- Department of Evolutionary Genetics, Cavanilles Institute, University of Valencia, Paterna, Valencia, Spain
New pathogenic bacteria belonging to the genus Erwinia associated with pome fruit trees (Erwinia, E. piriflorinigrans, E. uzenensis) have been increasingly described in the last years, and comparative analyses have found that all these species share several genetic characteristics. Studies at different level (whole genome comparison, virulence genes, plasmid content, etc.) show a high intraspecies homogeneity (i.e., among E. amylovora strains) and also abundant similarities appear between the different Erwinia species: presence of plasmids of similar size in the pathogenic species; high similarity in several genes associated with exopolysaccharide production and hence, with virulence, as well as in some other genes, in the chromosomes. Many genetic similarities have been observed also among some of the plasmids (and genomes) from the pathogenic species and E. tasmaniensis or E. billingiae, two epiphytic species on the same hosts. The amount of genetic material shared in this genus varies from individual genes to clusters, genomic islands and genetic material that even may constitute a whole plasmid. Recent research on evolution of erwinias point out the horizontal transfer acquisition of some genomic islands that were subsequently lost in some species and several pathogenic traits that are still present. How this common material has been obtained and is efficiently maintained in different species belonging to the same genus sharing a common ecological niche provides an idea of the origin and evolution of the pathogenic Erwinia and the interaction with non-pathogenic species present in the same niche, and the role of the genes that are conserved in all of them.
Introduction
The genus Erwinia belongs to the Enterobacteriaceae family and essentially comprises plant-associated bacteria that are pathogenic and epiphytic to pome fruit trees (Palacio-Bielsa et al., 2011). The most important species is Erwinia amylovora, causal agent of fire blight on rosaceous hosts, which is present worldwide and produces very high economic losses (Bonn and van der Zwet, 2000). Other pathogenic Erwinia species have been described in the last decades: Erwinia pyrifoliae, a pathogen described in Asian pear isolated in South Korea (Kim et al., 1999; Rhim et al., 1999; Shrestha et al., 2003); E. piriflorinigrans, isolated in 1999 in Spain, causes necrosis of pear blossoms (López et al., 2011), and Erwinia uzenensis from Japan, which produces bacterial black shoot disease (BBSDP) on European pear (Matsuura et al., 2012). Other related Erwinia species, E. tasmaniensis and E. billingiae, are epiphytes in the same hosts. All these species are genetically and phenotypically closely related, although they can be distinguished by taxonomic criteria (Mergaert et al., 1999; Kube et al., 2008; Palacio-Bielsa et al., 2011).
In the last years, several sequencing projects have been carried out which included all the Erwinia species except E. uzenensis. All have provided interesting clues about the relationships among these organisms and the exchange of genetic material (Sundin, 2007; Kube et al., 2008, 2010; Sebaihia et al., 2010; Smits et al., 2010a,b; Powney et al., 2011; Smits et al., 2013; Ismail et al., 2014; Smits et al., 2014). Because E. amylovora is a very important pathogen, the majority of information is related to it. Genetic studies have divided E. amylovora strains into two major groups with different host range: strains isolated from Spiraeoideae and from Rosoideae (Rubus spp., Mann et al., 2013). The genomes of Spiraeoideae-infecting strains are highly homogeneous, and greater diversity was observed between Spiraeoideae- and Rubus-infecting strains, the majority of which was attributed to variable genomic islands (Triplett et al., 2006; Smits et al., 2010b).
Comparative genomics of E. amylovora strains from different origins showed that the pan-genome is highly conserved relative to other phytopathogenic bacteria species, with homogeneity of 99.99% identity at the nucleotide level (Smits et al., 2010b; Zhao and Qi, 2011; Mann et al., 2013). The genomes of two E. pyrifoliae strains sequenced (Ep1/96) and DSM 12163 (Ep16/99) are almost identical whereas the two saprophytic species are distantly related to the pathogenic species, with E. tasmaniensis more related than E. billingae (Kube et al., 2010; Smits et al., 2010a). Comparison of genomes of Japanese (Ejp617) and Korean (Ep1/96) E. pyrifoliae strains revealed a high level of genome conservation (more than 95% nucleotide sequence identity) despite the numerous insertion/deletion rearrangements and inversions associated with Insertion Sequences (IS). The differences are mainly based on transposases, phage-related genes, and single genes (Smits et al., 2010a; Thapa et al., 2013). The genes acquired by horizontal gene transfer (HGT) are introduced via mobile genetic elements (MGEs) and incorporated into the chromosome by homologous or illegitimate recombination.
Other characteristics observed are related to the genome size. Differences in size between E. pyrifoliae and E. tasmaniensis genomes are due to the insertion of MGEs in the E. pyrifoliae genome that code transposases, integrases, and phage-related proteins. The prevalence of a high number of mobile elements in Ep1/96 may suggest frequent genomic changes and a higher rate of evolution (Smits et al., 2010a; Thapa et al., 2013).
Ancestral origins of several virulence factors have been found, and the two major virulence determinants required for E. amylovora to infect and cause disease are the genes involved in amylovoran biosynthesis and the type III secretion systems (T3SS). Other genes that could have been acquired after a divergence of pathogenic species are flagellar genes (Zhao et al., 2011) and the type VI secretion systems (T6SS; Smits et al., 2011; Mann et al., 2013). In this review, I will discuss the presence of transfer elements, involving from individual genes to entire plasmids, and how these genetic transfers intervene in the emergence of characteristics like pathogenicity, virulence and the fitness of the pome fruit erwinias (Zhao and Qi, 2011).
Exopolysaccharide Biosynthesis
Exopolysaccharide (EPS) is a pathogenicity factor contributing to biofilm formation of E. amylovora (Koczan et al., 2009), encoded by the ams operon. This gene cluster is present in the genomes of E. amylovora, E. pyrifoliae, and E. piriflorinigrans (Bernhard et al., 1996; Kube et al., 2008). In the sequenced E. tasmaniensis and E. billingiae genomes these genes are present in a different cluster (cps) yielding an EPS more related to stewartan of Pantoea stewartii subsp. stewartii (Coplin et al., 1996; Kim et al., 2002; Smits et al., 2010a; Malnoy et al., 2012). The hypothesis could be that an Erwinia ancestor produced an EPS similar to stewartan of P. stewartii (Coplin et al., 1996; Kube et al., 2008), and the differentiation took place at or after the separation of the pathogenic Erwinia from E. tasmaniensis, and this would indicate that the genes involved in amylovoran production are probably acquired (Bernhard et al., 1996; Smits et al., 2011).
