
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 July 2015
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 6 - 2015 | https://doi.org/10.3389/fmicb.2015.00634
 Abdulrahman A. Al-Sultan1
Abdulrahman A. Al-Sultan1 Benjamin A. Evans2*
Benjamin A. Evans2* Elsayed Aboulmagd1†
Elsayed Aboulmagd1† Ahmed A. Al-Qahtani3,4,5
Ahmed A. Al-Qahtani3,4,5 Marie Fe F. Bohol3
Marie Fe F. Bohol3 Mohammed N. Al-Ahdal3,4,6
Mohammed N. Al-Ahdal3,4,6 Andres F. Opazo7
Andres F. Opazo7 Sebastian G. B. Amyes8
Sebastian G. B. Amyes8It has previously been shown that carbapenem-resistant Acinetobacter baumannii are frequently detected in Saudi Arabia. The present study aimed to identify the epidemiology and distribution of antibiotic resistance determinants in these bacteria. A total of 83 A. baumannii isolates were typed by pulsed-field gel electrophoresis (PFGE), and screened by PCR for carbapenemase genes and insertion sequences. Antibiotic sensitivity to imipenem, meropenem, tigecycline, and colistin were determined. Eight different PFGE groups were identified, and were spread across multiple hospitals. Many of the PFGE groups contained isolates belonging to World-wide clone 2. Carbapenem resistance or intermediate resistance was detected in 69% of isolates. The blaVIM gene was detected in 94% of isolates, while blaOXA–23–like genes were detected in 58%. The data demonstrate the co-existence and wide distribution of a number of clones of carbapenem-resistant A. baumannii carrying multiple carbapenem-resistance determinants within hospitals in the Eastern Region of Saudi Arabia.
A number of recent studies have identified high levels of carbapenem resistance in isolates of Acinetobacter baumannii from Saudi Arabia (Alsultan et al., 2013; Abdalhamid et al., 2014; Aly et al., 2014). These studies have demonstrated that a large number of different genetic lineages within a hospital are resistant to the carbapenems, rather than there being the predominance of a single epidemic clone (Abdalhamid et al., 2014; Aly et al., 2014). These isolates often belong to widespread clonal lineages (Zarrilli et al., 2013). However, due to examining only isolates obtained from a single hospital (Abdalhamid et al., 2014; Aly et al., 2014), or the absence of molecular typing data (Alsultan et al., 2013), it is still not clear what the epidemiological pattern of carbapenem-resistant A. baumannii is on a wider scale within Saudi Arabia.
It was previously shown in a study utilizing relatively few isolates that a number of different lineages of A. baumannii, defined by pulsed-field gel electrophoresis (PFGE), were associated with infections in diabetic patients in Saudi Arabia, with one of these lineages found in more than one hospital (Alsultan et al., 2009). A much larger study found that isolates from diabetic patients were more likely to be carbapenem resistant and carry an acquired OXA-type β-lactamase, but did not identify the molecular epidemiology of the isolates (Alsultan et al., 2013). A recent study carried out in Abu Dhabi, United Arab Emirates, that borders Saudi Arabia, identified a diverse range of isolates, with even the epidemic isolates belonging to multiple PFGE pulsotypes and genotypes (Sonnevend et al., 2013).
The high and increasing prevalence of type 2 diabetes is a very serious health concern in Saudi Arabia, and as these patients are particularly susceptible to bacterial infection, the prevalence of carbapenem-resistant A. baumannii in this group is of particular concern (Prata-Rocha et al., 2012). Here we have typed a larger collection of 83 A. baumannii isolates taken from a combination of diabetic and non-diabetic patients from the Eastern District of Saudi Arabia. We aim to determine the epidemiological relationships between isolates and the distribution of antibiotic resistance determinants in this bacterial population, in order to address the lack of molecular epidemiological data between hospitals in a wider geographic context.
A total of 83 isolates were collected between 2008 and 2012 from different specimens from patients from 7 major hospitals in the Eastern province of Saudi Arabia—23 isolates from patients with type 2 Diabetes Mellitus, and 60 isolates from patients without this condition. For the purpose of our study, patients were considered to be diabetic if they were using insulin and were undergoing hospital treatment for the condition. All isolates were identified presumptively by the Vitek compact II system as being A. baumannii.
