- Department of Applied Microbiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland
The mobilome is a pool of genes located within mobile genetic elements (MGE), such as plasmids, IS elements, transposons, genomic/pathogenicity islands, and integron-associated gene cassettes. These genes are often referred to as “flexible” and may encode virulence factors, toxic compounds as well as resistance to antibiotics. The phenomenon of MGE transfer between bacteria, known as horizontal gene transfer (HGT), is well documented. The genes present on MGE are subject to continuous processes of evolution and environmental changes, largely induced or significantly accelerated by man. For bacteria, the only chance of survival in an environment contaminated with toxic chemicals, heavy metals and antibiotics is the acquisition of genes providing the ability to survive in such conditions. The process of acquiring and spreading antibiotic resistance genes (ARG) is of particular significance, as it is important for the health of humans and animals. Therefore, it is important to thoroughly study the mobilome of Aeromonas spp. that is widely distributed in various environments, causing many diseases in fishes and humans. This review discusses the recently published information on MGE prevalent in Aeromonas spp. with special emphasis on plasmids belonging to different incompatibility groups, i.e., IncA/C, IncU, IncQ, IncF, IncI, and ColE-type. The vast majority of plasmids carry a number of different transposons (Tn3, Tn21, Tn1213, Tn1721, Tn4401), the 1st, 2nd, or 3rd class of integrons, IS elements (e.g., IS26, ISPa12, ISPa13, ISKpn8, ISKpn6) and encode determinants such as antibiotic and mercury resistance genes, as well as virulence factors. Although the actual role of Aeromonas spp. as a human pathogen remains controversial, species of this genus may pose a serious risk to human health. This is due to the considerable potential of their mobilome, particularly in terms of antibiotic resistance and the possibility of the horizontal transfer of resistance genes.
Introduction
Bacteria of the genus Aeromonas are common in a variety of environments. They have been isolated from water, mammals, fish, invertebrates, birds, insects, soil (Palumbo et al., 1985; Ceylan et al., 2009) as well as from food (Neyts et al., 2000; Kingombe et al., 2004). However, they mainly inhabit all kinds of aquatic environments, such as rivers, lakes, ponds, estuaries of marine waters, drinking water and groundwater as well as wastewater at various stages of purification (Gordon et al., 2008; Reith et al., 2008; Moura et al., 2012). The genus Aeromonas is composed of a large number of species (31 species and 12 subspecies Martin-Carnahan and Joseph, 2005; http://www.bacterio.net/aeromonas.html) but only a few of them have been found to be primarily pathogens of fish and warm-blooded animals, including humans. In fish, mainly mesophilic A. hydrophila, A. veronii bv. sobria and psychrophilic strains of A. salmonicida are predominantly responsible for fish infections, e.g., furunculosis (Burr et al., 2005; Dallaire-Dufresne et al., 2014), but A. caviae, A. jandaei, A. sobria, A. bestiarum have also been reported to cause several known types of diseases as well as unusual infections, e.g., epizootic ulcerative syndrome (Rahman et al., 2002). Mesophilic A. hydrophila, A. caviae, and A. veronii bv. sobria strains are important human pathogens, responsible for a variety of infectious complications in both immunocompetent and immunocompromised individuals. They cause various types of infections of the digestive system, such as gastroenteritis (Holmberg and Farmer, 1984; Figueras, 2005; Edberg et al., 2007), respiratory (Bossi-Küpfer et al., 2007) and genitourinary infections (Al-Benwan et al., 2007; Huang et al., 2007), wound infections, infections of skin and soft tissue (Jorge et al., 1998; Vally et al., 2004; Chim and Song, 2007), sepsis (Ko et al., 2000; Lau et al., 2000; Tsai et al., 2006), eye infections (Khan et al., 2007), and meningitis (Seetha et al., 2004).
The mechanisms of the pathogenicity of Aeromonas spp. are not yet well understood, this being a concern because of recent reports of antibiotic resistant clinical strains (Ghenghesh et al., 2008; Wu et al., 2015). Moreover, cases of isolation of pathogenic strains from the environment are increasingly frequent, which can pose a serious threat to public health during natural disasters (Lin et al., 2013). Taking into account the pathogenicity potential of Aeromonas spp. and differences in antibiotic resistance profiles it seems reasonable to study the genetic background of these phenomena. In our study we have decided to focus on the mobile part of the Aeromonas spp. genome, that is the mobilome, since this topic has not yet been comprehensively reviewed. Based on the current knowledge we have focused in this review on plasmids, which carry a number of transposons, integron-associated gene cassettes and IS elements, and encode such determinants as antibiotic and heavy metal resistance, as well as virulence factors.
Mobile Genetic Elements and Mechanisms of Horizontal Gene Transfer
The mobilome is the total pool of mobile genes in the genome and consists of mobile genetic elements (MGE), such as plasmids, insertion sequences (IS), transposons, integron-associated gene cassettes and bacteriophages. Plasmids are mostly double-stranded and circular independent replicons of extrachromosomal DNA. They cover a variety of sizes from small, often cryptic plasmids to large megaplasmids with many features allowing them to adapt to different environmental conditions (Table 1). Other MGE are transposable elements (TE) such as the insertion sequences (IS) and transposons (Tn) (Table 2). IS are the most simple TE that reach about 0.5–3 kb and very often are flanked by short sequences of inverted repeats (IR). A transposase gene, encoding the transposition of an IS, is usually located between IRs. Transposons have a more complex structure, because in addition to the transposase, they also harbor various genes responsible for specific phenotypes (Oliver et al., 2013). Integrons are non-replicative genetic elements, which are able to capture and incorporate gene cassettes by site-specific recombination. They are composed of three main elements: the intI gene, coding for a site-specific recombinase of the integrase family, specific recombination site attI, where a gene cassette may be inserted, and the Pc promoter, managing the transcription of the captured gene (Stokes and Hall, 1989).
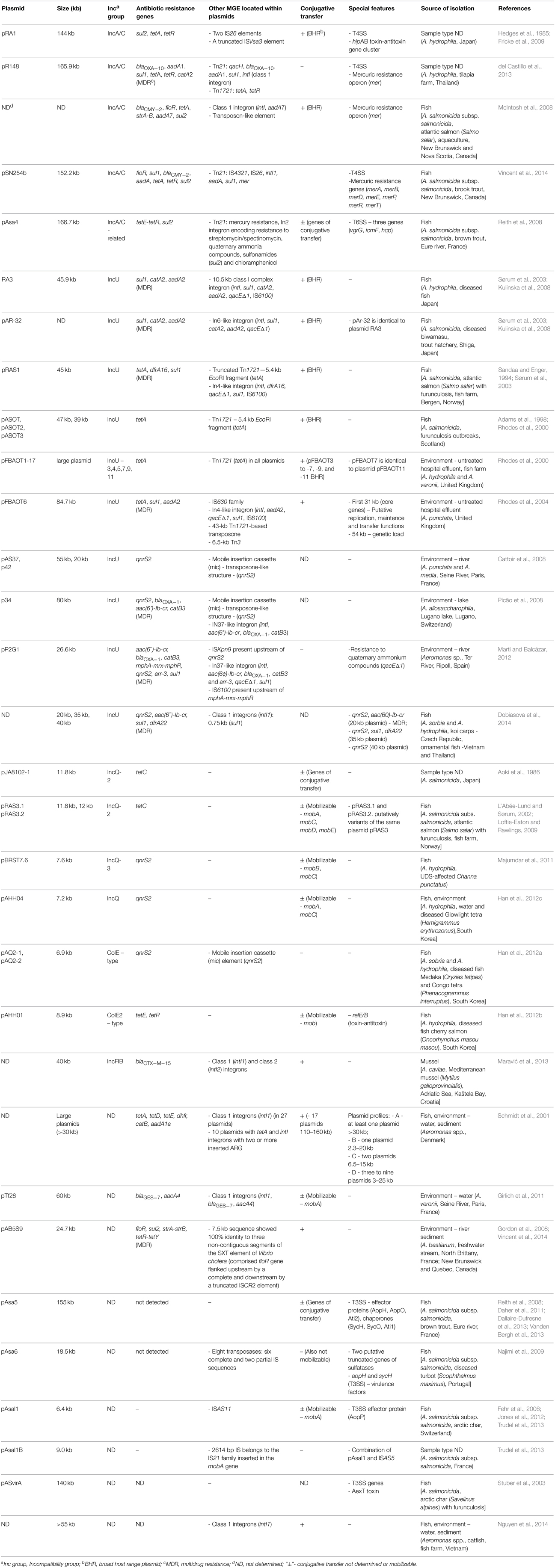
Table 1. Plasmids of Aeromonas spp., their origin and virulence features, and the presence of resistance determinants and mobile elements.
MGE are ubiquitous among all prokaryotes and play a significant role in horizontal gene transfer (HGT) and interspecies dissemination of resistance and virulence determinants (Brouwer et al., 2011; Oliveira et al., 2014). HGT occurs mainly by three mechanisms: DNA transformation, conjugative transfer involving plasmids, and other conjugative elements (conjugative transposons) and transduction by phages (Thomas and Nielsen, 2005). In the case of Aeromonas spp. until now gene transfer by transduction has never been observed. However, Aeromonas spp. phages have been identified in various environments, such as A. hydrophila phages Aeh1 and Aeh2 from sewage (Chow and Rouf, 1983), A. hydrophila phage CC2 from sewage in China (Shen et al., 2012), A. salmonicida phage phiAS4 from river in Korea (Kim et al., 2012a), and A. salmonicida phage PAS-1 from aquaculture in Korea (Kim et al., 2012b). Additionally, prophages have been also detected in Aeromonas spp.: Φ O18P (Myoviridae) in A. media isolated from a pond in Germany (Beilstein and Dreiseikelmann, 2008), AH1, AH2, AH3, AH4, and AH5 in A. hydrophila isolated from epidemic outbreak of catfish in the USA (Hossain et al., 2013). Hossain et al. (2013) detected five putative prophages (AH1-5) located in epidemic-associated regions in the genome. Unfortunately, their studies have not shown the ability of the prophages to transduce any bacterial genes. The other two mechanisms of HGT (DNA transformation and conjugative transfer by plasmids) are ubiquitous among Aeromonas spp. and will be the subject of this review. Various MGE, such as plasmids, transposons, or insertion sequences have been isolated from aeromonads, and many of them, regardless of the strain origin, have been found to carry resistance or virulence determinants (Sørum et al., 2003; Dallaire-Dufresne et al., 2014).
