- 1Laboratory of Microbiology, Wageningen University, Wageningen, Netherlands
- 2Centre of Biological Engineering, University of Minho, Braga, Portugal
Carbon monoxide can act as a substrate for different modes of fermentative anaerobic metabolism. The trait of utilizing CO is spread among a diverse group of microorganisms, including members of bacteria as well as archaea. Over the last decade this metabolism has gained interest due to the potential of converting CO-rich gas, such as synthesis gas, into bio-based products. Three main types of fermentative CO metabolism can be distinguished: hydrogenogenesis, methanogenesis, and acetogenesis, generating hydrogen, methane and acetate, respectively. Here, we review the current knowledge on these three variants of microbial CO metabolism with an emphasis on the potential enzymatic routes and bio-energetics involved.
Introduction
Carbon monoxide is a tasteless, odorless gas, best known for its toxic characteristics. It is part of the global carbon cycle, is involved in assimilatory and dissimilatory pathways of numerous microorganisms and was found to act as a signal molecule in mammals (Ryter and Otterbein, 2004). Additionally, it is speculated to be an important substrate for early life on earth (Miyakawa et al., 2002). CO is chemically formed during oxygen limited combustion of carbon materials, and can be biologically generated via cellular metabolism (Eikmanns et al., 1985; Ryter and Otterbein, 2004) or enzymatic degradation of heme (Chin and Otterbein, 2009). A large part of naturally generated CO is assumed to be formed via photochemical reactions (Weinstock and Niki, 1972). Other sources contributing to the atmospheric CO level are: volcanic activity, forest fires, and over the last two centuries industrial activity. Cumulative, these activities result in the production of approximately 2.6 petagram (Pg) CO per year (Khalil and Rasmussen, 1990).
Due to the foreseen depletion of fossil energy sources and consideration on environmental impact of current chemical industry, alternative sustainable technologies are being developed. A bio-based economy is considered one of the sustainable solutions for the growing resource depletion, and has potential to replace petroleum industries in the future. However, the hydrolysis of non-food-competing, ligno-cellulosic biomass limits the application of bio-based technologies, leaving a significant amount of the initial substrate unused (Hamelinck et al., 2005). Gasification of the carbohydrate material, forming syngas, is a potential way of gaining access to all material of the original source, mainly in the form of CO, H2, and CO2. A large spectrum of carbon sources can fuel this technology, from coal, tar, and gas to ligno-cellulosic biomass, all kinds of municipal waste, and digester sludge. As syngas mainly consists of CO, H2, and CO2, autotrophic, CO-tolerant microorganisms are required as biocatalysts for fermentation of this gas. Numerous microorganisms have shown to utilize CO as a substrate, producing organic compounds such as acetate, ethanol, 2,3-butanediol, and butyrate (Latif et al., 2014; Tiquia-Arashiro, 2014; Dürre and Eikmanns, 2015). In addition, methanogenesis and hydrogenogenesis using syngas as a substrate could have large implications for biofuel production. Even though production of interesting chemicals from syngas is possible, the exact metabolism of syngas conversion is not fully understood. This limits the optimization of potential production strains and, thus, the development of bio-based production processes. Different aspects of biological conversion of CO and syngas have been reviewed in the past decade (Henstra et al., 2007b; Oelgeschlager and Rother, 2008; Bengelsdorf et al., 2013; Sokolova and Lebedinsky, 2013; Latif et al., 2014; Dürre and Eikmanns, 2015). However, a large part of these reviews focus on industrially relevant species and their application potential, while only few address fundamental aspects.
Two types of CO metabolism can be distinguished: respiratory and fermentative (Table 1), the former involving an exogenous electron acceptor, whereas the latter utilizes internally generated intermediates as electron acceptor. A relatively well studied example of respiratory CO metabolism is CO oxidation coupled to oxygen reduction (Meyer and Schlegel, 1983; Meyer et al., 1986). Other, less characterized, electron acceptors that have been identified for carboxydotrophic growth are sulfate (Parshina et al., 2005a, 2010), anthroquinone disulfonate (AQDS; Henstra and Stams, 2004), fumarate (Henstra and Stams, 2004), and ferrihydrite (Slobodkin et al., 2006). The main focus of this review is on fermentative CO metabolism, distinguishing hydrogenogenesis, acetogenesis, and methanogenesis with a special emphasis on the potential enzymatic routes and bio-energetics involved. Classifying hydrogenogenesis as a fermentative process is debatable because protons can be considered exogenous acceptors. However, as protons are present in any microbial environment and are generated from the substrate water during hydrogenogenesis, hydrogenogenic metabolism is considered here as fermentative.
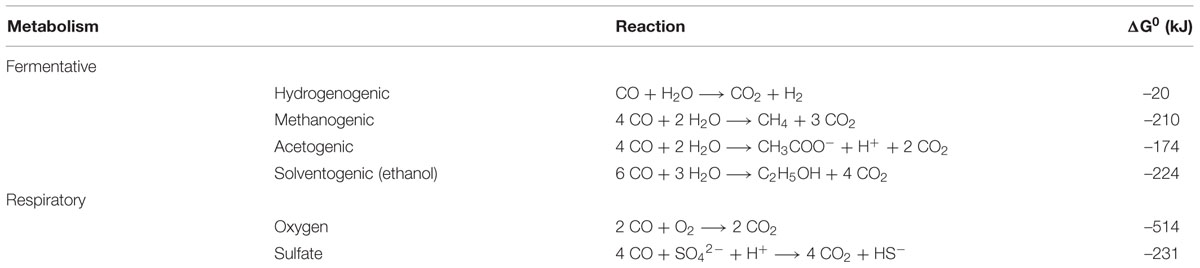
TABLE 1. Reaction equations and their standard Gibbs free energy (ΔG0) for several modes of carboxydotrophic growth.
General Microbial Co Metabolism
Both aerobic and anaerobic CO metabolism process the CO molecule via the enzyme: carbon monoxide dehydrogenase (CODH). Here, we only consider the anaerobic CODHs, which differ from the aerobic CODHs by structure and the presence of nickel–iron clusters in their active centers (Jeoung et al., 2014). About 6% of all known microbial genomes contain at least one [Ni–Fe] CODH gene sequence, out of which 43% contain at least two, suggesting a more widespread anaerobic CO-utilizing capability than assumed before (Techtmann et al., 2012). It has been shown that CODH genes cluster according to function, instead of clustering by phylogeny, suggesting horizontal gene transfer events have led to the establishment of the CODH gene in the different microbial genomes (Techtmann et al., 2012). CODHs from different organisms have been purified and characterized, including the ones from three relatively well studied anaerobic carboxydotrophic organisms: Rhodospirillum rubrum (Drennan et al., 2001), Carboxydothermus hydrogenoformans (Dobbek et al., 2001), and Moorella thermoacetica (Doukov et al., 2002; Darnault et al., 2003). All the structures of these anaerobic CODHs contain iron-sulfur center (Hu et al., 1996; Dobbek et al., 2001). Additionally, these CODHs contain nickel as a cofactor, for binding and coordinating CO in the active site (Dobbek et al., 2001; Drennan et al., 2001). Other divalent metals were found in the active center of anaerobic CODHs, however, only the nickel containing enzymes were observed to be active in CO conversion (Darnault et al., 2003). CODH can be mono- or bi-functional, both enabling the organism to utilize CO for the energy metabolism. The bifunctional CODH is associated with an acetyl-CoA synthase (ACS), and additionally has a role in carbon fixation, catalyzing the condensation of CO, CoA-SH, and a methyl-group into acetyl-CoA. The bacterial CODH and ACS are connected via a hydrophobic tunnel (Maynard and Lindahl, 1999; Seravalli and Ragsdale, 2000; Lindahl, 2002), preventing CO from being a toxic intermediate in the metabolism of the cell (Doukov et al., 2002).
The redox potential of the CO/CO2 pair (E0 = –520 mV), is lower than that of H2/H+ (E0 = –414 mV), which has significant implications for the metabolism. While a metabolism driven by hydrogen requires bifurcation mechanisms to reduce ferredoxin (E0 = –400 mV) (Buckel and Thauer, 2013), CO can solely drive this reaction. However, the more negative redox potential of CO poses a challenge for the redox balance of the organism. Therefore, efficient cofactor re-oxidizing pathways are required to avoid the cell from becoming completely reduced. Additionally, hydrogenases are considered to be a weak point in CO metabolism, as hydrogen metabolism is often observed to be rapidly inactivated upon CO exposure (Purec et al., 1962; Daniels et al., 1977; Genthner and Bryant, 1982; Bertsch and Müller, 2015). However, [Ni–Fe]-hydrogenases were found to be less sensitive to CO than [Fe–Fe] or iron-only hydrogenases (Adams, 1990b; De Lacey et al., 2007). Some microorganisms possess [Ni–Fe] hydrogenases that are highly tolerant to CO, such as Rhodospirillum rubrum (Fox et al., 1996b) and Pyrococcus furiosus (Adams, 1990a). CO is known to strongly bind to metals via a process called back bonding, which is also considered to be the mechanism of toxicity (Jeoung et al., 2014).
Co Driven Hydrogenogenic Metabolism
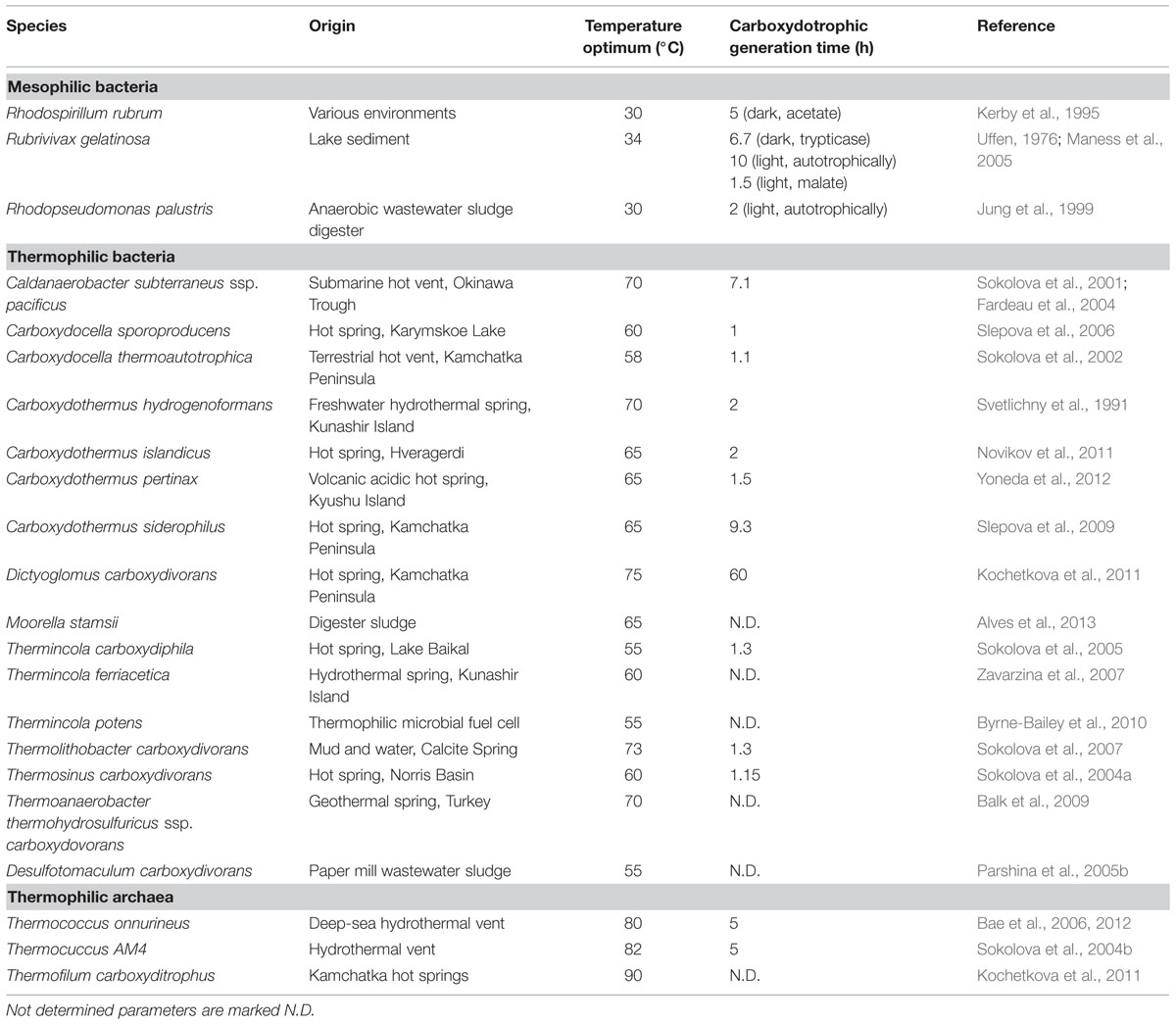
Coupling CO oxidation to proton reduction is, conceptually seen, one of the simplest mechanisms of biological energy conservation. This reaction, also known as the water–gas shift reaction, results in the formation of hydrogen and CO2 (Table 1). The reaction was found to be completed by three enzymes: CODH, an electron transfer protein and an energy converting hydrogenase (EcH). CO is oxidized via the CODH complex, and electrons are transferred to a “ferredoxin-like” electron carrier. Oxidation of this electron carrier can be coupled to proton reduction via an EcH complex, producing hydrogen and simultaneously generating an ion motive force (Hedderich and Forzi, 2004). Besides being involved in hydrogenogenic metabolism, EcH enzymes also play a role in sugar fermentation (Sapra et al., 2003) and methanogenesis (Thauer et al., 2010). Several microorganisms which hydrogenogenically metabolize CO have been isolated; most of them are thermophiles (Table 2). Two microorganisms conserving energy via the water–gas shift reaction have been rather well studied, the mesophilic Rhodospirillum rubrum (Kerby et al., 1995) and the thermophilic Carboxydothermus hydrogenoformans (Svetlichny et al., 1991).
