- 1Molecular and Translational Sciences Division, United States Army Medical Research Institute of Infectious Diseases, Frederick, MD, USA
- 2Department of Defense Biotechnology High Performance Computing Software Applications Institute, Telemedicine and Advanced Technology Research Center, United States Army Medical Research and Materiel Command, Frederick, MD, USA
- 3National Center for Biodefense and Infectious Diseases, and School of Systems Biology, George Mason University, Manassas, VA, USA
- 4PerkinElmer, Inc., Waltham, MA, USA
Burkholderia is a diverse genus of gram-negative bacteria that causes high mortality rate in humans, equines and cattle. The lack of effective therapeutic treatments poses serious public health threats. Developing insights toward host-Burkholderia spp. interaction is critical for understanding the pathogenesis of infection as well as identifying therapeutic targets for drug development. Reverse-phase protein microarray technology was previously proven to identify and characterize novel biomarkers and molecular signatures associated with infectious disease and cancer. In the present study, this technology was utilized to interrogate changes in host protein expression and phosphorylation events in macrophages infected with a collection of geographically diverse strains of Burkholderia spp. The expression or phosphorylation state of 25 proteins was altered during Burkholderia spp. infections of which eight proteins were selected for further characterization by immunoblotting. Increased phosphorylation of AMPK-α1, Src, and GSK3β suggested the importance of their roles in regulating Burkholderia spp. mediated innate immune response. Modulating the inflammatory response by perturbing their activities may provide therapeutic routes for future treatments.
Introduction
Burkholderia pseudomallei (Bp) and Burkholderia mallei (Bm) are facultative intracellular gram-negative bacterial pathogens that cause melioidosis and glanders, respectively. While genetically similar, Bp and Bm are associated with their own hallmarks. Bp is endemic to tropical regions of Southeast Asia, Northern Australia, and China, where it inhabits the soil and stagnant water. Routes of human infection include inhalation, ingestion, and contact with open wounds; however, human-to-human transmission is extremely rare (White, 2003; Gilad, 2007; Galyov et al., 2010). Symptomatic infection may present with flu-like symptoms such as fever and pulmonary distress, making accurate diagnosis of melioidosis difficult in the early stages (Dance, 1991; Leelarasamee, 2004; De Keulenaer and Cheng, 2006). Fatal infection is often due to progression to septicemia and acute pneumonia (Dance, 1991, 2000; Dharakul and Songsivilai, 1999; Galyov et al., 2010). By contrast, Bm is a non-motile obligate mammalian pathogen endemic among domestic animals in Africa, Asia, the Middle East, and Central and South America (Whitlock et al., 2007). Equine species are the natural reservoir for Bm and are responsible for transmission of glanders to humans through contact, leading to development of pneumonia and septicemia, often resulting in fatality (Whitlock et al., 2007; Galyov et al., 2010; Van Zandt et al., 2013). Given that both species are highly infectious via aerosolization, coupled with the lack of vaccines, Bp and Bm are considered category B bioterrorism agents by the U. S. Centers for Disease Control (Holden et al., 2004; Whitlock et al., 2007; Galyov et al., 2010).
Bp and Bm are resistant to many traditional antibiotics (Whitlock et al., 2007; Estes et al., 2010; Galyov et al., 2010). Although successful treatment is possible with several months of a combination antimicrobial regimen, relapse is common (Holden et al., 2004; Estes et al., 2010). Effort has been placed on evaluating the therapeutic potential of bacterial secretion systems and effectors as components of candidate vaccines (Whitlock et al., 2007); however, none are available at this time. Additional therapeutic strategies are required for controlling disease in endemic regions as well as for protection against potential bioterrorism threats. Immunotherapy in conjunction with antimicrobials is a new theme in the Burkholderia treatment paradigm. A promising study found that combining interferon (IFN)-γ with an antimicrobial showed synergistic inhibition of growth in Bp infected macrophages (Propst et al., 2010). Identification of additional host targets that function in resolving the overactive immune and inflammatory responses elicited by infection is an active area of research (Ulett et al., 2000).
Pattern recognition receptors (PRRs) are innate immune sensors that serve as the first line of defense against pathogen infections. Toll like receptor 4 (TLR4) is a PRR that detects lipopolysaccharide (LPS) expressed on the surface of Burkholderia spp. TLR4 activation recruits two adaptor proteins, myeloid differentiation primary response gene 88 (MyD88) and TIR-domain-containing adapter-inducing interferon-β (TRIF). The TRIF-dependent pathway mediates the upregulation of type I IFN and subsequent phosphorylation of Signal Transducers and Activators of Transcriptions (STATs). The MyD88-dependent pathway leads to the activation of IKKα and IKKβ, which specifically phosphorylate two serines on the cytoplasmic NF-κB inhibitor, IκB. Phosphorylation of IκB causes its ubiquitin-dependent degradation, leading to the activation and nuclear translocation of NF-κB (Akira et al., 2006). In addition to the NF-κB pathway, TLR4 also activates Stress Activated Protein Kinases (SAPK) such as p38 via apoptosis signal-regulating kinase 1 (ASK1), a MAP kinase kinase kinase (MEKK; Matsuzawa et al., 2005). Although surrogate ligand-mediated (e.g., LPS) pro-inflammatory pathway is extensively studied, the role of Bp and Bm LPS in human melioidosis or glanders is just beginning to unravel (Chantratita et al., 2013; Alam et al., 2014). Furthermore, larger molecular signaling networks that are critical for invasion, survival and replication of Burkholderia spp. in the host require characterization.
Reverse-phase protein microarray (RPMA) is a quantitative, high throughput tool that enables interrogation of changes in protein expression and post-translational modification in complex cellular signaling networks upon perturbation. This technology has been successfully utilized to profile molecular signatures and functional pathway activation associated with ovarian and breast cancer, respectively, (Sheehan et al., 2005; Wulfkuhle et al., 2012). Furthermore, phosphorylation states of key host signaling proteins were characterized in human samples infected with highly pathogenic agents such as Rift valley fever virus and Yersinia pestis (Popova et al., 2010; Alem et al., 2015). The goal of this study was to comprehensively examine whether diverse strains of Bp, Bm, and Burkholderia thailandensis (Bt) trigger signaling through common or distinct host pathways. To this end, we utilized a RPMA platform to simultaneously detect changes in the expression of host signaling proteins as well as alterations in phosphorylation-mediated host signaling caused by infection with Burkholderia spp. To generate host protein expression and phosphorylation profiles, RAW264.7 murine macrophages were infected with geographically diverse isolates of Bp and Bm, and at various time points protein lysates were examined using RPMA. This approach identified 25 candidates whose expression levels and/or phosphorylation states were altered, of which eight proteins were selected for further characterization through immunoblot analysis. Consistent with published studies, we identified phosphorylation of glycogen synthase kinase 3 beta (GSK3β) and components of the NF-κB and MAPK pathways, validating the RPMA approach. Moreover, we report phosphorylation of additional host proteins that were not previously linked to Burkholderia spp. infection, including AMP activated protein kinase (AMPK-α1) and Src. The signaling architecture is similar between Burkholderia spp. with minor distinctions in the magnitude of activation or induction for iNOS, phosphorylated GSK3β and STAT1 and c-Myc. Pharmacological perturbation of these critical cascades may serve as therapeutic routes for Burkholderia spp. infections.
Materials and Methods
Bacterial Strains
Bm ATCC 23344, NCTC 10247, NCTC 10229, NCTC 3708, NCTC 3709, 2002721278 (Burtnick et al., 2002); Bp E8, 576, MSHR305 (Bast et al., 2011; Van Zandt et al., 2012) and Bt DW503 were used in this study. Burkholderia spp. were maintained on Luria Broth (LB) plates with 1.5% agar or on 5% sheep blood agar (SBA) plates. Bp and Bt strains were cultured in LB whereas Bm strains were propagated in LB with 4% glycerol. Bacterial concentrations were quantified using optical density (OD) at 600 nm readings and diluted using a conversion factor of 5 × 108 CFU/ml per unit of OD. All studies using viable Bp and Bm isolates were performed using biosafety level three conditions.
Cell Lines
Mouse macrophage cell line RAW264.7 was obtained from ATCC (Manassas, VA). Cells were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Hyclone), 1% nonessential amino acids (Sigma-Aldrich), and 1% glutamax (Life Technologies), grown at 37°C in 5% CO2.
Antibodies
For the RPMA studies, only validated antibodies known to cross react with murine target proteins were selected (Supplementary Table S1). For immunoblotting, the following antibodies were purchased from Cell Signaling Technology: iNOS (#2977), IκBα (#9242), phospho STAT1 (#9171), phospho p38 (#9211), phospho ERK1/2 (#9101), phospho AMPK-α1 (#4184), total p38 (#2387), phospho Src (#6943), c-Myc (#9402), phospho GSK3β (#9331). GAPDH was purchased from Sigma-Alrich (G9545). Total GSK3α/β was purchased from Santa Cruz biotechnology (SC-56913). Total ERK1 was purchased from BD Transduction laboratories (M12320).
