- 1School of Life Sciences, University of Nevada, Las Vegas, Las Vegas, NV, USA
- 2Nevada Institute of Personalized Medicine, University of Nevada, Las Vegas, Las Vegas, NV, USA
- 3Biology Department and Center for Life in Extreme Environments, Portland State University, Portland, OR, USA
- 4State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Beijing, China
- 5Department of Biology, California State University San Bernardino, San Bernardino, CA, USA
- 6Department of Geology and Environmental Earth Science, Miami University,Oxford, OH, USA
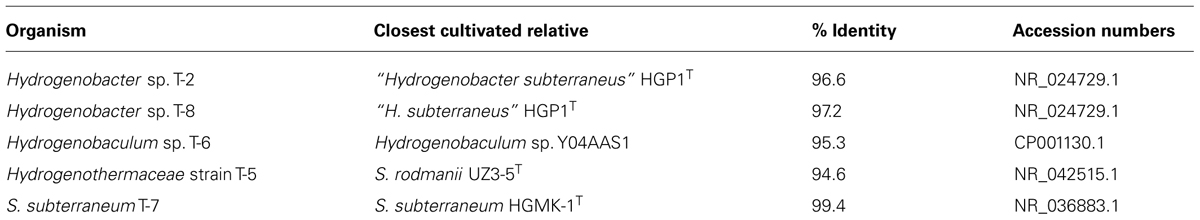
The order Aquificales (phylum Aquificae) consists of thermophilic and hyperthermophilic bacteria that are prominent in many geothermal systems, including those in Tengchong, Yunnan Province, China. However, Aquificales have not previously been isolated from Tengchong. We isolated five strains of Aquificales from diverse springs (temperature 45.2–83.3°C and pH 2.6–9.1) in the Rehai Geothermal Field from sites in which Aquificales were abundant. Phylogenetic analysis showed that four of the strains belong to the genera Hydrogenobacter, Hydrogenobaculum, and Sulfurihydrogenibium, including strains distant enough to likely justify new species of Hydrogenobacter and Hydrogenobaculum. The additional strain may represent a new genus in the Hydrogenothermaceae. All strains were capable of aerobic respiration under microaerophilic conditions; however, they had variable capacity for chemolithotrophic oxidation of hydrogen and sulfur compounds and nitrate reduction.
Introduction
The phylum Aquificae is composed of a single order, Aquificales, and three families, Aquificaceae, Hydrogenothermaceae, and Desulfurobacteriaceae (Reysenbach et al., 2005; L’Haridon et al., 2006). Aquificales are present in many terrestrial and marine geothermal systems where they often form multicellular “streamer” assemblages (Huber et al., 1998; Reysenbach et al., 2000, 2005; Takacs et al., 2001; Eder and Huber, 2002; Spear et al., 2005; Hou et al., 2013; Takacs-Vesbach et al., 2013) but can also be prominent members of planktonic microbial communities (Cole et al., 2013; Hou et al., 2013; Murphy et al., 2013). Most members of the Aquificales are obligate or facultative autotrophs (Kawasumi et al., 1984; Stohr et al., 2001; Takai et al., 2001; Eder and Huber, 2002; Aguiar et al., 2004; Caldwell et al., 2010), although at least one isolate was reported to be incapable of autotrophic growth under the conditions that were tested (Takai et al., 2001). Although very few studies have quantified autotrophy in terrestrial geothermal systems inhabited by Aquificales (Boyd et al., 2009), Aquificales are broadly hypothesized to be important primary producers and are capable of using a variety of inorganic compounds to fuel chemolithotrophy, including diverse electron donors (H2, S2-, S2O32-, SO32-, S0, Fe2+, AsO33-) and terminal electron acceptors (O2, NO3-, SO32-, Fe3+, AsO43-, SeO32-; Stohr et al., 2001; Takai et al., 2001; Eder and Huber, 2002; O’Neill et al., 2008).
Two families of Aquificales dominate in terrestrial geothermal systems, the Aquificaceae and Hydrogenothermaceae. The Aquificaceae includes three genera that are abundant in terrestrial systems: Hydrogenobacter, Thermocrinis, and Hydrogenobaculum (Reysenbach, 2001; Takacs-Vesbach et al., 2013). Hydrogenobacter and Thermocrinis are closely related and are capable of axenic growth at circumneutral pH to ≥85°C (Kawasumi et al., 1984; Takai et al., 2001; Eder and Huber, 2002) and ≥89°C (Huber et al., 1998; Eder and Huber, 2002; Caldwell et al., 2010), respectively. In contrast, known isolates of Hydrogenobaculum are acidophilic (optimum pH 3–4) and have lower growth temperature ranges, with optima between 60 and 70°C (Shima and Suzuki, 1993; D’Imperio et al., 2008). The family Hydrogenothermaceae includes a single genus that is prominent in many terrestrial geothermal systems, Sulfurihydrogenibium, with known isolates capable of growth to ≥75°C at circumneutral pH (5.0–8.8; O’Neill et al., 2008).
Yunnan Province, in southwest China, has a large number of geothermal springs, particularly in Tengchong County, which is located within the Indo-Burma Range along the central-western border between Yunnan Province and Myanmar. Geothermal activity in Yunnan Province is typically located along arched fault structures and circular depressions and is likely fueled by latent heat from tectonic activity associated with the subduction of Tethys Ocean lithosphere (Liao and Guo, 1986; Wang et al., 2008). The largest and best-known geothermal area in Tengchong is the Rehai (“Hot Sea”) Geothermal Field, with springs reaching the boiling point (∼95°C at ∼1,500 m elevation) and spanning a pH range of 2.5–9.4 at high temperature (>80°C; Figure 1; Table 1; Hedlund et al., 2012). A large number of Bacteria and Archaea have been isolated from Rehai springs, particularly thermophilic members of the Firmicutes (Bacillales, Thermoanaerobales, Clostritiales), Deinococcus-Thermus phylum (Thermales), and Crenarchaeota (Sulfolobales) (reviewed in Hedlund et al., 2012). However, despite recent cultivation-independent studies suggesting that Aquificales are abundant in nearly all high-temperature sites in Rehai (Pagaling et al., 2012; Hou et al., 2013; Song et al., 2013; Briggs et al., 2014), there are no published reports of the isolation or characterization of Aquificales from Rehai or anywhere in China.