Type III Secretion Systems
The T3SS is a protein complex found in several Gram-negative bacteria with a needle-like structure used as a sensory probe to detect the presence of eukaryotic organisms and to secrete and inject virulence factors into the host cells (Galan and Collmer, 1999) and are located in pathogenicity islands (PAIs) integrated into the genomes in the plant pathogens (Oh et al., 2005; Toth et al., 2006). The PAI in all isolates analyzed of E. amylovora is divided into four distinct DNA regions, and is delimited by genes suggesting horizontal gene transfer and the remnants of an integrative conjugative element (ICE) are present at the flank of the Hrp cluster (Wei and Beer, 1993; Oh and Beer, 2005; Oh et al., 2005; Mann et al., 2012, 2013; Vrancken et al., 2013). The Hrp region is conserved in the Spiraeoideae strains sequenced (CFBP 1430, ATCC 49946; Smits et al., 2010b) whereas the genomes of several Rubus strains (ATCC BAA-2158, Ea644, and MR-1; Powney et al., 2011) showed larger sizes. Similarly, the island transfer (IT) regions of Spiraeoideae-infecting strains of E. amylovora (IT: group of MGEs that reside in a host chromosome but retain the ability to excise and to transfer by conjugation) are highly conserved (>99% nucleotide identity and identical synteny), but the IT regions of the Rubus-infecting strains all vary in sequence identity and length. Comparative genome sequences revealed two additional T3SS PAIs (PAI2 and PAI3) and two flagellar T3SS systems (Fla1 and Fla2; Zhao et al., 2009; Smits et al., 2011; Zhao and Qi, 2011). PAI2 and PAI3 have a significantly lower G+C content and are closely related to those of Sodalis glossinidius (an endosymbiont of the tsetse fly) and to the pathogens Salmonella and Yersinia (Dale et al., 2001; Young and Young, 2002; Gendlina et al., 2007; Zhao et al., 2009; Smits et al., 2010b). Sequences upstream of PAI2 and PAI3 contained genes associated with MGEs, thus, the insertion of a mobile element deleted a part of PAI-2 in E. tasmaniensis Et1/99 (Kube et al., 2008), and PAI2 is lost in E. pyrifoliae (Smits et al., 2010a). It could be speculated that E. amylovora may have acquired these novel T3SS PAIs from other bacteria associated with their insect vectors during evolution, or that these novel T3SS PAIs may contribute to the association of E. amylovora with its insect vectors (Zhao and Qi, 2011). Two other sets of genes encoding for flagellar assembly and chemotaxis related proteins were found in the genome of E. pyrifoliae. One set is tightly clustered and the encoded proteins show high identity with the corresponding proteins of Salmonella and Escherichia spp. (Kube et al., 2008). This suggests that the entire region was acquired as a genomic island via horizontal genetic transfer (Smits et al., 2010a).
Type VI Secretion Systems
Type VI secretion systems (T6SS) have been identified in at least a quarter of the sequenced Gram-negative bacteria (Boyer et al., 2009; Records, 2011), and three gene clusters (T6SS1-3) have been found in the genome of E. amylovora CFBP 1430 (Smits et al., 2010b). Comparison of the T6SS clusters among Erwinia and Pantoea species has identified conserved core regions and variable islands (De Maayer et al., 2011). T6SS clusters 1 and 2 are highly similar to E. pyrifoliae DSM 12163T and E. tasmaniensis Et1/99, with the exception of some genes encoding hypothetical proteins that do not belong to the core genes of T6SS (Bingle et al., 2008). E. amylovora showed variations within non-conserved islands of T6SS-1 in regions that share high sequence similarity to bacteria of the genus Pantoea, including the plant pathogen Pantoea stewartii subsp. stewartii (Braun, 1982; Smits et al., 2010a; Figures 1A,B). The third T6SS cluster was identified only in E. amylovora CFBP 1430, located within a putative genomic island and, therefore, might be acquired by horizontal gene transfer (Smits et al., 2010b).
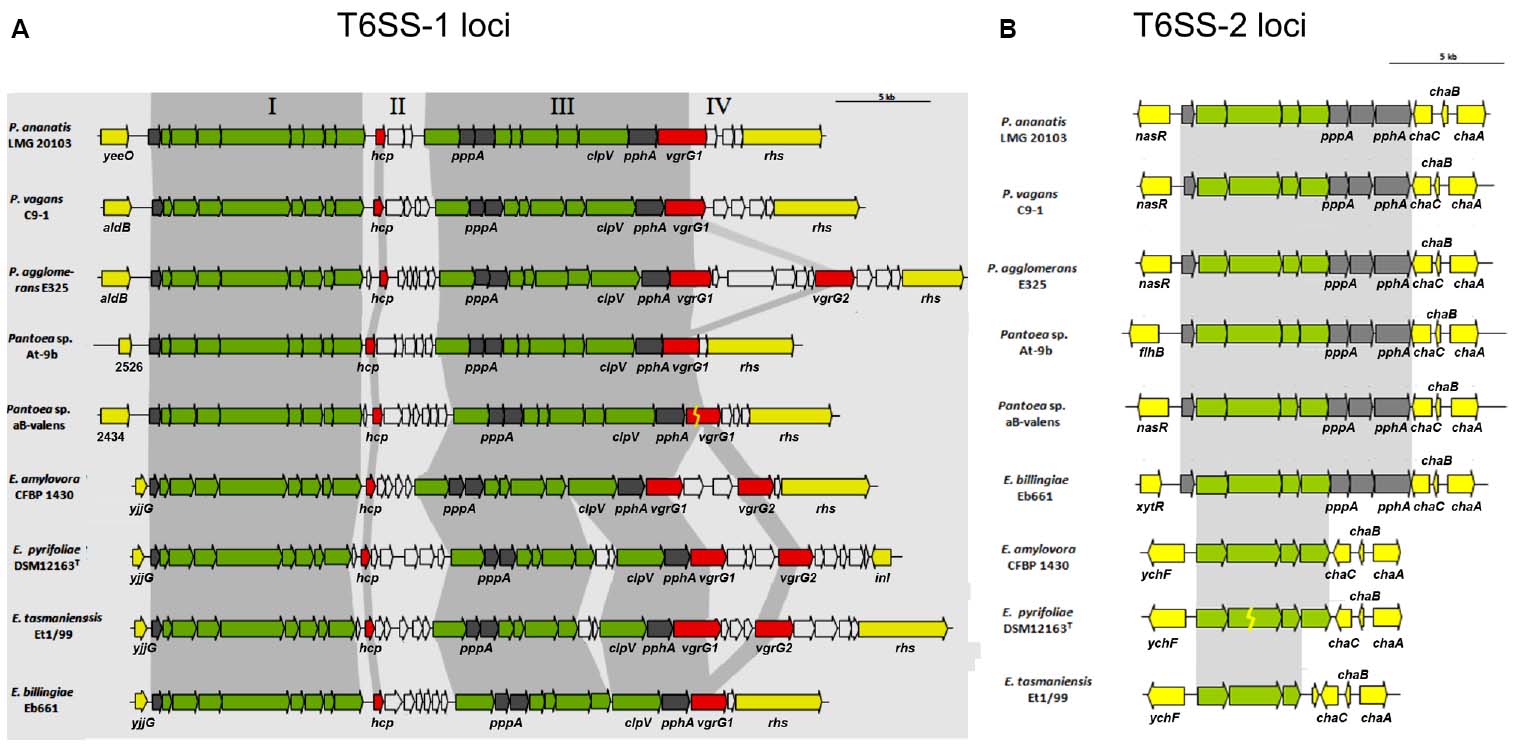
