- 1Department of Microbiology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
- 2Department of Microbiology, School of Medicine, Bushehr University of Medical Sciences, Bushehr, Iran
- 3Pediatrics Infectious Diseases Research Center, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
- 4Molecular Biology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran
- 5Anti-microbial Resistance Research Center, Iran University of Medical Sciences, Tehran, Iran
- 6Department of Microbiology, Faculty of Medicine, Hormozgan University of Medical Sciences, Bandar Abbas, Iran
Antimicrobial resistance in carbapenem non-susceptible Acinetobacter baumannii (CNSAb) is a major public health concern globally. This study determined the antibiotic resistance and molecular epidemiology of CNSAb isolates from a referral burn center in Tehran, Iran. Sixty-nine CNSAb isolates were tested for susceptibility to antimicrobial agents using the E test methodology. Multiple locus variable number tandem repeat analysis (MLVA), Multilocus sequence typing (MLST) and multiplex PCR were performed. PCR assays tested for ambler classes A, B, and D β-lactamases. Detection of ISAba1, characterization of integrons, and biofilm formation were investigated. Fifty-three (77%) isolates revealed XDR phenotypes. High prevalence of blaOXA-23-like (88%) and blaPER-1 (54%) were detected. ISAba1 was detected upstream of blaADC, blaOXA-23-like and blaOXA51-like genes in, 97, 42, and 26% of isolates, respectively. Thirty-one (45%) isolates were assigned to international clone (IC) variants. MLVA identified 56 distinct types with six clusters and 53 singleton genotypes. Forty previously known MLST sequence types forming 5 clonal complexes were identified. The Class 1 integron (class 1 integrons) gene was identified in 84% of the isolates. The most prevalent (33%) cassette combination was aacA4-catB8-aadA1. The IC variants were predominant in the A. baumannii lineage with the ability to form strong biofilms. The XDR-CNSAb from burned patients in Iran is resistant to various antimicrobials, including tigecycline. This study shows wide genetic diversity in CNSAb. Integrating the new Iranian A. baumannii IC variants into the epidemiologic clonal and susceptibility profile databases can help effective global control measures against the XDR-CNSAb pandemic.
Introduction
Acinetobacter baumannii is an important healthcare-associated pathogen that can cause of life threatening infections (Öncül et al., 2014; Girerd-Genessay et al., 2015). A. baumannii is the second most common multidrug-resistant cause of nosocomial infection in burn patients in Iran (Alaghehbandan et al., 2012; Azimi et al., 2015). Its infections have become critical challenge to health care systems due to increasing levels of resistance to antimicrobial agents in nosocomial isolates of A. baumannii (Cai et al., 2012; Moradi et al., 2015). In developing countries, including Iran, the occurrence of carbapenem non-susceptinle A. baumannii (CNSAb) infections is a growing problem in hospitalized burn patients (Mahdian et al., 2015; Zowawi et al., 2015). Inadequate management of the antibiotic therapy of CNSAb infections often leads to the emergence of extensive and pandrug resistant (XDR and PDR) CNSAb strains, which present significant health challenges by prolonging hospitalization, treatment failures, and increased mortality (Liu et al., 2014b). The most important mechanism of carbapenem resistance in A. baumannii is the production of carbapenem-hydrolyzing β-lactamases of Ambler classes A, B, and D. In addition to being resistant to all β-lactams available, carbapenemases have a high capacity to spread, since their genes have been commonly found in transferable plasmids containing integrons and insertion sequence (IS) elements (Karah et al., 2012).
Worldwide surveillance has shown that the global population structure of CNSAb isolates is diverse, but a small number of widespread clones, including the international clonal (IC) lineage I–III, also defined based on multilocus sequence typing (MLST) as clonal complexes (CC), may be predominant in healthcare settings (Jeannot et al., 2014). Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) has been considered as a rapid and cost-effective method for fine-scale typing of A. baumannii, enabling inter-laboratory comparison of typing data (Karah et al., 2012). A recent study found the high discriminatory power of MLVA, sufficient to detect diversity among isolates showing identical MLST types (Almeida and Araujo, 2013).
Up-to-date surveillance information regarding genotypic spread, plus local antimicrobial susceptibility patterns of clinical isolates of A. baumannii, are necessary for effective drug therapy and control of XDR and PDR A. baumannii hospital outbreaks (Fishbain and Peleg, 2010). Considering the points mentioned above, the aim of this study was to determine molecular epidemiology, to identify the dissemination of the most common resistance genes, to investigate the prevalence of integrons and arrangement of integron gene cassettes among CNSAb isolates from a referral burn center in Tehran, Iran.
Materials And Methods
Bacterial Isolates
A total of 92 non-repetitive A. baumannii strains were collected between January 2012 to May 2013, from the burn wound infections of hospitalized patients in Shahid Motahari hospital, the only referral burn center in Tehran, Iran. Species of these isolates were initially characterized using the API20NE system (bioMérieux, Marcy-l’Etoile, France) and then final identification of isolates were performed by multiplex PCR using gyrB-directed primers according to the study of Higgins et al. (2010b).
Antimicrobial Susceptibility Tests and Biofilm Formation Assay
The Clinical and Laboratory Standards Institute (CLSI) guideline (Clinical and Laboratory Standards Institute [CLSI], 2014) for minimum inhibitory concentrations (MICs) using the E test was used to assess the susceptibility of 92 A. baumannii isolates to imipenem (Ezy MICTM strips, Himedia, India). All isolates with MICs of imipenem >4 mg/L were defined as CNSAb isolates and were recruited for further screening (Clinical and Laboratory Standards Institute [CLSI], 2014).The susceptibility of CNSAb isolates to 17 other antimicrobial agents such as amikacin, ampicillin-sulbactam, cefepime, ceftazidime, ciprofloxacin, colistin, gentamicin, levofloxacin, meropenem, minocycline, piperacillin, piperacillin-tazobactam, rifampicin, tetracycline, tigecycline, tobramicin, and trimethoprim-sulfamethoxazole were also carried out using the E test (Ezy MICTM strips, Himedia, India). The phenotypic presence of Metallo-β-lactamase (MBL) enzymes was detected by imipenem/imipenem + EDTA strips (Ezy MICTM strips, Himedia, India). The MIC ratio of MBL-E test of ≥8 mg/L was interpreted as indicative of MBL activity, according to the manufacturer’s instructions. Since there is no breakpoint for tigecycline and rifampicin against A. baumannii strains in the CLSI guidelines; therefore, the criteria for interpretation of the MIC values of tigecycline (MIC of ≤1 mg/L defined as susceptible and >2 mg/L as resistant) were determined according to the European committee on antimicrobial susceptibility testing (EUCAST) (European Committee on Antimicrobial Susceptibility Testing, 2014) for members of the Enterobacteriaceae sp. and CLSI criteria, using breakpoint values suggested for Staphylococcus aureus applied to rifampicin (susceptible defined as ≤1 mg/L and resistant defined as ≥4 mg/L). The MIC50 and MIC90 of each antibiotic were calculated. Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and E. coli ATCC 35218 were used as quality control organisms. The phenotype of A. baumannii is defined as MDR and XDR according to the International Expert proposal for Interim Standards Guidelines (Magiorakos et al., 2012). The formation assay and quantitative analysis of biofilm were performed according to a previous study (Zhang et al., 2014).
