- 1State Key Laboratory of Forest and Soil Ecology, Institute of Applied Ecology, Chinese Academy of Sciences, Shenyang, China
- 2College of Resources and Environment, University of Chinese Academy of Sciences, Beijing, China
- 3Department of Biological Sciences, Institute for Bioinformatics and Evolutionary Studies, University of Idaho, Moscow, ID, USA
A self-transmissible broad-host-range (BHR) plasmid pSFA231 was isolated from petroleum-contaminated sediment in Shen-fu wastewater irrigation zone, China, using the triparental mating exogenous plasmid capture method. Based on its complete sequence the plasmid has a size of 41.5 kb and codes for 50 putative open reading frames (orfs), 29 of which represent genes involved in replication, partitioning and transfer functions of the plasmid. Phylogenetic analysis grouped pSFA231 into the newly defined PromA plasmid family, which currently includes five members. Further comparative genomic analysis shows that pSFA231 shares the common backbone regions with the other PromA plasmids, i.e., genes involved in replication, maintenance and control, and conjugative transfer. Nevertheless, phylogenetic divergence was found in specific gene products. We propose to divide the PromA group into two subgroups, PromA-α (pMRAD02, pSB102) and PromA-β (pMOL98, pIPO2T, pSFA231, pTer331), based on the splits network analysis of the RepA protein. Interestingly, a cluster of hypothetical orfs located between parA and traA of pSFA231 shows high similarity with the corresponding regions on pMOL98, pIPO2T, and pTer331, suggesting these hypothetical orfs may represent “essential” plasmid backbone genes for the PromA-β subgroup. Alternatively, they may also be accessory genes that were first acquired and then stayed as the plasmid diverged. Our study increases the available collection of complete genome sequences of BHR plasmids, and since pSFA231 is the only characterized PromA plasmid from China, our findings also enhance our understanding of the genetic diversity of this plasmid group in different parts of the world.
Introduction
Plasmids are extra-chromosomal self-replicating DNA elements within the microorganisms (Mela et al., 2008). They are important members of the mobile gene pool, and are among the most important contributors to horizontal gene transfer between bacteria (Frost et al., 2005). The broad-host-range (BHR) plasmids have been defined as those plasmids that can self-transfer themselves and can stably replicate and maintain in bacterial species from at least two subgroups within the Proteobacteria (e.g., between α- and β- Proteobacteria) (Szpirer et al., 1999; Sen et al., 2011). The BHR plasmids typically have mosaic genomes including two distinct regions (Thomas, 2000). The “plasmid backbone” genes encode proteins involved in the replication, maintenance, control and conjugative transfer of the BHR plasmid. Other plasmid regions are comprised of various “accessory” genes conferring important benefits to the host, including resistance to antibiotics (Rhodes et al., 2004), resistance to heavy metals (Schneiker et al., 2001), catabolic functions (Ono et al., 2007), and virulence determinants (Schlüter et al., 2008), etc.
Conjugative gene transfer mediated by BHR plasmids is generally believed to be a common and widespread mechanism for the transfer of genes across a broad phylogenetic range of bacteria (Top and Springael, 2003; Van der Auwera et al., 2009), and plays a crucial role in the adaptation of bacteria to environmental challenges and spread of antibiotic resistance (Jechalke et al., 2013). Despite the general agreement on the importance of BHR plasmids in the adaptive evolution of bacteria, the BHR plasmids being identified and completely sequenced are still few, initially limited by the high sequencing cost of first generation (Sanger) sequencing technology. To the best of our knowledge, no more than 15 BHR plasmids had been fully sequenced until 2006. With the development of next-generation sequencing methods, such as 454 pyrosequencing and Illumina high-throughput sequencing technology, more complete sequences of BHR plasmids have been added to the data pool in recent years, most of which were identified as IncP-1 plasmids (Sen et al., 2012; Brown et al., 2013). In spite of the increasing number of BHR plasmids identified, complete sequences of BHR plasmids available in GenBank are still not sufficient for systematically analyzing their genetic diversity. Thus, isolation and characterization of new BHR plasmids from environmental samples is still required to better understand the nature and evolutionary history of these important mobile genetic elements and their role in horizontal gene transfer.
Among the fully sequenced BHR plasmids, most are classified as the well-known incompatibility groups, such as IncP-1 (41 plasmids), IncW (5 plasmids), and IncU (4 plasmids), based on the backbone genes (Fernández-López et al., 2006; Norberg et al., 2011; Sen et al., 2011; Brown et al., 2013). Recently, a novel plasmid group, which could not be classified by the conventional inc/rep grouping system, was proposed by Van der Auwera et al. (2009) in terms of phylogenetic analysis of the complete genome sequence. This new BHR plasmid group was defined as PromA family, now including five members, namely plasmids pMOL98 (Gstalder et al., 2003), pIPO2 (Tauch et al., 2002), pSB102 (Schneiker et al., 2001), pTer331 (Mela et al., 2008), and pMRAD02 (Ito and Iizuka, 1971). Their RepA protein formed a separate cluster related to but distinct from the IncW plasmids, while several plasmid maintenance genes were most closely related to those of other incompatibility group, and the plasmid mating pair formation genes appeared similar to chromosomally encoded Brucella sp. virB genes (Schneiker et al., 2001; Tauch et al., 2002). Additionally, a putative PromA plasmid, defined as pMBUI6, was recently identified from freshwater sample (Brown et al., 2013). Important PromA-like features identified on pMBUI6 is the presence of topoisomerase gene (topA), the relaxase gene (mobB), and the long direct repeats in intergenic regions. Nevertheless, some of its backbone genes bear close similarity to pXF51 from Xylella fastidiosa but more distantly from PromA group.
In our present work, a self-transmissible BHR plasmid pSFA231 was isolated from petroleum-contaminated sediment in Shen-fu, China, by using the triparental mating method, selecting only for the plasmid's ability to mobilize a non-selftransmissible plasmid. Complete sequencing and phylogenetic analyses of this newly isolated plasmid revealed that it fell within the recently defined PromA plasmid group. Isolation of PromA plasmids with highly similar backbone sequences from different parts of the world provides further evidence for global spread of bacteria or their plasmids, and improves our understanding of the evolution of the PromA plasmid group.
Materials and Methods
Site Description and Sampling
The sampling site was located in Shen-fu wastewater irrigation zone (123°35′ E, 41°44′ N), the largest petroleum wastewater irrigation zone in Northeast of China. A 70-km irrigation channel was constructed in 1960's, and the wastewater mainly comes from an oil refinery. After 50-year exposure to petroleum-containing wastewater, soils in the irrigation zone have been seriously contaminated (Li et al., 2005; Zhou et al., 2012). Plasmid pSFA231 was isolated from a sediment sample collected from down-stream of the irrigation channel. Surface sediment sample (0~10 cm) was collected with a shovel as described by Cook et al. (2001). The collected sediment sample was placed in a plastic bag, and then was transported to the laboratory on ice. The fresh sediment sample was kept at 4°C for plasmid isolation, and was air-dried and sieved (2 mm) for the analysis of the basic physical and chemical properties. The total petroleum hydrocarbons (TPH) was determined as 760.1 mg/kg using gravimetric method (Villalobos et al., 2008), and the pH value (soil: water = 1: 5) was 6.5. The organic matter, total nitrogen and available nitrogen of the sample were 12.8 g/kg, 0.7 g/kg, 4.0 mg/kg, respectively.
