Corrigendum: Purification and Characterisation of Malate Dehydrogenase From Synechocystis sp. PCC 6803: Biochemical Barrier of the Oxidative Tricarboxylic Acid Cycle
- 1RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 2Department of Agricultural Chemistry, School of Agriculture, Meiji University, Kawasaki, Japan
- 3Biomass Engineering Program, RIKEN, Yokohama, Japan
- 4Department of Chemical Science and Engineering, Graduate School of Engineering, Kobe University, Kobe, Japan
Succinate is a building block compound that the U.S. Department of Energy (DOE) has declared as important in biorefineries, and it is widely used as a commodity chemical. Here, we identified the two genes increasing succinate production of the unicellular cyanobacterium Synechocystis sp. PCC 6803. Succinate was excreted under dark, anaerobic conditions, and its production level increased by knocking out ackA, which encodes an acetate kinase, and by overexpressing sigE, which encodes an RNA polymerase sigma factor. Glycogen catabolism and organic acid biosynthesis were enhanced in the mutant lacking ackA and overexpressing sigE, leading to an increase in succinate production reaching five times of the wild-type levels. Our genetic and metabolomic analyses thus demonstrated the effect of genetic manipulation of a metabolic enzyme and a transcriptional regulator on succinate excretion from this cyanobacterium with the data based on metabolomic technique.
Introduction
In 2004, the U.S. Department of Energy (DOE) selected the top 12 building block chemicals from a list of more than 300 candidates that were produced from biomass (Werpy and Petersen, 2004). Among these, four-carbon dicarboxylic acids, including succinate, were included. Succinate can be used as a precursor to numerous chemicals such as a biodegradable plastic like polybutylene succinate, fibers, and pigments (Zeikus et al., 1999; Hong and Lee, 2002; Werpy and Petersen, 2004). Succinate is currently derived from petroleum, but it could also be produced using bacteria (McKinlay et al., 2007).
Production of succinate by recombinant heterotrophic bacteria such as Escherichia coli, Corynebacterium glutamicum, Anaerobiospirillum succiniciproducens, Actinobacillus succinogenes, and Mannheimia succiniciproducens has been intensively studied (Samuelov et al., 1991; Guettler et al., 1999; Chatterjee et al., 2001; Hong and Lee, 2001; Hong et al., 2004; Lee et al., 2006). Succinate is an intermediate in the tricarboxylic acid (TCA) cycle and is excreted by succinate-producing cells during anaerobic fermentation (McKinlay et al., 2007). Succinate is produced from phosphoenolpyruvate via the reductive branch of the TCA cycle, in which phosphoenolpyruvate is converted to oxaloacetate by phosphoenolpyruvate carboxylase (PEPC) or phosphoenolpyruvate carboxykinase (PEPCK) under anaerobic conditions (McKinlay et al., 2007). Malate is produced from oxaloacetate when catalyzed by malate dehydrogenase, and fumarate is produced from malate when catalyzed by fumarase. This is followed by the production of succinate when catalyzed by succinate dehydrogenase (SDH; McKinlay et al., 2007). The overexpression of a gene encoding PEPC in E. coli increases succinate production 3.8-fold (Millard et al., 1996). The introduction of PEPC, PEPCK, or malic enzyme (catalyzing a reaction from pyruvate to malate), also enhances succinate production in E. coli (Hong and Lee, 2001; Kim et al., 2004; Lin et al., 2005; Zhang et al., 2009). The deletion of ldhA (encoding L-lactate dehydrogenase), adhE (encoding alcohol dehydrogenase), and ack-pta (encoding acetate kinase and phosphotransacetylase, respectively) prevented the production of L-lactate, ethanol, and acetate, that are by-products during anaerobic fermentation, also increases the production of succinate in E. coli (Sánchez et al., 2005a,b; Jantama et al., 2007). In addition, the activation of the glyoxylate pathway by the deletion of iclR, which encodes the transcriptional repressor of the genes related to glyoxylate pathway, increased the succinate productivity in E. coli (Sánchez et al., 2005a,b). Thus, inhibition of by-product formation combined with additional genetic engineering can up-regulate succinate productivity.
Cyanobacteria are a group of bacteria that fix carbon dioxide via oxygenic photosynthesis. The potential applications of cyanobacteria in providing renewable energy and resources may reduce the environmental burden. Genome information for cyanobacteria is available (Kanesaki et al., 2012), and genetic engineering is easily performed by homologous recombination in several cyanobacterial strains, including the non-nitrogen fixing cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803; Ikeuchi and Tabata, 2001). The genome of Synechocystis 6803 was the first sequenced among the cyanobacteria (Kaneko et al., 1996), and it has been used extensively in basic and applied sciences.
There are few reports of succinate production using cyanobacteria. McNeely et al. revealed that five fermentation products, lactate, acetate, succinate, alanine, and hydrogen, were produced under dark, anaerobic conditions by the marine cyanobacterium Synechococcus sp. PCC 7002 (hereafter Synechococcus 7002; McNeely et al., 2010). A knockout of ldhA increased acetate and hydrogen levels, and diminished lactate production (McNeely et al., 2010). Succinate was excreted from the ldhA knockout cells, but it was not detected from the wild-type cells of Synechococcus 7002 (McNeely et al., 2010). The filamentous, non-diazotrophic cyanobacteria Arthrospira maxima CS-328 cells produced lactate, acetate, ethanol, formate, and hydrogen under dark, anaerobic conditions, but succinate excretion was not detected (Carrieri et al., 2010, 2011). For Synechocystis 6803, hydrogen is generated under both light and dark, anaerobic conditions (Osanai et al., 2013). Organic acids, including D-lactates, were highly produced by genetically engineered Synechocystis 6803 cells, and succinate was also generated but its levels were only 3% of total carbon excreted, suggesting that genetic and metabolic engineering are necessary to increase succinate production (Angermayr et al., 2014; Hollinshead et al., 2014; McNeely et al., 2014). The genes encoding enzymes for organic acid production exist in the Synechocystis 6803 genome (Figure 1).
