- Centre of Microbial and Plant Genetics, Department of Molecular and Microbial Systems, KU Leuven, Leuven, Belgium*
Post-transcriptional regulation is a very important mechanism to control gene expression in changing environments. In the past decade, a lot of interest has been directed toward the role of small RNAs (sRNAs) in bacterial post-transcriptional regulation. However, sRNAs are not the only molecules controlling gene expression at this level, RNA-binding proteins (RBPs) play an important role as well. CsrA and Hfq are the two best studied bacterial proteins of this type, but recently, additional proteins involved in post-transcriptional control have been identified. This review focuses on the general working mechanisms of post-transcriptionally active RBPs, which include (i) adaptation of the susceptibility of mRNAs and sRNAs to RNases, (ii) modulating the accessibility of the ribosome binding site of mRNAs, (iii) recruiting and assisting in the interaction of mRNAs with other molecules and (iv) regulating transcription terminator/antiterminator formation, and gives an overview of both the well-studied and the newly identified proteins that are involved in post-transcriptional regulatory processes. Additionally, the post-transcriptional mechanisms by which the expression or the activity of these proteins is regulated, are described. For many of the newly identified proteins, however, mechanistic questions remain. Most likely, more post-transcriptionally active proteins will be identified in the future.
Introduction
Bacteria need to survive in constantly changing environments. Therefore, they must be able to alter their gene expression in response to environmental signals, causing protein levels to be adjusted according to the needs of the cell. This can be achieved by adjusting transcription initiation with sigma factors and proteins that activate or repress transcription. However, gene expression regulation also occurs after transcription is initiated (Perez-Rueda and Martinez-Nuñez, 2012). The importance of these post-transcriptional regulatory processes is highlighted by the weak correlation that has been observed between RNA and protein abundance (Picard et al., 2009).
Prokaryotic post-transcriptional regulators typically modulate RNA decay, translation initiation efficiency or transcript elongation. Different types of prokaryotic post-transcriptional regulators have been identified, including small RNAs (sRNAs) and RNA-binding proteins (RBPs). sRNAs are typically defined as non-coding RNA molecules that bind with limited complementarity near the ribosome binding site (RBS) of their target mRNA, causing competition with the ribosome for binding to this region. However, the number of sRNAs that deviate from this general definition is increasing (Liu and Camilli, 2010; Storz et al., 2011). The new insights into the post-transcriptional mechanisms of sRNAs and their role in gene expression regulation were reviewed recently (Desnoyers et al., 2013). Here, RBPs involved in post-transcriptional regulation are discussed. For some of these proteins, the mechanism of action and the targets are well described, as for CsrA and Hfq. Their post-transcriptional function in Escherichia coli was already reported almost 20 years ago (Liu et al., 1995; Muffler et al., 1996). Lately, more insight was gained into the diverse mechanisms these two well-studied proteins use to regulate the expression of their target genes and how they regulate their own expression or activity in E. coli and in other bacteria. Additional RBPs involved in post-transcriptional regulation have been identified only recently and not much is known about their post-transcriptional function. In this review, the general working mechanisms of RBPs are discussed first. Afterward, examples of well-known and recently identified proteins, from E. coli and from other bacteria, are described.
General Mechanisms of Regulatory Proteins that Act Post-Transcriptionally
Bacterial post-transcriptionally active regulatory proteins typically bind RNA molecules and regulate translation initiation, stability, and transcript elongation of their RNA targets, using different regulatory mechanisms. These mechanisms include (i) adaptation of the susceptibility of the target RNAs to RNases, (ii) modulation of the accessibility of the RBS of mRNA targets for ribosome binding, (iii) acting as a chaperone for the interaction of the RNA target with other effector molecules, and (iv) modulation of transcription terminator/antiterminator structure formation, and will be described hereafter.
Adaptation of the Susceptibility to RNases
Regulation of RNA stability is an important mechanism to post-transcriptionally control gene expression, as it affects the number of mRNAs that can be translated or the number of sRNAs that can execute their regulatory function. RNA stability is determined by intrinsic RNA elements, such as primary sequence and secondary structure, but can be affected by sRNAs or proteins that bind to the RNA molecule. These proteins are mainly ribonucleases (RNases). In E. coli, single-stranded RNA-specific endoribonucleases (e.g., RNaseE and RNaseG) or double-stranded RNA-specific endoribonucleases (e.g., RNaseIII) generally initiate mRNA decay by making endoribonucleolytic cleavages. This yields smaller products that are further degraded by a combination of endo- and exonucleases, like PNPase (polynucleotide phosphorylase), RNaseII, or occasionally RNaseR (reviewed in Kaberdin et al., 2011). sRNAs are mainly degraded by RNaseE and PNPase, or by RNaseIII if the sRNA is hybridized to an mRNA target (reviewed in Saramago et al., 2014). In addition to RNases, other RBPs can play a role in the regulation of RNA stability by modulating the susceptibility of mRNAs and sRNAs to these RNases. Regulatory RBPs can act by directly shielding the recognition sites of RNases involved in the decay of RNA molecules if they have a shared binding preference, e.g., proteins that bind to single stranded AU-rich regions which are also recognized by RNaseE in E. coli (Moll et al., 2003). Other regulatory RBPs are involved in the regulation of RNA stability and induce a change in the secondary structure of their mRNA targets upon binding. Consequently, RNase recognition sites become buried or more exposed in locally formed structures, which positively or negatively affects the RNA stability of these molecules, respectively (Barria et al., 2013; see Figure 1).
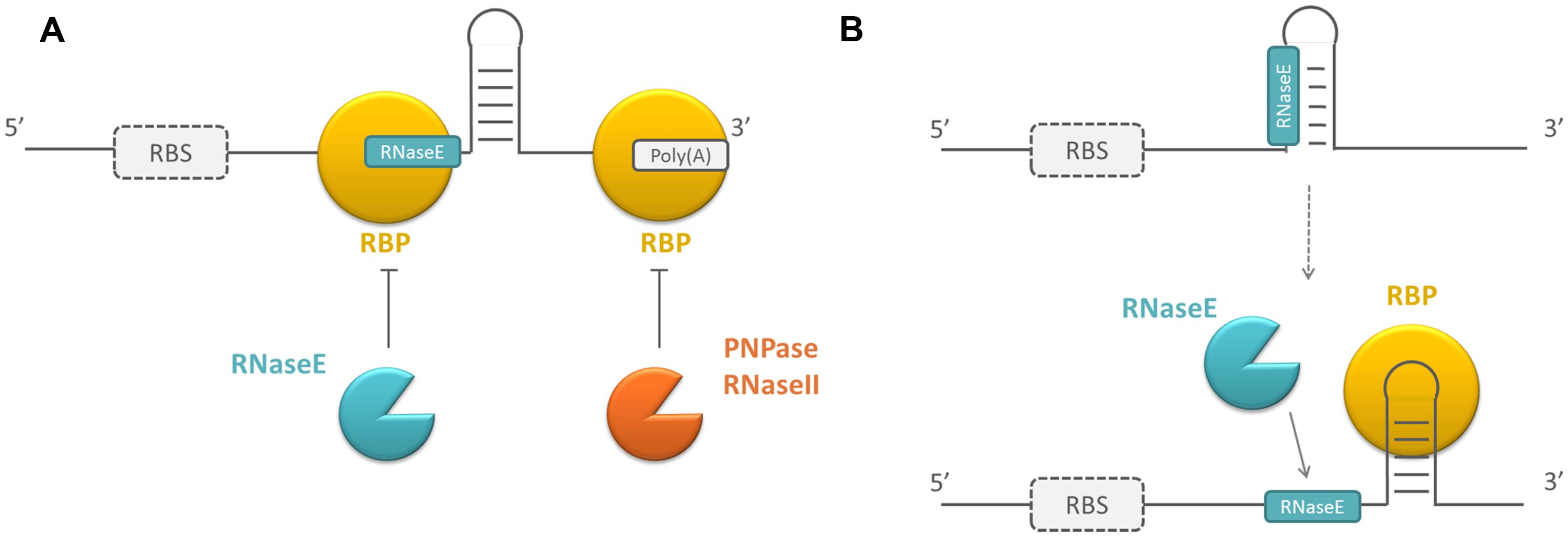
FIGURE 1. Schematic overview of the working mechanism of RNA-binding proteins (RBPs) influencing RNase susceptibility. (A) By directly blocking RNase recognition sites or (B) by changing the secondary structure of the mRNAs they bind. Because the targets of RBPs can be both mRNAs and small RNAs (sRNAs) and sRNAs are not translated, the ribosome binding site (RBS) is surrounded by a dotted line. RBPs are depicted in yellow, endoribonucleases in blue, exoribonucleases in orange.
Although RNases are the proteins that are mainly involved in RNA degradation, mRNA modifying enzymes can facilitate mRNA turnover as well. Pyrophosphate removal at the 5′ end by RppH (pyrophosphate hydrolase) and addition of a single stranded poly(A) extension at the 3′ end of the mRNA by PAPI [Poly(A) polymerase I] both promote mRNA degradation. Additionally, the exonucleolytic decay of highly structured mRNAs can be facilitated by RhlB, which unwinds RNA structures in an ATP-dependent way (reviewed in Kaberdin et al., 2011). E. coli regulatory RBPs can interfere with the poly(A)-assisted decay of mRNA molecules by binding to the poly(A) tail and protecting the mRNA to which it is bound from degradation (Folichon, 2003). Other regulatory RBPs facilitate mRNA degradation by recruiting RNases or RNA modifying enzymes, e.g., PAPI (De Lay et al., 2013). Because these proteins form a platform for the interaction of RNA molecules and proteins, their mechanism of action is assumed to be different and will be described later.
Many components of the mRNA decay machinery, like the RNases RNaseIII, PNPase, RNaseII, and RNaseR as well as the polymerase PAPI are well conserved across the bacterial kingdom (Kaberdin et al., 2011). This is not the case for the major endonucleases RNaseE/G in E. coli. However, functional homologs of RNaseE/G were identified, e.g., RNase J1/J2 or RNaseY in Bacillus subtilis (Even et al., 2005; Shahbabian et al., 2009). Therefore, it is likely that the regulatory mechanisms identified for regulatory RBPs influencing the susceptibility to degrading or modifying enzymes in E. coli are conserved in other bacteria.
