- 1Institute of Biosciences, University Putra Malaysia, Selangor, Malaysia
- 2School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 4Department of Pharmacognosy, Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran
Screening of lactic acid bacteria (LAB) isolated from ewe colostrum led to the identification and isolation of Enterococcus faecium CM33 with interesting features like high survival rates under acidic or bile salts condition, high tolerance for the simulated gastrointestinal condition, and high adhesive potential to Caco-2 cells. According the inhibition of pathogen adhesion test results, this strain can reduce more than 50% adhesion capacity of Escherichia coli, Shigella flexneri, Klebsiella pneumoniae, Listeria monocytogenes, and Staphylococcus aureus to Caco-2 cells. Based on the antibiotic sensitivity test findings, E. faecium CM33 was susceptible to gentamycin, vancomycin, erythromycin, ampicillin, penicillin, tetracycline, and rifampicin, but resistant to chloramphenicol, clindamycin, and kanamycin. Upon assessment of the virulence determinants for E. faecium CM33, this strain was negative for all tested virulence genes. Furthermore, the genome of this strain was evaluated for the incidence of the known enterocin genes by specific PCR amplification and discovered the genes encoding enterocins A, 31, X, and Q. Based on this study findings, the strain E. faecium CM33 can be considered as a valuable nutraceutical and can be introduced as a new potential probiotic.
Introduction
The health promoting effects of probiotic bacteria are strain-specific, hence their discrimination and identification is very important and crucial under strain level by applying an efficient and valid strain-identification techniques such as 16S rRNA sequencing. Besides, some identification methods can be utilized for assessment on the safety aspect and ecological properties of probiotics (Pereira and Gibson, 2002; Senok, 2009). The choice of appropriate probiotics is the most important base for enhancing the bio-therapeutic achievement in nutraceutical/pharmaceutical products. Several important characters of bacteria used as probiotics are the ability to survive in gastric conditions and adhere to the host intestinal cells (Kirjavainen et al., 1998; Dunne et al., 2001). However, in vitro procedures for choice of promising probiotics are necessary because in vivo methods are costly and need agreement by ethical committees.
Caco-2 cells have been effectively applied to find the mechanism of cellular adhesion of non-pathogenic lactic acid bacteria (LAB; Tuomola and Salminen, 1998). It has also been explained that enterococci produce antimicrobial substances such as bacteriocins, hydrogen peroxide, and lactic acid (Sparo et al., 2006; Franz et al., 2007), which could elucidate their ability against several pathogens such as Staphylococcus aureus and Listeria monocytogenes. The connection of Enterococcus bacteria with human diseases has raised concern regarding their use as probiotic microorganisms (Franz et al., 2011). Several enterococcal strains are able to have antiviral activity (Wang et al., 2013), beneficial effects in anti-tumoral protective responses (Thirabunyanon and Hongwittayakorn, 2013) and can restore the microbiota balance in antibiotic-induced dysbiosis (Tarasova et al., 2010). Enterococcus bacteria, utilized in fermentation of vegetable products, meat, and dairy, are the reason of clinical infections. Therefore, it is vital to evaluate the virulence genes as well the antibiotic resistance profile.
Although the safety aspect of this genus is still blurred, members of enterococci are now engaged as probiotic bacteria to improve the immunity and human health (Brandão et al., 2010). The capability of these bacteria to make enterocins is remarkable and can be applied as food bio-preservatives (Khan et al., 2010). Bacteriocins are small, ribosomally synthesized and heat-stable peptides that apply antimicrobial activity against pathogenic bacteria or food spoilage, including Staphylococcus aureus, Clostridium spp. Bacillus spp., L. monocytogenes, and Campylobacter spp. (Nes et al., 2007; Line et al., 2008). Franz et al. (2007) categorized bacteriocins produced by Enterococcus bacteria into four different classes: Class I is lantibiotic enterocins such as cytolysin (CylLL/S; Gilmore et al., 1994); Class II is small, non-lantibiotic enterocins; Class III is cyclic enterocins for instance enterocin AS-48 (Eijsink et al., 2002); and Class IV is large proteins like enterolysin A (Hickey et al., 2003). There are three subclasses within class II including subclass IIa such as enterocin A and P (Ent A and Ent P), which are pediocin-like bacteriocins; subclass IIb like enterocin L50 and Q (Ent L50A/B and Ent Q), which required two peptides for full antimicrobial activity and subclass IIc like enterocin B (Ent B), which are other linear and non-pediocin-type enterocins (Casaus et al., 1997).
Although there are rising concern about the safety of Enterococcus bacteria because of their relationship with several human infections (Brandão et al., 2010), a lot of works specify that enterococci play a key function in the improvement of the sensory properties of fermented foods like olives, sausages and cheese (Moreno et al., 2006). Carlos et al. (2010) elucidated a lot of virulence factors and their actions in Enterococcus bacteria (Carlos et al., 2010). These include the cylA which is in charge of the transportation and activation of the cytolisin; cylB and cylM involved in the post-translational modification; esp gene in charge of a cell wall protein involved in immune evasion; agg encodes an aggregation protein which is responsible for adherence to eukaryotic cells; gelE is in charge of toxin production that hydrolyzes gelatine and other compounds, and finally cpd, ccf, and cad genes encode sex pheromones, which are in charge of facilitating conjugation.
Because of the high potential of Enterococcus bacteria in health and food, the purpose of current study was to find out the antimicrobial activity and the presence of bacteriocin structural genes and virulence factors in Enterococcus faecium CM33 isolated from ewe colostrum to assess the potential probiotic properties and safety aspect of use of this strain.
Materials and Methods
Bacteria Isolation
Enterococcus faecium CM33 was isolated from ewe colostrum by culturing in de Man Rogosa and Sharpe broth (MRS) and following incubation of 50 μl of cultured medium onto MRS agar supplemented with 6.5% (w/v) NaCl (MRS-NaCl) for 18 h at 37°C in aerobic conditions. After enrichment, five passages by isolated colony, the pure bacterium isolate was obtained.
PCR Amplification of Isolate
The amplification of 16S rRNA gene (1500 bp) of the isolate was performed by the primers (F 5′-AGAGTTTGATC CTGGCTCAG-3′ and R 5′-GGCTGCTGGCACGTAGTTAG-3′), which those primarily used by Kullen et al. (2000). The amplification reaction was performed in a thermocycler (Thermo Electron Corporation, Waltham, MA, USA) with the following thermocycle program: Four min at 96°C as an initial denaturation; 30 s at 96°C as a denaturation for 30 cycles; 30 s at 48°C as an annealing and 45 s at 72 C as an extension, with a final extension step for 4 min at 72°C. The total volume of reaction was 50 μl. The PCR product was visualized through 0.8% (w/v) agarose gel (Sigma Chemical Co., Poole, UK) electrophoresis by using ethidium bromide staining (Kullen et al., 2000). PCR product was sequenced by the Macrogen DNA Sequencing Service (Korea). The sequence was interrogated through the BLAST search of the NCBI database. The isolate was identified on account of the highest matching score.
