- 1Laboratory of Molecular Microbiology and Biotechnology, Department of Medical Biotechnologies, University of Siena, Siena, Italy
- 2Unit of Endodontology, UCL Eastman Dental Institute, University College London, London, UK
- 3Department of Microbial Diseases, UCL Eastman Dental Institute, University College London, London, UK
The oral and nasopharyngeal streptococci are a major part of the normal microbiota in humans. Most human associated streptococci are considered commensals, however, a small number of them are pathogenic, causing a wide range of diseases including oral infections such as dental caries and periodontitis and diseases at other body sites including sinusitis and endocarditis, and in the case of Streptococcus pneumoniae, meningitis. Both phenotypic and sequence based studies have shown that the human associated streptococci from the mouth and nasopharynx harbor a large number of antibiotic resistance genes and these are often located on mobile genetic elements (MGEs) known as conjugative transposons or integrative and conjugative elements of the Tn916/Tn1545 family. These MGEs are responsible for the spread of the resistance genes between streptococci and also between streptococci and other bacteria. In this review we describe the resistances conferred by, and the genetic variations between the many different Tn916-like elements found in recent studies of oral and nasopharyngeal streptococci and show that Tn916-like elements are important mediators of antibiotic resistance genes within this genus. We will also discuss the role of the oral environment and how this is conducive to the transfer of these elements and discuss the contribution of both transformation and conjugation on the transfer and evolution of these elements in different streptococci.
Introduction
The oral microbiota is one of the most diverse bacterial populations found in the human body (Topazian et al., 2002) with the total number of distinct taxonomic units found in the oral cavity in the 100s, if not the 1000s (Aas et al., 2005; Keijser et al., 2008; Wade, 2013), however, these are not all found in every mouth as individuals have distinct bacterial populations, and distinct bacterial populations are found in different habitats within each mouth (Eren et al., 2014 ). The vast majority of the oral inhabitants are commensal species and Streptococcus spp. are the most diverse and one of the most abundant genera present in saliva of both dentulous and edentulous individuals (Keijser et al., 2008; Ealla et al., 2013). The oral streptococci are pioneers in the colonization of the oral cavity soon after birth with the numbers and complexity of the oral bacterial species increasing gradually from exposure with microbial sources from the external environment and also following tooth eruption. Streptococcus salivarius, S. mitis, and S. oralis have been identified as the first and most dominant oral microbes in the oral cavities of newborns (Pearce et al., 1995).
Most Streptococcus spp. are non-pathogenic, forming part of the commensal human microbiome present in the mouth, intestine, and upper respiratory tract and they are also present on the skin. However, in the presence of host defense imbalance, many of these species can cause disease, such as dental caries and other infections (Topazian et al., 2002).
Diseases Caused by Oral Streptococci
Streptococcus spp. are major residents of oral biofilms (dental plaque). Due to ecological shifts in the biofilm ecology a more lactic acid rich environment can result causing demineralization and destruction of the hard tissues of the teeth (e.g., enamel, dentin, and acellular cementum). S. mutans and S. sanguinis are strongly associated with early childhood caries, suggesting that the relative levels of these two microorganisms in the oral cavity play an important role in caries development in deciduous teeth (Hardie, 1992; Ge et al., 2008). The adhesion mechanisms of various Streptococcus spp. enable multiple intimate contacts and interactions between the bacterial cells and the host, which is crucial in biofilm formation, caries, and periodontal disease development. In vitro and in vivo studies have demonstrated direct roles for many streptococcal adhesins as colonization factors (Nobbs et al., 2009).
Periodontal disease corresponds to an inflammatory pathosis of bacterial origin resulting in the loss of tooth attachment and as a final consequence, the loss of the tooth (Nguyen and Martin, 2008). Although not the primary candidates for periodontal pathogens (Hardie, 1992), S. mutans, S. sanguinis, and S. mitis are the primary colonizers of the tooth surface and subsequent attachment of other bacteria results in a multispecies biofilm which can lead to inflammation (Ochiai et al., 1993). These species are also found in the colonized periodontal pockets (Edwardsson et al., 1999).
There is also evidence for the involvement of Streptococcus spp. in root canal infections with periapical disease, S. gordonii, S. anginosus, and S. oralis were the most frequently isolated streptococci from infected root canals, however, S. mutans, S. intermedius, S. parasanguinis have also been isolated (Chávez De Paz et al., 2003). Additionally other Streptococcus spp. have been detected in human infected root canals using checkerboard DNA–DNA hybridization including S. mitis, S. intermedius, and S. constellatus (Vianna et al., 2008). A clinical development of periapical disease is the dental abscess containing pus composed of dead host and bacterial cells. The causative microbiota is comprised of a mix of strict anaerobes in association with facultative anaerobes such as viridans and anginosus groups (Robertson and Smith, 2009). S. anginosus, S. intermedius, and S. constellatus have been identified from abscess cases in association with other species (Siqueira and Rôças, 2013).
A potential life-threatening complication of oral infections, involves the spreading of invasive microorganisms through connective tissue and fascial planes (cellulitis), brain, maxillary and cavernous sinus, eye, or mediastinum. The microbiology of the severe spreading odontogenic infection seems to differ from the more localized dental abscess with anginosus group streptococci and Fusobacterium spp. often isolated (Han and Kerschner, 2001).
Less specific, in terms of infected tissues is Noma, a gangrenous disease that affects maxillary of children with compromised immune function. This is most probably a polymicrobial infection, however, it has been demonstrated that S. pyogenes and S. anginosus (among other non-streptococcal species) are more abundant in the gingival microbiota of Noma patients with acute necrotizing gingivitis compared to healthy controls (Huyghe et al., 2013).
The spread of oral microorganisms by the blood circulation has been reported. Bacteraemia can occur as a result of tooth brushing (Lucas et al., 2008) and invasive procedures, however, these are usually transient in normal individuals and the exact definition of bacteraemia remains a topic of discussion (Bergstrom, 2009). However, in compromised hosts (e.g., patients with cancer, untreated diabetes or immunodeficiency) it can result in a generalized fatal infection (sepsis). The magnitude of bacteraemia of oral origin seems to be directly proportional to the degree of oral inflammation and infection (Bender et al., 1984), S. viridans (Heimdahl et al., 1990), S. intermedius and S. sanguinis (Debelian et al., 1995) have been isolated from blood samples. Bacteraemia is also considered a risk factor for the development of infective endocarditis, an infection of the heart valves (Starkebaum et al., 1977).
Antibiotics are administered as part of the clinical management of life-threatening infections and for the prevention of infective endocarditis in patients undergoing dental treatment, patients with prosthetic heart valves and/or patients with previous history of endocarditis and a cardiac transplant that develops a problem in a heart valve. The usual recommended antibiotics for these cases are: (1) oral amoxicillin, intravenous or intramuscular ampicillin, where patients are unable to take oral medication; (2) oral clindamycin, oral cephalexin, oral azithromycin or oral clarithromycin where patients are allergic to penicillin or ampicillin; (3) clindamycin or cefazolin (intravenous or intramuscular) where patients are allergic to penicillin or ampicillin and unable to take oral medication (Wilson et al., 2008). Although the use of systemic antibiotics is not a recommended routine to treat oral infections, its benefit is debatable when used in conjunction with non-surgical periodontal therapy (Keestra et al., 2014). The antibiotic resistance of isolates from dental abscess has increased over the years (Kuriyama et al., 2007) which is extremely concerning taking into consideration that general practitioners knowledge about antibiotics may not always be sufficient (e.g., Nabavizadeh et al., 2011) and its use can be indiscriminate.