Rhodospirillum rubrum, Rubrivivax gelatinosa, and Rhodopseudomonas palustris are photosynthetic bacteria and the only known mesophiles capable of efficiently conserving energy from the water–gas shift reaction (Table 2). In R. rubrum the CO-dependent metabolism is regulated via a heme-protein, which acts as CO sensor (CooA) and controls transcription of the enzymatic machinery required for CO dependent growth (Roberts et al., 2001). The genes controlled by CooA in R. rubrum are arranged in two gene clusters: cooFSCTJ and cooMKLXUH. The first gene cluster codes for the active CODH (cooS), electron carrier (cooF) and a nickel inserting complex (cooCTJ) (Kerby et al., 1997), whereas the latter codes for a six subunit EcH complex. The CODH structure of R. rubrum has been resolved to 2.8Å, and is similar to the CODH of anaerobes such as C. hydrogenoformans and M. thermoacetica (Dobbek et al., 2001; Drennan et al., 2001). Electrons from CO oxidation are transferred to an iron–sulfur protein (CooF), which shuttles the electrons to the EcH complex. The CooF complex is tightly associated with the CODH, and was shown to be reduced upon CO exposure (Ensign and Ludden, 1991). Other electron carriers, such as other native ferredoxins from R. rubrum, were ineffective in mediating electron transfer from CODH to the hydrogenase (Ensign and Ludden, 1991). This suggests that the CooF subunit is highly specific for electron transfer from CODH to the hydrogenase. Not only is the R. rubrum CODH efficient in converting CO to CO2, also its CO-induced hydrogenase is well adapted to CO dependent growth (Bonam et al., 1989). The EcH of R. rubrum consists of 6 subunits, of which two subunits, CooH and CooL, are similar to the ones found in some [Ni–Fe] hydrogenases. In addition, all six subunits show high similarity with complex I NADH:oxidoreductases, which are involved in proton translocation coupled to NADH oxidation (Fox et al., 1996a,b). During activity assays, the EcH is found to function optimally in presence of CODH:CooF, which is theorized to promote forming and maintaining a stable complex (Singer et al., 2006). The CO-induced hydrogenase of R. rubrum is highly CO tolerant, and only shows signs of inhibition above 60% CO in the headspace (Fox et al., 1996b). Despite the seemingly efficient water–gas shift metabolism in R. rubrum, autotrophic growth on solely CO as a carbon source is very slow (Dashekvicz and Uffen, 1979). R. rubrum requires small amounts of yeast extract and acetate as a carbon source to grow efficiently. R. gelatinosus and R. palustris exhibit a similar hydrogenogenic CO metabolism as R. rubrum. However, in contrast to R. rubrum, these bacteria were able to perform the water–gas shift reaction and grow on CO as a sole carbon source, but merely in presence of light (Jung et al., 1999; Maness et al., 2005). Growth was significantly slowed down for R. gelatinosus in the dark, which was not assessed for R. palustris. The growth rate of R. gelatinosus increased significantly after addition of malate as a carbon source (Maness et al., 2005). So, despite efficient energy conservation via the water–gas shift reaction, as shown in the presence of organic carbon sources, autotrophic growth seems very energy intensive for these phototrophic bacteria. The relatively slow growth on CO as a sole carbon source is likely due to the use of the energy demanding Calvin-cycle, which can be considered the main carbon-fixation mechanism in these phototrophic bacteria. All three isolated mesophilic, hydrogenogenic carboxydotrophs are phototrophs. However, it is unclear why the trait of hydrogenogenic CO-utilization among mesophiles is exclusive to this group.
Among thermophilic hydrogenogens, C. hydrogenoformans is one of the best studied. This bacterium was first thought to only grow fermentatively on CO or pyruvate as substrate, but was later shown to be capable of respiratory growth with CO as well (Henstra and Stams, 2004). With five different CODHs encoded in its genome, it is one of the few organisms known to have multiple CODH types, which is likely related to its exceptional growth capabilities on CO (Wu et al., 2005). C. hydrogenoformans uses a CODH–CooF–EcH complex, which is highly similar to the system found in R. rubrum (Soboh et al., 2002). In contrast to the mentioned mesophilic phototrophs, C. hydrogenoformans is capable of efficient autotrophic growth, using solely CO as energy and carbon source. This characteristic might be assigned to the presence of the Wood–Ljungdahl pathway, which in contrast to the Calvin-cycle is not as energy demanding. Additionally, a turnover rate of 31000 s-1 was found for the CODHII of C. hydrogenoformans (Svetlitchnyi et al., 2001), allowing for fast generation of reduction equivalents and thus a quick energy metabolism. Upon increased hydrogen and carbon dioxide pressure, acetate is produced from CO by C. hydrogenoformans. This suggests acetogenic use of the Wood–Ljungdahl pathway could act as a backup for its hydrogenogenic metabolism (Henstra and Stams, 2011). C. hydrogenoformans and related thermophilic species are suggested to fulfill an important role in the volcanic environments they originate from, ensuring CO concentrations are kept below toxic levels, making life of other non-CO tolerant organisms possible (Techtmann et al., 2009). Furthermore, it seems horizontal gene transfer events have played an important role in the establishment of CO-driven hydrogenogenic metabolism in these environments (Techtmann et al., 2012; Sant’Anna et al., 2015).
When assessing the distribution of isolated hydrogenogenic carboxydotrophic microorganisms (Table 2), thermophilic isolates seem to be more prevalent than mesophilic ones, which contrasts with the solubility of gaseous substrates at elevated temperatures. Temperature increase has two effects on dissolved gases: decreased gas solubility and increased gas diffusion rates. In a hydrogenogenic metabolism the microorganisms use a gaseous substrate, subsequently producing a gaseous product. The thermodynamics of this metabolism thus relies on the concentration of two gases, which is indirectly related to the diffusion rate of these gases. The Km values of the CODH in R. rubrum, C. hydrogenoformans, and the acetogen M. thermoacetica, are in the order of 0.032, 0.018, and 0.01 mM, respectively (Raybuck et al., 1988; Jeon et al., 2005; Seravalli and Ragsdale, 2008). The maximal solubility of CO in water is approximately 1.6 to 0.38 mM, in the range of 273 to 353 K, respectively. Assuming a Km of 0.03 mM and applying simple Michaelis–Menten kinetics, the associated CODH reaction rate at these CO concentrations goes from 98 to 93% of Vmax (Figure 1). This suggests that carboxydotrophic microorganisms are not significantly limited by the maximal solubility of CO at elevated temperatures. The CO diffusion coefficient, an indication of the diffusion rate of the gas, is 2.0 × 10-5 cm2/s in water at 298 K. Compared to 298 K, the estimated diffusion constant at 333 K is two times larger and three times larger at 353 K (approximated by the Stokes–Einstein equation, using the dynamic viscosity of water, Figure 1). This suggests that at increased temperatures, CO is more rapidly supplied to the microorganisms. Via the same mechanism, the temperature indirectly affects the degree of accumulation of hydrogen in the near vicinity of the microorganism, allowing two times faster removal of H2 at 333 K, and three times at 353 K. This suggests thermophilic hydrogenogenic metabolism suffers less from hydrogen accumulation to thermodynamically unfavorable levels when compared to mesophilic conditions. Therefore, we hypothesize that CO driven hydrogenogenic metabolism is more favorable at higher temperatures when compared to lower temperatures, giving rise to the currently observed temperature distribution of carboxydotrophic hydrogenogenic isolates (Table 2). If this potential advantage also translates into an increased energy yield, and thus a higher growth rate with increasing temperatures is unclear as numerous factors influence the growth rate.
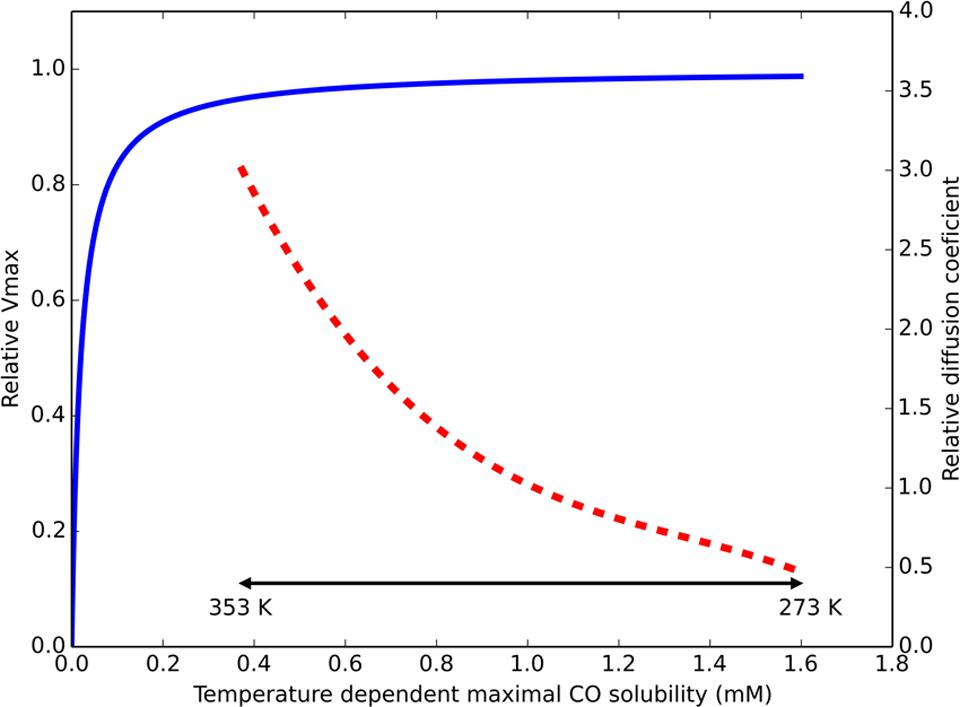
FIGURE 1. Temperature effect on parameters for CO driven hydrogenogenic growth. Estimated relative Vmax of a CO dehydrogenase (CODH), displayed by the blue continuous line, assuming Michaelis–Menten kinetics (Km = 0.03 mM) based on maximal solubilized CO. The approximated ratio of the diffusion constant for any dissolved gas (at atmospheric pressure) in water compared to its diffusion constant at 298 K is displayed by the red dashed line. The physiologically relevant temperature range is displayed by the double headed black arrow. The displayed temperature scale is not linear due to the non-linear relation between temperature and CO solubility.
Co Driven Acetogenesis and Related Metabolisms
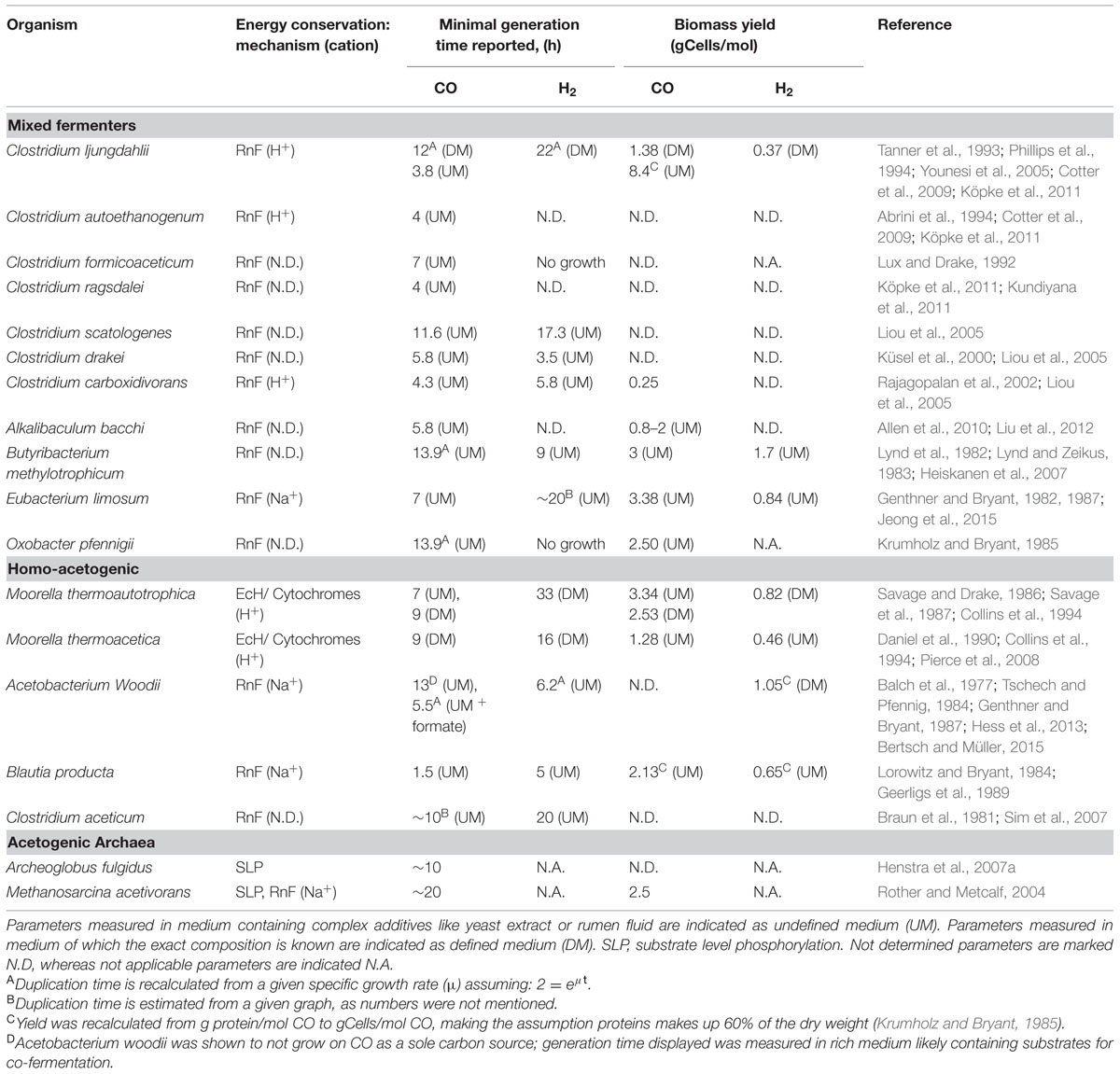
Formation of acetate from CO was first reported by Lynd et al. (1982). Since then many acetogens have been discovered to utilize CO, both homo-acetogenic organisms (i.e., generating solely acetate) and mixed-product acetogenic organisms (i.e., generating a mixture of end-products; Table 3). Large differences can be observed between the growth rates and yields of CO-grown acetogens, indicating differences in their ways of energy conservation and pathways utilized (Table 3). The Wood–Ljungdahl pathway is the central metabolism for acetogenic growth (Ragsdale and Pierce, 2008). In contrast to the Calvin-cycle, the reductive TCA-cycle or the 3-hydroxypropionate cycle, the Wood–Ljungdahl pathway can be used to conserve energy. The Wood–Ljungdahl pathway consists of two branches, which in total require eight reduction equivalents and one ATP to form acetyl-CoA from two CO2. During formation of acetate the ATP investment is regained by substrate level phosphorylation, but no net ATP is gained, thus requiring an ion motive force to conserve net energy. Complex I related RnF complexes have been identified as the cation extruding mechanism in many acetogens, linking ferredoxin oxidation to NAD reduction, simultaneously forming either a sodium or proton gradient (Biegel et al., 2011). Upon consumption of this gradient, ATP is formed from ADP and phosphate via an ATPase (Müller, 2003). The availability of ferredoxin can thus be considered the driving force of energy generation in the autotrophic acetogenic metabolism. It is assumed that in hydrogenotrophic acetogens electron-bifurcating hydrogenases are required to generate reduced ferredoxin (Poehlein et al., 2012; Buckel and Thauer, 2013). As CO can directly reduce ferredoxin, these bifurcating systems seem redundant during CO-driven growth, but might be utilized to correctly distribute reduction equivalents over the metabolism. Four steps in the Wood–Ljungdahl pathway require input of electrons for fixation of two CO2. The type of cofactors utilized in each of these steps differs per enzyme and per microorganism, making it impossible to propose a general metabolism for acetogenic CO metabolism. This distinct use of cofactors can explain part of the differences observed in yield and growth rate of different acetogens (Table 3). Recently, the energy metabolism for autotrophic growth on H2/CO2 for Acetobacterium woodii, M. thermoacetica, and Clostridium ljungdahlii was reviewed (Schuchmann and Müller, 2014). Here, we assess and compare the CO metabolism of these three species, which are the current model organisms for acetogenic metabolism.
