- 1Department of Bioengineering and Biotechnology, Huaqiao University, Xiamen, China
- 2State Key Laboratory Breeding Base of Marine Genetic Resource, Third Institute of Oceanography, State Oceanic Administration, Xiamen, China
- 3Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI, USA
Arsenic (As) is widespread in the environment and causes numerous health problems. Rhodopseudomonas palustris has been regarded as a good model organism for studying arsenic detoxification since it was first demonstrated to methylate environmental arsenic by conversion to soluble or gaseous methylated species. However, the detailed arsenic resistance mechanisms remain unknown though there are at least three arsenic-resistance operons (ars1, ars2, and ars3) in R. palustris. In this study, we investigated how arsenic multi-operons contributed to arsenic detoxification in R. palustris. The expression of ars2 or ars3 operons increased with increasing environmental arsenite (As(III)) concentrations (up to 1.0 mM) while transcript of ars1 operon was not detected in the middle log-phase (55 h). ars2 operon was actively expressed even at the low concentration of As(III) (0.01 μM), whereas the ars3 operon was expressed at 1.0 μM of As(III), indicating that there was a differential regulation mechanism for the three arsenic operons. Furthermore, ars2 and ars3 operons were maximally transcribed in the early log-phase where ars2 operon was 5.4-fold higher than that of ars3 operon. A low level of ars1 transcript was only detected at 43 h (early log-phase). Arsenic speciation analysis demonstrated that R. palustris could reduce As(V) to As(III). Collectively, strain CGA009 detoxified arsenic by using arsenic reduction and methylating arsenic mechanism, while the latter might occur with the presence of higher concentrations of arsenic.
Introduction
Arsenic (As) is a highly toxic, carcinogenic, clastogenic and teratogenic metalloid (Slyemi and Bonnefoy, 2012). Arsenic occurs primarily as inorganic forms of pentavalent arsenate As(V) and trivalent arsenite As(III), with the latter being regarded as the most mobile and toxic form (Yang et al., 2012). Arsenicals generated from natural and anthropogenic sources are the widely distributed contaminants of freshwater, groundwater and seawater (Stolz et al., 2010; Slyemi and Bonnefoy, 2012; Rodríguez-Lado et al., 2013). Combustion of fossil fuels, mining, and applications of arsenic-containing pesticides/herbicides account for most of the arsenic pollution sources (Stolz and Oremland, 1999). Microorganisms are the principal drivers of arsenic chemical speciation by redox transformations and influence arsenic mobility and toxicity. Most of bacteria and archaea virtually carry arsenic-resistance (ars) genes that potentially confer resistance to As(V) and/or As(III) (Jackson and Dugas, 2003). The phenomenon (widespread occurrence of ars genes) indicates the ubiquitous distribution of this toxic metalloids in nature.
Some of microorganisms have a significant impact on the biogeochemical transformations of arsenic and are considered as bioremediation reagents for arsenic contamination (Zhang et al., 2015). Increasing interests have been drawn to investigate microbial detoxification of arsenic compounds, particularly to study the arsenic metabolic mechanisms. Furthermore, some microorganisms such as chemoautotrophic arsenite oxidizers oxidize As(III) to As(V) to gain energy for cell growth when fixing inorganic carbon (CO2). Instead, microbes that use As(V) as an electron acceptor in anaerobic respiration lead to the production of As(III), indicating that arsenic plays various roles in microbial metabolism (Heinrich-Salmeron et al., 2011; Kruger et al., 2013). Arsenic metabolism and its genetic determinants have been investigated in many Gram-positive and Gram-negative bacteria (Kruger et al., 2013). Bacteria acquired several different arsenic-metabolic strategies including arsenite oxidation (aio system) in arsenite oxidizers which oxidize As(III) to As(V) (Oremland and Stolz, 2003; Heinrich-Salmeron et al., 2011), anaerobic arsenate respiration (arr system) in arsenate-reducing microbes which respire and reduce As(V) to As(III) (Oremland and Stolz, 2003; Páez-Espino et al., 2009), arsenate system (ars system) in arsenate-resistant microbes which reduce cytoplasmic toxic As(V) to As(III) (Oremland and Stolz, 2003; Kruger et al., 2013), and arsenic methylation (arsM system) in arsenic methylating bacteria which convert inorganic arsenic into methylated arsenic (Qin et al., 2006; Kruger et al., 2013; Zhang et al., 2015). Arsenic-metabolic genes are assembled with multiple arsenic operons in some bacteria. For example, both arr and ars operons are found in Shewanella sp. (Saltikov and Newman, 2003); Thiomonas sp. possess two operons (aio and ars system) (Arsène-Ploetze et al., 2010); Herminiimonas arsenicoxydans has aio and ars (Muller et al., 2007); Cyanidioschyzon sp. 5508 has aio, arr, and ars operons (including arsM) (Qin et al., 2009). Arsenic-resistant microorganisms possibly benefited from multiple arsenic-resistance operons, e.g., they can utilize different detoxification strategies under the complex environments.
