- State Key Laboratory Breeding Base of Eco-Environment and Bio-Resource of the Three Gorges Area, Key Laboratory of Eco-environments in Three Gorges Reservoir Region, Ministry of Education, School of Life Sciences, Institute of Modern Biopharmaceuticals, Southwest University, Chongqing, China
PE/PPE family proteins, named after their conserved PE (Pro-Glu) and PPE (Pro-Pro-Glu) domains of N-terminal, are most intriguing aspects of pathologic mycobacterial genome. The roles of most members of this family remain unknown, although selected genes of this family are related to the virulence of Mycobacterium tuberculosis. In order to decipher the role of Rv1169c, the Mycobacterium smegmatis strain heterologous expressed this ORF was constructed and identified that Rv1169c was a cell wall associated protein with a novel function in modifying the cell wall fatty acids. The growth of Rv1169c expressing strain was affected under surface stress, acidic condition and antibiotics treatment. M. smegmatis expressing Rv1169c induced necrotic cell death of macrophage after infection and significantly decreased interlukin-6 production compared to controls. In general, these results underscore a proposing role of Rv1169c in virulence of M. tuberculosis, as it's role in the susceptibility of anti-mycobacteria factors caused by modified cell wall fatty acid, and the induced necrotic cell death by Rv1169c is crucial for M. tuberculosis virulence during infection.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis infection, remains a formidable threat to global public health. M. tuberculosis can modulate and elude host immune responses and persist for prolonged periods (Lin and Flynn, 2010). One hallmark of the M. tuberculosis genome is the presence of the multi-genic PE/PPE family proteins, which consist of PPE, PE, and PE_PGRS subfamilies and account for about 10% of the coding capacity of the M. tuberculosis genome. There are 69 genes encoding PPE subfamily which are named after its N terminal Pro(P)-Pro(P)-Glu(E) motif (Cole and Barrell, 1998; Cole et al., 1998) and about 100 genes encoding PE subfamily, which harbor a conserved N-terminal domain with 110 amino acid residues with the Pro-Glu (PE) motif, while the C-termini vary significantly in size and protein-specific polymorphic GC-rich repeats PGRS (Cole et al., 1998; Akhter et al., 2012). The 69 PPE proteins are classified into PPE_SVP with typical G-X-S-V-P-X-X-W repeats, PPE_PPW with special G-F-X-G-T and Pro-X-X-P-X-X-W sequences and PPE_MPTR with N-X-G-X-G-N-A-G major polymorphic tandem motifs in their C terminal (Cole and Barrell, 1998; Cole et al., 1998). The PE subfamily contains 37 PE genes with a conserved N terminal and 61 PE_PGRS genes with G-G-A and G-G-N tandem repeats in their C terminal (Brennan and Delogu, 2002; Fleischmann et al., 2002; Voskuil et al., 2004; Dheenadhayalan et al., 2006a). The unique sequences of these proteins might underlie the specific physiological role of this family during M. tuberculosis infection.
The exclusive presence of PE/PPE family among pathogenic mycobacteria (Gey van Pittius et al., 2006) has attracted many researchers. The primary origin, regulation and physiological role of some PE/PPE family proteins have been well characterized and reviewed (Brennan and Delogu, 2002; Tian and Jian-Ping, 2010; Mohareer et al., 2011; Sampson, 2011; Akhter et al., 2012; Kohli et al., 2012; Vordermeier et al., 2012; Fishbein et al., 2015). The origin of the PE/PPE genes is associated with the Type VII secretion system (T7S) (Abdallah et al., 2009). Several transcriptional regulators involved in the regulation of PE/PPE family proteins have been characterized, including the stringent response mediator RelA (Dahl et al., 2003), ESX-1 secreted protein regulator EspR (Blasco et al., 2012) and global nucleoid-associated transcriptional inhibitor Lsr2 (Gordon et al., 2010) and sigma factors, such as sigF (Williams et al., 2007; Humpel et al., 2010), sigB (Dahl et al., 2003; Fontan et al., 2009), and sigD (Raman et al., 2004; Calamita et al., 2005). It has been shown that several members of PE/PPE proteins are immunogenic (Delogu and Brennan, 2001; Chaitra et al., 2005; Campuzano et al., 2007) and might contribute to the antigenic diversity and immune evasion of mycobacteria (Cole et al., 1998; Brennan and Delogu, 2002). The C-terminal fragments of the PE_PGRS protein Rv1759c (Espitia et al., 1999) or the PGRS domain of Rv3367 (Singh et al., 2001) can react with TB patients sera. Other PE/PPE proteins such as PE25/PPE41 complex, Rv1169c, Rv0978c as well as Rv1818c showed same characteristics (Delogu and Brennan, 2001; Narayana et al., 2007; Tundup et al., 2008). Some PE/PPE proteins are implicated in immune evasion and antigenic variation (Banu et al., 2002; Brennan and Delogu, 2002) or may be linked to virulence and responsible for its ability to grow in a macrophage (Jha et al., 2010; Dong et al., 2012; Iantomasi et al., 2012; Tiwari et al., 2012; Thi et al., 2013). PE/PPE family proteins are cell wall associated (Delogu et al., 2004; Cascioferro et al., 2007, 2011; Dona et al., 2013; Chatrath et al., 2014; Deng et al., 2014), suggesting that a role in directly interaction with host targets such as the cell surface receptor TLR2, or even interfering the host immunity (Nair et al., 2009; Bansal et al., 2010; Tiwari et al., 2012; Zumbo et al., 2013; Deng et al., 2014). Macrophages are the first line of defense against bacteria infection, which can secrete various cytokines to mediate the inflammatory response. The varied transcription level of several PE/PPE genes within macrophages or mouse during M. tuberculosis infection (Dheenadhayalan et al., 2006b) suggested a role in manipulation the host macrophage activity. PE_PGRS62 inhibited the maturation of phagosome (Huang et al., 2012), PE_PGRS30 (Iantomasi et al., 2012), as well as PPE25 (Jha et al., 2010) disturbed the phagolysosomal fusion. In brief, mounting evidences showed that PE/PPE family proteins contribute significantly to the successful immune evasion of M. tuberculosis.
M. tuberculosis Rv1169c encodes for PE11, a prototypical member of PE/PPE family protein, was characterized in this study. The up-regulation of Rv1169c after 24 h of starvation (Betts et al., 2002) or transient acid exposure (Fisher et al., 2002), suggested a role in pathogen persistence or dormancy, in particular, in fatty acid metabolism. To study the role of Rv1169c and underlying mechanism, M. smegmatis expressing Rv1169c and M. smegmatis harboring the vector only were constructed. Our studies demonstrated that M. smegmatis expressing Rv1169c has a modified cell wall fatty acid consonant, and its ability to against anti-tuberculosis factors was abated. Moreover, Rv1169c was also found to affect the pleiotropic pro-inflammatory cytokine IL-6 secretion and manuscript cell death of macrophage.
Methods and Materials
Construction Recombinant M. smegmatis Strains
Mycobacterial expression vector pALACE used in this study has been described previously (Lakshminarayan et al., 2008). The full-length of Rv1169c gene was amplified from M. tuberculosis genome using gene-specific primers listed in Table 1 (pALACE-Rv1169c-F and pALACE-Rv1169c-R). The EcoR I-BamH I-digested PCR product was cloned into pALACE to generate pALACE-Rv1169c. The plasmids (pALACE and pALACE-Rv1169c) were electroporated into fast-growing non-pathogenic M. smegmatis mc2155 according standard procure (Lakshminarayan et al., 2008). The recombinant M. smegmatis strains were selected on MB 7H10 medium containing 100 μg/ml hygromycin (Hyg). The constructs harboring Rv1169c gene were confirmed by PCR amplification, and the positive recombinant strains were stored with sterile 20% glycerol at −80°C for further use. Escherichia coli DH5a strains using for gene cloning were grown at 37°C using Luria–Bertani (LB) broth and LB agar with the addition of appropriate antibiotics. M. smegmatis mc2155 were grown in 7H9 broth medium or on 7H10 agar supplemented with 0.05% (v/v) Tween 80, 0.2% (w/v) glucose, and 0.5% (v/v) glycerol.