Moorella thermoacetica is one of the best studied homo-acetogenic bacteria able to utilize CO. This organism differs from other acetogenic strains in the sense that it does not possess an RnF complex, leaving the mechanism of cation transport unknown (Pierce et al., 2008). Either an EcH complex or cytochromes are proposed to perform the build-up of an ion motive force. Several cytochromes have been found in Moorella species that are potentially active in proton transport (Gottwald et al., 1975). However, the role of these electron carrier proteins in acetogenic metabolism has never been experimentally shown. Additionally, a role of these cytochromes in respiratory metabolism is likely to exist, as growth with nitrate and nitrite was shown to be possible (Drake and Daniel, 2004). An EcH complex is coded for in the genome of M. thermoacetica (Huang et al., 2012), making cation export via this enzyme a possibility. Based on presence of an EcH complex, an energy metabolism for growth on H2/CO2 has been proposed for M. thermoacetica (Schuchmann and Müller, 2014). The plausible metabolism is based on the assumption that methylenetetrahydrofolate reductase, responsible for the exergonic reduction of methylenetetrahydrofolate to methyltetrahydrofolate, is somehow coupled to energy conservation. As all enzymes of the Wood–Ljungdahl pathway are considered soluble, none of these is expected to be involved in generation of an ion motive force. Crude membrane extraction methods were thought to be the cause of finding all the enzymes in the soluble fraction, and more gentle extraction methods confirmed membrane attachment of methylenetetrahydrofolate reductase (Hugenholtz and Ljungdahl, 1989). Also in the acetogen Blautia producta, methylenetetrahydrofolate reductase was found to be loosely attached to the cellular membrane, supporting a potential role in energy conservation (Wohlfarth et al., 1990). Further experimental evidence for a direct role of this enzyme in energy conservation has never been found though. A recent theory is that the methylenetetrahydrofolate reductase has bifurcation activity, coupling the oxidation of two NADH molecules to the reduction of ferredoxin and methylenetetrahydrofolate (Huang et al., 2012). The hypothesis for a bifurcating function of this enzyme in M. thermoacetica is supported by the following arguments: i) the enzyme was found to be associated with subunits similar to F420 non-reducing hydrogenases, also found in archaea for bifurcational reduction of CoM–CoB (Huang et al., 2012), ii) in several acetogens the complex was found to contain flavin (Clark and Ljungdahl, 1984; Wohlfarth et al., 1990), which is thought to be essential for bifurcation of the electrons over the two acceptors (Herrmann et al., 2008; Thauer et al., 2008; Kaster et al., 2011; Nitschke and Russell, 2012; Buckel and Thauer, 2013). The flavin is theorized to donate one electron to a high potential acceptor, leaving the flavin at a “red hot” flavosemiquinone state, capable of reducing a low potential acceptor (Buckel and Thauer, 2013). This can be repeated for another two electrons, obtaining two fully reduced products. In case of methylenetetrahydrofolate reductase, the high potential acceptor would be methylenetetrahydrofolate (E0 = –117 mV) whereas ferredoxin (E0 = –400 mV) would be the low potential acceptor. Assuming a bifurcating function of methylenetetrahydrofolate reductase and a proton translocation ratio of 1:1 per hydrogen formed by the EcH complex, the energy yield of H2/CO2 grown M. thermoacetica was suggested to be 0.5 ATP per acetate formed (Schuchmann and Müller, 2014). Applying a model using similar assumptions, the metabolism on CO is expected to yield 1.5 ATP per acetate formed (Figure 2). The suggested three times increase in energy yield matches with the increased observed growth yield of M. thermoacetica with CO (Table 3). The same yield increase is observed in the related organism M. thermoautotrophica, which is thought to exhibit a similar metabolism (Table 3).
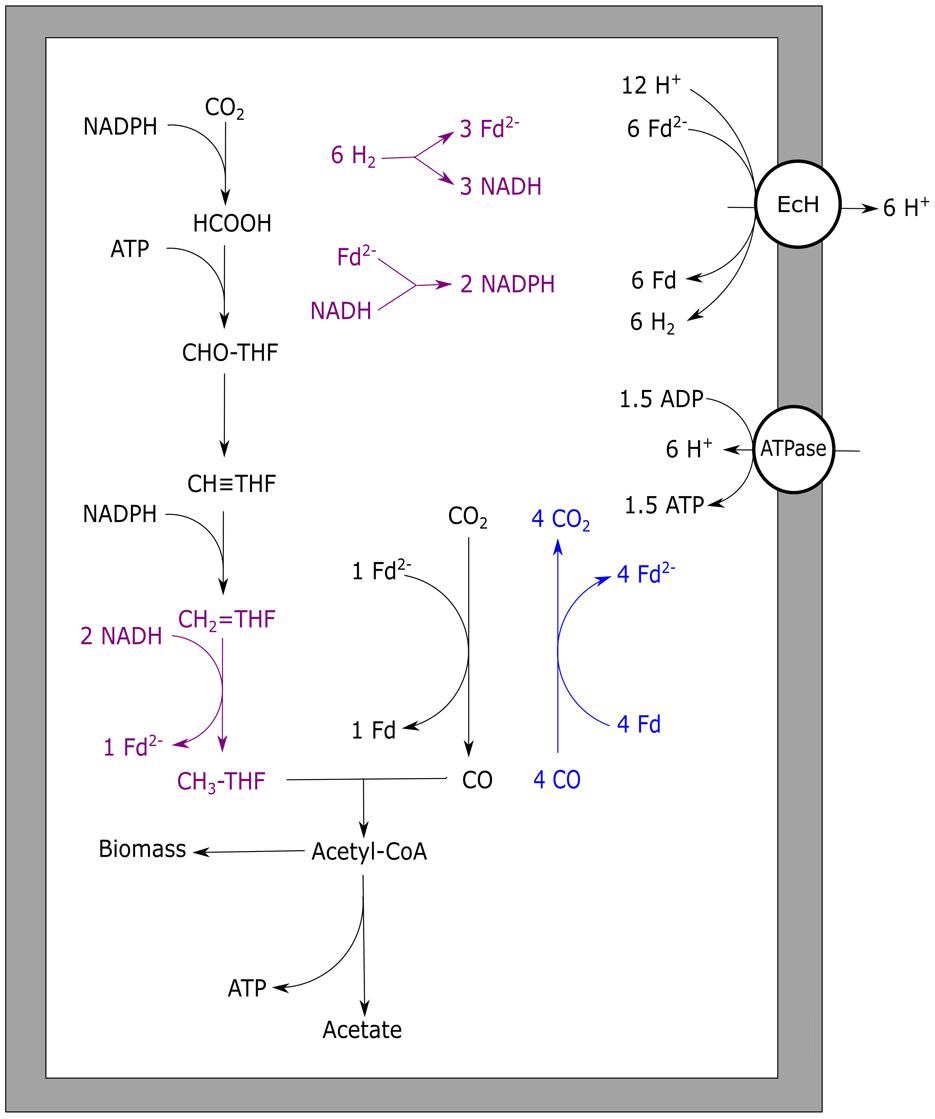
FIGURE 2. Carbon monoxide metabolism of Moorella thermoacetica. Stoichiometric conversion of CO to acetate by M. thermoacetica is disaplayed. Reactions marked blue indicate CO oxidizing activity by CODH, bifurcating reactions are marked purple. The EcH is assumed to transport one proton per hydrogen formed whereas the ATPase is assumed to generate one ATP per four protons translocated. EcH, energy converting hydrogenase; ATPase, ATP synthase; Fd, ferredoxin; THF, tetrahydrofolate.
Initially, the acetogen Acetobacterium woodii was reported to grow homo-acetogenically on CO as a sole energy source (Genthner and Bryant, 1987). However, recently it was shown that the organism can only utilize CO in co-fermentation with either hydrogen or formate (Bertsch and Müller, 2015). Additionally, A. woodii was shown to produce ethanol when the pressure of CO in the headspace was over 25 kPa (Bertsch and Müller, 2015). These contradictory observations might be explained by the rich undefined medium (UM) used in the initial study, which makes it possible that the organism has co-fermented CO with other substrates, such as formate (Bertsch and Müller, 2015). This is further supported by findings of the initial study that it was not possible to grow A. woodii with solely CO on defined medium (Genthner and Bryant, 1987). For A. woodii, a metabolism on H2/CO2 was proposed based on genomic data (Poehlein et al., 2012), and was later adapted suggesting a gain of ∼0.3 ATP per acetate formed (Schuchmann and Müller, 2014). A. woodii is suggested to contain a non-bifurcating methylenetetrahydrofolate reductase, using only NADH to form methyltetrahydrofolate (Schuchmann and Müller, 2014). Growth on H2/CO2 for A. woodii was reported with a generation time of approximately 6 h. Presence of increasing levels of CO negatively affected the growth rates on H2/CO2, and became almost fully inhibited above 15 kPa CO (Bertsch and Müller, 2015). A. woodii efficiently co-fermented CO (25 kPa) with formate at a generation time of approximately 5.5 h. Levels up to 50 kPa CO stimulated growth when co-fermenting formate, however, higher CO pressures caused a decrease in growth rate. Growth remained possible up to a maximally tested pressure of 100 kPa CO (Bertsch and Müller, 2015). The inhibitory effect of CO on the A. woodii metabolism is suggested to be related to its formate dehydrogenase (Bertsch and Müller, 2015), which is associated with a [Fe–Fe] hydrogenase (Schuchmann and Müller, 2013). Additionally, bifurcational [Fe–Fe] hydrogenases present in A. woodii can be a bottleneck in utilization of CO (Figure 3A). In the presence of formate, CO-inhibited enzymes are expected not to be required, facilitating the use of CO as a substrate (Figure 3B).
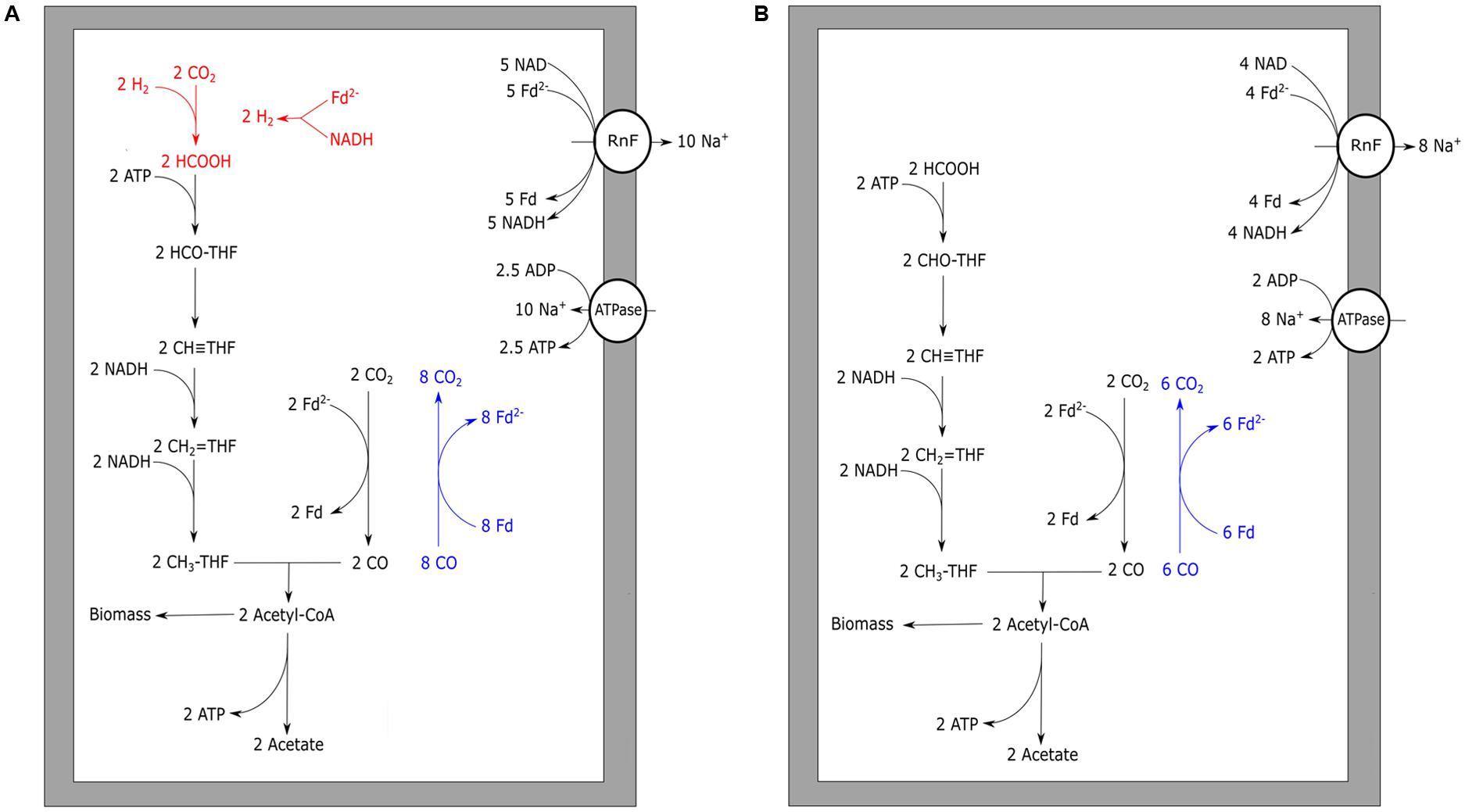
FIGURE 3. Carbon monoxide metabolism of Acetobacterium woodii. Acetogenic CO metabolism of A. woodii is displayed. (A) The proposed theoretical pathway and energetic yield of CO conversion to acetate. Pathways prone to CO inhibition are shown in red. (B) Proposed metabolism and energetic yield of acetogenic metabolism driven by CO co-fermented with formate. Reactions marked blue indicate CO oxidizing activity by CODH. The RnF complex is assumed to transport two sodium ions per Fd oxidized whereas the ATPase is assumed to generate one ATP per four sodium ion translocated. Rnf, RnF complex; ATPase, ATP synthase; Fd, ferredoxin; THF, tetrahydrofolate.