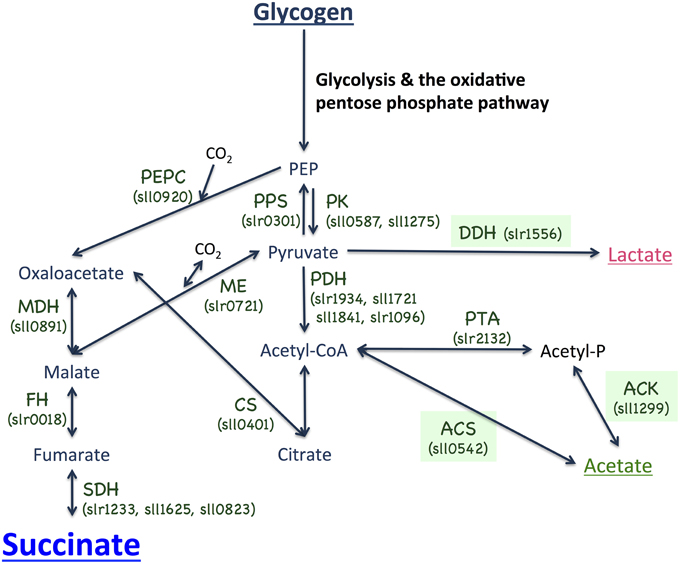
Figure 1. Metabolic map surrounding succinate biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase; PPS, phosphoenolpyruvate synthase; PK, pyruvate kinase; MDH, malate dehydrogenase; ME, malic enzyme; FH, fumarate hydratase; SDH, succinate dehydrogenase; PDH, pyruvate dehydrogenase; DDH, D-lactate dehydrogenase, CS, citrate synthase; PTA, phosphotransacetylase; ACK, acetate kinase; and ACS, acetyl-CoA synthetase.
We reveal here that succinate was excreted from Synechocystis 6803 cells under dark, anaerobic conditions and the succinate levels were enhanced by reducing acetate biosynthesis and overexpressing sigE encoding a sigma factor. These results demonstrated the genetic manipulation of two types of genes increasing the succinate excretion from this cyanobacterium.
Materials and Methods
Bacterial Strains and Culture Conditions
The glucose-tolerant strain of Synechocystis sp. PCC 6803, isolated by Williams (Williams, 1988), was grown in modified BG-11 medium, consisting of BG-110 liquid medium (Rippka, 1988) supplemented with 5 mM NH4Cl (buffered with 20 mM HEPES–KOH, pH 7.8). The GT-I strain, among GT substrains, was used in the current study (Kanesaki et al., 2012). Liquid cultures were bubbled with 1% (v/v) CO2 in air and incubated at 30°C under continuous white light (~50–70 μmol photons m−2 s−1). For the mutant strains, 10, 0.3, and 10 μg/mL of kanamycin, gentamycin and chloramphenicol, respectively, were added for preculturing. Modified BG-11 medium (containing 10 mM NH4Cl in liquid medium) was solidified with agar (1.5% w/v) for plate cultures, and similarly incubated in air at 30°C under continuous white light (~50–70 μmol photons m−2 s−1). Cell densities were measured at A730 using a Hitachi U-3310 spectrophotometer (Hitachi High-Tech., Tokyo, Japan).
For succinate production, cells grown in 70 mL modified BG-11 medium (started from A730 = 0.4) for 3 days were concentrated into 10 mL HEPES buffer (20 mM HEPES–KOH, pH 7.8) or modified BG-11 medium to A730 = 20 in a GC vial. The vial was sealed using butyl rubber, and N2 gas was introduced using syringes for 1 h to produce anaerobic conditions. After removing the syringes, the vial was wrapped with aluminum foil and shaken at 30°C. Cell cultures were then centrifuged at 5800 × g for 2 min, the supernatant was filtrated, and 1 mL supernatant was freeze-dried for 1 day. The dried sample was used for high-performance liquid chromatography analysis.
Plasmid Construction of Knock-in Vectors pTCP1556, pTCP0542, and pTCP1299
The kanamycin resistance cassette of the pTKP2031V vector (Osanai et al., 2011) was removed by digestion with XhoI and AatII (Takara Bio, Shiga, Japan). The chloramphenicol resistance cassette from pKRP10 (Reece and Phillips, 1995) was amplified by PCR with KOD polymerase (Toyobo, Osaka, Japan) and the specific primers in Table S2, digested with XhoI and AatII, and inserted into the XhoI-AatII sites of pTKP2031V. The resultant plasmid was named pTCP2031. Regions of ddh (slr1556), from −297 to +800 bp, acs (sll0542), from +921 to +1962 bp, and ackA (sll1299), from +270 to +1238 bp, based on the translation initiation codons, were amplified by PCR with KOD plus neo polymerase (Toyobo) and the specific primers in Table S2. The fragments amplified by PCR were digested with SphI and EcoRV (Takara Bio) and inserted into the SphI-SmaI sites of the pUC119 vector (Takara Bio). The resultant plasmid was digested with HincII (for ddh and ackA) or ApaI (for acs), and the region including the chloramphenicol resistance cassette, psbAII promoter, and NdeI-HpaI cloning sites of pTCP2031 was amplified with KOD plus neo polymerase and the specific primers 5′-TTTGCTTCATCGCTCGAG-3′ and 5′-ATCCAATGTGAGGTTAAC-3′, and integrated into the HincII or ApaI site of the plasmid. The resultant plasmids were named pTCP1556, pTCP0542, and pTCP1299 for knockouts of ddh, acs, and ackA, respectively. The sigE ORF was obtained by digestion with NdeI and HpaI from pTGP0945-sigE plasmid (Osanai et al., 2014a) and cloned into the NdeI-HpaI sites of pTCP1556, pTCP0542, and pTCP1299. The plasmids were integrated into the GT-I strain by natural transformation as described previously (Osanai et al., 2011). Knockouts and the insertion of the sigE ORF were confirmed by PCR using GoTaq (Promega, Fitchburg, WI, USA) with the primers in Table S2.
