- 1Laboratory of Molecular Microbiology and Biotechnology, Department of Medical Biotechnologies, University of Siena, Siena, Italy
- 2Section of Microbiology, Department of Biomedical and Biotechnological Sciences, University of Catania, Catania, Italy
Genetic element Φ1207.3 (formerly Tn1207.3) is a prophage of Streptococcus pyogenes which carries the macrolide efflux resistance genes mef(A)/msr(D) and is capable of conjugal transfer among streptococci. Complete nucleotide sequence showed that Φ1207.3 is 52,491 bp in length and contained 58 open reading frames (ORFs). A manual homology-based annotation with functional prediction of the hypothetical gene product was possible only for 34 out of 58 ORFs. Φ1207.3 codes for two different C-methylation systems, several phage structural genes, a lysis cassette (composed by a holin and a peptidoglycan hydrolase), and three site-specific resolvases of the serine recombinase family.
Introduction
In Streptococcus pyogenes, the mef (A)/msr(D) pair of genes encoding efflux resistance to 14- and 15-membered macrolides is carried by a mobile genetic element originally described as a conjugative transposon which was called Tn1207.3 (Santagati et al., 2003; Pozzi et al., 2004). This element was found to be 52.5 kb in size, and to contain a complete copy of Tn1207.1, a 7,244-bp defective transposon previously found to carry mef(A)/msr(D) in S. pneumoniae (Santagati et al., 2000). Integration of the element into the S. pyogenes chromosome occurred at a specific GA dinucleotide target site located into the comEC coding sequence, with integration producing a duplication of the GA site. Upon conjugal transfer to S. pneumoniae, chromosomal integration occurred in celB, the pneumococcal homolog of comEC, at the same insertion site of Tn1207.1 (Santagati et al., 2003; Pozzi et al., 2004). A copy of Tn1207.1 was also found integrated in the 58,761-bp genetic element Φ10394.4, described as a prophage integrated at the same GA site within the comEC coding sequence of an erythromycin resistant clinical strain of S. pyogenes (Banks et al., 2003, 2004).
Here we report the manually annotated DNA sequence of Tn1207.3 which indicates that the element is in fact a prophage, identical to the right end of Φ10394.4. For this reason the element was renamed Φ1207.3.
Materials and Methods
Streptococcus pyogenes Strains and Growth Conditions
2812A is an erythromycin-resistant italian clinical isolate containing Φ1207.3 (Santagati et al., 2003). Bacteria were routinely grown in tryptic soy broth or tryptic soy agar (Difco) supplemented with 3% horse blood and, where appropriate, in presence of 1 μg/ml erythromycin.
PCR and Sequencing
Long PCR fragments were obtained with Takara LA Taq (Takara) following essentially the protocol suggested by the manufacturer. Briefly, the 25-μl reaction mixture was in 1X LA PCR Buffer II Taq buffer and contained: (i) 2.5 mM MgCl2, (ii) 200 μM dNTPs, (iii) 10 pmol of each primer, (iv) 0.25 units of Takara LA Taq, (v) 1 μl of liquid bacterial culture. Thermal cycling profile was as follows: 1 cycle at 92°C for 2 min, then 30 cycles at 50°C for 10 s, 68°C for 15 min, 92°C for 10 s, and 1 cycle at 50°C for 1 min and 68°C for 20 min. A primer walking approach (Santoro et al., 2010) was used to sequence the PCR products. The Expand High Fidelity PCR System (Roche) was used to produce PCR fragments of about 1,000 bp in size which were used as sequencing starting template to confirm sequence on the other strand. Four primer pairs were used to amplify the four fragments, ranging from 10,358 to 16,223 bp in size, containing Φ1207.3 and its chromosomal junction fragments.
DNA Sequence Analysis
DNA sequence analysis was performed with the software Artemis version 111. Manual gene annotation was carried out conducting BLAST homology searches of the databases available at the National Center for Biotechnology Information2. Protein domains were identified searching the protein family database Pfam available at the Wellcome Trust Sanger Institute3. The nucleotide sequence of Φ1207.3 is assigned GenBank accession no. AY657002.
