- 1Department of Microbiology and Immunology, Dalhousie University, Halifax, NS, Canada
- 2Department of Medicine (Infectious Diseases), Dalhousie University, Halifax, NS, Canada
Vibrio parahaemolyticus (Vp) is a marine halophilic bacterium that is commonly associated with oysters and shrimp. Human consumption of contaminated shellfish can result in Vp mediated gastroenteritis and severe diarrheal disease. Vp encodes two type 3 secretion systems (T3SS-1 and T3SS2) that have been functionally implicated in cytotoxicity and enterotoxicity respectively. In this study, we profiled protein secretion and temporal promoter activities associated with exsA and exsB gene expression. exsA is an AraC-like transcriptional activator that is critical for activating multiple operons that encode T3SS-1 genes, whereas exsB is thought to encode an outer membrane pilotin component for T3SS-1. The exsBA genetic locus has two predicted promoter elements. The predicted exsB and exsA promoters were individually cloned upstream of luxCDABE genes in reporter plasmid constructs allowing for in situ, real-time quantitative light emission measurements under many growth conditions. Low calcium growth conditions supported maximal exsB and exsA promoter activation. exsB promoter activity exhibited high basal activity and resulted in an exsBA co-transcript. Furthermore, a separate proximal exsA promoter showed initial low basal activity yet eventually exceeded that of exsB and reached maximal levels after 2.5 h corresponding to an entry into early log phase. exsA promoter activity was significantly higher at 30°C than 37°C, which also coincided with increased secretion levels of specific T3SS-1 effector proteins. Lastly, bioinformatic analyses identified a putative expanded ExsA binding motif for multiple transcriptional operons. These findings suggest a two wave model of Vp T3SS-I induction that integrates two distinct promoter elements and environmental signals into a complex ExsA activation framework.
Introduction
Vibrio parahaemolyticus (Vp) is a significant food borne human pathogen associated with severe diarrhea arising from enteric infections (Ceccarelli et al., 2014). It has also been associated to a lesser extent with wound infections and septicaemia. A pandemic serotype Vp O3:K6 has emerged, reaching many continents of the world, suggesting that this serotype has adapted to efficiently establish disease in humans (Velazquez-Roman et al., 2014). Sequence data from various Vp isolates indicate that many but not all human pathogenic strains contain two type III secretion systems (T3SS) (Makino et al., 2003; Banerjee et al., 2015). Studies using a small animal infection model with Vp strain RIMD2216330 (O3:K6) have demonstrated that both secretion systems function to deliver effector proteins into host cells, with T3SS-1 likely contributing to host cell cytotoxicity and T3SS-2 being involved in intestinal disruption and acute diarrhea (Ritchie et al., 2012). Therefore, it has been established that type III secretion is a key facet of Vp infection biology. In contrast, little is known regarding what environmental or host factors contribute to regulating T3SS genes and only a few studies have evaluated Vp T3SS gene regulation (Kodama et al., 2010a; Zhou et al., 2010a; Livny et al., 2014).
The majority of T3SS-1 associated genes within Vp are found in operon or co-transcribed units, similar to the situation found in other pathogens that express T3SS. Regulation of T3SS-1 genes in V. parahaemolyticus occurs in part through its transcriptional activator ExsA and a cognate anti-activator ExsD (Zhou et al., 2008), presumably functioning in a manner similar to the homologs that are found in Pseudomonas aeruginosa (King et al., 2013). ExsA is considered a member of the AraC/XylR transcriptional activator family and physically binds and bends DNA to activate gene transcription as demonstrated in both Vp and P. aeruginosa (Zhou et al., 2008; Brutinel et al., 2009). ExsA family members are thought to recognize and bind DNA via two intrinsic helix turn helix (HTH) domains. In the case of P. aeruginosa, two adjacent DNA binding sites are thought to support binding of two ExsA monomers (Diaz et al., 2011). Moreover, the monomers interact with their respective amino-terminal domains to form a functional ExsA homodimer. ExsD acts as an anti-activator, binding to an ExsA monomer within the cell, thereby preventing ExsA from binding to DNA and limiting ExsA dependent gene transcription (Zhou et al., 2010a; Kodama et al., 2010b). ExsA is likely involved in transcriptional activation of 8 or more operons which contain over 40 T3SS-1 associated genes, thereby placing it as a key activator of T3SS-1 expression.
Specific environmental signals that are sensed by pathogens often result in temporal and coordinated virulence gene expression. Temperature and low calcium has been implicated in inducing active type III secretion for a variety of pathogens (Fields et al., 1994; Thomas et al., 2005; Diaz et al., 2011), although the regulatory mechanisms that promote secretion events remain poorly understood. Many studies have demonstrated that low calcium growth conditions support the expression of the P. aeruginosa T3SS gene repertoire in an ExsA dependent manner (Diaz et al., 2011). Recently, a proteomic study revealed that low calcium growth conditions supported protein expression and secretion for many of the Vp T3SS-1 translocators and effectors (Sarty et al., 2012). It has also been reported that Vp exsA gene expression was upregulated when Vp was in contact with HeLa cells, indicating that T3SS-1 gene expression is induced during infection-like conditions (Zhou et al., 2008). Moreover, low cell density has been observed to promote Vp and V. harveyi T3SS-1 gene expression via quorum sensing mechanisms (Henke and Bassler, 2004). In contrast, exsA gene expression is inhibited by the global negative regulator H-NS (Sun et al., 2015). Still, environmental signals and requisite genetic mechanisms that result in exsA gene transcription, and hence T3SS-1 activation, remain unresolved.
In this study we set out to evaluate what factors contribute to T3SS-1 expression. Based on previous studies that evaluated T3SS-1 protein expression (Park et al., 2004b; Ono et al., 2006; Sarty et al., 2012), we assayed in situ real-time exsB and exsA promoter activity under various culture conditions which revealed differential activation patterns. Total secreted protein profiles for tested cultured conditions further revealed differential secretion of translocators and effectors. The data suggest a two wave model of Vp T3SS-1 induction that integrates environmental signals into a complex ExsA activation framework.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains generated and used in this study are listed in Table 1. Bacteria were routinely cultured in Luria broth (LB) [Sigma, L3522] at 37°C with shaking at 200 r.p.m., or 30°C at 250 r.p.m. for specific assays. Antibiotics (Sigma) were added when appropriate, to a final concentration of 100 μg kanamycin ml−1 (for Vp strains) or 50 μg for Escherichia coli DH5α. Sterile media supplements (MgSO4, EGTA, CaCl2) were added to basal LB media from concentrated stock solutions followed by a brief 10 min equilibration prior to use in assays.
