- 1Cancer Center, School of Life Sciences, School of Advanced Computing, Cooperative Innovation Center for High Performance Computing, Sun Yat-sen University, Guangzhou, China
- 2Orthopaedic Department of Anhui Medical University Affiliated Provincial Hospital, Hefei, China
Eukaryotic cells may divide via the critical cellular process of cell division/mitosis, resulting in two daughter cells with the same genetic information. A large number of dedicated proteins are involved in this process and spatiotemporally assembled into three distinct super-complex structures/organelles, including the centrosome/spindle pole body, kinetochore/centromere and cleavage furrow/midbody/bud neck, so as to precisely modulate the cell division/mitosis events of chromosome alignment, chromosome segregation and cytokinesis in an orderly fashion. In recent years, many efforts have been made to identify the protein components and architecture of these subcellular organelles, aiming to uncover the organelle assembly pathways, determine the molecular mechanisms underlying the organelle functions, and thereby provide new therapeutic strategies for a variety of diseases. However, the organelles are highly dynamic structures, making it difficult to identify the entire components. Here, we review the current knowledge of the identified protein components governing the organization and functioning of organelles, especially in human and yeast cells, and discuss the multi-localized protein components mediating the communication between organelles during cell division.
Introduction
Cell division/mitosis is a precisely modulated process of chromosome segregation and nuclear division in which one eukaryotic cell divides into two daughter cells with identical chromosomes in order to produce more cells for growth and replace any damaged, dying or senescent cells (Sancar et al., 2004). Mitosis is always accompanied by a separation of the cell cytoplasm, known as cytokinesis, in which the daughter cells become completely separated (Wheatley et al., 2001; Straight et al., 2003). Mitosis (nuclear division) and cytokinesis (cytoplasmic division), which define the mitotic (M) phase, are the most crucial and fundamental activities of the eukaryotic cell cycle. Before entering the M phase of the cell cycle, the cell undergoes a period of growth and maturation during the interphase, duplicating genetic materials and organelles for the performance of cell division (Heun et al., 2001). The interphase and M phase of the cell cycle are complex and highly regulated by numerous proteins which are spatially and temporally organized as protein super-complexes. The super-complexes carry out chromosome replication and alignment, sister chromatid separation and cytoplasm division (Straight et al., 2003).
Chromosome must be precisely replicated once per cell cycle to maintain genome integrity. Eukaryotic cells may use multiple proteins, many of which are also involved in super-complex formation to regulate chromosome alignment, separation and cytoplasm division, to control the origins of chromosome replication. During the interphase, the origin recognition complex (ORC), a six-subunit complex comprised of ORC1-6, binds to chromosomes at the replication origin sites and acts as a central component for eukaryotic chromosome replication initiation (Bell and Dutta, 2002). As the initiation of replication is a central event in cell cycle, the identification of replication origin sites and its binding proteins is essential to the understanding of DNA replication. Benefit from recent genome-wide approaches, a huge number of replication origins and ORC proteins were identified. Also, several specialized databases, such as DeOri (Gao et al., 2012), have been developed to assist the comprehensive study on eukaryotic DNA replication. Over the years, new roles for many ORC proteins were revealed in cells. Unlike their regular function that controls the initiation of DNA replication, a fair amount of ORC proteins also binds to other cell cycle-related organelles, including centrosome, kinetochore and midbody. Evidences have shown that ORC1 and ORC2 can regulate centrosome duplication and a depletion of them resulted in abnormal centrosome copy number (Prasanth et al., 2004; Hemerly et al., 2009). Coincidentally, researches also demonstrated that ORC6 and ORC2 can localize to kinetochore. The absence of ORC proteins may lead to kinetochore dysfunction (Shimada and Gasser, 2007). Furthermore, in anaphase, the ORC6 may target to the midbody in controlling of chromosome segregation (Prasanth et al., 2002). In the process of chromosome replication, the enzymes that catalyze DNA duplication are unable to reach the very end of the chromosome. Chromosome has a special DNA structure named telomere at the end. Thus, in the course of each replication, the length of the telomeres is shortened (Von Zglinicki, 2002). Once the telomeres shrink to a critical minimum size, the cells no longer divide and ultimately become senescent or die (Hahn et al., 1999; Henson et al., 2002). However, telomerase, a unique protein-RNA complex that is activated in certain cells (Rudolph et al., 1999; Hanahan and Weinberg, 2000), such as yeast cells, stem cells, reproductive cells and cancer cells, is responsible for elongating telomeres (Herbert et al., 1999; Dunham et al., 2000). It thus prevents the chromosome degradation, maintains the stability of the genome and assists cells to escape the fate of being unable to continue division (Hoeijmakers, 2001).
In the M phase of the cell cycle, multiple proteins assemble in the three distinct regions of the centrosome/spindle pole body, kinetochore/centromere and cleavage furrow/midbody/bud neck, directing the process of cell division. The centrosomes in animal cells, spindle pole bodies (SPB) in budding yeast and related/homologous structures in other organisms have been characterized as the microtubule organizing centers (MTOCs) (Veith et al., 2005), which participate in the organization and orientation of the mitotic spindle apparatus, and thus direct the chromosome alignment and sister chromatids segregation during cell division. In addition, the kinetochore, a specialized protein complex which is dynamically assembled around the centromere of chromosomes (Ditchfield et al., 2003), acts as the “handle” of the chromosome and specifies the attachments between the chromosomes and spindle to ensure accurate chromosome segregation (Hauf et al., 2003). Dysfunction of the centrosome/spindle pole body and kinetochore/centromere is catastrophic for cells and contributes to aberrant division and chromosome instability (Fodde et al., 2001), both of which are hallmarks of cancer cells (Schuyler et al., 2012). The chromosome separation in animal cells is always accompanied by cytokinesis, which begins with ingression of the cleavage furrow mediated by the actomyosin ring (Somers and Saint, 2003), followed by the formation dense structure of the midbody (Gromley et al., 2005) which is also known as the phragmoplast in plants (Van Damme et al., 2004) and the bud neck in budding yeast (Vallen et al., 2000; Caviston et al., 2003). Numerous proteins are recruited to the midbody and form a super-complex which mediates the midbody abscission in order to perform cytokinesis, with complete separation of the two daughter cells (Adams et al., 2001; Wheatley et al., 2001; Mollinari et al., 2002).
Although the importance of organelles to cell biology has been repeatedly demonstrated by multiple reports over the past decades, many aspects of their function, structure and composition are still largely unknown. In this regard, comprehensive identification of the protein components of the super-complex structures will be one of the keys to understanding the mechanisms of chromosome segregation and cytokinesis, and may provide important clues for the discovery and validation of new therapeutic targets. Recently, many protein components have been identified, but according to a combination of proteomic analysis, biochemical studies and genetic screening, there still remain a large number of proteins that are predicted to be associated with these organelles (Table 1). In this review, we will present a general overview of the identified components of the super-complexes involved in the mitosis and cytokinesis with the aim of integrating the relevant information of organelles and thus broadening the knowledge of cell division. Remarkably, the process of the cell division is highly conserved in eukaryotic cells, we therefore briefly review the two commonly studied systems, human and yeast cells.