Immunoblotting
Cells were collected by centrifugation (5800 × g for 2 min), and the supernatant was removed and cells were frozen by liquid nitrogen. Then, cells were dissolved in PBS-T and disrupted by sonication as described previously (Osanai et al., 2014a). Immunoblotting was performed as described previously (Osanai et al., 2014a). Antisera against SigE were generated previously (Osanai et al., 2009).
Glycogen Measurement
Glycogen levels were measured at the Biotechnology Center of Akita Prefectural University (Akita, Japan), as described in Osanai et al. (2014a).
LC-MS/MS Analysis
Equal amounts of cells (10 mL cell culture with A730 = 1.0) were harvested by rapid filtration, and metabolites were extracted using a previously described method (Osanai et al., 2014b). Briefly, the cells were filtrated, and then the intermediate metabolites were quenched and extracted in 1.2 mL of solvent mixture (CHCl3:CH3OH:H2O, 2.5:2.5:1, v/v/v) containing 10 μg/L D-(+)-camphor-10-sulfonic acid as an internal standard. After centrifugation at 15,000 × g at 4°C for 5 min, 400 μL of the upper phase was transferred to a new tube and vacuum-dried.
GC-MS Analysis
Equal amounts of cells (10 mL cell culture with A730 = 1.0) were harvested by rapid filtration as mentioned above. GC-MS was carried out using a GCMS-QP2010 Ultra, and the detailed protocol is described in Osanai et al. (2015).
Measurement of Organic Acids by High-Performance Liquid Chromatography (HPLC)
Freeze-dried supernatants were resolved in 100 μL of filtered 3 mM perchloric acid. The resolved samples were analyzed by HPLC using a LC-2000Plus Systems (JASCO, Tokyo, Japan) with a photodiode array detector and two RSpak KC-811 columns (Showa Denko, Tokyo, Japan). Organic acids were quantified with 0.2 mM bromothymol blue in 15 mM sodium phosphate buffer; peaks were detected at 445 nm. The column temperature was 60°C, and the flow rates of 3 mM perchloric acid and 0.2 mM bromothymol blue solutions were 1.0 and 1.5 mL/min, respectively.
Results
ackA Knockout and sigE Overexpression Enhanced Succinate Production
The identities of the excreted organic acids from the wild-type Synechocystis 6803 (GT) during anaerobic conditions were determined first. After cultivation for 3 days under light, aerobic conditions (1% CO2 in the air), cells were concentrated into 10 mL BG-110 medium or HEPES buffer with or without nitrogen sources (5 mM NH4Cl) in a GC-vial, subjected to anaerobic conditions by introducing N2 gas, and incubated for 3 days under dark conditions with shaking at 30°C. Organic acids excreted into the medium or buffer were analyzed by HPLC. Succinate, lactate and acetate were detected, and the succinate levels were highest in HEPES buffer with nitrogen source among the four conditions tested (Figure 2). Lactates were not detected in HEPES buffer (Figure 2). Acetate levels were higher in BG-11 or HEPES buffer without nitrogen sources than those in BG-11 or HEPES buffer with nitrogen sources (Figure 2). To reduce the cost of succinate production, subsequent experiments were performed using HEPES buffer without nitrogen sources.
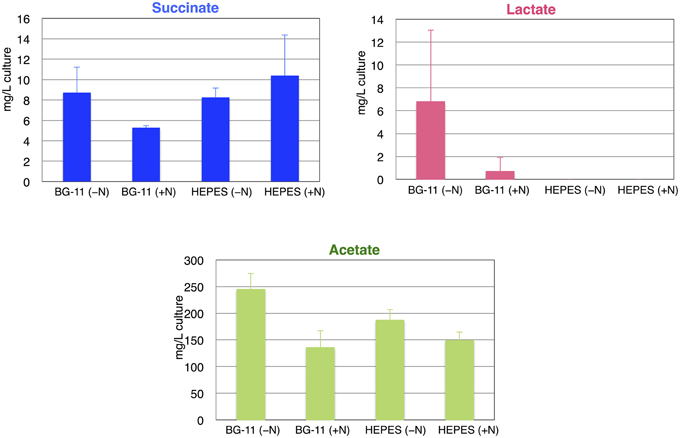
Figure 2. Levels of succinate, lactate, and acetate from the wild-type cyanobacterium Synechocystis 6803 using different media or buffers during anaerobic conditions. Organic acids excreted from 3 days under dark, anaerobic cultivation were quantified by HPLC. Data represent means ± SD from three independent experiments. +N designates 5 mM NH4Cl was added to the BG-11 medium or 20 mM HEPES–KOH (pH 7.8) buffer.
To increase succinate production, we applied two strategies, decreasing lactate and acetate by knocking out each of the three genes (ddh, acs, or ackA; Figure 1) and promoting the sugar catabolic pathway by overexpressing sigE, encoding an RNA polymerase sigma factor, which activates the expression of sugar catabolic enzymes (Osanai et al., 2011). Knock-in vectors, which integrate the region containing the chloramphenicol resistance cassette, the psbAII promoter from the D1 protein of Photosystem II, and NdeI-HpaI cloning sites, were constructed to generate the knockout mutants of ddh, acs, and ackA (Figure 3A). The sigE open reading frame (ORF) was cloned into the NdeI-HpaI sites to generate the sigE overexpression strain combined with the ddh, acs, or ackA knockout, and the resultant strains were designated as 1556E, 0542E, and 1299E, respectively (Figure 3A). The insertion of these DNA fragments was confirmed by PCR (Figure 3B). Immunoblotting confirmed that SigE proteins in the three sigE-overexpressing strains were higher than in GT after 3 days of cultivation under dark, anaerobic conditions (Figure 3C).