Strains, Plasmids, and Media
A list of the strains and plasmids used in this study is provided in Table 1. Luria-Bertani (LB) broth was generally used to culture the strains. Solid medium was prepared by addition of 1.7% agar. When necessary for selection, antibiotics were added to the medium at the following concentrations: rifampicin, 50 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml. Mueller–Hinton (Becton, Dickinson, and Co., Franklin Lakes, NJ) agar was used for detection of antibiotic resistance. Cycloheximide (300 μg/ml) was added to LB agar (LBA) to prevent growth of fungus during screening of transconjugants.
Exogenous Isolation of the Plasmid
Triparental exogenous isolation of plasmids was performed as described by Hill et al. (1992). A rifampicin-resistant strain E. coli MG1685 (K12 RifR) (Fox et al., 2008) was used as the recipient to capture the plasmid from the sediment sample. E. coli JM109 (pBBR1MCS-5) (Yanischperron et al., 1985) was used as the donor, with the mobilizable plasmid pBBR1MCS-5 conveying resistance to gentamicin. Cultures of recipient and donor cells were grown overnight in LB broth containing corresponding antibiotics at 37°C.
Five grams of air-dried and sieved sediment sample was shaken for 1 h in 45 ml of sterile saline. The previously reported BHR plasmid pB10 (Schlüter et al., 2003) was added as a positive control. In the positive control flask, 5 g of soil sample was mixed with 100 μl of a 10−1 dilution (in saline) of a fully grown E. coli DH5α (pB10) culture (approximately 107 CFU/g soil). The suspension was allowed to settle for approximately 30 min. The supernatant was transported to the Eppendorf tube and then centrifuged (4°C, 10,000 rpm) for 10 min. After centrifugation, the supernatant was discarded and 3 ml of LB solid medium was added to resuspend the pellet. Individually, 500 μl of donor, recipient, and sediment were dispensed into 1.5 ml Eppendorf tubes as controls for the mating. For every mating, 500 μl portions of overnight grown cultures of the donor and the recipient were mixed in an Eppendorf tube with 500 μl of sediment supernatant. All the mating and control preparations were centrifuged (4°C, 10,000 rpm) for 5 min, and the pellet was resuspended in 50 μl of LB broth. Then 30 μl of this cell suspension was spotted onto an LB agar plate. After overnight of mating at 30°C, using a sterile loop a portion of the biomass (~1/4–1/3 of the mating “spot”) from each droplet was resuspended in 500 μl of saline, and then agitated vigorously with a Vortex mixer. The cell suspension was serially diluted in saline, and 0.1 ml samples were plated on LB agar supplemented with rifampicin (50 μg/ml) and gentamicin (10 μg/ml). Transconjugant colonies were picked up after 2 days incubation and purified on the same agar medium. Finally, cultures of transconjugant cells were grown overnight in LB medium containing rifampicin (50 μg/ml) and gentamicin (10 μg/ml) at 37°C. Physical evidence that mobilizing plasmids were present was obtained by plasmid extraction by using the alkaline lysis method, followed by agarose gel electrophoresis.
Detection of Antibiotic Resistance
To test antibiotic resistance of the isolated BHR plasmid, 1.8 ml overnight culture was centrifuged at 10,000 rpm for 10 min, and the cell pellets were washed with sterile saline for three times. The cells were resuspended in 500 μl sterile saline (CFU approximately 108), and 250 μl bacterial suspension was added into 150 ml pre-heated Mueller–Hinton agar, mixed by inversion and quickly poured to make an inoculated plate. Paper discs (6 mm in diameter) containing different antibiotics of known amounts (Oxoid Ltd) were placed on the inoculated plates with sterile forceps. The type and content of antibiotics are the following: kanamycin (K30, 30 μg/slice), chloramphenicol (C30), Ciprofloxacin (CIP5), erythromycin (E15), amoxicillin (AMC30), rifampicin (RD5), macrodantin(F300), nalidixic acid (NA30), imipenem (IPM10), gentamycin (CN10), carbenicillin (CAR100), sulfamethoxazole (W5), ceftazidime (CAZ30), polymyxin B (PB300), miramycin (SH100), tetracycline (TE30). After 24-h incubation at 37°C, antibiotic resistance was determined by measuring the inhibition zones around the antibiotic paper discs compared with those of the recipient strain E. coli MG1685 and the donor plasmid pBBR1MCS-5. When the diameter of the inhibition zone was 8 mm, the transconjugant strain was considered as resistant to the antibiotic.
Tagging the Plasmid with mini-Tn5 Transposon
Antibiotic resistance test revealed that plasmid pSFA231showed no resistance to any antibiotics. To facilitate selection in further analysis, pSFA231 was tagged with a miniTn5::Km1 transposon (De Lorenzo et al., 1990) using a biparental mating/ mobilization strategy. E. coli MG1685 (pSFA231) served as the recipient and the transposon pUTminiTn5::Km1 was provided by donor strain E. coli S17-1. Transconjugants were picked and streaked on LB agar supplemented with kanamycin (50 μg/ml). The tagged plasmid (KmR) was eventually transferred to E. coli EC100 (SmR) in a second round of biparental mating, at which point the captured plasmid was separated from the donor plasmid pBBR1MCS-5.
Host Range Test
To determine the self-transferability and the host range, plasmid pSFA231 was transferred from E. coli EC100 (SmR) to bacterial species from the other two Proteobacteria subgroups. Rifampicin-resistant (RifR) strains, Agrobacterium tumefaciens C58 (α-Proteobacteria) (Wood et al., 2001) and Cupriavidus necator JMP228 (β-Proteobacteria) (Top et al., 1995), were used as recipients in biparental matings, respectively (Heuer et al., 2012). The plasmids were considered to transfer successfully if colonies could grow on medium added with kanamycin and rifampicin. Finally, plasmid was extracted by alkaline lysis method (Feliciello and Chinali, 1993) to confirm the presence of the plasmid in transconjugants.
Sequencing and Annotation
Plasmid DNA for sequencing analyses was prepared using the QIAGEN Plasmid Midi Kit (QIAGEN GmbH, Germany) according to the protocols provided by manufacturer. The whole genome of plasmid pSFA231 was sequenced by Illumina Hiseq 2000 high-throughput sequencing platform at the Majorbio Bioinformatics Technology Co. Ltd (Shanghai, China). The paired-end library was generated for high-throughput sequencing. Sequence assembly was primarily done with the SOAPdenovo (http://soap.genomics.org.cn/, version: v1.05) and GapCloser software (Li et al., 2010). To acquire the complete sequence of plasmid pSFA231, gaps in the plasmid sequence were closed using the general PCR method. Gene prediction was done using the Glimmer 3.0 (http://www.cbcb.umd.edu/software/glimmer). The annotation information of the predicted genes was obtained through blastp alignment between the amino acid sequences of the predicted genes and the Nr database information using BLAST 2.2.24+. The annotated nucleotide sequence of plasmid pSFA231 was submitted to the GenBank database under the accession number KJ850907.