Polymicrobial Biofilms of the Oral Cavity: A “Gene Transfer Prone” Environment
The oral cavity, and to a lesser extent the nasopharyngeal cavity, are physicochemically complex environments, with many distinct habitats, in which different bacterial species coexist. Different bacteria are found in close contact on the mouth surfaces where they exist as polymicrobial biofilms. These are complex communities of bacteria adhering to a surface, embedded in a polymeric extracellular matrix consisting of polysaccharides, proteins and nucleic acids. Extracellular DNA (eDNA) is a key component (Whitchurch et al., 2002) of the biofilm matrix produced by many bacterial species (Roberts and Kreth, 2014). This eDNA has structural, nutritional, and informational value to the cells within the biofilm. Many of the species living in the oral cavity are naturally transformable, especially among streptococci, which are the most relevant genus of culturable microorganisms in the oral microflora (Cvitkovitch, 2001), these species can be transformed by eDNA present in the biofilm (e.g., Hannan et al., 2010). Beyond transformation, the close contact between bacteria in biofilms allows genetic exchange by means of conjugation, which requires cell-to-cell contact. Conjugation has been proven to happen in oral biofilms and in biofilms derived from different environments, with a frequency usually higher compared to conjugation between planktonically growing cells (Roberts et al., 1999, 2001a; Ready et al., 2006; Cook et al., 2011). There is a wealth of evidence suggesting that the oral biofilm environment is a suitable environment for extensive horizontal gene transfer and the readers are directed to recent reviews on this topic (Roberts and Mullany, 2006, 2010; Olsen et al., 2013; Roberts and Kreth, 2014).
Mobile Genetic Elements – Tn916
Mobile genetic elements (MGEs) are regions of DNA that encode the necessary proteins in order to catalyze their own movement. MGEs are capable of movement within a bacterial cell, either between different sites within the same replicon or between replicons (e.g., from a chromosome to a plasmid). There are many different families of MGEs with the families being delineated by their structure or the type of recombinase responsible for their integration and excision from a replicon (Roberts et al., 2008). One of the types of elements which are often associated with the carriage of antibiotic resistance genes are the conjugative transposons, also known as integrative and conjugative elements (ICEs; Mullany et al., 2002; Roberts and Mullany, 2011). These MGEs are capable of both intracellular transfer between sites within a cell and intercellular conjugation using their self-encoded conjugation apparatus. There are different families of conjugative transposons, some of which seem to be confined to a particular species or genera of bacteria. Members of the Tn916/Tn1545 family of conjugative transposons have, however, been found in, or introduced into, over 30 different genera of bacteria. The basic biology of Tn916/Tn1545 like elements has been reviewed recently and readers are directed to these reviews (Roberts and Mullany, 2009, 2011; Ciric et al., 2011a).
Tn916, encoding tet(M) for the ribosomal protection protein Tet(M), was discovered and reported around 1980 (Franke and Clewell, 1981) when plasmid independent transfer of tetracycline resistance was demonstrated between strains of Enterococcus faecalis. Since then it, or variants of it (called here Tn916-like), has been found in many different organisms including important human pathogens such as E. faecalis, Clostridium difficile, Staphylococcus aureus, and S. pneumoniae (reviewed in Roberts and Mullany, 2011). Of these pathogens S. pneumoniae deserves particular attention in this review as its natural habitat is contiguous with the oral cavity and it is apparent that the influence of Tn916-like elements is similar in many respects to the activity and incidence of Tn916-like elements in the oral streptococci. In addition many of the elements found in the oral streptococci were initially discovered in S. pneumoniae.
Streptococcus pneumoniae: Disease, Competence, and Antibiotic Resistance
Streptococcus pneumoniae is a member of the normal human nasopharyngeal microflora; it is present in a significant portion of the general population, with prevalence in children peaking at <5 years of age (Le Polain de Waroux et al., 2014). From the nasal cavity where it resides, S. pneumoniae can invade contiguous sites such as the ears and frontal sinus causing otitis media and sinusitis, which are mild but common diseases, and it can be aspirated in the lungs where it causes pneumonia. S. pneumoniae is also a causative agent of meningitis. It can reach meninges from the middle ear, the frontal sinus or from the bloodstream, which can be transiently invaded during pneumonia (Mook-Kanamori et al., 2011). The genome of S. pneumoniae shows high plasticity, with evidence of extensive horizontal gene transfer mediated by transformation and, to a lesser extent, conjugation and transduction. Natural genetic transformation was discovered in S. pneumoniae (Griffith, 1928) and since then has been intensively studied. In the process of transformation, a bacterium can internalize exogenous naked DNA and integrate it into its chromosome by homologous recombination. The acquisition, processing and integration of DNA are mediated by an array of proteins which are produced only transiently during the growth of the microorganism, in a phase called competence. The entrance of a pneumococcal population into the competence phase is regulated by a quorum sensing peptide called competence stimulating peptide (CSP), which is encoded by comC, synthesized as a 41 amino acid precursor, cleaved to 17 amino acids and secreted by the proteolytic ABC-transporter ComAB. CSP subsequently binds its receptor, which is a transmembrane histidine protein kinase coded by comD. Upon binding of CSP, ComD autophosphorylates, and donates the phosphoryl group to its cognate ComE response regulator (Martin et al., 2013). The phosphorylated ComE is able to bind to the promoter region of the early competence genes and activate their transcription: the activated operons include comAB and comCDE in an auto-induction loop (Peterson et al., 2004). The induction of competence by CSP is also essential for S. pneumoniae biofilm formation, suggesting that the two processes are intertwined and that competence has a deep effect on the cellular physiology (Oggioni et al., 2006). Antibiotic treatment has been demonstrated to induce competence in S. pneumoniae, the bacterium is therefore able to modify its genetic milieu and to acquire new phenotypes which might help its survival under stress conditions (Prudhomme et al., 2006). DNA can be acquired by transformation from co-colonizing pneumococci as well as from other commensal bacteria, which might be present in the same ecological niche, given enough homology between the transforming and recipient DNAs. Among the new phenotypes acquired by transformation there is increased resistance to antibiotics such as penicillin and cotrimoxazole (Chewapreecha et al., 2014).
Antibiotic resistance in S. pneumoniae is becoming more and more common, but has been known for a long time. Increased MIC for penicillin in a clinical strain of S. pneumoniae was first detected in 1967 in Australia (Hansman and Bullen, 1967), since then isolates with decreased susceptibility to penicillin have arisen and spread worldwide. An international network (Pneumococcal Molecular Epidemiology Network, PMEN) was established in 1997 in order to characterize and classify the international spread antibiotic resistant pneumococcal clones (McGee et al., 2001). The first identified clone is the Spain23F-1, also known as PMEN-1, resistant to penicillin, tetracycline, chloramphenicol, and cotrimoxazole.
In S. pneumoniae, resistance to tetracycline is often due to Tn916-like MGEs, which usually carry the tet(M) gene. Resistance to macrolides can be due to erm genes, coding for rRNA modifying methyltransferase, or to mef/msr genes which code for an efflux pump. The erm genes usually confer the MLSB phenotype, resistance to macrolides, lincosamides and streptogramin B with high MIC values (>64 mg/L for erythromycin), while mef/msr genes confer the M phenotype, resistance to macrolides albeit with low MIC values (1–16 mg/L for erythromycin). In S. pneumoniae, both classes of resistance genes are carried by Tn916-like elements.