Multiplex polymerase chain reactions (PCRs) using primers for the blaOXA–51–like, blaOXA–23–like, blaOXA–40–like, and blaOXA–58–like genes were performed (Woodford et al., 2006), with species identification being confirmed by the production of a blaOXA–51–like amplicon. Amplification and sequencing of complete blaOXA–51–like alleles was performed using the primers OXA-69A and OXA-69B (Heritier et al., 2005), and the allele identity used to determine the epidemic lineage to which the isolate belonged as first described by Evans et al. (2008) and later confirmed by Pournaras et al. (2014). Isolates were screened by PCR for the presence of acquired metallo-β-lactamase genes (MBLs) of the blaIMP, blaVIM, blaNDM, blaSIM, and blaGIM (Ellington et al., 2007; Poirel et al., 2011) families using consensus primers, and for the insertion sequences ISAba1, ISAba2, ISAba3, and IS18 (Poirel and Nordmann, 2006).
The minimum inhibitory concentrations (MICs) of imipenem, meropenem, tigecycline and colistin were determined by the agar plate doubling-dilution method according to the British Society for Antimicrobial Chemotherapy (BSAC) guidelines (Andrews, 2010).
Isolates were typed by PFGE using the method described by Miranda et al. (1996) as previously described (Evans et al., 2008). PFGE profiles were analyzed using the Bionumerics software version 6.5. All data were analyzed in SPSS version 20 (SPSS Inc.,). Following Kruskal Wallis tests, differences between groups were determined by pairwise Mann-Whitney U tests. Correction for multiple testing was performed using the method of Benjamini and Hochberg (1995).
