- 1Department of Biological Sciences, Northern Illinois University, DeKalb, IL, USA
- 2Southern Regional Research Center, United States Department of Agriculture – Agricultural Research Service, New Orleans, LA, USA
Fungal secondary metabolism and morphological development have been shown to be intimately associated at the genetic level. Much of the literature has focused on the co-regulation of secondary metabolite production (e.g., sterigmatocystin and aflatoxin in Aspergillus nidulans and Aspergillus flavus, respectively) with conidiation or formation of sexual fruiting bodies. However, many of these genetic links also control sclerotial production. Sclerotia are resistant structures produced by a number of fungal genera. They also represent the principal source of primary inoculum for some phytopathogenic fungi. In nature, higher plants often concentrate secondary metabolites in reproductive structures as a means of defense against herbivores and insects. By analogy, fungi also sequester a number of secondary metabolites in sclerotia that act as a chemical defense system against fungivorous predators. These include antiinsectant compounds such as tetramic acids, indole diterpenoids, pyridones, and diketopiperazines. This chapter will focus on the molecular mechanisms governing production of secondary metabolites and the role they play in sclerotial development and fungal ecology, with particular emphasis on Aspergillus species. The global regulatory proteins VeA and LaeA, components of the velvet nuclear protein complex, serve as virulence factors and control both development and secondary metabolite production in many Aspergillus species. We will discuss a number of VeA- and LaeA-regulated secondary metabolic gene clusters in A. flavus that are postulated to be involved in sclerotial morphogenesis and chemical defense. The presence of multiple regulatory factors that control secondary metabolism and sclerotial formation suggests that fungi have evolved these complex regulatory mechanisms as a means to rapidly adapt chemical responses to protect sclerotia from predators, competitors and other environmental stressors.
Introduction
Fungal species are able to develop specialized structures allowing them to disseminate and survive adverse environmental conditions. Aspergilli differentiate by forming conidiophores, structures that produce conidiospores. Some Aspergillus species, such as the model species Aspergillus nidulans, also produce sexual fruiting bodies known as cleistothecia where meiospores (i.e., ascospores) are generated. Both reproductive processes, asexual and sexual development, are controlled by temporal and spatial genetic regulation (Adams and Yu, 1998; Calvo et al., 2002; Fischer and Kues, 2006). Other species, such as Aspergillus flavus or Aspergillus parasiticus, form resting structures capable of surviving environmental extremes termed sclerotia that represent vestiges of fruiting bodies incapable of producing ascospores (Coley-Smith and Cooke, 1971; Malloch and Cain, 1972; Wicklow, 1987). Initial evidence presented by Geiser et al. (1996) supported that asexual Aspergilli are often derived from meiotic lineages and postulated for the first time that sclerotia might be vestigial cleistothecia that lost the capacity to produce ascospores. In more recent years, the complementary alpha- and HMG-domain MAT genes have been characterized from A. flavus and A. parasiticus (Ramirez-Prado et al., 2008). Presence and functionality of mating type genes in Aspergillus oryzae was also found, supporting a possible heterothallic breeding system in this fungus (Wada et al., 2012). Furthermore, Horn et al. (2009b, 2014) reported ascospore-bearing ascocarps embedded within sclerotia of A. flavus and A. parasiticus. The proposed common origin between cleistothecia and sclerotia suggested that conserved genetic regulatory pathways controlling cleistothecia formation could also control sclerotial production. Rapid progress on studies of the cleistothecium-producing model fungus A. nidulans and other related fungi [i.e., Dyer and O’Gorman (2012) and references therein] has facilitated uncovering regulatory pathways controlling sclerotial production in other fungi, particularly in A. flavus.
Studies have found that a number of genetic regulators controlling the formation of developmental structures, including sclerotia, also govern the production of secondary metabolites (Calvo et al., 2002; Calvo, 2008). While some of these compounds, also termed natural products, are beneficial (e.g., penicillin and lovastatin), other secondary metabolites are deleterious, such as mycotoxins [reviewed in Gloer (2007)]. Among fungal secondary metabolites, aflatoxins (AFs) are probably the most well known and studied. These compounds were discovered after the United Kingdom’s outbreak of Turkey X disease in 1962, caused by consumption of A. flavus-contaminated feed and resulted in the deaths of numerous turkey poults (Bennett and Klich, 2003). A. flavus is capable of colonizing economically important crops such as peanut, cotton, maize and other oilseed crops both pre- and post-harvest. In the U.S. alone, A. flavus costs 100s of millions USD annually due to market losses from AF contaminated crops (Wu, 2004). In addition to AFs, A. flavus produces other secondary metabolites and many of them have been found in sclerotia (Gloer, 1995). Genetic regulation of development and secondary metabolism has been intensely studied in the Aspergilli, and in particular A. flavus and A. nidulans. In this review we focus on the association between secondary metabolism and sclerotial formation in this fungal genus, including genetic co-regulatory patterns leading to the activation of the secondary metabolic gene clusters and formation of sclerotia. Important components of this shared regulatory mechanism are the global regulatory proteins VeA and LaeA, part of the velvet complex (Bayram et al., 2008a; Bayram and Braus, 2012). Additionally, we also discuss the possible roles of secondary metabolites associated with sclerotia, particularly in Aspergilli.
Secondary Metabolites Present in Fungal Sclerotia
It is difficult to determine just how many species of fungi exist, but estimates have suggested that the fungal kingdom is very diverse having anywhere from 1.0 to 2.7 million species with only a fraction of these having been isolated and described Hawksworth and Rossman (1997) and Mueller and Schmit (2007). One common theme of many of the described species is that they are prolific producers of biologically active secondary metabolites. The diversity of these natural products rivals that of the fungal kingdom. Fungal secondary metabolites have garnered much attention for their beneficial impact as therapeutic agents (e.g., lovastatin and penicillin) and continue to be mined as a source of important end products and building blocks for pharmaceutical development. On the other hand, secondary metabolites have also received considerable attention for their adverse impact of humans and animals due to their widespread occurrence as mycotoxins (e.g., AFs and fumonisins) on food and feed crops as well as indoor environments. Fungi produce a number of structural classes of secondary metabolites including polyketides (PKs), non-ribosomal peptides (NRPs), hybrid PK-NRPs, indole alkaloids, and terpenes (Keller et al., 2005). In almost all cases the genes responsible for the production of these classes of secondary metabolites are organized as a gene cluster (discussed below). Secondary metabolites of this type that have been identified in sclerotia will be the main focus of this section.
Though many of the recognized biological activities of important secondary metabolites relate to their direct influence on humans and other vertebrates, it is generally accepted that these natural products play key roles in the ecology of the fungus as well. Over the course of evolution, secondary metabolites have been fashioned for numerous biological functions in microorganisms, as chemical messengers between microbes and as a means of defense from predation and competing microbes (Wicklow, 1988; Yim et al., 2007; Rohlfs and Churchill, 2011; Yin et al., 2012). Fungi are much like plants in that; in general, they are static organisms incapable of readily escaping from encroaching predators and competing microbes. In spite of this, fungi are quite successful at inhabiting and surviving for long periods of time in highly competitive environments. It has been hypothesized that these competitive environments have provided considerable selective pressure for fungi to produce an array of antagonistic secondary metabolites as part of their “chemical” defense against numerous fungivores and competitors (Gloer, 2007; Rohlfs et al., 2007; Rohlfs and Churchill, 2011). A recent study showed that arthropod grazing induces a “resistance” phenotype in A. nidulans to fungivory that coincided with elevated levels of secondary metabolite and sexual fruiting body formation (Rohlfs and Churchill, 2011). Plants also tend to concentrate secondary metabolites in reproductive structures (e.g., seeds) as a means of defense against herbivores; as well, herbivores tend to avoid feeding or oviposition on plants or plant tissues that contain high levels of secondary metabolites (Rhoades, 1985). In an analogous fashion, various fungi are known to sequester secondary metabolites in asexual conidia and sexual fruiting structures that are critical to survival and which often results in reduced incidences of insect fungivory (Doll et al., 2013).
In addition to conidia and fruiting bodies, numerous fungi also produce structures termed sclerotia. Sclerotia are compacted mats of hyphae produced by certain fungi that allow survival for long periods of time under adverse environmental conditions (Coley-Smith and Cooke, 1971). Upon onset of favorable conditions, sclerotia can germinate to produce large quantities of either hyphae or conidia, and as such they represent a primary source of fungal inoculum in the field. Sclerotia are commonly produced on plant tissues during fungal invasion and eventually end up in soil, or on decaying plant tissues, in the field where they are exposed to predation by insects. In addition to serving as survival structures, in many Aspergillus species (e.g., A. flavus and A. nomius), with proper environmental conditions and mating pair interactions, sclerotia can serve as a substrate (termed stromata) for the formation of sexual structures (Horn et al., 2009a, 2011). The stromata harbor ascospore-bearing cleistothecia, similar to cleistothecia of other ascomycetous species that have a sexual cycle (Horn et al., 2009a, 2014). Many of the genetic mechanisms that connect secondary metabolism to morphogenesis of sexual fruiting bodies have also been shown to control sclerotial production (discussed below). Production of sclerotia represents a substantial metabolic investment by the fungus that is warranted based on the critical role of these structures in reproduction and survival. The importance of sclerotia to fungal biology combined with their high nutrient value to insects would justify the existence of considerable selective pressure on the fungus to produce antiinsectan/antifeedant secondary metabolites as part of their chemical defenses. In fact, this appears to be the case as numerous studies have shown sclerotia to be veritable storehouses of a diversity of secondary metabolites with antiinsectan properties [reviewed in Wicklow (1988) and Gloer (1995, 1997, 2007)]. The fungus’ need for a diverse array of defensive secondary metabolites may be a reflection of the ability of the target organism to develop resistance to specific inhibitory agents. One would predict that the presence of a number of secondary metabolites in sclerotia, many of which may have different modes of action, would make it more difficult for the target organism to evolve resistance either through mutation or acquisition of resistance genes than if it were faced with having to overcome just one inhibitory metabolite.
Perhaps the quintessential example of sclerotia-based chemical defense is that of Claviceps purpurea. This ascomycetous fungus produces a group of indole-derived secondary metabolites known as ergot alkaloids (EAs) during growth on a number of plants including many cereal crops (Haarmann et al., 2009). Consumption of food and feeds contaminated with the alkaloid-containing sclerotia (ergot) resulted in vast epidemics of human and animal disease that were reported as early as 600 BC. In addition to C. purpurea, a number of chemically diverse EAs are produced by other fungi including many grass endophytes, as well as strains of Penicillium and Aspergillus, though most of these strains are not known to produce sclerotia (Gloer, 2007). The proposed ecological role of EAs is to protect the fungus by reducing consumption of the host crop by herbivores or from direct consumption by fungivorous insects (Schardl et al., 2006). The remainder of this section will focus on secondary metabolites identified in sclerotia, primarily of Aspergillus species, though a few examples will be provided for other fungi. A list of secondary metabolites found in sclerotia from Aspergillus species is presented in Table 1. Reports by Gloer (1995, 1997, 2007) provide an excellent source of information on the chemistry and biological function of fungal metabolites associated with sclerotia. This review will only touch on new findings since the (Gloer, 2007) publication and provide a few examples of interest. In many cases, previous literature on fungal secondary metabolites describe whole culture extracts and fail to specify if the metabolite(s) was present in sclerotia. In some instances, the investigators report on secondary metabolites that were extracted from isolated sclerotia but fail to indicate if they were also present in other fungal structures such as mycelia and conidia. This review is focused on secondary metabolites of sclerotial origin, but in some cases information will be presented on metabolites that are present in sclerotia as well as other fungal structures, or secreted outside of the cell.
One of the most intensely studied fungal genera with respect to production of secondary metabolites is Aspergillus. Members of this genus of fungi are ubiquitous in nature and are capable of living as saprophytes in soils or as opportunistic pathogens of humans, plants and animals. With well over 250 identified species of Aspergillus (Geiser et al., 2007), probably the best known members of this genus are A. flavus and A. parasiticus, that produce carcinogenic and toxigenic AFs. Many species of Aspergillus produce both sclerotia and the polyketide-derived AFs, however, the majority of the literature has focused on AF production in A. flavus as it is most commonly associated with contamination of food and feed crops (Payne and Brown, 1998; Cary et al., 2000; Bhatnagar et al., 2002). AFs are produced during growth of the fungus on oilseed crops such as corn, peanuts, cottonseed, and treenuts and they can also contaminate many additional crops during storage. Production of AFs in A. flavus and A. parasiticus is known to occur in specialized endosomes of mycelia and subsequently secreted into the environment (Chanda et al., 2009). AFs have also been found in all fungal cell structures including mycelia, conidia, and sclerotia (Wicklow and Cole, 1982; Wicklow and Shotwell, 1983). Though the exact role of AFs in the biology of producing species remains elusive, evidence suggests that they are produced in response to oxidative stress and may also be endowed with antiinsectan properties (Chinnici and Bettinger, 1984; Narasaiah et al., 2006; Grintzalis et al., 2014). In addition, AF production and sclerotial development may be closely related, as increased production of AF precursors was associated with a decrease in sclerotial size (Chang et al., 2002). It was suggested that this may be due to common substrates like acetate being directed toward AF production resulting in lowered availability for biogenesis of sclerotia.
