- Department of Food, Environmental and Nutritional Sciences, University of Milan, Milan, Italy
Beneficial microorganisms are increasingly used in agriculture, but their efficacy often fails due to limited knowledge of their interactions with plants and other microorganisms present in rhizosphere. We studied spatio-temporal colonization dynamics of lettuce roots and rhizosphere by genetically modified Streptomyces spp. Five Streptomyces strains, strongly inhibiting in vitro the major soil-borne pathogen of horticultural crops, Sclerotinia sclerotiorum, were transformed with pIJ8641 plasmid harboring an enhanced green fluorescent protein marker and resistance to apramycin. The fitness of transformants was compared to the wild-type strains and all of them grew and sporulated at similar rates and retained the production of enzymes and selected secondary metabolites as well as in vitro inhibition of S. sclerotiorum. The tagged ZEA17I strain was selected to study the dynamics of lettuce roots and rhizosphere colonization in non-sterile growth substrate. The transformed strain was able to colonize soil, developing roots, and rhizosphere. When the strain was inoculated directly on the growth substrate, significantly more t-ZEA17I was re-isolated both from the rhizosphere and the roots when compared to the amount obtained after seed coating. The re-isolation from the rhizosphere and the inner tissues of surface-sterilized lettuce roots demonstrated that t-ZEA17I is both rhizospheric and endophytic.
Introduction
Roots anchor plants in soil, provide uptake of water and nutrients, and mediate numerous interactions with soil organisms. The interface between roots and soil – where most of these interactions take place – is called rhizosphere. This narrow and specific zone is distinct from bulk soil in terms of nutrient availability, pH and presence of a wide variety of microorganisms and invertebrates attracted and influenced by root exudates and rhizodeposits (Hinsinger et al., 2009; Compant et al., 2010; Philippot et al., 2013). Many microbes present in rhizosphere have neutral effect on plants, while others positively or negatively affect host development and health via complex interactions, which we are only beginning to understand (Raaijmakers et al., 2009; Compant et al., 2010; Glick, 2012). Some microorganisms are deleterious as they compete with plants for nutrients or cause disease (soil borne plant pathogens), while others support their hosts by mobilizing nutrients, stimulating growth, and increasing yield or reducing biotic and abiotic stresses, such as mycorrhizal fungi and plant growth promoting bacteria (PGPB; Compant et al., 2010; Aeron et al., 2011; Smith and Smith, 2011).
Plant growth promoting bacteria are gaining more and more attention in modern agriculture, where sustainable and environmentally friendly strategies of crop cultivation increasingly rely on their use as biofertilizers, phytostimulants, or biopesticides. They employ several mechanisms to improve the plant growth, such as synthesis of phytohormones, nitrogen fixation and increasing availability of nutrients by production of siderophores and solubilization of phosphates (Lugtenberg et al., 2002; Compant et al., 2010). Furthermore, special attention is dedicated to biological control agents (BCAs), a group of microorganisms producing a wide variety of biologically active molecules potentially able to inhibit plant pathogens. Antagonism is one of the most common modes of action; here the BCA inhibits or kills pathogens via production of diffusible or volatile antimicrobial compounds and cell wall degrading enzymes. Antagonism is widespread in Bacillus, Pseudomonas, and Streptomyces spp., from which a wide range of biologically active secondary metabolites were isolated (Raaijmakers et al., 2002; Compant et al., 2005).
Despite the optimal performance at laboratory-scale screening tests, PGPB often fail to demonstrate their potential or show inconsistent results in greenhouse and field trials. This variable performance may have different causes, such as reduced or delayed expression of bioactive molecules in the presence of competing microorganisms or lower rhizosphere competence, i.e., poor colonization of root tissues and rhizosphere of the host plant (Lugtenberg et al., 2001; Compant et al., 2010; Ghirardi et al., 2012). To overcome these obstacles, it is essential to understand how PGPB interact with the host plant and with other microorganisms present in soil. Several studies have demonstrated better plant protection when bioactive Pseudomonas spp. strains with improved rhizosphere-competence were used (Ghirardi et al., 2012). In recent years, several characters essential for rhizosphere colonization were identified in Pseudomonas spp. (Latour et al., 2003; Lugtenberg and Kamilova, 2009), however, similar studies are missing for other beneficial genera of bacteria.
One of such genera is Streptomyces, filamentous Gram-positive bacteria commonly inhabiting soil and rhizosphere and renowned for the production of a variety of bioactive secondary metabolites (Loria et al., 2006; Hopwood, 2007). They have been largely exploited in pharmaceutical industry since 1940s (Watve et al., 2001; Lucas et al., 2013), whereas only a few have been developed as commercial products for plant application in agriculture (Yuan and Crawford, 1995; Minuto et al., 2006; Berg, 2009). Streptomycetes have been long considered simply as free-living soil inhabitants, but recently the importance of their complex interactions with plants and other organisms is being uncovered (Seipke et al., 2012). Some of them, such as S. scabies or S. turgidiscabies, are plant pathogens with broad host range, causing important economic losses especially on tap root and tuber crops, such as potatoes, sweet potatoes, carrots, or beet (Loria et al., 2006; Seipke et al., 2012). On the contrary, many others establish beneficial relationships with host plants as endophytes (Sardi et al., 1992; Coombs and Franco, 2003; Cao et al., 2004). Auxin production was described for endophytic and free living Streptomyces in rhizosphere (Coombs et al., 2004; Khamna et al., 2009), while S. lydicus augmented the nodulation by Rhizobium species in pea plants, increasing iron and molybdenum assimilation as well as root growth (Tokala et al., 2002; Seipke et al., 2012).
Several markers have been developed and adopted to study localization and quantification of PGPB in the rhizosphere; among these, antibiotic resistance has been widely used (Prosser, 1994; Gamalero et al., 2003). Because many of soil microorganisms produce a variety of different antibiotics, it is necessary to determine the specificity of the antibiotic marker selected for the identification of PGPB before its use. Currently, fluorescent markers are gaining increasing popularity for colonization studies (Lu et al., 2004; Cao et al., 2011; Krzyzanowska et al., 2012). Various derivatives of green fluorescent protein (GFP) have been engineered to increase the fluorescence and to overcome the variable expression of the original marker in different species (Errampalli et al., 1999; Gamalero et al., 2003). Enhanced GFP (EGFP) contains numerous silent nucleotide changes in comparison to GFP to maximize its expression in mammalian cells (Haas et al., 1996), and was adopted for use in Streptomyces spp., which have a similar codon usage (Sun et al., 1999).
Green fluorescent protein has been utilized to study PGPB colonization of roots and rhizosphere in sterile conditions (Coombs and Franco, 2003; Weyens et al., 2012). These studies provide a basic understanding of the interactions between PGPB and the host plant, but they do not consider the complex interactions in vivo. In non-sterile conditions with high microbial diversity, PGPB have to compete with other microorganisms present in the rhizosphere, and in some cases the competition reduced the colonization ability of PGPB (Cao et al., 2011; Hohmann et al., 2012; Weyens et al., 2012). Moreover, the activity and the fitness of the transformed strain need to be controlled following the transformation, as it has been observed that the presence of the transgene may interfere with the biological activity of the studied organism (Nigro et al., 1999; Lübeck et al., 2002; Weyens et al., 2012).
The objective of this work was to get insight into the localization and colonization of a genetically modified Streptomyces strain, selected as potential BCA, in lettuce roots and rhizosphere. First, we compared the fitness of the transformed and the corresponding wild-type strains, then we studied the colonization dynamics of the most promising transformed strain in rhizosphere and roots of lettuce plants in non-sterile growth substrate. Finally, we compared the effect of two inoculation methods on the ability of the Streptomyces strain to differentially colonize rhizosphere and roots.
Materials and Methods
Transformation of Streptomyces spp.