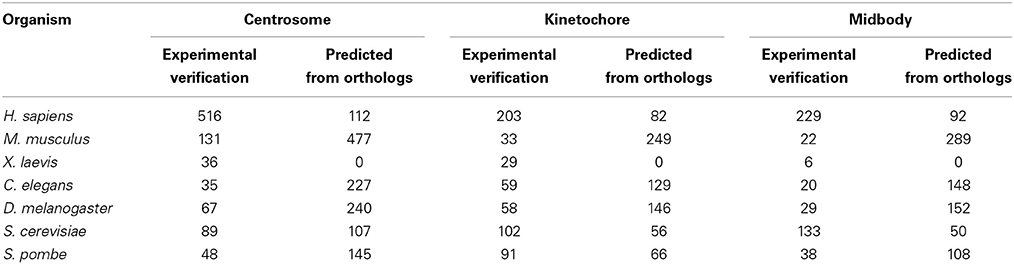
Table 1. The number of proteins located in centrosome, kinetochore, midbody with experimental verification and predicted in 7 different species from MiCroKiTS (Updated June 27, 2014).
The Centrosome
As a complex and dynamic organelle, the MTOC contributes to both microtubule organization and nucleation, which are important for chromosomes separation during mitosis (Brinkley, 1985; Luders and Stearns, 2007). Multiple proteins must be involved in manipulating MTOC functions, controlling its duplication and driving maturation. To further clarify the functional processes of the organization and regulation of the MTOC, the protein components must be identified. Recently, evidence from a combination of genetic and biochemical studies has revealed many important MTOC-associated proteins in a variety of species (Masuda et al., 2013). However, according to the MiCroKiTS database (Ren et al., 2010) (http://microkit.biocuckoo.org/), an integrated database of the midbody, centrosome and kinetochore most recently updated in June 27, 2014, a large number of proteins that are predicted to be located on the MTOC are still not well validated (Table 1). Confirmation of the functions of these predicted proteins has broad implications for the understanding of the MTOC.
The centrosome is the primary MTOC, which contains two orthogonally arranged centrioles and the surrounding pericentriolar material (PCM) (Nigg and Raff, 2009). The centriole, composed mainly of tublin, is a typically cylindrical organelle made up of nine triplets of microtubules in most animal eukaryotic cells (Kitagawa et al., 2011), although absent in most fungi and high plant cells (Bell and Dutta, 2002; Gao et al., 2012). In the G1 phase of the cell cycle, the paired centrioles, termed the mother and the daughter centrioles, are connected via interconnecting fibers. Morphologically distinct from the daughter centriole, the mother centriole has both distal and subdistal appendages that serve to anchor the centrioles to the plasma membrane (Bettencourt-Dias and Glover, 2009). Recently, several components of the centriole appendages have been described, such as the distal appendage proteins CEP164 and CEP89, as well as three novel components of CEP83, the Sodium channel and clathrin linker 1 (SCLT1) and the Fas-binding factor 1 (FBF1) (Tanos et al., 2013; Kloc et al., 2014). The subdistal appendage proteins include Outer dense fiber 2 (ODF2; also known as cenexin) (Chang et al., 2003), ninein (Graser et al., 2007), epsilon-tubulin (Chang et al., 2003), Centriolin (Gromley et al., 2003), and CC2D2A (Veleri et al., 2014). However, the molecular composition and the exact functions of the appendages remain largely unclear. The mechanisms underlying the assembly of the centriole are still poorly understood. In recent years, identification of the proteins that are responsible for centriole formation has advanced the understanding of the assembly mechanisms. In human cells, spindle assembly abnormal 6 (HsSAS-6), Polo-like kinase 4 (PLK4), SCL/TAL1 interrupting locus (STIL), centrosomal P4.1-associated protein (CPAP) (Brownlee and Rogers, 2013), located at the centriole, have been identified as the core components required for centriole assembly. Using proteomic and biochemical analysis as well as genetic screening, a list of the proteins associated with centriole, such as centrosomal protein of 135 kDa (CEP135), CEP152, CEP63, spindle and centriole-associated protein (SPICE), CP110, centrobin, CEP120, and CEP192 (Gonczy, 2012), are considered to govern the centriole assembly. The maintenance of a constant centriole number is critical for the progression of the cell cycle, and precisely controlled by numerous proteins which are involved in regulating centrosome duplication in the G1 and S phases, centrosome maturation in the G2/M phase and separation in the mitotic phase (Brownlee and Rogers, 2013). In human cells, PLK4, hsSAS-6 and STIL are three regulators necessary for centrosome duplication (Vulprecht et al., 2012). Following the activation of PLK4 and accumulation of STIL around the mother centriole, F-box protein FBXW5 stabilizes HsSAS-6 (Puklowski et al., 2011). In addition, several other proteins, such as CEP135, CPAP (Tang et al., 2009), γ-tubulin, CEP192, BRCA2, CP110 and its interaction protein USP33 (Li et al., 2013) that are essential for centrosome duplication, are recruited to the centriole, thus orchestrating centrosome duplication (Brownlee and Rogers, 2013). Additionally, cell cycle kinase CDK2 as well as potential partners of cyclin A and cyclin E are required for the two centrioles to split during centrosome duplication (Stearns, 2001). At the onset of mitosis, NEK2 and centrin are required for the sister centrosome disjunction as well as the formation of the two spindle poles during mitosis (Hinchcliffe and Sluder, 2001). Centrosome maturation is accompanied by the recruitment of many proteins to the centrioles and a dramatic expansion of the pericentriolar matrix (PCM) (Mennella et al., 2014). Phosphorylation is considered to be a key mechanism underlying centrosome maturation. The Polo-like kinases1 (PLK1) (Barr et al., 2004; Conduit et al., 2014) and Aurora kinases (Carmena and Earnshaw, 2003) have been identified as two important regulators of centrosome maturation. The specific phosphorylation of pericentrin (PCNT) by PLK1 results in the recruitment of many centrosomal proteins, such as γ-tubulin, Aurora A, PLK1, CEP192, and GCP-WD (γ-complex protein with WD repeats), to the centrosome during mitosis (Lee and Rhee, 2011). PCM is a matrix of proteins involved in centrosomal organization, microtubule nucleation and anchoring. The main components of PCM exist in the form of two proteins layers, one comprising a large number of coiled-coil proteins, such as pericentrin/pericentrin-like protein (PLP) and CEP152, with the other one including CEP215, γ-tubulin and CEP192 (Mennella et al., 2014). In PCM, γ-tubulin and other proteins such as γ tubulin complex protein (GCP) family can be assembled as γ-tubulin ring complexes (γ-TuRCs) for microtubule nucleation. The GCP family is also involved in the γ-TuRCs function, regulation and localization of γ-TuRCs (Kollman et al., 2011). The centrosomal protein pericentrin and the ninein-like protein (NLP) have been shown to anchor γ-TuRCs at the spindle poles (Zimmerman et al., 2004). Meanwhile, the precise components and regulators of γ-TuRCs remains incompletely understood.
