- 1School of Biotechnology, Banaras Hindu University, Varanasi, India
- 2Botany Section, Mahila Maha Vidyalaya, Banaras Hindu University, Varanasi, India
For examining how UV-B radiation alters the proteome of the N2-fixing cyanobacterium, Anabaena L31, we extracted proteins from cultures irradiated with UV-B + white light and controls (white light irradiated) and analyzed the proteins using two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). Twenty one proteins, including two hypothetical proteins (HPs) were identified and placed in eight functional categories. However several of the proteins were housekeeping proteins involved in key metabolic processes such as carbon, amino acid biosynthesis and energy metabolism, certain proteins seem to have a role in stress (antioxidative enzymes), translation, cellular processes and reductases. Two novel HPs (all3797 and all4050) were characterized in detail. These two were over-expressed after UV-B irradiation and characterized as FAS 1 (all3797) and PRC barrel-like (all4050) proteins. Bioinformatics analysis revealed that the genes of both the HPs have promoter regions as well as transcription binding sites in their upstream region (UTR). Promoters present in all3797 genes suggest their crucial role against UV-B and certain other abiotic stresses. To our knowledge these novel proteins have not been previously reported in any Anabaena strains subjected to UV-B stress. Although we have focused our study on a limited number of proteins, results obtained shed light on the highly complicated but poorly studied aspect of UV-B radiation-mediated changes in the proteome and expression of proteins in cyanobacteria.
Introduction
While sunlight provides the energy for life, the ultraviolet-B (UV-B; 280–315 nm) and long-wavelength UV light (UV-A; 315–400 nm) components of the solar spectrum are harmful to many organisms. The rise of solar UV-B radiation reaching the earth’s surface due to the depletion of the stratospheric ozone layer caused by the anthropogenic inputs of chlorinated fluorocarbons has become an important issue over the past two decades (Crutzen, 1992; Kerr and McElroy, 1993). UV-B radiation can damage key biological molecules such as proteins, DNA and lipids (Häder et al., 2007; Castenholz and Garcia-Pichel, 2012). Cyanobacteria are photosynthetic prokaryotes with an evolutionary history that precedes the development of atmospheric ozone protection. They grow in a wide range of diverse and extreme habitats such as hot springs, arctic regions, desert soils, and rocky surfaces (Whitton and Potts, 2000). They show considerable metabolic plasticity and, besides assimilating CO2 and O2, certain species also assimilate N2 and H2 (Stewart, 1980). Studies conducted so far suggest that the deleterious effects of UV-B radiation in cyanobacteria are in part due to the direct effects on membrane components, growth and survival, motility, pigmentation, uptake of CO2, photosynthetic apparatus particularly the reaction center of photosystem II, RuBisCO, N2 fixation, DNA, proteins, and enzymes (Wilson et al., 1995; Kumar et al., 2003, 2004; Singh et al., 2010, 2013).
The molecular mechanisms of various physiological responses in cyanobacteria under UV-B radiation stress are poorly explored (Häder et al., 2007; Singh et al., 2010). In general, studies mainly focused on DNA repair and D1 reaction center protein of photosystem II (Vass et al., 2005; Castenholz and Garcia-Pichel, 2012). SDS-PAGE analysis of whole cell protein of certain cyanobacteria under the stress of UV-B radiation showed negligible change in the protein pattern (Chauhan et al., 1998). However, studies on the global gene expression profile under UV-B stress revealed significant alteration within 2 h of exposure (Huang et al., 2002). Transcriptome analysis pointed to changes of several antioxidative genes but the mRNA abundance did not match with the protein level (Gygi et al., 1999). While these reports do shed light on molecular response, there is a general consensus that the study of differential gene expression is only an indirect approach to understanding the molecular mechanisms of stress response including UV-B radiation (Vass et al., 2005; Fulda et al., 2006; Kurian et al., 2006). During the last two decades, proteome analysis employing 2-dimensional gel electrophoresis (2-DE) has become a powerful tool for visualizing many proteins synthesized in the cell and paved the way for understanding global changes in the gene expression (Godon et al., 1998; Hecker and Volker, 2004; Lei et al., 2005; Kumar et al., 2014). A few workers have studied proteomic changes following UV-B irradiation to cyanobacteria and reported significant alterations in the number and expression of proteins (Ehling-Schulz et al., 2002; Gao et al., 2009; Rai et al., 2013). Induction of early shock proteins and late acclimation proteins following UV-B exposure to Nostoc commune has been reported (Ehling-Schulz et al., 2002). Similarly, short-term and long-term exposure of UV-B to Synechocystis sp. PCC 6803 showed two groups of proteins, namely short-term and long-term responsive proteins on the basis of their expression (Gao et al., 2009). Till date the best characterized cyanobacterial response to UV-B stress is the cyanobacterium Synechocystis sp. PCC 6803, and only sporadic reports are available for the N2-fixing species from tropical rice paddy fields. It is also pertinent to mention that in most previous studies, the response of cyanobacteria was tested by exposing cultures to UV-B radiation alone, little, if any, attempt has been made to examine such effects in combination with white light.
Prompted by the above lacuna, we examined the effects of UV-B radiation in combination with white (visible) light in the N2-fixing cyanobacterium Anabaena strain L31. We aimed to (a) examine the changes in proteome under UV-B + white light stress, (b) identify and characterize the novel proteins, and (c) assess the possible role of identified proteins. This study may provide new insights for understanding how Anabaena L31 cells can induce certain novel proteins and adapt to UV-B stress via expression of UV-B responsive genes.
Materials and Methods
Test Organism and Growth Conditions
The test organism, a filamentous heterocystous cyanobacterium, Anabaena strain L31 was obtained from Dr. S. K. Apte, Bhabha Atomic Research Centre, Trombay, Mumbai, India. Axenic cultures were routinely grown diazotrophically in BG 11 medium (Rippka et al., 1979) in a culture room at 27 ± 2°C and illuminated with white light from Sylvania 40 W T12 cool fluorescent tubes at an intensity of 14.4 W m-2 for a 14/10 h light/dark cycle. Unless otherwise stated, all the experiments were performed with exponentially grown cultures having an initial dry weight of 0.1 mg mL-1.
Source and Mode of UV Radiation
The source of artificial UV-B irradiation was a UV-B lamp (Cat No. 3-4408, Fotodyne Inc., USA) giving its main output at 312.67 nm. The desired intensity of UV-B reaching the test samples was obtained by adjusting the distance between the UV-B lamp and the test samples. UV-B intensity was measured by Black-Ray J-221, Long Wave Ultraviolet Intensity Meter (UVP Inc., San Gabriel, CA, USA). Percent survival following exposure of cultures to UV-B + white light or UV-B alone was determined as described previously (Singh et al., 2013). For proteome analysis, 200 mL of homogeneous culture suspension was equally divided into two sets and transferred to two sterile “Corning” make glass Petri dishes (120 mm). Set one was placed in a specially fabricated UV-chamber and irradiated with UV-B radiation (1 W m-2) together with white light (14.4 W m-2) for 6, 9, and 12 h. Set two (served as control) was transferred to culture room and exposed to white light (14.4 W m-2). The culture suspension was gently stirred magnetically during UV-B irradiation to facilitate uniform exposure. Thirty mL culture suspensions were removed at indicated time intervals from both the sets and immediately processed for protein extraction. Unless otherwise stated, cultures were always exposed to UV-B radiation in combination with white light.
Protein Extraction
Whole cell protein was extracted as described previously (Kurian et al., 2006). Cells were harvested from UV-B treated and control (untreated) cultures by centrifugation at 10,000 ×g for 10 min at 4°C in a Sorvall RC-5B Refrigerated Superspeed Centrifuge (DuPont Instruments, USA). The resulting pellet was suspended in 10 mM HEPES buffer (pH 7.2) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and cells were broken by vortexing with glass beads (0.17–0.18 mm) at 4°C. Glass beads and unbroken cells were removed by centrifugation at 10,000 ×g for 20 min. The supernatant obtained mainly contained cytoplasmic proteins. Salts and insoluble impurities present, if any, were removed by a 2-D Clean-Up kit (GE Healthcare, UK). The protein content in each sample was measured by Bradford (1976) method.
Protein Separation by 2-Dimensional Gel Electrophoresis
The first dimension isoelectric focusing (IEF) was performed using Immobiline DryStrip (pH 4–7, 13 cm; GE Healthcare Bio-Sciences AB, Sweden). Protein sample (500 μg) dissolved in rehydration buffer (7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 0.3% (w/v) DTT, and 0.5% (v/v) IPG buffer) was loaded onto each IPG strip and rehydrated overnight at 4°C in the dark. After rehydration, IEF was performed using the Ettan IPGphor 3 IEF system (GE Healthcare, UK) using the following steps; 2 h at 100 V, 2 h at 200 V, 2 h at 500 V, 2 h at 1500 V, 2 h at 3000 V, 2 h at 6000 V, and 8000 V until a total of 60000 Vh was reached. Following the two steps of equilibrations (Nouwens et al., 2002), the second dimension separation was carried out on 12% SDS-PAGE gel at 20 mA/gel using SE 600 Ruby multiple gel-electrophoresis system (GE Healthcare, UK). Three gels were run for each protein sample of UV-B treated and untreated control cultures. Gels were stained with Colloidal Coomassie Blue G-250 (Candiano et al., 2004) and images were captured using the AlphaImager Gel Documentation unit (Alpha Innotech, USA).