Results
Φ1207.3 Nucleotide Sequence
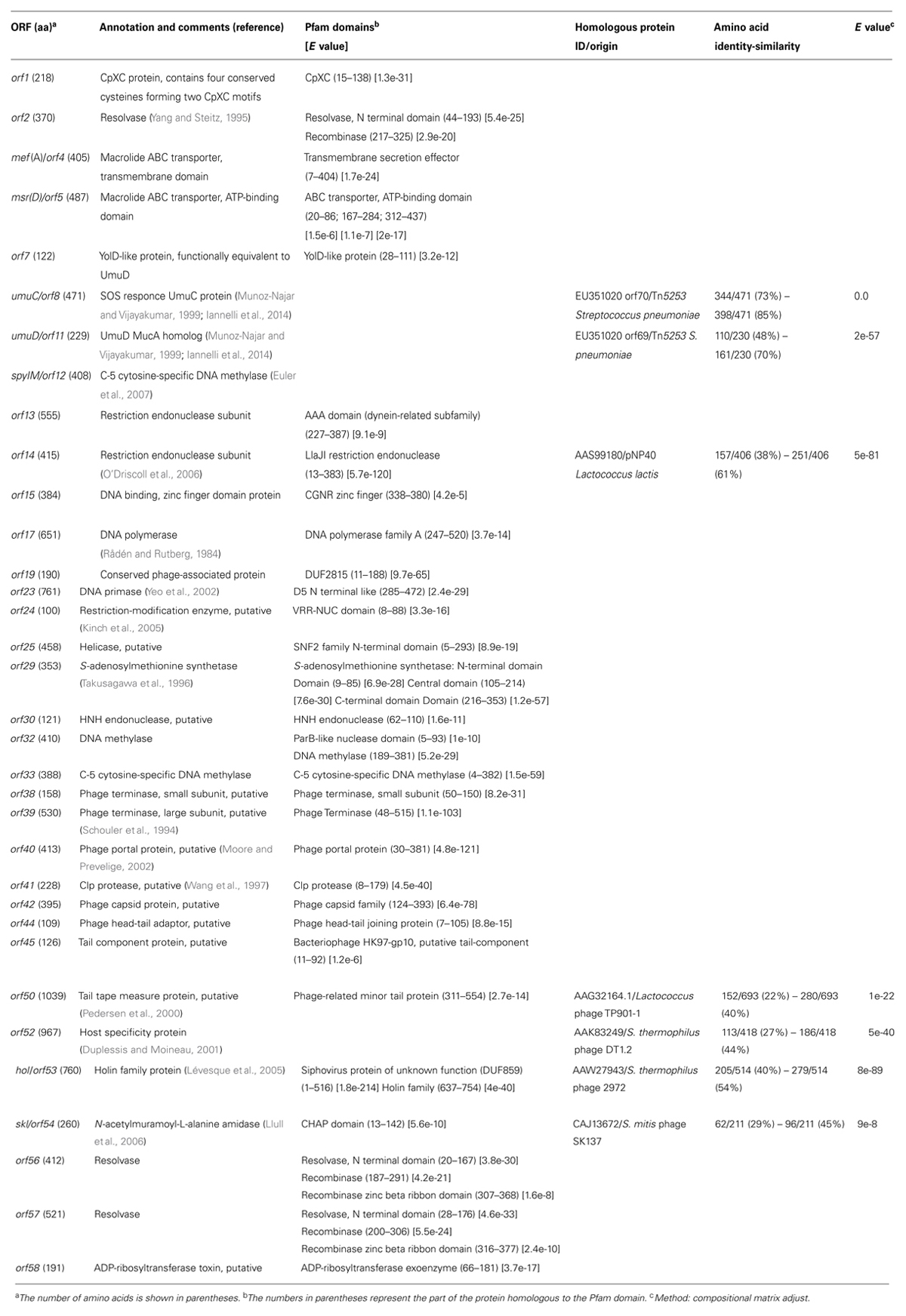
The complete nucleotide sequence of Φ1207.3 was obtained by primer walking on four long PCR fragments spanning the whole element. Inverse PCR on genomic DNA of S. pyogenes strain 2812A was carried out with divergent primers matching the already known 7,244-bp sequence of Φ1207.3 and its chromosomal junction sequences (Santagati et al., 2003). Amplicons obtained were used as sequencing template to determine internal sequences of Φ1207.3 on which primers were designed for the amplification of the four long overlapping fragments. The nucleotide sequence of Φ1207.3 was confirmed on the other strand using short PCR fragments as sequencing templates. Φ1207.3 was found to be 52,491 bp in length and DNA sequence analysis showed the presence of 58 open reading frames (ORFs), 44 of which have the same direction of transcription (Figure 1). Regions with different GC content can be could be identified in the DNA sequence of Φ1207.3: (i) the left end, containing ORFs 1–11, with 36.4% GC; (ii) ORFs 12–16 with 31.6% GC; (iii) ORFs 17–54 with 41,4% GC; (iv) the right end, corresponding to ORFs 55–58, with 37,8% GC (Figure 1). Comparison of Φ1207.3 with sequences present in public