- Department of Food and Bioproduct Sciences, University of Saskatchewan, Saskatoon, SK, Canada
Probiotic bacteria offer a number of potential health benefits when administered in sufficient amounts that in part include reducing the number of harmful organisms in the intestine, producing antimicrobial substances and stimulating the body’s immune response. However, precisely elucidating the probiotic effect of a specific bacterium has been challenging due to the complexity of the gut’s microbial ecosystem and a lack of definitive means for its characterization. This review provides an overview of widely used and recently described probiotics, their impact on the human’s gut microflora as a preventative treatment of disease, human/animal models being used to help show efficacy, and discusses the potential use of probiotics in gastrointestinal diseases associated with antibiotic administration.
Microbial Ecology of the Human Gastrointestinal Tract
The human intestinal microbiota is a complex ecosystem with considerable impact on human health and well-being, contributing to maturation of the immune system and providing a direct barrier against pathogen colonization (Doré and Corthier, 2010). It consists of bacteria, archaea, some protozoa, anaerobic fungi, and different bacteriophages and viruses, and it has been estimated that more than 1000 species of microbes inhabit the human intestine (Tuohy et al., 2012). The presence of a great number of microbes (up to 5 × 1011 bacterial cells per gram of intestinal contents) suggests strong regulatory effects on the human host, and recent findings suggest that gut microbiota can have a considerable impact on both our weight and mood (Duca et al., 2014; Naseribafrouei et al., 2014). The composition and function of human microbial populations associated with various body sites have been studied with the help of metagenomic tools as part of two recent initiatives – the NIH Human Microbiome Project (HMP) and the European Metagenomics of the Human Intestine (metaHIT) project (NIH HMP Working Group et al., 2009; Dusko Ehrlich and MetaHIT Consortium, 2011). These massive molecular approaches have already revealed the presence of three different clusters, or enterotypes, which correspond to one of three most abundant genera of human intestine – Bacteroides, Prevotella, and Ruminococcus (Arumugam et al., 2013).
Bacteria that initially colonize the large gut of an infant are facultative anaerobes, such as Escherichia coli and Streptococcus sp. These species metabolize oxygen in the gut, thereby creating anaerobic conditions. Subsequent colonization largely depends on food profile and environmental factors (i.e., sanitary conditions). After the full formation of the gastrointestinal microflora, its composition has been shown to include such genera as Bacteroides, Bifidobacterium, Eubacterium, Clostridium, Lactobacillus, Fusobacterium, and various Gram-positive cocci (Fooks et al., 1999; Wallace et al., 2011).
Within the gastrointestinal tract (GIT), the microbiota provides various functions, such as digestion of essential nutrients and maturation of intestinal epithelial cells. Studies on mice have shown a number of significant effects of microbiota on the host: in ex-germ-free reconventionalized mice, their intestinal epithelium was thicker, kinetics of enterocytes – faster, short-chain fatty acids were produced at significantly higher concentrations, and there was a normal level of immunological activity present, compared to germ-free animals (Aureli et al., 2011). Microbes also have the ability to affect physiologic parameters, providing systemic effects on blood lipids and generally influencing the immune system, as well as inhibiting harmful bacteria (Mikelsaar, 2011). Pathogen inhibition by human intestinal microbiota may provide significant human health benefits through protection against infection as a natural barrier against pathogen exposure in the GIT (Wallace et al., 2011). Factors such as food contamination by pathogens, as well as the high load of antibiotics in soil and animal feed, can influence the microbial ecology of human GIT (Sapkota et al., 2007). Using molecular genetic tools, it has been shown that antibiotics could induce significant alterations in the dominant colonic microbiota that are not detectable using bacteriological (culture-based) techniques, with effects lasting for up to 2 months (Mangin et al., 1999). Several more specific disorders involve disruption of the human microflora ecology: acute gastroenteritis, Clostridium difficile infection (CDI), necrotising enterocolitis in neonates, irritable bowel syndrome and Helicobacter pylori infection (Kotzampassi and Giamarellos-Bourboulis, 2012). Probiotics are currently being examined for their potential treatments of these aforementioned disorders.
Probiotic Bacteria
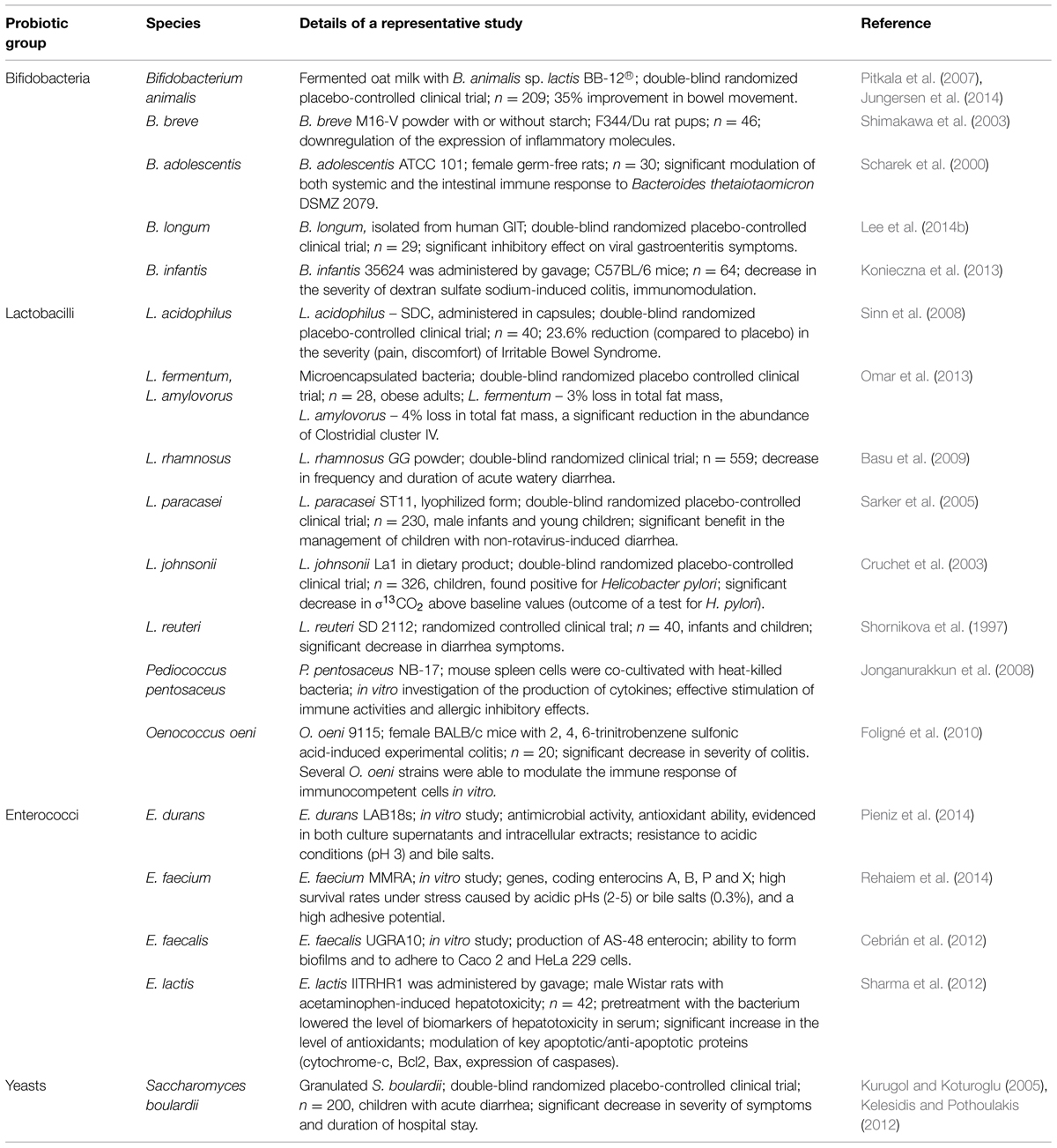
According to the popularized definition by the Food and Agriculture Organization/World Health Organization, and as grammatically modified by Hill et al. (2014), probiotics are defined as “Live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (FAO/WHO, 2001). The most common probiotics include representatives of lactobacilli, enterococci, bifidobacteria, and yeasts (Table 1). In addition, bacterial mixtures may be used to achieve the complex beneficial effect of probiotics (Caballero-Franco et al., 2007).
Presumed health benefits of probiotics include reducing harmful organisms in the intestine, producing antimicrobial factors, and stimulating the body’s immune response (Collado et al., 2007; Foligné et al., 2010; Konieczna et al., 2013). Some of the beneficial effects of probiotics (e.g., lowering of cholesterol level) are yet to be substantiated by well-controlled clinical trials. However, there are a growing number of studies providing data on effects of probiotic bacteria on the human immune system and on microflora of the GIT (Holzapfel and Schillinger, 2002; Foligne et al., 2007; Verdú et al., 2009; Wen et al., 2012). Increasingly, reports of the human/animal microbiome playing a central role in other key aspects of health functionality are emerging, including beneficial impacts on the treatment of metabolic disorders, such as obesity and type 2 diabetes, improvement of bowel function in patients with colorectal cancer, potential cognitive, and mood-enhancing benefits, antidepressant, and anxiolytic (antianxiety) activity (Desbonnet et al., 2008; Bravo et al., 2011; DiBaise et al., 2012; Lee et al., 2014a; Owen et al., 2014). The latter anxiolytic effect has even led to the emergence of the new term, psychobiotic, coined by Dinan et al. (2013) as a “live organism that, when ingested in adequate amounts, produces a health benefit in patients suffering from psychiatric illness.”
Products containing probiotic bacteria generally include supplements and foods. Live probiotics are commonly available in fermented dairy products and probiotic-fortified foods. These bacteria are added into numerous foods and beverages, ranging from yogurts to breakfast cereals. There are also tablets, capsules, powders, and sachets containing probiotics in freeze-dried form. Functional foods, defined as food preparations with various health-related properties, often include bacterial strains with declared probiotic properties (Turroni et al., 2011). The scientific interest in probiotics is growing exponentially: the search for published papers featuring the keyword “probiotic” in NIH PubMed database revealed 7265 articles for the period from 2000 to 2010, with 953 of them being clinical trials. Within the following 5 years (up to May 20th 2015), the frequency of publications doubled with 7979 papers being published, including 778 clinical trials.