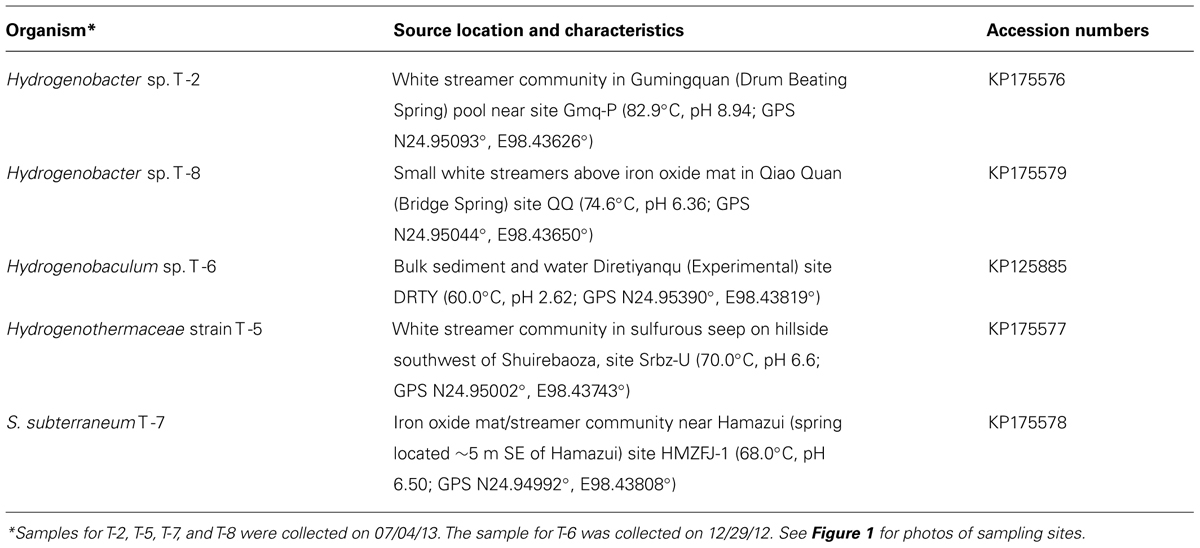
TABLE 1. Sources of Aquificales strains isolated from Tengchong hot springs and their 16S rRNA gene sequences.
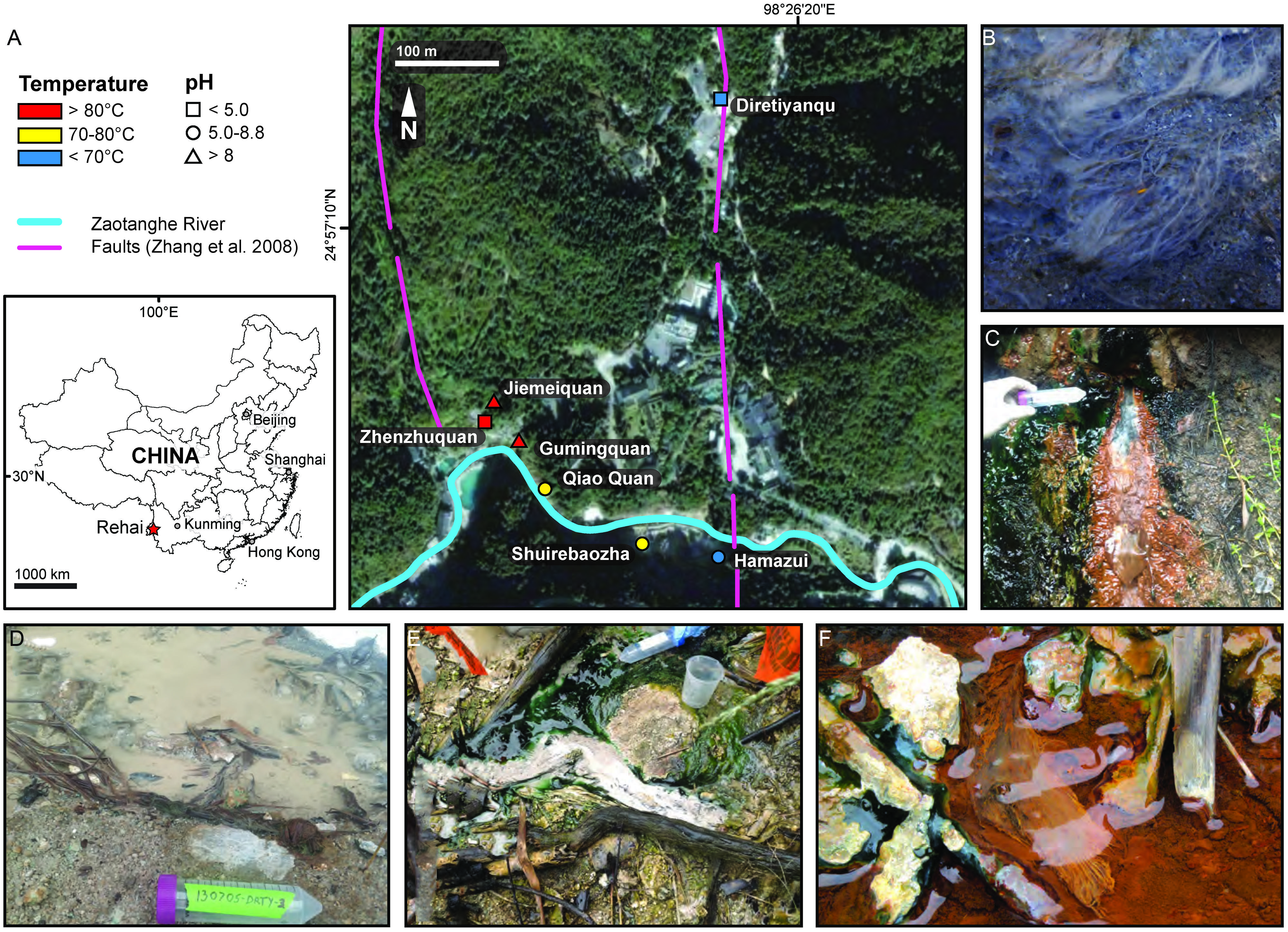
FIGURE 1. Study area map and sample locations. Strains were isolated from five hot spring locations in the Rehai Geothermal Field (A) in Yunnan, China: (B) Hydrogenobacter sp. T-2 from a white streamer community in Gumingquan (Drum Beating Spring) pool site Gmq-P (82.9°C, pH 8.94), (C) Hydrogenobacter sp. T-8 from Qiao Quan (Bridge Spring) site QQ (74.6°C, pH 6.36), (D) Hydrogenobaculum sp. T-6 from Diretiyanqu (Experimental) site DRTY (60.0°C, pH 2.62), (E) Hydrogenothermaceae strain T-5 from a white/yellow streamer community in a sulfurous seep on a hillside south of Shuirebaoza, site Srbz-U (70.0°C, pH 6.6), (F) Sulfurihydrogenibium subterraneum T-7 from a white/yellow streamer community in Hamazui-3 (spring located ∼5 m SE of Hamazui), site HMZFJ-3 (68.0°C, pH 6.50). (A) is modified from Hedlund et al. (2012). Fifty mL conical tubes used for scale are 11.7 cm long.