Effects of Heat, pH, and Hydrolytic Enzymes
This strain was cultured in MRS broth for 18 h at 37°C. After 10 min centrifugation at 12000 g, cells were removed and the cell free supernatant (metabolites) was utilized for additional characterization. To determine the effect of pH on the metabolites action, pH of the crude supernatant was adjusted in the range between 1.5 and 9 by adding sterile 1 M HCl or 1 M NaOH. Sample was incubated at room temperature for 4 h. Thermostability of metabolites was determined at diverse temperatures. Crude supernatant was autoclaved at 121°C for 15 min and then incubated at different temperatures (4, -20, and -70C) for 24 h, 48 h, 2 weeks, and 1 month.
The sensitivity of cell free supernatant toward different proteolytic enzymes, amylase, and catalase was investigated. One mg/ml of enzymes including proteinase K, pronase E, trypsin, and chymotrypsin were prepared in 15 mM sodium phosphate buffer (pH 7.2) and pepsin was prepared in glycine buffer pH 2. Before incubating for 2 h at 37°C, proteases at a final concentration of 0.1 mg/ml were added. The crude cell free supernatant was treated with catalase at 0.1 mg/ml to remove potential inhibitory effects because of production of hydrogen peroxide.
Targeting Bacteriocin Genes
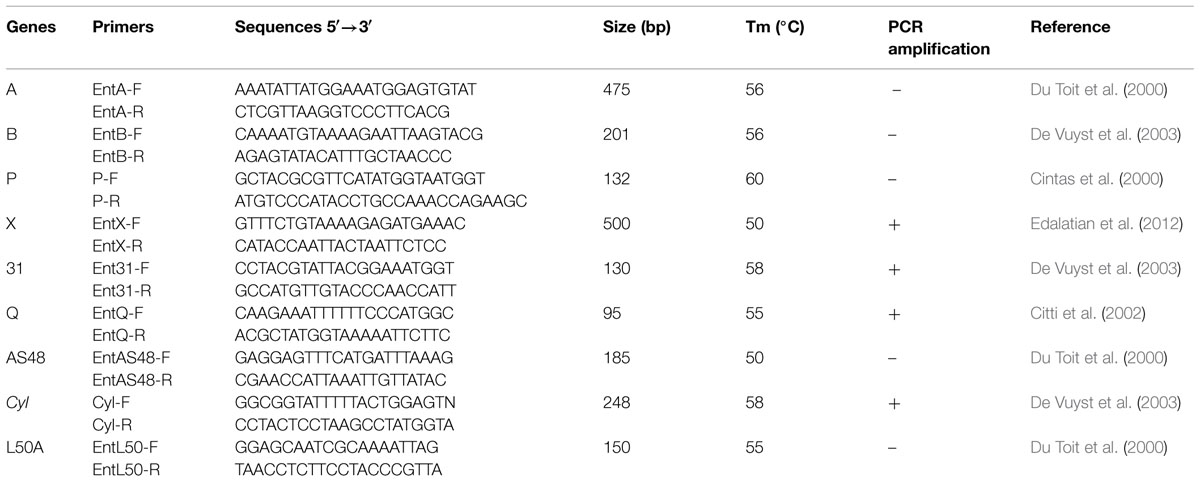
To detect genes encoding the known enterocins, genomic DNA of strain CM33 was used as template. PCR were performed in a Hybrid PCR Sprint thermocycler (Thermo Electron Corporation, Waltham, MA, USA) in 20 ml reaction mixture (10 pmoles of each primer, 1 mL DNA template, and 1.25 U Taq polymerase). The PCR conditions for enterocin-encoded genes are illustrated in Table 1. The PCR products were visualized by electrophoresis in 1.5% agarose gels.
Bile Tolerance Test
The method previously described by Pereira and Gibson (2002) was used to measure the viability of strain in bile salt. The isolate was grown in MRS medium supplemented with 0.5% (w/v) oxgall then was incubated for 4 h at 37°C. To determine of cell count, aliquots were plated onto MRS-agar medium at times 0, 1, 3, and 4 h. Control tube was without bile salt (Pereira and Gibson, 2002).
Survivability in Simulated In Vitro Digestion
Simulated in vitro digestion was carried out to evaluate the survival rate of strain CM33 in similar condition of the human gastrointestinal tract. To simulate the stomach digestion, samples (pH 3.0) were treated with 5% (w/v) pepsin (Sigma Chemical Co., Poole, UK) then were incubated for 120 min at 37°C with gentle agitation. The intestinal digestion was simulated by inoculating solutions of 0.1% (w/v) pancreatin (Sigma Chemical Co., Poole, UK) and 0.3% (w/v) bile oxgall. The samples were incubated for 180 min at 37°C with gentle shaking. Samples were removed before and after gastric and intestinal digestion to measure cell count. The serially diluted aliquots were cultured on MRS agar medium and were incubated for 24 h under anaerobic conditions (Seiquer et al., 2001).
Adhesion Ability to Caco-2 Cells
Caco-2, the colon adenocarcinoma cell line, was used to evaluate the adhesion ability of E. faecium CM33 to human epithelial cells. RPMI medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin mixture (Sigma Chemical Co., Poole, UK) was used to culture human colon cancer cells. Cells were seeded on 24-well tissue culture plates and incubated at 37°C in 5% CO2 in a humidified atmosphere. Before the adhesion assay, fresh antibiotic-free RPMI was used to wash wells, which have a monolayer of Caco-2 cells. Then, each well was inoculated with 1 × 107 cfu/ml of E. faecium in a total volume of 1 ml and incubated at 37°C for 3 h in an atmosphere of 5% (v/v) CO2. The wells were washed thrice with a pre-warmed phosphate buffer saline (Sigma Chemical Co., Poole, UK) to remove non-attached cells of E. faecium. To detach the cells, 1 ml of 1% Triton X-100 (Sigma Chemical Co., Poole, UK) was added to each well with gently stirring for 5–10 min and then the viable human colon cancer cells, Caco-2, were counted. Finally, bacteria and Caco-2 cells were cultured onto MRS agar by the pure plate method and incubated at 37°C anaerobically. The total number of bacteria attached to viable Caco-2 cells expressed as bacterial adhesion and carried out in triplicate.
Inhibition of Pathogen Adhesion to Caco-2 cells
Caco-2, human colon cells, were used to assess the effect of E. faecium treatment on the pathogen interaction with cells. E. faecium and pathogens (Escherichia coli, S. flexneri, K. pneumonia, and S. aureus) were cultured in MRS and LB, respectively. Overnight culture of E. faecium and pathogens were harvested and washed thrice with phosphate buffer saline. Bacteria were diluted in antibiotic-free RPMI and 0.5 ml/well (108 CFU per well) of E. faecium suspension was inoculated to 24-well tissue culture plates. To evaluate the capability of E. faecium to restrain pathogenic bacteria adhesion to Caco-2 cells, simultaneous addition of 108 CFU per well of tested pathogens was performed separately. Plates were incubated 1 h at 37°C and washed twice with sterile PBS then 1% (v/v) Triton X-100 (Sigma Chemical Co., Poole, UK) in deionized water for 5 min was used to lyse the cell-associated pathogens. To find out the number of viable cell-linked bacteria by colony forming counts, the suitable dilutions of the lysate were placed on LB agar. The inhibition analysis was calculated in three independent trials and each experiment was carried out in triplicate.
