- Department of Molecular Microbiology, John Innes Centre, Norwich, UK
PII proteins are pivotal players in the control of nitrogen metabolism in bacteria and archaea, and are also found in the plastids of plants. PII proteins control the activities of a diverse range of enzymes, transcription factors and membrane transport proteins, and their regulatory effect is achieved by direct interaction with their target. Many, but by no means all, PII proteins are subject to post-translational modification of a residue within the T-loop of the protein. The protein’s modification state is influenced by the cellular nitrogen status and in the past this has been considered to regulate PII activity by controlling interaction with target proteins. However, the fundamental ability of PII proteins to respond to the cellular nitrogen status has been shown to be dependent on binding of key effector molecules, ATP, ADP, and 2-oxoglutarate which brings into question the precise role of post-translational modification. In this study we have used the Escherichia coli PII protein GlnK to examine the influence of post-translational modification (uridylylation) on the interaction between GlnK and its cognate target the ammonia channel protein AmtB. We have compared the interaction with AmtB of wild-type GlnK and a variant protein, GlnKTyr51Ala, that cannot be uridylylated. This analysis was carried out both in vivo and in vitro and showed that association and dissociation of the GlnK–AmtB complex is not dependent on the uridylylation state of GlnK. However, our in vivo studies show that post-translational modification of GlnK does influence the dynamics of its interaction with AmtB.
Introduction
Proteins of the PII signal transduction superfamily play a major role in coordinating the regulation of nitrogen metabolic processes, and have recently also been implicated in regulation of at least one facet of carbon metabolism (Huergo et al., 2013; Rodrigues et al., 2014). They mediate their effects by protein–protein interaction and their targets include key metabolic and regulatory enzymes, transcription factors, and nutrient transporters. The PII proteins are the most widely distributed signalling proteins in nature, with ubiquitous members among prokaryotes, and representatives among nitrogen-fixing Archaea and in the chloroplasts of eukaryotic phototrophs (Sant’Anna et al., 2009; Huergo et al., 2013). They are highly conserved homotrimeric proteins consisting of 12–13 kDa subunits that form a cylindrical body with three protruding T-loops have been shown to be capable of adopting a variety of conformations (Conroy et al., 2007; Truan et al., 2010; Huergo et al., 2013). In the majority of cases where the structure of a PII protein bound to one of its targets has been determined the T-loops play a key role in that interaction.
The universally conserved mechanism by which PII activity is regulated involves binding of the effector molecules 2-oxoglutarate (2-OG) and Mg-ATP or ADP in the lateral clefts formed between the momomers (Radchenko et al., 2010; Radchenko and Merrick, 2011). When the intracellular nitrogen status is low, and 2-OG levels are high, PII proteins bind 2-OG and Mg-ATP in the lateral clefts and in this condition the T-loops are relatively unstructured. However when the intracellular nitrogen status rises 2-OG levels fall and 2-OG dissociates from the effector binding site. Studies of E. coli GlnK indicate that in the absence of 2-OG the bound ATP is subject to PII-mediated hydrolysis to ADP (Radchenko et al., 2013). This in turn leads to a rearrangement of particular residues in the GlnK binding pocket, most notably Gln39 and Lys58, and a concomitant change in the T-loop to form a more defined structure such that the apex of the loop projects very markedly above the protein’s surface (Conroy et al., 2007; Truan et al., 2010). Hence the switch from the MgATP, 2-OG form of PII in N-limited conditions to the ADP-bound form in N-sufficient conditions is reflected in a concomitant change in T-loop structure which could potentially be sufficient to regulate the ability of PII proteins to interact with their cognate targets.
Nevertheless, in addition to this conserved mode of regulation, some PII proteins have another regulatory mechanism that involves post-translational modification of the T-loop. The E. coli PII proteins, GlnB and GlnK, are subject to uridylylation of the conserved tyrosine residue (Tyr51) in the T-loop by a uridylyltransferase/uridylyl-removing enzyme (UTase/UR or GlnD; Jiang et al., 1998; Atkinson and Ninfa, 1999). In N-limiting conditions UTase activity is predominant and GlnB, and GlnK are uridylylated, whereas in N-sufficient conditions the intracellular glutamine concentration rises, glutamine binds to GlnD and the UR activity deuridylylates the PII proteins. Uridylylation of PII proteins is widespread in the Proteobacteria (Jiang et al., 1998) and a very similar GlnD-mediated adenylylation of PII proteins occurs in some Actinobacteria (Hesketh et al., 2002; Strosser et al., 2004). Furthermore, in some Cyanobacteria PII proteins are subject to phosphorylation of the T-loop (Forchhammer et al., 2004). It is, however, notable that in many organisms PII is not subject to any form of post-translational modification (Huergo et al., 2013) which raises the question of the biological role of this modification in those organisms where it occurs. We have chosen to investigate this in a model PII interaction, namely that of E. coli GlnK with the integral membrane ammonia transporter protein AmtB.