In this study, we isolated Aquificales from sites in Tengchong known to host abundant Aquificales populations and sites with abundant streamer growth that were deemed likely to host Aquificales. The strains belong to the genera Hydrogenobacter, Hydrogenobaculum, and Sulfurihydrogenibium, and possibly a new genus within the Hydrogenothermaceae. Although most of the strains likely represent new taxa, their general physiological traits are similar to known members of these genera, including variable capacity for aerobic hydrogen oxidation via the “knallgas reaction,” chemolithotrophic oxidation of sulfur compounds, and anaerobic respiration of nitrate.
Materials and Methods
Sample Collection, Enrichment, and Isolation
Sediment, streamer, and mat samples were collected from five hot springs located in the Rehai Geothermal Field in Tengchong County, Yunnan Province, China (Figure 1). Prior to sampling, the temperature and pH were measured at the precise sampling location with a field-calibrated pH probe with temperature correction (LaMotte five Series, Chestertown, MD, USA). Detailed water chemistry and microbial community composition at most of the sampling locations on several previous sampling trips has been reported elsewhere (Hou et al., 2013; Briggs et al., 2014).
Samples from which strains T-2, T-5, T-7, and T-8 were isolated were collected aseptically and transferred into 25 mL Balch tubes containing 5 mL modified MSH medium (Caldwell et al., 2010) containing S0 and S2O32- and adjusted to pH 8.0 (T-2), 6.5 (T-5 and T-7), or 7.5 (T-8). Tube headspace was either H2:CO2 (80:20) for strains T-2, T-7, and T-8, or N2:CO2 (80:20) for strain T-5, supplemented with 4% v/v O2. The tubes were stored and transported at room temperature. Once in the lab, the Balch tubes were incubated at 80°C (T-2) or 70°C (T-5, T-7, and T-8) and passaged in the same medium under the same conditions. To obtain pure cultures of strains T-2, T-7, and T-8, positive enrichments were streaked for isolation onto plates containing GBS salts medium (Dodsworth et al., 2014) containing thiosulfate (1 mM added as Na2S2O3⋅5H2O) and solidified with Gelrite [0.8% mass/vol, supplemented with 4 g/L MgCl2⋅6H2O (Serva, Heidelberg)] and incubated at 70°C in modified two quart Bandit pressure pots (C.A. Technologies). Pressure pot headspace consisted of ∼2 L anaerobic chamber gas (N2:CO2:H2 at 90:5:5) supplemented with 200 mL H2:CO2 (80:20) and 100 mL air. Isolated colonies were re-streaked two times to ensure purity. Strain T-5 was isolated using an extinction-to-dilution method that was repeated at least seven times. For all strains, purity was confirmed through microscopic observation and sequencing of the 16S rRNA gene, following the general approach described previously (Nakagawa et al., 2005).
The sample from which strain T-6 was isolated was aseptically transferred in the field into a 25 ml Balch tube containing 10 mL of DSMZ medium 743 (modified by replacing S0 with 30 μM Na2S, pH 3), given a headspace of N2/CO2/H2/air (30:40:20:10), and incubated in the spring. Following growth, the tube was transported to the lab without temperature control. For isolation, 1 mL of the enrichment culture was inoculated into 10 ml of the same medium with the same headspace as in initial enrichment. A pure isolate was obtained by three rounds of dilution to extinction and verified through microscopic observation and sequencing of the 16S rRNA gene.
Growth Characteristics
The capacity for growth of the strains on electron donors and electron acceptors commonly used by Aquificales was determined by growing each strain under conditions that permitted good growth, as determined by phase-contrast microscopy. In all cases, growth was determined by direct cell counts using a Petroff–Hausser counting chamber and a phase-contrast microscope. All experiments were performed in triplicate along with positive and negative controls. Strains T-2, T-7, and T-8 were routinely grown at 70°C in 5 mL volumes of GBS salts medium (Dodsworth et al., 2014) with an N2/H2/CO2/air (75:17:4:4) headspace or in 25 mL Balch tubes without shaking. The medium was adjusted to pH 8.0, 7.2, and 6.6 for T-2, T-7, and T-8, respectively. Strain T-5 was routinely grown in a modified MSH medium (Caldwell et al., 2010) containing S0 and S2O32- at 70°C in 5 mL volumes with a headspace of N2:CO2 (80:20). For testing electron donors, H2 was replaced with N2 in the headspace (for T-2, T-7, and T-8) and the following compounds were added as sources of possible electron donors, each tested at 1 mM final concentration: Na2S2O3⋅5H2O, sodium pyruvate, sodium formate, and sodium acetate; additionally, S0 was tested at 0.1 and 1.0% (w/vol). For testing terminal electron acceptors, air was replaced with N2 and the following possible electron acceptors were tested at 1 mM final concentration: NaNO3, NaNO2, Na2S2O3⋅5H2O, and Na2HAsO4.
Strain T-6 was routinely grown at 60°C in 10 mL volume of a modified DSMZ 743 medium with a N2/CO2/H2/air (30:40:20:10) headspace in 25 mL Balch tubes with no shaking. The following compounds were tested as possible electron donors under aerobic conditions with 5 mM citric acid as a buffer (pH 3.0; D’Imperio et al., 2008): S0 (w/vol 0.1%), Na2S (3 mM), Na2S2O3⋅5H2O (100 μM), sodium lactate (1 g/L), sodium pyruvate (1 g/L), sodium formate (1 g/L), and sodium acetate (1 g/L; Shima and Suzuki, 1993). The following compounds were tested as possible terminal electron acceptors in the same medium with H2 as the electron donor with an atmosphere of N2/CO2/H2 (40:40:20): NaNO3 (100 μM), NaNO2 (100 μM), Na2S2O3⋅5H2O (100 μM), FeCl3 (100 μM), and Na2SO4 (100 μM; Shima and Suzuki, 1993).
Identification of Nitrate Reduction Products
Nitrate and nitrite were measured colorimetrically using reagents from LaMotte (LaMotte, Chesterton, MD, USA). Nitrate plus nitrite was determined by cadmium reduction of nitrate and subsequent diazotization of nitrite. Nitrite was determined by diazotization without reduction of nitrate. Nitrous oxide was measured by gas chromatography-electron capture detection on a GC-2014 Nitrous Oxide Analyzer (Shimadzu, Moorpark, CA, USA), modified and operated as described (Dodsworth et al., 2011). Production of N2 was tested by using Durham vials.
