- 1School of Public Health, University of Alberta, Edmonton, AB, Canada
- 2Department of Food and Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul, South Korea
Campylobacter jejuni is an important foodborne pathogen. The molecular mechanisms for the regulation of oxidative stress resistance have not yet been understood fully in this bacterium. In this study, we investigated how PerR (peroxide stress regulator) modulates the transcriptional regulation of both peroxide and superoxide resistance genes in C. jejuni, particularly under oxidative stress conditions. The transcriptional levels of ahpC, katA, and sodB were substantially increased by aeration and oxidant exposure. Interestingly, a perR mutation completely abrogated the transcriptional response of ahpC, katA and sodB to oxidants. Furthermore, we demonstrated that perR transcription was reduced by aeration and oxidant exposure. In contrast to the unique role of PerR homologs in peroxide stress regulation in other bacteria, C. jejuni PerR directly regulates the transcription of sodB, the most important gene in superoxide defense, as evidenced by the alteration of sodB transcription by the perR mutation and direct binding of rPerR to the sodB promoter. In addition, we also observed notable morphological changes in C. jejuni from spiral rods to cocoid morphology under aerobic conditions. Based on the intracellular ATP levels, C. jejuni entered a viable-but-non-culturable (VBNC) state under aerobic conditions. These findings clearly demonstrate that C. jejuni possesses a unique regulatory mechanism of oxidative stress defense that does not specifically distinguish between peroxide and superoxide defense, and PerR plays a pivotal role in this non-selective regulation of oxidative stress resistance in C. jejuni.
Introduction
Campylobacter jejuni is one of the leading bacterial causes of gastroenteritis worldwide (Young et al., 2007). C. jejuni infection also accounts for the majority (approximately one quarter) of disease cases of Guillain-Barré syndrome, an acute peripheral neuropathy, as a post-infection complication (Hughes and Cornblath, 2005). C. jejuni is a commensal bacterium in the intestines of poultry that do not develop any clinical symptoms (Moore et al., 2005); thus, human infections with C. jejuni are frequently caused by the consumption of contaminated poultry (Allos, 2001). C. jejuni requires low oxygen concentrations (3 ~ 15%) for growth and is sensitive to high oxygen tension under normal atmospheric conditions. C. jejuni favorably inhabits the gastrointestinal tracts in poultry that provide optimal growth temperatures (about 42°C), nutrients, and low oxygen levels. Once excreted from animals, however, C. jejuni encounters various harsh environmental stress, such as high oxygen tension and nutrient starvation. Particularly, increased oxidative stress in the atmosphere is a critical barrier that C. jejuni should overcome during its zoonotic transmission from animals (i.e., poultry) to humans via food. Therefore, a better understanding of the molecular mechanisms of oxidative stress is critical for elucidating the infection process of this microaerophilic foodborne pathogen.
C. jejuni has a unique oxidative stress defense system. For the detoxification of reactive oxygen species (ROS), for example, C. jejuni possesses only single copies of alkyl hydroperoxide reductase (ahpC), catalase (katA), and superoxide dismutase (sodB) (Atack and Kelly, 2009), whereas other bacteria often harbor redundant types of these antioxidant genes. Escherichia coli encodes three sod genes (sodA, sodB, and sodC) and two catalase genes (katE and katG) (Imlay, 2008). Alkyl hydroperoxide reductase in E. coli consists of two subunits, AhpC and AhpF (Poole et al., 2000), whereas ahpF homologs are absent from the C. jejuni genome (Baillon et al., 1999). C. jejuni also has unique regulatory mechanisms for oxidative stress resistance. In E.coli and Salmonella, OxyR and SoxRS regulate expression of peroxide and superoxide defense regulons, respectively (Imlay, 2008; Chiang and Schellhorn, 2012). However, C. jejuni lacks homologs of SoxRS and OxyR (Atack and Kelly, 2009). Instead, C. jejuni posseses PerR, a substitute for OxyR found in many Gram-positive bacteria and some Gram-negative bacteria; in fact, C. jejuni is the first Gram-negative bacterium that is known to harbor PerR (Van Vliet et al., 1999). In C. jejuni, PerR inactivation significantly increases the expression of peroxide resistance enzymes, such as AhpC and KatA, and renders C. jejuni hyper-resistant to H2O2 (Van Vliet et al., 1999).