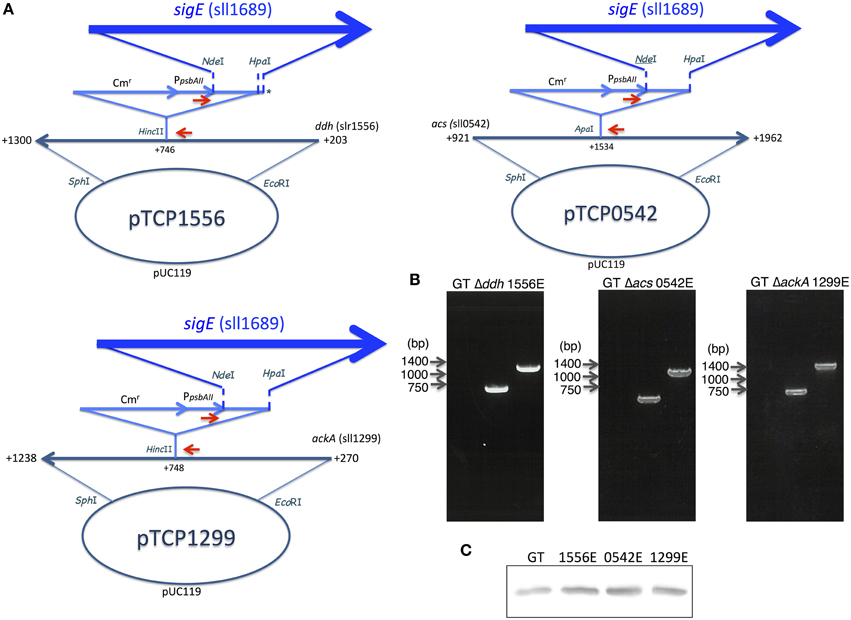
Figure 3. (A) Vector maps for three vectors disrupting ddh, acs, and ackA with or without sigE overexpression. Red arrows indicate the primer sets to confirm the insertion of the vectors in the non-nitrogen fixing cyanobacterium Synechocystis sp. PCC 6803 genome. Asterisk (*) in pTCP1556 indicates that the sigE ORF was inserted into the region from NdeI to 55-bp downstream of the HpaI site because of the star activity of HpaI. (B) The right bottom panels show DNA fragments amplified by PCR with the primers in agarose gel electrophoresis as visualized by ethidium bromide. (C) Protein levels of SigE in GT, 1556E, 0542E, and 1299E strains. Immunoblotting was performed with 12 μg of total protein from cells grown under dark, anaerobic conditions for 3 days.
Although the knockouts of ddh and acs did not increase the succinate levels, the ackA knockout increased the succinate level to 34.8 mg/L compared with the 13.9 mg/L produced by the parental wild-type strain under the same conditions. sigE overexpression (GOX50) alone increased the succinate level to 20.3 mg/L, and an additional knockout of ddh, acs, or ackA enhanced the levels to ~35.6, 29.4, or 71.5 mg/L, respectively (Figure 4A). The wild-type cells produced less than 10 mg/L lactate, while the lactate levels increased in the ackA knockout to 51.7 mg/L, and sigE overexpression with an acs or ackA knockout enhanced the levels to 30.0 or 93.5 mg/L, respectively (Figure 4A). Acetate levels were decreased to 62.0 mg/L by the ackA knockout, compared with 294.3 mg/L acetate produced by wild-type cells, and the ratio of succinate and lactate to acetate increased in the ackA knockout mutant (Figures 4A,B). The strain lacking ackA and overexpressing sigE (1299E) had the highest succinate levels and ratios among the eight strains (Figures 4A,B). A time-course experiment analyzed different lengths of dark, anaerobic incubations and showed that a 3- or 4-day incubation period was long enough to produce sufficient quantities and ratios of succinate in 1299E (Figure 5). The succinate production rates from 1299E were 1.38 and 1.18 mg/L/h for 3- and 4-day incubation, respectively (Figure 5). Therefore, subsequent experiments were performed using a 3-day incubation period under dark, anaerobic conditions.
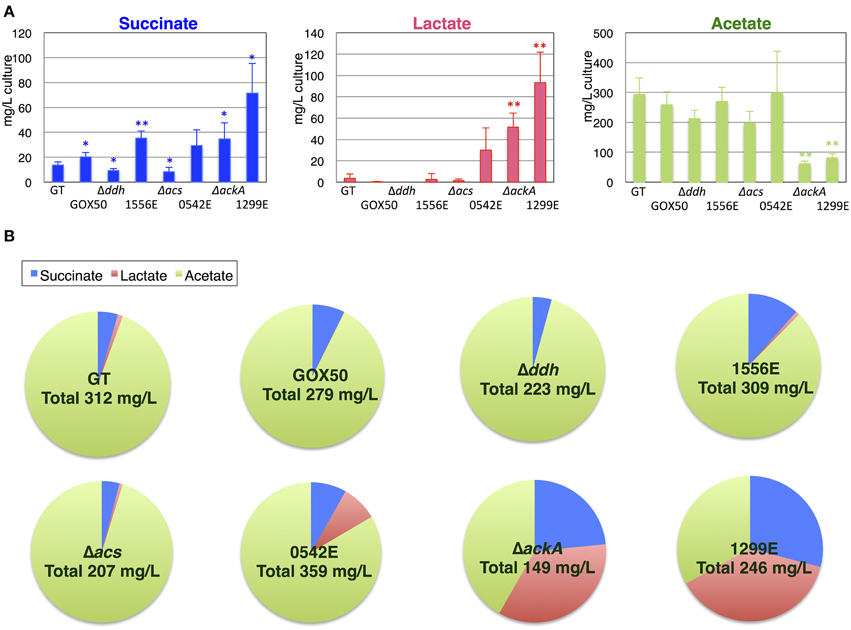
Figure 4. Production of organic acids from the cyanobacterium Synechocystis 6803 strain overexpressing sigE and lacking ddh (slr1556), acs (sll0542), or ackA (sll1299). (A) Levels of organic acids excreted during 3 days of dark, anaerobic cultivation were quantified by HPLC. Δddh, Δacs, and Δddh indicate the knockout of mutants of each gene. 1556E, 0542E, and 1299E represent the strains overexpressing sigE and lacking ddh, acs, or ackA, respectively. GOX50 designates the sigE-overexpressing strain. Data represent means ± SD from three or four independent experiments. Asterisks indicate statistically significant differences between GT and the mutant strains (Student's t-test; *P < 0.05, **P < 0.005). (B) The pie chart shows the ratio of succinate, lactate, and acetate excreted from the cells under anaerobic conditions. Total organic acid acids are sum of succinate, lactate, and acetate amounts excreted from the cells. Ratio is calculated by dividing the amount of each organic acid by the amounts of total organic acids.