Bioinformatic Analyses
GC contents were calculated using the BioXM software. GenBank was searched for similar sequences using BLAST (Altschul et al., 1997). The PromA plasmids used in the comparative analysis are listed in Table 2. Due to the unclear classification, PromA-like plasmid pMBUI6 was not included in the comparison. Nucleotide sequences and amino acid (AA) sequences translated from the coding sequences were aligned using ClustalX (Thompson et al., 1997), then Mega 6 (Tamura et al., 2013) was used to infer the phylogenetic trees using the neighbor joining algorithm with the best-fit model. The SplitsTree program was used to infer the phylogenetic network (Huson and Bryant, 2006). The Pairwise genetic distance based on each backbone protein was calculated by Mega 6 using the Jones–Taylor–Thornton method (Norberg et al., 2011). A circular plasmid map was generated using the SnapGene Viewer (http://www.snapgene.com/products/snapgene_viewer). Schematic diagrams of multiple alignments of plasmids were produced by manually realigning the linear plasmid maps generated by the SnapGene Viewer. The identity scores of translated DNA sequences were calculated by the BLAST program, bl2seq (Tatusova and Madden, 1999).
Results
Isolation and General Characterization of the Plasmid pSFA231
A BHR plasmid, named here as pSFA231, was isolated from the petroleum-contaminated sediment. Antibiotic resistance test indicated that plasmid pSFA231 carried no additional antibiotic resistance compared to the recipient strain and donor plasmid. For further analysis, the plasmid was marked by a mini-Tn5 transposon carrying a kanamycin resistance gene cassette. The tagged plasmid was eventually transferred to E. coli EC100 (SmR) by a second round of biparental mating.
To investigate the transferability of the exogenously isolated plasmid pSFA231, the selected clone of E. coli EC100 (pSFA231) was used as donors in biparental matings with rifampicin-resistant strains of Agrobacterium tumefaciens C58 (α-Proteobacteria) and C. necator JMP228 (β-Proteobacteria). Results showed that plasmid pSFA231 can self-transfer and replicate in representative strains from three different subgroups of Proteobacteria.
Basic Genome Sequence Information of the BHR Plasmid pSFA231
The complete nucleotide sequence of the wild-type plasmid pSFA231 is determined to be 41518 bp with a GC content of 60.54%. Annotation of the sequence revealed a total of 50 orfs, of which 22 are transcribed on one strand and 28 on the other (Figure 1). The sequence has about 84% coding ratio with an average orf length of 705 bp. The closest relatives (with highest amino acid identity scores) of these orfs in GenBank are summarized in Table 3. Among the 50 predicted orfs, 29 were attributed to certain biological functions, 19 orfs coding for conserved hypothetical proteins, and the remaining 2 predicted genes do not have any known homologs. The putative known coding regions of pSFA231 are dominated by essential plasmid backbone genes involved in plasmid replication, maintenance and control, and conjugative transfer (Table 3, Figure 1).
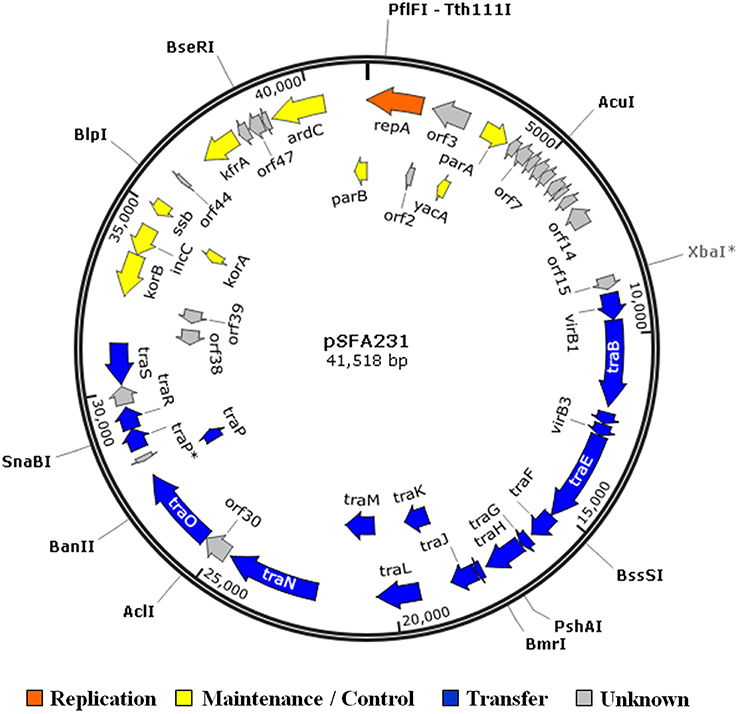
Figure 1. Circular map of plasmid pSFA231. The 50 orfs identified in the nucleotide sequence of pSFA231 are located on a circular map. The orfs are shown by arrows indicating the direction of transcription. Different colors indicate replication (orange) and partitioning functional regions (yellow), and the transfer functional regions (blue). Hypothetical coding regions are shown in gray. The genes transcribed in clockwise orientation are in the outer perimeter and those transcribed in anti-clockwise orientation are in the inner perimeter. The restriction enzyme cutting sites are shown as a filled circle.