2-De Gel Analysis
The gels were scanned and protein spots analyzed by PDQuest software version 8.0.1 (BioRad Laboratories, USA) for spot detection, quantification, background substraction, and spot matching between various gels. Spot quantification in the control and treated gels (UV-B exposed cultures) was performed by spot volumes (intensity × mm2) as described by Agrawal et al. (2014). The relative spot volume with respect to the UV-B treated and untreated control samples at desired time were compared using Student’s t-test. P values less than 0.05 were considered statistically significant.
Protein Spot Digestion and Mass Spectrometry
Desired spots were excised from the gel by manual picking using sterile OneTouch spot picker and placed into separate microcentrifuge tubes. MALDI-TOF MS analysis of protein spots was got done commercially from the Centre for Genomic Application (TCGA), New Delhi.
Database Search and Bioinformatics Analysis
The proteins were identified by comparing peptide mass fingerprints at the NCBInr database using the Mascot search engine1. Search parameters allowed for oxidized methionines, carbamidomethylation of cysteines, and one missed cleavage site of trypsin with mass accuracy of ±30 ppm. Identification was based on the first ranking result and Mascot scores of >74 which indicated that the hits were significant. Physico-chemical properties of proteins were determined using the ProtParam tool2 and database of Nostoc sp. PCC 7120 as reference (Kaneko et al., 2001). KEGG BLAST search3,4 ,5 was used to identify the signal transduction pathway for hypothetical proteins (HPs). SignalP 4.0 server6 was used for the prediction of putative signal peptides and their cleavage sites. The transmembrane helices and topology of proteins were predicted using the HMMTop7 server. InterProScan (version 4.8) server8 was used for the elucidation of protein family. Similarity search of HPs against different species from NCBI (National Center for Biotechnology Information) database was done using BLAST (Basic Local Alignment Search Tool) server9. Tree was constructed by UPGMA (Unweighted Pair Group Method with Arithmetic Mean) method. Proteins similar to the HPs were used as input for motif elucidation (highly conserved regions) by MEME tool10. For the prediction of promoter/cis-acting elements for HPs (designated as all3797 and all4050), upstream sequences were retrieved using genome blast11. Untranslated upstream region was taken as input to detect the promoter/cis-acting elements using BPROM tool from Softberry server12. Post-translational modification sites were detected by CBS prediction server13. Protein structure modeling and stereochemical properties of HP were done by CPHModels14 and PDBSum15.
Results
Effects of UV-B Radiation on Growth and Survival
Before assessing the effects of UV-B radiation + white light on proteome changes, we examined its effect on percentage survival and chlorophyll-a content in Anabaena L31. Cells exposed to UV-B (1 W m-2) + white light for 9 h showed 61% survival, and decreased to 4% after continuous exposure for 24 h (Figure 1). Chlorophyll-a content also decreased by 35% after 9 h of treatment but there was a steep decline after 12 h. This suggests that the cells of Anabaena L31 exposed to UV-B radiation (1 W m-2) + white light for 9 h do show significant physiological changes but retain more than 60% survival at this dose. Accordingly, all the following experiments were performed after 9 h employing 1 W m-2 of UV-B intensity together with white light.
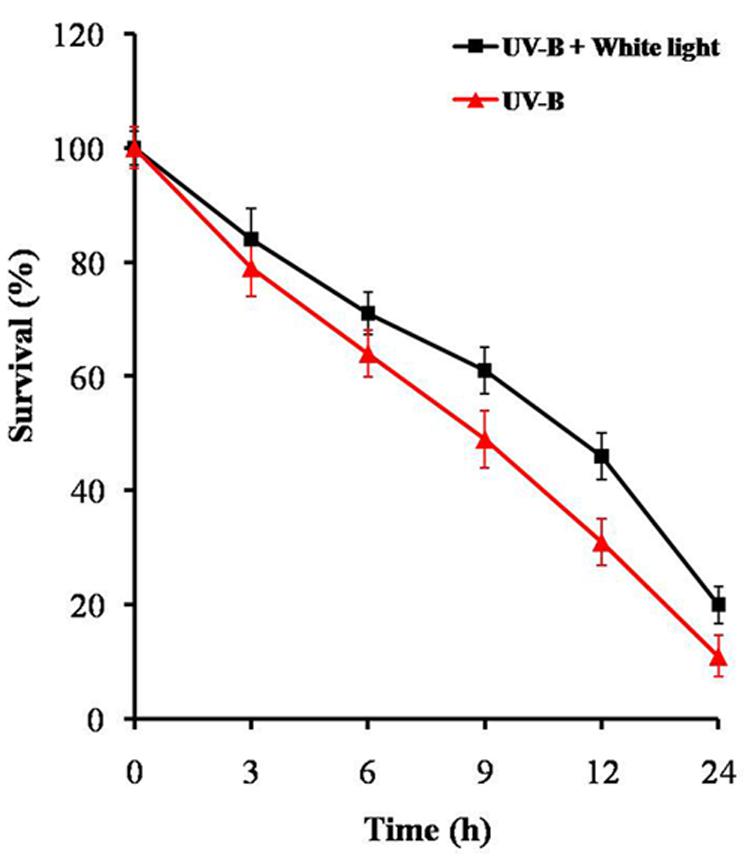
FIGURE 1. Percent survival of Anabaena L31. An exponentially grown culture was exposed to UV-B (1 W m-2) + white light or UV-B (1 W m-2) alone for desired time period and thereafter survival was determined. Results are based on the average of three experiments performed independently under identical conditions.
Effects of UV-B Radiation on Proteome
Once it became evident that UV-B (1 W m-2) + white light treatment do affect survival of Anabaena L31, we determined its possible effects on total proteome by employing 2-D gel electrophoresis. Analysis of whole cell protein in gels revealed 210 spots in untreated control and 223 in UV-B-treated Anabaena L31 (Figures 2A,B). Besides some changes in total number of spots, analysis of gels showed differential expression of several spots in UV-B treated cultures. Henceforth, attention was focused on the protein spots which were showing up-or down regulation on the basis of spot-to-spot comparison. Twenty one protein spots showing more than 4-fold higher and/or lower level of expression in UV-B-treated cultures as compared to untreated control (p < 0.05) were selected for MALDI-TOF MS analysis. Of the 21 identified proteins, 11 belonged to up-regulated group including two HPs (Figure 2C) and 10 to down-regulated group. Figure 2D represents expression level of all the above proteins.
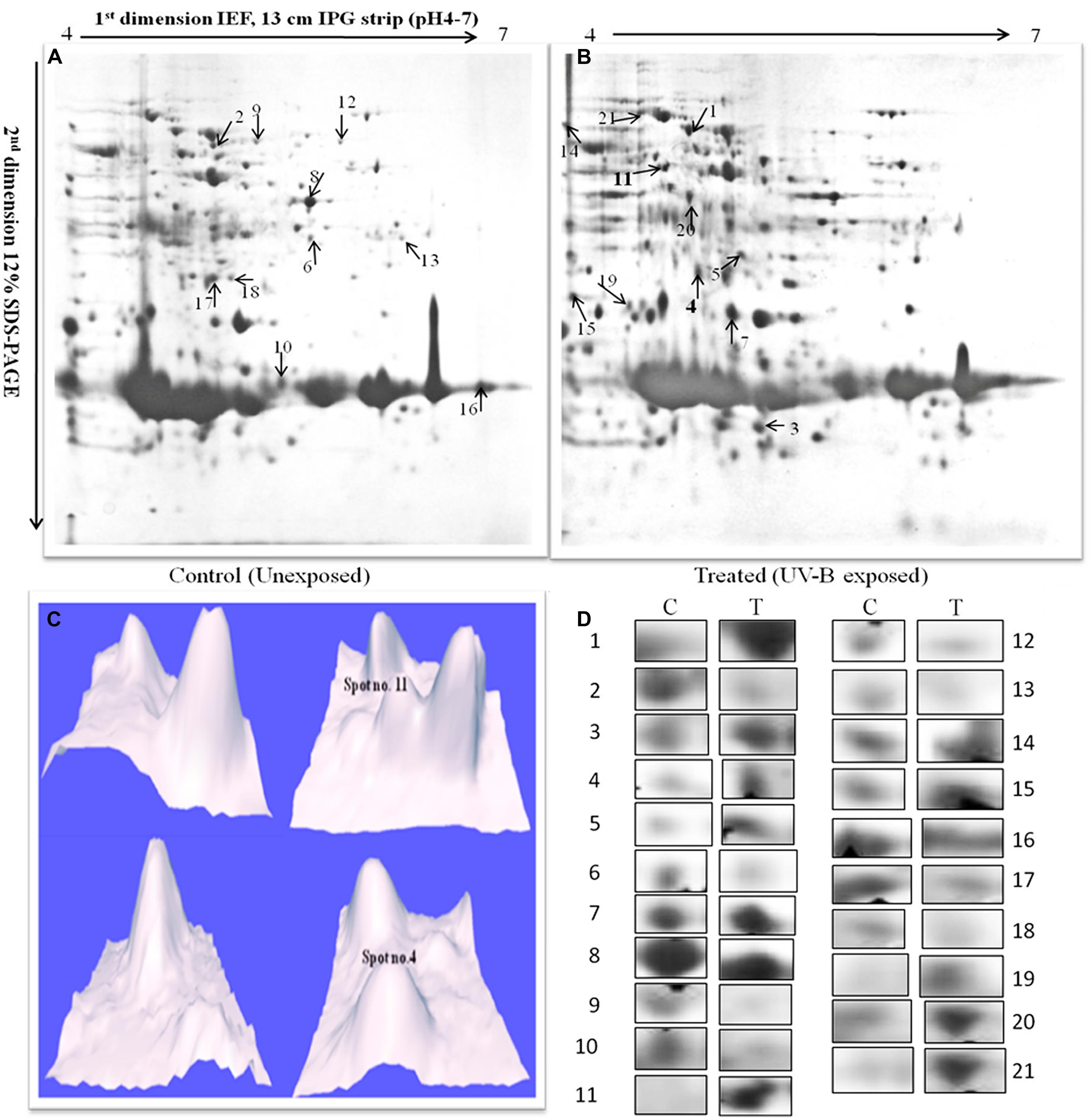
FIGURE 2. 2-DE images of cytosolic protein from Anabaena L31. (A)-control, and (B)-irradiated with UV-B (1 W m-2) + white light for 9 h. Proteins were extracted and separated by 2-DE and visualized by CBB staining. 500 μg proteins were loaded on to pH 4–7 IPG dry strips for first dimension IEF followed by 12% linear vertical SDS-PAGE as the second dimension. (C)-3-D images of the hypothetical protein spots shown in bold numbers on 2-DE gels, and (D)-expression pattern of selected proteins. Lane: C-control, and T- 9 h of UV-B treatment. Numbers represent the spots as indicated on 2-DE gels. Proteins showing significant and reproducible changes were subjected to MALDI-TOF-MS.