PerR belongs to the ferric uptake regulator class of metal-responsive repressor proteins (Van Vliet et al., 1999; Lee and Helmann, 2007). PerR uses either manganese or iron as a regulatory metal-cofactor to detect oxidative stress in Bacillus subtilis (Lee and Helmann, 2006; Jacquamet et al., 2009), whereas iron, but not manganese, affects perR transcription by PerR autoregulation in C. jejuni (Kim et al., 2011). Although PerR is the key regulator of oxidative stress defense in C. jejuni, the regulatory mechanism of PerR has not been fully defined in this microaerophilic foodborne pathogen, particularly under oxidative stress (e.g., aerobic conditions). In this study, we demonstrated that PerR controls the transcription of sodB, the most important gene in superoxide resistance, and plays a critical role in non-selective regulation of peroxide and superoxide resistance genes in C. jejuni.
Materials and Methods
Bacterial Strains and Growth Conditions
C. jejuni NCTC 11168 and its derivatives were grown at 42°C on Mueller-Hinton media (Difco) under microaerobic conditions (5% O2, 10% CO2, 85% N2). Kanamycin (50 μg/ml) was occasionally added to the culture media where required. Broth cultures were grown by shaking at 200 rpm aerobically or microaerobically.
Quantitation of Intracellular ATP Levels
The intracellular ATP levels were measured by using the ATP Bioluminescent Assay Kit (Sigma) according to the manufacturer's instructions. Briefly, after washing with PBS (pH 7.4) twice, the C. jejuni suspension was mixed with the same volume of the ATP assay solution in a 96-well plate. After incubation at room temperature for 3 min, the luminescence was measured with FLUOstar Omega (BMG Labtech). The total protein concentrations of the sample were measured by using the Bradford assay (Bradford, 1976) to normalize the ATP levels to the protein amounts. The experiments were repeated three times, and each set was run in triplicate.
Fluorescence Microscopic Analysis
C. jejuni was harvested after 4 h, 8 h, and 24 h culture under aerobic or microaerobic conditions and was stained with the LIVE/DEAD BacLight™ bacteria viability kit (Life Technologies). Images were visualized with a fluorescence microscope (Carl Zeiss M1) and were analyzed with Axio Vision LE (Zeiss).
Construction of lacZ Transcriptional Fusion and β-Galactosidase Assay
The promoters and partial coding regions of ahpC, katA, and sodB were PCR-amplified with the primer pairs of ahpC_PF_F(XbaI) and ahpC_PF_R(XbaI), katA_PF_F(XbaI) and katA_PF_R(XbaI), and sodB_PF_F(XbaI) and sodB_PF_R(XbaI), respectively (Table 1). The PCR products were cloned into an XbaI site of pMW10 (Wösten et al., 1998) that contains a promoterless lacZ gene. Each fusion plasmid was mobilized to C. jejuni NCTC 11168 by conjugation. C. jejuni strains harboring a PperR-lacZ plasmid and a perR complementation strain were described in our previous study (Kim et al., 2011). Occasionally, the assays were performed with Minimum Essential Medium alpha (MEMα) (Life Technologies), which does not contain iron, in the presence or absence of 40 μM FeSO4 (Sigma). To assess the effect of oxidative stress on the promoter activities of ahpC, katA, and sodB, β-galactosidase assays were carried out with oxidants, such as H2O2, cumene hydroperoxide (CHP, an organic peroxide), and menadione (MND, a superoxide generator). Briefly, C. jejuni in the exponential phase was harvested and resuspended in fresh MH broth to a cell concentration of 109 CFU/ml, and was exposed to oxidants for 2 h prior to the assay.
Purification of Recombinant PerR Protein (rPerR)
Purification of rPerR was performed as described in our pervious study (Kim et al., 2011). Briefly, an E. coli strain carrying pET15b::perR was grown in Luria-Bertani (LB) broth to an optical density at 600 nm of 1.0 at 37°C, and 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) was then added. After 5 h induction at 37°C, cells were harvested and resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 300 mM NaCl). Bacterial cells were lysed by sonication and purified under native conditions using Ni2+ affinity chromatography (Qiagen). The His-tag was removed from rPerR with the Thrombin Cleavage Capture Kit (Novagen) according to the manufacturer's instructions.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as described previously with some modifications (Jeon and Zhang, 2007). The Cy3-labeled DNA probes were PCR-generated with the primers of SB-P and SB-R (Table 1). The DNA probe was incubated with rPerR at different concentrations at 37°C for 15 min in 10 μl of gel-shift assay buffer (20 mM HEPES [pH 7.6], 1 mM EDTA, 10 mM (NH4)2SO4, 5 mM DTT, 0.2% Tween 20, 30 mM KCl, 0.1 μg poly dI-dC). The reaction mixtures were resolved in a 6% polyacrylamide gel, and the Cy3-labeled DNA fragments were visualized with FluorChem™-R (ProteinSimple, USA).