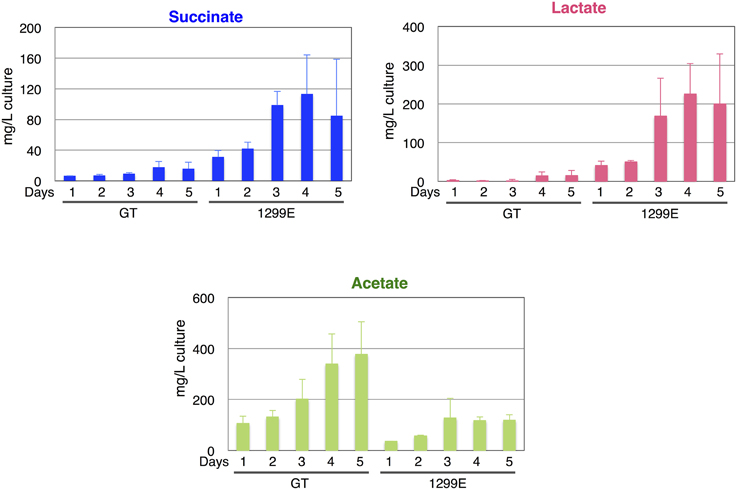
Figure 5. Time-course analysis of levels of succinate, lactate, and acetate from the wild-type cyanobacterium Synechocystis 6803 and the 1299E strain, which overexpresses sigE and lacks ackA. Organic acids excreted from 1 to 5 days under dark, anaerobic cultivation were quantified by HPLC. Data represent means ± SD from three independent experiments.
A Metabolome Analysis
A metabolome analysis using the GT and 1299E strains grown under aerobic and anaerobic conditions was then performed to clarify the metabolic profiles. After dark, aerobic cultivation, ADP-glucoses disappeared and the levels of several sugar phosphates increased (Figure 6 and Table S1). The fructose-1, 6-bisphoshate and dihydroxyacetone phosphate levels increased more than 10 times under anaerobic conditions in the wild-type strain (Figure 6). Phosphoenolpyruvate, pyruvate and acetyl-CoA decreased greatly under anaerobic conditions (Figure 6). The levels of sugar phosphates, such as glucose-1-phosphate, glucose-6-phosphate, ribulose-5-phosphate, 6-phosphogluconate, fructose-6-phosphate, and fructose-1, 6-bisphosphate, in the 1299E strain under anaerobic conditions, were higher than those in the wild-type strain (Figure 6). Phosphoenolpyruvate and acetyl-CoA were lower in the 1299E strain than in the wild-type strain, and organic acids, such as succinate, lactate, malate, and fumarate, were higher in the 1299E strain than in the wild-type strain under anaerobic conditions (Figure 6).
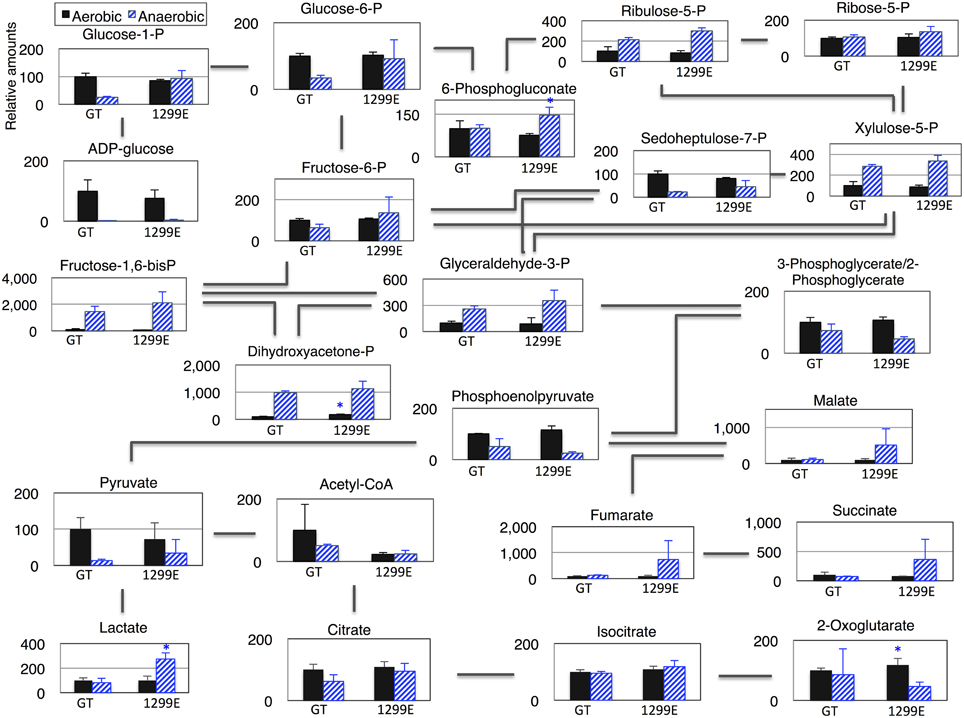
Figure 6. Levels of metabolites in primary metabolism of the cyanobacterium Synechocystis 1299E strain, which overexpresses sigE and lacks ackA. Data represent means ± SD from three independent experiments. Metabolite levels were calibrated relative to that of corresponding metabolites in the wild-type strain (GT) under aerobic conditions (set at 100%). P designates phosphate. Asterisks indicate the statistically significant differences between GT and 1299E (Student's t-test; *P < 0.05).