Lactic Acid Bacteria
Lactic acid bacteria (LAB) are Gram-positive, non-spore forming cocci, coccobacilli, or rods, which generally have non-respiratory (fermentative) metabolism and lack true catalase. Unlike bifidobacteria, which are active in lower parts of the colon, lactobacilli are prevalent in the upper GIT (Turroni et al., 2011). This group is also a normal member of the human microflora, found in the oral cavity, the small intestine, and the vaginal epithelium, where it is thought to play beneficial roles (Gomes and Malcata, 1999). Among the beneficial effects, lactobacilli can improve digestion, absorption, and availability of nutrients (Wallace et al., 2011). Furthermore, LAB are capable of hydrolyzing compounds that limit the bioavailability of minerals, like tannin and phytate, due to tannin acylhydrolase and phytase activities (Turpin et al., 2010). In addition, it was shown that some lactobacilli strains could enhance mineral absorption in Caco-2 cells and improve the nutritional status of the host by producing B-group vitamins. More recently, the role of lactobacilli in energy homeostasis, particularly in obese patients, has been the object of an increased interest (Guo et al., 2010; Mikelsaar, 2011). A further potential positive impact of LAB is their ability to inhibit or kill H. pylori, which is now regarded as the major cause of gastritis and peptic ulcers and is a risk factor for gastric malignancy (Hamilton-Miller, 2003). In addition, both Lactobacillus sp. and Bifidobacterium sp. strains can reduce the side effects of H. pylori eradication therapy (Canducci et al., 2002).
Pediococci are also related to the LAB group and are utilized in industrial fermentations of foods and silage (Raccach, 2014). Pediocin-producing Pediococcus sp. strains are of potential interest to food safety (Raccach, 2014), with three of them potentially possessing probiotic properties – Pediococcus pentosaceus, P. parvulus, and P. acidilactici. Osmanagaoglu et al. (2010) comprehensively studied the potential of a human P. pentosaceus isolate for probiotic use, and reported that the strain produced an anti-Listerial bacteriocin, had excellent autoaggregation characteristics and was also able to co-aggregate with Salmonella enterica serotype typhimurium and enterotoxigenic Escherichia coli (Osmanagaoglu et al., 2010). Antagonistic activity against Listeria monocytogenes was also discovered in P. acidilactici (Guerra and Pastrana, 2002). Clinical trials employing another strain of Pediococcus sp. revealed that the administration of P. parvulus decreased the serum cholesterol levels and increased counts of fecal Bifidobacterium sp. (Mårtensson et al., 2005).
Another group of LAB promoted as probiotics are enterococci, which reportedly help in the maintenance of normal intestinal microflora and stimulate the immune system (Bhardwaj et al., 2008). Studies of potential probiotic properties of E. faecium showed its efficacy in reducing the recovery period of acute diarrhea (Benyacoub et al., 2003). Another study by Pieniz et al. (2014) showed that E. durans possessed antimicrobial activity and antioxidant ability and was resistant to simulated gastric juice and bile salts. Though enterococci have probiotic potential, they are considered opportunistic pathogens for humans as they might cause nosocomial infection and are also known to possess resistance to vancomycin (Tambyah et al., 2004). Due to these controversial properties, the use of enterococci as probiotics remains under debate.
Bifidobacteria
Bifidobacteria are major constituents of the GIT microbiota of animals and humans. They are Gram-positive, non-motile anaerobic saccharolytic bacteria (Gomes and Malcata, 1999). In the gut environment, bifidobacteria have a commensal relationship with their hosts, and contribute to host nutrition by utilizing complex carbohydrates, which are important sources of carbon and energy, but are not degraded in the stomach or intestine (Biavati, 1994). These substances include plant-derived dietary fiber and diet-related carbohydrates, such as starch, galactan, sucrose, amylopectin, and pullulan (Ventura et al., 2007, 2012). The capacity of bifidobacteria to metabolize non-digestible host dietary carbohydrates (prebiotics) can be used for selective stimulation of certain strains colonizing the intestinal tract. Bifidobacteria used as probiotics include strains belonging to species of Bifidobacterium lactis, B. bifidum, B. animalis, B. thermophilum, B. breve, B. longum, B. infantis, and B. adolescentis (Table 1). These bacteria have been shown to inhibit the adherence of enterotoxigenic E. coli, enteropathogenic E. coli, and C. difficile to intestinal epithelial cells, an important trait for use of these bacteria as probiotics (Tsai et al., 2008). Additional beneficial effects of bifidobacterial strains include the prevention or alleviation of infectious diarrhea and the improvement of inflammatory bowel disease symptoms (Sanz, 2007). Bifidobacteria have also been shown to modulate the host’s immune response against other indigenous microflora (e.g., B. adolescentis down-regulates humoral immunity to Bacteroides thetaiotaomicron; Scharek et al., 2000). Some bifidobacterial strains suppress H. pylori-induced genes in human epithelial cells (Shirasawa et al., 2010) while other Bifidobacterium sp. cells and culture supernatants exerted inhibitory effects against Streptococcus mutans and Streptococcus sobrinus, important etiological agents in human dental caries (Lee et al., 2011).
Yeasts
Saccharomyces boulardii is one of the best-studied probiotic species, with a long history of successful use in treatment of multiple gastrointestinal disorders. The administration of this probiotic in lyophilized form was found effective in cases of diarrhea by decreasing the duration of the disease, regardless of its cause (McFarland, 2007; Dinleyici et al., 2012; Shan et al., 2013). It has also been reported that S. boulardii prevented and treated relapses of inflammatory bowel disease, including moderate cases of ulcerative colitis (Guslandi et al., 2000; Guslandi et al., 2003; Choi et al., 2011). Interesting results have also been reported by Lim et al. (2015), suggesting that yeasts can enhance the growth of other probiotics under acidic conditions: Saccharomyces cerevisiae EC-1118 was found to significantly enhance the viability of the probiotic strain Lactobacillus rhamnosus HN001 at pH 2.5–4.0. The use of S. boulardii in reduction of C. difficile infection relapse is still under debate due to controversial results of clinical trials (Flatley et al., 2015). Among other yeasts species, Torulaspora delbrueckii, Debaromyces hansenii, Yarrowia lipolytica, Kluyveromyces lactis, Kluyveromyces marxianus, and Kluyveromyces lodderae have shown strong antagonistic effect against pathogenic bacteria and high acid tolerance (Kumura et al., 2004; Psani and Kotzekidou, 2006; Chen et al., 2010). Despite an excellent record of safe use, yeasts may still be the cause of localized infections in immunocompromised patients (Thygesen et al., 2012).
Akkermansia muciniphila
Another recently described microorganism with possible probiotic potential is Akkermansia muciniphila – a mucin-degrading bacterium that resides within intestinal mucus layers (Derrien et al., 2004). According to several studies, obese patients have significantly lower amounts of this bacterium in their GIT (Collado et al., 2008; Karlsson et al., 2012). The genome sequence of A. muciniphila suggests the ability of this bacterium to metabolize a variety of complex carbohydrates, as well as synthesize multiple amino acids, vitamins, and cofactors (van Passel et al., 2011). Its influence on metabolic processes in the GIT is not fully investigated; however, it has already been shown that this bacterium may be a potential treatment for type II diabetes. Shin et al. (2014) have shown that oral administration of A. muciniphila to mice induced Foxp3 regulatory T cells in the visceral adipose tissue, which attenuated adipose tissue inflammation. Based on these results it has been suggested that pharmacological manipulation of the gut microbiota in favor of A. muciniphila might be beneficial in the treatment of diabetes.
Faecalibacterium prausnitzii and Other Clostridia
Another bacterium that has been demonstrated to have a considerable impact on human gastrointestinal microbiota is Faecalibacterium prausnitzii of the Clostridium sp. cluster IV. This microorganism accounts for 5–15% of the total fecal microbiota, making it one of the most abundant butyrate-producing bacteria in the GIT (Hold et al., 2003; Flint et al., 2012). Since butyrate is a primary energy source for intestinal epithelial cells, it is essential for maintenance of epithelial barrier integrity. Multiple beneficial effects of butyrate for health also include reduction of cancer progression, protection against pathogens, and stimulation of the immune system (Macfarlane and Macfarlane, 2011). The reduction of F. prausnitzii counts in fecal and biopsy samples has been observed in multiple studies of inflammatory bowel disease (especially, ileal Crohn’s disease and ulcerative colitis), suggesting that the presence of this species is important for normal GIT function (Wang et al., 2007; Swidsinski et al., 2008; Andoh et al., 2012). The first gnotobiotic rodent model with F. prausnitzii showed that it could influence gut physiology through the production of mucus O-glycans, thereby affecting the quality and quantity of produced mucus (Wrzosek et al., 2013). Though F. prausnitzii dysbiosis might be an important marker in the development of disease, routine diagnostic tools have not been developed mainly due to the extreme sensitivity of this species to oxygen.
Other bacteria of the class Clostridia might also find use as potential probiotics, since they are highly abundant in human GIT microbiota and may play an important role in metabolism and immune system function. Atarashi et al. (2013) have shown that a mixture of 17 strains of Clostridium sp., belonging to clusters IV, XIV, and XVIII, were able to suppress experimental colitis in mice through induction of interleukin-10-producing regulatory T cells. A similar mechanism of colitis suppression, via IL-10 production by induced macrophages, was observed using strain C. butyricum MIYAIRI 588 (Hayashi et al., 2013). According to another recent study, when mixed with B. infantis, C. butyricum was effective in treatment of experimentally-induced antibiotic-associated diarrhea in mice, and the beneficial effect of the mixture was superior to single strains (Ling et al., 2015). However, though clostridia have potential for use as probiotics, there is still not enough evidence to support their medical efficacy and safety for humans.
Use of Probiotics in Prevention and Treatment of Antibiotic-Associated Diseases
Although most antibiotics are generally safe, some have the potential to cause life-threatening side effects. Antimicrobial side effects are adverse drug reactions involving one or more organ systems. Moreover, even a short-term course of antibiotics may have a long-term negative impact on the normal human gut microbiota (Jernberg et al., 2010). The most commonly used classes of antibiotics include penicillins, cephalosporins, aminoglycosides, fluoroquinolones, macrolides, and tetracyclines; each of these compounds can cause their own specific side-effects (Cunha, 2001). In fact, most traditionally used antibiotics are able to cause health problems in the GIT, and are commonly related to disturbances in microflora composition caused by survival and spread of resistant strains. For instance, penicillins, which are known for having the least-frequent and -severe side effects, may cause diarrhea, and nausea, vomiting, and upset stomach. Fluoroquinolones are also considered relatively safe, but may similarly induce nausea, vomiting, diarrhea, and abdominal pain (Bertino and Fish, 2000). Side-effects of macrolides include GIT-associated nausea, vomiting, and diarrhea, whereas adverse effects of the tetracyclines depend on the concentration of the antibiotic in the affected organ. Their common side-effects include cramps or burning of the stomach, diarrhea, sore mouth, or tongue (Rubinstein, 2001). Research in this field is ongoing and has already provided evidence for efficacy of probiotic use for prevention of health problems emerging as a result of antibiotic use. Examples of such diseases are antibiotic-associated diarrhea (AAD) and C. difficile-associated diarrhea (CDAD; pseudomembranous colitis).