Recombinant DNA Techniques
All PCR, cloning, and recombinant DNA techniques were performed using standard protocols. Routine DNA cloning steps were performed in DH5α, followed by introducing relevant recombinant plasmids into Vp using electroporation. Restriction endonucleases and DNA polymerases were purchased from New England Biolabs (NEB). Oligonucleotide primers were obtained from Integrated DNA Technologies (IDT) (Table 2). pJW15 contains a promoter-less luxCDABE cassette (Macritchie et al., 2008) and served as a vector for cloning exsA, exsB, exsD, and rplN promoter fragments. To generate a recombinant ExsA-FLAG expression plasmid, Vp exsA (without a stop codon) was amplified from chromosomal DNA by PCR using primers NT387 and NT388 followed by cloning into pFLAG-CTC. A PCR using primers NT139 and NT140 and exsA/pFLAG-CTC as DNA template yielded a product containing a 5′ tac promoter and a 3′ FLAG tag fused to exsA. The product was finally blunt end cloned into pVSV105 and the resulting plasmid was introduced into Vp by conjugal mating. All plasmid constructs were subjected to DNA sequence analyses to verify cloned DNA.
Reverse Transcriptase (RT)-PCR
Total RNA was isolated from Vp using a Qiagen RNeasy kit. RNA preparations were treated with DNase and then column purified. Reverse transcription reactions using primer AL400 were performed with Omniscript reverse transcriptase (Qiagen) using the protocol described by the manufacturer. A PCR reaction using cDNA template generated from the reverse step was performed using primers AL399 and AL400. A separate PCR using Vp genomic DNA was used to generate the predicted amplicon control of 1.5 kb. The amplicon spans the partial open reading frame of exsB to within exsA, including the intergenic region.
Luciferase (lux) Reporter Assays
Luciferase reporter assays were performed similar to a previous promoter study (Brouwers et al., 2012). End-point or multiple point assays were performed with Vp strains harboring lux plasmid constructs. For end-point assays, starting cultures (OD595 0.025) were grown for 2.5 h in LB supplemented with magnesium and ethylene glycol tetracetic acid (Mg+EGTA) followed by a OD595 measurement by a Biophotometer Plus (Eppendorf) at a path length of 10 mm. The samples were then aliquoted into clear bottom white walled multiwell plates (Costar #3610) and measured for light emission by counts per second (c.p.s.) using a Victor X5 instrument (Perkin Elmer). For multiple time point assays, the relevant samples were initially prepared in the same way as for the end-point assays, followed by immediate OD595 and cps reading at each time point. The data were subjected to a Two Way ANOVA with a standard Tukey test. Plotted data is the average from three experiments. Data was plotted using GraphPad Prism5 software.
Protein Secretion Assays
A secretion assay protocol using syringe end filters was used to produce total secreted protein fractions from Vp culture supernatants (Sarty et al., 2012). Briefly, bacterial cultures (starting OD595 0.025, 30°C, 250 rpm) were harvested after 4 h of growth. Three milliliters of culture was centrifuged to remove whole cells, followed by filtering the clarified supernatant through a 0.22 μm PVDF syringe-end filter (Millipore). The resulting culture supernatant was subjected to trichloracetic acid precipitation [10% (vol/vol)] on ice. The sample was centrifuged at 16,000 × g followed by resuspending the pellet in 10 μl electrophoretic sample loading buffer (2XESB) as previously described (Sarty et al., 2012).
Protein Electrophoresis
All protein samples were separated by SDS-PAGE as described elsewhere (Laemmli, 1970). Separated polypeptides were visualized by Coomassie G-250 blue staining.
Results
Evaluation of exsA and exsB Promoter Activities
The Vp exsBA genetic locus has two promoter elements that support transcription initiation upstream of exsB and exsA respectively (Sun et al., 2015) (Figure 1A). In V. harveyi, a multigene exsBA transcript has been detected, presumably driven from the exsB promoter (Waters et al., 2010). This observation is somewhat surprising given the long intergenic region between exsB and exsA, in addition to the presence of gene specific promoters. Therefore, we set out to determine if Vp exsBA are co-transcribed. As shown in Figure 1B, Vp generated an exsBA mRNA co-transcript as determined by reverse transcription PCR (RT-PCR).
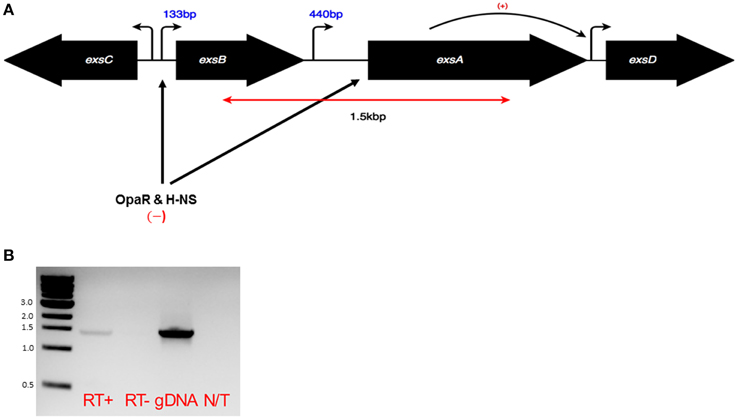
Figure 1. (A) Schematic of the exsA gene locus and adjacent genes. Bent arrows indicate promoter elements. In the case of exsB and exsA mapped transcriptional start sites are known, with numbers indicating mRNA leader lengths. The double headed red arrow corresponds to the predicted product size of the co-transcript as shown in a RT-PCR experiment (B). Black arrows indicate the roles of specific transcriptional regulators acting at indicated sites. (B) RT-PCR using sequence specific primers and purified mRNA was carried out to evaluate the existence of an exsBA co-transcript. Genomic DNA was used as a template to show primer specificity and expected amplicon length. The RT-PCR amplified 1.5 kb region is denoted in (A).