The quantification of the glycogen levels before and after anaerobic cultivation for 3 days revealed that 33% of glycogen was consumed in the wild-type strain; however, 70% of glycogen were consumed in the 1299E strains (Table 1).
Discussion
Fermentation is closely related to sugar metabolism. In Synechococcus 7002, the levels of excreted fermentation products were altered by the disruption of glgC, which encodes ADP-glucose pyrophosphorylase (Guerra et al., 2013). Lactate production in the glgC knockout strain was approximately half of that in the wild-type, while acetate and alanine production were not significantly affected (Guerra et al., 2013). The reason for the decreased lactate excretion may be the slower catabolic rate of reduced sugars in the glgC knockout mutant (Guerra et al., 2013). The rate of sugar catabolism, not the amount of total reduced sugars, was important for the increased lactate production (Guerra et al., 2013). This finding was consistent with our results that sigE overexpression, which accelerated glycogen degradation and glucose catabolism (Osanai et al., 2011), increased succinate production in Synechocystis 6803 (Figures 4A, 7). Glycogen degradation was enhanced by sigE overexpression during dark, anaerobic conditions (Table 1), possibly leading to increases in the biosynthesis of intracellular organic acids (Figure 6 and Table S1), which in turn led to the production of extracellular succinate at a higher level (Figures 4A, 7).
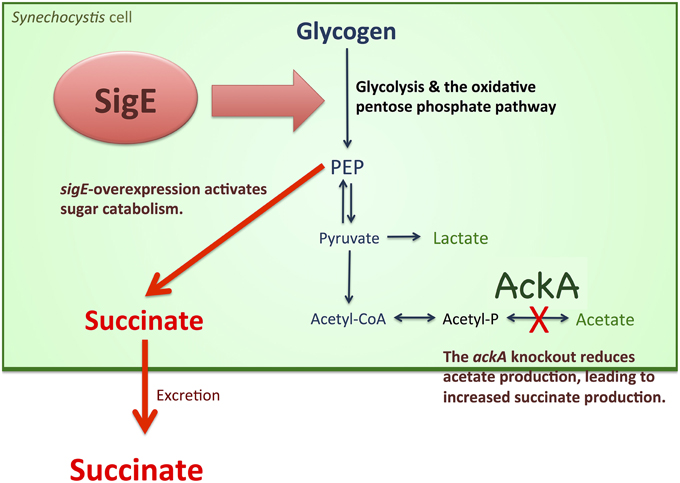
Figure 7. Schematic model of succinate production from Synechocystis 6803. sigE overexpression activates sugar catabolism, possibly leading to increased succinate production. The ackA knockout reduces acetate biosynthesis, resulted in increased production of succinate and lactate production.
A metabolome analysis revealed that the levels of fructose-1,6-bisphosphate and dihydroxyacetone phosphate increased more than 10 times after dark, anaerobic cultivation in the wild-type strain (Figure 6). Glyceraldehyde-3-phosphate production also increased, but 3-phosphoglycerate/2-phosphoglycerate, phosphoenolpyruvate, pyruvate, and acetyl-CoA decreased after dark, anaerobic cultivation in the wild-type strain (Figure 6). These results suggest that important enzymatic reactions exist downstream of glyceraldehyde-3-phosphate under dark, anaerobic conditions. Glyceraldehyde-3-phosphate dehydrogenase, which produces glycerate-1,3-bisphosphate from glyceraldehyde-3-phosphate, is encoded by gap1 and gap2, and the reactions were uniquely catalyzed by the two enzymes in Synechocystis 6803. Gap1 catalyzes the catabolic reactions and Gap2 catalyzes the anabolic reactions (Koksharova et al., 1998). In addition, the gap1 transcript levels are regulated by at least two transcriptional regulators, SigE and a response regulator Rre37 (Osanai et al., 2005; Azuma et al., 2011), indicating the importance of gap1 in the sugar catabolism of Synechocystis 6803. The flux into the TCA cycle in Synechocystis is relatively low compared with other heterotrophic bacteria (You et al., 2015). Current results demonstrate that the flux into succinate can be up-regulated by our genetic modification under dark, anaerobic conditions.