Identification and Functional Classification of Differentially Expressed Proteins
In functional studies we compared the protein sequences with the secondary (or derived) protein databases that contain information on motifs, signature, and protein domains. Highly significant hits against different protein databases allowed us to derive the biochemical functions of query protein. Table 1 summarizes the key findings related to cyanobacterial processes responding to UV-B treatment based on the identification of representative proteins. All the 21 proteins identified in this study were sorted according to the functional categories defined by cyanobase. On the basis of physiological functions, these proteins were placed in eight functional groups which included carbohydrate metabolism, photosynthesis and respiration, energy metabolism, translation and cellular processes, amino acid biosynthesis, hypothetical, reductases, chaperone, and antioxidative enzymes (Table 1). Notably, two HPs namely, all3797 and all4050 were found among the 21 identified proteins.
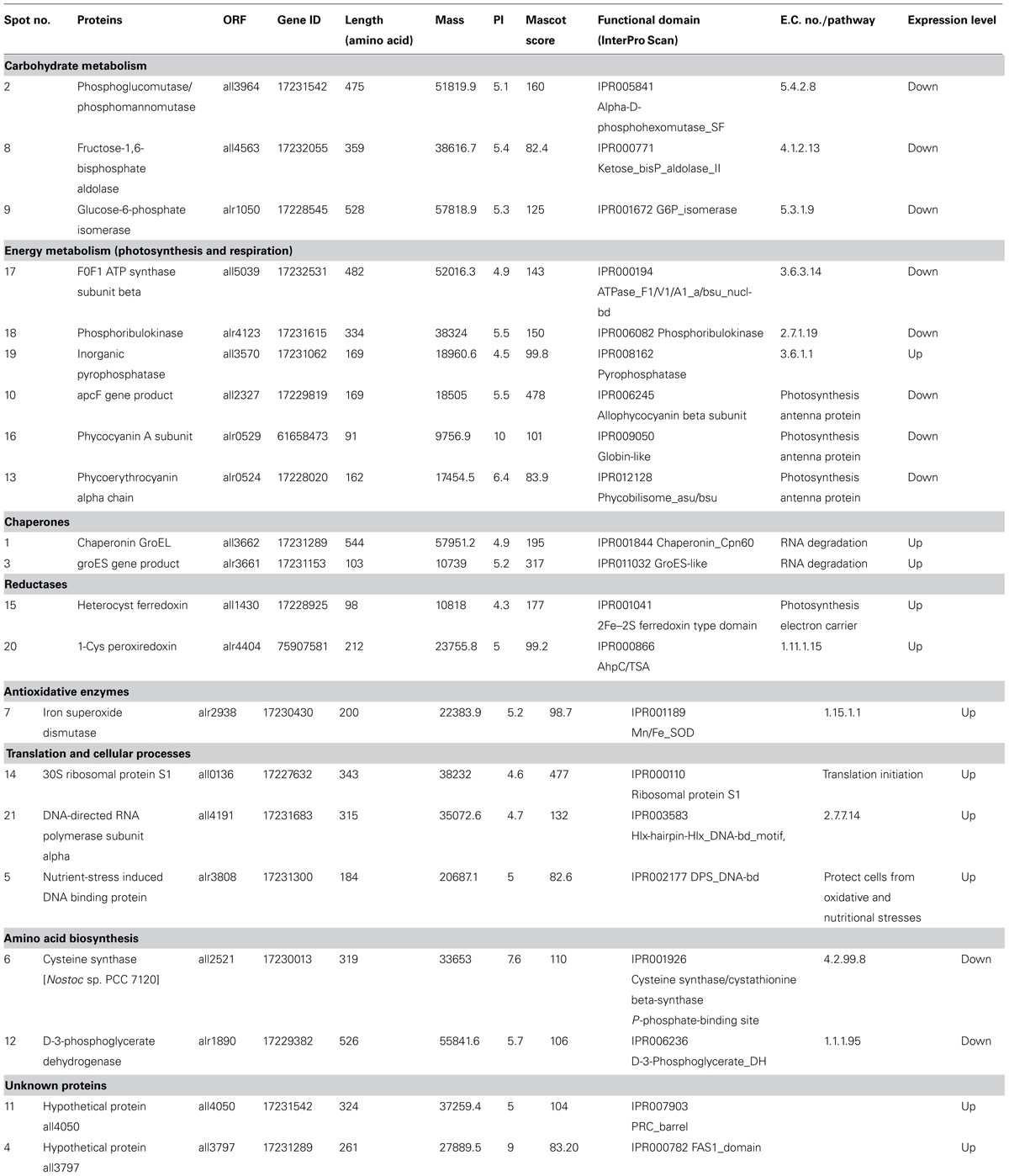
TABLE 1. Differentially expressed proteins identified in the cellular soluble fraction of Anabaena L31 by MALDI-TOF MS.
Prediction and Localization of Functional Domain, Motif Analysis, and Sequence Homology of Hypothetical Proteins
As both the HPs (all3797 and all4050) were always induced under UV-B stress and such proteins have not been reported previously from cyanobacteria, we focused our studies on these two proteins. Accordingly, with a view to obtaining preliminary functional information of these two, analysis of functional domain and motif analysis was made. Data obtained revealed that both the proteins possess functional domain as well as regulatory motifs. The protein all3797 encoded by ORF all3797 showed similarity with the FAS 1 like domain (IPR000782; Figure 3A) and all4050 encoded by ORF all4050 and having four domains (IPR007903, IPR011033, IPR011756, and IPR019060; Figure 3B) resembled with the photosynthetic reaction center (PRC).

FIGURE 3. Model showing the functional domains of hypothetical proteins. (A)- all3797, and (B)- all4050 obtained from (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
Prediction of localization, putative signal peptides and transmembrane helices of the proteins revealed that the protein all3797 has N terminal (inside) with single transmembrane helices from positions 8 to 27 and is extracellularly localized, indicating that it may be a member of type II transmembrane protein family (Figure 4A). Protein all4050 showed N terminal (outside) with single transmembrane helices from positions 65 to 83 and is localized in the cytoplasm, implying that it probably be a member of the type I transmembrane protein family (Figure 4B).