16S rRNA Gene PCR, Sequencing, and Phylogenetic Analysis
DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) and 16S rRNA genes were amplified by PCR using primers 9 bF (Eder et al., 1999) and 1512uR (Eder et al., 2001) and sequenced as previously described (Costa et al., 2009). Reads were trimmed to remove bases with quality scores of less than 20 and aligned against the SILVA alignment in the program mothur v1.20.2 (Schloss et al., 2009; Quast et al., 2013). Near full-length 16S rRNA gene sequences were aligned along with reference sequences of Aquificales, including the closest BLASTN matches, using the SILVA alignment in mothur v1.33.3. The alignment was curated manually using Bioedit v7.0.5.3 (Hall, 1999) and analyzed with and without the Lane mask (Lane, 1991) using maximum likelihood in RAxML v7.2.6 (100 replicates, GTR+CAT model of nucleotide substitution; Stamatakis, 2006) and neighbor joining in PHYLIP v3.69 (1,000 replicates; Felsenstein, 2005). Trees were visualized and manipulated using Dendroscope v2 (Huson et al., 2007). Distances shown in Table 2 were derived by applying the dist.seqs to the curated Silva alignment. Pairwise comparisons between 16S rRNA gene sequences from isolates and previously published sequence tags (Hou et al., 2013) were computed using DNAMAN software (Lynnon LLC, San Ramon, CA, USA).
Nucleotide Accession Numbers
Near full-length 16S rRNA gene sequences have been deposited in GenBank with the following accession numbers: KP125885 and KP175576–KP175579.
Results
Isolation and Phylogenetic Analysis
Chemolithotrophic isolates were obtained from five geochemically diverse sites in the Rehai Geothermal field (Table 1), which were chosen based on previous reports of Aquificales in Rehai (Hou et al., 2013; Song et al., 2013; Briggs et al., 2014) and identification of additional streamer communities deemed likely to host Aquificales. Sample sites included white streamer material and sediment collected from a high pH, high temperature site (Figure 1B), a small geothermal seep hosting small white streamers (Figure 1C), sediments in a small acidic pool dominated by silicate sand (Figure 1D; Hou et al., 2013; Briggs et al., 2014), a white streamer community in a sulfurous seep (Figure 1E), and a biofilm and streamer community encrusted with iron oxide (Figure 1F).
Phylogenetic analysis based on near-complete 16S rRNA genes showed that the strains belonged to the families Aquificaceae and Hydrogenothemaceae. Two Hydrogenobacter strains were isolated, designated T-2 and T-8, from sites differing in pH by > 2.5 units. They were grown in media with pH similar to their environmental source, although both were closely related to “Hydrogenobacter subterraneus” (Table 2; Figure 2). Both strains belonged to a species-level (98.65% identity; Kim et al., 2014) operational taxonomic unit (OTU) that comprised >50% of 16S rRNA gene sequence tags in either sediments or the bulk water in most circumneutral geothermal sites in both Rehai and Ruidian (Dientan) geothermal fields (Hou et al., 2013), including streamer and sediment communities in Guminquan, from which strain T-2 was isolated. However, the most abundant sequence within that OTU shared only 98.84% identity with strains T-2 and T-8, whereas the identical sequence to T-2 and T-8 was a rare variant in the cultivation-independent datasets. The other isolate belonging to the Aquificaceae, strain T-6, was isolated from the acidic pool, Diretiyanqu. Strain T-6 branched within the genus Hydrogenobaculum but was distant from the only validly described species, Hydrogenobaculum acidophilus, as well as other isolates from Yellowstone National Park (Table 2; Figure 2). Strain T-6 belonged to an OTU that comprised 31 to 66% of 16S rRNA gene sequence tags from pools ranging from 55 to 65°C from Diretiyanqu, the system of small acidic pools from which the strain was isolated (Hou et al., 2013). Within these systems, the dominant OTU was identical to T-6. Sulfurihydrogenibium strain T-7 was closely related to Sulfurihydrogenibium subterraneum and tentatively identified as a member of that species. Within the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum HGMK-1T, and strain T-7 branched with 16S rRNA gene clones from Asia (Japan and Taiwan), potentially representing a species exclusive to Asia (Figure 2). T-5, the other strain that branched from within the Hydrogenothermaceae, was only distantly related to cultivated strains of Sulfurihydrogenibium and Venenivibrio and branched with 16S rRNA gene clones from hot springs in China and Thailand (Table 2; Figure 2; JX298759 and KC831413, unpublished). Aside from strain T-7, the 16S rRNA gene identity between each new isolate and the most closely related species was well below the 16S rRNA gene identity threshold suggested to delimit bacterial species (98.65%; Kim et al., 2014), and strains T-6 and T-5 were also below the median 16S rRNA gene identity circumscribing bacterial genera (Yarza et al., 2014). Formal taxonomic treatment of these isolates will be determined pending detailed physiological and genomic analysis.