Infection of RAW264.7 Macrophages
RAW264.7 macrophages (1 × 106 cells/well) were seeded overnight in six well plates. The next day, cells were infected with one Bt, five Bm, and three Bp strains at multiplicity of infection (MOI) of 1 and 10. In parallel, cells were stimulated with Escherichia coli LPS at 100 ng/ml. Uninfected and unstimulated cells were used as negative controls. RAW264.7 cell samples were collected at 30 min, 1, 4, and 8 h post Burkholderia spp. infection, washed with phosphate buffered saline (PBS) and lysed. For the longer incubation time points of 4 and 8 h, extracellular bacteria were removed 2 h post infection by washing the cells three times with PBS and then further incubated with pre-warmed DMEM containing 10% fetal bovine serum and 250 μg/ml of kanamycin (Km) (Brett et al., 2008) to reduce extracellular bacterial growth. All experiments were performed on two independent days and each biological sample lysate was printed in triplicate on nitrocellulose coated glass slides.
Reverse Phase Antibody Array
For RPMA assays, cells were harvested, washed with PBS, and then lysed in a mixture of T-PER Reagent (Thermo Scientific) and 2X Tris-Glycine SDS sample buffer (Life Technologies), complete protease inhibitor cocktail (Roche), Na3VO4, NaF, EDTA, and DTT. RPMAs were constructed and analyzed as previously described (Improta et al., 2011; Federici et al., 2013). Samples from duplicate sets of Burkholderia spp. infections were printed in triplicate spots on nitrocellulose coated glass slides (GRACE Bio-Labs, Inc.) using an Aushon 2470 microarrayer equipped with 185 μm pins (Aushon Biosystems), according to manufacturer’s instructions. A subset of the printed array slides was stained with Spyro Ruby Protein Blot Stain (Life Technologies) to estimate sample total protein concentration. Before staining, proteins array slides were treated with 1X ReBlot Mild Solution (Chemicon) for 15 min, washed two times for 5 min in PBS (Life Technologies), and incubated for 1 h in blocking solution (2% I-Block, 0.1% Tween-20 in PBS; Life Technologies). Immunostaining was performed on an automated slide stainer using a signal amplification kit (DAKO). The arrays were probed with a collection of 114 antibodies. Primary antibody binding was detected using a biotinylated goat-anti rabbit immunoglobulin G (IgG) H + L at 1:7,500 dilution (Vector Laboratories) or rabbit anti-mouse IgG at 1:10 dilution (DAKO) followed by streptavidin-conjugated IRDye680 fluorophore (LI-COR Biosciences). All slides were scanned using a Revolution 4550 scanner (Vidar Systems Corporation), and acquired images were analyzed with MicroVigene v.4.0.0.0 (VigeneTech Inc.), which conducted spot detection, local background subtraction, negative control subtraction, replicate averaging, and total protein normalization, producing a single value for each sample.
RPMA Data Analysis
Reverse-phase protein microarray data for 15 out of the 114 antibodies used in the study had expression values around zero. As we could not assess whether these values represent true biological values or are a consequence of experimental and array processing procedures, data from these 15 antibodies was not included in further analyses. Technical replicates were independently averaged to generate a value for each biological sample. Expression values for individual biological replicates were corrected by normalizing to the value of the uninfected control condition. Fold-change values were generated for each time point. Biological replicates were averaged and those for which the average fold change remained twofold or larger (i.e., fold-change ≤ 0.5 for under-expressed proteins or fold change ≥2 for over-expressed proteins) were included in further analysis.
Immunoblotting
For immunoblot analysis, RAW264.7 macrophages infected at MOI 10 were pelleted and lysed in radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific) containing complete protease inhibitor cocktail (Roche), and phosphatase inhibitors (Roche). The amount of total protein in each sample was quantified by Bradford assay (Bio-Rad). RAW264.7 cell lysate preparations were boiled for 10 min with NuPAGE LDS Sample Buffer (Life Technologies) containing 10% 2-Mercaptoethanol. Equivalent amounts of total protein were loaded onto 4–12% Bis-Tris gradient gels (Life Technologies) and electrophoresed. Following electrophoresis, proteins were transferred to a nitrocellulose membrane (Life Technologies) and then blocked in 5% bovine serum albumin for 1 h prior to incubation with primary antibodies overnight. Blots were washed, then incubated with appropriate HRP-conjugated secondary antibodies (Jackson ImmunoResearch). Signal was detected by ECL Prime (GE Healthcare Life Sciences). Image acquisition was performed with ChemiDocTM XRS+ system (Bio-Rad). Relative protein levels were normalized with endogenous GAPDH as a loading control.
Immuno-Fluorescence Staining
RAW264.7 macrophages (4 × 104 cells/well) were seeded in a Falcon 96 well flat bottom plate (REF353219). Cells were stimulated with 1 μg/ml of LPS or infected with Bm ATCC23344, Bm 2002721278, and Bp E8 at MOI of 10. At 30 min, 1, 4 and 8 h post infection, cells were washed and fixed with formaldehyde. Immuno-fluorescent staining of phospho ERK1/2 (Cell Signaling Technology #9101) was performed according to the manufacturer’s protocol. Images were acquired by Opera confocal reader (model 3842) using a 20X water objective. Analysis of the images was accomplished within the Opera system using standard Acapella scripts.
Colony Forming Unit (CFU) Assay to Quantify Bacterial Uptake and Intracellular Replication
RAW264.7 macrophages (2 × 105 cells/well) were seeded in 24 well tissue culture plates and incubated overnight at 37°C with 5% CO2. Cells were infected with Burkholderia spp. at an MOI of 10, and after 1 h infected cells were treated with or without aminoguandine (200 μg/ml, Sigma-Aldrich; Brett et al., 2008). Two hours post-infection, cells were washed three times with PBS to remove extracellular bacteria and then further incubated with pre-warmed DMEM containing 10% fetal bovine serum and 250 μg/ml of Km. At 3 and 24 h post-infection, supernatants were harvested and macrophage monolayers were washed two times with PBS and lysed with 0.1% (vol/vol) Triton X-100. Serial dilutions of the lysates were plated onto SBA plates. After incubation for 48 h at 37°C, colonies were counted and CFU/ml was computed.
Quantification of Nitrite and Murine IFN-β
Supernatants harvested from the above assay were evaluated for nitrite and IFN-β production using a Nitric Oxide Assay Kit (Thermo Scientific) and mouse IFN-β ELISA kit (Thermo Scientific), respectively.
Results
Identification of Host Factors that were Differentially Expressed upon Burkholderia spp. Infection
To gain insight into host proteins that are differentially expressed or post-translationally modified following Burkholderia spp. infection, a high throughput RPMA-based screening platform was utilized. RAW264.7 macrophage were adopted due to its ability to phagocytose Burkholderia spp., which in turn exploits the host’s cellular systems by promoting host actin polymerization on the bacterial surface and inducing multinucleated giant cells (MNGCs) formation (Galyov et al., 2010; Pegoraro et al., 2014). Host responses to a collection of nine Burkholderia spp. originating from various geographical locations isolated from human, animals, or environmental sources were investigated. The ancestry for the majority of the strains is known along with their virulence profiles and genome sequences. RAW264.7 macrophage lysates harvested at various time points post Burkholderia spp. infection were individually arrayed onto nitrocellulose coated slides and probed with a total of 114 well characterized antibodies (Figure 1A). Statistical analysis revealed 25 candidates whose change in expression was twofold or higher in both replicates in any strain, at any time point and at MOI of 10 (Figure 1B; Supplementary Figure S1). At MOI of 1, very few robust changes were observed in host signaling molecules. Hence, subsequent studies were performed using MOI of 10. RPMA data analysis did not identify any proteins whose expression was considerably down regulated compared to uninfected and untreated control. The selected candidates were further characterized by performing traditional immunoblots using cell lysates collected from RAW264.7 macrophage treated with LPS, or infected with Bm ATCC23344, and Bp E8. Although not included in the initial RPMA, Bm 2002721278, an avirulent strain (personal communication), was included for purposes of comparative analyses of host signaling dynamics between virulent and avirulent Bm.