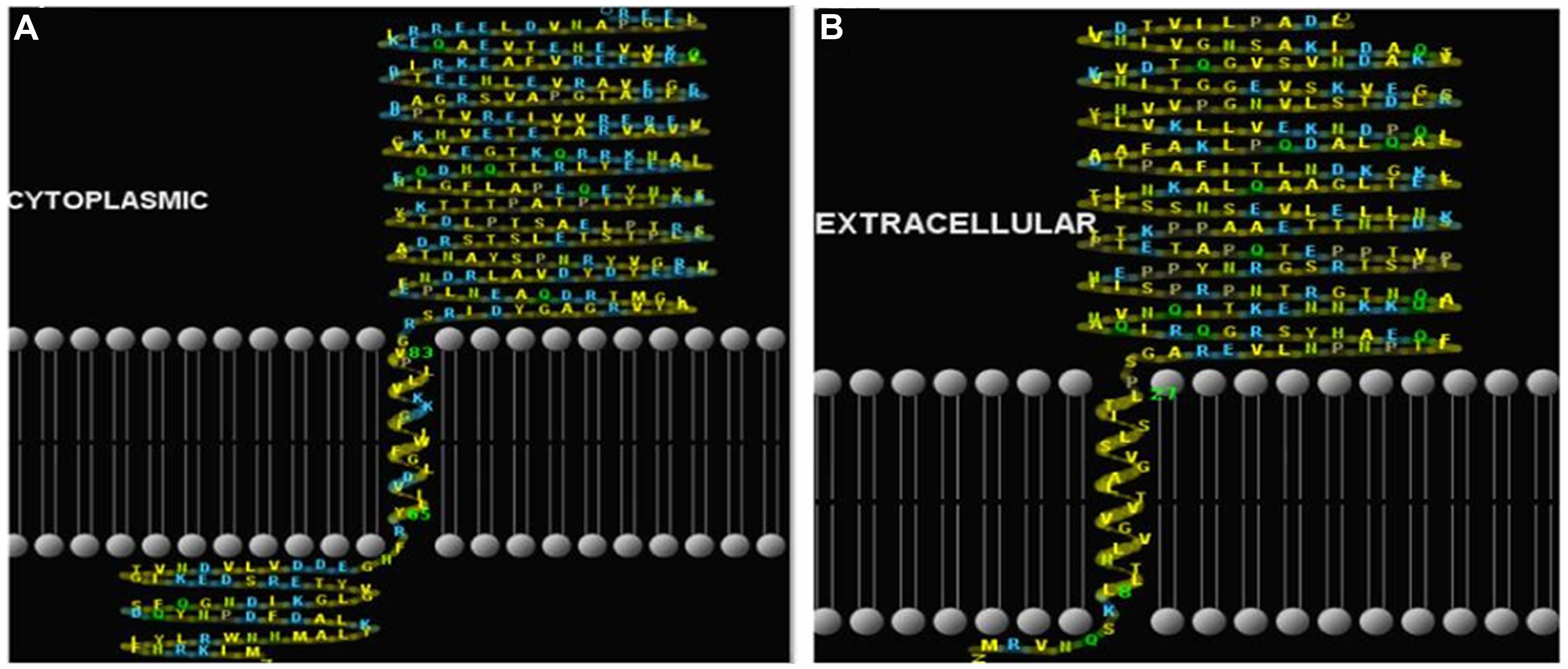
FIGURE 4. Prediction of putative signal peptides and transmembrane helices of hypothetical proteins. (A)-all3797, and (B)-all4050 (http://www.cbs.dtu.dk/services/SignalP/) and HMMTop (http://www.enzim.hu/hmmtop/).
With a view to revealing homology of the proteins all3797 and all4050 with proteins of other species of cyanobacteria (available in data base), a phylogenetic tree was constructed using reference strain Nostoc sp. PCC 7120 (also known as Anabaena sp. PCC 7120) because of its whole genome sequence availability in database. Accordingly, various cyanobacterial proteins showing more than 30% homology with reference strain were selected for multiple sequence alignment. From the tree it is evident that the protein all3797 shares 100% similarity with the protein of Anabaena variabilis (Figure 5A). However, a copy of identical ORF is also present in Nostoc PCC 7120 but grouped in a separate cluster which might be responsible for certain other functions in this species. HP all4050 showed 99% similarity with the protein of Nostoc punctiforme (Figure 5C). Analysis of different motifs present in the protein sequences of all3797 suggests that the first four motifs are highly conserved in all the selected species. Furthermore, all the motifs present in all3797 were also present in A. variabilis which is in accordance with the similarity index noted in phylogenetic tree (Figure 5B). In the case of protein all4050, seven motifs were found to be highly conserved in all the selected species. Interestingly, these motifs were also noted in three species of the bacterium Deinococcus (an extreme radioresistant organism) but they also had certain other motifs which resulted in separate cluster formation (Figure 5D). It is well-known that the multifunctional consensus sequences corresponding to the motif are useful in understanding the functional aspect of motifs. The sequence analysis shows that in the case of all3797, phenylalanine at position numbers (138, 163, 170), leucine (141,150,154,173,182,183,191), asparagine (246,186), serine (205,245), glycine (247,218,211), threonine (161,166), alanine (144,147,148,164,169), proline (165,174,184), histidine (197,250), glutamate (167), and tyrosine (196) are totally conserved in the sequence alignment. Similarly, in the case of protein all4050, the core conserved residues, namely leucine (16), glycine (50, 61, and 84), arginine (64), and proline (82) were noted.
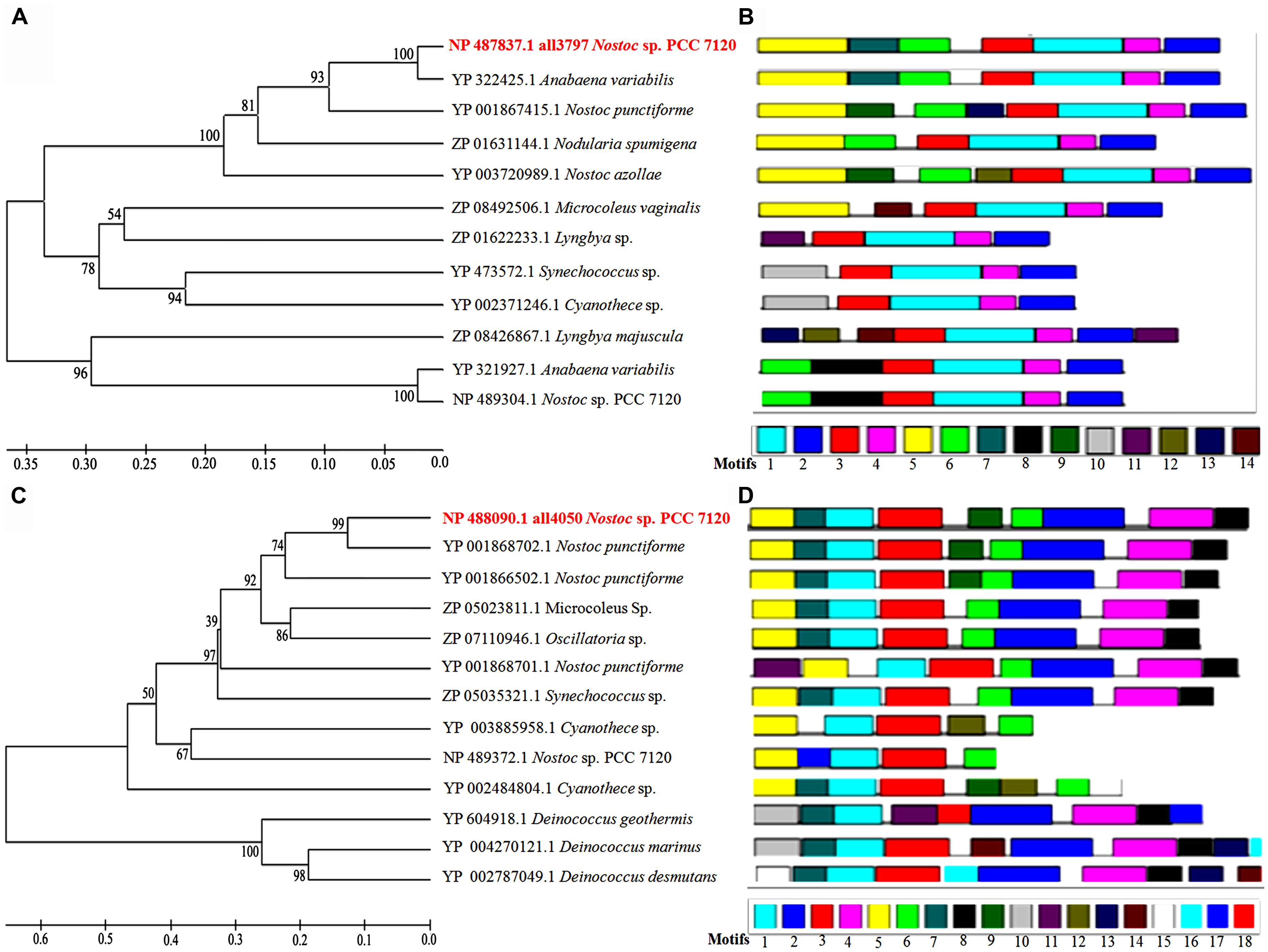
FIGURE 5. Phylogenetic tree showing homology with proteins of other species of cyanobacteria and bacteria available in the database. (A)-phyologenetic tree, and (B)-motif analysis of all3797; (C)-phyologenetic tree, and (D)-motif analysis of all4050 (http://www.icp.ucl.ac.be/∼opperd/private/upgma.html) and MEME tool (http://meme.nbcr.net/meme/).