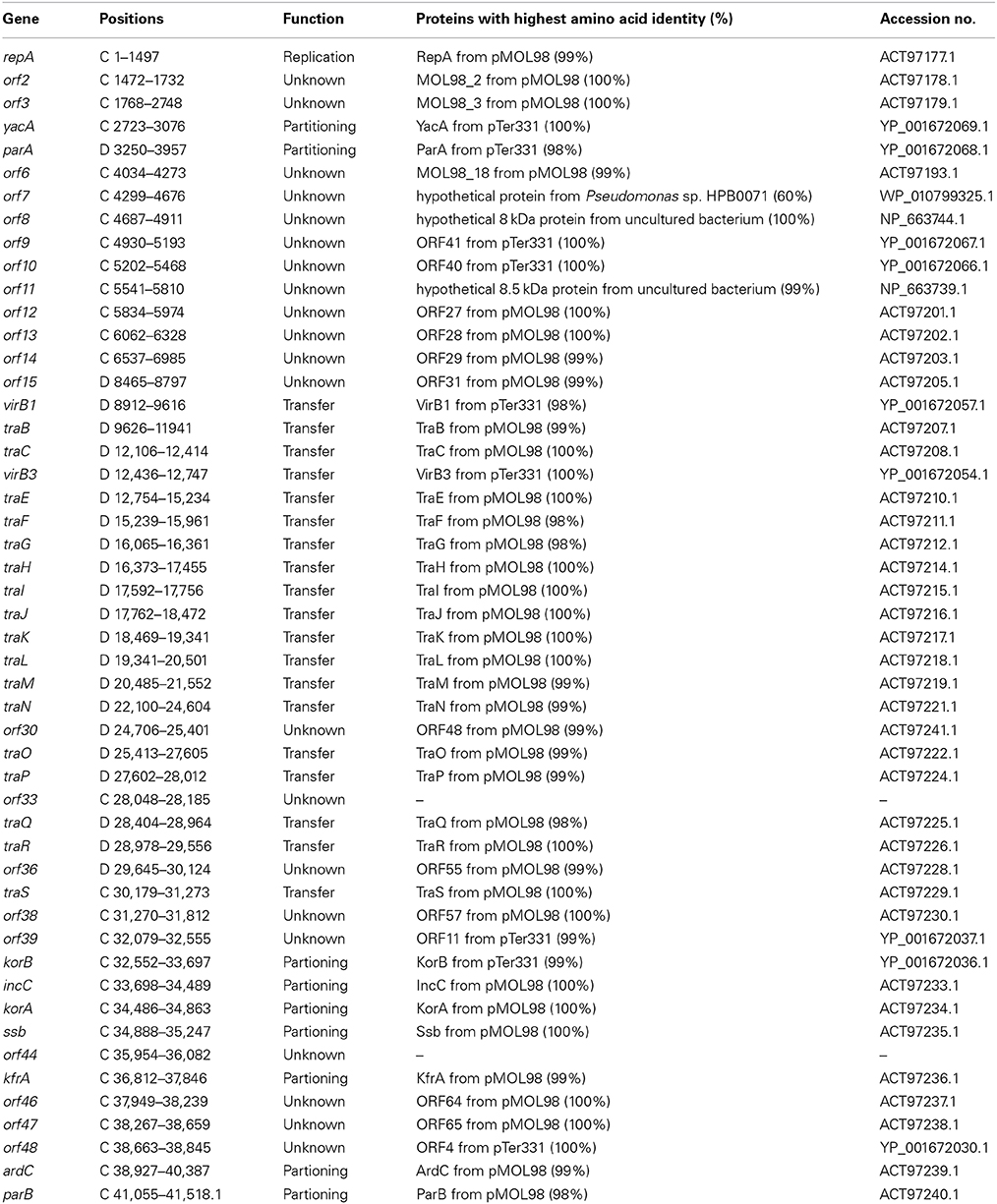
Table 3. Location of putative coding regions on plasmid pSFA231 and the closest relatives of the deduced proteins.
Similarity searches showed that most of the predicted orfs in plasmid pSFA231 coding for proteins are highly similar to those from plasmid pMOL98, and the remaining orfs were annotated to the proteins from plasmid pTer331 (Table 3). Both pMOL98 and pTer331 are listed as the members of the recently defined PromA plasmid family (Van der Auwera et al., 2009), leading to the conclusion that plasmid pSFA231 is a member of the PromA group.
Plasmid pSFA231 Harbors a Replication Module Unique to the PromA Family
Before PromA plasmids were recommended as a new family, their RepA proteins were reported to show some degree of identity with the RepA proteins of IncW plasmids. To further reveal the phylogenetic relationship of the PromA plasmids with IncW plasmids, a splits network (Figure 2) was constructed for 1000 bootstrap replicates of the RepA protein phylogeny of the currently reported PromA members and the selected IncW plasmids, R388 [GenBank accession: BR000038], R7K [GenBank accession: NC_010643.1], pPAES01 [GenBank accession: CP001109], pSa [GenBank accession: U3071.1], pXV2 [GenBank accession: AF201825.1], pIE321 [GenBank accession: NC_010716.1], pRM21 [GenBank accession: NC_001755.1], and pPRO2 [GenBank accession: NC_008608] (Fernández-López et al., 2006). The network, which presents a combinatorial generalization of phylogenetic trees, presents a star-like topology with six main clades (Figure 2). It can be visualized that the six PromA plasmids formed a cluster distinct from IncW plasmids, and the PromA-clade was clearly divided into two sub-clades. Plasmids pMRAD02 and pSB102 were clustered separately from the other four members, and was suggested here as the PromA-α. The initiator protein RepA of plasmid pSFA231 had 99, 91, and 90% similarity to the corresponding protein of pMOL98, pTer331, and pIPO2T, respectively, and thus, they all together were proposed to be grouped into the PromA-β.
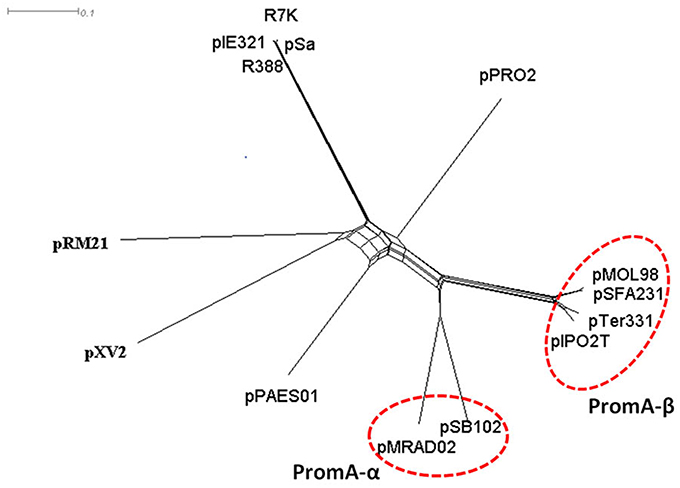
Figure 2. Phylogenetic network of RepA proteins of selected IncW and PromA plasmids, using the neighbor joining algorithm on protein distances with Poisson correction. Phylogenetic distance (amino acid difference percentage) was indicated by the length of the tree branches and the scale bars.
Comparison of the oriV region of plasmid pSFA231 and its closest homolog, pMOL98, further verified that the replication module of pSFA231 was similar to PromA plasmids. Like other members of the PromA family, pSFA231 was characterized as θ-type mode of replication, with an oriV-like region being located at 5.1 kb downstream of the repA gene. Within this region, we identified four putative iterons (RepA binding site), which are identical to the iteron sequences from pMOL98. Furthermore, a putative DnaA-binding site, a potential integration host factor (IHF) site and an AT-rich (86.67%) region were also found in this region (Figure 3).