Collectively, the identification of the structural and functional proteins of centrosome is clearly crucial for elucidating the structure of the centrosome and uncovering the underlying mechanisms in centrosome organization and regulation. Up to now, only a portion of the centrosome components have been detected (Tables S1, S2), and more efforts are required for the experimental validation of the remaining components. The centrosome in yeast cells is termed the spindle pole body (SPB), which is composed of a half-bridge for new SPB assembly, and three plaques, including an inner plaque for nuclear microtubules forms as the mitotic spindles originate, a central plaque spanning the nuclear membrane, and an outer plaque for cytoplasmic microtubules that used for karyogamy, nuclear positioning and spindle orientation (Seybold and Schiebel, 2013). The identified SPB proteins involved in the organization and regulation of SPB are listed in Table S2. However, there is still a large number of proteins located on the SPB that need to be further validated (Table 1).
The centrosome is a complex and precisely regulated organelle for bipolar spindle assembly, primary cilia formation, cell division and certain other cellular processes, including cell migration, protein degradation and axonal growth in human cells. More recent studies have shown that aberrant organization of centrosome resulting from defects in structural and functional proteins of the centrosome (Ganem et al., 2009; Nigg and Raff, 2009) is linked to neurodegenerative, Bardet–Biedl syndrome (Swaminathan, 2004), microcephaly (Marthiens et al., 2013), cystic kidney disease (Ong and Wheatley, 2003) and tumorigenesis (Marina and Saavedra, 2014). Thus, identification of the centrosomal proteins and clarification of the mechanisms underlying the centrosome assembly and regulation may lead to new drug targets, diagnostics or therapeutic approaches.
The Kinetochore
During mitosis in eukaryotic cells, a large number of proteins are assembled as a unique protein complex called the kinetochore, at the surface of the centromeric chromatin/centromere. The kinetochore functions as the binding site of the spindle microtubules to chromatin and directs sister chromatid segregation (Cheeseman and Desai, 2008). The protein components of the kinetochore modulate the connection between the centromeric chromatin and microtubules from the mitotic spindle to facilitate the proper segregation of the chromosomes during cell division (Gonen et al., 2012). According to the MiCroKiTS database and a comprehensive literature review (Cheeseman and Desai, 2008; Gonen et al., 2012), many kinetochore proteins have been identified in different species (Table 1). However, there are still a number of proteins localized at the kinetochore without any functional validation, as shown in the MiCroKiTS database.
The kinetochore is a complex and dynamic structure of variable size and shape. It is difficult to obtain the structural information on the complete kinetochore, so the structure is still not entirely clear. Previous studies have revealed that the overall positioning, main components and architecture of kinetochore are highly conserved from yeast to human (Quarmby and Parker, 2005). Many copies of centromeric proteins are assembled as a trilaminar kinetochore structure with the inner layer, a platform for kinetochore assembly that is located on the centromeric chromatin, the outer layer, responsible for the interaction with spindle microtubules, and the central layer, a region that links the inner and outer layers. In vertebrate cells, the inner layer consists of at least 18 centromeric proteins (Santaguida and Musacchio, 2009). Histone H3 variant centromeric protein A (CENP-A), also known as Cse4 in budding yeast, is one inner layer protein that may function as an early epigenetic marker for centromere localization and formation by making the centromeres distinct from the rest of the chromosome (Barnhart et al., 2011; Guse et al., 2011; Henikoff et al., 2014). CENP-A, together with CENP-B and CENP-C, are three main auto-antigens recognized by anticentromeric antibodies (Masumoto et al., 1989). Many other CENPs are also included in the inner layer, such as CENP-H, CENP-I, and CENP-K–W, all of which along with CENP-C colocalize with CENP-A and constitute the constitutive centromere-associated network (CCAN) (Cheeseman and Desai, 2008) (Table S3). Most of the components of the inner layer are evolutionarily conserved. They are responsible for keeping the kinetochore tethered to the centromere throughout the cell cycle and are essential for outer layer assembly (Carroll and Straight, 2006; Okada et al., 2006; Tanaka et al., 2009). The outer layer of the kinetochore is composed of several super-complexes, including Mis12, Ndc80 and Ska. The Mis12 complex provides the main platform for outer layer assembly, and consists of MIS12, NSL1, NNF1, and DSN1 (Screpanti et al., 2011). The Knl1 complex, which consists of KNL-1 and ZWINT, has been shown to recruit other outer layer proteins, such as spindle assembly checkpoint (SAC) proteins, CENP-F and the Rod–ZW10–Zwilch (RZZ) complex. The Ndc80 complex (NDC80, NUF2, SPC24, and SPC25) is one of the core binding sites of kinetochore-microtubules (kMTs) (Malvezzi et al., 2013). The Ska complex, composed of SKA1, SKA2, and SKA3, is essential for stabilizing kMT attachement (Welburn et al., 2009). In addition, the Knl1 complex, together with the Mis12 and Ndc80 complexes, forms the core of a highly conserved KMN network. This network is required for effective kMT attachment and force generation, and regulated by the Ska complex (the Dam/Dash complex in yeast) (Varma and Salmon, 2012). During mitosis, the components of the SAC, a mechanism that acts in response to unattached kinetochores, are recruited to the kinetochore monitor the correct kMT attachment by inhibiting the polyubiquitylation activities of the anaphase promoting complex (APC) (Peters, 2006). Several SAC components have been identified to date, including the non-kinase components Mad1, Mad2 and Bub3, the kinase components BubR1 (Mad3 in budding yeast), Bub1 and Mps1, the RZZ complex and other proteins (Lara-Gonzalez et al., 2012), as shown in Table S4. Among these components, Mad2 can interact with the APC activator of CDC20 to negatively regulate its function for the purpose of APC inhibition (Yu, 2002). In recent studies, several other mitotic protein kinases, including Aurora B (Chan et al., 2012) and PLK1 (Kang et al., 2006), PP2A phosphatase (Schmitz et al., 2010) and a number of nuclear pore proteins, including the Nup107–160 complex and SEH1 (D'angelo and Hetzer, 2008), have also been shown to transiently localize to the kinetochore during mitosis. They are involved in accurate segregation of chromosomes and controlling kinetochore function (Cheeseman and Desai, 2008), possibly by modulating checkpoint signaling. As the main structural features of kinetochores are conserved from yeast to human, the kinetochore also consists of the inner and outer layers in yeast, and the kMT attachment is regulated by numerous SAC proteins (Tables S3, S4). Among the components of the kinetochore found in yeast and human, the Ndc80 complex and some of the SAC proteins are highly conserved and exist in both species, indicating the importance of these proteins for correct chromosome segregation during cell division.
A combination of biochemical, fluorescence-microscopy and electron microscopy (EM) studies has led to the proposal of several structural models of the kinetochore with only weak supporting evidence. However, in recent studies, the first three-dimensional images of the kinetochore core structure have been obtained from budding yeast. These images show that the size of the kinetochore is approximately 126 nm, with a large central hub surrounded by multiple outer globular domains that form a ring-like structure around the microtubules (Gonen et al., 2012). This finding is important and extends the knowledge of the kinetochore. To further the understanding of the assembly process of the kinetochore and the mechanisms underlying chromosome segregation, additional kinetochore components and higher resolution images of kinetochore are needed to assist the elucidation of the structure and regulatory network. These are key elements in advancing our understanding of the mechanisms of the kinetochore-associated diseases, such as cancer, and may contribute to the development of early-stage clinical treatments (Gonen et al., 2012).