Antimicrobial Activity
To determine and identify inhibitory substances of metabolites produced by E. faecium CM33, well-diffusion method was performed (Hechard et al., 1990). Overnight cultures of the indicator strains were cultured in MRS agar at 37°C for 18 h. 50 μl of filtered crude supernatant of CM33 were dropped to each well (5 mm) into agar plates. The suspensions were inoculated to the wells and permitted to diffuse for 4 h at room temperature into MRS agar. The inhibition zone produced around the wells was determined after 24 h incubation at the optimum growth temperature of the indicator strains. The diameter of the produced inhibition zones around the wells was determined by digital caliper. These diameters were classified to four different groups including no inhibition (–), weak (+), good (++), and strong (+++). All trials were carried out in triplicate.
PCR Detection of Virulence Factors in Enterococci
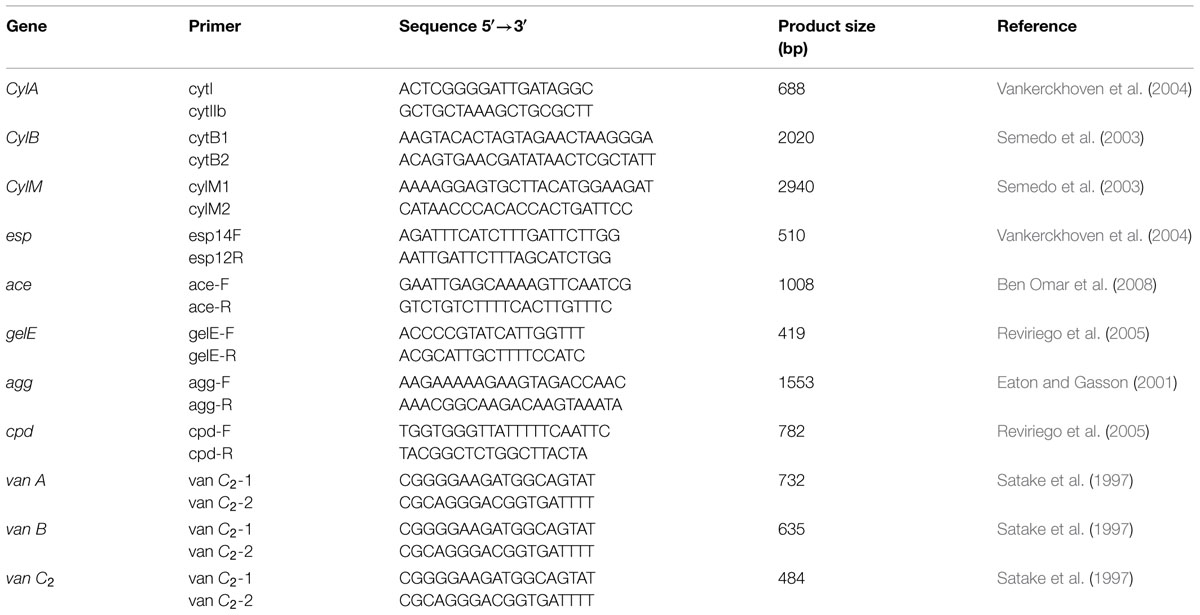
Detection of genes encoding potential virulence factors in E. faecium CM33 was performed by PCR. Total bacterial DNA used as template in PCR reactions was isolated by the alkaline lysis method (Baele et al., 2001). Virulence genes evaluated in this study include: cytolysin (CylA, CylB, CylM; Semedo et al., 2003; Vankerckhoven et al., 2004), enterococcal surface protein (esp; Vankerckhoven et al., 2004), collagen-binding protein (ace; Omar et al., 2004), gelatinase (gelE), aggregation substance (agg; Eaton and Gasson, 2001; Ahmed et al., 2012), sex pheromone peptides (cpd; Abriouel et al., 2008), cell-wall anchored collagen adhesion (acm; Cariolato et al., 2008; Nallapareddy et al., 2008), and van A, van B, van C2 (Satake et al., 1997). E. faecium ATCC 51299 and ATCC 29212 were controls for this test. PCR was used to detect of genes encoding these factors using several primers (Table 2) which have been described by Eaton and Gasson (2001).
Interpretation of Results
Data were analyzed using SPSS 19.0 software (SPSS Inc., an IBM Company, Schaumburg, IL, USA). One-way ANOVA followed by multiple mean comparisons Duncan’s test were used to assess statistical differences in multiple groups. Values are mean ± SE and p ≤ 0.05 is considered statistically significant.
Results
Bacteria Isolation
Screening of different dairy products like colostrum, yogurt, curd, cheese, and yogurt drink led to the isolation of E. faecium CM33 from ewe colostrum. After DNA extraction, the 16S rRNA fragments were amplified by PCR. According to 16S rRNA identification, 50 of the isolated bacteria were classified into three major groups of LAB, namely, 14% enterococci, 66% lactobacilli, and 20% lactococci. The isolates classified as LAB were also separated and identified by sequencing. After sequencing, the Enterococcus strains were categorized into E. lactis, E. hirae, E. avium, E. durans, E. faecalis, and E. faecium. The Lactobacillus strains representing 66% were classified into L. acidophilus and L. plantarum. The Lactococcus species were classified only into one species (Lactococcus lactis). Probiotic characterization of these isolates showed that E. faecium CM33 has the best properties such as tolerance to acid and bile, antimicrobial activity and antibiotic resistance thus it was selected for further analysis.
Effects of pH, Heat, and Hydrolytic Enzymes on E. faecium Bacteriocin
The stability and activity of E. faecium CM33 bacteriocins has been shown in Table 3. Bacteriocins were active over broad ranges of pH from 1.5 to 9. It was also stable after autoclaving at 121°C for 15 min. The treatment of cell free supernatant of E. faecium CM33 with amylase, catalase, and proteolytic enzymes showed that amylase and catalase enzymes do not influence on the bacteriocin activity, while proteolysis enzymes affect on the bacteriocin activity after 2 h.

TABLE 3. Effects of pH, heat, and enzymes (concentration 1 mg/ml) on the antibacterial activity present in E. faecium CM33 culture metabolites.
PCR Analysis of Bacteriocin Genes
PCR amplification using specific primers as shown in Table 1 was used to screen DNA of E. faecium CM33 for the existence of the known enterocin genes. This study focused on enterocin genes including Cyl, A, P, 31, Q, X, L50A, and AS-48. E. faecium CM33 showed four bands: 95, 130, 248, and 500 bp corresponding to the specific amplification of enterocins Q, 31, Cyl, and X, respectively.