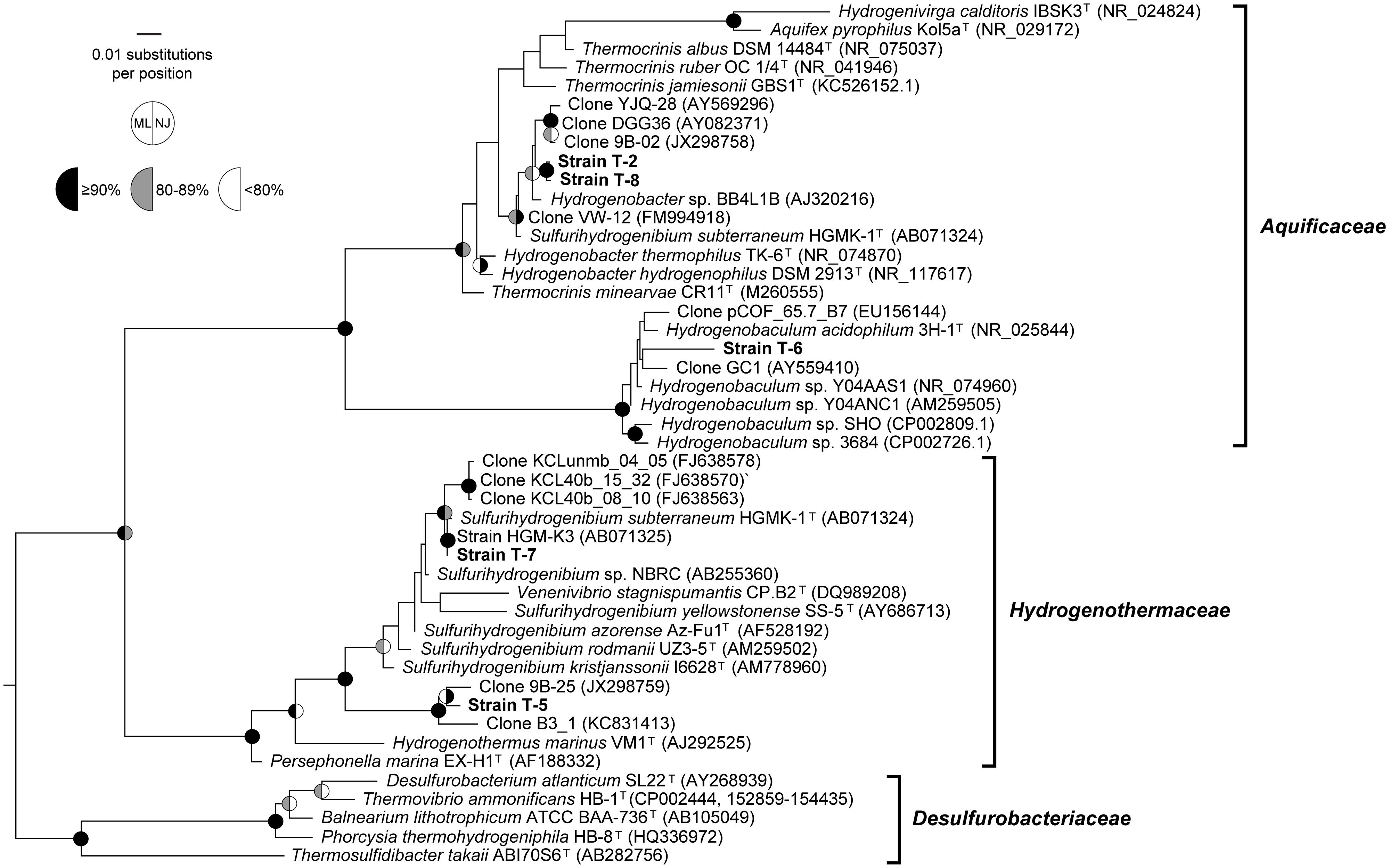
FIGURE 2. Phylogenetic analysis. Maximum-likelihood (ML) phylogeny of the Aquificales including all genera and type strains of all species in the genera Hydrogenobacter, Hydrogenobaculum, and Sulfurihydrogenibium, as well as closely related clones from cultivation-independent studies. Bootstrap values represent 100 replicates for ML and 1,000 replicates for neighbor joining (NJ). Similar analyses with a Lane mask or without an outgroup sequence yielded similar results. Bootstrap support for nodes supported by<80% recovery from both methods is not shown. Bar, 0.01 changes per nucleotide. The outgroup was Methanocaldococcus jannaschii (AB603516).
With the exception of T-5, all strains were capable of chemotrophic growth with H2 as the electron donor under microaerophilic conditions (Table 3). Both Hydrogenobacter strains also used S2O32- as an electron donor and Hydrogenobacter sp. T-2 additionally used S0 and acetate as electron donors. Hydrogenobacter sp. T-8 grew anaerobically by reducing nitrate. Neither nitrous oxide nor dinitrogen were identified as products of nitrate reduction. Hydrogenobaculum strain T-6 was capable of microaerobic growth with S2- and S0 as alternative electron donors. S. subterraneum T-7 was capable of growth with S2- and S2O32- as alternative electron donors. T-5 could only use sulfur or thiosulfate as electron donors and O2 as the electron acceptor. All strains could grow autotrophically, with the exception of Hydrogenobaculum strain T-6, which required or was greatly stimulated by citrate, which is the buffer for DSM medium 743.
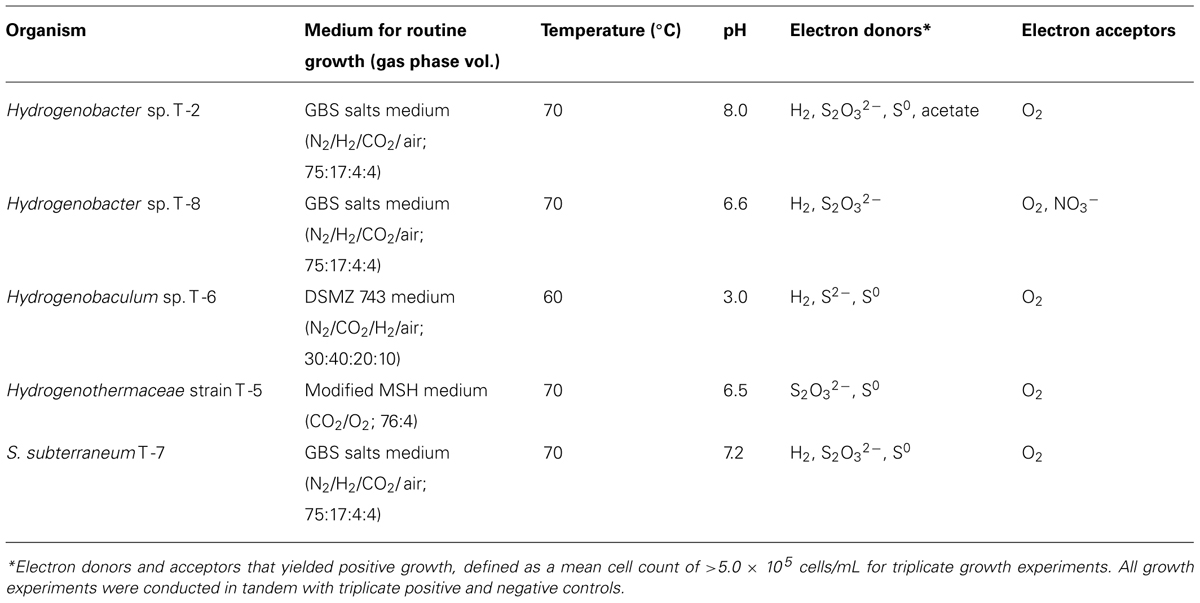
TABLE 3. Media for routine growth and growth characteristics for Aquificales strains from Tengchong hot springs.