Isolates were grouped according to their PFGE profiles (Figure 1). Sixty-two isolates were assigned to one of eight different PFGE groups, defined as having PFGE profiles with 85% similarity or greater, and consisting of at least three isolates. The remaining 21 isolates that did not meet these criteria were considered to be ungrouped. There was a significant difference in the diabetic status of the patients between the individual PFGE groups (Kruskal Wallis, χ2 = 26.32, d.f. = 7, p = 0.0024 following correction), with isolates from diabetic patients being in the majority in PFGE groups B, D, and H (Figure 1).
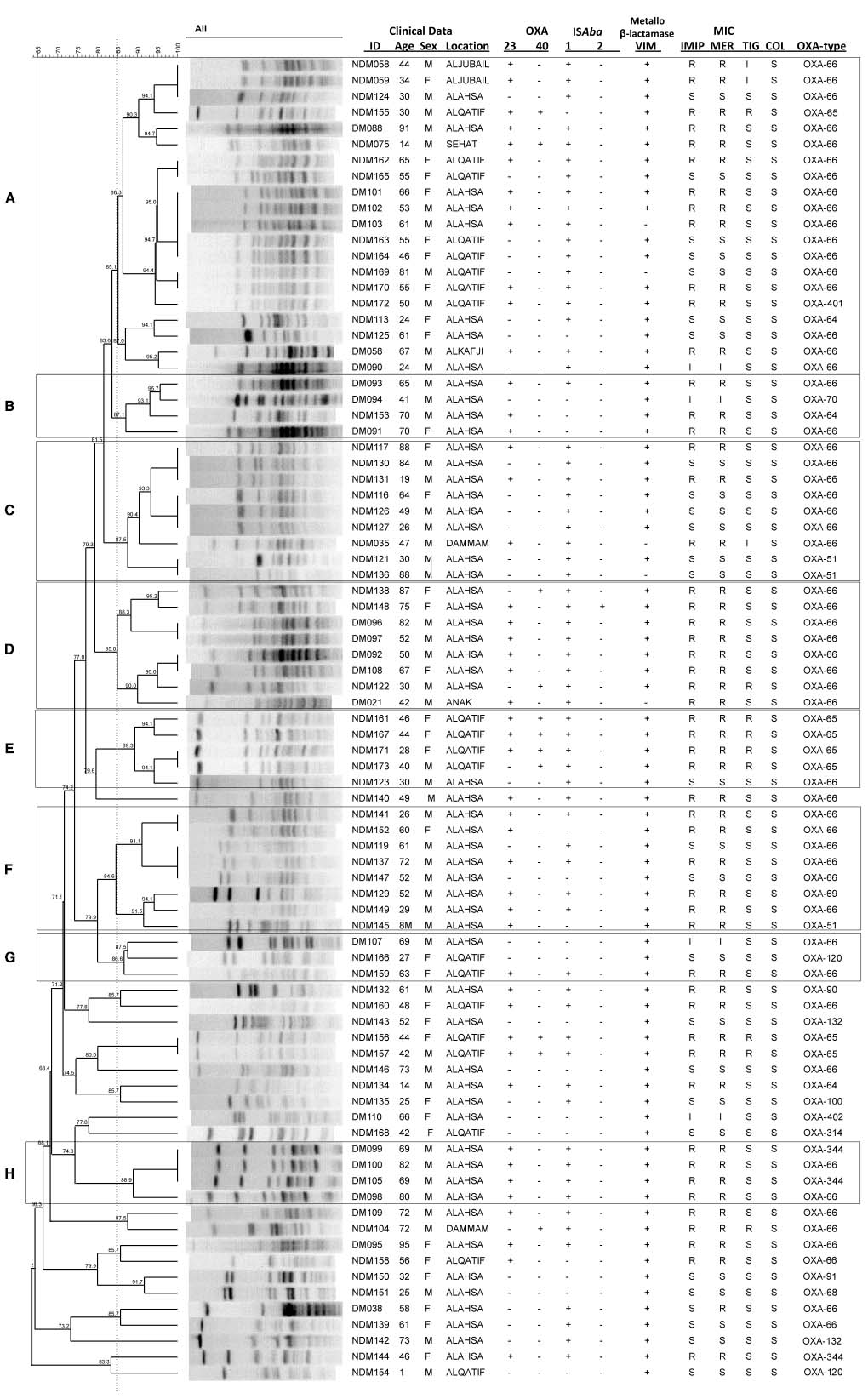
Figure 1. Isolate relatedness, antibiotic resistances and molecular screening of isolates. In the ID column, DM stands for Diabetes Mellitus, NDM for non-Diabetes Mellitus. IMIP, imipenem; MER, meropenem; TIG, tigecycline; COL, colistin. (A–H) Indicate PFGE groups assigned as described in the text.
The majority of the isolates (67%) were from Al Ahsa, with most of the remaining isolates (24%) originating from Al Qatif. In Al Ahsa, isolates representing all eight of the PFGE groups were identified (Figure 2). Three of these groups were also identified in Al Qatif. Additionally, isolates from one or more of the PFGE groups were identified at the five other locations sampled, despite the geographic distance between these hospitals (Figure 2).
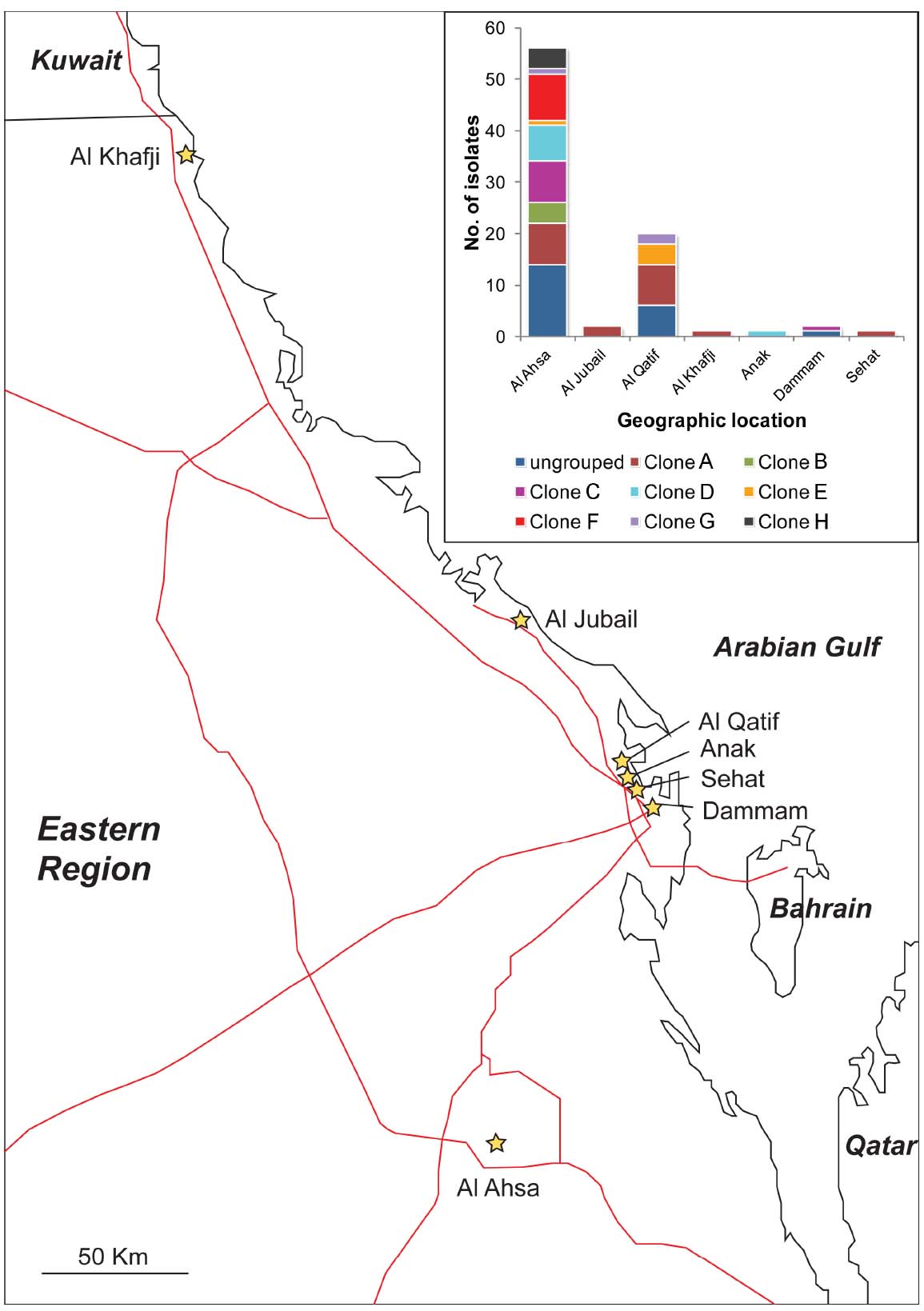