The Midbody
During cytokinesis, many proteins promote furrow ingression, dividing one cell into two daughter cells still connected by midbody, a cellular substructure contains many transient protein complexes formed at the narrow intracellular bridge (Steigemann and Gerlich, 2009). The midbody is generally considered to be an important structure for directing the abscission and completely separating the two daughter cells at the final stage of cytokinesis (Pohl and Jentsch, 2008). However, more functions of the midbody are still unclear. According to recent studies, the midbody may also be involved in cell-fate determination. Morphologically, the midbody is a dense structure formed by a tightly packed anti-parallel microtubule array, and many proteins are recruited to this site to assist in the cytokinesis process (Mullins and Biesele, 1977; Steigemann and Gerlich, 2009). However, the current knowledge of the midbody components and the way the midbody proteins are organized is limited. To further clarify the functions and the processes of assembly and regulation of the midbody, the primary task is to identify its protein components. Although there are approximately 229 proteins identified as being associated with the midbody in human cells, and 133 proteins in yeast cells (Table 1), there are still many remaining components that urgently need to be uncovered and validated.
Previous studies have shown that the midbody proteins are organized in three parts, the bulge, the dark zone and the flanking zone (Mullins and Biesele, 1977; Steigemann and Gerlich, 2009). The bulge is at the center of the midbody, containing few bundled anti-parallel microtubules and various proteins. In human cells, centralspindlin, a key component of the bulge, is a complex of the human GTPase-activating protein MgcRacGAP and Mitotic kinesin-like protein 1 (MKLP1). Centralspindlin is essential for the midbody formation and links the midbody to the plasma membrane. Many of the identified bulge proteins are associated with centralspindlin. The ADP-ribosylation factor 6 (ARF6) GTPase can interact with centralspindlin and may be respectively responsible for midbody stabilization (Joseph et al., 2012). The Rho guanine nucleotide exchange factor (RhoGEF) Ect2 is also a centralspindlin-interacting protein and localizes at the bulge to facilitate midbody abscission (Yuce et al., 2005). The coiled-coil protein centriolin, recruited to the midbody by centralspindlin, is important for integrating the process of membrane-vesicle fusion with abscission by interacting with the exocyst components and SNARE complexes (Gromley et al., 2005). Another centralspindlin binding protein is a centrosomal protein of 55 kDa (CEP55) that is persistently localized at the midbody bulge during cytokinesis. The tumor-susceptibility gene 101 (TSG101) has been observed at the bulge. TSG101 and another midbody protein called Alg2-interacting protein X (ALIX) are associated with CEP55, and are proposed to be responsible for recruiting ESCRT-III components to the dark zone and thus assisting with the midbody abscission (Morita et al., 2007; Lee et al., 2008; Elia et al., 2011). The dark zone is a narrow region in the center of the midbody where antiparallel microtubules overlap. The microtubule-associated protein regulator of cytokinesis 1 (PRC1), in association with a microtubule-based motor protein of kinesin superfamily protein member 4 (KIF4), colocalizes at the midbody dark zone and together they are essential for cytokinesis (Kurasawa et al., 2004). Wnt5a signaling is important for stabilization. In recent studies, Wnt receptor Frizzled 2 (FZD2), which has been observed in the dark zone and has a similar localization pattern as the ESCRT-III subunit of CHMP4B, may regulate ESCRT-III localization via a Wnt5a-mediated β-catenin-independent signaling pathway (Fumoto et al., 2012). The midbody flanking zone resides outside of the dark zone, containing multi-proteins (Hu et al., 2012), such as the negative cytokinesis-regulator of centromere protein E (CENPE) (Liu et al., 2006), mitotic kinesin-like protein 2 (MKLP2) that regulates the localization of the chromosomal passenger complex (CPC) during cytokinesis (Gruneberg et al., 2004), and a CPC subunit of the Aurora B kinase-mediated abscission checkpoint (Steigemann et al., 2009). In yeast cells, the bud neck, which is analogous to the midbody, is responsible for cytokinesis and abscission (Guertin et al., 2002). And the main components of the organism are evolutionarily conserved from yeast to vertebrate (Otegui et al., 2005).
Actually, many components of each substructure of the midbody and bud neck subregions listed in Tables S5, S6 display a dynamic localization pattern, but the detailed composition of midbody and bud neck is still not known. In human cells, the midbody contains secretory or membrane-trafficking proteins, actin-associated proteins, microtubule-associated proteins, kinases proteins, and other uncharacterized or other function proteins, involved in many processes, such as the cytoskeleton, lipid rafts and vesicle trafficking (Skop et al., 2004). In addition, recent studies indicate that the functions of the midbody are not only related to abscission, but also involved in patterning, morphogenesis and development during embryogenesis (Chai et al., 2012). The accumulation of midbodies has been shown to correlate with the pluripotency of stem cells and to increase the tumorigenicity of cancer cells, while in differentiated cells, the midbody is degraded through an autophagy pathway (Ettinger et al., 2011; Kuo et al., 2011; Schink and Stenmark, 2011). Thus, identification of the midbody components is essential for advancing our knowledge of midbody and cell-fate determination, and also for exploring new therapeutic strategies for midbody related diseases treatment, such as cancer.
Discussion
A large number of proteins have been shown to participate in the process of cell division and spatiotemporally assemble as super-complexes at defined subcellular localizations, such as kinetochores at the centromeric chromatin, the centrosome near the nucleus, and the midbody between two daughter cells. According to the MiCroKiTS database search, during cell division, there are a total of approximately 754 identified proteins localized at the organelles of the centrosome, kinetochore and midbody in Homo sapiens, and 278 in Saccharomyces cerevisiae (Figure 1). Despite the fact that the protein components of each organelle are recruited to a specific subcellular localization, some proteins exhibit multi-localization in various species. Collectively, there are approximately 165 proteins which have more than one subcellular localizations in Homo sapiens, while there are 41 proteins in S. cerevisiae.
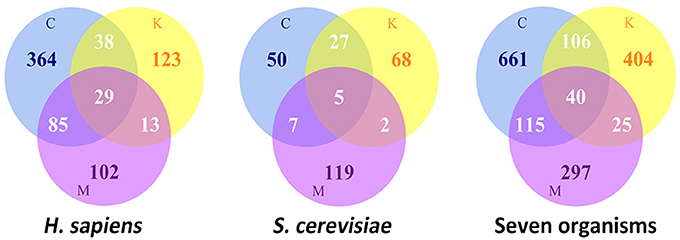
Figure 1. The statistics of Location distributions of MicroKiTS proteins. C refers to Centrosome. K refers to Kinetochore. M refers to Midbody. Seven organisms include of H. sapiens, S. cerevisiae, C. elegans, D. melanogaster, X. laevis, M. musculus, and S. prombe.