Discussion
Aquificales are globally distributed and often abundant in both marine and terrestrial geothermal systems where they likely play important roles in C, N, H, and S cycles. Recent cultivation-independent censuses of Bacteria and Archaea in hot springs in Tengchong County, China suggested the wide distribution of Aquificales in the region, particularly in the Rehai Geothermal System, where Aquificales dominated many 16S rRNA gene pyrotag datasets generated using a few different primer sets and on several different sampling campaigns (Pagaling et al., 2012; Hou et al., 2013; Song et al., 2013). Both 16S rRNA gene pyrotag data and phylochip data suggest that Hydrogenobacter is a dominant member of most circumneutral to alkaline springs in Rehai (pH 8.1–9.4; Hou et al., 2013; Song et al., 2013; Briggs et al., 2014), including large growths of white streamer material in springs Gumingquan and Jiemeiquan (Figure 1A; Hou et al., 2013; Briggs et al., 2014). Strains T-2 and T-8 shared 98.84% 16S rRNA gene sequence identity across the V4 region, suggesting they may belong to the same species as the dominant Hydrogenobacter OTU in the springs. However, extrapolation of physiological traits of these strains to the abundant natural populations must be done with caution, since even T-2 and T-8 were different with regard to both electron donor and acceptor use, despite being nearly identical across the near-complete 16S rRNA gene. High intra-species variation in respiratory capacity may be a common feature in the Aquificales (D’Imperio et al., 2008). The electron donors and acceptors used by the Hydrogenobacter isolates are similar to those described for other members of the genus (Kawasumi et al., 1984; Takai et al., 2001; Eder and Huber, 2002). “H. subterraneus,” the most closely related isolate described in detail, is similar in its ability to use reduced sulfur compounds as electron donors; however, “H. subterraneus” is unable to use H2 as an electron donor and appears to be incapable of autotrophic growth (Takai et al., 2001). The genus Thermocrinis, which often forms conspicuous streamer growth (Reysenbach et al., 1994; Huber et al., 1998; Eder and Huber, 2002), has only been detected in one 16S rRNA gene census at Rehai (Song et al., 2013) and was not detected by other 16S rRNA gene PCR censuses and phylochip analysis (Pagaling et al., 2012; Hou et al., 2013; Briggs et al., 2014). The sporadic detection of Thermocrinis in Rehai may explain why cultivation experiments described here did not yield Thermocrinis cultures.
Cultivation-independent surveys in Tengchong also identified abundant Hydrogenobaculum populations in Rehai springs with pH < 4, particularly within silica sand-dominated acidic pools in Diretiyanqu and Zhenzuquan (Hou et al., 2013; Song et al., 2013; Briggs et al., 2014). Hydrogenobaculum strain T-6, used both H2 and reduced S compounds as electron donors (S2-, S0, and S2O32-). These compounds are widely used by Hydrogenobaculum isolates from different locations (Shima and Suzuki, 1993; Donahoe-Christiansen et al., 2004; D’Imperio et al., 2008), although isolates from Yellowstone National Park are heterogeneous with regard to their ability to oxidize H2 (D’Imperio et al., 2008). Arsenite oxidation and the encoding structural genes, aioBA, have been documented for some Hydrogenobaculum isolates (Donahoe-Christiansen et al., 2004; Clingenpeel et al., 2009; Romano et al., 2013). The aioBA genes have also been cloned from natural geothermal environments, including those inhabited by Hydrogenobaculum (Clingenpeel et al., 2009; Hamamura et al., 2009) that are similar to those in our study site with regard to temperature and pH. However, strain T-6 was unable to oxidize arsenite under the conditions tested and so perhaps it is similar in this regard to Yellowstone strain Y04AAS1, which lacks recognizable aioBA (Romano et al., 2013) and has not been reported to oxidize arsenite. Lack of arsenite oxidation capability may reflect the relatively low concentrations of total arsenic at this site [50–200 ppb (Hou et al., 2013)].
In contrast to the Aquificaceae, cultivation-independent surveys have suggested a low abundance of Hydrogenothermaceae, including sequences that were related to Hydrogenothermus, Persephonella, and Sulfirihydrogenibium (Pagaling et al., 2012; Song et al., 2013; Briggs et al., 2014). However, two springs not included in published cultivation-independent studies of Rehai yielded strains related to Sulfurihydrogenibium. Strain T-7 was very closely related to S. subterraneum HGMK-1T. The electron donors used by strain T-7 were identical to those used by S. subterraneum HGMK-1T (Nakagawa et al., 2005), although T-7 appeared to be more restricted in its use of terminal electron acceptors. Strain T-5 could not grow on any complex organics, and could only use sulfur or thiosulfate as electron donors in the presence of oxygen as an electron acceptor.
Conclusion
This study expands both the geographic and phylogenetic coverage of Aquificales cultivated from terrestrial geothermal springs. This study is particularly important within the context of the study of thermophilic microbial communities in Tengchong County because abundant evidence from cultivation-independent studies implicate the Aquificales as widely distributed and abundant microorganisms with potential roles in several biogeochemical cycles. Known phenotypic variability within the Aquificales notwithstanding, these studies provide a strong foundation for understanding the potential roles of these organisms in C, N, S, and H cycles in the Rehai Geothermal System. The Aquificales isolates described here likely represent novel species of Hydrogenobacter (strains T-2 and T-8) and Hydrogenobaculum (strain T-6) and a new genus in the Hydrogenothermaceae (strain T-5). Further work is underway to thoroughly taxonomically describe these novel organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Tengchong PIRE team and the staff from the Yunnan Tengchong Volcano and Spa Tourist Attraction Development Corporation for their support and assistance. We thank Chrisabelle Cempron for assistance with determination of nitrate reduction products, Senthil Murugapiran and Eric Boyd for advice and assistance with phylogenetics, and Kristen Brileya and Annie Lindgren for assistance in sequencing the 16S rRNA genes. We thank Hilairy Hartnett for a photograph of the streamer community in Guminquan. This research was supported by National Science Foundation grant OISE-0968421 and the National Natural Science Foundation of China grant 40972211 and National Science Foundation DEB-1134877 funding to Reysenbach. BH acknowledges the generous support of Greg Fullmer through the UNLV Foundation.