Antibiotic-associated diarrhea is defined as “otherwise unexplained diarrhea that occurs in association with the administration of antibiotics” (Friedman, 2012). However, mild cases of C. difficile infection are sometimes also considered as the cause of AAD (Kelly et al., 1994). The disease comes as one of the most frequent side effects of antibiotic use: 5–39% of patients, depending on the type of antibiotic (e.g., certain β-lactam antibiotics are more likely to cause diarrheal side-effects than cephalosporins) and is associated with increased length and cost of hospitalization (Videlock and Cremonini, 2012). There are several mechanisms of antibiotic effect on humans that can result in AAD. These include osmotic diarrhea, caused by suppression of anaerobic bacteria and a reduction in carbohydrate metabolism, disruption of protective effect of commensal bacteria and reduction of colonic mucosal resistance to pathogenic opportunistic bacteria. Full restoration of the normal gut microbiota may take several weeks or even months (Friedman, 2012; Kaier, 2012).
Many studies have been conducted assessing the efficacy of probiotics in the treatment of AAD and have provided data supporting the usage of both single-strain and mixed-probiotics for diarrhea treatment (Surawicz, 2003; Szajewska et al., 2006; McFarland, 2009). A meta-analysis by Hempel et al. (2012) revealed 82 studies that provided evidence of probiotic efficiency in treatment of AAD. Microorganisms used in these studies included the genera Lactobacillus, Bifidobacterium, Saccharomyces, Streptococcus, Enterococcus, and Bacillus. According to Friedman (2012), several mechanisms of action of probiotics contribute to the prevention and treatment of diarrhea: enhancing mucosal barrier function by secreting mucins, increasing tight junctions in epithelial cells, providing colonization resistance, producing bacteriocins, increasing production of secretory lgA, producing balanced T-helper cell response, increasing production of IL-10 and transforming growth factor beta. Collectively, these factors contribute to the restoration of a normal gastrointestinal balance following damage by antibiotics (Friedman, 2012).
Clostridium difficile-associated diarrhea or pseudomembranous colitisis is an inflammation of the intestine walls caused by toxins produced by C. difficile. CDAD is one of the most common hospital-acquired infections and is a frequent cause of morbidity and mortality among elderly hospitalized patients. Complications include shock, need for colectomy, toxic megacolon, and in severe cases, perforation of the colon wall. C. difficile colonizes the GIT after the alteration of normal gut flora by antibiotic therapy (Bergogne-Bérézin, 2000; Ndegwa and Nkansah, 2008). Extremely high rates of CDAD have been reported in Quebec from 2002 to 2005, totaling 14000 cases (a 4.5-fold increased incidence compared with 1991), with evidence suggesting the emergence of a highly-virulent strain of C. difficile (Pepin et al., 2004, 2005).
Several studies have shown that probiotics aid in prevention and treatment of CDAD. Gao et al. (2010) reported lower risk of disease occurrence after intake of a preparation based on two Lactobacillus strains. S. boulardii has also been successfully used for treatment of CDAD (McFarland et al., 1994). However, a large multi-center study is needed to build sufficient evidence in support of probiotic use as a treatment for C. difficile-associated infections.
Problems Associated with Transfer of Antibiotic Resistance Determinants
Many probiotic strains have naturally acquired resistance toward one or several antimicrobial agents (Table 2). Though intrinsic resistance of probiotic bacteria to certain antibiotics might offer benefits for their use in the prevention and treatment of AAD, the issue of possible transfer of resistance determinants has been raised (Pflughoeft and Versalovic, 2012), particularly for strains that carry plasmids.
Courvalin (2006) specified two distinct types of acquired antibiotic resistance in bacteria: (i) initially non-transferred resistance that occurred as a result of one or several mutations in indigenous gene(s), and (ii) transferred resistance, acquired from a different organism by horizontal gene transfer. Antibiotic resistance (both intrinsic and acquired) can occur as a result of three major mechanisms: (i) altering the outer- and/or inner membrane permeability and transport activity, which leads to lower accumulation of the antibiotic within the cell, (ii) using enzymes to detoxify the antibiotic, and (iii) modifying the antibiotic target site (Guardabassi and Courvalin, 2006). The gene responsible for acquisition of antibiotic resistance often resides on a plasmid or transposon, which might be easily transferred (Bennett, 2008). In fact, transposon-mediated transfer of genetic material between species was recently described as the most frequent mechanism contributing to the spread of antibiotic resistance in bacteria (Wozniak and Waldor, 2010).
Multiple studies have already shown that antibiotic resistance can be transferred between different bacterial species that reside in the human GIT. For instance, it has been reported that both Lactobacillus lactis and Streptococcus thermophilus are able to transfer erythromycin resistance [erm(B) gene, located on a plasmid] to L. monocytogenes under in vitro conditions (Toomey et al., 2009). Another study provided the evidence of in vivo transfer of ampicillin resistance between two strains of E. coli co-residing in human gut: it was demonstrated that a plasmid carrying a β-lactamase gene had been transferred from an ampicillin- resistant E. coli strain to an initially susceptible strain (Karami et al., 2007). Devirgiliis et al. (2009) reported the transfer of a tet(M) gene (tetracycline resistance; located on broad host range Tn916 transposon) from L. paracasei to E. faecalis in vitro. In another set of experiments, the erythromycin resistance pLFE1 plasmid of L. plantarum strain M345 was successfully transferred to five different species: L. rhamnosus, Lc. lactis, Listeria innocua, E. faecalis, and L. monocytogenes (Feld et al., 2009). These and other examples raise a safety concern; strains to be used as probiotics should be carefully selected, and only those free of transferrable antibiotic-resistance determinants ought to be considered safe (Radulovic et al., 2012).
In Vitro and In Vivo Systems Used to Study Probiotic Effects
Novel probiotic-based strategies for therapeutic and prophylactic use against multiple GIT diseases are gaining popularity worldwide. Their effectiveness has been predicted by numerous animal model studies and proven by extensive research involving humans. However, the initial step in confirming probiotic effects is the extensive characterization of a bacterial strain to be used as a probiotic, which is usually performed under in vitro conditions by studying bacterial acid resistance, bile resistance, carbon source utilization, and aggregative properties, or ex vivo for their ability to adhere to mammalian cells (Kotikalapudi et al., 2010; Wood et al., 2012). Similarly, probiotic delivery methods, such as lyophilization or encapsulation, are also tested for their protective potential in vitro under simulated gastric conditions (Klemmer et al., 2011; Wood et al., 2012; Khan et al., 2013; Wang et al., 2014). The most popular materials used for encapsulation of bacteria are alginate, carrageenans and gums, since they are easy to process, resistant to low pH and freezing, and are generally recognized as safe (Gbassi and Vandamme, 2012). We have recently reported the efficient delivery of B. adolescentis, encapsulated for this purpose in an alginate-pea protein protective matrix, into the lower gut of rats (Varankovich et al., 2015).
Apart from basic synthetic gastric juice solutions (low pH, 37°C), more complex systems have been developed, such as SHIME (Simulator of the Human Intestinal Microbial Ecosystem), designed to simulate different parts of the human GIT (Cook et al., 2012). Probiotic strains and methods for their delivery, preselected in vitro, are subsequently tested in animal models.
Traditionally used animal models include mice and rats. Larger animals like rabbits, dogs, and pigs are generally considered to have more common features with the physiology and microflora of the human GIT (Kararli, 1995). However, rodents are cheap, standardized, and have short life-cycles; thus, their extensive use in large-scale research. Investigation of probiotic effects on animal microflora may be approached by: (i) examining the quantitative and qualitative characteristics of bacterial microflora in animals using cultivation and/or molecular biology techniques, such as real-time polymerase chain reaction (qPCR), next-generation sequencing (NGS), and fluorescence in situ hybridization (FISH), or (ii) evaluating treatment efficiency indirectly by using it to cure an artificially induced disease.
Distribution of specific species of microorganisms is still being studied in healthy humans and compared with those of patients with various gastrointestinal diseases. Perturbations of microbiota, even in case of alterations in numbers of a single species (i.e., A. muciniphila), might be a cause (and an indicator) of the development of disease (Karlsson et al., 2012). In this case, probiotic treatment might be useful in restoring microbiota balance in the gut. An interesting example of quantitative/qualitative analysis of animal gut microbiota after probiotic administration can be found in the study by Wang et al. (2015b): 454 pyrosequencing of fecal bacterial 16S rRNA genes in obese vs. lean mice showed that the probiotic strains shifted the overall structure of the gut microbiota of obese animals toward that of lean mice fed a normal diet, with significant changes observed in 83 operational taxonomic units. Due to complicated analyses required to understand specific mechanisms of disease development, as well as the mode of action of a certain probiotic microorganism, the use of disease models is generally more widespread.
Rodent Models of GIT Diseases
Generally, in order to establish a disease model, mice are infected with the pathogen or irritant either one time or continuously (Pawlowski et al., 2010; Bhinder et al., 2013). Subsequently, animals are treated with probiotics with concomitant monitoring of the disease symptoms and evaluation of changes in the gut microflora. Following this approach, Verdú et al. (2008) infected mice with H. pylori for 4–6 months to investigate the effect of probiotic therapy on upper gastrointestinal dysfunction induced by chronic H. pylori infection. The authors reported that with probiotic treatment delayed gastric emptying in mice normalized significantly faster post-eradication, compared to control groups, where the dysfunction was observed during 2 months after pathogen administration was ceased. Mice and rats have also been used to evaluate the efficacy of probiotics for the treatment of Salmonella and E. coli O157:H7 infections (Asahara et al., 2001, 2004), inflammatory bowel disease (Shiba et al., 2003) and immune suppression (Lollo et al., 2012). Asahara et al. (2001) showed that intestinal growth and subsequent extra-intestinal translocation of orally-infected Salmonella typhimurium in mice were inhibited during administration of probiotic B. breve. Later, the same group reported B. breve was also effective in protecting mice against Shiga toxic-producing E. coli 0157:H7 (Asahara et al., 2004). Extrapolation of results achieved in animal studies and in vitro experiments to humans remains a difficult challenge. Many factors, such as differences in physiology and microflora composition of respective gastrointestinal systems, must be considered before interpreting the outcome.