Proteins with multiple localizations are the key factors for mediating the communication between the organelles. In human cells, Ndc80 complex dynamically localizes at centrosome, and then concentrates at centromere and becomes a stable component of kinetochore until completion of the mitosis (Hori et al., 2003). Ndc80 complex is required for the stable kinetochore-spindle microtubule attachments, which controls the chromosome alignment and segregation in mitosis (Wei et al., 2005). The kinetochore protein components of INCENP (Cooke et al., 1987), CENP-A(Liu et al., 2013) and Aurora B (Kimura and Okano, 2005) for the chromosome biorientation, and the centrosome proteins of BARD1 (Ryser et al., 2009), BRCA2 (Daniels et al., 2004) and CEP55 (Fabbro et al., 2005), can be recruited to the midbody for the progression of cytokinesis. PLK1 (Cdc5 in yeast), a key mitotic regulator that phosphorylates substrate proteins on several different mitotic structures in human cells, first localizes at the centrosome before associating with kinetochore, and then is recruited to the midbody (Petronczki et al., 2008). The dynamic localization of PLK1, mediated by the polo-box domain (PBD) and kinase activity, is critical for chromosome alignment, spindle assembly and cytokinesis (Petronczki et al., 2008; Liu et al., 2012). A ubiquitin-ligase complex of APC and the HECT E3 ligase Smurf2, both of which control the progression of mitosis and cytokinesis through ubiquitin modification of substrate proteins and thus altering the protein localization and stability, have also been found to be dynamically localized at the centrosome, kinetochore and midbody (Kurasawa and Todokoro, 1999; Osmundson et al., 2008). In yeast cells, 5 proteins, including Cdc5 (Snead et al., 2007), protein phosphatase 2A regulatory subunit RTS1 (Gentry and Hallberg, 2002) and TPD3 (Gentry and Hallberg, 2002), Casein kinase I homolog HRR25 (Lusk et al., 2007) and protein phosphatase PP1-2 (Bloecher and Tatchell, 2000), are spatiotemporally recruited to the SPB, kinetochore and bud neck, and precisely regulate the cell division progression by altering the phosphorylation state of the substrates proteins. The subcellular localization determines the biological activities of multi-localized proteins through controlling the access of these proteins to different interaction partners, and is critical for the formation of the dynamic protein-protein interaction network to govern the process of the cell division/mitosis. Meanwhile, the posttranslational modifications (PTMs), including phosphorylation and ubiquitylation, as well as altering of the subcellular localizations, are essential mechanisms used by multi-localized proteins to diversify function and regulate cell division. A latest analysis of the dynamics of proteome and phosphoproteome during the cell division of the fission yeast revealed that changes of proteome level are weak, whereas changes of protein phosphorylation states are the predominant events occurred in mitosis, indicating that phosphorylation is probably associated with the functions and localizations of the proteins, which are involved in regulating mitotic progression and completion(Carpy et al., 2014). Additionally, the progresses in proteome-wide analysis of ubiquitination modifications in cell division demonstrated that ubiquitination, which affect protein stability, activity, and localization, plays an important role in regulating the mitotic progression (Chuang et al., 2010; Merbl et al., 2013). Certainly, the current understanding of the mechanisms used by multi-localized proteins to dynamically control the formation and functions of subcellular structures is still limited. Future studies are needed to identify the components of the subcellular structures as well as the multi-localized proteins, and also to characterize their functions, on–off mechanisms and crosstalk.
Author's Contributions
Yueyuan zheng, Junjie Guo, and Xu Li contribute to the literature collection and help to draft the manuscript. Mingming Hou, Yubin Xie, Xuyang Fu, Shengkun Dai, and Rucheng Diao participate in drafting the manuscript and revising it critically for important intellectual content. Yanyan Miao writes the manscript and interprets the data. Jian Ren contributes to the conception and design of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Basic Research Program (973 project) [2013CB933900, 2012CB911201]; National Natural Science Foundation of China [31471252, 81201383]; Guangdong Natural Science Funds for Distinguished Young Scholar [S20120011335]; Zhujiang Nova Program of Guangzhou [2011J2200042]; Program of International S&T Cooperation [2014DFB30020]; Fundamental Research Funds for the Central Universities [14lgjc14, 14lgpy02]; Program for New Century Excellent Talents in University [NCET-13-0610]; China Postdoctoral Science Foundation [2014M562238].
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00573/abstract
References
Adams, R. R., Maiato, H., Earnshaw, W. C., and Carmena, M. (2001). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865–879. doi: 10.1083/jcb.153.4.865
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barnhart, M. C., Kuich, P. H. J. L., Stellfox, M. E., Ward, J. A., Bassett, E. A., Black, B. E., et al. (2011). HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194, 229–243. doi: 10.1083/jcb.201012017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barr, F. A., Sillje, H. H., and Nigg, E. A. (2004). Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440. doi: 10.1038/nrm1401
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bell, S. P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. doi: 10.1146/annurev.biochem.71.110601.135425
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bettencourt-Dias, M., and Glover, D. M. (2009). SnapShot: centriole biogenesis. Cell 136, 188–188.e1. doi: 10.1016/j.cell.2008.12.035
Bloecher, A., and Tatchell, K. (2000). Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 149, 125–140. doi: 10.1083/jcb.149.1.125
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brinkley, B. R. (1985). Microtubule organizing centers. Annu. Rev. Cell Biol. 1, 145–172. doi: 10.1146/annurev.cb.01.110185.001045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brownlee, C. W., and Rogers, G. C. (2013). Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification. Cell. Mol. Life Sci. 70, 1021–1034. doi: 10.1007/s00018-012-1102-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carmena, M., and Earnshaw, W. C. (2003). The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854. doi: 10.1038/nrm1245
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carpy, A., Krug, K., Graf, S., Koch, A., Popic, S., Hauf, S., et al. (2014). Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast). Mol. Cell. Proteomics 13, 1925–1936. doi: 10.1074/mcp.M113.035824
Carroll, C. W., and Straight, A. F. (2006). Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 16, 70–78. doi: 10.1016/j.tcb.2005.12.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Caviston, J. P., Longtine, M., Pringle, J. R., and Bi, E. (2003). The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066. doi: 10.1091/mbc.E03-04-0247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chai, Y., Tian, D., Yang, Y., Feng, G., Cheng, Z., Li, W., et al. (2012). Apoptotic regulators promote cytokinetic midbody degradation in C. elegans. J. Cell Biol. 199, 1047–1055. doi: 10.1083/jcb.201209050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chan, Y. W., Jeyaprakash, A. A., Nigg, E. A., and Santamaria, A. (2012). Aurora B controls kinetochore–microtubule attachments by inhibiting Ska complex–KMN network interaction. J. Cell Biol. 196, 563–571. doi: 10.1083/jcb.201109001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P., Giddings, T. H. Jr., Winey, M., and Stearns, T. (2003). Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat. Cell Biol. 5, 71–76. doi: 10.1038/ncb900
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheeseman, I. M., and Desai, A. (2008). Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46. doi: 10.1038/nrm2310
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chuang, C., Lin, S. H., Huang, F., Pan, J., Josic, D., and Yu-Lee, L. Y. (2010). Acetylation of RNA processing proteins and cell cycle proteins in mitosis. J. Proteome Res. 9, 4554–4564. doi: 10.1021/pr100281h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conduit, P. T., Feng, Z., Richens, J. H., Baumbach, J., Wainman, A., Bakshi, S. D., et al. (2014). The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell 28, 659–669. doi: 10.1016/j.devcel.2014.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cooke, C. A., Heck, M. M., and Earnshaw, W. C. (1987). The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105, 2053–2067. doi: 10.1083/jcb.105.5.2053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
D'angelo, M. A., and Hetzer, M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466. doi: 10.1016/j.tcb.2008.07.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Daniels, M. J., Wang, Y., Lee, M., and Venkitaraman, A. R. (2004). Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science 306, 876–879. doi: 10.1126/science.1102574
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ditchfield, C., Johnson, V. L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., et al. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280. doi: 10.1083/jcb.200208091