The primary function of E. coli GlnK is to regulate the activity of AmtB which is a homotrimer with an ammonia conduction channel through each subunit (Thomas et al., 2000; Coutts et al., 2002; Zheng et al., 2004). In E. coli an increase in the cellular N status promotes binding of GlnK to AmtB and the crystal structure of the complex shows that GlnK has a molecule of ADP bound to each monomer. GlnK interacts with AmtB almost exclusively via the T-loops the tips of which insert deeply into the cytoplasmic pore exits of AmtB such that residue Arg47 blocks ammonia conduction into the cell (Conroy et al., 2007). Initial studies of GlnK–AmtB interaction noted that complex formation in response to an ammonium shock occurred synchronously with deuridylylation of GlnK (Coutts et al., 2002) suggesting that association/dissociation of the complex was regulated by the post-translational state of GlnK.
The GlnK–AmtB interaction is highly conserved in bacteria and Archaea as reflected by conserved linkage between the structural genes (Thomas et al., 2000). Indeed it has been proposed that regulation of AmtB is the ancestral function of GlnK (Sant’Anna et al., 2009). However the glnK amtB operon is found in many organisms where there is no evidence of post-translational modification of PII and it was noted by Coutts et al. (2002) that this suggested that regulation of the complex in those organisms would require an alternative mechanism. In this study we describe experiments to examine this topic and show that association/dissociation of the GlnK–AmtB complex can indeed respond to the cellular N status in the absence of post-translational modification of GlnK.
Materials and Methods
Strains, Plasmids, and Growth Media
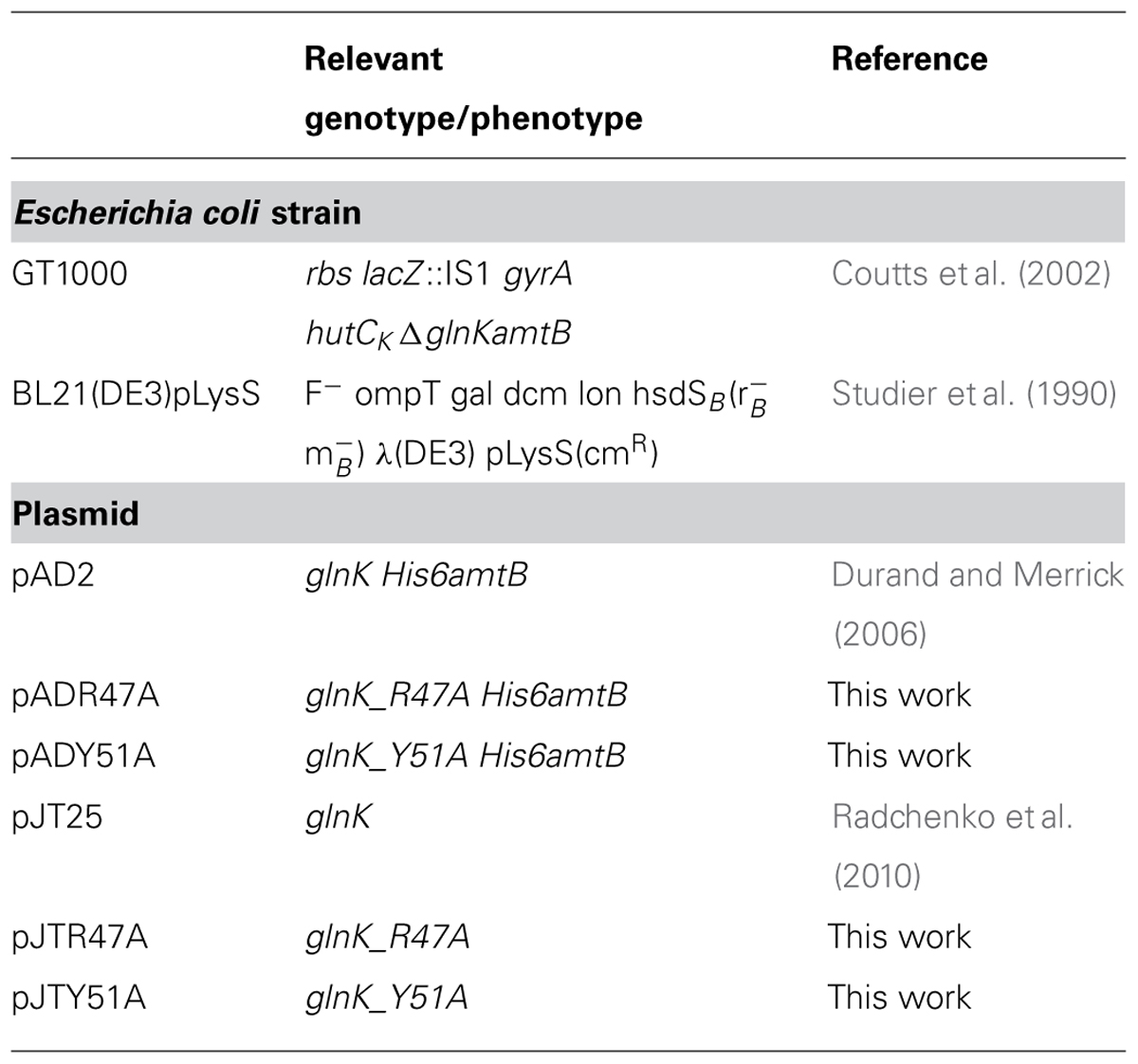
The strains and plasmids used are listed in Table 1. Plasmid pAD2 and its derivatives were expressed in GT1000 and used for His6AmtB–GlnK (wild-type or variants) complex purification. Plasmid pJT25 or its derivatives in E. coli BL21(DE3)pLysS were used for overexpression of GlnK (wild type or variants). Derivatives of pAD2, plasmids pADR47A, and pADY51A, and derivatives of pJT25, plasmids pJTR47A, and pJTY51A, were generated using the QuickChange Lightning Site-directed Mutagenesis Kit (Agilent Technologies). E. coli strains were routinely grown in Luria Bertani medium, and for nitrogen-limited growth M9Gln medium (Javelle et al., 2005) was used. Ampicillin (100 μg ml-1), chloramphenicol (30 μg ml-1) were included as appropriate.