Detection the Expression of Rv1169c in M. smegmatis
The recombinant M. smegmatis strains harboring His-tagged Rv1169c (Ms_Rv1169c) and vector pALACE (Ms_Vec) were cultured in MB 7H9 broth medium supplemented with 100 μg/ml Hyg. At the OD600-value of 0.8, the recombinant strains were subjected to 28 mM acetamide (Aladdin, China) for protein expression. M. smegmatis cell fractionation was carried out essentially as described earlier, with minor modifications. In general, the recombinants including Ms_Vec and Ms_Rv1169c were harvested after 16 h acetamide induction using centrifugation at the speed of 3000 × g for 10 min, 4°C. The collected cells were washed and then sonicated in cold PBS supplemented with protease inhibitor P-8849 (Sigma-Aldrich). After sonication, the prepared whole-cell lysate were centrifuged at the speed of 20,000 × g for separating the insoluble (pellets in the bottom) and the soluble (supernatant in the upper layer) fractions. The separated fractions were loaded to SDS-PAGE and further detected by Western blot analysis with using specific anti-His monoclonal antibody (TIANGEN, China). The blots were formed when incubation with IgG-HRP, an anti-mouse IgG monoclonal antibody labeled with horseradish peroxidase (TIANGEN, China).
Subcellular Fractionation of Recombinant M. smegmatis
Recombinant Ms_Rv1169c and Ms_Vec constructs were grown and subjected to cell fractionation separation as described previously (Deng et al., 2014), with minor modification. Generally, the acetamide-induced recombinant Ms_Vec and Ms_Rv1169c were subjected to sonication. The whole lysates were centrifuged at the speed of 3000 × g for 5 min at 4°C for removing un-lysed cells and cell debris. The supernatants were ultra-centrifuged at the speed of 27,000 × g for 30 min, at 4°C. After ultra-centrifugation, the pellets were considered the cell wall fraction, and the supernatants were supposed to cell membrane and cytosol fractions. The pellets were further suspended in PBS. Equal amounts of protein from pellets and supernatants fraction were subjected to Western blot as described previously for analysis the expression of Rv1169c.GroEL2 protein served as cytosol marker protein of mycobacteria.
Analysis in Vitro Survival under Different Stress Conditions
Recombinant M. smegmatis strains were grown into optimal concentration in 7H9 medium containing 100 μg/ml Hyg. Ms_Vec and Ms_Rv1169c were performed in presence of stress condition after 16 h induction by acetamide. Ms_Vec and Ms_Rv1169c were treated by 0.05% SDS for 1, 2, 3, and 4 h. In addition, pH gradient was generated by adding HCl into 7H9 medium, and sterilized by passing through a 2 μm filter. After the treatment, the recombinant strains were ten-fold dilution spotted onto MB 7H10 agar containing Hyg and bacteria numbers were counted after 3 days.
Anti-Tuberculosis Drug Sensitivity Assays
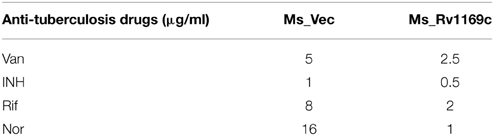
Four antibiotics were used in this study, including vancomycin (Van), isoniazid (INH), norfloxacin (Nor) and rifampicin (Rif). Acetamide induced Ms_Vec and Ms_Rv1169c strains were prepared for treatment with these four different antibiotics. The original concentration for Van is 320, 512 ug/ml for INH, 512 ug/m for Nor and 128 ug/m for Rif, and different concentration of each antibiotic was made prepared by 2-fold dilution. MIC values of each antibiotic were determined when the bacterial activity was killed at least 99% on liquid medium. For bactericidal ability test, Acetamide induced Ms_Vec and Ms_Rv1169c strains were exposed to Van (5, 10, 20, 80, 160 μg/ml), INH (4, 16, 64, 128 μg/ml), Nor (1, 2, 4, 8, 32, 64 μg/ml) and Rif (4, 16, 64, 128 μg/ml) for 24 h, respectively. After treatment with these four antibiotics in different concentration, the recombinant strains were diluted by 10-fold and plated into 7H10 agar medium and counted the bacterial number after 3 days culture. The medium without any antibiotics serves as the control to make sure the normal growth of bacteria.
Macrophages Infection by Recombinant M. smegmatis
The human monocyte cell line U-937 was maintained in RPMI 1640 medium (Invitrogen) supplemented with 2 mM L-glutamine, 10% (v/v) heat inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen) and cultured in humidified incubator supplemented with 5% CO2 at in 37°C. Macrophages were seeded at 1 × 106 cells per well in 12-well tissue culture plates or at 5 × 105 cells per well in 24-well tissue culture plates. The suspension cell line U-937 cells were transformed into adherent macrophage after 48 h treatment with 100 μg/ml phorbol 12-myristate 13-acetate (PMA, Sigma). Cells were infected with Ms_Vec or Ms_Rv1169c at MOI of 10. Gentamicin was added into culture at the concentration of 150 ug/ml after 4 h infection to remove the bacteria outside the macrophages. After 6, 24, 48, and 72 h infection, the culture supernatants were collected for detecting lactate dehydrogenase (LDH) activities. The LDH activities were detected by commercially LDH cytotoxicity kit (Takara Bio) according to standard procedure. For the detection of bacteria survival within macrophage, the infected macrophages were washed by PBS for 3 times and lysed by 0.05% SDS. The cell lysates were serially ten-fold diluted and then spotted on 7H10 agar containing Hyg. The numbers of bacteria were enumerated after 3 days.
Assay for Cytokines Production
Culture supernatants were harvested after infection of macrophages with Ms_Vec or Ms_Rv1169c for 6, 24, 48, and 72 h. The RNA was extracted from the infected cells using RNA extraction kit (TIANGEN) and mRNA level of cytokines were detected by RT-PCR using gene specific PCR primers listed in Table 1. The cytokines in the culture supernatants were detected using commercially ELISA kits of interleukin 1β(IL-1β), interleukin 6 (IL-6).
GC-MS Analysis of Fatty Acid Components
The samples used in GC-MS analysis were extracted according the standard procedure (http://www.midi-inc.com). The determination of fatty acids was performed on an Agilent 7890A gas chromatograph with GC-MSD (5975C mass selective detector) equipped with an Agilent 7693A automatic liquid sampler and a DB-5MS capillary column (30 m, 0.25 mm i.d, 0.25 μm film thickness). High purity (99.999%) helium served as the carrier gas in constant-flow mode at a column flow rate of 1 mL/min. 150, 230, 250, and 280°C were the optimal temperature of quadrupole, ion source, injector and transfer line temperatures, respectively. The oven temperature program was as follows: initial temperature 60°C, held for 1 min; increased to 160°C; later increased to 290°C held for 10 min. 70eV was set for the electron impact energy. One microliter of each sample was infused in splitless mode. PAHs were identified by comparing the relative RT (retention time) with IS. Those peaks were integrated for quantification and qualification when their location was within the right range (2%) of RT.