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dunham, M. A., Neumann, A. A., Fasching, C. L., and Reddel, R. R. (2000). Telomere maintenance by recombination in human cells. Nat. Genet. 26, 447–450. doi: 10.1038/82586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elia, N., Sougrat, R., Spurlin, T. A., Hurley, J. H., and Lippincott-Schwartz, J. (2011). Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U.S.A. 108, 4846–4851. doi: 10.1073/pnas.1102714108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ettinger, A. W., Wilsch-Brauninger, M., Marzesco, A. M., Bickle, M., Lohmann, A., Maliga, Z., et al. (2011). Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat. Commun. 2, 503. doi: 10.1038/ncomms1511
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fabbro, M., Zhou, B. B., Takahashi, M., Sarcevic, B., Lal, P., Graham, M. E., et al. (2005). Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 9, 477–488. doi: 10.1016/j.devcel.2005.09.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fodde, R., Kuipers, J., Rosenberg, C., Smits, R., Kielman, M., Gaspar, C., et al. (2001). Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 3, 433–438. doi: 10.1038/35070129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fumoto, K., Kikuchi, K., Gon, H., and Kikuchi, A. (2012). Wnt5a signaling controls cytokinesis by correctly positioning ESCRT-III at the midbody. J. Cell Sci. 125, 4822–4832. doi: 10.1242/jcs.108142
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ganem, N. J., Godinho, S. A., and Pellman, D. (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. doi: 10.1038/nature08136
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, F., Luo, H., and Zhang, C. T. (2012). DeOri: a database of eukaryotic DNA replication origins. Bioinformatics 28, 1551–1552. doi: 10.1093/bioinformatics/bts151
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gentry, M. S., and Hallberg, R. L. (2002). Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol. Biol. Cell 13, 3477–3492. doi: 10.1091/mbc.02-05-0065
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gonczy, P. (2012). Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13, 425–435. doi: 10.1038/nrm3373
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gonen, S., Akiyoshi, B., Iadanza, M. G., Shi, D., Duggan, N., Biggins, S., et al. (2012). The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 19, 925–929. doi: 10.1038/nsmb.2358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Graser, S., Stierhof, Y. D., Lavoie, S. B., Gassner, O. S., Lamla, S., Le Clech, M., et al. (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330. doi: 10.1083/jcb.200707181
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gromley, A., Jurczyk, A., Sillibourne, J., Halilovic, E., Mogensen, M., Groisman, I., et al. (2003). A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161, 535–545. doi: 10.1083/jcb.200301105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gromley, A., Yeaman, C., Rosa, J., Redick, S., Chen, C. T., Mirabelle, S., et al. (2005). Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75–87. doi: 10.1016/j.cell.2005.07.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gruneberg, U., Neef, R., Honda, R., Nigg, E. A., and Barr, F. A. (2004). Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166, 167–172. doi: 10.1083/jcb.200403084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guertin, D. A., Trautmann, S., and McCollum, D. (2002). Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66, 155–178. doi: 10.1128/MMBR.66.2.155-178.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guse, A., Carroll, C. W., Moree, B., Fuller, C. J., and Straight, A. F. (2011). In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–U136. doi: 10.1038/nature10379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hahn, W. C., Stewart, S. A., Brooks, M. W., York, S. G., Eaton, E., Kurachi, A., et al. (1999). Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 5, 1164–1170. doi: 10.1038/13495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanahan, D., and Weinberg, R. A. (2000). The hallmarks of cancer. Cell 100, 57–70. doi: 10.1016/S0092-8674(00)81683-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hauf, S., Cole, R. W., Laterra, S., Zimmer, C., Schnapp, G., Walter, R., et al. (2003). The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294. doi: 10.1083/jcb.200208092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hemerly, A. S., Prasanth, S. G., Siddiqui, K., and Stillman, B. (2009). Orc1 controls centriole and centrosome copy number in human cells. Science 323, 789–793. doi: 10.1126/science.1166745
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henikoff, S., Ramachandran, S., Krassovsky, K., Bryson, T. D., Codomo, C. A., Brogaard, K., et al. (2014). The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. Elife 3:e01861. doi: 10.7554/eLife.01861
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henson, J. D., Neumann, A. A., Yeager, T. R., and Reddel, R. R. (2002). Alternative lengthening of telomeres in mammalian cells. Oncogene 21, 598–610. doi: 10.1038/sj.onc.1205058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herbert, B. S., Pitts, A. E., Baker, S. I., Hamilton, S. E., Wright, W. E., Shay, J. W., et al. (1999). Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. U.S.A. 96, 14276–14281. doi: 10.1073/pnas.96.25.14276
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heun, P., Laroche, T., Shimada, K., Furrer, P., and Gasser, S. M. (2001). Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181–2186. doi: 10.1126/science.1065366
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hinchcliffe, E. H., and Sluder, G. (2001). “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15, 1167–1181. doi: 10.1101/gad.894001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoeijmakers, J. H. J. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374. doi: 10.1038/35077232
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hori, T., Haraguchi, T., Hiraoka, Y., Kimura, H., and Fukagawa, T. (2003). Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116, 3347–3362. doi: 10.1242/jcs.00645
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, C. K., Coughlin, M., and Mitchison, T. J. (2012). Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell 23, 1024–1034. doi: 10.1091/mbc.E11-08-0721
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joseph, N., Hutterer, A., Poser, I., and Mishima, M. (2012). ARF6 GTPase protects the post-mitotic midbody from 14-3-3-mediated disintegration. EMBO J. 31, 2604–2614. doi: 10.1038/emboj.2012.139
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kang, Y. H., Park, J.-E., Yu, L.-R., Soung, N.-K., Yun, S.-M., Bang, J. K., et al. (2006). Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell 24, 409–422. doi: 10.1016/j.molcel.2006.10.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kitagawa, D., Vakonakis, I., Olieric, N., Hilbert, M., Keller, D., Olieric, V., et al. (2011). Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375. doi: 10.1016/j.cell.2011.01.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloc, M., Kubiak, J. Z., Li, X. C., and Ghobrial, R. M. (2014). The newly found functions of MTOC in immunological response. J. Leukoc. Biol. 95, 417–430. doi: 10.1189/jlb.0813468
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kollman, J. M., Merdes, A., Mourey, L., and Agard, D. A. (2011). Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721. doi: 10.1038/nrm3209
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuo, T. C., Chen, C. T., Baron, D., Onder, T. T., Loewer, S., Almeida, S., et al. (2011). Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13, 1214–1223. doi: 10.1038/ncb2332
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kurasawa, Y., Earnshaw, W. C., Mochizuki, Y., Dohmae, N., and Todokoro, K. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248. doi: 10.1038/sj.emboj.7600347
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kurasawa, Y., and Todokoro, K. (1999). Identification of human APC10/Doc1 as a subunit of anaphase promoting complex. Oncogene 18, 5131–5137. doi: 10.1038/sj.onc.1203133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lara-Gonzalez, P., Westhorpe, F. G., and Taylor, S. S. (2012). The spindle assembly checkpoint. Curr. Biol. 22, R966–R980. doi: 10.1016/j.cub.2012.10.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, H. H., Elia, N., Ghirlando, R., Lippincott-Schwartz, J., and Hurley, J. H. (2008). Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 322, 576–580. doi: 10.1126/science.1162042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, K., and Rhee, K. (2011). PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 195, 1093–1101. doi: 10.1083/jcb.201106093
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, J., D'angiolella, V., Seeley, E. S., Kim, S., Kobayashi, T., Fu, W., et al. (2013). USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 495, 255–259. doi: 10.1038/nature11941
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, D., Davydenko, O., and Lampson, M. A. (2012). Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J. Cell Biol. 198, 491–499. doi: 10.1083/jcb.201205090
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, D., Zhang, N., Du, J., Cai, X., Zhu, M., Jin, C., et al. (2006). Interaction of Skp1 with CENP-E at the midbody is essential for cytokinesis. Biochem. Biophys. Res. Commun. 345, 394–402. doi: 10.1016/j.bbrc.2006.04.062
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, R. M., Tian, X. Y., Huang, X. T., and Zhou, H. (2013). Transfer of Ser7 phosphorylated CENP-A from centromere to midbody during mitosis in MCF-7 cells. Folia Biol. (Praha.) 59, 105–109.