Modulating RBS Accessibility
Besides their involvement in the regulation of RNA stability, RBPs can post-transcriptionally control gene expression by altering the efficiency of translation initiation. Translation initiation of an mRNA requires ribosome binding to the RBS of the mRNA. This RBS contains the Shine–Dalgarno sequence, which is a sequence complementary to the 3′ end of the 16S rRNA. This sequence is important for the recruitment and the correct positioning of the ribosome on the mRNA (Shine and Dalgarno, 1974). The more efficient the pairing between the Shine–Dalgarno region of the mRNA and the 16S rRNA, the more efficient ribosomes are recruited. Although the Shine–Dalgarno region is very important, the interaction region of an initiating ribosome is larger than the Shine–Dalgarno sequence alone and comprises nucleotides -20 to +19 relative to the start codon of mRNAs broadening the region that needs to be accessible in order for ribosome binding to occur (Beyer et al., 1994; Huttenhofer and Fnolier, 1994; Mackay et al., 2011; Desnoyers and Masse, 2012). Regulatory RBPs can modulate the efficiency of translation initiation by directly competing with ribosomes for binding to the ribosome interaction region or by initiating a change in the secondary structure of the mRNA sequence near this region (see Figure 2; Dubey et al., 2005; Baker et al., 2007; Irie et al., 2010). The resulting reduction in translation initiation efficiency often causes mRNA stability to be decreased as well. This can be explained by two mechanisms. Firstly, RNaseE can bind internally to a transcript (Mackie, 1998; Kime et al., 2010), but it can also interact with 5′ monophosphorylated transcripts with its 5′ binding pocket (Callaghan et al., 2005). When there is no ribosome bound, the mRNA is not protected from this kind of interaction with RNaseE. Secondly, when translation initiation efficiency is reduced, the spacing of the translating ribosomes on the mRNA is less compact. Consequently, it is more likely that RNase recognition sites in the mRNA become exposed causing transcript decay (Deana and Belasco, 2005).

FIGURE 2. Schematic overview of the working mechanisms of RBPs influencing the accessibility of the RBS. (A) By directly blocking this region and (B) by changing the secondary structure of the region surrounding the RBS. RBPs are depicted in yellow, the ribosomes in black, CDS, coding sequence.
Recruiting and Assisting in the Interaction with Other Molecules
RNA stability or translation initiation efficiency can also be affected by RBPs that form a platform to assist in the interaction of other molecules, which consequently affect RNA stability or translation efficiency (see Figure 3). The mechanism of action of these RBPs is described here as different compared to the previous two, because they typically bind simultaneously to an RNA target and an effector molecule. The effector molecule bound by the regulatory protein can be an sRNA or a protein. As previously mentioned, sRNAs typically regulate translation efficiency and RNA stability by binding near the RBS of their mRNA targets. Intermolecular basepairing between the sRNAs and mRNAs is facilitated by regulatory RBPs that function as a chaperone (Herschlag, 1995; Soper et al., 2011). The proteins recruited by regulatory RBPs can be proteins facilitating mRNA degradation, e.g., PAPI, RNases, or the degradosome (De Lay et al., 2013). The degradosome is a multiprotein complex in which different components cooperate during mRNA decay. Often, it contains RNaseE as a scaffolding protein and the protein partners PNPase, enolase, and RhlB. However, its assembly is not essential for RNA decay in E. coli (Carpousis, 2007). The recruitment of these proteins by other RBPs negatively affects transcript stability.
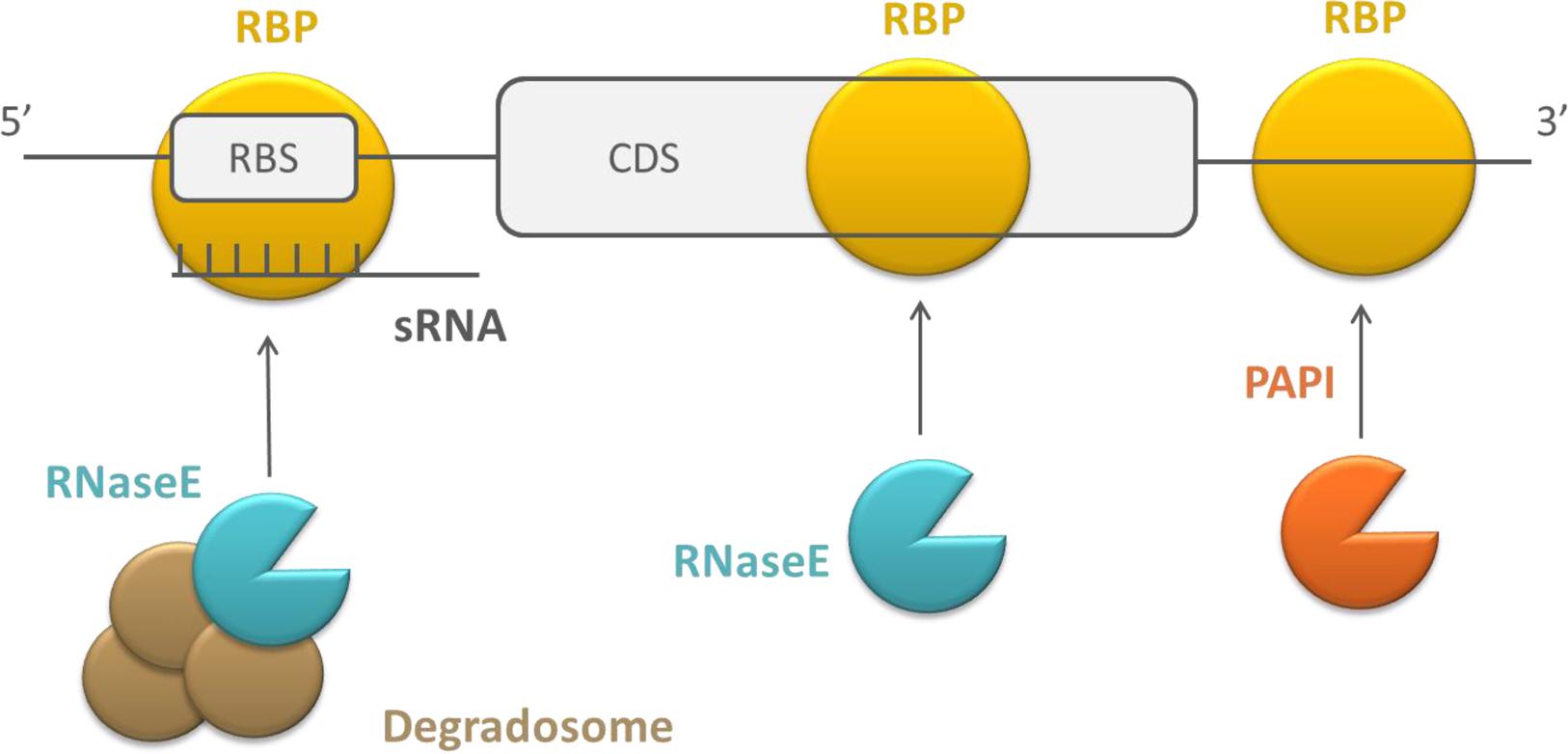
FIGURE 3. Schematic overview of the working mechanism of RBPs that recruit and assist in the interaction with sRNAs and proteins. RBPs are depicted in yellow, endoribonucleases in blue, exoribonucleases in orange, and auxiliary factors of the degradosome in brown, CDS, coding sequence.
Modulation of Transcription Terminator/Antiterminator Structure Formation
A last mechanism by which RBPs can post-transcriptionally affect gene expression is by modulating transcription elongation. After RNA polymerases initiate transcription, transcripts are elongated until a terminator is reached. There are two classes of terminators: intrinsic and factor-dependent terminators. At intrinsic terminators, dissociation of the elongation complex is dependent on the nucleic acid sequence and structure, while factor-dependent termination is dependent on the action of a protein factor, like the Rho-protein (Santangelo and Artsimovitch, 2011). Typically, these terminators are present at the end of the operon. However, some also exist within the 5′ leader region of the transcript. The presence of a terminator at this site prevents transcript elongation to full length. Premature termination can be abrogated by proteins that bind to the polymerase and allow transcription beyond the terminator signals or by the formation of an alternative secondary structure the enables transcription to progress (Stülke, 2002). In the latter process, RBPs can play a role. In the case of intrinsic termination, the RBPs can stabilize either the terminator structure or an alternative secondary structure, the antiterminator, which prevents the terminator form forming. Often the formation of both structures are mutually exclusive (Santangelo and Artsimovitch, 2011; see Figure 4). In general, the activity of this type of RBPs is controlled by their phosphorylation state or by a bound ligand, which induces major conformational changes in the proteins (Santangelo and Artsimovitch, 2011), although there are exceptions (Tortosa et al., 1997; Bachem and Stülke, 1998). Furthermore, RBPs can play a role in rho-dependent termination by inducing a secondary structure change, exposing a rho utilization (rut) sequence that is normally inaccessible for the rho-factor. Protein binding enables access to this region and rho-dependent transcription termination takes place (Figueroa-Bossi et al., 2014).
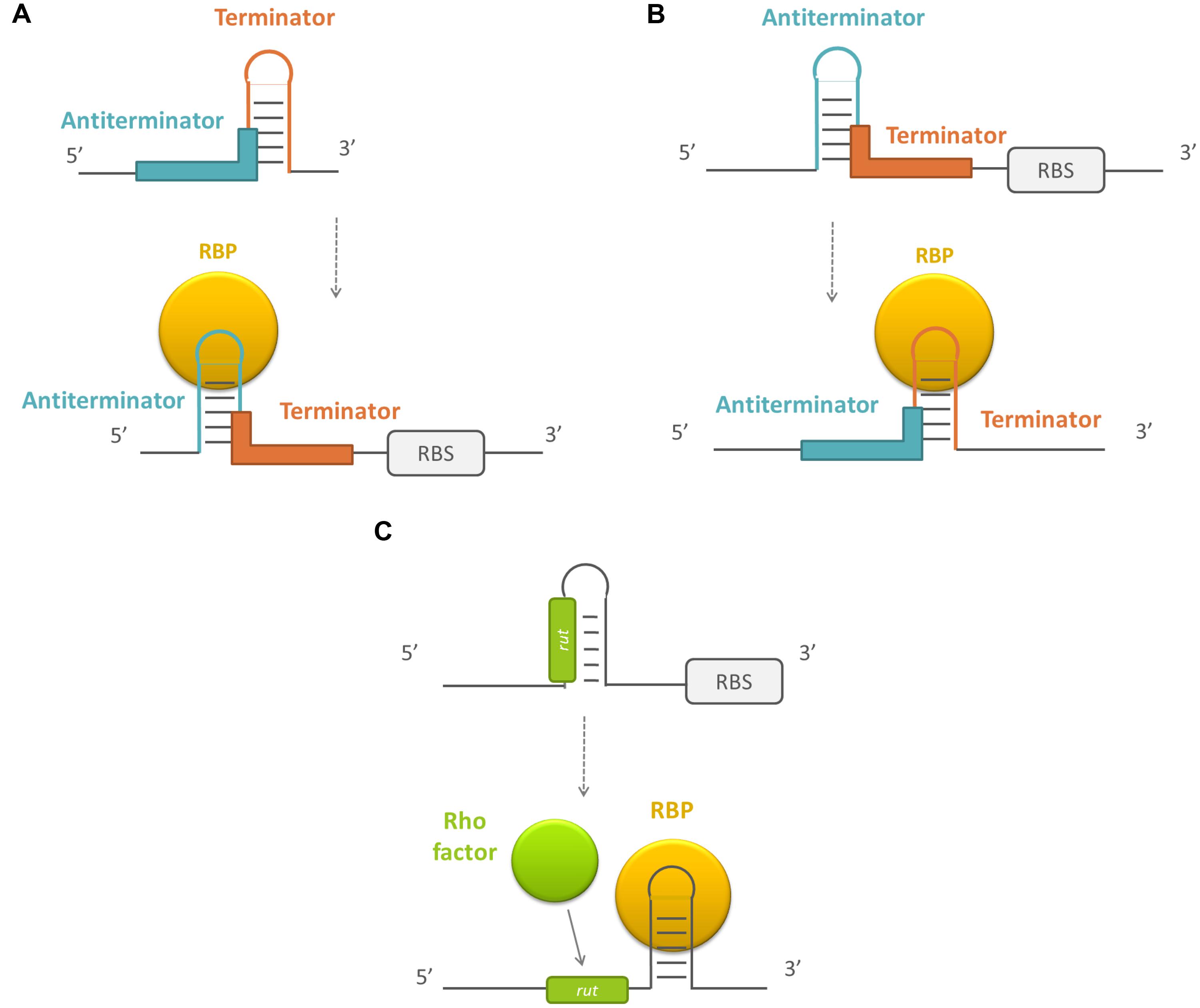
FIGURE 4. Schematic overview of the working mechanism of RBPs that modulate transcription terminator/antitermator structure formation. (A) By stabilizing the antiterminator structure, (B) by stabilizing the terminator structure, and (C) by exposing a rho utilization site. RBPs are depicted in yellow, the sequences forming the terminator in red, the sequences forming the antiterminator in blue, and the rho utilization sequences (rut) en rho factor in green. RBS, ribosome binding site.
