- 1Department of Biological Sciences, University of Calgary, Calgary, AB, Canada
- 2Geological Survey of Canada, Calgary, AB, Canada
Carbon monoxide (CO) is a potential energy and carbon source for thermophilic bacteria in geothermal environments. Geothermal sites ranging in temperature from 45 to 65°C were investigated for the presence and activity of anaerobic CO-oxidizing bacteria. Anaerobic CO oxidation potentials were measured at up to 48.9 μmoles CO g−1 (wet weight) day−1 within five selected sites. Active anaerobic carboxydotrophic bacteria were identified using 13CO DNA stable isotope probing (SIP) combined with pyrosequencing of 16S rRNA genes amplified from labeled DNA. Bacterial communities identified in heavy DNA fractions were predominated by Firmicutes, which comprised up to 95% of all sequences in 13CO incubations. The predominant bacteria that assimilated 13C derived from CO were closely related (>98% 16S rRNA gene sequence identity) to genera of known carboxydotrophs including Thermincola, Desulfotomaculum, Thermolithobacter, and Carboxydocella, although a few species with lower similarity to known bacteria were also found that may represent previously unconfirmed CO-oxidizers. While the distribution was variable, many of the same OTUs were identified across sample sites from different temperature regimes. These results show that bacteria capable of using CO as a carbon source are common in geothermal springs, and that thermophilic carboxydotrophs are probably already quite well known from cultivation studies.
Introduction
Carbon monoxide (CO) is an odorless gas toxic to many animals due to its competitive binding to hemoglobin (Haab, 1990). It has been estimated that about 3.3 × 109 metric tons of CO are released annually to the atmosphere (Conrad, 1996). There are numerous natural biogenic and abiogenic sources of CO. Thermal decomposition and photochemical degradation of organic compounds are important sources of abiotic CO (Sipma et al., 2006). CO is also a component of volcanic emissions, which may contain as much as 1–2% of CO per volume of total gas (Giggenbach, 1980; Svetlichny et al., 1991a; Sokolova et al., 2009 and references therein). Biogenic CO may also be produced in microbial ecosystems, and net CO production has been reported for marine algae (Conrad, 1988) and hypersaline cyanobacterial mats (Hoehler et al., 2001) during photosynthesis. Sulfate-reducing bacteria (SRB) have also been shown to produce CO during fermentation (Voordouw, 2002). Some microbes growing in high temperature environments are likely capable of growth at low concentrations of CO (Sokolova et al., 2009). It has also been suggested that CO-oxidizing microbes may occupy micro-niches in which biogenic CO locally accumulates to high levels (Techtmann et al., 2009).
Microorganisms that have the ability to oxidize CO are termed “carboxydotrophs” (King and Weber, 2007). A number of aerobic and anaerobic bacteria as well as some anaerobic archaea (e.g., methanogens) are capable of using CO as a source of energy and/or carbon (e.g., Mörsdorf et al., 1992; Oelgeschläger and Rother, 2008; Sokolova et al., 2009). Carboxydotrophic energy generation employs the enzyme CO dehydrogenase (CODH) that oxidizes CO to CO2, generating electrons. The aerobic and anaerobic versions of this enzyme differ. Anaerobic CODH in bacteria is encoded by cooS genes (Techtmann et al., 2011) and contains nickel in the active site, while aerobic CODH is encoded by cox genes and contains molybdenum (e.g., Dobbek et al., 2001; King and Weber, 2007). Within the Domain Bacteria, anaerobic carboxydotrophs are typically found within the phylum Firmicutes and some in the class Alphaproteobacteria of the Proteobacteria (see Techtmann et al., 2009). Purple non-sulfur bacteria (i.e., phototrophic Alphaproteobacteria) exist among the known anaerobic CO oxidizers and were among the first discovered (Uffen, 1981; Kerby et al., 1995). However, an increasing number have been identified that are strictly anaerobic thermophiles belonging to the phylum Firmicutes (e.g., Svetlichny et al., 1991b; Sokolova et al., 2002). Hydrothermal systems have been proposed as early ecosystems supporting chemolithotrophic life, including thermophilic anaerobic bacteria and archaea using CO as an energy and carbon source (e.g., Cavicchioli, 2002; Wächtershäuser, 2006; King and Weber, 2007). Examples of thermophilic archaea that use CO include Thermococcus sp. NA1, capable of both heterotrophic and carboxydotrophic growth (Lee et al., 2008) and Archaeoglobus fulgidus capable of using CO as an autotrophic growth substrate (Henstra et al., 2007a).
While the anaerobic oxidation of CO may be coupled to a variety of respiratory processes such as sulfate reduction and acetogenesis (Oelgeschläger and Rother, 2008), hydrogenogenic carboxydotrophs make up the majority of thermophilic CO oxidizing microbes that have been identified in geothermal environments (e.g., Svetlichny et al., 1991a,b; Sokolova et al., 2004, 2005; Slepova et al., 2006). These bacteria oxidize CO via the water-gas-shift reaction (Uffen, 1981; Sipma et al., 2006):
Thermophilic bacteria and archaea with the capacity for hydrogenogenic carboxydotrophy have been isolated from various locations around the world including the Kunashir Island, Russia (Svetlichny et al., 1991a), Kamchatka (Sokolova et al., 2002; Slepova et al., 2006), Yellowstone National Park (Sokolova et al., 2004), and Iceland (Novikov et al., 2011). The isolates share similar ranges of optimal pH (ca. 6.8–7.0) and temperature (ca. 55–83°C) (see Henstra et al., 2007b; Techtmann et al., 2009) but are phylogenetically divergent (Techtmann et al., 2009). Unlike mesophilic carboxydotrophs, the thermophilic hydrogenogenic species isolated so far do not show growth inhibition by high levels of CO. In fact, most grow under atmospheres of 100% CO, far above natural CO concentrations in geothermal systems (e.g., Svetlichny et al., 1991a,b).
CO oxidizing thermophiles are of potential biotechnological interest for the anaerobic fermentation of synthesis gas (“syngas”). Syngas is a product comprised mostly of H2, CO, and CO2 resulting from the high temperature gasification of waste biomass, into higher-value bioalcohol fuel (Henstra et al., 2007b). As syngas is produced at high temperatures, bacteria from geothermal sites are of particular interest due to the expected high rates of substrate conversion at high temperatures. Characterization of anaerobic CO-oxidizing bacteria in geothermal systems therefore could provide fundamental information about the natural diversity of thermophilic carboxydotrophs available for these biotechnological applications. DNA-Stable Isotope Probing (SIP) is a valuable tool in assessment of functional bacterial groups. It has been used in a variety of environments to identify active consumers of substrates such as methane (He et al., 2012; Sharp et al., 2012, 2014b). There are however some difficulties in applying SIP to identify carboxydotrophs. Firstly, while some carboxydotrophic bacteria directly incorporate CO-carbon into the carboxyl group of acetate via acetyl-CoA synthases (ACSs) using the Wood-Ljungdahl pathway (e.g., Henstra et al., 2007b), others incorporate the CO2 produced from CO oxidation as the direct source of cellular carbon, using reductive CO2 pathways such as the Calvin-Benson-Bassham (CBB) Cycle, or the reverse tricarboxylic acid (TCA) cycle (Uffen, 1981; Ragsdale, 1991, 2004; Berg, 2011). As such, CO2 present in the atmosphere may be incorporated rather than the CO2 produced directly from the oxidation of CO diluting the labeling effect. Another issue with CO-SIP is that the products of CO oxidation, i.e., H2 and CO2, may result in labeling of other autotrophs. Cross-feeding is a caveat in any SIP experiment, but the severity of the cross-feeding, especially via CO2, can often be assessed with controls such as the addition of exogenous 12CO2 (e.g., Sharp et al., 2012). CO-SIP will also not detect carboxydoheterotrophs that may oxidize CO but use a different carbon source. Therefore, in this study we assessed the value of CO-SIP to identify anaerobic thermophilic carboxydoautotrophic bacteria in some geothermal springs.