Results
Differential Induction of ahpC, katA, and sodB Transcription Under Aerobic Conditions
AhpC, SodB, and KatA are the sole alkyl hydroperoxide reductase, superoxide dismutase (SOD), and catalase in C. jejuni, respectively, significantly contributing to oxidative stress defense in C. jejuni (Atack and Kelly, 2009). Although C. jejuni is a microaerophilic bacterium, it harbors a certain level of aerotolerance (Gundogdu et al., 2011). Due to the importance of ahpC, katA, and sodB in ROS detoxification, we hypothesized that C. jejuni would differentially regulate ahpC, katA, and sodB expression to scavenge toxic ROS under aerobic conditions. To measure the transcriptional levels of ahpC, katA, and sodB, we constructed transcriptional fusions of the ahpC, katA, and sodB promoters to a promoterless lacZ in pMW10 (Wösten et al., 1998). Although C. jejuni growth was significantly decreased under aerobic conditions (Figure 1A), interestingly, the transcriptional levels of ahpC and sodB were increased under aerobic conditions (Figure 1B). After 24 h culture under aerobic conditions, the promoter activities of ahpC and sodB were increased by approximately 62% and 89%, respectively, compared to microaerobic conditions (Figure 1B).
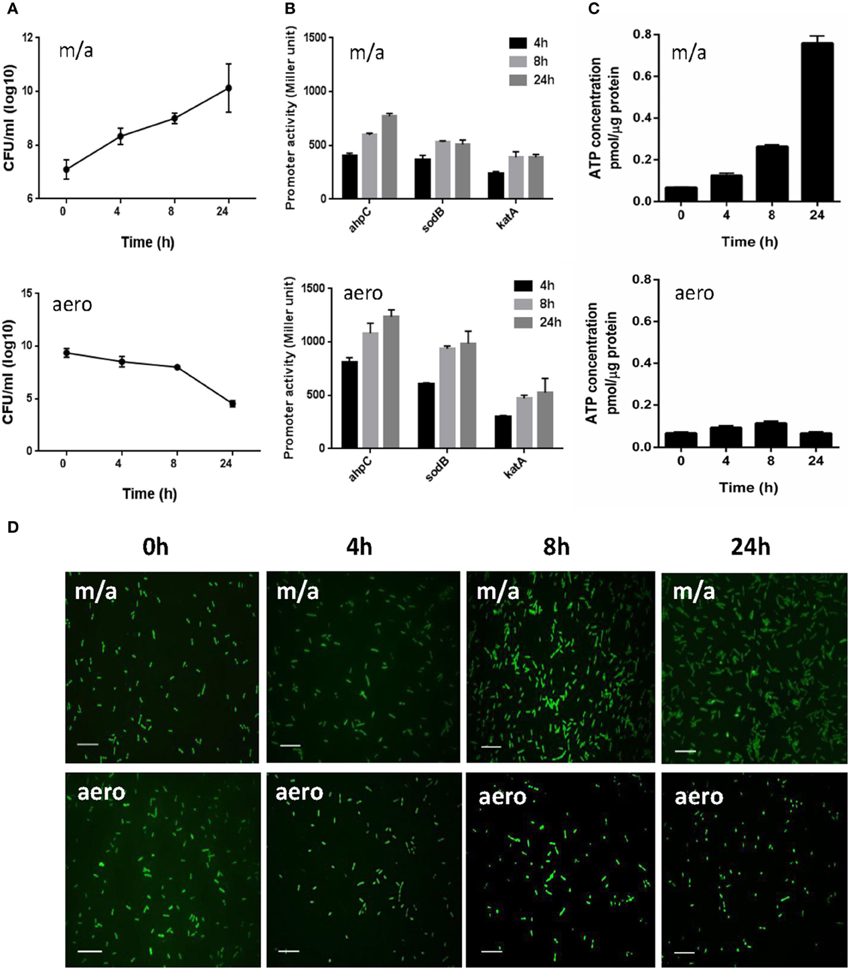
Figure 1. Bacterial growth, transcriptional levels of ROS-detoxification genes, intracellular ATP levels, and morphological changes in C. jejuni under microaerobic and aerobic conditions. (A) The growth kinetics of C. jejuni under microaerobic and aerobic culture conditions. The means and standard deviations of triplicate samples in a single experiment are indicated. The experiments were repeated three times. (B) Transcriptional levels of ahpC, katA, and sodB genes under microaerobic and aerobic culture conditions. C. jejuni strains harboring PahpC-lacZ, PkatA-lacZ, and PsodB-lacZ were grown in MH medium microaerobically or aerobically. The results are representative of at least three independent experiments. Each experiment was performed with triplicate samples and all the repeated experiments exhibited similar results. (C) Intracellular ATP levels in C. jejuni under microaerobic and aerobic conditions. The ATP concentrations were normalized to the total protein concentrations. The experiment was performed in triplicate and was repeated three times. Error bars indicate the standard deviations. (D) Fluorescent microscopic images of C. jejuni with LIVE/DEAD BacLight™ staining under microaerobic and aerobic conditions. The scale bar shows 10 μm. “m/a” and “aero” indicate microaerobic and aerobic conditions, respectively.