Different RNA-Binding Proteins that Act Post-Transcriptionally in Bacteria
A number of proteins involved in post-transcriptional regulation have been identified in E. coli as well as in other bacteria. They are listed in Table 1. From this list, the global regulatory proteins CsrA and Hfq are best described in literature. CsrA works predominantly by competing with the ribosome for binding to the RBS of its mRNA targets. Hfq is best known for its role in assisting interacting sRNAs and mRNAs, but the protein applies a variety of other mechanisms to post-transcriptionally control gene expression. Besides Hfq and CsrA, there are other proteins that regulate the expression of their mRNA targets using similar working mechanisms. Moreover, additional RBPs have been identified that specifically regulate sRNA stability. For other RBPs affecting translation efficiency or RNA stability, the exact working mechanism is still unknown. Possibly they do use analogous mechanisms compared to those described for the well-known proteins.
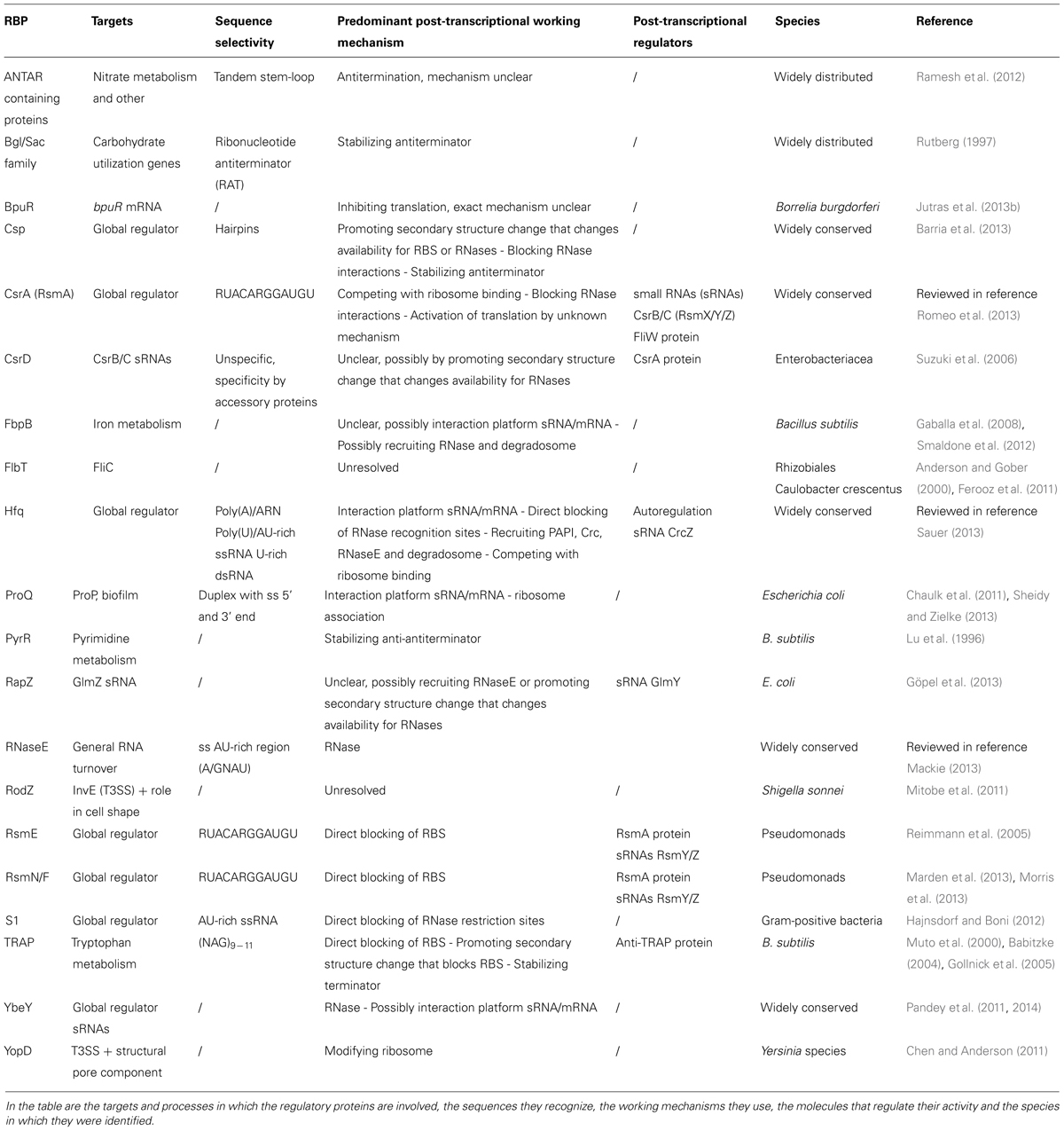
TABLE 1. Bacterial RNA-binding proteins (RBPs) that influence gene expression post-transcriptionally.
CsrA, an RNA-Binding Protein Predominantly Acting by Changing RBS Accessibility for Ribosomes
CsrA from E. coli and its orthologs in other bacteria, are RBPs that predominantly regulate gene expression by competing with the ribosome for binding to the RBS. CsrA is a widely conserved protein that has been annotated in over 1500 species (Finn et al., 2014). The protein is a global regulator, as illustrated by the changed expression level of approximately 10% of the genes in a csrA mutant in different bacteria (Lawhon et al., 2003; Burrowes et al., 2006; Brencic and Lory, 2009). In general, CsrA activates exponential phase functions and represses stationary phase processes (reviewed in Romeo et al., 2013). This is caused by direct and indirect regulatory events, as some CsrA targets encode regulatory proteins themselves (Jonas et al., 2008; Edwards et al., 2011).
To determine the selectivity of the CsrA protein, a SELEX experiment was conducted, enabling the identification of the RNA ligands binding with the highest affinity to CsrA. The CsrA-binding consensus sequence was shown to be RUACARGGAUGU; with the ACA and GGA being 100% conserved. Besides by the presence of this sequence in the mRNA target, CsrA specificity is additionally determined by the secondary structure of the target, as CsrA preferentially binds to RNA molecules that have the binding motif in a hairpin structure, with GGA in the loops of the hairpin (Dubey et al., 2005). The similarity between the CsrA recognition sequence and the consensus sequence of the Shine–Dalgarno region, i.e., AGGAGG, explains why there can be competition between CsrA and the ribosome for binding at this region. However, there are examples of CsrA targets that have CsrA binding sites that do not overlap with the Shine–Dalgarno region. In these cases, one of the CsrA binding sites overlaps with the translation initiation codon (Jonas et al., 2008) or the binding sites are solely present within the coding region (Yakhnin et al., 2011a). In the latter case, CsrA still competes with ribosome binding (Yakhnin et al., 2011a). Generally, the reduced translation initiation efficiency that results from CsrA binding to an mRNA target, leads to mRNA degradation as well (Baker et al., 2002; Dubey et al., 2003; Wang et al., 2005), although there are exceptions (Baker et al., 2007).
While the predominant regulatory mechanism of CsrA is direct competition with the ribosome for RBS binding, the protein can also use other mechanisms. One example is the binding of RsmA, the CsrA ortholog in Pseudomonas aeruginosa, to one of its targets, psl, which causes a stabilization of a hairpin structure in the region spanning the RBS, blocking the Shine–Dalgarno region and preventing ribosome binding (Irie et al., 2010). Secondly, binding of CsrA can have a positive effect on gene expression by blocking RNaseE interaction sites at the 5′ region of the mRNA of flhDC in E. coli which has a positive effect on RNA stability (Yakhnin et al., 2014). Thirdly, CsrA has been implicated recently in promoting the translation of the moaA mRNA. CsrA binding influences the structure of the moaA mRNA, however, this does not affect moaA mRNA levels. The exact mechanism of translational activation therefore remains to be unraveled. Remarkably, the moaA mRNA region that contains one of the CsrA binding sites, can also form a molybdenum cofactor (MOCO) binding riboswitch. It is unclear whether MOCO and CsrA can bind simultaneously or whether MOCO prevents CsrA from binding (Patterson-Fortin et al., 2013). Lastly, CsrA induces premature transcription termination of the pgaA mRNA in E. coli by unfolding a secondary structure sequestering an entry site for transcription terminator factor Rho thereby regulating transcription elongation (Figueroa-Bossi et al., 2014). Although CsrA is predominantly involved in post-transcriptional regulation, the protein has recently been shown to affect transcription as well. In P. protegens, RsmA represses lipA transcription, although the mechanism remains unclear (Zha et al., 2014). Remarkably, lipA is additionally regulated by RsmE, which is one of the paralogs of RsmA in this species. RsmE blocks ribosome access by binding to the RBS of lipA.
Different Pseudomonas species indeed have three non-identical copies of RsmA, the CsrA ortholog in these bacteria. Although the expression profile of RsmE is slightly different from the one of RsmA, both proteins function in a largely redundant way (Reimmann et al., 2005). Another RsmA paralog is RsmN (also called RsmF). RsmN has a different structural organization of α-helices and β-sheets compared to RsmA and RsmE, but the tertiary structure is similar. This results in a conserved spatial organization of key residues within the dimeric structure, which is necessary for RNA-hairpin recognition (Marden et al., 2013; Morris et al., 2013). RsmA can bind to the mRNAs of its paralogs, thereby negatively influencing RsmE and RsmN protein expression (Reimmann et al., 2005; Marden et al., 2013). Altogether, these elements indicate that these proteins have a unique but overlapping regulatory role compared to RsmA. It has been suggested that variations in sequence, structure, RNA-binding affinities and specificities between these different paralogs facilitate tight gene-specific control at the global post-transcriptional level for Pseudomonads. The question remains why E. coli does not possess this wide array of CsrA paralogs (Morris et al., 2013).