To investigate individual exsB and exsA promoters, the relevant chromosomal DNA fragments were fused to a promoterless luxCDABE gene cassette within pJW15 (Table 1). Plasmids pexsA-lux, pexsB-lux and pJW15 were each electroporated into wild type Vp. Notably, the pexsB-lux and pexsA-lux transformant colonies recovered post- electroporation were observed to emit light when grown on solid LB agar, whereas the transformants harboring pJW15 did not emit light (data not shown). This indicated that the cloned exsB and exsA promoters were functional and supported transcriptional activity when Vp was grown on LB.
ExsA is known to act at many Vp T3SS-1 operons, including those involved in forming secretion apparatus components (Zhou et al., 2008). Using Vp harboring pexsA-lux, we set out to evaluate exsA gene expression under a variety of media conditions at 30°C. This growth temperature was previously shown to support high levels of translocon and effector protein secretion via T3SS-I (Sarty et al., 2012). We show here that LB supports low level exsB and exsA promoter activity therefore we used it as the derivation point for the basal growth media. In luciferase (lux) assay experiments, real-time in situ exsA promoter activation is directly proportional to light emitted from the bacterial culture (Meighen, 1993). When Vp with pexsA-lux was grown in LB for 1 h, a basal level of exsA promoter activation was observed (Figure 2), consistent with the light emitting colonies observed when Vp was grown on LB agar. Vp exsA promoter activity for LB supplemented with magnesium or calcium was comparable to LB alone, however, LB supplemented with magnesium and EGTA produced significantly elevated exsA promoter activation (Figure 2). exsA promoter activation was even more pronounced after 3 h. Bacteria continued to demonstrate exsA promoter activation in LB supplemented with magnesium and EGTA after 4 h of incubation, although promoter activity exhibited a decreasing trend from that observed after 3 h. The data indicate that LB containing magnesium and EGTA supported maximal exsA and exsB promoter activation. LB that contained magnesium, calcium, or both elements, all supported basal level activation comparable to LB (only). Notably, the addition of calcium to LB media containing magnesium and EGTA lowered exsA and exsB promoter activation (over all time points evaluated) implicating calcium sensing in T3SS-1 gene expression (see Discussion).
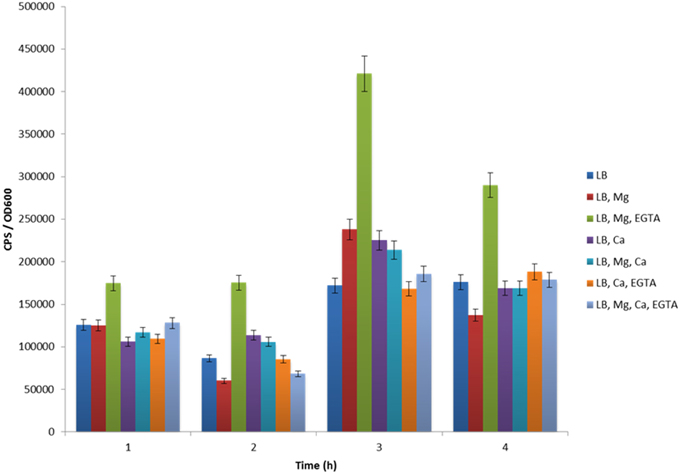
Figure 2. The effect of various growth media formulations on exsA promoter activity. Maximal exsA promoter activity was evident when Vp was grown in LB+Mg+EGTA for all time points assessed.
Experiments to evaluate exsB promoter activation revealed a similar trend to that observed for the exsA promoter (Figure 3). One notable difference was that exsB promoter activity was higher (10000–12000 units) than that of exsA (5000–7000 units) at early time points (0–30 min). Maximal exsA and exsB promoter activation occurred at 2.5 h in growth media containing Mg+EGTA. This time point also corresponded to when the bacteria were entering into early log phase growth (Figure 3). The effect of growth media on exsB promoter activation was similar to that observed for exsA promoter activation (Figure S1). Collectively, these data quantitatively demonstrate differential activity profiles for exsB and exsA.
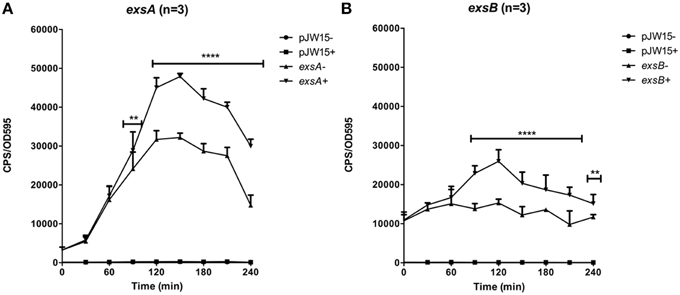
Figure 3. Temporal promoter profiles for (A) exsA and (B) exsB were generated by diluting overnight cultures containing the reporter plasmids in LB medium (–) and LB supplemented with EGTA, MgSO4 (+). Real time measurements were collected at 30 min intervals and normalized to OD595. The averaged data values from three experiments are shown. Error bars represent S.E.M. **p < 0.01, ****p < 0.0001. Representative growth curves for each strain from a single experiment are shown below the relevant data set.
T3SS-1 Activity Is Not Required for Vp exsA Gene Expression
For many pathogens with a T3SS, a negative regulator protein is often secreted through the assembled T3SS apparatus. This action typically leads to effector gene expression. For Vp, the negative regulator ExsE interacts with ExsC within the cytoplasm, which allows ExsD to inhibit ExsA activity. Upon secretion of ExsE by the T3SS-1, ExsC interacts with ExsD, thereby allowing ExsA to activate T3SS-1 associated genes (Kodama et al., 2010b). To assess whether ExsE secretion impacts on exsA promoter activity, the pexsA-lux construct was introduced into Vp ΔvscN and Vp ΔvscN1ΔvscN2. These mutants are defective for T3SS-1 and T3SS-1 and T3SS-2 respectively due to the absence of ATPases normally required for function. As shown in Figure 4A, exsA promoter activity was unchanged in ΔvscN and ΔvscN1ΔvscN2 compared to the activity observed for wild type. Furthermore, we profiled exsA promoter activity within ΔvscN over 4 h and did not see any change compared to wild type Vp (Figure S1). Furthermore, exsB promoter activity was unchanged in ΔvscN (Figure 4B). These data indicate that under the conditions tested, the absence of T3SS-1 or T3SS-2 activity does not significantly impact on exsA promoter activation. ExsB is thought to encode a T3SS-1 pilotin and the data indicates that T3SS-1 activity does not impact on exsB promoter activity.