Antibiotic Resistance Genes on MGE
Over the years, there has been very little research on the in vitro susceptibility of Aeromonas bacteria isolated from clinical material to various chemotherapeutic agents. Most of the available information is focused on antibiotics used to treat infections caused by A. hydrophila, A. caviae, and A. veronii bv. sobria, and it is not yet certain whether these results can be extrapolated to other species of this genus (Fricke et al., 2009; Girlich et al., 2011; Maravić et al., 2013). The Clinical and Laboratory Standards Institute (CLSI) has recently published guidelines for the assessment of the sensitivity of clinical isolates of Aeromonas spp. using disk diffusion and MIC tests, but these data are based on testing of the three above- mentioned-, most clinically relevant species of Aeromonas (Jorgensen and Hindler, 2007) plus A. jandaei and A. schubertii (Clinical and Laboratory Standards Institute, 2006). The most commonly administered antibiotics In the treatment of Aeromonas infections are ciprofloxacin, levofloxacin, sulfamethoxazole/trimethoprim, amikacin, gentamicin, ciprofloxacin, and trimethoprim (Jones and Wilcox, 1995). Sensitivity analysis of clinical strains demonstrated that more than half of the strains tested were resistant to antibiotics of the following groups: antifolates (sulfamethoxazole), cephalosporins, penicillins (amoxicillin, ampicillin, ampicillin-sulbactam, oxacillin, penicillin, ticarcillin. The susceptibilities were determined by agar dilution or disc diffusion method, respectively, according to CLSI guidelines, but the ARG were not pin-pointed (Lamy et al., 2009; Aravena-Román et al., 2012). However, the susceptibility profile of individual strains can also vary depending on the particular species, different geographical localization and environment in which they occur (Ghenghesh et al., 2008). Such differences could be related to the recommended approach for the treatment of Aeromonas infections in different countries.
On the basis of fairly abundant literature data concerning the antibiotic resistance of environmental Aeromonas strains, it can be concluded that this phenomenon mostly concerns strains isolated from various water environments, including wastewater (Figueira et al., 2011), natural waters such as rivers (Girlich et al., 2011), lakes (Picão et al., 2008), and estuaries (Fiorentini et al., 1998; Henriques et al., 2006), aquacultures (Schmidt et al., 2001; Jacobs and Chenia, 2007; Yi et al., 2014a,b), and urban drinking water (Carvalho et al., 2012). ARG recently found in water strains encoded resistance to four major groups of antibiotics: β-lactams, quinolones, aminoglycosides, tetracyclines and less frequently to sulfonamides and trimethoprim, chloramphenicol, florfenicol, macrolides, streptogramins, streptothricin, and ansamycins. In general, the resistance profile and the presence of specific resistance genes depends on the particular aquatic environment (Piotrowska and Popowska, 2014). Given the risk to human health, the incidence of ARG is alarming, particularly among A. hydrophila, A. caviae, and A. sorbia, which are considered opportunistic pathogens responsible for infections in both fish and humans (Alcaide et al., 2010; Ottaviani et al., 2011; Shak et al., 2011; Dias et al., 2012; Yi et al., 2014b). The localization of ARG and virulence determinants of Aeromonas spp. on MGE such as plasmids, insertion sequences, transposons and mobile integron gene cassettes have been determined by many environmental studies. There is some literature data on the localization of ARG among clinical Aeromonas strains. However, in the recent publications, the presence of ARG on plasmids (e.g., MOX, TEM, PSE, and CTX-M β-lactamase genes, sul1 and sul2) has been confirmed, but no characteristics have been provided (Ye et al., 2010; Puah et al., 2013). Moreover, among clinical strains three cases of β-lactamases genes located within integrons: blaVIM from A. caviae (Adler et al., 2014), blaVIM−4 from A. hydrophila (Libisch et al., 2008) blaIMP which also was located on 35-kb plasmid from A. caviae have been identified (Neuwirth et al., 2007). Also (Wu et al., 2011) identified the ESBL gene blaPER−3 in two A. caviae isolates. The gene was located in both chromosomes and plasmids. Additionally, there is only one clinical report of gene qnrS2 that has been found on a plasmid isolated from A. veronii (Sánchez-Céspedes et al., 2008). However, all these reports are still sufficient enough to look for any gene-MGE correlation and consequently research on a larger scale should be conducted.
Plasmids
Analysis of a number of studies on Aeromonas spp. showed the prevalence of different incompatibility groups of plasmids, i.e., IncA/C, IncU, IncQ, IncF, IncI, and ColE-type, with the greatest frequency of the first two groups (Table 1). The vast majority of plasmids carry a number of different determinants such as antibiotic and metal resistance genes or virulence factors. Aeromonas spp. very often carry resistance plasmids (R-plasmids) of various length belonging to different incompatibility groups and of worldwide spread. Furthermore, numerous R-plasmids contain multidrug-resistance (MDR) to three or more antimicrobial classes (according to the European Centre of Disease Prevention and Control, Magiorakos et al., 2012). Of particular concern is the fact that most of the isolated plasmids are broad-host-range (BHR), capable of conjugative transfer (tra genes) or capable of mobilization (mob genes).
Plasmids of the IncA/C incompatibility group have been described as conjugative and BHR, and capable of spreading multidrug-resistance. A variety of isolates among different bacterial genera, such as Escherichia, Salmonella, Vibrio, or Yersinia (Winokur et al., 2001; Pan et al., 2008; Guo et al., 2014) of environmental, animal and human origin have been reported to carry these plasmids, which increases public health concerns worldwide (Mataseje et al., 2010). The first member of the IncA/C family was the pRA1 plasmid isolated in 1971 from A. hydrophila derived from Japan (Hedges et al., 1985). The complete DNA sequence of this large plasmid (144 kb) revealed that pRA1 and other members of the IncA/C family shared 100 kb of a highly conserved plasmidic backbone with more than 80% nucleotide sequence identity. Among the most important core genes are those that encode type IV secretion-like conjugative transfer operons. A complete set of the type IV secretion system operons was also found on pR148 (MDR plasmid) of IncA/C group isolated from diseased fish from Thailand (del Castillo et al., 2013). Another interesting feature was the hipAB toxin-antitoxin gene cluster, which was also partially (hipAB-related gene cluster) described in the IncA/C-related plasmid pAsa4, isolated from the fish pathogen A. salmonicida subsp. salmonicida (Fricke et al., 2009). According to the BLAST database, integrating conjugative elements (ICE) were identified as the closest relatives of IncA/C plasmids (Fricke et al., 2009). Antimicrobial resistance profile of pRA1 is reduced compared with all other IncA/C plasmids sequenced of Aeromonas spp., as it is limited to tetracyclines (tetRA cluster) and sulfonamides (sul2) (McIntosh et al., 2008; del Castillo et al., 2013). The sul2 gene was located next to the truncated ISVsa3 element, which has been observed previously in an IncA/C plasmid isolated from a Spanish S. enterica strain (García et al., 2011). Class D tetRA gene cluster was found within two IS26 elements that played a key role in the distribution of ARG on different plasmids (Cullik et al., 2010). In addition to antibiotics, heavy metals are also implicated as potential substances that can co-select antibiotic resistance in the environment, resulting in a frequent presence of heavy metal resistance genes on the same MGE as ARG (Lazar et al., 2002). Mercury resistance operons (mer) have been found on IncA/C plasmids such as pR148 or pAsa4 in Aeromonas spp. The plasmid isolated from A. salmonicida subsp. salmonicida showed 100% nucleotide sequence homology to the mer operon carried by S. enterica IncA/C plasmid pSN254 (McIntosh et al., 2008; del Castillo et al., 2013). The pR148 plasmid also carries genes encoding resistance to quaternary ammonium compounds. Moreover, phylogenetic comparative studies revealed that pR148 is the most closely related to human pathogenic E. coli and Acinetobacter baumanii. This similarity indicates that the IncA/C group of plasmids was transferred between different genera (McIntosh et al., 2008; Moura et al., 2012). Furthermore, the IncA/C MDR plasmid isolated by McIntosh et al., 2008 carried floR, tetA, sul2, and strA/strB sequences on a cassette that had 99.9% nucleotide sequence homology to that of the pSN254 plasmid isolated from S. enterica. Recently Vincent et al. (2014) identified a 152-kb pSN254b plasmid which is a different variant of pSN254. This MDR plasmid provides resistance to chloramphenicol (floR), florfenicol (floR), streptomycin (aadA), spectinomycin (aadA), tetracycline (tet), sulfonamide (sul1), beta-lactam antibiotics (blaCMY−2), quaternary ammonium compounds (sugE2), and mercury (merA, merB, merD, merE, merP, merR, merT). There is no strong correlation between IncA/C plasmids and other MGE, but suggestive associations have been observed and will be described in the next chapter.
Plasmids of the IncU group have been isolated from many clinical and environmental strains of Esherichia coli and Aeromonas spp. Conjugative and BHR plasmids are members of this group and are also involved in the dissemination of antibiotic resistance among Aeromonas spp. This group of plasmids is widely distributed around the world (Table 1) and it has been postulated that they share a conserved backbone structure with a variable region limited to resistance-determining genes (Rhodes et al., 2000; Sørum et al., 2003). The RA3 plasmid (45.9 kb) was isolated from A. hydrophila in Japan and serves as the reference plasmid of the IncU group (Kulinska et al., 2008). Functional analysis demonstrated that RA3, as a BHR plasmid, could self-transfer, replicate and be stably maintained in Alpha-, Beta-, and Gammaproteobacteria. RA3, similarly to other members of the IncU group, is a MDR plasmid and contains a 10.5-kb antibiotic resistance region that comprises class I integron with sul1, catA2, aadA2, qacE resistance genes. The pAr-32 plasmid, isolated from A. salmonicida in Japan in 1970 is very similar to the RA3 plasmid and carries the same integron cassette, which is highly similar to the In6 integron of the pSa plasmid (Sørum et al., 2003). Another IncU plasmid (pRAS1) was isolated from A. salmonicida from Norway in 1989 and had the same backbone structure as pAR-32. The region controlling drug resistance in pRAS1 contains two main elements: the complete class 1 In4-like integron with dfrA16, qacE, sul1 gene cassette and a fragment of the Tn1721 transposon carrying tetA resistance gene. The study of Schmidt et al. (2001) demonstrated a positive correlation between oxytetracycline resistant strains of Aeromonas spp. containing large plasmids, and the presence of tetA genes. Among IncU R-plasmids, such as pRAS1, pASOT, or pFBOAT, tetracycline resistance determinants were observed in the complete or truncated Tn1721 (Adams et al., 1998; Rhodes et al., 2000). In all cases the TetA determinant was located within a 5.5-kb EcoRI restriction fragment. Based upon RFLP assessment, antibiotic resistance, and frequency of transfer all these tetracycline resistance encoding plasmids are considered to be closely related to plasmid pIE420 isolated from a German hospital strain of E. coli (Rhodes et al., 2000). The study of Rhodes et al. (2004) showed that plasmids pRAS1 and pIE420 are probably identical. These results support the hypothesis that IncU is an evolutionarily narrow group. However, Rhodes et al. (2004) also characterized plasmid pFBOAT6 (84.7 kb), which had a 31-kb region of core genes and a 54-kb region of genetic load, which made this plasmid almost twice as large as the other IncU plasmids. This was due to the presence of a 43-kb resistance region flanked by Tn1721. This region is highly similar to those of the pXF51, pIPO2, and pSB102 plasmids found in a plant-associated bacterial hosts. Nevertheless, only several nucleotide differences in the core genes were found between RA3 and pFBAOT6 plasmids.