Hfq Uses Different Mechanisms to Post-Transcriptionally Regulate Target Gene Expression
Hfq is another very well-studied post-transcriptional regulator. The protein is widespread but not ubiquitously present throughout the bacterial kingdom (Sobrero and Valverde, 2012). In general, a knockout in hfq reduces the fitness of bacteria to survive in stressful environments (Tsui et al., 1994; Christiansen et al., 2004; Liu et al., 2010; Wang et al., 2014). Its role in RNA metabolism is more limited in Gram-positive bacteria compared to Gram-negative bacteria, which is illustrated by the fact that an hfq deletion does not have the same global effect on the transcriptome of Bacillus, like it has on the transcriptome of E. coli or Salmonella (Hämmerle et al., 2014).
Hfq has three different sites that can bind RNA: the distal, proximal and lateral site (Sauer, 2013). The binding preferences of these different RNA-binding sites are probably not that strict, as a study performed in Listeria monocytogenes shows that RNA-binding sites of Hfq have the potential to bind a wider variety of RNA sequences than was previously thought (Kovach et al., 2014). However, in E. coli, the distal site of Hfq does have a preference for repetitions of ARN-triplets or poly(A) stretches (Mikulecky et al., 2004; Link et al., 2009; Lorenz et al., 2010). The proximal site, on the other hand, has a preference for AU-rich single stranded sequences or poly(U) stretches (Schumacher et al., 2002; Moll et al., 2003). Finally, the lateral site of Hfq binds U-rich sequences and double stranded elements (Sauer et al., 2012), although the function of this lateral site in RNA-binding is controversial (Sauer, 2013). Some of the Hfq binding specificities overlap with the binding preferences of certain RNases. Therefore, Hfq can influence the RNase susceptibility of an mRNA or an sRNA. The AU-rich binding preference of the proximal binding site of Hfq, for example, is similar to the sequence that is recognized by RNaseE. Therefore, Hfq and RNaseE can compete for binding to the same region, reducing RNA decay (Massé et al., 2003; Moll et al., 2003; Mohanty et al., 2004). In the same way, Hfq plays a role in poly(A)-assisted RNA degradation. With its distal binding site, Hfq can bind to the poly(A) tail of mRNAs. Consequently, this region becomes inaccessible for exonucleases, like PNPase and RNaseII. Hfq binding at the poly(A) tail also impairs RNaseE processing. Both processes increase mRNA stability. However, binding of Hfq can also promote polyadenylation and thus promote poly(A)-assisted decay (Hajnsdorf and Régnier, 2000; Le Derout, 2003; Régnier and Hajnsdorf, 2013). Altogether, these mechanisms indicate how Hfq is important for the stability of sRNAs and mRNAs. Although Hfq is present at high levels in the cell, there is not enough Hfq to stabilize all sRNAs and mRNAs. Therefore, there is constant competition amongst sRNAs and mRNAs for Hfq binding (Vogel and Luisi, 2011).
Next to its role in regulating RNA stability, Hfq can function as a chaperone to stimulate the interaction of mRNAs and sRNAs (Møller et al., 2002; Zhang et al., 2002; Geissmann and Touati, 2004; Rasmussen et al., 2005; Kawamoto et al., 2006). Because there are different RNA-binding sites present in a hexameric Hfq molecule, it is possible that an sRNA and an mRNA are simultaneously bound to one Hfq molecule. Such binding to Hfq brings sRNAs and mRNAs in close proximity, enhancing the likelihood of interaction (Soper et al., 2011). However, cobinding of sRNAs and mRNAs to Hfq is transient and insufficient for sRNA-dependent regulation (Hopkins et al., 2011). Therefore RNA restructuring is also an important function of Hfq in this process (Henderson et al., 2013). The protein can change the secondary structure of the RNA molecules, making some regions in the mRNA more accessible for base pairing (Soper et al., 2011).
In addition to assisting sRNA/mRNA interactions, Hfq can also form a platform for the interaction of these RNA molecules with other proteins, e.g., RNaseE. This RNA/protein complex can then further interact with the other subunits of the RNaseE-based degradosome, causing degradation of the mRNA, and often also of the sRNA, in the complex (Morita et al., 2005; Aiba, 2007). The interaction between Hfq and RNaseE is most likely a combination of direct protein interactions between RNaseE and Hfq, which occur at the RhlB recognition region of RNaseE in the canonical degradosome (Ikeda et al., 2011), and indirect interactions via the RNA molecules they bind (De Lay et al., 2013). Hfq can indirectly interact in this way with a number of other proteins, like the cold shock protein CspC in E. coli and RsmA in P. aeruginosa, a protein that was discussed previously in this review (Sorger-Domenigg et al., 2007; Cohen-Or et al., 2010). Another protein that may be recruited by Hfq is Crc, a protein involved in catabolite repression control in P. fluorescens and P. aeruginosa. Crc was originally identified as an RBP, able to bind short unpaired A-rich motifs (AAnAAnAA) at or near the RBS, thereby inhibiting translation initiation (Moreno et al., 2009; Sonnleitner et al., 2009; Browne et al., 2010). However, a recent study shows that Crc has no RNA-binding capacity and previous results on RNA-binding rely on contaminations of Crc protein samples with Hfq (Milojevic et al., 2013). Hfq and Crc are now assumed to cooperate for binding to RNAs that contain an A-rich motif, because both proteins form a cocomplex and are both necessary for catabolite repression. Possibly Hfq recruits Crc or Crc modifies Hfq in such a way that it can more efficiently bind to the A-rich stretches with its distal site, preventing translation (Moreno et al., 2014).
Next to its functions as a (de)stabilizing factor for RNA molecules and as an interaction platform for RNA/RNA an RNA/protein interactions, Hfq carries out other functions. One of these functions is competing with initiating ribosomes for access to the RBS by binding AU-rich regions close to the RBS which are acting as translational enhancers (Vytvytska et al., 2000; Desnoyers and Masse, 2012). These translational enhancers facilitate the interaction of an mRNA with protein S1 near the RBS, but this interaction is impaired when Hfq is bound. S1 is a protein, weakly associated to the 30S subunit of the ribosome that facilitates the recognition of mRNAs by ribosomes at the initial step of translation (Subramanian, 1983). Sometimes, sRNAs are involved in this regulatory process. Spot42, for example, recruits Hfq at the enhancer region (Desnoyers and Masse, 2012). In another case, Hfq binding takes place without an sRNA as a recruiting molecule. Oppositely, binding of the sRNA RyhB can prevent Hfq from binding to the enhancer region (Salvail et al., 2013). In addition, Hfq has a role in transcription regulation, by inhibiting the function of the Rho protein, which is involved in transcription termination (Le Derout et al., 2010; Rabhi et al., 2011).
While CsrA and Hfq use different mechanisms to globally regulate gene expression, there are proteins that have only been shown to use a confined number of these mechanisms or regulate a more limited number of genes. They will be described hereafter.
RNA-Binding Proteins Adapting the Susceptibility to RNases
In addition to its role in facilitating ribosome interactions with the mRNA, protein S1 also has a function in post-transcriptional regulation. The protein can stabilize RNA molecules by directly shielding RNase recognition sites (Hajnsdorf and Boni, 2012). Although S1 has no strict sequence specificity, it does have a higher affinity for AU-rich mRNA sites. As RNaseE preferentially binds AU-rich single stranded regions as well, S1 can shield RNaseE recognition sites and protect mRNAs against cleavage (Komarova et al., 2005). Because S1 is capable of binding to sRNAs with the same affinity as Hfq, it has been suggested that S1 can theoretically regulate sRNA stability as well. However, the biological relevance of this suggestion still needs to be elucidated (Koleva et al., 2006; Windbichler et al., 2008).
S1 directly competes with RNases for binding to specific sequences. However, other proteins affect mRNA stability by promoting a change in the secondary structure of their mRNA targets. Consequently, RNase recognition sites become more or less available for RNases. An example of proteins that use this mechanism are cold shock proteins. These proteins are induced when bacteria encounter a temperature downshift. One of the physiological effects of cold is the stabilization of secondary structures that make RNase recognition sites inaccessible, which likewise impairs RNA degradation. Binding of the cold shock protein, CspA, to these mRNAs causes, together with the induced helicases, cold-induced secondary structures in the mRNA to be melted. Adversely, cold shock proteins can also prevent RNA degradation. CspE can bind poly(A) sites and can consequently interfere with either binding of PNPase or with internal cleavage by RNaseE (Feng et al., 2001). Additionally, cold shock proteins can assist in unwinding of secondary structures that sequester the RBS, which enhances translation efficiency (Barria et al., 2013).
RNA-Binding Proteins that Affect sRNA Stability
Hfq is very well known for its role in regulating sRNA and mRNA stability. However, recently, other RBPs were identified that regulate the stability of specific sRNAs. A first example is CsrD (Suzuki et al., 2006). In E. coli, this protein is involved in the turnover of the sRNAs CsrB and CsrC. These sRNAs regulate the activity of CsrA, which is an RBP that was described earlier. Although CsrD destabilizes the sRNAs CsrB and CsrC in an RNaseE-dependent way, these sRNAs have no obvious RNaseE recognition sites and CsrD has no RNase activity itself. Therefore, it was suggested that CsrD might induce structural changes in the sRNA, making it more susceptible for RNaseE (Suzuki et al., 2006). CsrD does not bind specifically to CsrB or CsrC, however, the action of CsrD seems to be specific. This indicates that there are additional factors in the cell that determine the specificity of the process (Suzuki et al., 2006). A second example of an RBP specifically regulating sRNA stability is RapZ. RapZ is a protein identified in E. coli that functions as an adaptor protein guiding the processing of the sRNA, GlmZ. The protein has been reported to recruit RNaseE to the sRNA. It has been hypothesized that this occurs through changing the structure of the sRNA so it can be recognized by RNaseE, or by functioning as an interaction platform by delivering the sRNA to RNaseE (Göpel et al., 2013). Most likely, there are more proteins that bind sRNAs and target them for degradation, just like RapZ and CsrD.
RNA-Binding Proteins that Modulate RBS Accessibility
TRAP (trp RNA-binding attenuation protein), a protein involved in the regulation of tryptophan metabolism of B. subtilis, acts through modulating RBS accessibility for ribosomes. The protein specifically binds multiple (9–11) NAG repeats, separated by non-conserved spacers. Because of the extended recognition sequence, TRAP can regulate a small subset of genes, all involved in tryptophan metabolism. TRAP can act as a post-transcriptional regulator by directly blocking ribosome access to the RBS (Babitzke et al., 1994; Yang et al., 1995; Du et al., 1997; Sarsero et al., 2000; Yakhnin et al., 2004). Although the protein binds multiple NAG repeats, it is sufficient that one repeat overlaps with the RBS to block ribosome access to this region (Babitzke et al., 1994; Babitzke et al., 1995). TRAP can also promote a change in secondary structure which influences the availability of the RBS for a subset of genes (Merino et al., 1995; Du and Babitzke, 1998). Additionally, the protein is involved in the regulation of transcription elongation, which will be described later.