Statistical Analysis
Graphpad Prism 6 software was used for analysis of differences between experimental and control group. Statistical significance (P-value) was decided using Student's t-test. P-values were less than 0.05 were supposed to be statistically significant. *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
M. tuberculosis Rv1169c Can Be Expressed in M. smegmatis
M. tuberculosis PE/PPE family Rv1169c gene encodes 12 kDa protein with about 300 bp in size. In this study, we constructed two recombinant M. smegmatis strains to probe the role of Rv1169c, recombinant Ms_Vec and Ms_Rv1169c strains were constructed. Ms_Rv1169c expressed a His-tagged Rv1169c protein harboring pALACE-Rv1169c, while Ms_Vec only harbored the vector with Hyg resistant marker gene (Lakshminarayan et al., 2008). Both Ms_Vec and Ms-Rv1169c were grown in MB 7H9 medium supplemented with Hyg. PCR amplification identified there was about 300 bp of Rv1169c gene in Ms_Rv1169c strain by specific primers (Figure 1A). Western Blot confirmed only Ms_ Rv1169c strain expressed ~12 kDa Rv1169c-His protein, while its absence in Ms_Vec strain (Figure 1B). These results identify that M. tuberculosis Rv1169c gene was constructed and expressed in M. smegmatis strains.
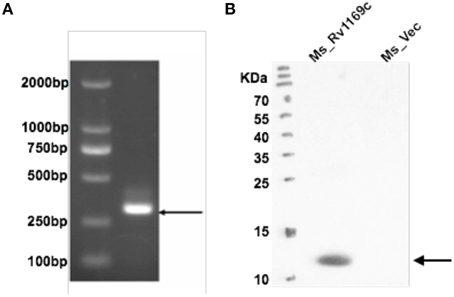
Figure 1. Heterologous expression of M. tuberculosis Rv1169c in M. smegmatis. (A) Ms_Vec and Ms_Rv1169c were grown into an OD600 of 0.8 and then were subjected to PCR amplification to detect Rv1169c gene. (B) The cell lysates were prepared from acetamide-induced Ms_Vec and Ms_Rv1169c were subjected to Western blot to determine the expression of His-tagged Rv1169c protein in M. smegmatis by mouse anti-His antibody.
Rv1169c is Associated with Mycobacterial Cell Wall
Bioinformatics approach suggested that at least 29 PE/PPE proteins are associated with the “cell wall and cell processes” (Mazandu and Mulder, 2012). Selected PE/PPE genes were experimentally identified localized to cell wall of mycobacteria (Fishbein et al., 2015). We hypothesized that Rv1169c might be also localized to the cell wall of M. smegmatis. In order to further identify this hypothesis, M. smegmatis expression His-tagged Rv1169c strain was constructed. Recombinant M. smegmatis strains including Ms_Rv1169c and Ms_Vec were subjected to cell fractionation experiment, their subcellular localization was finally determined by Western blot. The Rv1169c protein was present in the cell wall fraction and was not found in cytoplasm fraction (Figure 2), suggesting Rv1169c is a cell wall-associated protein. As expected, cytoplasmic heat-shock protein GroEL2 was detected only in the cytoplasm of M. smegmatis.
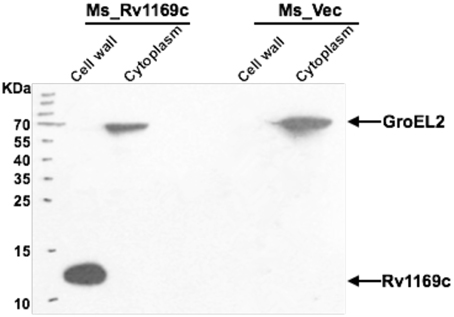
Figure 2. Rv1169c is associated with Mycobacterium cell wall. Cell fractionation experiments were performed to detect detection the expression of Rv1169c protein, and further confirmed by Western blot by specific anti-His antibody. The cytoplasm marker of Mycobacterium GroEL2 served as a control.
Rv1169c Reprograms the Cell Wall Components
The cell wall associated Rv1169c protein would provide a proof for the observed phenotypes related to the characteristic of cell surface, as another PE/PPE member PE30 (also designated lipase LipY) involved in lipids metabolism and was mainly present in the cell wall (Mishra et al., 2008). Rv1169c was predicted as lipase LipX (Deb et al., 2006). We therefore tested whether Rv1169c was involved in lipids metabolism, recombinant Ms_Rv1169c, Ms_Vec and wild type M. smegmatis strains were subjected to GC-MS to determine the fatty acids components of these strains. A novel unsaturated fatty acid C17: 1w7c was found in the cell wall component of recombinant Ms_Rv1169c while another saturated 9MeC19:0 was disappeared compared with Ms_Vec and wild type M. smegmatis strains (Figure 3). The difference of saturability of fatty acid between Ms_Rv1169c and wild type M. smegmatis might be due to the directly and indirectly effect of Rv1169c on the expression of Mycobacterium fatty acid desaturase. On the other hand, the modified components in cell wall of Ms_Rv1169c may affect their permeability to antimicrobial factors and their survival in the host.
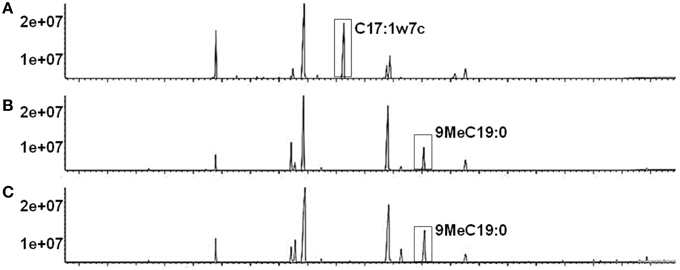
Figure 3. The fatty acid component in cell wall of recombinant Ms_Rv1169c (A), Ms_Vec (B), and wild-type M. smegmatis (C).
Ms_Rv1169c is Highly Sensitive to Anti-Tuberculosis Drugs
The cell wall serves as an effective permeable barrier of M. tuberculosis for diverse antibiotics (Brennan and Nikaido, 1995). Therefore, the alterations of cell wall fatty acid component in Ms_Rv1169c might be highlighted a role of Rv1169c in cell wall permeability. To study whether Rv1169c function in cell wall permeability, Ms_Vec and Ms_Rv1169c were treated with four anti-tuberculosis drugs as described in method. Both Ms_Vec and Ms_Rv1169c displayed comparable capacity to Van and INH. The MIC values of Van and INH for Ms_Vec were 5 and 1 μg/ml, and 2.5 and 0.5 μg/ml for Ms_Rv1169c, respectively. However, Ms_Rv1169c was highly susceptible to Rif and especially sensitive to Nor. The MIC values of Rif and Nor for Ms_Vec were 8 and 16 μg/ml, while 2 and 1 μg/ml for Ms_Rv1169c, respectively. The MIC values of Ms_Rv1169c for Rif and Nor were 4 and 16 fold lower than Ms_Vec, respectively (Table 2). In addition, the Ms_Rv1169c was more sensitive than Ms_Vec to Rif, especially Nor, while there was no difference in INH and Van (Figure 4). These results thus suggest a novel function of PE family protein Rv1169c in the susceptibility of antibiotic.