Materials and Methods
Geothermal Spring Sample Collection
Five geothermal springs were selected for CO-SIP investigations from among geothermal sites in Western Canada (Grasby et al., 2000; Sharp et al., 2012). Soil or sediment or biomat samples were collected at various times of the year between fall 2010 and fall 2012 into sterile screw-cap tubes (Table 1). Samples were kept cold as soon as possible to minimize changes in the microbial community during transport. Collected material was sub-sampled within approximately 5 days of sampling for DNA extraction, and the remainder was stored at 4°C for 1–2 d prior to incubation studies.
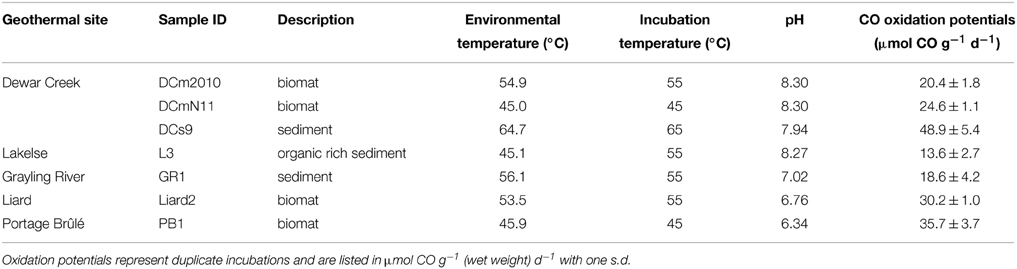
Table 1. Name of geothermal spring, measured in situ pH and temperature, and anaerobic CO-oxidation potentials for sites included in the current study.
Soil Microcosms and CO Oxidation
Approximately 2–5 g of sample material (wet weight) was added to 120-ml serum bottles and crimp-sealed with sterile blue butyl rubber stoppers. An anoxic environment was created by repeated (3 ×) evacuation and refilling with N2 gas. CO was added to final mixing ratios of 5–10% in the headspace to assess oxidation potential and act as a 12C incubation for stable isotope probing (SIP) experiments. For SIP experiments, labeled gases 13CO (99 atom % 13C, Sigma Aldrich) and 13CO2 (99 atom % 13C, Sigma Aldrich) were used at mixing ratios of 10% v/v, both separately and in combination with non-labeled CO and CO2 gases in different SIP trials. “Control” is used to refer to the un-incubated (i.e., no CO or CO2 added) environmental samples. Microcosms were incubated at close to environmental temperatures (Table 1). Headspace CO was monitored at ca. 1-d intervals using a Varian 450-Gas Chromatograph equipped with a 0.5-m Hayseep N and a 1.2-m Mol Sieve 16X column in series coupled to a Thermal Conductivity Detector (GC/TCD). Potential production of methane was monitored using a GC-Flame Ionization Detector (FID). Incubations proceeded until approximately 95–100% of the added CO had been consumed, typically within a week. Samples were then harvested and frozen immediately at −85°C. CO consumption rates were based on duplicates of any sample (12CO and 13CO only incubations), while duplicate SIP experiments were performed only in some cases.
DNA Extraction and Density Fractionation
DNA was extracted from approximately 500 mg of sample using the FastDNA Spin Kit (MP Biomedicals) with the addition of washing steps using guanidine thiocyanate (Knief et al., 2003). Quantification was performed using the Quant-iT™ dsDNA HS Assay Kit (Invitrogen) and extracted DNA was stored at −20°C prior to ultracentrifugation separation. Heavy and light DNA were separated by density gradient ultracentrifugation using cesium chloride (CsCl) as described by Neufeld et al. (2007), with minor modifications. Five hundred nanograms to one microgram of total DNA was typically used for each SIP assay. To account for any variability in DNA distribution patterns that may arise due to using inconsistent amounts of DNA in CsCl gradients, within an individual sample set (i.e., all trials from the same sample site) similar total DNA amounts were used. Centrifugation and gradient fractionation were performed as described by Sharp et al. (2014b). DNA was precipitated from each fraction using polyethylene glycol (PEG) and glycogen as in Neufeld et al. (2007). DNA present in each density gradient was quantified using the Quant-iT™ dsDNA HS Assay Kit (Invitrogen). As the focus of this paper is assessing bacterial rather than archaeal carboxydotrophy, 16S rRNA gene PCR assays for each fraction were set-up using a QIAgility (v. 4.13.5) with bacterial specific primers 519f and 907r (Stubner, 2002). 16S rRNA gene copies were quantified on a Rotor-Gene Q (Qiagen) as in Sharp et al. (2012).
Microbial Community Analysis
The density gradients of DNA extracted from incubations with 13C-labeled vs. 12C-labeled substrates were compared. Heavy SIP fractions with increases in the relative amounts of DNA and/or 16S rRNA gene copies were selected for pyrosequencing analysis of the 16S rRNA gene. “Light” fractions (density ca. 1.690 g ml−1) from 13CO incubated samples were also analyzed in some cases for comparison to heavy fractions (Table S1). Where possible, DNA was also amplified from the corresponding heavy fractions from untreated controls (Figure 2, Table S1). However, in some cases the amount of DNA present in the fractions was below detection, or too low to obtain enough for pyrosequencing (e.g., DCm2010) (Supplementary Figure S1). Samples were prepared for sequencing analysis as described previously (Grasby et al., 2013; Sharp et al., 2014a) using FLX Titanium amplicon primers 454T_RA_X and 454T_F, which contain 16S rRNA gene targeted primers 926fw (5′-aaactYaaaKgaattgRcgg-3′) and 1392r (5′-acgggcggtgtgtRc-3′) designed to target both bacteria and archaea (Ramos-Padrón et al., 2011). PCR reactions and purification were performed as described in Sharp et al. (2014a). Purified PCR products (ca. 150 ng total DNA) were analyzed at the Genome Quebec and McGill University Innovation Centre, Montreal, Quebec on a 454 Life Sciences Genome Sequencer FLX (Roche) machine running the Titanium chemistry.