A knockout of ackA reduced the acetate level but a knockout of acs did not affect the acetate level (Figure 4A), suggesting the major route of acetate biosynthesis under dark, anaerobic conditions is through an AckA-dependent pathway in this cyanobacterium (Figure 1). Prochlorococcus species lack ackA in their genomes (KEGG database URL: http://www.genome.jp/kegg-bin/show_pathway?syn00620), and thus, acetate biosynthesis pathway may be diverse among cyanobacteria. The mutant lacking ddh showed diminished lactate production, but lactate was produced by sigE overexpression even in the ddh knockout (Figure 4A). These results indicate another pathway for lactate biosynthesis exists in this cyanobacterium. Lactate can be synthesized from lactoylglutathione, which is derived from dihydroxyacetone phosphate (KEGG database URL: http://www.genome.jp/kegg-bin/show_pathway?syn00620), and the pathway may be activated by sigE overexpression. Our metabolome analysis revealed that NADPH disappeared, and that malate, fumarate, and succinate generally increased in the strains producing more succinate (Figure 6 and Table S1). These data suggest that succinate is produced through the reverse TCA cycle, as shown in Figure 1, as in other heterotrophic bacteria (Lee et al., 2006). Thus, the metabolic flux toward succinate production has been clarified in this cyanobacterium by the metabolome analysis. Pyruvate and phosphoenolpyruvate were severely reduced by light-to-dark transition (Iijima et al., 2015), and thus, the provision of these metabolites may be important to increase organic acid production under dark, anaerobic conditions. The current results demonstrated that a combination of the genetic manipulation of genes encoding a metabolic enzyme and a sigma factor succeeded in up-regulating the succinate levels. Future study about the detailed metabolic regulation will contribute to further understanding of the mechanistic implication of succinate excretion from this cyanobacterium.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan, by a grant to TO from ALCA (Project name “Production of cyanobacterial succinate by the genetic engineering of transcriptional regulators and circadian clocks”) from the Japan Science and Technology Agency. All authors contributed to the work and approved the submission of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01064
References
Angermayr, S. A., van der Woude, A. D., Correddu, D., Vreugdenhil, A., Verrone, V., and Hellingwerf, K. J. (2014). Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 7, 99. doi: 10.1186/1754-6834-7-99
Azuma, M., Osanai, T., Hirai, M. Y., and Tanaka, K. (2011). A response regulator Rre37 and an RNA polymerase sigma factor SigE represent two parallel pathways to activate sugar catabolism in a cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 52, 404–412. doi: 10.1093/pcp/pcq204
Carrieri, D., Ananyev, G., Lenz, O., Bryant, D. A., and Dismukes, G. C. (2011). Contribution of a sodium ion gradient to energy conservation during fermentation in the cyanobacterium Arthrospira (Spirulina) maxima CS-328. Appl. Environ. Microbiol. 77, 7185–7194. doi: 10.1128/AEM.00612-11
Carrieri, D., Momot, D., Brasg, I. A., Ananyev, G., Lenz, O., Bryant, D. A., et al. (2010). Boosting autofermentation rates and product yields with sodium stress cycling: application to production of renewable fuels by cyanobacteria. Appl. Environ. Microbiol. 76, 6455–6462. doi: 10.1128/AEM.00975-10
Chatterjee, R., Millard, C. S., Champion, K., Clark, D. P., and Donnelly, M. I. (2001). Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl. Environ. Microbiol. 67, 148–154. doi: 10.1128/AEM.67.1.148-154.2001
Guerra, L. T., Xu, Y., Bennette, N., McNeely, K., Bryant, D. A., and Dismukes, G. C. (2013). Natural osmolytes are much less effective substrates than glycogen for catabolic energy production in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Biotechnol. 166, 65–75. doi: 10.1016/j.jbiotec.2013.04.005
Guettler, M. V., Rumler, D., and Jain, M. K. (1999). Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int. J. Syst. Bacteriol. 49, 207–216. doi: 10.1099/00207713-49-1-207
Hollinshead, W. D., Varman, A. M., You, L., Hembree, Z., and Tan, Y. J. (2014). Boosting D-lactate production in engineered cyanobacteria using sterilized anaerobic digestion effluents. Bioresour. Technol. 169, 464–467. doi: 10.1016/j.biortech.2014.07.003
Hong, S. H., Kim, J. S., Lee, S. Y., In, Y. H., Choi, S. S., Rih, J. K., et al. (2004). The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat. Biotechnol. 22, 1275–1281. doi: 10.1038/nbt1010
Hong, S. H., and Lee, S. Y. (2001). Metabolic flux analysis for succinic acid production by recombinant Escherichia coli with amplified malic enzyme activity. Biotechnol. Bioeng. 74, 89–95. doi: 10.1002/bit.1098
Hong, S. H., and Lee, S. Y. (2002). Importance of redox balance on the production of succinic acid by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 58, 286–290. doi: 10.1007/s00253-001-0899-y
Iijima, H., Shirai, T., Okamoto, M., Kondo, A., Hirai, M. Y., and Osanai, T. (2015). Changes in primary metabolism under light and dark conditions in response to overproduction of a response regulator RpaA in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 6:888. doi: 10.3389/fmicb.2015.00888
Ikeuchi, M., and Tabata, S. (2001). Synechocystis sp. PCC 6803 – a useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 70, 73–83. doi: 10.1023/A:1013887908680
Jantama, K., Haupt, M. J., Svoronos, S. A., Zhang, X., Moore, J. C., Shanmugam, K. T., et al. (2007). Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99, 1140–1153. doi: 10.1002/bit.21694
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. doi: 10.1093/dnares/3.3.109
Kanesaki, Y., Shiwa, Y., Tajima, N., Suzuki, M., Watanabe, S., Sato, N., et al. (2012). Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 19, 67–79. doi: 10.1093/dnares/dsr042
Kim, P., Laivenieks, M., Vieille, C., and Zeikus, J. G. (2004). Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl. Microbiol. Biotechnol. 70, 1238–1241. doi: 10.1128/aem.70.2.1238-1241.2004
Koksharova, O., Schubert, M., Shestakov, S., and Cerff, R. (1998). Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 36, 183–194. doi: 10.1023/A:1005925732743
Lee, S. J., Song, H., and Lee, S. Y. (2006). Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl. Environ. Microbiol. 72, 1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006
Lin, H., San, K. Y., and Bennett, G. N. (2005). Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli. Appl. Microbiol. Biotechnol. 67, 515–523. doi: 10.1007/s00253-004-1789-x
McKinlay, J. B., Vieille, C., and Zeikus, J. G. (2007). Prospects for a bio-based succinate industry. Appl. Microbiol. Biotechnol. 76, 727–740. doi: 10.1007/s00253-007-1057-y
McNeely, K., Kumaraswamy, G. K., Guerra, T., Bennette, N., Ananyev, G., and Dismukes, G. C. (2014). Metabolic switching of central carbon metabolism in response to nitrate: application to autofermentative hydrogen production in cyanobacteria. J. Biotechnol. 182–183, 83–91. doi: 10.1016/j.jbiotec.2014.04.004
McNeely, K., Xu, Y., Bennette, N., Bryand, D. A., and Dismukes, G. C. (2010). Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 76, 5032–5038. doi: 10.1128/AEM.00862-10
Millard, C. S., Chao, Y. P., Liao, J. C., and Donnelly, M. I. (1996). Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl. Environ. Microbiol. 62, 1808–1810.