Figure 1. The orthologous T6SS-1 (A) and T6SS-2 (B) loci in Pantoea and Erwinia species. (A) The conserved regions (block I and III) are shaded in gray, while the non-conserved hcp and vgrG islands are not shaded. Genes encoding conserved domain proteins identified by Boyer et al. (2009) are represented by green arrows, and gray arrows indicate other genes conserved among the Pantoea and Erwinia T6SS-1 loci which were not identified as part of the conserved core described by Boyer et al. (2009). Red arrows represent the hcp and vgrG genes while genes not conserved among the Pantoea and Erwinia species are colored in white. (B) The orthologous T6SS-2 loci in Pantoea and Erwinia species. Genes encoding proteins with the conserved domains identified by Boyer et al. (2009) are represented by green arrows while the gray arrows indicate other genes conserved among the Pantoea and Erwinia T6SS-2 loci, which were not identified as part of the conserved core. White arrows represent the genes not conserved among the Pantoea and Erwinia species (Illustration from De Maayer et al., 2011).
Gene Transfer in Related Plasmids in Erwinia spp
One of the most obvious differences among strains of Erwinia spp is the presence of different plasmids in all genome-sequenced Erwinia spp. New plasmids have been described during the latest genome sequencing projects, and is the largest factor influencing the pan-genome size of E. amylovora (McGhee et al., 2002; Maxson-Stein et al., 2003; Foster et al., 2004; Sebaihia et al., 2010; Powney et al., 2011; Kamber et al., 2012; Llop et al., 2012; Ismail et al., 2014). The analyses performed have revealed a strong relationship with other plant and human pathogenic and non-pathogenic bacteria, and constitute the widest host range of the genetic exchange in Erwinia spp. Some of the genetic material exchanged involving different plasmids are described below.
Streptomycin Resistance in pEA34
Streptomycin resistance (SmR) E. amylovora strains were found harboring transposon Tn5393 including the strA-strB gene pair (Chiou and Jones, 1993; McGhee and Sundin, 2011). Tn5393 was introduced to E. amylovora on the conjugative plasmid pEa34. This transposon is also present in Pantoea agglomerans and is thought to have been transferred to E. amylovora on pEA34 (Chiou and Jones, 1991). Plasmid pEA34 could have originated from the insertion of Tn5393 into a 28 kb plasmid no related with pEA29 (Chiou and Jones, 1993; Jones and Schnabel, 2000), and the transposon probably moved from pEa34 to pEA29 because this transposon was found in pEA29 (Burr et al., 1988; Chiou and Jones, 1993; McManus and Jones, 1994; Sundin and Bender, 1995, 1996; McGhee and Sundin, 2011). An additional insertion of Tn5393 was found into the thiO gene of pEA29 (McGhee and Jones, 2000).
Other E. amylovora isolates were found harboring genes strA-strB within the same Tn5393 transposon in a different plasmid (pEA8.7; Sundin and Bender, 1996). This plasmid is identical to the SmR plasmid RSF1010 that has a broad distribution among enteric bacteria and also encodes the strA-strB gene pair (Palmer et al., 1997). These genes are identical to the streptomycin resistance genes found in at least 14 genera of gram-negative animal and human pathogens. In the clinical strains, the genes reside on small broad-host-range plasmids like RSF1010 and pBP1 or large self-transmissible R plasmids like pGS05 and pCJ004 (Sundin and Bender, 1996; McGhee and Sundin, 2011).
Plasmids and Genetic Transfer Elements
Erwinia amylovora isolates from Poland and Belgium showed the presence of a new plasmid of 68 kb (pEA68). It is closely related to other plasmids from different E. amylovora strains, pEA72 from USA (Sebaihia et al., 2010) and pEA78 in E. amylovora from Mexico (Smits et al., 2014). The amino acid sequence identity between these plasmids range between 60 and 90% for the CDS shared, and include genes of transfer and mobilization (mob, tra, and trb). Large regions between the repA gene and the mobABC gene cluster are divergent in all three plasmids, indicating that these regions are highly variable likely due to horizontal gene transfer (Ismail et al., 2014).
Plasmid pEI70 was found in Spanish strains of E. amylovora (Llop et al., 2006). It is conjugative and widespread in European countries, and similarly to pEA29, it induces an increase in symptoms development but with no specific pathogenicity genes (Llop et al., 2008, 2011). pEI70 is almost entirely included in plasmid pEB102 from E. billingiae, with nucleotide sequence identities superior to 98%. The organization is identical as well, with only a 36-kb region in pEB102 absent in pEI70. Another major feature of pEI70 is the presence of an Integrating Conjugative Element (ICE) that shares similarities to specific regions of Pseudomonas fluorescens Pf-5 (Mavrodi et al., 2009) and Pectobacterium atrosepticum SCRI 1043 (Toth et al., 2006; Llop et al., 2011; Figure 2A) and its possible role in the fitness of the bacteria (Llop et al., 2011).