Molecular Detection of β-lactamases-encoding Genes and Characterization of Integrons
A series of PCR amplifications were done for detection of different Ambler class bla gene groups: class A; blaPER-1, blaV EB-1, blaCTX-M, blaKPC, blaGES, blaTEM and blaSHV -1 (Poirel et al., 2003; Ellington et al., 2007; Bonnin et al., 2011a,b; Potron et al., 2011; Robledo et al., 2011; Climaco et al., 2013); Class B, blaIMP-1, blaV IM-2, blaGIM-1, blaSPM-1 blaSIM-1 and blaNDM-1 (Chen et al., 2011); Class C, blaADC (Bou and Martinez-Beltran, 2000) and Class D, blaOXA-23-, blaOXA-40-, blaOXA-51-, blaOXA-58 (Woodford et al., 2006) and blaOXA-143-like (Higgins et al., 2010a) encoding genes were detected. To determine whether ISAba1 was present upstream of blaOXA-23-like, blaOXA-51-like (Turton et al., 2006) and blaADC (Heritier et al., 2006) genes, PCR mapping experiments were performed. Detection and characterization of class 1, 2, and 3 integrons, as well as gene cassette mapping and sequencing of class 1 integrons were carried out using previously described PCR assay (Dillon et al., 2005).
Genetic Diversity and Population Structure
The IC lineage so-called PCR-based group were identified using three-locus dual assay multiplex PCR as described previously (Turton et al., 2007). Isolates pertaining to the novel variant of the PCR-based group, according to the new combination of amplified products obtained from the two separate multiplex PCRs, did not correspond to those previously defined. Moreover, the CNSAb isolates were genotyped using the MLVA-8 scheme method developed by Pourcel et al. (2011). For clustering analysis, the allele strings were entered into the BioNumerics software v.7.0 (Applied Maths, Sint-Martens-Latem, Belgium) as character values. A cut-off value of 90% similarity was applied to define clusters (Nhu et al., 2014). MLST was performed according to the Bartual method described previously (Bartual et al., 2005). Allele sequences, sequence types (STs), primer sequences and other details are available from the MLST website at http://pubmlst.org. The minimum spanning tree (MSTree) algorithm was used to predict the clonal relationship between all CNSAb isolates. MSTree algorithm was constructed with the BioNumerics software v.7.0 (Applied Maths, Sint-Martens-Latem, Belgium) using a categorical coefficient according to a previous report (Ruan et al., 2013).
Statistical Analysis
Statistical analyses were performed by Student’s t-test, Fisher’s exact test and the chi-square test with the SPSS software package (version 16). P-values <0.05 in all experiments were considered as significant.
Results
Antibiotic Susceptibility Testing and Biofilm Assay
In this study, from the 92 A. baumannii isolates, 69 isolates were confirmed as CNSAb. The MIC determination of all CNSAb isolates, exhibited non-susceptibility rates ≥95% to cefepime, ceftazidime, levofloxacin, meropenem, piperacillin, piperacillin-tazobactam, rifampicin and trimethoprim-sulfamethoxazole (Table 1). CNSAb isolates showed high susceptibility to colistin (100%), tigecycline (87%) and minocycline (69%) with MIC50 values of 0.01, 0.25, and 5 mg/L, respectively and MIC90 values of 1, 3, and 10 mg/L, respectively (Table 1). Fifty-three (77%) CNSAb isolates revealed XDR phenotypes. In all XDR isolates following colistin with 100% sensitivity, tigecycline (83%) and minocycline (67%) were the most effective antibiotics. Fifty-six (81%) of all CNSAb isolates formed strong biofilm whereas 13 (19%) of these isolates were considered as weak biofilm forming strains. Our study revealed a significant association of strong biofilm formation with antimicrobial resistance (P = 0.0001).
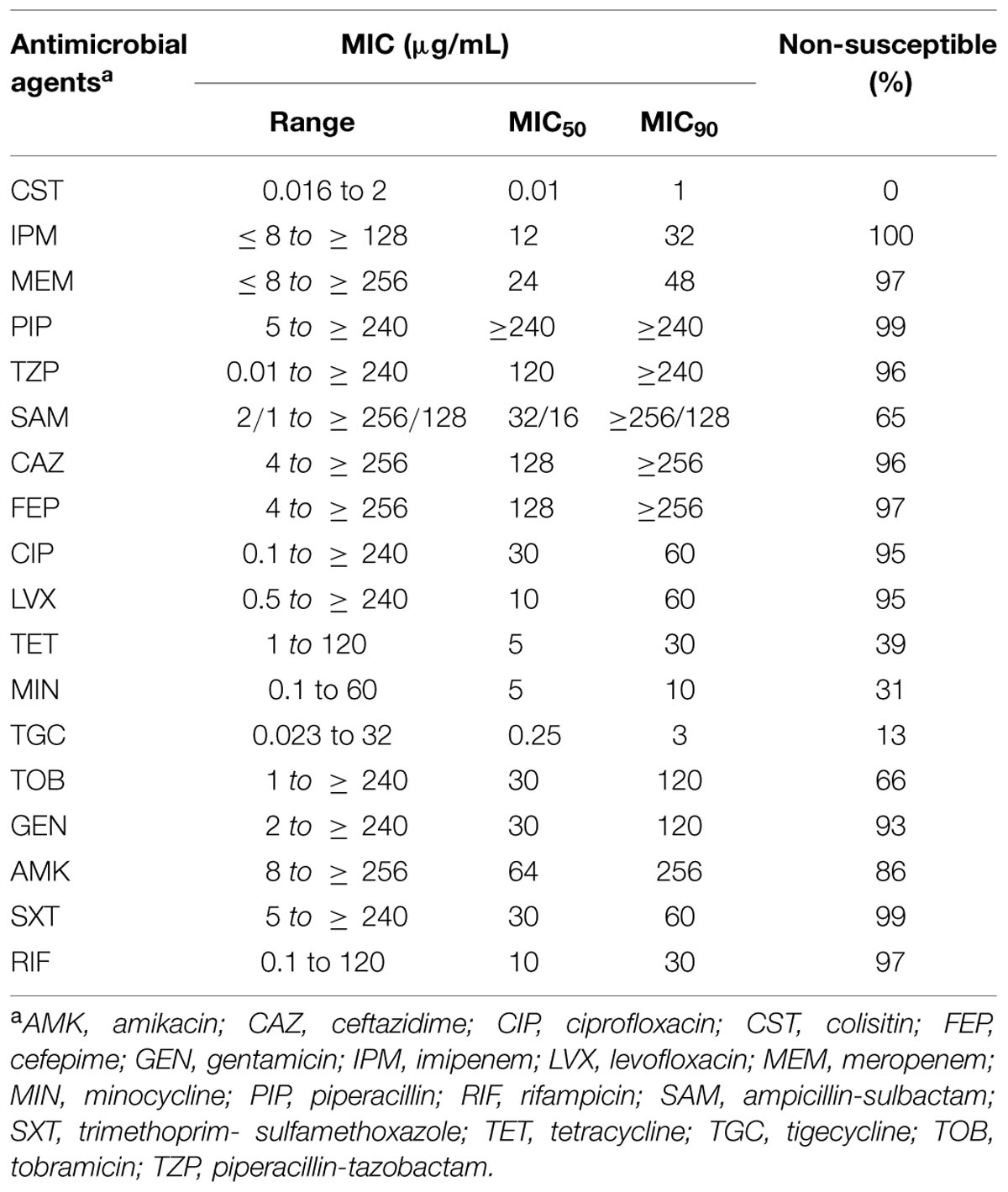
TABLE 1. The minimum inhibitory concentration (MIC) distribution of 18 antimicrobial agents for the 69 CNSAb isolates as determined by E test.