Survivability in Bile Salt and Simulated Digestion
Enterococcus faecium CM33 was incubated in 0.3% bile salts for 4 h. It showed the survival rate of >93% after this incubation period. One of the most desirable features needed for probiotics is the ability to survive in the GI tract. For further analysis, E. faecium CM33 was evaluated using simulated digestion test. This test revealed that this strain had 57.6% digestion survivability.
Adhesion Assay to Caco-2 Cells
The strain was tested further for its capability to colonize Caco-2 cells. E. faecium CM33 showed the acceptable adhesion ability with an adhesion value of 32 × 104 cfu/ml. E. faecium ATCC 29212, which used as a control to compare their adhesion ability, could not significantly adhere to Caco-2 cells.
Inhibition of Pathogens Adhesion
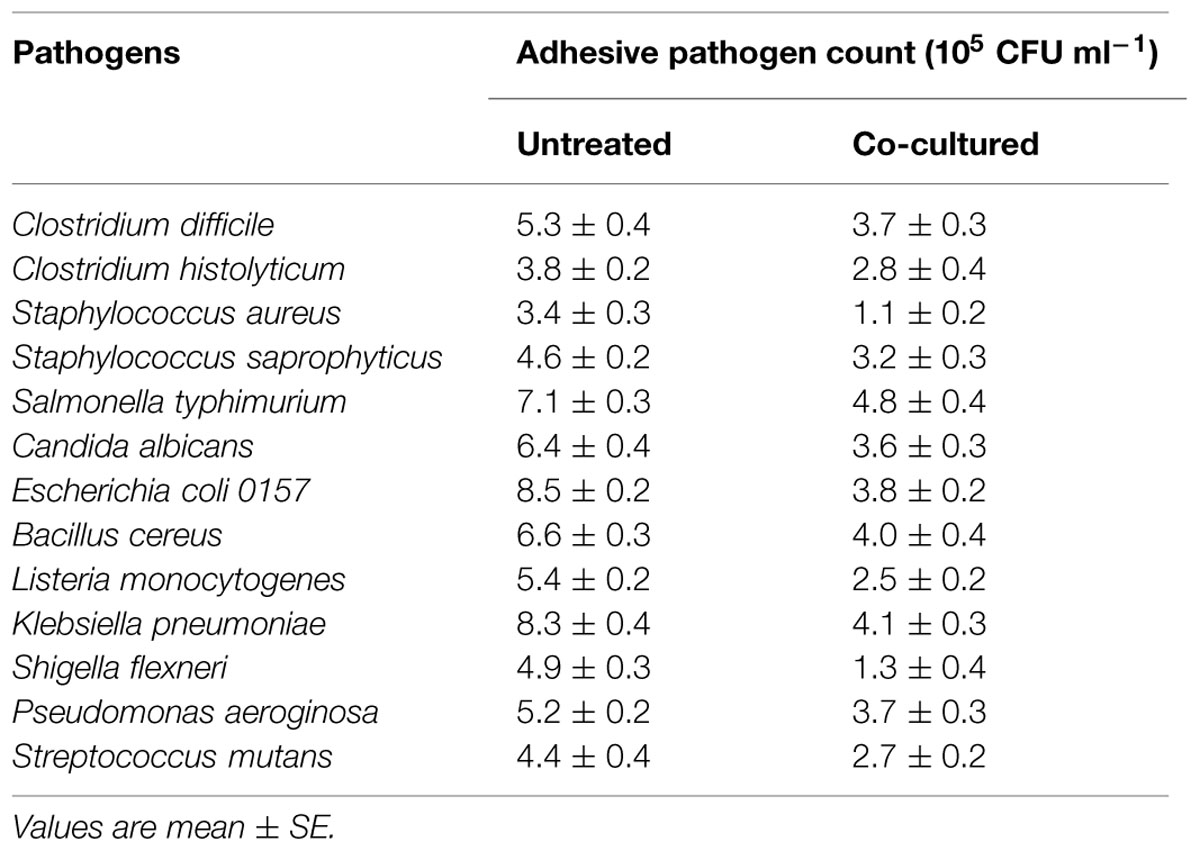
In the presence of E. faecium CM33, the inhibition of pathogens adhesion was presented in Table 4. All the adherent pathogens were clearly reduced by co-culture with E. faecium. The reduction of E. coli, S. flexneri, K. pneumoniae, and S. aureus adhesion to Caco-2 cells was more than 50% added with E. faecium 108 CFU per well.
Antimicrobial Activity
The strain CM33 isolated from colostrum displayed antimicrobial activity against all the tested indicator microorganisms (Table 5). The most sensitive indicators were L. monocytogenes, followed by Candida albicans. On the contrary, Clostridium difficile, Clostridium histolyticum, Salmonella typhimurium, and Streptococcus mutans were the less sensitive indicator microorganisms.
Virulence Genes
The presence of virulence genes in genomic DNA of E. faecium CM33 was screened by PCR. Currently several enterococci are being used as probiotics but they should be carefully assessed to ensure that they are not pathogenic. E. faecium CM33 was found to be negative for all tested virulent genes (Table 6). In the case of van A, van B and van C2 genes, it is essential to note that this result associate with the lack of resistance to vancomycin.
Discussion
The food industry is requested to present increasingly functional foods containing beneficial components because of the great interest on health-oriented nutritional habits. Bacteria generally recognize as probiotic commonly belong to LAB. These microorganisms, as commensals in the human gastrointestinal tract, show a long history of use in foods and fermented products (Haghshenas et al., 2014a,b, 2015; Nami et al., 2014a,b,c). Enterococcus bacteria have various helpful functions in the dairy industry (Haghshenas et al., 2014c; Nami et al., 2014d). As starters, enterococci fulfill a considerable function in developing flavor progress and quality of cheeses. As probiotics, LAB are able to be used for healing of gut disorders in both humans and animals since these bacteria contribute to the intestinal health of the host by the development of gut microbial balance (Rehaiem et al., 2014). On the other hand, some requirements should be fulfilled to introduce a certain strain as a probiotic (Collins et al., 1998) and probiotic activities should be confirmed in well-made human studies. The aim of this paper was to evaluate safety and the potential beneficial properties of E. faecium CM33 isolated from ewe colostrum.
The most important characteristics required for microorganisms to be probiotics is their ability to survive while passing through the upper digestive tract to reach the large intestine, where their positive functions are predictable (Bezkorovainy, 2001; Tuomola et al., 2001; Nueno-Palop and Narbad, 2011). Probiotics can adhere to epithelial cells and should tolerate high acidic pH and high bile salt conditions in the gastric and intestinal fluid. Resistance to acid is of the popular properties used during the screening of potential probiotic strains. Another important mechanism in resisting intestinal pathogens and preventing diseases is the competition of probiotic bacteria with pathogens to adhere and colonize (Pascual et al., 2008). In this study, E. faecium CM33 isolated from ewe coloctrum dairy products was capable to survive both in bile salts and the gastric simulated digestion condition. This strain was further evaluated for their adhesion ability to Caco-2 cells. Data showed that E. faecium CM33 possessed an ability to resist the digestion conditions and was able to adhere to Caco-2 cells. These results show that E. faecium CM33 could be a suitable candidate to be as potential new probiotic.