databases showed that: (i) the first 7,244 bp of the Φ1207.3 sequence are identical to the 7,244-bp element Tn1207.1 of S. pneumoniae (Santagati et al., 2000, 2003); (ii) the whole Φ1207.3 is identical to the 52,491-bp right end sequence of S. pyogenes Φ10394.4 (Banks et al., 2003; Pozzi et al., 2004); (iii) Seven sequence fragments, ranging in size from 0.2 to 9.5 kb, show 76–97% nucleotide identity to the S. pyogenes prophage Φm46.1 (Brenciani et al., 2010); (iv) homologous sequence fragments are present in the genetic elements from the genomes of S. dysgalactiae AC-2713 (GenBank no. HE858529), S. suis JS14 (GenBank no. CP002465), and S. agalactiae A909 (GenBank no. CP000114).
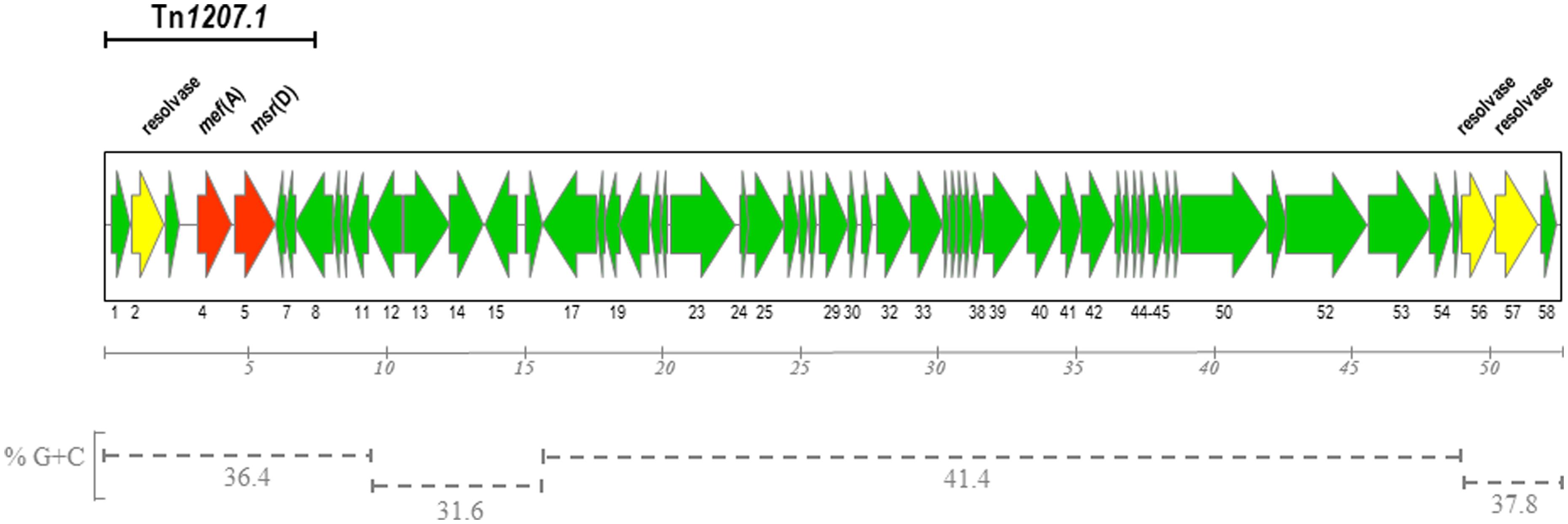
FIGURE 1. Schematic diagram of Streptococcus pyogenes Φ1207.3. The element is 52,491 bp in size and contains 58 open reading frames (ORFs). ORFs and their direction of transcription are represented by arrows and the annotated ORFs are indicated only by their numbers. Macrolide resistance genes and resolvase genes are reported as red and yellow arrows respectively. The region homologous to Tn1207.1 is indicated by a solid bar. The different GC content of the various regions are indicated with dotted bars. Scale is in kilobases.