Tn916 Variants Detected in S. pneumoniae Genomes
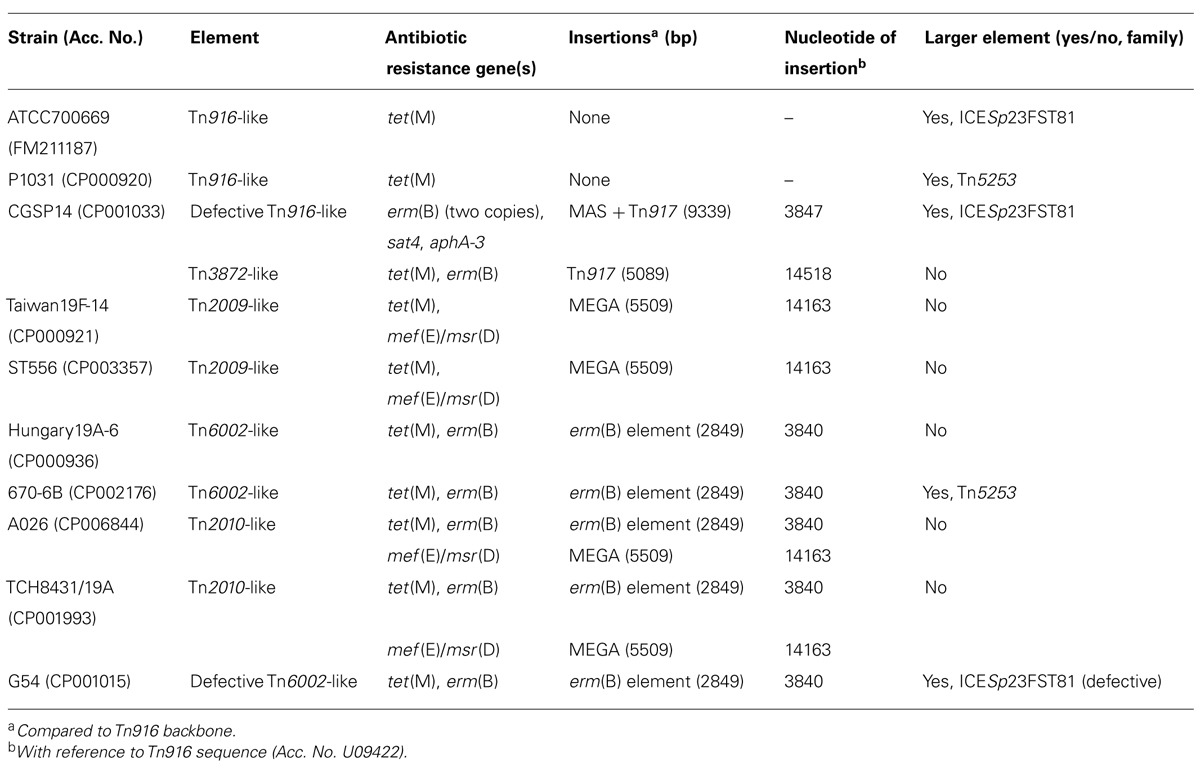
Many Tn916-family elements were originally detected and sequenced in clinical strains of S. pneumoniae. Eleven Tn916-like elements are present in 10 out of 25 complete pneumococcal genome sequences present in GenBank (as to 20th May 2014); strain CGSP14 harbors two different Tn916-like CTns (Table 1). In five out of 11 of these genomes, Tn916-like elements are integrated into a larger composite Tn5253 family or ICESp23FST81 family element (see below), while in the remaining six cases they are present as individual elements. Nine out of the 11 Tn916-like elements are in fact composite elements themselves, as there are insertions of other DNA within the Tn916 sequence backbone, two of these elements are also presumed defective, as they lack some of the Tn916 sequences. The DNA sequences that can be found inserted within the Tn916 backbone are: (i) Tn917, 5,089 bp, carrying the erm(B) gene, (ii) the “MEGA” element (macrolide efflux genetic assembly), 5,511 bp, carrying mef(E)/msr(D), (iii) the MAS (macrolide–aminoglycoside–streptothricin) element, 4,225 bp, carrying erm(B), aadE, sat4, and aphA-3 genes, and (iv) a 2,849 bp fragment containing erm(B). It is worth noting that the 2,849 bp erm(B) fragment derives from a 5,288 bp omega element (GenBank accession no. FR671418) which carries two copies of an omega repressor encoding gene and the aphA-3 gene (for a diagram of these elements and their location within Tn916 readers a referred to Ciric et al., 2011a). The deletion of the aphA-3 fragment happens through a recombination event between the omega-encoding genes, and results in the fusion of the 3′ end of Tn916 orf20 with the omega repressor coding sequence (Croucher et al., 2011).
As reported in Table 1, the Tn916-like elements present in pneumococcal complete genome strains are: (i) Tn3872, a Tn916 carrying Tn917, present in one strain (McDougal et al., 1998), (ii) Tn6002 carrying the 2,849 bp erm(B) fragment, present in two strains (Warburton et al., 2007), (iii) Tn2009, carrying “MEGA,” in two strains (Del Grosso et al., 2004), and (iv) Tn2010 carrying both “MEGA” and the 2,849 bp erm(B) fragment, in two strains (Del Grosso et al., 2006). Two more strains carry Tn916-like elements: one has a defective version of Tn6002, with a deletion from the end of tet(M) to the end of the element, and another resembles the structure of SpnRierm(B) (GenBank accession no. AM490850, Cochetti et al., 2007), where the MAS element is fused to Tn917 replacing the portion from orf19 to tet(M).
Tn6003 and Tn1545 are other Tn916-family elements which were first detected in S. pneumoniae and both of them contain the MAS element and the erm(B) fragment, the latter also has the insertion of a transposase at another site (Cochetti et al., 2008). It has recently been shown that the MAS element is able to excise and circularize using a 1,166 bp direct repeat containing erm(B), thus turning Tn6003 into Tn6002 (Palmieri et al., 2012).
A few epidemiological studies have reported the frequencies of the different elements in clinical strains of S. pneumoniae. A Chinese study performed on 328 invasive isolates from children <5 years in 2005 reported that 31% of the isolates were macrolide resistant, and among these, 74% were also tetracycline resistant (Xu et al., 2010). Investigation of the genetic elements present in the tetracycline and macrolide resistant strains was performed by PCR detection of integrase, excisionase, transposase, and resistance genes carried by Tn916-like elements, with subsequent reverse blot hybridization of the PCR product with specific probes and assignment to a category of genetic elements based on hybridization results. The study might have therefore underestimated the presence of new Tn916 variants. Data analysis revealed that 49 strains carried Tn6002, 15 strains carried Tn3872 and three carried Tn1545/Tn6003; variants of Tn6002 and Tn3872 lacking the tet(M) gene were also detected. A later study performed in China in 2010 on 135 macrolide resistant pneumococci from nasal swabs of children, confirmed the high prevalence of Tn6002-like elements (56.3% of the isolates), followed by Tn2010 (28.9%), Tn1545/Tn6003 (5.9%), and Tn3872 (5.2%; Zhou et al., 2012). Tn2010 was first detected in an Italian isolate (Del Grosso et al., 2006) and has later been completely sequenced (Li et al., 2011). In most strains analyzed, it was found to be inserted between bases 1,731,928 and 1,743,232 of the R6 genome (GenBank accession no. NC_003098), flanked at both ends by short sequence stretches (24 and 66 nucleotides) that did not show homology with the R6 genome nor with the ends of Tn916, in one strain it has also been found inserted between nucleotides 1,820,138 and 1,820,139 of the R6 genome, with only a 7 bp stretch at the left end (Li et al., 2011).
A PCR study performed on multidrug resistant pneumococcal strains colonizing children in Venezuela reported a high prevalence of Tn3872 (44% of isolates) and the presence of Tn6002 (10%), Tn2009 (10%), Tn2010 (4%), and an element related to SpnRierm(B) (only one isolate; Quintero et al., 2011). The elements were detected in strains of different serotypes, suggesting horizontal gene transfer of the resistance determinants, Tn916-family element are therefore widespread among multidrug resistant pneumococcal clinical isolates. Two studies on macrolide resistant pneumococci and their association with the presence of Tn916 family of genetic elements were also performed in Spain (Calatayud et al., 2007, 2010), both of them reported the high prevalence of Tn6002-like elements (about 50% of macrolide resistant strains examined), a significant prevalence of Tn3872 (about 25% of the strains) with a lower prevalence of the other elements. Interestingly, both the studies detected a number of strains (about 16%) harboring erm(B), tet(M), and tndX, suggesting the presence of an ICESp1116 related element (GenBank accession no. HE802677; Brenciani et al., 2012). This element was originally described in S. pyogenes as a defective Tn5397 with an erm(B) gene and an Insertion Sequence encoded transposase disrupting tet(M), with a recombination module including tndX, a large serine-recombinase (Wang and Mullany, 2000; Roberts et al., 2001b) and orf45 coding for a DDE transposase.