Unlike M. thermoacetica, which contains an EcH, RnF-utilizing acetogens cannot directly couple ferredoxin oxidation to proton reduction. Therefore, to prevent a completely reduced state of the cell, RnF-containing acetogens need to stoichiometrically couple CO oxidation to the Wood–Ljungdahl pathway. When compared to hydrogenotrophic acetogenesis, acetogenic growth on CO often leads to the formation of additional alcohols, hydrogen, or fatty acids. These by-products are likely formed due to the more strongly reduced environment created by CO. An example is the CO metabolism of E. limosum, which produced solely acetate when grown on H2/CO2, but generates a mixture of acetate and butyrate when grown on CO (Jeong et al., 2015). The best studied pathway for maintaining redox balance during acetogenic growth on CO is solventogenesis. The model organism for this type of fermentation is C. ljungdahlii, which is known for its fast growth rate and solventogenic production characteristics on CO (Köpke et al., 2010). In contrast to A. woodii and M. thermoacetica, the enzymes from the Wood–Ljungdahl pathway in C. ljungdahlii have not been purified and tested for cofactor specificity. Therefore, the proposed metabolism of C. ljungdahlii on H2/CO2 was partly based on genomic data (Schuchmann and Müller, 2014). The proposed metabolism on H2 is to yield a minimum of 0.13 ATP per acetate formed, and can go up to 0.63 ATP per acetate, depending on which cofactors are utilized in each of the steps. Based on the assumptions of the minimal H2 metabolism, a model for CO driven acetogenic growth can be proposed for C. ljungdahlii, and related bacteria (Figure 4). The model shows a yield of about 1.125 ATP per acetate formed (Figure 4), which is further reduced when formation of side products is taken into account. Despite the fact that the energy yield per acetate formed is less as proposed for M. thermoacetica, the generation time of C. ljungdahlii is shorter. Two mechanisms might contribute to this enhanced growth rate: a bifurcational formate dehydrogenase, and the up-regulation of re-oxidizing reactions. A related bacterium, Clostridium autoethanogenum, was found to highly express a formate dehydrogenase associated with a [Fe–Fe]-bifurcating hydrogenase. This formate dehydrogenase is suggested to use one mole of NADPH and one mole of ferredoxin to reduce two moles of CO2 to formate (Wang et al., 2013). Despite the sensitivity of [Fe–Fe]-hydrogenases to CO, levels in the cell are assumed to be kept low enough for the hydrogenase to function. The utilization of both NADPH and ferredoxin results in re-oxidation of these two important cofactors, and prevents over-reduction of the cell. However, loss of ferredoxin in re-oxidizing reactions reduces its capacity to act as a driving force for cation export via the RnF complex, lowering the energy yield of the overall metabolism. C. ljungdahlii codes for a similar formate dehydrogenase complex in its genome, and might thus utilize a similar system during growth on CO. Additionally, solventogenic reactions play a role in maintaining redox balance. Ethanol production in C. ljungdahlii occurs via two pathways: a direct or an indirect pathway (Köpke et al., 2010, 2011). The direct pathway forms ethanol via acetaldehyde directly from acetyl-CoA, utilizing an acetaldehyde/alcohol dehydrogenase complex. This pathway is expected to largely reduce the overall energy yield as no ATP is generated via acetate formation. The indirect pathway does not omit the ADP phosphorylation step, maintaining the energy conservation via acetate synthesis. Acetate is theorized to subsequently be reduced with ferredoxin to acetaldehyde via an aldehyde oxidoreductase. This enzyme was shown to be expressed in CO grown C. ljungdahlii, and its expression is stimulated by the addition of external acids (Xie et al., 2015). Ethanol is subsequently formed from acetaldehyde via an alcohol dehydrogenase, utilizing additional reducing equivalents such as NADH or NADPH (Köpke et al., 2010). Energy conservation linked to ethanol formation from H2/CO2 in C. autoethanogenum is also expected to run via this indirect pathway (Mock et al., 2015). Additionally, a similar pathway could be responsible for conversion of different carboxylic acids into alcohols, as observed in mixed cultures exposed to syngas (Liu et al., 2014).
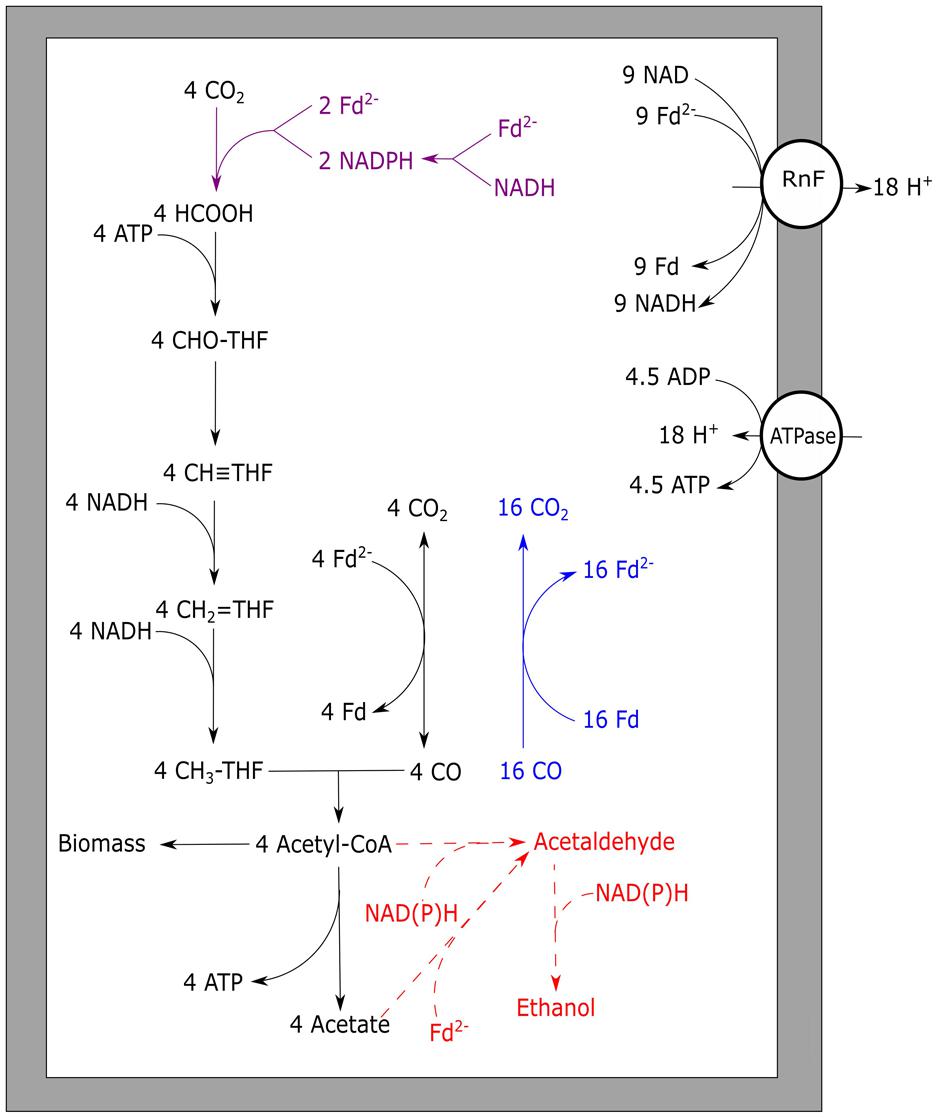
FIGURE 4. Carbon monoxide metabolism of Clostridium ljungdahlii. Stoichiometric conversion of CO to acetate for C. ljungdahlii is displayed. The pathways of ethanol formation are indicated by the dotted line in red, and are not taken into account for the energy yield displayed. Reactions marked blue indicate CO oxidizing activity by CODH, bifurcating reactions are marked purple. The RnF complex is assumed to transport two protons per Fd oxidized whereas the ATPase is assumed to generate one ATP per four protons translocated. Rnf, RnF complex; ATPase, ATP synthase; Fd, ferredoxin; THF, tetrahydrofolate.
As the suggested energy yield per acetate formed from CO for C. ljungdahlii is 1.125 ATP (Figure 4), 0.125 ATP is generated up to acetyl-CoA formation. This suggests net energy can be yielded by this organism from any product formed from acetyl-CoA which does not require further investment of ATP. This could include compounds such as C4-carboxylic acids, lactate, fatty acids, and a variety of alcohols, which are considered interesting end products of bio-based processes (Dürre and Eikmanns, 2015).
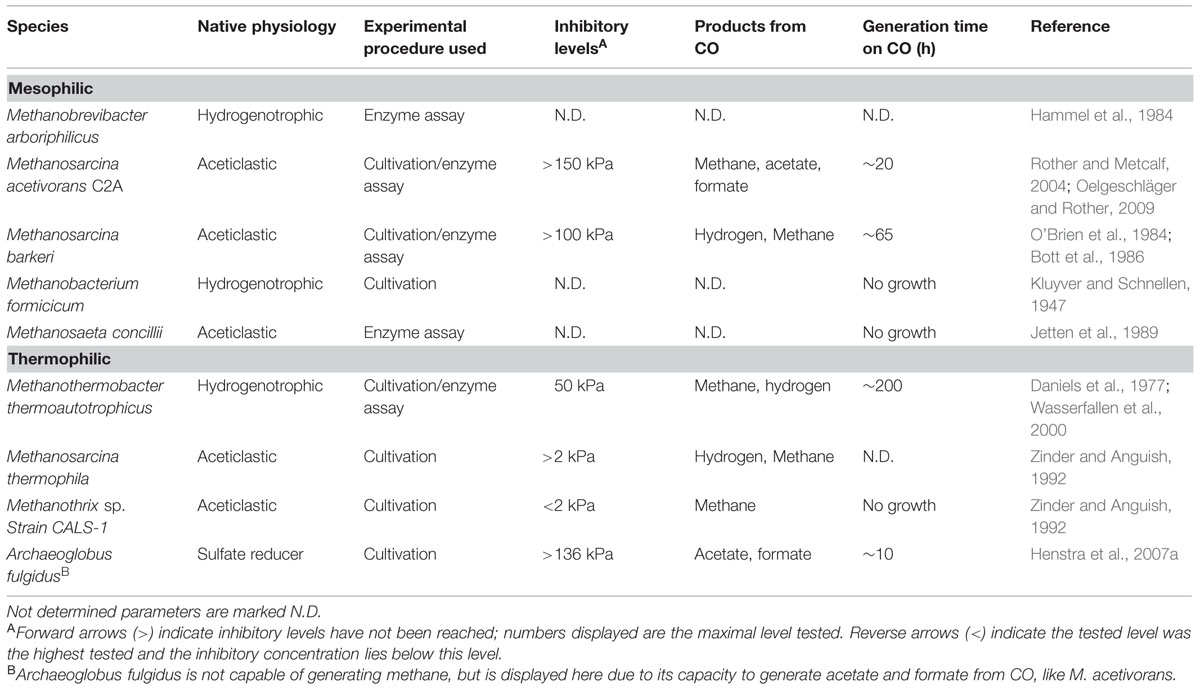
Carboxydotrophic Methanogenic Metabolism
Methane can be formed anaerobically from different substrates, such as H2/CO2, formate, methanol, acetate, or methylamines. The physiology and bioenergetics of different types of methanogenic metabolism have been reviewed before (Thauer et al., 2008). CO is a less studied substrate of methanogenesis, and was first reported by Fischer et al. (1931). It has thereafter been shown to be utilized in the metabolism of several methanogenic strains (Table 4). Methanogenic growth on CO as sole substrate appears not to be very efficient, as only three species have been reported to grow while producing methane: Methanobacterium thermoautotrophicus, Methanosarcina acetivorans, and Methanosarcina barkeri. To gain insight in CO utilization by different groups of methanogens, two main types of methanogenic metabolism are of interest: hydrogenotrophic and aceticlastic methanogenesis. CO is an intermediate in both types of methanogenic metabolism: playing a role in anabolism or catabolism of acetyl-CoA. It is therefore not surprising that genes coding for CODH in methanogens are mainly located in the genome as neighbor of an ACS (Techtmann et al., 2012). Some methanogens do, however, possess more than one CODH, which are not all associated with an ACS complex. Examples are Methanothermobacter marburgensis and Methanococcus jannaschii. M. jannaschii has a CODH which is in located the same operon as a hydrogenase, suggesting that hydrogenogenic carboxydotrophic metabolism is possible. However, many methanogenic strains have never been tested for utilization of CO as a substrate, and their growth potential on CO is therefore unknown. Unlike in many bacteria, the routes for electron transfer from CODH to the rest of the metabolism are not well established for archaea. In general, “ferredoxin-like” proteins are proposed as the acceptor molecules for CODH complexes. This is confirmed for some methanogens, such as Methanosarcina thermophila (Terlesky and Ferry, 1988; Abbanat and Ferry, 1991) and Methanosarcina barkeri (Fischer and Thauer, 1990), which require ferredoxin to perform CO-dependent reactions. However, cell-free extract of M. thermoautotrophicus exhibited F420-reducing activity in presence of CO, while ferredoxin of Clostridium pasteurianum was not reduced (Daniels et al., 1977). F420 was also observed to be reduced by purified CODH from Methanosaeta concilii, indicating potential ability of this enzyme to reduce this cofactor (Jetten et al., 1989). However, ferredoxin was not tested as acceptor for the CODH complex of M. concilii, and thus cannot be excluded as acceptor. When assuming E = –500 mV for ferredoxin under physiological conditions (Buckel and Thauer, 2013), it can be expected that ferredoxin is an ideal acceptor for electrons from CO (E0 = –520 mV). Transfer of electrons to F420 (E0 = –380 mV) would result in additional energy loss, and would require bifurcation processes to generate reduced ferredoxin. Ferredoxins have in general been shown to be interchangeable over long phylogenetic ranges, as indicated by the similar characteristics of plant and bacterial ferredoxins (Tagawa and Arnon, 1968). However, ferredoxin from spinach did not form a complex with the CODH of M. thermoacetica and was not reduced by this bacterial CODH (Drake et al., 1980; Shanmugasundaram and Wood, 1992). Additionally, the observation that the CODH from R. rubrum could only effectively donate electrons to the tightly associated CooF complex (Ensign and Ludden, 1991), suggests not all ferredoxins are efficient in receiving in electrons from a CODH complex. Therefore, the observation that some archaeal CODHs do not transfer electrons to non-native ferredoxin might be related to a difference in characteristics, such as structure and location of the special nickel–iron–sulfur clusters present in the CODH and the possibly different mid-point redox potentials of the ferredoxin.