The regulatory RBP BpuR from Borrelia burgdorferi possibly acts by competing with ribosome binding as well, as the protein has been shown to bind to the 5′ region of its mRNA target, thereby blocking translation. However, the only identified RNA target of this protein this far is its own mRNA. Most likely, BpuR is a post-transcriptional regulator of other genes in the Borrelia genome (Jutras et al., 2013b). BpuR can also act as a DNA binding protein, but it binds RNA with higher affinity (Jutras et al., 2013a).
Proteins Acting as an Interaction Platform with Other Molecules
YbeY is a widely conserved protein, known to influence the maturation of rRNAs and to be involved in the quality control of 70S ribosomes (Davies et al., 2011; Jacob et al., 2014). Although it is an essential RNase in some bacteria like Vibrio cholera, the protein is not essential and has weak RNase activity in E. coli and Sinorhizobium meliloti (Vercruysse et al., 2014). In these bacteria, both in silico and phenotypic indications exist, supporting the hypothesis that YbeY plays a role in sRNA regulation, although YbeY has not yet been shown to actually bind sRNAs in vivo in these species. In silico analyses show that YbeY displays high sequence and structural similarities to MID domains of Argonaut proteins, the central component of sRNA-mediated gene silencing in eukaryotes. Argonaut proteins bind sRNAs that function as sequence-specific guides to lead the Argonaut proteins to perfectly or partially complementary sequences (Mallory and Vaucheret, 2010). Additionally, structural models assigned a probable RNA-binding site for YbeY. Phenotypically, there are striking similarities between an smc01113 mutant, the ybeY ortholog in S. meliloti, and an hfq mutant (Pandey et al., 2011). Mutated YbeY indeed causes an increased sensitivity to various stresses, similarly to when Hfq is mutated. Moreover, YbeY modulates the levels of both already identified Hfq-dependent and Hfq-independent sRNAs and their targets in E. coli (Pandey et al., 2014), which suggests that YbeY has a central role in RNA metabolism. The exact working mechanism of the YbeY protein remains unknown. Besides the suggested role in the interaction of mRNAs and sRNAs, YbeY might still have a catalytic role as an RNase as a functional equivalent of RNaseE (Vercruysse et al., 2014).
While YbeY may play a central role in the general sRNA metabolism, there are some RBPs that form a platform of interaction for a more limited number of sRNAs. One example is the FinO family of bacterial chaperones. FinO is involved in the regulation of gene expression from the F-plasmid by facilitating the interaction between the sRNA FinP and the mRNA of the F plasmid transcription factor traJ. The protein facilitates sRNA-mRNA interactions by destabilizing internal hairpins in target RNAs (Arthur et al., 2003) and protects them from RNaseE (Jerome et al., 1999). However, contrary to Hfq, FinO does not simultaneously bind mRNA and sRNA molecules (Chaulk et al., 2010). Other proteins assigned to this FinO-family include NMB1681 in Neisseria meningitides and ProQ in E. coli. Although NMB1681 has been shown to restore phenotypes in an E. coli finO mutant, the role of this protein in Neisseria has not been unraveled (Chaulk et al., 2010). ProQ has an N-terminus homologous to FinO and a C-terminus homologous to the C-terminus of Hfq. Both the N- and the C-terminus facilitate sRNA-mRNA interactions. However, the Hfq-like domain most likely assists in sRNA–mRNA interaction, while the FinO-like domain confers sequence-specific properties to the protein (Chaulk et al., 2011; Sheidy and Zielke, 2013). ProQ also associates with the ribosome, which appears to be mediated by an interaction between ProQ and its targets being translated (Sheidy and Zielke, 2013).
A second example of an RBP possibly involved in assisting a limited number of sRNA–mRNA interactions is FbpB. FbpB is a small protein involved in the regulation of iron metabolism in B. subtilis. The protein is suggested to function as a coregulator of the translational repressor FsrA, by targeting the sRNA FsrA to specific transcripts and increasing the effectiveness of the sRNA. The phenotype of an fbpB mutant can indeed be restored by an upregulation of FsrA. FbpB is possibly involved in the recruitment of the degradation machinery of B. subtilis as well, thereby causing degradation of the sRNA–mRNA complex (Smaldone et al., 2012). Although this is still speculative, this would be consistent with the functions of Hfq in E. coli.
RNA-Binding Proteins that Modulate Transcription Terminator/Antiterminator Formation
TRAP and cold shock proteins have been described earlier as they can induce a change in the secondary structure of their RNA target which modulates RNA stability or translation initiation efficiency. Additionally, these proteins can modulate transcription elongation by stabilizing a transcription terminator or antiterminator structure upon binding. TRAP, activated by tryptophan, binds to the Bacillus trp 5′ leader transcript and occludes the formation of the antiterminator. This antiterminator is located upstream of a terminator and the formation of both structures is mutually exclusive. Thereby, TRAP binding enables the terminator to be formed and transcription is prematurely stopped (Babitzke, 2004). The cold shock proteins CspA and homologs CspC and CspE from E. coli, work in the opposite way. These proteins prevent the formation of transcription terminators by stabilizing an antiterminator structure (Bae et al., 2000; Phadtare et al., 2002).
Another example of RBPs that stabilize an antiterminator structure upon binding is the Bgl/Sac family. These proteins are widely distributed and recognize a 23–30 nucleotide stretch called ribonucleotide antiterminator (RAT) that partially overlaps with the terminator sequence (Aymerich and Steinmetz, 1992). Similarly to cold shock proteins, binding of an RBP of the Bgl/Sac family causes this antiterminator region to fold in a stem-loop structure that occludes the formation of the terminator. The Bgl sytem of E. coli was the first mechanism that involves protein-mediated antitermination (Mahadevan and Wright, 1987; Schnetz and Rak, 1988). Members of this family have been identified in different bacteria such as E. coli, B. subtilis, Lactococcus lactis, and Erwinia chrysanthemi. They control the expression of genes required for the utilization of carbohydrates (Rutberg, 1997).
Lastly, PyrR of B. subtilis is involved in modulating transcription terminator/antiterminator formation, although its mechanism of action is a little different compared to the examples described above. PyrR, activated in the presence of uridine, stabilizes an anti-antiterminator structure. This structure sequesters nucleotides of the antiterminator by basepairing with sequences that lie further upstream, inducing terminator formation and preventing gene expression. Only in the absence of uridine the antiterminator can form and expression of the RNA target is possible (Lu et al., 1996).
Post-Transcriptionally Active Regulatory Proteins with an Unknown Mechanism
Other post-transcriptionally active regulatory proteins have been identified. However, often their exact mechanism of action remains unclear. For example, YopD is a component of the type III secretion system (T3SS) of Yersinia species and is conserved in pathogens with a T3SS (Schiano and Lathem, 2012). It translocates virulence factors across the cell membrane of the host. Simultaneously, YopD post-transcriptionally regulates genes of the T3SS directly and specifically in complex with LcrH, a secretion chaperone (Schiano and Lathem, 2012). The specificity of YopD is thought to be based on the interaction of the protein with short AU-rich sequences, both up- and downstream of the start codon of the target genes. However, the interaction is more complex than YopD/LcrH and AU sequences alone. Other interaction partners are involved. The post-transcriptionally active complex of YopD, LcrH and other interacting components binds to the 5′UTR of its targets and represses their translation, but the mechanism remains unclear. YopD has been suggested to facilitate degradation of these targets by directly competing with ribosome binding or promoting degradation (Chen and Anderson, 2011). However, a recent report shows that YopD affects translation by modifying the ribosome itself (Kopaskie et al., 2013). It is remarkable that a protein with a structural function has a regulatory role as well.
In Shigella sonnei, RodZ was identified as a membrane-localized cytoskeletal protein that retains the rod-shaped morphology of the bacterium. Later, this protein was also shown to be involved in post-transcriptional regulation, as the protein has RNA-binding capacity and its expression leads to repression of InvE protein synthesis by means of a decreased stability of the mRNA. However, the working mechanism still needs to be resolved. Possibly it forms a platform where mRNAs and other putative regulatory factors coincide (Mitobe et al., 2011).
FlbT has been identified in Caulobacter crescentus and Brucella melitensis, where it is proposed to bind to the 5′UTR of the fliC mRNA. However, it is not clear how it post-transcriptionally regulates gene expression. In Caulobacter binding of FlbT promotes degradation of its mRNA targets, whereas in Brucella FlbT is proposed to be an activator of gene expression (Anderson and Gober, 2000; Ferooz et al., 2011).
The family of AmiR and NasR transcriptional antiterminator regulator (ANTAR) domain proteins is a last example of RBPs acting at the post-transcriptional level with an undefined regulatory mechanism. They are widely distributed among different species and are involved in transcription antitermination. These proteins recognize an RNA motif consisting of two tandem stem-loops. However the exact molecular mechanism of antitermination has not been determined (Ramesh et al., 2012). For NasR of Klebsiella oxytoca, it has been suggested that it does not involve the formation of an antitermination structure (Chai and Stewart, 1999).
Post-Transcriptional Regulation of Post-Transcriptionally Active Regulatory Proteins
To ensure that gene expression is adjusted according to the needs of the cell, it is crucial that the expression and the activity of the regulatory RBPs themselves are tightly controlled as well. Remarkably, protein expression or protein activity is regulated itself by post-transcriptional regulatory processes. Proteins that modulate RBS accessibility, like CsrA and BpuR, have their recognition site present in the 5′UTR of their own mRNA. Protein binding to the mRNA consequently competes with ribosome binding, reducing translation initiation efficiency and the expression of the protein (Yakhnin et al., 2011b; Jutras et al., 2013b). Similarly, Hfq expression is autoregulated. The protein inhibits ribosome binding to its own mRNA, making it at the same time more vulnerable for cleavage by RNaseE. Two Hfq binding sites were identified upstream of the start codon, with one overlapping with the RBS. However, the other binding site is also necessary for translational repression together with a hairpin structure in the coding region. These elements possibly function as stabilizing elements for RNA/protein interaction (Vecerek et al., 2005).
Secondly, the activity of different RBPs is regulated with sRNAs. These sRNAs carry multiple high affinity sequences that are specifically recognized by a regulatory RBP. Binding of the sRNA sequesters the protein, resulting in a lower number of proteins available for binding to its mRNA targets. The best studied examples of sRNAs that regulate protein activity by mimicking the protein binding sequence are the sRNAs CsrB and CsrC (Liu et al., 1997; Weilbacher et al., 2003). They bind to the global regulator CsrA, which was discussed previously. CsrB and CsrC carry 18 and 9 CsrA binding sequences, respectively. Recently, another E. coli sRNA, McaS, was shown to bind CsrA. This sRNA has at least two CsrA binding sites (Jørgensen et al., 2013). The different sRNA molecules that regulate CsrA activity are differentially expressed in some conditions. Consequently, the activity of the RBP can be regulated in response to different environmental conditions (Jørgensen et al., 2013). Remarkably, CsrA activity is not only regulated by sRNAs. In Salmonella Typhimurium, for example, the fim mRNA can inhibit CsrA function as well. This mRNA carries the CsrA recognition sequence but the stability or the translation of fim mRNA is not affected by CsrA binding, excluding it from being a regulated CsrA target (Sterzenbach et al., 2013).