Detection of Antibiotic Resistance Determinants
Phenotypic detection of MBL showed that none of the isolates were positive for MBL. The presence of blaOXA51-like and blaADC were confirmed in all CNSAb isolates. The high prevalence of blaOXA-23-like (53; 77%) and blaPER-1 (37; 54%) were seen in CNSAb isolates. Co-existence of blaOXA-23-like/blaOXA-40-like and blaOXA-23-like/blaOXA-58-like genes were detected in 6 (9%) and 2 (3%) of the isolates, respectively. The PCR results were negative for the other β-lactamase encoding genes including blaOXA-143-like, bla MBL (blaIMP-1, blaV IM-2, blaGIM-1, blaSPM-1 blaSIM-1 and blaNDM-), blaCTX-M, blaKPC, blaGES blaTEM and blaSHV -1. ISAba1 was detected upstream of blaADC, blaOXA-23-like, and blaOXA51-like genes in 67 (97%), 29(42%), and 18 (26%) of CNSAb isolates, respectively (Figure 1). A significant correlation was observed between the presence of ISAba1 in upstream of blaOXA-23-like and increasing MIC of imipenem (MIC ≥ 16 mg/L) (P = 0.042). In the presence of ISAba1 in upstream of blaOXA-51-like and increasing imipenem MICs, there was no significant correlation (P = 0.347).
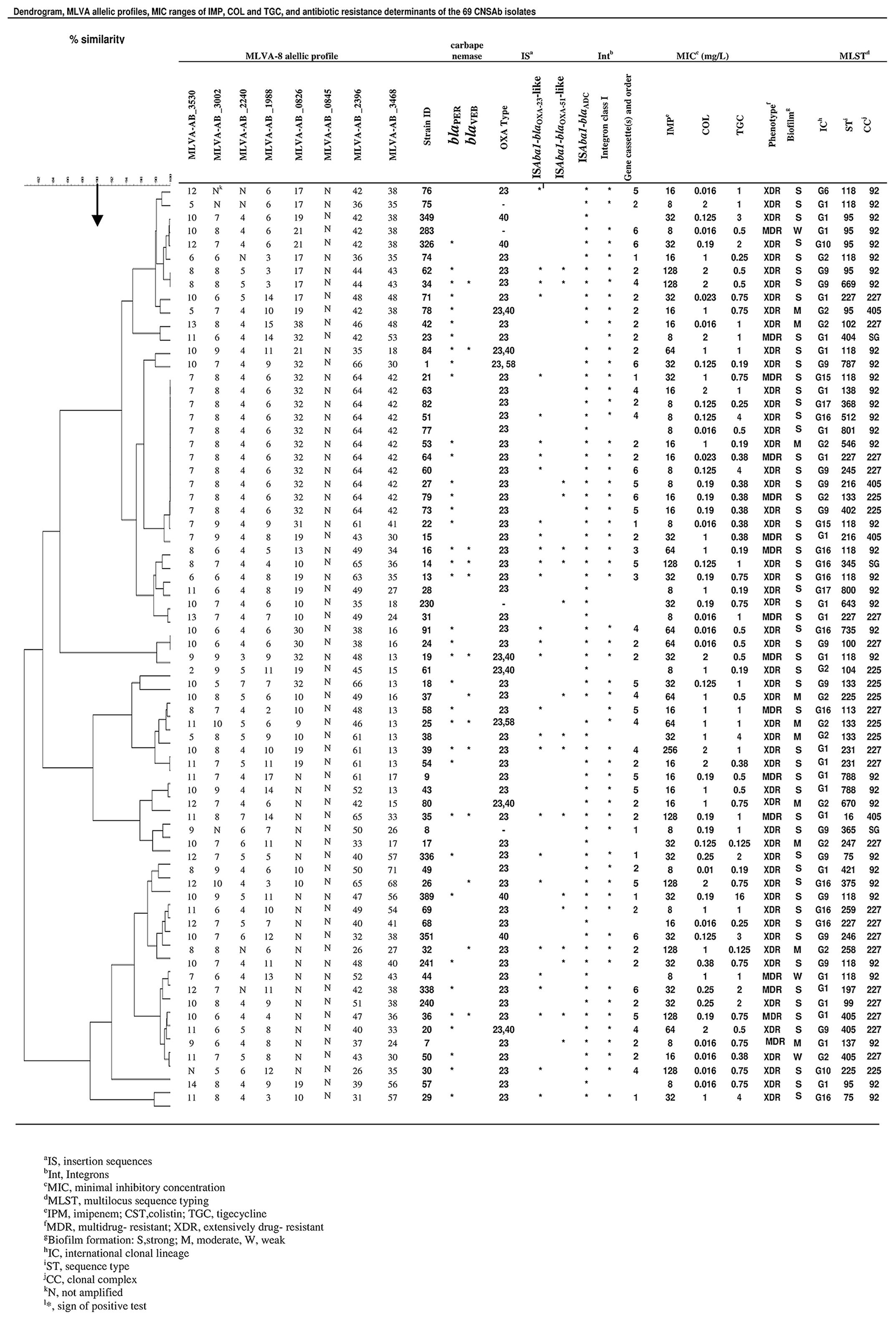
FIGURE 1. Dendogram shows the genetic diversity of 69 carbapenem non-susceptible Acinetobacter baumannii isolates by MLVA and MLST; carbapenemase encoding genes; MIC ranges of colisitin, imipenem, and tigecycline; resistance phenotype; biofilm formation and international clonal lineage.
Class 1 integron gene cassettes were identified in 58 (84%) CNSAb isolates while class 2 and 3 integrons were not found. Six different cassette combinations were detected within the class 1 integrons. The most prevalent cassette combination, aacA4-catB8-aadA1 (2.3 Kb), were detected in 23 (39%) of class 1 integrons. The other cassette combinations including arr3-aacA4 (1.3 Kb), aacC1-orfX-orfX’-aadA1 (2.5 Kb), orfI-aadA1 (1.5 Kb), dfrVII (0.7 Kb) and dfrXII-orfF-aadA2 (1.7 Kb) were detected in 10 (14.5%), 9 (13%), 7 (10%), 7 (10%), and 2 (3%) CNSAb isolates, respectively. The gene cassette arrangement showed that the class 1 integrons harbor genes encoding resistance to aminoglycosides, chloramphenicol, rifampicin and trimethoprim/sulfamethoxazole. Also, the results of this study show that integron-associated β-lactamases encoding genes were not detected in class 1 integrons-harboring CNSAb isolates.