Figure 2. Location of hospitals from where isolates were obtained, and (insert) the distribution of PFGE clones (as defined in Figure 1) by their geographic location. Hospitals are shown as stars, and major roads as red lines.
Of the 83 isolates, 57 (69%) were resistant or had intermediate resistance to at least one carbapenem. In agreement with previous findings (Alsultan et al., 2013), isolates from diabetic patients were more likely to be resistant to a carbapenem than those from non-diabetic patients (diabetic status vs meropenem resistance, Mann Whitney U test, Z = –3.787, p = 0.001 following correction). A total of 13 isolates (16%) were resistant to tigecycline, and none to colistin. A large proportion of isolates were found to carry resistance genes, with 48 isolates (58%) carrying blaOXA–23–like genes, 11 isolates (13%) carrying blaOXA–40–like genes, and 78 isolates (94%) carrying a blaVIM gene. PCRs for blaOXA–58–like, blaIMP, blaNDM, blaSIM, and blaGIM genes were all negative. There was a significant difference in the presence of blaOXA–40 genes between the different PFGE groups (Kruskal Wallis, χ2 = 25.12, d.f. = 7, p = 0.003 following correction), with the gene being detected with particularly high frequency in PFGE group E (Figure 1). Phenotypic resistance to the carbapenems was associated with the presence of a blaOXA–23–like gene rather than the presence of a blaVIM–like gene (meropenem resistance vs blaOXA–23–like carriage: Mann Whitney U test, Z = –7.162, p < 0.00001 following correction; meropenem resistance vs blaVIM carriage: Mann Whitney U test, Z = –0.429, p = 0.713 following correction).