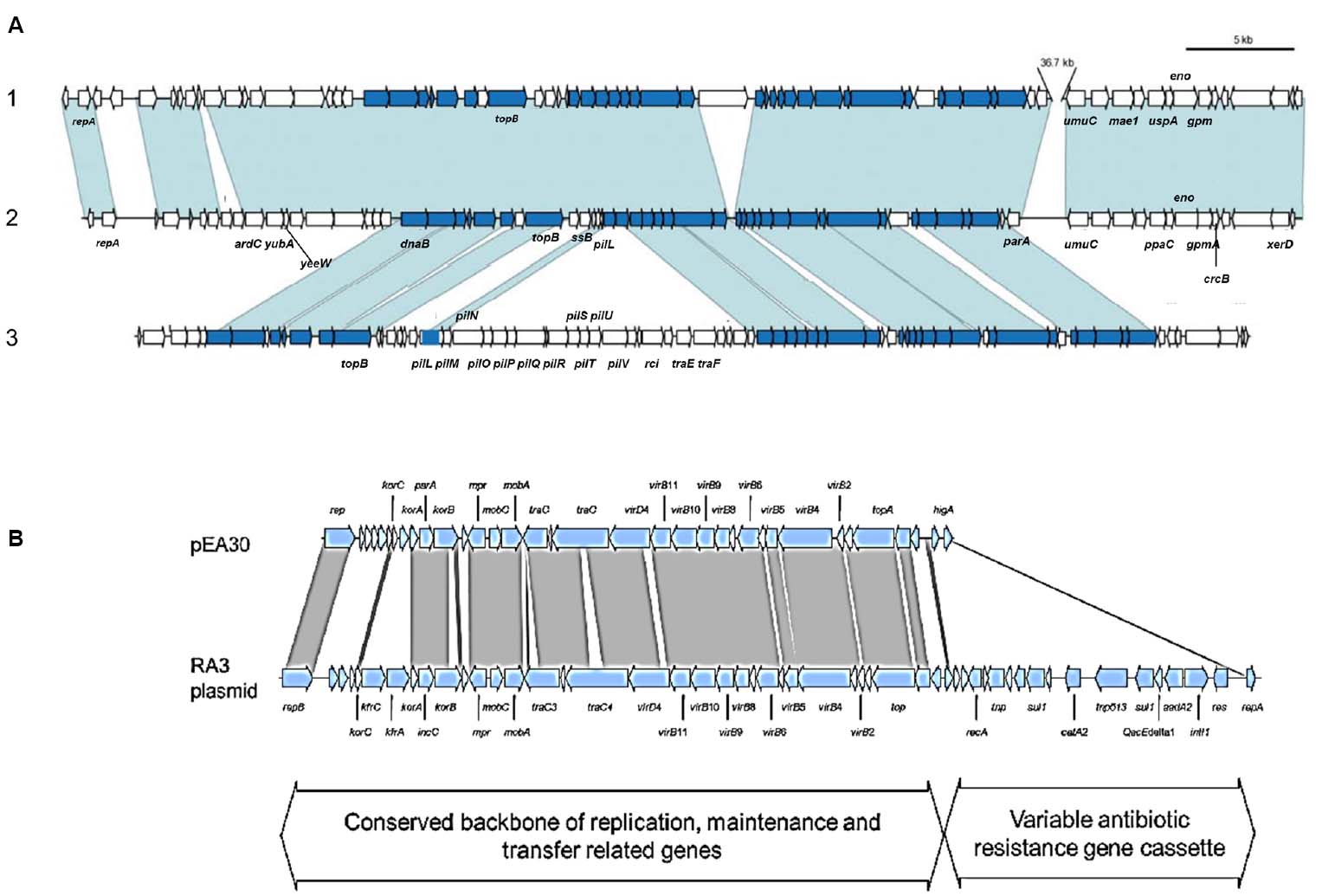
Figure 2. (A) Comparison of plasmid pEB102 of E. billingiae Eb661 (1) with E. amylovora ACW 56400 plasmid pEI70 (2), and the conserved region of GAI-2 of Pectobacterium atrosepticum SCRI 1043 (3). Orthologous genes are indicated by blue shading (conserved ICE element genes) and shading. Genes in white do not have orthologs in these regions (Illustration from Llop et al., 2011). (B) Comparison of plasmid pEA30 of CFBP 2585 (Ea495) to the RA3 plasmid of Aeromonas hydrophila. The RA3 plasmid is the archetype of the IncU plasmids which are a distinct group of mobile elements with highly conserved backbones and variable antibiotic resistance gene cassettes. Conservation between pEA30 and RA3 (represented by the gray shaded lines) is limited to the conserved backbone of replication, maintenance and transfer related genes. Nucleotide similarity searches to known sequences in GenBank indicate that pEA30 has 70% total sequence coverage and 64–81% identity of all high-scoring segment pair matches related to the RA3 plasmid (Illustration from Mann et al., 2013).
Plasmid pEL60 was reported in E. amylovora strains from Lebanon in half of the isolates analyzed. pEL60 has strong relationships with other enterobacterial plasmids, with a high similarity to the Citrobacter freundii plasmid pCTX-M3, sharing 66 of the 68 ORFs it contains (Foster et al., 2004). Transfer genes are similar to genes on plasmid pACM1 from Klebsiella oxytoca (Preston et al., 2000) and to tra genes from plasmids isolated from other enteric bacteria and from the plant pathogen P. syringae pv. tomato DC3000. These observations suggest that the environmental enteric plant pathogen E. amylovora can access the horizontal gene pool shared among clinical enteric bacteria.