The production of antimicrobial compounds like bacteriocins, organic acids, and hydrogen peroxide is of the well-designed properties applied to characterize probiotics (Rehaiem et al., 2014). Production of bacteriocins (enterocins) is the useful biotechnological trait of Enterococcus bacteria (Franz et al., 2011). The PCR results (Table 1; Figure 1) showed that E. faecium CM33 carried enterocins A, 31, Q, and X structural genes, while the other primers used in this test did not yield any visible band. Franz et al. (2007) classified enterocins A and 31 in the class IIa bacteriocins (pediocin-like bacteriocins). This group of bacteriocins is cationic and shows very effectual anti-Listeria activity and permeabilizes the cell membrane to kills target cells. Moreover, results showed that E. faecium CM33 has the greatest inhibitory function against L. monocytogenes. This fact could be described by the capability of Enterococcus bacteria to obtain and replace genetic information by conjugation (Brandão et al., 2010), not only between strains of Enterococcus genus, but also with other genera (Strompfová et al., 2008).
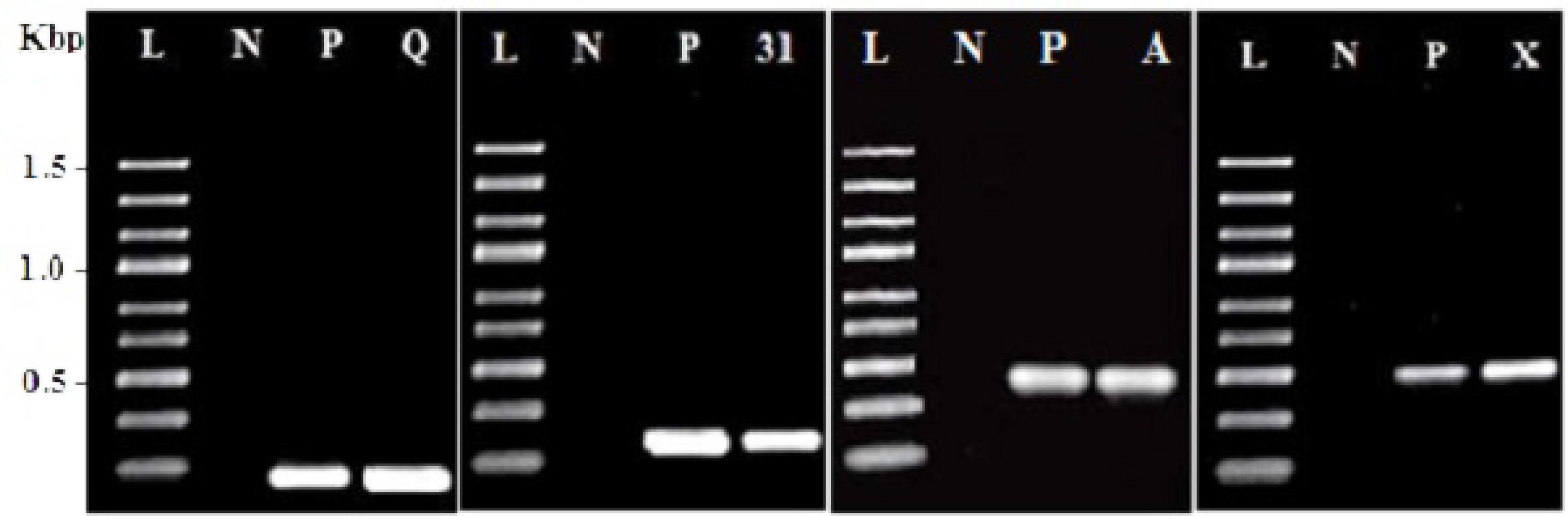
FIGURE 1. Amplification results for screening enterocin genes in Enterococcus faecium CM33. L, molecular weight ladder (kbp); N, negative control; P, positive control by using purified DNA from a producer strain; Q, enterocin Q; 31: enterocin 31; A: enterocin A and X: enterocin X.
The class IIb bacteriocins, two-peptide bacteriocins, also need two diverse peptides (enterocins Q and X) for activity. The production of multiple bacteriocins is not unusual and could be a general aspect of enterococci. Many Enterococcus bacteria are able to produce multiple bacteriocins, such as E. faecium WHE81 (Ennahar et al., 2001), E. faecium KV-B5 (Hu et al., 2010), E. faecium NKR-5-3A (Ishibashi et al., 2012), E. faecium JCM 5804T (Park et al., 2003), and E. faecium DAC2 (Sánchez et al., 2007). Nevertheless, the incidence of numerous enterocin genes in enterococci does not forever link with an advanced bacteriocin activity in their supernatants and not all enterocin genes should be expressed at the same time (Casaus et al., 1997).
Enterococcus faecium is the most normally occurring enterococcal species in dairy industry (Morandi et al., 2006) and fermented vegetable (Landeta et al., 2013). This species is also found in raw fruits (Abriouel et al., 2008) and probiotic preparations (Senok, 2009). However the nosocomial infections related to wound, bacteraemia, urinary-tract infections, and abdominal infections are caused by many enterococcal species (Thurlow et al., 2009). Enterococcus bacteria, as pathogens, have intrinsic and acquired resistance to many antibiotics that were considered as a major concern. The resistance against antibiotics and the production of virulence factors are two significant factors for the assessment of safety in enterococci. Probiotic strains may harbor antibiotic resistance genes that are able to be transferred to pathogenic bacteria. In particular, the most significant concern among antibiotic resistance is vancomycin resistance since it is the last antibiotics largely efficient against clinical infections by multidrug-resistant pathogens (Klein, 2003). Several studies elucidated the incidence of vancomycin-resistant enterococci in food of animal origin, mainly in E. faecium and E. faecalis species; although the isolation frequency seems to be lower than in clinical samples (Klein, 2003). In our case, CM33 strain was found to be susceptible to vancomycin, and this lack of resistance associated with the absence of van A, van B and van C2 genes. According to the EFSA, if the strain has one or more these genetic elements, this strain is unsafe and could not be applied as a feed additive (Ogier and Serror, 2008). Moreover, no resistance to other antibiotics such as gentamycin, erythromycin, tetracycline, and rifampicin was detected in this colostrum-isolated Enterococcus.
The potential pathogenicity of CM33 strain was evaluated by assessing the existence of genes encoding for other virulence factors, as determined by studies in various enterococcal species (Omar et al., 2004). Genes encoding for cell-wall anchored collagen adhesion (acm), aggregation substance (agg), cytolysin (cylA), and gelatinase (gelE) were not found in our isolated strain. Besides, the amplification for a sex pheromones gene (ccf), which could be involved in initiating conjugation processes (Clewell, 2011) was negative.
One of important properties of strain to be as a potential probiotic is anti-pathogen activity. In vitro test under neutralized pH was performed using 13 pathogenic microorganisms and supernatants (metabolites) obtained from the exponential E. faecium CM33 culture. The well diffusion technique was performed to evaluate the inhibitory effect of the CM33 strain against 13 pathogenic microorganisms. The E. faecium supernatant demonstrated the strong inhibition function against indicator pathogens (Table 5). This strain was found to have the strongest inhibition activity against L. monocytogenes and Candida albicans. It was recommended that E. faecium CM33 produced bacteriocins (enterocin A and 31) to inhibit these pathogens.