With extraordinary metabolic versatility, R. palustris has been widely studied for wastewater treatment, bioremediation, hydrogen production and electricity generation (Zhao et al., 2011; Fu et al., 2014; Liu et al., 2015). R. palustris is possibly exposed to a high concentration of arsenic under the above field application conditions. Despite considerable research on the arsenic-metabolic mechanisms in chemotrophic bacteria, only few studies have been conducted in anoxygenic phototrophic bacteria. R. palustris CGA009 has three arsenic-resistance operons (ars1, ars2, and ars3) in the chromosome (Qin et al., 2006). Deciphering the arsenic resistance mechanism(s) in anoxygenic phototrophic bacteria may facilitate engineering stronger arsenic resistant strains with desirable features in industrial applications. The objectives of the present study were as follows: (i) to examine the arsenic resistance capacity; (ii) to exam the differential regulation of the three arsenic operons; (iii) to analyze the arsenic speciation in anoxygenic phototrophic bacteria by using R. palustris CGA009 as a model organism; (iv) to propose a working model for arsenic resistance in R. palustris CGA009.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions
R. palustris CGA009 (ATCC BAA-98) was obtained from the American Type Culture Collection (ATCC, USA). Bacteria were anaerobically grown in modified Ormerod medium at 30°C with continuous illumination (Zhao et al., 2011). Briefly, ammonium sulfate and DL-malic acid were substituted by 10 mM of ammonium chloride, 30 mM of sodium acetate, 10 mM of sodium succinate, 10 mM of sodium pyruvate, 10 mM of sodium malate, 10 mM of sodium bicarbonate and 0.1% yeast extract (Oxoid, UK), pH 6.8.
Arsenic Resistance Determination
Cells in the log-phase were dispensed into 20-ml screw cap test tube. Various concentrations of Na3AsO4·12H2O and NaAsO2 (Merck, Germany) were added with final concentrations ranging from 0.5 to 6.0 mM and ranging from 0.5 to 2.5 mM, respectively. Cell growth was estimated by measuring the optical density at 660 nm (OD660) after incubation anaerobically at 30°C and with 2500 lux light for 4 days (Carius et al., 2013). The starting OD660 was 0.09. The growth index was defined as the median effective concentration (EC50) and was used to assess the arsenic tolerance in R. palustris.
Determination of Arsenic Speciation
The process of determination of arsenic speciation as described previously (Lin et al., 2014). Freeze-dried samples were extracted with 10 ml of 1% nitric acid (Merck, Germany) in a microwave-accelerated reaction system (CEM Microwave Technology, UK). This system was provided with a stably increasing temperature from 55 to 75°C within 10 min. Then the extracts were heated at 95°C for 30 min. Finally, the extracted solutions were centrifuged and passed through a nylon filter with a size of 0.22 μm. Arsenic speciation of cells was determined using the high-performance liquid chromatography (HPLC) (Agilent 1200, Japan) coupled with inductively-coupled plasma mass spectrometry (ICP-MS) (Agilent 7500cx, USA) as described previously (Lin et al., 2014). The mobile phases consisted of 6.67 mM of NH4H2PO4 (Merck, Germany) and 6.67 mM of NH4NO3 (Merck, Germany) at pH 6.2. Arsenic speciation in the samples were identified by comparing their retention times to the standards including arsenite, arsenate, dimethylarsinic acid (DMA) (Chem Service, PA, USA) and monomethylarsonic acid (MMA) (Beijing Chemicals, China).
Molecular Manipulation
Genomic DNA was extracted using the One-4-all Genomic DNA Mini-Preps Kit (Sangon, China). Total RNA was extracted and purified by using an EasyPure™ RNA Kit (TransGen, China). The contaminating DNA was removed by DNase I procedure (Takara, China) if necessary. DNA-free RNA samples were confirmed by PCR amplification of the house-keeping gene gyrB (encoding DNA gyrase subunit B) with the forward primer gyrBF (AACTGAACGGCATTATGG) and the reverse primer gyrBR (GGGATGTTGTTGGTGAAG). cDNAs were synthesized using an PrimeScript™ II 1st strand cDNA synthesis kit (Takara, China) following the protocol. Prior to quantitative PCR, cDNAs were diluted 10-fold as template in nuclease-free water.