Many IncU plasmids also harbor quinolone resistance determinants–qnrS2 and aac(6′)-Ib-cr (Table 1). Plasmid-mediated qnr genes have been identified in many Enterobacteriaceae isolates (Nordmann and Poirel, 2005; Kehrenberg et al., 2006; Pasom et al., 2013) and recently also in Pseudomonas spp. (Cayci et al., 2014). In addition, the qnrS2 gene has been recently detected in A. caviae clinical strain isolated from a stool sample collected from a patient with gastroenteritis (Arias et al., 2010). Among the environmental isolates of Aeromonas spp., the qnrS2 gene was found in the following plasmids: pAS37 and p42 from French strains of A. punctata and A. media (Cattoir et al., 2008), p34 from a Swiss strain of A. allasacharophila (Picão et al., 2008), pP2G1 from a Spanish strain (Marti and Balcázar, 2012) and recently isolated unnamed plasmids from Thai strains of A. sorbia and A. hydrophila (Dobiasova et al., 2014). All of the described plasmids are medium to large in size (20–80 kb) and have an interesting genetic descent. The first two plasmids, pAS37 and p34, contain the qnrS2 gene as a part of a novel transposon-like genetic structure called the mobile insertion cassette (mic) instead of the transposase gene. Moreover, this specific mobile element has been previously found in a Bacillus cereus strain, which makes mic a possible vector of ARG between environmental and clinical pathogens (de Palmenaer et al., 2004). Furthermore, the study of Han et al. (2012a) revealed two small (6.9 kb) plasmids (pAQ2-1 and pAQ2-2) carrying the mic-qnr2S structure. These ColE-type plasmids were 99% identical and genes for plasmid replication were organized in a similar way to ColE2-type cryptic plasmids pAsa1, pAsa2, and pAsa3, isolated from A. salmonicida subsp. salmonicida (Boyd et al., 2003). This observation suggests that these mic-type structures are potential vehicles of plasmid-mediated quinolone resistance determinants among different groups of plasmids in various geographical locations, and more importantly in clinically relevant strains.
Plasmids of the IncQ group are also strongly associated with quinolone resistance that have been identified among Aeromonas spp. These small, mobilizable plasmids (5.1–14.2-kb) are BHR and are found in many bacterial species worldwide (Loftie-Eaton and Rawlings, 2009). Among Aeromonas spp., two plasmids (pBRST7.6 and pAHH04) isolated from A. hydrophila strains from diseased fish and water samples harbored qnrS2 genes (Majumdar et al., 2011; Han et al., 2012c). In addition, the exogenous pGNB2 plasmid obtained from the wastewater treatment plant in Germany also harbored the qnrS2 gene (Bönemann et al., 2006). In contrast to IncU plasmid-mediated qnr genes, quinolone determinants of the IncQ plasmids were not associated with any mic or integron, and did not harbor any additional resistance determinants. Moreover, the pJA8102-1 plasmid found in A. salmonicida from Japan, and pRAS3.1 and pRAS3.2 of Norwich A. salmonicida strains carried tetAR(C) genes (L'Abée-Lund and Sørum, 2002). However, it is worth to emphasize that pRAS3.1 and pRAS3.2 are considered variants of the same plasmid pRAS3, which was also identified in a Scottish A. salmonicida strain and appears to be identical to the R-plasmid pJA8102-2.
In addition to the frequently occurring BHR plasmids that were discussed earlier (IncA/C, IncU, IncQ), plasmids belonging to IncFrepB, IncFIB, IncFIC and IncI groups were also observed in the genus Aeromonas (Han et al., 2012b; Moura et al., 2012; Maravić et al., 2013). The study of Maravić et al. (2013) found 40-kb conjugative plasmids described as narrow host range IncFIB group in 11 A. caviae strains isolated from Croatian mussels. All the vectors carried the blaCTX−M−15 gene, encoding ESBL β-lactamase. The same β-lactamase was also identified in IncFIB plasmids isolated from E. coli, and a large (210 kb), non-conjugative IncFIA plasmid identified in an A. hydrophila strain (Dolejska et al., 2011; Gómez-Garcés et al., 2011).
Insertion Sequences and Transposons
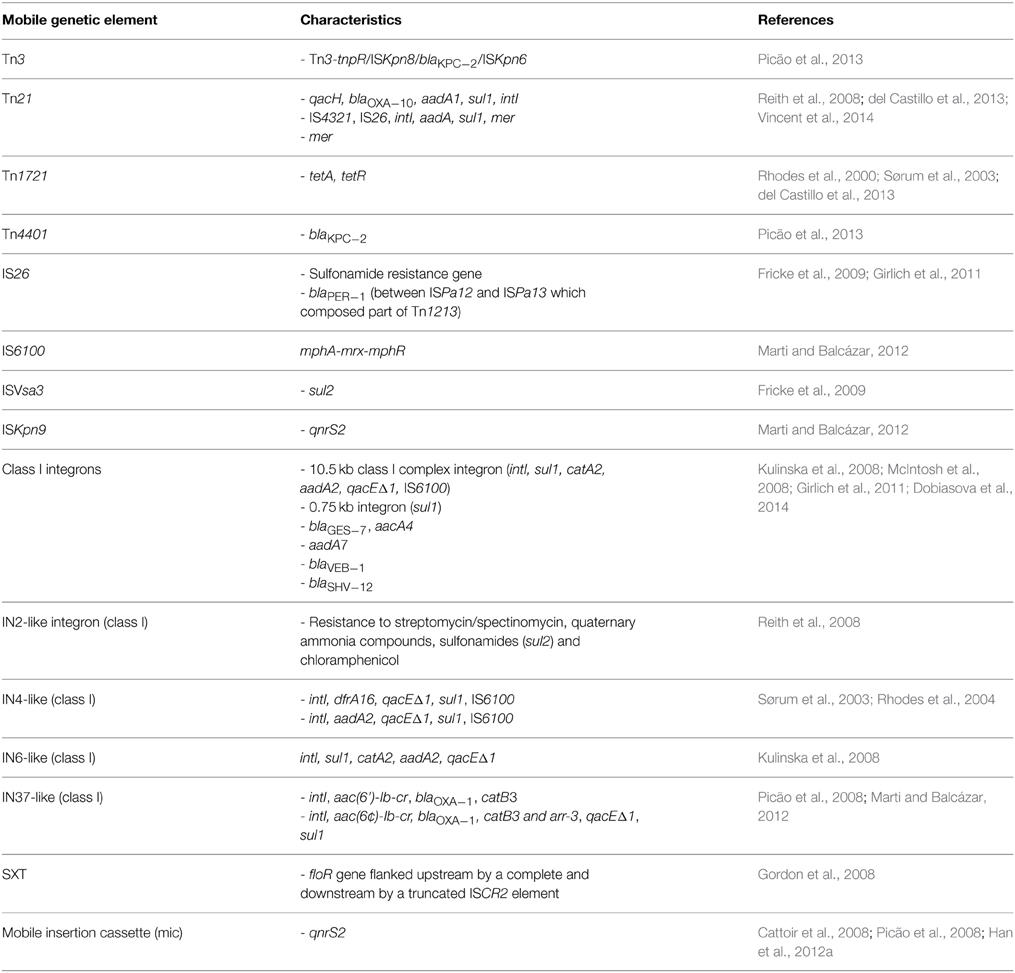
Aeromonas spp. often contain transposons located on plasmids and chromosomes (Tn3, Tn21, Tn1213, Tn1721, Tn4401) as well as insertion sequences (e.g., IS26, ISPa12, ISPa13, ISKpn8, ISKpn6) (Tables 1, 2). This makes the mobilome an even more complex structure that likely plays an important role in the dissemination of various resistance and virulence determinants.
McIntosh et al. (2008) found a transposon-like element that contained the blaCMY−2 β-lactamase gene. This element is known to be widely distributed among foodborne and clinical Salmonella strains as well as other Enterobacteriaceae in Asia and the United States. Other transposons (e.g., Tn21) are involved in the dissemination of ARG and mercury resistance genes (as in pAsa4) between gram-negative bacteria (Liebert et al., 1999). Transposon Tn21 located on the pR148 plasmid carried qacH, blaOXA−10, aadA1, and sul1 cassette, which showed 100% similarity (when the last gene is excluded) to Acinetobacter baumanii AYE genome. A Tn1721-like transposon, conferring tetracycline resistance via tetA/R genes, was identified on the same plasmid. Tn1721 belongs to the Tn501 subfamily and the Tn3 family of transposons. The involvement of Tn1721 and Tn1721-like elements in the dissemination of the tetA gene has been observed in many other studies (Rhodes et al., 2000; Pasquali et al., 2005; Girlich et al., 2010). This transposon is also ubiquitous among IncU plasmids, which form another important group within Aeromonas spp.