Sequence Data Processing
Quantitative Insights Into Microbial Ecology (QIIME) pipeline version 1.8 (Caporaso et al., 2010) was used to process raw sequence data as in Sharp et al. (2014a). A minimum quality score of 25 was used and sequences were screened using ChimeraSlayer (Haas et al., 2011). Taxonomic identification of a representative sequence (most common) for each phylotype (clustered at 97% similarity) was determined using nucleotide Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990) against the Silva 111 reference database (Pruesse et al., 2007). Eukaryotic and chloroplast sequences were removed from further analysis. Final numbers ranged from 2195 to 21118 sequences per sample (Table S1). A phylogenetic tree was constructed using the parsimony-add function in ARB (Ludwig et al., 2004).
16S rRNA gene sequences obtained from this study have been deposited in the SRA database under accession numbers SRP028305 and SRP059036. Representative sequences of identified OTUs present at 25 fold enrichment are provided in the Supplementary Material.
Results
Biodegradation of Carbon Monoxide
Anaerobic oxidation of carbon monoxide was detected via monitoring of the CO mixing ratio in serum bottle headspaces after adding CO. Communities that showed evidence for CO consumption were selected for further investigation using stable isotope probing (SIP). The average values for anaerobic CO oxidation potentials for each sample is listed in Table 1. Rates ranged from 13.6 to 48.9 μmol CO g−1 d−1. The fastest rates were observed in samples from the Dewar Creek hot spring. Despite the anaerobic conditions, no methane production was observed in sample sites GraylingRiver1, Liard2, and PortageBrûlé1 during any incubation and only minor amounts were observed in other Dewar Creek and Lakelse samples, with estimated CH4 production rates of 0.013–11.4 nmol mol day−1 g−1. The amount of CH4 produced corresponds to ca. < 0.1% of the added CO being converted to methane. One exception was a Lakelse sample that produced more total CH4 (~ 18 μmol) than CO added, indicating other substrates were present for methanogenesis.
Identification of Active CO Consuming Bacteria Using SIP Combined with 16S rRNA Gene Sequencing
Density profiles of DNA extracted from samples after incubation with 13CO generally ranged from 1.660 to 1.800 g ml−1. Shifts in DNA density compared to un-amended control samples were often subtle (Supplementary Figure S1). Therefore, quantitative real-time PCR (qPCR) counts of total bacterial 16S rRNA gene abundance were used to identify density fractions that had an increase in the relative number of gene copies (Figure 1, Supplementary Figure S2).
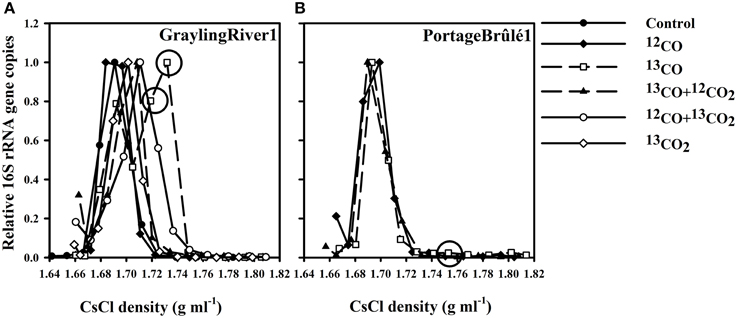
Figure 1. (A) Representative 16S rRNA gene counts vs. DNA density for SIP experiments using GraylingRiver1 sediment, illustrating the shifts in relative 16S rRNA gene copies associated with different incubations. The incubations using only 13CO as the sole carbon and energy source showed the greatest increase in the relative number of 16S rRNA gene copies in heavy fractions at densities of approximately 1.730 g ml−1. DNA from these heavy fractions was used for 16S rRNA gene pyrosequencing. (B) Relative 16S rRNA gene copies from PB1 sediment showing no density shift with 13CO incubations despite active CO oxidation. Circles indicate 13CO incubation fractions used for 16S rRNA gene pyrosequencing analyses. “Control” represents the un-incubated (no CO or CO2 added) environmental sample.
Representative profiles of 16S rRNA gene copies vs. DNA density for GraylingRiver1 (GR1) and PortageBrûlé1 (PB1) SIP experiments are shown in Figure 1. The number of 16S rRNA gene copies in the control environmental communities peaked in the density range of 1.690–1.700 g ml−1. Incubations using 12CO showed the same peaks, demonstrating that incubation with CO itself had little effect on the overall DNA density profile of the community. However, incubation of GR1 with 13CO resulted in a marked shift in 16S rRNA gene copies toward heavier fractions, indicating assimilation of the 13C from 13CO into DNA. In contrast, no significant shift in DNA density or 16S rRNA gene copies was observed over multiple incubations of the PB1 sample under 13CO. Although this sample also oxidized CO, no assimilation of 13C was evident. These two samples were representative of the two major patterns observed. Other samples analyzed are shown in Supplementary Figures S1, S2.
A fundamental issue with interpreting SIP results is cross-labeling of other bacteria via metabolic products, especially CO2. The severity of this problem can be estimated via several controls. In GR1 (Figure 1A), as well as in several other samples tested (Supplementary Figures S1, S2) incubations with only 13CO2 and no added CO always showed very minor density shifts in DNA and 16S rRNA gene counts. This indicated that other autotrophs growing on substrates such as sulfur and ammonia were of minor importance. CO was therefore the primary energy source in the incubations, and the food webs detected were ultimately based on CO oxidation.
In samples like GR1, microcosms containing 13CO as the sole carbon and energy source (with no CO2 addition) displayed the greatest shift in density (Figure 1). Less of a shift in density was observed in incubation with 13CO + 12CO2, probably because assimilation of 12C from the added CO2 diluted the labeling effect. Incubation with 12CO + 13CO2 showed some increase in 16S rRNA gene copies compared to the un-incubated control, albeit to a lesser extent than in 13CO. The apparent assimilation of C preferably from 13CO but also from 13CO2 indicates that CO2 was probably the primary C source of the CO oxidizers, but that there may be a diffusion effect whereby the 13CO2 produced directly by a carboxydotroph from 13CO is preferentially assimilated compared to exogenous 12CO2 supplied in the atmosphere.
In the majority of cases, the “heavy” fractions that showed the greatest increase in 16S rRNA gene copies were at a density of ca. 1.730 g ml−1 (Figure 1, Supplementary Figure S2). Heavy fractions that contained a large increase in DNA amounts and/or 16S rRNA gene copies as compared to an un-incubated, non-labeled control were selected for 16S rRNA gene sequencing. In some cases (e.g., DCm2010), the amount of DNA present in the corresponding high-density fractions of the unlabeled controls was too low to obtain enough for successful pyrotag sequencing despite inputs of similar total amounts of DNA (see Supplementary Figure S1).
