- 1STELA Dairy Research Center, Nutrition and Functional Foods Institute, Université Laval, Québec, QC, Canada
- 2Centre de Recherche en Infectiologie de l’Université Laval, Axe Maladies Infectieuses et Immunitaires, Centre de Recherche du CHU de Québec, Québec, QC, Canada
- 3Département de Microbiologie-Infectiologie et d’Immunologie, Faculté de Médecine, Université Laval, Québec, QC, Canada
Clostridium difficile is the most frequently identified enteric pathogen in patients with nosocomially acquired, antibiotic-associated diarrhea and pseudomembranous colitis. Although metronidazole and vancomycin were effective, an increasing number of treatment failures and recurrence of C. difficile infection are being reported. Use of probiotics, particularly metabolically active lactic acid bacteria, was recently proposed as an alternative for the medical community. The aim of this study was to assess a probiotic candidate, nisin Z-producer Lactococcus lactis UL719, competitivity and nisin (Nisaplin®) capacity to inhibit C. difficile in a model of human colon. Bacterial populations was enumerated by qPCR coupled to PMA treatment. L. lactis UL719 was able to survive and proliferate under simulated human colon, did not alter microbiota composition, but failed to inhibit C. difficile. While a single dose of 19 μmol/L (5× the MIC) was not sufficient to inhibit C. difficile, nisin at 76 μmol/L (20×the MIC) was effective at killing the pathogen. Nisin (at 76 μmol/L) caused some temporary changes in the microbiota with Gram-positive bacteria being the mostly affected. These results highlight the capacity of L. lactis UL719 to survive under simulated human colon and the efficacy of nisin as an alternative in the treatment of C. difficile infections.
Introduction
Clostridium difficile is a Gram-positive anaerobic sporulating pathogen causing intestinal infections following disturbance of the human and animal gut microbiota, usually subsequent to an antibiotic therapy. C. difficile is now thought to be responsible for a wide range of diseases including acute diarrhea and pseudomembranous colitis, and could lead to colonic perforation and death if untreated (Borriello et al., 1990). Although metronidazole and vancomycin are well-established treatments for C. difficile infections (CDI) (Surowiec et al., 2006; Kelly and LaMont, 2008), an increasing number of treatment failures with these antibiotics and recurrence of C. difficile infection are being reported, reviewed in Vardakas et al. (2012). Vancomycin is also losing its attractiveness for CDI treatment with emergence of vancomycin-resistant enterococci and dissemination of antibiotic-resistance determinants within the hospital environment (Lagrotteria et al., 2006). The emergence of C. difficile isolates with multiple-drug resistance is rarely explicitly mentioned (Peláez et al., 2002; Mutlu et al., 2007), but constitutes further a serious public health threat that urges the need of novel antimicrobial treatments.
Previously, a large number of clinical trials highlighted the positive role of probiotics in the treatment of diarrhea by either shortening its duration and/or preventing its complications in infants and young children, reviewed in Guandalini (2011). In instance, a yogurt containing a combination of Lactobacillus rhamnosus GG, L. acidophilus La-5, and Bifidobacterium lactis Bb12 was shown to be an effective method for reducing the incidence of antibiotic-associated diarrhea in children (Fox et al., 2015). Moreover, different probiotics (Saccharomyces boulardii, L. casei DN114001, a mixture of L. acidophilus and B. bifidum, and a mixture of L. acidophilus, L. casei and L. rhamnosus) significantly improved CDI prevention, reviewed in McFarland (2015). Although several meta-analyses pointed the positive effect of probiotics, their role in the prevention of CDI remains unclear. The health-promoting properties of probiotics are numerous and their effects on host include competition with pathogens for adhesion sites and nutrients, stimulation of immunity/immunomodulation, and production of inhibitory substances such as bacteriocins (Fliss et al., 2011). Bacteriocins have been suggested as promising alternative to conventional antibiotics (Rea et al., 2007, 2010), and their production is being considered as a probiotic trait although not clearly demonstrated in vivo (Dobson et al., 2012). While several bacteriocins including nisin (Le Blay et al., 2007; Le Lay et al., under revision), Microbisporicin (Castiglione et al., 2008), Lacticin 3147 (Rea et al., 2007) and thuricin CD (Rea et al., 2010) were shown effective against C. difficile, to date only nisin is approved by the American Food and Drug Administration, the World Health Organization, and the European Union as natural food additive (Delves-Broughton, 1990). Nisin displays high antibacterial activity against multi-resistant Streptococcus pneumoniae, methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium, E. faecalis, and C. difficile (Severina et al., 1998; Le Blay et al., 2007).
Previously, we have observed that potential probiotic Lactococcus lactis UL719, a nisin Z producer, was able to survive through the gastrointestinal tract (unpublished data). The strain L. lactis UL719 was able to grow and inhibit Listeria in a medium simulating the nutrient composition of the human colon (Fernandez et al., 2013). The aim of this study was to evaluate the capacity of L. lactis UL719 and nisin (Nisaplin®) to inhibit C. difficile in a model of the colon mimicking physiological and microbiological conditions of the large intestine. In addition, impact of both strain and its bacteriocin on the gut microbiota composition were also investigated.
Materials and Methods
Bacterial Strains and Growth Conditions
Lactococcus lactis sp. lactis biovar. diacetylactis UL719, a nisin Z-producer, was isolated from raw milk cheese (Ali et al., 1995; Meghrous et al., 1997). C. difficile ATCC43255 was purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). L. lactis UL719 was reactivated in De Man Rogosa Sharpe (MRS) broth (Difco Laboratories, Sparks, MD, USA) and cultivated for 24 h at 30°C. C. difficile was reactivated in Brain Heart Infusion (BHI) broth (Difco laboratories, Sparks, MD, USA) supplemented with 0.05% L-cysteine-HCl (Sigma chemicals). C. difficile culture was cultivated in an anaerobic chamber (Forma scientific anaerobic system Model 1025; Forma Scientific, Marietta, OH, USA) at 37°C for 24 h. Escherichia coli ATCC25922 and E. faecalis ATCC27275 were grown aerobically at 37°C for 24 h in BHI and TSBYE, respectively. B. adolescentis ATCC15703, Bacteroides thetaiotaomicron ATCC29741, Blautia coccoides ATCC29236, and C. leptum ATCC29065 were, respectively, grown in MRS broth (0.05% L-cysteine-HCl), BHI (0.05% L-cysteine-HCl) and a modified chopped meat medium with maltose (ATCC medium 2751) under anaerobic conditions at 37°C. All bacterial strains were maintained in 20% glycerol at -80°C. Prior to each experiment, each bacterial strain was subcultured at least three times (inoculation at 1%, v/v) at 24 h intervals.
Development of Large Intestine Fermentation Model
Feces Collection and Immobilization in Gel Beads
A fresh fecal sample was obtained from one 27 years old healthy donor who had not taken antibiotics for the previous 3 months. The collected fecal sample was used for immobilization following procedure described by Le Blay et al. (2012). The entire process was completed aseptically under anaerobic conditions within 1 h after sample collection.