Five Streptomyces strains, potential BCAs against Sclerotinia sclerotiorum, were used in this study: CX14W, CX16W, FT05W, SW06W, and ZEA17I. They were maintained at the Plant Pathology Laboratory, Department of Food, Environmental and Nutritional Sciences (DeFENS), University of Milan, and selected previously from a wide collection of actinomycetes isolated from roots of different plants (Sardi et al., 1992; Petrolini et al., 1996; Bonaldi et al., 2011, 2014). Escherichia coli strain ET12567 (harboring the helper plasmid pUZ8002), was provided by prof. Flavia Marinelli, University of Insubria, Italy, and was used as donor strain for conjugation. Plasmid pIJ8641, obtained from prof. Mervyn Bibb, John Innes Centre, UK, was maintained in E. coli strain DH5α. It carries the EGFP gene under the constitutive ermE promoter, an apramycin resistance marker [aac(3)IV], an oriT/RK2 region, and a lambda phage chromosomal integration sequence (IntC31; Sun et al., 1999). The strain S. coelicolor A3(2) was obtained from F. Marinelli, and used as a reference strain to evaluate transformation efficiency.
Plasmid pIJ8641 was transformed into the donor strain E. coli ET12567 (pUZ8002) by rubidium chloride method (Hanahan, 1983) and conjugated into recipient Streptomyces strains as previously described (Kieser et al., 2000). Prior to conjugation, the concentration of the E. coli donor strain ET12567 containing plasmid pIJ8641 was adjusted to 1 × 107 CFU/mL. The ex-conjugants were selected on the basis of apramycin resistance. The conjugation efficiency was calculated as number of ex-conjugant colonies per number of recipient spores.
Genomic DNA of wild-type and transformed (t-) Streptomyces strains was extracted by the CTAB method (Kieser et al., 2000). The amplification of 16S rDNA fragment (expected size 1500 bp) was used to evaluate the quality of DNA in all samples, using PCR primers fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and fD2 (5′-ACGGCTACCTTGTTACGACTT-3′; Weisburg et al., 1991), and the following thermal cycling conditions: initial denaturation at 94°C for 1 min, 30 cycles of denaturation at 92°C for 45 s, annealing at 56°C for 30 s and extension at 72°C for 2 min, a final extension at 72°C for 5 min. PCR primers rEGFP-N (5′-CTGGTCGAGCTGGACGGCGACG-3′) and rEGFP-C (5′-CACGAACTCCAGCAGGACCA TG-3′) were designed to amplify the EGFP gene fragment (expected fragment size 700 bp), using the following thermal cycling conditions: initial denaturation at 94°C for 1 min, 30 cycles of denaturation at 92°C for 45 s, annealing at 60°C for 45 s and extension at 72°C for 2 min, a final extension at 72°C for 2 min. The DNA amplification was carried out using PCR thermal cycler (BioRad, USA), performed in a total volume of 25 μL containing 30–50 ng DNA, 0.25 μM each primer, 1 U/μL Go Taq DNA polymerase (Promega, USA), 5 μL of 5x Go Taq buffer (Promega, USA), and 0.2 mM of each dNTP. The PCR products were visualized under Gel Doc transilluminator (BioRad, USA) following electrophoresis in 1%(w/v) agarose gel.
For microscopic observations, the transformed Streptomyces strains were inoculated on Czapek yeast extract agar (CZY; 35 g/L Czapek-Dox Broth Difco, 2 g/L Yeast Extract Difco, 15 g/L agar). A microscopic cover slide was partially inserted in the medium at the edge of the inoculated strain under the 45° angle to allow the strain to grow on the cover slide. The plates were incubated at 24°C for 5 days. Subsequently, cover slides were removed from the medium and observed by brightfield and epifluorescence microcopy using Olympus BX51 with the FITC filter set (467–498 nm excitation and 513–556 nm emission) to confirm the expression of EGFP in transformants.
Inhibition of Sclerotinia sclerotiorum Growth In Vitro
The antagonistic activity of wild-type and transformed (t-) Streptomyces strains against S. sclerotiorum was determined by dual culture assay on CZY agar as described (Bonaldi et al., 2014). Briefly, 10 μL of Streptomyces spore suspension (1 × 107 CFU/mL) were inoculated on a 40 mm line two days prior to S. sclerotiorum inoculation. An agar-mycelium plug (5 mm diameter), obtained from the edge of an actively growing colony of S. sclerotiorum grown on Malt Extract Agar (MEA; 20 g/L Malt Extract, Difco, and 15 g/L agar), was placed at 25 mm distance from the growing Streptomyces colony and the plates were incubated for 72 h at 24°C. Plates inoculated with S. sclerotiorum only were used as a control. The antagonistic activity was determined by calculating the percentage of growth inhibition of S. sclerotiorum compared to the control. The experiment was repeated twice in three replicates.
Mycelium Growth and Sporulation
The mycelium growth curve of transformed and wild-type strains was determined daily as follows: 20 μL of Streptomyces spore suspension (1 × 107 CFU/mL) were transferred into a 50 mL tube containing 20 mL of CZY broth, and incubated at 30°C with 200 rpm constant shaking for 8 days. Each day, 2 mL of liquid culture were removed and spun at 10600 g for 10 min and the dry weight of the pellet was repeated twice in three replicates and expressed in g/L.
The sporulation of the strains was measured by plating 1 mL of spore suspension (1 × 107 CFU/mL) on a CZY agar plate and determining the number of spores produced after 6 days of incubation at 30°C (Grantcharova et al., 2005). Five mL of sterile water were added to the Petri plate and the surface of colonies was gently scraped to release the newly formed spores (Kieser et al., 2000). The spore suspension was filtrated through two layers of sterile gauze and the spore concentration (CFU/mL) was quantified by plating serial dilutions of the spore suspension and counting the number of colonies grown after 4 days of incubation at 30°C. The experiment was repeated twice in three replicates.
Production of Secondary Metabolites
Siderophore production
Ten milliliter of Fe-free Czapek solution (300 g/L NaNO3, 50 g/L KCl, 50 g/L MgSO4 ⋅ 7H2O) were mixed with 15 g/L agar, 30 g/L sucrose, 1 g/L K2HPO4, and 5 g/L yeast extract to prepare the Fe-free Czapek agar medium. Ten microliter of Streptomyces spore suspension (1 × 107 CFU/mL) were inoculated in the center of a Fe-free Czapek agar plate and incubated at 30°C for two weeks. Subsequently, the Streptomyces colony was overlaid by 15 mL of the Chrome azurol S (CAS) agar (Schwyn and Neilands, 1987; Perez-Miranda et al., 2007). The siderophore production was determined as color change in the overlay medium (from blue to orange) after 24 h of incubation at room temperature. The experiment was repeated twice in three replicates.
Chitinase production
The colloidal chitin and the colloidal chitin agar were prepared as described previously (Saima et al., 2013). Ten microliter of Streptomyces spore suspension (1 × 107 CFU/mL) were inoculated in the center of colloidal chitin agar plate (chitin as single carbon sources) as a 40 mm line and incubated at 30°C for 10 days. The production of chitinase was determined based on the presence of a clear hydrolysis zone on the agar plate below the colony. The experiment was repeated twice in three replicates.
Phosphate solubilization
The phosphate solubilization activity of the Streptomyces strains was assessed using a plate assay with National Botanical Research Institute’s Phosphate (NBRIP) medium (Nautiyal, 1999), in which Ca3(PO4)2 is the only phosphate source. Ten microliter of Streptomyces spore suspension (1 × 107 CFU/mL) were inoculated in the center of a NBRIP-medium Petri plate and incubated at 30°C for 2 weeks. The phosphate solubilization was determined based on the presence of a clear hydrolysis zone on the agar plate below the colony. The test was repeated twice in three replicates.
Indole-3-acetic acid (IAA) synthesis
The IAA production was determined as described previously (Bric et al., 1991; Bano and Musarrat, 2003). In brief, 10 μL of Streptomyces spore suspension (1 × 107 CFU/ml) were incubated with constant shaking at 5 g in 5 mL CZY broth added with 500 μg/mL tryptophan (Sigma, USA) in the dark at 30°C for 10 days. Two mL of the liquid culture were centrifuged for 10 min at 18000 g. One mL of the supernatant was mixed with 50 μL 10 mM orthophosphoric acid and 2 mL of Salkowski reagent (1 mL of 0.5M FeCl3 in 50 mL of 35% HClO4). The tubes were incubated at room temperature for 30 min. The development of pink color indicated the IAA production, which was quantified by spectrophotometer at 530 nm. The experiment was repeated twice in three replicates.