Despite a significant growth defect under aerobic conditions, C. jejuni exhibited a substantial increase in the transcriptional levels of the antioxidant genes. The intracellular ATP levels were measured to examine the physiological activity of C. jejuni under aerobic conditions. The ATP concentrations were approximately 16-fold higher under microaerobic conditions than aerobic cultures after 24 h (Figure 1C), indicating that C. jejuni is not physiologically active under aerobic conditions. Microscopic observation with the LIVE/DEAD staining revealed that most viable C. jejuni cells were in coccoid forms under aerobic conditions (Figure 1D). Based on the morphology and ATP levels, C. jejuni is likely to enter a viable-but-non-culturable (VBNC) state under aerobic conditions.
Abrogation of Oxidant-Mediated Induction of ahpC, katA, and sodB by a perR Mutation
Since the transcriptional levels of ahpC, katA, and sodB were increased under aerobic conditions (Figure 1C) and the common biological role of the antioxidant genes is ROS detoxification, we hypothesized the augmented transcriptional levels in ahpC, katA, and sodB under aerobic conditions would result from increased oxidative stress in C. jejuni. Thus, we investigated whether oxidants would increase the promoter activity of the ROS-detoxification genes. The promoter activities of ahpC, katA, and sodB were monitored in the presence of oxidizing agents, such as H2O2, CHP (an organic peroxide), and MND (a superoxide anion generator). Regardless of the oxidant type, whether peroxide or superoxide, the transcription of ahpC, katA, and sodB was induced by oxidant exposure (Figure 2). The transcriptional induction by oxidants occurred in a concentration-dependent manner, and antioxidant treatment removed the oxidant-mediated induction of ahpC, katA, and sodB transcription (Supplementary Figure 1).
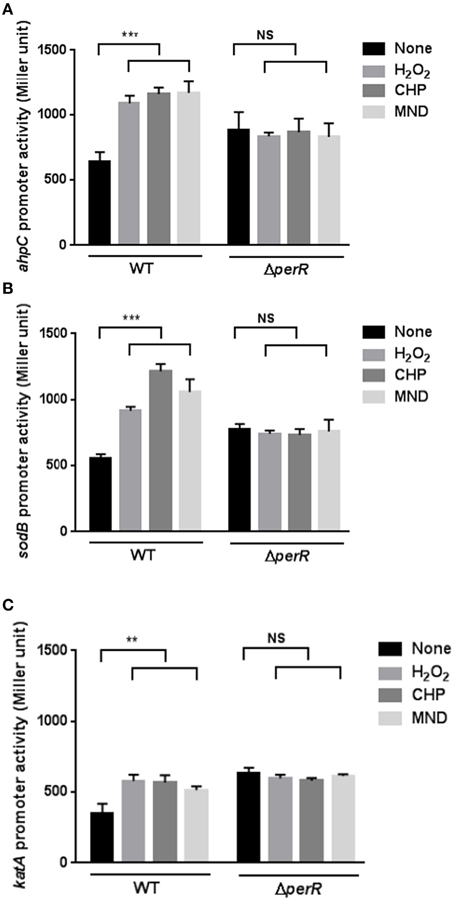
Figure 2. Effects of a perR mutation on the transcription of ahpC (A), sodB (B), and katA (C) under oxidative stress. β-Galactosidase assays of ahpC, sodB, and katA expression after exposure to 1 mM hydrogen peroxide (H2O2), 100 μM cumene hydroperoxide (CHP), and 100 μM menadione (MND). The promoter fusion assays were performed in both wild type and a perR mutant. After 8 h culture under microaerobic conditions, C. jejuni was exposed to oxidants for 2 h prior to an assay. The values represent the means and standard deviations of three independent experiments. Statistical analysis was performed with the Student's t-test using GraphPad Prism 6 (GraphPad Software Inc., USA). **, P < 0.01; ***, P < 0.001; NS, not significant.