Furthermore, many transposons and various insertion sequences were identified among Aeromonas spp. in association with β-lactamases. The study of Girlich et al. (2011) revealed many ESBL β-lactamases of different genetic backgrounds. The blaSHV−12 gene was preceded by IS26 while the blaPER−1 gene was located between ISPa12 and ISPa13, thus forming a part of a composite transposon Tn1213. It is also alarming that blaKPC−2 genes encoding carbapenemases have been isolated from Aeromonas spp. recovered from hospital sewage (Picão et al., 2013). The blaKPC−2 gene was found on the Tn4401 transposon and Tn3-tnpR/ISKpn8/blaKPC−2/ISKpn6 array.
Integrons
Integrons are widely distributed bacterial genetic elements that are able to acquire gene cassettes frequently containing ARG. Most of them belong to the 1st, 2nd, or 3rd class of integrons and contain intI1, intI2, or intI3 integrase genes, respectively (Hall et al., 1999). Integrons harbored by plasmids, transposons and other mobile structures are called “mobile integrons” (MI), because MGE promote their dissemination. For this reason, MI are also involved in spreading antibiotic resistance in the environment (Laroche et al., 2009). Integrons found in Aeromonas spp. mainly belong to the class 1 and carry a number of antibiotic resistance gene cassettes (Tables 1, 2). Schmidt et al. (2001) demonstrated that class 1 integrons frequently occurred on oxytetracycline resistance plasmids (most often tetA), but they did not observe any strong correlations between integrons and tet genes or any other group of plasmids. Similar results were reported by Jacobs and Chenia (2007), who observed class 1 integrons and tet genes in 68.4% isolates that also harbored different types of plasmid profiles. However, the study of Moura et al. (2012) demonstrated a positive correlation between integrons and FrepB and I1 plasmids isolated from Aeromonas spp. from wastewater. Furthermore, Schmidt et al. (2001) reported a close association of sulfadiazine/trimethoprim resistance and class 1 integrons, which manifested in the presence of sul1 and dfr gene cassette inserts in class 1 integrons. Henriques et al. (2006), detected intI genes in 21% of Aeromonas and 29.6% of Enterobacteriaceae isolates. The most often found resistance gene cassettes contained various aadA genes, which were also observed in later studies (Moura et al., 2007; Koczura et al., 2014; Sarria-Guzmán et al., 2014). Integrons are also correlated with β-lactamase genes, e.g., genes blaVEB−1 and blaSHV−12were located in class 1 integrons on plasmids of different sizes (30–170 kb) (Jacobs and Chenia, 2007; Carvalho et al., 2012). Integrons of class 2 were found in a couple of studies that indicated putative chromosomal location of these integrons (Carvalho et al., 2012). Other integrons belonging to class 1, such as IN2-like, IN4-like, IN6-like, IN37-like, or SXT, as well as many undescribed integrons, have been also reported on Aeromonas spp. plasmids (Table 1).
Virulence Factors on MGE
The role of potential virulence factors in the pathogenesis of Aeromonas spp. is not yet fully understood. However, several factors probably play important roles in the host infection process. In addition to adhesive factors, the capsular polysaccharide of Aeromonas (Khan et al., 2008), bacterial flagella and pili are needed for the first stage of infection. Lysis proteases, which are involved in the second step (metalloproteases, serine proteases, and aminopeptidases) are capable of degrading complex proteins present in the serum and connective tissue (Merino et al., 1996; Han et al., 2008; Imamura et al., 2008; Ottaviani et al., 2011; Puthucheary et al., 2012; John and Hatha, 2013). Many other factors are also likely to play important roles in infections, including specific proteins required for adaptive acid tolerance, biofilm formation and synthesis of autoinducers (e.g., acyl-homoserine lactone) in the quorum sensing process (Jangid et al., 2007) and type three secretion systems (Vanden Bergh et al., 2013). S-layers are also important, as they have the ability to bind different proteins, such as the extracellular matrix proteins fibronectin, laminin, and vitronectin, which provide a defense against the components of the serum and protease digestion (Noonan and Trust, 1997). The main pathogenic factors of Aeromonas spp. have also been observed on several plasmids (Table 1). Genes coding for the AexT toxin and three types of secretion systems—type III (T3SS), type IV (T4SS), and type VI (T6SS) have been identified on virulence plasmids (Table 1).
Six virulence plasmids were isolated from strains of the fish pathogen A. salmonicida i.e., pASvirA from diseased fish, pAsa4 and pAsa5 from the same French strain, pAsa6 from diseased fish in Portugal (Stuber et al., 2003; Reith et al., 2008; Najimi et al., 2009) and pAsal1 and pAsal1B (Fehr et al., 2006; Jones et al., 2012; Trudel et al., 2013). Three plasmids were large (140–166.7 kb), i.e., pASvirA, pAsa4 and pAsa5, and in addition the latter two harbored conjugative transfer genes. Plasmid pAsa5 contained most of the T3SS genes that have been shown to be required for virulence in A. salmonicida. Three putative effector proteins (AopH, AopO, Ati2) and their associated chaperones (SycH, SycO, Ati1) were identified in it. The recommended temperature for growth of A. salmonicida by Bergey's Manual of Systematic Bacteriology is 22–28°C. However, several studies demonstrated that culturing at 25°C resulted in a lack of virulence due to the loss of virulence factors, such as the A-layer (Ishiguro et al., 1981), T3SS region (Stuber et al., 2003), or AexT toxine (Najimi et al., 2009). The mechanism of this phenomenon is not fully understood, but Stuber et al. (2003) explained it as a result of the loss of a virulence plasmid. As an example, the loss of plasmid pASvirA is accompanied by the inability of A. salmonicida to secrete AexT and loss of virulence toward RTG-2 cells. However, Tanaka et al. (2013) proposed a mechanism of the loss of virulence mediated by IS, wherein rearrangements are caused by recombination of three IS from thermolabile plasmids, e.g., pAsa5 or pASvirA. A consequence of the recombination of ISAS1, ISAS2, and ISAS11 is the deletion of the T3SS region in A. salmonicida, resulting in the loss of virulence. This is consistent with a previous study that showed a large number of IS in A. salmonicida genome being involved in virulence gene disruption with the formation of pseudogenes (Reith et al., 2008). Sequencing and analysis studies of the total genome of A. salmonicida subsp. salmonicida A449 revealed the occurrence of 88 complete IS sequences of different types: ISAs1, ISAs2, ISAs3, ISAs4, ISAs5, ISAs6, ISAs7, ISAs8, ISAs9, ISAs10, ISAs11 and a significant number of pseudogenes (170). Many putative transposons and IS sequences, as well as AopH and SycH proteins are also present on pAsa6. This vector is a non-mobilizable 18-kb plasmid with characteristic strong homology to many pAsa5 genes (Najimi et al., 2009). Comparative analyses suggested that pAsa6 might be derived from pAsa5 through a deletion of numerous genes, or conversely, pAsa5 might have been formed as a fusion of a pAsa6-like plasmid with another megaplasmid. This is even more interesting when one considers the fact that both plasmids were isolated in different countries. Three genes of T6SS (vgrG, icmF, hcp) were found on pAsa4, but the majority of them were located on the chromosome of A. salmonicida subs. salmonicida or A. hydrophila (Reith et al., 2008). It has been demonstrated that T6SS plays an important role in the pathogenesis of Aeromonas strains, in the translocation of hemolysin protein (Hcp) into the host cells (Suarez et al., 2008). In addition, two small mobilizable plasmids, pAsal1 and pAsal1B (6.7 and 9.0 kb, respectively) encoding T3SS effector protein AopP, were recently discovered (Trudel et al., 2013). AopP has been reported to have a inhibitory activity against the NF-κB pathway in cultured cells (Fehr et al., 2006) and has potent pro-apoptotic activity when expressed in cultured mammalian macrophage or epithelial cells (Jones et al., 2012). Trudel et al. (2013) showed that pAsalB is a combination of pAsal1 and ISAS5, where this 2614 bp IS belongs to the IS21 family inserted in the mobA gene sequence of the pAsal1 plasmid.
Conclusions
Bacteria from the genus Aeromonas have a complex mobilome consisting of many different MGE. This review presents the characteristics of more than 26 plasmids belonging to different incompatibility groups, all of which were isolated from environmental strains. Resistance genes have been detected in 21 of them, and 7 meet the MDR criteria for the isolated Aeromonas strains. Of particular note are the conjugative broad-host-range plasmids, belonging to the incompatibility group IncA/C, IncU, IncQ, IncF, IncI, and ColE-type. These plasmids are primarily responsible for multi-drug resistance among bacteria both in clinical and natural environments. They harbor resistance genes against antibiotics of key importance in clinical therapy, such as the quinolones, β-lactams, aminoglycosides, tetracyclines and sulfonamide. Aeromonas strains causing infections in humans may transfer MGEs carrying resistance genes to pathogenic or opportunistic bacteria in the human microbiome, and thus pose a threat to public health. This in vivo transfer has been reported from two clinical outbreaks in France where a 180-kb plasmid carrying the blaTEM−24 gene has been isolated from Enterobacter aerogenes and two Aeromonas species: A. hydrophila and A. caviae (Marchandin et al., 2003; Fosse et al., 2004). The same plasmid has been previously characterized in the case of Klebsiella pneumonia and Pseudomonas aeruginosa (Giraud-Morin and Fosse, 2003; Marchandin et al., 2000) indicative of their broad host range potential among pathogenic bacterial species. Studies of antibiotic resistance in clinical strains focus solely on the determination of susceptibility using disk diffusion tests and subsequent classification to the R (resistant) or S (susceptible) groups according to the guidelines of such organizations as CLSI (Clinical and Laboratory Standards Institute, 2006). Publications concerning such strains rarely explain the molecular mechanisms of resistance, i.e., the identification of specific genes by PCR amplification or hybridization. There are also no studies on the correlation between the presence of these genes and MGE, in contrast to studies on environmental Aeromonas strains. Hence, the comparison of the mobilome of environmental and clinical isolates of Aeromonas at this stage of research is virtually impossible. One can only compare the phenotypic profiles of resistance. The resistance profile of Aeromonas clinical and environmental strains is very similar, but additional resistance to chloramphenicol and florfenicol can be found in the latter (Michel et al., 2003). However, resistant profiles differ depending on species and geographical localization. The main reason for this is the serious lack of sufficient data about the contribution of antibiotic resistant clinical strains of Aeromonas that are not under epidemiological surveillance in most parts of the world. At this point it is worthwhile to refer to the term “clinical strain” itself, and answer the question what the difference between “clinical” and “environmental” strain is. While in the case of environmental strains an explanation arises spontaneously, it is no longer so obvious for clinical strains. Based on an analysis of the literature data, it can be said that the site of isolation is the essence of the definition of a clinical strain and the ability to cause disease in humans. Thus, clinical strains may be the same as environmental ones, while the opposite is not always true. The aquatic environment seems to be a “hot spot” for the transmission of antibiotic resistance caused by the selective pressure associated with excessive use of antimicrobial compounds. A wide range of ARG have been found in Aeromonas spp., as described in the preceding chapters. Although numerous ARG have been found on plasmids and other MGE, sulfonamide (sul), tetracycline (tet), quinolone (qnr), and β-lactam (bla) resistance genes are most common. Nevertheless, there is no correlation between one definite group of plasmids and any particular ARG. In addition, among heavy metal resistance genes, only mercury resistance genes (mer) have been found on MGE. They were identified among several R-plasmids that belong to IncA/C. It is worth noticing that the co-localization of heavy metal resistance and ARG on the same MGE can promote a co-selection mechanism (Pérez-Valdespino et al., 2014; Yi et al., 2014b).