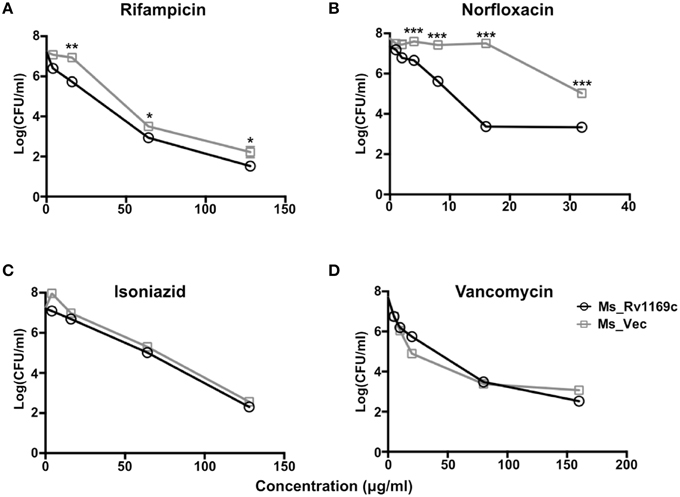
Figure 4. The survival of Ms_Vec and Ms_Rv1169c after treatment with different antibiotics, including Rif (A), Nor (B), INH (C), and Van (D). Ms_Vec and Ms_Rv1169c were exposed to Rif, Nor, INH and Van, and then ten-fold dilution spotted the bacteria on MB 7H10 supplemented with Hyg, the bacterial number of Ms_Rv1169c and Ms_Vec were counted after 3 days cultivation. *P < 0.05, **P < 0.01, and ***P < 0.001.
Ms_Rv1169c is More Susceptible to Antimicrobial Factors
To gain further insight into whether Rv1169c will affect the permeability to antimicrobial factors, the growth characteristics of recombinant Ms_Vec and Ms_Rv1169c under different acid condition and surface stress were analyzed. As shown in the Figure 5A, there was no significant difference between Ms_Vec and Ms_Rv1169c under in vitro acid stress at early time points (0, 3, 6 h after treatment). The survival percentage of Ms_Rv1169c was significant lower than Ms_Vec after 9 h treatment with acid stress (pH = 3 and 5). The acid sensitive mutants of M. tuberculosis were observed also hypersensitive to antibiotics, surface stress, heat shock, oxidative stress, oxygen and nitrogen intermediates (Vandal et al., 2009). Whether Ms_Rv1169c was hypersensitive to surface stress, Ms_Vec and Ms_Rv1169c were exposed to 0.05% SDS that mimicked surface stress. Although there was a rapid decrease in the bacterial numbers for all tested strains exposed to the detergent SDS (Figure 5B), Ms_Rv1169c was more sensitive to SDS compared to Ms_Vec: the survival percentage was 0.5% for Ms_Rv1169c while 3% for Ms_Vec. The sensitivity of Ms_Rv1169c to SDS was validated when bacterial survival was tested after 1 h of incubation with SDS (Figure 5B). These results suggest that Rv1169c promotes the susceptibility of M. smegmatis to surface and acid stress.
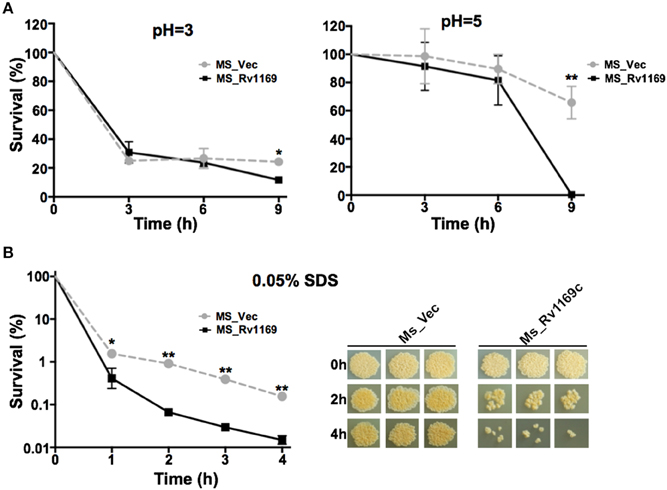
Figure 5. The sensitivity of Ms_Vec and Ms_1169c strains to acid and surface stress. (A) In vitro growth of recombinant Ms_Vec and Ms_Rv1169c after treatment with different pH gradient for 0, 3, 6, and 9 h. The Ms_Vec and Ms_Rv1169c strains were centrifuged, re-suspended to 5 ml MB 7H9 at an OD600 of 0.5, 10-fold serial dilutions of Ms_Vec and Ms_Rv1169c were spotted on MB 7H10 containing Hyg. (B) Survival of Ms_Vec and Ms_Rv1169c after exposure to 0.05% SDS. Re-suspended 5 ml recombinant MS_Vec and MS_Rv1169c (OD600 = 0.5) were exposed to 0.05% SDS for 1, 2, 3, and 4 h. And then the recombinant strains were plated onto 7H10 plates by serially ten-fold dilution, the bacterial numbers were counted after 3–4 days of cultivation at 37°C. *P < 0.05 and ** P < 0.01.
Intracellular Survival of Recombinant M. smegmatis within Macrophages
Several PE/PPE proteins are responsible for the virulence M. tuberculosis and contribute to its ability to grow in a macrophage (Bottai et al., 2012; Tiwari et al., 2012; Singh et al., 2013). Whether Rv1169c will affect the intrecellur survival of M. smegmatis in host macrophage, as selected PE/PPE proteins were reported to mediate bacterial survival within macrophage through different mechanisms (Tiwari et al., 2012; Singh et al., 2013). We compared the survival rate of recombinant Ms_Rv1169c and Ms_Vec to determine whether Rv1169c can enhance the intracellular survival of M. smegmatis within macrophages. PMA-differentiated U-937 macrophages were infected with Ms_Vec and MS_Rv1169c at an MOI of 10. There was a significant difference in the percent survival of bacteria in macrophages between Ms_Rv1169c and Ms_Vec at 24 h after infection (Figure 6), implying the presence of Rv1169c can enhance the intracellular survival of M. smegmatis within macrophages at early stage of infection.
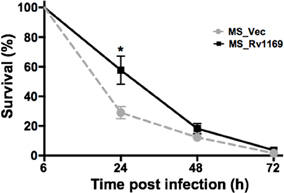
Figure 6. Intracellular survival of recombinant Ms_Vec and Ms_Rv1169c within macrophages. PMA-differentiated macrophages were infected with Ms_Vec or Ms_Rv1169c at an MOI of 10. At 6, 24, 48, and 72 h after infection, the macrophages were washed and lysed using 0.01% SDS. Lysates were plated on MB 7H10 medium containing 100 μg/ml Hyg to determine the bacterial number. *P < 0.05.
Ms_Rv1169c Promotes the Death of Macrophage
Infection of macrophages with M. tuberculosis can induce necrosis, defined by cell lysis. M. tuberculosis might manipulate host cell death to cause disease. Alternatively, infection can result in macrophages apoptosis to maintain an intact plasma membrane, to diminish pathogen viability and enhance host immunity. To determine the effect of Rv1169c on viability of macrophages, PMA-differentiated macrophages were infected with Ms_Vec or Ms_Rv1169c, LDH release into the culture supernatants was determined after Ms_Vec and Ms_Rv1169c infection. LDH release from infected macrophage was increased following 6 h infection with both Ms_Rv1169c and the control strain Ms_Vec, Macrophages infected with Ms_Rv1169c released more LDH compared to Ms_Vec after 24 h infection and later time points (Figure 7), suggesting Ms_Rv1169c has can induce the cell death of macrophage.