Composite Elements Carrying Tn916-Family Elements in S. pneumoniae
It is well known that Tn916 is a promiscuous element, as it can integrate at different sites not only into the host chromosome, but also into plasmids and into other larger ICEs. In S. pneumoniae at least two families of composite elements have integrated copies of Tn916; these are the Tn5253 family and ICESp23FST81 family. The prototype elements are large (64.5 and 81 kbps respectively), share some sequence homology but have two different recombination modules and consequently different, specific integration sites in the S. pneumoniae chromosome. The insertion of Tn5253 into S. pneumoniae chromosome leads the duplication of an 83 bp target site located between spr1042 and the 5′ end of spr1043 (rbgA; GenBank EU351020), while ICESpn23FST81 is inserted at the 3′ end of spr1211 (rplL), with its insertion causing a duplication of 16 bp (Croucher et al., 2009; insertion sites refer positions in the R6 genome). Both integration sites lie at one end of a pneumococcal conserved gene, whose coding sequence is not disrupted by the insertion of the element, this site selection strategy guarantees that the element always has a “safe site” to integrate in. These larger ICEs show a typical modular structure in which Tn916-like elements are inserted. The insertion site of Tn916 within the larger element is variable, for instance in strains CGSP14 (GenBank CP001033), ATCC700619 (GenBank FM211187), and P1031 (GenBank CP000920) it integrates downstream Tn5253 orf8 homolog; in strains 670-6B (GenBank CP002176) and G54 (GenBank CP001015) it integrates in a site not present in Tn5253, while in Tn5253 it is integrated downstream orf20, coding for a truncated transposase of the IS110 family. The presence of Tn5253-family and ICESp23FST81-family elements has been investigated in clinical isolates of S. pneumoniae and proven to be frequent, especially among multidrug resistant strains (Henderson-Begg et al., 2009; Mingoia et al., 2011); moreover ICESp23FST81-family elements are a distinctive feature of the isolates belonging to the multidrug resistant clone PMEN-1 (Croucher et al., 2011).
Biology of Tn916-Like Elements in S. pneumoniae: Is there any Evidence for Conjugation?
Tn916 has limited requirements for target site selection in that it recognizes an A:T rich region (Scott et al., 1988, 1994; Mullany et al., 2012), consequently its host range is very broad (Bertram et al., 1991). However, it has been reported that Tn916 integration can also occur at some specific or preferred sites, as in Clostridium difficile strain CD37 (Mullany et al., 1991; Wang et al., 2000) or in the enterococcal plasmid pAD1 (Jaworski and Clewell, 1994). In S. pneumoniae sequenced genomes, the insertions of Tn916-family elements can be mapped at a few hotspots (Santoro et al., 2010), suggesting a target site specificity. On the other hand, transconjugants generated in the lab harboring Tn5251 (the Tn916 component of the composite Tn5253 element) seem to have multiple insertions at A:T rich sites within the chromosome (Santoro et al., 2010) and transconjugants generated in a serotype 3 S. pneumoniae strain, with E. faecalis as a Tn916 donor, harbored multiple and widespread insertions (Watson and Musher, 1990). The actual functionality of Tn916-family elements in S. pneumoniae has been investigated in very few studies. Tn3872, originally described in 1998, was not capable of conjugal transfer to the standard pneumococcal recipient R6 (McDougal et al., 1998), this was later confirmed by Cochetti et al. (2007) who also showed that Tn6002 and Tn6003 are able to conjugate, albeit at low frequency and in a recipient dependent manner (transfer frequency of 1.1 × 10-8 to S. pyogenes 12RF and of 1.7 × 10-7 to E. faecalis JH2-2 CFU transconjugants/CFU donors, respectively). The “MEGA” carrying elements Tn2009 and Tn2010 are not conjugative using pneumococcal recipients and selection for either tetracycline or erythromycin resistance (transfer frequency<10-9 per donor cell), while both elements are transferable by transformation to standard pneumococcal recipients (Del Grosso et al., 2006). The first step in the conjugal transfer of Tn916 family of genetic elements is the excision from the chromosome with formation of a circular intermediate (CI). A CI has been detected in strains carrying Tn2010 (Zhou et al., 2014), indicating that XisTn and IntTn are probably functional, therefore the conjugal transfer may either happen at frequencies below the detection threshold or be impaired because of the insertion of “MEGA” within the regulatory region. The insertion of the erm(B) element in the conjugal transfer region does not seem to be detrimental for conjugation, as Tn1545, Tn6002, and Tn6003 are conjugative. CIs have also been detected for Tn5251 which is integrated in the composite element Tn5253 (Provvedi et al., 1996), subsequently the autonomous transfer of Tn5251 has been demonstrated, even if, again, at low frequencies and in a recipient dependent manner (Santoro et al., 2010). The fact that Tn916-family elements in S. pneumoniae tend to be either inserted in composite elements or at the same hotspots, suggests that their autonomous conjugation in vivo might not be frequent, even if there is evidence that this event is possible, and that they may rely on the conjugation machinery of larger elements such as ICESp23FST81 or Tn5253 which are able to transfer at frequencies about three orders of magnitude higher (Iannelli et al., 2014).
Transformability of S. pneumoniae with Respect to Conjugative Transposons: From Pioneering Studies to the Genomic ERA
Tn5253 had originally been identified as a chromosomal genetic element bearing resistance to chloramphenicol and tetracycline in the clinical strain BM6001. Work performed in the late seventies in the laboratories of Walter Guild investigated the transformation properties of pneumococcal lysates containing Tn5253 (Shoemaker et al., 1979). First, isogenic strains with or without Tn5253 were constructed, and then DNA contained in the lysates was used in transformation experiments. Different shearing treatments of donor DNA were used to infer the length of the heterologous insertions, the whole Tn5253 was predicted to be longer than 30 kb and the cat containing fragment between 4 and 8 kb, which is consistent with the actual sizes; 64,528 and 7,627 bp, respectively. Transformation has been used to study the transferability of Tn916 elements which could not be transferred by conjugation, the transformation frequencies of Tn2009 and Tn2010 have been shown to be 4 × 10-7 and 3 × 10-7 CFU/ml of recipient, respectively (Del Grosso et al., 2004). Whole genome sequencing of three Tn2010 bearing transformants showed that multiple SNPs had been acquired together with the element; moreover the presence of the element did not involve any fitness cost (Zhou et al., 2014). A transforming pneumococcal DNA containing a “MEGA” carrying Tn916-like has been used to assay the in vitro transformation frequencies of S. pneumoniae isolates belonging to different serotypes. All the serotypes investigated had similar transformation frequencies, ranging from 2.6 × 10-5 to 7.9 × 10-8 transformants per recipient cell, with no significant difference between drug-resistance and drug-susceptibility associated serotypes (Joloba et al., 2010). Nowadays, next generation sequencing technologies allow the rapid and parallel sequencing of 100s of genomes and bring new knowledge on the evolution of S. pneumoniae genome. Many of these studies are aimed at finding recombination events due to transformation and therefore exclude the recombination events due to MGEs (Mostowy et al., 2014). The seminal study by Croucher et al. (2011) examined the evolution of ICESp23FST81 in the PMEN1 clone and found that the acquisition of macrolide resistance genes on Tn916 occurred independently multiple times across the phylogeny. Another work was focused on PMEN2 and PMEN22 lineages, both carrying an independently acquired Tn5253 family ICE. In some PMEN2 isolates from Iceland, recombination events lead to inactivation or loss of resistance determinants, interestingly one of these events was the precise excision of the Tn916-like element from the Tn5253 backbone (Croucher et al., 2014). A recent sequencing study was performed on 3,085 nasopharyngeal pneumococcal isolates collected from infants and their mothers in a refugee camp over a period of 3 years (Chewapreecha et al., 2014). A total of 2,209 putative recombination events were detected among strains of different lineages, out of these 191 (8.5%) were plausible capsular switches while 132 (6%) were possibly due to MGEs. These data cannot confirm whether the transfer is due to conjugation or transformation, but underline how horizontal transfer of MGEs contributes to pneumococcal genome plasticity and likely utilizes at least two different mechanisms for horizontal gene transfer.