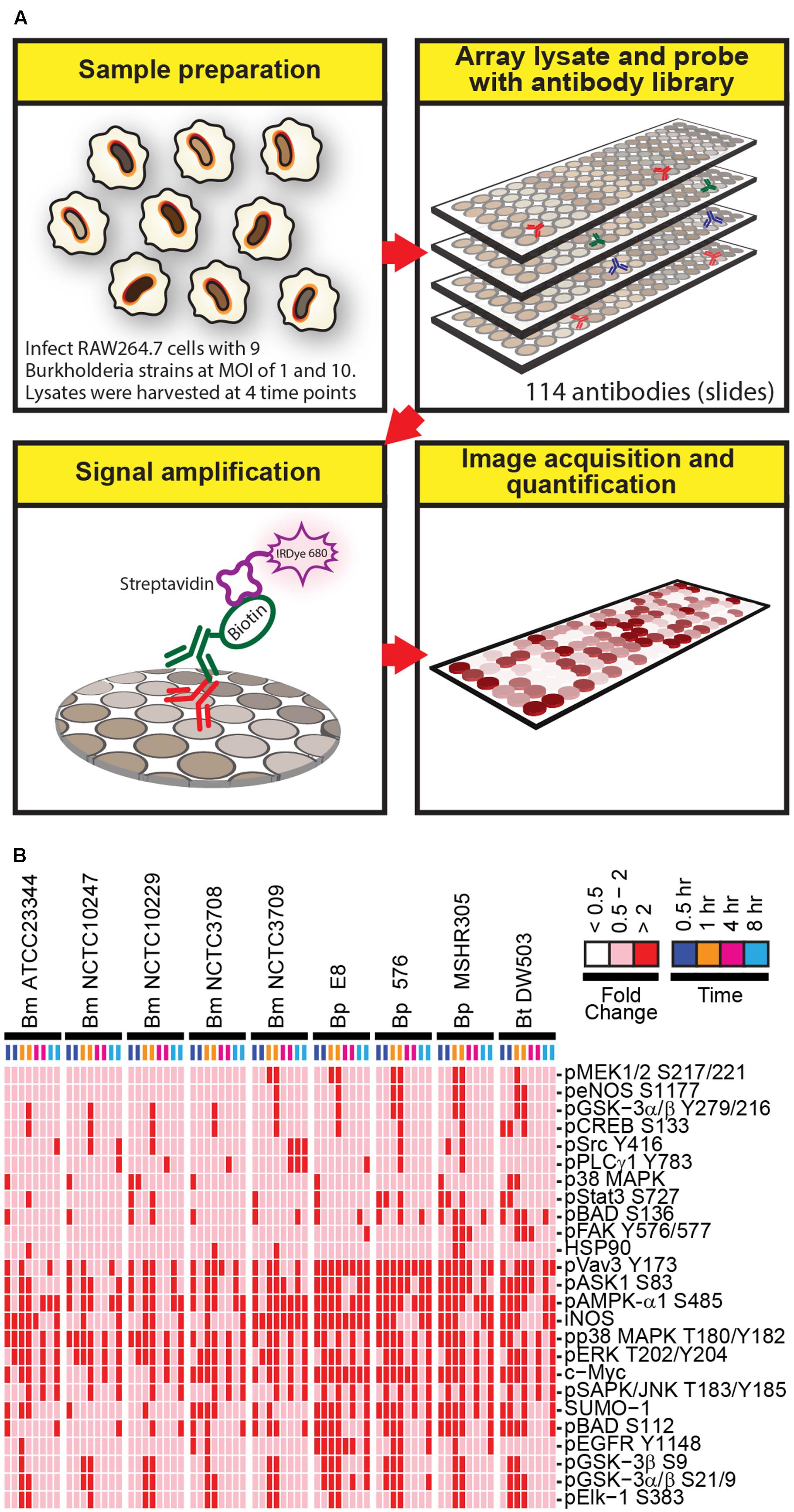
FIGURE 1. A schematic diagram of the experimental design and overview of the results. (A) Lysates harvested from Burkholderia spp. infected RAW264.7 macrophages were arrayed on nitrocellulose coated slides. Each slide was incubated with one primary antibody with a total of 114 antibodies in the library. The primary antibody was detected by biotin-labeled secondary antibody, which, in turn, was recognized by IRDye680 fluorophore conjugated streptavidin. Signals were acquired, quantified, and analyzed according to materials and methods section. (B) RAW264.7 macrophages were infected at multiplicity of infection (MOI) of 10 with indicated Burkholderia spp. for 0.5, 1, 4, and 8 h. Lysates were harvested and subjected to reverse-phase protein microarray (RPMA) studies. A heat map of fold changes over untreated samples is depicted. Experiments were performed on two independent days and data from these repeat studies are shown.
Burkholderia spp. Infection Induces iNOS Expression and Activation of STAT1
RPMA revealed iNOS expression was elevated 1 h post Burkholderia spp. infection, then reduced at 4 h and restored by 8 h (Figure 2A). Immunoblotting of independently prepared lysates samples revealed robust up-regulation of iNOS 8 h post Bm ATCC2334 and Bm 2002721278 infection. Furthermore, Bp E8 was a weaker inducer of iNOS expression when compared to both Bm strains (Figures 2B,C). Aminoguanidine (AG), an iNOS inhibitor, was utilized to correlate the importance of iNOS activity with clearance of intracellular Burkholderia spp. in RAW264.7 macrophages. The colony formation assay conducted 3 h post Burkholderia spp. infection suggested AG did not affect the uptake of bacteria. Treatment with AG augmented or restored replication of Bm strains in RAW264.7 macrophages 24 h post infection. Conversely, AG did not impact bacterial replication in Bp strains (Figure 2D). The efficacy of AG mediated inhibition of iNOS was further confirmed by quantifying nitrite production. AG inhibited Bm mediated nitrite production by approximately 80% whereas only basal levels of nitrite production was observed in Bp (Figure 2E). This is consistent with reported observations that iNOS expression was strongly induced when infected by Bm but not Bp (Utaisincharoen et al., 2001; Brett et al., 2008).
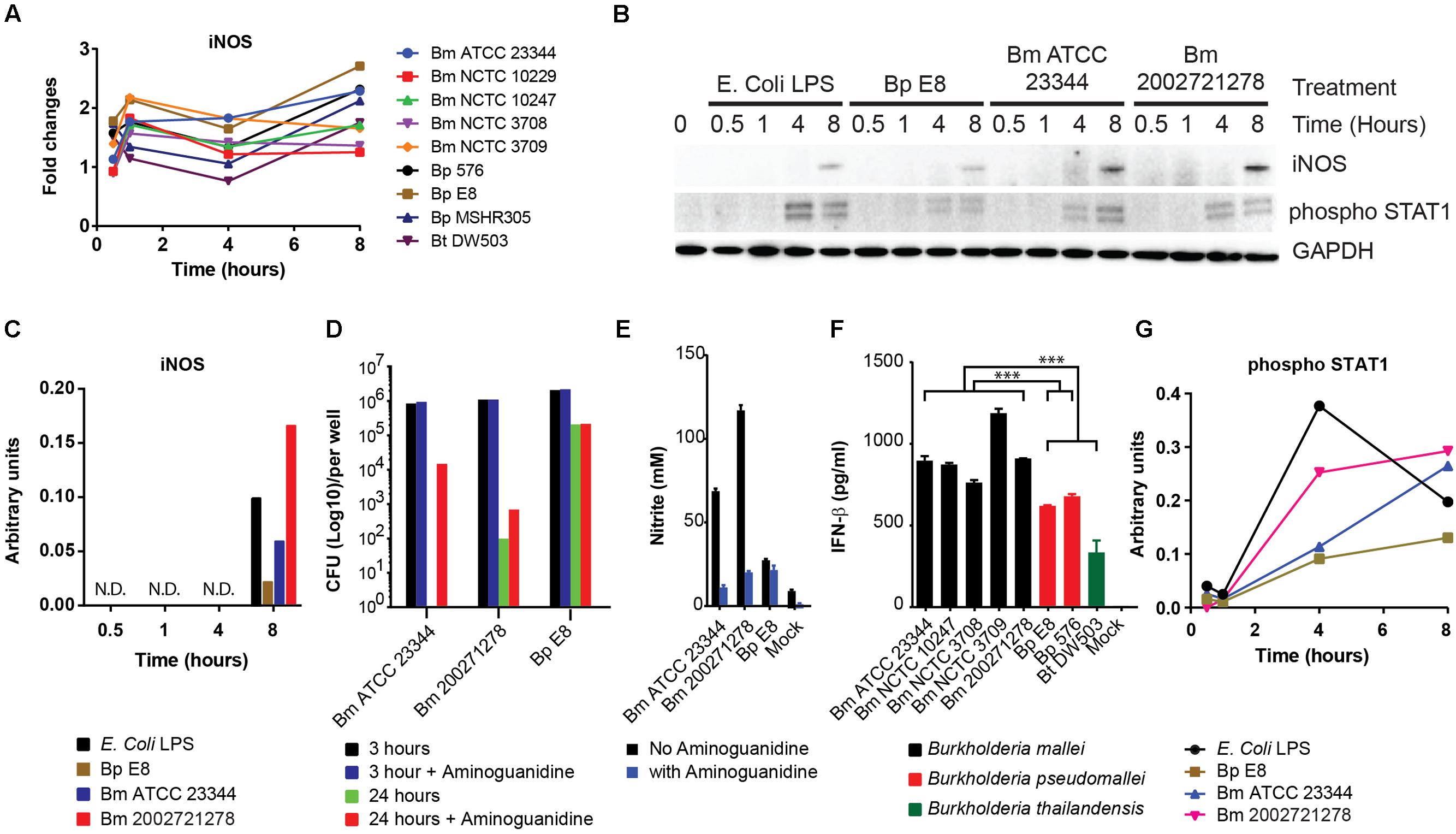
FIGURE 2. Induction of iNOS and activation of STAT1 in response to Burkholderia spp. infection. (A) The expression pattern of iNOS obtained from RPMA study. (B) RAW264.7 macrophages were infected with Burkholderia pseudomallei (Bp) E8, Burkholderia mallei (Bm) 23344, and Bm 2002721278 at MOI of 10, or treated with LPS at 100 ng/ml. Samples were collected at 0.5, 1, 4, and 8 h post treatments. The amount of iNOS and phosphorylated STAT1 was evaluated by immunoblotting, followed by (C) densitometric quantification of iNOS. N.D. indicates signal was not detectable. (D) Colony forming unit (CFU) was determined 3 and 24 h post Burkholderia spp. infection of RAW264.7 cells, with and without aminoguanidine (AG) treatment. Culture supernatants harvested at 24 h post infection from mock-infected and Burkholderia spp. infected RAW264.7 macrophages were assayed for the production of (E) nitrite and (F) IFN-β. P-values between species were calculated by Mann–Whitney U test. ∗∗∗p-value < 0.001. (G) densitometric quantification of phosphorylated STAT1. Immunoblots in Figures 2–4 were performed concurrently from the same samples and loading control, GAPDH. The immunoblot data is representative of three independent trials. Bacterial CFUs represent the average of two biological replicates. Nitrite and IFN-β levels were computed by averaging three biological replicates.