ORFs of Φ1207.3
For 34 out of 58 ORFs it was possible a manual homology-based annotation with functional prediction of the hypothetical gene product (Table 1). Public protein databases and the Pfam protein family database were used for Blast searches of predicted gene products, taking into account significant homologies with functionally characterized proteins or good matches with Pfam domains. A putative ribosome binding sequence preceded all ORFs except orf36 and orf37. The alternative start codon TTG was present in orf34, orf36, orf43, and orf56, whereas orf37, orf38, and orf51 started with GTG. In the left arm of the element, the gene product of orf2 was predicted to be a site-specific resolvase of the serine recombinase family, mef(A) (orf4) and msr(D) (orf5) encoded respectively the transmembrane domains and the ATP-binding domains of the ABC transporter responsible for the M-type resistance to macrolides (Iannelli et al., 2014, Abstract C1-1188, 44th Interscience Conference Antimicrobial Agents Chemotherapy, 2004), orf8 and orf11 were homologous to the Tn5253 umuC/umuD operon conferring UV resistance by activation of the SOS repair system (Munoz-Najar and Vijayakumar, 1999). SpyIM, encoded by orf12, is a C-5 cytosine-specific DNA methylase belonging to the SpyI type II restriction-modification cassette, and responsible for inhibition of restriction by SmaI, whereas orf13 and orf14 encoded the two subunits of SpyI restriction endonuclease (Euler et al., 2007). Other genes coding for putative restriction-modification proteins include orf24 (restriction enzyme), orf30 (endonuclease), orf33 (cytosine-specific DNA methylase), while orf29 gene product presented an S-adenosylmethionine synthetase domain which may act as a methyl group donor for Orf33. In the central region, orf17, orf23, and orf25 encoded a DNA polymerase, a DNA primase, and a DNA helicase, possibly involved in phage DNA replication, whereas orf38 and orf39 encoded for the small and large subunit of the phage terminase, which, together with the portal protein encoded by orf40, could be involved in phage DNA packaging. orf42, orf44, orf45, and orf50 code for putative structural phage proteins. A putative lysis cassette, which is typically composed by a holin and an endolysin (Young et al., 2000) was encoded by hol (orf53) and skl (orf54). At the right end of the element, orf56 and orf57 gene products were homologous to resolvases of the serine recombinase family, possibly involved in excision, circularization, and site specific integration of Φ1207.3, whereas orf58 encoded a putative ADP-ribosyltransferase toxin.
Discussion
The complete and annotated DNA sequence of the mobile genetic element previously called Tn1207.3 clearly shows that the element is a prophage which we renamed Φ1207.3. At the sequence level, Φ1207.3 (52,491 bp) shows homology to two S. pyogenes prophages: (i) Φ10394.4 (58,761 bp), integrated at the same chromosomal site of Φ1207.3 (Banks et al., 2003; Pozzi et al., 2004); (ii) Φm46.1 (55,172 bp), integrated in the rum gene encoding an RNA uracil methyltransferase (Brenciani et al., 2010). The whole Φ1207.3 is identical to the right end of Φ10394.4, whereas high homology (>70%) to Φm46.1 is limited to 57% of the Φ1207.3 genome. Prophages similar to Φ1207.3 were also described by Giovanetti et al. (2005).
The recombination machinery of Φ1207.3 consists of three resolvases of the serine recombinase family. Serine recombinases are less common than tyrosine recombinases in prophage genomes and are usually present as a single large recombinase gene (Smith and Thorpe, 2002). Most of the S. pyogenes prophages present in sequenced genomes have a tyrosine recombinase (integrase) as the recombination module (Beres and Musser, 2007). The two resolvase genes at the right end of Φ1207.3 are probably transcribed as a single unit, as their coding sequences overlap by one nucleotide. It is likely that their gene products cooperate in mediating excision and integration of the Φ1207.3 DNA. This arrangement in tandem of two resolvase genes is also found in SSCmec elements (types I to IV) of Staphylococcus aureus (Wang and Archer, 2010), and in streptococcal prophage Φm46.1 (Giovanetti et al., 2005; Brenciani et al., 2010).