Evidence for Tn916-Like Elements in Members of the Oral Streptococci – A Diversity of Resistance Genes
Following the original description of Tn916 (Franke and Clewell, 1981) reports of transferable tetracycline resistance in oral streptococci were published soon after. The host species of these Tn916-like elements were S. mutans U202 (Hartley et al., 1984), S. anginosus (Clermont and Horaud, 1990), and S. mitis (Bentorcha et al., 1992). The majority of these early studies were based on Southern blots being probed with wildtype Tn916 sequences, however, these techniques, while confirming the presence of Tn916-like structures were not sophisticated enough to show all the details of the elements involved. Following the advent of modern day sequencing and improved PCR techniques it became possible to gain a better understanding of the genetic context of the resistances found in the oral streptococci. Two recent studies have focussed on characterizing the whole element responsible for tetracycline resistance in oral streptococci. In the first study (Ciric et al., 2012) the authors used a PCR based method followed by RFLP profiling of large amplicons spanning the entire length of Tn916 (18,032 bp). They showed that wildtype Tn916 elements were not the most common element found in the tetracycline resistant streptococci cultured from the saliva of healthy humans. Rather the majority of elements were Tn6002 and Tn3872 which were found in strains where these elements have never been reported before; Tn3872 in S. salivarius and S. sanguinis and Tn6002 in S. australis, S. infantis, S. mitis, S. oralis, S. parasanguinis, S. salivarius, and S. sanguinis (Ciric et al., 2012). In a larger, more recent study 263 resistant viridans group streptococci were analyzed in order to elucidate the genetic basis of resistance (Brenciani et al., 2014). The authors showed the presence of multiple new structures of MGEs based on Tn916-like elements and, similarly to both the previous study (Ciric et al., 2012), and the situation previously described for S. pneumoniae (see above) showed the co-carriage of multiple resistance genes, e.g., tet(M) and erm(B) on Tn916-like MGEs in different oral streptococci. The architecture of these different accessory elements responsible for incoming resistances has been described above and reviewed relatively recently (Ciric et al., 2011a; Roberts and Mullany, 2011).
This acquisition of multiple resistance genes by Tn916-like elements is an emerging theme in studies analysing the entire element present in the oral and nasopharyngeal streptococci, and the authors suggest that researchers analyse the entire element before reporting the presence of Tn916 based simply on the detection or one or two genes by PCR, such as the integrase gene. In support of this suggestion it is also worth mentioning that the presence of intTn has been detected in excess of tet(M) in oral metagenomic DNA samples which strongly suggests there are elements containing the integrase gene which do not contain tet(M; Seville et al., 2009) and indeed a small number of these tet(M)-free Tn916-like elements have been reported from oral streptococci, e.g., Tn916S from S. intermedius (see below; Lancaster et al., 2004; Novais et al., 2012a).
Antiseptic Resistance is Co-Localized on Tn6087 with Tetracycline Resistance in an Oral S. oralis
During the previously described survey of Tn916-like elements in oral tetracycline resistant streptococci (Ciric et al., 2012) a strain of S. oralis was isolated which contained a novel element; detected by showing a different RFLP pattern following digestion of the long amplicons derived from Tn916. Upon further analysis and subsequent sequencing the authors demonstrated the presence of additional DNA inserted within orf15 of Tn916 (Ciric et al., 2011b). Analysis of this region showed that there were two almost identical copies of IS1216, an insertion sequence which is commonly associated with tet(S) containing Tn916-like elements in the enterococci (Novais et al., 2012b). Between these two copies of IS1216 there are two ORFs, one predicted to encode a hypothetical protein of unknown function and the other a small multi-drug resistance protein which was shown, by mutational analysis to encode resistance to cetyltrimethylammonium bromide (CTAB). CTAB is an antiseptic which is effective against bacteria and fungi and is one of the components in the topical antiseptic cetrimide used widely to treat minor skin injuries, minor burns, scalds, and nappy rash. In order to determine how this gene cassette can associate and insert within Tn916, PCRs were carried out across the region of insertion with primers designed to amplify a circular form of the small antiseptic resistance composite transposons and ligated target sites in Tn916. Multiple different products were isolated which showed that the small composite transposon consisting of the two IS elements and the intervening DNA could be detected within its target site, the antiseptic resistance gene and one copy of IS1216 could be detected in a circular form, one copy of a chimeric IS1216 could be detected in the target site within orf15 and finally the restored target site was detected by PCR. These products suggest that there are multiple mobility mechanisms being utilized by this element, the activity of the IS1216 encoded transposase could explain the excision reactions leading to the circular form detected, conversely homologous recombination between the two copies of the element must have led to the formation of the chimeric version of IS1216 which was amplified. This redundancy in the mobility of the DNA gives a possible scenario for the continued isolation of novel variants of Tn916-like elements. Insertion into a replicon, due to homologous recombination presumably negates the need for transposition reactions to occur in order to obtain successful integration into the Tn916 backbone. An interesting example of a recombination within Tn916 is seen within the conjugative transposon Tn916S from an oral S. intermedius. It was originally (and erroneously) described as a Tn916-like element containing the tet(S) gene in place of tet(M) (Lancaster et al., 2004). More recent analysis of the sequence of this element, however, has shown that it is infact a hybrid element containing a mosaic tetracycline resistance gene composed of tet(S) (encoding 599 amino acids of the gene) from a related element from Enterococcus casseliflavus designated Tn6000 (Roberts et al., 2006; Brouwer et al., 2010) and the Tn916 wildtype tet(M) gene (encoding 61 amino acids of the hybrid gene) and designated tet(S/M) (GenBank accession no. AY534326.1; Novais et al., 2012a).
Interestingly with Tn6087 the target site for the small composite transposon encoding qrg was within orf15. Despite repeated attempts the authors could not detect transfer of tetracycline and CTAB resistance by conjugation, however, they did show transformation into an oral S. australis (Ciric et al., 2011b).
Conclusion
The repeatedly observed redundancy in transfer mechanisms (conjugation and transformation) of many of the Tn916-like elements described in this review will likely lead to a higher chance of successful transfer than if these elements only relied on conjugation in order to spread. This may be one of the reasons why the Tn916-like elements are so successful within this genera and why there is so much variation of these elements in oral and nasopharyngeal streptococci. The elements themselves; both Tn916 and the smaller elements found within it, may well have exploited the competence inherent in many species within this genera.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in the laboratories of Francesco Santoro and Adam P. Roberts was funded by the Commission of the European Communities, specifically the Infectious Diseases research domain of the Health theme of the seventh Framework Program, contract 241446, “The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiotas at various body sites.”
References
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., and Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bender, I. B., Naidorf, I. J., and Garvey, G. J. (1984). Bacterial endocarditis: a consideration for physician and dentist. J. Am. Dent. Assoc. 109, 415–420.
Bentorcha, F., Clermont, D., de Cespédès, G., and Horaud, T. (1992). Natural occurrence of structures in oral streptococci and enterococci with DNA homology to Tn916. Antimicrob. Agents Chemother. 36, 59–63. doi: 10.1128/AAC.36.1.59
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergstrom, J. (2009). It is unclear if toothbrushing is a risk factor for infective endocarditis in children. J. Evid. Based Dent. Pract. 9, 219–220. doi: 10.1016/j.jebdp.2009.06.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bertram, J., Strätz, M., and Dürre, P. (1991). Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J. Bacteriol. 173, 443–448.
Brenciani, A., Tiberi, E., Morici, E., Oryasin, E., Giovanetti, E., and Varaldo, P. E. (2012). ICESp1116, the genetic element responsible for erm(B)-mediated, inducible resistance to erythromycin in Streptococcus pyogenes. Antimicrob. Agents Chemother. 56, 6425–6429. doi: 10.1128/AAC.01494-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Tiberi, E., Tili, E., Mingoia, M., Palmieri, C., Varaldo, P. E.,et al. (2014). Genetic determinants and elements associated with antibiotic resistance in viridans group streptococci. J. Antimicrob. Chemother. 69, 1197–1204. doi: 10.1093/jac/dkt495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brouwer, M. S., Mullany, P., and Roberts, A. P. (2010). Characterization of the conjugative transposon Tn6000 from Enterococcus casseliflavus 664.1H1 (formerly Enterococcus faecium 664.1H1). FEMS Microbiol. Lett. 309, 71–76.