Cyclopiazonic acid (CPA) is an indole-tetramic acid mycotoxin that is produced by a number of species of Aspergillus and Penicillium (Burdock and Flamm, 2000; Vinokurova et al., 2007). It is a common contaminate of a number of food commodities and is often present as a co-contaminate with AFs (Martins and Martins, 1999). CPA has been found in sclerotia of A. flavus, however, it was also detected in mycelia (Wicklow and Cole, 1982). Though its role in the ecology of the fungus is not known, CPA has been shown to be an inhibitor of calcium-dependent ATPase in the sarcoplasmic reticulum with exposure in some animals leading to organ necrosis and death (Riley et al., 1992).
Another mycotoxin of importance to human health is ochratoxin A (OTA; El Khoury and Atoui, 2010). OTA is produced by several species of Aspergillus and Penicillium via a pentaketide that is derived from a dihydrocoumarin coupled to β-phenylalanine. It is detected worldwide in various food and beverage sources. OTA can have several toxicological effects such as nephrotoxic, hepatotoxic, neurotoxic, teratogenic, and immunotoxic. OTA has been isolated from sclerotia of Aspergillus ochraceus, Aspergillus sclerotioniger, and Aspergillus carbonarius, with OTA isolated from the latter shown to have antiinsectan properties (Paster et al., 1984; Wicklow et al., 1996; Frisvad et al., 2014). Only a few strains of Aspergillus section Nigri have been reported to produce sclerotia, but when cultured in artificial media supplemented with natural substrates such as fruits and grains, sclerotial production was induced along with numerous sclerotial secondary metabolites (Frisvad et al., 2014). In addition to detection of OTA, some isolates were found to produce apolar indoloterpenes of the aflavinine type and okaramines (Frisvad et al., 2014; Petersen et al., 2014).
A number of fungi produce polyketide-derived melanins which are the black or near-black pigments formed by oxidative polymerization of phenolic compounds produced by the dihydroxynaphthalene (DHN)-melanin pathway (Wheeler, 1983; Butler and Day, 1998). Melanin has been shown to be a virulence factor in plant, animal, and human pathogenic fungi and it also functions in survival and longevity in nature of fungal propagules such as sclerotia (Butler and Day, 1998). Sclerotial DHN-melanins have been reported as a component of black sclerotia of Sclerotinia sclerotiorum and S. trifoliorum (Butler et al., 2009). Recently, an A. flavus gene cluster was found to be responsible for the production of a sclerotia-specific pigment identified as the polyketide, asparasone (discussed below; Cary et al., 2014). Sclerotia produced by mutants of the asparasone polyketide synthase (PKS) lacked dark pigmentation, were significantly less resistant to insect predation than wild-type sclerotia and were more susceptible to the deleterious effects of ultraviolet light and heat. Fungal sclerotia and conidia were previously thought to be mostly resistant to this type of damage due to the presence of DHN-melanins. The study of Cary et al. (2014) showed that the dark brown pigments in A. flavus sclerotia derive from anthraquinones produced by the asparasone cluster rather than from the typical DHN-melanin pathway.
Global Genetic Regulatory Mechanisms Governing Production of Secondary Metabolites That Influence Sclerotia
The global regulatory proteins VeA and LaeA, components of the velvet nuclear protein complex, control both development and secondary metabolism in numerous fungi, including Aspergillus species. The characterization of the veA gene began more than 60 years ago, when Kafer (1965) generated the first veA mutant in A. nidulans, veA1, a mutant with partial loss-of-function. However, its characterization was delayed for many years due to the fact that the VeA predicted protein did not demonstrate homology with any other proteins of known function. Further studies with veA deletion mutants in A. nidulans and in other fungi provided valuable insight into the role of this regulator. veA is known to have a role in activating sexual development and inhibiting asexual development (Champe et al., 1981; Yager, 1992; Kim et al., 2002). Interestingly the role of veA in the regulation of morphogenesis is light-dependent; light reduces and delays cleistothecial formation and promotes conidiation in A. nidulans strains with a veA wild-type allele, while in the dark the fungus develops fruiting bodies (Yager, 1992; Stinnett et al., 2007). Deletion of veA in A. nidulans resulted in hyperconidiating strains unable to produce cleistothecia (Kim et al., 2002; Kato et al., 2003). Similarly, deletion of veA increases conidiation and completely blocks sclerotial formation in A. flavus and A. parasiticus (Calvo et al., 2004; Duran et al., 2007).
Another major breakthrough contributing to the understanding of veA function was the discovery of its regulatory role in secondary metabolism in A. nidulans (Kato et al., 2003). veA was shown to control the biosynthesis of several compounds including antibiotics and mycotoxins, specifically sterigmatocystin (ST), the penultimate intermediate in the AF biosynthetic pathway (Kato et al., 2003). Further studies revealed this regulatory role to be conserved in many other fungi. Importantly, veA was demonstrated to be required for the production of AFs in A. parasiticus and A. flavus, as well as CPA and aflatrem in A. flavus (Calvo et al., 2004; Duran et al., 2007, 2009). Studies of the veA-dependent transcriptome in Aspergillus fumigatus indicated that veA affects the expression of 100s of genes (Dhingra et al., 2013), while studies in A. flavus and Fusarium verticillioides demonstrated a role for veA in response to oxidative stress (Baidya et al., 2014; Lan et al., 2014) and hydrolytic activity (Duran et al., 2014). However, in this review we will mainly focus on the role of veA and known veA-related regulatory factors in the control of morphogenesis, particularly in the formation of sclerotia and in the biosynthesis of secondary metabolites.
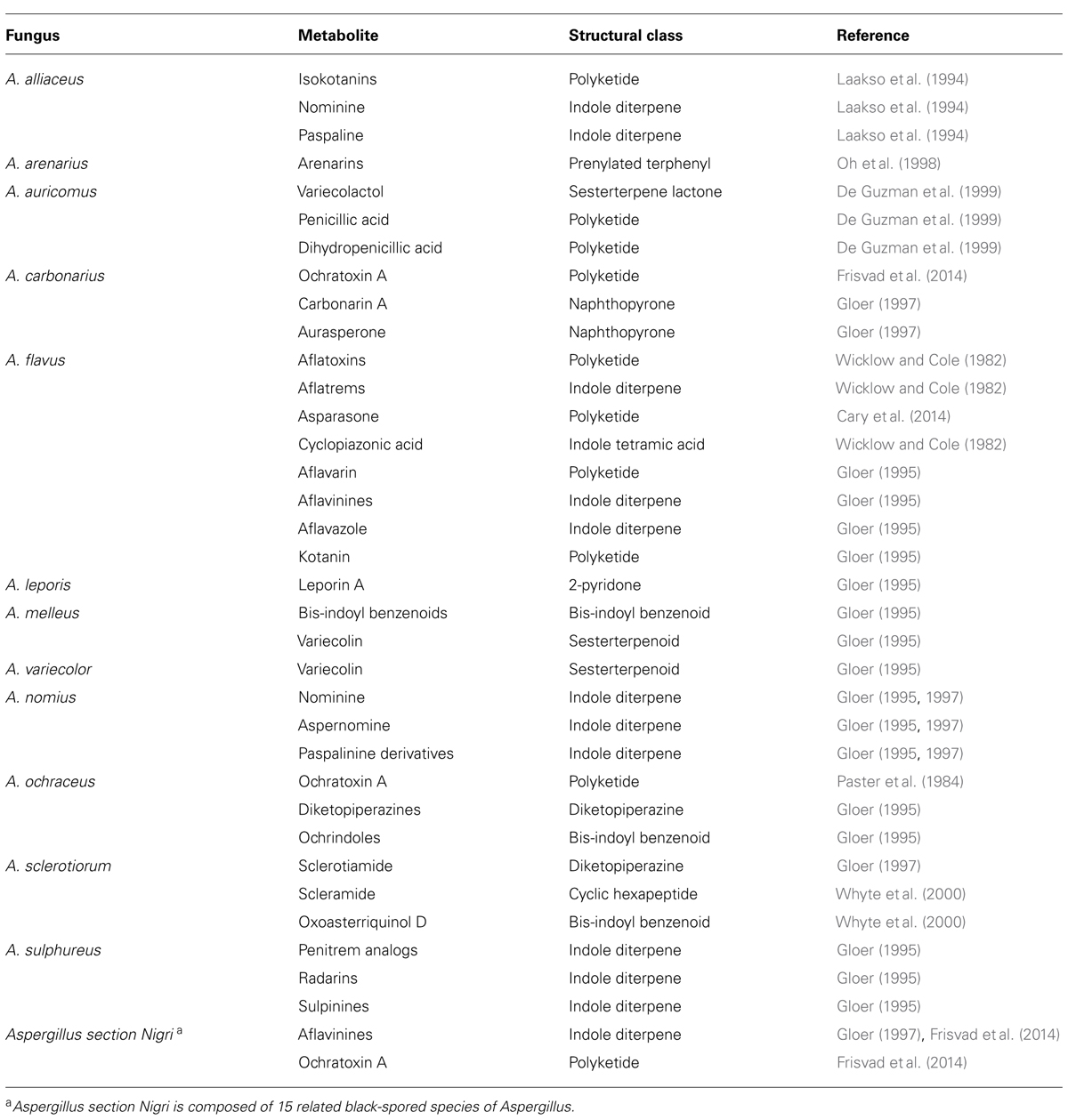
Numerous putative veA homologs have been identified in other fungal species and many of them have been experimentally characterized (Li et al., 2006; Dreyer et al., 2007; Bayram et al., 2008b; Chettri et al., 2012; Dhingra et al., 2012; Laskowski-Peak et al., 2012; Myung et al., 2012; Lopez-Berges et al., 2013). In A. nidulans the study of possible veA-interacting proteins revealed that VeA forms complexes with other proteins (Bayram et al., 2008a; Calvo, 2008; Purschwitz et al., 2008; Figure 1). After its transport to the nucleus by the alpha-importin KapA (Stinnett et al., 2007; Araujo-Bazan et al., 2009) VeA forms a complex with the red phytochrome FphA (Purschwitz et al., 2008). This interaction is dependent on the presence of the tetrapyrrole chromophore. LreA and LreB, blue sensing proteins, do not interact directly with VeA, but through FphA association; the FphA protein was found to interact with LreB, which interacts with LreA. Deletion of either fphA or lreA/lreB genes affected sexual development as well as secondary metabolism in A. nidulans where they play antagonistic functions (Purschwitz et al., 2008). FphA also negatively affects VeA transport to the nucleus in the presence of light. It is likely that a similar regulatory output of the light-sensing proteins is also conserved in A. flavus.
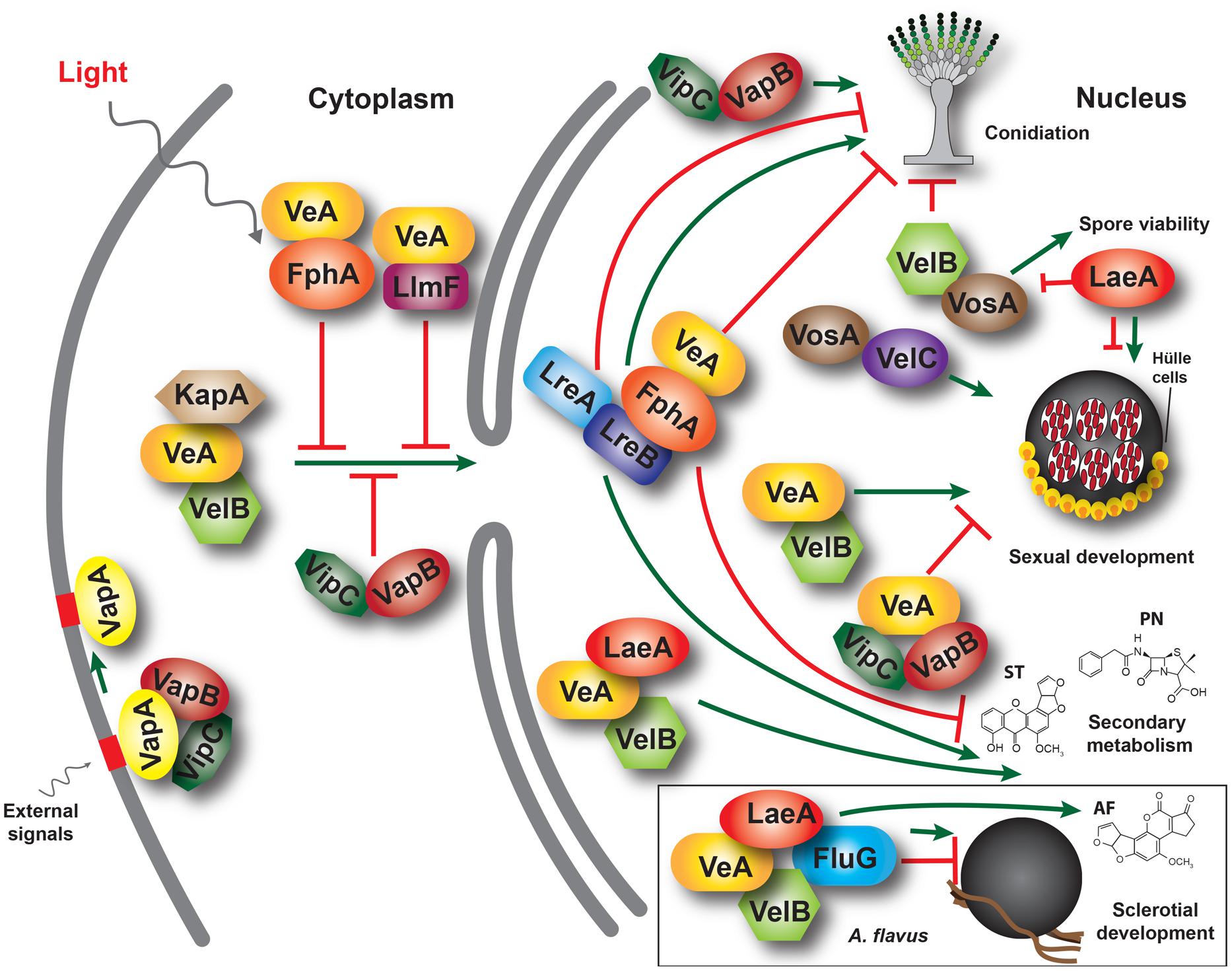
FIGURE 1. A model illustrating interactions between velvet family proteins, LaeA and other associated proteins in the model fungus Aspergillus nidulans. The α-importin KapA transports the VeA-VelB dimer from the cytoplasm to the nucleus, particularly in the dark. This transport is negatively influenced by other proteins, such as FphA, LlmF and Vip-VapB dimer in the light. In the nucleus, VelB-VeA activates sexual development and can interact with LaeA, forming the velvet complex. VeA also interacts with FphA, which is associated with LreB-LreA forming a light-sensing protein complex. VelB, repressor of asexual development, also forms homodimers and heterodimers with VosA, a protein required for spore viability activating trehalose biosynthesis. VosA also interacts with VelC, which positively regulates sexual development. Additionally, VipC and VapB associate with VeA in the nucleus repressing cleistothecial formation. These complexes regulate development and secondary metabolism in a coordinated manner. VeA, LaeA, and VelB have also been shown to control sclerotial and AF production in A. flavus, where they also form a protein complex, together with FluG (box). ST, sterigmatocystin. PN, penicillin; AF, aflatoxin B1.