Purification of His6-AmtB and GlnK Proteins
GlnK–AmtB complexes and Histidine-tagged AmtB (His6-AmtB) were obtained by purifying the AmtB–GlnK complex as described previously (Durand and Merrick, 2006; Radchenko et al., 2010). GlnK wild-type and variants were purified from whole cell extracts of BL21(DE3)pLysS carrying pJT25, pJTR47A, or pJTY51A that were heated to 80°C for 4 min and centrifuged at 30,000 g for 30 min to remove debris (Moure et al., 2012). Supernatants were processed by affinity chromatography on a Heparin column (GE Healthcare), using a step gradient of 5, 10, 20, 40, and 100% elution buffer (protein eluted at 20% step). Equilibration buffer was 50 mM Tris, 0.1 M KCl, pH 7.5, and elution buffer was 50 mM Tris, 1.0 M KCl, pH 7.5.
Analysis of GlnK–AmtB Interaction in Vitro
The ability of the variant GlnK proteins to interact with AmtB was assessed by in vitro binding assays as described previously (Radchenko et al., 2010). For assessment of association His6-AmtB (400 μg) was immobilized on a HIS-Select spin column previously equilibrated with buffer A [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10% (v/v) glycerol, 0.05% (w/v) n-dodecyl-N,N-dimethylamine-N-oxide (LDAO)] containing 5 mM imidazole. The flow through was collected, and unbound proteins were removed by two washes with the same buffer. Three additional washes were carried out in buffer A containing 5 mM imidazole, 300 μg GlnK protein and various combinations of effectors. The effector concentrations used were reflective of those measured in vivo under conditions of N-limitation and N-sufficiency (Radchenko et al., 2010). As a control, buffer A containing 5 mM imidazole, 300 μg GlnK protein and 0.6 M NaCl was used. The molar ratio of AmtB to GlnK was approximately 1:3. A single column was used for each condition tested. Elution of His6AmtB or His6AmtB–GlnK complex was performed by the addition of buffer A containing 500 mM imidazole. The eluted fractions were analyzed on 15% SDS-PAGE. For assessment of dissociation, complexes of AmtB with bound GlnK (wild-type or variants) were applied to a HIS-Select spin column in the presence of different combinations of effectors and washed and eluted fractions were analyzed by SDS–PAGE.