Because the analysis of transmembrane helices revealed that both the HPs have one transmembrane helix each, we attempted to detect post-translational modification sites present, if any, in the functional region. Based on the prediction of serine and threonine phosphorylation sites, a total of eight sites were identified in all3797 and three in all4050 (Figures 6A,B).
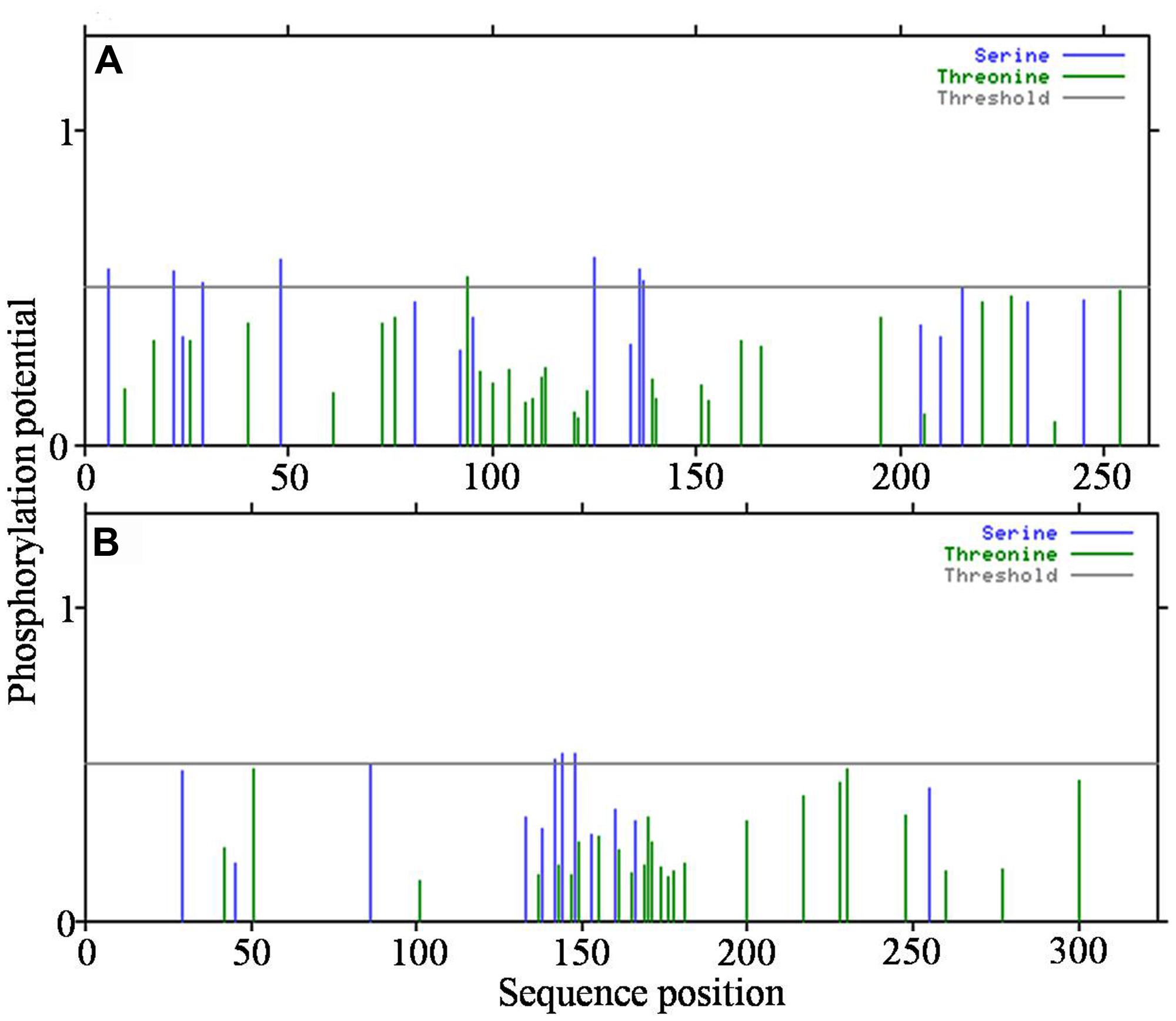
FIGURE 6. Detection of post-translational modification sites in hypothetical proteins. (A)-all3797, and (B)-all4050 (obtained from http://www.cbs.dtu.dk/services/NetPhosBac-1.0/).
Prediction of UTR and Promoter Regions
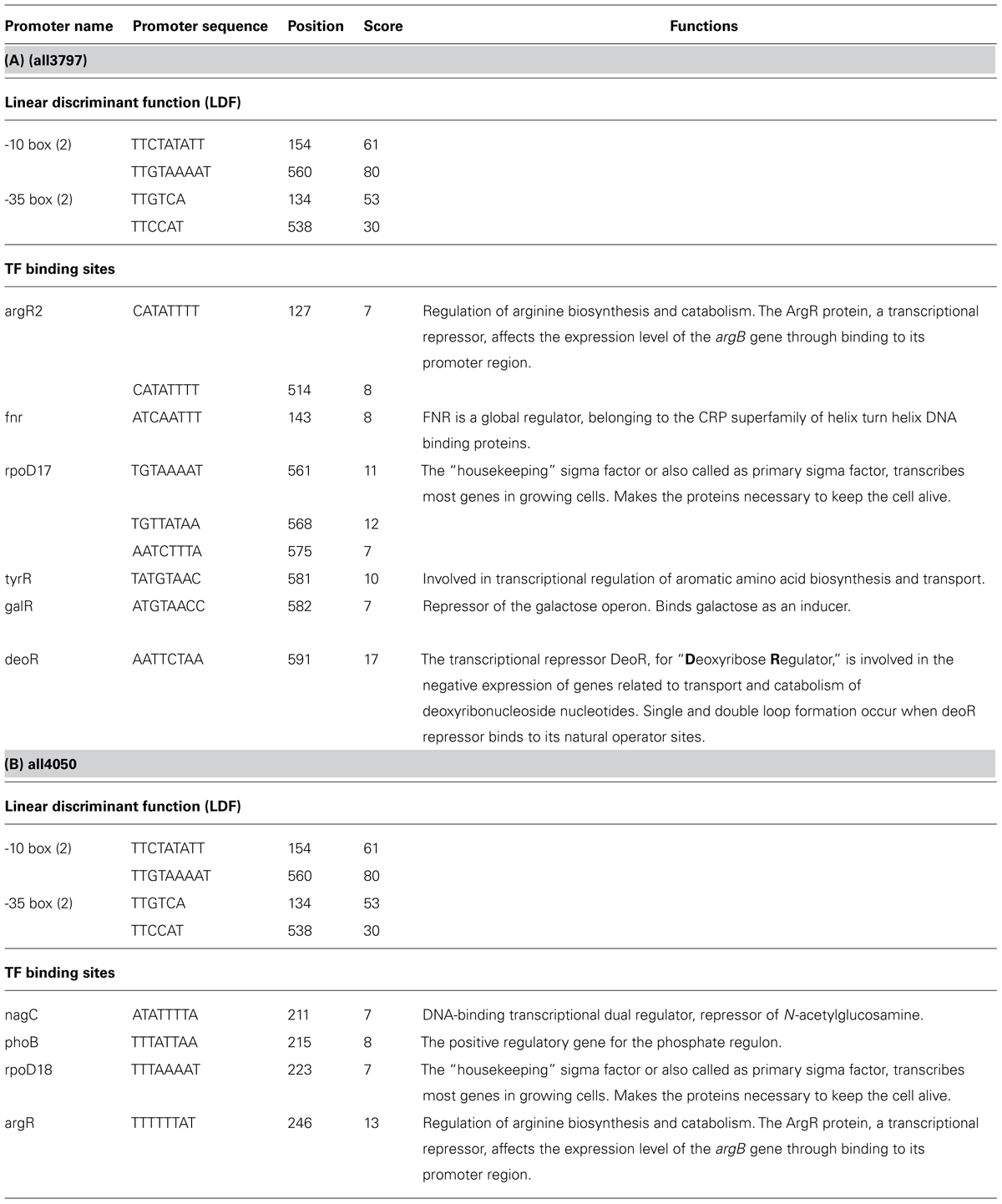
As the full genome sequence of Anabaena PCC7120 is available in the database16, an attempt was made to retrieve upstream region (UTR) of putative genes of both the HPs to make prediction of promoter binding sites and cis-regulating elements. Analysis showed that the genes of both the UV-B responsive HPs have promoter regions as well as transcription binding sites in their UTR. Details of promoters and transcription binding sites are presented in Table 2. The protein all3797 had promoters argR2 and rpoD17 which are known to play important role against UV radiation and certain other abiotic stresses (Table 2A). Likewise, protein all4050 showed the presence of promoters nagC, phoB, rpoD18, and argR whose roles in the regulation of vital metabolic processes are well-documented (Table 2B).
Structure Modeling and Stereochemical Properties
A 3-D model of both the HPs was constructed using sequence similarity and available structures in the data base. Validity of the structure was checked by the Ramchandran plot (Figures 7A,B). Such study of the all3797 revealed 75.2% residues in the most favored region, 23.1% in additional allowed region and 0.8% in disallowed region (Figure 7A). Similarly, 3-D model of all4050 showed 90.1% residues in the most favored region, 9.9% in additional allowed region and no residue in disallowed region (Figure 7B). The values obtained are well within normal range for a high quality model. The best validated structures of all3797 and all4050 have been deposited in the PMDB database with PMDB-IDs; PM0077736 and PM0077737 respectively.