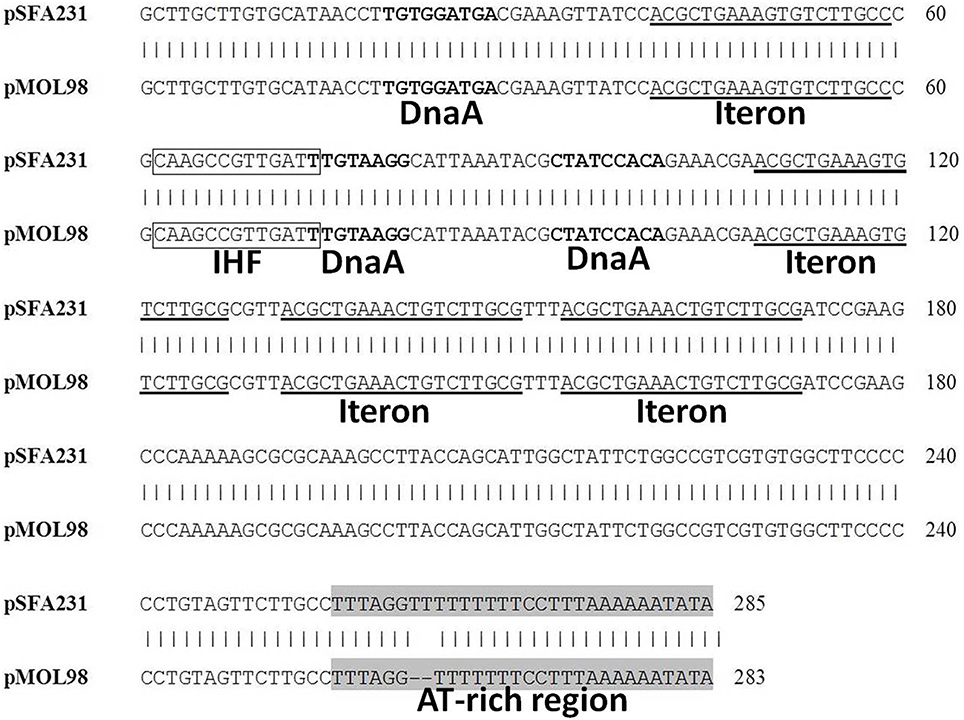
Figure 3. Alignment of the pSFA231 replicative origin region with the putative oriV regions of pMOL98. DnaA refers to DnaA boxes, IHF refers to putative Integration Host Factor binding sites.
Comparison of the Backbone Structure of PromA Plasmids
Comparison of the genome sequence between plasmid pSFA231 and the other five PromA plasmids, pMOL98, pIPO2, pTer331, pSB102, and pMRAD02, revealed a high level of structural similarity. They shared the common backbone regions including functional modules for replication (repA, oriV), conjugative transfer (tra), and maintenance/control (yacA, parA, korB, incC, korA, ssb, kfrA, ardC, and parB) (Figure 4). Nevertheless, phylogenetic divergences were found in specific loci indicating that PromA plasmids may have complex evolutionary histories. For instance, traO and traO* present in pMOL98 provide evidence for duplication (Figure 4). The putative relaxase gene traS locus on pSFA231, pMOL98, pSB102, and pMRAD02 is not visible on the pTer331 and pIPO2T, additionally, the krfA locus and parB locus located on pSFA231, pMOL98, pIPO2T, and pTer331, are almost entirely unrecognizable on pSB102 and pMRAD02. The presence and absence of specific backbone genes are most likely the result of insertions and/or deletions.
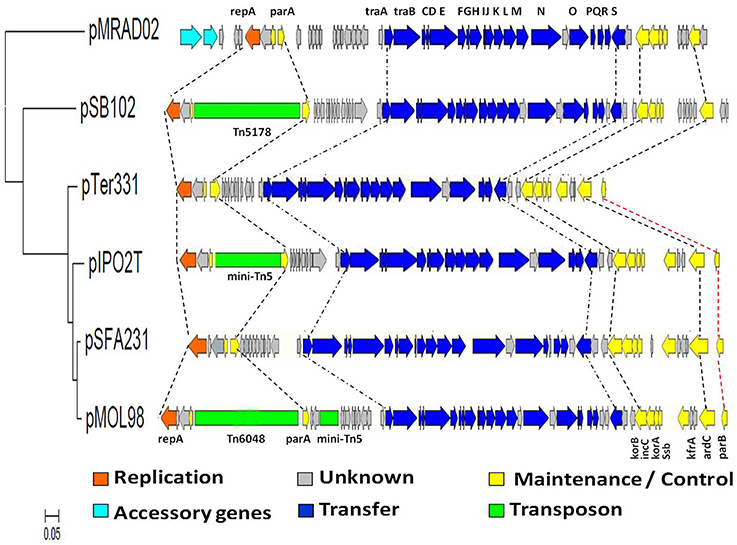
Figure 4. Schematic diagram of linear alignment of the 6 PromA plasmids. Phylogenetic tree was constructed based on the DNA sequences of concatenated 25 backbone genes using the Tamura- Nei model. The orfs are represented by block arrows. Predicted functions are indicated by the color key featured below the figure. Key backbone genes and accessory genes are annotated in the corresponding regions.
To evaluate the evolutionary history of the backbone regions, the pairwise genetic distances among the PromA plasmids were calculated based on the amino acid (AA) sequences of each backbone protein using the Jones-Taylor-Thornton method (Table S1). The AA similarity differed across the 6 plasmids. Among all the 5 known PromA plasmids, pMOL98 had the closest genetic distance to pSFA231 with respect genes of traC, virB3, traE, traH, traI, traJ, traK, traR, incC, korA, and ssb. In contrast, almost all the pMRAD02 backbone proteins showed the greatest genetic distance to our newly isolated plasmid pSFA231, except the traQ and traR. As for the plasmid pTer331, genes yacA, parA, virB1, traC, traM, traO, korB, and kfrA presented the closest, while genes traP, traQ, traR, and parB presented the greatest genetic distance to pSFA231.
To further reconstruct their evolutionary history, the phylogenetic analysis of the whole backbone regions, which are conserved and present in all 6 plasmids, was performed. It was observed that the gene coding for protein TraS is absent in plasmids pTer331 and pIPO2T, and plasmids pSB102 and pMRAD02 are lacking in the genes coding for proteins KfrA and ParB (Table S1). Thus, the traS gene sequence and the DNA sequences from kfrA to parB were excluded from the alignment, leaving two large backbone regions. One region contains 21 continuous genes, organized from repA to traR, while the other region includes genes korB, incC, korA, and ssb. Although the amino acid sequences translated from the counterparts of backbone genes in selected PromA plasmids (gene yacA in pMOL98, pSB102, and pMRAD02; parA in pSB102; traG in pMRAD02; traP in pMRAD02; korB in pIPO2T; incC in pIPO2T; and korA in pTer331, pIPO2T, and pMRAD02) were defined as hypothetical proteins, the corresponding genes are present in these plasmids, allowing us to include them in our comparative genomic analysis. A phylogenetic tree constructed from the concatenated DNA sequence showed that pSFA231 is most similar to pMOL98, then to pIPO2T and pTer331, but is phylogenetically divergent from pSB102 and pMRAD02 (Figure 4).