While CsrA is the best studied example of a protein that is regulated by sRNAs, more of them are known. The Hfq protein, which was extensively discussed above, is regulated by the sRNA CrcZ in E. coli. The sRNA has multiple A-rich stretches to which Hfq can potentially bind with its distal RNA-binding region. As Hfq has other RNA-binding sites it is possible that the protein can still bind and regulate other RNA molecules when CrcZ is bound to this distal site. However, this remains to be resolved (Sonnleitner and Bläsi, 2014). A last example is RapZ, which is sequestered by binding to the sRNA GlmY. This sRNA resembles the mRNA target of RapZ, i.e., GlmZ, by a conserved central stem-loop structure (Göpel et al., 2013). This shows that not only the global regulators, like CsrA and Hfq, are regulated through sRNA mimicry.
Although the activity of some RBPs is regulated by sRNAs, the activity of others is still regulated by proteins. This is the case for the TRAP protein, which is regulated by an anti-TRAP protein that binds near the RNA-binding pocket of the TRAP protein, preventing TRAP from binding to its mRNA targets (Snyder et al., 2004). CsrA from B. subtilis is another RBP that is regulated by a protein, FliW. This antagonistic protein binds near the active site of the protein. Remarkably, a CsrB-like sRNA has also been identified in B. subtilis (Mukherjee et al., 2011). Its regulatory role and the importance compared to FliW has not been unraveled yet (Kulkarni et al., 2006).
Conclusion and Perspectives
Regulation of gene expression is very complex and takes place at different regulatory levels. At the post-transcriptional level, RBPs are important gene expression regulators. These proteins generally act by adapting RNA stability and mRNA translation efficiency. Hereto, they use different mechanisms, including (i) adaptation of the susceptibility of the target RNAs for RNases, (ii) modulation of the accessibility of the RBS for ribosome binding, (iii) acting as a chaperone for the interaction of the RNA target with other effector molecules, and (iv) modulation of transcription terminator/antiterminator formation. Different post-transcriptionally active RBPs have been identified in E. coli and in other bacteria, with Hfq and CsrA being the two best studied examples. These proteins apply different mechanisms to regulate the expression of their target genes. Recently, additional RBPs that affect mRNA and/or sRNA stability or translation efficiency have been identified. Although many mechanistic questions remain, they use mostly similar regulatory mechanisms to regulate the expression of their targets. Additionally, they are post-transcriptionally regulated themselves through autoregulation and regulation by sRNA mimicry.
A number of post-transcriptionally active RBPs have been identified. However, it is very likely that more RBPs are active as post-transcriptional regulators, given that some known regulatory RBPs have another function, e.g., as a transcription regulator or a structural protein. Moreover, proteins that specifically regulate sRNA stability have only recently been discovered. Therefore, most likely, more proteins of this class will be identified in the future. Additionally, relatively little is known about this type of regulatory proteins in other bacteria than E. coli. Therefore, methods have been developed to identify new RBPs. One of these methods is the in vitro or in vivo assembly of RNA and RBPs, followed by mass spectrometry (Tsai et al., 2011). This method has already been optimized for Helicobacter pylori, E. coli, Salmonella Typhimurium and P. aeruginosa (Windbichler et al., 2008; Said et al., 2009; Rieder et al., 2012; Osborne et al., 2014).
Further studies on well-known and newly identified post-transcriptionally active proteins will lead to a better understanding of how bacteria use this type of gene regulation to respond to changes in the environment and how different post-transcriptional networks interact with transcriptional regulons and with each other. This knowledge will create opportunities for new or improved biotechnological applications, e.g., in synthetic biology as a tool to control gene expression, complementing the current approaches of transcription control. Additionally, as many of these proteins play a central role in RNA metabolism, interfering with the expression or the function of these proteins can be interesting as an alternative antimicrobial strategy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the funding received by the Centre of Excellence SymBioSys (Research Council K.U.Leuven EF/05/007), the Strategic Basic Research of the Institute for the Promotion of Innovation through Science and Technology in Flanders under grant IWT-SBO 120050 (NEMOA) and the FWO [Fund for Scientific Research – Flanders (Belgium)] through a postdoctoral fellowship to HS.
References
Aiba, H. (2007). Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10, 134–139. doi: 10.1016/j.mib.2007.03.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anderson, P. E., Gober J. W. (2000). FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5’ untranslated region of flagellin mRNA. Mol. Microbiol. 38, 41–52. doi: 10.1046/j.1365-2958.2000.02108.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arthur, D. C., Ghetu, A. F., Gubbins, M. J., Edwards, R. A., Frost, L. S., and Glover, J. N. M. (2003). FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 22, 6346–6355. doi: 10.1093/emboj/cdg607
Aymerich, S., and Steinmetz, M. (1992). Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG / SacY family. Proc. Natl. Acad. Sci. U.S.A. 89, 10410–10414. doi: 10.1073/pnas.89.21.10410
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Babitzke, P. (2004). Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr. Opin. Microbiol. 7, 132–139. doi: 10.1016/j.mib.2004.02.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Babitzke, P., Bear, D. G., and Yanofsky, C. (1995). TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc. Natl. Acad. Sci. U.S.A. 92, 7916–7920. doi: 10.1073/pnas.92.17.7916
Babitzke, P., Stults, J. T., Shire, S. J., and Yanofsky, C. (1994). TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J. Biol. Chem. 269, 16597–16604.
Bachem, S., and Stülke, J. (1998). Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J. Bacteriol. 180, 5319–5326.
Bae, W., Xia, B., Inouye, M., and Severinov, K. (2000). Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U.S.A. 97, 7784–7789. doi: 10.1073/pnas.97.14.7784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baker, C. S., Eöry, L. A., Yakhnin, H., Mercante, J., Romeo, T., and Babitzke, P. (2007). CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J. Bacteriol. 189, 5472–5481. doi: 10.1128/JB.00529
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baker, C. S., Morozov, I., Suzuki, K., Romeo, T., and Babitzke, P. (2002). CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44, 1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barria, C., Malecki, M., and Arraiano, C. M. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. doi: 10.1099/mic.0.052209-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beyer, D., Skripkin, E., Wadzack, J., and Nierhaus, K. H. (1994). How the ribosome moves along the mRNA during protein synthesis. J. Biol. Chem. 269, 30713–30717.
Brencic, A., and Lory, S. (2009). Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632. doi: 10.1111/j.1365-2958.2009.06670.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Browne, P., Barret, M., O’Gara, F., and Morrissey, J. P. (2010). Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 10:300. doi: 10.1186/1471-2180-10-300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burrowes, E., Baysse, C., Adams, C., and O’Gara, F. (2006). Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152, 405–418. doi: 10.1099/mic.0.28324-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Callaghan, A. J., Marcaida, M. J., Stead, J. A., McDowall, K. J., Scott, W. G., and Luisi, B. F. (2005). Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437, 1187–1191. doi: 10.1038/nature04084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carpousis, A. J. (2007). The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87. doi: 10.1146/annurev.micro.61.080706.093440
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chai, W., and Stewart, V. (1999). RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader. J. Mol. Biol. 292, 203–216. doi: 10.1006/jmbi.1999.3084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chaulk, S., Lu, J., Tan, K., Arthur, D. C., Edwards, R. A., Frost, L. S.,et al. (2010). N. meningitidis 1681 is a member of the FinO family of RNA chaperones. RNA Biol. 7, 812–819. doi: 10.4161/rna.7.6.13688
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chaulk, S. G., Smith-Frieday, M. N., Arthur, D. C., Culham, D. E., Edwards, R. A., Soo, P.,et al. (2011). ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry 50, 3095–3106. doi: 10.1021/bi101683a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Y., and Anderson, D. M. (2011). Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol. 80, 966–980. doi: 10.1111/j.1365-2958.2011.07623.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christiansen, J. K., Larsen, M. H., Ingmer, H., Søgaard-andersen, L., and Kallipolitis, B. H., (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186, 3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cohen-Or, I., Shenhar, Y., Biran, D., and Ron, E. Z. (2010). CspC regulates rpoS transcript levels and complements hfq deletions. Res. Microbiol. 161, 694–700. doi: 10.1016/j.resmic.2010.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davies, B. W., Köhrer, C., Jacob, A. I., Simmons, L. A., Zhu, J., Aleman, L. M.,et al. (2011). Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol. Microbiol. 78, 506–518. doi: 10.1111/j.1365-2958.2010.07351.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deana, A., and Belasco, J. G. (2005). Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19, 2526–2533. doi: 10.1101/gad.1348805
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Lay, N., Schu, D. J., and Gottesman, S. (2013). Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288, 7996–8003. doi: 10.1074/jbc.R112.441386
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Desnoyers, G., Bouchard, M. P., and Massé, E. (2013). New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 29, 92–98. doi: 10.1016/j.tig.2012.10.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Desnoyers, G., and Masse, E. (2012). Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. 42, 726–739. doi: 10.1101/gad.182493.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, H., and Babitzke, P. (1998). trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 273, 20494–20503. doi: 10.1074/jbc.273.32.20494
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, H., Tarpey, R., and Babitzke, P. (1997). The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 179, 2582–2586.
Dubey, A. K., Baker, C. S., Romeo, T., and Babitzke, P. (2005). RNA sequence and secondary structure participate in high-affinity CsrA – RNA interaction. RNA 11, 1579–1587. doi: 10.1261/rna.2990205.3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dubey, A. K., Baker, C. S., Suzuki, K., Jones, A. D., Pandit, P., Romeo, T.,et al. (2003). CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 185, 4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Edwards, A. N., Patterson-Fortin, L. M., Vakulskas, C. A., Mercante, J. W., Potrykus, K., Vinella, D.,et al. (2011). Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol. 80, 1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Even, S., Pellegrini, O., Zig, L., Labas, V., Vinh, J., Bréchemmier-Baey, D.,et al. (2005). Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33, 2141–2152. doi: 10.1093/nar/gki505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feng, Y., Huang, H., Liao, J., and Cohen, S. N. (2001). Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 276, 31651–31656. doi: 10.1074/jbc.M102855200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferooz, J., Lemaire, J., and Letesson, J.-J. (2011). Role of FlbT in flagellin production in Brucella melitensis. Microbiology 157, 1253–1262. doi: 10.1099/mic.0.044867-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Figueroa-Bossi, N., Schwartz, A., Guillemardet, B., D’Heygère, F., Bossi, L., and Boudvillain, M. (2014). RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 28, 1239–1251. doi: 10.1101/gad.240192.114
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R.,et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D2230. doi: 10.1093/nar/gkt1223
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Folichon, M. (2003). The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 31, 7302–7310. doi: 10.1093/nar/gkg915
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaballa, A., Antelmann, H., Aguilar, C., Khakh, S. K., Song, K.-B., Smaldone, G. T.,et al. (2008). The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 11927–11932. doi: 10.1073/pnas.0711752105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geissmann, T. A., and Touati, D. (2004). Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23, 396–405. doi: 10.1038/sj.emboj.7600058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gollnick, P., Babitzke, P., Antson, A., and Yanofsky, C. (2005). Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu. Rev. Genet. 39, 47–68. doi: 10.1146/annurev.genet.39.073003.093745
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Göpel, Y., Papenfort, K., Reichenbach, B., Vogel, J., and Görke, B. (2013). Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 27, 552–564. doi: 10.1101/gad.210112.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hajnsdorf, E., and Boni, I. V. (2012). Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94, 1544–1553. doi: 10.1016/j.biochi.2012.02.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hajnsdorf, E., and Régnier, P. (2000). Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. U.S.A. 97, 1501–1505. doi: 10.1073/pnas.040549897
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hämmerle, H., Amman, F., Veèerek, B., Stülke, J., Hofacker, I., and Bläsi, U. (2014). Impact of Hfq on the Bacillus subtilis transcriptome. PLoS ONE 9:e98661. doi: 10.1371/journal.pone.0098661
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henderson, C. A., Vincent, H. A., Casamento, A., Stone, C. M., Phillips, J. O., Cary, P. D.,et al. (2013). Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA 19, 1089–1104. doi: 10.1261/rna.034595.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herschlag, D. (1995). RNA chaperones and the RNA folding problem. J. Biol. Chem. 270, 20871–20874. doi: 10.1074/jbc.270.36.20871
Hopkins, J. F., Panja, S., and Woodson, S. A. (2011). Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 39, 5193–5202. doi: 10.1093/nar/gkr062
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huttenhofer, A., and Fnolier, H. (1994). Footprinting mRNA- ribosome complexes with chemical probes. EMBO J. 13, 3892–3901.