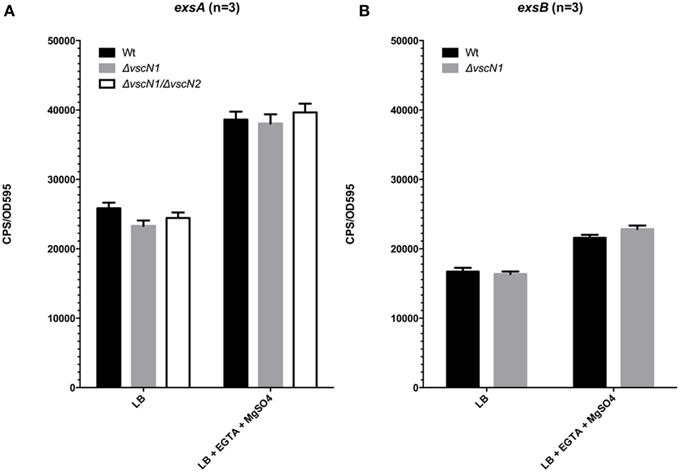
Figure 4. T3SS-1 and T3SS-2 activity are not required for exsA or exsB promoter activation. (A) exsA promoter activity in wild type Vp, ΔvscN1 (T3SS-1 ATPase-deficient mutant), and ΔvscN1ΔvscN2. (B) exsB promoter activity in wild type Vp and ΔvscN1. Endpoint measurements were taken at 2.5 h and compared. The averaged data values from three experiments are shown. Error bars represent S.E.M.
exsA Promoter Activity is Auto-activated
A common feature of AraC family regulators is the ability to auto-activate gene expression. To investigate this possibility for Vp ExsA, we generated a plasmid encoded ExsA-FLAG expression construct, with its transcription regulated by a tac promoter. This plasmid is expected to deliver ExsA overexpression without influences of feedback regulation or quorum sensing (due to the absence of the native exsA promoter). The ExsA-FLAG expression plasmid or empty control plasmid was transferred to Vp harboring pexsA-lux via conjugation. The resulting bacterial strains were then subjected to lux assays. ExsA overexpression did not impact exsA promoter activity when bacteria were grown in LB supplemented with magnesium +EGTA (data not shown). When bacteria overexpressing ExsA were grown in LB only, exsA promoter activity increased after 2.5 h compared to bacteria expressing native levels of ExsA. The increase reached statistical significance after 3.5 h (p = 0.015) and continued to 4.5 h (Figure 5A). These data indicate that ExsA overexpression results in higher levels of exsA promoter activity. Furthermore, in conditions that typically support low levels of type III protein secretion (LB growth), ExsA overexpression was observed to upregulate exsA promoter activity. This result is in complete agreement with a previous observation showing increased T3SS-1 associated gene expression upon ExsA overexpression (see Discussion).
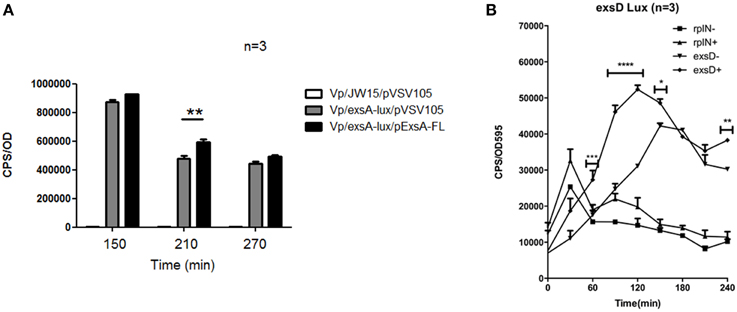
Figure 5. ExsA is involved in auto-activation and mediates exsD expression. (A) The effect of ExsA overexpression on exsA promoter activity was assessed using a lux assay. Error bars represent S.E.M, **p = 0.015. (B) Temporal promoter activity profiles for exsD and rplN were generated by diluting overnight cultures containing the reporter plasmids in LB medium (−), and LB supplemented with EGTA, MgSO4 (+). Real time measurements were collected at 30 min intervals and normalized to OD595. The averaged data values from three experiments are shown. Error bars represent S.E.M. *p < 0.05, **p < 0.01, ****p < 0.0001 for comparison of exsD-lux conditions (±).
exsD Promoter Activity Is Higher in Conditions That Support Maximal exsA Gene Expression
In Vp, ExsD binds to ExsA within the cell, thereby preventing ExsA binding to DNA (Zhou et al., 2010a). This regulatory paradigm is also observed in P. aeruginosa, where exsACDE are co-transcribed as a single operon. Hence, in P. aeruginosa the transcriptional activator ExsA is co-regulated with its own inhibitor. In Vp, the gene organization is different from P. aeruginosa. Specifically, exsD has its own promoter (Figure 1). Therefore, we set out to investigate whether the exsD promoter would show a similar activity profile to that of the exsA promoter. To address this we generated a pexsD-lux plasmid. We also generated a prplN-lux plasmid as a control since rplN is a ribosomal protein gene that is not expected to be regulated by ExsA. These plasmids were added to wild type Vp and the bacteria were subjected to lux assays. As shown in Figure 5B, exsD promoter activity reached a maximal level at 2 h. Importantly, this occurred in growth conditions that supported maximal exsA promoter activity (LB containing Mg+EGTA, refer to Figure 2). When bacteria were grown in LB only, exsD promoter activity increased, although at a slower rate than when bacteria were grown in LB supplemented with Mg+EGTA. The rplN promoter showed an early sharp increase in activity irrespective of growth media (Figure 5B). This was expected since the bacteria had been introduced into fresh growth media and ribosomal protein gene expression would be required to support de novo protein synthesis. More importantly, rplN promoter activity showed a steady state temporal activity trend between 60 and 240 min which was different to the activity profile for the exsD promoter. These data reveal that exsD promoter activity correlates with that of exsA.