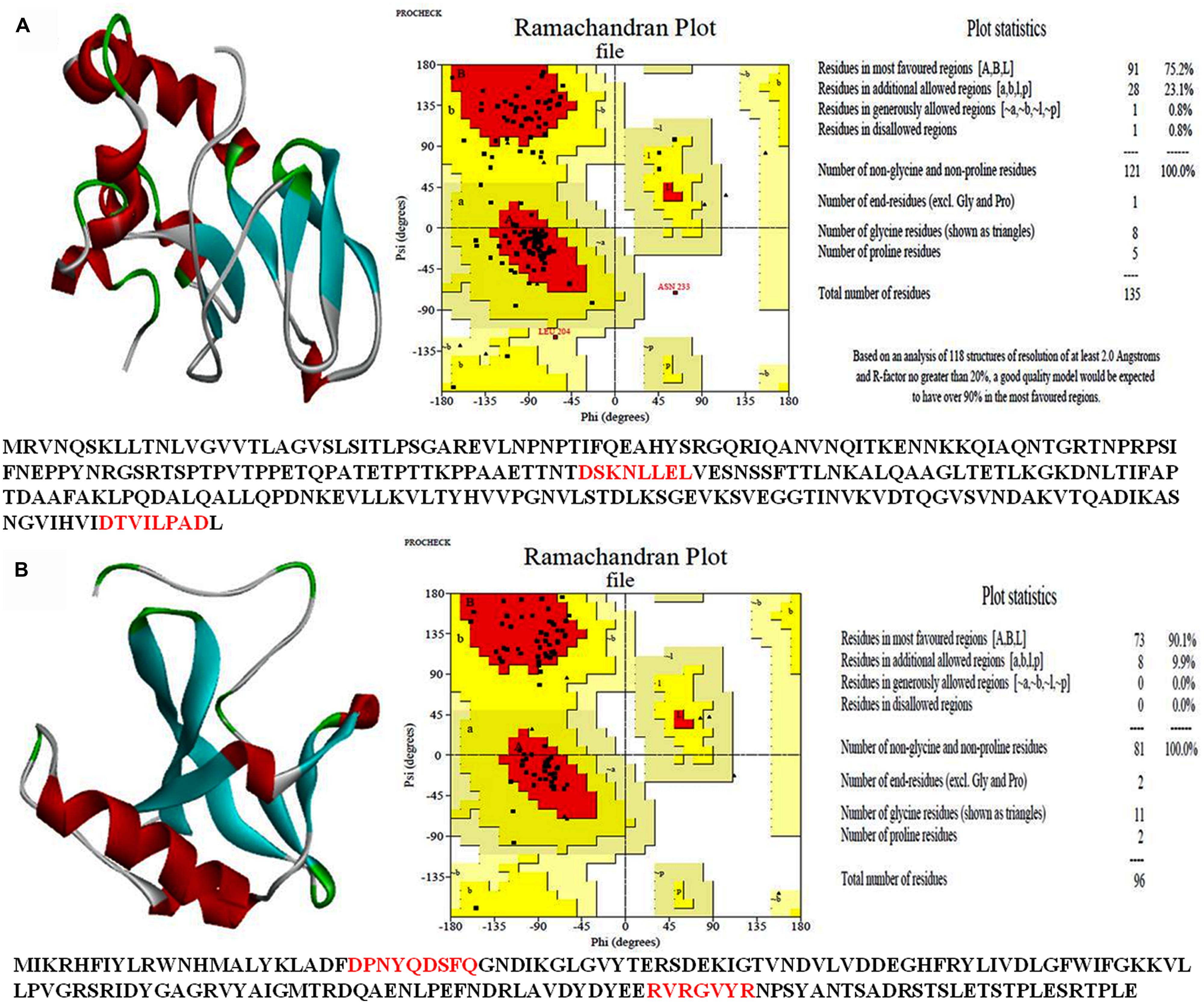
FIGURE 7. 3-D structure of hypothetical proteins. (A)-all3797 (PDB ID PM0077736), and (B) all4050 (PDB ID PM0077737) obtained from CPHModels (http://www.cbs.dtu.dk/services/CPHmodels/) and PDBSum (http://www.ebi.ac.uk/pdbsum/).
Discussion
Damaging effects of UV-B radiation on various metabolic processes of different species of cyanobacteria are well-documented (Kumar et al., 2003; Häder et al., 2007; Babele et al., 2012; Singh et al., 2013). In the present study, results obtained showed that UV-B radiation exposure to Anabaena L31 in the presence of white light also causes adverse effects on various physiological processes and brings about marked changes in the metabolic machinery. This finding is important and pertinent in view of the fact that living organisms are never exposed to UV-B radiation alone under ambient conditions. Accordingly, to gain better understanding of the metabolic events affected by UV-B stress, we targeted comparative proteomics study of the Anabaena L31 cells grown under UV-B stress and normal conditions. As alteration was noted in the expression of several proteins, we selected and identified 21 UV-B responsive proteins whose expression was drastically up- or down- regulated. Alteration in quantity and quality of proteins is expected as it is an essential step of adaptive mechanism developed by cyanobacteria under UV-B stress (Gao et al., 2009). As reported previously (Häder et al., 2007; Singh et al., 2013), that major part of the UV-B energy is absorbed by phycobiliproteins and chlorophyll, these act as the main targets of UV-B action. That phycobiliproteins are indeed the target of UV-B action is supported from our finding wherein the proteins, apcF gene product (all2327), phycocyanin A subunit (alr0529), and phycoerythrocyanin alpha chain (alr0524) are down regulated following UV-B exposure of Anabaena L31 cells. This clearly suggests that UV-B absorbed by the cells may directly affect the stability of pigment complex, and prolonged exposure may inhibit the synthesis of phycobiliproteins. Altogether, the damage caused to photosynthetic pigments may lead to diminished PS-II activity resulting in subtle changes in enzymes of Calvin cycle. This is in agreement with an earlier report (Huang et al., 2002) wherein operon composed of genes encoding carbon dioxide-concentrating mechanism proteins (fructose-1, 6-bis phosphate aldolase, glucose 6-phosphate isomerase, and phosphoglucomutase/phosphomannomutase) were found repressed under UV-B stress. Similar to above report, enzymes such as fructose-1, 6-bisphosphate aldolase (all4563), and glucose 6-phosphate isomerase (alr1050) were down regulated under UV-B stress in our study. Fructose-1, 6-bis phosphate aldolase is an important enzyme of glycolysis and Calvin cycle that converts fructose-1, 6-bis phosphate to glyceraldehyde 3-phosphate and dihydroxy acetone 3-phosphate. Under stress conditions glyceraldehyde 3-phosphate and fructose-1, 6-bis phosphate may be converted to glucose 6-phosphate to carry out pentose phosphate pathway for NADPH synthesis (Nakahara et al., 2003). This is an important step as it may partly meet the reductant requirement for vital metabolic processes. Down regulation of phosphoglucomutase (all3964) following exposure of Anabaena L31 cells to UV-B is an interesting observation and to our knowledge not reported from any other species of cyanobacteria. Phosphoglucomutase may have significant role under UV-B stress as it converts glucose-1-phosphate to glucose -6-phosphate which is an important reversible reaction of glycolysis (Fridlyand and Scheibe, 1999).
That energy metabolic proteins were one of the smallest fractions of the identified proteome in this study is in agreement with the report that majority of these proteins remain embedded into integral membranes and only a few occur in soluble fraction (Anderson et al., 2006). Consistent with the down regulation of proteins of carbohydrate metabolism, the transcript allowing ATP synthesis by F0F1 ATP synthase (all5039) was down regulated in the presence of UV-B stress. This enzyme is of great significance to the test cyanobacterium as it uses a proton gradient to drive ATP synthesis and hydrolyzes ATP to build the proton gradient. Another important protein identified as phosphoribulokinase (alr4123) which catalyzes the ATP-dependent phosphorylation of ribulose-5-phosphate to ribulose-1, 5-phosphate during CO2 assimilation by autotrophic organisms was also down regulated under UV-B stress. This may also adversely affect pentose phosphate pathway and growth of the test organism.
Amino acids are essential components of proteins with a key role in protein biosynthesis and regulation of growth and development of all living organisms. In this context, our data show that D-3-phosphoglycerate dehydrogenase (alr1890) and cysteine synthase (all2521) are down regulated under UV-B stress and thus may adversely affect the amino acid biosynthesis pathway. Down regulation of D-3-phosphoglycerate dehydrogenase which catalyzes the formation of 3-phosphonooxypyruvate from 3-phospho-D-glycerate in the serine biosynthesis may reduce the level of α-ketoglutarate and affect the formation of 2-hydroxyglutarate. Likewise, down regulation of cysteine synthase may also affect the synthesis and cellular level of L-cysteine as it converts O3-acetyl-L-serine into L-cysteine. Conceivably, diminished growth of Anabaena L31 under UV-B stress could be due to reduction in the level of enzymes of amino acid biosynthesis.