The majority of in vivo experiments investigating the effects of probiotics on pathogenic bacterial populations use gnotobiotic mice (usually with human microflora systems in their GIT; Bernet-Camard et al., 1997; Aiba et al., 1998; Gill et al., 2001; Pawlowski et al., 2010). For instance, in a study by Shiba et al. (2003), probiotic B. infantis 1222 was found to significantly suppress the systemic antibody response raised by Bacteroides vulgates, a representative pathogenic Bacteroides sp. species, in a gnotobiotic mice model of inflammatory bowel disease. The use of conventional mice as a model for investigating human diseases is more problematic due to significant differences in animal and human gut microflora. Nevertheless, it is possible to use murine-specific organisms as models for the study of human pathogens. For instance, Ge et al. (2001) used H. hepaticus infection as an animal model for examining the pathogenesis of gastrointestinal diseases in humans caused by H. pylori. More recently, Bhinder et al. (2013) described the Citrobacter rodentium mouse model for the study of pathogen and host contributions during infectious colitis. C. rodentium is a murine-specific bacterial pathogen, closely related to enteropathogenic and enterohaemorrhagic strains of E. coli (Borenstein et al., 2008). Several C. rodentium infection studies involving mice models have shown probiotics to reduce the severity of symptoms and prevent death caused by the pathogenic agent (Chen et al., 2005; Gareau et al., 2010; Mackos et al., 2013). Chen et al. (2005) successfully treated C. rodentium-induced murine colitis with probiotic L. acidophilus. Gareau et al. (2010) similarly reported that L. rhamnosus, combined with L. helveticus, were effective in prevention and treatment of the same disease state in mice. Later, another group showed that L. reuteri was able to attenuate the severity of murine colitis caused by C. rodentium (Mackos et al., 2013). Further investigation of host–pathogen and probiotic–pathogen interactions will likely provide better insight into treatment of C. rodentium infection in mice, and possibly E. coli infections in humans. However, confirmation of probiotic benefits and possible side effects will ultimately require human trials.
Human Clinical Trials
Human studies generally take the form of randomized clinical trials involving participants with some type of intestinal disorder. After assessment of eligibility and recruitment, participants are given either probiotic treatment or a placebo as a control. Results of these experiments have provided enough evidence for considering probiotics an efficient treatment for multiple GIT-associated diseases, such as acute gastroenteritis (Huang et al., 2002), irritable bowel syndrome (Nikfar et al., 2008) and necrotizing enterocolitis (Alfaleh and Anabrees, 2014). Some trials showing the efficacy of bacteria of interest in the treatment of specific gastrointestinal disorders are listed in Table 3. In one recent trial aimed to assess the efficiency of S. cerevisiae in treatment of irritable bowel syndrome, 179 adults, diagnosed with this condition, were randomized to receive once-daily 500 mg of S. cerevisiae or placebo for 8 weeks. Cardinal symptoms (abdominal pain/discomfort, bloating/distension, bowel movement difficulty) were recorded daily after a 2-week run-in period. The results showed that abdominal pain/discomfort scores were significantly reduced during probiotic intake (Pineton de Chamburn et al., 2015). A major trial involving 362 participants was conducted by Whorwell et al. (2006) in order to study the effect of B. infantis on symptoms of irritable bowel syndrome: probiotic administration lead to improvements in the majority of symptoms by more than 20%, compared to placebo. Another human clinical trial proved the efficacy of Lactobacillus GG in treatment of H. pylori infection: daily administration of the probiotic led to significant reduction in disease symptoms (diarrhea, nausea, and taste disturbances; Armuzzi et al., 2001). In general, data from multiple lines of research involving humans suggests that probiotic bacteria suppress gastrointestinal pathogens by simple competition by prevailing in numbers, and by producing antibacterial factors (bacteriocins and small organic molecules, such as fatty acids). Though more details into the mechanisms of action of probiotics on gut microbiota are essential, the large base of evidence already collected has proven the beneficial role in prevention and treatment of various GIT diseases in humans.
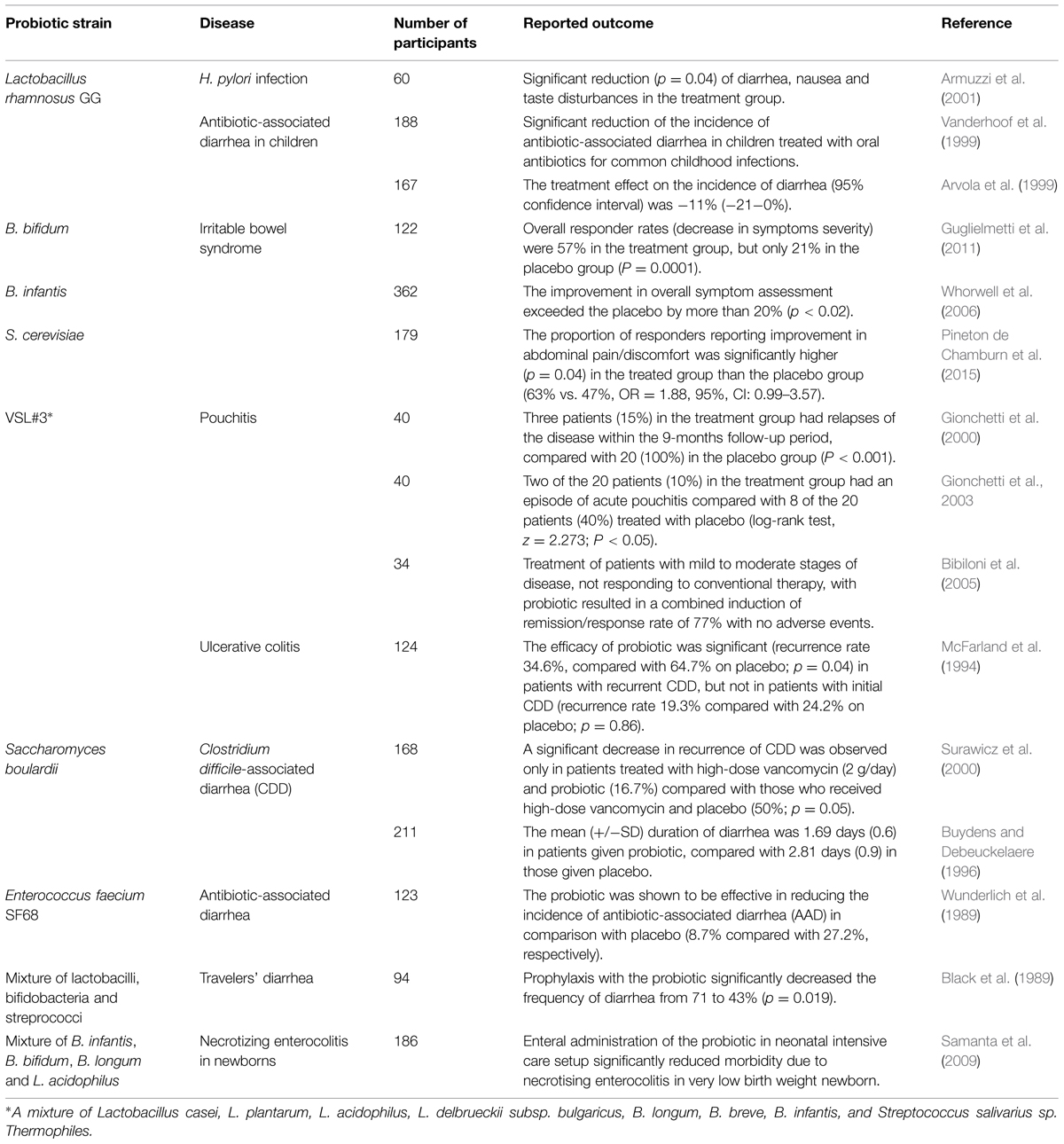
TABLE 3. Some of the major human trials of probiotics for the treatment of gastrointestinal diseases.
Conclusion
Many strains of genera Lactobacillus and Bifidobacterium, as well as some enterococci and yeasts, have been shown to possess probiotic properties with potential for prophylaxis and treatment of a range of gastrointestinal disorders. The effectiveness of probiotic bacteria in the treatment of these conditions is supported by many clinical trials involving patients of all ages and probiotic organisms chosen based on laboratory research trials. Notably, most of the work in the probiotic field has been conducted in vitro, as it is an essential step in the investigation of bacterial growth, metabolite production, ability to form biofilms, compete with pathogens, co-aggregate, and produce antimicrobials. All of these characteristics are important factors for identification of potential probiotic strains that possess desirable properties along with the ability to establish itself in the human gut. Intrinsic antibiotic resistance and transferability of genetic determinants are two additional factors to account for at the initial stage of a probiotic study. Novel putative probiotic species, such as A. muciniphila, are yet to be tested in both animal and human trials; however, the results achieved to date suggest that they might be beneficial in treatment or diagnosis of GIT diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Financial support for this project was provided by the Saskatchewan Ministry of Agriculture Development Fund.
References
Aiba, Y., Suzuki, N., Kabir, A. M., Takagi, A., and Koga, Y. (1998). Lactic acid-mediated suppression of Helicobacter pylori by the oral administration of Lactobacillus salivarius as a probiotic in a gnotobiotic murine model. Am. J. Gastroenterol. 93, 2097–2101. doi: 10.1111/j.1572-0241.1998.00600.x
AlFaleh, K., and Anabrees, J. (2014). Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst. Rev. 2014:CD005496. doi: 10.1002/14651858.CD005496.pub4
Andoh, A., Kuzuoka, H., Tsujikawa, T., Nakamura, S., Hirai, F., Suzuki, Y., et al. (2012). Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J. Gastroenterol. 47, 1298–1307. doi: 10.1007/s00535-012-0605-0
Armuzzi, A., Cremonini, F., Bartolozzi, F., Canducci, F., Candelli, M., Ojetti, V., et al. (2001). The effect of oral administration of Lactobacillus GG on antibiotic-associated gastrointestinal side-effects during Helicobacter pylori eradication therapy. Aliment. Pharmacol. Ther. 15, 163–169. doi: 10.1046/j.1365-2036.2001.00923.x
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2013). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Arvola, T., Laiho, K., Torkkeli, S., Mykkanen, H., Salminen, S., Maunula, L., et al. (1999). Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: a randomized study. Pediatrics 104, e64. doi: 10.1542/peds.104.5.e64
Asahara, T., Nomoto, K., Shimizu, K., Watanuki, M., and Tanaka, R. (2001). Increased resistance of mice to Salmonella enterica serovar Typhimurium infection by synbiotic administration of Bifidobacteria and transgalactosylated oligosaccharides. J. Appl. Microbiol. 91, 985–996. doi: 10.1046/j.1365-2672.2001.01461.x
Asahara, T., Shimizu, K., Nomoto, K., Asahara, T., Shimizu, K., and Nomoto, K. (2004). Probiotic Bifidobacteria protect mice from lethal infection with shiga toxin-producing Escherichia coli O157: H7. Infect. Immun. 72, 2240–2247. doi: 10.1128/IAI.72.4.2240-2247.2004
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 500, 232–236. doi: 10.1038/nature12331
Aureli, P., Capurso, L., Castellazzi, A. M., Clerici, M., Giovannini, M., Morelli, L., et al. (2011). Probiotics and health: an evidence-based review. Pharmacol. Res. 63, 366–376. doi: 10.1016/j.phrs.2011.02.006
Basu, S., Paul, D. K., Ganguly, S., Chatterjee, M., and Chandra, P. K. (2009). Efficacy of high-dose Lactobacillus rhamnosus GG in controlling acute watery diarrhea in Indian children: a randomized controlled trial. J. Clin. Gastroenterol. 43, 208–213. doi: 10.1097/MCG.0b013e31815a5780
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153, S347–S357. doi: 10.1038/sj.bjp.0707607
Benyacoub, J., Czarnecki-Maulden, G. L., Cavadini, C., Sauthier, T., Anderson, R. E., Schiffrin, E. J., et al. (2003). Supplementation of food with Enterococcus faecium (SF68) stimulates immune functions in young dogs. J. Nutr. 133, 1158–1162.