Genetic Diversity and Population Structure
Three-locus dual assay multiplex PCR showed eight different PCR-based groups (G1, G2, G6, G9, G10, and G15–G17) among the 69 CNSAb isolates. Twenty-five (36%) and 13 (19%) CNSAb isolates belonged to G1 (IC II) and G2 (IC I), respectively. Based on the variations in combination of PCR amplicons of the blaOXA-51-like, csuE and ompA genes, 31(45%) of the CNSAb isolates belonged to 6 IC variants PCR-based groups (Table 2). G9 (14; 45%) was the most common IC variant (Table 2).

TABLE 2. Combinations of amplicons obtained in the dual multiplex PCRs used to describe 31 A. baumannii IC variants.
The MSTree algorithm of the MLST data of this study suggests that 40 previously known STs combined into four CC (Figure 2). Thirty-six (52%) and 13 (19%) of all 69 CNSAb isolates belonging to CC-92 and CC-405, respectively. According to the MSTree algorithm, only one singleton ST (ST345) was identified (Figure 2). ST118 was the predominant ST, comprising 17.3% of isolates.
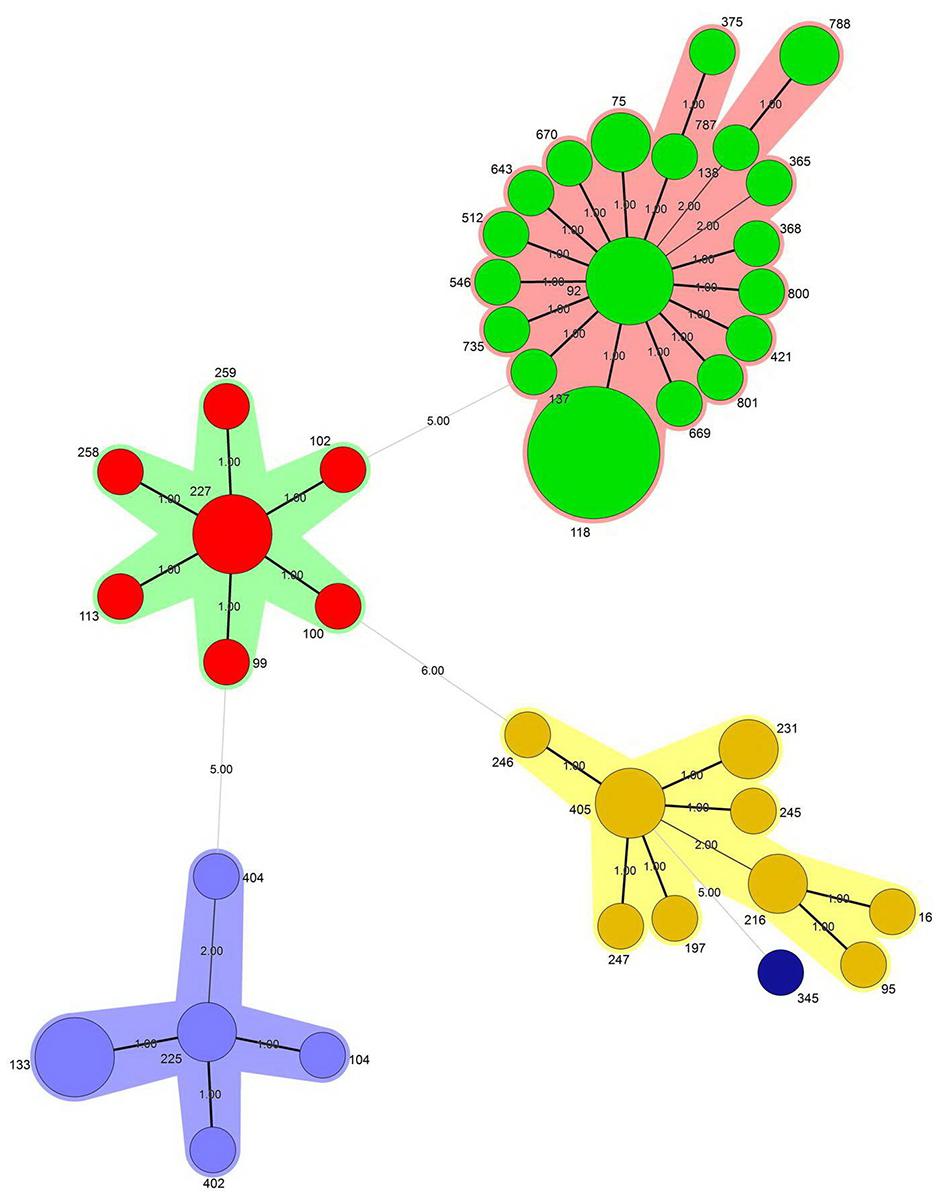
FIGURE 2. The minimum spanning tree (MStree) was constructed using a categorical coefficient based on multilocus sequence typing (MLST) data. Each circle stands for a different sequence type (ST). The color of a circle and the line clustering the ST with the same color represents an identical ST. Size of the circle corresponds to the number of isolates. The width of the line reflects the genetic distance between ST (heavy and thin lines represent a difference in one allele and two or more alleles, respectively).
By the MLVA typing method, the 69 CNSAb isolates were grouped into 56 distinct MLVA types with 6 clusters and 53 singleton genotypes. Loci MLVA-AB_0845 was absent (N) in all studied isolates. In this study, the VNTR loci MLVA-AB_3002 and MLVA-AB_2240 displayed lower diversity, whereas VNTR loci MLVA-AB_2396 and MLVA-AB_3468 showed higher level of diversity.
Thirty STs corresponded to 30 MLVA types; other STs (118, 95, 75, 227, 405, 231, 788, 225, 227, and 133) included heterogeneous MLVA profiles (Figure 1).
In this study, IC variants were classified into 23 STs belonging to CC92, CC405, CC227, CC225 and one singleton (Figure 1).
Relationship between IC Lineages with Antibiotic Resistance Determinants
Table 3 shows the frequency of antibiotics resistance in two major epidemic lineages and IC variants. Susceptibility to all antimicrobial agents, except tobramicin, in isolates belonged to IC II was higher than the IC I and IC variants (Table 3). It is noteworthy that the IC variants had the highest resistance rate to tigecycline (7; 22.5%) with MIC50 and MIC90 of 0.25 and 3 mg/L, respectively (Tables 1 and 3). Among the 13 carbapenem non-susceptible IC I, 12 (92%) were XDR while this rate was 14 (56%) and 27 (87%) in the isolates belonging to the IC variants and IC II, respectively. The resistance to beta lactam antibiotics in IC I and IC variant isolates was higher than that in IC II. Thirty-one (100%) IC variants and 22 (88%) of the IC II population formed strong biofilm but only 3 (23%) of IC I had this characteristic. In the present study, resistance to antimicrobials was comparatively higher among IC variant than IC I and IC II CNSAb isolates (P = 0.0012 and P = 0.0001, respectively). A slightly higher susceptibility to antimicrobials was observed in IC II than IC variants and IC I (P > 0.05). Our analysis revealed a significant correlation (P = 0.003) between the presence of blaPER1and biofilm formation in the CNSAb isolates. The CNSAb isolates belonging to all clones had the blaOXA-23-like gene. All CNSAb isolates classified in IC I, were positive for the blaOXA-23-like gene (Table 4). According to Table 4, the frequency of ISAba1/blaOXA-23-like and blaPER-1 genes in a population belonging to IC variants were significantly higher than IC I and IC II (P = 0.027 and P = 0.003, respectively). Table 4 shows that the distribution of different types of class 1 integrons is related to major epidemic lineages with predominance of class 1 integrons belonging to type 2.