According to previous studies, the multiple enterocins-producing isolate is likely more capable and could display a wider range of inhibition in inhibiting the growth of undesirable bacteria than a simple bacteriocin producer (Ishibashi et al., 2012). E. faecium DAC2 (producer both Ent A and Ent P) produces a higher antagonistic activity than those of the controls E. faecium T136 and E. faecium P13 (producer of enterocin A and enterocin P, respectively; Sánchez et al., 2007). The production of bacteriocins by LAB helps LAB to be colonized in their habitats and to compete with other bacteria (Garriga et al., 1993). Many factors including decreased pH levels, competition for substrates, and the production of substances such as bacteriocins are caused the antimicrobial activity of LAB (Parente and Ricciardi, 1999).
The competitive inhibition of enteropathogen attachment to epithelial cells by LAB is another important factor. So, the competitive inhibition of adherence of pathogenic bacteria to Caco-2 cells by adhering E. faecium CM33 cells was evaluated. This strain was able to strongly inhibit the adhesion of many tested pathogens (Table 4). However, the mechanisms by which LAB inhibit pathogen adhesion to human cell lines in vitro are not fully understood. Steric hindrance rather than blockage of specific receptors may be involved (Bernet et al., 1993). Additional trials are required to discover the exact mechanism of inhibition observed in this study.
Conclusion
An identified strain of E. faecium from colostrum can present interesting probiotic characteristics. It can stay alive during passing the digestive system and has ability to be colonized on the intestinal epithelial cells. In addition, this strain showed a significant enteropathogen growth inhibiting activity and interference with pathogens adhesion to human colon Caco-2 cells. These features could enable this strain to establish itself in the intestinal tract and to compete with other bacterial species. However, the safety and implications for public health of individual enterococcal strains must be carefully evaluated to fully exploit their industrial potential.
Ethical Issues
No ethical issues to be promulgated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The financial support of the University Putra Malaysia and the Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran are gratefully acknowledged.
References
Abriouel, H., Omar, N. B., Molinos, A. C., López, R. L., Grande, M. J., Martínez-Viedma, P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 123, 38–49. doi: 10.1016/j.ijfoodmicro.2007.11.067
Ahmed, W., Sidhu, J., and Toze, S. (2012). Speciation and frequency of virulence genes of Enterococcus spp. isolated from rainwater tank samples in Southeast Queensland, Australia. Environ. Sci. Technol. 46, 6843–6850. doi: 10.1021/es300595g
Baele, M., Storms, V., Haesebrouck, F., Devriese, L. A., Gillis, M., Verschraegen, G., et al. (2001). Application and evaluation of the interlaboratory reproducibility of tRNA intergenic length polymorphism analysis (tDNA-PCR) for identification of Streptococcus species. J. Clin. Microbiol. 39, 1436–1442. doi: 10.1128/JCM.39.4.1436-1442.2001
Ben Omar, N., Abriouel, H., Keleke, S., Sánchez Valenzuela, A., Martínez-Cañamero, M., Lucas López, R., et al. (2008). Bacteriocin-producing Lactobacillus strains isolated from poto poto, a Congolese fermented maize product, and genetic fingerprinting of their plantaricin operons. Int. J. Food microbiol. 127, 18–25. doi: 10.1016/j.ijfoodmicro.2008.05.037
Bernet, M.-F., Brassart, D., Neeser, J., and Servin, A. (1993). Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl. Environ. Microbiol. 59, 4121–4128.
Bezkorovainy, A. (2001). Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73, 399s–405s.
Brandão, A., Almeida, T., Muñoz-Atienza, E., Torres, C., Igrejas, G., Hernández, P., et al. (2010). Antimicrobial activity and occurrence of bacteriocin structural genes in Enterococcus spp. of human and animal origin isolated in Portugal. Arch. Microbiol. 192, 927–936. doi: 10.1007/s00203-010-0619-z
Cariolato, D., Andrighetto, C., and Lombardi, A. (2008). Occurrence of virulence factors and antibiotic resistances in Enterococcus faecalis and Enterococcus faecium collected from dairy and human samples in North Italy. Food Control 19, 886–892. doi: 10.1016/j.foodcont.2007.08.019
Carlos, A., Semedo-Lemsaddek, T., Barreto-Crespo, M., and Tenreiro, R. (2010). Transcriptional analysis of virulence-related genes in enterococci from distinct origins. J. Appl. Microbiol. 108, 1563–1575. doi: 10.1111/j.1365-2672.2009.04551.x
Casaus, P., Nilsen, T., Cintas, L. M., Nes, I. F., Hernández, P. E., and Holo, H. (1997). Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143, 2287–2294. doi: 10.1099/00221287-143-7-2287
Cintas, L. M., Casaus, P., Herranz, C., Hâvarstein, L. S., Holo, H., Hernández, P. E., et al. (2000). Biochemical and genetic evidence that Enterococcus faecium L50 produces enterocins L50A and L50B, the sec-dependent enterocin P, and a novel bacteriocin secreted without an N-terminal extension termed enterocin Q. J. Bacteriol. 182, 6806–6814. doi: 10.1128/JB.182.23.6806-6814.2000
Citti, L., Rovero, P., Colombo, M. G., Mariani, L., Poliseno, L., and Rainaldi, G. (2002). Efficacy of an amphipathic oligopeptide to shuttle and release a cis-acting DNA decoy into human cells. Biotechniques 32, 172–177.
Clewell, D. B. (2011). Tales of conjugation and sex pheromones: a plasmid and enterococcal odyssey. Mob. Genet. Elements 1, 38–54. doi: 10.4161/mge.1.1.15409
Collins, J., Thornton, G., and Sullivan, G. (1998). Selection of probiotic strains for human applications. Int. Dairy J. 8, 487–490. doi: 10.1016/S0958-6946(98)00073-9
De Vuyst, L., Foulquié Moreno, M. R., and Revets, H. (2003). Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 84, 299–318. doi: 10.1016/S0168-1605(02)00425-7
Dunne, C., O’mahony, L., Murphy, L., Thornton, G., Morrissey, D., O’halloran, S., et al. (2001). In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73, 386s–392s.