In autotrophic, hydrogen-utilizing methanogens, CO is an intermediate of the anabolic reductive acetyl-CoA pathway, a pathway functionally similar to the Wood–Ljungdahl pathway in bacteria (Berg et al., 2010). The archaeal CODH required for growth is a nickel-dependent enzyme, just as observed in bacteria (Hammel et al., 1984). For CO to be utilized as energy source by methanogens, a CODH has to be present that can function in the CO oxidizing direction, and suitable cofactors for electron transfer to the methanogenic metabolism should be available. Thermophilic Methanothermobacter thermoautotrophicus is capable of growing on CO, but slowly; at CO pressures up to 50 kPa at a rate of 1% compared to its growth rate on H2/CO2 (Daniels et al., 1977). The genome of M. thermoautotrophicus (Smith et al., 1997) codes for a single CODH enzyme with ACS as a neighboring gene, which are used for anabolism during growth on hydrogen (Stupperich et al., 1983). Growing M. thermoautotrophicus on CO as sole electron donor, results in methane formation and small amounts of H2, suggesting hydrogen is an intermediate or side product of the metabolism. In hydrogenotrophic methanogens an EcH is present, which is involved in the reduction of ferredoxin with H2, driven by a proton gradient (Thauer et al., 2010). Generation of reduced ferredoxin by CODH allows for the reverse reaction to take place, generating a proton gradient. This additionally results in the formation of hydrogen, which subsequently can be used for reduction of CO2 to methane (Figure 5). Metabolic activity using CO, with H2 as intermediate, is also observed in Methanosarcina barkeri (Fischer and Thauer, 1990) and Methanosarcina thermophila (Terlesky and Ferry, 1988; Zinder and Anguish, 1992). Experiments with cell extracts and washed cells of M. thermophila show increased hydrogenogenic activity upon exposure to CO (Zinder and Anguish, 1992). When growing M. thermophila on acetate, hydrogen production was detected besides methane formation, suggesting coupling of CO oxidation to proton reduction (Terlesky and Ferry, 1988). M. barkeri also has the capability of oxidizing CO coupled to formation of hydrogen, resulting in the formation of a proton gradient (Bott and Thauer, 1989). Upon addition of methanogenic inhibitors, cell suspensions of M. barkeri were still capable of utilizing CO, producing hydrogen and additionally resulting in formation of ATP (Bott et al., 1986). If a hydrogenogenic metabolism can sustain growth in M. barkeri is unclear. Without methanogenic inhibitors, the formed hydrogen is further oxidized to form methane, allowing for growth (O’Brien et al., 1984). Judging from the carboxydotrophic generation time of M. barkeri (∼65 h) and M. thermoautotrophicus (∼200 h), the methanogenic metabolism on CO is not very efficient. For both strains, hydrogen accumulation is observed during methanogenic carboxydotrophic growth (Daniels et al., 1977; O’Brien et al., 1984). This suggests hydrogenases, required for the formation of methane from the intermediately formed hydrogen, are inhibited by CO. The involved hydrogenases, such as the heterodisulfide reductase-associated hydrogenase and the F420-reducing hydrogenase, are of the [Ni–Fe] type (Thauer et al., 2010) and can be expected to be relatively resistant, but not insensitive, to CO (De Lacey et al., 2007). Additionally, M. thermoautotrophicus contains an iron-dependent methylenetetrahydromethanopterin dehydrogenase, which is involved in the conversion of methenyltetrahydromethanopterin to methylenetetrahydromethanopterin directly using hydrogen as a donor (Zirngibl et al., 1990). This hydrogenase is found in several hydrogenotrophic methanogens, and is mainly expressed under nickel-deprived conditions (Afting et al., 1998; Afting et al., 2000). The enzyme is susceptible to CO (Lyon et al., 2004) and is therefore a potential target for inhibition during carboxydotrophic growth. Despite the indication that hydrogenases are a limiting factor for carboxydotrophic methanogenesis, inhibition of other enzymes by CO cannot be ruled out as only limited information is available on the mechanisms of CO toxicity in methanogens.
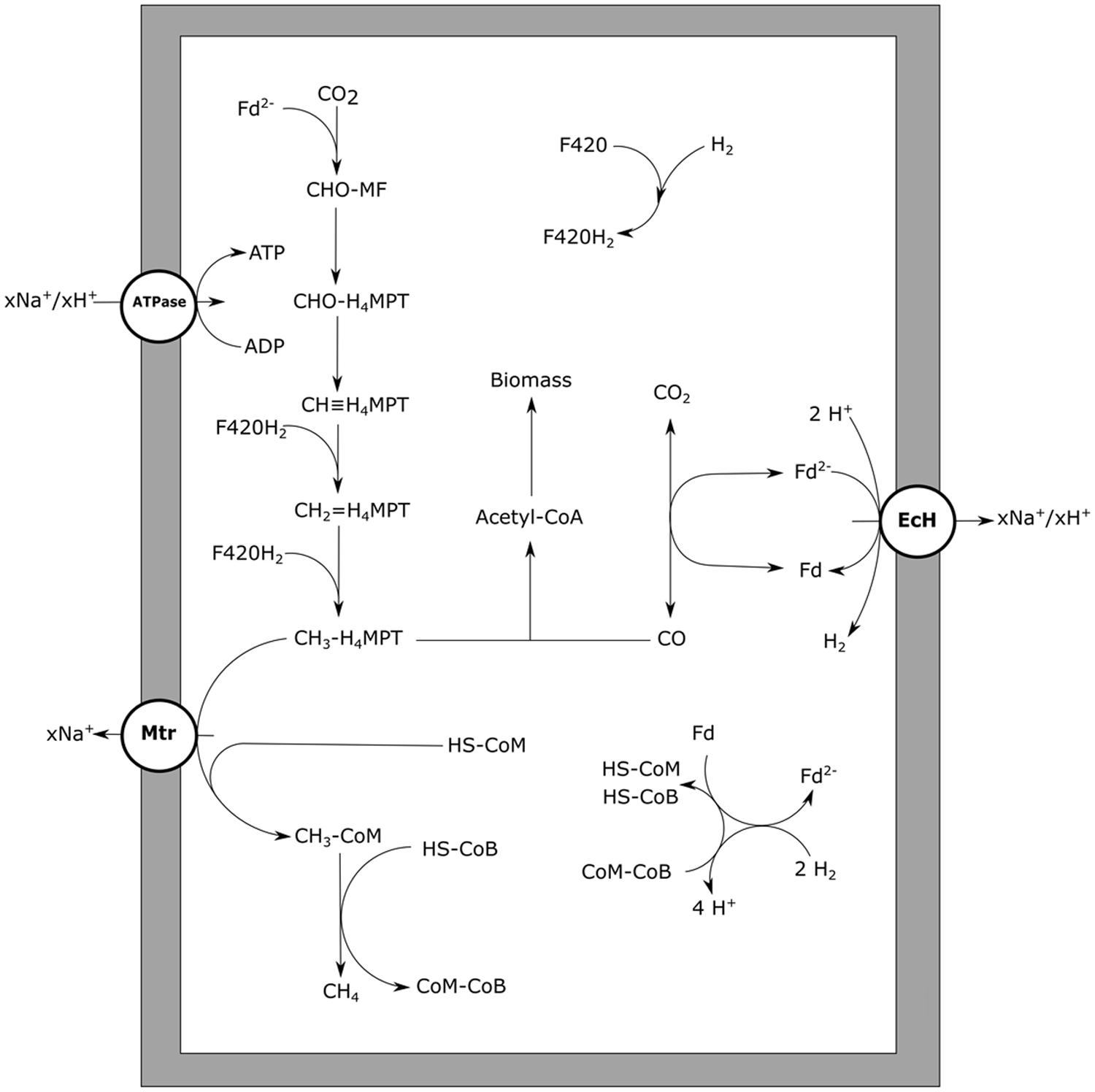
FIGURE 5. Carbon monoxide metabolism of hydrogenotrophic methanogens. CO driven methanogenesis with hydrogen as an intermediate is displayed. xH+ or xNa+ indicate translocation of an undefined number of protons or sodium ions, respectively. Reactions are not displayed stoichiometrically. EcH, energy converting hydrogenase; Mtr, methyl-H4MPT:HS-CoM methyltransferase; ATPase, ATP synthase; Fd, ferredoxin; MF, methanofuran; H4MPT, tetrahydromethanopterin; HS-CoM, coenzyme M; HS-CoB, coenzyme B.
During aceticlastic methanogenesis, CO is an intermediate originating from the splitting of acetyl-CoA by the CODH/ACS complex (Grahame, 2003). Subsequently, CODH is used to further oxidize CO and provide electrons for reduction of the methyl-group to methane. Methanogenic bioreactors fed with acetate accumulated CO to levels up to 0.25 Pa, likely resulting from its role as intermediate in aceticlastic methanogenesis (Hickey and Switzenbaum, 1990). Obligate aceticlastic methanogens such as Methanothrix sp. strain CALS-1, were observed to accumulate CO to low partial pressures while metabolizing acetate (Zinder and Anguish, 1992). Upon addition of low amounts of CO, it was consumed till equilibrium levels of 0.16 Pa. Addition of CO to levels of 2 kPa caused inhibition of growth on acetate. Obligate aceticlastic methanogens contain a low level of hydrogenases and are in general assumed not to utilize these enzymes for their energy metabolism (Deppenmeier et al., 1996; Smith and Ingram-Smith, 2007). This makes it unlikely that inhibition of hydrogenases results in CO toxicity for these methanogens. However, in contrast to EcH containing methanogens, the electrons released from CO oxidation cannot be coupled to proton reduction, requiring other pathways to re-oxidize formed reduction equivalents. The methyl-branch of the reductive acetyl-CoA pathway could theoretically fulfill this role, as the genes are present in the genome of aceticlastic Methanosaeta/Methanothrix species (Zhu et al., 2012). All of these organisms, however, lack the ability to grow on H2/CO2 or formate (Thauer et al., 2008), suggesting no activity of this pathway in the CO2-reducing direction. The genes of the methyl-branch coded for in these species are phylogenetically similar to genes present in methylotrophic methanogens, which use this branch in the oxidative direction, in order to generate reduction equivalents for biosynthesis (Zhu et al., 2012). This is further supported by C13 labeling studies in Methanosaeta harundinacea which confirms activity in the oxidizing but not in the reducing direction (Zhu et al., 2012). This suggests the methyl-branch of the reductive acetyl-CoA pathway in aceticlastic methanogens is not optimal for re-oxidation of reduced cofactors, as is suggested for hydrogenotrophic methanogens (Figure 5). When exposed to elevated levels of CO this could result in an over-reduced state of the cell, making it difficult for obligate aceticlastic methanogens to utilize it as a substrate.
The only methanogen which appears to deal quite well with CO is Methanosarcina acetivorans, which was initially isolated from marine sediments (Sowers et al., 1984). In addition to methane, acetate and formate were observed to be the main end products from CO (Rother and Metcalf, 2004). Additionally, M. acetivorans was found to produce methylated-thiols from CO (Oelgeschläger and Rother, 2009). The M. acetivorans genome codes for two isoforms of CODH/ACS, Cdh1, and Cdh2, which are both considered to be functional in acetyl-CoA anabolism and catabolism (Matschiavelli et al., 2012). The expression levels of the two isoforms are theorized to be regulated on transcriptional and posttranscriptional level by a CdhA subunit (CdhA3), which is suggested to act as a CO sensor (Matschiavelli et al., 2012). In addition to the CODH/ACS complex, two monofunctional CODH, CooS1F, and CooS2, were found to assist in removal of the CO at high CO partial pressures (Rother et al., 2007). For M. acetivorans no hydrogen formation is observed during growth on CO, which is supported by the fact that it is devoid of any significant hydrogen metabolism (Sowers et al., 1984). Despite the inability of M. acetivorans to grow on H2/CO2, its genome codes for homologs of the methyl-branch of the reductive acetyl-CoA pathway (Galagan et al., 2002). Proteomic analysis shows that these genes are more abundantly expressed during CO-dependent growth when compared to growth on acetate or methanol (Lessner et al., 2006). This suggests that, in contrast to what is proposed for Methanosaeta/Methanotrix species, M. acetivorans uses the methyl-branch of the reductive acetyl-CoA pathway to regenerate its reduction equivalents. However, due to its lack of hydrogenases it is unclear how the organism couples oxidation of CO to reduction of F420, which is required to operate this pathway. M. acetivorans was shown to express a sodium dependent “RnF-like” complex when metabolizing acetate (Li et al., 2007; Schlegel et al., 2012). It is speculated that this RnF complex couples ferredoxin oxidation to reduction of methanophenazine, subsequently passing on the electrons to the heterodisulfide reductase (Hdr) complex, involved in HS-CoM/HS-CoB regeneration (Li et al., 2007; Figure 6). Proteomic data of M. acetivorans show that in cells grown on CO the F420-oxidizing:Fpo complex is relatively more abundant, which suggests a role in CO metabolism (Lessner et al., 2006). This protein complex possibly operates in combination with the RnF complex, to catalyze ferredoxin-dependent F420 reduction (Lessner et al., 2006; Figure 6). An alternative for coupling ferredoxin oxidation to F420 reduction is via a subunit from the Fpo complex which partly resides in the cytoplasm: FpoF. This subunit was found to catalyze ferredoxin:F420 reduction in EcH knockout mutants of Methanosarcina mazei, whereas knockout mutants of the FpoF subunit did not show this activity (Welte and Deppenmeier, 2011). Despite the lack of genes coding for formate dehydrogenases in the genome of M. activorans (Galagan et al., 2002), formate is produced in addition to acetate and methane during carboxydotrophic growth. Formate is suggested to originate from activity of formyl-methanofuran dehydrogenase and is theorized to act as a redox exhaust of the cell during CO driven growth (Matschiavelli and Rother, 2015).