Infection of macrophages by Gram-negative bacteria induces the production of IFN-β (Utaisincharoen et al., 2003), which acts in autocrine or paracrine fashion and triggers subsequent STAT1 phosphorylation (Gao et al., 1998). Attenuation of iNOS gene expression in LPS stimulated, IFN-β deficient macrophages underscores the importance of IFN-β in modulating host innate immune responses (Thomas et al., 2006). To examine whether a relationship exists between IFN-β production and the induction of iNOS in the strains included in our study, culture supernatants were assayed for IFN-β by ELISA. When grouped, Bm infected RAW264.7 macrophages showed significant elevation of IFN-β production when compared to the Bp group (Bp E8 and Bp 576; p-value < 0.001) and Bp and Bt (Bp E8, Bp 576, Bt DW503) as a group (p-value < 0.00013). Furthermore, Bt was the least potent inducer among all the strains inspected in this study (Figure 2F). IFN-β mediated signal activation was further confirmed by immunoblotting for levels of STAT1 phosphorylation. Quantification of the phospho-STAT1 immunoblot shows increased activation of STAT1 in Bm strains versus Bp, which demonstrates distinct signaling dynamics between species (Figures 2B,G).
Burkholderia spp. Induced NF-κB Mediated Responses
NF-κB pathway constitutes an important part of the host defense against pathogens (Akira et al., 2006). Several key regulators of NF-κB signaling pathway were also identified by RPMA. These include the phosphorylated forms of Src and GSK3β (Sasaki et al., 2005; Steinbrecher et al., 2005; Han et al., 2010). RPMA detected changes in Src and GSK3β phosphorylation (Figure 3A). The activities of these proteins were later characterized by immunoblotting. Phosphorylated Src, an integrin associated non-receptor tyrosine kinase, accumulated over time throughout the course of the study (Figures 3B,C). Furthermore, the level of phosphorylated GSK3β at serine 9 and 21, conferring the inactive form, increased over time in RAW264.7 macrophages treated with LPS, infected by Bm ATCC23344 and Bp E8. No apparent accumulation of phosphorylated GSK3β was observed in Bm 2002721278 infected RAW264.7 macrophages (Figures 3B,C).
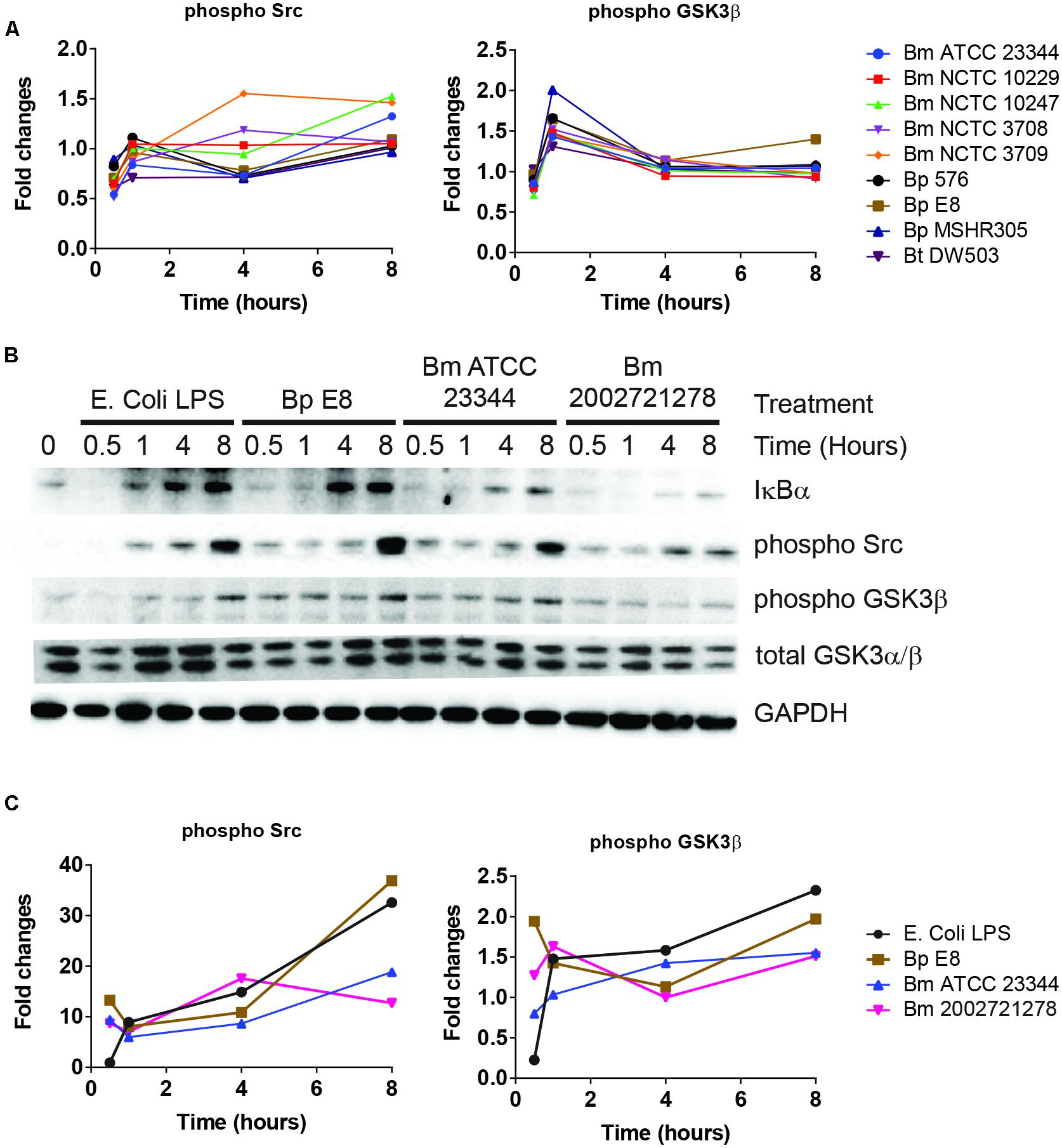
FIGURE 3. Burkholderia spp. induces NF-κB mediated responses. (A) The phosphorylation state of indicated proteins acquired by RPMA. (B) Samples were prepared as described in Figure 2B. The expression or phosphorylation state of indicated proteins was evaluated by immunoblotting, followed by (C) densitometry quantification. Immunoblots in Figures 2–4 were performed concurrently with the same sample and loading control, GAPDH. The immunoblot data is representative of three independent trials.
Activation of NF-κB first requires phosphorylation of IκBα, which primes IκBα for ubiquitination and proteasome-mediated degradation. NF-κB subsequently translocates to the nucleus and activates a series of genes involved in the inflammatory response (Thanos and Maniatis, 1995). Although no dramatic changes in the expression level of total IκBα or phosphorylated IκBα were observed in the RPMA dataset, immunoblot analysis showed that cell lysates from LPS treated RAW264.7 macrophages were void of IκBα at 30 min post stimulation; however, it was quickly resynthesized by the 1 h time point. The expression level of IκBα remained intact at 4 and 8 h under LPS stimulation. After Bp and Bm infections, IκBα expression was absent at 30 and 60 min but was detected by 4 h, and maintained at 8 h. The avirulent Bm 2002721278 strain showed reduced expression of IκBα compared to the virulent strains (Figure 3B).
Alternation of Intracellular Energy Level upon Burkholderia spp. Infection
AMPK-α1 is a serine/threonine protein kinase that has emerged as a master sensor of cellular energy balance in mammalian cells (Carling et al., 2011). However, post-translational modification of AMPK-α1 has not been implicated in the pathogenesis of Burkholderia spp. infections. The abundance of AMPK-α1 phosphorylation at an inhibitory site was altered in RPMA (Figure 4A). Immunoblot analyses confirmed increased expression of this phosphorylated form of AMPK-α1 after 1 h, which remained elevated throughout LPS stimulation or Burkholderia spp. infections (Figures 4B,C).