Du Toit, M., Franz, C. M., Dicks, L. M., and Holzapfel, W. H. (2000). Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J. Appl. Microbiol. 88, 482–494. doi: 10.1046/j.1365-2672.2000.00986.x
Eaton, T. J., and Gasson, M. J. (2001). Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67, 1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001
Edalatian, M. R., Habibi Najafi, M. B., Mortazavi, S. A., Alegría, Á., Delgado, S., Bassami, M. R., et al. (2012). Production of bacteriocins by Enterococcus spp. isolated from traditional, Iranian, raw milk cheeses, and detection of their encoding genes. Eur. Food Res. Technol. 234, 789–796. doi: 10.1007/s00217-012-1697-8
Eijsink, V. G., Axelsson, L., Diep, D. B., Håvarstein, L. S., Holo, H., and Nes, I. F. (2002). Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Van Leeuwenhoek 81, 639–654. doi: 10.1023/A:1020582211262
Ennahar, S., Asou, Y., Zendo, T., Sonomoto, K., and Ishizaki, A. (2001). Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. Int. J. Food Microbiol. 70, 291–301. doi: 10.1016/S0168-1605(01)00565-7
Franz, C. M., Huch, M., Abriouel, H., Holzapfel, W., and Gálvez, A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151, 125–140. doi: 10.1016/j.ijfoodmicro.2011.08.014
Franz, C. M., Van Belkum, M. J., Holzapfel, W. H., Abriouel, H., and Gálvez, A. (2007). Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 31, 293–310. doi: 10.1111/j.1574-6976.2007.00064.x
Garriga, M., Hugas, M., Aymerich, T., and Monfort, J. (1993). Bacteriocinogenic activity of Lactobacilli from fermented sausages. J. Appl. Bacteriol. 75, 142–148. doi: 10.1111/j.1365-2672.1993.tb02759.x
Gilmore, M. S., Segarra, R. A., Booth, M. C., Bogie, C. P., Hall, L. R., and Clewell, D. B. (1994). Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176, 7335–7344.
Haghshenas, B., Abdullah, N., Nami, Y., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2014a). Different effects of two newly-isolated probiotic Lactobacillus plantarum 15HN and Lactococcus lactis subsp. Lactis 44Lac strains from traditional dairy products on cancer cell lines. Anaerobe 30, 51–59. doi: 10.1016/j.anaerobe.2014.08.009
Haghshenas, B., Nami, Y., Abdullah, N., Radiah, D., Rosli, R., Barzegari, A., et al. (2014b). Potentially probiotic acetic acid bacteria isolation and identification from traditional dairies microbiota. Int. J. Food Sci. Technol. 50, 1056–1064. doi: 10.1111/ijfs.12718
Haghshenas, B., Nami, Y., Abdullah, N., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2014c). Anti-proliferative effects of Enterococcus strains isolated from fermented dairy products on different cancer cell lines. J. Funct. Foods 11, 363–374. doi: 10.1016/j.jff.2014.10.002
Haghshenas, B., Nami, Y., Abdullah, N., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2015). Anticancer impacts of potentially probiotic acetic acid bacteria isolated from traditional dairy microbiota. LWT Food Sci. Technol. 60, 690–697. doi: 10.1016/j.lwt.2014.09.058
Hechard, Y., Dherbomez, M., Cenatiempo, Y., and Letellier, F. (1990). Antagonism of lactic acid bacteria from goats’ milk against pathogenic strains assessed by the ‘sandwich method.’ Lett. Appl. Microbiol. 11, 185–188. doi: 10.1111/j.1472-765X.1990.tb00156.x
Hickey, R. M., Twomey, D. P., Ross, R. P., and Hill, C. (2003). Production of enterolysin A by a raw milk enterococcal isolate exhibiting multiple virulence factors. Microbiology 149, 655–664. doi: 10.1099/mic.0.25949-0
Hu, C.-B., Malaphan, W., Zendo, T., Nakayama, J., and Sonomoto, K. (2010). Enterocin X, a novel two-peptide bacteriocin from Enterococcus faecium KU-B5, has an antibacterial spectrum entirely different from those of its component peptides. Appl. Environ. Microbiol. 76, 4542–4545. doi: 10.1128/AEM.02264-09
Ishibashi, N., Himeno, K., Fujita, K., Masuda, Y., Perez, R. H., Zendo, T., et al. (2012). Purification and characterization of multiple bacteriocins and an inducing peptide produced by Enterococcus faecium NKR-5-3 from Thai fermented fish. Biosci. Biotechnol. Biochem. 76, 947–953. doi: 10.1271/bbb.110972
Khan, H., Flint, S., and Yu, P.-L. (2010). Enterocins in food preservation. Int. J. Food Microbiol. 141, 1–10. doi: 10.1016/j.ijfoodmicro.2010.03.005
Kirjavainen, P. V., Ouwehand, A. C., Isolauri, E., and Salminen, S. J. (1998). The ability of probiotic bacteria to bind to human intestinal mucus. FEMS Microbiol. Lett. 167, 185–189. doi: 10.1111/j.1574-6968.1998.tb13226.x
Klein, G. (2003). Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 88, 123–131. doi: 10.1016/S0168-1605(03)00175-2
Kullen, M., Sanozky-Dawes, R., Crowell, D., and Klaenhammer, T. (2000). Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89, 511–516. doi: 10.1046/j.1365-2672.2000.01146.x
Landeta, G., Curiel, J. A., Carrascosa, A. V., Muñoz, R., and De Las Rivas, B. (2013). Technological and safety properties of lactic acid bacteria isolated from Spanish dry-cured sausages. Meat Sci. 95, 272–280. doi: 10.1016/j.meatsci.2013.05.019
Line, J., Svetoch, E., Eruslanov, B., Perelygin, V., Mitsevich, E., Mitsevich, I., et al. (2008). Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 52, 1094–1100. doi: 10.1128/AAC.01569-06
Morandi, S., Brasca, M., Andrighetto, C., Lombardi, A., and Lodi, R. (2006). Technological and molecular characterisation of enterococci isolated from north–west Italian dairy products. Int. Dairy J. 16, 867–875. doi: 10.1016/j.idairyj.2005.09.005
Moreno, M. F., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Nallapareddy, S. R., Singh, K. V., Okhuysen, P. C., and Murray, B. E. (2008). A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect. Immun. 76, 4110–4119. doi: 10.1128/IAI.00375-08
Nami, Y., Abdullah, N., Haghshenas, B., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2014a). Assessment of probiotic potential and anticancer activity of newly isolated vaginal bacterium Lactobacillus plantarum 5BL. Microbiol. Immunol. 58, 492–502. doi: 10.1111/1348-0421.12175
Nami, Y., Abdullah, N., Haghshenas, B., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2014b). Probiotic assessment of Enterococcus durans 6HL and Lactococcus lactis 2HL isolated from vaginal microflora. J. Med. Microbiol. 63, 1044–1051. doi: 10.1099/jmm.0.074161-0
Nami, Y., Abdullah, N., Haghshenas, B., Radiah, D., Rosli, R., and Khosroushahi, A. Y. (2014c). Probiotic potential and biotherapeutic effects of newly isolated vaginal Lactobacillus acidophilus 36YL strain on cancer cells. Anaerobe 28, 29–36. doi: 10.1016/j.anaerobe.2014.04.012