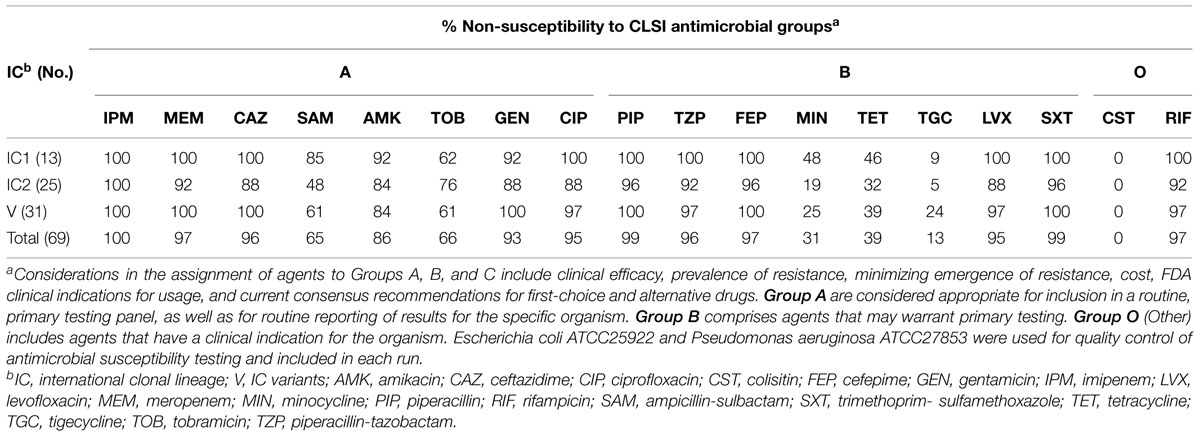
TABLE 3. Frequency of antimicrobial non-susceptibility in three major epidemic lineages in the 69 CNSAb isolates.
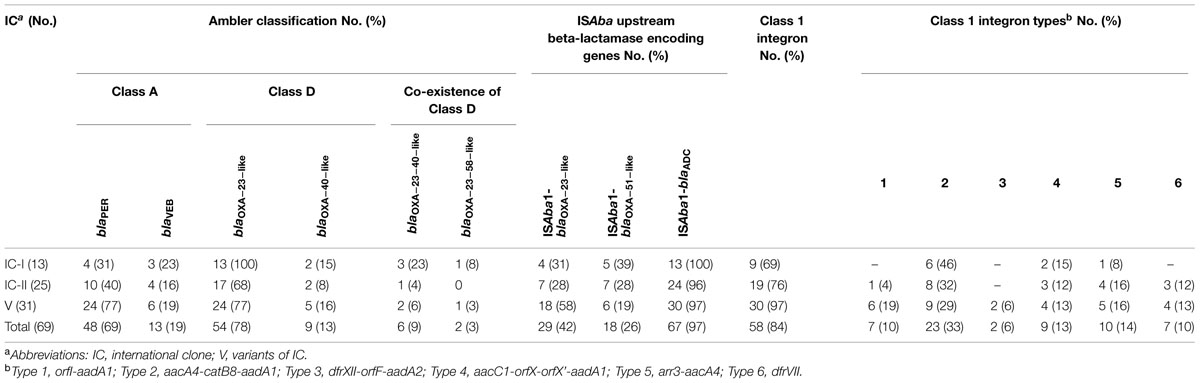
TABLE 4. Frequency of antimicrobials resistance determinants in epidemic clonal lineages of the CNSAb isolates.
Discussion
Nosocomial infections caused by CNSAb are currently among the most difficult to treat. The control of CNSAb infections among hospitalized patients continues to present serious challenges to patient management in developing countries like Iran (Diancourt et al., 2010; Giannouli et al., 2010; Bahador et al., 2013). The main challenge remains choosing the most likely effective antibiotics as determined by in vitro testing of antibiotics, based on the local susceptibility patterns (Fishbain and Peleg, 2010). Overall, the findings of this study are consistent with recent studies from Iran that show an alarming trend of CNSAb increase and in MDR A. baumannii resistance against a wide spectrum of antimicrobial agents (Moradi et al., 2015). However, in the present study colistin, tigecycline, and tetracyclines exhibited a potent activity against CNSAb isolates. Our data revealed a decrease (10%) of resistance in CNSAb isolates to tigecycline (Bahador et al., 2013), likely due to the implementation of restriction policy on the empirical prescription of tigecycline. Decrease in antibiotic resistance after implementation of restriction policies have been shown in several reports (Altunsoy et al., 2011; Sistanizad et al., 2013). Our finding of positive association in strong biofilm formation and resistance to antimicrobial is consistent with a recent report (Badave and Kulkarni, 2015). This could be due to inadequate penetration of the antimicrobials into the biofilms, and the emergence of persister cells in the biofilms (Penesyan et al., 2015).
The findings of our study on ISAba1 elements upstream of blaOXA genes and their role as a promoter to enhance OXA–enzymes expression and MIC of imipenem confirms previous reports (Liu et al., 2014a); however, this finding are in contrast to a recent report by Peymani et al. (2012b) from Iran. Not only did our research find that 42% of CNSAb isolates were the ISAba1-blaOXA-23-like gene; but also, a significant correlation was observed between the presence of ISAba1 upstream of blaOXA-23-like gene and increasing imipenem MICs (≥16 mg/L). This dissimilarity might be due to differences in the A. baumannii strains available for study. Additionally, our finding of high prevalence of ISAba1 sequences upstream of the blaADC among 97% of CNSAb isolates is consistent with previous report from Iran; and indicate that presence of ISAba1 sequences upstream of blaADC played an important role in increasing resistance to cephalosporins (MIC50 = 128 mg/L).
However, IC II is the most common strain in the world (Liu et al., 2014a); and majority of the clinical strains (45%) in this study belong to IC variants. The incidence rate of IC variants observed in this study is similar to previously reported rates of 46% from Sweden and 51% from Iran, and is considerably higher than that in other recent reports, including Iran (3%), Latvia (6%), Romania (15%), Italy (23%) and Norway (28%) (Fishbain and Peleg, 2010; Hojabri et al., 2014; Liu et al., 2014a). With regard to a previous study (Fishbain and Peleg, 2010) and the study of Karah et al. (2012), 14 CNSAb isolates showed a new combination of amplicons and were considered in this article as the PCR-based group (G) 15–17. Three of the 6 PCR-based groups in the present study, namely G6, G9, and G10, have been documented in previous studies (Karah et al., 2012; Bahador et al., 2014). Although there are differences among the IC variant types in this study and previous studies in Iran (Peymani et al., 2012b; Rezaee et al., 2013; Hojabri et al., 2014), the IC variants are highly resistant to antibiotics, which suggests that it is conceivable that the MDR phenotype has substantially contributed to their spread in Iran (Bahador et al., 2014).