Since PerR is a major regulator of oxidative stress in C. jejuni (Van Vliet et al., 1999), we investigated if PerR could be associated with the transcriptional changes in the mutants under oxidative stress. Interestingly, the transcriptional induction of ahpC, katA, and sodB by oxidants completely disappeared in the perR mutant (Figure 2), indicating that PerR is associated with the transcriptional changes in ahpC, katA, and sodB under oxidative stress. In addition, the perR mutation increased the transcriptional levels of ahpC, katA, and sodB (Figure 2), suggesting that the perR mutation de-repressed the antioxidant genes, even including sodB. These results strongly suggest that PerR plays a role in the transcriptional control of both peroxide- and superoxide- detoxification genes in C. jejuni under oxidative stress conditions.
PerR Regulation of sodB Transcription
Since a perR mutation affected sodB transcription (Figure 2B), we further investigated if PerR regulates sodB transcription by performing the PsodB-lacZ promoter fusion assay. A perR mutation de-repressed sodB transcription, regardless of the presence or absence of iron, and perR complementation restored sodB transcription to the wild-type level (Figure 3A). The increase in sodB transcription by the perR mutation was more significant in MEMα than MH broth (data not shown). To examine if PerR regulates sodB by binding to the sodB promoter, a gel-shift assay was performed with rPerR. Interestingly, rPerR directly bound to the sodB promoter (Figure 3B). A putative PerR binding site was predicted in the sodB promoter region based on the conserved PerR binding site in C. jejuni (Kim et al., 2011; Figure 3C). Taken together, these data suggest that PerR regulates sodB transcription by binding to the sodB promoter.
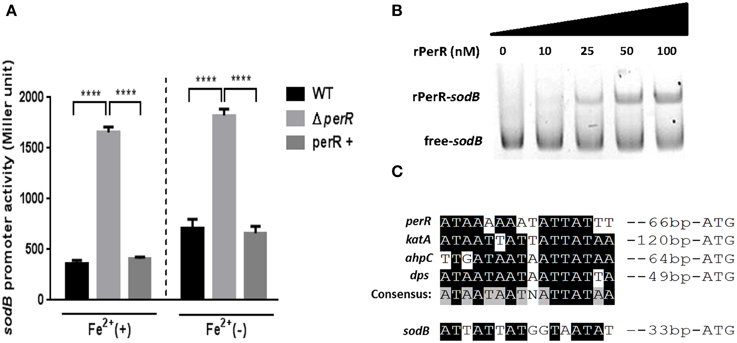
Figure 3. PerR regulation of sodB in C. jejuni. (A) De-repression of sodB by a perR mutation. A perR mutation de-repressed sodB transcription, and the level of sodB transcription was completely restored by complementation. The sodB promoter activity was determined in MEMα with or without 40μM FeSO4. The results show the standard deviations of three different experiments. Statistical analysis was performed with a Student's t-test using GraphPad Prism 6. ****P < 0.0001; perR+, a perR complementation strain. (B) rPerR binding to the sodB promoter. Cy3-labeled PCR probes of a sodB promoter region were incubated with rPerR at different concentrations that are indicated above the gel. The assay was conducted in a 6% polyacrylamide gel under native conditions. (C) A putative PerR-binding site in the sodB promoter. The binding site was predicted based on the consensus PerR-binding sequence in C. jejuni (Kim et al., 2011).
Alteration of perR Transcription by Oxidants and Aeration
Since the perR mutation completely removed the oxidant-mediated induction of ahpC, sodB, and katA (Figure 2), we investigated if perR transcription is affected by oxidants. The PperR-lacZ fusion assay was conducted with MEMα, because iron reduces perR transcription in C. jejuni (Kim et al., 2011). Oxidative stress, including oxidants (i.e., H2O2, organic peroxide, and superoxide) and aeration, substantially decreased the levels of perR transcription (Figure 4). The reduction of perR transcription by oxidative stress was not influenced by iron, although iron decreased the levels of perR transcription (Figure 4). The reduction of perR transcription by aeration would result from the increased ROS accumulation under aerobic conditions. The results clearly showed that perR transcription is affected by oxidative stress in C. jejuni.
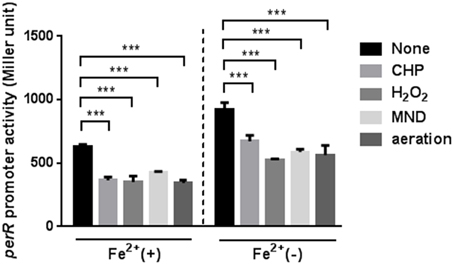
Figure 4. Down-regulation of perR transcription under oxidative stress. The perR promoter activity was evaluated after 2 h exposure to H2O2, CHP, MND, and aeration in MEMα with or without 40 μM FeSO4. The assays were repeated three times. Statistical analysis was performed with a Student's t-test. ***, P < 0.001.