A total of 65 isolates (78%) were positive by PCR for ISAba1, while only two isolates were positive for ISAba2. The insertion sequences ISAba3 and IS18 were not detected. The different PFGE groups differed significantly in the presence of ISAba1 (Kruskal Wallis, χ2 = 23.06, d.f. = 7, p = 0.004 following correction) with higher frequencies of detection of this element in PFGE groups A, C, D, E, and H (Figure 1).
Sequencing of the blaOXA–51–like alleles identified the presence of the blaOXA–66 allele in 54 isolates (65%), with the remaining 29 isolates encoding one of 15 other OXA-51-like enzymes (Figures 1 and 3). Isolates carrying a blaOXA–66 allele were considered to belong to the World-wide clone 2 (WW2) lineage (Evans et al., 2008; Higgins et al., 2009). Isolates belonging to one of the PFGE groups were more likely to encode a blaOXA–66 allele, while isolates that were not grouped were more likely to encode a non-blaOXA–66 allele (Mann Whitney U test, Z = –2.981, p = 0.012 following correction). In addition there was a significant difference in the association of ISAba1 with blaOXA–51–like alleles, with the insertion sequence found significantly more often upstream of blaOXA–66 alleles than non-blaOXA–66 alleles (Mann Whitney U test, Z = –3.171, p = 0.011 following correction).
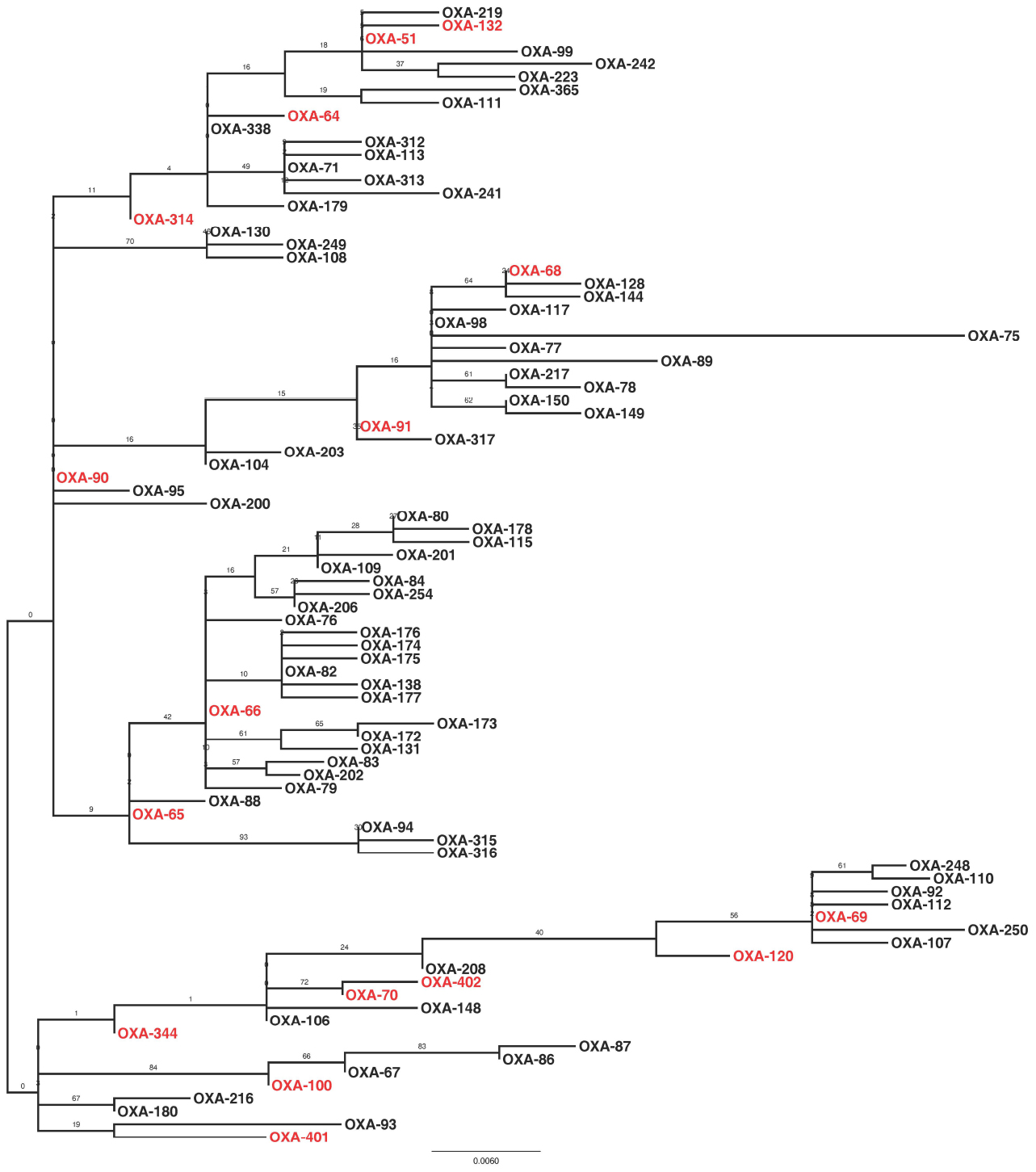
Figure 3. Maximum likelihood phylogeny based upon amino-acid sequences of OXA-51-like enzymes. Enzymes that were encoded by isolates in the present study are highlighted in red. The phylogeny is mid-point rooted, with support for the branches estimated from 100 bootstraps.
Carbapenem-resistant A. baumannii pose a significant challenge to the treatment of infected patients. These patients are usually immunocompromised, and the elimination of the carbapenem antibiotics as an effective therapy leaves very few treatment options open. In the current study, A. baumannii isolates from hospitals in the Eastern District of Saudi Arabia were examined to identify their mechanisms of resistance to the carbapenems and their local epidemiology, both within the hospitals and the District.