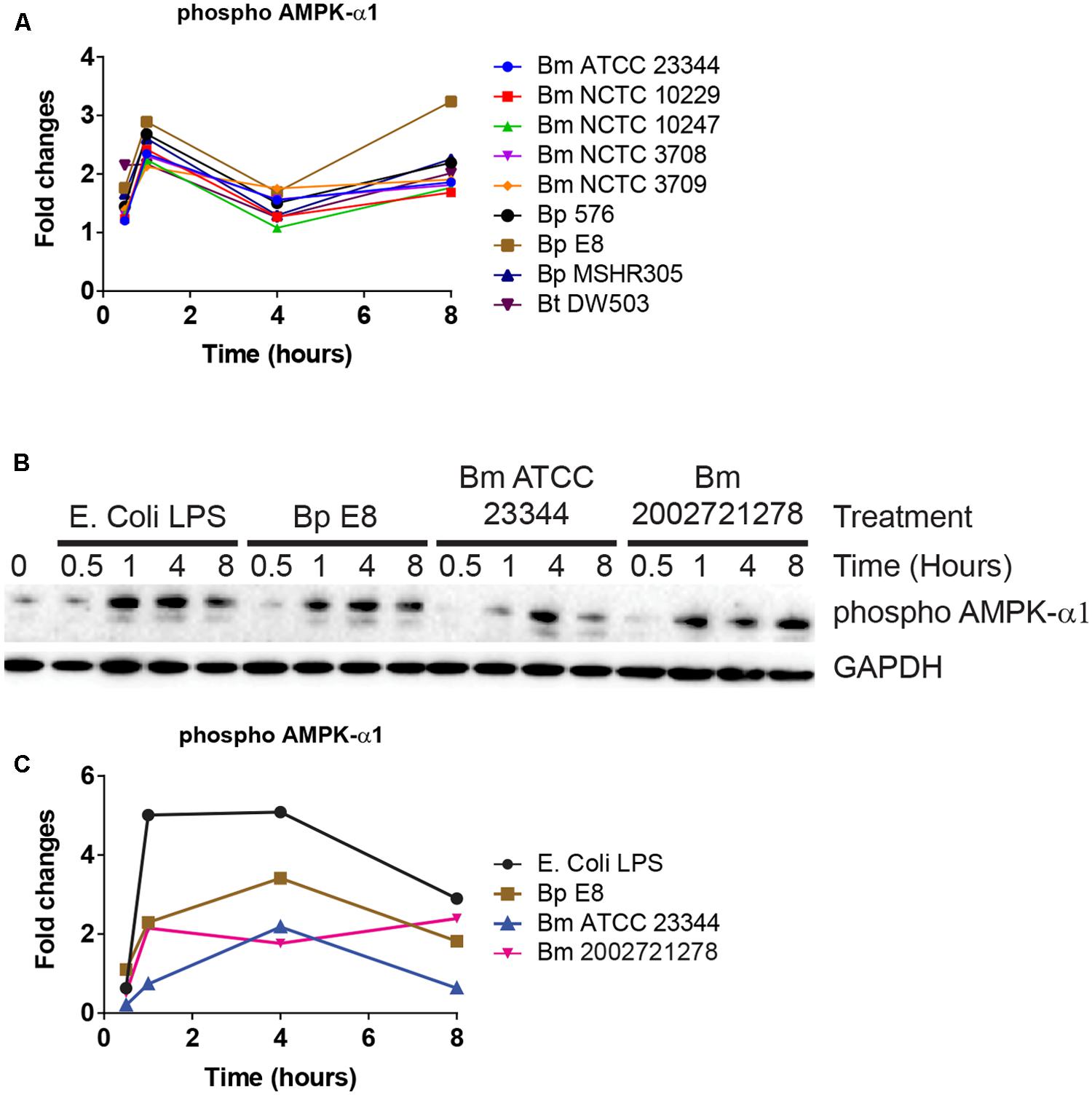
FIGURE 4. AMPK-α1 is phosphorylated upon Burkholderia spp. infection. (A) The phosphorylation state of AMPK-α1 acquired by RPMA. (B) Samples were prepared as described in Figure 2B. The phosphorylation state of AMPK-α1 was evaluated by immunoblotting, followed by (C) densitometry quantification. Immunoblots in Figures 2–4 were performed concurrently with the same sample and loading control, GAPDH. The immunoblot data is representative of three independent trials.
Activation of MAPK Pathway in Response to Burkholderia spp. Infections
Reverse-phase protein microarray identified multiple components of MAPK pathway. These included c-Myc and phosphorylated forms of ASK1, p38, and ERK (Figures 1 and 5A). Immunoblot analyses revealed elevated expression of phosphorylated p38 over the time course of treatments (Figures 5B,C). Both immunoblotting and immuno-fluorescent assay showed an increase in phosphorylated ERK at 30 min and 1 h post treatment (Figure 5D). This expression pattern matched the RPMA result where the phosho-ERK1/2 signal peaked at 1 h. c-Myc, a downstream transcription factor of p38, was robustly up-regulated 4 h post LPS stimulation and Burkholderia spp. infection. c-Myc expression was sustained 8 h post Bm ATCC23344 and Bp E8 infection but was attenuated in macrophages infected with Bm 2002721278 or treated with LPS (Figures 5B,C).
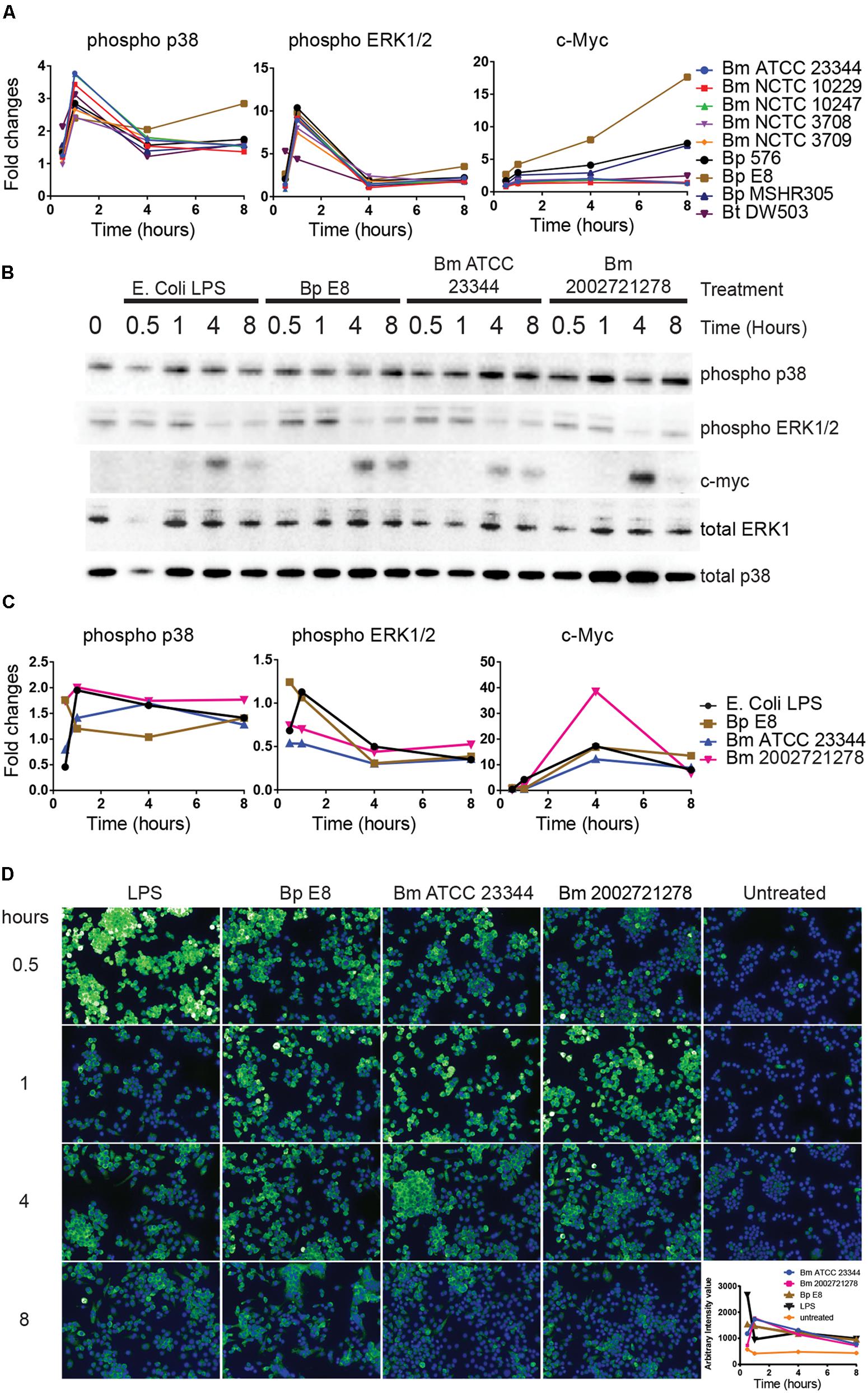
FIGURE 5. Activation of MAPK pathway components upon Burkholderia spp. infection. (A) The expression and phosphorylation states of indicated proteins acquired by RPMA. (B) Samples were prepared as described in Figure 2B. The expression and phosphorylation states of indicated proteins were evaluated by immunoblotting, followed by (C) densitometry quantification. (D) RAW264.7 macrophages were stimulated with lipopolysaccharide (LPS) or infected with Burkholderia spp. at indicated time points. Phosphorylation of ERK1/2 was visualized by indirect immuno-fluorescent staining. Expression of phosphorylated ERK1/2 was subsequently quantified using the Opera confocal system. Phosphorylated ERK1/2 is pseudocolored green and nuclear stain is colored blue. The immunoblot data is representative of three independent trials.