Bergogne-Bérézin, E. (2000). Treatment and prevention of antibiotic associated diarrhea. Int. J. Antimicrob. Agents 16, 521–526. doi: 10.1016/S0924-8579(00)00293-4
Bernet-Camard, M. F., Lievin, V., Brassart, D., Neeser, J. R., Servin, A. L., and Hudault, S. (1997). The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl. Environ. Microbiol. 63, 2747–2753.
Bertino, J., and Fish, D. (2000). The safety profile of the fluoroquinolones. Clin. Ther. 22, 798–817. doi: 10.1016/S0149-2918(00)80053-3
Bhardwaj, A., Malik, R. K., and Chauhan, P. (2008). Functional and safety aspects of enterococci in dairy foods. Indian J. Microbiol. 48, 317–325. doi: 10.1007/s12088-008-0041-2
Bhinder, G., Sham, H. P., Chan, J. M., Morampudi, V., Jacobson, K., and Vallance, B. A. (2013). The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. J. Vis. Exp. 72:e50222. doi: 10.3791/50222
Biavasco, F., Vignaroli, C., Lupidi, R., Manso, E., Facinelli, B., and Varaldo, P. E. (1997). In vitro antibacterial activity of LY333328, a new semisynthetic glycopeptide. Antimicrob. Agents Chemother. 41, 2165–2172.
Biavati, B. (1994). Degradation of complex carbohydrates. Int. J. Food Microb. 24, 199–210. doi: 10.1016/0168-1605(94)90119-8
Bibiloni, R., Fedorak, R. N., Tannock, G. W., Madsen, K. L., Gionchetti, P., Campieri, M., et al. (2005). VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am. J. Gastroenterol. 100, 1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x
Black, F. T., Andersen, P. L., Ørskov, J., Ørskov, F., Gaarslev, K., and Laulund, S. (1989). Prophylactic efficacy of lactobacilli on traveler’s diarrhea. Trav. Med. 7, 333–335. doi: 10.1007/978-3-642-73772-5_70
Borenstein, D., McBee, M. E., and Schauer, D. B. (2008). Utility of the Citrobacter rodentium infection model in laboratory mice. Curr. Open. Gastroenterol. 24, 32–37. doi: 10.1097/MOG.0b013e3282f2b0fb
Bravo, J. A., Forsythe, P., Chew, M. V., Escaravage, E., Savignac, H. M., Dinan, T. G., et al. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 108, 16050–16055. doi: 10.1073/pnas.1102999108
Buydens, P., and Debeuckelaere, S. (1996). Efficacy of SF 68 in the treatment of acute diarrhea. A placebo-controlled trial. Scand J. Gastroenterol. 31, 887–891. doi: 10.3109/00365529609051997
Caballero-Franco, C., Keller, K., De Simone, C., and Chadee, K. (2007). The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastroint. Liver Physiol. 292, 315–322. doi: 10.1152/ajpgi.00265.2006
Canducci, F., Cremonini, F., Armuzzi, A., Di Caro, S., Gabrielli, M., Santarelli, L., et al. (2002). Probiotics and Helicobacter pylori eradication. Digest Liver Dis. 34, S81–S83. doi: 10.1016/S1590-8658(02)80172-4
Cebrián, R., Baños, A., Valdivia, E., Pérez-Pulido, R., Martínez-Bueno, M., and Maqueda, M. (2012). Characterization of functional, safety, and probiotic properties of Enterococcus faecalis UGRA10, a new AS-48-producer strain. Food Microbiol. 30, 59–67. doi: 10.1016/j.fm.2011.12.002
Charteris, W. P., Kelly, P. M., Morelli, L., and Collins, J. K. (1998). Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 61, 1636–1643.
Chen, C.-C., Louie, S., Shi, H. N., and Walker, W. A. (2005). Preinoculation with the probiotic Lactobacillus acidophilus early in life effectively inhibits murine Citrobacter rodentium colitis. Pediatr. Res. 58, 1185–1191. doi: 10.1203/01.pdr.0000183660.39116.83
Chen, L.-S., Ma, Y., Maubois, J.-L., He, S.-H., Chen, L.-J., and Li, H.-M. (2010). Screening for the potential probiotic yeast strains from raw milk to assimilate cholesterol. Dairy Sci. Technol. 90, 537–548. doi: 10.1051/dst/2010001
Choi, C. H., Jo, S. Y., Park, H. J., Chang, S. K., Byeon, J. S., and Myung, S. J. (2011). A randomized, double-blind, placebo-controlled multicenter trial of Saccharomyces boulardii in irritable bowel syndrome: effect on quality of life. J. Clin. Gastroenterol. 45, 679–683. doi: 10.1097/MCG.0b013e318204593e
Collado, M. C., Isolauri, E., Laitinen, K., and Salminen, S. (2008). Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 88, 894–899.
Collado, M. C., Meriluoto, J., and Salminen, S. (2007). Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett. Appl. Microbiol. 45, 454–460. doi: 10.1111/j.1472-765X.2007.02212.x
Cook, M. T., Tzortzis, G., Charalampopoulos, D., and Khutoryanskiy, V. V. (2012). Microencapsulation of probiotics for gastrointestinal delivery. J. Control. Release 162, 56–67. doi: 10.1016/j.jconrel.2012.06.003
Courvalin, P. (2006). Antibiotic resistance: the pros and cons of probiotics. Dig. Liver Dis. 38(Suppl. 2), S261–S265. doi: 10.1016/S1590-8658(07)60006-1
Cruchet, S., Obregon, M. C., Salazar, G., Diaz, E., and Gotteland, M. (2003). Effect of the ingestion of a dietary product containing Lactobacillus johnsonii La1 on Helicobacter pylori colonization in children. Nutrition 19, 716–721. doi: 10.1016/S0899-9007(03)00109-6
Cunha, B. A. (2001). Antibiotic therapy, Part II. Int. Med. Clin. North Am. 85, 149–185. doi: 10.1016/S0025-7125(05)70309-6
Danielsen, M., and Wind, A. (2003). Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 82, 1–11. doi: 10.1016/S0168-1605(02)00254-4
Derrien, M., Vaughan, E. E., Plugge, C. M., and de Vos, W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476. doi: 10.1099/ijs.0.02873-0
Desbonnet, L., Garrett, L., Clarke, G., Bienenstock, J., and Dinan, T. G. (2008). The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J. Psychiatr. Res. 43, 164–174. doi: 10.1016/j.jpsychires.2008.03.009
Devirgiliis, C., Coppola, D., Barile, S., Colonna, B., and Perozzi, G. (2009). Characterization of the Tn916 conjugative transposon in a food-borne strain of Lactobacillus paracasei. Appl. Environ. Microbiol. 75, 3866–3871. doi: 10.1128/AEM.00589-09
DiBaise, J. K., Frank, D. N., and Mathur, R. (2012). Impact of the gut microbiota on the development of obesity: current concepts. Am. J. Gastroenterol. Suppl. 1, 22–27. doi: 10.1038/ajgsup.2012.5
Dinan, T. G., Stanton, C., and Cryan, J. F. (2013). Psychobiotics: a novel class of psychotropic. Biol. Psych. 74, 720–726. doi: 10.1016/j.biopsych.2013.05.001
Dinleyici, E. C., Eren, M., Ozen, M., Yargic, Z. A., and Vandenplas, Y. (2012). Effectiveness and safety of Saccharomyces boulardii for acute infectious diarrhea. Expert Opin. Biol. Ther. 12, 395–410. doi: 10.1517/14712598.2012.664129
Doré, J., and Corthier, G. (2010). The human intestinal microbiota. Gastroentérol. Clin. Biol. 34, S7–S15. doi: 10.1016/S0399-8320(10)70002-6
Duca, F., Sakar, Y., Lepage, P., Devime, F., Langelier, B., Doré, J., et al. (2014). Replication of obesity and associated signaling pathways through transfer of microbiota from obese-prone rats. Diabetes Metab. Res. Rev. 63, 1624–1636. doi: 10.2337/db13-1526
Dusko Ehrlich, S., and MetaHIT Consortium. (2011). “MetaHIT: the European Union Project on metagenomics of the human intestinal tract,” in Metagenomics of the Human Body, Chap. 15. ed. K. Nelson (Berlin: Springer).
FAO/WHO. (2001). Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health, and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria (Córdoba: Food and Agriculture Organization of the United Nations, World Health Organization).