Additional studies in A. nidulans showed that VeA also interacts with LaeA, forming part of the velvet complex (Bayram et al., 2008a). LaeA encodes a putative methyl transferase involved in chromatin remodeling (Keller et al., 2005; Bok et al., 2006b; Reyes-Dominguez et al., 2010). In addition, LaeA influences VeA post-translational modifications and inhibits sexual development in A. nidulans in response to light (Sarikaya Bayram et al., 2010). Moreover, laeA has been shown to be a positive regulator of gene clusters involved in secondary metabolism in this model organism (Keller et al., 2005; Bok et al., 2006a). In A. flavus the laeA homolog is necessary for production of AFs and sclerotial formation (Kale et al., 2008). Additionally, it has been shown that laeA is a negative regulator of veA expression in A. flavus. Transcriptome analysis of A. flavus wild-type and laeA deletion strains indicated that laeA not only regulates AF production but also controls the expression of other secondary metabolic gene clusters (Georgianna et al., 2010). Similar to FphA, an A. nidulans LaeA-like putative methyltransferase, designated LlmF, also interacts with VeA, negatively affecting VeA transport to the nucleus and acting as negative regulator of ST production and sexual development (Palmer et al., 2013).
Another component of the velvet complex interacting directly with VeA is VelB (Bayram et al., 2008a; Park et al., 2012), a member of the velvet protein family together with VosA and VelC (Ni and Yu, 2007; Park et al., 2014). In A. nidulans, VelB binds to VeA in the cytoplasm and they are co-transported to the nucleus (Bayram et al., 2008a). Similar to the veA deletion mutant, deletion of velB results in a strain unable to display a light-dependent developmental pattern and it is unable to form cleistothecia (Bayram et al., 2008a). However, dissimilar from the veA deletion, absence of velB only showed reduced and delayed production of ST. VelB also interacts with VosA (Bayram et al., 2010). The velvet domain in these two proteins has been shown to bind DNA in A. nidulans (Ahmed et al., 2013) and in Histoplasma capsulatum, where there are involved in the activation of the yeast-phase specific gene expression program (Beyhan et al., 2013). In addition, the VelC velvet protein functions as a positive regulator of sexual development in A. nidulans (Park et al., 2014). Homologs of A. nidulans VelB, and VelC have also been characterized in A. flavus (Chang et al., 2013). Deletion of A. flavus velB (but not velC), similar to the case of veA (Duran et al., 2007), prevents sclerotial formation and AF biosynthesis.
In addition to the interaction between A. flavus LaeA and VelB with VeA, Chang et al. (2013) also described interactions between LaeA and VelB with FluG (Figure 1), a known developmental regulator previously characterized in A. nidulans. FluG contributes to the inactivation of the FadA G-protein signaling pathway in the model fungus, leading to ST production and allowing sexual and asexual development. Mutations in A. nidulans fluG result in proliferation of undifferentiated vegetative hyphae that yield fluffy cotton-like colonies lacking the capacity to produce ST (Adams et al., 1992; Wieser et al., 1994). FadA function was also conserved in the AF-producer A. parasiticus (Hicks et al., 1997). Evidence for a connection between fluG and veA was previously provided by Mooney and Yager (1990) and Yager et al. (1998). Mooney et al. (1990) found three extragenic veA1 suppressor mutations that restored light-dependent conidiation in A. nidulans corresponded to different fluG alleles. This suggested that veA light-dependent activities are related to fluG function. fluG is involved in the synthesis of a diffusible compound that triggers the FluG signaling pathway directing conidiation and mycotoxin biosynthesis while reducing vegetative growth (Lee and Adams, 1996). The diffusible molecule was determined to be dehydroaustinol (Rodriguez-Urra et al., 2012). Two gene clusters in A. nidulans have been found to encode the complete dehydroaustinol pathway (Lo et al., 2012). However, co-culturing experiments did not show a similar diffusible secondary metabolite produced by A. flavus. These results suggest that the function of fluG and the signaling pathways related to conidiation might be different in these two related Aspergilli (Chang et al., 2012). Based on A. flavus studies, Chang et al. (2013) postulated that a delicate balance in the interaction between VeA, VelB, FluG, and LaeA is necessary to maintain normal sclerotiogenesis, conidiogenesis and secondary metabolism, where FluG plays an antagonistic role with respect to VeA, VelB, and LaeA regarding sclerotia formation (Chang et al., 2012). Deletion of fluG resulted in a notable increase in sclerotial formation but did not affect AF production. This also differs from the role of fluG in A. nidulans, where this gene is necessary for ST biosynthesis.
Other characterized VeA-interacting proteins include VipC-VapB methyltransferases, released from the VapA-VipC-VapB membrane-bound complex (Sarikaya Bayram et al., 2014). Presence of VipC-VapB reduces the abundance of the nuclear VelB-VeA-LaeA complex resulting in decreased sexual development. Additionally, VapB also decreases histone 3 lysine 9 trimethylation favoring asexual development.
Post-translational modification of VeA, such as that detected in LaeA-dependent modification in A. nidulans, could also have an effect on the velvet complex function (Sarikaya Bayram et al., 2010). Purschwitz et al. (2009) demonstrated that VeA is phosphorylated. Later Bayram et al. (2012) showed that MpkB phosphorylates VeA. The MAP-kinase mpkB, homolog of FUS3 in Saccharomyces cerevisiae, was first characterized in A. nidulans by Paoletti et al. (2007) and Atoui et al. (2008). MpkB transcription increased during sexual development and deletion of the mpkB gene resulted in sterility (Paoletti et al., 2007), as well as in a decreased in the expression of ST biosynthetic genes and concomitant ST biosynthesis (Atoui et al., 2008). Furthermore, the absence of mpkB also decreased the expression of genes in the penicillin and terrequinone A gene clusters (Atoui et al., 2008). mpkB is also necessary for normal expression of laeA, that as discussed above, is a global regulator of secondary metabolism (Atoui et al., 2008). The mpkB homolog is present in the A. flavus genome, however, its possible function in sclerotial development and secondary metabolism has not yet been characterized experimentally in this AF producer.
Both sclerotial and conidial development and secondary metabolism have been shown to be modulated by A. flavus oxylipins as well as by endogenous plant oxylipins that interact with the infecting fungus (Burow et al., 1997; Calvo et al., 1999; Brown et al., 2008; Affeldt et al., 2012; Scarpari et al., 2014). The A. flavus genome harbors four dioxygenase genes, ppoA, ppoB, ppoC, and ppoD, and one lipoxygenase gene, loxA (Brown et al., 2008, 2009). In the model fungus A. nidulans it has been shown that veA is important for ppo-dependent regulation of development. For instance, veA regulates ppoA expression (Tsitsigiannis et al., 2004). Furthermore, the triple mutant ppoA/B/C showed an increase in veA expression suggesting a regulatory loop between ppo genes and the master regulator veA (Tsitsigiannis et al., 2005). Absence of these genes results in alteration in morphological and chemical development in A. flavus [review by Amaike and Keller (2011)]. For example, strains with deletion of these five genes showed high levels of AF production and sclerotial formation (Brown et al., 2009). The antagonistic roles of different types of oxylipins appear to contribute to a balance between conidiation and sclerotial formation.
The necessity of veA for sclerotial production and AF biosynthesis could also be related to the requirement of veA for a proper oxidative stress response in A. flavus (Baidya et al., 2014). Using modulators that inhibit oxidative stress as well as thiol redox state, Grintzalis et al. (2014) demonstrated that oxidative stress regulates both AF biosynthesis and sclerotial development. Several research groups have also provided evidence of the association between AF production and oxidative stress in Aspergilli (Chang et al., 2011; Reverberi et al., 2012; Hong et al., 2013; Roze et al., 2013).
Recently other regulatory genes have been found to affect development and secondary metabolism in A. flavus, specifically nsdD and nsdC (Cary et al., 2012). The nsdD gene, first described in A. nidulans, encodes a GATA-type zinc finger transcription factor necessary for cleistothecia formation (Han et al., 2001), while nsdC encodes a C2H2 zinc finger-type transcription factor shown to negatively regulate asexual sporulation (Kim et al., 2009). veA only slightly affects nsdD expression (Kato et al., 2003), and has no effect on nsdC expression (Kim et al., 2009), suggesting that the role of these genes is independent of veA in A. nidulans. In A. flavus, both nsdC and nsdD are necessary for sclerotial production and normal levels of AF biosynthesis (Figure 2; Cary et al., 2012).
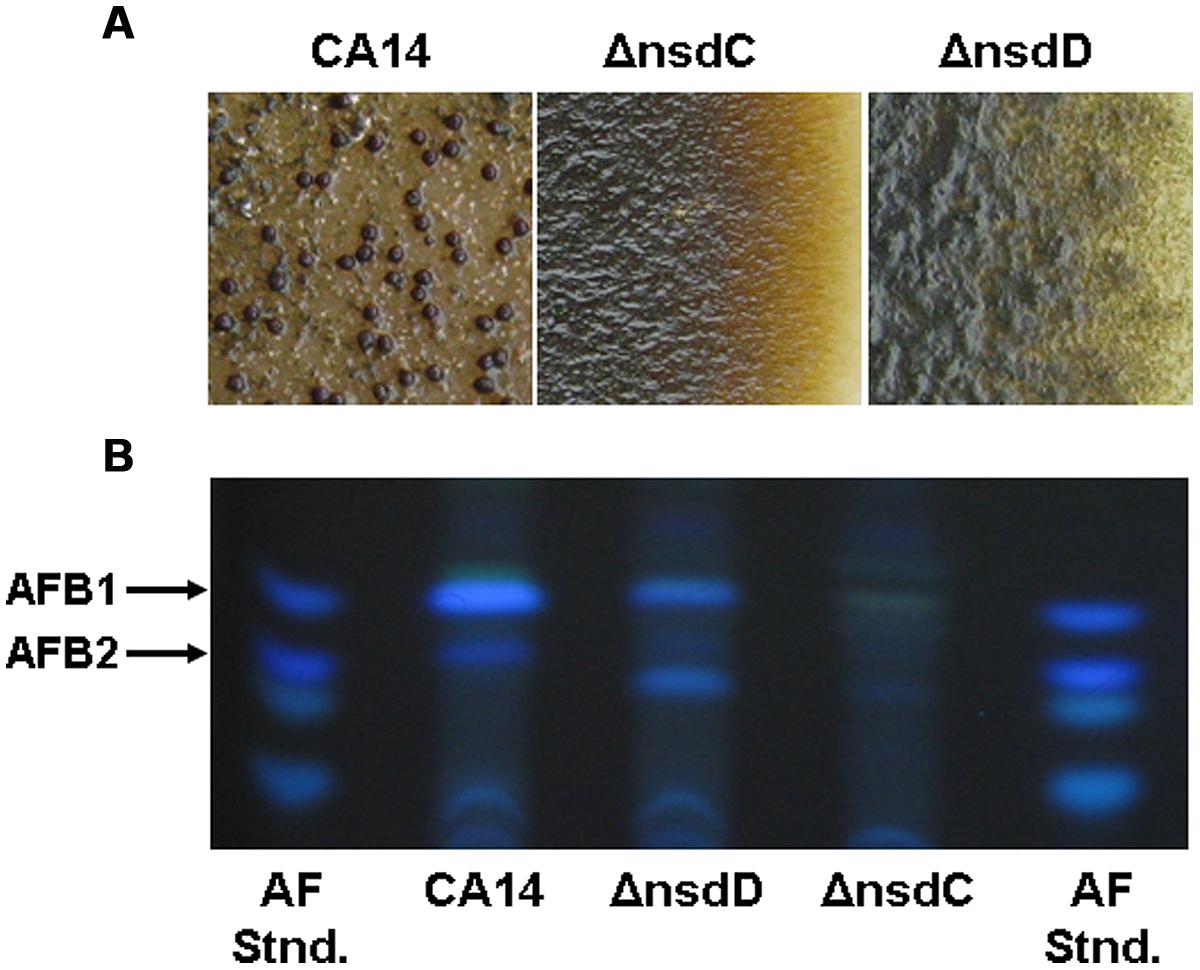
FIGURE 2. Production of sclerotia and aflatoxins in A. flavus CA14 ΔnsdC and ΔnsdD mutants. (A) Surface of colonies demonstrating sclerotial production after 14 days growth in the dark. Sclerotia were absent in the ΔnsdC and ΔnsdD mutants and were produced in the wild-type CA14 (dark structures). (B) TLC analysis of aflatoxin production from the wild-type CA14, ΔnsdC, and ΔnsdD mutants. Extracts (5 ul) were spotted onto 250 um silica gel TLC plates and metabolites were separated in ethyl acetate: methanol: water (40:1:1). Aflatoxin standards were also spotted on the plate. Adapted and modified from Cary et al. (2012).