Nami, Y., Abdullah, N., Haghshenas, B., Radiah, D., Rosli, R., and Yari Khosroushahi, A. (2014d). A newly isolated probiotic Enterococcus faecalis strain from vagina microbiota enhances apoptosis of human cancer cells. J. Appl. Microbiol. 117, 498–508. doi: 10.1111/jam.12531
Nes, I. F., Diep, D. B., and Holo, H. (2007). Bacteriocin diversity in Streptococcus and Enterococcus. J. Bacteriol. 189, 1189–1198. doi: 10.1128/JB.01254-06
Nueno-Palop, C., and Narbad, A. (2011). Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int. J. Food Microbiol. 145, 390–394. doi: 10.1016/j.ijfoodmicro.2010.12.029
Ogier, J.-C., and Serror, P. (2008). Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126, 291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017
Omar, N. B., Castro, A., Lucas, R., Abriouel, H., Yousif, N. M., Franz, C. M., et al. (2004). Functional and safety aspects of enterococci isolated from different Spanish foods. Syst. Appl. Microbiol. 27, 118–130. doi: 10.1078/0723-2020-00248
Parente, E., and Ricciardi, A. (1999). Production, recovery and purification of bacteriocins from lactic acid bacteria. Appl. Biochem. Biotechnol. 52, 628–638. doi: 10.1007/s002530051570
Park, S., Itoh, K., and Fujisawa, T. (2003). Characteristics and identification of enterocins produced by Enterococcus faecium JCM 5804T. J. Appl. Microbiol. 95, 294–300. doi: 10.1046/j.1365-2672.2003.01975.x
Pascual, L. M., Daniele, M. B., Ruiz, F., Giordano, W., Pájaro, C., and Barberis, L. (2008). Lactobacillus rhamnosus L60, a potential probiotic isolated from the human vagina. J. Gen. Appl. Microbiol. 54, 141–148. doi: 10.2323/jgam.54.141
Pereira, D. I., and Gibson, G. R. (2002). Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human gut. Appl. Environ. Microbiol. 68, 4689–4693. doi: 10.1128/AEM.68.9.4689-4693.2002
Rehaiem, A., Belgacem, Z. B., Edalatian, M. R., Martínez, B., Rodríguez, A., Manai, M., et al. (2014). Assessment of potential probiotic properties and multiple bacteriocin encoding-genes of the technological performing strain Enterococcus faecium MMRA. Food Control 37, 343–350. doi: 10.1016/j.foodcont.2013.09.044
Reviriego, C., Eaton, T., Martín, R., Jiménez, E., Fernández, L., Gasson, M. J., et al. (2005). Screening of virulence determinants in Enterococcus faecium strains isolated from breast milk. J. Hum. Loct. 21, 131–137. doi: 10.1177/0890334405275394
Sánchez, J., Basanta, A., Gómez-Sala, B., Herranz, C., Cintas, L., and Hernández, P. (2007). Antimicrobial and safety aspects, and biotechnological potential of bacteriocinogenic enterococci isolated from mallard ducks (Anas platyrhynchos). Int. J. Food Microbiol. 117, 295–305. doi: 10.1016/j.ijfoodmicro.2007.04.012
Satake, S., Clark, N., Rimland, D., Nolte, F. S., and Tenover, F. C. (1997). Detection of vancomycin-resistant enterococci in fecal samples by PCR. J. Clin. Microbiol. 35, 2325–2330.
Seiquer, I., Aspe, T., Vaquero, P., and Navarro, P. (2001). Effects of heat treatment of casein in the presence of reducing sugars on calcium bioavailability: in vitro and in vivo assays. J. Agric. Food Chem. 49, 1049–1055. doi: 10.1021/jf001008v
Semedo, T., Santos, M. A., Martins, P., Lopes, M. F. S., Marques, J. J. F., Tenreiro, R., et al. (2003). Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J. Clin. Microbiol. 41, 2569–2576. doi: 10.1128/JCM.41.6.2569-2576.2003
Senok, A. C. (2009). Probiotics in the arabian gulf region. Food Nutr. Res. 53. doi: 10.3402/fnr.v53i0.1842
Sparo, M., Castro, M., Andino, P., Lavigne, M., Ceriani, C., Gutiérrez, G., et al. (2006). Partial characterization of enterocin MR99 from a corn silage isolate of Enterococcus faecalis. J. Appl. Microbiol. 100, 123–134. doi: 10.1111/j.1365-2672.2005.02752.x
Strompfová, V., Lauková, A., Simonová, M., and Marciňáková, M. (2008). Occurrence of the structural enterocin A, P, B, L50B genes in enterococci of different origin. Vet. Microbiol. 132, 293–301. doi: 10.1016/j.vetmic.2008.05.001
Tarasova, E., Yermolenko, E., Donets, V., Sundukova, Z., Bochkareva, A., Borshev, I., et al. (2010). The influence of probiotic Enterococcus faecium strain L5 on the microbiota and cytokines expression in rats with dysbiosis induced by antibiotics. Benef. Microbes 1, 265–270. doi: 10.3920/BM2010.0008
Thirabunyanon, M., and Hongwittayakorn, P. (2013). Potential probiotic lactic acid bacteria of human origin induce antiproliferation of colon cancer cells via synergic actions in adhesion to cancer cells and short-chain fatty acid bioproduction. Appl. Biochem. Biotechnol. 169, 511–525. doi: 10.1007/s12010-012-9995-y
Thurlow, L. R., Thomas, V. C., Fleming, S. D., and Hancock, L. E. (2009). Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect. Immun. 77, 5551–5557. doi: 10.1128/IAI.00576-09
Tuomola, E., Crittenden, R., Playne, M., Isolauri, E., and Salminen, S. (2001). Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr. 73, 393s–398s.
Tuomola, E. M., and Salminen, S. J. (1998). Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 41, 45–51. doi: 10.1016/S0168-1605(98)00033-6
Vankerckhoven, V., Van Autgaerden, T., Vael, C., Lammens, C., Chapelle, S., Rossi, R., et al. (2004). Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42, 4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004
Keywords: antimicrobial activity, Enterococcus faecium, enterocins, probiotic, virulence genes
Citation: Nami Y, Haghshenas B, Haghshenas M and Yari Khosroushahi A (2015) Antimicrobial activity and the presence of virulence factors and bacteriocin structural genes in Enterococcus faecium CM33 isolated from ewe colostrum. Front. Microbiol. 6:782. doi: 10.3389/fmicb.2015.00782
Received: 27 May 2015; Accepted: 16 July 2015;
Published: 29 July 2015.
Edited by:
Jean-Christophe Augustin, Ecole Nationale Vétérinaire d’Alfort, FranceReviewed by:
Alexandra Lianou, Agricultural University of Athens, GreeceEva-Guadalupe Lizárraga-Paulín, Instituto Tecnológico y de Estudios Superiores de Monterrey, Mexico
Copyright © 2015 Nami, Haghshenas, Haghshenas and Yari Khosroushahi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmad Yari Khosroushahi, Drug Applied Research Center, and Department of Pharmacognosy, Faculty of Pharmacy, Tabriz University of Medical Sciences, P.O. Box 51664-14766, Daneshgah Street, Tabriz, Iran, yarikhosroushahia@tbzmed.ac.ir
 Yousef Nami
Yousef Nami Babak Haghshenas
Babak Haghshenas Minoo Haghshenas2
Minoo Haghshenas2 Ahmad Yari Khosroushahi
Ahmad Yari Khosroushahi