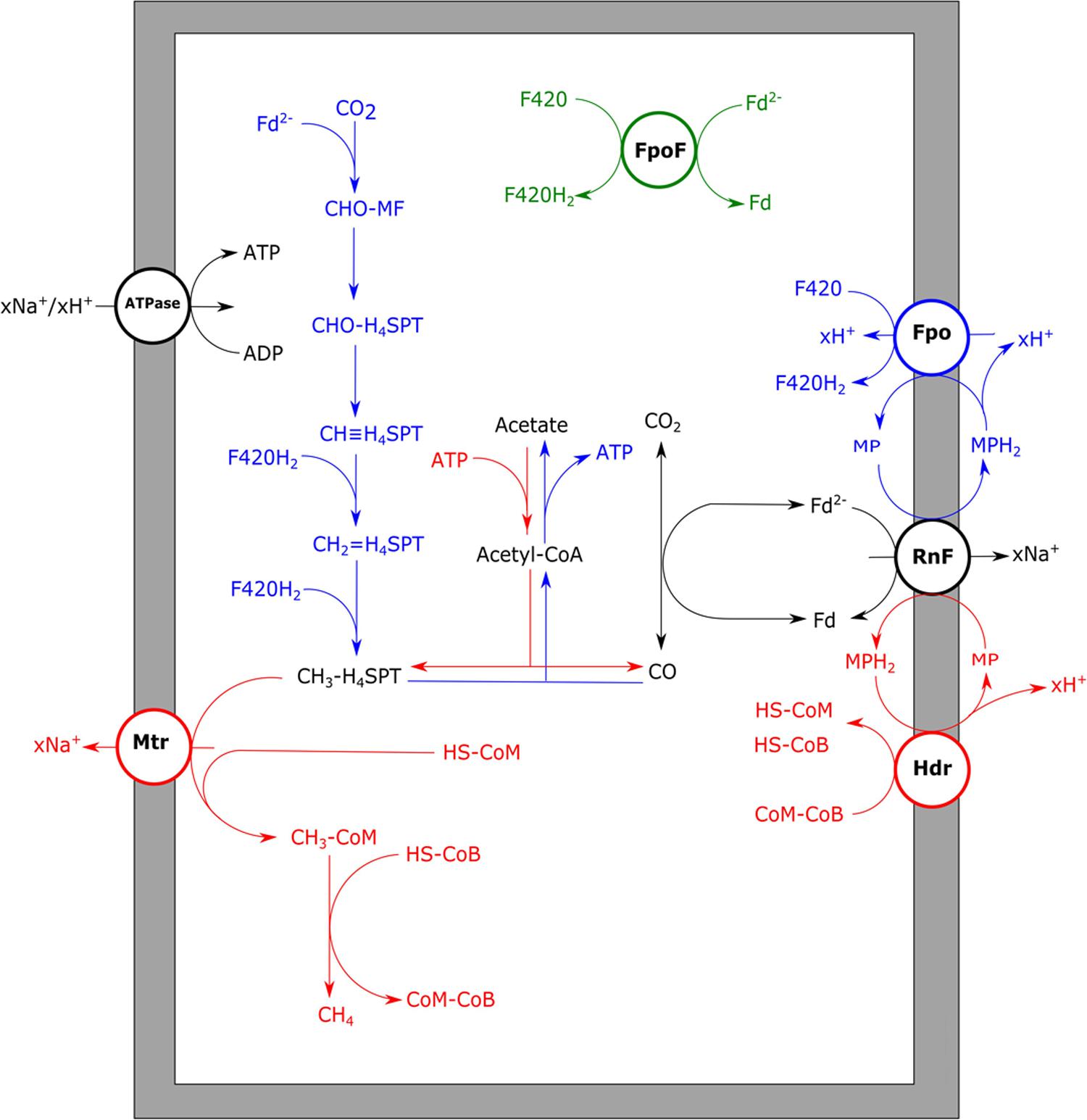
FIGURE 6. Aceticlastic and carboxydotrophic metabolism of Methanosarcina acetivorans. Aceticlastic methanogenic pathway proposed for M. acetivorans (red) and carboxydotrophic pathway proposed for M. acetivorans (blue). For carboxydotrophic growth either RnF/Fpo (blue) or FpoF (green) are proposed to generate reduced F420 from ferredoxin. xH+ or xNa+ indicate translocation of an undefined number of protons or sodium ions, respectively. RnF, RnF-like complex; ATPase, ATP synthase; Mtr, methyl-H4SPT:HS-CoM methyltransferase; Fpo, F420 dehydrogenase complex; FpoF, F420 dehydrogenase subunit F; Hdr, heterodisulfide reductase; Fd, ferredoxin; MF, methanofuran; H4SPT, tetrahydrosarcinapterin; HS-CoM, coenzyme M; HS-CoB, coenzyme B.
Methanosarcina acetivorans is not the only archaeon producing acetate and formate from CO, as also the hyperthermophilic, sulfate-reducing Archaeoglobus fulgidus generates these products during carboxydotrophic growth in absence of sulfate. In A. fulgidus, however, both the RnF and EcH complexes are not coded for in the genome (Hocking et al., 2014). A FpoF homolog, FqoF, is encoded for in the genome of A. fulgidus (Brüggemann et al., 2000), which could take part in the coupling of ferredoxin oxidation to F420 reduction. The absence of an EcH complex poses also a question for the redox balance of the organism, as hydrogen cannot be used as a redox exhaust. It is possible that formate is formed in a similar way as theorized for M. acetivorans, but it can also be formed via F420-dependent formate dehydrogenases, allowing favorable redox balance during growth.
Comparison and Application of Fermentative Carboxydotrophic Metabolism
Comparing the three main variants of fermentative CO metabolism, it is obvious that similar sets of enzymes and pathways are used. Hydrogenogenic microorganisms seem to be the most efficient CO-utilizers due to their relatively simple and “redox-closed” energy metabolism. This allows the water–gas shift mechanism to operate at a rate independent of the rest of the metabolism, minimizing metabolic stress. To maximize the energetic yield on CO, a methanogenic or acetogenic microorganism would first have to perform hydrogenogenesis, conserving energy via an EcH complex, subsequently gaining energy from the use of hydrogen for methanogenesis or acetogenesis. Hydrogenotrophic methanogens and acetogens like M. thermoacetica seem to make use of this strategy, maximizing the ATP yield per CO utilized. However, despite these optimized energy yields, these organisms have no outstanding growth performance on CO. This is likely related to the CO sensitivity of hydrogenases, essential for this type of metabolism. Acetogens which employ RnF complexes, seem less prone to inhibition by CO, but need to couple CO oxidation stoichiometrically to the Wood–Ljungdahl pathway in order to prevent an over-reduced state of the cell. In relatively fast growing acetogenic bacteria, this is prevented by utilizing alternative pathways to re-oxidize cofactors faster. Loss of “energy-rich” reduction equivalents in these re-oxidizing reactions lowers the overall energy yield of the cell. However, this is expected to allow for better maintenance of redox balance, resulting in reduced stress for the overall metabolism. Still net energy is assumed to be conserved by CO-utilizing acetogens, such as C. ljungdahlii, during acetyl-CoA formation. This suggests a large and diverse range of products can be formed from carboxydotrophic metabolism, making it interesting for future research and bio-based applications.
Author Contributions
MD, Drafted and wrote the manuscript; AS, reviewed and revised the manuscript; DS, reviewed and revised the manuscript. All authors gave approval for publication of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors involved were financially supported by an ERC grant (project 323009) and the Gravitation grant (project 024.002.002) of the Netherlands Ministry of Education, Culture and Science and the Netherlands Science Foundation (NWO).
References
Abbanat, D. R., and Ferry, J. G. (1991). Resolution of component proteins in an enzyme complex from Methanosarcina thermophila catalyzing the synthesis or cleavage of acetyl-CoA. Proc. Natl. Acad. Sci. U.S.A. 88, 3272–3276. doi: 10.1073/pnas.88.8.3272
Abrini, J., Naveau, H., and Nyns, E.-J. (1994). Clostridium autoethanogenum, sp. nov., an anaerobic bacterium that produces ethanol from carbon monoxide. Arch. Microbiol. 161, 345–351. doi: 10.1007/BF00303591
Adams, M. W. (1990a). The metabolism of hydrogen by extremely thermophilic, sulfur-dependent bacteria. FEMS Microbiol. Rev. 75, 219–237. doi: 10.1111/j.1574-6968.1990.tb04096.x
Adams, M. W. (1990b). The structure and mechanism of iron-hydrogenases. Biochim. Biophys. Acta 1020, 115–145. doi: 10.1016/0005-2728(90)90044-5
Afting, C., Hochheimer, A., and Thauer, R. (1998). Function of H2-forming methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum in coenzyme F420 reduction with H2. Arch. Microbiol. 169, 206–210. doi: 10.1007/s002030050562
Afting, C., Kremmer, E., Brucker, C., Hochheimer, A., and Thauer, R. K. (2000). Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch. Microbiol. 174, 225–232. doi: 10.1007/s002030000197
Allen, T. D., Caldwell, M. E., Lawson, P. A., Huhnke, R. L., and Tanner, R. S. (2010). Alkalibaculum bacchi gen. nov., sp. nov., a CO-oxidizing, ethanol-producing acetogen isolated from livestock-impacted soil. Int. J. Syst. Evol. Microbiol. 60, 2483–2489. doi: 10.1099/ijs.0.018507-0
Alves, J. I., Van Gelder, A. H., Alves, M. M., Sousa, D. Z., and Plugge, C. M. (2013). Moorella stamsii sp. nov., a new anaerobic thermophilic hydrogenogenic carboxydotroph isolated from digester sludge. Int. J. Syst. Evol. Microbiol. 63, 4072–4076. doi: 10.1099/ijs.0.050369-0
Bae, S. S., Kim, T. W., Lee, H. S., Kwon, K. K., Kim, Y. J., Kim, M.-S., et al. (2012). H2 production from CO, formate or starch using the hyperthermophilic archaeon, Thermococcus onnurineus. Biotechnol. Lett. 34, 75–79. doi: 10.1007/s10529-011-0732-3
Bae, S. S., Kim, Y. J., Yang, S. H., Lim, J. K., Jeon, J. H., Lee, H. S., et al. (2006). Thermococcus onnurineus sp. nov., a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent area at the PACMANUS field. J. Microbiol. Biotechnol. 16, 1826–1831.
Balch, W. E., Schoberth, S., Tanner, R. S., and Wolfe, R. (1977). Acetobacterium, a new genus of hydrogen-oxidizing, carbon dioxide-reducing, anaerobic bacteria. Int. J. Syst. Bacteriol. 27, 355–361. doi: 10.1099/00207713-27-4-355
Balk, M., Heilig, H. G., Van Eekert, M. H., Stams, A. J., Rijpstra, I. C., Sinninghe-Damsté, J. S., et al. (2009). Isolation and characterization of a new CO-utilizing strain, Thermoanaerobacter thermohydrosulfuricus subsp. carboxydovorans, isolated from a geothermal spring in Turkey. Extremophiles 13, 885–894. doi: 10.1007/s00792-009-0276-9
Bengelsdorf, F. R., Straub, M., and Durre, P. (2013). Bacterial synthesis gas (syngas) fermentation. Environ. Technol. 34, 1639–1651. doi: 10.1080/09593330.2013.827747
Berg, I. A., Kockelkorn, D., Ramos-Vera, W. H., Say, R. F., Zarzycki, J., Hügler, M., et al. (2010). Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 8, 447–460. doi: 10.1038/nrmicro2365
Bertsch, J., and Müller, V. (2015). CO metabolism in the acetogen Acetobacterium woodii. Appl. Environ. Microbiol. 81, 5949–5956. doi: 10.1128/AEM.01772-15
Biegel, E., Schmidt, S., González, J. M., and Müller, V. (2011). Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 68, 613–634. doi: 10.1007/s00018-010-0555-8
Bonam, D., Lehman, L., Roberts, G., and Ludden, P. (1989). Regulation of carbon monoxide dehydrogenase and hydrogenase in Rhodospirillum rubrum: effects of CO and oxygen on synthesis and activity. J. Bacteriol. 171, 3102–3107.
Bott, M., Eikmanns, B., and Thauer, R. K. (1986). Coupling of carbon monoxide oxidation to CO2 and H2 with the phosphorylation of ADP in acetate-grown Methanosarcina barkeri. Eur. J. Biochem. 159, 393–398. doi: 10.1111/j.1432-1033.1986.tb09881.x
Bott, M., and Thauer, R. K. (1989). Proton translocation coupled to the oxidation of carbon monoxide to CO2 and H2 in Methanosarcina barkeri. Eur. J. Biochem. 179, 469–472. doi: 10.1111/j.1432-1033.1989.tb14576.x
Braun, M., Mayer, F., and Gottschalk, G. (1981). Clostridium aceticum (Wieringa), a microorganism producing acetic acid from molecular hydrogen and carbon dioxide. Arch. Microbiol. 128, 288–293. doi: 10.1007/BF00422532
Brüggemann, H., Falinski, F., and Deppenmeier, U. (2000). Structure of the F420H2: quinone oxidoreductase of Archaeoglobus fulgidus. Eur. J. Biochem. 267, 5810–5814. doi: 10.1046/j.1432-1327.2000.01657.x
Buckel, W., and Thauer, R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. doi: 10.1016/j.bbabio.2012.07.002
Byrne-Bailey, K. G., Wrighton, K. C., Melnyk, R. A., Agbo, P., Hazen, T. C., and Coates, J. D. (2010). Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J. Bacteriol. 192, 4078–4079. doi: 10.1128/JB.00044-10
Chin, B. Y., and Otterbein, L. E. (2009). Carbon monoxide is a poison… to microbes! CO as a bactericidal molecule. Curr. Opin. Pharmacol. 9, 490–500. doi: 10.1016/j.coph.2009.06.025
Clark, J., and Ljungdahl, L. (1984). Purification and properties of 5, 10-methylenetetrahydrofolate reductase, an iron-sulfur flavoprotein from Clostridium formicoaceticum. J. Biol. Chem. 259, 10845–10849.
Collins, M., Lawson, P., Willems, A., Cordoba, J., Fernandez-Garayzabal, J., Garcia, P., et al. (1994). The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44, 812–826. doi: 10.1099/00207713-44-4-812
Cotter, J. L., Chinn, M. S., and Grunden, A. M. (2009). Influence of process parameters on growth of Clostridium ljungdahlii and Clostridium autoethanogenum on synthesis gas. Enzyme Microb. Technol. 44, 281–288. doi: 10.1016/j.enzmictec.2008.11.002
Daniel, S. L., Hsu, T., Dean, S., and Drake, H. (1990). Characterization of the H2-and CO-dependent chemolithotrophic potentials of the acetogens Clostridium thermoaceticum and Acetogenium kivui. J. Bacteriol. 172, 4464–4471.
Daniels, L., Fuchs, G., Thauer, R., and Zeikus, J. (1977). Carbon monoxide oxidation by methanogenic bacteria. J. Bacteriol. 132, 118–126.
Darnault, C., Volbeda, A., Kim, E. J., Legrand, P., Vernède, X., Lindahl, P. A., et al. (2003). Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] clusters in closed and open α subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase. Nat. Struct. Mol. Biol. 10, 271–279. doi: 10.1038/nsb912
Dashekvicz, M., and Uffen, R. (1979). Identification of a carbon monoxide-metabolizing bacterium as a strain of Rhodopseudomonas gelatinosa (Molisch) van Niel. Int. J. Syst. Bacteriol. 29, 145–148. doi: 10.1099/00207713-29-2-145
De Lacey, A. L., Fernandez, V. M., Rousset, M., and Cammack, R. (2007). Activation and inactivation of hydrogenase function and the catalytic cycle: spectroelectrochemical studies. Chem. Rev. 107, 4304–4330. doi: 10.1021/cr0501947
Deppenmeier, U., Müller, V., and Gottschalk, G. (1996). Pathways of energy conservation in methanogenic archaea. Arch. Microbiol. 165, 149–163. doi: 10.1007/BF01692856
Dobbek, H., Svetlitchnyi, V., Gremer, L., Huber, R., and Meyer, O. (2001). Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293, 1281–1285. doi: 10.1126/science.1061500
Doukov, T. I., Iverson, T. M., Seravalli, J., Ragsdale, S. W., and Drennan, C. L. (2002). A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science 298, 567–572. doi: 10.1126/science.1075843
Drake, H., Hu, S., and Wood, H. (1980). Purification of carbon monoxide dehydrogenase, a nickel enzyme from Clostridium thermocaceticum. J. Biol. Chem. 255, 7174–7180.