Functional genomic analysis is a powerful approach that has helped to elucidate the genetic connections between sclerotial formation and secondary metabolism. For instance, Wu et al. (2014) compared the transcriptome of mycelium and sclerotium developmental stages and found that backbone genes of 38 secondary metabolite pathways were transcribed in both the mycelial and sclerotial cultures, including the AF biosynthetic pathway. A transcriptome study by Lin et al. (2013) of A. flavus cultures treated with 5-azacytidine, an inactivator of DNA methyltransferase, provided further evidence that secondary metabolism and development are co-regulated. Addition of 5-azacytidine altered the expression of backbone genes of two identified secondary metabolite gene clusters, #35 and also #27, both of which have been demonstrated experimentally to be associated to sclerotial biology in either a veA- or laeA-dependent manner (Forseth et al., 2012; Cary et al., 2014). Additionally, Chang et al. (2014) studied the transcriptome of cultures treated with decanal and observed that this volatile compound halted development at the vegetative state rendering the fungus unable to produce sclerotia. This coincided with early transcriptional activation of AF and kojic acid biosynthesis gene clusters as well as subtle altered timing of other secondary metabolite gene clusters.
Gene Clusters Producing Secondary Metabolites Associated with Sclerotia
Rapid progress in sequencing of fungal genomes, coupled with bioinformatics, has provided researchers with an in silico approach for identifying potential secondary metabolic gene clusters (Bergmann et al., 2007; Winter et al., 2011; Ehrlich and Mack, 2014). Many of the prediction algorithms (e.g., SMURF, antiSMASH, and MIDDAS-M) in use are based on identification of core or “backbone” genes that encode enzymes such as a PKSs, NRPSs, or dimethylallyltryptophans (DMATs) as well as closely allied genes encoding “decorating” enzymes (e.g., dehydrogenases, methyltransferases, and oxidases), transcription factors and transporters (Khaldi et al., 2010; Medema et al., 2011). The MIDDAS-M algorithm has been used to identify potential secondary metabolic gene clusters that may not contain common core genes such as that for ustiloxin B, an A. flavus secondary metabolite produced by a ribosomal peptide synthetic pathway (Umemura et al., 2014). These types of algorithms have resulted in the identification of numerous putative secondary metabolic gene clusters in fungi, typically between 30 and 40 in Aspergillus species sequenced (Brakhage and Schroeckh, 2011; Andersen et al., 2013; Inglis et al., 2013) including as many as 55 in A. flavus (Georgianna et al., 2010). Some of the products of these clusters have been verified based on prior knowledge of genes and metabolites that constitute the cluster (e.g., AF and CPA). In other cases, the products have been predicted in one species based on homology to genes known to produce the metabolite in another fungal species; for example, the identification of the penicillin gene cluster in A. flavus based on amino acid identity to the known penicillin biosynthetic genes from A. nidulans. However, in most instances, the identity of the metabolites encoded by predicted secondary metabolic gene clusters remains unknown. In these instances the clusters have been termed “orphans.” In a number of cases, these orphan clusters can be “cryptic” or silent when the conditions required to activate expression of the cluster genes have not been determined (Brakhage and Schroeckh, 2011; Brakhage, 2013). Once a putative secondary metabolic gene cluster has been identified, a number of techniques can then be utilized to aid in identification of the cluster metabolite (Brakhage and Schroeckh, 2011; Sanchez et al., 2012).
When genes from orphan clusters are actively transcribed under laboratory growth conditions, standard gene-inactivation techniques can be applied, coupled with comparative metabolic profiling of the pathway mutant and the control strain using LC–MS. A common method used for the identification of cryptic gene cluster metabolites is to overexpress the pathway-specific transcriptional activator (if known) by placing it under the control of a strong inducible or constitutive promoter. For example, normally silent genes of the A. nidulans aspyridone (apd) gene cluster were activated by coupling of the apdR transcriptional activator to the inducible alcohol dehydrogenase promoter of A. nidulans, thus allowing identification of aspyridones A and B (Bergmann et al., 2007). In the absence of a pathway-specific transcription factor, it may be possible to activate gene expression of cryptic clusters by overexpressing global regulatory factors. This is exemplified by the use of overexpressing and deletion mutants of the global regulator, laeA, to identify the terrequinone A gene cluster in A. nidulans (Bok et al., 2006a). In keeping with epigenetic regulation of secondary metabolite production, a number of chemical agents (e.g., histone deacetylase or DNA methyltransferase inhibitors) or genes (e.g., inactivation of a histone deacetylase or sumoylation gene) that modulate chromatin structure have been used to successfully induce expression of cryptic clusters [reviewed in Sanchez et al. (2012) and Brakhage (2013)].
A recent study indicated the presence of secondary metabolite-mediated crosstalk between two separate gene clusters (Forseth et al., 2012). Comparative metabolomics of gene knockout, knockdown (RNAi-based), and overexpression strains of A. flavus were used to identify a group of secondary metabolites derived from two laeA-regulated orphan gene clusters, designated lna and lnb. The lna cluster is located on chromosome 6 and lnb on chromosome 8. The two clusters harbor non-canonical NRPS genes (lnaA and lnbA) with high sequence identity as well as associated genes encoding tailoring enzymes that are involved in the production of a group of piperazines. It was shown that addition of the one of the piperazine metabolites, produced almost exclusively by the lnaA cluster, to wild-type cultures greatly increased expression of the lnaB NRPS. The apparent “sensing” of a metabolite produced by a separate but related gene cluster may represent another layer in the complex regulation of secondary metabolism in fungi. Interestingly, loss of these lnaA- and lnaB-derived piperazines resulted in a significant reduction in sclerotial formation in the mutant strains, thereby demonstrating a role of these secondary metabolites in fungal development.
Lastly, the most ecologically based of all secondary metabolite induction techniques, is the simulation of interactions in nature between the fungus and other resident microbes. This technique is based on the premise that microorganisms share ecological niches; and as such, produce secondary metabolites as a means of intra- and interspecies communication or as defense mechanisms. By simulating these interactions in culture, using two or more organisms, there is a chance that the organism of interest will respond by eliciting production of a secondary metabolite. For example, utilizing microarray technology with co-cultivation techniques, the interaction of A. nidulans with the soil-dwelling actinomycete, Streptomyces rapamycinicus, induced the expression of a cryptic gene cluster in A. nidulans involved in the production of the polyketide, orsellinic acid (Schroeckh et al., 2009).
Sclerotia represent a means by which fungi maintain a quiescent viable state in the absence of a suitable host or of conditions favoring active growth (Coley-Smith and Cooke, 1971). As such, mature sclerotia are essentially dormant metabolically, and therefore would not be amenable to any of the methods discussed above for activation of cryptic secondary metabolic pathways. However, it is probable that many of the secondary metabolites present in sclerotia are produced in the hyphae that coalesce during the early phases of sclerotial morphogenesis. Most sclerotial metabolites identified so far in fungi have been identified from extracts of sclerotia generated under laboratory conditions on artificial media. It is likely that sclerotia found in nature harbor many additional as of yet unidentified secondary metabolites. Below we describe a number of genetically and biochemically well characterized secondary metabolite gene clusters whose products have been found in sclerotia of filamentous fungi. As most of these clusters have been thoroughly reviewed in the literature, only a brief synopsis with references will be provided here.
Ergot Alkaloids
Ergot alkaloids represent a complex family of indole derivatives with diverse structures and broad biological and pharmacological activities. The genetics and enzymology of EA biosynthesis is detailed in reviews by Wallwey and Li (2011) and Jakubczyk et al. (2014). Chemically, EAs can be divided into three groups: ergoamides, ergopeptines, and clavines. The biosynthetic gene clusters responsible for the production of each of these types of EAs have been identified in a number of fungal species. The gene cluster in C. purpurea leading to the formation of complex ergopeptines consists of 14 genes spanning about 68.5 kb of the genome. The Claviceps fusiformis cluster is responsible for the production of the clavines, agroclavine and elymoclavine, that lack the peptide moieties present on the ergoline ring of ergopeptines. The C. purpurea and C. fusiformis gene clusters are homologous with the exception of three genes that are lacking in C. fusiformis. These genes (lpsA1, lpsA2, and lpsC) encode NRPSs that are responsible for biosynthesis of the peptide moieties present in the ergopeptines. A. fumigatus also produces the clavine-type metabolites, fumigaclavines, but these have only been associated with conidiation. No genes encoding a putative pathway-specific transcriptional activator or transporter have been identified in EA gene clusters.
The genes involved in EA biosynthesis are divided into early and late pathway steps. The first step of the early pathway is catalyzed by the dimethylallyl prenyltransferases (DmaW) that prenylates L-tryptophan in the presence of dimethylallyl pyrophosphate (DMAPP) to form DMAT. Subsequent methylation (EasF) and two successive oxidations (EasC and EasE) produce chanoclavine-I, the ergoline ring C structure. Chanoclavine-I is then oxidized by EasD to generate the aldehyde form which in Claviceps is subsequently cyclized by EasA and EasG reductases to form the unsaturated ergoline ring D structure, agroclavine, that represents the last intermediate of the early pathway. The late step pathway genes encode an oxidase (CloA) responsible for the formation of paspalic acid which, either spontaneously or via an isomerase, forms lysergic acid. Three NRPSs (Lps1, Lps2, and lpsC) activate lysergic acid and form the tripeptide moiety of the ergopeptine end products.
Asparasone A
Expression of genes present in an A. flavus cluster, designated #27 based on SMURF analysis by Georgianna et al. (2010) was found to be significantly downregulated in a veA mutant (Cary et al., 2014). The cluster was predicted to consist of a Zn(2)-Cys(6)-type transcription factor, PKS, two putative transporters and a gene encoding a hypothetical protein. A schematic depiction of the cluster is shown in Figure 3A. Expression of the pks27 gene was first observable at 48 h, was maximal at 120 h, and decreased by 144 h (Cary et al., 2014). Transcription paralleled sclerotial development and pigmentation which appeared to be maximal at 120 h in wild-type A. flavus. The transcription factor, znf27, was required for wild-type levels of expression of the other three cluster genes but not for the gene encoding the hypothetical protein. The putative high-affinity glucose (mfs1) and MFS transporter (mfs2) genes showed an expression profile similar to that observed for the pks27 and the znf27 genes. qRT-PCR of RNA isolated from mycelia, conidia, and sclerotia of the A. flavus wild-type showed that expression of pks27 and znf27 was specific to the sclerotium.