In E. pyrifoliae strains, plasmids pEP36 and pEJ30 were found in South Korean and Japan isolates, respectively. They are nearly identical, both contain the element IS285 and the transposon (Tn5394) is missing in pEJ30. Other significant similarities of pEP36 were found in Yersinia pestis genome and Shigella flexneri SHI-2 PAIs (Mann et al., 2013). In E. piriflorinigrans strains, sequencing of a common 37-kb plasmid (pEPIR37) revealed high similarity to plasmids pEA29 of E. amylovora, and plasmids pEP36 and pEJ30 of E. pyrifoliae (McGhee et al., 2002; Maxson-Stein et al., 2003; Barbé et al., 2012). The replication origin has high homology with plasmids pET46 of E. tasmaniensis and pPag3 of P. vagans (Kube et al., 2008; Smits et al., 2010c) but also, a fragment of RepA protein (94% sequence identity) present in pEA29, pEJ30, and pEP36 plasmids, is present in pEPIR37. 12 CDS are similar to genes present in the genomes of E. pyrifoliae, E. tasmaniensis, and E. billingiae (Barbé et al., 2012).
A novel plasmid pEA30 was found in E. amylovora strain CFBP 2585, and nucleotide searches showed that is closely related to the RA3 plasmid of Aeromonas hydrophila (64–81% identity). Its high genetic similarity to RA3, a broad host range and self-transmissible plasmid, stably maintained in Alpha-, Beta-, and Gammaproteobacteria, is another example of the possible generation of the mobilome in pome fruit erwinias by means of plasmids present in the environment (Kulinska et al., 2008; Mann et al., 2013; Figure 2B).
In the plasmid pEA29 from E. amylovora, apart from the similarities with plasmids pEP36, pEJ30, and pEPIR37, remnants of several IS detected resembled insertion elements identified previously in other unrelated bacteria. A vestige of Tn2501 and direct repeats found in the IS911 were detected in all derivatives of pEA29 (McGhee and Jones, 2000). A 108 aa CDS with similarity to ParA from Agrobacterium tumefaciens was also found (Kim and Geider, 1999). The presence of a partial parA gene may explain the occurrence of the 8-bp repeats found consistently in this plasmid.
An striking feature of E. pyrifoliae strain Ep1/96 is the presence of an assumed non-ribosomal peptide synthetase EppT (NRPS) with high similarity with a protein of unknown function from Photorhabdus luminescens (syn. Xenorhabdus luminescens), an enterobacterial pathogen of insects, and an NRPS of the distantly related soft rot pathogen P. atrosepticum encoded in an island typical for horizontal gene transfer (Duchaud et al., 2003; Bell et al., 2004; Kube et al., 2010). Similar NRPS proteins are also encoded in the genome of E. tasmaniensis strain Et1/99 and E. billingiae strain Eb661, but differ in size and domain content. The presence of the eppT gene in this strain may be a result of an integration event as indicated by a phage integrase located upstream (Kube et al., 2010).
Discussion
Many genomic studies on almost all the pome fruit Erwinia species performed in the last years have unveiled the occurrence of transposition events related to HGT, and the presence of different genetic elements. They have allowed inferring the evolution and relatedness of the species within this genus and offer broad information about the mobilome of the plant host erwinias, both pathogenic and epiphytic (Smits et al., 2011). The pathogenic species show the acquisition of a large range of pathogenicity and virulence factors and a reduction of chromosome size by a significant gene loss. The important pathogenicity factor EPS, is present only in the genomes of E. amylovora, E. pyrifoliae, and E. piriflorinigrans (Smits et al., 2010b, 2011; Kamber et al., 2012). All these features demonstrate that horizontal gene transfer is responsible for many of the differential features between these species and may have led to the emergence of pathogenic species from the non-pathogenic (Smits et al., 2011). It is interesting to point out that the largest factor accounting for the genetic variability influencing the pan-genome of this genus is due to the presence of plasmids (Smits et al., 2011; Malnoy et al., 2012). Plasmid sequencing has uncovered the relationship observed in some of the medium and small plasmids in Erwinia species. Thus, partial sequences of plasmids present in some E. amylovora strains are most probably originated from other plasmids from human and animal pathogens (plasmids pEA30 and RA3, Figure 2B) and also the high genetic identity between plasmids pEI70 from E. amylovora and pEB102 from the epiphytic species E. billingiae indicates that lateral transfer of almost entire extrachromosomal material could take place between species sharing host and niche and be stably maintained (Llop et al., 2011; Figure 2A). Several of the plasmids reported show the potential for conjugal transfer, such as pEL60 and pEI70 from E. amylovora, pEB170 of E. billingiae, and several plasmids from E. tasmaniensis, and others carry mob genes and may contain an oriT to be mobilized by Tra proteins of other plasmids (plasmids pEP05 and pEt46 from E. pyrifoliae and E. tasmaniensis, Kube et al., 2010). The chromosome of E. tasmaniensis Et1/99 encodes part of the central region of E. pyrifoliae plasmid pEP36 carrying the thiOSGF and the betB genes, but not the entire plasmid (Smits et al., 2010a). The most recent sequencing projects performed are still unveiling new plasmids that are related to other plasmids or/and genomes from other genera, indicating that in this genus the main genetic source of variability is this extrachromosomal material (Smits et al., 2011, 2014; Ismail et al., 2014). Then, as reflected in this review, the mobilome in this genus shows an extended presence of transferable elements of different sizes, affecting all the species and situated mainly in PAIs related to virulence determinants, but also to the fitness of the bacterium, and all this information allows the knowledge about how the plant pathogens of the Erwinia pome fruits could access the gene pools of other enteric bacteria through horizontal transfer. As special features of the HGT in this genus are the role of MGEs based on plasmids in the acquisition of new traits related not only on classical aspects (i.e., antibiotic resistance) but in bacterial fitness that leads to a better survival in epiphytic species or increased aggressiveness in pathogenic ones. Also, the spread and stability of entire plasmids in both pathogenic and non-pathogenic species is remarkable. Many different factors can influence the prevalence of HGT in a community, but we do not know how the composition of the community shapes the likelihood of HGT events. In this case, knowledge of the bacterial community composition of the different species that are present in a particular site would be of great interest because HGT might be easier in a community composed of closely related species, and some species seem to be more prone to HGT events than others are. This would provide useful information for illuminating patterns of gene transfer in the microbial world.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
I want to thank MM. López for the opportunity to prepare this publication.