Drake, H. L., and Daniel, S. L. (2004). Physiology of the thermophilic acetogen Moorella thermoacetica. Res. Microbiol. 155, 869–883. doi: 10.1016/j.resmic.2004.10.002
Drennan, C. L., Heo, J., Sintchak, M. D., Schreiter, E., and Ludden, P. W. (2001). Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 98, 11973–11978. doi: 10.1073/pnas.211429998
Dürre, P., and Eikmanns, B. J. (2015). C1-carbon sources for chemical and fuel production by microbial gas fermentation. Curr. Opin. Biotechnol. 35, 63–72. doi: 10.1016/j.copbio.2015.03.008
Eikmanns, B., Fuchs, G., and Thauer, R. K. (1985). Formation of carbon monoxide from CO2 and H2 by Methanobacterium thermoautotrophicum. Eur. J. Biochem. 146, 149–154. doi: 10.1111/j.1432-1033.1985.tb08631.x
Ensign, S. A., and Ludden, P. (1991). Characterization of the CO oxidation/H2 evolution system of Rhodospirillum rubrum. Role of a 22-kDa iron-sulfur protein in mediating electron transfer between carbon monoxide dehydrogenase and hydrogenase. J. Biol. Chem. 266, 18395–18403.
Fardeau, M.-L., Salinas, M. B., L’haridon, S., Jeanthon, C., Verhé, F., Cayol, J.-L., et al. (2004). Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int. J. Syst. Evol. Microbiol. 54, 467–474. doi: 10.1099/ijs.0.02711-0
Fischer, F., Lieske, R., and Winzer, K. (1931). Die umsetzungen des kohlenoxyds. Biochem. Z. 236, 247–267.
Fischer, R., and Thauer, R. (1990). Ferredoxin-dependent methane formation from acetate in cell extracts of Methanosarcina barkeri (strain MS). FEBS Lett. 269, 368–372. doi: 10.1016/0014-5793(90)81195-T
Fox, J. D., He, Y., Shelver, D., Roberts, G. P., and Ludden, P. W. (1996a). Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J. Bacteriol. 178, 6200–6208.
Fox, J. D., Kerby, R. L., Roberts, G. P., and Ludden, P. W. (1996b). Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J. Bacteriol. 178, 1515–1524.
Galagan, J. E., Nusbaum, C., Roy, A., Endrizzi, M. G., Macdonald, P., Fitzhugh, W., et al. (2002). The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12, 532–542. doi: 10.1101/gr.223902
Geerligs, G., Schönheit, P., and Diekert, G. (1989). Sodium dependent acetate formation from CO2 in Peptostreptococcus productus (strain Marburg). FEMS Microbiol. Lett. 57, 353–357. doi: 10.1111/j.1574-6968.1989.tb03363.x
Genthner, B. S., and Bryant, M. (1982). Growth of Eubacterium limosum with carbon monoxide as the energy source. Appl. Environ. Microbiol. 43, 70–74.
Genthner, B. S., and Bryant, M. (1987). Additional characteristics of one-carbon-compound utilization by Eubacterium limosum and Acetobacterium woodii. Appl. Environ. Microbiol. 53, 471–476.
Gottwald, M., Andreesen, J. R., Legall, J., and Ljungdahl, L. G. (1975). Presence of cytochrome and menaquinone in Clostridium formicoaceticum and Clostridium thermoaceticum. J. Bacteriol. 122, 325–328.
Grahame, D. A. (2003). Acetate C–C bond formation and decomposition in the anaerobic world: the structure of a central enzyme and its key active-site metal cluster. Trends Biochem. Sci. 28, 221–224. doi: 10.1016/S0968-0004(03)00063-X
Hamelinck, C. N., Van Hooijdonk, G., and Faaij, A. P. (2005). Ethanol from lignocellulosic biomass: techno-economic performance in short-, middle-and long-term. Biomass Bioenergy 28, 384–410. doi: 10.1016/j.biombioe.2004.09.002
Hammel, K. E., Cornwell, K., Diekert, G. B., and Thauer, R. K. (1984). Evidence for a nickel-containing carbon monoxide dehydrogenase in Methanobrevibacter arboriphilicus. J. Bacteriol. 157, 975–978.
Hedderich, R., and Forzi, L. (2004). Energy-converting [NiFe] hydrogenases: more than just H2 activation. J. Mol. Microbiol. Biotechnol. 10, 92–104. doi: 10.1159/000091557
Heiskanen, H., Virkajärvi, I., and Viikari, L. (2007). The effect of syngas composition on the growth and product formation of Butyribacterium methylotrophicum. Enzyme Microb. Technol. 41, 362–367. doi: 10.1016/j.enzmictec.2007.03.004
Henstra, A. M., Dijkema, C., and Stams, A. J. (2007a). Archaeoglobus fulgidus couples CO oxidation to sulfate reduction and acetogenesis with transient formate accumulation. Environ. Microbiol. 9, 1836–1841. doi: 10.1111/j.1462-2920.2007.01306.x
Henstra, A. M., Sipma, J., Rinzema, A., and Stams, A. J. (2007b). Microbiology of synthesis gas fermentation for biofuel production. Curr. Opin. Biotechnol. 18, 200–206. doi: 10.1016/j.copbio.2007.03.008
Henstra, A. M., and Stams, A. J. (2004). Novel physiological features of Carboxydothermus hydrogenoformans and Thermoterrabacterium ferrireducens. Appl. Environ. Microbiol. 70, 7236–7240. doi: 10.1128/AEM.70.12.7236-7240.2004
Henstra, A. M., and Stams, A. J. (2011). Deep conversion of carbon monoxide to hydrogen and formation of acetate by the anaerobic thermophile Carboxydothermus hydrogenoformans. Int. J. Microbiol. 2011, 641582. doi: 10.1155/2011/641582
Herrmann, G., Jayamani, E., Mai, G., and Buckel, W. (2008). Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791. doi: 10.1128/JB.01422-07
Hess, V., Schuchmann, K., and Müller, V. (2013). The ferredoxin: NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J. Biol. Chem. 288, 31496–31502. doi: 10.1074/jbc.M113.510255
Hickey, R. F., and Switzenbaum, M. S. (1990). Behavior of carbon monoxide as a trace component of anaerobic digester gases and methanogenesis from acetate. Environ. Sci. Technol. 24, 1642–1648. doi: 10.1021/es00081a003
Hocking, W. P., Stokke, R., Roalkvam, I., and Steen, I. H. (2014). Identification of key components in the energy metabolism of the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus by transcriptome analyses. Front. Microbiol. 5:95. doi: 10.3389/fmicb.2014.00095
Hu, Z., Spangler, N. J., Anderson, M. E., Xia, J., Ludden, P. W., Lindahl, P. A., et al. (1996). Nature of the C-cluster in Ni-containing carbon monoxide dehydrogenases. J. Am. Chem. Soc. 118, 830–845. doi: 10.1021/ja9528386
Huang, H., Wang, S., Moll, J., and Thauer, R. K. (2012). Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J. Bacteriol. 194, 3689–3699. doi: 10.1128/JB.00385-12
Hugenholtz, J., and Ljungdahl, L. G. (1989). Electron transport and electrochemical proton gradient in membrane vesicles of Clostridium thermoautotrophicum. J. Bacteriol. 171, 2873–2875.
Jeon, W. B., Singer, S. W., Ludden, P. W., and Rubio, L. M. (2005). New insights into the mechanism of nickel insertion into carbon monoxide dehydrogenase: analysis of Rhodospirillum rubrum carbon monoxide dehydrogenase variants with substituted ligands to the [Fe3S4] portion of the active-site C-cluster. J. Biol. Inorg. Chem. 10, 903–912. doi: 10.1007/s00775-005-0043-z
Jeong, J., Bertsch, J., Hess, V., Choi, S., Choi, I.-G., Chang, I. S., et al. (2015). A model for energy conservation based on genomic and experimental analyses in a carbon monoxide-utilizing, butyrate-forming acetogen, Eubacterium limosum KIST612. Appl. Environ. Microbiol. 81, 4782–4790. doi: 10.1128/AEM.00675-15
Jeoung, J.-H., Fesseler, J., Goetzl, S., and Dobbek, H. (2014). “Carbon monoxide. toxic gas and fuel for anaerobes and aerobes: carbon monoxide dehydrogenases,” in The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment, eds P. M. H. Kroneck and M. E. Sosa Torres (Dordrecht: Springer), 37–69.
Jetten, M. S., Stams, A. J., and Zehnder, A. J. (1989). Purification and characterization of an oxygen-stable carbon monoxide dehydrogenase of Methanothrix soehngenii. Eur. J. Biochem. 181, 437–441. doi: 10.1111/j.1432-1033.1989.tb14744.x
Jung, G. Y., Jung, H. O., Kim, J. R., Ahn, Y., and Park, S. (1999). Isolation and characterization of Rhodopseudomonas palustris P4 which utilizes CO with the production of H2. Biotechnol. Lett. 21, 525–529. doi: 10.1023/A:1005560630351
Kaster, A.-K., Moll, J., Parey, K., and Thauer, R. K. (2011). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 2981–2986. doi: 10.1073/pnas.1016761108
Kerby, R., Ludden, P., and Roberts, G. (1995). Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 177, 2241–2244.
Kerby, R., Ludden, P., and Roberts, G. (1997). In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J. Bacteriol. 179, 2259–2266.
Khalil, M., and Rasmussen, R. (1990). The global cycle of carbon monoxide: trends and mass balance. Chemosphere 20, 227–242. doi: 10.1016/0045-6535(90)90098-E
Kluyver, A., and Schnellen, C. G. (1947). On the fermentation of carbon monoxide by pure cultures of methane bacteria. Arch. Biochem. 14, 57–70.
Kochetkova, T. V., Rusanov, I. I., Pimenov, N. V., Kolganova, T. V., Lebedinsky, A. V., Bonch-Osmolovskaya, E. A., et al. (2011). Anaerobic transformation of carbon monoxide by microbial communities of Kamchatka hot springs. Extremophiles 15, 319–325. doi: 10.1007/s00792-011-0362-7
Köpke, M., Held, C., Hujer, S., Liesegang, H., Wiezer, A., Wollherr, A., et al. (2010). Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U.S.A. 107, 13087–13092. doi: 10.1073/pnas.1004716107
Köpke, M., Mihalcea, C., Bromley, J. C., and Simpson, S. D. (2011). Fermentative production of ethanol from carbon monoxide. Curr. Opin. Biotechnol. 22, 320–325. doi: 10.1016/j.copbio.2011.01.005
Krumholz, L., and Bryant, M. (1985). Clostridium pfennigii sp. nov. uses methoxyl groups of monobenzenoids and produces butyrate. Int. J. Syst. Bacteriol. 35, 454–456. doi: 10.1099/00207713-35-4-454
Kundiyana, D. K., Huhnke, R. L., and Wilkins, M. R. (2011). Effect of nutrient limitation and two-stage continuous fermentor design on productivities during “Clostridium ragsdalei” syngas fermentation. Bioresour. Technol. 102, 6058–6064. doi: 10.1016/j.biortech.2011.03.020
Küsel, K., Dorsch, T., Acker, G., Stackebrandt, E., and Drake, H. (2000). Clostridium scatologenes strain SL1 isolated as an acetogenic bacterium from acidic sediments. Int. J. Syst. Evol. Microbiol. 50, 537–546. doi: 10.1099/00207713-50-2-537
Latif, H., Zeidan, A. A., Nielsen, A. T., and Zengler, K. (2014). Trash to treasure: production of biofuels and commodity chemicals via syngas fermenting microorganisms. Curr. Opin. Biotechnol. 27, 79–87. doi: 10.1016/j.copbio.2013.12.001
Lessner, D. J., Li, L., Li, Q., Rejtar, T., Andreev, V. P., Reichlen, M., et al. (2006). An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U.S.A. 103, 17921–17926. doi: 10.1073/pnas.0608833103
Li, L., Li, Q., Rohlin, L., Kim, U., Salmon, K., Rejtar, T., et al. (2007). Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6, 759–771. doi: 10.1021/pr060383l
Lindahl, P. A. (2002). The Ni-containing carbon monoxide dehydrogenase family: light at the end of the tunnel? Biochemistry 41, 2097–2105. doi: 10.1021/bi015932+
Liou, J. S.-C., Balkwill, D. L., Drake, G. R., and Tanner, R. S. (2005). Clostridium carboxidivorans sp. nov., a solvent-producing Clostridium isolated from an agricultural settling lagoon, and reclassification of the acetogen Clostridium scatologenes strain SL1 as Clostridium drakei sp. nov. Int. J. Syst. Evol. Microbiol. 55, 2085–2091. doi: 10.1099/ijs.0.63482-0
Liu, K., Atiyeh, H. K., Stevenson, B. S., Tanner, R. S., Wilkins, M. R., and Huhnke, R. L. (2014). Mixed culture syngas fermentation and conversion of carboxylic acids into alcohols. Bioresour. Technol. 152, 337–346. doi: 10.1016/j.biortech.2013.11.015
Liu, K., Atiyeh, H. K., Tanner, R. S., Wilkins, M. R., and Huhnke, R. L. (2012). Fermentative production of ethanol from syngas using novel moderately alkaliphilic strains of Alkalibaculum bacchi. Bioresour. Technol. 104, 336–341. doi: 10.1016/j.biortech.2011.10.054
Lorowitz, W. H., and Bryant, M. P. (1984). Peptostreptococcus productus strain that grows rapidly with CO as the energy source. Appl. Environ. Microbiol. 47, 961–964.
Lux, M. F., and Drake, H. L. (1992). Re-examination of the metabolic potentials of the acetogens Clostridium aceticum and Clostridium formicoaceticum: chemolithoautotrophic and aromatic-dependent growth. FEMS Microbiol. Lett. 95, 49–56. doi: 10.1111/j.1574-6968.1992.tb05341.x
Lynd, L., Kerby, R., and Zeikus, J. (1982). Carbon monoxide metabolism of the methylotrophic acidogen Butyribacterium methylotrophicum. J. Bacteriol. 149, 255–263.
Lynd, L., and Zeikus, J. (1983). Metabolism of H2-CO2, methanol, and glucose by Butyribacterium methylotrophicum. J. Bacteriol. 153, 1415–1423.