Soil, Root, and Rhizosphere Colonization by t-ZEA17I
The transformed ZEA17I strain (t-ZEA17I) was grown on CZY medium containing 50 mg/L of apramycin at 24°C for 10 days. Spores were collected in 10% sterile glycerol and filtered through two layers of gauze. The concentration was determined and the spore suspension was stored at -20°C.
Bulk soil colonization
Prior to colonization studies, the presence of naturally occurring apramycin-resistant streptomycetes in non-sterilized Irish and Baltic peat-based growth substrate (Vigorplant, Italy) was assessed. A portion of the substrate was resuspended in sterile water and plated on water agar medium (WA) containing 15 g/L agar, 25 mg/L nalidixic adic, 50 mg/L apramycin, 50 mg/L nystatin, and 50 mg/L cycloheximide. Plates were incubated for 7 days at 24°C and the presence of apramycin-resistant streptomycete colonies was visually checked.
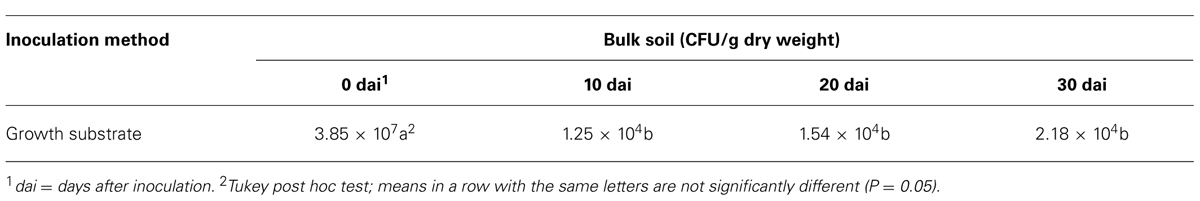
The growth substrate was placed in a polystyrene seed tray (48 cm3/cell) and watered with tap water. In every cell, t-ZEA17I was uniformly distributed on the top of the substrate adding 1 mL spore suspension (1 × 107 CFU/mL). The growth substrate was incubated in a growth chamber (24°C, 55% relative humidity and 15 h photoperiod) and watered every 2–3 days with tap water. t-ZEA17I was re-isolated 4 h (day 0), 10, 20, and 30 days after inoculation (dai) in six replicates. The entire amount of growth substrate in the cell was collected and weighed. The substrate was mixed to homogenize the inoculum and divided in two identical parts. One part was incubated in the oven at 50°C and the dry weight was determined. The other part was stirred in sterilized water (1:10 substrate fresh w/v) for one hour and serial dilutions were plated on WA. Plates were incubated for 7 days at 24°C and streptomycete colonies were counted. The t-ZEA17I concentration was expressed as CFU/g of growth substrate dry weight.
Plant inoculation
Ice queen lettuce seedlings (Lactuca sativa var. capitata, Iceberg group, Semeurop, Italy) were grown in polystyrene seed trays, as described previously. Seeds were surface sterilized in 0.7% sodium hypochlorite for 5 min and rinsed three times in sterile water. Two methods were used to inoculate the t-ZEA17I strain. In the growth substrate inoculation method, 1 mL spore suspension (1 × 107 CFU/mL) was uniformly distributed in every cell on the top of the growth substrate. In the seed coating method, 50 seeds were soaked in 1 mL of t-ZEA17I spore suspension (1 × 107 CFU/mL) and left to dry under the laminar flow hood. One seed for each cell of the tray was sown and the seedlings were incubated and watered as described previously.
To determine the inoculum load t-ZEA17I was re-isolated from seeds and growth substrate after inoculum application. In case of the growth substrate inoculation method, the t-ZEA17I strain was re-isolated four hours after soil inoculation as described above for bulk soil, and its amount was expresses as CFU/g of growth substrate dry weight. For the seed coating method, 10 randomly collected seeds were incubated for 30 min in 1 mL of sterile 0.9% NaCl and serial dilutions were plated on WA medium in six replicates. Following incubation at 24°C for 7 days the t-ZEA17I colonies were counted and the amount was expressed first as CFU/seed and then recalculated as CFU/g of growth substrate dry weight.
t-ZEA17I re-isolation from rhizosphere and root tissues
The t-ZEA17I strain was re-isolated 10, 20, and 30 days after sowing from rhizosphere and root tissues of six lettuce seedlings, equal to number of replicates. Seedlings with root system were carefully taken off the cell and the bulk soil was removed by gently shaking the plants (Bulgarelli et al., 2012).
For the rhizosphere analysis, each seedling was cut at base and the roots were vortexed two times for 15 s in 1–3 mL (volume varying according to period of sampling) of sterilized 0.9% NaCl and 0.02% Silwet L-77 (washing solution). The roots were removed and the suspension was filtered through a 300 μm nylon mesh to obtain the rhizosphere soil and its dry weight was determined. The suspension was centrifuged at 10600 g for 10 min and the pellet was resuspended in 0.5–1.5 mL of washing solution and plated in serial dilutions on WA medium. The plates were incubated at 24°C for 7 days. The t-ZEA17I colonies were counted and the concentration was expressed as CFU/g of rhizosphere dry weight.
For inner root tissues analysis, the roots were surface sterilized with propylene oxide for one hour (Sardi et al., 1992). Then, they were washed in washing solution and 1/10 of the total volume was plated on WA medium to verify the absence of contaminants. Subsequently the roots were finely homogenized in 1–3 mL washing solution, let to macerate for one hour and the suspension was plated in serial dilutions. The t-ZEA17I concentration was determined as described before and expressed as CFU/g of roots dry weight.
Statistical Analyses
All analyses were done using R software, version R3.0.2. (R Core Team, 2013). The statistical differences between data of transformed and wild-type strains in S. sclerotiorum growth inhibition, sporulation, and IAA production were compared by a Student’s t-test (P = 0.05). The percent data were arcsine root-squared transformed. The soil, root, and rhizosphere colonization data were submitted to ANOVA, followed by a Tukey post hoc test for multiple comparison (P = 0.05), using the TukeyC package (Faria et al., 2013).
Results
Transformation of Streptomyces spp. With Plasmid pIJ8641
All six strains, including S. coelicolor A3(2) were transformed with the pIJ8641 plasmid harboring the EGFP gene under a constitutive promoter and apramycin resistance. Conjugation efficiency varied among strains: four strains, CX14W, SW06W, CX16W, and FT05W showed conjugation efficiency similar to S. coelicolor A3(2), while one strain, ZEA17I conjugated with lower efficiency (Table 1). The EGFP gene was detected in transformed strains (data not shown), and its expression was confirmed by fluorescence microscopy observing a strong green fluorescence in all transformants following exposition to fluorescent light (Figure 1), while the corresponding wild-type strains did not fluoresce.