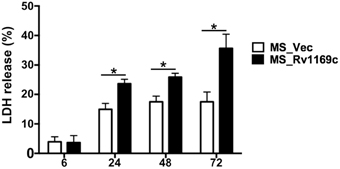
Figure 7. Assay of cell death in macrophages infected with Ms_Vec or Ms_Rv1169c. Macrophages were infected with Ms_Vec (white bars) or Ms_Rv1169c (black bars) at an MOI of 10. After 6, 24, 48, and 72 h infection, culture supernatants were collected and the release of LDH was measured. Data are shown as means ± SEM of triplicate wells. *P < 0.05.
Rv1169c Selectively Regulates the Expression of Pleiotropic Pro-Inflammatory Cytokine IL-6
To explore whether Rv1169c helps M. smegmatis subvert the early immune responses, PMA-differentiated U937 macrophages were infected with Ms_Vec and Ms_Rv1169c for 6, 24, 48, and 72 h. The infected macrophages were collected to detect the relative expression of cytokines by RT-PCR, total RNA was extracted from the infected macrophages at different intervals (6, 24, 48, and 72 h). The supernatants were harvested for cytokines production using ELISA. Interestingly, macrophages infected with Ms_Rv1169c secreted significantly lower amounts of the pro-inflammatory cytokine IL-6 than Ms_Vec (Figure 8C), while IL-6 mRNA level mRNA of Ms_Rv1169c infected macrophages was higher than Ms_Vec (Figure 8A). These suggest that Rv1169c serves as a double-edged sword in regulation of the transcription and translation of host cytokine IL-6. The relative expression of IL-1β was induced following 6 h infection with both Ms_Rv1169c and Ms_Vec strains and their expression was decreased quickly at later time points (Figure 8B), while the secretion of IL-1β was increased after 6 h infection in both bacterial infected macrophages (Figure 8D).
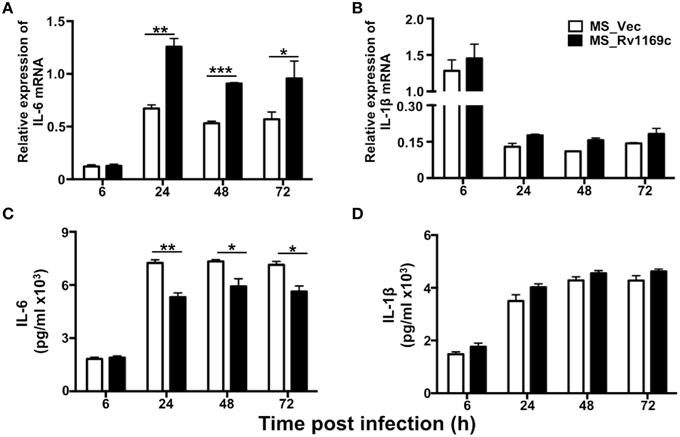
Figure 8. The expression of cytokines in macrophage infected by Ms_Vec and Ms_Rv1169c. PMA-differentiated U937 macrophages (2 × 106/well/2 ml) were infected with Ms_Vec and Ms_Rv1169c strains. After 6, 24, 48, and 72 h infection, the infected macrophage were collected and the transcription of IL-6 (A) and IL-1β mRNA (B) was detected by RT-PCR. Culture supernatants were harvested, the secretion of IL-6 (C) and IL-1β (D) were detected by ELISA analysis. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
There are increasing evidences suggesting M. tuberculosis PE/PPE proteins represent one of the most intriguing aspects of the M. tuberculosis genome (Fishbein et al., 2015). Selected members of this family have been reported to be associated with cell wall and serve as virulence factors to disturb the function of macrophage (Jha et al., 2010; Dong et al., 2012; Iantomasi et al., 2012; Tiwari et al., 2012; Thi et al., 2013). Rv1169c was found in the membrane protein fraction of M. tuberculosis H37Rv but absent in the culture filtrate (Malen et al., 2010). Experimental evidence for its cell wall localization remains absent. In this study, we successfully constructed the recombinant Ms_Rv1169c and confirmed Rv1169c is a cell wall associated protein.
As a predicted vaccine candidate (Narayana et al., 2007), the function of Rv1169c remains largely unknown. Rv1169c is a member of the M. tuberculosis PE/PPE family, also a predicted lipase lipX (http://tuberculist.com). It has been shown that PE_PGRS63 (encodes for LipY) hydrolyzes triacylglycerol stored in lipid inclusion bodies, which provides energy for the dormant Mycobacteria (Saxena et al., 2013). We found that a distinct cell wall fatty acid component in Ms_Rv1169c that is significantly different from Ms_Vec and wild type M. smegmatis strain, suggesting a potential role of Rv1169c in cell wall fatty acid metabolism. The changes in cell wall fatty acid component of recombinant Ms_Rv1169c might affect the function of mycobacterial cell wall, which is an important defense against stresses from environment. Many acid-sensitive mutants showed defect cell wall function, and also sensitive to antibiotics and other stresses (Vandal et al., 2009). We identified recombinant Ms_Rv1169c is more susceptible to surface stress SDS and acid stress compared to Ms_Vec, implicating a role of Rv1169c in stress response. Further evidence proved that Ms_Rv1169c is more sensitive to antibiotics including Rif and Nor. These results indicate that Rv1169c may increase the cell wall permeability of mycobacteria by affecting their stability.
Macrophage is the primary residence of infected M. tuberculosis, which sought to contain the invaded pathogens via multiple mechanisms (Leemans et al., 2005). The interaction between the M. tuberculosis and macrophage is crucial for the outcome of infection (Loeuillet et al., 2006). The evidence showed M. tuberculosis can escape from the attack of host macrophages and persist in hostile environment (Flynn and Chan, 2003), including reactive oxygen reactive nitrogen compounds, iron-deprived conditions and low pH conditions. We found recombinant Ms_Rv1169c is more sensitive to in vitro acid condition and SDS than Ms_Vec. Moreover, this phenotype is not correlated with increased survival within macrophage, as Rv1169 enhanced the intracellular survival of M. smegmatis. Pervious study has shown that necrotic cell death induced by bacterial infection is one strategy that promotes bacterial dissemination, while blocking apoptotic cell death, which serves as an innate defense against M. tuberculosis infection (Behar et al., 2011). It is smeared how M. tuberculosis operates host cell necrosis and manages its growth especially during persistent infection, although necrotic cell death commonly observed in mycobacterial lesion and granulomas is driven by bacterial factors (Dobos et al., 2000). We demonstrated that Rv1169c helps M. smegmatis enhance cell death with unknown mechanism. This is an important virulence mechanisms of M. tuberculosis appears to be their ability to induce necrosis of host immune cells, as not only Rv1169c, ESAT-6 and PE25/PPE41 have also been demonstrated to induce necrosis of immune cells via an unknown mechanism (Welin et al., 2011). The cell death induced by Rv1169c might not be apoptosis but necrosis since there is no difference in mRNA level of well-known apoptotic cell death related proteins such as bcl2 and caspase-1 (Figure S1) and cytokines TNF-α (Figure S2) and IL1β (Figure 8B) after both Ms_Rv1169c and Ms_Vec infection. This needs to be further verified since a new strategy has been recently found that IL-32-mediated apoptotic pathway is involved in controlling M. tuberculosis infection (Bai et al., 2015).