We also performed a phylogenetic analysis using six concatenated backbone gene products (Figure S1), namely, RepA, TraB, TraE, TraN, TraO, and KorB. Not surprisingly, the results are consistent with those based on the entire backbone regions, indicating that to simplify the process of comparative genomic analysis we can choose gene products with relatively large size and high level of synteny as targets for comparison.
The Accessory Regions and Transposons of the PromA Plasmids
One cluster of hypothetical orfs (orf6-orf15) with unknown functions was detected between parA and traA of the PromA members. Interestingly, we found that this gene cluster shows high DNA similarity with the corresponding regions on pMOL98 (97%), pIPO2T (95%), and pTer331 (95%), inferring that these genes (or some of them) may also be part of the common backbone of PromA-β sub-clade, although we still lack direct evidence. While transposons Tn5178 and Tn6048 were found to be inserted between yacA and parA gene on plasmids pSB102 and pMOL98, respectively. No transposon was detected in the corresponding region on our plasmid pSFA231, nor on pTer331.
Discussion
In the present study, a new BHR plasmid pSFA231 was isolated from a sediment sample collected from a petroleum-wastewater irrigation channel, using the triparental exogenous plasmid capture method. It was recommended that choosing donor and recipient strains that are phylogenetically distinct will increase the possibility of obtaining plasmids with a broad host range (Top et al., 1994). In our analysis, we also tried the donor/recipient system of E. coli DH5α (pSU4814)/C. necator JMP228 (β-Proteobacteria, RifR), but no self-transmissible BHR plasmids were isolated from the same sediment sample (data not shown). Although the plasmid pSFA231 was isolated by using a donor and recipient that both belonged to the same subgroup of Proteobacteria, further host range test showed that pSFA231 could successfully self-transfer and replicate in α-, β -, and γ-Proteobacteria.
Sediments are likely important reservoirs of BHR plasmid (Paul et al., 1991; Sobecky et al., 1997; Smalla et al., 2006). The rich biofilm structures in sediments (Westall and Rincé, 1994; Sutherland et al., 1998) may provide the bacteria more opportunities for cell contact and therefore transfer of mobile genetic elements. Moreover, BHR plasmids are frequently captured from contaminated environmental samples (Gstalder et al., 2003; Schlüter et al., 2007). BHR plasmids play an important role in the adaptation of bacterial populations to pollution stress, and long-term contamination may induce horizontal gene exchange and reshuffling of genetic information between phylogenetically distinct prokaryotes (Top and Springael, 2003). With long-term exposure to petroleum contamination, the Shen-fu wastewater irrigation zone likely provides a natural pool of BHR plasmids. Actually, a set of diverse BHR plasmids were captured from dozens of samples collected from Shen-fu irrigation zone in our experiment (unpublished data), including 4 IncP-1ε plasmids, 2 unknown plasmids, together with the PromA plasmid reported in this study. We expect an increasing number of phylogenetically diverse self-transmissible plasmids would be identified from this region by trying different capturing methods, such as biparental mating method or endogenous plasmid isolation (Top et al., 1994). Unexpectedly, in contrast to the BHR plasmids isolated from sludge collected from wastewater treatment plant (WWTP) (Schlüter et al., 2003, 2007; Wibberg et al., 2013), plasmid pSFA231 carried no antibiotic resistance genes. This phenomenon may not be abnormal. In a previous comparison of antibiotic resistance profiles of plasmids captured from non-polluted creek and WWTP effluent, no clear difference was found in the proportions of resistant plasmids captured from the two sites (Brown et al., 2013).
Based on a similarity search for putative orfs and subsequent comparative analysis, plasmid pSFA231 was proposed as a new member of the recently defined PromA plasmid family. Compared with the other incompatibility groups, PromA members were symbolized by a distinct replication initiation module, which contains a specific oriV-like region and a RepA protein (Gstalder et al., 2003; Van der Auwera et al., 2009). While the typical replication module of the IncP-1 group, into which most BHR plasmids have been classified, consists of trfA and ssb genes, and also an oriV region (Brown et al., 2013). Splits network analysis of the RepA protein clearly separated PromA members into two sub-divisions, illustrating the slight difference in their replication modules. Thus, in this study, we proposed to divide PromA family into two sub-groups, though there were only six members available. We believe that with more BHR plasmids being added into this recently defined group, new subgroups may be recommended in the future.
The phylogenetic information of backbone genes provides fundamental information on the “long-term” evolutionary history of BHR plasmids. In this study, the concatenated backbone regions of PromA family were compared between plasmid pSFA231 and five previously reported members. Although concatenation of backbone genes is problematic when there are distinct evolutionary histories of different functional regions of the plasmids (Sen et al., 2012), it is still a recommended method for inferring the evolutionary history of plasmids with higher backbone similarity (Norberg et al., 2011). Also, the SplitsTree algorithm allows us to discern the presence of divergent histories. Here, we use gene products for comparison rather than DNA sequence, because proteins are built from 20 amino acids while DNA only contains four different bases, meaning that the “signal-to-noise ratio” in protein sequence alignments is much better than in alignments of DNA (Wernersson, 2003). We found that all 6 PromA members share the highly conserved backbone regions, comprising replication, maintenance and control, and conjugative transfer functions. These plasmids were isolated from a variety of habitats, such as sediment, rhizosphere and soils, distributed in different locations in The Netherlands, Germany, Japan, and in this study, China (Table 2). It is of great interest that these geographically distinct BHR plasmids harbor backbone genes of high similarity. This fact suggests the wide distribution of PromA members.
Despite members of PromA family sharing a common backbone structure, phylogenetic analysis of the complete backbone regions still revealed significant divergence among the PromA members. Obviously, pSFA231 is more diverged from plasmids pMRAD02 and pSB102 than from pMOL98 and pTer331 (Figures 2, 4), which supports our recommendation on dividing the PromA family into two sub-divisions. During the process of evolution, the genetic organization of the backbone regions can rearrange via inversion, transposition, and duplication/loss processes (Price et al., 2001; Darling et al., 2004; Joshi et al., 2005), leading to the presence of diverse plasmids belonging to the same incompatibility group. For example, plasmid pMOL98 has two traO genes, providing evidence for duplication (Figure 4). In addition, lack of the traS genes in the backbone regions of the plasmids pSFA231, pMOL98, pSB102, and pMRAD02 indicated an indel during evolution.
Gains and losses of transposons and other MGEs often happen in BHR plasmid evolution (Kamachi et al., 2006). It was observed that transposons are embedded in the backbone of the plasmids pSB102 and pMOL98. The Tn5178 located on plasmid pSB102 confers mercury resistance (Schneiker et al., 2001), and Tn6048 carried on plasmid pMOL98 was characterized as multiple metal response (Van der Auwera et al., 2009). Because the lack of inserted elements is considered to be a sign of ancestry (Norberg et al., 2011), plasmid pSFA231 and pTer331 without any transposons are most likely to be closely related to the ancient ancestor of the PromA-β sub-clade.