Functional genes arsB, arsC2 and arsM were chosen as representative genes for ars1, ars2, and ars3 operons in this study, respectively. arsB was amplified with arsBF (GCTGAT CGTTTCCAACCT) and arsBR (ACCATCACCGAGGCATAA); arsC2 was amplified with arsCF (CGGGCACTTCAGATACTC) and arsCR (CGTCGTCATCTATCACAAC); arsM was amplified with arsMF (CCGACAGGTTGATGACGCAGT) and arsMR (CGTCGTCAT CTATCACAAC). gyrB gene was used to normalize the expression for arsB, arsC2, and arsM as described by Saltikov et al. (2005). General PCR experiments were conducted in a T-100 Thermal Cycler (Bio-Rad). Quantitative real-time PCR was carried out in the Applied Biosystems 7300 Real-Time PCR System (Life Technologies). The cycle thresholds (CT) were determined for samples and genomic DNA standards. Diluted CGA009 genomic DNA was used as standard for quantification. For each transcript, the CT value was converted to a genomic DNA equivalent in copies by comparing the CT of an unknown sample to standard curves (prepared by using CGA009 genomic DNA).
Results and Discussion
Effects of As(V) (0.5~6.0 mM) and As(III) (0.5~2.5 mM) on the bacterial growth of R. palustris were tested in modified Ormerod medium and under anaerobic–light conditions (Figure 1). Negligible difference was observed between the culture supplemented with 0.5 mM of As(V) and the control (no AS(V)), indicating that a low concentration of As(V) was not toxic to R. palustris. The cells could retain at least 60, 70, and 80% of the control growth when 3.0 mM, 2.0 mM and 1.0 mM of As(V) were added to the culture respectively, indicating a good resistance to As(V). However, higher concentrations of As(V) (i.e., 4.0 mM to 6.0 mM) severely inhibited the bacterial growth with the most significant inhibition (nearly 100%) at the concentration of 6.0 mM. R. palustris was more sensitive to environmental As(III) than that in As(V) as shown in Figure 1B. For example, 2.0 mM and 2.5 mM of As(III) inhibited up to 90 and 100% of the bacterial growth, respectively. However, cells retained at least 70 and 60% of the control growth when 1.0 and 1.5 mM of the As(III) were added, respectively. Furthermore, we studied the relationship between the growth rate and the arsenic concentrations (Figure 2). Median effective concentration (EC50) values for As(V) and As(III) were estimated to be 2.44 and 1.55 mM, respectively.
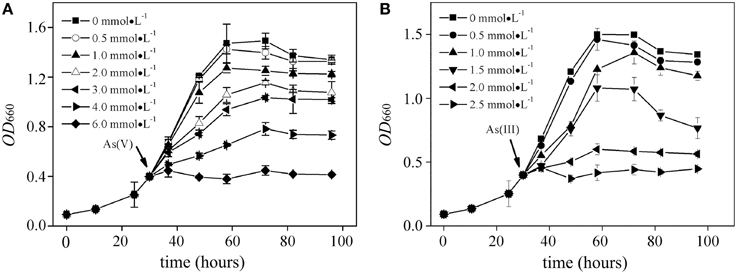
Figure 1. Growth curves of R. palustris CGA009 in the presence of different concentrations of arsenate (A) and arsenite (B), respectively. Strain CGA009 was anaerobically grown at 30°C with continuous illumination. Error bars indicate the standard deviation from three independent experiments.
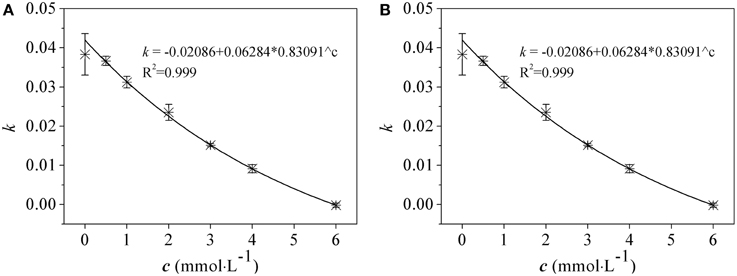
Figure 2. The relationship between the growth rate and arsenate (A) and arsenite (B) concentrations in R. palustris CGA009. Growth rate (k) was calculated from the increased OD660 values in log phase cultures grown in different concentrations of arsenic (c). The equations were obtained by nonlinear fitting. EC50 was obtained from c value which corresponded to a half of maximum of k value by equations.
LV et al. investigated the arsenic resistance in three purple non-sulfur bacteria and reported the EC50 for As(V) and As(III) Rhodopseudomona palustris CQV97 (2.4 and 0.9 mM, respectively), Rhodobacter azotoformans 134K20 (1.6 and 0.2 mM, respectively) and Rhodobacter capsulatus XJ-1 (1.9 and 0.3 mM, respectively) (Lv et al., 2012a,b). The EC50-value for arsenic in strain CQV97 was higher or comparable to E. coli, P. aeruginosa, Rhodococcus equi, and Methylosinus trichosporium. However, compared to those bacteria with a “high” arsenic resistance capability (>100 mM), there is still considerable room for further improvement.