Nutritive Medium
The culture medium used for colonic fermentation was the same as described by Macfarlane et al. (1998) with some modifications. Briefly, 0.5 mL of a vitamin solution (mg/L: pyridoxine–HCl 20; p-aminobenzoic acid 10; nicotinic acid 10; biotin 4; folic acid 4; vitamin B12 1; thiamine 8; riboflavin 10; menadione 2; vitamin K1 0.005; pantothenate 20) described by Gibson and Wang (1994) was added to each liter of the culture medium. The vitamin solution was sterilized by filtration (0.2 μm, VWR) and added to the autoclaved medium (15 min, 121°C) after cooling at room temperature.
Experimental Setup and Sampling
The colonic fermentation was based on the model described by Cinquin et al. (2004). A single-stage reactor (Bioflo III, New Brunswick Scientific Inc., Edison, NJ, USA) with 1 L working volume containing 30% (v/v) of freshly prepared beads was used to mimic the microbial ecosystem of adult distal colon. The colonization of beads with fecal microbiota was carried out during 2 days, and the nutritive medium was aseptically replaced by fresh culture medium every 12 h. pH (6.2) and anaerobic and temperature (37°C) conditions were maintained during the whole fermentation by addition of 5 M NaOH and a continuous flow of pure CO2 in the headspace. The continuous fermentation was carried out in the same reactor connected to a stirred feedstock vessel containing the sterile culture medium at 4°C under a CO2 atmosphere and to an effluent-receiving vessel. Feed flow rate was adjusted to 83.3 mL/h to mimic a mean retention time of 12 h encountered in adult distal colon.
The fermentation process was carried out for a total of 82 days and microbiota was stabilized 2 weeks before challenging tests. First, a cell suspension of L. lactis UL719 (at final concentration 109 CFU/mL in the reactor) was added twice to the reactor (day 17 and 22) (Figure 1). Then, nisin A (Nisaplin®, Danisco, Copenhagen, Denmark) was added to the reactor at 5× (at day 27 and 32) and 20× (at day 37 and 42) the MIC (3.8 μmol/L vs. C. difficile) to measure the impact of high doses of nisin on the intestinal flora. Next, challenges with C. difficile ATCC43255 (at a final concentration of 5 × 106 CFU/mL in the reactor) in absence (day 47 and 52) or in presence of L. lactis (added at a final concentration of 109 CFU/mL in the reactor; day 57 and 62) or in presence of different concentrations of nisin A [5× (day 67 and 72) or 20× (day 77 and 82) the MIC] were performed. Samples were collected for bacterial enumeration by qPCR. After each addition, samples were hourly taken during first 4 and at 8 h.
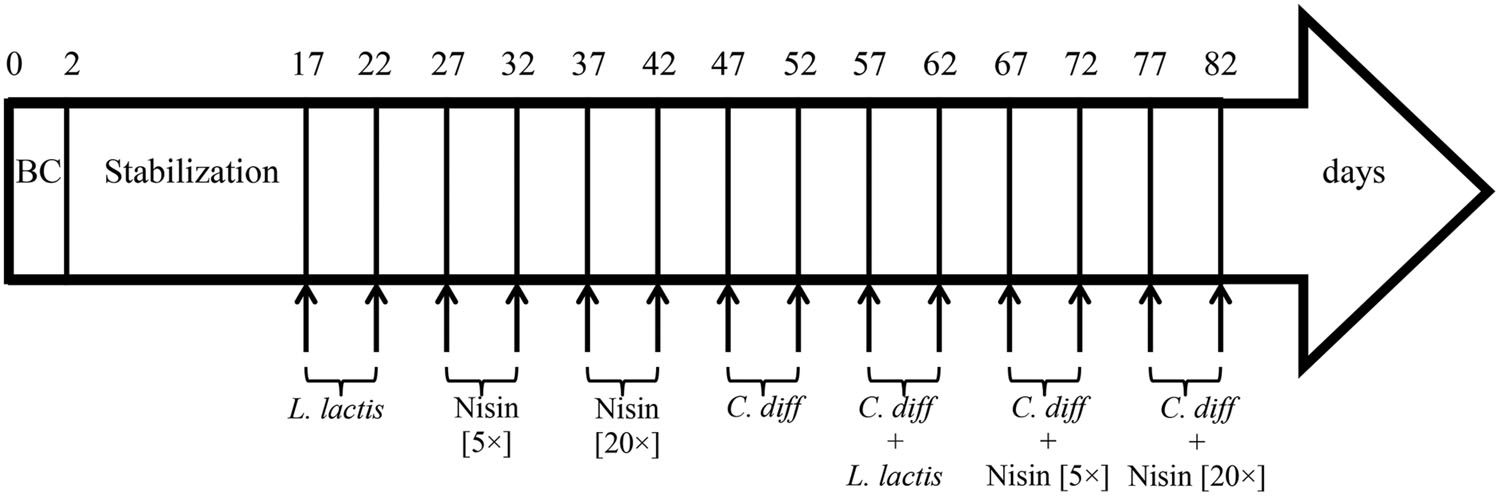
FIGURE 1. Time schedule of continuous intestinal fermentation during the different treatment periods. BC, bead colonization. Lactococcus lactis UL719 was added at final concentration of 109 CFU/mL in the reactor. Clostridium difficile ATCC43255 was added at a final concentration of 5 × 106 CFU/mL.
Microbiota Composition Analysis using q-PCR Coupled to PMA Treatment
Standard curve for the qPCR quantification was done using the following strains: E. coli ATCC 25922, B. adolescentis ATCC15703, B. thetaiotaomicron ATCC29741, C. leptum ATCC29065, B. coccoides ATCC29236, and E. faecalis ATCC27275. Samples were collected from the reactor and treated with propidium monoazide (Biotium, Inc., Hayward, CA, USA) prior enumeration of viable bacteria, as described in Fernandez et al. (2015). The DNA from fecal and fermentation samples were then extracted following the protocol of Ahlroos and Tynkkynen (2009) using the Wizard®genomic DNA Purification Kit (Promega, Madison, WI, USA) with some modifications (Fernandez et al., 2015). Real-time PCR was carried out on an ABI 7500 real-time PCR system (Applied biosystem, Streetsville, ON, Canada) with the iTaqTM Universal SYBR®Green supermix (Biorad, Oakville, ON, Canada) in 96-well plates following method described in Fernandez et al. (2015). Primers used in this study are summarized in Table 1. Each sample was done in triplicate.