References
Barbé, S., Llop, P., Blom, J., Cabrefiga, J., Goesmann, A., Duffy, B., et al. (2012). Complete sequence of Erwinia piriflorinigrans plasmids pEPIR37 and pEPIR5 and role of pEPIR37 in pathogen virulence. Plant Pathol. 62, 786–798. doi: 10.1111/ppa.12002
Bell, K. S., Sebaihia, M., Pritchard, L., Holden, M. T., Hyman, L. J., Holeva, M. C., et al. (2004). Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. U.S.A. 101, 11105–11110. doi: 10.1073/pnas.0402424101
Bernhard, F., Schullerus, D., Bellemann, P., Geider, K., Nimtz, M., Majerczak, D. R., et al. (1996). Genetics and complementation of DNA regions involved in amylovoran synthesis of Erwinia amylovora and stewartan synthesis of Erwinia stewartii. Acta Hortic. 411, 269–274. doi: 10.17660/actahortic.1996.411.53
Bingle, L. E. H., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Bonn, W. G., and van der Zwet, T. (2000). Distribution and Economic Importance of Fire Blight. Fire Blight: the Disease and its Causative Agent, Erwinia amylovora. London: CABI Publishing. 37–53 doi: 10.1079/9780851992945.0037
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104–111. doi: 10.1186/1471-2164-10-104
Braun, E. J. (1982). Ultrastructural investigation of resistant and susceptible maize inbreeds infected with Erwinia stewartii. Phytopathology 72, 159–166. doi: 10.1094/Phyto-72-159
Burr, T. J., Norelli, J. L., Katz, B., Wilcox, W. F., and Hoying, S. A. (1988). Streptomycin resistance of Pseudomonas syringae pv. papulans in apple orchards and its association with a conjugative plasmid. Phytopathology 78, 410–413. doi: 10.1094/Phyto-78-410
Chiou, C. S., and Jones, A. L. (1991). The analysis of plasmid-mediated streptomycin resistance in Erwinia amylovora. Phytopathology 81, 710–714. doi: 10.1094/Phyto-81-710
Chiou, C. S., and Jones, A. L. (1993). Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 175, 732–740.
Coplin, D. L., Majerczak, D. R., Bugert, P., and Geider, K. (1996). Nucleotide sequence analysis of the Erwinia stewartii cps gene cluster for synthesis of stewartan and comparison to the Erwinia amylovora ams cluster for synthesis of amylovoran. Acta Hortic. 411, 251–5717. doi: 10.17660/actahortic.1996.411.49
Dale, C., Young, S. A., Haydon, D. T., and Welburn, S. C. (2001). The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 98, 1883–1888. doi: 10.1073/pnas.98.4.1883
De Maayer, P., Venter, S. N., Kamber, T., Duffy, B., Coutinho, T. A., and Smits, T. H. M. (2011). Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 12:576. doi: 10.1186/1471-2164-12-576
Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., et al. (2003). The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21, 1307–1313. doi: 10.1038/nbt886
Foster, G. C., McGhee, G. C., Jones, A. L., and Sundin, G. W. (2004). Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70, 7539–7544. doi: 10.1128/AEM.70.12.7539-7544.2004
Galan, J. E., and Collmer, A. (1999). Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284, 1322–1328. doi: 10.1126/science.284.5418.1322
Gendlina, I., Held, K. G., Bartra, S. S., Gallis, B. M., Doneanu, C. E., Goodlett, D. R., et al. (2007). Identification and type III-dependent secretion of the Yersinia pestis insecticidal-like proteins. Mol. Microbiol. 64, 1214–1227. doi: 10.1111/j.1365-2958.2007.05729.x
Ismail, E., Blom, J., Bultreys, A., Ivanovic, M., Obradovic, A., van Doorn, J., et al. (2014). A novel plasmid pEA68 of Erwinia amylovora and the description of a new family of plasmids. Arch. Microbiol. 196, 891–899. doi: 10.1007/s00203-014-1028-5
Jones, A. L., and Schnabel, E. L. (2000). “The development of streptomycin resistant strains of Erwinia amylovora,” in Fire Blight: the Disease and its Causative Agent Erwinia amylovora, ed. J. L. Vanneste (Wallingford: CAB International), 235–251. doi: 10.1079/9780851992945.0235
Kamber, T., Smits, T. H. M., Rezzonico, F., and Duffy, B. (2012). Genomics and current genetic understanding of Erwinia amylovora and the fire blight antagonist Pantoea vagans. Trees Struct. Funct. 26, 227–238. doi: 10.1007/s00468-011-0619-x
Kim, W. S., and Geider, K. (1999). Analysis of variable short-sequence DNA repeats on the 29 kb plasmid of Erwinia amylovora strains. Eur. J. Plant Pathol. 105, 703–713. doi: 10.1023/A:1008723717211
Kim, W. S., Gardan, L., Rhim, S. L., and Geider, K. (1999). Erwinia pyrifoliae sp. nov., a novel pathogen that affects Asian pear trees (Pyrus pyrifolia Nakai). Int. J. Syst. Bacteriol. 49, 899–906. doi: 10.1099/00207713-49-2-899
Kim, W. S., Schollmeyer, M., Nimtz, M., Wray, V., and Geider, K. (2002). Genetics of biosynthesis and structure of the capsular exopolysaccharide from the Asian pear pathogen Erwinia pyrifoliae. Microbiology 148, 4015–4024.