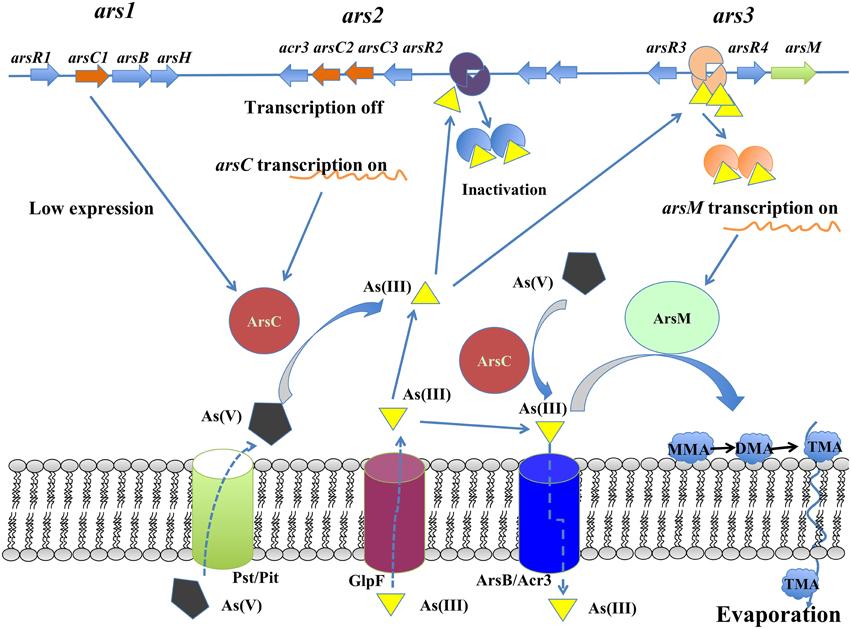
The genome-mining analysis showed that arsRBC operon existed in seven R. palustris strains (CGA009, HaA2, TIE-1, DX-1, BisB5, BisA53, and BisB18) (Figure 3), while additional arsRM operon was only found in strains CGA009, HaA2, TIE-1, DX-1, and BisB5 (Figure 3) (Lv et al., 2012a). The first operon resembles ars operon (named ars1 operon, arsRCBH type), consisting of arsenite-responsive transcriptional regulator gene (arsR1), arsenate reductase gene (arsC1), arsenite efflux pump gene (arsB) and NADPH-dependent flavin mononucleotide reductase gene (arsH). The second operon [named ars2 operon, arsRRCC (acr3) type] contains two arsR genes (arsR2 and arsR3), two As(V) reductases genes (arsC2 and arsC3), one arsenite permease gene (acr3), and two genes encoding the hypothetical proteins. Both ars1 and ars2 operons encode these proteins that perform As(V) reduction and As(III) extrusion mechanisms. The third operon (named ars3 operon, arsRM type) consists of arsenite-responsive transcriptional regulator gene (arsR4) and As(III)-methyltransferase gene (arsM). ars3 operon encode methylation protein that methylate As(III) to a number of methylated intermediates such as onomethylarsenite [MMA(III)] and dimethylarsenite [DMA(III)]. It should be note that arsenic resistance in microorganisms did not show a direct correlation with arsenic operon number(s) (Kruger et al., 2013). For example, at least five arsenic operons were found in Herminiimonas arsenicoxydans while its tolerance to arsenic was much lower than that in Corynebacterium glutamicum which has only two arsenic operons. Despite of their widespread distribution in various bacteria, the arsenic multi-operons were understudied. Due to arsenic multi-operons co-existing in one microorganism, it is difficult to define the arsenic resistance mechanisms. Therefore, it is necessary to further investigate how these arsenic multi-operons were differentially regulated and their contributions to arsenic speciation.
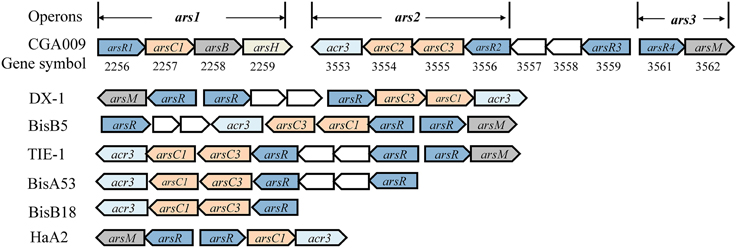
Figure 3. Schematic comparison of arsenic gene clusters from seven R. palustris strains. Genes were shown as different color and arrows indicated the direction of transcription. arsR was arsenite-responsive transcriptional regulator gene. arsC/arsC' was As (V) reductases gene, arsC was As (V) reductases genes, arsC1 and arsC2 were glutathione (GSH)/glutaredoxin (Grx)-coupled reductases genes, arsC3 was thioredoxin (Trx)/thioredoxin reductase (TrxR)-dependent reductases gene; arsB was arsenite efflux pump gene, acr3 was arsenite permease (ACR3) gene, they came from unrelated As(III) transporter families; arsH was NADPH-dependent flavin mononucleotide reductase gene; arsM was As(III)-methyltransferase gene. Other genes found in or adjacent to the ars clusters were shown as empty boxes.