Typically, the isolation of many carbapenem-resistant bacteria within a hospital may be explained by two epidemiological patterns: the emergence and spread of a particularly successful clone, or the persistence and co-existence of many clonal lineages, potentially with importation from the community. Our data are most consistent with the latter scenario for two reasons. Firstly, many different PFGE groups were identified both within Al Ahsa and Al Qatif. Moreover, isolates from the same PFGE groups were identified in different hospitals. It is also noteworthy that 25% of the isolates were not assigned to a PFGE group. The distribution of isolates from the same PFGE group across the Eastern District (Al Kafji and Al Ahsa are ∼430 km apart by road, Figure 2) and the identification of many ungrouped carbapenem-resistant isolates suggest the persistent co-existence of many lineages across the region. Secondly, there is a large degree of variation in antibiotic resistance phenotypes and genotypes within PFGE groups. The only features that were significantly associated with PFGE groups were the presence of blaOXA–40–like genes, and the presence of ISAba1. However, these genes were still identified in isolates belonging to PFGE groups that were not significantly associated with their carriage. Antibiotic resistance phenotypes and other genes associated with resistance were distributed amongst PFGE groups and ungrouped isolates, with no statistically significant positive or negative association with any PFGE group. This suggests that these genes are endemic within the general A. baumannii population where they have either spread amongst the many genetic lineages, or they have existed within the A. baumannii population for a substantial period of time as the different genetic lineages have diverged, with some genes being lost over time. Taken together, these data are consistent with the endemicity of a polyclonal carbapenem-resistant or carbapenemase-encoding population of A. baumannii in the Eastern District of Saudi Arabia.