Most predominant operational taxonomic units (OTUs) in the heavy fractions of all samples (i.e., most putative carboxydotrophs) belonged to the phylum Firmicutes, in particular to the class Clostridia. In heavy fractions recovered from 13CO incubations that showed observable shifts in 16S rRNA gene copies, members of the phylum Firmicutes accounted for 31–95% of the reads (Table S1). One of the lowest % of Firmicutes was from GR1 which had a very high proportion of Crenarchaeota in both the original community and in all heavy fractions. However, of bacterial sequences only, Firmicutes accounted for 81.5% in the heavy fraction of GR1_13CO. The proportions of top phyla (>1% of sequences) of both the un-amended environmental samples and the heavy fractions for two sites that showed strong DNA labeling are shown in Figure 2.
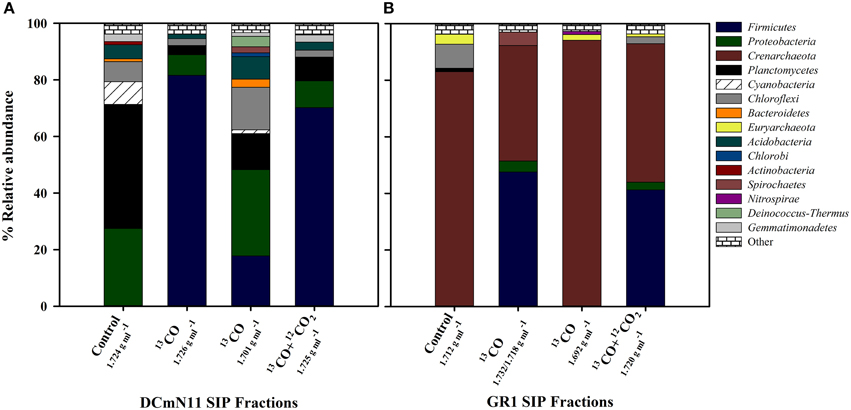
Figure 2. Relative abundance (%) of different phyla for SIP incubations showing results from heavy (density ca. 1.730 g ml−1) fractions and light fractions (density ca. 1.700 g ml−1) of samples incubated with 13CO or 13CO + 12CO2 compared to the community detected in heavy fractions of un-incubated Control samples (i.e., no CO or CO2 added). (A) DCmN11 showing significant increase in the proportion of Firmicutes in heavy fractions and (B) GR1 showing an increase in Firmicutes but also the large proportion of Crenarchaeota present in both the un-incubated environmental control sample and SIP fractions. 16S rRNA gene sequences were clustered at 97% similarity and classified using QIIME. “Other” includes phyla present at < 1%.
The taxonomic identifications of OTUs recovered from13CO microcosm heavy fractions that were present at 25 fold enrichment compared to the original environmental sample are shown in Table 2. A 16S rRNA gene phylogenetic tree was constructed using the top OTUs from each 13CO heavy fraction showing an observable shift compared to reference sequences (Figure 3). Taxonomic identifications of OTUs present at >1% of all sequences are presented in Supplementary Table S2. Most of the putative carboxydotrophs identified belonged with >98% sequence identity to genera that include known CO-oxidizers, such as Thermincola, Desulfotomaculum, Carboxydocella, and Thermolithobacter (Sokolova et al., 2002, 2005, 2007; Parshina et al., 2005a,b). For example, the most abundant OTU (OTU_17948) recovered from 13CO heavy fractions from Dewar Creek (DCm2010 and DCmN11) and Lakelse springs showed 99% sequence identity to both Thermincola potens and to Thermincola carboxydiphila, known CO-oxidizing bacteria (Sokolova et al., 2005; Byrne-Bailey et al., 2010) (Table 2). Members of the genus Carboxydocella were also detected in the majority of heavy fractions across all sites. This genus was represented by multiple OTUs, however the top OTU_7600 identified in most samples corresponded to Carboxydocella thermautotrophica (98% similarity). Members of the genus Desulfotomaculum were most predominant in DCs9, a sediment sample collected from Dewar Creek. In DCs9_13CO incubations, 31.9% of sequences were attributed to the genus Desulfotomaculum. 30.0% of sequences were in OTU_3148, which showed 99% sequence identity to D. kuznetsovii and D. luciae.
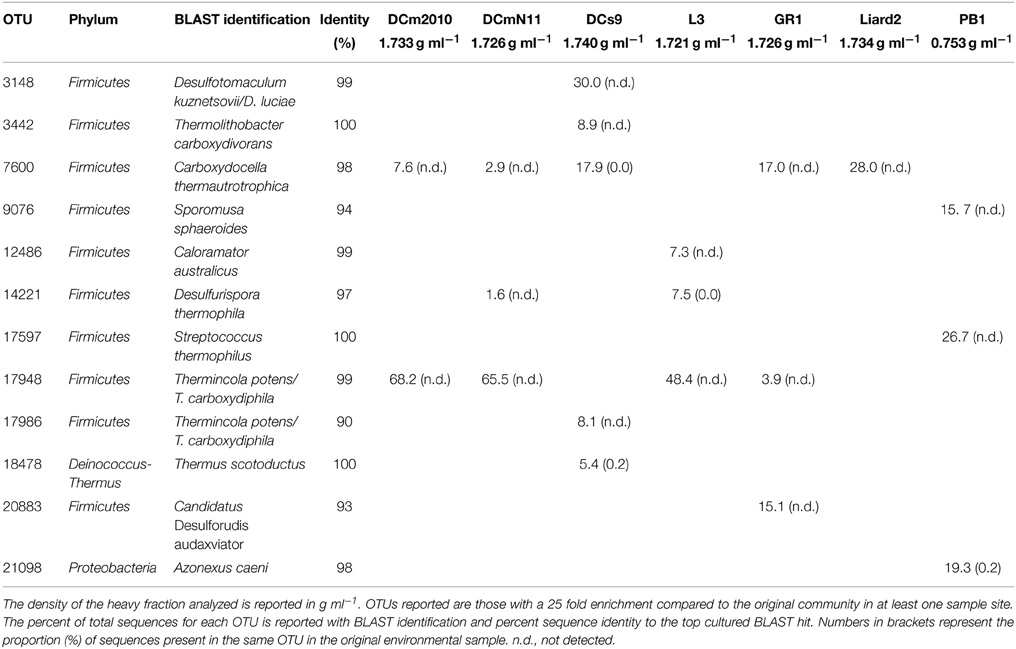
Table 2. The percent of total sequences associated with top OTUs identified in heavy fractions of 13CO incubations.
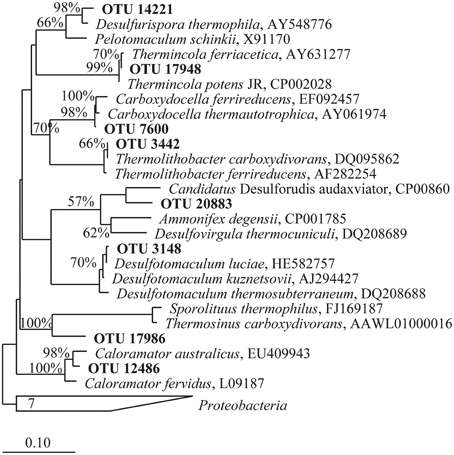
Figure 3. Phylogenetic tree of partial 16S rRNA gene sequences belonging to OTUs identified in heavy fractions from 13CO SIP incubations present at a minimum of 25 fold enrichment. OTUs presented are from those incubations that showed an observable shift in density and are hypothesized to represent microbes that oxidized CO. A skeleton tree was constructed from near complete 16S rRNA gene sequences (>1400 bp) via Neighbor-joining with a Jukes-Cantor correction and 10,000 bootstraps. Shorter sequences produced via 454 pyrosequencing obtained in this study were added by parsimony using ARB (in bold). The scale bar represents 0.1 change per nucleotide position. Bootstrap support values greater than 55% for the major nodes are given. The tree was rooted using 7 Proteobacteria 16S rRNA gene sequences.