Discussion
PerR is a primary regulator of peroxide stress defense in many bacteria. In order for PerR to control the expression of peroxide stress defense genes, bacteria should control the intracellular level of PerR in response to peroxide stress. However, previous observations in other bacteria have shown that perR transcription is not affected by H2O2. For example, a transcriptomic analysis in Staphylococcus aureus demonstrated that the transcription of perR and its regulatory genes is not altered by H2O2 exposure (Chang et al., 2006). In B. subtilis, perR transcription is not changed by H2O2 (Fuangthong et al., 2002); instead, PerR regulates peroxide resistance genes by protein conformational changes under peroxide stress (Herbig and Helmann, 2001). B. subtilis PerR senses H2O2 by oxidation of one of two histidine residues (i.e., H37 and H91) that coordinate the bound Fe2+; this modification results in the dissociation of Fe2+ from PerR. The demetallated PerR cannot bind to DNA, and this conformational changes in PerR induces gene expression (Lee and Helmann, 2006). Interestingly, in this study, we demonstrated that oxidative stress, such as oxidant exposure and aerobiosis, directly reduced the levels of perR transcription (Figure 4). To the extent of our knowledge, C. jejuni perR is the only perR homolog whose transcription is known to be affected by oxidative stress. Although PerR is considered as a peroxide resistance regulator, perR transcription was altered by exposure to both peroxides (H2O2 and CHP) and superoxide (MND; Figure 4). Since PerR represses ahpC and katA (Van Vliet et al., 1999) and sodB (Figure 3), down-regulation of perR transcription under oxidative stress will de-repress ahpC, katA, and sodB. Consistently, we observed that the transcriptional levels of ahpC, katA, and sodB were increased by aeration and oxidant exposure (Figures 1, 2), and a perR mutation abrogated the transcriptional response of ahpC, katA, and sodB to oxidants (Figure 2), suggesting that PerR is a key player in the induction of both peroxide- and superoxide-detoxification genes under oxidative stress.
In this study, we demonstrated that PerR negatively regulates sodB expression (Figure 3). Based on the promoter sequence of sodB in C. jejuni (Pesci et al., 1994), a predicted PerR-binding site overlaps with the -10 region of the sodB promoter (data not shown); thus, PerR binding would interfere with sodB transcription. PerR is known as a regulator of peroxide resistance. However, our findings revealed that PerR also regulates sodB transcription in C. jejuni. To the best of our knowledge, this report is first describing direct regulation of sodB transcription by PerR. As a key enzyme of superoxide resistance, SOD catalyzes the dismutation of superoxide to H2O2. Due to its physiological importance in oxidative stress defense, bacteria often harbor redundant types of SOD. For example, E. coli contains three types of SOD, including two cytoplasmic SOD isoenzymes [SodA (Mn-cofactored SOD) and SodB (Fe-cofactored SOD)] and a periplasmic SOD [SodC (Cu/Zn-cofactored SOD)](Imlay, 2008). However, SodB is the sole SOD present in Campylobacter (Pesci et al., 1994; Purdy and Park, 1994), significantly contributing to Campylobacter's stress resistance and colonization of chicken intestines (Pesci et al., 1994; Palyada et al., 2009). Various mechanisms for the regulation of sod genes have been reported in different bacteria. In E. coli and S. enterica, exposure to superoxide-generating agents increases SodA expression through positive regulation by SoxRS (Greenberg et al., 1990; Pomposiello et al., 2001); however, C. jejuni lacks the SoxRS system (Parkhill et al., 2000). In E. coli, Fur indirectly regulates sodB expression by RhyB small RNA (Masse and Gottesman, 2002), while apo-Fur represses sodB by directly binding to the sodB promoter in Helicobacter pylori (Ernst et al., 2005); however, sodB is not found in the Fur regulon of C. jejuni (Butcher et al., 2012). In C. jejuni, the sodB regulation relies on the two-component regulatory systems, such as CosR and CprS (Svensson et al., 2009; Hwang et al., 2011). In this study, we have demonstrated another novel mechanism of sodB regulation by PerR in C. jejuni.