In this study, we preliminary investigated their respective gene expression in the three ars operons by using quantitative real-time PCR in order to understand which one was actively involved in arsenic resistance in R. palustris CGA009. It should be noted that studying ars operon regulation by As(V) is difficult because it can be quickly reduced to As(III) by As(V) reductase in vivo. Therefore, we first examined the effect of As(III) (rather than As(V)) on the expression of ars1, ars2 and ars3 operons. To do that, we selected the functional genes arsB, arsC2, and arsM as marker genes for ars1, ars2, and ars3 operons, respectively (Figure 4). Gene expression of arsB, arsC2, and arsM were undetectable when As(III) was absent, indicating the three operons were not expressed without arsenic induction. arsB was not transcribed when As(III) was added to the culture at concentrations ranging from 0 to 1.0 mM. Remarkably, the expression of ars2 operon (arsC2) was readably detected when cells were even exposed to a low level of As(III) (0.01 μM). Compared to the control, its transcript level was increased 9.5, 23.8, and 126.8–fold when 0.01, 0.1, and 1.0 mM of As(III) were added respectively, demonstrating that ars2 expression was up-regulated by As(III). However, arsM was not transcribed when the cells were exposed to 1.0 μM of As(III), indicating that gene product of arsM was not critical for detoxifying As(III) under low environmental As(III) conditions (<1.0 μM). However, its expression level was 3.86-fold higher than the control when 1.0 μM concentration of As(III) was present in the medium (Figure 4). Furthermore, arsM transcript level increased with the increasing environmental As(III) concentration (Figure 4). It should be noted that the expression levels of arsM and arsC2 were almost equal when As(III) concentration reached a 1.0 mM, showing that R. palustris CGA009 probably required expression of ars2 and ars3 operons to detoxify the high concentrations of As(III).
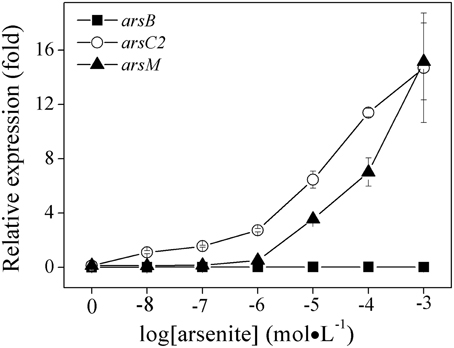
Figure 4. Effects of As (III) concentrations on the expressions of ars1, ars2 and ars3 operons in R. palustris CGA009. The expressions of arsB(■), arsC2(○), and arsM(▴) represent the expression of ars1, ars2, and ars3 operons, respectively. The expression of genes was calculated by determing the content ratio of functional genes to house-keeping gene (gyrB). Cultures were harvested in the middle-log phase (50 h). Error bars indicate the standard deviation from three independent experiments.
The expression dynamics of arsB, arsC2 and arsM (ars1, ars2, and ars3 operons) were investigated in R. palustris CGA009 at different phases of growth (Figure 5). arsC2 expression was highly induced by arsenate at the log-phase (between 43 and 60 h); the expression level decreased when cells entered the stationary phase (67–80 h). A similar expression pattern was found in arsM; however, a relatively low level of expression of arsM was recorded during the whole growth phase, compared to that in arsC2. ars1 transcript level was much lower than those in ars2 and ars3 at different phases of growth.
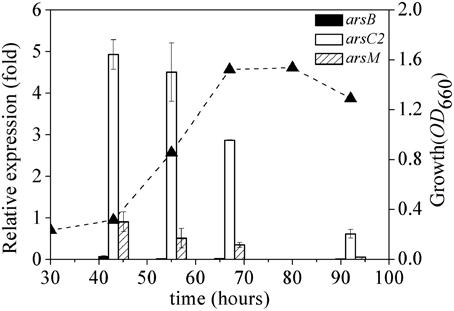
Figure 5. Dynamics of expression of ars1, ars2 and ars3 operons. The expression of arsB (black bars), arsC2 (white bars) and arsM (bias bars) was normalized to the expression of gyrB. The growth curve(▴) of R. palustris CGA009 grown on 0.5 mM arsenate is shown as dash line. Error bars indicate the standard deviation from three independent experiments.