Discussion
Identification of Gross Changes in Host Protein Expression and Phosphorylation During Burkholderia spp. Infection
In this study, we undertook quantitative analysis of phosphoprotein signaling induced by Burkholderia spp. in RAW264.7 macrophages using an RPMA platform. This approach allowed us to simultaneously analyze changes in phosphorylation states or total protein levels for 114 protein species, across nine Burkholderia isolates, at two MOI and four different time points. We did not observe any strain specific up/down regulation or activation of proteins queried at either MOI (Supplementary Figure S1). RPMA screening identified 25 candidate proteins whose expression or phosphorylation states were altered during Burkholderia spp. infections. The observed differences in the host signaling are not a result of defects in bacterial entry or replication within macrophages, as all the Bp, Bm, and Bt strains were able to efficiently replicate within macrophages (data not shown; Pilatz et al., 2006; Brett et al., 2008; Wand et al., 2011). Eight proteins were selected for further characterization by immunoblotting. Although both RPMA and immunoblotting approaches revealed changes in protein expression levels after Burkholderia spp. infection, the kinetic expression profiles of these proteins were slightly different. For example, phosphorylated AMPK-α1 signal obtained from RPMA peaked at 1 h post infection, attenuated by 4 h and restored by 8 h. On the contrary, immunoblot analyses showed a time dependent increase of phosphorylated AMPK-α1 with a peak at 4 h. The observed difference may be attributed to the existence of variations in the microenvironment of the immobilized cell extracts within and across arrays, such as possible spatial inconsistencies on the nitrocellulose surface, non-uniform reagent treatment, or printing inconsistencies (Anderson et al., 2009). Furthermore, fundamental differences exist between the RPMA and immunoblot sample preparation and experimental methods. For example, the fluorophore used in the RPMA platform is highly sensitive with a broad dynamic range compared to the chemiluminescence detection system, resulting in different relative signal outputs. These variations may contribute to the disparity we observe in peaks of expression between the two assays. Despite the variation between the observed protein expression kinetics, our goal to identify critical host factors is not undermined.
The signaling pathways altered in Burkholderia spp. infected RAW264.7 macrophages include AMPK-α1, regulators of NF-κB signaling pathway (IκBα, GSK3β, Src, STAT1) and MAPK (p38, ERK1/2, c-Myc). Notably, RPMA platform study by Molero et al. (2009) revealed both MAPK and GSK-3β activities are exploited by Salmonella enterica Serovar Typhimurium (S. Typhimurium) encoded type III secretion systems (T3SS) during the internalization process. Internalization of S. Typhimurium is dependent on the function of Cdc42- and Rac1-activating factors SopE/E2, which promotes actin rearrangement. A third effector protein, SigD, which also contributes to bacterial internalization, triggered Akt activation which subsequently led to GSK-3β inhibition by phosphorylation on serine 9. Cells pretreated with a p38 inhibitor showed significantly reduced Bp entry; however, the exact component of the T3SS that mediates this process has yet to be identified (Hii et al., 2008). The ability of Burkholderia spp. to alter phosphorylation states of several MAPK family members and GSK-3β (Serine 9) suggests both MAPK and GSK-3β are within a conserved axis exploited during the Burkholderia pathogenesis.
Manipulation of host kinase signaling by bacterial effectors is a major mechanism in the pathogenesis of infection (Krachler et al., 2011). It was reported that Bp T3SS contribute to activation of host proteins such as NF-κB and JNK in a TLR-independent manner although the specific bacterial effectors have not been identified (Hii et al., 2008). Presently, there is limited information linking the specific bacterial effectors to host pathways. While it is beyond the scope of this study, elucidating the bacterial-host networks will be critical for identification of host proteins that could be targeted for development of host-based therapeutics.
Induction of iNOS in Response to Burkholderia spp. Infection
Induction of iNOS upon bacterial infection of macrophages is a host response reported to enhance antimicrobial activity through activation of the TLR4 pathway as well as IFN signaling (Jacobs and Ignarro, 2001; Utaisincharoen et al., 2004; Brett et al., 2008). iNOS catalyzes production of nitric oxide (NO), which is oxidized to reactive nitrogen oxide species that play an important role in the clearance of intracellular bacteria (Coleman, 2001). RAW264.7 cells infected with Bm ATCC23344 and Bm 2002721278 showed reduced levels of total IκBα in comparison with Bp E8, suggesting elevated NF-κB activity upon Bm infection. Moreover, production of IFN-β was elevated in Bm infected RAW264.7 cells compared to Bp strains. This suggests upregulation of iNOS protein expression observed in Bm strains may thus be a result of the activity orchestrated by NF-κB and IFN-β. In fact, multiple transcription factors such as NF-κB, STAT1, and interferon regulatory factor (IRF) are demonstrated to regulate the iNOS promoter (Taylor and Geller, 2000). Notably, our results demonstrate that differential expression of iNOS in Bm vs. Bp strains plays a role in bacterial clearance as evidenced by bacterial survival in the presence of AG.
Potential Targets for Anti-Inflammatory Therapeutics in Burkholderia spp. Infection
Balancing the inflammatory network may direct the host responses toward an anti-inflammatory state and may represent a more effective means of treatment for postsymptomatic infections (D’Elia et al., 2013). Phosphorylation of AMPK-α1, Src, and GSK3β upon Burkholderia spp. infection suggested their involvement in modulating host inflammatory response. First, suppression of AMPK-α1 activation via Serine 485 phosphorylation was observed in Burkholderia spp. infected cells. This suggested that a pro-inflammatory mechanism was quickly mounted in response to the infection. Potentiating AMPK-α1 activation may mitigate the inflammatory response through promoting oxidative metabolism, which is connected to an anti-inflammatory state, rather than the glycolytic state associated with inflammation. Secondly, the tyrosine kinase Src mediated integrin signaling pathway not only modulates leukocyte adhesion and migration but also negatively regulates TLR4 activation (Han et al., 2010). In this context, activation of Src implicates initiation of a TLR4 mediated negative feedback mechanism. Attenuating activation of NF-κB through potentiating Src mediated integrin signal cascade may thus serve as a therapeutic route for Burkholderia spp. infection. Thirdly, reduced transcriptional activity of NF-κB was associated with GSK3β inhibition and this resulted in attenuation of TLR mediated inflammatory cytokine production (Martin et al., 2005). Bp infected mice pretreated with GSK3β inhibitor showed reduced levels of pro-inflammatory cytokines (TNF-α, IFN-γ, IL-1β) and elevated levels of anti-inflammatory cytokines (IL-10 and IL1Ra). Strikingly, Bp infected mice showed improved survival when treated with the GSK3β inhibitor (Tay et al., 2012). This selective action of GSK3β on NF-κB-induced gene expression will likely facilitate the therapeutic anti-inflammatory uses of GSK3β inhibitors.
Burkholderia spp. Host Signal Transduction Network
Empirical studies have characterized the function of numerous host proteins in the context of Burkholderia spp. infection; however, they have not been queried systematically against multiple Burkholderia spp. Our high throughput RPMA approach successfully identified several previously published protein targets as well as new, unreported host proteins that were altered in response to Burkholderia spp. infection. To the best of our knowledge, activation of Src, inactivation of AMPK-α1 or expression of c-Myc has not been implicated as altered in Burkholderia spp. infection. We have constructed a representative network that encompasses critical signaling axes based on current framework of signal transduction pathways and our survey of host protein expression and phosphorylation profiles. Canonical pathways downstream of TLR4 include several host proteins from our studies, namely IκBα, active Src, ERK1/2, p38, and inactive AMPK-α1. The aforementioned kinases ultimately activate transcription factors (e.g., c-Myc, STAT1, and NF-κB) which serve to regulate gene expression (e.g., iNOS) during Burkholderia spp. infection (Figure 6). Based on our results, induction of phosphorylated forms of AMPK-α1, GSK3β, and Src in the context of canonical pathways suggest that they play a role in regulating the inflammatory response of Burkholderia spp. infections. Inhibiting AMPK-α1 (by phosphorylating Serine 485) triggers a pro-inflammatory mechanism whereas inhibiting GSK3β (by phosphorylating Serine 9) may result in suboptimal NF-κB mediate inflammatory response. In addition, activating integrin associated Src kinase may negatively regulate MyD88 dependent NF-κB activation (Figure 6). Importantly, perturbing the nodes in these pathways by antisense technology and bioactive small molecules will serve to validate these host factors in the pathogenesis of Burkholderia spp. infections.