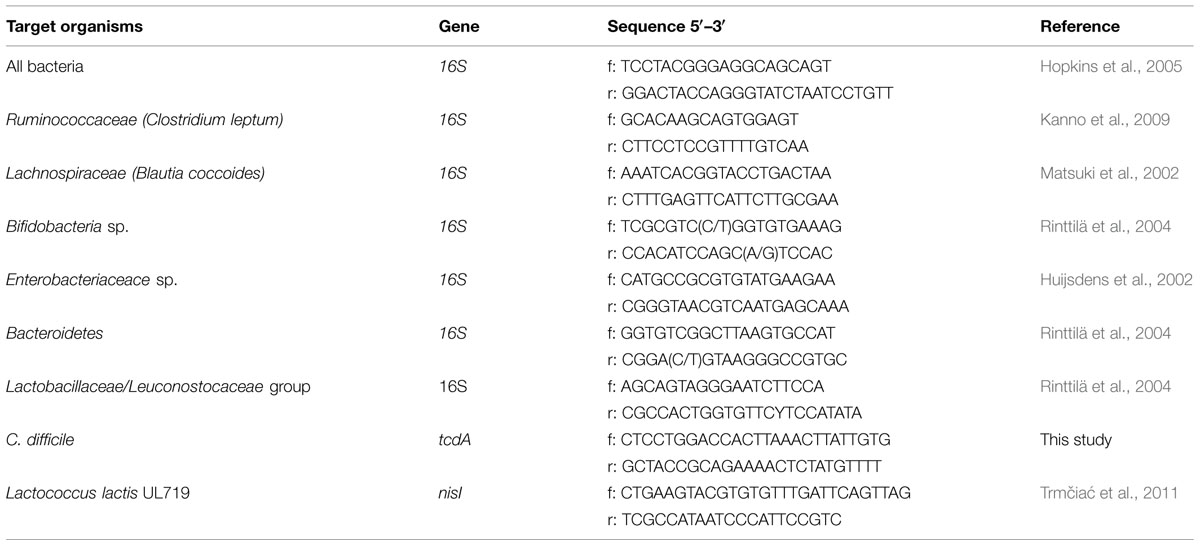
TABLE 1. Primers used for the detection of different bacterial groups in inoculum or fermentation samples by real-time qPCR analysis.
Analyses of Metabolites
Short-chain fatty acids (SCFA: acetate, propionate, butyrate, and valerate) and isoacids (isobutyrate and isovalerate) were determined by high-performance liquid chromatography (HPLC) analysis (Waters, Milford, MA, USA) equipped with an Ion 300 column (Transgenomic, San Jose, CA, USA), a differential refractometer (Model R410, Waters) as previously described by Cinquin et al. (2004). The analysis was performed at a flow rate of 0.4 mL/min at 37°C, with an injection volume of 100 μL. Each analysis was done in duplicate. The mean metabolite concentrations were expressed in mmol/L.
Statistical Analysis
Data are presented as means ± SD. Cell counts values were log10-transformed and analyzed for repeated measures using the PROC MIXED procedure of SAS v9.2 statistical package (SAS Institute Inc., Cary, NC, USA). The statistical differences in metabolites contents between treatments were evaluated using a one-way ANOVA t-test. The level of significance was P ≤ 0.05.
Results
Microbiota Composition during Stabilization Period
Bacterial populations enumerated by qPCR coupled to PMA treatment in the fecal inoculum and effluent samples at the end of stabilization period are summarized in Table 2. The fecal inoculum presented a total bacterial cell counts of 11.84 ± 0.04 log10 CFU/g, which was dominated by Bacteroidetes (10.85 ± 0.02 log10 CFU/g), clostridia (10.55 ± 0.02 log10 CFU/g), and bifidobacteria (10.16 ± 0.15 log10 CFU/g). At the end of the stabilization period (16 days) under simulated colon conditions, the microbiota population reached a pseudo steady state in which a slight change was observed in the microbial balance, compared to the initial fecal inoculum. The microbiota decreased by -0.7 log10 CFU/mL at this stage and was dominated by Bacteroidetes group with 10.52 ± 0.08 log10 CFU/mL. While Enterobacteriaceae group increased by 1.49 log10 and reached 8.73 ± 0.01 log10 CFU/mL, bifidobacteria, and Lactobacillaceae/Leuconostocaceae group populations dropped to 6.14 ± 0.08 and 3.82 ± 0.14 log10 CFU/mL, respectively. Nevertheless, the obtained results are in accordance to those previously reported for colonic fermentation models (Brück et al., 2002; Probert and Gibson, 2004; Cleusix et al., 2008; Le Blay et al., 2012).
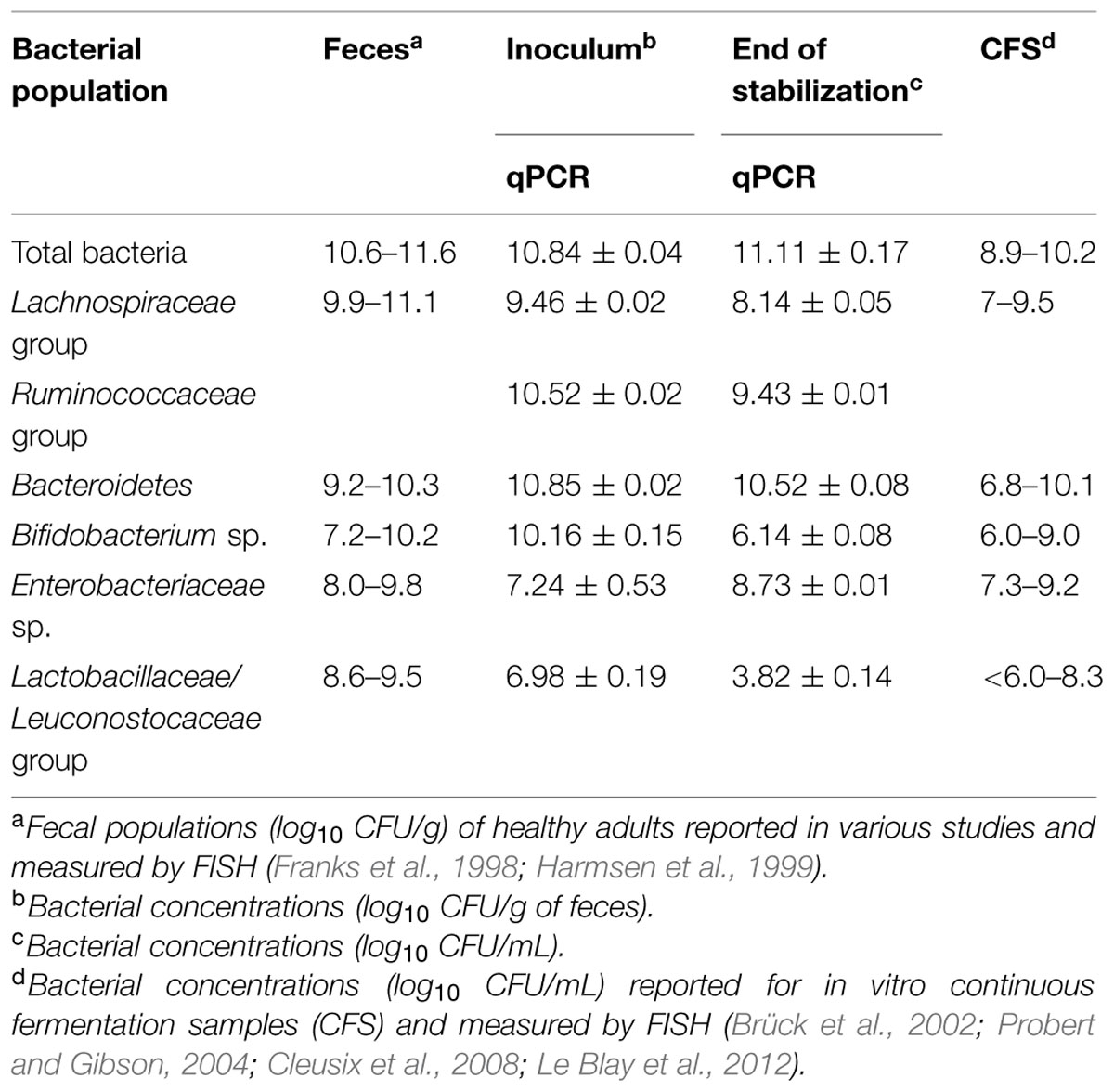
TABLE 2. Bacterial cell counts in the fecal inoculum and during the fermentation at the end of the stabilization period of the continuous culture measured by qPCR.