Luders, J., and Stearns, T. (2007). Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167. doi: 10.1038/nrm2100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lusk, C. P., Waller, D. D., Makhnevych, T., Dienemann, A., Whiteway, M., Thomas, D. Y., et al. (2007). Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic 8, 647–660. doi: 10.1111/j.1600-0854.2007.00559.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malvezzi, F., Litos, G., Schleiffer, A., Heuck, A., Mechtler, K., Clausen, T., et al. (2013). A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32, 409–423. doi: 10.1038/emboj.2012.356
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marina, M., and Saavedra, H. I. (2014). Nek2 and Plk4: prognostic markers, drivers of breast tumorigenesis and drug resistance. Front. Biosci. (Landmark Ed.) 19, 352–365. doi: 10.2741/4212
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marthiens, V., Rujano, M. A., Pennetier, C., Tessier, S., Paul-Gilloteaux, P., and Basto, R. (2013). Centrosome amplification causes microcephaly. Nat. Cell Biol. 15, 731–740. doi: 10.1038/ncb2746
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Masuda, H., Mori, R., Yukawa, M., and Toda, T. (2013). Fission yeast MOZART1/Mzt1 is an essential gamma-tubulin complex component required for complex recruitment to the microtubule organizing center, but not its assembly. Mol. Biol. Cell 24, 2894–2906. doi: 10.1091/mbc.E13-05-0235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Masumoto, H., Masukata, H., Muro, Y., Nozaki, N., and Okazaki, T. (1989). A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973. doi: 10.1083/jcb.109.5.1963
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mennella, V., Agard, D. A., Huang, B., and Pelletier, L. (2014). Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 24, 188–197. doi: 10.1016/j.tcb.2013.10.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merbl, Y., Refour, P., Patel, H., Springer, M., and Kirschner, M. W. (2013). Profiling of ubiquitin-like modifications reveals features of mitotic control. Cell 152, 1160–1172. doi: 10.1016/j.cell.2013.02.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mollinari, C., Kleman, J. P., Jiang, W., Schoehn, G., Hunter, T., and Margolis, R. L. (2002). PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157, 1175–1186. doi: 10.1083/jcb.200111052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, E., Sandrin, V., Chung, H. Y., Morham, S. G., Gygi, S. P., Rodesch, C. K., et al. (2007). Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227. doi: 10.1038/sj.emboj.7601850
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mullins, J. M., and Biesele, J. J. (1977). Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 73, 672–684. doi: 10.1083/jcb.73.3.672
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nigg, E. A., and Raff, J. W. (2009). Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678. doi: 10.1016/j.cell.2009.10.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okada, M., Cheeseman, I. M., Hori, T., Okawa, K., McLeod, I. X., Yates, J. R. 3rd., et al. (2006). The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446–457. doi: 10.1038/ncb1396
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ong, A. C., and Wheatley, D. N. (2003). Polycystic kidney disease–the ciliary connection. Lancet 361, 774–776. doi: 10.1016/S0140-6736(03)12662-1
Osmundson, E. C., Ray, D., Moore, F. E., Gao, Q., Thomsen, G. H., and Kiyokawa, H. (2008). The HECT E3 ligase Smurf2 is required for Mad2-dependent spindle assembly checkpoint. J. Cell Biol. 183, 267–277. doi: 10.1083/jcb.200801049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Otegui, M. S., Verbrugghe, K. J., and Skop, A. R. (2005). Midbodies and phragmoplasts: analogous structures involved in cytokinesis. Trends Cell Biol. 15, 404–413. doi: 10.1016/j.tcb.2005.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peters, J. M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656. doi: 10.1038/nrm1988
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Petronczki, M., Lenart, P., and Peters, J. M. (2008). Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev. Cell 14, 646–659. doi: 10.1016/j.devcel.2008.04.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pohl, C., and Jentsch, S. (2008). Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell 132, 832–845. doi: 10.1016/j.cell.2008.01.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prasanth, S. G., Prasanth, K. V., Siddiqui, K., Spector, D. L., and Stillman, B. (2004). Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23, 2651–2663. doi: 10.1038/sj.emboj.7600255
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prasanth, S. G., Prasanth, K. V., and Stillman, B. (2002). Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297, 1026–1031. doi: 10.1126/science.1072802
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Puklowski, A., Homsi, Y., Keller, D., May, M., Chauhan, S., Kossatz, U., et al. (2011). The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13, 1004–1009. doi: 10.1038/ncb2282
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Quarmby, L. M., and Parker, J. D. (2005). Cilia and the cell cycle? J. Cell Biol. 169, 707–710. doi: 10.1083/jcb.200503053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ren, J., Liu, Z., Gao, X., Jin, C., Ye, M., Zou, H., et al. (2010). MiCroKit 3.0: an integrated database of midbody, centrosome and kinetochore. Nucleic Acids Res. 38, D155–D160. doi: 10.1093/nar/gkp784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rudolph, K. L., Chang, S., Lee, H. W., Blasco, M., Gottlieb, G. J., Greider, C., et al. (1999). Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712. doi: 10.1016/S0092-8674(00)80580-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ryser, S., Dizin, E., Jefford, C. E., Delaval, B., Gagos, S., Christodoulidou, A., et al. (2009). Distinct roles of BARD1 isoforms in mitosis: full-length BARD1 mediates Aurora B degradation, cancer-associated BARD1beta scaffolds Aurora B and BRCA2. Cancer Res. 69, 1125–1134. doi: 10.1158/0008-5472.CAN-08-2134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K., and Linn, S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85. doi: 10.1146/annurev.biochem.73.011303.073723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santaguida, S., and Musacchio, A. (2009). The life and miracles of kinetochores. EMBO J. 28, 2511–2531. doi: 10.1038/emboj.2009.173