Growth of Vp in Media Supplemented with Magnesium, Calcium, or EGTA Produce Differential Secreted Protein Profiles
We previously reported growth conditions that support maximal levels of Vp translocator and effector secretion (Sarty et al., 2012). Based on the differential exsA promoter activation patterns in several distinct media (Figure 2), we reassessed the data and provide new observations. For these experiments, Vp was grown for 4 h and filtered culture supernatants were processed for SDS-PAGE and Coomassie staining (see Materials and Methods).
Vp was grown in LB media with different supplementations, similar to the lux based exsA promoter activation studies. It was noted that each media produced differential total secreted protein profiles (Figure 6). The addition of EGTA to LB reduced Vp growth rate (data not shown), probably as a result of cell envelope stress and hence the secreted protein profile was too complex to interpret. Vp grown in LB had a complex secreted protein profile, however, the addition of magnesium to LB resulted in the appearance of 4 dominant protein species. Furthermore, growth of Vp in LB media containing magnesium and EGTA produced additional dominant secreted proteins (Figure 6). In contrast, growth of Vp in LB supplemented with (i) magnesium and calcium or (ii) magnesium, calcium and EGTA, did not yield abundant levels of these proteins.
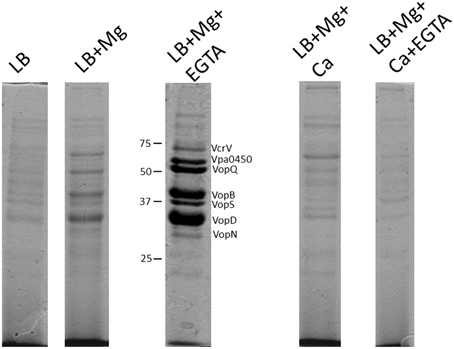
Figure 6. Total protein secretion profiles of Vp grown for 4 h in the indicated growth media conditions. Samples were normalized for cell density to account for growth rate differences. All protein samples were stained with Coomassie G-250. The protein species have been previously identified using mass spectrometry approaches. Note the specific appearance of Vpa0450 and VopN under LB+Mg+EGTA conditions only. The addition of calcium to media containing Mg+EGTA reduced the secretion of all T3SS-1 associated proteins.
The secreted proteins produced from Vp growth in LB+Mg+EGTA were previously subjected to mass spectrometry analyses and identified as type III translocators and effector proteins (Sarty et al., 2012). Notably, VopN and Vpa450 secretion were readily apparent in Vp cultures supplemented with Mg+EGTA yet not detectable from Vp cultures supplemented with magnesium (only) or any media supplemented with calcium (Figure 6). This observation appeared unique to VopN and Vpa0450. These data indicate that defined growth conditions significantly impact the T3SS-1 associated secretion profile of Vp and that certain effectors are efficiently secreted under low calcium conditions.
Growth Temperature Impacts on exsA Promoter Activity and T3SS-1 Secretion Levels
Vp is commonly found in brackish estuarine waters that range in temperature from 15 to 30°C (Ceccarelli et al., 2014). However, during human infection, Vp encounters a body temperature of 37°C. We set out to assess exsA promoter activity at 30°C and 37°C. It was observed that exsA promoter activity was significantly higher at 30°C when compared to 37°C (Figure 7A). The culture supernatants from these samples were subjected to SDS-PAGE to assess total protein secretion under the different temperature conditions. Coomassie staining revealed that effector protein secretion was more abundant at 30°C than 37°C (Figure 7B). These data correlated with the elevated exsA promoter activity observed at 30°C.
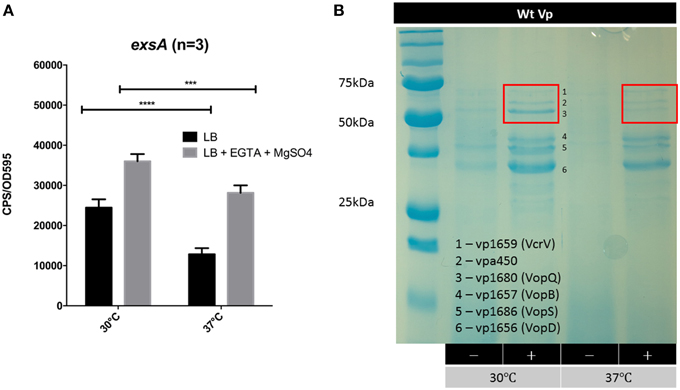
Figure 7. The effect of growth temperature on exsA promoter activity and Vp total secretion profiles. (A) exsA promoter activity measured in LB (−) or LB+Mg+EGTA (+) grown at 30°C or 37°C. Measurements were taken at 2.5 h post inoculation and normalized to OD595. ***p < 0.001, ****p < 0.0001. (B) Total secretion profiles revealed by Coomassie G-250 staining. Vp was grown in LB (−) or LB+Mg+EGTA (+), at 30°C or 37°C. Culture supernatants were harvested 4 h post inoculation. The red boxes highlight different banding intensities of effector proteins between the two conditions.
Identification of an Expanded Putative Vp ExsA Binding Motif for 11 Genetic Loci
We noted that elevated Vpa0450 and VopN secretion levels exclusively correlated with growth conditions that supported elevated exsA gene expression (Figures 2, 3). This observation is in direct agreement with a RNA-seq study showing ExsA overexpression upregulated vpa0450 and vopN gene expression (Nydam et al., 2014). Therefore, we sought to discover an explanation that could link elevated Vpa0450 and VopN protein secretion to exsA expression.
We hypothesized that ExsA could activate vpa0450 and vopN gene expression by recognizing the respective promoter regions of these genes. For Vp, a previous study using two promoter DNA fragments identified an ExsA DNA binding motif (TTTAGN4TT) (Zhou et al., 2008). This DNA binding motif is similar to one identified for VirF, an AraC-type regulator of Yersinia species (Wattiau and Cornelis, 1994). Notably, Vp exsA has been shown to complement a P. aeruginosa exsA mutant for transcriptional activation (King et al., 2013) and detailed mutagenesis studies have implicated an A-box and a separate G/C box motif in P. aeruginosa ExsA binding to DNA (King et al., 2012). Therefore, we predicted that the identified DNA motifs for both Vp and P. aeruginosa ExsA DNA binding would be present as a “hybrid” or expanded motif upstream of Vp exsA-dependent promoters. Sequence analysis of the DNA region upstream of vopN (vp1667) and vpa0451 (a chaperone gene co-transcribed with vpa0450), revealed DNA sequences consistent with our prediction. Specifically, considerable identity to the binding motif (TTTAGN4TT), along with an A-box and a G/C box (Figure 8).