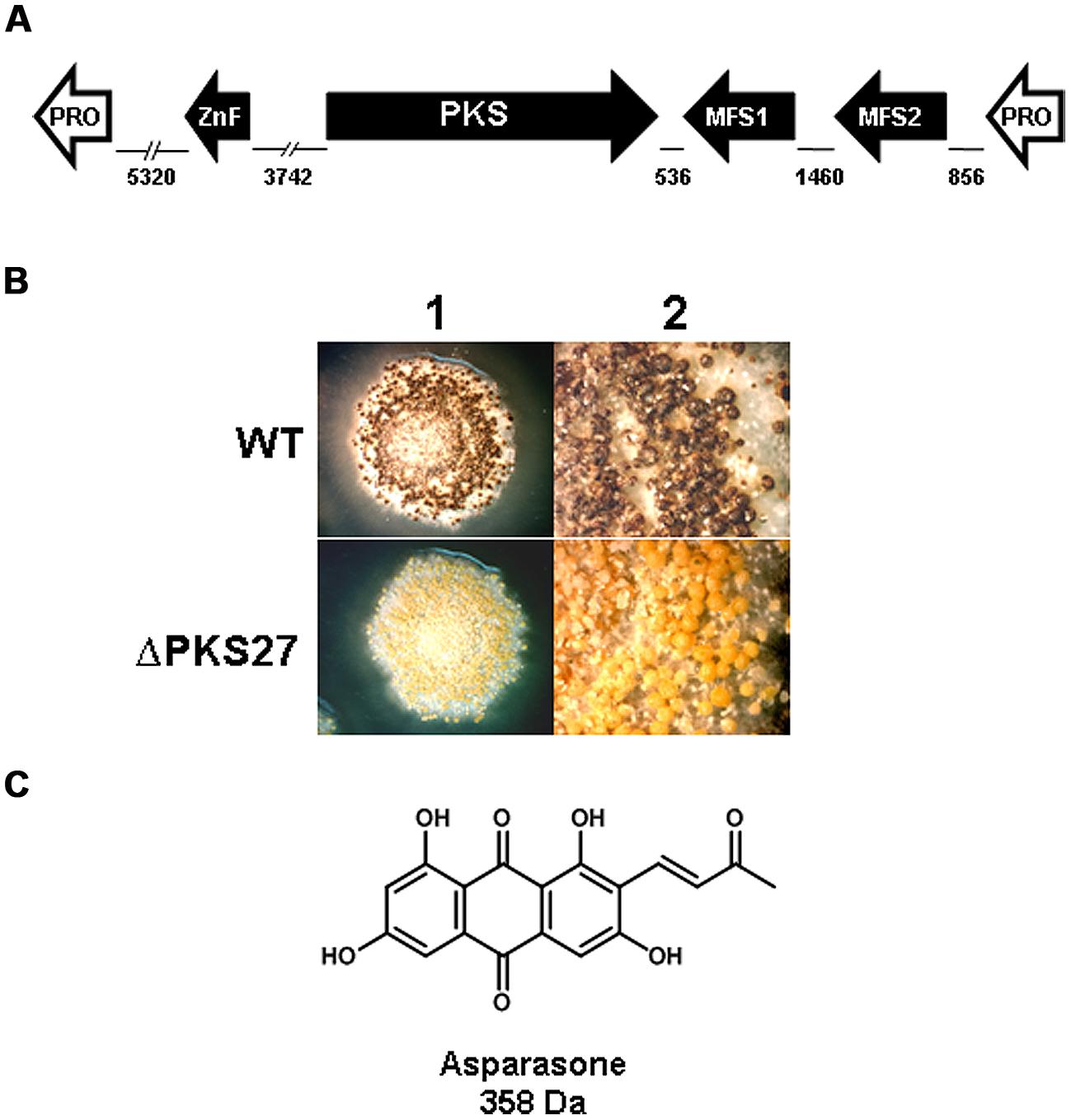
FIGURE 3. (A) Schematic representation of the A. flavus asparasone gene cluster. The cluster is composed of four genes [putative Gal4-type transcription factor (ZnF); polyketide synthase (PKS); putative high-affinity glucose transporter (MFS1); and a second putative MFS transporter (MFS2)]. The two hypothetical protein encoding non-cluster genes (Pro) flanking the asparasone cluster are shown in white. The size in bp of intergenic regions are shown. Arrowheads denote direction of transcription. (B) Microscopic examination of sclerotia. Sclerotium production in Af70 pyrG-1 (WT) and an Af70 Δpks27 mutant was observed using an Olympus SZH10 stereomicroscope and Nikon DS-Qi1 camera. Panel: (1) colony surface, X10 magnification; (2) colony surface, X25 magnification. (C) Chemical structure of asparasone A. Adapted and modified from Cary et al. (2014).
Inactivation of pks27 resulted in A. flavus colonies that produced only grayish-yellow sclerotia, instead of the characteristic dark brown color of the wild-type, indicating that the mutational defect was only in pigmentation and not in sclerotial maturity (Figure 3B). Comparison of extracts of the wild-type and the Δpks27 mutant by ultra-high performance liquid chromatography and mass spectrometry revealed a metabolite of mass 358 Da that was present in the wild-type but missing in the mutant. Based on this mass, the metabolite was putatively identified as the anthraquinone asparasone A [1,3,6,8-tetrahydroxy-2-(1′-hydroxy-3′-oxobutyl)-anthraquinone; M = 358 Da; Figure 3C]. It was previously reported to be produced by A. parasiticus which is a close relative of A. flavus (Sobolev et al., 1997). The identification of the 358 Da metabolite as asparasone A was confirmed by LC–MS comparison with an authentic asparasone A standard. It was hypothesized that dehydration of asparasone would result in conjugated olefins which, like styrene, might rapidly polymerize in the presence of metal dioxygenases such as laccases to form the dark sclerotial pigment (Cary et al., 2014).
Aflatoxin
The genetics, molecular biology, and biochemistry of AF biosynthesis in A. flavus and A. parasiticus have been the focus of a number of reviews, and we direct the reader to these references for more detailed information (Yu et al., 2004; Ehrlich et al., 2005; Georgianna and Payne, 2009; Yu, 2012). Many of the contributions to our understanding of AF biosynthesis and its regulation have come from studies in the model fungus, A. nidulans, on the biosynthesis of ST. ST a precursor of AF, and the genes for ST biosynthesis in A. nidulans are highly homologous to those required for the production of ST in the AF gene cluster. The AF biosynthetic gene cluster of A. flavus spans ∼70 kb of chromosome 3 and consists of 28 genes including two regulatory factors, aflR and aflS (aflJ). AflR is a Zn(2)-Cys(6)-type, pathway-specific transcriptional activator while AflS does not share any significant identity with other fungal proteins but has been shown to be required for AF production. AflS has been shown to interact with AflR and facilitate the activation of other AF biosynthetic genes (Du et al., 2007). AflR protein binds to the palindromic sequence 5′-TCGN5CGA-3′ (or deviations thereof) found in the promoter region of all AF biosynthetic genes. The AF gene cluster in A. flavus is under the control of the global regulators, VeA, NsdD, NsdC, and LaeA (Duran et al., 2007; Kale et al., 2008; Cary et al., 2012).
Aflatoxins are a group of polyketide-derived furanocoumarins that are produced from acetate via a PKS (AflC) and two fatty acid synthetases (AflA and AflB), and a number of tailoring enzymes. AF biosynthesis requires at least 18 known enzymatic reactions to catalyze synthesis of the four major AFs, AFB1, AFB2, AFG1, and AFG2 found in A. parasiticus. In general, A. flavus only produces AFB1 and AFB2. Just outside of the distal end of the AF gene cluster in A. parasiticus and A. flavus is a conserved sugar utilization gene cluster. However, the genetic composition at the proximal end (the end closest to the telomere) of the AF cluster is not conserved in these two species. A. flavus strains contain a deletion at the proximal end of the cluster that result in functional loss of aflU (cypA) and aflF (norB) genes. The inability of A. flavus to produce the G toxins is due to the partial deletions of aflU and aflF, which encode a P450 monooxygenase and a putative aryl alcohol dehydrogenase, respectively, and are required for conversion of hydroxymethyl-ST to AFG1 in A. parasiticus (Zeng et al., 2011).
Aspergillus flavus as a species contains two morphotypes that differ in sclerotial size and in their ability to produce AFs. Large (L) and small (S) sclerotial strains are often found in soils from agricultural fields, and the S strains are generally found to produce higher levels of AF than L strains (Zhang and Cotty, 2006; Horn, 2007). A. flavus is a genetically diverse species and, unlike other aflatoxigenic Aspergillus species, a portion of A. flavus populations has lost the ability to produce AFs (Cotty and Bhatnagar, 1994). A survey of 38 non-aflatoxigenic A. flavus strains, isolated from across the Southern United States, identified eight patterns of gene deletion present in the AF gene cluster (Chang et al., 2005). There is evidence that gene loss in the AF gene cluster of non-aflatoxigenic A. flavus isolates is irreversible, and that balancing selection maintains non-aflatoxigenicity and lineage-specific gene loss in A. flavus populations (Moore et al., 2009, 2011).
Aflatrems
Both aflatrem and its isomer, β-aflatrem (502 Da), are indole-diterpenes that have been isolated from the sclerotia of A. flavus (TePaske et al., 1992). Aflatrems are tremorigenic mycotoxins that have been shown to cause neurological disorders in mammals, including muscle tremors and hyperexcitability in livestock, that have consumed feed contaminated with A. flavus (Valdes et al., 1985). β-aflatrem displayed significant activity against corn earworm in feeding studies (TePaske et al., 1992). Biosynthesis of aflatrems proceeds much like that of paxilline in Penicillium paxilli (Parker and Scott, 2004), in that aflatrem consists of a paxilline-like core, with an additional prenyl group on the indole moiety and an acetyl group on the diterpene skeleton (Nicholson et al., 2009). Utilizing sequence information for genes involved in paxilline biosynthesis in P. paxilli and the genome sequence of A. flavus, the genes required for aflatrem biosynthesis were found to be present on two separate loci in A. flavus (Zhang et al., 2004; Nicholson et al., 2009). Expression of aflatrem cluster genes and concomitant production of aflatrem has been shown to require the presence of veA and laeA (Duran et al., 2007; Georgianna et al., 2010).
Two gene clusters involved in aflatrem biosynthesis have been described. The ATM1 locus, present on chromosome 5 in A. flavus, harbors a gene cluster consisting of the atmG, atmC, and atmM genes. These encode the geranylgeranyl pyrophosphate (GGPP) synthase, prenyltransferases, and monoxygenase, respectively, that are involved in synthesis of paspaline, the first stable intermediate in paxilline and aflatrem biosynthesis. The ATM2 locus, located on chromosome 7, contains atmD, encoding an aromatic prenyltransferse; atmQ and atmP, both encoding P450 monooxygenases; and atmA and atmB, both encoding predicted membrane proteins believed (but not proven) to be transporters required for paxilline biosynthesis. The exact functions of atmG, atmC, atmM, and atmB and their pax orthologs in paspaline biosynthesis are not clear. It is believed that AtmG catalyzes the condensation of indole-3-glycerol phosphate and DMAPP to generate GGPP, which is then epoxidated by AtmM and cyclized by AtmC to form paspaline (Saikia et al., 2006). AtmP converts paspaline to 13-desoxypaxilline via removal of the C-30 methyl group and oxidation at C-10. AtmQ catalyzes the oxidation of 13-desoxypaxilline at C-7 then C-13 to form paspalinine. Finally, AtmD prenylates paspalinine on the indole moiety to form aflatrem. No pathway-specific transcription activator gene was identified in the clusters.
Cyclopiazonic Acid
It was noted that A. flavus strains unable to form AFs, due to deletions that extended from the adjacent subtelomeric region to within the AF gene cluster, were often unable to produce CPA (Chang et al., 2009). A region spanning about 30 kb from the subtelemeric end of the AF cluster was shown to harbor genes encoding a putative DMAT (dmtA), PKS-NRPS (pks-nrps), and FAD-dependent oxidoreductase (moaA) that were considered candidates for CPA production based on enzymes identified in biosynthesis of EAs. Inactivation of these three genes resulted in loss of CPA production. Orthologous genes (cpaD = dmtA; cpaA = pks-nrps; cpaO = moaA) have been identified in A. oryzae and also shown, by gene disruption, to be required for biosynthesis of CPA (Shinohara et al., 2011). Interestingly, the CPA cluster in both of these fungi also contained a putative transcription factor (cpaR = ctfR1), however, disruption of this gene in both A. flavus and A. oryzae did not affect CPA production. Production of CPA has been shown to require the presence of the veA gene (Duran et al., 2007).
In the initial step in CPA biosynthesis, the PKS-NRPS catalyzes the condensation of L-tryptophan and two molecules of acetyl-CoA to generate cycloacetoacetyl-L-tryptophan (cAATrp) which is then converted by the DMAT to β-CPA. The FAD-dependent oxidoreductase is then responsible for the cyclization of β-CPA to CPA (Shinohara et al., 2011). Interestingly, A. oryzae RIB40, which does not make CPA, was found to have a truncated version of the PKS-NRPS (cpaA) gene.
Conclusion and Future Perspectives
It will be difficult to ascertain the exact role of individual sclerotial secondary metabolites in fungal biology. However, observations such as the induction of orsellinic acid production in A. nidulans upon co-culture with a soil microbe provide strong support for a role of these natural products in cross-species communication or defense against competing microbes. The potential role of sclerotial secondary metabolites as a chemical defense against insect predators is supported by the plethora of studies that have demonstrated their antiinsectan/antifeedant properties (Gloer, 1995, 2007). The finding of Cary et al. (2014) of preferential feeding by insects on sclerotia collected from a mutant A. flavus no longer producing asparasone A represents the first in vivo experimental evidence of the contribution of a secondary metabolite to sclerotial chemical defense. These types of gene knockout experiments should prove invaluable in identifying the contribution of individual secondary metabolites to fungal chemical defense. This will be important as many of the secondary metabolites identified in A. flavus have not been assigned to a predicted gene cluster, and it is highly probable that in the near future many of these orphan clusters will be found to produce compounds that are associated with the sclerotium. Advances in functional genomics and metabolomics will invariably accelerate the pace in the identification of secondary metabolic gene clusters associated with the synthesis of sclerotial compounds. Accordingly, these studies will also provide relevant information on the genetic regulatory networks governing activation and modulation of secondary metabolic gene clusters that play a role in sclerotial biology as well as other cellular processes. In this regard, continued structural and comparative analyses of sequenced fungal genomes, coupled with ever-increasing understanding of the molecular and functional biology of secondary metabolites in the Aspergilli, will undoubtedly accelerate the identification and functional characterization of secondary metabolite gene clusters in other filamentous fungi.