Koczan, J. M., McGrath, M. J., Zhao, Y., and Sundin, G. W. (2009). Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99, 1237–44 doi: 10.1094/PHYTO-99-11-1237
Kube, M., Migdoll, A. M., Gehring, I., Heitmann, K., Mayer, Y., Kuhl, H., et al. (2010). Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae. BMC Genomics 11:393. doi: 10.1186/1471-2164-11-393
Kube, M., Migdoll, A. M., Müller, I., Kuhl, H., Beck, A., Reinhardt, R., et al. (2008). The genome of Erwinia tasmaniensis strain Et1/99, a non-pathogenic bacterium in the genus Erwinia. Environ. Microbiology 10, 2211–2222. doi: 10.1111/j.1462-2920.2008.01639.x
Kulinska, A., Czeredys, M., Hayes, F., and Jagura-Burdzy, G. (2008). Genomic and functional characterization of the modular broad-host-range RA3 plasmid, the archetype of the IncU group. Appl. Environ. Microbiol. 74, 4119–4132. doi: 10.1128/AEM.00229-08
Llop, P., Barbé, S., and López, M. M. (2012). Functions and origin of plasmids in Erwinia species that are pathogenic to or epiphytically associated with pome fruit trees. Trees Struct. Funct. 26, 31–46. doi: 10.1007/s00468-011-0630-2
Llop, P., Cabrefiga, J., Smits, T. H. M., Dreo, T., Barbé, S., Pulawska, J., et al. (2011). Erwinia amylovora novel plasmid pEI70: complete sequence, biogeography, and role in aggressiveness in the fire blight phytopathogen. PLoS ONE 6:e28651. doi: 10.1371/journal.pone.0028651
Llop, P., Donat, V., Rodríguez, M., Cabrefiga, J., Ruz, L., Palomo, J. L., et al. (2006). An indigenous virulent strain of Erwinia amylovora lacking the ubiquitous plasmid pEA29. Phytopathology 96, 900–907. doi: 10.1094/PHYTO-96-0900
Llop, P., González, R., Pulawska, J., Bultreys, A., Dreo, T., and López, M. M. (2008). The new plasmid pEI70 is present in Erwinia amylovora European strains. Acta Hortic. 793, 131–136. doi: 10.17660/actahortic.2008.793.15
López, M. M., Roselló, M., Llop, P., Ferrer, S., Christen, R., and Gardan, L. (2011). Erwinia piriflorinigrans sp. nov., a novel pathogen that causes necrosis of pear blossoms. Int. J. Syst. Evol. Microbiol. 61, 561–567. doi: 10.1099/ijs.0.020479-0
Malnoy, M., Martens, S., Norelli, J. L., Barny, M., Sundin, G. W., Smits, T. H. M., et al. (2012). Fire Blight: applied genomic insights of the pathogen and host. Ann. Rev. Phytopathol. 50, 475–494. doi: 10.1146/annurev-phyto-081211-172931
Mann, R. A., Blom, J., Bühlmann, A., Plummer, K. M., Beer, S. V., Luck, J. E., et al. (2012). Comparative analysis of the Hrp pathogenicity island of Rubus- and Spiraeoideae-infecting Erwinia amylovora strains identifies the IT region as a remnant of an integrative conjugative element. Gene 504, 6–12. doi: 10.1016/j.gene.2012.05.002
Mann, R. A., Smits, T. H. M., Bühlmann, A., Blom, J., Goesmann, A., Frey, J. E., et al. (2013). Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core. PLoS ONE 8:e55644. doi: 10.1371/journal.pone.0055644
Matsuura, T., Mizuno, A., Tsukamoto, T., Shimizu, Y., Saito, N., Sato, S., et al. (2012). Erwinia uzenensis sp. nov., a novel pathogen that affects European pear trees (Pyrus communis L.) Int. J. Syst. Evol. Microbiol. 62, 1799–1803. doi: 10.1099/ijs.0.032011-0
Mavrodi, D. V., Loper, J. E., Paulsen, I. T., and Thomashow, L. S. (2009). Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol. 9:8. doi: 10.1186/1471-2180-9-8
Maxson-Stein, K., McGhee, G. C., Smith, J. J., Jones, A. L., and Sundin, G. W. (2003). Genetic analysis of a pathogenic Erwinia sp. isolated from pear in Japan. Phytopathology 93, 1393–1399. doi: 10.1094/PHYTO.2003.93.11.1393
McGhee, G. C., and Jones, A. L. (2000). Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl. Environ. Microbiol. 66, 4897–4907. doi: 10.1128/AEM.66.11.4897-4907.2000
McGhee, G. C., Schnabel, E. L., Maxson-Stein K, Jones, B., Stromberg, V. K., Lacy, G. H., et al. (2002). Relatedness of chromosomal and plasmid DNAs of Erwinia pyrifoliae and Erwinia amylovora. Appl. Environ. Microbiol. 68, 6182–6192. doi: 10.1128/AEM.68.12.6182-6192.2002
McGhee, G. C., and Sundin, G. W. (2011). Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology 101, 192–204. doi: 10.1094/PHYTO-04-10-0128
McManus, P. S., and Jones, A. L. (1994). Epidemiology and genetic analysis of streptomycin-resistant Erwinia amylovora from Michigan and evaluation of oxytetracycline for control. Phytopathology 84, 627–633. doi: 10.1094/Phyto-84-627
Mergaert, J., Hauben, L., Cnockaert, M. C., and Swings, J. (1999). Reclassification of non-pigmented Erwinia herbicola strains from trees as Erwinia billingiae sp. nov. Int. J. Syst. Bacteriol. 49, 377–383. doi: 10.1099/00207713-49-2-377
Oh, C-S., and Beer, S. V. (2005). Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192. doi: 10.1016/j.femsle.2005.09.051
Oh, C. -S., Kim, J. F., and Beer, S. V. (2005). The Hrp pathogenicity island of Erwinia amylovora and the identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6, 125–138. doi: 10.1111/j.1364-3703.2005.00269.x
Palacio-Bielsa, A., Roselló, M., Llop, P., and López, M. M. (2011). Erwinia spp. from pome fruit trees: similarities and differences among pathogenic and non-pathogenic species. Trees Struct. Funct. 26, 13–29. doi: 10.1007/s00468-011-0644-9
Palmer, E. L., Teviotdale, B. L., and Jones, A. L. (1997). A relative of the broad-host-range plasmid RSF1010 detected in Erwinia amylovora. Appl. Environ. Microbiol. 63, 4604–4607.