In Vivo Detection of GlnK Membrane Sequestration Following Ammonium Shock
Escherichia coli cultures of GT1000 carrying plasmids pAD2, pADR47A, and pADY51A were grown in M9Gln medium at 30°C and subjected to ammonium shock by the addition of NH4Cl to a final concentration of 200 μM. Samples were taken prior to ammonium addition and then at 0.5, 1, 2, 3, 5, 10, and 15 mins after addition. Whole cell protein extracts, membrane and cytoplasmic protein fractions were prepared as described previously (Coutts et al., 2002). Membrane fractions were subjected to two high-salt washes (600 mM NaCl) prior to analysis. Protein quantification of the fractions was determined with Coomassie PlusTM protein assay reagent (Thermo Scientific) using an albumin standard (Thermo Scientific).
To determine the cellular location of GlnK membrane protein fractions were separated by 12.5% SDS-PAGE (5–10 μg/lane), analyzed by Western blotting and detected as described elsewhere (Radchenko et al., 2010). The uridylylation state of GlnK was also assessed for each sample by separation of the different GlnK states (UMP0–UMP3) using native PAGE followed by Western blotting. All experiments were replicated three times.
Results
In order to investigate the influence of GlnK uridylylation on the interaction between GlnK and AmtB we constructed two T-loop variants for comparison with the wild-type. Residue Tyr51 was changed to Ala thereby generating a variant that would not be subject to uridylylation. As a control residue Arg 47 was also changed to Ala, generating a variant that would still be subject to modification but lacked the charged residue at the tip of the T-loop in the GlnK–AmtB complex. The variants were analyzed in two assays. Firstly the purified GlnK proteins were analyzed in vitro for their ability to interact with AmtB in response to the effector molecules ATP, 2-OG, and ADP. Secondly the ability of GlnK wild-type and the variants to be sequestered to the membrane by AmtB was analyzed in vivo during a transition from N-limited to N-sufficient conditions.
Association and Dissociation of GlnK–AmtB Complexes in Vitro
The ability of the variant GlnK proteins to interact with AmtB was assessed by in vitro binding assays as described previously (Radchenko et al., 2010). For assessment of association His6-AmtB was bound to a HIS-Select spin column and the ability of GlnK wild-type and variants to bind to AmtB in the presence of a variety of effector concentrations was studied. For assessment of dissociation complexes of AmtB with bound GlnK (wild-type or variants) were bound to a HIS-Select spin column and the dissociation of GlnK in the presence of a variety of effector concentrations was studied. The effectors concentrations used were reflective of those measured in vivo under conditions of N-limitation and N-sufficiency (Radchenko et al., 2010).
In all conditions tested the GlnK variants, Arg47Ala, and Tyr51Ala, behaved in the same way as wild-type GlnK. In N-limited conditions in vivo the intracellular 2-OG pool is around 1.5 mM and this falls rapidly following ammonium shock to 0.3 mM (Radchenko et al., 2010). GlnK behavior in vitro reflects this, in that association with AmtB requires a low 2-OG level (Figure 1). Following ammonium shock intracellular ADP levels rise transiently to around 0.75 mM (Radchenko et al., 2010) and this level of ADP promotes complex formation (Figure 1). Dissociation of the complex in vitro (Figure 2) only occurred in a combination of low ADP (0.35 mM) and high ATP (4.5 mM) and 2-OG (1.5 mM) and these are conditions comparable to those found in vivo when cells that are subject to N-limitation (Radchenko et al., 2010).