References
Aguiar, P., Beveridge, T. J., and Reysenbach, A. L. (2004). Sulfurihydrogenibium azorense sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int. J. Syst. Evol. Microbiol. 54, 33–39. doi: 10.1099/ijs.0.02790-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boyd, E. S., Leavitt, W. D., and Geesey, G. G. (2009). CO2 uptake and fixation by a thermoacidophilic microbial community attached to sulfur flocs in a geo-thermal spring. Appl. Environ. Microbiol. 75, 4289–4296. doi: 10.1128/AEM.02751-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Briggs, B. R., Brodie, E. L., Tom, L. M., Dong, H., Jiang, H., Huang, Q.,et al. (2014). Seasonal patterns in microbial communities inhabiting the hot springs of Tengchong, Yunnan Province, China. Environ. Microbiol. 16, 1579–1591. doi: 10.1111/1462-2920.12311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Caldwell, S. L., Liu, Y., Ferrera, I., Beveridge, T., and Reysenbach, A. L. (2010) Thermocrinis minervae sp. nov., a hydrogen- and sulfur-oxidizing, thermophilic member of the Aquificales from a costa rican terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 60, 338–343. doi: 10.1099/ijs.0.010496–10490
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clingenpeel, S. R., D’Imperio, S., Oduro, H., Druschel, G. K., and McDermott, T. R. (2009). Cloning and in situ expression studies of the Hydrogenobaculum arsenite oxidase genes. Appl. Environ. Microbiol. 75, 3362–3365. doi: 10.1128/AEM.00336-339
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cole, J. K., Peacock, J. P., Dodsworth, J. A., Williams, A. J., Thompson, D. B., Dong, H.,et al. (2013). Sediment microbial communities in great boiling spring are controlled by temperature and distinct from water communities. ISME J. 7, 718–729. doi: 10.1038/ismej.2012.157
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Costa, K., Navarro, J., Shock, E., Zhang, C., Soukup, D., and Hedlund, B. (2009). Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13, 447–459. doi: 10.1007/s00792-009-0230-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
D’Imperio, S., Lehr, C. R., Oduro, H., Druschel, G., Kühl, M., and McDermott, T. R. (2008). Relative importance of H2 and H2S as energy sources for primary production in geothermal springs. Appl. Environ. Microbiol. 74, 5802–5808. doi: 10.1128/AEM.00852-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodsworth, J. A., Gevorkian, J., Despujos, F., Cole, J. K., Murugapiran, S. K., Ming, H.,et al. (2014). Thermoflexus hugenholtzii gen. nov., sp. nov., a thermophilic, microaerophilic, filamentous bacterium representing a novel class in the Chloroflexi, Thermoflexia classis nov., and description of Thermoflexaceae fam. nov. and Thermoflexales ord. nov. Int. J. Syst. Evol. Microbiol. 64, 2119–2127. doi: 10.1099/ijs.0.055855-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dodsworth, J. A., Hungate, B., de la Torre, J. R., Jiang, H., and Hedlund, B. P. (2011). Measuring nitrification, denitrification, and related biomarkers in continental geothermal ecosystems. Meth. Enzymol. 486, 171–203. doi: 10.1016/S0076-6879(11)86008-8
Donahoe-Christiansen, J., D’Imperio, S., Jackson, C. R., Inskeep, W. P., and McDermott, T. R. (2004). Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in yellowstone national park. Appl. Environ. Microbiol. 70, 1865–1868. doi: 10.1128/AEM.70.3.1865-1868.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eder, W., and Huber, R. (2002). New isolates and physiological properties of the Aquificales and description of Thermocrinis albus sp. nov. Extremophiles 6, 309–618. doi: 10.1007/s00792-001-0259-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eder, W., Jahnke, L. L., Schmidt, M., and Huber, R. (2001). Microbial diversity of the brine-seawater interface of the kebrit deep, red sea, studied via 16S rRNA gene sequences and cultivation methods. Appl. Environ. Microbiol. 67, 3077–3085. doi: 10.1128/AEM.67.7.3077-3085.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eder, W., Ludwig, W., and Huber, R. (1999). Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of kebrit deep, red sea. Arch. Microbiol. 172, 213–218. doi: 10.1007/s002030050762
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Felsenstein, J. (2005). PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the Author. Seattle, WA: Department of Genome Sciences, University of Washington.
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hamamura, N., Macur, R. E., Korf, S., Ackerman, G., Taylor, W. P., Kozubal, M.,et al. (2009). Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ. Microbiol. 11, 421–431. doi: 10.1111/j.1462-2920.2008.01781.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hedlund, B. P., Cole, J. K., Hou, W., Zhou, E., Li, W., and Dong, H. (2012). A review of the microbiology of the Rehai geothermal field in Tengchong, Yunnan Province, China. Geosci. Front. 3:273–288. doi: 10.1016/j.gsf.2011.12.006
Hou, W., Wang, S., Dong, H., Jiang, H., Briggs, B. R., Peacock, J. P., et al. (2013). A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS ONE 8:e53350. doi: 10.1371/journal.pone.0053350
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huber, R., Eder, W., Heldwein, S., Wanner, G., Huber, H., Rachel, R.,et al. (1998). Thermocrinis ruber gen. nov., sp. nov., A pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl. Environ. Microbiol. 64, 3576–3583.
Huson, D. H., Richter, D. C., Rausch, C., Dezulian, M. F., and Rupp, R. (2007). An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. doi: 10.1186/1471-2105-8-460
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawasumi, T., Igarashi, Y., Kodama, T., and Minoda, Y. (1984). Hydrogenobacter thermophilus gen. nov., sp. nov., an extremely thermophilic, aerobic, hydrogen-oxidizing bacterium. Int. J. Syst. Evol. Bacteriol. 34, 5–10. doi: 10.1099/00207713-34-1-5
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014.) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-59770
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
L’Haridon, S., Reysenbach, A. L., Tindall, B. L., Schönheit, P., Banta, A., Johnsen, U.,et al. (2006). Desulfurobacterium atlanticum sp. nov., Desulfurobacterium pacificum sp. nov. and Thermovibrio guaymasensis sp. nov., three thermophilic members of the Desulfurobacteriaceae fam. nov., a deep branching lineage within the Bacteria. Int. J. Syst. Evol. Microbiol. 56, 2843–2852. doi: 10.1099/ijs.0.63994-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lane, D. J. (1991). “Nucleic acid techniques in bacterial systematics,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Hoboken, NJ: Wiley), 115–174.
Liao, Z., and Guo, G. (1986). Geology of the tengchong geothermal field and surrounding area, West Yunnan, China. Geothermics 15, 339–345. doi: 10.1016/0375-6505(86)90110-0
Murphy, C. N., Dodsworth, J. A., Babbitt, A. B., and Hedlund, B. P. (2013). Community microrespirometry and molecular analyses reveal a diverse energy economy in Great Boiling Spring and Sandy’s Spring West in the U.S. Great Basin. Appl. Environ. Microbiol. 79, 3306–3310. doi: 10.1128/AEM.00139-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakagawa, S., Shtaih, Z., Banta, A., Beveridge, T. J., Sako, Y., and Reysenbach, A. L. (2005). Sulfurihydrogenibium yellowstonense sp. nov., an extremely thermophilic, facultatively heterotrophic, sulfur-oxidizing bacterium from Yellowstone National Park, and emended descriptions of the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int. J. Syst. Evol. Microbiol. 55, 2263–2268. doi: 10.1099/ijs.0.63708-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O’Neill, A. H., Liu, Y., Ferrera, I., Beveridge, T. J., and Reysenbach, A. L. (2008). Sulfurihydrogenibium rodmanii sp. nov., a sulfur-oxidizing chemolithoautotroph from the Uzon Caldera, Kamchatka Peninsula, Russia, and emended description of the genus Sulfurihydrogenibium. Int. J. Syst. Evol. Microbiol. 58, 1147–1152. doi: 10.1099/ijs.0.65431-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pagaling, E., Grant, W. D., Cowan, D. A., Jones, B. E., Ma, Y., Ventosa, A.,et al. (2012). Bacterial and archaeal diversity in two hot spring microbial mats from the geothermal region of Tengchong, China. Extremophiles 16, 607–618. doi: 10.1007/s00792-012-0460-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P.,et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 4, D590–D596. doi: 10.1093/nar/gks1219
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reysenbach, A.-L., (2001). “Aquificales ord. nov,” in Bergey’s Manual of Systematic Bacteriology, 2nd Edn, Vol. 1, The Archaea and the Deeply Branching and Phototrophic Bacteria, eds D. R. Boone, R. W. Castenholz, and G. M. Garrity (New York, NY: Springer-Verlag), 359.