Since Φ1207.3 can move among streptococcal species (S. pyogenes, S. pneumoniae, S. gordonii) by a mechanism which fits the operational definition of conjugation (Santagati et al., 2003), this conjugative prophage may represent a novel class of genetic elements with a molecular mechanism of transfer that still needs to be elucidated. It is entirely possible that assembly of a complete phage particle may not be essential for the observed interspecific DNA transfer, and we hypothesize that the lysis cassette of Φ1207.3, rather than producing the bacterial cell burst after intracellular phage expansion (Young et al., 2000), could contribute to the formation of cytoplasmic bridges between donor and recipient cell.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This study was supported by the European Commission grant ANTIRESDEV HEALTH-F3-2009-241446.
Footnotes
- ^http://www.sanger.ac.uk/Software/Artemis/website
- ^http://www.ncbi.nlm.nih.gov/sites/gquery
- ^http://pfam.sanger.ac.uk
References
Banks, D. J., Porcella, S. F., Barbian, K. D., Beres, S. B., Philips, L. E., Voyich, J. M.,et al. (2004). Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190, 727–738. doi: 10.1086/422697
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Banks, D. J., Porcella, S. F., Barbian, K. D., Martin, J. M., and Musser, J. M. (2003). Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phenotypically diverse clones of group A Streptococcus. J. Infect. Dis. 188, 1898–1908. doi: 10.1086/379897
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beres, S. B., and Musser, J. M. (2007). Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS ONE 8:e800. doi: 10.1371/journal.pone.0000800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Bacciaglia, A., Vignaroli, C., Pugnaloni, A., Varaldo, P. E., and Giovanetti, E. (2010). Φm46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob. Agents Chemother. 54, 221–229. doi: 10.1128/AAC.00499-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duplessis, M., and Moineau, S. (2001). Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41, 325–336. doi: 10.1046/j.1365-2958.2001.02521.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Euler, C. W., Ryan, P., Martin, J. M., and Fischetti, V. A. (2007). M.SpyI, a DNA methyltransferase encoded on a mefA chimeric element, modifies the genome of Streptococcus pyogenes. J. Bacteriol. 189, 1044–1054. doi: 10.1128/JB.01411-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giovanetti, E., Brenciani, A., Vecchi, M., Manzin, A., and Varaldo, P. E. (2005). Prophage association of mef(A) elements encoding efflux-mediated erythromycin resistance in Streptococcus pyogenes. J. Antimicrob. Chemother. 55, 445–451. doi: 10.1093/jac/dki049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iannelli, F., Santoro, F., Oggioni, M. R., and Pozzi, G. (2014). Nucleotide sequence analysis of integrative conjugative element Tn5253 of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 1235–1239. doi: 10.1128/AAC.01764-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kinch, L. N., Ginalski, K., Rychlewski, L., and Grishin, N. V. (2005). Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 33, 3598–3605. doi: 10.1093/nar/gki676
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lévesque, C. M., Duplessis, M., Labonté, J., Labrie, S., Fremaux, C., Tremblay, D.,et al. (2005). Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 71, 4057–4068. doi: 10.1128/AEM.71.7.4057-4068.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Llull, D., López, R., and García, E. (2006). Skl, a novel choline-binding N-acetylmuramoyl-L-alanine amidase of Streptococcus mitis SK137 containing a CHAP domain. FEBS Lett. 580, 1959–1964. doi: 10.1016/j.febslet.2006.02.060
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, S. D., and Prevelige, P. E. J. (2002). DNA packaging: a new class of molecular motors. Curr. Biol. 12, R96–R98. doi: 10.1016/S0960-9822(02)00670-X
Munoz-Najar, U., and Vijayakumar, M. N. (1999). An operon that confers UV resistance by evoking the SOS mutagenic response in streptococcal conjugative transposon Tn5252. J. Bacteriol. 181, 2782–2788.