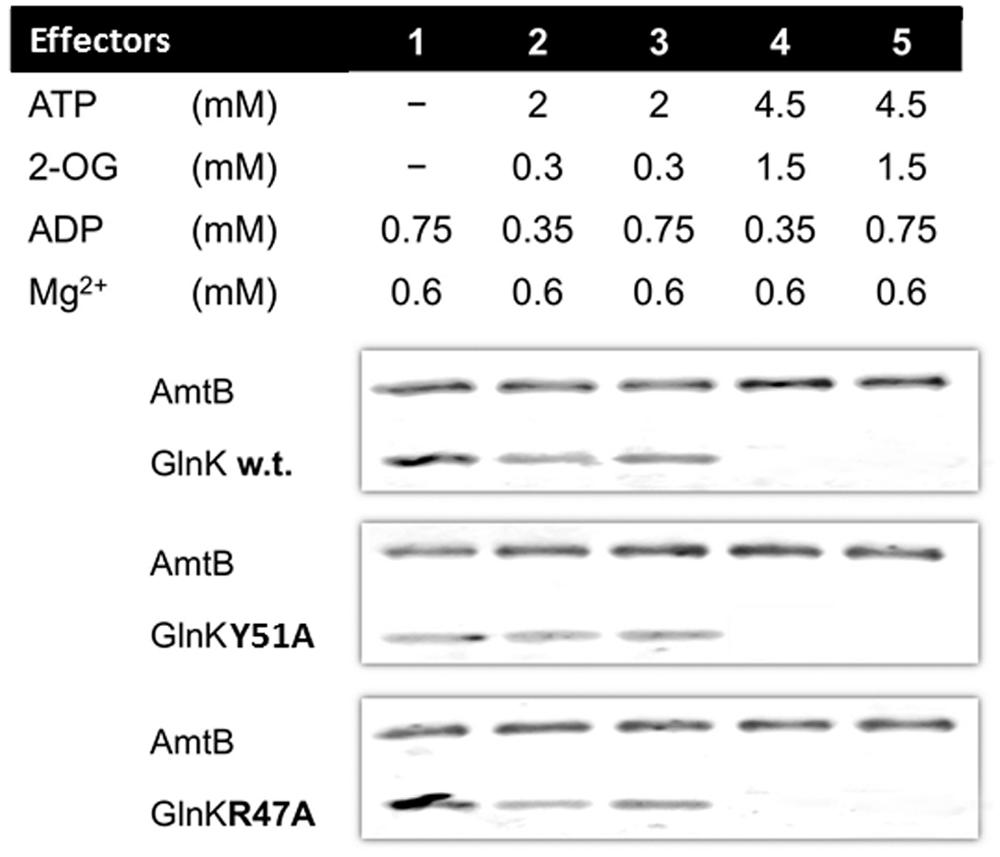
FIGURE 1. GlnK–AmtB association in vitro. His6-AmtB was bound to a HIS-Select spin column and GlnK proteins were then applied to the column in the presence of various combinations of effectors. The His6-AmtB or the GlnK–His6AmtB complex were then eluted with 500 mM imidazole and analyzed by SDS-PAGE. The presence of GlnK in the eluted fraction indicates association to AmtB.

FIGURE 2. GlnK–AmtB dissociation in vitro. GlnK–AmtB complexes containing wild type or variant GlnK were bound to a HIS-Select spin column and then subject to washes with buffer containing various combinations of effectors. The His6-AmtB or the GlnK–His6AmtB complex were then eluted with 500 mM imidazole and analyzed by SDS-PAGE. The absence of GlnK in the eluted fraction indicates dissociation of the complex.
GlnK–AmtB Complex Formation in Vivo
Cells were grown in N-limitation and then subject to an ammonium shock by addition of NH4Cl to the culture medium. Aliquots of the culture were taken prior to ammonium addition and then periodically over a period of 15 mins following the ammonium shock. The interaction of GlnK with AmtB was monitored by detection of GlnK association with the membrane fraction and the uridylylation state of GlnK was monitored by native PAGE as described previously (Radchenko et al., 2010).
As reported previously (Radchenko et al., 2010), sequestration of wild-type GlnK to the membrane was maximal 2 mins after ammonium shock and corresponded exactly with the time taken for the protein to become fully deuridylylated (Figure 3). As the added ammonium was metabolised the cells returned to N-limitation and 15 mins after ammonium addition GlnK was again dissociated from the membrane and fully uridylylated (Figure 3). The GlnK Arg47Ala variant showed an identical membrane sequestration profile to the wild-type. It was also deuridylylated at an identical rate to wild-type but took very slightly longer to be reuridylylated (Figure 3). As expected, the Tyr51Ala variant was fully deuridylylated regardless of the N-status of the cells. Nevertheless it still responded to ammonium shock by becoming rapidly sequestered to the membrane. However, in this case the sequestration rate was significantly more rapid than observed with wild-type GlnK or the Arg47Ala variant such that sequestration to the membrane was maximal only 1 min after ammonium shock (Figure 3). These experiments were repeated three times with essentially identical results on each occasion.