Calatayud, L., Ardanuy, C., Cercenado, E., Fenoll, A., Bouza, E., Pallares, R.,et al. (2007). Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in spain. Antimicrob. Agents Chemother. 51, 3240–3246. doi: 10.1128/AAC.00157-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Calatayud, L., Ardanuy, C., Tubau, F., Rolo, D., Grau, I., Pallarés, R.,et al. (2010). Serotype and genotype replacement among macrolide-resistant invasive pneumococci in adults: mechanisms of resistance and association with different transposons. J. Clin. Microbiol. 48, 1310–1316. doi: 10.1128/JCM.01868-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chávez De Paz, L. E., Dahlén, G., Molander, A., Möller, A., and Bergenholtz, G. (2003). Bacteria recovered from teeth with apical periodontitis after antimicrobial endodontic treatment. Int. Endod. J. 36, 500–508. doi: 10.1046/j.1365-2591.2003.00686.x
Chewapreecha, C., Harris, S. R., Croucher, N. J., Turner, C., Marttinen, P., Cheng, L.,et al. (2014). Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46, 305–309. doi: 10.1038/ng.2895
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ciric, L., Ellatif, M., Sharma, P., Patel, R., Song, X., Mullany, P.,et al. (2012). Tn916-like elements from human, oral, commensal streptococci possess a variety of antibiotic and antiseptic resistance genes. Int. J. Antimicrob. Agents 39, 360–361. doi: 10.1016/j.ijantimicag.2011.12.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ciric, L., Jasni, A., de Vries, L. E., Agersø, Y., Mullany, P., and Roberts, A. P. (2011a). “The Tn916/Tn1545 family of conjugative transposons,” in Bacterial Integrative Mobile Genetic Elements, eds A. P. Roberts and P. Mullany (Texas, TX: Landes Bioscience).
Ciric, L., Mullany, P., and Roberts, A. P. (2011b). Antibiotic and antiseptic resistance genes are linked on a novel mobile genetic element: Tn6087. J. Antimicrob. Chemother. 66, 2235–2239. doi: 10.1093/jac/dkr311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clermont, D., and Horaud, T. (1990). Identification of chromosomal antibiotic resistance genes in Streptococcus anginosus (“S. milleri”). Antimicrob. Agents Chemother. 34, 1685–1690. doi: 10.1128/AAC.34.9.1685
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cochetti, I., Tili, E., Mingoia, M., Varaldo, P. E., and Montanari, M. P. (2008). erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 52, 1285–1290. doi: 10.1128/AAC.01457-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cochetti, I., Tili, E., Vecchi, M., Manzin, A., Mingoia, M., Varaldo, P. E.,et al. (2007). New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J. Antimicrob. Chemother. 60, 127–131. doi: 10.1093/jac/dkm120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cook, L., Chatterjee, A., Barnes, A., Yarwood, J., Hu, W. S., and Dunny, G. (2011). Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol. Microbiol. 81, 1499–1510. doi: 10.1111/j.1365-2958.2011.07786.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croucher, N. J., Hanage, W. P., Harris, S. R., McGee, L., van der Linden, M., de Lencastre, H. M.,et al. (2014). Variable recombination dynamics during the emergence, transmission and “disarming” of a multidrug-resistant pneumococcal clone. BMC Biol. 1:49. doi: 10.1186/1741-7007-12-49
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croucher, N. J., Harris, S. R., Fraser, C., Quail, M. A., Burton, J., van der Linden, M.,et al. (2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434. doi: 10.1126/science.1198545
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croucher, N. J., Walker, D., Romero, P., Lennard, N., Paterson, G. K., Bason, N. C.,et al. (2009). Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191, 1480–1489. doi: 10.1128/JB.01343-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cvitkovitch, D. G. (2001). Genetic competence and transformation in oral Streptococci. Crit. Rev. Oral Biol. Med. 12, 217–243. doi: 10.1177/10454411010120030201
Debelian, G. J., Olsen, I., and Tronstad, L. (1995). Bacteremia in conjunction with endodontic therapy. Endod. Dent. Traumatol. 11, 142–149. doi: 10.1111/j.1600-9657.1995.tb00476.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Del Grosso, M., Camilli, R., Iannelli, F., Pozzi, G., and Pantosti, A. (2006). The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50, 3361–3366. doi: 10.1128/AAC.00277-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Del Grosso, M., Scotto d’Abusco, A., Iannelli, F., Pozzi, G., and Pantosti, A. (2004). Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48, 2037–2042. doi: 10.1128/AAC.48.6.2037-2042.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ealla, K. K., Ghanta, S. B., Motupalli, N. K., Bembalgi, M., Madineni, P. K., and Raju, P. K. (2013). Comparative analysis of colony counts of different species of oral streptococci in saliva of dentulous, edentulous and in those wearing partial and complete dentures. J. Contemp. Dent. Pract. 14, 601–604. doi: 10.5005/jp-journals-10024-1371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Edwardsson, S., Bing, M., Axtelius, B., Lindberg, B., Söderfeldt, B., and Attström, R. (1999). The microbiota of periodontal pockets with different depths in therapy-resistant periodontitis. J. Clin. Periodontol. 26, 143–152. doi: 10.1034/j.1600-051X.1999.260303.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eren, A. M., Borisy, G. G., Huse, S. M., and Mark Welch, J. L. (2014). Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. U.S.A. 111, E2875–E2884. doi: 10.1073/pnas.1409644111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Franke, A. E., and Clewell, D. B. (1981). Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145, 494–502.
Ge, Y., Caufield, P. W., Fisch, G. S., and Li, Y. (2008). Streptococcus mutans and Streptococcus sanguinis colonization correlated with caries experience in children. Caries Res. 42, 444–448. doi: 10.1159/000159608
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Griffith, F. (1928). The significance of pneumococcal types. J. Hyg. (Lond.) 27, 113–159. doi: 10.1017/S0022172400031879
Han, J. K., and Kerschner, J. E. (2001). Streptococcus milleri: an organism for head and neck infections and abscess. Arch. Otolaryngol. Head Neck Surg. 127, 650–654. doi: 10.1001/archotol.127.6.650
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hannan, S., Ready, D., Jasni, A. S., Rogers, M., Pratten, J., and Roberts, A. P. (2010). Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Microbiol. 59, 345–349. doi: 10.1111/j.1574-695X.2010.00661.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansman, D., and Bullen, M. M. (1967). A resistant pneumococcus. Lancet 290, 264–265. doi: 10.1016/S0140-6736(67)92346-X
Hardie, J. M. (1992). Oral microbiology: current concepts in the microbiology of dental caries and periodontal disease. Br. Dent. J. 172, 271–278. doi: 10.1038/sj.bdj.4807849
Hartley, D. L., Jones, K. R., Tobian, J. A., LeBlanc, D. J., and Macrina, F. L. (1984). Disseminated tetracycline resistance in oral streptococci: implication of a conjugative transposon. Infect. Immun. 45, 13–17.
Heimdahl, A., Hall, G., Hedberg, M., Sandberg, H., Söder, P. O., Tunér, K.,et al. (1990). Detection and quantitation by lysis-filtration of bacteremia after different oral surgical procedures. J. Clin. Microbiol. 28, 2205–2209.
Henderson-Begg, S. K., Roberts, A. P., and Hall, L. M. (2009). Diversity of putative Tn5253-like elements in Streptococcus pneumoniae. Int. J. Antimicrob. Agents 33, 364–367. doi: 10.1016/j.ijantimicag.2008.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huyghe, A., François, P., Mombelli, A., Tangomo, M., Girard, M., Baratti-Mayer, D.,et al. (2013). Microarray analysis of microbiota of gingival lesions in noma patients. PLoS Negl. Trop. Dis. 7:e2453. doi: 10.1371/journal.pntd.0002453
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iannelli, F., Santoro, F., Oggioni, M. R., and Pozzi, G. (2014). Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 1235–1239. doi: 10.1128/AAC.01764-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jaworski, D. D., and Clewell, D. B. (1994). Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J. Bacteriol. 176, 3328–3335.