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schink, K. O., and Stenmark, H. (2011). Cell differentiation: midbody remnants - junk or fate factors? Curr. Biol. 21, R958–960. doi: 10.1016/j.cub.2011.10.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmitz, M. H., Held, M., Janssens, V., Hutchins, J. R., Hudecz, O., Ivanova, E., et al. (2010). Live-cell imaging RNAi screen identifies PP2A-B55 [alpha] and importin-[beta] 1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12, 886–893. doi: 10.1038/ncb2092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schuyler, S. C., Wu, Y. F., and Kuan, V. J. W. (2012). The Mad1-Mad2 balancing act - a damaged spindle checkpoint in chromosome instability and cancer. J. Cell Sci. 125, 4197–4206. doi: 10.1242/jcs.107037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Screpanti, E., De Antoni, A., Alushin, G. M., Petrovic, A., Melis, T., Nogales, E., et al. (2011). Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391–398. doi: 10.1016/j.cub.2010.12.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seybold, C., and Schiebel, E. (2013). Spindle pole bodies. Curr. Biol. 23, R858–R860. doi: 10.1016/j.cub.2013.07.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shimada, K., and Gasser, S. M. (2007). The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell 128, 85–99. doi: 10.1016/j.cell.2006.11.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Skop, A. R., Liu, H., Yates, J. 3rd., Meyer, B. J., and Heald, R. (2004). Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305, 61–66. doi: 10.1126/science.1097931
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Snead, J. L., Sullivan, M., Lowery, D. M., Cohen, M. S., Zhang, C., Randle, D. H., et al. (2007). A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem. Biol. 14, 1261–1272. doi: 10.1016/j.chembiol.2007.09.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Somers, W. G., and Saint, R. (2003). A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubles at the onset of cytokinesis. Dev. Cell 4, 29–39. doi: 10.1016/S1534-5807(02)00402-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stearns, T. (2001). Centrosome duplication. a centriolar pas de deux. Cell 105, 417–420. doi: 10.1016/S0092-8674(01)00366-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Steigemann, P., and Gerlich, D. W. (2009). Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 19, 606–616. doi: 10.1016/j.tcb.2009.07.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Steigemann, P., Wurzenberger, C., Schmitz, M. H., Held, M., Guizetti, J., Maar, S., et al. (2009). Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484. doi: 10.1016/j.cell.2008.12.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Straight, A. F., Cheung, A., Limouze, J., Chen, I., Westwood, N. J., Sellers, J. R., et al. (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 299, 1743–1747. doi: 10.1126/science.1081412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Swaminathan, S. (2004). Human disease: the centrosome connection. Nat. Cell Biol. 6, 383. doi: 10.1038/ncb0504-383
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tanaka, K., Li Chang, H., Kagami, A., and Watanabe, Y. (2009). CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343. doi: 10.1016/j.devcel.2009.08.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tang, C. J. C., Fu, R. H., Wu, K. S., Hsu, W. B., and Tang, T. K. (2009). CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11, 825–U103. doi: 10.1038/ncb1889
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tanos, B. E., Yang, H. J., Soni, R., Wang, W. J., Macaluso, F. P., Asara, J. M., et al. (2013). Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168. doi: 10.1101/gad.207043.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vallen, E. A., Caviston, J., and Bi, E. (2000). Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 593–611. doi: 10.1091/mbc.11.2.593
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Damme, D., Bouget, F. Y., Van Poucke, K., Inze, D., and Geelen, D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 40, 386–398. doi: 10.1111/j.1365-313X.2004.02222.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varma, D., and Salmon, E. D. (2012). The KMN protein network–chief conductors of the kinetochore orchestra. J. Cell Sci. 125, 5927–5936. doi: 10.1242/jcs.093724
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Veith, D., Scherr, N., Efimov, V. P., and Fischer, R. (2005). Role of the spindle-pole-body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans. J. Cell Sci. 118, 3705–3716. doi: 10.1242/jcs.02501
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Veleri, S., Manjunath, S. H., Fariss, R. N., May-Simera, H., Brooks, M., Foskett, T. A., et al. (2014). Ciliopathy-associated gene Cc2d2a promotes assembly of subdistal appendages on the mother centriole during cilia biogenesis. Nat. Commun. 5, 4207. doi: 10.1038/ncomms5207
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Von Zglinicki, T. (2002). Oxidative stress shortens telomeres. Trends Biochem. Sci. 27, 339–344. doi: 10.1016/S0968-0004(02)02110-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vulprecht, J., David, A., Tibelius, A., Castiel, A., Konotop, G., Liu, F., et al. (2012). STIL is required for centriole duplication in human cells. J. Cell Sci. 125, 1353–1362. doi: 10.1242/jcs.104109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wei, R. R., Sorger, P. K., and Harrison, S. C. (2005). Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. U.S.A. 102, 5363–5367. doi: 10.1073/pnas.0501168102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Welburn, J. P., Grishchuk, E. L., Backer, C. B., Wilson-Kubalek, E. M., Yates, J. R. 3rd., and Cheeseman, I. M. (2009). The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell 16, 374–385. doi: 10.1016/j.devcel.2009.01.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wheatley, S. P., Carvalho, A., Vagnarelli, P., and Earnshaw, W. C. (2001). INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 11, 886–890. doi: 10.1016/S0960-9822(01)00238-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, H. (2002). Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14, 706–714. doi: 10.1016/S0955-0674(02)00382-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yuce, O., Piekny, A., and Glotzer, M. (2005). An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170, 571–582. doi: 10.1083/jcb.200501097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zimmerman, W. C., Sillibourne, J., Rosa, J., and Doxsey, S. J. (2004). Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657. doi: 10.1091/mbc.E03-11-0796
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: super-complex structures, cell division/mitosis, protein components, centrosome, kinetochore, midbody
Citation: Zheng Y, Guo J, Li X, Xie Y, Hou M, Fu X, Dai S, Diao R, Miao Y and Ren J (2014) An integrated overview of spatiotemporal organization and regulation in mitosis in terms of the proteins in the functional supercomplexes. Front. Microbiol. 5:573. doi: 10.3389/fmicb.2014.00573
Received: 05 September 2014; Accepted: 11 October 2014;
Published online: 29 October 2014.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Zhixiang Zuo, University of Chicago, USAZiding Zhang, China Agricultural University, China
Penghui Zhou, Dana Farber Cancer Institute, USA
Copyright © 2014 Zheng, Guo, Li, Xie, Hou, Fu, Dai, Diao, Miao and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Miao and Jian Ren, Cancer Center, School of Life Sciences, School of Advanced Computing, Cooperative Innovation Center for High Performance Computing, Sun Yat-sen University, 135 West Xingang Road, Guangzhou 510275, China e-mail: myy@mail.ustc.edu.cn; renjian.sysu@gmail.com
†The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
 Yueyuan Zheng
Yueyuan Zheng Junjie Guo1†
Junjie Guo1† Yubin Xie
Yubin Xie Xuyang Fu
Xuyang Fu Shengkun Dai
Shengkun Dai Rucheng Diao
Rucheng Diao Yanyan Miao
Yanyan Miao Jian Ren
Jian Ren