The results of this study indicate that, in spite of the high rate of antibiotic resistance determinants in the IC I stains in comparison with the IC variant strains, the IC I strains had lower prevalence. This dissimilarity might be due to the higher frequency of strong biofilm formers in the IC variant strains than in the IC I strains. Several studies proved that, cell adhesiveness and ability to form biofilm were higher in the blaPER-1 producing isolates (Longo et al., 2014), and this is in agreement with the result of this study. The capability to form biofilm and presence of the blaPER gene were significantly higher in IC variants. In this study, the IC II had slightly higher susceptibility to antimicrobial agents than other clonal lineage population. This might be due to low-frequency of carbapenem-hydrolyzing class D β-lactamase (CHDL) genes and ISAba1 elements, as well as low ability of biofilm formation in IC II strain as compared to IC I and IC variant stains.
It has been clearly shown that, the high prevalence of class 1 integrons among MDR A. baumannii clinical isolates is responsible for several nosocomial outbreaks globally (Liu et al., 2014b). The resistance rates to all the tested antibiotics, except colistin, tigecycline, minocycline, and tetracycline, of the collected CNSAb isolates were >65%. However, the presence of integron cassette arrays cannot be responsible for all these resistance phenotypes. In Iran, class 1 integrons have been reported previously in >11% of A. baumannii isolates (Peymani et al., 2012a; Japoni-Nejad et al., 2013). In the current study, 84% of isolates contain class 1 integrons, but no class 2 and class 3 integrons was detected. This result shows that class 2 and class 3 integrons are not the major resistant determinants in CNSAb isolates in this study, which is consistent with previous studies (Peymani et al., 2012a; Japoni-Nejad et al., 2013).
Only six categories of gene cassettes, including aminoglycoside-modifying enzymes, chloramphenicol-resistant, trimethoprim/sulfamethoxazole-resistant, and rifampicin-resistant were detected in spite of previous studies reporting integrons carrying carbepenemase genes (Japoni-Nejad et al., 2013). This result indicates that acquisitions of carbapenem-hydrolyzing β-lactamase genes plays more important role than the class 1 integrons in carbapenem resistance in CNSAb. The class 1 integrons rate (84%) here defined for a referral burn center in central Iran is higher than the rates found in other geographical regions, including Western Iran (11%) (Salimizand et al., 2014), China (74%) (Wu et al., 2012) and European countries in general (43%) (Castanheira et al., 2014). This result is similar to that of previous studies from the Northwest and center of Iran (Peymani et al., 2012a; Japoni-Nejad et al., 2013). From literature survey, this is the first study to report carriage of class 1 integrons and associated arrays in A. baumannii isolates from burn patients in Iran.
Analysis of the integron cassette arrays shows that most class 1 integrons include the globally distributed aacA4-catB8-aadA1 array (Liu et al., 2014a), and the cassette array aacC1-orfX-orfX’-aadA1 has been documented in previous studies in Iran (Japoni-Nejad et al., 2013). Moreover, gene cassettes encoding aminoglycoside- resistance are present in the majority of CNSAb isolates in this study, suggesting a close relationship between high-level resistance rate to aminoglycoside compounds such as gentamicin and amikacin and the A. baumannii isolates. The different types of gene cassette array in this study and previous studies in Iran (Japoni-Nejad et al., 2013; Salimizand et al., 2014), suggests that a geographical feature plays a major role in MDR isolates formation, and it could be due to different volumes and patterns of antibiotic consumption in distinct areas. In addition, the results of this study indicate that strains indistinguishable from each other by MLST and MLVA typing can have different cassette arrays, indicating that some of these may be transmitted very efficiently. In this study, amikacin resistance recorded in 10% of the CNSAb isolates was not cassette-encoded. Analysis of the integron cassette arrays show that 50% of these isolates harbor an integron without aminoglycoside resistance genes. In the cases where the class 1 integrons could not be amplified, there were no integrons and alteration of the primer binding site or a large size of the Integron are the most likely explanations for the negative result of PCRs.
According to the findings of this research, MLVA could identify closely epidemiologically related isolates clustered by MLST in some cases (Figure 1). In this study, isolates with the same ST, such as ST118, have closely related MLVA type (MTs) (Figure 1). This result suggests that the evaluation of MLVA in CNSAb isolates shows its great ability for discrimination of genetically closely related isolates.
In agreement with previous studies (Hojabri et al., 2014), the present results substantiated that CC92 represented the recent widely distributed clone in Iran, accounting for 52.2% of CNSAb isolates examined in this study. This most likely indicates that the isolates belonging to CC92 might benefit with respect to acquiring resistance determinants and surviving in the health care environment, when encountering selection by antibiotics such as carbapenems. The international spread of the CC92 isolates has been reported from all around the world (Karah et al., 2012). This study revealed that the 28 isolates belonging to this lineage were shown to be heterogeneous by MLVA (22 MTs), followed by MLST (13 STs). The 69 CNSAb isolates were shown to be heterogeneous by MLVA (56 MTs) and MLST (40 STs). Taken together, genetic diversity and wide dissemination clones among CNSAb isolates is consistent with recent studies from Iran that show high diversity of STs in A. baumannii isolates from burned patients (Hojabri et al., 2014), which are the evidence of dynamic population structure and evolution in progress. The referral of patients from other medical centers of Iran to the hospital in Tehran could be among the possible reasons for this diversity and failure to colonize a specific type. Spread of specific clonally related isolates could have been prevented by the success of infection control programs in the teaching hospital in Iran.
Conclusion
Evidence that XDR-CNSAb from burned patients in Iran is rapidly changing toward growing resistance to various antimicrobials, including tigecycline was presented. Despite the increasing resistance to several first-line antimicrobials, all resistant CNSAb isolates remain sensitive to the antimicrobial colistin, a viable agent in controlling XDR-CNSAb outbreaks, especially in developing countries. This is the first study to perform IC analysis and gene cassette mapping of class integrons of A. baumannii, obtained from burned patients in Iran. It can be confirmed that the dominant local IC is an IC variant, with the likelihood that G9 is the predominant member. In the current study, the aacA4-catB8-aadA1 gene cassette array which confers resistance to aminoglycoside and chloramphenicol was identified as the predominant cassette array of class 1 integrons in CNSAb. This study described the widest distribution of blaOXA-23-like-horbering IC ST118 (CC92) and ST405 (CC405) were the principal reason for the rapid increase in the carbapenem resistance rate in burned patients in Iran. The findings highlight the importance of an international drug resistance and molecular epidemiology monitoring network of A. baumannii isolates to effect global control measures against XDR-CNSAb.
Funding
This research has been supported by Tehran University of Medical Sciences and health Services grant No. 92- 03-30-23186.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors would like to thank the dedicated laboratory personnel at the Shahid Motahari hospital (Iran University of Medical Sciences Complex), who helped collect specimens for this study.