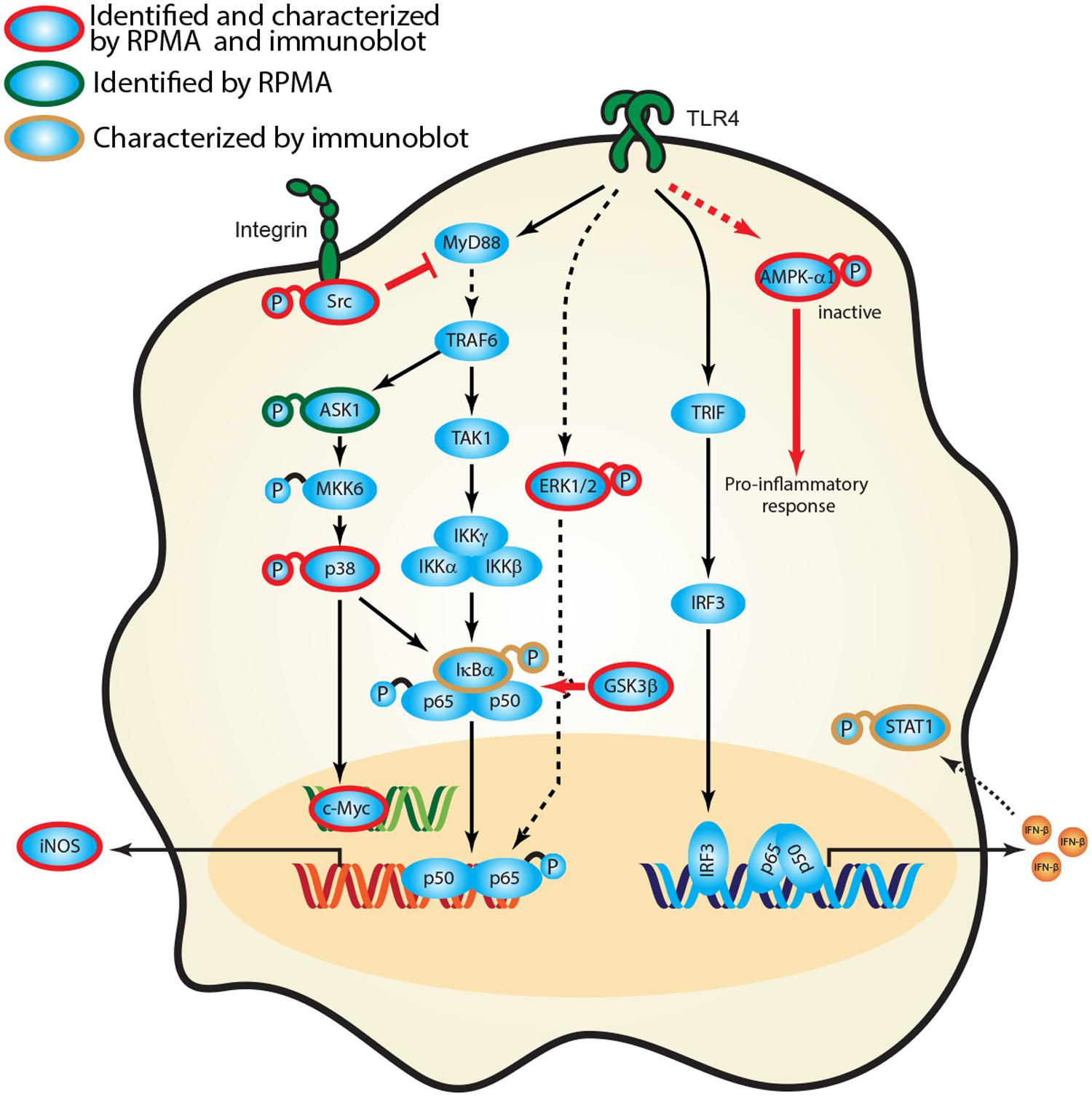
FIGURE 6. Activation of signaling cascades in response to Burkholderia spp. infection. A signaling network was constructed based on current knowledge of signal transduction pathways and our survey of host protein expression and phosphorylation profiles in response to Burkholderia spp. infection. Proteins identified in the RPMA screen (green circle) and/or characterized by immunoblotting are designated by color coding (gold circle, confirmed by immunoblot; red, confirmed by RPMA and immunoblot). Red bold arrow represents potential therapeutic routes.
Conflict of Interest Statement
The Associate Editor Kurt John Langenbach declares that, despite being affiliated to the same institution as author Ramin M. Hakami, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the Department of Defense Chemical Biological Defense Program through the Defense Threat Reduction Agency (DTRA) JSTO-CBS.MEDBIO.02.10.RD.010 (to RP), and by funding awarded by the U.S. Army Medical Research and Materiel Command (W81XWH-11-P-0310) and George Mason University (to RMH). We would like to thank Oak Ridge Institute for Science and Education for participating in the Postgraduate Research Program at the U.S. Army Medical Research and Materiel Command. We also thank Professor Emanuel F. Petricoin for invaluable advice regarding the RPMA data analysis and assistance with performance of the RPMA experiments. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00683
FIGURE S1 | Heat map of fold changes over mock treated samples. RAW264.7 macrophages were infected with indicated Burkholderia spp. for 0.5, 1, 4, 8 h. Lysates were harvested and subjected to RPMA methodology. The expression patterns and phosphorylation states of indicated proteins after infection with an MOI of 10 (A) or MOI of 1 (B) are depicted.
TABLE S1 | Antibodies used in RPMA experiment. Explanations of column headings are: Name of antibody: name of the proteins that antibodies recognize; Vendor: the vendor of the antibodies; Catalog #: the catalog number associated with the vendor; Molecular Weight: the size of the protein in kilo-Dalton; Host: the source of the antibodies; Dilution: dilutions used in the PRMA.
References
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Alam, S., Amemiya, K., Bernhards, R. C., Ulrich, R. G., Waag, D. M., and Saikh, K. U. (2014). Characterization of cellular immune response and innate immune signaling in human and nonhuman primate primary mononuclear cells exposed to Burkholderia mallei. Microb. Pathog. 78, 20–28. doi: 10.1016/j.micpath.2014.11.009
Alem, F., Yao, K., Lane, D., Calvert, V., Petricoin, E. F., Kramer, L., et al. (2015). Host response during Yersinia pestis infection of human bronchial epithelial cells involves negative regulation of autophagy and suggests a modulation of survival-related and cellular growth pathways. Front. Microbiol. 6:50. doi: 10.3389/fmicb.2015.00050
Anderson, T., Wulfkuhle, J., Liotta, L., Winslow, R. L., and Petricoin E. III, (2009). Improved reproducibility of reverse-phase protein microarrays using array microenvironment normalization. Proteomics 9, 5562–5566. doi: 10.1002/pmic.200900505
Bast, A., Schmidt, I. H., Brauner, P., Brix, B., Breitbach, K., and Steinmetz, I. (2011). Defense mechanisms of hepatocytes against Burkholderia pseudomallei. Front. Microbiol. 2:277. doi: 10.3389/fmicb.2011.00277
Brett, P. J., Burtnick, M. N., Su, H., Nair, V., and Gherardini, F. C. (2008). iNOS activity is critical for the clearance of Burkholderia mallei from infected RAW 264.7 murine macrophages. Cell. Microbiol. 10, 487–498. doi: 10.1111/j.1462-5822.2007.01063.x
Burtnick, M. N., Brett, P. J., and Woods, D. E. (2002). Molecular and physical characterization of Burkholderia mallei O antigens. J. Bacteriol. 184, 849–852. doi: 10.1128/JB.184.3.849-852.2002
Carling, D., Mayer, F. V., Sanders, M. J., and Gamblin, S. J. (2011). AMP-activated protein kinase: nature’s energy sensor. Nat. Chem. Biol. 7, 512–518. doi: 10.1038/nchembio.610
Chantratita, N., Tandhavanant, S., Myers, N. D., Seal, S., Arayawichanont, A., Kliangsa-Ad, A., et al. (2013). Survey of innate immune responses to Burkholderia pseudomallei in human blood identifies a central role for lipopolysaccharide. PLoS ONE 8:e81617. doi: 10.1371/journal.pone.0081617
Coleman, J. W. (2001). Nitric oxide in immunity and inflammation. Int. Immunopharmacol. 1, 1397–1406. doi: 10.1016/S1567-5769(01)00086-8
Dance, D. A. (2000). Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human-animal hosts. Acta Trop. 74, 159–168. doi: 10.1016/S0001-706X(99)00066-2
De Keulenaer, B. L., and Cheng, A. C. (2006). Severe sepsis due to melioidosis. Chest 130, 1282. doi: 10.1378/chest.130.4.1282
D’Elia, R. V., Harrison, K., Oyston, P. C., Lukaszewski, R. A., and Clark, G. C. (2013). Targeting the “cytokine storm” for therapeutic benefit. Clin. Vaccine Immunol. 20, 319–327. doi: 10.1128/CVI.00636-12
Dharakul, T., and Songsivilai, S. (1999). The many facets of melioidosis. Trends Microbiol. 7, 138–140. doi: 10.1016/S0966-842X(99)01477-8
Estes, D. M., Dow, S. W., Schweizer, H. P., and Torres, A. G. (2010). Present and future therapeutic strategies for melioidosis and glanders. Expert Rev. Anti Infect. Ther. 8, 325–338. doi: 10.1586/eri.10.4
Federici, G., Gao, X., Slawek, J., Arodz, T., Shitaye, A., Wulfkuhle, J. D., et al. (2013). Systems analysis of the NCI-60 cancer cell lines by alignment of protein pathway activation modules with “-OMIC” data fields and therapeutic response signatures. Mol. Cancer Res. 11, 676–685. doi: 10.1158/1541-7786.MCR-12-0690
Galyov, E. E., Brett, P. J., and Deshazer, D. (2010). Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 64, 495–517. doi: 10.1146/annurev.micro.112408.134030
Gao, J. J., Filla, M. B., Fultz, M. J., Vogel, S. N., Russell, S. W., and Murphy, W. J. (1998). Autocrine/paracrine IFN-alphabeta mediates the lipopolysaccharide-induced activation of transcription factor Stat1alpha in mouse macrophages: pivotal role of Stat1alpha in induction of the inducible nitric oxide synthase gene. J. Immunol. 161, 4803–4810.