A cluster of hypothetical orfs located between the parA and traA gene of plasmid pSFA231 catches our attention, due to its striking high similarity with the corresponding regions on pMOL98, pIPO2T, or pTer331. The conservation demonstrated by these hypothetical proteins indicates that these hypothetical ORFs may contain “essential” backbone proteins of PromA-β subgroup, although we still have no further direct evidence. Another hypothesis is that this orfs cluster is accessory genes that were first acquired and then stayed as the plasmid diverged. We made such a hypothesis just because this orfs cluster was exactly situated near the parA locus, which was usually considered as a “hot-spot” in PromA plasmids for insertion of accessory elements (Minakhina et al., 1999; Van der Auwera et al., 2009). The reason why the accessory genes insert into or near the region of parA locus so frequently is that this site contains a consensus AT-palindromic sequence and topology of the target DNA (Liu et al., 2005; Tobes and Pareja, 2006). Further analysis is required to confirm the structure and function of these hypothetical proteins in future studies.
In the present study, a new self-transmissible BHR plasmid pSFA231 was isolated from the petroleum-contaminated sediment and was recommended as a new member of the recently defined PromA family, based on phylogenetic and comparative genomic analysis. The present work is of great significance to add new information to the BHR plasmid sequence database that now exists. We believe that in this era of high-throughput sequencing, more members of BHR plasmids would be fully sequenced, and the extension of the database will improve our understanding of the genetic diversity of this important mobile genetic element.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Program of the National Science Foundation of China (31070102) for Hui Li, the US National Science Foundation Grant EF-0627988 to Eva M. Top, Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15010103) for Yong Jiang, and also the Idaho INBRE Program, NIH grants P20RR016454 and P20GM103408 through support for Celeste J. Brown. We also thank for Dr. Hao Sun from Institute of Applied Ecology, Chinese Academy of Sciences for helpful discussion in bioinformatics analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00777/abstract
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, C. J., Sen, D., Yano, H., Bauer, M. L., Rogers, L. M., Van der Auwera, G. A., et al. (2013). Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl. Environ. Microbiol. 79, 7684–7695. doi: 10.1128/AEM.02252-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cook, M. A., Osborn, A. M., Bettandorff, J., and Sobecky, P. A. (2001). Endogenous isolation of replicon probes for assessing plasmid ecology of marine sediment microbial communities. Microbiology 147, 2089–2101.
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Lorenzo, V., Herrero, M., Jakubzik, U., and Timmis, K. N. (1990). Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172, 6568–6572.
Feliciello, I., and Chinali, G. (1993). A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia Coli. Anal. Biochem. 212, 394–401. doi: 10.1006/abio.1993.1346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fernández-López, R., Garcillán-Barcia, M. P., Revilla, C., Lázaro, M., Vielva, L., and De La Cruz, F. (2006). Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30, 942–966. doi: 10.1111/j.1574-6976.2006.00042.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fox, R. E., Zhong, X., Krone, S. M., and Top, E. M. (2008). Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2, 1024–1039. doi: 10.1038/ismej.2008.53
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gstalder, M. E., Faelen, M., Mine, N., Top, E. M., Mergeay, M., and Couturier, M. (2003). Replication functions of new broad host range plasmids isolated from polluted soils. Res. Microbiol. 154, 499–509. doi: 10.1016/S0923-2508(03)00143-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hale, T., Guerry, P., Seid, R., Kapfer, C., Wingfield, M., Reaves, C., et al. (1984). Expression of lipopolysaccharide O antigen in Escherichia coli K-12 hybrids containing plasmid and chromosomal genes from Shigella dysenteriae 1. Infect. Immun. 46, 470–475.
Heuer, H., Binh, C. T., Jechalke, S., Kopmann, C., Zimmerling, U., Krögerrecklenfort, E., et al. (2012). IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front. Microbiol. 3:2. doi: 10.3389/fmicb.2012.00002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hill, K., Weightman, A., and Fry, J. (1992). Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl. Environ. Microbiol. 58, 1292–1300.
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ito, H., and Iizuka, H. (1971). Taxonomic studies on a radio-resistant Pseudomonas. XII. Studies on the microorganisms of cereal grain. Agric. Biol. Chem. 35, 1566–1571. doi: 10.1271/bbb1961.35.1566
Jechalke, S., Dealtry, S., Smalla, K., and Heuer, H. (2013). Quantification of IncP-1 plasmid prevalence in environmental samples. Appl. Environ. Microbiol. 79, 1410–1413. doi: 10.1128/AEM.03728-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joshi, B. D., Berg, M., Rogers, J., Fletcher, J., and Melcher, U. (2005). Sequence comparisons of plasmids pBJS-O of Spiroplasma citri and pSKU146 of S. kunkelii: implications for plasmid evolution. BMC Genomics 6:175. doi: 10.1186/1471-2164-6-175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kamachi, K., Sota, M., Tamai, Y., Nagata, N., Konda, T., Inoue, T., et al. (2006). Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1β plasmids without accessory mobile elements. Microbiology 152, 3477–3484. doi: 10.1099/mic.0.29056-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, H., Zhang, Y., Zhang, C., and Chen, G. (2005). Effect of petroleum-containing wastewater irrigation on bacterial diversities and enzymatic activities in a paddy soil irrigation area. J. Environ. Qual. 34, 1073–1080. doi: 10.2134/jeq2004.0438
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, G., Geurts, A. M., Yae, K., Srinivasan, A. R., Fahrenkrug, S. C., Largaespada, D. A., et al. (2005). Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 346, 161–173. doi: 10.1016/j.jmb.2004.09.086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mazodier, P., Petter, R., and Thompson, C. (1989). Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171, 3583–3585.
Mela, F., Fritsche, K., Boersma, H., Van Elsas, J. D., Bartels, D., Meyer, F., et al. (2008). Comparative genomics of the pIPO2/pSB102 family of environmental plasmids: sequence, evolution, and ecology of pTer331 isolated from Collimonas fungivorans Ter331. FEMS Microbiol. Ecol. 66, 45–62. doi: 10.1111/j.1574-6941.2008.00472.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Minakhina, S., Kholodii, G., Mindlin, S., Yurieva, O., and Nikiforov, V. (1999). Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33, 1059–1068. doi: 10.1046/j.1365-2958.1999.01548.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Norberg, P., Bergstrom, M., Jethava, V., Dubhashi, D., and Hermansson, M. (2011). The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2, 268. doi: 10.1038/ncomms1267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ono, A., Miyazaki, R., Sota, M., Ohtsubo, Y., Nagata, Y., and Tsuda, M. (2007). Isolation and characterization of naphthalene-catabolic genes and plasmids from oil-contaminated soil by using two cultivation-independent approaches. Appl. Microbiol. Biotechnol. 74, 501–510. doi: 10.1007/s00253-006-0671-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paul, J. H., Frischer, M. E., and Thurmond, J. M. (1991). Gene transfer in marine water column and sediment microcosms by natural plasmid transformation. Appl. Environ. Microbiol. 57, 1509–1515.