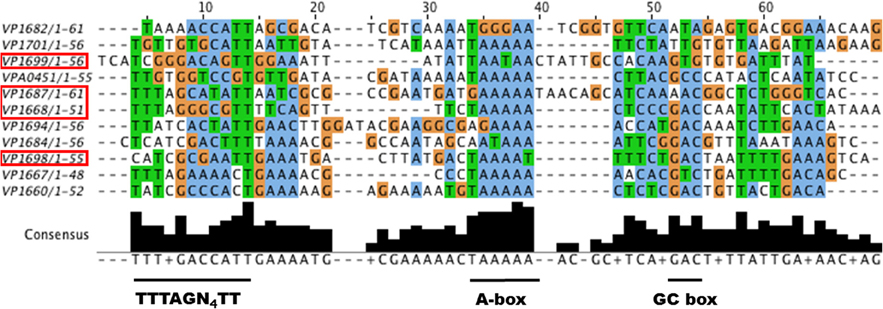
Figure 8. A putative expanded ExsA binding motif is found for many Vp T3SS-1 associated genes. Vp T3SS-1 promoter elements were retrieved from the genome sequence of Vp RIMD2210633 within the NCBI database, aligned using MUSCLE (Edgar, 2004), and visualized with JALView (Waterhouse et al., 2009). Boxed genes have been validated for either ExsA binding or ExsA mediated gene regulation in this study. The underlined regions below the sequences represent conserved DNA motifs. The TTTAGN4TT motif has been shown to support ExsA binding to DNA in Vp. The A-box and GC box motifs are known to support DNA binding for P. aeruginosa ExsA.
Next, we considered that the observed increase in T3SS-1 associated proteins and effectors under LB+Mg+EGTA growth conditions (Figure 6) likely required increased expression of specific T3SS-1 apparatus genes, presumably achieved through ExsA. We evaluated all the annotated T3SS-1 genes and identified 11 putative expanded ExsA binding sites (Figure 8). Two of these are known to bind ExsA (vp1668 and vp1687) and we provide evidence in this study that exsA (vp1699) and exsD (vp1698) are regulated by ExsA. Putative expanded ExsA binding sites are located upstream of chaperone and effector gene pairs (vp1684-vp1683, vp1687-vp1686, vp1682-vp1680).
Discussion
In this study, we profiled two distinct promoter elements that control exsA gene expression. Specific growth conditions produced differential exsA promoter activity. Maximal exsA gene expression correlated with elevated T3SS-1 protein secretion levels when Vp was grown in media supplemented with Mg+EGTA at 30°C. In contrast, exsA gene expression and T3SS-1 activity was minimal (baseline) when Vp was cultured in the presence of calcium. We identify that ExsA serves as an auto-activator, while at the same time positively regulates gene expression of exsD, an inhibitor of ExsA activity. Lastly, combining known features of DNA binding motifs from Vp ExsA, P. aeruginosa ExsA, and Yersinia VirF, fresh bioinformatic analyses identified a putative expanded Vp ExsA DNA binding motif linked to multiple T3SS-1 genetic operons. The findings indicate that Vp ExsA is a key entry level regulator of T3SS-1 gene expression and that multiple genetic and environmental factors contribute to exsA gene expression levels.
Genetic analyses have identified a transcriptional start site and putative promoter upstream of exsA (Sun et al., 2015). In V. harveyi, an exsBA co-transcript has been identified (Waters et al., 2010), which is somewhat surprising given the large exsB-exsA intergenic region [~600bp, also present in Vp (Figure 1)]. This prompted us to investigate whether exsBA are co-transcribed in Vp. Using RT-PCR, we detected a Vp exsBA transcript (Figure 1). This finding indicates that exsA gene expression can be initiated by two distinct promoter elements, similar to V. harveyi. Quantitative activity profiling of the distinct exsB and exsA promoters revealed differential temporal features and activity levels (Figure 3). Collectively, the promoter profiling data indicates that for the growth conditions tested, Vp can initiate exsBA co-transcription followed by high level exsA gene expression from a proximal promoter.
A clear finding from this study is that high T3SS-1 secreted protein levels correlate with an increase in exsA promoter activity (Figures 6, 7). We had previously shown that growth media with calcium reduced Vp T3SS-1 activity and that Mg+EGTA growth media promoted T3SS-1 activity (Sarty et al., 2012), although the mechanisms behind those observations were not evident. Multiple growth conditions were used to profile light emission of an exsA promoter-lux fusion which indicated that when Vp is grown in the presence of calcium, exsA promoter activity (and by extension exsA gene expression) was at low baseline levels. In sharp contrast, Vp growth in Mg+EGTA significantly upregulated exsA promoter activity. Hence, growth conditions can impact exsA gene expression levels with consequences for T3SS-1 activity.
Temperature is known to have a regulating impact on gene expression and T3SS activity for many pathogens. In the case of Vp, it appears that exsA gene expression is elevated at 30°C compared to 37°C (Figure 7). This observation extends to protein secretion levels where translocon and effectors were relatively more abundant at 30°C. It is important to note that T3SS-1 associated proteins are expressed and secreted at 37°C, which agrees with various lines of evidence from cytotoxicity and infection assays (Park et al., 2004a; Broberg et al., 2010; Zhou et al., 2010b; Sarty et al., 2012). While T3SS-1 is required for full virulence in a small animal model of infection (Ritchie et al., 2012), it's role in human diarrheal disease has yet to be confirmed. A recent transcriptomic screen of Vp during infection suggests that ExsA levels need to reach a threshold to mediate downstream events (Livny et al., 2014). Our data supports this view, particularly with respect to differential exsA promoter activation.