Lactococcus lactis UL719 Alone or in Presence of C. difficile ATCC43255 have no Perturbing Impact on Intestinal Microbiota under Simulated Colonic Conditions
After the stabilization period, L. lactis UL719, C. difficile ATCC43255, and their combination were successively added to the bioreactor and the microbiota populations were monitored by qPCR (Table 3). Interestingly, the addition of L. lactis UL719 at 1 × 109 CFU/mL to the bioreactor, did not induce any significant change neither in the intestinal microbiota composition nor in metabolites production (Table 4). Since the last addition of L. lactis UL719 to the reactor, the strain was detected at about 0.1 - 1 × 109 CFU/mL during the remaining 20 days of fermentation (Figure 2). While the infection of the bioreactor with 5 × 106 CFU/mL of C. difficile did not affect the microbiota composition, a slight but significant decrease (p < 0.05) of acetate and butyrate was detected (from 76.24 to 72.59 mmol/L and from 32.13 to 29.54 mmol/L, respectively) (Table 4). Simultaneous addition of C. difficile and L. lactis UL719 had no impact on the microbiota cell counts but a significant decrease (p < 0.05) of butyrate (from 32.13 to 28.40 mmol/L). Under these conditions, L. lactis has no inhibitory effect on C. difficile (Figure 3).
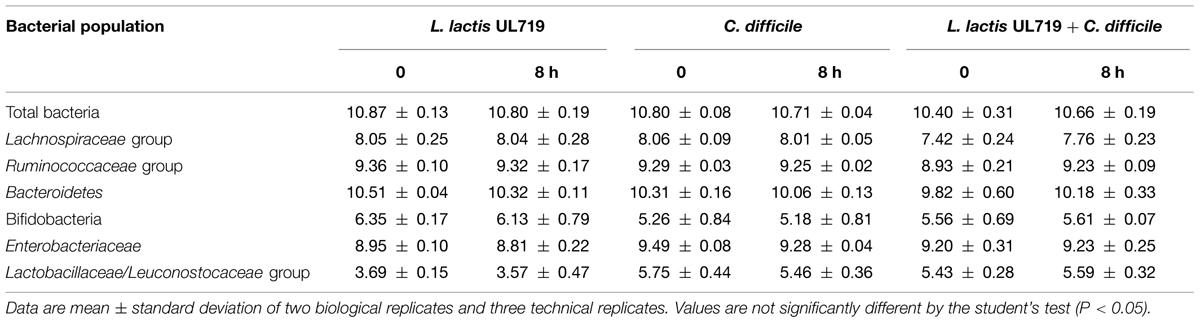
TABLE 3. Impact of L. lactis UL719 (109 CFU/mL) and/or C. difficile ATCC43255 (5 × 106 CFU/mL) addition on the microbiota.
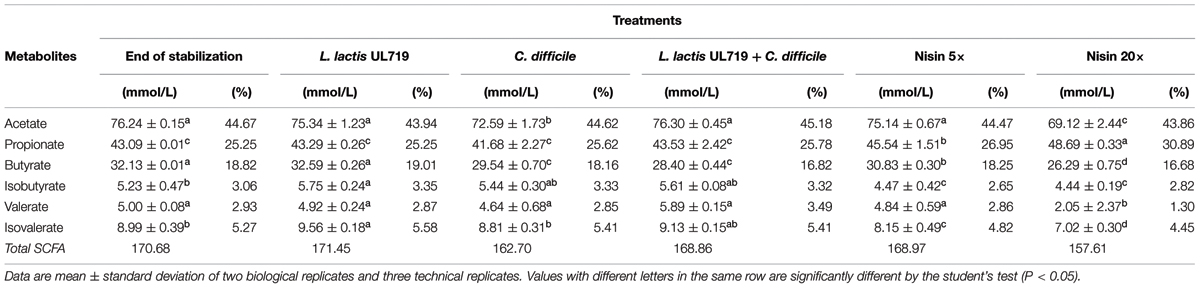
TABLE 4. Concentration of short chain fatty acids (SFCA) in effluent samples at 4 h following various treatments.
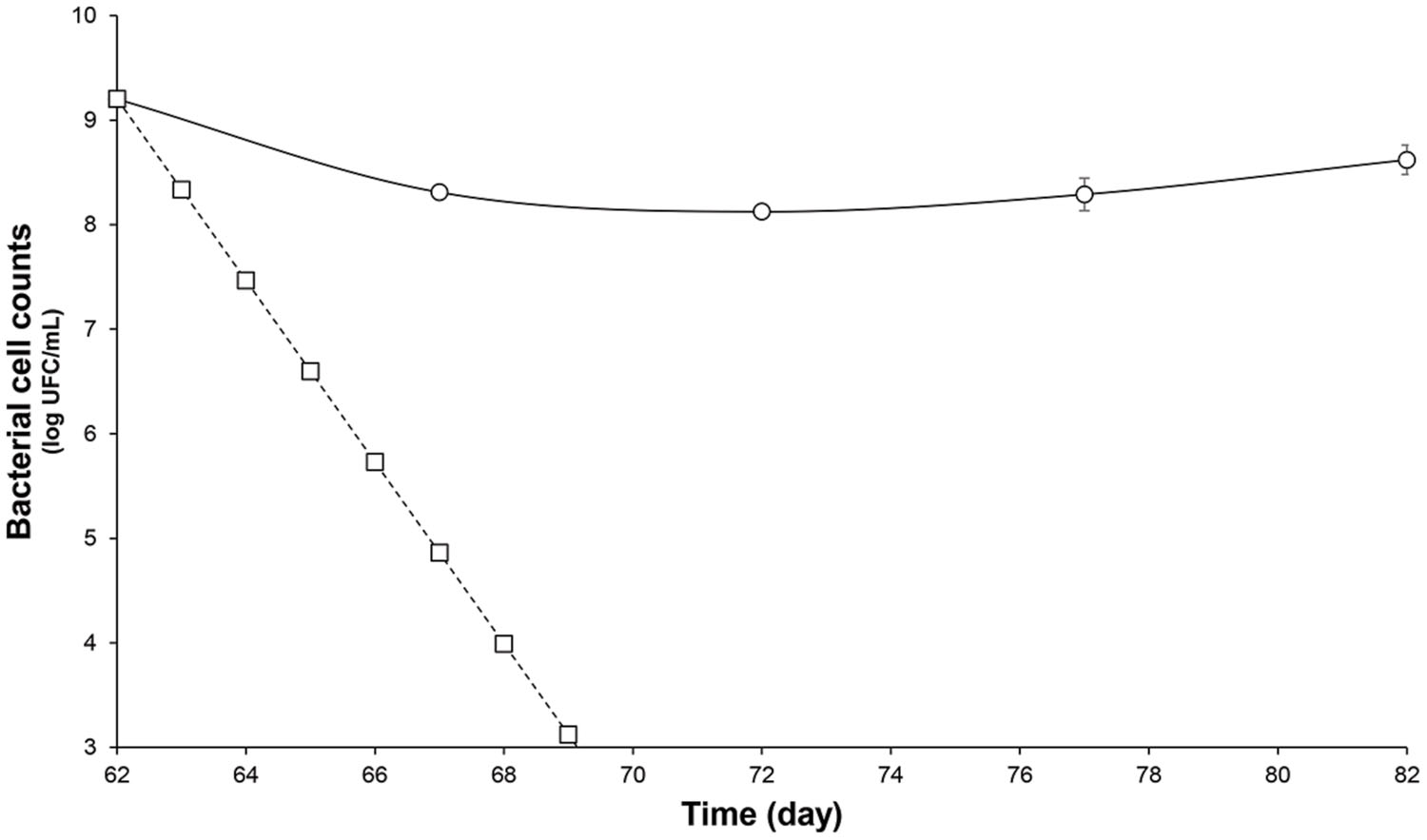
FIGURE 2. Survival of L. lactis UL719 after its last addition (day 62) in a human colon model. L. lactis UL719 (circle); theoretical washout (square).