It should also be noted that other MGE, such as IS, transposons or mobile integrons form a complex mobilome and may play a significant role in the dissemination of ARG. This can be explained by natural transformation, which is a general property of Aeromonas spp. in the environment (Huddleston et al., 2013). Frequent transformation of exogenous DNA may indicate different genetic structures of Aeromonas populations, including the participation of various MGE. This is consistent with previous observations in that there is no clear, detectable association between Aeromonas species, virulence pattern, source or origin (Tanaka et al., 2013; Martino et al., 2014). However, a review of the literature data shows a clear association between mobilome and ARG. This makes the genus Aeromonas a complex one and highlights the fact that there are many mechanisms of antibiotic resistance dissemination among prokaryotes. The situation is different for virulence factors. As knowledge about virulence factors and infection is incomplete, there is no clear evidence of their association with MGE. However, it has been demonstrated that the genomic plasticity of A. salmonicida is dependent on various IS. The majority of clinical strains, especially of A. caviae are considered pathogenic to humans, but they did not present all of the main known virulence factors (Janda and Kokka, 1991; Khajanchi et al., 2010; Ottaviani et al., 2011). The low prevalence of these factors suggests that pathogenicity may not depend on these virulence markers, but primarily on adaptation toward specific habitats. It should be noted that these processes also play a significant role in the distribution of strains in the environment. Originally, Aeromonas spp. were described as fish pathogens. Currently, these bacteria are considered emerging human pathogens, but their effective role in virulence toward humans remains controversial. This does not change the fact that the mobilome of Aeromonas has a considerable potential, particularly in terms of antibiotic resistance, the possibility of horizontal transfer of resistance genes, and the threat it may pose to humans.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors' research was financed by a grant from the National Center of Science, Poland (741/N-COST/2010/0). The authors would like to thank EU for the support provided through COST Action ES1403 “New and emerging challenges and opportunities in wastewater reuse (NEREUS).”
References
Adams, C. A., Austin, B., Meaden, P. G., and McIntosh, D. (1998). Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl. Environ. Microbiol. 64, 4194–4201.
Adler, A., Assous, M. V., Paikin, S., Shulman, A., Miller-Roll, T., Hillel, S., et al. (2014). Emergence of VIM-producing Aeromonas caviae in Israeli hospitals. J. Antimicrob. Chemother. 69, 1211–1214. doi: 10.1093/jac/dkt505
Al-Benwan, K., Abbott, S., Janda, J. M., Huys, G., and Albert, M. J. (2007). Cystitis caused by Aeromonas caviae. J. Clin. Microbiol. 45, 2348–2350. doi: 10.1128/JCM.00480-07
Alcaide, E., Blasco, M. D., and Esteve, C. (2010). Mechanisms of quinolone resistance in Aeromonas species isolated from humans, water and eels. Res. Microbiol. 161, 40–45. doi: 10.1016/j.resmic.2009.10.006
Aoki, T., Mitoma, Y., and Crosa, J. H. (1986). The characterization of a conjugative R-plasmid isolated from Aeromonas salmonicida. Plasmid 16, 213–218. doi: 10.1016/0147-619X(86)90059-4
Aravena-Román, M., Inglis, T. J. J., Henderson, B., Riley, T. V., and Chang, B. J. (2012). Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob. Agents Chemother. 56, 1110–1112. doi: 10.1128/AAC.05387-11
Arias, A., Seral, C., Navarro, F., Miró, E., Coll, P., and Castillo, F. J. (2010). Plasmid-mediated QnrS2 determinant in an Aeromonas caviae isolate recovered from a patient with diarrhoea. Clin. Microbiol. Infect. 16, 1005–1007. doi: 10.1111/j.1469-0691.2009.02958.x
Beilstein, F., and Dreiseikelmann, B. (2008). Temperate bacteriophage PhiO18P 790 from an Aeromonas media isolate: characterization and complete genome sequence. Virology 30, 25–29. doi: 10.1016/j.virol.2007.11.016
Bönemann, G., Stiens, M., Pühler, A., and Schlüter, A. (2006). Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 50, 3075–3080. doi: 10.1128/AAC.00378-06
Bossi-Küpfer, M., Genini, A., Peduzzi, R., and Demarta, A. (2007). Tracheobronchitis caused by Aeromonas veronii biovar sobria after near-drowning. J. Med. Microbiol. 56, 1563–1564. doi: 10.1099/jmm.0.47202-0
Boyd, J., Williams, J., Curtis, B., Kozera, C., Singh, R., and Reith, M. (2003). Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid 50, 131–144. doi: 10.1016/S0147-619X(03)00058-1
Brouwer, M. S., Warburton, P. J., Roberts, A. P., Mullany, P., and Allan, E. (2011). Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS ONE 6:e23014. doi: 10.1371/journal.pone.0023014
Burr, S. E., Pugovkin, D., Wahli, T., Segner, H., and Frey, J. (2005). Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbowtrout model. Microbiology 151, 2111–2118. doi: 10.1099/mic.0.27926-0
Carvalho, M. J., Martínez-Murcia, A., Esteves, A. C., Correia, A., and Saavedra, M. J. (2012). Phylogenetic diversity, antibiotic resistance and virulence traits of Aeromonas spp. from untreated waters for human consumption. Int. J. Food Microbiol. 159, 230–239. doi: 10.1016/j.ijfoodmicro.2012.09.008
Cattoir, V., Poirel, L., Aubert, C., Soussy, C. J., and Nordmann, P. (2008). Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14, 231–237. doi: 10.3201/eid1402.070677
Cayci, Y. T., Coban, A. Y., and Gunaydin, M. (2014). Investigation of plasmid-mediated quinolone resistance in Pseudomonas aeruginosa clinical isolates. Indian J. Med. Microbiol. 32, 285–289. doi: 10.4103/0255-0857.136567
Ceylan, E., Berktas, M., and Ağaoğlu, Z. (2009). The occurrence and antibiotic resistance of motile Aeromonas in livestock. Trop. Anim. Health Prod. 41, 199–204. doi: 10.1007/s11250-008-9175-9
Chim, H., and Song, C. (2007). Aeromonas infection in critically ill burn patients. Burns 33, 756–759. doi: 10.1016/j.burns.2006.10.389
Chow, M. S., and Rouf, M. A. (1983). Isolation and partial characterization of two aeromonas hydrophila bacteriophages. Appl. Environ. Microbiol. 45, 1670–1676.
Clinical and Laboratory Standards Institute. (2006). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline. M45-A. Wayne, PA: Clinical and Laboratory Standards Institute.