The data indicate that in vitro low calcium conditions are associated with elevated exsA promoter activation. This is similar to many observations from different pathogens that express a T3SS, including Yersinia, Pseudomonas, and EPEC (DeBord et al., 2003; Deng et al., 2005; Dasgupta et al., 2006). However, we are cautious with our interpretations since the experiments used EGTA, which can chelate a variety of divalent cations with varying efficiency. The affinity of EGTA for calcium is known to be high, although other cations can be bound including Fe2+. We also note that the inclusion of calcium to Vp growth media containing magnesium and EGTA was sufficient to limit exsA promoter activity (Figure 2), although one cannot rule out the possibility that other chelated cations are freed up due to EGTA competitively binding calcium. For example, low soluble iron availability has been implicated in Vp T3SS induction (Gode-Potratz et al., 2010). Under certain in vitro conditions tested here, soluble iron cations may have come available when EGTA competitively bound to calcium, potentially limiting T3SS-1 activity. Therefore, it remains to be determined what exact environmental signals induce Vp T3SS-1, although low amounts of free divalent cations could be involved. During Vp infection of human cells, bacterial contact with target cells also is believed to have a role in T3SS-1 induction (Zhou et al., 2008), although the detailed mechanisms have not been elucidated to date.
A secretion hierarchy in T3SS occurs with translocator proteins being secreted ahead of effector proteins (Tomalka et al., 2012). Interestingly, different total secreted protein profiles were observed when Vp was cultured in specific growth media (Figure 6). The inclusion of Mg+EGTA to LB resulted in higher levels of effectors along with the specific appearance of VopN and Vpa450 effector proteins in secreted fractions. exsA promoter activity levels were significantly different for LB+Mg and LB+Mg+EGTA conditions especially at 3 h post inoculation (Figure 2). This prompted us to examine if ExsA could be involved in activating gene expression of vopN and vpa0450. In both cases, we identified DNA sequences that strongly match an expanded Vp ExsA binding motif (containing TTTAGN4TT, A-box, and G/C box motifs) located upstream of these genes (Figure 8). Furthermore, our bioinformatic analyses reveal putative expanded ExsA binding motifs upstream of 11 genetic loci associated with T3SS-1 operons. Two of these genetic loci have been shown to support ExsA binding, and we provide promoter activity data to implicate the exsA and exsD promoters as being regulated by ExsA (Figure 5).
The 11 identified genetic loci in Figure 8 are arranged in transcriptional operons and collectively encode 41 genes. This number of ExsA regulated genes is in agreement with a recent Vp RNA-Seq study (Nydam et al., 2014) and also closely resembles the 10 ExsA regulated operons within P. aeruginosa (Diaz et al., 2011). The expanded Vp ExsA binding motif identified here combines DNA elements that bring together data from various systems in Yersinia (VirF), Pseudomonas (ExsA) and Vp (ExsA) (Wattiau and Cornelis, 1994; Brutinel et al., 2008; Zhou et al., 2008). The fact that Vp exsA can complement a P. aeruginosa exsA mutant (King et al., 2013) further indicates common functionality at a DNA binding level. The binding of AraC/XylR/ExsA family members to DNA is complex, in some cases involving sequential steps. Therefore, we propose that further DNA binding assays and detailed mutagenesis investigations are required to experimentally validate and delineate the expanded Vp ExsA binding motif presented in Figure 8.
In summary, Vp was found to have two distinct mechanisms to initiate exsA expression. Growth media and temperature were found to impact on T3SS-1 activity, which was linked to exsA expression levels. ExsA auto-activates is own expression, whereas its negative regulator ExsD is expressed under similar conditions. ExsA is therefore a central transcriptional activator that responds to environmental stimuli to regulate T3SS-1 associated genes. The results suggest that temporal ExsA cellular levels exhibit fine tuning regulatory roles resulting in differential T3SS-1 activity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Thomas laboratory for their comments toward the manuscript. Lauren Campbell provided support for some of the lux experiments. This work was funded by a grant awarded to NT. from the Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN/342111-2013). The Canadian Foundation for Innovation (CFI) provided infrastructure support in a CFI-LOF grant to NT.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01089
Figure S1. (A) exsA promoter activity profiles within ΔvscN grown in LB (−) or LB+Mg+EGTA (+). (B) exsB promoter activities for various growth media conditions. Samples were measured at 2.5 h post inoculation. A representative data set is shown.
References
Banerjee, S., Petronella, N., Chew Leung, C., and Farber, J. (2015). Draft Genome Sequences of Four Vibrio parahaemolyticus Isolates from Clinical Cases in Canada. Genome Announc 3:e01482-14. doi: 10.1128/genomeA.01482-14
Broberg, C. A., Zhang, L., Gonzalez, H., Laskowski-Arce, M. A., and Orth, K. (2010). A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 329, 1660–1662. doi: 10.1126/science.1192850
Brouwers, E., Ma, I., and Thomas, N. A. (2012). Dual temporal transcription activation mechanisms control cesT expression in enteropathogenic Escherichia coli. Microbiology 158(Pt 9), 2246–2261. doi: 10.1099/mic.0.059444-0
Brutinel, E. D., Vakulskas, C. A., Brady, K. M., and Yahr, T. L. (2008). Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 68, 657–671. doi: 10.1111/j.1365-2958.2008.06179.x
Brutinel, E. D., Vakulskas, C. A., and Yahr, T. L. (2009). Functional domains of ExsA, the transcriptional activator of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 191, 3811–3821. doi: 10.1128/JB.00002-09
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2014). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Dasgupta, N., Ashare, A., Hunninghake, G. W., and Yahr, T. L. (2006). Transcriptional induction of the Pseudomonas aeruginosa type III secretion system by low Ca2+ and host cell contact proceeds through two distinct signaling pathways. Infect. Immun. 74, 3334–3341. doi: 10.1128/IAI.00090-06
DeBord, K. L., Galanopoulos, N. S., and Schneewind, O. (2003). The ttsA gene is required for low-calcium-induced type III secretion of Yop proteins and virulence of Yersinia enterocolitica W22703. J. Bacteriol. 185, 3499–3507. doi: 10.1128/JB.185.12.3499-3507.2003
Deng, W., Li, Y., Hardwidge, P. R., Frey, E. A., Pfuetzner, R. A., Lee, S., et al. (2005). Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73, 2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005
Diaz, M. R., King, J. M., and Yahr, T. L. (2011). Intrinsic and extrinsic regulation of type III secretion gene expression in pseudomonas aeruginosa. Front. Microbiol. 2:89 doi: 10.3389/fmicb.2011.00089
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fields, K. A., Plano, G. V., and Straley, S. C. (1994). A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J. Bacteriol. 176, 569–579.