Since C. jejuni is a microaerophilic bacterium, prolonged exposure to aerobic conditions substantially reduced the growth of C. jejuni; aerobic growth for 24 h resulted in an approximate 5-log reduction in colony forming units (Figure 1A). However, the transcriptional levels of ahpC, katA, and sodB were higher under aerobic conditions than microaerobic conditions (Figure 1B). C. jejuni exhibited aerotolerance but was not physiologically active based on the intracellular ATP levels (Figure 1C). In addition, the morphology of C. jejuni changed from spiral rods to coccoid forms under aerobic conditions (Figure 1D). A previous study demonstrated that C. jejuni is able to survive under prolonged exposure to aerobic stress by forming VBNC cells with a typical morphological change to coccoid forms (Rollins and Colwell, 1986). Harvey and Leach reported the formation of coccoid forms of C. jejuni is enhanced by high oxygen tension, presumably as a result of oxidative damage, and coccoid forms of C. jejuni may regain normal spiral morphology following adaptation to oxidative stress (Harvey and Leach, 1998). In response to various harsh stress conditions, many bacterial species are known to enter a VBNC state with significant dwarfing in size (Oliver, 2010). Although a few stress conditions have been reported to induce a VBNC state in Campylobacter, including nutrient starvation (e.g., incubation in water), cold stress (e.g., 4°C), and organic acids (e.g., formic acid) (Rollins and Colwell, 1986; Harvey and Leach, 1998; Kassem et al., 2013), mechanisms for the VBNC formation remain largely unknown. Based on the findings in this study, C. jejuni enters a VBNC state under aerobic conditions. Compared with microaerobic conditions, interestingly, the transcriptional levels of ahpC, katA, and sodB were higher under aerobic conditions, in which the majority of C. jejuni cells were in coccoid forms (Figures 1B,D). At this stage, it is not clear whether the up-regulation of ahpC, katA, and sodB occurred in viable-and-culturalble or VBNC cells or both. However, increased expression of the antioxidant genes will alleviate oxidative stress under aerobic conditions and may contribute to the protection of C. jejuni from increased oxidative stress during the physiological transition of C. jejuni to a VBNC state under aerobic conditions.
In this study, we presented a unique regulatory mechanism of oxidative stress defense in C. jejuni that non-selectively regulates superoxide and peroxide stress via PerR. Such an integrative regulatory system of PerR will help C. jejuni to survive in oxygen-rich conditions during transmission from food to humans by allowing for effective coordination of the expression of relatively few ROS-detoxification enzymes in this microaerophilic foodborne pathogen. Future studies will include the investigation of the role of the unique stress response mechanism in the survival of C. jejuni under stress (i.e., oxygen-rich) conditions, and the interplay between PerR and other oxidative stress response regulators in C. jejuni.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), the Alberta Livestock and Meat Agency (ALMA), and the Canada Foundation for Innovation (CFI).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00126/abstract
References
Allos, B. M. (2001). Campylobacter jejuni Infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206. doi: 10.1086/319760
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Atack, J. M., and Kelly, D. J. (2009). Oxidative stress in Campylobacter jejuni: responses, resistance and regulation. Future Microbiol. 4, 677–690. doi: 10.2217/fmb.09.44
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baillon, M. L., Van Vliet, A. H., Ketley, J. M., Constantinidou, C., and Penn, C. W. (1999). An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181, 4798–4804.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Butcher, J., Sarvan, S., Brunzelle, J. S., Couture, J. F., and Stintzi, A. (2012). Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 10047–10052. doi: 10.1073/pnas.1118321109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, W., Small, D. A., Toghrol, F., and Bentley, W. E. (2006). Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188, 1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, S. M., and Schellhorn, H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169. doi: 10.1016/j.abb.2012.02.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ernst, F. D., Homuth, G., Stoof, J., Mader, U., Waidner, B., Kuipers, E. J., et al. (2005). Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J. Bacteriol. 187, 3687–3692. doi: 10.1128/JB.187.11.3687-3692.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fuangthong, M., Herbig, A. F., Bsat, N., and Helmann, J. D. (2002). Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184, 3276–3286. doi: 10.1128/JB.184.12.3276-3286.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Greenberg, J. T., Monach, P., Chou, J. H., Josephy, P. D., and Demple, B. (1990). Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 6181–6185. doi: 10.1073/pnas.87.16.6181
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gundogdu, O., Mills, D. C., Elmi, A., Martin, M. J., Wren, B. W., and Dorrell, N. (2011). The Campylobacter jejuni transcriptional regulator Cj1556 plays a role in the oxidative and aerobic stress response and is important for bacterial survival in vivo. J. Bacteriol. 193, 4238–4249. doi: 10.1128/JB.05189-11