Among the up-regulated proteins, those involved in DNA repair, chaperonins, and antioxidative defense mechanism are the most prominent. Induction of proteins involved in DNA repair such as nutrient stress-induced DNA-binding protein (alr3808) and DNA-directed RNA polymerase alpha subunit (all4191) suggests that DNA could be a target of UV-B effects in Anabaena sp. This is also supported from our earlier conclusion where we reported that that UV-B affects the DNA of cyanobacteria and the killing of these microbes might be due to the irreversible damage caused to DNA by this high energy radiation (Kumar et al., 2004). To this effect, up-regulation of DNA-binding protein is expected as DNA-binding protein from stationary phase (DpsA) of the caynobacterium Synechococcus sp. PCC7942 is known to confer resistance to oxidative stress and long-term nutrient starvation (Pena and Bullerjahn, 1995; Sen et al., 2000). A DpsA homolog has also been reported from Nostoc (Anabaena) sp. PCC7120 (Hernandez et al., 2007). To our knowledge, there is no report showing induction of DNA-binding protein under UV-B stress in any cyanobacteria; so identification of alr3808 protein in this study may be a novel finding. RNA polymerases that make up transcription machinery routinely traverse through many parts of the genome; this transcription machinery seems to have an important role in sensing DNA repair and stress responsive pathway (Ljungman, 2007). As DNA-directed RNA polymerase and nutrient-induced DNA-binding proteins are regulated by the transcription machinery, their up regulation may be a common event under different abiotic stresses.
Various abiotic stresses can induce chaperonin GroEL, including heat, acid, radicals, and UV radiation (Frydman, 2001; Jang et al., 2004; Bukau et al., 2006; Gao et al., 2009). We also observed four-fold up regulation of GroEL (all3662) and GroES (alr3661) in response to UV-B stress in Anabaena L31. Similar response of above proteins in plants and bacteria under stress condition has been reported (Jang et al., 2004). It has been suggested that proteomic mechanisms act as a check point inside the cell under stress conditions with crucial roles in modifications and survival of cyanobacteria (Balogi et al., 2008; Gao et al., 2009). The most striking feature of the proteins involved in protection against oxidative stress is their expression and maintenance of homeostasis in a stressed environment. In this context the activated antioxidative systems under stress play key role in the protection mechanism of any organism. In the present study, iron superoxide dismutase (SOD; alr2938) and 1-cys peroxiredoxin (alr4404) increased fourfold following UV-B stress and could possibly mitigate the damaging effect of increased ROS level in the cells. SOD effectively reduces superoxide radicals to hydrogen peroxide and peroxiredoxins metabolize peroxides. Peroxiredoxins have multiple roles including reduction of H2O2 and organic hydrogen peroxides (Dietz et al., 2006), and as molecular chaperones similar to HSPs with roles in cell signaling (Jang et al., 2004).
Inorganic-pyrophosphatase plays a critical role in lipid metabolism (including lipid synthesis and degradation) and DNA synthesis, as well as in other biochemical transformations. By promoting rapid hydrolysis of pyrophosphate (PPi), inorganic pyrophosphatase activates oxidation of fatty acids. As UV-B radiation is reported to cause lipid peroxidation (Rai et al., 2013), over-expression of inorganic pyrophosphatase in this study might be due to this stress.
One novel finding from this study is the identification and characterization of two proteins (all3797 and all4050) annotated as HPs which are new and so far not reported from any cyanobacteria subjected to UV-B stress. In silico analysis revealed that all3797 is related to the FAS1 (fasciclin-like) domain and is an extracellular module of about 140 amino acid residues of fasciclin superfamily protein. It seems to be involved in cell adhesion with extracellular matrix and may be responsible for aggregation of cyanobacterial filaments to protect them from UV radiation.
Another HP all4050 showed similarity to PRC (barrel)-like protein. The PRC-barrel, approximately 80 residues long, is widely distributed in bacteria, archaea and plants. Its role in a variety of biological systems, ranging from RNA processing to photosynthesis is well-documented. Several researchers have reported drastic effects of UV-B irradiation on the water soluble pigment phycobiliprotein, including disintegration of phycobilisomes and loss of α and β monomers of phycocyanin. Additionally, phycobiliproteins act as photosensitizers and produce ROS under UV-B stress. Most probably enhanced expression of PRC-barrel protein all4050 may be the result of stress caused by UV-B and may partially restore light-driven electron transfer reactions, further study is needed to reveal the exact function of all4050 under UV-B stress. However, we have provided sufficient data related to localization, functional domain, signal peptide, sequence similarity, motif types and UTR and promoter regions of both the HPs by applying in silico approaches. Prediction of functional domains, UTR, motif analysis, and putative model of these HPs could be useful in future studies dealing with unknown proteins specifically induced under other types of abiotic stress. In summary, the protein profiling and expression data obtained in this study suggest that UV-B stress is quite effective in the presence of white light but more work is warranted to unravel the complexity of the proteome following growth of cyanobacteria under different light regimes with UV-B radiation.
Conclusion
A few researchers have reported changes in the proteome of cyanobacteria under UV-B radiation stress but those studies were mostly conducted on the unicellular cyanobacterium, Synechocystis sp. PCC 6803 but only in the presence of UV-B light. Our study is the first of its kind that demonstrates changes in the proteome of the N2-fixing Anabaena strain L31 following UV-B radiation stress in the presence of white light. Analysis of 223 proteins on 2-D gels revealed several differentially expressed proteins of which 21 spots were identified by mass spectrometry. Among the identified proteins, chaperonin GroEL and groES gene product, 30S ribosomal protein S1, and iron SOD are the well-documented UV-B induced proteins but induction of certain novel proteins including HPs (all3797 and all4050) under UV-B stress is new and not reported so far. It would be worthwhile to make detailed and comparative analysis of UV-B responsive proteins from both non-N2-fixing and N2-fixing cyanobacteria to better understand the stress tolerance mechanism against UV-B as well as the roles of different classes of proteins in this important process. Further study is needed to assign the exact function of HPs which are specifically induced under UV-B stress. Nevertheless, identification of differentially expressed proteins in this study may prove useful in future studies especially for assessing their significance in the adaptation mechanism of cyanobacteria under UV-B radiation stress.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the Department of Science and Technology, Govt. of India, New Delhi, project (No. SR/SO/PS-49/09) sanctioned to AK. PKB and GS worked as Junior Research Fellows in the project. We thank Prof. H. D. Kumar, ex-professor and coordinator, Department of Botany and School of Biotechnology, Banaras Hindu University, Varanasi, for his suggestions in improving the manuscript.
Footnotes
- ^http://www.matrixscience.com
- ^http://web.expasy.org/protparam/
- ^http://www.genome.jp/tools/blast/
- ^http://biocyc.org/blast.html
- ^http://www.genome.jp/kegg/genes.html
- ^http://www.cbs.dtu.dk/services/SignalP/
- ^http://www.enzim.hu/hmmtop/
- ^http://www.ebi.ac.uk/Tools/pfa/iprscan/
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^http://meme.sdsc.edu/meme/intro.html
- ^http://genome.kazusa.or.jp/cyanobase
- ^http://linux1.softberry.com/berry.phtml
- ^http://www.cbs.dtu.dk/services/NetPhosBac-1.0/
- ^http://www.cbs.dtu.dk/services/CPHmodels/
- ^http://www.ebi.ac.uk/pdbsum/
- ^http://genome.kazusa.or.jp/cyanobase/Anabaena
References
Agrawal, C., Sen, S., Singh, S., Rai, S., Singh, P. K., Singh, V. K.,et al. (2014). Comparative proteomics reveals association of early accumulated proteins in conferring butachlor tolerance in three N2-fixing Anabaena spp. J. Proteomics 96, 271–290. doi: 10.1016/j.jprot.2013.11.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anderson, D. C., Campbell, E. L., and Meeks, J. C. (2006). A soluble 3D LC-2S-2S proteome of the filamentous cyanobacterium Nostoc punctiforme. J. Proteom. Res. 5, 3096–3104. doi: 10.1021/pr060272m
Babele, P. K., Singh, G., Tyagi, M. B., Sinha, R. P., and Kumar, A. (2012). Ultraviolet-B radiation effects on cyanobacteria and the role of sunscreen pigments in its protection. Phykos 42, 1–13.