Osanai, T., Imashimizu, M., Seki, A., Sato, S., Tabata, S., Imamura, S., et al. (2009). ChlH, the H subunit of the Mg-chelatase, is an anti-sigma factor for SigE in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 106, 6860–6865. doi: 10.1073/pnas.0810040106
Osanai, T., Kanesaki, Y., Nakano, T., Takahashi, H., Asayama, M., Shirai, M., et al. (2005). Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor SigE. J. Biol. Chem. 280, 30653–30659. doi: 10.1074/jbc.M505043200
Osanai, T., Kuwahara, A., Iijima, H., Toyooka, K., Sato, M., Tanaka, K., et al. (2013). Pleiotropic effect of sigE over-expression on cell morphology, photosynthesis and hydrogen production in Synechocystis sp. PCC 6803. Plant J. 76, 456–465. doi: 10.1111/tpj.12310
Osanai, T., Oikawa, A., Azuma, M., Tanaka, K., Saito, K., Hirai, M., et al. (2011). Genetic engineering of group 2 sigma factor SigE widely activates expressions of sugar catabolic genes in Synechocystis species PCC 6803. J. Biol. Chem. 286, 30962–30971. doi: 10.1074/jbc.M111.231183
Osanai, T., Oikawa, A., Numata, K., Kuwahara, A., Iijima, H., Doi, Y., et al. (2014a). Pathway-level acceleration of glycogen catabolism by a response regulator in the cyanobacterium Synechocystis species PCC 6803. Plant Physiol. 164, 1831–1841. doi: 10.1104/pp.113.232025
Osanai, T., Oikawa, A., Shirai, T., Kuwahara, A., Iijima, H., Tanaka, K., et al. (2014b). Capillary electrophoresis-mass spectrometry reveals the distribution of carbon metabolites during nitrogen starvation in Synechocystis sp. PCC 6803. Environ. Microbiol. 16, 512–524. doi: 10.1111/1462-2920.12170
Osanai, T., Shirai, T., Iijima, H., Kuwahara, A., Suzuki, I., Kondo, A., et al. (2015). Alteration of cyanobacterial sugar and amino acid metabolism by overexpression hik8, encoding a KaiC-associated histidine kinase. Environ. Microbiol. 17, 2430–2440. doi: 10.1111/1462-2920.12715
Reece, K. S., and Phillips, G. J. (1995). New plasmids carrying antibiotic-resistance cassettes. Gene 165, 141–142. doi: 10.1016/0378-1119(95)00529-F
Rippka, R. (1988). Isolation and purification of cyanobacteria. Meth. Enzymol. 167, 3–27. doi: 10.1016/0076-6879(88)67004-2
Samuelov, N. S., Lamed, R., Lowe, S., and Zeikus, J. G. (1991). Influence of CO2-HCO3 levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl. Environ. Microbiol. 57, 3013–3019.
Sánchez, A. M., Bennett, G. N., and San, K. Y. (2005a). Efficient succinic acid production from glucose through overexpression of pyruvate carboxylase in an Escherichia coli alcohol dehydrogenase and lactate dehydrogenase mutant. Biotechnol. Prog. 21, 358–365. doi: 10.1021/bp049676e
Sánchez, A. M., Bennett, G. N., and San, K. Y. (2005b). Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab. Eng. 7, 229–239. doi: 10.1016/j.ymben.2005.03.001
Werpy, T., and Petersen, G. (2004). Top Value Added Chemicals from Biomass, Vol. 1, Results of Screening for Potential Candidates from Sugars and Synthesis Gas. Oak Ridge, TN: U.S. Department of Energy.
Williams, J. G. K. (1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Meth. Enzymol. 167, 766–778. doi: 10.1016/0076-6879(88)67088-1
You, L., He, L., and Tang, Y. J. (2015). Photoheterotrophic fluxome in Synechocystis sp. strain PCC 6803 and its implications for cyanobacterial bioenergetics. J. Bacteriol. 197, 943–950. doi: 10.1128/JB.02149-14
Zeikus, J. G., Jain, M. K., and Elankovan, P. (1999). Biotechnology of succinic acid production and markets for derived industrial products. Appl. Microbiol. Biotechnol. 51, 545–552. doi: 10.1007/s002530051431
Keywords: cyanobacteria, metabolism, metabolomics, sigma factor, succinate
Citation: Osanai T, Shirai T, Iijima H, Nakaya Y, Okamoto M, Kondo A and Hirai MY (2015) Genetic manipulation of a metabolic enzyme and a transcriptional regulator increasing succinate excretion from unicellular cyanobacterium. Front. Microbiol. 6:1064. doi: 10.3389/fmicb.2015.01064
Received: 21 August 2015; Accepted: 15 September 2015;
Published: 06 October 2015.
Edited by:
Weiwen Zhang, Tianjin University, ChinaCopyright © 2015 Osanai, Shirai, Iijima, Nakaya, Okamoto, Kondo and Hirai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Osanai, Department of Agricultural Chemistry, School of Agriculture, Meiji University, 1-1-1 Higashimita, Tama-ku, Kawasaki, Kanagawa 214-8571, Japan, tosanai@meiji.ac.jp
Masami Y. Hirai, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan, masami.hirai@riken.jp
 Takashi Osanai
Takashi Osanai Tomokazu Shirai3
Tomokazu Shirai3 Yuka Nakaya
Yuka Nakaya Akihiko Kondo
Akihiko Kondo Masami Y. Hirai
Masami Y. Hirai