Ikeda, Y., Yagi, M., Morita, T., and Aiba, H. (2011). Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 79, 419–432. doi: 10.1111/j.1365-2958.2010.07454.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Irie, Y., Starkey, M., Edwards, A. N., Wozniak, D. J., Romeo, T., and Parsek, M. R. (2010). Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78, 158–172. doi: 10.1111/j.1365-2958.2010.07320.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacob, A. I., Köhrer, C., Davies, B. W., Rajbhandary, U. L., and Walker, G. C. (2014). Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol. Cell 49, 427–438. doi: 10.1016/j.molcel.2012.11.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jerome, L. J., van Biesen, T., and Frost, L. S. (1999). Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol. 285, 1457–1473. doi: 10.1006/jmbi.1998.2404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jonas, K., Edwards, A. N., Simm, R., Romeo, T., Römling, U., and Melefors, O. (2008). The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70, 236–257. doi: 10.1111/j.1365-2958.2008.06411.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jørgensen, M. G., Thomason, M. K., Havelund, J., Valentin-Hansen, P., and Storz, G. (2013). Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 27, 1132–1145. doi: 10.1101/gad.214734.113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jutras, B. L., Chenail, A. M., Carroll, D. W., Miller, M. C., Zhu, H., Bowman A.,et al. (2013a). Bpur, the Lyme disease spirochete’s PUR domain protein: identification as a transcriptional modulator and characterization of nucleic acid interactions. J. Biol. Chem. 288, 26220–26234. doi: 10.1074/jbc.M113.491357
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jutras, B. L., Jones, G. S., Verma, A., Brown, N. A., Antonicello, A. D., Chenail, A. M.,et al. (2013b). Posttranscriptional self-regulation by the Lyme disease bacterium’s BpuR DNA/RNA-binding protein. J. Bacteriol. 195, 4915–4923. doi: 10.1128/JB.00819-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaberdin, V. R., Singh, D., and Lin-Chao, S. (2011). Composition and conservation of the mRNA-degrading machinery in bacteria. J. Biomed. Sci. 18, 23. doi: 10.1186/1423-0127-18-23
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawamoto, H., Koide, Y., Morita, T., and Aiba, H. (2006). Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 61, 1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kime, L., Jourdan, S. S., Stead, J. A., Hidalgo-Sastre, A., and McDowall, K. J. (2010). Rapid cleavage of RNA by RNase E in the absence of 5’ monophosphate stimulation. Mol. Microbiol. 76, 590–604. doi: 10.1111/j.1365-2958.2009.06935.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koleva, R. I., Austin, C. A., Kowaleski, J. M., Neems, D. S., Wang, L., Vary, C. P. H.,et al. (2006). Interactions of ribosomal protein S1 with DsrA and rpoS mRNA. Biochem. Biophys. Res. Commun. 348, 662–668. doi: 10.1016/j.bbrc.2006.07.102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Komarova, A. V., Tchufistova, L. S., Dreyfus, M., and Boni, I. V. (2005). AU-Rich sequences within 5’ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 187, 1344–1349. doi: 10.1128/JB.187.4.1344
Kopaskie, K. S., Ligtenberg, K. G., and Schneewind, O. (2013). Translational regulation of Yersinia enterocolitica mRNA encoding a type III secretion substrate. J. Biol. Chem. 288, 35478–35488. doi: 10.1074/jbc.M113.504811
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kovach, A. R., Hoff, K. E., Canty, J. T., Orans, J., and Brennan, R. G. (2014). Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA 20, 1548–1559. doi: 10.1261/rna.044032.113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kulkarni, P. R., Cui, X., Williams, J. W., Stevens, A. M., and Kulkarni, R. V. (2006). Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 34, 3361–3369. doi: 10.1093/nar/gkl439
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lawhon, S. D., Frye, J. G., Suyemoto, M., Porwollik, S., McClelland, M., and Altier, C. (2003). Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48, 1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Le Derout, J. (2003). Hfq affects the length and the frequency of short oligo(A) tails at the 3’ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 31, 4017–4023. doi: 10.1093/nar/gkg456
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Le Derout, J., Boni, I. V., Régnier, P., and Hajnsdorf, E. (2010). Hfq affects mRNA levels independently of degradation. BMC Mol. Biol. 11:17. doi: 10.1186/1471-2199-11-17.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Link, T. M., Valentin-Hansen, P., and Brennan, R. G. (2009). Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 106, 19292–19297. doi: 10.1073/pnas.0908744106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, J. M., and Camilli, A. (2010). A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13, 18–23. doi: 10.1016/j.mib.2009.11.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, M. Y., Gui, G., Wei, B., Preston, J. F., Oakford, L., Yuksel, U.,et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. doi: 10.1074/jbc.272.28.17502
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, M. Y., Yang, H., and Romeo, T. (1995). The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177, 2663–2672.
Liu, Y., Wu, N., Dong, J., Gao, Y., Zhang, X., Mu, C.,et al. (2010). Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS ONE 5:13069. doi: 10.1371/journal.pone.0013069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lorenz, C., Gesell, T., Zimmermann, B., Schoeberl, U., Bilusic, I., Rajkowitsch, L.,et al. (2010). Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 38, 3794–3808. doi: 10.1093/nar/gkq032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lu, Y., Turner, R., and Switzer, R. (1996). Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl. Acad. Sci. U.S.A. 93, 14462–14467. doi: 10.1073/pnas.93.25.14462
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mackay, J. P., Font, J., and Segal, D. J. (2011). The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 18, 256–261. doi: 10.1038/nsmb.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mackie, G. A. (1998). Ribonuclease E is a 5’-end-dependent endonuclease. Nature 395, 720–723. doi: 10.1038/27246
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mackie, G. A. (2013). RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11, 45–57. doi: 10.1038/nrmicro2930
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mahadevan, S., and Wright, A. (1987). A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell 50, 485–494. doi: 10.1016/0092-8674(87)90502-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mallory, A., and Vaucheret, H. (2010). Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22, 3879–3889. doi: 10.1105/tpc.110.080671
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marden, J. N., Diaz, M. R., Walton, W. G., Gode, C. J., Betts, L., Urbanowski, M. L.,et al. (2013). An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 110, 15055–15060. doi: 10.1073/pnas.1307217110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Massé, E., Escorcia, F. E., and Gottesman, S. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 19, 2374–2383. doi: 10.1101/gad.1127103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merino, E., Babitzke, P., Yanofsky, C., Merino, E., Babitzke, P., and Yanofsky, C. (1995). trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 177, 6362–6370.
Mikulecky, P. J., Kaw, M. K., Brescia, C. C., Takach, J. C., Darren, D., and Feig, A. L. (2004). Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 11, 1206–1214. doi: 10.1038/nsmb858
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Milojevic, T., Grishkovskaya, I., Sonnleitner, E., Djinovic-Carugo, K., and Bläsi, U. (2013). The Pseudomonas aeruginosa catabolite repression control protein Crc is devoid of RNA binding activity. PLoS ONE 8:e64609. doi: 10.1371/journal.pone.0064609
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitobe, J., Yanagihara, I., Ohnishi, K., Yamamoto, S., Ohnishi, M., Ishihama, A.,et al. (2011). RodZ regulates the post-transcriptional processing of the Shigella sonnei type III secretion system. EMBO Rep. 12, 911–916. doi: 10.1038/embor.2011.132
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mohanty, B. K., Maples, V. F., and Kushner, S. R. (2004). The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54, 905–920. doi: 10.1111/j.1365-2958.2004.04337.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moll, I., Afonyushkin, T., Vytvytska, O., and Kaberdin, V. R. (2003). Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 11, 1308–1314. doi: 10.1261/rna.5850703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Møller, T., Franch, T., Højrup, P., Keene, D. R., Ba, H. P., Brennan, R. G.,et al. (2002). Hfq: a bacterial sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9, 23–30. doi: 10.1016/S1097-2765(01)00436-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moreno, R., Hernández-Arranz, S., La Rosa, R., Yuste, L., Madhushani, A., Shingler, V.,et al. (2014). The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ. Microbiol. 17, 105–118. doi: 10.1111/1462-2920.12499
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moreno, R., Marzi, S., Romby, P., and Rojo, F. (2009). The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 37, 7678–7690. doi: 10.1093/nar/gkp825
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, T., Maki, K., and Aiba, H. (2005). RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19, 2176–2186. doi: 10.1101/gad.1330405.1996
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morris, E. R., Hall, G., Li, C., Heeb, S., Kulkarni, R. V., Lovelock, L.,et al. (2013). Structural rearrangement in an RsmA/CsrA Ortholog of pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21, 1659–1671. doi: 10.1016/j.str.2013.07.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muffler, A., Fischer, D., and Hengge-Aronis, R. (1996). The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10, 1143–1151. doi: 10.1101/gad.10.9.1143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mukherjee, S., Yakhnin, H., Kysela, D., Sokoloski, J., Babitzke, P., and Kearns, D. B. (2011). CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 82, 447–461. doi: 10.1111/j.1365-2958.2011.07822.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muto, Y., Oubridge, C., and Nagai, K. (2000). RNA-binding proteins: trapping RNA bases. Curr. Biol. 10, 19–21. doi: 10.1016/S0960-9822(99)00250-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osborne, J., Djapgne, L., Tran, B. Q., Goo, Y. A., and Oglesby-Sherrouse, A. G. (2014). A method for in vivo identification of bacterial small RNA-binding proteins. Microbiologyopen 3, 950–960. doi: 10.1002/mbo3.220
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pandey, P, S., Winkler, J. A., Li, H., Camacho, D. M., Collins, J. J., and Walker, G. C. (2014). Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics 15:121. doi: 10.1186/1471-2164-15-121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pandey, S. P., Minesinger, B. K., Kumar, J., and Walker, G. C. (2011). A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 39, 4691–4708. doi: 10.1093/nar/gkr060
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patterson-Fortin, L. M., Vakulskas, C. A., Yakhnin, H., Babitzke, P., and Romeo, T. (2013). Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J. Mol. Biol. 425, 3662–3677. doi: 10.1016/j.jmb.2012.12.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perez-Rueda, E., and Martinez-Nuñez, M. A. (2012). The repertoire of DNA-binding transcription factors in prokaryotes: functional and evolutionary lessons. Sci. Prog. 95, 315–329. doi: 10.3184/003685012X13420097673409