Cullik, A., Pfeifer, Y., Prager, R., von Baum, H., and Witte, W. (2010). A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J. Med. Microbiol. 59, 580–587. doi: 10.1099/jmm.0.016188-0
Daher, R. K., Filion, G., Tan, S. G., Dallaire-Dufresne, S., Paquet, V. E., and Charette, S. J. (2011). Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 152, 353–360. doi: 10.1016/j.vetmic.2011.04.034
Dallaire-Dufresne, S., Barbeau, X., Sarty, D., Tanaka, K. H., Denoncourt, A. M., Lagüe, P., et al. (2013). Aeromonas salmonicida Ati2 is an effector protein of the type three secretion system. Microbiology 159, 1937–1945. doi: 10.1099/mic.0.067959-0
Dallaire-Dufresne, S., Tanaka, K. H., Trudel, M. V., Lafaille, A., and Charette, S. J. (2014). Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 21, 1–7. doi: 10.1016/j.vetmic.2013.06.025
del Castillo, C. S., Hikima, J., Jang, H. B., Nho, S. W., Jung, T. S., Wongtavatchai, J., et al. (2013). Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob. Agents Chemother. 57, 120–129. doi: 10.1128/AAC.01239-12
de Palmenaer, D., Vermeiren, C., and Mahillon, J. (2004). IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol. Microbiol. 53, 457–467. doi: 10.1111/j.1365-2958.2004.04146.x
Dias, C., Mota, V., Martinez-Murcia, A., and José Saavedra, M. (2012). Antimicrobial resistance patterns of Aeromonas spp. Isolated from ornamental fish. J. Aquacult. Res. Dev. 3:3. doi: 10.4172/2155-9546.1000131
Dobiasova, H., Kutilova, I., Piackova, V., Vesely, T., Cizek, A., and Dolejska, M. (2014). Ornamental fish as a source of plasmid-mediated quinolone resistance genes and antibiotic resistance plasmids. Vet. Microbiol. 171, 413–421. doi: 10.1016/j.vetmic.2014.02.011
Dolejska, M., Duskova, E., Rybarikova, J., Janoszowska, D., Roubalova, E., Dibdakova, K., et al. (2011). Plasmids carrying blaCTX-M- 1 and qnr genes in Escherichia coli isolates from an equine clinic and a horseback riding centre. J. Antimicrob. Chemother. 66, 757–764. doi: 10.1093/jac/dkq500
Edberg, S. C., Browne, F. A., and Allen, M. J. (2007). Issues for microbial regulation: Aeromonas as a model. Crit. Rev. Microbiol. 33, 89–100. doi: 10.1080/10408410601172180
Fehr, D., Casanova, C., Liverman, A., Blazkova, H., Orth, K., Dobbelaere, D., et al. (2006). AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-κB signalling pathway. Microbiology 152, 2809–2818. doi: 10.1099/mic.0.28889-0
Figueira, V., Vaz-Moreira, I., Silva, M., and Manaia, C. M. (2011). Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 45, 5599–5611. doi: 10.1016/j.watres.2011.08.021
Figueras, M. J. (2005). Clinical relevance of Aeromonas sM503. Rev. Med. Microbiol. 16, 145–153. doi: 10.1097/01.revmedmi.0000184410.98677.8a
Fiorentini, C., Barbieri, E., Falzano, L., Baffone, W., Pianetti, A., Katouli, M., et al. (1998). Occurrence, diversity and pathogenicity of mesophilic Aeromonas in estuarine waters of the Italian coast of the Adriatic Sea. J. Appl. Microbiol. 85, 501–511. doi: 10.1046/j.1365-2672.1998.853517.x
Fosse, T., Giraud-Morin, C., Madinier, I., Mantoux, F., Lacour, J. P., and Ortonne, J. P. (2004). Aeromonas hydrophila with plasmid-borne class A extended-spectrum β-lactamase TEM-24 and three chromosomal class B, C, and D β-lactamases, isolated from a patient with necrotizing fasciitis. Antimicrob. Agents Chemother. 48, 2342–2343. doi: 10.1128/AAC.48.6.2342-2343.2004
Fricke, W. F., Welch, T. J., McDermott, P. F., Mammel, M. K., LeClerc, J. E., White, D. G., et al. (2009). Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191, 4750–4757. doi: 10.1128/JB.00189-09
García, P., Guerra, B., Bances, M., Mendoza, M. C., and Rodicio, M. R. (2011). IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging Salmonella enterica serotype 4,[5],12:i:-. J. Antimicrob. Chemother. 66, 543–549. doi: 10.1093/jac/dkq481
Ghenghesh, K. S., Ahmed, S. F., El-Khalek, R. A., Al-Gendy, A., and Klena, J. (2008). Aeromonas-associated infections in developing countries. J. Infect. Dev. Ctries. 2, 81–98. doi: 10.3855/jidc.277
Giraud-Morin, C., and Fosse, T. (2003). A seven-year survey of Klebsiella pneumoniae producing TEM-24 extended-spectrum β-lactamase in Nice University Hospital (1994–2000). J. Hosp. Infect. 54, 25–31. doi: 10.1016/S0195-6701(03)00038-0
Girlich, D., Poirel, L., and Nordmann, P. (2010). PER-6, an extended-spectrum beta-lactamase from Aeromonas allosaccharophila. Antimicrob. Agents Chemother. 54, 1619–1622. doi: 10.1128/AAC.01585-09
Girlich, D., Poirel, L., and Nordmann, P. (2011). Diversity of clavulanic acid-inhibited extended-spectrum β-lactamases in Aeromonas spp. from the Seine River, Paris, France. Antimicrob. Agents Chemother. 55, 1256–1261. doi: 10.1128/AAC.00921-10
Gómez-Garcés, J. L., Saéz, D., Almagro, M., Fernández-Romero, S., Merino, F., Campos, J., et al. (2011). Osteomyelitis associated to CTX-M-15-producing Aeromonas hydrophila: first description in the literature. Diagn. Microbiol. Infect. Dis. 70, 420–422. doi: 10.1016/j.diagmicrobio.2011.03.004
Gordon, L., Cloeckaert, A., Doublet, B., Schwarz, S., Bouju-Albert, A., Ganière, J. P., et al. (2008). Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J. Antimicrob. Chemother. 62, 65–71. doi: 10.1093/jac/dkn166
Guo, Y. F., Zhang, W. H., Ren, S. Q., Yang, L., Lü, D. H., Zeng, Z. L., et al. (2014). IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS ONE 9:e96738. doi: 10.1371/journal.pone.0096738
Hall, R. M., Collis, C. M., Kim, M. J., Partridge, S. R., Recchia, G. D., and Stokes, H. W. (1999). Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870, 68–80. doi: 10.1111/j.1749-6632.1999.tb08866.x
Han, H. J., Taki, T., Kondo, H., Hirono, I., and Aoki, T. (2008). Pathogenic potential of a collagenase from Aeromonas veronii. Can. J. Microbiol. 54, 1–10. doi: 10.1139/W07-109
Han, J. E., Kim, J. H., Choresca, C. H. Jr., Shin, S. P., Jun, J. W., Chai, J. Y., et al. (2012a). First description of ColE-type plasmid in Aeromonas spp. carrying quinolone resistance (qnrS2) gene. Lett. Appl. Microbiol. 55, 290–294. doi: 10.1111/j.1472-765X.2012.03293.x
Han, J. E., Kim, J. H., Choresca, C. H. Jr., Shin, S. P., Jun, J. W., Chai, J. Y., et al. (2012b). Prevalence of tet gene and complete genome sequencing of tet gene-encoded plasmid (pAHH01) isolated from Aeromonas species in South Korea. J. Appl. Microbiol. 112, 631–638. doi: 10.1111/j.1365-2672.2012.05237.x
Han, J. E., Kim, J. H., Choresca, C. H. Jr., Shin, S. P., Jun, J. W., Chai, J. Y., et al. (2012c). A small IncQ-type plasmid carrying the quinolone resistance (qnrS2) gene from Aeromonas hydrophila. Lett. Appl. Microbiol. 54, 374–376. doi: 10.1111/j.1472-765X.2012.03208.x
Hedges, R. W., Smith, P., and Brazil, G. (1985). Resistance Plasmids of Aeromonads. J. Gen. Microbiol. 131, 2091–2095. doi: 10.1099/00221287-131-8-2091
Henriques, I. S., Fonseca, F., Alves, A., Saavedra, M. J., and Correia, A. (2006). Occurrence and diversity of integrons and β-lactamase genes among ampicillin-resistant isolates from estuarine waters. Res. Microbiol. 157, 938–947. doi: 10.1016/j.resmic.2006.09.003
Holmberg, S. D., and Farmer, J. J. III. (1984). Aeromonas hydrophila and Plesiomonas shigelloides as causes of intestinal infection. Rev. Infect. Dis. 6, 633–639. doi: 10.1093/clinids/6.5.633
Hossain, M. J., Waldbieser, G. C., Sun, D., Capps, N. K., Hemstreet, W. B., Carlisle, K., et al. (2013). Implication of lateral genetic transfer in the emergence of Aeromonas hydrophila isolates of epidemic outbreaks in channel catfish. PLoS ONE 20:e80943. doi: 10.1371/journal.pone.0080943
Huang, H. C., Yu, W. L., Huan, K. H., Cheng, K. C., and Chuang, Y. C. (2007). Aeromonas sobria prostatitis and septic shock in a healthy man with chronic alcoholic consumption. Jpn. J. Infect. Dis. 60, 400–401.
Huddleston, J. R., Brokaw, J. M., Zak, J. C., and Jeter, R. M. (2013). Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst. Appl. Microbiol. 36, 224–234. doi: 10.1016/j.syapm.2013.01.004
Imamura, T., Nitta, H., Wada, Y., Kobayashi, H., and Okamoto, K. (2008). Impaired plasma clottability induction through fibrinogen degradation by ASP, a serine protease released from Aeromonas sobria. FEMS. Microbiol. Lett. 284, 35–42. doi: 10.1111/j.1574-6968.2008.01184.x
Ishiguro, E. E., Kay, W. W., Ainsworth, T., Chamberlain, J. B., Austen, R. A., Buckley, J. T., et al. (1981). Loss of virulence during culture of Aeromonas salmonicida at high temperature. J. Bacteriol. 148, 333–340.
Jacobs, L., and Chenia, H. Y. (2007). Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int. J. Food Microbiol. 114, 295–306. doi: 10.1016/j.ijfoodmicro.2006.09.030
Janda, J. M., and Kokka, R. P. (1991). The pathogenicity of Aeromonas strains relative to genospecies and phenospecies identification. FEMS. Microbiol. Lett. 90, 29–34. doi: 10.1111/j.1574-6968.1991.tb05120.x
Jangid, K., Kong, R., Patole, M. S., and Shouche, Y. S. (2007). luxRI homologs are universally present in the genus Aeromonas. BMC. Microbiol. 7:93. doi: 10.1186/1471-2180-7-93
John, N., and Hatha, A. A. M. (2013). Distribution, extracellular virulence factors and drug resistance of motile aeromonads in fresh water ornamental fishes and associated carriage water. Int. J. Aquacult. 3, 92–100. doi: 10.5376/ija.2013.03.0017
Jones, B. L., and Wilcox, M. H. (1995). Aeromonas infections and their treatment. J. Antimicrob. Chemother. 35, 453–461. doi: 10.1093/jac/35.4.453
Jones, R. M., Luo, L., and Moberg, K. H. (2012). Aeromonas salmonicida-secreted protein AopP is a potent inducer of apoptosis in a mammalian and a Drosophila model. Cell Microbiol. 14, 274–285. doi: 10.1111/j.1462-5822.2011.01717.x
Jorge, M. T., Nishioka Sde, A., de Oliveirá, R. B., Ribeiro, L. A., and Silveira, P. V. (1998). Aeromonas hydrophila soft-tissue infection as a complication of snake bite: report of three cases. Ann. Trop. Med. Parasitol. 92, 213–217. doi: 10.1080/00034989860067
Jorgensen, J. H., and Hindler, J. F. (2007). New consensus guidelines from the Clinical and Laboratory Standards Institute for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria. Clin. Infect. Dis. 44, 280–286. doi: 10.1086/510431
Kehrenberg, C., Friederichs, S., de Jong, A., Michael, G. B., and Schwarz, S. (2006). Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis. J. Antimicrob. Chemother. 58, 18–22. doi: 10.1093/jac/dkl213
Khajanchi, B. K., Fadl, A. A., Borchardt, M. A., Berg, R. L., Horneman, A. J., Stemper, M. E., et al. (2010). Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl. Environ. Microbiol. 76, 2313–2325. doi: 10.1128/AEM.02535-09
Khan, M. I., Walters, G., and Metcalfe, T. (2007). Bilateral endogenous endophthalmitis caused by Aeromonas hydrophila. Eye. 21, 1244–1245. doi: 10.1038/sj.eye.6702926
Khan, R., Takahashi, E., Nakura, H., Ansaruzzaman, M., Banik, S., Ramamurthy, T., et al. (2008). Toxin production by Aeromonas sobria in natural environments: river water vs. seawater. Acta. Med. Okayama 62, 363–371.