References
Alaghehbandan, R., Azimi, L., and Rastegar Lari, A. (2012). Nosocomial infections among burn patients in Teheran, Iran: a decade later. Ann. Burns Fire Disasters 25, 3–7.
Almeida, L. A., and Araujo, R. (2013). Highlights on molecular identification of closely related species. Infect. Genet. Evol. 13, 67–75. doi: 10.1016/j.meegid
Altunsoy, A., Aypak, C., Azap, A., Ergönül,Ö, and Balik, I. (2011). The impact of a nationwide antibiotic restriction program on antibiotic usage and resistance against nosocomial pathogens in Turkey. Int. J. Med. Sci. 8, 339–344.
Azimi, L., Talebi, M., Pourshafie, M. R., Owlia, P., and Rastegar Lari, A. (2015). Characterization of carbapenemases in extensively drug resistance Acinetobacter baumannii in a burn care center in Iran. Int. J. Mol. Cell Med. 4, 46–53.
Badave, G. K., and Kulkarni, D. (2015). Biofilm producing multidrug resistant Acinetobacter baumannii: an emerging challenge. J. Clin. Diagn. Res. 9, 8–10. doi: 10.7860/JCDR/2015/11014.5398
Bahador, A., Raoofian, R., Taheri, M., Pourakbari, B., Hashemizadeh, Z., and Hashemi, F. B. (2014). Multidrug resistance among Acinetobacter baumannii isolates from Iran: changes in antimicrobial susceptibility patterns and genotypic profile. Microb. Drug Resist. 20, 632–640. doi: 10.1089/mdr.2013.0146
Bahador, A., Taheri, M., Pourakbari, B., Hashemizadeh, Z., Rostami, H., Mansoori, N., et al. (2013). Emergence of rifampicin, tigecycline, and colistin-resistant Acinetobacter baumannii in Iran; spreading of MDR strains of novel International Clone variants. Microb. Drug Resist. 19, 397–406. doi: 10.1089/mdr.2012.0233
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A., Wisplinghoff, H., and Rodríguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005
Bonnin, R. A., Nordmann, P., Potron, A., Lecuyer, H., Zahar, J. R., and Poirel, L. (2011a). Carbapenem-hydrolyzing ges-type extended-spectrum β-Lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 349–354. doi: 10.1128/AAC.00773-10
Bonnin, R. A., Potron, A., Poirel, L., Lecuyer, H., Neri, R., and Nordmann, P. (2011b). PER-7, an extended-spectrum β-lactamase with increased activity toward broad-Spectrum cephalosporins in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 2424–2427. doi: 10.1128/AAC.01795-10
Bou, G., and Martinez-Beltran, J. (2000). Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44, 428–432. doi: 10.1128/AAC.44.2.428-432.2000
Cai, Y., Chai, D., Wang, R., Liang, B., and Bai, N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615. doi: 10.1093/jac/dks084
Castanheira, M., Costello, S. E., Woosley, L. N., Deshpande, L. M., Davies, T. A., and Jones, R. N. (2014). Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and Enterobacteriaceae isolates collected in European and Mediterranean countries and detection of two novel β-lactamases, GES-22 and VIM-35. Antimicrob. Agents Chemother. 58, 7358–7366. doi: 10.1128/AAC.03930-14
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
Climaco, E. C., Oliveira, M. L., Pitondo-Silva, A., Oliveira, M. G., Medeiros, M., Lincopan, N., et al. (2013). Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect. Genet. Evol. 19, 127–133. doi: 10.1016/j.meegid.2013.06.024
Clinical and Laboratory Standards Institute [CLSI] (2014). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty fourth Informational Supplement M100-S24CLSI. Wayne, PA: CLSI.
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 5:e10034. doi: 10.1371/journal.pone.0010034
Dillon, B., Thomas, L., Mohmand, G., Zelynski, A., and Iredell, J. (2005). Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods 62, 221–232. doi: 10.1016/j.mimet.2005.02.007
Ellington, M. J., Kistler, J., Livermore, D. M., and Woodford, N. (2007). Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59, 321–322. doi: 10.1093/jac/dkl481
Fishbain, J., and Peleg, A. Y. (2010). Treatment of Acinetobacter infections. Clin. Infect. Dis. 51, 79–84. doi: 10.1086/653120
Giannouli, M., Cuccurullo, S., Crivaro, V., Di Popolo, A., Bernardo, M., Tomasone, F., et al. (2010). Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J. Clin. Microbiol. 48, 1223–1230. doi: 10.1128/JCM.02263-09
Girerd-Genessay, I., Bénet, T., and Vanhems, P. (2015). Multidrug-resistant bacterial outbreaks in burn units: a synthesis of the literature according to the orion statement. J. Burn Care Res. [Epub ahead of print]
Heritier, C., Poirel, L., and Nordmann, P. (2006). Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12, 123–130. doi: 10.1111/j.1469-0691.2005.01320.x
Higgins, P. G., Lehmann, M., and Seifert, H. (2010a). Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 35:305. doi: 10.1016/j.ijantimicag.2009.10.014
Higgins, P. G., Lehmann, M., Wisplinghoff, H., and Seifert, H. (2010b). gyrB Multiplex PCR To differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J. Clin. Microbiol. 48, 4592–4594. doi: 10.1128/JCM.01765-10
Hojabri, Z., Pajand, O., Bonura, C., Aleo, A., Giammanco, A., and Mammina, C. (2014). Molecular epidemiology of Acinetobacter baumannii in Iran: endemic and epidemic spread of multiresistant isolates. J. Antimicrob. Chemother. 69, 2383–2387. doi: 10.1093/jac/dku045
Japoni-Nejad, A., Farshad, S., van Belkum, A., and Ghaznavi-Rad, E. (2013). Novel cassette array in a class 1 integron in clinical isolates of Acinetobacter baumannii from central Iran. Int. J. Med. Microbiol. 303, 645–650. doi: 10.1016/j.ijmm.2013.09.005
Jeannot, K., Diancourt, L., Vaux, S., Thouverez, M., Ribeiro, A., Coignard, B., et al. (2014). Molecular epidemiology of carbapenem non-susceptible Acinetobacter baumannii in France. PLoS ONE 9:e115452. doi: 10.1371/journal.pone.0115452
Karah, N., Sundsfjord, A., Towner, K., and Samuelsen, Ø (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001
Liu, C. C., Tang, C. Y., Chang, K. C., Kuo, H. Y., and Liou, M. L. (2014a). A comparative study of class 1 integrons in Acinetobacter baumannii. Gene 544, 75–82. doi: 10.1016/j.gene.2014.04.047
Liu, C. P., Shih, S. C., Wang, N. Y., Wu, A. Y., Sun, F. J., Chow, S. F., et al. (2014b). Risk factors of mortality in patients with carbapenem-resistant Acinetobacter baumannii bacteremia. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2014.10.006 [Epub ahead of print].
Longo, F., Vuotto, C., and Donelli, G. (2014). Biofilm formation in Acinetobacter baumannii. N. Microbiol. 37, 119–127.