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Phadtare, S., Inouye, M., and Severinov, K. (2002). The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277, 7239–7245. doi: 10.1074/jbc.M111496200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Picard, F., Dressaire, C., Girbal, L., and Cocaign-Bousquet, M. (2009). Examination of post-transcriptional regulations in prokaryotes by integrative biology. C. R. Biol. 332, 958–973. doi: 10.1016/j.crvi.2009.09.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rabhi, M., Espéli, O., Schwartz, A., Cayrol, B., Rahmouni, A. R., Arluison, V.,et al. (2011). The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 30, 2805–2816. doi: 10.1038/emboj.2011.192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ramesh, A., DebRoy, S., Goodson, J. R., Fox, K. A., Faz, H., Garsin, D. A.,et al. (2012). The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 8:e1002666. doi: 10.1371/journal.pgen.1002666
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rasmussen, A. A., Eriksen, M., Gilany, K., Udesen, C., Franch, T., Petersen, C.,et al. (2005). Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 58, 1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Régnier, P., and Hajnsdorf, E. (2013). The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3’ ends of RNAs resulting from Rho-independent termination: a tentative model. RNA Biol. 10, 602–609. doi: 10.4161/rna.23664
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reimmann, C., Valverde, C., Kay, E., and Haas, D. (2005). Posttranscriptional repression of GacS / GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187, 276–285. doi: 10.1128/JB.187.1.276
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rieder, R., Reinhardt, R., Sharma, C., and Vogel, J. (2012). Experimental tools to identify RNA-protein interactions in Helicobacter pylori. RNA Biol. 9, 520–531. doi: 10.4161/rna.20331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Romeo, T., Vakulskas, C. A., and Babitzke, P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324. doi: 10.1111/j.1462-2920.2012.02794.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rutberg, B. (1997). MicroReview antitermination of transcription of catabolic operons. Mol. Microbiol. 23, 413–421. doi: 10.1046/j.1365-2958.1997.d01-1867.x
Said, N., Rieder, R., Hurwitz, R., Deckert, J., Urlaub, H., and Vogel, J. (2009). In vivo expression and purification of aptamer-tagged small RNA regulators. Nucleic Acids Res. 37, e133. doi: 10.1093/nar/gkp719
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Salvail, H., Caron, M.-P., Bélanger, J., and Massé, E. (2013). Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J. 32, 2764–2778. doi: 10.1038/emboj.2013.205
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santangelo, T. J., and Artsimovitch, I. (2011). Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 9, 319–329. doi: 10.1038/nrmicro2560
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saramago, M., Bárria, C., Dos Santos, R. F., Silva, I. J., Pobre, V., Domingues, S.,et al. (2014). The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 18, 105–115. doi: 10.1016/j.mib.2014.02.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sarsero, J. P., Merino, E., and Yanofsky, C. (2000). A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 182, 2329–2331. doi: 10.1128/JB.182.8.2329-2331.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sauer, E. (2013). Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 10, 610–618. doi: 10.4161/rna.24201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sauer, E., Schmidt, S., and Weichenrieder, O. (2012). Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 9396–9401. doi: 10.1073/pnas.1202521109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schiano, C. A., and Lathem, W. W. (2012). Post-transcriptional regulation of gene expression in Yersinia species. Front. Cell. Infect. Microbiol. 2:129. doi: 10.3389/fcimb.2012.00129
Schnetz, K., and Rak, B. (1988). Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 7, 3271–3277.
Schumacher, M. A., Pearson, R. F., Møller, T., Valentin-Hansen, P., and Brennan, R. G. (2002). Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21, 3546–3556. doi: 10.1093/emboj/cdf322
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shahbabian, K., Jamalli, A., Zig, L., and Putzer, H. (2009). RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28, 3523–3533. doi: 10.1038/emboj.2009.283
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sheidy, D. T., and Zielke, R. A. (2013). Analysis and expansion of the role of the Escherichia coli protein ProQ. PLoS ONE 8:e79656. doi: 10.1371/journal.pone.0079656
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shine, J., and Dalgarno, L. (1974). The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 71, 1342–1346. doi: 10.1073/pnas.71.4.1342
Smaldone, G. T., Antelmann, H., Gaballa, A., and Helmann, J. D. (2012). The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J. Bacteriol. 194, 2586–2593. doi: 10.1128/JB.05567-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Snyder, D., Lary, J., Chen, Y., Gollnick, P., and Cole, J. L. (2004). Interaction of the trp RNA-binding attenuation protein (TRAP) with anti-TRAP. J. Mol. Biol. 338, 669–682. doi: 10.1016/j.jmb.2004.03.030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sobrero, P., and Valverde, C. (2012). The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 38, 276–299. doi: 10.3109/1040841X.2012.664540
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sonnleitner, E., Abdou, L., and Haas, D. (2009). Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 106, 21866–21871. doi: 10.1073/pnas.pnas.0910308106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sonnleitner, E., and Bläsi, U. (2014). Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 10:e1004440. doi: 10.1371/journal.pgen.1004440
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Soper, T. J., Doxzen, K., and Woodson, S. A. (2011). Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA 17, 1544–1550. doi: 10.1261/rna.2767211
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sorger-Domenigg, T., Sonnleitner, E., Kaberdin, V. R., and Bläsi, U. (2007). Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 352, 769–773. doi: 10.1016/j.bbrc.2006.11.084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sterzenbach, T., Nguyen, K. T., Nuccio, S.-P., Winter, M. G., Vakulskas, C. A., Clegg, S.,et al. (2013). A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. EMBO J. 32, 2872–2883. doi: 10.1038/emboj.2013.206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stülke, J. (2002). Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch. Microbiol. 177, 433–440. doi: 10.1007/s00203-002-0407-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Subramanian, A. R. (1983). Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28, 101–142. doi: 10.1016/S0079-6603(08)60085-9
Suzuki, K., Babitzke, P., Kushner, S. R., and Romeo, T. (2006). Identification of a novel regulatory protein ( CsrD ) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20, 2605–2617. doi: 10.1101/gad.1461606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tortosa, P., Lindner, C., Saier, M. H., Reizer, J., and Le Coq, D. (1997). Multiple Phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system. J. Biol. Chem. 272, 17230–17237. doi: 10.1074/jbc.272.27.17230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsai, B. P., Wang, X., Huang, L., and Waterman, M. L. (2011). Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell Proteomics 10, M110.007385. doi:10.1074/mcp.M110.007385
Tsui, H. C., Leung, H. C., and Winkler, M. E. (1994). Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K–12. Mol. Microbiol. 13, 35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vecerek, B., Moll, I., and Bläsi, U. (2005). Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA 11, 976–984. doi: 10.1261/rna.2360205
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vercruysse, M., Köhrer, C., Davies, B. W., Arnold, M. F. F., Mekalanos, J. J., RajBhandary, U. L.,et al. (2014). The highly conserved bacterial RNase YbeY is essential in vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog. 10:e1004175. doi: 10.1371/journal.ppat.1004175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vytvytska, O., Moll, I., Kaberdin, V. R., Von Gabain, A., and Bläsi, U. (2000). binding Hfq ( HF1 ) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14, 1109–1118. doi: 10.1101/gad.14.9.1109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, M.-C., Chien, H.-F., Tsai, Y.-L., Liu, M.-C., and Liaw, S.-J. (2014). The RNA chaperone Hfq is involved in stress tolerance and virulence in uropathogenic Proteus mirabilis. PLoS ONE 9:e85626. doi: 10.1371/journal.pone.0085626
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Dubey, A. K., Suzuki, K., Baker, C. S., Babitzke, P., and Romeo, T. (2005). CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56, 1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weilbacher, T., Suzuki, K., Dubey, A. K., Wang, X., Gudapaty, S., Morozov, I.,et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. doi: 10.1046/j.1365-2958.2003.03459.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Windbichler, N., von Pelchrzim, F., Mayer, O., Csaszar, E., and Schroeder, R. (2008). Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol. 5, 30–40. doi: 10.4161/rna.5.1.5694
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yakhnin, A. V, Baker, C. S., Vakulskas, C. A., Yakhnin, H., Berezin, I., Romeo, T.,et al. (2014). CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol. Cell 87, 851–866. doi: 10.1111/mmi.12136.CsrA
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yakhnin, H., Baker, C. S., Berezin, I., Evangelista, M. A., Rassin, A., Romeo, T.,et al. (2011a). CsrA represses translation of sdiA, which encodes the N-acylhomoserine-L-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J. Bacteriol. 193, 6162–6170. doi: 10.1128/JB.05975-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yakhnin, H., Yakhnin, A. V., Baker, C. S., Sineva, E., Berezin, I., Romeo, T.,et al. (2011b). Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Esigma70 andEsigmaS, and indirect transcriptional activation by CsrA. Mol. Microbiol. 81, 689–704. doi: 10.1111/j.1365-2958.2011.07723.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yakhnin, H., Zhang, H., Yakhnin, A. V., and Babitzke, P. (2004). The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP ( yhaG ) by blocking ribosome binding. J. Bacteriol. 186, 278–286. doi: 10.1128/JB.186.2.278
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, M., de Saizieu, A., van Loon, A. P., and Gollnick, P. (1995). Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP). J. Bacteriol. 177, 4272–4278.
Zha, D., Xu, L., Zhang, H., and Yan, Y. (2014). The two-component GacS-GacA System activates lipA translation by RsmE but not RsmA in Pseudomonas protegens Pf-5. Appl. Environ. Microbiol. 80, 6627–6637. doi: 10.1128/AEM.02184-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, A., Wassarman, K. M., Ortega, J., Steven, A. C., and Storz, G. (2002). The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9, 11–22. doi: 10.1016/S1097-2765(01)00437-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: post-transcriptional regulation, RNA-binding proteins, bacteria, working mechanisms, biotechnological applications, regulation of translation, stability regulation
Citation: Van Assche E, Van Puyvelde S, Vanderleyden J and Steenackers HP (2015) RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 6:141. doi: 10.3389/fmicb.2015.00141
Received: 24 December 2014; Paper pending published: 24 January 2015;
Accepted: 06 February 2015; Published online: 03 March 2015.
Edited by:
Martin G. Klotz, University of North Carolina at Charlotte, USACopyright © 2015 Van Assche, Van Puyvelde, Vanderleyden and Steenackers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hans P. Steenackers, Centre of Microbial and Plant Genetics, Department of Molecular and Microbial Systems, KU Leuven, Kasteelpark Arenberg 20 bus 2460, 3000 Leuven, Belgium e-mail: hans.steenackers@biw.kuleuven.be
 Elke Van Assche
Elke Van Assche Sandra Van Puyvelde
Sandra Van Puyvelde Jos Vanderleyden
Jos Vanderleyden Hans P. Steenackers
Hans P. Steenackers