Price, C. W., Fawcett, P., Ceremonie, H., Su, N., Murphy, C. K., and Youngman, P. (2001). Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41, 757–774. doi: 10.1046/j.1365-2958.2001.02534.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rhodes, G., Parkhill, J., Bird, C., Ambrose, K., Jones, M. C., Huys, G., et al. (2004). Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 70, 7497–7510. doi: 10.1128/AEM.70.12.7497-7510.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning. New York, NY: Cold spring harbor laboratory press.
Schlüter, A., Heuer, H., Szczepanowski, R., Forney, L., Thomas, C., Pühler, A., et al. (2003). The 64508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149, 3139–3153. doi: 10.1099/mic.0.26570-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schlüter, A., Krause, L., Szczepanowski, R., Goesmann, A., and Pühler, A. (2008). Genetic diversity and composition of a plasmid metagenome from a wastewater treatment plant. J. Biotechnol. 136, 65–76. doi: 10.1016/j.jbiotec.2008.03.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schlüter, A., Szczepanowski, R., Pühler, A., and Top, E. M. (2007). Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31, 449–477. doi: 10.1111/j.1574-6976.2007.00074.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schneiker, S., Keller, M., Dröge, M., Lanka, E., Pühler, A., and Selbitschka, W. (2001). The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucl. Acids Res. 29, 5169–5181. doi: 10.1093/nar/29.24.5169
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sen, D., Brown, C. J., Top, E. M., and Sullivan, J. (2012). Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol. Biol. Evol. 30, 154–166. doi: 10.1093/molbev/mss210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sen, D., Van der Auwera, G. A., Rogers, L. M., Thomas, C. M., Brown, C. J., and Top, E. M. (2011). Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl. Environ. Microbiol. 77, 7975–7983. doi: 10.1128/AEM.05439-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smalla, K., Haines, A. S., Jones, K., Krögerrecklenfort, E., Heuer, H., Schloter, M., et al. (2006). Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1β plasmids with a complex mer transposon as the sole accessory element. Appl. Environ. Microbiol. 72, 7253–7259. doi: 10.1128/AEM.00922-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sobecky, P. A., Mincer, T. J., Chang, M. C., and Helinski, D. R. (1997). Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl. Environ. Microbiol. 63, 888–895.
Sutherland, T., Amos, C., and Grant, J. (1998). The effect of buoyant biofilms on the erodibility of sublittoral sediments of a temperate microtidal estuary. Limnol. Oceanogr. 43, 225–235. doi: 10.4319/lo.1998.43.2.0225
Szpirer, C., Top, E. M., Couturier, M., and Mergeay, M. (1999). Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology 145, 3321–3329.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tatusova, T. A., and Madden, T. L. (1999). BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174, 247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tauch, A., Schneiker, S., Selbitschka, W., Pühler, A., van Overbeek, L. S., Smalla, K., et al. (2002). The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere. Microbiology 148, 1637–1653.
Thomas, C. M. (2000). Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491. doi: 10.1046/j.1365-2958.2000.02006.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tobes, R., and Pareja, E. (2006). Bacterial repetitive extragenic palindromic sequences are DNA targets for insertion sequence elements. BMC Genomics 7:62. doi: 10.1186/1471-2164-7-62
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Top, E. M., De Smet, I., Verstraete, W., Dijkmans, R., and Mergeay, M. (1994). Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl. Environ. Microbiol. 60, 831–839.
Top, E. M., Holben, W. E., and Forney, L. J. (1995). Characterization of diverse 2, 4- dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl. Environ. Microbiol. 61, 1691–1698.
Top, E. M., and Springael, D. (2003). The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotech. 14, 262–269. doi: 10.1016/S0958-1669(03)00066-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van der Auwera, G. A., Król, J. E., Suzuki, H., Foster, B., Van Houdt, R., Brown, C. J., et al. (2009). Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie Van Leeuwenhoek 96, 193–204. doi: 10.1007/s10482-009-9316-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Villalobos, M., Avila-Forcada, A. P., and Gutierrez-Ruiz, M. (2008). An improved gravimetric method to determine total petroleum hydrocarbons in contaminated soils. Water Air Soil Pollut. 194, 151–161. doi: 10.1007/s11270-008-9704-1
Wernersson, R. (2003). RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic. Acids Res. 31, 3537–3539. doi: 10.1093/nar/gkg609
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Westall, F., and Rincé, Y. (1994). Biofilms, microbial mats and microbe−particle interactions: electron microscope observations from diatomaceous sediments. Sedimentology 41, 147–162. doi: 10.1111/j.1365-3091.1994.tb01396.x
Wibberg, D., Szczepanowski, R., Eikmeyer, F., Puhler, A., and Schluter, A. (2013). The IncF plasmid pRSB225 isolated from a municipal wastewater treatment plant's on-site preflooder combining antibiotic resistance and putative virulence functions is highly related to virulence plasmids identified in pathogenic E. coli isolates. Plasmid 69, 127–137. doi: 10.1016/j.plasmid.2012.11.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323. doi: 10.1126/science.1066804
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yanischperron, C., Vieira, J., and Messing, J. (1985). Improved M13 phage cloning vectors and host strains-nucleotide-sequences of the M13MP18 and PUC19 vectors. Gene 33, 103–119. doi: 10.1016/0378-1119(85)90120-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, L., Li, H., Zhang, Y., Wang, Y., Han, S., and Xu, H. (2012). Abundance and diversity of Sphingomonas in Shenfu petroleum-wastewater irrigation zone, China. Environ. Sci. Pollut. Res. Int. 19, 282–294. doi: 10.1007/s11356-011-0552-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: broad-host-range plasmid, PromA plasmid family, complete sequence, plasmid backbone regions, comparative genomic analysis
Citation: Li X, Top EM, Wang Y, Brown CJ, Yao F, Yang S, Jiang Y and Li H (2015) The broad-host-range plasmid pSFA231 isolated from petroleum-contaminated sediment represents a new member of the PromA plasmid family. Front. Microbiol. 5:777. doi: 10.3389/fmicb.2014.00777
Received: 07 November 2014; Accepted: 18 December 2014;
Published online: 12 January 2015.
Edited by:
Holger Heuer, Julius Kühn-Institut, GermanyReviewed by:
Kok Gan Chan, University of Malaya, MalaysiaMichael Gillings, Macquarie University, Australia
Oleg Reva, University of Pretoria, South Africa
Copyright © 2015 Li, Top, Wang, Brown, Yao, Yang, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, State Key Laboratory of Forest and Soil Ecology, Institute of Applied Ecology, Chinese Academy of Sciences, No. 72 Wenhua Road, Shenyang 110164, China e-mail: huili@iae.ac.cn
 Xiaobin Li
Xiaobin Li Eva M. Top
Eva M. Top Yafei Wang1
Yafei Wang1 Celeste J. Brown
Celeste J. Brown Fei Yao
Fei Yao Yong Jiang
Yong Jiang Hui Li
Hui Li