Unfortunately, only few studies have been conducted to exam the differential regulation of arsenic multi-operons by arsenic. In bacteria and archaea, ars operons are often controlled by ArsR (Figure 3). When As(III) is absent, ArsR binds to the operator/promoter region of the operon and thus repressed the downstream genes' transcription. When As(III) is available, ArsR binds to it and goes through conformational changes, resulting in dissociation from the operator/promoter region. In our transcript analysis experiments, we could not detect a significant ars1 expression (Figure 4). However, this observation was not new. For example, in C. glutamicum, only arsC1' was constitutively transcribed though there were two arsenic resistance operons (ars1 and ars2) (Villadangos et al., 2011). Furthermore, the expression of ars3 operon in R. palustris CGA009 was induced with the presence of 1.0 μM of As(III), indicating it may only contribute to the arsenic detoxification under the higher concentrations of As(III) (see next). However, the expression pattern was not similar to those previously observed in Synechocystis sp. PCC 6803 and P. alcaligenes NBRC14159 (López-Maury et al., 2003; Zhang et al., 2015). Synechocystis sp. PCC 6803 has at least two operons (ars and arsM) on its chromosome. ars operon was regulated by ArsR though arsR was located far away from ars. DNase I footprinting experiments indicated that ArsR binds to two 17-bp direct repeats (ATCAA(N)6TTGAT) in the promoter-operator region. However, the upstream sequence of arsM does not have the 17-bp repeats (López-Maury et al., 2003). Authors proposed that ArsM is constitutively expressed whilst how ArsM expression is regulated has yet to be investigated. In P. alcaligenes NBRC14159, PaarsM was expressed in the absence of As(III) and the expression was further enhanced by As(III) exposure (Zhang et al., 2015). Our results revealed that the expression of the different arsenic operons were affected by growth cycle: the maximal expressions of both ars2 and ars3 operons in CGA009 appeared at early log-phase (43 h) and maintained at high level during the growth of middle log-phase (55 h). It is different from expression patterns reported in the earlier studies in Shewanella sp. ANA-3 (Saltikov et al., 2005). The highest arr expression appeared at the log-phase while peak expression level of ars was observed in the stationary phase, respectively. That transcription of the ars operon in Shewanella sp. ANA-3 could involve other factors, such as those related to quorum sensing and energy production. In fact, a quorum-sensing-based response was shown to be a second regulatory circuit for aio transcription in A. tumefaciens (Kashyap et al., 2006).
HPLC-ICP-MS analysis in middle log-phase demonstrated that As(V) was reduced to As(III), indicating that strain CGA009 detoxified arsenate by reducing As(V) to As(III) by ArsC (Figure 6). However, we failed to detect dimethylarsine (DMA), monomethylarsine (MMA) even in the growth stage where arsM transcript approached to the highest level (Figure 6). DMA and MMA, the intermediates produced in As(III) methylation process did not accumulate in the cells and were immediately converted into volatile trimethylarsine (TMA). Thereafter, TMA were rapidly expelled to extracellular space (Qin et al., 2006). In the same study, Qin et al. heterologously expressed the arsM gene from R. palustris CGA009 in an arsenic-sensitive strain of E. coli. Their results showed that ArsM catalyzed the formation of a number of methylated intermediates from As(III), with TMA as the end product and increased the arsenic resistance, indicating that it was very possible for arsM to be functional in vivo (Qin et al., 2006).
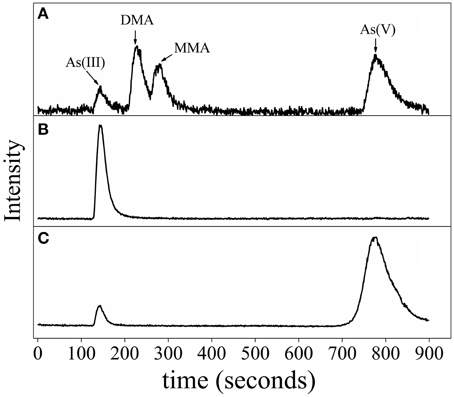
Figure 6. Arsenic speciation of R. palustris CGA009 determined by anion exchange HPLC-ICP-MS. (A) Chromatogram of the standards, including As (III), dimethylarsine (DMA), monomethylarsine (MMA) and As (V). (B) Speciation of arsenic of R. palustris CGA009 grown on 1.0 mM arsenite in the middle-log phase. (C) Speciation of arsenic of R. palustris CGA009 grown on 0.5 mM arsenate in the middle-log phase.