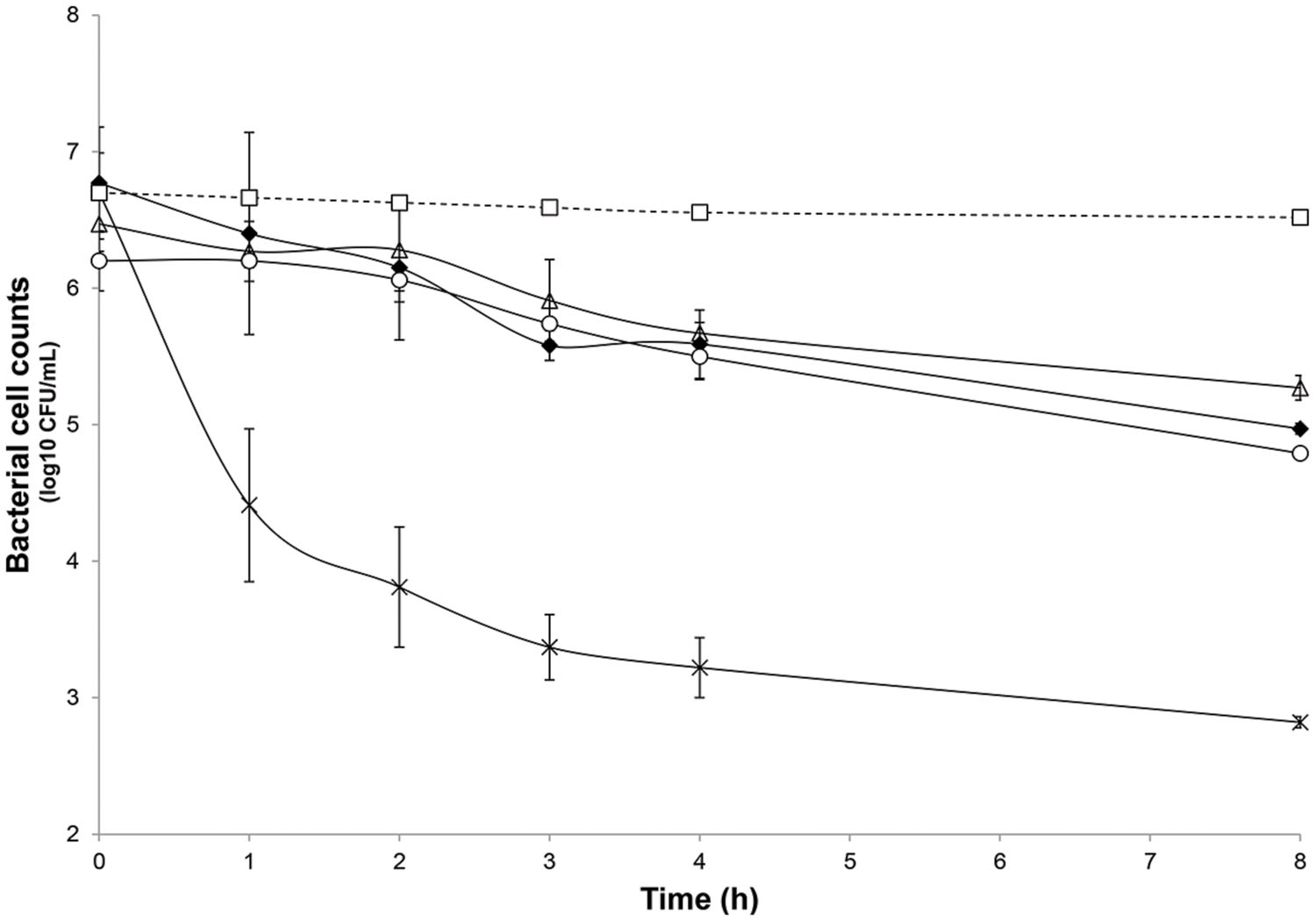
FIGURE 3. Inhibitory activity of nisin at 5× and 20× the MIC (3.8 μmol/L) and L. lactis UL719 (109 CFU/mL) against C. difficile ATCC43255 in a human colon model. C. difficile alone (black diamond); C. difficile plus nisin 5× (white triangle); C. difficile plus nisin 20× (cross); C. difficile plus L. lactis UL719 (white circle); theoretical washout (white square).
A Nisin Concentration of 20× the MIC is Required to Effective Inhibition of C. difficile ATCC43255 in a Model of Human Colon
The microbiota was challenged by 5× and 20× the MIC vs. C. difficile ATCC43255. Nisin at 5× the MIC did not alter the microbiota which remained stable (data not shown) although minor variations in the metabolite production profile (Table 4). At a nisin concentration of 20× the MIC, total microbiota significantly decreased by 0.7 log10 (p < 0.008), as shown in Figure 4. Gram-positive bacteria were affected by this higher amount of nisin, with Ruminococcaceae group being the mostly altered (-3.7 log10) after 24 h. In a lesser extent, a reduction of 1.5 log10, 1.3 log10, and 1 log10 were recorded for Lachnospiraceae group, Lactobacillaceae/Leuconostocaceae group and bifidobacteria, respectively. After 24 h of nisin administration, all bacterial populations recovered their initial counts except Ruminococcaceae group which dropped to its minimum counts. While acetate and butyrate significantly decreased (p < 0.05) from 76.24 and 32.13 mmol/L to 69.12 and 26.29 mmol/L, propionate production increased by 13% (Table 4). Besides, a nisin concentration of 5× did not inhibit C. difficile, which counts remained close to control (C. difficile alone) (Figure 3). Conversely, nisin at 20× was effective at inhibiting C. difficile with a significant reduction (p < 0.001) of 2.3 log10 at 1 h that lasted for 8 h (Figure 3). C. difficile was not detected after 24 h in this model (data not shown).