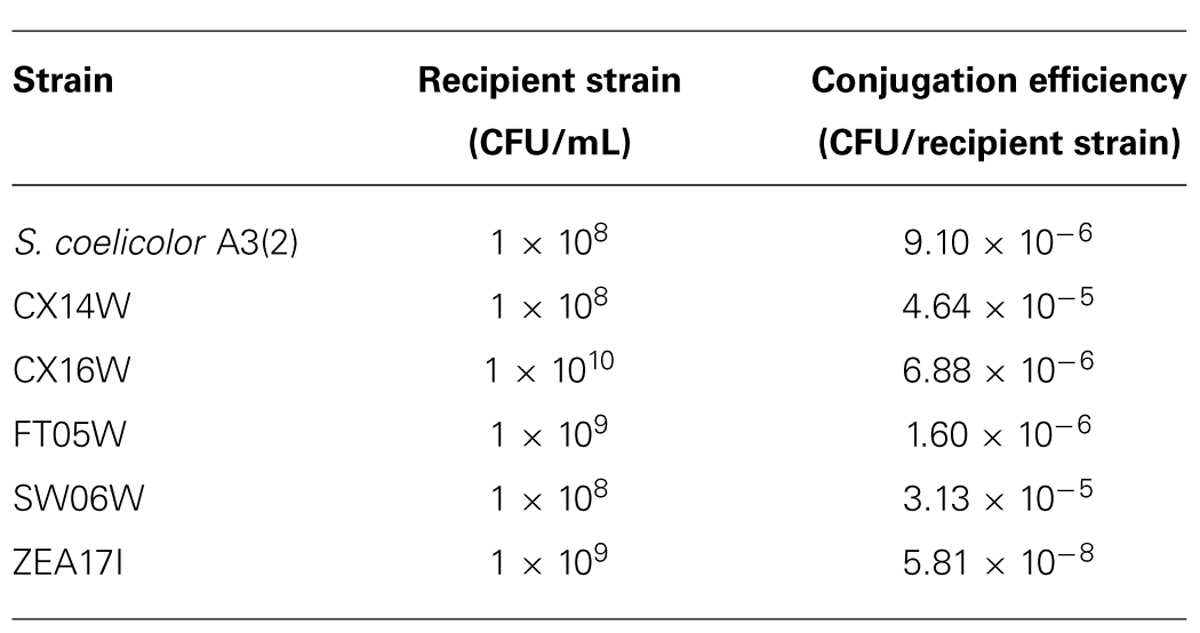
TABLE 1. Conjugation efficiencies of five Streptomyces spp. strains and S. coelicolor A3(2) with the pIJ8641 plasmid.
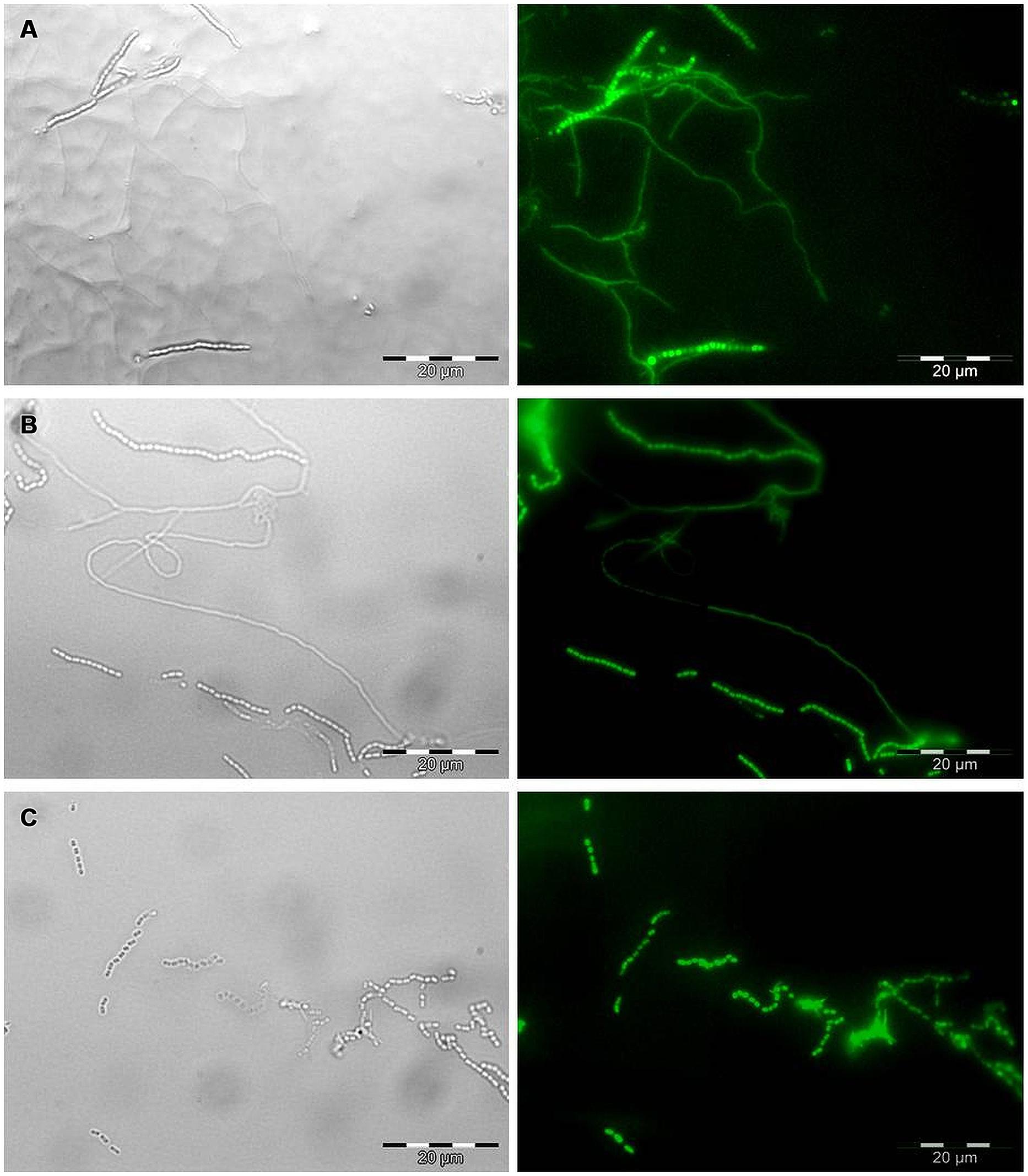
FIGURE 1. Microscopic observation of EGFP expression in transformed Streptomyces strains by bright field (left) and epifluorescence microscopy (right). Mycelium and spore chains of (A) the strain t-ZEA17I, (B) the strain t-FT05W, and (C) spore chains of the strain t-CX16W. Scale bar, 20 μm.
Effect of the Transformation on Strain Fitness
Following the transformation, the fitness of transformants was evaluated in terms of mycelium growth and sporulation, inhibition of S. sclerotiorum mycelium growth, and production of selected secondary metabolites. All the transformed strains showed similar mycelium growth and sporulation to their corresponding wild-type strains (Figure 2 – for simplicity, the growth curves for only two strains were reported, Table 2). The transformants inhibited S. sclerotiorum radial growth from 66 to 81%, and no significant differences were observed between wild-type and transformed strains. Finally, no significant differences were detected in siderophore, auxin, chitinase production, and phosphate solubilization (Tables 2 and 3).
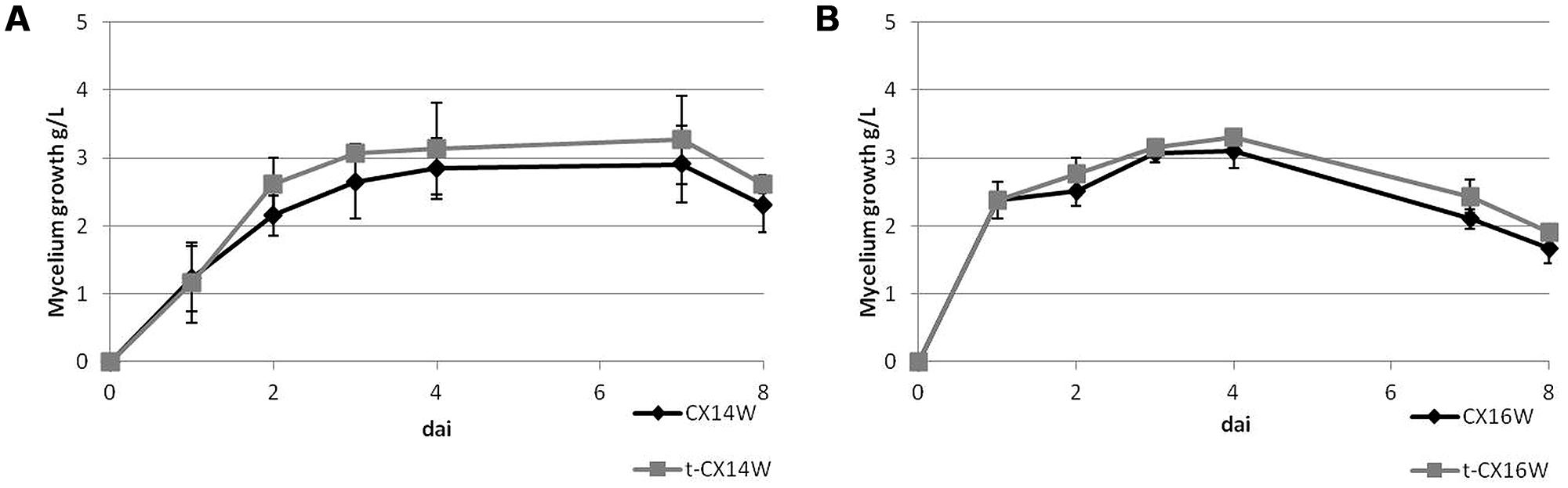
FIGURE 2. Mycelium growth of the wild-type and transformed Streptomyces spp. strains, (A) CX14W and (B) CX16W. Vertical bars represent SE (N = 3).