IL-6, a pleiotropic pro-inflammatory cytokine, is an indicator of many human chronic inflammatory diseases. Pervious studies have shown that IL-6 is crucial for the control of Mycobacterial infections due to its roles in producing protective Th1 immune responses against M. tuberculosis after subunit vaccine vaccination (Leal et al., 1999). In addition, IL-6 KO mice became more susceptible to very high doses of intravenously infected M. tuberculosis (Ladel et al., 1997) and lower the protective effect against M. tuberculosis infection (Leal et al., 1999). Moreover, other groups have proved IL-6 is critical for controlling infection in mice challenged with high doses of intravenously delivered M. tuberculosiss (Ladel et al., 1997; Saunders et al., 2000; Nagabhushanam et al., 2003). The depletion of IL-6 can aggravate other mycobacterial infection, such as M. avium (Appelberg et al., 1994). In this study, we demonstrated Rv1169c controls the expression of IL-6. This modulation is selective, since no discernable effect on IL-1β and other cytokines including IL12p40, TNF-α and IL-10 (Figure S2). However, how M. tuberculosis Rv1169c selectively regulates host pleiotropic pro-inflammatory cytokine IL-6 remains to be determined, as Rv1169c promotes the relative expression of IL6 mRNA while decreases the IL-6 secretion. As we noticed that Rv1169c contains a transcriptional regulator characterized helix-turn-helix motif from amino acid 88 to 109 in its C terminal. Therefore, we suspected that Rv1169c, like other regulators, might regulate the IL-6 transcription, and this might be responsible for the highly IL-6 mRNA level detected in Ms_Rv1169c infected cells. On the other hand, like other well-documented PE/PPE proteins, Rv1169c has the ability to control the secretion of cytokine, and this might be the reason for the reduced IL-6 secretion in Ms_Rv1169c infected macrophage.
Conclusion
In summary, despite intense studies on the PE and PPE families of mycobacterial proteins, conclusive evidences for their roles remain to be found. We provide here evidence that M. tuberculosis Rv1169c regulates macrophage IL-6 secretion and plays an important role in cell death of macrophage. It's first reported that Rv1169c may directly or indirectly involves in fatty acid metabolism, resulting in the remarkable changes in fatty acid component of mycobacterial cell wall. Moreover, a role of Rv1169c in stability of mycobacterial cell wall integrity was confirmed as evidenced by the declined resistance of recombinant M. smegmatis to anti-mycobacterial factors including SDS, acid stress and antibiotics.
Author Contributions
These experiments were conceived and designed by WD and JX. Most experiments performed by WD and JZ, they contributed equally to this paper. XX and PL conducted several experiments. This paper wrote by WD and JX. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation [grant numbers 81371851, 81071316, 81271882, 81301394], New Century Excellent Talents in Universities [grant number NCET-11-0703], National Megaprojects for Key Infectious Diseases [grant numbers 2008ZX10003-006], Excellent PhD thesis Fellowship of Southwest University [grant numbers kb2010017, ky2011003], The Fundamental Research Funds for the Central Universities [grant numbers XDJK2011D006, XDJK2012D011, XDJK2012D007,XDJK2013D003, XDJK2014D040], The Chongqing Municipal Committee of Education for Postgraduates Excellence Program [grant numbers YJG123104], The Undergraduatesc Teaching Reform Program [grant numbers 2013JY201]. Graduates Innovative Project of Chongqing Municipal Education Commission [grant numbers CYS14044].
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00613
Abbreviations
TB, Tuberculosis; M.tuberculosis (Schrager and Vaccine Prevention of Sustained Mycobacterium tuberculosis Infection Summary, 2015), M. smegmatis, Mycobacterium smegmatis mc2155; Hyg, hygromycin; Van, vancomycin; Rif, rifampicin; INH, isoniazide; Nor, norfloxacin; SDS, Sodium Dodecyl Sulfate; IL-6, interleukin-6; IL-1β, interleukin-1β; IL-10, interleukin-10; TNF-α, tumor necrosis factor-α; LDH, lactate dehydrogenase.
References
Abdallah, A. M., Verboom, T., Weerdenburg, E. M., Gey van Pittius, N. C., Mahasha, P. W., Jimenez, C., et al. (2009). PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol. Microbiol. 73, 329–340. doi: 10.1111/j.1365-2958.2009.06783.x
Akhter, Y., Ehebauer, M. T., Mukhopadhyay, S., and Hasnain, S. E. (2012). The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie 94, 110–116. doi: 10.1016/j.biochi.2011.09.026
Appelberg, R., Castro, A. G., Pedrosa, J., and Minoprio, P. (1994). Role of interleukin-6 in the induction of protective T cells during mycobacterial infections in mice. Immunology 82, 361–364.
Bai, X., Kinney, W. H., Su, W. L., Bai, A., Ovrutsky, A. R., Honda, J. R., et al. (2015). Caspase-3-independent apoptotic pathways contribute to interleukin-32gamma-mediated control of Mycobacterium tuberculosis infection in THP-1 cells. BMC Microbiol. 15:39. doi: 10.1186/s12866-015-0366-z
Bansal, K., Sinha, A. Y., Ghorpade, D. S., Togarsimalemath, S. K., Patil, S. A., Kaveri, S. V., et al. (2010). Src homology 3-interacting domain of Rv1917c of Mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-kappaB signaling and drives Th2 immune responses. J. Biol. Chem. 285, 36511–36522. doi: 10.1074/jbc.M110.158055
Banu, S., Honore, N., Saint-Joanis, B., Philpott, D., Prevost, M. C., and Cole, S. T. (2002). Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 44, 9–19. doi: 10.1046/j.1365-2958.2002.02813.x
Behar, S. M., Martin, C. J., Booty, M. G., Nishimura, T., Zhao, X., Gan, H. X., et al. (2011). Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 4, 279–287. doi: 10.1038/mi.2011.3
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Blasco, B., Chen, J. M., Hartkoorn, R., Sala, C., Uplekar, S., Rougemont, J., et al. (2012). Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 8:e1002621. doi: 10.1371/journal.ppat.1002621
Bottai, D., Di Luca, M., Majlessi, L., Frigui, W., Simeone, R., Sayes, F., et al. (2012). Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol. Microbiol. 83, 1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x
Brennan, M. J., and Delogu, G. (2002). The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 10, 246–249. doi: 10.1016/S0966-842X(02)02335-1
Brennan, P. J., and Nikaido, H. (1995). The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63. doi: 10.1146/annurev.bi.64.070195.000333
Calamita, H., Ko, C., Tyagi, S., Yoshimatsu, T., Morrison, N. E., and Bishai, W. R. (2005). The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell. Microbiol. 7, 233–244. doi: 10.1111/j.1462-5822.2004.00454.x
Campuzano, J., Aguilar, D., Arriaga, K., Leon, J. C., Salas-Rangel, L. P., Gonzalez-y-Merchand, J., et al. (2007). The PGRS domain of Mycobacterium tuberculosis PE_PGRS Rv1759c antigen is an efficient subunit vaccine to prevent reactivation in a murine model of chronic tuberculosis. Vaccine 25, 3722–3729. doi: 10.1016/j.vaccine.2006.12.042
Cascioferro, A., Daleke, M. H., Ventura, M., Dona, V., Delogu, G., Palu, G., et al. (2011). Functional dissection of the PE domain responsible for translocation of PE_PGRS33 across the mycobacterial cell wall. PLoS ONE 6:e27713. doi: 10.1371/journal.pone.0027713
Cascioferro, A., Delogu, G., Colone, M., Sali, M., Stringaro, A., Arancia, G., et al. (2007). PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol. Microbiol. 66, 1536–1547. doi: 10.1111/j.1365-2958.2007.06023.x
Chaitra, M. G., Hariharaputran, S., Chandra, N. R., Shaila, M. S., and Nayak, R. (2005). Defining putative T cell epitopes from PE and PPE families of proteins of Mycobacterium tuberculosis with vaccine potential. Vaccine 23, 1265–1272. doi: 10.1016/j.vaccine.2004.08.046
Chatrath, S., Gupta, V. K., and Garg, L. C. (2014). The PGRS domain is responsible for translocation of PE_PGRS30 to cell poles while the PE and the C-terminal domains localize it to the cell wall. FEBS Lett. 588, 990–994. doi: 10.1016/j.febslet.2014.01.059
Cole, S. T., and Barrell, B. G. (1998). Analysis of the genome of Mycobacterium tuberculosis H37Rv. Novartis Found. Symp. 217, 160–72.