Some OTUs identified may reflect bacteria with as yet unknown or unconfirmed CO-oxidizing capabilities. One such cluster, OTU_20883, present in both the GR1_13CO and GR1_13CO + 12CO2 heavy fraction at 15.1 and 5.3% of sequences respectively, is 93% similar to Candidatus “Desulforudis audaxviator.” Neither this OTU nor any others that showed any similarity to this bacterium were detected in the control environmental sample from this site. BLAST results for OTU_17986 representing 8.1% of the total for DCs9_13CO returned equal results for Thermincola potens and Thermincola carboxydiphila. However, the sequence identity to both was only 90%, and it branches distantly from Thermincola in the phylogenetic tree (Figure 3). This may represent another related CO-oxidizing genus, the exact nature of which requires further study.
While PB1 and Liard2 sediments did oxidize CO, incubations with 13CO did not show any observable shift in 16S rRNA gene density profiles. Nevertheless, we did for comparison analyze the heavy fractions from these incubations. The heavy fraction of PB1 showed a slight increase in Firmicutes as compared to the proportion present in the original community (Table S1), however no OTUs with sequences >1% were associated with known CO-oxidizing bacteria (Table S2). In comparison, the heavy fraction from Liard2_13CO had a number of OTUs associated with CO-oxidizing bacteria. Only one OTU was present at 25 fold enrichment and represented the top OTU of this fraction (Table 2). OTU_7600 had a 98% BLAST identity to Carboxydocella thermoautotrophica. It represented 28.0% of sequences and was the same Carboxydocella OTU found in other sites at relatively high abundance. Therefore, growth of carboxydotrophs was probably occurring in these samples as well, albeit at slow rates.
High G + C content may be responsible for the observation of some organisms in heavy fractions, including uncultured Crenarchaeota present in heavy fractions recovered from GR1_13CO (Table 2). The G + C content of previously identified CO-oxidizing thermophiles ranges from ca. 40–48% (Svetlichny et al., 1991b; Sokolova et al., 2002, 2004, 2005), corresponding to a density range of 1.698–1.705 g ml−1. Increased G + C content results in a higher buoyant density that may initially suggest 13C incorporation (Schildkraut et al., 1962). However, the high proportions in un-amended and 12C controls of Crenarchaeota in the case of GR1 suggest that these microbes were not carboxydotrophs but rather are naturally present at that density due to relatively high G + C content.
Discussion
In this study, bacteria potentially involved in the anaerobic oxidation of carbon monoxide were identified from hot spring environments using DNA-SIP. Five geographically diverse geological settings were identified in which potential anaerobic CO-oxidation was detectable. These five locations are widely dispersed geographically over an area of approximately 1 million km2. The measured CO-oxidation potentials were variable between geothermal springs. Comparative rates from other environments are rarely reported, however CO-oxidation rates of 120 μmol l−1 of sediment d−1 were estimated in slurries from Uzon Caldera, Kamchatka (Kochetkova et al., 2011), and a 14CO tracer was used to estimate a rate of 40.75 nmol CO cm−3 sediment d−1 for another anaerobic hot spring community in Kamchatka (Slepova et al., 2007). While obtained following different methodology, these in vitro rate estimates are 2–4 orders of magnitude lower than those measured in the present study, and show that potential rates of CO oxidation may vary greatly between environments. The observation of little to no methane production in most samples was consistent with the negligible proportions of Euryarchaeota detected in the original environmental communities and the lack of archaeal sequences detected in heavy SIP fractions (e.g., Figure 2A).
Distinct differences were noted between the original microbial communities and the communities detected in 13C-labeled heavy DNA fractions for each sample. In most cases, Firmicutes were present at < 1% in the original communities but increased in abundance in 13CO incubated heavy fractions, reaching up to ca. 95% of all 16S rRNA gene reads (Table S1). These results indicate that CO-metabolizing bacteria make up a relatively minor component of the overall population within these geothermal systems but are still present and may become active if CO is provided. Most of the bacteria identified in heavy fractions showed high identities (>98%) to known CO-oxidizing bacteria described from other geothermal springs, particularly Carboxydocella and Thermincola species (Table 2). This finding suggests not only that the CO-SIP procedure was successful in identifying primary carboxydotrophs without cross-feeding artifacts, but also that the predominant carboxydotrophic bacteria in geothermal environments may in fact already be well described from cultivation studies. This is a rather unusual finding for SIP experiments (e.g., Redmond et al., 2010). Geographically, it also indicates that anaerobic thermophilic carboxydotrophic bacteria are highly cosmopolitan (at least at the species/genus level), since most described isolates have been obtained from Russian geothermal sites. Aerobic carobyxdotrophy is taxonomically diverse (e.g., King and Weber, 2007). And while the presence of CO in geothermal spring emissions may suggest the potential for wide-spread CO metabolism, the current study supports the notion that the capacity for anaerobic carboxydotrophy among thermophiles is more limited.
While the most predominant bacteria identified via SIP were similar to carboxydotrophs isolated from other geothermal springs, there were a few exceptions. Among the predominant OTUs detected in heavy DNA fractions (Table 2), three OTUs showed < 95% 16S rRNA gene sequence identity to any described species. For example, an OTU making up 15% of the heavy DNA fraction in sample GR1 had only a moderate similarity to the proposed genus “Desulforudis.” Candidatus “Desulforudis audaxviator” was identified in the fracture water of a South African gold mine. This isolate has components of the Wood-Ljungdahl pathway and may be capable of CO oxidation and assimilation (Chivian et al., 2008). However, the low identity (93%) of our OTU indicates a genus-level divergence to Ca. “D. audaxviator.”