The majority of studies on the biological functions of sclerotial secondary metabolites have focused on their role in chemical defense against insect predators and other competing organisms. More research is needed to investigate other possible roles for these metabolites in sclerotial biology. For example, no information exists as to why A. flavus produces two morphotypes of sclerotia and if there is any difference in the secondary metabolic profiles of these morphotypes. If a metabolite(s) is consistently present in one sclerotial morphotype versus the other this may indicate a role for the metabolite(s) in sclerotial morphogenesis. Not only are S morphotype sclerotia smaller than L morphotype, but they are almost always produced in greater numbers. The ability to produce greater numbers of S morphotype sclerotia may represent an adaptive response by the fungus to survive insect predation compared to that of L strains. A correlation between selective pressure, due to predation, and sclerotial size has been suggested (Wicklow, 1988), in which long-term survival of a fungus is improved by the production of larger, better chemically defended sclerotia compared to those of fungi that produce numerous small sclerotia. However, it can also be argued that smaller size may increase the ease with which S morphotype sclerotia are damaged/consumed by predators and therefore the fungus has evolved to produce increased numbers as a means to ensure dissemination and survival of the species. During the course of evolution, selective pressure from increased predation on S morphotype sclerotia may have led to an increase in the levels/classes of antiinsectan secondary metabolites present in small sclerotia. The study of Chang et al. (2002) demonstrated that an increase in AF intermediates led to smaller sclerotial size in A. flavus. A similar correlation may be used to explain the existence of the S morphotype. If sclerotia of S morphotype A. flavus strains have increased levels of secondary metabolites compared to L morphotype, the increased demand for carbon building blocks (e.g., acetate) for biosynthesis of the additional secondary metabolites would result in less availability of carbon for sclerotial biogenesis, resulting in the observed small sclerotial morphotype. Chemical analysis of sclerotial extracts coupled with insect feeding studies should be able to shed some light on the relationship of sclerotial morphotype and fungivory.
Another unexplored area is the potential role of secondary metabolites in mating of normally asexual species of Aspergilli. It will be of interest to determine if secondary metabolite profiles differ in the stromata generated from the mating of two A. flavus strains compared to that present in the sclerotia produced during normal growth of each strain. Perhaps a chemical signal produced by hyphae of the interacting mating types can induce production of novel secondary metabolites in the sexual stromata that are not present in sclerotia of the individual mating partners. The chemical signal itself could be the product of a secondary metabolic gene cluster that is activated upon interaction of hyphae of opposite mating types. The presence of novel secondary metabolites in stromata would indicate that these compounds may play a role in the recognition and initiation of sexual reproduction by strains of opposite mating type, or they may be produced as a means of expanding the chemical arsenal of antiinsectan agents present in the fruiting structures.
Sclerotia are very important to the survival and dissemination of fungi in nature, and as such should be the target of strategies for control of fungal contamination of food and feed crops. As presented in this review, a number of global regulators that control production of secondary metabolites also control sclerotial formation. Novel technologies such as host-induced gene silencing can take advantage of host plant-derived siRNAs that target expression of these global regulators in the invading fungus. For example, maize can be transformed with RNAi-based constructs that generate siRNAs targeting veA or nsdC gene transcripts of A. flavus. This approach, in theory, would reduce both AF and sclerotial production in the invading fungus (Nunes and Dean, 2012). The soundness of this concept has already been demonstrated for control of several cereal pathogens, including barley powdery mildew (Nowara et al., 2010), wheat leaf rust (Panwar et al., 2013) and maize ear-rot caused by F. verticillioides (Tinoco et al., 2010). Continued study of the biogenesis and function of fungal secondary metabolites and their association with development, as well as elucidation of the regulatory mechanisms controlling production of these natural products, will facilitate the design of additional strategies to reduce the detrimental effects of pathogenic fungi.
Conflict of Interest Statement
The Review Editor Geromy G. Moore declares that, despite being affiliated to the same institution as author Jeffrey W. Cary, the review process was handled objectively and no conflict of interest exists.
References
Adams, T. H., Hide, W. A., Yager, L. N., and Lee, B. N. (1992). Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 12, 3827–3833.
Adams, T. H., and Yu, J. H. (1998). Coordinate control of secondary metabolite production and asexual sporulation in Aspergillus nidulans. Curr. Opin. Microbiol. 1, 674–677. doi: 10.1016/S1369-5274(98)80114-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Affeldt, K. J., Brodhagen, M., and Keller, N. P. (2012). Aspergillus oxylipin signaling and quorum sensing pathways depend on g protein-coupled receptors. Toxins (Basel) 4, 695–717. doi: 10.3390/toxins4090695
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ahmed, Y. L., Gerke, J., Park, H. S., Bayram, O., Neumann, P., Ni, M.,et al. (2013). The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-kappaB. PLoS Biol. 11:e1001750. doi: 10.1371/journal.pbio.1001750
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Andersen, M. R., Nielsen, J. B., Klitgaard, A., Petersen, L. M., Zachariasen, M., Hansen, T. J.,et al. (2013). Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. U.S.A. 110, E99–E107. doi: 10.1073/pnas.1205532110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Araujo-Bazan, L., Dhingra, S., Chu, J., Fernandez-Martinez, J., Calvo, A. M., and Espeso, E. A. (2009). Importin alpha is an essential nuclear import carrier adaptor required for proper sexual and asexual development and secondary metabolism in Aspergillus nidulans. Fungal Genet. Biol. 46, 506–515. doi: 10.1016/j.fgb.2009.03.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Atoui, A., Bao, D., Kaur, N., Grayburn, W. S., and Calvo, A. M. (2008). Aspergillus nidulans natural product biosynthesis is regulated by mpkB, a putative pheromone response mitogen-activated protein kinase. Appl. Environ. Microbiol. 74, 3596–3600. doi: 10.1128/AEM.02842-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baidya, S., Duran, R. M., Lohmar, J. M., Harris-Coward, P. Y., Cary, J. W., Hong, S. Y.,et al. (2014). VeA is associated with the response to oxidative stress in the aflatoxin producer Aspergillus flavus. Eukaryot. Cell 13, 1095–1103. doi: 10.1128/EC.00099-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bayram, O., Bayram, O. S., Ahmed, Y. L., Maruyama, J., Valerius, O., Rizzoli, S. O.,et al. (2012). The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 8:e1002816. doi: 10.1371/journal.pgen.1002816
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bayram, O., and Braus, G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24. doi: 10.1111/j.1574-6976.2011.00285.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bayram, O., Braus, G. H., Fischer, R., and Rodriguez-Romero, J. (2010). Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet. Biol. 47, 900–908. doi: 10.1016/j.fgb.2010.05.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bayram, O., Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O.,et al. (2008a). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bayram, O., Krappmann, S., Seiler, S., Vogt, N., and Braus, G. H. (2008b). Neurospora crassa ve-1 affects asexual conidiation. Fungal Genet. Biol. 45, 127–138. doi: 10.1016/j.fgb.2007.06.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bennett, J. W., and Klich, M. (2003). Mycotoxins. Clin. Microbiol. Rev. 16, 497–516. doi: 10.1128/CMR.16.3.497-516.2003
Bergmann, S., Schumann, J., Scherlach, K., Lange, C., Brakhage, A. A., and Hertweck, C. (2007). Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3, 213–217. doi: 10.1038/nchembio869
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beyhan, S., Gutierrez, M., Voorhies, M., and Sil, A. (2013). A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 11:e1001614. doi: 10.1371/journal.pbio.1001614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhatnagar, D., Brown, R., Ehrlich, K., and Cleveland, T. E. (2002). “Mycotoxins contaminating cereal grain crops: their occurrence and toxicity,” in Applied Mycology and Biotechnology, eds G. G. Khachatourians and D. K. Arora (New York, NY: Elsevier B.V.), 171–196.
Bok, J. W., Hoffmeister, D., Maggio-Hall, L. A., Murillo, R., Glasner, J. D., and Keller, N. P. (2006a). Genomic mining for Aspergillus natural products. Chem. Biol. 13, 31–37. doi: 10.1016/j.chembiol.2005.10.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bok, J. W., Noordermeer, D., Kale, S. P., and Keller, N. P. (2006b). Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 61, 1636–1645. doi: 10.1111/j.1365-2958.2006.05330.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brakhage, A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. doi: 10.1038/nrmicro2916
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brakhage, A. A., and Schroeckh, V. (2011). Fungal secondary metabolites – strategies to activate silent gene clusters. Fungal Genet. Biol. 48, 15–22. doi: 10.1016/j.fgb.2010.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, S. H., Scott, J. B., Bhaheetharan, J., Sharpee, W. C., Milde, L., Wilson, R. A.,et al. (2009). Oxygenase coordination is required for morphological transition and the host-fungus interaction of Aspergillus flavus. Mol. Plant Microbe Interact. 22, 882–894. doi: 10.1094/MPMI-22-7-0882
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, S. H., Zarnowski, R., Sharpee, W. C., and Keller, N. P. (2008). Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl. Environ. Microbiol. 74, 5674–5685. doi: 10.1128/AEM.00565-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burdock, G. A., and Flamm, W. G. (2000). Safety assessment of the mycotoxin cyclopiazonic acid. Int. J. Toxicol. 19, 195–218. doi: 10.1080/10915810050074964
Burow, G. B., Nesbitt, J. D., and Keller, N. P. (1997). Seed lipoxygenase products modulate Aspergillus mycotoxin synthesis. Mol. Plant Microbe Interact. 10, 380–387. doi: 10.1094/MPMI.1997.10.3.380
Butler, M. J., and Day, A. W. (1998). Fungal melanins: a review. Can. J. Microbiol. 44, 1115–1136. doi: 10.1139/w98-119
Butler, M. J., Gardiner, R. B., and Day, A. W. (2009). Melanin synthesis by Sclerotinia sclerotiorum. Mycologia 101, 296–304. doi: 10.3852/08-120
Calvo, A. M. (2008). The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 45, 1053–1061. doi: 10.1016/j.fgb.2008.03.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Calvo, A. M., Bok, J.-W., Brooks, W., and Keller, N. P. (2004). VeA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70, 4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Calvo, A. M., Hinze, L. L., Gardner, H. W., and Keller, N. P. (1999). Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65, 3668–3673.
Calvo, A. M., Wilson, R. A., Bok, J. W., and Keller, N. P. (2002). Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459. doi: 10.1128/MMBR.66.3.447-459.2002
Cary, J. W., Harris-Coward, P. Y., Ehrlich, K. C., Di Mavungu, J. D., Malysheva, S. V., De Saeger, S.,et al. (2014). Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet. Biol. 64, 25–35. doi: 10.1016/j.fgb.2014.01.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cary, J. W., Harris-Coward, P. Y., Ehrlich, K. C., Mack, B. M., Kale, S. P., Larey, C.,et al. (2012). NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 11, 1104–1111. doi: 10.1128/EC.00069-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cary, J. W., Linz, J. E., and Bhatnagar, D. (2000). “Aflatoxins: biological significance and regulation of biosynthesis,” in Microbial Foodborne Diseases: Mechanisms of Pathogenesis and Toxin Synthesis, eds J. W. Cary, J. E. Linz, and D. Bhatnagar (Lancaster, PA: Technomic Publishing Co.), 317–361.
Champe, S. P., Kurtz, M. B., Yager, L. N., Butnick, N. J., and Axelrod, D. E. (1981). “Spore formation in Aspergillus nidulans: competence and other developmental processes,” in The Fungal Spores: Morphogenic Controls, eds H. R. Hohl and G. Turian (New York, NY: Academic Press), 255–276.
Chanda, A., Roze, L. V., Kang, S., Artymovich, K. A., Hicks, G. R., Raikhel, N. V.,et al. (2009). A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 106, 19533–19538. doi: 10.1073/pnas.0907416106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P.-K., Bennett, J. W., and Cotty, P. J. (2002). Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 153, 41–48. doi: 10.1023/A:1015211915310
Chang, P. K., Horn, B. W., and Dorner, J. W. (2005). Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 42, 914–923. doi: 10.1016/j.fgb.2005.07.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P. K., Horn, B. W., and Dorner, J. W. (2009). Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet. Biol. 46, 176–182. doi: 10.1016/j.fgb.2008.11.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P. K., Scharfenstein, L. L., Li, P., and Ehrlich, K. C. (2013). Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 58–59, 71–79. doi: 10.1016/j.fgb.2013.08.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P. K., Scharfenstein, L. L., Luo, M., Mahoney, N., Molyneux, R. J., Yu, J.,et al. (2011). Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins (Basel) 3, 82–104. doi: 10.3390/toxins3010082
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P. K., Scharfenstein, L. L., Mack, B., and Ehrlich, K. C. (2012). Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl. Environ. Microbiol. 78, 7557–7563. doi: 10.1128/AEM.01241-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, P. K., Scharfenstein, L. L., Mack, B., Yu, J., and Ehrlich, K. C. (2014). Transcriptomic profiles of Aspergillus flavus CA42, a strain that produces small sclerotia, by decanal treatment and after recovery. Fungal Genet. Biol. 68, 39–47. doi: 10.1016/j.fgb.2014.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chettri, P., Calvo, A. M., Cary, J. W., Dhingra, S., Guo, Y., Mcdougal, R. L.,et al. (2012). The veA gene of the pine needle pathogen Dothistroma septosporum regulates sporulation and secondary metabolism. Fungal Genet. Biol. 49, 141–151. doi: 10.1016/j.fgb.2011.11.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chinnici, J. P., and Bettinger, D. A. (1984). Effects of aflatoxin B1 and caffeine on viability in natural strains of Drosophila melanogaster. J. Invertebr. Pathol. 44, 263–266. doi: 10.1016/0022-2011(84)90023-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coley-Smith, J. R., and Cooke, R. C. (1971). Survival and germination of fungal sclerotia. Annu. Rev. Phytopathol. 9, 65–92. doi: 10.1146/annurev.py.09.090171.000433
Cotty, P. J., and Bhatnagar, D. (1994). Variability among atoxigenic Aspergillus flavus strains in ability to prevent aflatoxin contamination and production of aflatoxin biosynthetic pathway enzymes. Appl. Environ. Microbiol. 60, 2248–2251.
De Guzman, F. S., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1999). Variecolactol: a new sesterterpene lactone from the sclerotia of Aspergillus auricomus (Gueguen) Saito. Sci. Diliman 11, 1–5.