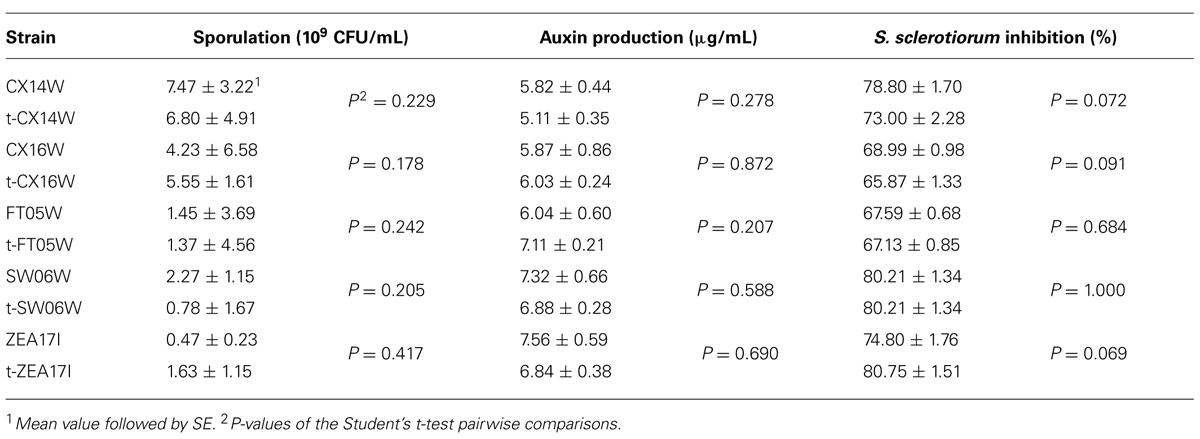
TABLE 2. Sporulation, auxin production, and inhibition of Sclerotinia sclerotiorum mycelium growth of the wild-type and transformed (t-) Streptomyces spp.
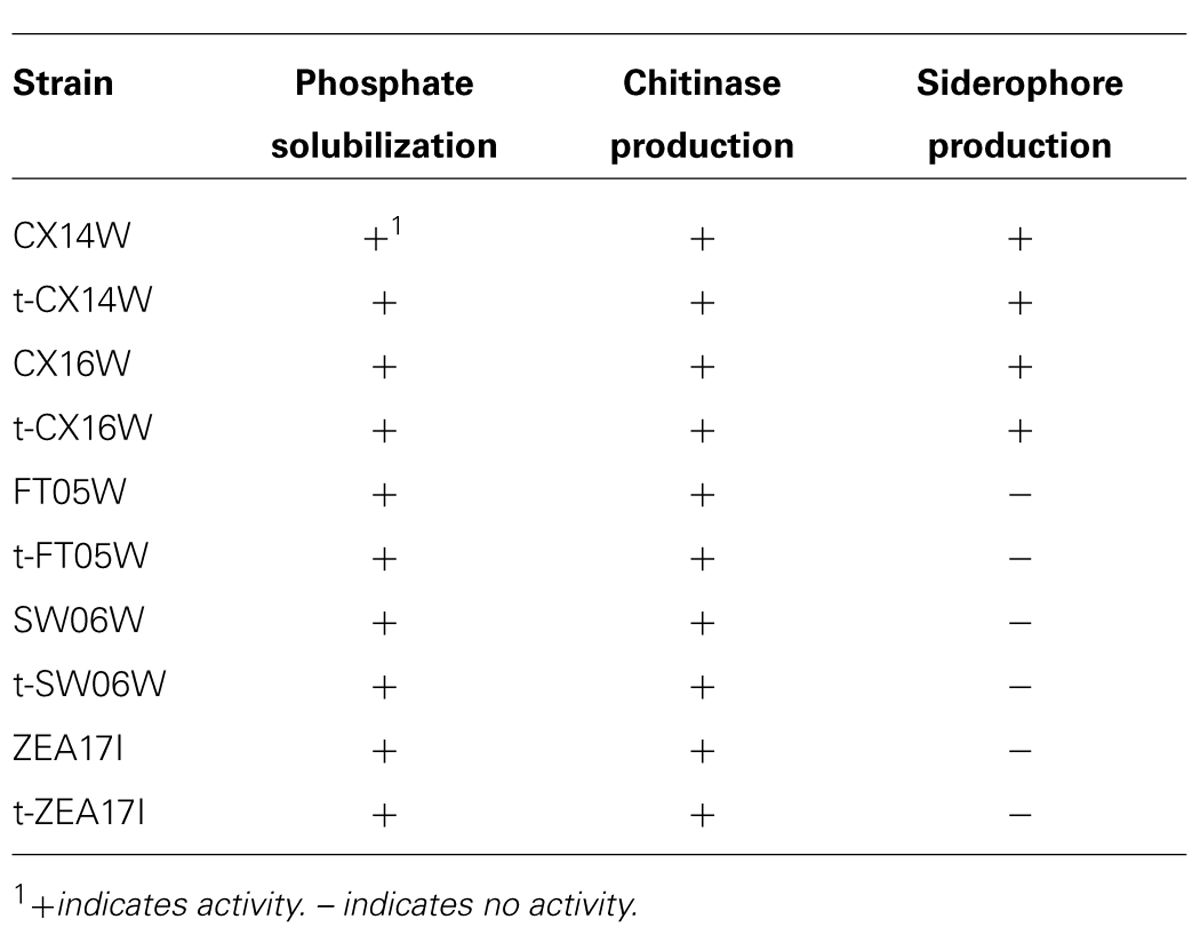
TABLE 3. Phosphate solubilization, chitinase, and siderophore production of the wild-type and transformed (t-) Streptomyces spp.
Colonization Dynamics of the Transformed ZEA17I in Bulk Soil
In this study, apramycin was used as a marker to identify the transformed t-ZEA17I strain. However, to be able to use it in greenhouse experiments, we first checked for the presence of naturally occurring apramycin-resistant Streptomyces spp. in the growth substrate, but none were detected. Therefore non-sterilized growth substrate was used in following experiments.
The colonization dynamics of t-ZEA17I in bulk soil showed that the initial inoculum, 1.16 × 108 CFU/g dry weight added to the soil, was recovered at 3.85 × 107 CFU/g dry weight four hours after inoculation (0 dai). The t-ZEA17I amount decreased significantly within the first 10 days and thereafter it remained stable up to 30 days at 2.18 × 104 CFU/g dry weight (Table 4).
Colonization of Lettuce Rhizosphere and Inner Root Tissues by the Transformed ZEA17I
The Streptomyces strain t-ZEA17I was inoculated with two different methods: as a spore suspension distributed on soil surface and as seed coating. The colonization dynamics of rhizosphere and inner root tissues of lettuce seedlings differed between the two methods.
In the rhizosphere, the concentration of the t-ZEA17I strain remained similar to the inoculated amount during the first 20 dai with either method. When t-ZEA17I was distributed on top of the growth substrate, a significant increase in concentration 30 dai was observed. In the case of the seed coating, after a slight increase within the first 10 days, the final amount was not significantly different from the initial inoculum (Table 5).