While the use of 16S rRNA gene qPCR greatly improved the detection of shifts in density within the CsCl gradients, in general the observed shifts were subtle. Approximately 0.47 mmol of total 13C was added to each of the 13CO SIP incubations. However, previous studies show that relatively little of the CO oxidized microbially in geothermal habitats is incorporated into biomass. Using radioisotope tracers to examine a hot spring community from Kamchatka, it was estimated that 85% of the 14CO was oxidized to CO2 while only 0.5% was used for cell biomass production. The remainder was distributed between dissolved organic matter and minor (0.001%) amounts of methane (Slepova et al., 2007). At 0.5% incorporation, a maximum of ca. 2.35 μmol of 13C would have been incorporated into the bacteria identified in the current study and may explain why shifts in the density of labeled DNA were minor compared to SIP experiments using substrates such as 13CH4 where more C is incorporated into the cells over a short incubation period (Dumont et al., 2011). The use of qPCR and the examination of the 16S rRNA gene copy profiles provided a means by which to identify subtle shifts in DNA density (Lueders et al., 2004; Sharp et al., 2012). The lack of observable shifts in 16S rRNA gene copies in one of our samples (13CO-incubated PortageBrûlé) indicated that some geothermal communities may contain bacteria that are using CO as an energy source to maintain the population but are perhaps growing slowly, maybe due to other nutrient limitations, or more likely are incorporating other, possibly organic, C sources into biomass (Figure 1B). This does indicate limitations in the 13CO-SIP technique. While it appeared to be effective in identifying some carboxydotrophs, it cannot identify all of these metabolically diverse microorganisms including potential carboxydoheterotrophs.
The 13CO-SIP technique is also challenging because carboxydotrophs may incorporate CO2 rather than CO directly, and because the initial products of CO-oxidation, H2 and CO2, may lead to labeling of other autotrophs. Our experiments included controls suggesting that these problems were minor. The greatest shifts in density were observed when 13CO was provided as the sole carbon source (i.e., no extra CO2 was added), but incubations with 13CO + 12CO2 showed smaller shifts in DNA density compared to 13CO alone. Most likely, an initial oxidation of 13CO to 13CO2 via the gas-water shift reaction is followed by assimilation of the produced 13CO2. Incubations with 13CO2 alone (and no CO added) showed little or no apparent shifts in DNA density, indicating that labeling of autotrophs growing on substrates already present in the samples, such as sulfur or ammonia, was not an issue, and that CO was ultimately the primary energy source in the incubations. However, there is the distinct possibility of hydrogenotrophic organisms using the H2 and 13CO2 produced via the gas-water shift reaction. This particular form of cross-feeding cannot be eliminated from the current results, but lines of evidence suggest that it was minor: (1) Cross-feeding with H2 may occur only in addition to primary CO oxidation- i.e., it can only be a secondary process given that CO was the major energy source available; (2) almost all of the detected bacteria in the present study were closely related to known carboxydotrophs; and (3) potentially hydrogenotrophic but non-carboxydotrophic bacteria such as the members of the phylum Deinococcus-Thermus were detected in some heavy DNA fractions, but were always of minor importance compared to known carboxydotrophs (e.g., Thermus scotoductus in DCs9, Table 2; Table S2). Complete genomes of both T. scotoductus and T. antranikianii (Table S2) lack genes related to CO metabolism (http://img.jgi.doe.gov/).
Many bacterial species that possess cooS genes have a primary metabolism that does not focus on CO, however their presence may imply a potential underlying or backup CO-dependent physiology should conditions vary and become more optimal for CO-oxidation (Techtmann et al., 2011). While challenging to measure in situ, localized accumulations of CO may create microniches within geothermal systems in which low abundance carboxydotrophic population members may thrive. For example, the species composition of the heavy fractions was similar for two biomat samples DCm2010 and DCmN11 with a dominance of Thermincola potens in both cases. The presence of CO oxidizing bacteria in these samples is perhaps not surprising given the observation of net CO production within microbial mats. Saline and intertidal sand flat photosynthetic microbial mats exhibited a net production of CO (3.1–5.4 μmol m−2 d−1) during daylight hours and were also observed to have a net production of H2 (Hoehler et al., 2001). As many of the Dewar Creek samples that showed evidence of CO-oxidation were comprised of biomats with relatively high proportions of cyanobacteria (Figure 2B, Table S1), these results support CO as a potentially important carbon and energy source in microenvironments within geothermal systems. Variation in CO metabolizing bacteria was observed between samples collected from the same geothermal system but under different temperature regimes. OTU_3148 was 99% similar to both Desulfotomaculum kuznetsovii and Desulfotomaculum luciae, detected in 13CO incubations of DCs9, another site from the Dewar Creek hot spring. Desulfotomaculum kuznetsovii is an obligate anaerobe and is capable of growth with CO as the sole carbon and energy source (Parshina et al., 2005a). Thermolithobacter carboxydivorans was also detected in this particular sample site but was not detected in any other microcosm. The optimum growth temperature for T. carboxydivorans of 70°C may also explain the presence of this bacterium in DCs9 with an environmental and incubation temperature of 65°C as opposed to other Dewar Creek samples with lower in situ temperatures incubated at 55°C (Sokolova et al., 2005).
The dominance of Firmicutes in heavy-density DNA fractions of geothermal samples incubated under 13CO confirms that representatives of this phylum may play a predominant role relative to other phyla in anaerobic oxidation of CO in geothermal environments. In particular, they may reflect minor populations within geothermal microenvironments where localized CO concentrations may be high. While the detection of such microenvironments in situ is challenging, the hypothesized presence of these localized CO-rich niches (e.g., Techtmann et al., 2009) suggests a mechanism by which these carboxydotrophs may exist. Despite comprising a relatively small proportion of in situ communities, the CO oxidizing bacteria are active and show some variation across geothermal environments. Oxidation potentials are higher than the few previously reported rates for mixed geothermal communities. Despite geographical differences, thermophilic bacteria associated with anaerobic CO-oxidation are widely distributed geographically and the predominant species are well-described from cultivation studies. The presence of a few OTUs that do not show high degrees of similarity to any known cultured representatives indicates that a few new lithoautotrophs that have not been previously identified as CO-oxidizers may also be present in the geothermal springs tested and require further study.
The detection of sequences associated with known CO-oxidizing bacteria in high abundance supports the applicability of the CO-SIP technique. SIP can be applied to target autotrophs by adding an energy substrate along with 13CO2. This works as long as the added substrate is the primary energy source for the autotrophic community. We have previously used this approach to identify autotrophic methanotrophs (Sharp et al., 2012). We therefore conclude that the CO-SIP technique, which works in a similar way to identify autotrophic carboxydotrophs, does have some value, although of course the results still need to be interpreted with caution. Controls are necessary to demonstrate a low rate of assimilation of 13CO2 by other autotrophs. It should also be stressed that heterotrophic carboxydotrophs will be missed- this was a possible explanation for the failure of some of the samples assayed in this study. Cross feeding via H2 + CO2 produced via the gas-water shift reaction is also a potential issue, although this appeared to be minor in this particular study. This study appeared to work because the bacteria identified were primarily known carboxydotrophs, however identification of a new potential carboxydotroph should only be taken as initial evidence that requires verification with other methods including sequencing of potential cooS genes.