A working model for the arsenic detoxification by the arsenic multi-operons was proposed in R. palustris (Figure 7). In this model, the As(V) enters into cells through inorganic phosphate (Pit) or phosphate specific transport (Pst) systems (Kruger et al., 2013); once As(V) arrives inside the cells, it is reduced to As(III) by ArsC produced from ars1 and/or ars2 operon (possible at a low expression) (Figure 3). As(III) formed in the reaction then inactivates ArsR, initiating the transcription of the arsRRCC (acr3) operon (ars2). Due to the two copies of arsC in ars2, it allows to reduce arsenate more promptly, leading to As(III) accumulation in the cells. If the accumulated As(III) could not be expelled out of the cells by arsenite permease (Acr3), the increasing As(III) triggers the ars3 transcription by releasing ArsR which originally binds arsM promoter/operator. Arsenic resistance genetic units ars2 and ars3 in R. palustris contribute to detoxifying arsenic at a high dose because their transcription level is comparable when cells are treated with 1.0 mM arsenite. With cooperation of ars2 and ars3, R. palustris detoxifying As(III) by extruding it out of the cell by As(III) transporter (Acr3) or transforming As(III) to volatile methylated As(III) (TMA) by ArsM. It is reasonable to assume that ars2 is more important when cells are exposed to lower levels of arsenic due to its activity expression. However, a relatively complex arsenate detoxification system described here suggests that R. palustris growing in the natural environment must be equipped to deal with rapidly changing and perhaps relatively high levels of arsenic.
Conclusion
This study provided a novel insight into arsenic resistance mechanisms in R. palustris CGA009, a member of anoxygenic phototrophic bacteria. R. palustris possessed good arsenic resistance which possibly linked to the arsenic multi-operons in the chromosome. Our results showed ars2 and ars3 operons were upregulated by increasing As(III) concentrations while ars3 operon was only expressed when exposed to the arsenic concentrations more than 1.0 μM. However, the expression of ars1 operon was very low, indicating that ars1 may not be actively used. Collectively, our preliminary data showed that arsenic was possibly transformed by the combination mechanisms of the cytoplasmic As(V) reduction, As(III) extrusion and arsenic methylation when exposed to arsenic at a high concentration.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been supported by the National Natural Science Foundation of China (No. 31070054), by National Marine Public Industry Research (No. 201505026), by Natural Science Foundation of Fujian Province (No. 2015J01137).
References
Arsène-Ploetze, F., Koechler, S., Marchal, M., Coppée, J. Y., Chandler, M., Bonnefoy, V., et al. (2010). Structure, function, and evolution of the Thiomonas spp. Genome. PLoS Genet. 6:e1000859. doi: 10.1371/journal.pgen.1000859
Carius, L., Carius, A. B., McIntosh, M., and Grammel, H. (2013). Quorum sensing influences growth and photosynthetic membrane production in high-cell-density cultivations of Rhodospirillum rubrum. BMC Microbiol. 13:189. doi: 10.1186/1471-2180-13-189
Fu, Q. M., Zhao, C. G., Yang, S. P., and Wu, J. H. (2014). The photoelectric performance of dye-sensitized solar cells fabricated by assembling pigment–protein complexes of purple bacteria on nanocrystalline photoelectrode. Mater. Lett. 129, 195–197. doi: 10.1016/j.matlet.2014.05.054
Heinrich-Salmeron, A., Cordi, A., Brochier-Armanet, C., Halter, D., Pagnout, C., Abbaszadeh-fard, E., et al. (2011). Unsuspected diversity of arsenite-oxidizing bacteria as revealed by widespread distribution of the aoxB gene in Prokaryotes. Appl. Environ. Microbiol. 77, 4685–4692. doi: 10.1128/AEM.02884-10
Jackson, C. R., and Dugas, S. L. (2003). Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 3:18. doi: 10.1186/1471-2148-3-18
Kashyap, D. R., Botero, L. M., Franck, W. L., Hassett, D. J., and McDermott, T. R. (2006). Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188, 1081–1088. doi: 10.1128/JB.188.3.1081-1088.2006
Kruger, M. C., Bertin, P. N., Heipieper, H. J., and Arsène-Ploetze, F. (2013). Bacterial metabolism of environmental arsenic–mechanisms and biotechnological applications. Appl. Microbiol. Biotechnol. 97, 3827–3841. doi: 10.1007/s00253-013-4838-5
Lin, Z. H., Yue, H. Y., Lü, C. J., Zhao, C. G., and Yang, P. S. (2014). Variation in composition and relative content of accumulated photopigments in a newly isolated Rhodobacter capsulatus strain XJ-1 in response to arsenic. J. Environ. Sci. Health A 49, 1493–1500. doi: 10.1080/10934529.2014.937168
Liu, Y., Ghosh, D., and Hallenbeck, P. C. (2015). Biological reformation of ethanol to hydrogen by Rhodopseudomonas palustris CGA009 Bioresource Technology 176, 189–195. doi: 10.1016/j.biortech.2014.11.047
López-Maury, L., Florencio, F. J., and Reyes, J. C. (2003). Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185, 5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003
Lv, C. J., Zhang, Y. Y., Zhao, C. G., Guo, S. W., Yang, S. P., and Chen, S. H. (2012b). Arsenic resistance mechanisms in Rhodopseudomonas palustris under anaerobic and light conditions. Acta Sci. Circumstantiae 32, 2375–2383.