Dhingra, S., Andes, D., and Calvo, A. M. (2012). VeA regulates conidiation, gliotoxin production, and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 11, 1531–1543. doi: 10.1128/EC.00222-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dhingra, S., Lind, A. L., Lin, H. C., Tang, Y., Rokas, A., and Calvo, A. M. (2013). The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE 8:e77147. doi: 10.1371/journal.pone.0077147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doll, K., Chatterjee, S., Scheu, S., Karlovsky, P., and Rohlfs, M. (2013). Fungal metabolic plasticity and sexual development mediate induced resistance to arthropod fungivory. Proc. Biol. Sci. 280, 20131219. doi: 10.1098/rspb.2013.1219
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dreyer, J., Eichhorn, H., Friedlin, E., Kurnsteiner, H., and Kuck, U. (2007). A homologue of the Aspergillus velvet gene regulates both cephalosporin C biosynthesis and hyphal fragmentation in Acremonium chrysogenum. Appl. Environ. Microbiol. 73, 3412–3422. doi: 10.1128/AEM.00129-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, W., Obrian, G. R., and Payne, G. A. (2007). Function and regulation of aflJ in the accumulation of aflatoxin early pathway intermediate in Aspergillus flavus. Food Addit. Contam. 24, 1043–1050. doi: 10.1080/02652030701513826
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duran, R. M., Cary, J. W., and Calvo, A. M. (2007). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. doi: 10.1007/s00253-006-0581-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duran, R. M., Cary, J. W., and Calvo, A. M. (2009). The role of veA in Aspergillus flavus infection of peanut, corn and cotton. Open Mycol. J. 3, 27–36. doi: 10.2174/1874437000903010027
Duran, R. M., Gregersen, S., Smith, T. D., Bhetariya, P. J., Cary, J. W., Harris-Coward, P. Y.,et al. (2014). The role of Aspergillus flavus veA in the production of extracellular proteins during growth on starch substrates. Appl. Microbiol. Biotechnol. 98, 5081–5094. doi: 10.1007/s00253-014-5598-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dyer, P. S., and O’Gorman, C. M. (2012). Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36, 165–192. doi: 10.1111/j.1574-6976.2011.00308.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ehrlich, K. C., and Mack, B. M. (2014). Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins (Basel) 6, 1916–1928. doi: 10.3390/toxins6061916
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ehrlich, K. C., Yu, J., and Cotty, P. J. (2005). Aflatoxin biosynthesis gene clusters and flanking regions. J. Appl. Microbiol. 99, 518–27. doi: 10.1111/j.1365-2672.2005.02637.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
El Khoury, A., and Atoui, A. (2010). Ochratoxin a: general overview and actual molecular status. Toxins (Basel) 2, 461–493. doi: 10.3390/toxins2040461
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fischer, R., and Kues, U. (2006). “Asexual sporulation in mycelial fungi,” in The Mycota: Growth, Differentiation and Sexuality, 2nd Edn, eds U. Kues and R. Fischer (Berlin: Springer), 263–292. doi: 10.1007/3-540-28135-5_14
Forseth, R. R., Amaike, S., Schwenk, D., Affeldt, K. J., Hoffmeister, D., Schroeder, F. C.,et al. (2012). Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. Engl. 51, 1–7. doi: 10.1002/anie.201207456
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frisvad, J. C., Petersen, L. M., Lyhne, E. K., and Larsen, T. O. (2014). Formation of sclerotia and production of indoloterpenes by Aspergillus niger and other species in section Nigri. PLoS ONE 9:e94857. doi: 10.1371/journal.pone.0094857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geiser, D. M., Klich, M. A., Frisvad, J. C., Peterson, S. W., Varga, J., and Samson, R. A. (2007). The current status of species recognition and identification in Aspergillus. Stud. Mycol. 59, 1–10. doi: 10.3114/sim.2007.59.01
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geiser, D. M., Timberlake, W. E., and Arnold, M. L. (1996). Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13, 809–817. doi: 10.1093/oxfordjournals.molbev.a025641
Georgianna, D. R., Fedorova, N. D., Burroughs, J. L., Dolezal, A. L., Bok, J. W., Horowitz-Brown, S.,et al. (2010). Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11, 213–226. doi: 10.1111/j.1364-3703.2009.00594.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Georgianna, D. R., and Payne, G. A. (2009). Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet. Biol. 46, 113–125. doi: 10.1016/j.fgb.2008.10.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gloer, J. B. (1995). Antiinsectan natural products from fungal sclerotia. Acc. Chem. Res. 28, 343–350. doi: 10.1021/ar00056a004
Gloer, J. B. (1997). “Applications of fungal ecology in the search for new bioactive natural products,” in The Mycota, 1st Edn, eds D. T. Wicklow and B. Soderstrom (Berlin: Springer), 249–268.
Gloer, J. B. (2007). “Applications of fungal ecology in the search for new bioactive natural products,” in The Mycota, Vol. IV, 2nd Edn, eds C. P. Kubicek and I. S. Druzhinina (New York, NY: Springer-Verlag).
Grintzalis, K., Vernardis, S. I., Klapa, M. I., and Georgiou, C. D. (2014). Role of oxidative stress in sclerotial differentiation and aflatoxin B1 biosynthesis in Aspergillus flavus. Appl. Environ. Microbiol. 80, 5561–5571. doi: 10.1128/AEM.01282-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haarmann, T., Rolke, Y., Giesbert, S., and Tudzynski, P. (2009). Ergot: from witchcraft to biotechnology. Mol. Plant Pathol. 10, 563–577. doi: 10.1111/j.1364-3703.2009.00548.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, K. H., Han, K. Y., Yu, J. H., Chae, K. S., Jahng, K. Y., and Han, D. M. (2001). The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41, 299–309. doi: 10.1046/j.1365-2958.2001.02472.x
Hawksworth, D. L., and Rossman, A. Y. (1997). Where are all the undescribed fungi? Phytopathology 87, 888–891. doi: 10.1094/PHYTO.1997.87.9.888
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hicks, J. K., Yu, J. H., Keller, N. P., and Adams, T. H. (1997). Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16, 4916–4923. doi: 10.1093/emboj/16.16.4916
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hong, S. Y., Roze, L. V., Wee, J., and Linz, J. E. (2013). Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. Microbiologyopen 2, 144–160. doi: 10.1002/mbo3.63
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horn, B. W. (2007). Biodiversity of Aspergillus section Flavi in the United States: a review. Food Addit. Contam. 24, 1088–1101. doi: 10.1080/02652030701510012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horn, B. W., Moore, G. G., and Carbone, I. (2009a). Sexual reproduction in Aspergillus flavus. Mycologia 101, 423–429. doi: 10.3852/09-011
Horn, B. W., Ramirez-Prado, J. H., and Carbone, I. (2009b). The sexual state of Aspergillus parasiticus. Mycologia 101, 275–280. doi: 10.3852/08-205
Horn, B. W., Moore, G. G., and Carbone, I. (2011). Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia 103, 174–183. doi: 10.3852/10–115.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horn, B. W., Sorensen, R. B., Lamb, M. C., Sobolev, V. S., Olarte, R. A., Worthington, C. J.,et al. (2014). Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 104, 75–85. doi: 10.1094/PHYTO-05-13-0129-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Inglis, D. O., Binkley, J., Skrzypek, M. S., Arnaud, M. B., Cerqueira, G. C., Shah, P.,et al. (2013). Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 13:91. doi: 10.1186/1471-2180-13-91
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jakubczyk, D., Cheng, J. Z., and O’connor, S. E. (2014). Biosynthesis of the ergot alkaloids. Nat. Prod. Rep. 31, 1328–1338. doi: 10.1039/c4np00062e
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kale, S. P., Milde, L., Trapp, M. K., Frisvad, J. C., Keller, N. P., and Bok, J. W. (2008). Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45, 1422–1429. doi: 10.1016/j.fgb.2008.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kato, N., Brooks, W., and Calvo, A. M. (2003). The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2, 1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism – from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khaldi, N., Seifuddin, F. T., Turner, G., Haft, D., Nierman, W. C., Wolfe, K. H.,et al. (2010). SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. doi: 10.1016/j.fgb.2010.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, H. R., Chae, K. S., Han, K. H., and Han, D. M. (2009). The nsdC gene encoding a putative C2H2-type transcription factor is a key activator of sexual development in Aspergillus nidulans. Genetics 182, 771–783. doi: 10.1534/genetics.109.101667
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, H., Han, K., Kim, K., Han, D., Jahng, K., and Chae, K. (2002). The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37, 72–80. doi: 10.1016/S1087-1845(02)00029-4
Laakso, J. A., Narske, E. D., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1994). Isokotanins A-C: new bicoumarins from the sclerotia of Aspergillus alliaceus. J. Nat. Prod. 57, 128–133. doi: 10.1021/np50103a018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lan, N., Zhang, H., Hu, C., Wang, W., Calvo, A. M., Harris, S. D.,et al. (2014). Coordinated and distinct functions of velvet proteins in Fusarium verticillioides. Eukaryot. Cell 13, 909–918. doi: 10.1128/EC.00022-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laskowski-Peak, M. C., Calvo, A. M., Rohrssen, J., and Smulian, A. G. (2012). VEA1 is required for cleistothecial formation and virulence in Histoplasma capsulatum. Fungal Genet. Biol. 49, 838–846. doi: 10.1016/j.fgb.2012.07.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, B. N., and Adams, T. H. (1996). FluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA beta activation. EMBO J. 15, 299–309.
Li, S., Myung, K., Guse, D., Donkin, B., Proctor, R. H., Grayburn, W. S.,et al. (2006). FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol. Microbiol. 62, 1418–1432. doi: 10.1111/j.1365-2958.2006.05447.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, J. Q., Zhao, X. X., Zhi, Q. Q., Zhao, M., and He, Z. M. (2013). Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 56, 78–86. doi: 10.1016/j.fgb.2013.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lo, H. C., Entwistle, R., Guo, C. J., Ahuja, M., Szewczyk, E., Hung, J. H.,et al. (2012). Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 134, 4709–4720. doi: 10.1021/ja209809t
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lopez-Berges, M. S., Hera, C., Sulyok, M., Schafer, K., Capilla, J., Guarro, J.,et al. (2013). The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Microbiol. 87, 49–65. doi: 10.1111/mmi.12082
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malloch, D., and Cain, R. F. (1972). The Trichocomataceae: ascomycetes with Aspergillus, Paeciloyces and Penicillium imperfect states. Can. J. Bot. 50, 2613–2628. doi: 10.1139/b72-335
Martins, M. L., and Martins, H. M. (1999). Natural and in vitro coproduction of cyclopiazonic acid and aflatoxins. J. Food Prot. 62, 292–294.
Medema, M. H., Blin, K., Cimermancic, P., De Jager, V., Zakrzewski, P., Fischbach, M. A.,et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. doi: 10.1093/nar/gkr466
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mooney, J. L., Hassett, D. E., and Yager, L. N. (1990). Genetic analysis of suppressors of the veA1 mutation in Aspergillus nidulans. Genetics 126, 869–874.
Mooney, J. L., and Yager, L. N. (1990). Light is required for conidiation in Aspergillus nidulans. Genes Dev. 4, 1473–1482. doi: 10.1101/gad.4.9.1473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, G. G., Beltz, S. B., Carbone, I., Ehrlich, K. C., and Horn, B. W. (2011). “The population dynamics of aflatoxigenic Aspergilli,” in Aflatoxins-Biochemistry and Molecular Biology, eds R. G. Guevara-Gonzalez (Rijeka: InTech Open Access Publisher), 347–366.
Moore, G. G., Singh, R., Horn, B. W., and Carbone, I. (2009). Recombination and lineage-specific gene loss in the aflatoxin gene cluster of Aspergillus flavus. Mol. Ecol. 18, 4870–4887. doi: 10.1111/j.1365-294X.2009.04414.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mueller, G. M., and Schmit, J. P. (2007). Fungal biodiversity: what do we know? What can we predict? Biodivers. Conserv. 16, 1–5. doi: 10.1007/s10531-006-9117-7
Myung, K., Zitomer, N. C., Duvall, M., Glenn, A. E., Riley, R. T., and Calvo, A. M. (2012). The conserved global regulator VeA is necessary for symptom production and mycotoxin synthesis in maize seedlings by Fusarium verticillioides. Plant Pathol. 61, 152–160. doi: 10.1111/j.1365-3059.2011.02504.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Narasaiah, K. V., Sashidhar, R. B., and Subramanyam, C. (2006). Biochemical analysis of oxidative stress in the production of aflatoxin and its precursor intermediates. Mycopathologia 162, 179–189. doi: 10.1007/s11046-006-0052-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ni, M., and Yu, J. H. (2007). A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS ONE 2:e970. doi: 10.1371/journal.pone.0000970
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nicholson, M. J., Koulman, A., Monahan, B. J., Pritchard, B. L., Payne, G. A., and Scott, B. (2009). Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75, 7469–7481. doi: 10.1128/AEM.02146-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D.,et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nunes, C. C., and Dean, R. A. (2012). Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. doi: 10.1111/j.1364-3703.2011.00766.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oh, H., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1998). Arenarins A-C: new cytotoxic fungal metabolites from the sclerotia of Aspergillus arenarius. J. Nat. Prod. 61, 702–705. doi: 10.1021/np980001t
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmer, J. M., Theisen, J. M., Duran, R. M., Grayburn, W. S., Calvo, A. M., and Keller, N. P. (2013). Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet. 9:e1003193. doi: 10.1371/journal.pgen.1003193
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. doi: 10.1007/s11103-013-0022-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paoletti, M., Seymour, F. A., Alcocer, M. J., Kaur, N., Calvo, A. M., Archer, D. B.,et al. (2007). Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17, 1384–1389. doi: 10.1016/j.cub.2007.07.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, H. S., Nam, T. Y., Han, K. H., Kim, S. C., and Yu, J. H. (2014). VelC positively controls sexual development in Aspergillus nidulans. PLoS ONE 9:e89883. doi: 10.1371/journal.pone.0089883
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, H. S., Ni, M., Jeong, K. C., Kim, Y. H., and Yu, J. H. (2012). The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS ONE 7:e45935. doi: 10.1371/journal.pone.0045935
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parker, E. J., and Scott, D. B. (2004). “Indole-diterpene biosynthesis in ascomycetous fungi,” in Handbook of Industrial Mycology, ed. Z. An (New York, NY: Marcel Dekker), 405–426.