Joloba, M. L., Kidenya, B. R., Kateete, D. P., Katabazi, F. A., Muwanguzi, J. K., Asiimwe, B. B.,et al. (2010). Comparison of transformation frequencies among selected Streptococcus pneumoniae serotypes. Int. J. Antimicrob. Agents 36, 124–128. doi: 10.1016/j.ijantimicag.2010.03.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
KeestraJ. A., Grosjean, I., Coucke, W., Quirynen, M., and Teughels, W. (2014). Non-surgical periodontal therapy with systemic antibiotics in patients with untreated chronic periodontitis: a systematic review and meta-analysis. J. Periodontal Res. doi: 10.1111/jre.12221 [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keijser, B. J., Zaura, E., Huse, S. M., van der Vossen, J. M., Schuren, F. H., Montijn, R. C.,et al. (2008). Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87, 1016–1020. doi: 10.1177/154405910808701104
Kuriyama, T., Williams, D. W., Yanagisawa, M., Iwahara, K., Shimizu, C., Nakagawa, K.,et al. (2007). Antimicrobial susceptibility of 800 anaerobic isolates from patients with dentoalveolar infection to 13 oral antibiotics. Oral Microbiol. Immunol. 22, 285–288. doi: 10.1111/j.1399-302X.2007.00365.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lancaster, H., Roberts, A. P., Bedi, R., Wilson, M., and Mullany, P. (2004). Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet(S). J. Bacteriol. 186, 4395–4398. doi: 10.1128/JB.186.13.4395-4398.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Le Polain de Waroux, O., Flasche, S., Prieto-Merino, D., and Edmunds, W. J. (2014). Age-dependent prevalence of nasopharyngeal carriage of Streptococcus pneumoniae before conjugate vaccine introduction: a prediction model based on a meta-analysis. PLoS ONE 9:e86136. doi: 10.1371/journal.pone.0086136
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, Y., Tomita, H., Lv, Y., Liu, J., Xue, F., Zheng, B.,et al. (2011). Molecular characterization of erm(B)- and mef(E)-mediated erythromycin-resistant Streptococcus pneumoniae in China and complete DNA sequence of Tn2010. J. Appl. Microbiol. 110, 254–265. doi: 10.1111/j.1365-2672.2010.04875.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lucas, V. S., Gafan, G., Dewhurst, S., and Roberts, G. J. (2008). Prevalence, intensity and nature of bacteraemia after toothbrushing. J. Dent. 36, 481–487. doi: 10.1016/j.jdent.2008.03.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, B., Soulet, A. L., Mirouze, N., Prudhomme, M., Mortier-Barrière, I., Granadel, C.,et al. (2013). ComE/ComEP interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87, 394–411. doi: 10.1111/mmi.12104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McDougal, L. K., Tenover, F. C., Lee, L. N., Rasheed, J. K., Patterson, J. E., Jorgensen, J. H.,et al. (1998). Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42, 2312–2318.
McGee, L., McDougal, L., Zhou, J., Spratt, B. G., Tenover, F. C., George, R.,et al. (2001). Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39, 2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mingoia, M., Tili, E., Manso, E., Varaldo, P. E., and Montanari, M. P. (2011). Heterogeneity of Tn5253-like composite elements in clinical Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 55, 1453–1459. doi: 10.1128/AAC.01087-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mook-Kanamori, B. B., Geldhoff, M., van der Poll, T., and van de Beek, D. (2011). Pathogenesis and pathophysiology of pneumococcal meningitis. Clin. Microbiol. Rev. 24, 557–591. doi: 10.1128/CMR.00008-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mostowy, R., Croucher, N. J., Hanage, W. P., Harris, S. R., Bentley, S., and Fraser, C. (2014). Heterogeneity in the frequency and characteristics of homologous recombination in pneumococcal evolution. PLoS Genet. 10:e1004300. doi: 10.1371/journal.pgen.1004300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mullany, P., Roberts, A. P., and Wang, H. (2002). Mechanism of integration and excision in conjugative transposons. Cell. Mol. Life Sci. 59, 2017–2022. doi: 10.1007/s000180200001
Mullany, P., Wilks, M., and Tabaqchali, S. (1991). Transfer of Tn916 and Tn916 delta E into Clostridium difficile: demonstration of a hot-spot for these elements in the C. difficile genome. FEMS Microbiol. Lett. 63, 191–194.
Mullany, P., Williams, R., Langridge, G. C., Turner, D. J., Whalan, R., Clayton, C.,et al. (2012). Behavior and target site selection of conjugative transposon Tn916 in two different strains of toxigenic Clostridium difficile. Appl. Environ. Microbiol. 78, 2147–2153. doi: 10.1128/AEM.06193-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nabavizadeh, M. R., Sahebi, S., and Nadian, I. (2011). Antibiotic prescription for endodontic treatment: general dentist knowledge + practice in shiraz. Iran. Endod. J. 6, 54–59.
Nguyen, D. H., and Martin, J. T. (2008). Common dental infections in the primary care setting. Am. Fam. Physician 77, 797–802.
Nobbs, A. H., Lamont, R. J., and Jenkinson, H. F. (2009). Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450. doi: 10.1128/MMBR.00014-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Novais, C., Freitas, A. R., Silveira, E., Baquero, F., Peixe, L., Roberts, A. P.,et al. (2012a). A tet(S/M) hybrid from CTn6000 and CTn916 recombination. Microbiology 158, 2710–2711. doi: 10.1099/mic.0.062729-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Novais, C., Freitas, A. R., Silveira, E., Baquero, F., Peixe, L., Roberts, A. P.,et al. (2012b). Different genetic supports for the tet(S) gene in enterococci. Antimicrob. Agents Chemother. 56, 6014–6018. doi: 10.1128/AAC.00758-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ochiai, K., Kurita-Ochiai, T., Kamino, Y., and Ikeda, T. (1993). Effect of co-aggregation on the pathogenicity of oral bacteria. J. Med. Microbiol. 39, 183–190. doi: 10.1099/00222615-39-3-183
Oggioni, M. R., Trappetti, C., Kadioglu, A., Cassone, M., Iannelli, F., Ricci, S.,et al. (2006). Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 61, 1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olsen, I., Tribble, G. D., Fiehn, N. E., and Wang, B. Y. (2013). Bacterial sex in dental plaque. J. Oral Microbiol. 3, 5. doi: 10.3402/jom.v5i0.20736
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmieri, C., Mingoia, M., Massidda, O., Giovanetti, E., and Varaldo, P. E. (2012). Streptococcus pneumoniae transposon Tn1545/Tn6003 changes to Tn6002 due to spontaneous excision in circular form of the erm(B)- and aphA3-containing macrolide-aminoglycoside-streptothricin (MAS) element. Antimicrob. Agents Chemother. 56, 5994–5997. doi: 10.1128/AAC.01487-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pearce, C., Bowden, G. H., Evans, M., Fitzsimmons, S. P., Johnson, J., Sheridan, M. J.,et al. (1995). Identification of pioneer viridans streptococci in the oral cavity of human neonates. J. Med. Microbiol. 42, 67–72. doi: 10.1099/00222615-42-1-67
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peterson, S. N., Sung, C. K., Cline, R., Desai, B. V., Snesrud, E. C., Luo, P.,et al. (2004). Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51, 1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Provvedi, R., Manganelli, R., and Pozzi, G. (1996). Characterization of conjugative transposon Tn 5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 135, 231–236. doi: 10.1111/j.1574-6968.1996.tb07994.x
Prudhomme, M., Attaiech, L., Sanchez, G., Martin, B., and Claverys, J. P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92. doi: 10.1126/science.1127912
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Quintero, B., Araque, M., van der Gaast-de Jongh, C., and Hermans, P. W. (2011). Genetic diversity of Tn916-related transposons among drug-resistant Streptococcus pneumoniae isolates colonizing healthy children in Venezuela. Antimicrob. Agents Chemother. 55, 4930–4932. doi: 10.1128/AAC.00242-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ready, D., Pratten, J., Roberts, A. P., Bedi, R., Mullany, P., and Wilson, M. (2006). Potential role of Veillonella spp. as a reservoir of transferable tetracycline resistance in the oral cavity. Antimicrob. Agents Chemother. 50, 2866–2868. doi: 10.1128/AAC.00217-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., Chandler, M., Courvalin, P., Guédon, G., Mullany, P., Pembroke, T.,et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., Cheah, G., Ready, D., Pratten, J., Wilson, M., and Mullany, P. (2001a). Transfer of Tn916-like elements in microcosm dental plaques. Antimicrob. Agents Chemother. 45, 2943–2946. doi: 10.1128/AAC.45.10.2943-2946.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., Johanesen, P. A., Lyras, D., Mullany, P., and Rood, J. I. (2001b). Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147, 1243–1251.