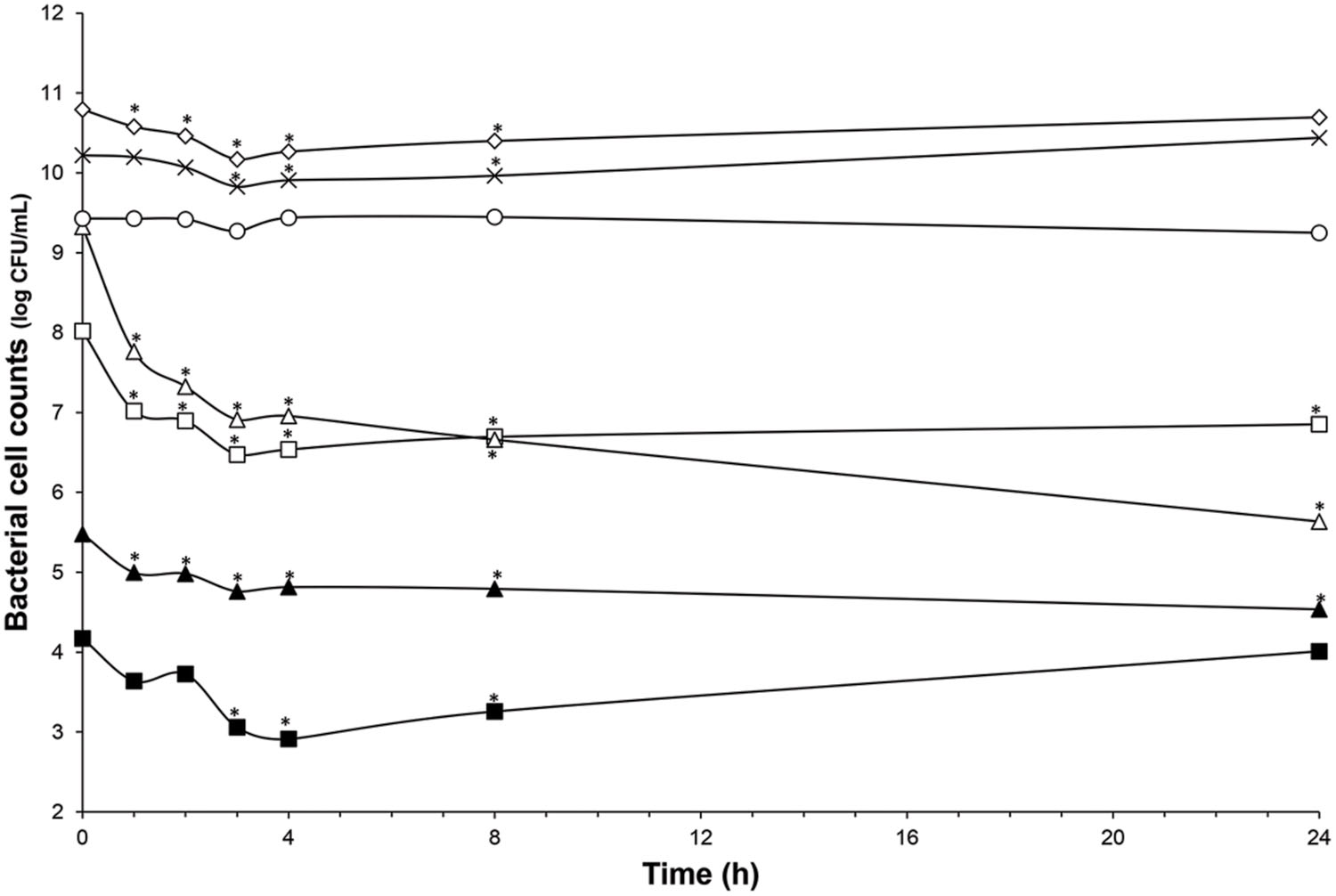
FIGURE 4. Impact of nisin addition at 76 μmol/L (20× the MIC vs. C. difficile ATCC43255) on microbiota population enumerated by qPCR. Total bacteria (white diamond); Lachnospiraceae group. (white square); Ruminococcaceae group. (white triangle); Bacteroidetes (cross); Bifidobacteria (black triangle); Enterobacteriaceae (white circle); Lactobacillaceae/Leuconostocaceae group. (black square); Values with asterisk are significantly different (P < 0.05).
Discussion
Previously, we have demonstrated the nisin efficacy against several clinical isolates of C. difficile vegetative cells and spores (Le Lay et al., under revision). In addition, we have observed that L. lactis UL719, a nisin Z producer, was able to survive these GIT stressful conditions, to keep ability to produce its bacteriocin, and to reach the colon in large enough numbers (>108 CFU) to comply with the recommended daily dose of 108–109 cells delivery to exert a beneficial effect on the host (unpublished data). The aim of this study was to assess L. lactis UL719 competitivity and nisin capacity to inhibit C. difficile ATCC43255 in a model of human colon. In this study, L. lactis UL719 at 109 CFU/mL did not induce any significant change neither in the intestinal microbiota composition nor in metabolites production. The strain was monitored by quantification of nisI gene by PMA-qPCR, and found able to survive and proliferate up to 108 CFU/mL in our colonic model during the 82 days of fermentation (Figure 2). Unlikely, L. lactis DPC6520 was shown more susceptible to GIT conditions, which cell counts were reduced by 10 000-fold 24 h after its inoculation into a colon model (Dobson et al., 2011). Likewise, a 19 μmol/L concentration of nisin (corresponding to 5× the MIC vs. C. difficile ATCC43255) did not alter microbiota levels. At a higher concentration of 76 μmol/L (20×), Gram-positive bacteria were affected and Ruminococcaceae group was the mostly altered (-3.7 log10), while increase in Gram-negative population (Bacteroidetes and Enterobacteriaceae) were observed. Nevertheless, the initial bacterial balance was quickly restored within 24 h after the addition of 20× nisin. Previously, we have shown in vitro the sensitivity of colonic Gram-positive bacteria such as B. bifidum DSM 20456, L. fermentum ETHZ, C. clostridioforme DSM933, Eubacterium biforme DSM3989 to nisin (Le Blay et al., 2007). Recently, Rea et al. (2011) reported that lacticin 3147 induce similar variations in microbiota composition, with a decrease in Firmicutes abundance in favor of Proteobacteria. Broad-spectrum antibiotics like vancomycin and metronidazole seems to induce also decrease of Firmicutes and an increase in Enterobacteriaceae and Proteobacteria (Antonopoulos et al., 2009; Rea et al., 2011). More recently, thuricin CD, a narrow spectrum bacteriocin produced by Bacillus thuringiensis, was used in the distal colon model and had no significant impact on the composition of the microbiota (Rea et al., 2011).
Although its capacity to survive colonic conditions, L. lactis UL719 had no significant effect on C. difficile. Similar results were previously reported with L. lactis DPC6520 (a lacticin 3147 producer) and L. lactis DPC6519 (lacticin non-producer) in an ex vivo human colonic model (Dobson et al., 2011). Although L. lactis UL719 is able to produce nisin in a Macfarlane medium simulating the nutrient composition of the colon (Fernandez et al., 2013), the lack of effectiveness observed here is likely due to no or a low production of nisin, not sufficient to inhibit C. difficile. Conversely, L. salivarius UCC118 has demonstrated its capacity to produce the Abp118 bacteriocin in vivo and to protect mice against infection with the invasive foodborne pathogen Listeria monocytogenes. This protection was related to bacteriocin production, and mutant of L. salivarius UCC118 lacking the bacteriocin gene failed to protect mice against infection (Corr et al., 2007). Some similar results were obtained with human L. lactis and Pediococcus acidilactici nisin- and pediocin-producing strains that were able to reduce vancomycin-resistant enterococci intestinal colonization in a mouse model (Millette et al., 2008).