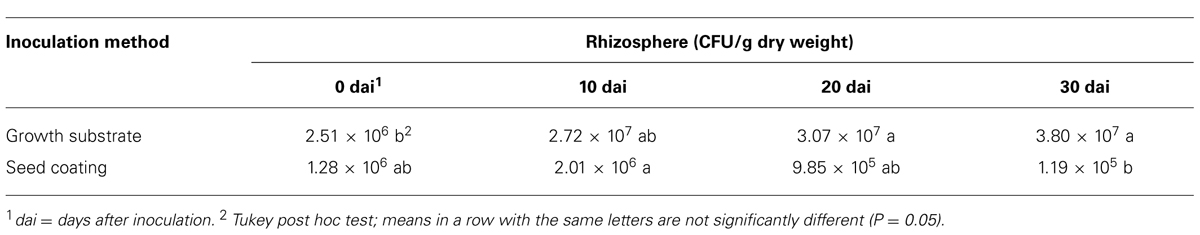
TABLE 5. Colonization dynamics of Lactuca sativa var. capitata rhizosphere by transformed Streptomyces ZEA17I strain according to two inoculation methods.
Similarly, we studied the dynamics of t-ZEA17I colonization in the inner tissues of lettuce roots. First, we ruled out the possible external root contamination due to ineffective sterilization, and no Streptomyces colonies were detected. The t-ZEA17I strain was re-isolated from inner root tissues of surface-sterilized roots independently of the inoculation method, confirming its ability to endophytically colonize lettuce roots. The concentration of t-ZEA17I declined steadily through time, however, this reduction was not significant with either inoculation method (Table 6).
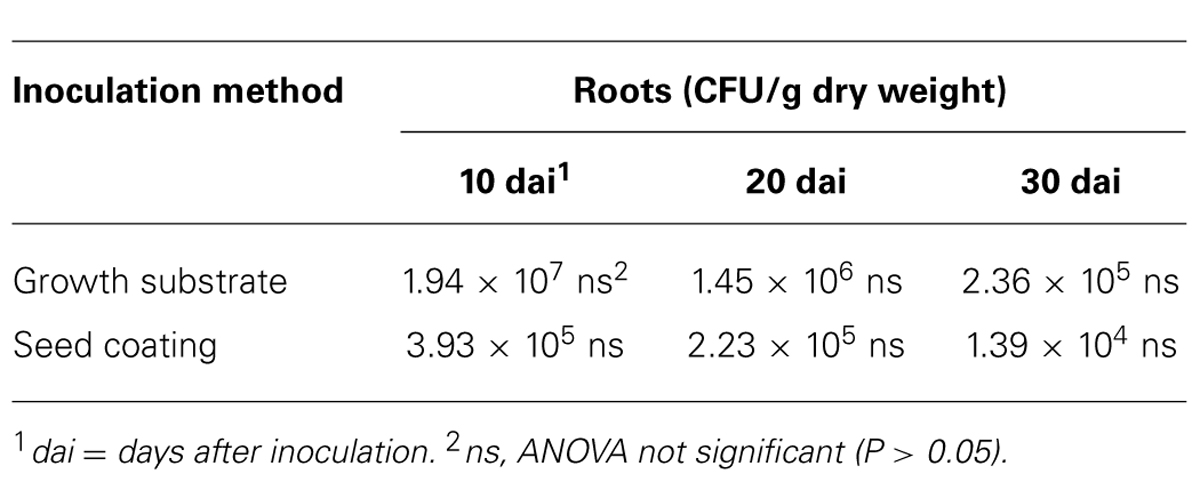
TABLE 6. Colonization dynamics of L. sativa var. capitata inner root tissues by transformed Streptomyces ZEA17I strain according to two inoculation methods.
Finally, we compared the two inoculation methods to get a further insight into whether one of them could improve the survival and colonization rates of t-ZEA17I in lettuce rhizosphere and roots. In the rhizosphere, significantly more t-ZEA17I was re-isolated at all sampling times using the growth substrate inoculation rather than seed coating (P = 0.0037, 0.0389, and 0.0005, for sampling time 10, 20, and 30 dai, respectively). Similarly, in roots, significantly higher concentration of t-ZEA17I was re-isolated using the growth substrate inoculation at 20 and 30 dai (P = 0.0415 and P = 0.0604, respectively). However, in spite of higher strain amounts present in roots using the growth substrate inoculation method, not all seedlings were colonized. Indeed, we failed to re-isolate t-ZEA17I from roots of three seedlings (one seedling at 20 dai and two seedlings at 30 dai), whereas, using the seed coating method, all roots were endophytically colonized.
Discussion
Plant beneficial bacteria have a great potential in agriculture as PGPB and BCAs and reports about successful control of plant diseases are increasing. However, application of these microbial agents in field often fails to achieve the expected results, which could be due to lack of knowledge about their biology and interactions with the host plant, the pathogens, and other microorganisms in the rhizosphere. Therefore, there are increasing attempts to study these complex interactions that take place in the rhizosphere (Gamalero et al., 2003; Compant et al., 2010).
Our aim was to study spatio-temporal dynamics of colonization of lettuce roots and rhizosphere by Streptomyces spp. with biological control potential, to better understand if and how they inhabit the rhizosphere and colonize plant roots. We selected five Streptomyces strains on the basis of their strong in vitro antagonism against the major soil-borne pathogen of horticultural crops, S. sclerotiorum (Bonaldi et al., 2014), and we transformed them with the pIJ8641 plasmid harboring apramycin resistance marker and EGFP gene under a strong constitutive promoter (Sun et al., 1999). The conjugation efficiency varied, and for most strains it was comparable to the reference strain S. coelicolor A3(2). The pIJ8641 plasmid integrates at the chromosomal attachment site for the temperate phage φC31, which may result in disruption or alteration of fitness and biological activity of the transformed strains. Indeed, decrease or loss of biological activity was detected after GFP-transformation of various BCAs, e.g., Pseudomonas putida, Metschnikowia pulcherrima, or Clonostachys rosea (Nigro et al., 1999; Lübeck et al., 2002; Weyens et al., 2012). We compared several traits important for biological control and plant growth promotion of transformed and wild-type strains, before studying their interactions with the host plant. None of the transformed strains showed altered growth or sporulation, which could have conferred a disadvantage in plant root and rhizosphere colonization. All transformants retained the ability to suppress growth of S. sclerotiorum in vitro, therefore they will also be used for studying their interactions with the pathogen and the mechanisms of biological control in vivo. Moreover, we compared the expression of some of the most common traits involved in plant growth promotion and biological control (Brader et al., 2014), such as production of auxins, siderophores and lytic enzymes, and no change in performance between the wild type and the transformants was observed.
We chose the most promising transformed strain, t-ZEA17I, for pilot studies of lettuce roots and rhizosphere colonization. We intentionally used a non-sterile growth substrate to simulate competition with natural microflora and evaluate the competitiveness of the inoculated Streptomyces strain exploiting the apramycin resistance for its identification among soil microorganisms. In absence of the host plant, we confirmed that t-ZEA17I freely survives in soil, although we observed a sharp decrease in its density within the first 10 days after bulk soil inoculation. Similar dynamics for introduced microbial population in non-sterile soil are already known, attributed to scarcity of available nutrients and adverse biotic and abiotic factors (van Veen et al., 1997). However, following the initial fall in population density, the t-ZEA17I population remained stable for up to 30 days, probably establishing an equilibrium with the indigenous microflora as described previously (Yuan and Crawford, 1995; Merzaeva and Shirokikh, 2006). In the presence of the lettuce plant, we did not detect in the rhizosphere the initial rapid decrease in t-ZEA17I amount that was observed in bulk soil. On the contrary, its concentration augmented when applied directly on the growth substrate. It is possible that t-ZEA17I was chemoattracted to the rhizosphere of the growing seedling, where it quickly established a stable interaction with the host plant roots. Indeed, the presence of a host plant may greatly affect the survival of PGPB, as was observed, i.e., for the sharp decline in Azospirillum brasilense population after removal of inoculated plants (Bashan et al., 1995).