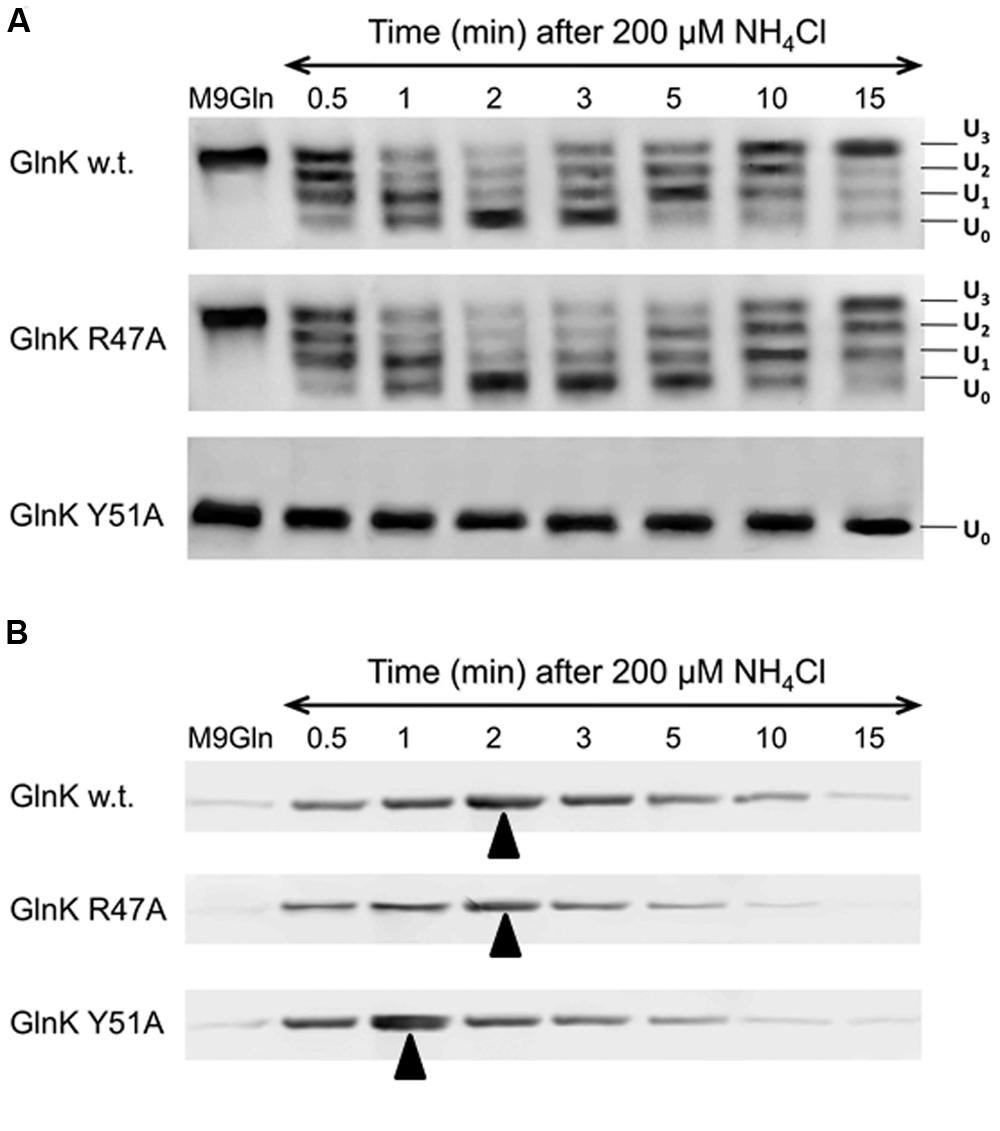
FIGURE 3. GlnK–AmtB complex formation in vivo. (A) Changes in the uridylylation state of GlnK wild-type and variants R47A and Y51A following ammonium shock. Culture samples were taken prior to ammonium addition and then at a series of time points and the four possible uridylylation states (UMP0–UMP3) of the protein were visualized by native PAGE followed by Western blotting. (B) Binding of GlnK wild-type and variants R47A and Y51A to AmtB was assessed by Western blotting to detect GlnK association with the cell membrane prior to and following ammonium shock. Arrows indicate point of maximum complex formation.
Discussion
We have employed a combination of in vitro and in vivo studies to investigate the role of GlnK uridylylation with respect to the interaction of GlnK with its primary target, the ammonium transport protein AmtB. Prior to the recognition of the role that the effector molecules ATP, ADP, and 2-OG play in influencing the conformation of the T-loop in PII proteins it seemed possible that post-translational modification of the T-loop could be the primary factor controlling the interaction of PII proteins with their targets (Coutts et al., 2002). In such a model, uridylylation of the GlnK T-loop in N-limiting conditions would prevent interaction with AmtB and deuridylylation in response to an elevated cellular N-status would leave the T-loops free to effect complex formation. However, in contradiction of such a model many organisms that encode a glnK amtB operon and are predicted to regulate AmtB function by GlnK–AmtB interaction do not have mechanisms for post-translational modification of GlnK.