Although L. lactis UL719 had no significant effect on C. difficile in this model of human colon, addition of nisin (in Nisaplin®form) at 76 μmol/L induced a significant reduction of C. difficile. The observed efficacy of Nisaplin®against C. difficile could be due to a synergy between nisin and salt present in the commercial product. At lower concentration of nisin (19 μmol/L), we did not show any significant effect on C. difficile, its rapid adsorption on the surface of the colonic microbiota or its inactivation due to enzymatic activities (proteolysis mainly) could explain this lack of activity (Dobson et al., 2011). Rea et al. (2011) have reported on the effectiveness of other bacteriocins such as lacticin 3147 and thuricin CD against C. difficile in a distal colon model. Lacticin 3147 (270 μmol/L) and thuricin CD (90 μmol/L) affected the viability of C. difficile (106 CFU/mL) with a loss of detection after 12 h and three log10 reduction after 24 h, respectively (Rea et al., 2011). After respective addition of lacticin 3147 (270 μmol/L) and thuricin CD (90 μmol/L), authors have shown a CFU reduction of 4 log10 and 1.2 log10, but lacticin at 90 μmol/L had no significant effect on the C. difficile viability (Rea et al., 2011). In this study, nisin was as effective as lacticin 3147 and more efficient than thuricin CD with a CFU reduction of 3.23 log10 with nisin (76 μmol/L) compared to initial time. Besides, three times addition of vancomycin (90 μmol/L) or metronidazole (90 μmol/L) is required to induce a significant effect on C. difficile after 24 h (Rea et al., 2011). A single dose of nisin (76 μmol/L) was as effective as antibiotics traditionally used to treat CDIs.
With increase of failures and recurrences in the treatment of CDIs, development of alternative treatments has become necessary. In recent years, use of probiotic bacteria producing antimicrobial molecules (such as bacteriocins) constitute a promising alternative for prevention and treatment of C. difficile related diseases. In the study, we have shown that nisin-producer L. lactis UL719 was able to survive and proliferate in the human colon model. Although L. lactis UL719 failed to inhibit C. difficile in this model, L. lactis UL 719 had not affected the microbiota. Others studies aiming to increase competitivity and nisin production will be necessary and could include the addition of prebiotics or carbohydrate which stimulate nisin production. Nisin (Nisaplin®) causes some temporary changes in the microbiota but is effective at killing C. difficile in the human colon model.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science and Engineering Research Council of Canada (NSERC) and the Fond de Recherche Nature et Technologies from the province of Quebec (FQRNT).
References
Ahlroos, T., and Tynkkynen, S. (2009). Quantitative strain-specific detection of Lactobacillus rhamnosus GG in human faecal samples by real-time PCR. J. Appl. Microbiol. 106, 506–514. doi: 10.1111/j.1365-2672.2008.04018.x
Ali, D., Lacroix, C., Thuault, D., Bourgeois, C. M., and Simard, R. E. (1995). Characterization of diacetin B, a bacteriocin from Lactococcus lactis subsp. lactis bv. diacetylactis UL720. Can. J. Microbiol. 41, 832–841. doi: 10.1139/m95-114
Antonopoulos, D. A., Huse, S. M., Morrison, H. G., Schmidt, T. M., Sogin, M. L., and Young, V. B. (2009). Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77, 2367–2375. doi: 10.1128/IAI.01520-08
Borriello, S. P., Davies, H. A., Kamiya, S., Reed, P. J., and Seddon, S. (1990). Virulence factors of Clostridium difficile. Rev. Infect. Dis. 12, S185–S191. doi: 10.1093/clinids/12.Supplement_2.S185
Brück, W. M., Graverholt, G., and Gibson, G. R. (2002). Use of batch culture and a two-stage continuous culture system to study the effect of supplemental α-lactalbumin and glycomacropeptide on mixed populations of human gut bacteria. FEMS Microbiol. Ecol. 41, 231–237. doi: 10.1111/j.1574-6941.2002.tb00984.x
Castiglione, F., Lazzarini, A., Carrano, L., Corti, E., Ciciliato, I., Gastaldo, L., et al. (2008). Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem. Biol. 15, 22–31. doi: 10.1016/j.chembiol.2007.11.009
Cinquin, C., Le Blay, G., Fliss, I., and Lacroix, C. (2004). Immobilization of infant fecal microbiota and utilization in an in vitro colonic fermentation model. Microb. Ecol. 48, 128–138. doi: 10.1007/s00248-003-2022-7
Cleusix, V., Lacroix, C., Vollenweider, S., and Le Blay, G. (2008). Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiol. Ecol. 63, 56–64. doi: 10.1111/j.1574-6941.2007.00412.x
Corr, S. C., Li, Y., Riedel, C. U., O’toole, P. W., Hill, C., and Gahan, C. G. M. (2007). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621. doi: 10.1073/pnas.0700440104
Delves-Broughton, J. (1990). Nisin and its application as a food preservative. Int. J. Dairy Technol. 43, 73–76. doi: 10.1111/j.1471-0307.1990.tb02449.x
Dobson, A., Cotter, P. D., Ross, R. P., and Hill, C. (2012). Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78, 1–6. doi: 10.1128/AEM.05576-11
Dobson, A., Crispie, F., Rea, M. C., O’sullivan, O., Casey, P. G., Lawlor, P. G., et al. (2011). Fate and efficacy of lacticin 3147-producing Lactococcus lactis in the mammalian gastrointestinal tract. FEMS Microbiol. Ecol. 76, 602–614. doi: 10.1111/j.1574-6941.2011.01069.x
Fernandez, B., Le Lay, C., Jean, J., and Fliss, I. (2013). Growth, acid production and bacteriocin production by probiotic candidates under simulated colonic conditions. J. Appl. Microbiol. 114, 877–885. doi: 10.1111/jam.12081
Fernandez, B., Savard, P., and Fliss, I. (2015). Survival and metabolic activity of pediocin producer Pediococcus acidilactici UL5: its impact on intestinal microbiota and Listeria monocytogenes in a model of the human terminal ileum. Microb. Ecol. doi: 10.1007/s00248-015-0645-0 [Epub ahead of print].
Fliss, I., Hammami, R., and Le Lay, C. (2011). Biological Control of Human Digestive Microbiota using Antimicrobial Cultures and Bacteriocins. Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation. Cambridge: Woodhead Publishing Ltd, 240–263.
Fox, M. J., Ahuja, K. D. K., Robertson, I. K., Ball, M. J., and Eri, R. D. (2015). Can probiotic yogurt prevent diarrhoea in children on antibiotics? A double-blind, randomised, placebo-controlled study. BMJ Open 5, e006474. doi: 10.1136/bmjopen-2014-006474
Franks, A. H., Harmsen, H. J. M., Raangs, G. C., Jansen, G. J., Schut, F., and Welling, G. W. (1998). Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64, 3336–3345.