Different strategies are being used for studying BCAs and PGPB in the rhizosphere. Their localization in roots and seeds rely on microscopic tools exploiting fluorescent markers, which give a fundamental insight into the spatial distribution of the microorganism along and inside the growing root, but do not quantify the microbial amounts and their dynamics (Coombs and Franco, 2003; Olivain et al., 2006; Compant et al., 2010). Additionally, studying the dynamics of colonization by beneficial microorganisms exploits the strain identification mostly by natural or introduced antibiotic resistance and its quantification by dilution plating (Gamalero et al., 2003, 2005). Here, we quantified the t-ZEA17I in roots and rhizosphere through the introduced antibiotic resistance for its identification, to understand if it can inhabit soil in competitive concentrations in comparison to the indigenous microflora. t-ZEA17I was detected in the inner root tissues of growing seedlings already 10 days after inoculation at high concentrations. Indeed, Coombs and Franco (2003) demonstrated that the EGFP-tagged endophytic Streptomyces sp. strain EN27 rapidly colonizes the wheat embryo, as it was detected in developing roots as early as 24 h after inoculation. Re-isolation of t-ZEA17I from the rhizosphere and the inner tissues of surface-sterilized roots indicates that it is both rhizospheric and endophytic, although it is not known if its localization affects its potential for biocontrol and plant growth promotion. It has been hypothesized that endophytic bacteria form more stable interactions with plants, rather than rhizospheric or epiphytic bacteria (Compant et al., 2010; Malfanova et al., 2011).
Finally, we tested how different methods of inoculation influence the t-ZEA17I colonization ability. When it was distributed directly on the growth substrate, higher concentration was detected in roots, however, we could not re-isolate the strain from all inoculated plants. On the contrary, when the seed coating method was used, less propagules were recovered but all plants were endophytically colonized. It is possible that in case of seed coating, t-ZEA17I is more closely associated with the growing seedling, which increases its probability to internally colonize root tissues. In roots, we observed progressive decline in its concentration using either inoculation method. Although the total amount of t-ZEA17I increased at different sampling times (data not shown), the increase in lettuce root biomass was probably higher than the strain growth, thus resulting in lower strain concentration per g of root. To ensure that t-ZEA17I colonizes roots in effective concentrations, and to prevent its decline to undetectable levels, studies assessing optimal amount of inoculum are needed. Moreover, it is possible that strains colonize only certain root zones (Gamalero et al., 2004, 2005). Therefore, further studies are needed to establish which zones of the plant roots are colonized by t-ZEA17I and ultimately how it interacts with the plant in presence of S. sclerotiorum, to evaluate its biological control activity in vivo.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank prof. Flavia Marinelli (University of Insubria, Italy), for donor strain E. coli ET12567 and reference strain S. coelicolor A3(2) and Prof. Mervyn Bibb (John Innes Centre, UK), for kindly providing the plasmid pIJ8641. The authors also thank the reviewers for their constructive and useful comments that greatly improved the manuscript. This research was supported in part by research program “Dote ricerca applicata” funded by Lombardy Region and Sipcam Italia Spa.
References
Aeron, A., Kumar, S., Pandey, P., and Maheshwari, D. K. (2011). “Emerging role of plant growth promoting rhizobacteria in agrobiology,” in Bacteria in Agrobiology: Crop Ecosystems, ed. D. K. Maheshwari (Berlin: Springer), 1–36. doi: 10.1007/978-3-642-18357-7_1
Bano, N., and Musarrat, J. (2003). Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential biocontrol agent. Curr. Microbiol. 46, 0324–0328. doi: 10.1007/s00284-002-3857-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bashan, Y., Puente, M. E., Rodriguezmendoza, M. N., Toledo, G., Holguin, G., Ferreracerrato, R.,et al. (1995). Survival of Azospirillum brasilense in the bulk soil and rhizosphere of 23 soil types. Appl. Environ. Microbiol. 61, 1938–1945.
Berg, G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bonaldi, M., Kunova, A., Saracchi, M., Sardi, P., and Cortesi, P. (2014). Streptomycetes as biological control agents against basal drop. Acta Hortic. 1044, 313–318.
Bonaldi, M., Kunova, A., Sardi, P., Cortesi, P., and Saracchi, M. (2011). Streptomycetes as possible biocontrol agents against soil-borne pathogens. J. Plant Pathol. 93, S4.26–S4.27.
Brader, G., Compant, S., Mitter, B., Trognitz, F., and Sessitsch, A. (2014). Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 27, 30–37. doi: 10.1016/j.copbio.2013.09.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bric, J. M., Bostock, R. M., and Silverstone, S. E. (1991). Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538.
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren Van Themaat, E., Ahmadinejad, N., Assenza, F.,et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, L., Qiu, Z., You, J., Tan, H., and Zhou, S. (2004). Isolation and characterization of endophytic Streptomyces strains from surface-sterilized tomato (Lycopersicon esculentum) roots. Lett. Appl. Microbiol. 39, 425–430. doi: 10.1111/j.1472-765X.2004.01606.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, Y., Zhang, Z., Ling, N., Yuan, Y., Zheng, X., Shen, B.,et al. (2011). Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertility Soils 47, 495–506. doi: 10.1007/s00374-011-05562
Compant, S., Clement, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved, and prospects for utilization. Soil. Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Compant, S., Duffy, B., Nowak, J., Clement, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/Aem.71.9.4951-4959.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coombs, J. T., and Franco, C. M. M. (2003). Visualization of an endophytic Streptomyces species in wheat seed. Appl. Environ. Microbiol. 69, 4260–4262. doi: 10.1128/Aem.69.7.4260-4262.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coombs, J. T., Michelsen, P. P., and Franco, C. M. (2004). Evaluation of endophytic actinobacteria as antagonists of Gaeumannomyces graminis var. tritici in wheat. Biol. Control 29, 359–366. doi: 10.1016/j.biocontrol.2003.08.001
Errampalli, D., Leung, K., Cassidy, M. B., Kostrzynska, M., Blears, M., Lee, H.,et al. (1999). Applications of the green fluorescent protein as a molecular marker in environmental microorganisms. J. Microbiol. Methods 35, 187–199. doi: 10.1016/S0167-7012(99)00024-X
Faria, J. C., Jelihovschi, E. G., and Allaman, I. B. (2013). Conventional Tukey Test. Ilheus: Test.UESC.
Gamalero, E., Lingua, G., Berta, G., and Lemanceau, P. (2003). Methods for studying root colonization by introduced beneficial bacteria. Agronomie 23, 407–418. doi: 10.1051/Agro:2003014
Gamalero, E., Lingua, G., Capri, F. G., Fusconi, A., Berta, G., and Lemanceau, P. (2004). Colonization pattern of primary tomato roots by Pseudomonas fluorescens A6RI characterized by dilution plating, flow cytometry, fluorescence, confocal, and scanning electron microscopy. FEMS Microbiol. Ecol. 48, 79–87. doi: 10.1016/j.femsec.2003.12.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gamalero, E., Lingua, G., Tombolini, R., Avidano, L., Pivato, B., and Berta, G. (2005). Colonization of tomato root seedling by Pseudomonas fluorescens 92rkG5: spatio-temporal dynamics, localization, organization, viability, and culturability. Microb. Ecol. 50, 289–297. doi: 10.1007/s00248-004-0149-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ghirardi, S., Dessaint, F., Mazurier, S., Corberand, T., Raaijmakers, J. M., Meyer, J. M.,et al. (2012). Identification of traits shared by rhizosphere-competent strains of fluorescent pseudomonads. Microb. Ecol. 64, 725–737. doi: 10.1007/s00248-012-0065-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012, 15. doi: 10.6064/2012/963401
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grantcharova, N., Lustig, U., and Flardh, K. (2005). Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 187, 3227–3237. doi: 10.1128/Jb.187.3227.3237.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haas, J., Park, E.-C., and Seed, B. (1996). Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6, 315–324. doi: 10.1016/S0960-9822(02)00482-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152. doi: 10.1007/s11104-008-9885-9889
Hohmann, P., Jones, E. E., Hill, R. A., and Stewart, A. (2012). Ecological studies of the bio-inoculant Trichoderma hamatum LU592 in the root system of Pinus radiata. FEMS Microbiol. Ecol. 80, 709–721. doi: 10.1111/j.1574-6941.2012.01340.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hopwood, D. A. (2007). Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford: Oxford University Press New York.