Balogi, Z., Cheregi, O., Giese, K. C., Juhasz, K., Vierling, E., Vass, I.,et al. (2008). A mutant small heat shock protein with increased thylakoid association provides an elevated resistance against UV-B damage in Synechocystis 6803. J. Biol. Chem. 83, 22983–22991. doi: 10.1074/jbc.M710400200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantity of proteins utilising the principle of protein dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Bukau, B., Weissman, J., and Horwich, A. (2006). Molecular chaperones and protein quality control. Cell 125, 443–451. doi: 10.1016/j.cell.2006.04.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Candiano, G., Bruschi, M., Musante, L., Santucci, L., Ghiggeri, G. M., Carnemolla, B.,et al. (2004). Blue silver: a very sensitive colloidal coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333. doi: 10.1002/elps.200305844
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Castenholz, R. W., and Garcia-Pichel, F. (2012). “Cyanobacterial responses to UV radiation,” in The Ecology of Cyanobacteria II: Their Diversity in Time and Space, ed. B. A. Whitton (Dordrecht: Springer), 481–502. doi: 10.1007/978-94-007-3855-3_19
Chauhan, S., Pandey, R., and Singhal, G. S. (1998). Ultraviolet-B induced changes in ultrastructure and D1/D2 proteins in cyanobacteria Synechococcus sp. PCC 7942. Photosynthetica 35, 161–167. doi: 10.1023/A:1006939003814
Dietz, K. J., Jacob, S., Oelze, M. L., Laxa, M., Tognattiv, D., Miranda, S. M. N.,et al. (2006). The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57, 1697–1709. doi: 10.1093/jxb/erj160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ehling-Schulz, M., Schulz, S., Wait, R., Görg, A., and Scherer, S. (2002). The UV-B stimulon of the terrestrial cyanobacterium Nostoc commune comprises early shock proteins and late acclimation proteins. Mol. Microbiol. 46, 827–843. doi: 10.1046/j.1365-2958.2002.03209.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fridlyand, L. E., and Scheibe, R. (1999). Regulation of the calvin cycle for CO2 fixation as an example for general control mechanism in metabolic cycles. Biosystems 51, 79–93. doi: 10.1016/S0303-2647(99)00017-9
Frydman, J. (2001). Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647. doi: 10.1146/annurev.biochem.70.1.603
Fulda, S., Mikkat, S., Huang, F., Huckauf, J., Marin, K., Norling, B.,et al. (2006). Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6, 2733–2745. doi: 10.1002/pmic.200500538
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, Y., Xiong, W., Li, X., Gao, C., Zhang, Y., Li, H.,et al. (2009). Identification of the proteomic changes in Synechocystis sp. PCC 6803 following prolonged UV-B irradiation. J. Exp. Bot. 60, 1141–1154. doi: 10.1093/jxb/ern356
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M.,et al. (1998). The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273, 22480–22489. doi: 10.1074/jbc.273.35.22480
Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. (1999). Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730.
Häder, D. P., Kumar, H. D., Smith, R. C., and Worrest, R. C. (2007). Effects of solar UV radiation on aquatic ecosystems and interactions with climate change. Photochem. Photobiol. Sci. 6, 267–285. doi: 10.1039/B700020K
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hecker, M., and Volker, U. (2004). Towards a comprehensive understanding of Bacillus subtilis cell physiology by physiological proteomics. Proteomics 4, 3727–3750. doi: 10.1002/pmic.200401017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hernandez, J. A., Pellicer, S., Huang, L., Peleato, M. L., and Fillat, M. F. (2007). FurA modulates gene expression of alr3808, a DpsA homologue in Nostoc (Anabaena) sp. PCC7120. FEBS Lett. 581, 1351–1356. doi: 10.1016/j.febslet.2007.02.053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, L., McCluskey, M. P., Ni, H., and LaRossa, R. A. (2002). Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 184, 6845–6858. doi: 10.1128/JB.184.24.6845-6858.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jang, H. H., Lee, K. O., Chi, Y. H., Jung, B. G., Park, S. K., and Park, J. H. (2004). Two enzymes is one: two yeast peroxiredoxins display oxidative stress dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625–635. doi: 10.1016/j.cell.2004.05.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaneko, T., Nakamura, Y. C., Wolk, P., Kuritz, T., Sasamoto, S., and Watanabe, A. (2001). Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8, 205–213. doi: 10.1093/dnares/8.5.205
Kerr, J. B., and McElroy, C. T. (1993). Evidence for large upward trends of ultraviolet-B radiation linked to ozone depletion. Science 262, 1032–1034. doi: 10.1126/science.262.5136.1032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, A., Tyagi, M. B., and Jha, P. N. (2004). Evidences showing ultraviolet-B radiation-induced damage of DNA in cyanobacteria and its detection by PCR assay. Biochem. Biophys. Res. Commun. 318, 1025–1030. doi: 10.1016/j.bbrc.2004.04.129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, A., Tyagi, M. B., Jha, P. N., Srinivas, G., and Singh, A. (2003). Inactivation of cyanobacterial nitrogenase after exposure to ultraviolet-B radiation. Curr. Microbiol. 46, 380–384. doi: 10.1007/s00284-001-3894-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, K., Prakash, A., Tasleem, M., Islam, A., Ahmad, F., and Hassan, M. I. (2014). Functional annotation of putative hypothetical proteins from Candida dubliniensis. Gene 543, 93–100. doi: 10.1016/j.gene.2014.03.060
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kurian, D., Phadwal, K., and Maenpaa, P. (2006). Proteomic characterization of acid stress response in Synechocystis sp. PCC 6803. Proteomics 6, 3614–3624. doi: 10.1002/pmic.200600033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lei, Z., Elmer, A. M., Watson, B. S., Dixon, R. A., Mendes, P. J., and Sumner, L. W. (2005). A two-dimensional electrophoresis proteomic reference map and systematic identification of 1367 proteins from a cell suspension culture of the model legume Medicago truncatula. Mol. Cell Proteomics 4, 1812–1825. doi: 10.1074/mcp.D500005-MCP200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ljungman, M. (2007). The transcription stress response. Cell Cycle 6, 2252–2257. doi: 10.4161/cc.6.18.4751
Nakahara, K., Yamamoto, H., Miyake, C., and Yokota, A. (2003). Purification and characterization of class-I and class-II fructose-1, 6-bisphosphate aldolase from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 44, 326–333. doi: 10.1093/pcp/pcg044
Nouwens, A. S., Willcox, M. D. P., Walsh, B. J., and Cordwell, S. J. (2002). Proteomic comparison of membrane and extracellular proteins from invasive (PAO1) and cytotoxic (6206) strains of Pseudomonas aeruginosa. Proteomics 2, 1325–1346. doi: 10.1002/1615-9861(200209)2:9 < 1325
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pena, M. M., and Bullerjahn, G. S. (1995). The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J. Biol. Chem. 270, 22478–22482. doi: 10.1074/jbc.270.38.22478
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rai, S., Singh, S., Shrivastava, A. K., and Rai, L. C. (2013). Salt and UV-B induced changes in Anabaena PCC 7120: physiological, proteomic and bioinformatic perspectives. Photosynth. Res. 118, 105–114. doi: 10.1007/s11120-013-9931-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61. doi: 10.1099/00221287-111-1-1
Sen, A., Dwivedi, K., Rice, K. A., and Bullerjahn, G. S. (2000). Growth phase and metal-dependent regulation of the dpsA gene in Synechococcus sp. strain PCC 7942, USA. Arch. Microbiol. 173, 352–357. doi: 10.1007/s002030000153
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, G., Babele, P. K., Sinha, R. P., Tyagi, M. B., and Kumar, A. (2013). Enzymatic and non-enzymatic defense mechanisms against ultraviolet-B radiation in two Anabaena species. Proc. Biochem. 48, 796–802. doi: 10.1016/j.procbio.2013.04.022
Singh, S. P., Häder, D. P., and Sinha, R. P. (2010). Cyanobacteria and ultraviolet radiation (UVR) stress: mitigation strategies. Ageing Res. Rev. 9, 79–90. doi: 10.1016/j.arr.2009.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stewart, W. D. P. (1980). Some aspects of structure and function in N2 -fixing cyanobacteria. Annu. Rev. Microbiol. 34, 497–536. doi: 10.1146/annurev.mi.34.100180.002433
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vass, I., Szilard, A., and Sicora, C. (2005). “Adverse effects of UV-B light on the structure and function of the photosynthetic apparatus,” in Handbook of Photosynthesis, ed. M. Pessarakli (New York: Marcel Dekker Inc), 827–843.
Whitton, B. A., and Potts, M. (2000). The Ecology of Cyanobacteria: Their Diversity in Time and Space. Dordrecht: Kluwer Academic Publishers.
Keywords: UV-B radiation, cyanobacteria, Anabaena L31, 2-D gel electrophoresis, hypothetical proteins
Citation: Babele PK, Singh G, Kumar A and Tyagi MB (2015) Induction and differential expression of certain novel proteins in Anabaena L31 under UV-B radiation stress. Front. Microbiol. 6:133. doi: 10.3389/fmicb.2015.00133
Received: 11 November 2014; Accepted: 04 February 2015;
Published online: 24 February 2015.
Edited by:
Julie Anne Maupin-Furlow, University of Florida, USACopyright © 2015 Babele, Singh, Kumar and Tyagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashok Kumar, School of Biotechnology, Banaras Hindu University, Varanasi-221005, Uttar Pradesh, India e-mail: kasokbt@rediffmail.com
 Piyoosh K. Babele
Piyoosh K. Babele Garvita Singh
Garvita Singh Ashok Kumar
Ashok Kumar Madhu B. Tyagi
Madhu B. Tyagi