Kim, J. H., Son, J. S., Choi, Y. J., Choresca, C. H., Shin, S. P., Han, J. E., et al. (2012a). Isolation and characterization of a lytic Myoviridae bacteriophage PAS-1 with broad infectivity in Aeromonas salmonicida. Curr. Microbiol. 64, 418–426. doi: 10.1007/s00284-012-0091-x
Kim, J. H., Son, J. S., Choi, Y. J., Choresca, C. H., Shin, S. P., Han, J. E., et al. (2012b). Complete genomic sequence of a T4-like bacteriophage, phiAS4, infecting Aeromonas salmonicida subsp. salmonicida. Arch. Virol. 157, 391–395. doi: 10.1007/s00705-011-1175-9
Kingombe, C. I. B., Huys, G., Howald, D., Luthi, E., Swing, J., and Jemmi, T. (2004). The usefulness of molecular techniques to assess the presence of Aeromonas spp. harbouring virulence markers in foods. Int. J. Food Microbiol. 94, 113–121. doi: 10.1016/S0168-1605(03)00105-3
Ko, W. C., Lee, H. C., Chuang, Y. C., Liu, C. C., and Wu, J. J. (2000). Clinical features and therapeutic implications of 104 episodes of monomicrobial Aeromonas bacteraemia. J. Infect. 40, 267–273. doi: 10.1053/jinf.2000.0654
Koczura, R., Semkowska, A., and Mokracka, J. (2014). Integron-bearing Gram-negative bacteria in lake waters. Lett. Appl. Microbiol. 59, 514–519. doi: 10.1111/lam.12307
Kulinska, A., Czeredys, M., Hayes, F., and Jagura-Burdzy, G. (2008). Genomic and functional characterization of the modular broad-host-range RA3 plasmid, the archetype of the IncU group. Appl. Environ. Microbiol. 74, 4119–4132. doi: 10.1128/AEM.00229-08
L'Abée-Lund, T. M., and Sørum, H. (2002). A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47, 172–181. doi: 10.1016/S0147-619X(02)00001-X
Lamy, B., Kodjo, A. colBVH Study Group, and Laurent, F. (2009). Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 47, 1234–1237. doi: 10.1128/JCM.00155-09
Laroche, E., Pawlak, B., Berthe, T., Skurnik, D., and Petit, F. (2009). Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiol. Ecol. 68, 118–130. doi: 10.1111/j.1574-6941.2009.00655.x
Lau, S. M., Peng, M. Y., and Chang, F. Y. (2000). Outcomes of Aeromonas bacteremia in patients with different types of underlying disease. J. Microbiol. Immunol. Infect. 33, 241–247.
Lazar, V., Cernat, R., Balotsen, C., Cotar, A., Coipan, E., and Cojocaru, C. (2002). Correlation between multiple antibiotic resistance and heavy metal tolerance among some E. coli strains isolated from polluted waters. Bacteriol. Virusol. Parazitol. Epidemiol. 47, 155–160.
Libisch, B., Gisk, C. G., Kovács, B., Tóth, T. G., and Füzi, M. (2008). Identification of the first VIM metallo-beta-lactamase-producing multiresistant Aeromonas hydrophila strain. J. Clin. Microbiol. 46, 1878–1880. doi: 10.1128/JCM.00047-08
Liebert, C. A., Hall, R. M., and Summers, A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522.
Lin, W. T., Su, S. Y., Lai, C. C., Tsai, T. C., Gau, S. J., and Chao, C. M. (2013). Peritonitis caused by Aeromonas species at a hospital in southern Taiwan. Intern. Med. 55, 2517–2521. doi: 10.2169/internalmedicine.52.0180
Loftie-Eaton, W., and Rawlings, D. E. (2009). Comparative biology of two natural variants of the IncQ-2 family plasmids, pRAS3.1 and pRAS3.2. J. Bacteriol. 191, 6436–6446. doi: 10.1128/JB.00864-09
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Majumdar, T., Das, B., Bhadra, R. K., Dam, B., and Mazumder, S. (2011). Complete nucleotide sequence of a quinolone resistance gene (qnrS2) carrying plasmid of Aeromonas hydrophila isolated from fish. Plasmid 66, 79–84. doi: 10.1016/j.plasmid.2011.05.001
Maravić, A., Skoèibušić, M., Samanić, I., Fredotović, Z., Cvjetan, S., Jutronić, M., et al. (2013). Aeromonas spp. simultaneously harbouring bla(CTX-M-15), bla(SHV-12), bla(PER-1) and bla(FOX-2), in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food. Microbiol. 166, 301–308. doi: 10.1016/j.ijfoodmicro.2013.07.010
Marchandin, H., Godreuil, S., Darbas, H., Jean-Pierre, H., Jumas-Bilak, E., Chanal, C., et al. (2003). Extended-spectrum beta-lactamase TEM-24 in an Aeromonas clinical strain: acquisition from the prevalent Enterobacter aerogenes clone in France. Antimicrob. Agents Chemother. 47, 3994–3995. doi: 10.1128/AAC.47.12.3994-3995.2003
Marchandin, H., Jean-Pierre, H., De Champs, C., Sirot, D., Darbas, H., Perigault, P. F., et al. (2000). Production of a TEM-24 plasmid-mediated extended-spectrum β-lactamase by a clinical isolate of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 213–216. doi: 10.1128/AAC.44.1.213-216.2000
Marti, E., and Balcázar, J. L. (2012). Multidrug resistance-encoding plasmid from Aeromonas sp. strain P2G1. Clin. Microbiol. Infect. 18, E366–E368. doi: 10.1111/j.1469-0691.2012.03935.x
Martin-Carnahan, A., and Joseph, S. W. (2005). Genus I. Aeromonas Stanier 1943, 213AL, p. 557–578. Bergey's Manual of Systematic Bacteriology. New York, NY: Springer.
Martino, M. E., Fasolato, L., Montemurro, F., Novelli, E., and Cardazzo, B. (2014). Aeromonas spp.: ubiquitous or specialized bugs? Environ. Microbiol. 16, 1005–1018. doi: 10.1111/1462-2920.12215
Mataseje, L. F., Baudry, P. J., Zhanel, G. G., Morck, D. W., Read, R. R., Louie, M., et al. (2010). Comparison of CMY-2 plasmids isolated from human, animal, and environmental Escherichia coli and Salmonella spp. from Canada. Diagn. Microbiol. Infect. Dis. 67, 387–391. doi: 10.1016/j.diagmicrobio.2010.02.027
McIntosh, D., Cunningham, M., Ji, B., Fekete, F. A., Parry, E. M., Clark, S. E., et al. (2008). Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 61, 1221–1228. doi: 10.1093/jac/dkn123
Merino, S., Aguilar, A., Rubires, X., Simon-Pujol, D., Congregado, F., and Tomás, J. M. (1996). The role of the capsular polysaccharide of Aeromonas salmonicida in the adherence and invasion of fish cell lines. FEMS Microbiol. Lett. 142, 185–189. doi: 10.1111/j.1574-6968.1996.tb08428.x
Michel, C., Kerouault, B., and Martin, C. (2003). Chloramphenicol and florfenicol susceptibility of fish-pathogenic bacteria isolated in France: comparison of minimum inhibitory concentration, using recommended provisory standards for fish bacteria. J. Appl. Microbiol. 95, 1008–1015. doi: 10.1046/j.1365-2672.2003.02093.x
Moura, A., Henriques, I., Ribeiro, R., and Correia, A. (2007). Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 60, 1243–1250. doi: 10.1093/jac/dkm340
Moura, A., Oliveira, C., Henriques, I., Smalla, K., and Correia, A. (2012). Broad diversity of conjugative plasmids in integron-carrying bacteria from wastewater environments. FEMS Microbiol. Lett. 330, 157–164. doi: 10.1111/j.1574-6968.2012.02544.x
Najimi, M., Lemos, M. L., and Osorio, C. R. (2009). Identification of iron regulated genes in the fish pathogen Aeromonas salmonicida subsp. salmonicida: genetic diversity and evidence of conserved iron uptake systems. Vet. Microbiol. 133, 377–382. doi: 10.1016/j.vetmic.2008.07.008
Neuwirth, C., Siebor, E., Robin, F., and Bonnet, R. (2007). First occurrence of an IMP metallo-beta-lactamase in Aeromonas caviae: IMP-19 in an isolate from France. Antimicrob. Agents Chemother. 51, 4486–4488. doi: 10.1128/AAC.01462-06
Neyts, K., Huys, G., Uyttendaele, M., Swings, J., and Debevere, J. (2000). Incidence and identification of mesophilic Aeromonas spp. from retail foods. Lett. Appl. Microbiol. 31:359–363. doi: 10.1046/j.1472-765x.2000.00828.x
Nguyen, H. N., Van, T. T., Nguyen, H. T., Smooker, P. M., Shimeta, J., and Coloe, P. J. (2014). Molecular characterization of antibiotic resistance in Pseudomonas and Aeromonas isolates from catfish of the Mekong Delta, Vietnam. Vet. Microbiol. 171, 397–405. doi: 10.1016/j.vetmic.2014.01.028
Noonan, B., and Trust, T. J. (1997). The synthesis, secretion and role in virulence of the paracrystalline surface protein layers of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol. Lett. 154, 1–7. doi: 10.1111/j.1574-6968.1997.tb12616.x
Nordmann, P., and Poirel, L. (2005). Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56, 463–469. doi: 10.1093/jac/dki245
Oliveira, P. H., Touchon, M., and Rocha, E. P. (2014). The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 42, 10618–10631. doi: 10.1093/nar/gku734
Oliver, K. R., McComb, J. A., and Greene, W. K. (2013). Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol Evol. 5, 1886–1901. doi: 10.1093/gbe/evt141
Ottaviani, D., Parlani, C., Citterio, B., Masini, L., Leoni, F., Canonico, C., et al. (2011). Putative virulence properties of Aeromonas strains isolated from food, environmental and clinical sources in Italy: a comparative study. Int. J. Food Microbiol. 5, 538–545. doi: 10.1016/j.ijfoodmicro.2010.11.020
Palumbo, S. A., Maxino, F., Williams, A. C., Buchanan, R. L., and Thayer, D. T. W. (1985). Starch-ampicillin agar for the quantitative detection of Aeromonas hydrophila. Appl. Environ. Microbiol. 50, 1027–1030.