Roberts, A. P., Davis, I. J., Seville, L., Villedieu, A., and Mullany, P. (2006). Characterization of the ends and target site of a novel tetracycline resistance-encoding conjugative transposon from Enterococcus faecium 664.1 H1. J.Bacteriol. 188, 4356–4361. doi: 10.1128/JB.00129-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., and Kreth, J. (2014). The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front. Cell. Infect. Microbiol. 4:124. doi: 10.3389/fcimb.2014.00124
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., and Mullany, P. (2006). Genetic basis of horizontal gene transfer among oral bacteria. Periodontol. 2000 42, 36–46. doi: 10.1111/j.1600-0757.2006.00149.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., and Mullany, P. (2009). A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17, 251–258. doi: 10.1016/j.tim.2009.03.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., and Mullany, P. (2010). Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev. Anti Infect. Ther. 8, 1441–1450. doi: 10.1586/eri.10.106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., and Mullany, P. (2011). Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 35, 856–871. doi: 10.1111/j.1574-6976.2011.00283.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roberts, A. P., Pratten, J., Wilson, M., and Mullany, P. (1999). Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol. Lett. 177, 63–66. doi: 10.1111/j.1574-6968.1999.tb13714.x
Robertson, D., and Smith, A. J. (2009). Microbiology of acute dental abscess. J. Med. Microbiol. 58, 155–162. doi: 10.1099/jmm.0.003517-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santoro, F., Oggioni, M. R., Pozzi, G., and Iannelli, F. (2010). Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn 5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 308, 150–158. doi: 10.1111/j.1574-6968.2010.02002.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scott, J. R., Bringel, F., Marra, D., van Alstine, G., and Rudy, C. K. (1994). Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11, 1099–1108. doi: 10.1111/j.1365-2958.1994.tb00386.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scott, J. R., Kirchman, P. A., and Caparon, M. G. (1988). An intermediate in transposition of the conjugative transposon Tn916. Proc. Natl. Acad. Sci. U.S.A. 85, 4809–4813. doi: 10.1073/pnas.85.13.4809
Seville, L. A., Patterson, A. J., Scott, K. P., Mullany, P., Quail, M. A., Parkhill, J.,et al. (2009). Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb. Drug Resist. 15, 159–166. doi: 10.1089/mdr.2009.0916
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shoemaker, N. B., Smith, M. D., and Guild, W. R. (1979). Organization and transfer of heterologous chloramphenicol and tetracycline resistance genes in pneumococcus. J. Bacteriol. 139, 432–441.
Siqueira, J. F. Jr., and Rôças, I. N. (2013). Microbiology and treatment of acute apical abscesses. Clin. Microbiol. Rev. 26, 255–273. doi: 10.1128/CMR.00082-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Starkebaum, M., Durack, D., and Beeson, P. (1977). The “incubation period” of subacute bacterial endocarditis. Yale J. Biol. Med. 50, 49–58.
Topazian, R. G., Goldberg, M. H., and Hupp, J. R. (eds). (2002). Oral, and Maxillofacial Infections, 4th Edn. Philadelphia, PA: WB Saunders Company.
Vianna, M. E., Horz, H. P., Conrads, G., Feres, M., and Gomes, B. P. (2008). Comparative analysis of endodontic pathogens using checkerboard hybridization in relation to culture. Oral Microbiol. Immunol. 23, 282–290. doi: 10.1111/j.1399-302X.2007.00425.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wade, W. G. (2013). The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143. doi: 10.1016/j.phrs.2012.11.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., and Mullany, P. (2000). The large resolvase TndX is required and sufficient for integration and excision of derivatives of the novel conjugative transposon Tn5397. J. Bacteriol. 182, 6577–6583. doi: 10.1128/JB.182.23.6577-6583.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Roberts, A. P., and Mullany, P. (2000). DNA sequence of the insertional hot spot of Tn916 in the Clostridium difficile genome and discovery of a Tn916-like element in an environmental isolate integrated in the same hot spot. FEMS Microbiol. Lett. 192, 15–20. doi: 10.1111/j.1574-6968.2000.tb09352.x
Warburton, P. J., Palmer, R. M., Munson, M. A., and Wade, W. G. (2007). Demonstration of in vivo transfer of doxycycline resistance mediated by a novel transposon. J. Antimicrob. Chemother. 60, 973–980. doi: 10.1093/jac/dkm331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watson, D. A., and Musher, D. M. (1990). Interruption of capsule production in Streptococcus pneumoniae serotype 3 by insertion of transposon Tn 916. Infect. Immun. 58, 3135–3138.
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295, 1487. doi: 10.1126/science.295.5559.1487
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilson, W., Taubert, K. A., Gewitz, M., Lockhart, P. B., Baddour, L. M., Levison, M.,et al. (2008). American Heart Association. Prevention of infective endocarditis: guidelines from the American Heart Association: a guideline from the American Heart Association Rheumatic Fever, Endocarditis and Kawasaki Disease Committee, Council on Cardiovascular Disease in the Young, and the Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and the Quality of Care and Outcomes Research Interdisciplinary Working Group. J. Am. Dent. Assoc. 139(Suppl.), 3S–24S. doi: 10.14219/jada.archive.2008.0346
Xu, X., Cai, L., Xiao, M., Kong, F., Oftadeh, S., Zhou, F.,et al. (2010). Distribution of serotypes, genotypes, and resistance determinants among macrolide-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 54, 1152–1159. doi: 10.1128/AAC.01268-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, L., Ma, X., Gao, W., Yao, K. H., Shen, A. D., Yu, S. J.,et al. (2012). Molecular characteristics of erythromycin-resistant Streptococcus pneumoniae from pediatric patients younger than five years in Beijing, 2010. BMC Microbiol. 12:228. doi: 10.1186/1471-2180-12-228
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, W., Yao, K., Zhang, G., Yang, Y., Li, Y., Lv, Y.,et al. (2014). Mechanism for transfer of transposon Tn 2010 carrying macrolide resistance genes in Streptococcus pneumoniae and its effects on genome evolution. J. Antimicrob. Chemother. 69, 1470–1473. doi: 10.1093/jac/dku019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Streptococcus, conjugative transposon, antibiotic resistance, nasopharyngeal, oral cavity, transformation, conjugation, horizontal gene transfer
Citation: Santoro F, Vianna ME and Roberts AP (2014) Variation on a theme; an overview of the Tn916/Tn1545 family of mobile genetic elements in the oral and nasopharyngeal streptococci. Front. Microbiol. 5:535. doi: 10.3389/fmicb.2014.00535
Received: 03 September 2014; Paper pending published: 23 September 2014;
Accepted: 25 September 2014; Published online: 20 October 2014.
Edited by:
Bruna Facinelli, Università Politecnica delle Marche, ItalyReviewed by:
Andrea Brenciani, Polytechnic University of Marche, ItalySophie Payot, Institut National de la Recherche Agronomique, France
Copyright © 2014 Santoro, Vianna and Roberts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adam P. Roberts, Department of Microbial Diseases, UCL Eastman Dental Institute, University College London, 256 Gray’s Inn Road, London WC1X 8LD, UK e-mail: adam.roberts@ucl.ac.uk
 Francesco Santoro
Francesco Santoro Morgana E. Vianna
Morgana E. Vianna Adam P. Roberts
Adam P. Roberts