Feld, L., Bielak, E., Hammer, K., and Wilcks, A. (2009). Characterization of a small erythromycin resistance plasmid pLFE1 from the food-isolate Lactobacillus plantarum M345. Plasmid 61, 159–170. doi: 10.1016/j.plasmid.2009.01.002
Flatley, E. A., Wilde, A. M., and Nailor, M. D. (2015). Saccharomyces boulardii for the prevention of hospital onset Clostridium difficile infection. J. Gastrointestin. Liver Dis. 24, 21–24. doi: 10.15403/jgld.2014.1121.fly
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P., and Forano, E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Foligné, B., Dewulf, J., Breton, J., Claisse, O., Lonvaud-Funel, A., and Pot, B. (2010). Probiotic properties of non-conventional lactic acid bacteria: immunomodulation by Oenococcus oeni. Int. J. Food Microbiol. 140, 136–145. doi: 10.1016/j.ijfoodmicro.2010.04.007
Foligne, B., Zoumpopoulou, G., Dewulf, J., Ben Younes, A., Chareyre, F., Sirard, J.-C., et al. (2007). A key role of dendritic cells in probiotic functionality. PLoS ONE 2:e313. doi: 10.1371/journal.pone.0000313
Fooks, L. J., Fuller, R., and Gibson, G. R. (1999). Prebiotics, probiotics and human gut microbiology. Int. Dairy J. 9, 53–61. doi: 10.1016/S0958-6946(99)00044-8
Friedman, G. (2012). Preface: clinical applications of probiotics in gastroenterology: questions and answers. Gastroenterol. Clin. North Am. 41, 763–779. doi: 10.1016/j.gtc.2012.08.002
Gao, X. W., Mubasher, M., Fang, C. Y., Reifer, C., and Miller, L. E. (2010). Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am. J. Gastroenterol. 105, 1636–1641. doi: 10.1038/ajg.2010.11
Gareau, M. G., Wine, E., Reardon, C., and Sherman, P. M. (2010). Probiotics prevent death caused by Citrobacter rodentium infection in neonatal mice. J. Inf. Dis. 201, 81–91. doi: 10.1086/648614
Gbassi, G. K., and Vandamme, T. (2012). Probiotic encapsulation technology: from microencapsulation to release into the gut. Pharmaceutics 4, 149–163. doi: 10.3390/pharmaceutics4010149
Ge, Z., White, D. A., Whary, M. T., and Fox, J. G. (2001). Fluorogenic PCR-Based quantitative detection of a murine pathogen Helicobacter hepaticus. J. Clin. Microbiol. 39, 2598–2602. doi: 10.1128/JCM.39.7.2598-2602.2001
Gill, H. S., Shu, Q., Lin, H., Rutherfurd, K. J., and Cross, M. L. (2001). Protection against translocating Salmonella typhimurium infection in mice by feeding the immuno-enhancing probiotic Lactobacillus rhamnosus strain HN001. Med. Microbiol. Immunol. 190, 97–9104.
Gionchetti, P., Rizzello, F., Helwig, U., Venturi, A., Lammers, K. M., Brigidi, P., et al. (2003). Prophylactis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 124, 1202–1209. doi: 10.1016/S0016-5085(03)00171-9
Gionchetti, P., Rizzello, F., Venturi, A., Brigidi, P., Matteuzzi, D., Bazzocchi, G., et al. (2000). Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 119, 305–309. doi: 10.1053/gast.2000.9370
Gomes, A. M. P., and Malcata, F. X. (1999). Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci. Tech. 10, 139–157. doi: 10.1016/S0924-2244(99)00033-3
Guardabassi, L., and Courvalin, P. (2006). “Antibiotics: Mode of action and mechanisms of resistance,” in Antimicrobial Resistance in Bacteria of Animal Origin, eds F. Aarestrup and H. K. Wegener (Washington, DC: American Society for Microbiology), 1–18.
Guerra, N. P., and Pastrana, L. (2002). Modelling the influence of pH on the kinetics of both nisin and pediocin production and characterization of their functional properties. Proc. Biochem. 37, 1005–1015. doi: 10.1016/S0032-9592(01)00312-0
Guglielmetti, S., Mora, D., Gschwender, M., and Popp, K. (2011). Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life – a double-blind, placebo-controlled study. Alim. Pharmacol. Therapeu. 33, 1123–1132. doi: 10.1111/j.1365-2036.2011.04633.x
Guo, X. H., Kim, J. M., Nam, H. M., Park, S. Y., and Kim, J. M. (2010). Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe 16, 321–326. doi: 10.1016/j.anaerobe.2010.03.006
Guslandi, M., Giollo, P., and Testoni, P. A. (2003). A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur. J. Gastroenterol. Hepatol. 15, 697–698. doi: 10.1097/00042737-200306000-00017
Guslandi, M., Mezzi, G., Sorghi, M., and Testoni, P. A. (2000). Saccharomyces boulardii in maintenance treatment of Crohn’s disease. Dig. Dis. Sci. 45, 1462–1464. doi: 10.1023/A:1005588911207
Hamilton-Miller, J. M. T. (2003). The role of probiotics in the treatment and prevention of Helicobacter pylori infection. Int. J. Antimicrob. Agents 22, 360–366. doi: 10.1016/S0924-8579(03)00153-5
Hayashi, A., Sato, T., Kamada, N., Mikami, Y., Matsuoka, K., and Hisamatsu, T. (2013). A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe 13, 711–722. doi: 10.1016/j.chom.2013.05.013
Hempel, S., Maher, A. R., Wang, Z., Miles, J. N. V., Shanman, R., Johnsen, B., et al. (2012). Probiotics for the prevention and treatment of antibiotic-associated diarrhea. A systematic review and meta-analysis. J. Am. Med. Assoc. 307, 1959–1969. doi: 10.1001/jama.2012.3507
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hold, G. L., Schwiertz, A., Aminov, R. I., Blaut, M., and Flint, H. J. (2003). Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69, 4320–4324. doi: 10.1128/AEM.69.7.4320-4324.2003
Holzapfel, W. H., and Schillinger, U. (2002). Introduction to pre- and probiotics. Food Res. Int. 35, 109–116. doi: 10.1016/S0963-9969(01)00171-5
Huang, J. S., Bousvaros, A., Lee, J. W., Diaz, A., and Davidson, E. J. (2002). Efficacy of probiotic use in acute diarrhea in children: a meta-analysis. Diges Dis. Sci. 47, 2625–2634. doi: 10.1023/A:1020501202369
Jernberg, C., Löfmark, S., Edlund, C., and Jansson, J. K. (2010). Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156(Pt 11), 3216–3223. doi: 10.1099/mic.0.040618-0
Jonganurakkun, B., Wang, Q., Xu, S. H., Tada, Y., Minamida, K., Yasokawa, D., et al. (2008). Pediococcus pentosaceus NB-17 for probiotic use. J. Biosci. Bioeng. 106, 69–73. doi: 10.1263/jbb.106.69
Jungersen, M., Wind, A., Johansen, E., Christensen, J., Stuer-Lauridsen, B., and Eskesen, D. (2014). The science behind the probiotic strain Bifidobacterium animalis subsp. lactis BB-12. Microorganisms 2, 92–110. doi: 10.3390/microorganisms2020092
Kaier, K. (2012). Economic implications of the dynamic relationship between antibiotic use and hospital-acquired infections. Val. Health 15, 87–93. doi: 10.1016/j.jval.2011.09.005
Karami, N., Martner, A., Enne, V. I., Swerkersson, S., Adlerberth, I., and Wold, A. E. (2007). Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J. Antimicrob. Chemother. 60, 1142–1145. doi: 10.1093/jac/dkm327
Kararli, T. T. (1995). Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm. Drug Dispos. 16, 351–380. doi: 10.1002/bdd.2510160502
Karlsson, C. L., Onnerfält, J., Xu, J., Molin, G., Ahrné, S., and Thorngren-Jerneck, K. (2012). The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 20, 2257–2261. doi: 10.1038/oby.2012.110
Kelesidis, T., and Pothoulakis, C. (2012). Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therapeu. Adv. Gastroenterol. 5, 111–125. doi: 10.1177/1756283X11428502
Kelly, C. P., Pothoulakis, C., and Lamont, J. T. (1994). Clostridium difficile colitis. N. Engl. J. Med. 330, 257–262. doi: 10.1056/NEJM199401273300406
Khan, N. H., Korber, D. R., Low, N. H., and Nickerson, M. T. (2013). Development of extrusion-based legume protein isolate-alginate capsules for the protection and delivery of the acid sensitive probiotic, Bifidobacterium adolescentis. Food Res. Int. 54, 730–737. doi: 10.1016/j.foodres.2013.08.017
Kheadr, E., Bernoussi, N., Lacroix, C., and Fliss, I. (2004). Comparison of the sensitivity of commercial strains and infant isolates of bifidobacteria to antibiotics and bacteriocins. Int. Dairy J. 14, 1041–1053. doi: 10.1016/j.idairyj.2004.04.010
Klemmer, K. J., Korber, D. R., Low, N. H., and Nickerson, M. T. (2011). Pea protein-based capsules for probiotic and prebiotic delivery. Int. J. Food Sci. Tech. 46, 2248–2256. doi: 10.1111/j.1365-2621.2011.02743.x
Konieczna, P., Ferstl, R., Ziegler, M., Frei, R., Nehrbass, D., Lauener, R. P., et al. (2013). Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS ONE 8:e62617. doi: 10.1371/journal.pone.0062617
Kotikalapudi, B. L., Low, N. H., Nickerson, M. T., and Korber, D. R. (2010). In vitro characterization of probiotic survival, adherence and antimicrobial resistance: candidate selection for encapsulation in a pea protein isolate–alginate delivery system. Int. J. Probiotics Prebiotics 5, 1–12.
Kotzampassi, K., and Giamarellos-Bourboulis, E. J. (2012). Probiotics for infectious diseases: more drugs, less dietary supplementation. Int. J. Antimicrob. Agents 40, 288–296. doi: 10.1016/j.ijantimicag.2012.06.006
Kumura, H., Tanoue, Y., Tsukahara, M., Tanaka, T., and Shimazaki, K. (2004). Screening of dairy yeast strains for probiotic applications. J. Dairy Sci. 87, 4050–4056. doi: 10.3168/jds.S0022-0302(04)73546-8
Kurugol, Z., and Koturoglu, G. (2005). Effects of Saccharomyces boulardii in children with acute diarrhoea. Acta Paediatr. 94, 44–47. doi: 10.1080/08035250410022521
Lee, D. K., Park, S. Y., An, H. M., Kim, J. R., Kim, M. J., Lee, S. W., et al. (2011). Antimicrobial activity of Bifidobacterium spp. isolated from healthy adult Koreans against cariogenic microflora. Arch. Oral. Biol. 56, 1047–54. doi: 10.1016/j.archoralbio.2011.03.002
Lee, J.-Y., Chu, S.-H., Jeon, J. Y., Lee, M.-K., Park, J.-H., Lee, D.-C., et al. (2014a). Effects of 12 weeks of probiotic supplementation on quality of life in colorectal cancer survivors: a double-blind, randomized, placebo-controlled trial. Dig. Liver Dis. 46, 1126–1132. doi: 10.1016/j.dld.2014.09.004
Lee, D. K., Park, J. E., Kim, M. J., Seo, J. G., Lee, J. H., and Ha, N. J. (2014b). Probiotic bacteria, B. longum and L. acidophilus inhibit infection by rotavirus in vitro and decrease the duration of diarrhea in pediatric patients. Clin. Res. Hepatol. Gastroenterol. 39, 237–244. doi: 10.1016/j.clinre.2014.09.006
Lim, P. L., Toh, M., and Liu, S. Q. (2015). Saccharomyces cerevisiae EC-1118 enhances the survivability of probiotic Lactobacillus rhamnosus HN001 in an acidic environment. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-015-6560-y [Epub ahead of print].