O’Driscoll, J., Heiter, D. F., Wilson, G. G., Fitzgerald, G. F., Roberts, R., and van Sinderen, D. (2006). A genetic dissection of the LlaJI restriction cassette reveals insights on a novel bacteriophage resistance system. BMC Microbiol. 6:40. doi: 10.1186/1471-2180-6-40
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pedersen, M., Østergaard, S., Bresciani, J., and Vogensen, F. K. (2000). Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276, 315–328. doi: 10.1006/viro.2000.0497
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pozzi, G., Iannelli, F., Oggioni, M. R., Santagati, M., and Stefani, S. (2004). Genetic elements carrying macrolide efflux genes in streptococci. Curr. Drug Targets Infect. Disord. 4, 203–206. doi: 10.2174/1568005043340641
Rådén, B., and Rutberg, L. (1984). Nucleotide sequence of the temperate Bacillus subtilis bacteriophage SPO2 DNA polymerase gene L. J. Virol. 52, 9–15.
Santagati, M., Iannelli, F., Cascone, C., Campanile, F., Oggioni, M. R., Stefani, S.,et al. (2003). The novel conjugative transposon Tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb. Drug Resist. 9, 243–247. doi: 10.1089/107662903322286445
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santagati, M., Iannelli, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2000). Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44, 2585–2587. doi: 10.1128/AAC.44.9.2585-2587.2000
Santoro, F., Oggioni, M. R., Pozzi, G., and Iannelli, F. (2010). Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 308, 150–158. doi: 10.1111/j.1574-6968.2010.02002.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schouler, C., Ehrlich, S. D., and Chopin, M. C. (1994). Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140, 3061–3069. doi: 10.1099/13500872-140-11-3061
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, M. C., and Thorpe, H. M. (2002). Diversity in the serine recombinases. Mol. Microbiol. 44, 299–307. doi: 10.1046/j.1365-2958.2002.02891.x
Takusagawa, F., Kamitori, S., and Markham, G. D. (1996). Structure and function of S-adenosylmethionine synthetase: crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution. Biochemistry 35, 2586–2596. doi: 10.1021/bi952604z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., Hartling, J. A., and Flanagan, J. M. (1997). The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456. doi: 10.1016/S0092-8674(00)80431-6
Wang, L., and Archer, G. L. (2010). Roles of CcrA and CcrB in excision and integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J. Bacteriol. 12, 3204–3212. doi: 10.1128/JB.01520-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, W., and Steitz, T. A. (1995). Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell 82, 193–207. doi: 10.1016/0092-8674(95)90307-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yeo, H. J., Zieglin, G., Korolev, S., Calendar, R., Lanka, E., and Waksman, G. (2002). Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins. Mol. Microbiol. 43, 855–867. doi: 10.1046/j.1365-2958.2002.02796.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Young, R., Wang, I. N., and Roof, W. D. (2000). Phages will out: strategies of host cell lysis. Trends Microbiol. 8, 120–128. doi: 10.1016/S0966-842X(00)01705-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: prophage, ICE, Φ1207.3, Tn1207.3, Tn1207.1, S. pyogenes, macrolide resistance
Citation: Iannelli F, Santagati M, Santoro F, Oggioni MR, Stefani S and Pozzi G (2014) Nucleotide sequence of conjugative prophage Φ1207.3 (formerly Tn1207.3) carrying the mef(A)/msr(D) genes for efflux resistance to macrolides in Streptococcus pyogenes. Front. Microbiol. 5:687. doi: 10.3389/fmicb.2014.00687
Received: 04 October 2014; Accepted: 21 November 2014;
Published online: 09 December 2014.
Edited by:
Bruna Facinelli, Università Politecnica delle Marche, ItalyReviewed by:
Dmitri Debabov, NovaBay Pharmaceuticals, USAAndrea Brenciani, Marche Polytechnic University, Italy
Pietro Emanuele Varaldo, Marche Polytechnic University, Italy
Copyright © 2014 Iannelli, Santagati, Santoro, Oggioni, Stefani and Pozzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianni Pozzi, Laboratory of Molecular Microbiology and Biotechnology, Department of Medical Biotechnologies, University of Siena, Policlinico Le Scotte, V Lotto I Piano, Viale Bracci, Siena 53100, Italy e-mail: gianni.pozzi@unisi.it
†Present address: Marco R. Oggioni, Department of Genetics, University of Leicester, Leicester, UK
‡ Francesco Iannelli and Maria Santagati have contributed equally to this work.
 Francesco Iannelli
Francesco Iannelli Maria Santagati
Maria Santagati Francesco Santoro
Francesco Santoro Marco R. Oggioni
Marco R. Oggioni Stefania Stefani
Stefania Stefani Gianni Pozzi
Gianni Pozzi