Previous studies have identified carbapenem-resistant A. baumannii isolates belonging to the WW2 lineage as being widespread and responsible for outbreaks of infection in hospitals globally (Higgins et al., 2009; Karah et al., 2012). In the current study, the majority of isolates were considered to belong to WW2 by virtue of their specific blaOXA–51–like allele (Evans et al., 2008; Higgins et al., 2009). A previous study associated isolates carrying blaOXA–64 with multi-locus sequence typing (MLST) clonal complex (CC) 25, blaOXA–51 with CC15, blaOXA–100 with CC32, and blaOXA–65 with CC79 from the Pasteur MLST scheme (Pournaras et al., 2014). As such, isolates identified in the present study may also belong to these clonal complexes. Despite the fact that many different PFGE groups were identified, isolates belonging to one of these PFGE groups were statistically more likely to belong to WW2 than the ungrouped isolates. In addition, these WW2 isolates were statistically more likely to encode an ISAba1 element upstream of the blaOXA–51–like genes, which has been implicated in increased expression of the β-lactamase gene (Turton et al., 2006). As has been identified previously (Evans et al., 2008; Hamouda et al., 2010), PFGE profiles did not correlate perfectly with the blaOXA–51–like gene content of the isolates in a few cases, which may be explained by potential mobilization of the blaOXA–51–like genes (Chen et al., 2010). Together these data are consistent with previous studies, and demonstrate that the different PFGE groups identified in this study are subgroups of WW2, and therefore this global clone represents the majority of carbapenem-resistant isolates in the Eastern District.
The question arises as to the precise nature of the local epidemiology of carbapenem resistance in these isolates. There are two scenarios that can be explored. Firstly, a series of endemic clones may be circulating within and amongst hospitals in the Eastern District, with the transmission of plasmids carrying antibiotic resistance determinants responsible for a large degree of the variation in antibiotic resistance phenotype and genotype between strains. Secondly, carbapenem-resistant A. baumannii may be endemic within the community, with strains being imported into the hospital where they briefly spread, before being replaced by other strains. Most of the isolates (94%) in the current study were collected between October 2010 and April 2011, and as such an assessment cannot be made from these data of the changes in frequency of different strains over time. Establishing which of these scenarios reflect the situation in the Eastern District will enable the appropriate infection control advice to be given to hospitals to reduce the prevalence of carbapenem-resistant A. baumannii, and is the subject of our future research.
AAS, SA conceived and designed the study. AAS, EA, AAQ, MB, MA, AO acquired the data. AAS, BE, EA analyzed the data. AAS, BE, EA, SA drafted and critically evaluated the manuscript. All authors approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was funded by King Abdulaziz City for Science and Technology (KACST). The authors wish to thank King Faisal University (KFU) for its support, and Hani Alrasasi, Hani Alfarhan, Hajar Al-dohilan, Nof Alhomaini, Abdulqader Abouli, and Fatemah Al Najar for their technical assistance.
Abdalhamid, B., Hassan, H., Itbaileh, A., and Shorman, M. (2014). Characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates in a tertiary care hospital in Saudi Arabia. New Microbiol. 37, 65–73.
Alsultan, A. A., Evans, B. A., Elsayed, E. A., Al-Thawadi, S. I., Al-Taher, A. Y., Amyes, S. G., et al. (2013). High frequency of carbapenem-resistant Acinetobacter baumannii in patients with diabetes mellitus in Saudi Arabia. J. Med. Microbiol. 62, 885–888. doi: 10.1099/jmm.0.057216-0
Alsultan, A. A., Hamouda, A., Evans, B. A., and Amyes, S. G. (2009). Acinetobacter baumannii: emergence of four strains with novel blaOXA–51–like genes in patients with diabetes mellitus. J. Chemother. 21, 290–295. doi: 10.1179/joc.2009.21.3.290
Aly, M., Tayeb, H. T., Al Johani, S. M., Alyamani, E. J., Aldughaishem, F., Alabdulkarim, I., et al. (2014). Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1223–1228. doi: 10.1007/s10096-014-2068-0
Andrews, J. M. (2010). BSAC Methods for Antimicrobial Susceptibility Testing, Version 9.1. Available at: http://bsac.org.uk/wp-content/uploads/2012/02/Version_9.1_March_2010_final-v2.pdf [accessed August 30, 2012].
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300.