We have studied two GlnK T-loop variants; one (Tyr51Ala) that is unable to uridylylate GlnK and one (Arg47Ala) that is able to be uridylylated but that lacks the charged residue at the tip of the T-loop which interacts with the pore of AmtB. Both variants behaved in vitro in an identical manner to wild-type GlnK in that they showed association with AmtB in the presence of physiological concentrations of ADP (0.35–0.75 mM) excepting in those effector conditions seen in E. coli cells subject to N-limitation (4.5 mM ATP, 1.5 mM 2-OG, and 0.35 mM ADP). Hence in vitro the Tyr51Ala variant of GlnK was able to associate to and dissociate from AmtB independently of any post-translational modification.
When assessed in vivo, for both changes in uridylylation state and dynamics of interaction with AmtB in response to an ammonium shock, the Arg47Ala variant of GlnK behaved essentially as the wild-type. As expected the Tyr51Ala variant was not uridylylated under any of the conditions tested but was fully competent to associate with and dissociate from AmtB in response to the ammonium shock. This confirms that post-translational modification is not essential to regulate the interaction of GlnK with AmtB. Our in vitro studies indicate that binding of the effectors, MgATP/ADP, and 2-OG, in response to the metabolic state of the cell is sufficient to control the interaction of GlnK with AmtB through the effect on the conformation of the T-loop. Indeed it seems likely that in many, if not most, cases this could be the primary factor controlling association/dissociation of the PII-target complex.
However uridylylation of GlnK did influence the dynamics of GlnK–AmtB association, such that in the absence of the need to deuridylylate the protein GlnK was able to associate more rapidly with its target when the cellular N-status increased. Deuridylylation of GlnK is triggered by a rise in the intracellular glutamine pool and binding of glutamine to GlnD leading to enhancement of its uridyl-removing (UR) activity. Hence it would appear that the deuridylylation of GlnK prior to complex formation with AmtB acts as a check-point that integrates glutamine availability into the signal transduction network. In the proteobacteria and the actinobacteria glnD is widespread (Huergo et al., 2013) presumably reflecting a selective advantage of this enhanced signal transduction network. However, in organisms where post-translational modification of PII is apparently absent it seems that regulation of AmtB activity is controlled by the cellular 2-OG pool as the sole measure of N-status. Studies of the PII-PipX interaction in Synechococcus elongatus have similarly shown that, although PII is subject to phosphorylation of the T-loop, association/dissociation of the PII–PipX complex appears to be independent of PII modification (Llacer et al., 2010). Indeed to date there is little evidence of PII target interactions where post-translational modification of the T-loops plays an essential role (Merrick, 2015). There is therefore clearly a need for much more information on the function of post-translational modification of PII, both with respect to studies of many more PII interactions with different targets and in a wide range of physiological conditions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was funded by the Biotechnology and Biological Sciences Research Council Grant BB/E022308/1 to Mike Merrick.
References
Atkinson, M., and Ninfa, A. J. (1999). Characterization of the GlnK protein of Escherichia coli. Mol. Microbiol. 32, 301–313. doi: 10.1046/j.1365-2958.1999.01349.x
Conroy, M. J., Durand, A., Lupo, D., Li, X. D., Bullough, P. A., Winkler, F. K.,et al. (2007). The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. U.S.A. 104, 1213–1218. doi: 10.1073/pnas.0610348104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coutts, G., Thomas, G., Blakey, D., and Merrick, M. (2002). Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21, 536–545. doi: 10.1093/emboj/21.4.536
Durand, A., and Merrick, M. (2006). In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate. J. Biol. Chem. 281, 29558–29567. doi: 10.1074/jbc.M602477200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Forchhammer, K., Irmler, A., Kloft, N., and Ruppert, U. (2004). P signalling in unicellular cyanobacteria: analysis of redox-signals and energy charge. Physiol. Plant. 120, 51–56. doi: 10.1111/j.0031-9317.2004.0218.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hesketh, A., Fink, D., Gust, B., Rexer, H. U., Scheel, B., Chater, K.,et al. (2002). The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 46, 319–330. doi: 10.1046/j.1365-2958.2002.03149.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huergo, L. F., Chandra, G., and Merrick, M. (2013). PII signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. 37, 251–283. doi: 10.1111/j.1574-6976.2012.00351.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Javelle, A., Thomas, G., Marini, A. M., Kramer, R., and Merrick, M. (2005). In vivo functional characterisation of the E. coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem. J. 390, 215–222. doi: 10.1042/BJ20042094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, P., Peliska, J. A., and Ninfa, A. J. (1998). Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37, 12782–12794. doi: 10.1021/bi980667m