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Dahl, J. L., Kraus, C. N., Boshoff, H. I., Doan, B., Foley, K., Avarbock, D., et al. (2003). The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 10026–10031. doi: 10.1073/pnas.1631248100
Deb, C., Daniel, J., Sirakova, T. D., Abomoelak, B., Dubey, V. S., and Kolattukudy, P. E. (2006). A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 281, 3866–3875. doi: 10.1074/jbc.M505556200
Delogu, G., and Brennan, M. J. (2001). Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 69, 5606–5611. doi: 10.1128/IAI.69.9.5606-5611.2001
Delogu, G., Pusceddu, C., Bua, A., Fadda, G., Brennan, M. J., and Zanetti, S. (2004). Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52, 725–733. doi: 10.1111/j.1365-2958.2004.04007.x
Deng, W., Li, W., Zeng, J., Zhao, Q., Li, C., Zhao, Y., et al. (2014). Mycobacterium tuberculosis PPE family protein Rv1808 manipulates cytokines profile via co-activation of MAPK and NF-kappaB signaling pathways. Cell. Physiol. Biochem. 33, 273–288. doi: 10.1159/000356668
Dheenadhayalan, V., Delogu, G., and Brennan, M. J. (2006a). Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect. 8, 262–272. doi: 10.1016/j.micinf.2005.06.021
Dheenadhayalan, V., Delogu, G., Sanguinetti, M., Fadda, G., and Brennan, M. J. (2006b). Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J. Bacteriol. 188, 3721–3725. doi: 10.1128/JB.188.10.3721-3725.2006
Dobos, K. M., Spotts, E. A., Quinn, F. D., and King, C. H. (2000). Necrosis of lung epithelial cells during infection with Mycobacterium tuberculosis is preceded by cell permeation. Infect. Immun. 68, 6300–6310. doi: 10.1128/IAI.68.11.6300-6310.2000
Dona, V., Ventura, M., Sali, M., Cascioferro, A., Provvedi, R., Palu, G., et al. (2013). The PPE domain of PPE17 is responsible for its surface localization and can be used to express heterologous proteins on the mycobacterial surface. PLoS ONE 8:e57517. doi: 10.1371/journal.pone.0057517
Dong, D., Wang, D., Li, M., Wang, H., Yu, J., Wang, C., et al. (2012). PPE38 modulates the innate immune response and is required for Mycobacterium marinum virulence. Infect. Immun. 80, 43–54. doi: 10.1128/IAI.05249-11
Espitia, C., Laclette, J. P., Mondragon-Palomino, M., Amador, A., Campuzano, J., Martens, A., et al. (1999). The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology 145(Pt 12), 3487–3495.
Fishbein, S., van Wyk, N., Warren, R. M., and Sampson, S. L. (2015). Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol. Microbiol. 96, 901–916. doi: 10.1111/mmi.12981
Fisher, M. A., Plikaytis, B. B., and Shinnick, T. M. (2002). Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184, 4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002
Fleischmann, R. D., Alland, D., Eisen, J. A., Carpenter, L., White, O., Peterson, J., et al. (2002). Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184, 5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002
Flynn, J. L., and Chan, J. (2003). Immune evasion by Mycobacterium tuberculosis: living with the enemy. Curr. Opin. Immunol. 15, 450–455. doi: 10.1016/S0952-7915(03)00075-X
Fontan, P. A., Voskuil, M. I., Gomez, M., Tan, D., Pardini, M., Manganelli, R., et al. (2009). The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J. Bacteriol. 191, 5628–5633. doi: 10.1128/JB.00510-09
Gey van Pittius, N. C., Sampson, S. L., Lee, H., Kim, Y., van Helden, P. D., and Warren, R. M. (2006). Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 6:95. doi: 10.1186/1471-2148-6-95
Gordon, B. R., Li, Y., Wang, L., Sintsova, A., van Bakel, H., Tian, S., et al. (2010). Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 107, 5154–5159. doi: 10.1073/pnas.0913551107
Huang, Y., Zhou, X., Bai, Y., Yang, L., Yin, X., Wang, Z., et al. (2012). Phagolysosome maturation of macrophages was reduced by PE_PGRS 62 protein expressing in Mycobacterium smegmatis and induced in IFN-gamma priming. Vet. Microbiol. 160, 117–125. doi: 10.1016/j.vetmic.2012.05.011
Humpel, A., Gebhard, S., Cook, G. M., and Berney, M. (2010). The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J. Bacteriol. 192, 2491–2502. doi: 10.1128/JB.00035-10
Iantomasi, R., Sali, M., Cascioferro, A., Palucci, I., Zumbo, A., Soldini, S., et al. (2012). PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis. Cell. Microbiol. 14, 356–367. doi: 10.1111/j.1462-5822.2011.01721.x
Jha, S. S., Danelishvili, L., Wagner, D., Maser, J., Li, Y. J., Moric, I., et al. (2010). Virulence-related Mycobacterium avium subsp hominissuis MAV_2928 gene is associated with vacuole remodeling in macrophages. BMC Microbiol. 10:100. doi: 10.1186/1471-2180-10-100
Kohli, S., Singh, Y., Sharma, K., Mittal, A., Ehtesham, N. Z., and Hasnain, S. E. (2012). Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H(3)(7)Rv and H(3)(7)Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res. 40, 7113–7122. doi: 10.1093/nar/gks465
Ladel, C. H., Blum, C., Dreher, A., Reifenberg, K., Kopf, M., and Kaufmann, S. H. (1997). Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect. Immun. 65, 4843–4849.
Lakshminarayan, H., Narayanan, S., Bach, H., Sundaram, K. G., and Av-Gay, Y. (2008). Molecular cloning and biochemical characterization of a serine threonine protein kinase, PknL, from Mycobacterium tuberculosis. Protein Expr. Purif. 58, 309–317. doi: 10.1016/j.pep.2007.12.012
Leal, I. S., Smedegard, B., Andersen, P., and Appelberg, R. (1999). Interleukin-6 and interleukin-12 participate in induction of a type 1 protective T-cell response during vaccination with a tuberculosis subunit vaccine. Infect. Immun. 67, 5747–5754.