Reysenbach, A.-L., Banta, A., Civello, S., Daly, J., Mitchell, K., Lalonde, S.,et al. (2005). “The Aquificales in Yellowstone National Park,” in Geothermal Biology and Geochemistry in Yellowstone National Park, eds W. P. Inskeep and T. R. McDermott (Bozeman, MT: Thermal Biology Institute, Montana State University), 129–142.
Reysenbach, A. L., Ehringer, M., and Hershberger, K. (2000). Microbial diversity at 83 degrees C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 4,61–67.
Reysenbach, A. L., Wickham, G. S., and Pace, N. R. (1994). Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60, 2113–2119.
Romano, C., D’Imperio, S., Woyke, T., Mavromatis, K., Lasken, R., Shock, E. L.,et al. (2013). Comparative genomic analysis of phylogenetically closely related Hydrogenobaculum sp isolates from Yellowstone National Park. Appl. Environ. Microbiol. 79, 2932–2943. doi: 10.1128/AEM.03591-3512
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B.,et al. (2009). Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shima, S., and Suzuki, K. I. (1993). Hydrogenobacter acidophilus sp. nov., a thermoacidophilic, aerobic, hydrogen-oxidizing bacterium requiring elemental sulfur for growth. Int. J. Syst. Evol. Bacteriol. 43, 703–708. doi: 10.1099/00207713-43-4-703
Song, Z. Q., Wang, F. P., Zhi, X. Y., Chen, J. Q., Zhou, E. M., Liang, F.,et al. (2013). Bacterial and archaeal diversities in Yunnan and Tibetan hot springs, China. Environ. Microbiol. 15, 1160–1175. doi: 10.1111/1462-2920.12025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spear, J. R., Walker, J. J., McCollom, T. M., and Pace, N. R. (2005). Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. U.S.A. 102, 2555–2560. doi: 10.1073/pnas.0409574102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stohr, R., Waberski, A., Völker, H., Tindall, B. J., and Thomm, M. (2001). Hydrogenothermus marinus gen. nov., sp. nov., a novel thermophilic hydrogen-oxidizing bacterium, recognition of Calderobacterium hydrogenophilum as a member of the genus Hydrogenobacter and proposal of the reclassification of Hydrogenobacter acidophilus as Hydrogenobaculum acidophilum gen. nov., comb. nov., in the phylum ‘Hydrogenobacter/Aquifex’. Int. J. Syst. Evol. Microbiol. 51, 1853–1818. doi: 10.1099/00207713-51-5-1853
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takacs, C. D., Ehringer, M., Favre, R., Cermola, M., Eggertsson, G., Palsdottir, A.,et al. (2001). Phylogenetic characterization of the blue filamentous bacterial community from an Icelandic geothermal spring. FEMS Microbiol. Ecol. 35, 123–128. doi: 10.1111/j.1574-6941.2001.tb00795.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takacs-Vesbach, C., Inskeep, W. P., Jay, Z. J., Herrgard, M. J., Rusch, D. B., Tringe, S. G.,et al. (2013). Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three Aquificales lineages. Front. Microbiol. 4:84. doi: 10.3389/fmicb.2013.00084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takai, K., Komatsu, T., and Horikoshi, K. (2001). Hydrogenobacter subterraneus sp. nov., an extremely thermophilic, heterotrophic bacterium unable to grow on hydrogen gas, from deep subsurface geothermal water. Int. J. Syst. Evol. Microbiol. 51, 1425–1435.
Wang, C. S., Zhao, X. X., Liu, Z. F., Lippert, P. C., Graham, S. A., Coe, R. S.,et al. (2008). Constraints on the early uplift history of the Tibetan Plateau. Proc Natl. Acad. Sci. U.S.A. 105, 4987–4992. doi: 10.1073/pnas.0703595105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yarza, P., Yilmaz P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K. H.,et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330.
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Aquificales, Hydrogenobacter, Hydrogenobaculum, Sulfurihydrogenibium, hot springs, hydrogen oxidation, sulfide oxidation, thiosulfate oxidation
Citation: Hedlund BP, Reysenbach A-L, Huang L, Ong JC, Liu Z, Dodsworth JA, Ahmed R, Williams AJ, Briggs BR, Liu Y, Hou W and Dong H (2015) Isolation of diverse members of the Aquificales from geothermal springs in Tengchong, China. Front. Microbiol. 6:157. doi: 10.3389/fmicb.2015.00157
Received: 18 November 2014; Accepted: 11 February 2015;
Published online: 27 February 2015.
Edited by:
Jesse Dillon, California State University, USAReviewed by:
John R. Spear, Colorado School of Mines, USATim McDermott, Montana State University, USA
Copyright © 2015 Hedlund, Reysenbach, Huang, Ong, Liu, Dodsworth, Ahmed, Williams, Briggs, Liu, Hou and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian P. Hedlund, School of Life Sciences, University of Nevada, Las Vegas and Nevada Institute of Personalized Medicine, Las Vegas, NV 89154-4004, USA e-mail: brian.hedlund@unlv.edu; Anna-Louise Reysenbach, Biology Department and Center for Life in Extreme Environments, Portland State University, Portland, OR 89154-4004, USA e-mail: reysenbacha@pdx.edu; Hailiang Dong, State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Beijing 100083, China e-mail: dongh@miamioh.edu
 Brian P. Hedlund
Brian P. Hedlund Anna-Louise Reysenbach
Anna-Louise Reysenbach Liuquin Huang4
Liuquin Huang4 Zizhang Liu
Zizhang Liu Jeremy A. Dodsworth
Jeremy A. Dodsworth Amanda J. Williams
Amanda J. Williams Hailiang Dong
Hailiang Dong