Gode-Potratz, C. J., Chodur, D. M., and McCarter, L. L. (2010). Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J. Bacteriol. 192, 6025–6038. doi: 10.1128/JB.00654-10
Henke, J. M., and Bassler, B. L. (2004). Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186, 3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004
King, J. M., Brutinel, E. D., Marsden, A. E., Schubot, F. D., and Yahr, T. L. (2012). Orientation of Pseudomonas aeruginosa ExsA monomers bound to promoter DNA and base-specific contacts with the P(exoT) promoter. J. Bacteriol. 194, 2573–2585. doi: 10.1128/JB.00107-12
King, J. M., Schesser Bartra, S., Plano, G., and Yahr, T. L. (2013). ExsA and LcrF recognize similar consensus binding sites, but differences in their oligomeric state influence interactions with promoter DNA. J. Bacteriol. 195, 5639–5650. doi: 10.1128/JB.00990-13
Kodama, T., Gotoh, K., Hiyoshi, H., Morita, M., Izutsu, K., Akeda, Y., et al. (2010a). Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS ONE 5:e8678. doi: 10.1371/journal.pone.0008678
Kodama, T., Yamazaki, C., Park, K. S., Akeda, Y., Iida, T., and Honda, T. (2010b). Transcription of Vibrio parahaemolyticus T3SS1 genes is regulated by a dual regulation system consisting of the ExsACDE regulatory cascade and H-NS. FEMS Microbiol. Lett. 311, 10–17. doi: 10.1111/j.1574-6968.2010.02066.x
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Livny, J., Zhou, X., Mandlik, A., Hubbard, T., Davis, B. M., and Waldor, M. K. (2014). Comparative RNA-Seq based dissection of the regulatory networks and environmental stimuli underlying Vibrio parahaemolyticus gene expression during infection. Nucleic Acids Res. 42, 12212–12223. doi: 10.1093/nar/gku891
Macritchie, D. M., Ward, J. D., Nevesinjac, A. Z., and Raivio, T. L. (2008). Activation of the Cpx envelope stress response down-regulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect. Immun. 76, 1465–1475. doi: 10.1128/IAI.01265-07
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Meighen, E. A. (1993). Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7, 1016–1022.
Nydam, S. D., Shah, D. H., and Call, D. R. (2014). Transcriptome analysis of Vibrio parahaemolyticus in type III secretion system 1 inducing conditions. Front. Cell. Infect. Microbiol. 4:1. doi: 10.3389/fcimb.2014.00001
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74, 1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Iida, T., and Honda, T. (2004a). Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol. Immunol. 48, 313–318. doi: 10.1111/j.1348-0421.2004.tb03512.x
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T., et al. (2004b). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004
Ritchie, J. M., Rui, H., Zhou, X., Iida, T., Kodoma, T., Ito, S., et al. (2012). Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog 8:e1002593. doi: 10.1371/journal.ppat.1002593
Sarty, D., Baker, N. T., Thomson, E. L., Rafuse, C., Ebanks, R. O., Graham, L. L., et al. (2012). Characterization of the type III secretion associated low calcium response genes of Vibrio parahaemolyticus RIMD2210633. Can. J. Microbiol. 58, 1306–1315. doi: 10.1139/w2012-109
Sun, F., Zhang, Y., Qiu, Y., Yang, H., Yang, W., Yin, Z., et al. (2015). H-NS is a repressor of major virulence gene loci in Vibrio parahaemolyticus. Front. Microbiol. 5:675. doi: 10.3389/fmicb.2014.00675
Thomas, N. A., Deng, W., Puente, J. L., Frey, E. A., Yip, C. K., Strynadka, N. C., et al. (2005). CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57, 1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x
Tomalka, A. G., Stopford, C. M., Lee, P. C., and Rietsch, A. (2012). A translocator-specific export signal establishes the translocator-effector secretion hierarchy that is important for type III secretion system function. Mol. Microbiol. 86, 1464–1481. doi: 10.1111/mmi.12069
Velazquez-Roman, J., León-Sicairos, N., de Jesus Hernandez-Diaz, L., and Canizalez-Roman, A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 3:110. doi: 10.3389/fcimb.2013.00110
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Waters, C. M., Wu, J. T., Ramsey, M. E., Harris, R. C., and Bassler, B. L. (2010). Control of the type 3 secretion system in Vibrio harveyi by quorum sensing through repression of ExsA. Appl. Environ. Microbiol. 76, 4996–5004. doi: 10.1128/AEM.00886-10
Wattiau, P., and Cornelis, G. R. (1994). Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 176, 3878–3884.
Zhou, X., Konkel, M. E., and Call, D. R. (2010a). Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence 1, 260–272. doi: 10.3389/fcimb.2013.00110
Zhou, X., Konkel, M. E., and Call, D. R. (2010b). Vp1659 is a Vibrio parahaemolyticus type III secretion system 1 protein that contributes to translocation of effector proteins needed to induce cytolysis, autophagy, and disruption of actin structure in HeLa cells. J. Bacteriol. 192, 3491–3502. doi: 10.1128/JB.01493-09
Keywords: Type III secretion, transcription activator, gene regulation, environmental sensing
Citation: Liu AC and Thomas NA (2015) Transcriptional profiling of Vibrio parahaemolyticus exsA reveals a complex activation network for type III secretion. Front. Microbiol. 6:1089. doi: 10.3389/fmicb.2015.01089
Received: 30 June 2015; Accepted: 22 September 2015;
Published: 20 October 2015.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Qingping Zhong, South China Agricultural University, ChinaMatthew S. Francis, Umeå University, Sweden
Copyright © 2015 Liu and Thomas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nikhil A. Thomas, Dalhousie University, Tupper Medical Building, 7th floor, 5850 College Street, Halifax, NS B3H 4R2, Canada, n.thomas@dal.ca
 Aaron C. Liu
Aaron C. Liu Nikhil A. Thomas
Nikhil A. Thomas