Chen, T. L., Lee, Y. T., Kuo, S. C., Hsueh, P. R., Chang, F. Y., Siu, L. K., et al. (2010). Emergence and distribution of plasmids bearing the blaOXA–51–like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 54, 4575–4581. doi: 10.1128/aac.00764-10
Ellington, M. J., Kistler, J., Livermore, D. M., and Woodford, N. (2007). Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J. Antimicrob. Chemother. 59, 321–322. doi: 10.1093/jac/dkl481
Evans, B. A., Hamouda, A., Towner, K. J., and Amyes, S. G. (2008). OXA-51-like β-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin. Microbiol. Infect. 14, 268–275. doi: 10.1111/j.1469-0691.2007.01919.x
Hamouda, A., Evans, B. A., Towner, K. J., and Amyes, S. G. (2010). Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of blaOXA–51–like genes. J. Clin. Microbiol. 48, 2476–2483. doi: 10.1128/JCM.02431-09
Heritier, C., Poirel, L., Fournier, P. E., Claverie, J. M., Raoult, D., and Nordmann, P. (2005). Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005
Higgins, P., Dammhayn, C., Hackel, M., and Seifert, H. (2009). Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 233–238. doi: 10.1093/jac/dkp428
Karah, N., Sundsfjord, A., Towner, K., and Samuelsen, O. (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001
Miranda, G., Kelly, C., Solorzano, F., Leanos, B., Coria, R., and Patterson, J. E. (1996). Use of pulsed-field gel electrophoresis typing to study an outbreak of infection due to Serratia marcescens in a neonatal intensive care unit. J. Clin. Microbiol. 34, 3138–3141.
Poirel, L., and Nordmann, P. (2006). Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA–58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 1442–1448. doi: 10.1128/AAC.50.4.1442-1448.2006
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pournaras, S., Gogou, V., Giannouli, M., Dimitroulia, E., Dafopoulou, K., Tsakris, A., et al. (2014). Single-locus-sequence-based typing of blaOXA–51–like genes for rapid assignment of Acinetobacter baumannii clinical isolates to international clonal lineages. J. Clin. Microbiol. 52, 1653–1657. doi: 10.1128/JCM.03565-13
Prata-Rocha, M. L., Gontijo-Filho, P. P., and Melo, G. B. (2012). Factors influencing survival in patients with multidrug-resistant Acinetobacter baumannii infection. Braz. J. Infect. Dis. 16, 237–241. doi: 10.1590/s1413-86702012000300004
Sonnevend, A., Ghazawi, A., Al Munthari, N., Pitout, M., Hamadeh, M. B., Hashmey, R., et al. (2013). Characteristics of epidemic and sporadic strains of Acinetobacter baumannii isolated in Abu Dhabi hospitals. J. Med. Microbiol. 62, 582–590. doi: 10.1099/jmm.0.055681-0
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/j.1574-6968.2006.00195.x
Woodford, N., Ellington, M. J., Coelho, J. M., Turton, J. F., Ward, M. E., Brown, S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27, 351–353. doi: 10.1016/j.ijantimicag.2006.01.004
Keywords: Acinetobacter baumannii, carbapenem resistance, Saudi Arabia, PFGE, OXA, VIM
Citation: Al-Sultan AA, Evans BA, Aboulmagd E, Al-Qahtani AA, Bohol MFF, Al-Ahdal MN, Opazo AF and Amyes SGB (2015) Dissemination of multiple carbapenem-resistant clones of Acinetobacter baumannii in the Eastern District of Saudi Arabia. Front. Microbiol. 6:634. doi: 10.3389/fmicb.2015.00634
Received: 01 May 2015; Accepted: 11 June 2015;
Published: 02 July 2015.
Edited by:
Fiona Walsh, National University of Ireland Maynooth, IrelandReviewed by:
Raffaele Zarrilli, University of Naples Federico II, ItalyCopyright © 2015 Al-Sultan, Evans, Aboulmagd, Al-Qahtani, Bohol, Al-Ahdal, Opazo and Amyes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin A. Evans, Department of Biomedical and Forensic Sciences, Faculty of Science and Technology, Anglia Ruskin University, East Road, Cambridge, CB1 1PT, UK,benjamin.evans@anglia.ac.uk
†Present address: Elsayed Aboulmagd, Faculty of Pharmacy, Alexandria University, Alexandria, Egypt
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.