Gilad, J. (2007). Burkholderia mallei and Burkholderia pseudomallei: the causative micro-organisms of glanders and melioidosis. Recent Pat. Antiinfect. Drug Discov. 2, 233–241. doi: 10.2174/157489107782497335
Han, C., Jin, J., Xu, S., Liu, H., Li, N., and Cao, X. (2010). Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 11, 734–742. doi: 10.1038/ni.1908
Hii, C. S., Sun, G. W., Goh, J. W., Lu, J., Stevens, M. P., and Gan, Y. H. (2008). Interleukin-8 induction by Burkholderia pseudomallei can occur without Toll-like receptor signaling but requires a functional type III secretion system. J. Infect. Dis. 197, 1537–1547. doi: 10.1086/587905
Holden, M. T., Titball, R. W., Peacock, S. J., Cerdeno-Tarraga, A. M., Atkins, T., Crossman, L. C., et al. (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 101, 14240–14245. doi: 10.1073/pnas.0403302101
Improta, G., Zupa, A., Fillmore, H., Deng, J., Aieta, M., Musto, P., et al. (2011). Protein pathway activation mapping of brain metastasis from lung and breast cancers reveals organ type specific drug target activation. J. Proteome Res. 10, 3089–3097. doi: 10.1021/pr200065t
Jacobs, A. T., and Ignarro, L. J. (2001). Lipopolysaccharide-induced expression of interferon-beta mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J. Biol. Chem. 276, 47950–47957. doi: 10.1074/jbc.M106639200
Krachler, A. M., Woolery, A. R., and Orth, K. (2011). Manipulation of kinase signaling by bacterial pathogens. J. Cell Biol. 195, 1083–1092. doi: 10.1083/jcb.201107132
Leelarasamee, A. (2004). Recent development in melioidosis. Curr. Opin. Infect. Dis. 17, 131–136. doi: 10.1097/00001432-200404000-00011
Martin, M., Rehani, K., Jope, R. S., and Michalek, S. M. (2005). Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 6, 777–784. doi: 10.1038/ni1221
Matsuzawa, A., Saegusa, K., Noguchi, T., Sadamitsu, C., Nishitoh, H., Nagai, S., et al. (2005). ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat. Immunol. 6, 587–592. doi: 10.1038/ni1200
Molero, C., Rodriguez-Escudero, I., Aleman, A., Rotger, R., Molina, M., and Cid, V. J. (2009). Addressing the effects of Salmonella internalization in host cell signaling on a reverse-phase protein array. Proteomics 9, 3652–3665. doi: 10.1002/pmic.200800907
Pegoraro, G., Eaton, B. P., Ulrich, R. L., Lane, D. J., Ojeda, J. F., Bavari, S., et al. (2014). A high-content imaging assay for the quantification of the Burkholderia pseudomallei induced multinucleated giant cell (MNGC) phenotype in murine macrophages. BMC Microbiol. 14:98. doi: 10.1186/1471-2180-14-98
Pilatz, S., Breitbach, K., Hein, N., Fehlhaber, B., Schulze, J., Brenneke, B., et al. (2006). Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74, 3576–3586. doi: 10.1128/IAI.01262-05
Popova, T. G., Turell, M. J., Espina, V., Kehn-Hall, K., Kidd, J., Narayanan, A., et al. (2010). Reverse-phase phosphoproteome analysis of signaling pathways induced by Rift valley fever virus in human small airway epithelial cells. PLoS ONE 5:e13805. doi: 10.1371/journal.pone.0013805
Propst, K. L., Troyer, R. M., Kellihan, L. M., Schweizer, H. P., and Dow, S. W. (2010). Immunotherapy markedly increases the effectiveness of antimicrobial therapy for treatment of Burkholderia pseudomallei infection. Antimicrob. Agents Chemother. 54, 1785–1792. doi: 10.1128/AAC.01513-09
Sasaki, C. Y., Barberi, T. J., Ghosh, P., and Longo, D. L. (2005). Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J. Biol. Chem. 280, 34538–34547. doi: 10.1074/jbc.M504943200
Sheehan, K. M., Calvert, V. S., Kay, E. W., Lu, Y., Fishman, D., Espina, V., et al. (2005). Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol. Cell. Proteomics 4, 346–355. doi: 10.1074/mcp.T500003-MCP200
Steinbrecher, K. A., Wilson, W. III, Cogswell, P. C., and Baldwin, A. S. (2005). Glycogen synthase kinase 3beta functions to specify gene-specific, NF-kappaB-dependent transcription. Mol. Cell. Biol. 25, 8444–8455. doi: 10.1128/MCB.25.19.8444-8455.2005
Tay, T. F., Maheran, M., Too, S. L., Hasidah, M. S., Ismail, G., and Embi, N. (2012). Glycogen synthase kinase-3beta inhibition improved survivability of mice infected with Burkholderia pseudomallei. Trop. Biomed. 29, 551–567.
Taylor, B. S., and Geller, D. A. (2000). Molecular regulation of the human inducible nitric oxide synthase (iNOS) gene. Shock 13, 413–424. doi: 10.1097/00024382-200006000-00001
Thanos, D., and Maniatis, T. (1995). NF-kappa B: a lesson in family values. Cell 80, 529–532. doi: 10.1016/0092-8674(95)90506-5
Thomas, K. E., Galligan, C. L., Newman, R. D., Fish, E. N., and Vogel, S. N. (2006). Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J. Biol. Chem. 281, 31119–31130. doi: 10.1074/jbc.M604958200
Ulett, G. C., Ketheesan, N., and Hirst, R. G. (2000). Cytokine gene expression in innately susceptible BALB/c mice and relatively resistant C57BL/6 mice during infection with virulent Burkholderia pseudomallei. Infect. Immun. 68, 2034–2042. doi: 10.1128/IAI.68.4.2034-2042.2000
Utaisincharoen, P., Anuntagool, N., Arjcharoen, S., Limposuwan, K., Chaisuriya, P., and Sirisinha, S. (2004). Induction of iNOS expression and antimicrobial activity by interferon (IFN)-beta is distinct from IFN-gamma in Burkholderia pseudomallei-infected mouse macrophages. Clin. Exp. Immunol. 136, 277–283. doi: 10.1111/j.1365-2249.2004.02445.x
Utaisincharoen, P., Anuntagool, N., Limposuwan, K., Chaisuriya, P., and Sirisinha, S. (2003). Involvement of beta interferon in enhancing inducible nitric oxide synthase production and antimicrobial activity of Burkholderia pseudomallei-infected macrophages. Infect. Immun. 71, 3053–3057. doi: 10.1128/IAI.71.6.3053-3057.2003
Utaisincharoen, P., Tangthawornchaikul, N., Kespichayawattana, W., Chaisuriya, P., and Sirisinha, S. (2001). Burkholderia pseudomallei interferes with inducible nitric oxide synthase (iNOS) production: a possible mechanism of evading macrophage killing. Microbiol. Immunol. 45, 307–313. doi: 10.1111/j.1348-0421.2001.tb02623.x
Van Zandt, K. E., Greer, M. T., and Gelhaus, H. C. (2013). Glanders: an overview of infection in humans. Orphanet J. Rare Dis. 8, 131. doi: 10.1186/1750-1172-8-131
Van Zandt, K. E., Tuanyok, A., Keim, P. S., Warren, R. L., and Gelhaus, H. C. (2012). An objective approach for Burkholderia pseudomallei strain selection as challenge material for medical countermeasures efficacy testing. Front. Cell. Infect. Microbiol. 2:120. doi: 10.3389/fcimb.2012.00120
Wand, M. E., Muller, C. M., Titball, R. W., and Michell, S. L. (2011). Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 11:11. doi: 10.1186/1471-2180-11-11
Whitlock, G. C., Estes, D. M., and Torres, A. G. (2007). Glanders: off to the races with Burkholderia mallei. FEMS Microbiol. Lett. 277, 115–122. doi: 10.1111/j.1574-6968.2007.00949.x
Keywords: Burkholderia mallei, Burkholderia pseudomallei, lipopolysaccharide, reverse-phase protein microarrays
Citation: Chiang C-Y, Uzoma I, Lane DJ, Memišević V, Alem F, Yao K, Kota KP, Bavari S, Wallqvist A, Hakami RM and Panchal RG (2015) A reverse-phase protein microarray-based screen identifies host signaling dynamics upon Burkholderia spp. infection. Front. Microbiol. 6:683. doi: 10.3389/fmicb.2015.00683
Received: 06 October 2014; Accepted: 22 June 2015;
Published: 27 July 2015.
Edited by:
Kurt John Langenbach, American Type Culture Collection, USACopyright © 2015 Chiang, Uzoma, Lane, Memišević, Alem, Yao, Kota, Bavari, Wallqvist, Hakami and Panchal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rekha G. Panchal, Molecular and Translational Sciences Division, United States Army Medical Research Institute of Infectious Diseases, 1425 Porter Street, Frederick, MD 21702, USA, rekha.g.panchal.civ@mail.mil
†These authors have contributed equally to this work.
 Chih-Yuan Chiang
Chih-Yuan Chiang Ijeoma Uzoma
Ijeoma Uzoma Douglas J. Lane1
Douglas J. Lane1 Vesna Memišević
Vesna Memišević Farhang Alem
Farhang Alem Kuan Yao
Kuan Yao Anders Wallqvist
Anders Wallqvist Ramin M. Hakami
Ramin M. Hakami