Lv, C. J., Zhao, C. G., Yang, S. P., and Qu, Y. B. (2012a). Arsenic metabolism in purple nonsulfur bacteria. Acta Microbiol. Sinica 52, 1497–1507.
Muller, D., Médigue, C., Koechler, S., Barbe, V., Barakat, M., Talla, E., et al. (2007). A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genetics 3:e53. doi: 10.1371/journal.pgen.0030053
Oremland, R. S., and Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944. doi: 10.1126/science.1081903
Páez-Espino, D., Tamames, J., de Lorenzo, V., and Canovas, D. (2009). Microbial responses to environmental arsenic. Biometals 22, 117–130. doi: 10.1007/s10534-008-9195-y
Qin, J., Lehr, C. R., Yuan, C., Le, X. C., McDermott, T. R., and Rosen, B. P. (2009). Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc. Natl. Acad. Sci. U.S.A. 106, 5213–5217. doi: 10.1073/pnas.0900238106
Qin, J., Rosen, B. P., Zhang, Y., Wang, G., Franke, S., and Rensing, C. (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 2075–2080. doi: 10.1073/pnas.0506836103
Rodríguez-Lado, L., Sun, G., Berg, M., Zhang, Q., Xue, H., Zheng, Q., et al. (2013). Groundwater arsenic contamination throughout China. Science 341, 866–868. doi: 10.1126/science.1237484
Saltikov, C. W., and Newman, D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U.S.A. 100, 10983–10988. doi: 10.1073/pnas.1834303100
Saltikov, C. W., Wildman, R. A. Jr., and Newman, D. K. (2005). Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J. Bacteriol. 187, 7390–7396. doi: 10.1128/JB.187.21.7390-7396.2005
Slyemi, D., and Bonnefoy, V. (2012). How prokaryotes deal with arsenic. Environ. Microbiol. Reports 4, 571–586. doi: 10.1111/j.1758-2229.2011.00300.x
Stolz, J. F., Basu, P., and Oremland, R. S. (2010). Microbial arsenic metabolism: new twists on an old poison. Microbe 5, 53–59. doi: 10.1128/microbe.5.53.1
Stolz, J. F., and Oremland, R. S. (1999). Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23, 615–627. doi: 10.1111/j.1574-6976.1999.tb00416.x
Villadangos, A. F., Van Belle, K., Wahni, K., Dufe, V. T., Freitas, S., Nur, H., et al. (2011). Corynebacterium glutamicum survives arsenic stress with arsenate reductases coupled to two distinct redox mechanisms. Mol. Microbiol. 82, 998–1014. doi: 10.1111/j.1365-2958.2011.07882.x
Yang, H. C., Fu, H. L., Lin, Y. F., and Rosen, B. P. (2012). Pathways of arsenic uptake and efflux. Curr. Top. Membr. 69, 325–358. doi: 10.1016/B978-0-12-394390-3.00012-4
Zhang, J., Cao, T., Tang, Z., Shen, Q., Rosen, B. P., and Zhao, F. J. (2015). Arsenic methylation and volatilization by arsenite S-adenosylmethionine methyltransferase in Pseudomonas alcaligenes NBRC14159. Appl. Environ. Microbiol. 81, 2852–2860. doi: 10.1128/AEM.03804-14
Keywords: Rhodopseudomonas palustris, operons, arsenic resistance, regulation
Citation: Zhao C, Zhang Y, Chan Z, Chen S and Yang S (2015) Insights into arsenic multi-operons expression and resistance mechanisms in Rhodopseudomonas palustris CGA009. Front. Microbiol. 6:986. doi: 10.3389/fmicb.2015.00986
Received: 23 April 2015; Accepted: 04 September 2015;
Published: 17 September 2015.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Patrick Hallenbeck, University of Montreal, CanadaLei Chen, Tianjin University, China
Joseph Kuo-Hsiang Tang, Arizona State University, USA
Copyright © 2015 Zhao, Zhang, Chan, Chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chungui Zhao and Suping Yang, Department of Bioengineering and Biotechnology, College of Chemical Engineering, Huaqiao University, 668 Jimei Avenue, Xiamen 361021, China, chungui@hqu.edu.cn; yangsuping@hqu.edu.cn;
Shicheng Chen, Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI 48863, USA, shicheng@msu.edu
 Chungui Zhao1*
Chungui Zhao1* Shicheng Chen
Shicheng Chen