Harvey, P., and Leach, S. (1998). Analysis of coccal cell formation by Campylobacter jejuni using continuous culture techniques, and the importance of oxidative stress. J. Appl. Microbiol. 85, 398–404. doi: 10.1046/j.1365-2672.1998.00532.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herbig, A. F., and Helmann, J. D. (2001). Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41, 849–859. doi: 10.1046/j.1365-2958.2001.02543.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hughes, R. A., and Cornblath, D. R. (2005). Guillain-Barre syndrome. Lancet 366, 1653–1666. doi: 10.1016/S0140-6736(05)67665-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hwang, S., Kim, M., Ryu, S., and Jeon, B. (2011). Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS ONE 6:e22300. doi: 10.1371/journal.pone.0022300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776. doi: 10.1146/annurev.biochem.77.061606.161055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacquamet, L., Traore, D. A., Ferrer, J. L., Proux, O., Testemale, D., Hazemann, J. L., et al. (2009). Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73, 20–31. doi: 10.1111/j.1365-2958.2009.06753.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jeon, B., and Zhang, Q. (2007). Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol. 189, 7399–7407. doi: 10.1128/JB.01012-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kassem, I. I, Chandrashekhar, K., and Rajashekara, G. (2013). Of energy and survival incognito: a relationship between viable but non-culturable cells formation and inorganic polyphosphate and formate metabolism in Campylobacter jejuni. Front. Microbiol. 4:183. doi: 10.3389/fmicb.2013.00183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, M., Hwang, S., Ryu, S., and Jeon, B. (2011). Regulation of perR expression by iron and PerR in Campylobacter jejuni. J. Bacteriol. 193, 6171–6178. doi: 10.1128/JB.05493-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, J. W., and Helmann, J. D. (2006). The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367. doi: 10.1038/nature04537
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Masse, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, J. E., Corcoran, D., Dooley, J. S., Fanning, S., Lucey, B., Matsuda, M., et al. (2005). Campylobacter. Vet. Res. 36, 351–382. doi: 10.1051/vetres:2005012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oliver, J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34, 415–425. doi: 10.1111/j.1574-6976.2009.00200.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palyada, K., Sun, Y. Q., Flint, A., Butcher, J., Naikare, H., and Stintzi, A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. doi: 10.1186/1471-2164-10-481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pesci, E. C., Cottle, D. L., and Pickett, C. L. (1994). Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62, 2687–2694.
Pomposiello, P. J., Bennik, M. H., and Demple, B. (2001). Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183, 3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Poole, L. B., Reynolds, C. M., Wood, Z. A., Karplus, P. A., Ellis, H. R., and Li Calzi, M. (2000). AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267, 6126–6133. doi: 10.1046/j.1432-1327.2000.01704.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Purdy, D., and Park, S. F. (1994). Cloning, nucleotide sequence and characterization of a gene encoding superoxide dismutase from Campylobacter jejuni and Campylobacter coli. Microbiology 140, 1203–1208. doi: 10.1099/13500872-140-5-1203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rollins, D. M., and Colwell, R. R. (1986). Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52, 531–538.
Svensson, S. L., Davis, L. M., Mackichan, J. K., Allan, B. J., Pajaniappan, M., Thompson, S. A., et al. (2009). The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol. Microbiol. 71, 253–272. doi: 10.1111/j.1365-2958.2008.06534.x
Van Vliet, A. H., Baillon, M. L., Penn, C. W., and Ketley, J. M. (1999). Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181, 6371–6376.
Wösten, M. M., Boeve, M., Koot, M. G., Van Nuenen, A. C., and Van Der Zeijst, B. A. (1998). Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180, 594–599.
Young, K. T., Davis, L. M., and Dirita, V. J. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–679. doi: 10.1038/nrmicro1718
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Campylobacter, oxidative stress, PerR
Citation: Kim J-C, Oh E, Hwang S, Ryu S and Jeon B (2015) Non-selective regulation of peroxide and superoxide resistance genes by PerR in Campylobacter jejuni. Front. Microbiol. 6:126. doi: 10.3389/fmicb.2015.00126
Received: 15 January 2015; Accepted: 03 February 2015;
Published online: 17 February 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Lefu Lan, Chinese Academy of Sciences, ChinaCatherine M. Logue, Iowa State University, USA
Copyright © 2015 Kim, Oh, Hwang, Ryu and Jeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byeonghwa Jeon, School of Public Health, University of Alberta, 3-57 South Academic Building, Edmonton, AB T6G 2G7, Canada e-mail: bjeon@ualberta.ca
†Present address: Sunyoung Hwang, Nutrition Safety Policy Division, Food Nutrition and Dietary Safety Bureau, Ministry of Food and Drug Safety, Cheongwon-gun, Chungcheongbuk-do, South Korea
 Jong-Chul Kim
Jong-Chul Kim Euna Oh
Euna Oh Sunyoung Hwang2†
Sunyoung Hwang2† Byeonghwa Jeon
Byeonghwa Jeon