Powney, R., Smits, T. H. M., Sawbridge, T., Frey, B., Blom, J., Frey, J. E., et al. (2011). Genome sequence of an Erwinia amylovora strain with restricted pathogenicity to Rubus plants. J. Bacteriol. 193, 785–786. doi: 10.1128/JB.01352-10
Preston, K. E., Radomski, C. C. A., and Venezia, R. A. (2000). Nucleotide sequence of a 7-kb fragment of pACM1 encoding an IncM DNA primase and other putative proteins associated with conjugation. Plasmid 44, 12–23. doi: 10.1006/plas.2000.1472
Records, A. R. (2011). The type VI secretion system: a multi-purpose delivery system with a phage-like machinery. Mol Plant Microbe Interact. 24, 751–757. doi: 10.1094/MPMI-11-10-0262
Rhim, S. L., Völksch, B., Gardan, L., Paulin, J. P., Langlotz, C., Kim, W. S., et al. (1999). Erwinia pyrifoliae, an Erwinia species different from Erwinia amylovora, causes a necrotic disease of Asian pear trees. Plant Pathol. 48, 514–520. doi: 10.1046/j.1365-3059.1999.00376.x
Sebaihia, M., Bocsanczy, A. M., Biehl, B. S., Quail, M. A., Perna, N. T., Glasner, J. D., et al. (2010). Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J. Bacteriol. 192, 2020–2021. doi: 10.1128/JB.00022-10
Shrestha, R., Koo, J. H., Park, D. H., Hwang, I., Hur, J. H., and Lim, C. K. (2003). Erwinia pyrifoliae, a causal endemic pathogen of shoot blight of Asian pear tree in Korea. Plant Pathol. J. 19, 294–300. doi: 10.5423/PPJ.2003.19.6.294
Smits, T. H. M., Guerrero-Prieto, V. M., Hernández-Escarcega, G., Blom, J., Goesmann, A., Rezzonico, F., et al. (2014). Whole-Genome Sequencing of Erwinia amylovora strains from Mexico detects single nucleotide polymorphisms in rpsL conferring streptomycin resistance and in the avrRpt2 effector altering host interactions. Genome Announc. 2:e01229-13. doi: 10.1128/genomeA.01229-13
Smits, T. H. M., Jaenicke, S., Rezzonico, F., Kamber, T., Goesmann, A., Frey, J. E., et al. (2010a). Complete genome sequence of the fire blight pathogen Erwinia pyrifoliae DSM 12163T and comparative genomic insights into plant pathogenicity. BMC Genomics 11:2. doi: 10.1186/1471-2164-11-2
Smits, T. H. M., Rezzonico, F., Kamber, T., Blom, J., Goesmann, A., Frey, J. E., et al. (2010b). Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP1430 and comparison to other Erwinia spp. Mol. Plant Microbe Interact 23, 384–393. doi: 10.1094/MPMI-23-4-0384
Smits, T. H. M., Rezzonico, F., Kamber, T., Goesmann, A., Ishimaru, C. A., Stockwell, V. O., et al. (2010c). The genome sequence of the biocontrol agent Pantoea vagans strain C9-1. J. Bacteriol. 192, 6486–6487. doi: 10.1128/JB.01122-10
Smits, T.H.M., Rezzonico, F., and Duffy, B. (2011). Evolutionary insights from Erwinia amylovora genomics. J. Biotechnol. 155, 34–39. doi: 10.1016/j.jbiotec.2010.10.075
Smits, T. H. M., Rezzonico, F., López, M. M., Blom, J., Goesmann, A., Frey, J. E., et al. (2013). Phylogenetic position and virulence apparatus of the pear flower necrosis pathogen Erwinia piriflorinigrans CFBP 5888T as assessed by comparative genomics. Systematic Appl. Microbiol. 36, 449–456. doi: 10.1016/j.syapm.2013.04.003
Sundin, G. W. (2007). Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu. Rev. Phytopathol. 45, 129–151. doi: 10.1146/annurev.phyto.45.062806.094317
Sundin, G. W., and Bender, C. L. (1995). Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61, 2891–2897.
Sundin, G. W., and Bender, C. L. (1996). Dissemination of the strA-strB streptomycin resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 5, 133–143. doi: 10.1111/j.1365-294X.1996.tb00299.x
Thapa, S. P., Park, D. H., Kim, W. S., Choi, B. S., Lim, J. S., and Choi, I. Y. (2013). Comparative genomics of Japanese Erwinia pyrifoliae strain Ejp617 with closely related erwinias. Genome 56, 83–90. doi: 10.1139/gen-2012-0094
Toth, I. K., Pritchard, L., and Birch, P. J. R. (2006). Comparative genomics reveals what makes an enterobacterial plant pathogen. Annu. Rev. Phytopathol. 44, 305–336. doi: 10.1146/annurev.phyto.44.070505.143444
Triplett, L., Zhao, Y. F., and Sundin, G. W. (2006). Genetic differences among blight-causing Erwinia species with differing host specificities identified by suppression subtractive hybridization. Appl. Environ. Microbiol. 72, 7359–7364. doi: 10.1128/AEM.01159-06
Vrancken, K., Holtappels, M., Schoofs, H., Deckers, T., and Valcke, R. (2013). Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in Rosaceae: state of the art. Microbiology 159, 823–832. doi: 10.1099/mic.0.064881-0
Wei, Z. M., and Beer, S. V. (1993). HrpI of Erwinia amylovora functions in secretion of harpin and is a member of a new protein family. J Bacteriol. 175, 7958–7967.
Young, B. M., and Young, G. M. (2002). YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184, 1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002
Zhao, Y. F., and Qi, M. (2011). Comparative genomics of Erwinia amylovora and related Erwinia species—what do we learn? Genes 2, 627–639. doi: 10.3390/genes2030627
Zhao, Y. F., Qi, M., and Wang, D. (2011). Evolution and function of flagellar and non-flagellar type III secretion systems in Erwinia amylovora. Acta Hort. 896, 177–184. doi: 10.17660/actahortic.2011.896.23
Keywords: Erwinia genus, gene similarity, transfer elements, genetic diversity, gene interaction
Citation: Llop P (2015) Genetic islands in pome fruit pathogenic and non-pathogenic Erwinia species and related plasmids. Front. Microbiol. 6:874. doi: 10.3389/fmicb.2015.00874
Received: 31 October 2014; Accepted: 10 August 2015;
Published: 28 August 2015.
Edited by:
Kornelia Smalla, Julius Kühn-Institut, GermanyReviewed by:
Awdhesh Kalia, The University of Texas MD Anderson Cancer Center, USAKlaus Geider, Julius Kühn-Institut, Germany
Copyright © 2015 Llop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pablo Llop, Department of Evolutionary Genetics, Cavanilles Institute, University of Valencia, Calle Catedrático José Beltrán Martínez, nº 2, 46980 Paterna, Valencia, Spain, pablo.llop@uv.es
 Pablo Llop
Pablo Llop