Khamna, S., Yokota, A., and Lumyong, S. (2009). Actinomycetes isolated from medicinal plant rhizosphere soils: diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J. Microbiol. Biotechnol. 25, 649–655. doi: 10.1007/s11274-008-9933-x
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: The John Innes Foundation.
Krzyzanowska, D., Obuchowski, M., Bikowski, M., Rychlowski, M., and Jafra, S. (2012). Colonization of potato rhizosphere by GFP-tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 shown on large sections of roots using enrichment sample preparation and confocal laser scanning microscopy. Sensors 12, 17608–17619. doi: 10.3390/S121217608
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Latour, X., Delorme, S., Mirleau, P., and Lemanceau, P. (2003). Identification of traits implicated in the rhizosphere competence of fluorescent pseudomonads: description of a strategy based on population and model strain studies. Agronomie 23, 397–405. doi: 10.1051/Agro:2003015
Loria, R., Kers, J., and Joshi, M. (2006). Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 44, 469–487. doi: 10.1146/annurev.phyto.44.032905.091147
Lu, Z. X., Tombolini, R., Woo, S., Zeilinger, S., Lorito, M., and Jansson, J. K. (2004). In vivo study of Trichoderma-pathogen-plant interactions, using constitutive and inducible green fluorescent protein reporter systems. Appl. Environ. Microbiol. 70, 3073–3081. doi: 10.1128/Aem.70.5.3073-3081.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lübeck, M., Knudsen, I., Jensen, B., Thrane, U., Janvier, C., and Funck Jensen, D. (2002). GUS and GFP transformation of the biocontrol strain Clonostachys rosea IK726 and the use of these marker genes in ecological studies. Mycol. Res. 106, 815–826. doi: 10.1017/S095375620200607X
Lucas, X., Senger, C., Erxleben, A., Grüning, B. A., Döring, K., Mosch, J.,et al. (2013). StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 41, D1130–D1136. doi: 10.1093/nar/gks1253
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lugtenberg, B. J., Chin-A-Woeng, T. C., and Bloemberg, G. (2002). Microbe–plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek 81, 373–383. doi: 10.1023/a:1020596903142
Lugtenberg, B. J. J., Dekkers, L., and Bloemberg, G. V. (2001). Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39, 461–490. doi: 10.1146/annurev.phyto.39.1.461
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Malfanova, N., Kamilova, F., Validov, S., Shcherbakov, A., Chebotar, V., Tikhonovich, I.,et al. (2011). Characterization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microbial Biotechnol. 4, 523–532. doi: 10.1111/j.1751-7915.2011.00253.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merzaeva, O. V., and Shirokikh, I. G. (2006). Colonization of plant rhizosphere by actinomycetes of different genera. Microbiology 75, 226–230. doi: 10.1134/S0026261706020184
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Minuto, A., Spadaro, D., Garibaldi, A., and Gullino, M. L. (2006). Control of soilborne pathogens of tomato using a commercial formulation of Streptomyces griseoviridis and solarization. Crop Prot. 25, 468–475. doi: 10.1016/j.cropro.2005.08.001
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nigro, F., Finetti Sialer, M. M., and Gallitelli, D. (1999). Transformation of Metschnikowia pulcherrima 320, biocontrol agent of storage rot, with the green fluorescent protein gene. J. Plant Pathol. 205–208.
Olivain, C., Humbert, C., Nahalkova, J., Fatehi, J., L’haridon, F., and Alabouvette, C. (2006). Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl. Environ. Microbiol. 72, 1523–1531. doi: 10.1128/Aem.72.2.1523-1531.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perez-Miranda, S., Cabirol, N., George-Tellez, R., Zamudio-Rivera, L. S., and Fernandez, F. J. (2007). O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 70, 127–131. doi: 10.1016/j.mimet.2007.03.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Petrolini, B., Quaroni, S., Saracchi, M., and Sardi, P. (1996). Studies on the streptomycete population inhabiting plant roots. Actinomycetes 7, 66–78.
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Van Der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/Nrmicro3109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Prosser, J. I. (1994). Molecular marker systems for detection of genetically-engineered microorganisms in the environment. Microbiology 140, 5–17. doi: 10.1099/13500872-140-1-5
R Core Team. (2013). R: A Language and Environment for Statistical Computing. (Vienna: R Foundation for Statistical Computing).
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moenne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Raaijmakers, J. M., Vlami, M., and De Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek International J. Gen. Mol. Microbiol. 81, 537–547. doi: 10.1023/A:1020501420831
Saima, M., Kuddus, M., Roohi, I., and Ahmad, Z. (2013). Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J. Genet. Eng. Biotechnol. 11, 39–46. doi: 10.1016/j.jgeb.2013.03.001
Sardi, P., Saracchi, M., Quaroni, S., Petrolini, B., Borgonovi, G. E., and Merli, S. (1992). Isolation of endophytic Streptomyces strains from surface-sterilized roots. Appl. Environ. Microbiol. 58, 2691–2693.
Schwyn, B., and Neilands, J. B. (1987). Universal chemical-assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seipke, R. F., Kaltenpoth, M., and Hutchings, M. I. (2012). Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev. 36, 862–876. doi: 10.1111/j.1574-6976.2011.00313.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, S. E., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250. doi: 10.1146/annurev-arplant-042110-103846
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, J. H., Kelemen, G. H., Fernandez-Abalos, J. M., and Bibb, M. J. (1999). Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227.
Tokala, R. K., Strap, J. L., Jung, C. M., Crawford, D. L., Salove, M. H., Deobald, L. A.,et al. (2002). Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl. Environ. Microbiol. 68, 2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Veen, J. A., van Overbeek, L. S., and van Elsas, J. D. (1997). Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61, 121–135.
Watve, M., Tickoo, R., Jog, M., and Bhole, B. (2001). How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390. doi: 10.1007/s002030100345
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16s Ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Weyens, N., Boulet, J., Adriaensen, D., Timmermans, J.-P., Prinsen, E., Van Oevelen, S.,et al. (2012). Contrasting colonization and plant growth promoting capacity between wild type and a gfp-derative of the endophyte Pseudomonas putida W619 in hybrid poplar. Plant Soil 356, 217–230. doi: 10.1007/s11104-011-0831-x
Keywords: biocontrol, Lactuca sativa, Sclerotinia sclerotiorum, streptomycetes, rhizosphere competence
Citation: Bonaldi M, Chen X, Kunova A, Pizzatti C, Saracchi M and Cortesi P (2015) Colonization of lettuce rhizosphere and roots by tagged Streptomyces. Front. Microbiol. 6:25. doi: 10.3389/fmicb.2015.00025
Received: 13 November 2014; Accepted: 08 January 2015;
Published online: 06 February 2015.
Edited by:
Aurelio Ciancio, Consiglio Nazionale delle Ricerche – Istituto per la Protezione Sostenibile delle Piante, ItalyReviewed by:
Elena Prats, Spanish National Research Council, SpainStephane Compant, Austrian Institute of Technology GmbH, Austria
Copyright © 2015 Bonaldi, Chen, Kunova, Pizzatti, Saracchi and Cortesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paolo Cortesi, Department of Food, Environmental and Nutritional Sciences, University of Milan, Via Giovanni Celoria, 2 20133 Milano, Italy e-mail: paolo.cortesi@unimi.it
†These authors have contributed equally to this work.
 Maria Bonaldi
Maria Bonaldi Xiaoyulong Chen
Xiaoyulong Chen Andrea Kunova
Andrea Kunova Cristina Pizzatti
Cristina Pizzatti Marco Saracchi
Marco Saracchi Paolo Cortesi
Paolo Cortesi