Author Contributions
AB, CS and SG collected samples; AB performed SIP incubations with input from PD; AB with aid from CS extracted DNA, prepared samples for sequencing and carried out data processing. AB and PD wrote the initial draft of the paper; all authors designed the study, discussed the results and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding was provided by an Alberta Innovates Technology Futures (AITF) New Faculty Award grant to PD and in part by a Natural Science and Engineering Research Council of Canada (NSERC) Post-Doctoral Fellowship and PEO Scholar Award to AB. CS was supported by fellowships from NSERC and AITF. We thank B. C. Parks for permission to sample Dewar Creek.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00897
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Berg, I. A. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77, 1925–1936. doi: 10.1128/AEM.02473-10
Byrne-Bailey, K. G., Wrighton, K. C., Melnyk, R. A., Agbo, P., Hazen, T. C., and Coates, J. D. (2010). Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J. Bacteriol. 192, 4078–4079. doi: 10.1128/JB.00044-10
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cavicchioli, R. (2002). Extremophiles and the search for extraterrestrial life. Astrobiology 2, 281–292. doi: 10.1089/153110702762027862
Chivian, D., Brodie, E. L., Alm, E. J., Culley, D. E., Dehal, P. S., DeSantis, T. Z., et al. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322, 275–278. doi: 10.1126/science.1155495
Conrad, R. (1988). Biogeochemistry and ecophysiology of atmospheric CO and H2. Adv. Microb. Ecol. 10, 231–283. doi: 10.1007/978-1-4684-5409-3_7
Conrad, R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O and NO). Microbiol. Rev. 60, 609–640.
Dobbek, H., Svetlitchnyi, V., Gremer, L., Huber, R., and Meyer, O. (2001). Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Sci. New Ser. 293, 1281–1285. doi: 10.1126/science.1061500
Dumont, M. G., Pommerenke, B., Casper, P., and Conrad, R. (2011). DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ. Microbiol. 13, 1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x
Giggenbach, W. F. (1980). Geothermal gas equilibria. Geochim. Cosmochim. Acta 44, 2021–2032. doi: 10.1016/0016-7037(80)90200-8
Grasby, S. E., Hutcheon, I., and Krouse, H. R. (2000). The influence of water-rock interaction on the chemistry of thermal springs in western Canada. Appl. Geochem. 15, 439–454. doi: 10.1016/S0883-2927(99)00066-9
Grasby, S. E., Richards, B., Sharp, C. E., Brady, A. L., Jones, G. M., and Dunfield, P. F. (2013). The Paint Pots, Kootenay National Park, Canada – A natural acid spring analogue for Mars. Can. J. Earth Sci. 50, 94–108 doi: 10.1139/e2012-060
Haab, P. (1990). The effect of carbon monoxide on respiration. Experientia 46, 1202–1206. doi: 10.1007/BF01936937
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
He, R., Wooller, M. J., Pohlman, J. W., Quensen, J., Tiedje, J. M., and Leigh, M. B. (2012). Diversity of active aerobic methanotrophs along depth profiles of artic and subarctic lake water column and sediments. ISME J. 6, 1937–1948. doi: 10.1038/ismej.2012.34
Henstra, A. M., Dijkema, C., and Stams, A. J. M. (2007a). Archaeoglobus fulgidus couples CO oxidation to sulfate reduction and acetogenesis with transient formate accumulation. Environ. Microbiol. 9, 1836–1841. doi: 10.1111/j.1462-2920.2007.01306.x
Henstra, A. M., Sipma, J., Rinzema, A., and Stams, A. J. M. (2007b). Microbiology of synthesis gas fermentation for biofuel production. Curr. Opin. Biotechnol. 18, 200–206. doi: 10.1016/j.copbio.2007.03.008
Hoehler, T. M., Bebout, B. M., and Des Marais, D. J. (2001). The role of microbial mats in the production of reduced gases on early Earth. Nature 412, 324–327. doi: 10.1038/35085554
Kerby, R. L., Ludden, P. W., and Roberts, G. P. (1995). Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 177, 2241–2244.
King, G. M., and Weber, C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. 5, 107–118. doi: 10.1038/nrmicro1595
Knief, C., Lipski, A., and Dunfield, P. F. (2003). Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69, 6703–6714. doi: 10.1128/AEM.69.11.6703-6714.2003
Kochetkova, T. V., Rusanov, I. I., Pimenov, N. V., Kolganova, T. V., Lebedinsky, A. V., Bonch-Osmolovskaya, E. A., et al. (2011). Anaerobic transformation of carbon monoxide by microbial communities of Kamchatka hot springs. Extremophiles 15, 319–325. doi: 10.1007/s00792-011-0362-7
Lee, H. S., Kang, S. G., Bae, S. S., Lim, J. K., Cho, Y., Kim, Y. J., et al. (2008). The complete genome sequence of Thermococcus onnurineus NA1 reveals a mixed heterotrophic and carboxydotrophic metabolism. J. Bacteriol. 190, 7491–7499. doi: 10.1128/JB.00746-08
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, B. A., et al. (2004). Arb: a software environment for sequence data. Nucleic. Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Lueders, T., Manefield, M., and Friedrich, M. W. (2004). Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. doi: 10.1046/j.1462-2920.2003.00536.x
Mörsdorf, G., Frunzke, K., Gadkari, D., and Meyer, O. (1992). Microbial growth on carbon monoxide. Biodegradation 3, 61–82. doi: 10.1007/BF00189635
Neufeld, J. D., Vohra, J., Dumont, M. G., Lueders, T., Manefield, M., Friedrich, M. W., et al. (2007). DNA stable-isotope probing. Nature 2, 860–866. doi: 10.1038/nprot.2007.109
Novikov, A. A., Sokolova, T. G., Lebedinsky, A. V., Kolganova, T. V., and Bonch-Osmolovskaya, E. A. (2011). Carboxydothermus islandicus sp. nov., a thermophilic, hydrogenogenic, carboxydotrophic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 61, 2532–2537. doi: 10.1099/ijs.0.030288-0
Oelgeschläger, E., and Rother, M. (2008). Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch. Microbiol. 190, 257–269. doi: 10.1007/s00203-008-0382-6
Parshina, S. N., Kijlstra, S., Henstra, A. M., Sipma, J., Plugge, C. M., and Stams, A. J. M. (2005a). Carbon monoxide conversion by thermophilic sulfate-reducing bacteria in pure culture and in co-culture with Carboxydotherms hydrogenoformas. Appl. Microbiol. Biotechnol. 68, 390–396. doi: 10.1007/s00253-004-1878-x
Parshina, S. N., Sipma, J., Nakashimada, Y., Henstra, A. M., Smidt, H., Lysenko, A. M., et al. (2005b). Desulfotomaculum carboxydivorans sp. nov., a novel sulfate-reducing bacterium capable of growth at 100% CO. Int. J. Syst. Evol. Microbiol. 55, 2159–2165. doi: 10.1099/ijs.0.63780-0