Paster, N., Lisker, N., and Chet, I. (1984). Accumulation of ochratoxin A in sclerotia of Aspergillus ochraceus. Can. J. Bot. 63, 661–662. doi: 10.1139/b85-083
Payne, G. P., and Brown, M. P. (1998). Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 36, 329–362. doi: 10.1146/annurev.phyto.36.1.329
Petersen, L. M., Hoeck, C., Frisvad, J. C., Gotfredsen, C. H., and Larsen, T. O. (2014). Dereplication guided discovery of secondary metabolites of mixed biosynthetic origin from Aspergillus aculeatus. Molecules 19, 10898–10921. doi: 10.3390/molecules190810898
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Purschwitz, J., Muller, S., and Fischer, R. (2009). Mapping the interaction sites of Aspergillus nidulans phytochrome FphA with the global regulator VeA and the White Collar protein LreB. Mol. Genet. Genomics 281, 35–42. doi: 10.1007/s00438-008-0390-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Purschwitz, J., Muller, S., Kastner, C., Schoser, M., Haas, H., Espeso, E. A.,et al. (2008). Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 18, 255–259. doi: 10.1016/j.cub.2008.01.061
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ramirez-Prado, J. H., Moore, G. G., Horn, B. W., and Carbone, I. (2008). Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 45, 1292–1299. doi: 10.1016/j.fgb.2008.06.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reverberi, M., Punelli, M., Smith, C. A., Zjalic, S., Scarpari, M., Scala, V.,et al. (2012). How peroxisomes affect aflatoxin biosynthesis in Aspergillus flavus. PLoS ONE 7:e48097. doi: 10.1371/journal.pone.0048097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reyes-Dominguez, Y., Bok, J. W., Berger, H., Shwab, E. K., Basheer, A., Gallmetzer, A.,et al. (2010). Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76, 1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rhoades, D. F. (1985). Offensive-defensive interactions between herbivores and plants: their relevance in herbivore population dynamics and ecological theory. Am. Nat. 125, 205–238. doi: 10.1086/284338
Riley, R. T., Goeger, D. E., Yoo, H., and Showker, J. L. (1992). Comparison of three tetramic acids and their ability to alter membrane function in cultured skeletal muscle cells and sarcoplasmic reticulum vesicles. Toxicol. Appl. Pharmacol. 114, 261–267. doi: 10.1016/0041-008X(92)90076-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rodriguez-Urra, A. B., Jimenez, C., Nieto, M. I., Rodriguez, J., Hayashi, H., and Ugalde, U. (2012). Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans. ACS Chem. Biol. 7, 599–606. doi: 10.1021/cb200455u
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohlfs, M., Albert, M., Keller, N. P., and Kempken, F. (2007). Secondary chemicals protect mould from fungivory. Biol. Lett. 3, 523–525. doi: 10.1098/rsbl.2007.0338
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohlfs, M., and Churchill, A. C. (2011). Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 48, 23–34. doi: 10.1016/j.fgb.2010.08.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roze, L. V., Hong, S. Y., and Linz, J. E. (2013). Aflatoxin biosynthesis: current frontiers. Annu. Rev. Food Sci. Technol. 4, 293–311. doi: 10.1146/annurev-food-083012-123702
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saikia, S., Parker, E. J., Koulman, A., and Scott, B. (2006). Four gene products are required for the fungal synthesis of the indole-diterpene, paspaline. FEBS Lett. 580, 1625–1630. doi: 10.1016/j.febslet.2006.02.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sanchez, J. F., Somoza, A. D., Keller, N. P., and Wang, C. C. (2012). Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 29, 351–371. doi: 10.1039/c2np00084a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sarikaya Bayram, O., Bayram, O., Feussner, K., Kim, J. H., Kim, H. S., Kaever, A.,et al. (2014). Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev. Cell 29, 406–420. doi: 10.1016/j.devcel.2014.03.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sarikaya Bayram, O., Bayram, O., Valerius, O., Park, H. S., Irniger, S., Gerke, J.,et al. (2010). LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6:e1001226. doi: 10.1371/journal.pgen.1001226
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scarpari, M., Punelli, M., Scala, V., Zaccaria, M., Nobili, C., Ludovici, M.,et al. (2014). Lipids in Aspergillus flavus-maize interaction. Front. Microbiol. 5:74. doi: 10.3389/fmicb.2014.00074
Schardl, C. L., Panaccione, D. G., and Tudzynski, P. (2006). Ergot alkaloids – biology and molecular biology. Alkaloids Chem. Biol. 63, 45–86. doi: 10.1016/S1099-4831(06)63002-2
Schroeckh, V., Scherlach, K., Nutzmann, H. W., Shelest, E., Schmidt-Heck, W., Schuemann, J.,et al. (2009). Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 106, 14558–14563. doi: 10.1073/pnas.0901870106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shinohara, Y., Tokuoka, M., and Koyama, Y. (2011). Functional analysis of the cyclopiazonic acid biosynthesis gene cluster in Aspergillus oryzae RIB 40. Biosci. Biotechnol. Biochem. 75, 2249–2252. doi: 10.1271/bbb.110467
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sobolev, V. S., Cole, R. J., Dorner, J. W., Horn, B. W., Harrigan, G. G., and Gloer, J. B. (1997). Isolation and structure elucidation of a new metabolite produced by Aspergillus parasiticus. J. Nat. Prod. 60, 847–850. doi: 10.1021/np970131m
Stinnett, S. M., Espeso, E. A., Cobeno, L., Araujo-Bazan, L., and Calvo, A. M. (2007). Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 63, 242–255. doi: 10.1111/j.1365-2958.2006.05506.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
TePaske, M. R., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1992). Aflavarin and beta-aflatrem: new anti-insectan metabolites from the sclerotia of Aspergillus flavus. J. Nat. Prod. 55, 1080–1086. doi: 10.1021/np50086a008
Tinoco, M. L., Dias, B. B., Dall’Astta, R. C., Pamphile, J. A., and Aragao, F. J. (2010). In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 8:27. doi: 10.1186/1741-7007-8-27
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsitsigiannis, D. I., Bok, J. W., Andes, D., Nielsen, K. F., Frisvad, J. C., and Keller, N. P. (2005). Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect. Immun. 73, 4548–4559. doi: 10.1128/IAI.73.8.4548-4559.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsitsigiannis, D. I., Zarnowski, R., and Keller, N. P. (2004). The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 279, 11344–11353. doi: 10.1074/jbc.M310840200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Umemura, M., Nagano, N., Koike, H., Kawano, J., Ishii, T., Miyamura, Y.,et al. (2014). Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol. 68, 23–30. doi: 10.1016/j.fgb.2014.04.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valdes, J. J., Cameron, J. E., and Cole, R. J. (1985). Aflatrem: a tremorgenic mycotoxin with acute neurotoxic effects. Environ. Health Perspect. 62, 459–463. doi: 10.1289/ehp.8562459
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vinokurova, N. G., Ivanushkina, N. E., Khmel’nitskaia, I. I., and Arinbasarov, M. U. (2007). Synthesis of alpha-cyclopiazonic acid by fungi of the genus Aspergillus. Prikl. Biokhim. Mikrobiol. 43, 486–489. doi: 10.1134/S0003683807040138
Wada, R., Maruyama, J., Yamaguchi, H., Yamamoto, N., Wagu, Y., Paoletti, M.,et al. (2012). Presence and functionality of mating type genes in the supposedly asexual filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 78, 2819–2829. doi: 10.1128/AEM.07034-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wallwey, C., and Li, S. M. (2011). Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat. Prod. Rep. 28, 496–510. doi: 10.1039/c0np00060d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wheeler, M. H. (1983). Comparisons of fungal melanin biosynthesis in ascomycetous, imperfect and basidiomycetous fungi. Mycol. Res. 81, 29–36. doi: 10.1016/S0007-1536(83)80200-9
Whyte, A. C., Joshi, B. K., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (2000). New cyclic peptide and bisindolyl benzenoid metabolites from the sclerotia of Aspergillus sclerotiorum. J. Nat. Prod. 63, 1006–1009. doi: 10.1021/np000103v
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wicklow, D. T. (1988). “Metabolites in the coevolution of fungal chemical defence systems,” in Coevolution of Fungi with Plants and Animals, eds K. A. Pirozynski and D. Hawksworth (New York, NY: Academic Press), 173–201.
Wicklow, D. T., and Cole, R. J. (1982). Tremoragenic indole metabolites and aflatoxins in sclerotia of Aspergillus flavus link: an evolutionary perspective. Can. J. Bot. 60, 525–528. doi: 10.1139/b82-070
Wicklow, D. T., Dowd, P. F., Alfatafta, A. A., and Gloer, J. B. (1996). Ochratoxin A: an antiinsectan metabolite from the sclerotia of Aspergillus carbonarius NRRL 369. Can. J. Microbiol. 42, 1100–1103. doi: 10.1139/m96-141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wicklow, D. T., and Shotwell, O. L. (1983). Intrafungal distribution of aflatoxins among conidia and sclerotia of Aspergillus flavus and Aspergillus parasiticus. Can. J. Microbiol. 29, 1–5. doi: 10.1139/m83-001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wieser, J., Lee, B. N., Fondon, J. III, and Adams, T. H. (1994). Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27, 62–69. doi: 10.1007/BF00326580
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Winter, J. M., Behnken, S., and Hertweck, C. (2011). Genomics-inspired discovery of natural products. Curr. Opin. Chem. Biol 15, 22–31. doi: 10.1016/j.cbpa.2010.10.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, F. (2004). Mycotoxin risk assessment for the purpose of setting international regulatory standards. Environ. Sci. Technol. 38, 4049–4055. doi: 10.1021/es035353n
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, X., Zhou, B., Yin, C., Guo, Y., Lin, Y., Pan, L.,et al. (2014). Characterization of natural antisense transcript, sclerotia development and secondary metabolism by strand-specific RNA sequencing of Aspergillus flavus. PLoS ONE 9:e97814. doi: 10.1371/journal.pone.0097814
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yager, L. N. (1992). “Early developmental events during asexual and sexual sporulation in Aspergillus nidulans,” in Aspergillus: Biology and Industrial Applications, eds J. W. Bennet and M. A. Klich (Boston, MA: Butterworth-Heinemann), 19–41.
Yager, L. N., Lee, H. O., Nagle, D. L., and Zimmerman, J. E. (1998). Analysis of fluG mutations that affect light-dependent conidiation in Aspergillus nidulans. Genetics 149, 1777–1786.
Yim, G., Wang, H. H., and Davies, J. R. (2007). Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1195–1200. doi: 10.1098/rstb.2007.2044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yin, W. B., Amaike, S., Wohlbach, D. J., Gasch, A. P., Chiang, Y. M., Wang, C. C.,et al. (2012). An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol. 83, 1024–1034. doi: 10.1111/j.1365-2958.2012.07986.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, J. (2012). Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 4, 1024–1057. doi: 10.3390/toxins4111024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, J., Chang, P. K., Ehrlich, K. C., Cary, J. W., Bhatnagar, D., Cleveland, T. E.,et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70, 1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004
Zeng, H., Hatabayashi, H., Nakagawa, H., Cai, J., Suzuki, R., Sakuno, E.,et al. (2011). Conversion of 11-hydroxy-O-methylsterigmatocystin to aflatoxin G1 in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 90, 635–650. doi: 10.1007/s00253-010-2999-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, J. X., and Cotty, P. J. (2006). Diversity within communities of L and S strains of Aspergillus flavus in cotton fields in Texas and Arizona. Phytopathol. 97, S172.
Zhang, S., Monahan, B. J., Tkacz, J. S., and Scott, B. (2004). Indole-diterpene gene cluster from Aspergillus flavus. Appl. Environ. Microbiol. 70, 6875–6883. doi: 10.1128/AEM.70.11.6875-6883.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: secondary metabolism, genetic regulation, sclerotia, VeA, velvet, Aspergillus, gene cluster, morphogenesis
Citation: Calvo AM and Cary JW (2015) Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 6:62. doi: 10.3389/fmicb.2015.00062
Received: 06 October 2014; Accepted: 18 January 2015;
Published online: 16 February 2015.
Edited by:
Ozgur Bayram, Maynooth University, IrelandReviewed by:
Geromy G. Moore, United States Department of Agriculture – Agricultural Research Service, USAJae-Hyuk Yu, University of Wisconsin, USA
Copyright © 2015 Calvo and Cary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana M. Calvo, Department of Biological Sciences, Northern Illinois University, 155 Castle Road, DeKalb, IL 60115, USA e-mail: amcalvo@niu.edu
 Ana M. Calvo
Ana M. Calvo Jeffrey W. Cary
Jeffrey W. Cary