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Llacer, J. L., Espinosa, J., Castells, M. A., Contreras, A., Forchhammer, K., and Rubio, V. (2010). Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc. Natl. Acad. Sci. U.S.A. 107, 15397–15402. doi: 10.1073/pnas.1007015107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merrick, M. (2015). Post-translational modification of PII signal transduction proteins. Front. Microbiol. 5:763. doi: 10.3389/fmicb.2014.00763
Moure, V. R., Razzera, G., Araujo, L. M., Oliveira, M. A., Gerhardt, E. C., Muller-Santos, M.,et al. (2012). Heat stability of Proteobacterial PII protein facilitate purification using a single chromatography step. Protein Expr. Purif. 81, 83–88. doi: 10.1016/j.pep.2011.09.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radchenko, M., and Merrick, M. (2011). The role of effector molecules in signal transduction by PII proteins. Biochem. Soc. Trans. 39, 189–194. doi: 10.1042/BST0390189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radchenko, M. V., Thornton, J., and Merrick, M. (2010). Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP and 2-oxoglutarate. J. Biol. Chem. 285, 31037–31045. doi: 10.1074/jbc.M110.153908
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radchenko, M. V., Thornton, J., and Merrick, M. (2013). P(II) signal transduction proteins are ATPases whose activity is regulated by 2-oxoglutarate. Proc. Natl. Acad. Sci. U.S.A. 110, 12948–12953. doi: 10.1073/pnas.1304386110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rodrigues, T. E., Gerhardt, E. C., Oliveira, M. A., Chubatsu, L. S., Pedrosa, F. O., Souza, E. M.,et al. (2014). Search for novel targets of the PII signal transduction protein in Bacteria identifies the BCCP component of acetyl-CoA carboxylase as a PII binding partner. Mol. Microbiol. 91, 751–761. doi: 10.1111/mmi.12493
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sant’Anna, F. H., Trentini, D. B., de Souto,W. S., Cecagno, R., da Silva, S. C., and Schrank, I. S. (2009). The PII superfamily revised: a novel group and evolutionary insights. J. Mol. Evol. 68, 322–336. doi: 10.1007/s00239-009-9209-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Strosser, J., Ludke, A., Schaffer, S., Kramer, R., and Burkovski, A. (2004). Regulation of GlnK activity: modification, membrane sequestration and proteolysis as regulatory principles in the network of nitrogen control in Corynebacterium glutamicum. Mol. Microbiol. 54, 132–147. doi: 10.1111/j.1365-2958.2004.04247.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Studier, F. W., Rosenberg, A. H., Dunn, J. J., and Dubendorff, J. W. (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89. doi: 10.1016/0076-6879(90)85008-C
Thomas, G., Coutts, G., and Merrick, M. (2000). The glnKamtB operon: a conserved gene pair in prokaryotes. Trends Genet. 16, 11–14. doi: 10.1016/S0168-9525(99)01887-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Truan, D., Huergo, L. F., Chubatsu, L. S., Merrick, M., Li, X. D., and Winkler, F. K. (2010). A new PII protein structure identifies the 2-oxoglutarate binding site. J. Mol. Biol. 400, 531–539. doi: 10.1016/j.jmb.2010.05.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zheng, L., Kostrewa, D., Bernèche, S., Winkler, F. K., and Li, X.-D. (2004). The mechanism of ammonia transport based on the crystal structure of AmtB of E. coli. Proc. Natl. Acad. Sci. U.S.A. 101, 17090–17095. doi: 10.1073/pnas.0406475101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: PII protein, GlnK, post-translational modification, uridylylation, ammonium transport, AmtB
Citation: Radchenko MV, Thornton J and Merrick M (2014) Association and dissociation of the GlnK–AmtB complex in response to cellular nitrogen status can occur in the absence of GlnK post-translational modification. Front. Microbiol. 5:731. doi: 10.3389/fmicb.2014.00731
Received: 28 October 2014; Accepted: 04 December 2014;
Published online: 23 December 2014.
Edited by:
Jörg Stülke, Georg-August-Universität Göttingen, GermanyReviewed by:
Wolfgang Buckel, Philipps-Universität Marburg, GermanySven Halbedel, Robert Koch Institute, Germany
Copyright © 2014 Radchenko, Thornton and Merrick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mike Merrick, Department of Molecular Microbiology, John Innes Centre, Norwich Research Park, NR4 7UH Norwich, UK e-mail: mike.merrick@jic.ac.uk
†Present address: Martha V. Radchenko, Department of Biochemistry and Molecular Biology, Rosalind Franklin University of Medicine and Science, North Chicago, IL, USA
 Martha V. Radchenko†
Martha V. Radchenko† Jeremy Thornton
Jeremy Thornton Mike Merrick
Mike Merrick