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Ragsdale, S. W. (1991). Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit. Rev. Biochem. Mol. 26, 261–300. doi: 10.3109/10409239109114070
Ragsdale, S. W. (2004). Life with carbon monoxide. Crit. Rev. Biochem. Mol. 39, 165–195. doi: 10.1080/10409230490496577
Ramos-Padrón, E., Bordenave, S., Lin, S., Bhaskar, I. M., Dong, X., Sensen, C. W., et al. (2011). Carbon and sulfur cycling by microbial communities in a gypsum-treated oil sands tailings pond. Environ. Sci. Technol. 45, 439–446. doi: 10.1021/es1028487
Redmond, M. C., Valentine, D. L., and Sessions, A. L. (2010). Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl. Environ. Microbiol. 76, 6412–6422. doi: 10.1128/AEM.00271-10
Schildkraut, C. L., Marmu, J., and Doty, P. (1962). Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J. Mol. Biol. 4, 430–443. doi: 10.1016/S0022-2836(62)80100-4
Sharp, C. E., Brady, A. L., Sharp, G. H., Grasby, S. E., Stott, M. B., and Dunfield, P. F. (2014a). Humboldt's spa: microbial diversity is controlled by temperature in geothermal environments. ISME J. 8, 1166–1174. doi: 10.1038/ismej.2013.237
Sharp, C. E., Martínez-Lorenzo, A., Brady, A. L., Grasby, S. E., and Dunfield, P. F. (2014b). Methanotrophic bacteria in warm geothermal spring sediments identified using stable-isotope probing. FEMS Microbiol. Ecol. 90, 92–102. doi: 10.1111/1574-6941.12375
Sharp, C. E., Stott, M. B., and Dunfield, P. F. (2012). Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3:303. doi: 10.3389/fmicb.2012.00303
Sipma, J., Henstra, A. M., Parshina, S. N., Lens, P. N. L., Lettinga, G., and Stams, A. J. M. (2006). Microbial CO conversions with applications in synthesis gas purification and bio-desulfurization. Crit. Rev. Biotechnol. 26, 541–547. doi: 10.1080/07388550500513974
Slepova, T. V., Rusanov, I. I., Sokolova, T. G., Bonch-Osmolovskaya, E. A., and Pimenov, N. V. (2007). Radioisotopic tracing of carbon monoxide conversion by anaerobic thermophilic prokaryotes. Microbiology 76, 523–529. doi: 10.1134/S0026261707050025
Slepova, T. V., Sokolova, T. G., Lysenko, A. M., Tourova, T. P., Kolganova, T. V., Kamzolkina, O. V., et al. (2006). Carboxydocella sporoproducens sp. nov., a novel anaerobic CO-utilizing/H2-producing thermophilic bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 56, 797–800. doi: 10.1099/ijs.0.63961-0
Sokolova, T. G., González, J. M., Kostrikina, N. A., Chernyh, N. A., Slepova, T. V., Bonch-Osmolovskaya, E. A., et al. (2004). Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 54, 2353–2359. doi: 10.1099/ijs.0.63186-0
Sokolova, T. G., Henstra, A. M., Sipma, J., Parshina, S. N., Stams, A. J. M., and Lebedinsky, A. V. (2009). Diversity and ecophysiological features of thermophilic carboxydotrophic anaerobes. FEMS Microbiol. Ecol. 68, 131–141. doi: 10.1111/j.1574-6941.2009.00663.x
Sokolova, T. G., Kostrikina, N. A., Chernyh, N. A., Kolganova, T. V., Tourova, T. P., and Bonch-Osmolovskaya, E. A. (2005). Thermincola carboxydiphila gen. nov., sp. nov., a novel anaerobic, carboxydotrophic, hydrogenogenic bacterium from a hot spring of the Lake Baikal area. Int. J. Syst. Evol. Microbiol. 55, 2069–2073. doi: 10.1099/ijs.0.63299-0
Sokolova, T. G., Kostrikina, N. A., Chernyh, N. A., Tourova, T. P., Kolganova, T. V., and Bonch-Osmolovskaya, E. A. (2002). Carboxydocella thermautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring. Int. J. Syst. Evol. Microbiol. 52, 1961–1967. doi: 10.1099/ijs.0.02173-0
Sokolova, T., Hanel, J., Onyenwoke, R. U., Reysenbach, A.-L., Banta, A., Geyer, R., et al. (2007). Novel chemolithotrophic, thermophilic, anaerobic bacteria Thermolithobacter ferrireducens gen. nov., sp. nov. and Thermolithobacter carboxydivorans sp. nov. Extremophiles 11, 145–157. doi: 10.1007/s00792-006-0022-5
Stubner, S. (2002). Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J. Microbiol. Methods 50, 155–164. doi: 10.1016/S0167-7012(02)00024-6
Svetlichny, V. A., Sokolova, T. G., Gerhardt, M., Kostrikina, N. A., and Zavarzin, G. A. (1991a). Anaerobic extremely thermophilic carboxydotrophic bacteria in hydrotherms of Kuril Islands. Microb. Ecol. 21, 1–10. doi: 10.1007/BF02539140
Svetlichny, V. A., Sokolova, T. G., Gerhardt, M., Ringpfeil, M., Kostrikina, N. A., and Zavarzin, G. A. (1991b). Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir Island. Syst. Appl. Microbiol. 14, 254–260. doi: 10.1016/S0723-2020(11)80377-2
Techtmann, S. M., Colman, A. S., Murphy, M. B., Schackwitz, W. S., Goodwin, L. A., and Robb, F. T. (2011). Regulation of multiple carbon monoxide consumption pathways in anaerobic bacteria. Front. Microbiol. 2:147. doi: 10.3389/fmicb.2011.00147
Techtmann, S. M., Colman, A. S., and Robb, F. T. (2009). ‘That which does not kill us only makes us stronger’ the role of carbon monoxide in thermophilic microbial consortia. Environ. Microbiol. 11, 1027–1037. doi: 10.1111/j.1462-2920.2009.01865.x
Uffen, R. L. (1981). Metabolism of carbon monoxide. Enzyme Microb. Technol. 3, 197–206. doi: 10.1016/0141-0229(81)90086-7
Voordouw, G. (2002). Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184, 5903–5911. doi: 10.1128/JB.184.21.5903-5911.2002
Keywords: carboxydotrophs, stable isotope probing, geothermal, carbon monoxide (CO), thermophile
Citation: Brady AL, Sharp CE, Grasby SE and Dunfield PF (2015) Anaerobic carboxydotrophic bacteria in geothermal springs identified using stable isotope probing. Front. Microbiol. 6:897. doi: 10.3389/fmicb.2015.00897
Received: 30 May 2015; Accepted: 17 August 2015;
Published: 01 September 2015.
Edited by:
Steffen Kolb, Friedrich-Schiller-Universität Jena, GermanyReviewed by:
Alexander V. Lebedinsky, Winogradsky Institute of Microbiology, RussiaMartin Taubert, Friedrich-Schiller-Universität Jena, Germany
Copyright © 2015 Brady, Sharp, Grasby and Dunfield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter F. Dunfield, Department of Biological Sciences, University of Calgary, 2500 University Drive NW, Calgary, AB T2N 1N4, Canada, pfdunfie@ucalgary.ca
†Present Address: Allyson L. Brady, School of Geography and Earth Sciences, McMaster University, Hamilton, ON, Canada
 Allyson L. Brady
Allyson L. Brady Christine E. Sharp
Christine E. Sharp Stephen E. Grasby
Stephen E. Grasby Peter F. Dunfield
Peter F. Dunfield