Gibson, G. R., and Wang, X. (1994). Enrichment of bifidobacteria from human gut contents by oligofructose using continuous culture. FEMS Microbiol. Lett. 118, 121–127. doi: 10.1111/j.1574-6968.1994.tb06813.x
Guandalini, S. (2011). Probiotics for prevention and treatment of diarrhea. J. Clin. Gastroenterol. 45, S149–S153. doi: 10.1097/MCG.0b013e3182257e98
Harmsen, H. J. M., Elfferich, P., Schut, F., and Welling, G. (1999). A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11, 3–12. doi: 10.1080/089106099435862
Hopkins, M. J., Macfarlane, G. T., Furrie, E., Fite, A., and Macfarlane, S. (2005). Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol. Ecol. 54, 77–85. doi: 10.1016/j.femsec.2005.03.001
Huijsdens, X. W., Linskens, R. K., Mak, M., Meuwissen, S. G. M., Vandenbroucke-Grauls, C. M. J. E., and Savelkoul, P. H. M. (2002). Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40, 4423–4427. doi: 10.1128/jcm.40.12.4423-4427.2002
Kanno, T., Matsuki, T., Oka, M., Utsunomiya, H., Inada, K., Magari, H., et al. (2009). Gastric acid reduction leads to an alteration in lower intestinal microflora. Biochem. Biophys. Res. Commun. 381, 666–670. doi: 10.1016/j.bbrc.2009.02.109
Kelly, C. P., and LaMont, J. T. (2008). Clostridium difficile — more difficult than ever. N. Engl. J. Med. 359, 1932–1940. doi: 10.1056/NEJMra0707500
Lagrotteria, D., Holmes, S., Smieja, M., Smaill, F., and Lee, C. (2006). Prospective, randomized inpatient study of oral metronidazole versus oral metronidazole and rifampin for treatment of primary episode of Clostridium difficile–associated diarrhea. Clin. Infect. Dis. 43, 547–552. doi: 10.1086/506354
Le Blay, G., Hammami, R., Lacroix, C., and Fliss, I. (2012). Stability and inhibitory activity of pediocin PA-1 against Listeria sp. in simulated physiological conditions of the human terminal ileum. Probiotics Antimicrob. Proteins 4, 250–258. doi: 10.1007/s12602-012-9111-1
Le Blay, G., Lacroix, C., Zihler, A., and Fliss, I. (2007). In vitro inhibition activity of nisin A, nisin Z, pediocin PA-1 and antibiotics against common intestinal bacteria. Lett. Appl. Microbiol. 45, 252–257. doi: 10.1111/j.1472-765X.2007.02178.x
Macfarlane, G. T., Macfarlane, S., and Gibson, G. R. (1998). Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35, 180–187. doi: 10.1007/s002489900072
Matsuki, T., Watanabe, K., Fujimoto, J., Miyamoto, Y., Takada, T., Matsumoto, K., et al. (2002). Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68, 5445–5451. doi: 10.1128/aem.68.11.5445-5451.2002
McFarland, L. (2015). Probiotics for the primary and secondary prevention of C. difficile infections: a meta-analysis and systematic review. Antibiotics 4, 160–178. doi: 10.3390/antibiotics4020160
Meghrous, J., Lacroix, C., Bouksaïm, M., Lapointe, G., and Simard, R. E. (1997). Note: genetic and biochemical characterization of nisin Z produced by Lactococcus lactis ssp. lactis biovar. diacetylactis UL 719. J. Appl. Microbiol. 83, 133–138. doi: 10.1046/j.1365-2672.1997.00160.x
Millette, M., Cornut, G., Dupont, C., Shareck, F., Archambault, D., and Lacroix, M. (2008). Capacity of human nisin-and pediocin-producing lactic acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl. Environ. Microbiol. 74, 1997–2003. doi: 10.1128/AEM.02150-07
Mutlu, E., Wroe, A. J., Sanchez-Hurtado, K., Brazier, J. S., and Poxton, I. R. (2007). Molecular characterization and antimicrobial susceptibility patterns of Clostridium difficile strains isolated from hospitals in south-east Scotland. J. Med. Microbiol. 56, 921–929. doi: 10.1099/jmm.0.47176-0
Peláez, T., Alcalá, L., Alonso, R., Rodríguez-Créixems, M., García-Lechuz, J. M., and Bouza, E. (2002). Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob. Agents Chemother. 46, 1647–1650. doi: 10.1128/aac.46.6.1647-1650.2002
Probert, H. M., and Gibson, G. R. (2004). Development of a fermentation system to model sessile bacterial populations in the human colon. Biofilms 1, 13–19. doi: 10.1017/S1479050503001029
Rea, M. C., Clayton, E., O’Connor, P. M., Shanahan, F., Kiely, B., Ross, R. P., et al. (2007). Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56, 940–946. doi: 10.1099/jmm.0.47085-0
Rea, M. C., Dobson, A., O’sullivan, O., Crispie, F., Fouhy, F., Cotter, P. D., et al. (2011). Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. U.S.A. 108, 4639–4644. doi: 10.1073/pnas.1001224107
Rea, M. C., Sit, C. S., Clayton, E., O’connor, P. M., Whittal, R. M., Zheng, J., et al. (2010). Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U.S.A. 107, 9352–9357. doi: 10.1073/pnas.0913554107
Rinttilä, T., Kassinen, A., Malinen, E., Krogius, L., and Palva, A. (2004). Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97, 1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x
Severina, E., Severin, A., and Tomasz, A. (1998). Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J. Antimicrob. Chemother. 41, 341–347. doi: 10.1093/jac/41.3.341
Surowiec, D., Kuyumjian, A. G., Wynd, M. A., and Cicogna, C. E. (2006). Past, present, and future therapies for Clostridium difficile-associated disease. Ann. Pharmacother. 40, 2155–2163. doi: 10.1345/aph.1H332
Trmčiać, A., Monnet, C., Rogelj, I., and Matijašiać, B. B. (2011). Expression of nisin genes in cheese—a quantitative real-time polymerase chain reaction approach. J. Dairy Sci. 94, 77–85. doi: 10.3168/jds.2010-3677
Vardakas, K. Z., Polyzos, K. A., Patouni, K., Rafailidis, P. I., Samonis, G., and Falagas, M. E. (2012). Treatment failure and recurrence of Clostridium difficile infection following treatment with vancomycin or metronidazole: a systematic review of the evidence. Int. J. Antimicrob. Agents 40, 1–8. doi: 10.1016/j.ijantimicag.2012.01.004
Keywords: Clostridium difficile, probiotic, Lactococcus lactis UL719, bacteriocin, nisin, colon model
Citation: Le Lay C, Fernandez B, Hammami R, Ouellette M and Fliss I (2015) On Lactococcus lactis UL719 competitivity and nisin (Nisaplin®) capacity to inhibit Clostridium difficile in a model of human colon. Front. Microbiol. 6:1020. doi: 10.3389/fmicb.2015.01020
Received: 07 July 2015; Accepted: 08 September 2015;
Published: 25 September 2015.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Michael Gänzle, University of Alberta, CanadaMaria De Los Angeles Serradell, Universidad Nacional de La Plata, Argentina
Carolina Ibarguren, Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina
Copyright © 2015 Le Lay, Fernandez, Hammami, Ouellette and Fliss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ismail Fliss, STELA Dairy Research Center, Nutrition and Functional Foods Institute, Université Laval, G1V 0A6 Québec, QC, Canada, ismail.fliss@fsaa.ulaval.ca
 Christophe Le Lay
Christophe Le Lay Benoit Fernandez
Benoit Fernandez Riadh Hammami
Riadh Hammami Marc Ouellette2,3
Marc Ouellette2,3 Ismail Fliss
Ismail Fliss