Pan, J. C., Ye, R., Wang, H. Q., Xiang, H. Q., Zhang, W., Yu, X. F., et al. (2008). Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob. Agents Chemother. 52, 3829–3836. doi: 10.1128/AAC.00375-08
Pasom, W., Chanawong, A., Lulitanond, A., Wilailuckana, C., Kenprom, S., and Puang-Ngern, P. (2013). Plasmid-mediated quinolone resistance genes, aac(6′)-Ib-cr, qnrS, qnrB, and qnrA, in urinary isolates of Escherichia coli and Klebsiella pneumoniae at a teaching hospital, Thailand. Jpn. J. Infect. Dis. 66, 428–432. doi: 10.7883/yoken.66.428
Pasquali, F., Kehrenberg, C., Manfreda, G., and Schwarz, S. (2005). Physical linkage of Tn3 and part of Tn1721 in a tetracycline and ampicillin resistance plasmid from Salmonella Typhimurium. J. Antimicrob. Chemother. 55, 562–565. doi: 10.1093/jac/dkh553
Pérez-Valdespino, A., Celestino-Mancera, M., Villegas-Rodríguez, V. L., and Curiel-Quesada, E. (2014). Characterization of mercury-resistant clinical Aeromonas species. Braz. J. Microbiol. 10, 1279–1283.
Picão, R. C., Cardoso, J. P., Campana, E. H., Nicoletti, A. G., Petrolini, F. V., Assis, D. M., et al. (2013). The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 76, 80–85. doi: 10.1016/j.diagmicrobio.2013.02.001
Picão, R. C., Poirel, L., Demarta, A., Silva, C. S., Corvaglia, A. R., Petrini, O., et al. (2008). Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62, 948–950. doi: 10.1093/jac/dkn341
Piotrowska, M., and Popowska, M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64, 921–934. doi: 10.1007/s13213-014-0911-2
Puah, S. M., Puthucheary, S. D., Liew, F. Y., and Chua, K. H. (2013). Aeromonas aquariorum clinical isolates: antimicrobial profiles, plasmids and genetic determinants. Int. J. Antimicrob. Agents 41, 281–284. doi: 10.1016/j.ijantimicag.2012.11.012
Puthucheary, S. D., Puah, S. M., and Chua, K. H. (2012). Molecular characterization of clinical isolates of Aeromonas species from Malaysia. PLOS. ONE 7:e30205. doi: 10.1371/journal.pone.0030205
Rahman, M., Colque-Navarro, P., Kühn, I., Huys, G., Swings, J., and Möllby, R. (2002). Identification and characterization of pathogenic Aeromonas veronii biovar sobria associated with epizootic ulcerative syndrome in fish in Bangladesh. Appl. Environ. Microbiol. 68, 650–655. doi: 10.1128/AEM.68.2.650-655.2002
Reith, M. E., Singh, R. K., Curtis, B., Boyd, J. M., Bouevitch, A., Kimball, J., et al. (2008). The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. doi: 10.1186/1471-2164-9-427
Rhodes, G., Huys, G., Swings, J., McGann, P., Hiney, M., Smith, P., et al. (2000). Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tetA. Appl. Environ. Microbiol. 66, 3883–3890. doi: 10.1128/AEM.66.9.3883-3890.2000
Rhodes, G., Parkhill, J., Bird, C., Ambrose, K., Jones, M. C., Huys, G., et al. (2004). Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 70, 7497–7510. doi: 10.1128/AEM.70.12.7497-7510.2004
Sánchez-Céspedes, J., Blasco, M. D., Marti, S., Alba, V., Alcaide, E., Esteve, C., and Vila, J. (2008). Plasmid-mediated QnrS2 determinant from a clinical Aeromonas veronii isolate. Antimicrob. Agents Chemother. 52, 2990–2991. doi: 10.1128/AAC.00287-08
Sandaa, R. A., and Enger, O. (1994). Transfer in marine sediments of the naturally occurring plasmid pRAS1 encoding multiple antibiotic resistance. Appl. Environ. Microbiol. 60, 4234–4238.
Sarria-Guzmán, Y., López-Ramírez, M. P., Chávez-Romero, Y., Ruiz-Romero, E., Dendooven, L., and Bello-López, J. M. (2014). Identification of antibiotic resistance cassettes in class 1 integrons in Aeromonas spp. strains isolated from fresh fish (Cyprinus carpio L.). Curr. Microbiol. 68, 581–586. doi: 10.1007/s00284-013-0511-6
Schmidt, A. S., Bruun, M. S., Dalsgaard, I., and Larsen, J. L. (2001). Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67, 5675–5682. doi: 10.1128/AEM.67.12.5675-5682.2001
Seetha, K. S., Jose, B. T., and Jasthi, A. (2004). Meningitis due to Aeromonas hydrophila. Indian. J. Med. Microbiol. 22, 191–192.
Shak, J. R., Whitaker, J. A., Ribner, B. S., and Burd, E. M. (2011). Aminoglycoside-resistant Aeromonas hydrophila as part of a polymicrobial infection following a traumatic fall into freshwater. J. Clin. Microbiol. 49, 1169–1170. doi: 10.1128/JCM.01949-10
Shen, C. J., Liu, Y. J., and Lu, C. P. (2012). Complete genome sequence of Aeromonas hydrophila phage CC2. J. Virol. 86:10900. doi: 10.1128/JVI.01882-12
Sørum, H., L'Abée-Lund, T. M., Solberg, A., and Wold, A. (2003). Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47, 1285–1290. doi: 10.1128/AAC.47.4.1285-1290.2003
Stokes, H. W., and Hall, R. M. (1989). A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3, 1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x
Stuber, K., Burr, S. E., Braun, M., Wahli, T., and Frey, J. (2003). Type III secretion genes in Aeromonas salmonicida subsp salmonicida are located on a large thermolabile virulence plasmid. J. Clin. Microbiol. 41, 3854–3856. doi: 10.1128/JCM.41.8.3854-3856.2003
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Tanaka, K. H., Frenette, M., and Charette, S. J. (2013). IS-mediated loss of virulence by Aeromonas salmonicida: a tangible piece of an evolutionary puzzle. Mob. Genet. Elements 3:e23498. doi: 10.4161/mge.23498
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and Barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Trudel, M. V., Tanaka, K. H., Filion, G., Daher, R. K., Frenette, M., and Charette, S. J. (2013). Insertion sequence AS5 (IS AS5) is involved in the genomic plasticity of Aeromonas salmonicida. Mob. Genet. Elements 1:e25640. doi: 10.4161/mge.25640
Tsai, M. S., Kuo, C. Y., Wang, M. C., Wu, H. C., Chien, C. C., and Liu, J. W. (2006). Clinical features and risk factors for mortality in Aeromonas bacteremic adults with hematologic malignancies. J. Microbiol. Immunol. Infect. 39, 150–154.
Vally, H., Whittle, A., Cameron, S., Dowse, G. K., and Watson, T. (2004). Outbreak of Aeromonas hydrophila wound infections associated with mud football. Clin. Infect. Dis. 38, 1084–1089. doi: 10.1086/382876
Vanden Bergh, P., Heller, M., Braga-Lagache, S., and Frey, J. (2013). The Aeromonas salmonicida subsp. salmonicida exoproteome: determination of the complete repertoire of Type-Three Secretion System effectors and identification of other virulence factors. Proteome Sci. 11:42. doi: 10.1186/1477-5956-11-42
Vincent, A. T., Trudel, M. V., Paquet, V. E., Boyle, B., Tanaka, K. H., Dallaire-Dufresne, S., et al. (2014). Detection of variants of the pRAS3, pAB5S9, and pSN254 plasmids in Aeromonas salmonicida subsp. salmonicida: multidrug resistance, interspecies exchanges, and plasmid reshaping. Antimicrob. Agents Chemother. 58, 7367–7374. doi: 10.1128/AAC.03730-14
Winokur, P. L., Vonstein, D. L., Hoffman, L. J., Uhlenhopp, E. K., and Doern, G. V. (2001). Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45, 2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001
Wu, C. J., Chen, P. L., Hsueh, P. R., Chang, M. C., Tsai, P. J., Shih, H. I., et al. (2015). Clinical implications of species identification in monomicrobial Aeromonas bacteremia. PLoS ONE 10:e0117821. doi: 10.1371/journal.pone.0117821
Wu, C. J., Chuang, Y. C., Lee, M. F., Lee, C. C., Lee, H. C., Lee, N. Y., et al. (2011). Bacteremia due to extended-spectrum-β-lactamase-producing Aeromonas spp. at a medical center in Southern Taiwan. Antimicrob. Agents Chemother. 55, 5813–5818. doi: 10.1128/AAC.00634-11
Ye, Y., Xu, X. H., and Li, J. B. (2010). Emergence of CTX-M-3, TEM-1 and a new plasmid-mediated MOX-4 AmpC in a multiresistant Aeromonas caviae isolate from a patient with pneumonia. J. Med. Microbiol. 59, 843–847. doi: 10.1099/jmm.0.016337-0
Yi, S. W., Chung, T. H., Joh, S. J., Park, C., Park, B. Y., and Shin, G. W. (2014a). High Prevalence of blaCTX-M group Genes in Aeromonas dhakensis Isolated from Aquaculture Fish Species in South Korea. J. Vet. Med. Sci. 2014:3. doi: 10.1292/jvms.14-0274
Keywords: Aeromonas, mobilome, plasmid, transposon, integron, virulence factor, antibiotic resistance gene, horizontal gene transfer
Citation: Piotrowska M and Popowska M (2015) Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494. doi: 10.3389/fmicb.2015.00494
Received: 17 February 2015; Accepted: 05 May 2015;
Published: 27 May 2015.
Edited by:
Amy Pruden, Virginia Tech, USAReviewed by:
Peiying Hong, King Abdullah University of Science and Technology, Saudi ArabiaMichael J. Rothrock, United States Department of Agriculture – Agricultural Research Service, USA
Copyright © 2015 Piotrowska and Popowska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magdalena Popowska, Department of Applied Microbiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Ilji Miecznikowa 1, 02-096 Warsaw, Poland, magdapop@biol.uw.edu.pl
 Marta Piotrowska
Marta Piotrowska Magdalena Popowska
Magdalena Popowska