Ling, Z., Liu, X., Cheng, Y., Luo, Y., Yuan, L., Li, L., et al. (2015). Clostridium butyricum combined with Bifidobacterium infantis probiotic mixture restores fecal microbiota and attenuates systemic inflammation in mice with antibiotic-associated diarrhea. Biomed Res. Int. 2015:582048. doi: 10.1155/2015/582048 [Epub ahead of print].
Lollo, P. C. B., Cruz, G., Morato, P. N., Moura, C. S., Carvalho-Silva, L. B., Oliveira, C. F., et al. (2012). Probiotic cheese attenuates exercise-induced immune suppression in Wistar rats. J. Dairy Sci. 95, 3549–3558. doi: 10.3168/jds.2011-5124
Macfarlane, G. T., and Macfarlane, S. (2011). Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 45, S120–S127. doi: 10.1097/mcg.0b013e31822fecfe
Mackos, A. R., Eubank, T. D., Parry, N. M. A., and Bailey, M. T. (2013). Probiotic Lactobacillus reuteri attenuates the stressor-enhanced severity of Citrobacter rodentium infection. Infect. Immun. 81, 3253–3263. doi: 10.1128/IAI.00278-13
Mangin, I., Bouhnik, Y., Bisetti, N., and Decaris, B. (1999). Molecular monitoring of human intestinal Bifidobacterium strain diversity. Res. Microbiol. 150, 343–350. doi: 10.1016/S0923-2508(99)80060-6
Mårtensson, O., Biörklund, M., Lambo, A. M., Dueñas-Chasco, M., Irastorza, A., Holst, O., et al. (2005). Fermented, ropy, oat-based products reduce cholesterol levels and stimulate the bifidobacteria flora in humans. Nutr. Res. 25, 429–442. doi: 10.1016/j.nutres.2005.03.004
McFarland, L. V. (2007). Meta-analysis of probiotics for the prevention of traveler’s diarrhea. Travel. Med. Infect. Dis. 5, 97–105. doi: 10.1016/j.tmaid.2005.10.003
McFarland, L. V. (2009). Evidence-based review of probiotics for antibiotic-associated diarrhea and Clostridium difficile infections. Anaerobe 15, 274–280. doi: 10.1016/j.anaerobe.2009.09.002
McFarland, L. V., Surawicz, C. M., Greenberg, R. N., Fekety, R., Elmer, G. W., Moyer, K. A., et al. (1994). A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. J. Am. Med. Assoc. 271, 1913–1918. doi: 10.1001/jama.1994.03510480037031
Mikelsaar, M. (2011). Human microbial ecology: Lactobacilli, probiotics, selective decontamination. Anaerobe 17, 463–467. doi: 10.1016/j.anaerobe.2011.07.005
Naseribafrouei, A., Hestad, K., Avershina, E., Sekelja, M., Linløkken, A., Wilson, R., et al. (2014). Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26, 1155–1162. doi: 10.1111/nmo.12378
Ndegwa, S., and Nkansah, E. (2008). Vancomycin for C. difficile Pseudomembranous colitis: Guidelines and a Clinical Effectiveness Review. Ottawa, ON: CAN: Canadian Agency for Drugs and Technologies in Health, 2008.
NIH HMP Working Group, Peterson, J., Garges, S., Giovanni, M., McInnes, P., Wang, L., et al. (2009). The NIH human microbiome project. Genome Res. 19, 2317–2323. doi: 10.1101/gr.096651.109
Nikfar, S., Rahimi, R., Rahimi, F., Derakhshani, S., and Abdollahi, M. (2008). Efficacy of probiotics in irritable bowel syndrome: a meta-analysis of randomized, controlled trials. Dis. Colon. Rectum 5, 1775–1780. doi: 10.1007/s10350-008-9335-z
Omar, J. M., Chan, Y., Jones, M. L., Prakash, S., and Jones, P. J. H. (2013). Lactobacillus fermentum and Lactobacillus amylovorus as probiotics alter body adiposity and gut microflora in healthy persons. J. Funct. Foods 5, 116–123. doi: 10.1016/j.jff.2012.09.001
Osmanagaoglu, O., Kiran, F., and Ataoglu, H. (2010). Evaluation of in vitro probiotic potential of Pediococcus pentosaceus OZF isolated from human breast milk. Probiotics Antimicrobl. Prot. 2, 162–174. doi: 10.1007/s12602-010-9050-7
Owen, L., Reinders, M., Narramore, R., Marsh, A. M. R., Gar Lui, F., Baron, R., et al. (2014). A double blind, placebo controlled, randomised pilot trial examining the effects of probiotic administration on mood and cognitive function. Proceed Nutr. Soc. 73:e29. doi: 10.1017/S0029665114000433
Pawlowski, S. W., Calabrese, G., Kolling, G. L., Freire, R., Alcantara Warren, C., Liu, B., et al. (2010). Murine Model of Clostridium difficile infection with aged gnotobiotic C57BL/6 Mice and a BI/NAP1 strain. J. Infect. Dis. 202, 1708–1712. doi: 10.1086/657086
Pepin, J., Valiquette, L., Alary, M. E., Villemure, P., Pelletier, A., Forget, K., et al. (2004). Clostridium difficile - associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171, 466–472. doi: 10.1503/cmaj.1041104
Pepin, J., Valiquette, L., and Cossette, B. (2005). Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 173, 1037–1042. doi: 10.1503/cmaj.050978
Pflughoeft, K. J., and Versalovic, J. (2012). Human microbiome in health and disease. Annu. Rev. Pathol. 7, 99–122. doi: 10.1146/annurev-pathol-011811-132421
Pieniz, S., Andreazza, R., Anghinoni, T., Camargo, F., and Brandelli, A. (2014). Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 37, 251–256. doi: 10.1016/j.foodcont.2013.09.055
Pineton de Chambrun, G., Neut, C., Chau, A., Cazaubiel, M., Pelerin, F., Justen, P., et al. (2015). A randomized clinical trial of Saccharomyces cerevisiae versus placebo in the irritable bowel syndrome. Dig. Liver Dis. 47, 119–124. doi: 10.1016/j.dld.2014.11.007
Pitkala, K. H., Strandberg, T. E., Finne Soveri, U. H., Ouwehand, A. C., Poussa, T., and Salminen, S. (2007). Fermented cereal with specific bifidobacteria normalizes bowel movements in elderly nursing home residents. A randomized controlled trial. J. Nutr. Health Agents 11, 305–311.
Psani, M., and Kotzekidou, P. (2006). Technological characteristics of yeast strains and their potential as starter adjuncts in Greek-style black olive fermentation. World J. Microbiol. Biotechnol. 22, 1329–1336. doi: 10.1007/s11274-006-9180-y
Raccach, M. (2014). “Pediococcus,” in Encyclopedia of Food Microbiology, 2nd Edn, eds C. A. Batt and M. L. Tortorello (London: Academic Press Inc.), 1–5. doi: 10.1016/B978-0-12-384730-0.00247-0
Radulovic, Z., Petrovic, T., and Bulajic, S. (2012). “Antibiotic susceptibility of probiotic bacteria, antibiotic resistant bacteria,” A Continuous Challenge in the New Millennium, ed. M. Pana (Belgrade: University of Belgrade, Faculty of Agriculture, Republic of Serbia), 550–576.