Leemans, J. C., Thepen, T., Weijer, S., Florquin, S., van Rooijen, N., van de Winkel, J. G., et al. (2005). Macrophages play a dual role during pulmonary tuberculosis in mice. J. Infect. Dis. 191, 65–74. doi: 10.1086/426395
Lin, P. L., and Flynn, J. L. (2010). Understanding latent tuberculosis: a moving target. J. Immunol. 185, 15–22. doi: 10.4049/jimmunol.0903856
Loeuillet, C., Martinon, F., Perez, C., Munoz, M., Thome, M., and Meylan, P. R. (2006). Mycobacterium tuberculosis subverts innate immunity to evade specific effectors. J. Immunol. 177, 6245–6255. doi: 10.4049/jimmunol.177.9.6245
Malen, H., Pathak, S., Softeland, T., de Souza, G. A., and Wiker, H. G. (2010). Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol. 10:132. doi: 10.1186/1471-2180-10-132
Mazandu, G. K., and Mulder, N. J. (2012). Function prediction and analysis of Mycobacterium tuberculosis hypothetical proteins. Int. J. Mol. Sci. 13, 7283–7302. doi: 10.3390/ijms13067283
Mishra, K. C., de Chastellier, C., Narayana, Y., Bifani, P., Brown, A. K., Besra, G. S., et al. (2008). Functional role of the PE domain and immunogenicity of the Mycobacterium tuberculosis triacylglycerol hydrolase LipY. Infect. Immun. 76, 127–140. doi: 10.1128/IAI.00410-07
Mohareer, K., Tundup, S., and Hasnain, S. E. (2011). Transcriptional regulation of Mycobacterium tuberculosis PE/PPE genes: a molecular switch to virulence? J. Mol. Microbiol. Biotechnol. 21, 97–109. doi: 10.1159/000329489
Nagabhushanam, V., Solache, A., Ting, L. M., Escaron, C. J., Zhang, J. Y., and Ernst, J. D. (2003). Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 171, 4750–4757. doi: 10.4049/jimmunol.171.9.4750
Nair, S., Ramaswamy, P. A., Ghosh, S., Joshi, D. C., Pathak, N., Siddiqui, I., et al. (2009). The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. J. Immunol. 183, 6269–6281. doi: 10.4049/jimmunol.0901367
Narayana, Y., Joshi, B., Katoch, V. M., Mishra, K. C., and Balaji, K. N. (2007). Differential B-cell responses are induced by Mycobacterium tuberculosis PE antigens Rv1169c, Rv0978c, and Rv1818c. Clin. Vaccine Immunol. 14, 1334–1341. doi: 10.1128/CVI.00181-07
Raman, S., Hazra, R., Dascher, C. C., and Husson, R. N. (2004). Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol. 186, 6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004
Sampson, S. L. (2011). Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin. Dev. Immunol. 2011:497203. doi: 10.1155/2011/497203
Saunders, B. M., Frank, A. A., Orme, I. M., and Cooper, A. M. (2000). Interleukin-6 induces early gamma interferon production in the infected lung but is not required for generation of specific immunity to Mycobacterium tuberculosis infection. Infect. Immun. 68, 3322–3326. doi: 10.1128/IAI.68.6.3322-3326.2000
Saxena, A. K., Roy, K. K., Singh, S., Vishnoi, S. P., Kumar, A., Kashyap, V. K., et al. (2013). Identification and characterisation of small-molecule inhibitors of Rv3097c-encoded lipase (LipY) of Mycobacterium tuberculosis that selectively inhibit growth of bacilli in hypoxia. Int. J. Antimicrob. Agents 42, 27–35. doi: 10.1016/j.ijantimicag.2013.03.007
Schrager, L. Vaccine Prevention of Sustained Mycobacterium tuberculosis Infection Summary Group. (2015). Developing vaccines to prevent sustained infection with Mycobacterium tuberculosis: Conference proceedings: National Institute of Allergy and Infectious Diseases, Rockville, Maryland USA, November 7, 2014. Vaccine 33, 3056–3064. doi: 10.1016/j.vaccine.2015.03.061
Singh, K. K., Zhang, X., Patibandla, A. S., Chien, P. Jr., and Laal, S. (2001). Antigens of Mycobacterium tuberculosis expressed during preclinical tuberculosis: serological immunodominance of proteins with repetitive amino acid sequences. Infect. Immun. 69, 4185–4191. doi: 10.1128/IAI.69.6.4185-4191.2001
Singh, S. K., Tripathi, D. K., Singh, P. K., Sharma, S., and Srivastava, K. K. (2013). Protective and survival efficacies of Rv0160c protein in murine model of Mycobacterium tuberculosis. Appl. Microbiol. Biotechnol. 97, 5825–5837. doi: 10.1007/s00253-012-4493-2
Thi, E. P., Hong, C. J., Sanghera, G., and Reiner, N. E. (2013). Identification of the Mycobacterium tuberculosis protein PE-PGRS62 as a novel effector that functions to block phagosome maturation and inhibit iNOS expression. Cell. Microbiol. 15, 795–808. doi: 10.1111/cmi.12073
Tian, C., and Jian-Ping, X. (2010). Roles of PE_PGRS family in Mycobacterium tuberculosis pathogenesis and novel measures against tuberculosis. Microb. Pathog. 49, 311–314. doi: 10.1016/j.micpath.2010.07.004
Tiwari, B. M., Kannan, N., Vemu, L., and Raghunand, T. R. (2012). The Mycobacterium tuberculosis PE proteins Rv0285 and Rv1386 modulate innate immunity and mediate bacillary survival in macrophages. PLoS ONE 7:e51686. doi: 10.1371/journal.pone.0051686
Tundup, S., Pathak, N., Ramanadham, M., Mukhopadhyay, S., Murthy, K. J., Ehtesham, N. Z., et al. (2008). The co-operonic PE25/PPE41 protein complex of Mycobacterium tuberculosis elicits increased humoral and cell mediated immune response. PLoS ONE 3:e3586. doi: 10.1371/journal.pone.0003586
Vandal, O. H., Roberts, J. A., Odaira, T., Schnappinger, D., Nathan, C. F., and Ehrt, S. (2009). Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J. Bacteriol. 191, 625–631. doi: 10.1128/JB.00932-08
Vordermeier, H. M., Hewinson, R. G., Wilkinson, R. J., Wilkinson, K. A., Gideon, H. P., Young, D. B., et al. (2012). Conserved immune recognition hierarchy of mycobacterial PE/PPE proteins during infection in natural hosts. PLoS ONE 7:e40890. doi: 10.1371/journal.pone.0040890
Voskuil, M. I., Schnappinger, D., Rutherford, R., Liu, Y., and Schoolnik, G. K. (2004). Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis. 84, 256–262. doi: 10.1016/j.tube.2003.12.014
Welin, A., Eklund, D., Stendahl, O., and Lerm, M. (2011). Human macrophages infected with a high burden of ESAT-6-expressing M. tuberculosis undergo caspase-1- and cathepsin B-independent necrosis. PLoS ONE 6:e20302. doi: 10.1371/journal.pone.0020302
Williams, E. P., Lee, J. H., Bishai, W. R., Colantuoni, C., and Karakousis, P. C. (2007). Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 189, 4234–4242. doi: 10.1128/JB.00201-07
Keywords: Mycobacterium tuberculosis, PE family, Rv1169c, fatty acid components, cell wall integrality, cell death, interleukin-6
Citation: Deng W, Zeng J, Xiang X, Li P and Xie J (2015) PE11 (Rv1169c) selectively alters fatty acid components of Mycobacterium smegmatis and host cell interleukin-6 level accompanied with cell death. Front. Microbiol. 6:613. doi: 10.3389/fmicb.2015.00613
Received: 08 March 2015; Accepted: 02 June 2015;
Published: 23 June 2015.
Edited by:
Vishwanath Venketaraman, Western University of Health Sciences, USAReviewed by:
Hridayesh Prakash, University of Hyderabad, IndiaHeinrich Korner, Menzies Research Institute Tasmania, Australia
Copyright © 2015 Deng, Zeng, Xiang, Li and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Xie, State Key Laboratory Breeding Base of Eco-Environment and Bio-Resource of the Three Gorges Area, Key Laboratory of Eco-environments in Three Gorges Reservoir Region, Ministry of Education, School of Life Sciences, Institute of Modern Biopharmaceuticals, Southwest University, Chongqing 400715, China, georgex@swu.edu.cn
†These authors have contributed equally to this work.
‡Co-first authors.
 Wanyan Deng
Wanyan Deng Jie Zeng
Jie Zeng Xiaohong Xiang
Xiaohong Xiang Ping Li
Ping Li Jianping Xie
Jianping Xie