Magiorakos, A. P., Srinivasan, A., Carey, R., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrugresistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691
Mahdian, S., Sadeghifard, N., Pakzad, I., Ghanbari, F., Soroush, S., Azimi, L., et al. (2015). Acinetobacter baumannii clonal lineages I and II harboring different carbapenem-hydrolyzing-β-lactamase genes are widespread among hospitalized burn patients in Tehran. J. Infect. Public Health doi: 10.1016/j.jiph.2015.04.030 [Epub ahead of print].
Moradi, J., Hashemi, F. B., and Bahador, A. (2015). Antibiotic resistance of Acinetobacter baumannii in Iran: a systemic review of the published literature. Osong Public Health Res. Perspect. 6, 79–86. doi: 10.1016/j.phrp.2014.12.006
Nhu, N. T., Lan, N. P., Campbell, J. I., Parry, C. M., Thompson, C., Tuyen, H. T., et al. (2014). Emergence of carbapenem-resistant Acinetobacter baumannii as the major cause of ventilator-associated pneumonia in intensive care unit patients at an infectious disease hospital in southern Vietnam. J. Med. Microbiol. 63, 1386–1394. doi: 10.1099/jmm.0.076646-0
Öncül, O., Öksüz, S., Acar, A., Ülkür, E., Turhan, V., Uygur, F., et al. (2014). Nosocomial infection characteristics in a burn intensive care unit: analysis of an eleven-year active surveillance. Burns 40, 835–841. doi: 10.1016/j.burns
Penesyan, A., Gillings, M., and Paulsen, I. T. (2015). Antibiotic discovery: combatting bacterial resistance in cells, and in biofilm communities. Molecules 20, 5286–5298. doi: 10.3390/molecules20045286
Peymani, A., Farajnia, S., Nahaei, M. R., Sohrabi, N., Abbasi, L., Ansarin, K., et al. (2012a). Prevalence of class 1 integron among multidrug-resistant Acinetobacter baumannii in Tabriz, northwest of Iran. Pol. J. Microbiol. 61, 57–60.
Peymani, A., Higgins, P. G., Nahaei, M. R., Farajnia, S., and Seifert, H. (2012b). Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, northwest Iran. Int. J. Antimicrob. Agents 39, 526–528. doi: 10.1016/j.ijantimicag
Poirel, L., Menuteau, O., Agoli, N., Cattoen, C., and Nordmann, P. (2003). Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41, 3542–3547. doi: 10.1128/JCM.41.8.3542-3547.2003
Potron, A., Munoz-Price, L. S., Nordmann, P., Cleary, T., and Poirel, L. (2011). Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob. Agents Chemother. 55, 5946–5948. doi: 10.1128/AAC.05124-11
Pourcel, C., Minandri, F., Hauck, Y., D’Arezzo, S., Imperi, F., Vergnaud, G., et al. (2011). Identification of variable-number tandem-repeat (VNTR) sequences in Acinetobacter baumannii and interlaboratory validation of an optimized multiple-locus VNTR analysis typing scheme. J. Clin. Microbiol. 49, 539–548. doi: 10.1128/JCM.02003-10
Rezaee, M. A., Pajand, O., Nahaei, M. R., Mahdian, R., Aghazadeh, M., Ghojazadeh, M., et al. (2013). Prevalence of Ambler class A β-lactamases and ampC expression in cephalosporin-resistant isolates of Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 76, 330–334. doi: 10.1016/j.diagmicrobio
Robledo, I. E., Aquino, E. E., and Vazquez, G. J. (2011). Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 55, 2968–2970. doi: 10.1128/AAC.01633-10
Ruan, Z., Chen, Y., Jiang, Y., Zhou, H., Zhou, Z., Fu, Y., et al. (2013). Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int. J. Antimicrob. Agents 42, 322–328. doi: 10.1016/j.ijantimicag
Salimizand, H., Menbari, S., Ramazanzadeh, R., Khonsha, M., and Vahedi, M. S. (2014). DNA fingerprinting and antimicrobial susceptibility pattern of clinical and environmental Acinetobacter baumannii isolates: a multicentre study. J. Chemother. [Epub ahead of print].
Sistanizad, M., Kouchek, M., Miri, M., Goharani, R., Solouki, M., Ayazkhoo, L., et al. (2013). Carbapenem restriction and its effect on bacterial resistance in an intensive care unit of a teaching hospital. Iran J. Pharm. Res. 12, 503–509.
Turton, J. F., Gabriel, S. N., Valderrey, C., Kaufmann, M. E., and Pitt, T. L. (2007). Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13, 807–815. doi: 10.1111/j.1469-0691.2007.01759.x
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/j.1574-6968.2006.00195.x
Woodford, N., Ellington, M. J., Coelho, J. M., Turton, J. F., Ward, M. E., Brown, S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27, 351–353. doi: 10.1016/j.ijantimicag.2006.01.004
Wu, K., Wang, F., Sun, J., Wang, Q., Chen, Q., Yu, S., et al. (2012). Class 1 integron gene cassettes in multidrug-resistant Gram-negative bacteria in southern China. Int. J. Antimicrob. Agents 40, 264–267. doi: 10.1016/j.ijantimicag
Zhang, D., Xia, J., Xu, Y., Gong, M., Zhou, Y., Xie, L., et al. (2014). Biological features of biofilm-forming ability of Acinetobacter baumannii strains derived from 121 elderly patients with hospital-acquired pneumonia. Clin. Exp. Med. [Epub ahead of print].
Zowawi, H. M., Sartor, A. L., Sidjabat, H. E., Balkhy, H. H., Walsh, T. R., Al Johani, S. M., et al. (2015). Molecular epidemiology of carbapenem resistant Acinetobacter baumannii in the gulf cooperation council states. Dominance of OXA-23-type producers. J. Clin. Microbiol. 53, 896–903. doi: 10.1128/JCM.02784-14
Keywords: antimicrobial resistance, MLST, MLVA, international clone, integron
Citation: Farshadzadeh Z, Hashemi FB, Rahimi S, Pourakbari B, Esmaeili D, Haghighi MA, Majidpour A, Shojaa S, Rahmani M, Gharesi S, Aziemzadeh M and Bahador A (2015) Wide distribution of carbapenem resistant Acinetobacter baumannii in burns patients in Iran. Front. Microbiol. 6:1146. doi: 10.3389/fmicb.2015.01146
Received: 18 July 2015; Accepted: 05 October 2015;
Published: 20 October 2015.
Edited by:
Margaret Ip, Chinese University of Hong Kong, Hong KongReviewed by:
Benjamin Andrew Evans, University of Edinburgh, UKHenrietta Venter, University of South Australia, Australia
Copyright © 2015 Farshadzadeh, Hashemi, Rahimi, Pourakbari, Esmaeili, Haghighi, Majidpour, Shojaa, Rahmani, Gharesi, Aziemzadeh and Bahador. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abbas Bahador, abahador@tums.ac.ir; ab.bahador@gmail.com
 Zahra Farshadzadeh1
Zahra Farshadzadeh1 Farhad B. Hashemi
Farhad B. Hashemi Mohammad A. Haghighi
Mohammad A. Haghighi Abbas Bahador
Abbas Bahador