Rehaiem, A., Belgacem, Z., Ben Edalatian, M. R., Martínez, B., Rodríguez, A., Manai, M., et al. (2014). Assessment of potential probiotic properties and multiple bacteriocin encoding-genes of the technological performing strain Enterococcus faecium MMRA. Food Control. 37, 343–350. doi: 10.1016/j.foodcont.2013.09.044
Rubinstein, E. (2001). Comparative safety of the different macrolides. Int. J. Antimicrob. Agents 18(Suppl. 1), 71–76. doi: 10.1016/S0924-8579(01)00397-1
Samanta, M., Sarkar, M., Ghosh, P., Ghosh, J. K., Sinha, M. K., and Chatterjee, S. (2009). Prophylactic probiotics for prevention of necrotizing enterocolitis in very low birth weight newborns. J. Trop. Pediatr. 55, 128–131. doi: 10.1093/tropej/fmn091
Sanz, Y. (2007). Ecological and functional implications of the acid-adaptation ability of Bifidobacterium: a way of selecting improved probiotic strains. Int. Dairy J. 17, 1284–1289. doi: 10.1016/j.idairyj.2007.01.016
Sapkota, A. R., Lefferts, L. Y., Mckenzie, S., and Walker, P. (2007). What do we feed to food-production animals? A review of animal feed ingredients and their potential impacts on human health. Environ. Health Perspect. 115, 663–670. doi: 10.1289/ehp.9760
Sarker, S. A., Sultana, S., Fuchs, G. J., Alam, N. H., Azim, T., Brüssow, H., et al. (2005). Lactobacillus paracasei strain ST11 has no effect on rotavirus but ameliorates the outcome of nonrotavirus diarrhea in children from Bangladesh. Pediatrics 116:e221–e228. doi: 10.1542/peds.2004-2334
Scharek, L., Hartmann, L., Heinevetter, L., and Blaut, M. (2000). Bifidobacterium adolescentis modulates the specific immune response to another human gut bacterium, Bacteroides thetaiotaomicron, in gnotobiotic rats. Immunobiology 202, 429–441. doi: 10.1016/S0171-2985(00)80102-3
Shan, L. S., Hou, P., Wang, Z. J., Liu, F. R., Chen, N., Shu, L. H., et al. (2013). Prevention and treatment of diarrhoea with Saccharomyces boulardii in children with acute lower respiratory tract infections. Benef. Microbes 4, 329–334. doi: 10.3920/BM2013.0008
Sharma, S., Chaturvedi, J., Chaudhari, B. P., Singh, R. L., and Kakkar, P. (2012). Probiotic Enterococcus lactis IITRHR1 protects against acetaminophen-induced hepatotoxicity. Nutrition 28, 173–181. doi: 10.1016/j.nut.2011.02.012
Shiba, T., Aiba, Y., Ishikawa, H., Ushiyama, A., Takagi, A., Mine, T., et al. (2003). The suppressive effect of bifidobacteria on Bacteroides vulgatus, a putative pathogenic microbe in inflammatory bowel disease. Microbiol. Immunol. 47, 371–378. doi: 10.1111/j.1348-0421.2003.tb03368.x
Shimakawa, Y., Matsubara, S., Yuki, N., Ikeda, M., and Ishikawa, F. (2003). Evaluation of Bifidobacterium breve strain Yakult-fermented soymilk as a probiotic food. Int. J. Food Microbiol. 81, 131–136. doi: 10.1016/S0168-1605(02)00224-6
Shin, N. R., Lee, J. C., Lee, H. Y., Kim, M. S., Whon, T. W., Lee, M. S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735. doi: 10.1136/gutjnl-2012-303839
Shirasawa, Y., Shibahara-Sone, H., Lino, T., and Ishikawa, F. (2010). Bifidobacterium bifidum BF-1 suppresses Helicobacter pylori-induced genes in human epithelial cells. J. Dairy Sci. 93, 4526–4534. doi: 10.3168/jds.2010-3274
Shornikova, A. V., Casas, I. A., Isolauri, E., Mykkanen, H., and Vesikari, T. (1997). Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J. Ped. Gastroenterol. Nutr. 24, 399–404. doi: 10.1097/00005176-199704000-00008
Sinn, D. H., Song, J. H., Kim, H. J., Lee, J. H., Son, H. J., Chang, D. K., et al. (2008). Therapeutic effect of Lactobacillus acidophilus-SDC 2012, 2013 in patients with irritable bowel syndrome. Digest Dis. Sci. 53, 2714–2718. doi: 10.1007/s10620-007-0196-4
Surawicz, C. M. (2003). Probiotics, antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in humans. Best Pract. Res. Clin. Gastroenterol. 17, 775–783. doi: 10.1016/S1521-6918(03)00054-4
Surawicz, C. M., Mcfarland, L. V., Greenberg, R. N., Rubin, M., Fekety, R., Mulligan, M. E., et al. (2000). The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin. Infect. Dis. 31, 1012–1017. doi: 10.1086/318130
Swidsinski, A., Loening-Baucke, V., Vaneechoutte, M., and Doerffel, Y. (2008). Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Infl. Bowel. Dis. 14, 147–161. doi: 10.1002/ibd.20330
Szajewska, H., Ruszczynski, M., and Radzikowski, A. (2006). Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J. Pediatr. 149, 367–372. doi: 10.1016/j.jpeds.2006.04.053
Tambyah, P. A., Marx, J. A., and Maki, D. G. (2004). Nosocomial infection with vancomycin-dependent enterococci. Emerg. Infect. Dis. 10, 1277–1281. doi: 10.3201/eid1007.030993
Temmerman, R., Pot, B., Huys, G., and Swings, J. (2003). Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 81, 1–10. doi: 10.1016/S0168-1605(02)00162-9
Thygesen, J. B., Glerup, H., and Tarp, B. (2012). Saccharomyces boulardii fungemia caused by treatment with a probioticum. BMJ Case Rep. 2012. doi: 10.1136/bcr.06.2011.4412
Toomey, N., Monaghan, A., Fanning, S., and Bolton, D. J. (2009). Assessment of antimicrobial resistance transfer between lactic acid bacteria and potential foodborne pathogens using in vitro methods and mating in a food matrix. Foodborne Pathog. Dis. 6, 925–933. doi: 10.1089/fpd.2009.0278
Tsai, C.-C., Lai, C.-H., Yu, B., and Tsen, H.-Y. (2008). Use of specific primers based on the 16S-23S internal transcribed spacer (ITS) region for the screening Bifidobacterium adolescentis in yogurt products and human stool samples. Anaerobe 14, 219–223. doi: 10.1016/j.anaerobe.2008.05.001
Tuohy, K. M., Conterno, L., Gasperotti, M., and Viola, R. (2012). Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J. Agric. Food Chem. 60, 8776–8782. doi: 10.1021/jf2053959
Turpin, W., Humblot, C., Thomas, M., and Guyot, J.-P. (2010). Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int. J. Food Microbiol. 143, 87–102. doi: 10.1016/j.ijfoodmicro.2010.07.032
Turroni, F., Van Sinderen, D., and Ventura, M. (2011). Genomics and ecological overview of the genus Bifidobacterium. Int. J. Food Microbiol. 149, 37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010
van Passel, M. W., Kant, R., Zoetendal, E. G., Plugge, C. M., Derrien, M., Malfatti, S. A., et al. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6:e16876. doi: 10.1371/journal.pone.0016876
Vanderhoof, J. A., Whitney, D. B., Antonson, D. L., Hanner, T. L., Lupo, J. V., and Young, R. J. (1999). Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J. Pediatr. 135, 564–568. doi: 10.1016/S0022-3476(99)70053-3
Varankovich, N. V., Khan, N. H., Nickerson, M. T., Kalmokoff, M., and Korber, D. R. (2015). Evaluation of pea protein–polysaccharide matrices for encapsulation of acid-sensitive bacteria. Food Res. Int. 70, 118–124. doi: 10.1016/j.foodres.2015.01.028
Ventura, M., O’Connell Motherway, M., Leahy, S., Moreno-Munoz, J. A., Fitzgerald Gerald, F., and van Sinderen, D. (2007). From bacterial genome to functionality; case Bifidobacteria. Int. J. Food Microbiol. 120, 2–12. doi: 10.1016/j.ijfoodmicro.2007.06.011
Ventura, M., Turroni, F., Motherway, M. O. C., Macsharry, J., and Van Sinderen, D. (2012). Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20, 467–476. doi: 10.1016/j.tim.2012.07.002
Verdú, E. F., Bercik, P., and Collins, S. M. (2009). Effect of probiotics on gastrointestinal function: evidence from animal models. Therap. Adv. Gastroenterol. 2(Suppl. 4), 31–35. doi: 10.1177/1756283X09337645
Verdú, E. F., Bercik, P., Huang, X. X., Lu, J., Al-Mutawaly, N., Sakai, H., et al. (2008). The role of luminal factors in the recovery of gastric function and behavioral changes after chronic Helicobacter pylori infection. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G664–G670. doi: 10.1152/ajpgi.90316.2008
Videlock, E. J., and Cremonini, F. (2012). Meta-analysis: probiotics in antibiotic-associated diarrhoea. Aliment. Pharmacol. Ther. 35, 1355–1369. doi: 10.1111/j.1365-2036.2012.05104.x
Wallace, T. C., Guarner, F., Madsen, K., Cabana, M. D., Gibson, G., Hentges, E., et al. (2011). Human gut microbiota and its relationship to health and disease. Nutr. Rev. 69, 392–403. doi: 10.1111/j.1753-4887.2011.00402.x
Wang, J., Korber, D. R., Low, N. H., and Nickerson, M. T. (2014). Entrapment, survival and release of Bifidobacterium adolescentis within chickpea protein-based microcapsules. Food Res. Int. 55, 20–27. doi: 10.1016/j.foodres.2013.09.018
Wang, J., Korber, D. R., Low, N. H., and Nickerson, M. T. (2015a). Encapsulation of Bifidobacterium adolescentis with legume proteins and its survival under stimulated gastric conditions and during storage within commercial fruit juices. Food Sci. Biotechnol. 24, 383–391. doi: 10.1007/s10068-015-0051-x
Wang, J., Tang, H., Zhang, C., Zhao, Y., Derrien, M., Rocher, E., et al. (2015b). Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 9, 1–15. doi: 10.1038/ismej.2014.99
Wang, M., Molin, G., Ahrné, S., Adawi, D., and Jeppsson, B. (2007). High proportions of proinflammatory bacteria on the colonic mucosa in a young patient with ulcerative colitis as revealed by cloning and sequencing of 16S rRNA genes. Dig. Dis. Sci. 52, 620–627. doi: 10.1007/s10620-006-9461-1
Wei, Y.-X., Zhang, Z.-Y., Liu, C., Malakar, P. K., and Guo, X.-K. (2012). Safety assessment of Bifidobacterium longum JDM301 based on complete genome sequences. World J. Gastroenterol. 18, 479–488. doi: 10.3748/wjg.v18.i5.479
Wen, K., Li, G., Bui, T., Liu, F., Li, Y., Kocher, J., et al. (2012). High dose and low dose Lactobacillus acidophilus exerted differential immune modulating effects on T cell immune responses induced by an oral human rotavirus vaccine in gnotobiotic pigs. Vaccine 30, 1198–1207. doi: 10.1016/j.vaccine.2011.11.107
Werner, G., Coque, T. M., Hammerum, A. M., Hope, R., Hryniewicz, W., Johnson, A., et al. (2008). Emergence and spread of vancomycin resistance among enterococci in Europe. Euro. Surveill. 13, Pii: 19046.
Whorwell, P. J., Altringer, L., Morel, J., Bond, Y., Charbonneau, D., O’Mahony, L., et al. (2006). Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am. J. Gastroenterol. 101, 1581–1590. doi: 10.1111/j.1572-0241.2006.00734.x
Wood, K. A., Korber, D. R., Nickerson, M. T., and Low, N. H. (2012). Oligosaccharide utilization by and encapsulation of Bifidobacterium adolescentis in alginate and/or protein capsules. Int. J. Probiotics Prebiotics 7, 153–164.
Wozniak, R. A., and Waldor, M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. doi: 10.1038/nrmicro2382
Wrzosek, L. M. S., Noordine, M. L., Bouet, S., Joncquel Chevalier-Curt, M., Robert, V., Philippe, C., et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013:11. doi: 10.1186/1741-7007-11-61
Wunderlich, P. F., Braun, L., Fumagalli, I., D’Apuzzo, V., Heim, F., Karly, M., et al. (1989). Double-blind report on the efficacy of lactic acid-producing Enterococcus SF68 in the prevention of antibiotic-associated diarrhoea and in the treatment of acute diarrhoea. J. Int. Med. Res. 17,333–338.
Keywords: probiotic, human gut, gastrointestinal disorders, antibiotic resistance, gut microbiota
Citation: Varankovich NV, Nickerson MT and Korber DR (2015) Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front. Microbiol. 6:685. doi: 10.3389/fmicb.2015.00685
Received: 09 January 2015; Accepted: 22 June 2015;
Published: 14 July 2015.
Edited by:
Maria De Angelis, University of Bari Aldo Moro, ItalyReviewed by:
Maria Guadalupe Vizoso Pinto, National University of Tucumán, ArgentinaAldo Corsetti, University of Teramo, Italy
Copyright © 2015 Varankovich, Nickerson and Korber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darren R. Korber, Department of Food and Bioproduct Sciences, University of Saskatchewan, 51 Campus Drive, Saskatoon, SK S7N 5A8, Canada, darren.korber@usask.ca
 Natallia V. Varankovich
Natallia V. Varankovich Michael T. Nickerson
Michael T. Nickerson Darren R. Korber
Darren R. Korber