Lyon, E. J., Shima, S., Boecher, R., Thauer, R. K., Grevels, F.-W., Bill, E., et al. (2004). Carbon monoxide as an intrinsic ligand to iron in the active site of the iron-sulfur-cluster-free hydrogenase H2-forming methylenetetrahydromethanopterin dehydrogenase as revealed by infrared spectroscopy. J. Am. Chem. Soc. 126, 14239–14248. doi: 10.1021/ja046818s
Maness, P.-C., Huang, J., Smolinski, S., Tek, V., and Vanzin, G. (2005). Energy generation from the CO oxidation-hydrogen production pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 71, 2870–2874. doi: 10.1128/AEM.71.6.2870-2874.2005
Matschiavelli, N., Oelgeschlager, E., Cocchiararo, B., Finke, J., and Rother, M. (2012). Function and regulation of isoforms of carbon monoxide dehydrogenase/acetyl coenzyme A synthase in Methanosarcina acetivorans. J. Bacteriol. 194, 5377–5387. doi: 10.1128/JB.00881-12
Matschiavelli, N., and Rother, M. (2015). Role of a putative tungsten-dependent formylmethanofuran dehydrogenase in Methanosarcina acetivorans. Arch. Microbiol. 197, 379–388. doi: 10.1007/s00203-014-1070-3
Maynard, E. L., and Lindahl, P. A. (1999). Evidence of a molecular tunnel connecting the active sites for CO2 reduction and acetyl-CoA synthesis in acetyl-CoA synthase from Clostridium thermoaceticum. J. Am. Chem. Soc. 121, 9221–9222. doi: 10.1021/ja992120g
Meyer, O., Jacobitz, S., and Krüger, B. (1986). Biochemistry and physiology of aerobic carbon monoxide-utilizing bacteria. FEMS Microbiol. Rev. 2, 161–179. doi: 10.1111/j.1574-6968.1986.tb01858.x
Meyer, O., and Schlegel, H. G. (1983). Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 37, 277–310. doi: 10.1146/annurev.mi.37.100183.001425
Miyakawa, S., Yamanashi, H., Kobayashi, K., Cleaves, H. J., and Miller, S. L. (2002). Prebiotic synthesis from CO atmospheres: implications for the origins of life. Proc. Natl. Acad. Sci. U.S.A. 99, 14628–14631. doi: 10.1073/pnas.192568299
Mock, J., Zheng, Y., Mueller, A. P., Ly, S., Tran, L., Segovia, S., et al. (2015). Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J. Bacteriol. 197, 2965–2980. doi: 10.1128/JB.00399-15
Müller, V. (2003). Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 69, 6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003
Nitschke, W., and Russell, M. J. (2012). Redox bifurcations: mechanisms and importance to life now, and at its origin. Bioessays 34, 106–109. doi: 10.1002/bies.201100134
Novikov, A. A., Sokolova, T. G., Lebedinsky, A. V., Kolganova, T. V., and Bonch-Osmolovskaya, E. A. (2011). Carboxydothermus islandicus sp. nov., a thermophilic, hydrogenogenic, carboxydotrophic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 61, 2532–2537. doi: 10.1099/ijs.0.030288-0
O’Brien, J. M., Wolkin, R., Moench, T., Morgan, J., and Zeikus, J. (1984). Association of hydrogen metabolism with unitrophic or mixotrophic growth of Methanosarcina barkeri on carbon monoxide. J. Bacteriol. 158, 373–375.
Oelgeschläger, E., and Rother, M. (2008). Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch. Microbiol. 190, 257–269. doi: 10.1007/s00203-008-0382-6
Oelgeschläger, E., and Rother, M. (2009). In vivo role of three fused corrinoid/methyl transfer proteins in Methanosarcina acetivorans. Mol. Microbiol. 72, 1260–1272. doi: 10.1111/j.1365-2958.2009.06723.x
Parshina, S. N., Kijlstra, S., Henstra, A. M., Sipma, J., Plugge, C. M., and Stams, A. J. M. (2005a). Carbon monoxide conversion by thermophilic sulfate-reducing bacteria in pure culture and in co-culture with Carboxydothermus hydrogenoformans. Appl. Microbiol. Biotechnol. 68, 390–396. doi: 10.1007/s00253-004-1878-x
Parshina, S. N., Sipma, J., Nakashimada, Y., Henstra, A. M., Smidt, H., Lysenko, A. M., et al. (2005b). Desulfotomaculum carboxydivorans sp. nov., a novel sulfate-reducing bacterium capable of growth at 100% CO. Int. J. Syst. Evol. Microbiol. 55, 2159–2165. doi: 10.1099/ijs.0.63780-0
Parshina, S. N., Sipma, J., Henstra, A. M., and Stams, A. J. M. (2010). Carbon monoxide as an electron donor for the biological reduction of sulphate. Int. J. Microbiol. 2010, 319527. doi: 10.1155/2010/319527
Phillips, J. R., Clausen, E. C., and Gaddy, J. L. (1994). Synthesis gas as substrate for the biological production of fuels and chemicals. Appl. Biochem. Biotechnol. 45, 145–157. doi: 10.1007/BF02941794
Pierce, E., Xie, G., Barabote, R. D., Saunders, E., Han, C. S., Detter, J. C., et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x
Poehlein, A., Schmidt, S., Kaster, A.-K., Goenrich, M., Vollmers, J., Thürmer, A., et al. (2012). An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS ONE 7:e33439. doi: 10.1371/journal.pone.0033439
Purec, L., Krasna, A. I., and Rittenberg, D. (1962). The inhibition of hydrogenase by carbon monoxide and the reversal of this inhibition by light*. Biochemistry 1, 270–275. doi: 10.1021/bi00908a013
Ragsdale, S. W., and Pierce, E. (2008). Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898. doi: 10.1016/j.bbapap.2008.08.012
Rajagopalan, S., Datar, R. P., and Lewis, R. S. (2002). Formation of ethanol from carbon monoxide via a new microbial catalyst. Biomass Bioenergy 23, 487–493. doi: 10.1016/S0961-9534(02)00071-5
Raybuck, S. A., Bastian, N. R., Orme-Johnson, W. H., and Walsh, C. T. (1988). Kinetic characterization of the carbon monoxide-acetyl-CoA (carbonyl group) exchange activity of the acetyl-CoA synthesizing carbon monoxide dehydrogenase from Clostridium thermoaceticum. Biochemistry 27, 7698–7702. doi: 10.1021/bi00420a019
Roberts, G. P., Thorsteinsson, M. V., Kerby, R. L., Lanzilotta, W. N., and Poulos, T. (2001). Cook: a heme-containing regulatory protein that serves as a specific sensor of both carbon monoxide and redox state. Prog. Nucleic Acid Res. Mol. Biol. 67, 35–63. doi: 10.1016/S0079-6603(01)67024-7
Rother, M., and Metcalf, W. W. (2004). Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. U.S.A. 101, 16929–16934. doi: 10.1073/pnas.0407486101
Rother, M., Oelgeschläger, E., and Metcalf, W. W. (2007). Genetic and proteomic analyses of CO utilization by Methanosarcina acetivorans. Arch. Microbiol. 188, 463–472. doi: 10.1007/s00203-007-0266-1
Ryter, S. W., and Otterbein, L. E. (2004). Carbon monoxide in biology and medicine. Bioessays 26, 270–280. doi: 10.1002/bies.20005
Sant’Anna, F., Lebedinsky, A., Sokolova, T., Robb, F., and Gonzalez, J. (2015). Analysis of three genomes within the thermophilic bacterial species Caldanaerobacter subterraneus with a focus on carbon monoxide dehydrogenase evolution and hydrolase diversity. BMC Genomics 16:757. doi: 10.1186/s12864-015-1955-9
Sapra, R., Bagramyan, K., and Adams, M. W. (2003). A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. U.S.A. 100, 7545–7550. doi: 10.1073/pnas.1331436100
Savage, M. D., and Drake, H. L. (1986). Adaptation of the acetogen Clostridium thermoautotrophicum to minimal medium. J. Bacteriol. 165, 315–318.
Savage, M. D., Wu, Z., Daniel, S. L., Lundie, L., and Drake, H. L. (1987). Carbon monoxide-dependent chemolithotrophic growth of Clostridium thermoautotrophicum. Appl. Environ. Microbiol. 53, 1902–1906.
Schlegel, K., Welte, C., Deppenmeier, U., and Müller, V. (2012). Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279, 4444–4452. doi: 10.1111/febs.12031
Schuchmann, K., and Müller, V. (2013). Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342, 1382–1385. doi: 10.1126/science.1244758
Schuchmann, K., and Müller, V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821. doi: 10.1038/nrmicro3365
Seravalli, J., and Ragsdale, S. W. (2000). Channeling of carbon monoxide during anaerobic carbon dioxide fixation. Biochemistry 39, 1274–1277. doi: 10.1021/bi991812e
Seravalli, J., and Ragsdale, S. W. (2008). 13C NMR characterization of an exchange reaction between CO and CO2 catalyzed by carbon monoxide dehydrogenase. Biochemistry 47, 6770–6781. doi: 10.1021/bi8004522
Shanmugasundaram, T., and Wood, H. (1992). Interaction of ferredoxin with carbon monoxide dehydrogenase from Clostridium thermoaceticum. J. Biol. Chem. 267, 897–900.
Sim, J. H., Kamaruddin, A. H., Long, W. S., and Najafpour, G. (2007). Clostridium aceticum—a potential organism in catalyzing carbon monoxide to acetic acid: application of response surface methodology. Enzyme Microb. Technol. 40, 1234–1243. doi: 10.1016/j.enzmictec.2006.09.017
Singer, S. W., Hirst, M. B., and Ludden, P. W. (2006). CO-dependent H2 evolution by Rhodospirillum rubrum: role of CODH: CooF complex. Biochim. Biophys. Acta 1757, 1582–1591. doi: 10.1016/j.bbabio.2006.10.003
Slepova, T. V., Sokolova, T. G., Kolganova, T. V., Tourova, T. P., and Bonch-Osmolovskaya, E. A. (2009). Carboxydothermus siderophilus sp. nov., a thermophilic, hydrogenogenic, carboxydotrophic, dissimilatory Fe (III)-reducing bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 59, 213–217. doi: 10.1099/ijs.0.000620-0
Slepova, T. V., Sokolova, T. G., Lysenko, A. M., Tourova, T. P., Kolganova, T. V., Kamzolkina, O. V., et al. (2006). Carboxydocella sporoproducens sp. nov., a novel anaerobic CO-utilizing/H2-producing thermophilic bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 56, 797–800. doi: 10.1099/ijs.0.63961-0
Slobodkin, A., Sokolova, T., Lysenko, A., and Wiegel, J. (2006). Reclassification of Thermoterrabacterium ferrireducens as Carboxydothermus ferrireducens comb. nov., and emended description of the genus Carboxydothermus. Int. J. Syst. Evol. Microbiol. 56, 2349–2351. doi: 10.1099/ijs.0.64503-0
Smith, D. R., Doucette-Stamm, L. A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., et al. (1997). Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179, 7135–7155.
Smith, K. S., and Ingram-Smith, C. (2007). Methanosaeta, the forgotten methanogen? Trends Microbiol. 15, 150–155. doi: 10.1016/j.tim.2007.02.002
Soboh, B., Linder, D., and Hedderich, R. (2002). Purification and catalytic properties of a CO-oxidizing: H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur. J. Biochem. 269, 5712–5721. doi: 10.1046/j.1432-1033.2002.03282.x
Sokolova, T. G., Gonzalez, J. M., Kostrikina, N. A., Chernyh, N. A., Slepova, T. V., Bonch-Osmolovskaya, E. A., et al. (2004a). Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 54, 2353–2359. doi: 10.1099/ijs.0.63186-0
Sokolova, T. G., Jeanthon, C., Kostrikina, N. A., Chernyh, N. A., Lebedinsky, A. V., Stackebrandt, E., et al. (2004b). The first evidence of anaerobic CO oxidation coupled with H2 production by a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Extremophiles 8, 317–323. doi: 10.1007/s00792-004-0389-0
Sokolova, T., Gonzalez, J., Kostrikina, N., Chernyh, N., Tourova, T., Kato, C., et al. (2001). Carboxydobrachium pacificum gen. nov., sp. nov., a new anaerobic, thermophilic, CO-utilizing marine bacterium from Okinawa Trough. Int. J. Syst. Evol. Microbiol. 51, 141–149. doi: 10.1099/00207713-51-1-141
Sokolova, T., Hanel, J., Onyenwoke, R., Reysenbach, A.-L., Banta, A., Geyer, R., et al. (2007). Novel chemolithotrophic, thermophilic, anaerobic bacteria Thermolithobacter ferrireducens gen. nov., sp. nov. and Thermolithobacter carboxydivorans sp. nov. Extremophiles 11, 145–157. doi: 10.1007/s00792-006-0022-5
Sokolova, T. G., Kostrikina, N. A., Chernyh, N. A., Kolganova, T. V., Tourova, T. P., and Bonch-Osmolovskaya, E. A. (2005). Thermincola carboxydiphila gen. nov., sp. nov., a novel anaerobic, carboxydotrophic, hydrogenogenic bacterium from a hot spring of the Lake Baikal area. Int. J. syst. Evol. Microbiol. 55, 2069–2073. doi: 10.1099/ijs.0.63299-0
Sokolova, T., Kostrikina, N., Chernyh, N., Tourova, T., Kolganova, T., and Bonch-Osmolovskaya, E. (2002). Carboxydocella thermautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring. Int. J. Syst. Evol. Microbiol. 52, 1961–1967. doi: 10.1099/00207713-52-6-1961
Sokolova, T., and Lebedinsky, A. (2013). “CO-oxidizing anaerobic thermophilic prokaryotes,” in Thermophilic Microbes in Environmental and Industrial Biotechnology, eds T. Satyanarayana, J. Littlechild, J. A. Littlechild, and Y. Kawarabayasi (Dordrecht: Springer), 203–231. doi: 10.1007/978-94-007-5899-5_7
Sowers, K. R., Baron, S. F., and Ferry, J. G. (1984). Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47, 971–978.
Stupperich, E., Hammel, K., Fuchs, G., and Thauer, R. (1983). Carbon monoxide fixation into the carboxyl group of acetyl coenzyme A during autotrophic growth of Methanobacterium. FEBS Lett. 152, 21–23. doi: 10.1016/0014-5793(83)80473-6
Svetlichny, V., Sokolova, T., Gerhardt, M., Ringpfeil, M., Kostrikina, N., and Zavarzin, G. (1991). Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir Island. Syst. Appl. Microbiol. 14, 254–260. doi: 10.1016/S0723-2020(11)80377-2
Svetlitchnyi, V., Peschel, C., Acker, G., and Meyer, O. (2001). Two membrane-associated NiFeS-carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 183, 5134–5144. doi: 10.1128/JB.183.17.5134-5144.2001
Tagawa, K., and Arnon, D. I. (1968). Oxidation-reduction potentials and stoichiometry of electron transfer in ferredoxins. Biochim. Biophys. Acta 153, 602–613. doi: 10.1016/0005-2728(68)90188-6
Tanner, R. S., Miller, L. M., and Yang, D. (1993). Clostridium ljungdahlii sp. nov., an acetogenic species in clostridial rRNA homology group I. Int. J. Syst. Bacteriol. 43, 232–236. doi: 10.1099/00207713-43-2-232
Techtmann, S. M., Colman, A. S., and Robb, F. T. (2009). ‘That which does not kill us only makes us stronger’: the role of carbon monoxide in thermophilic microbial consortia. Environ. Microbiol. 11, 1027–1037. doi: 10.1111/j.1462-2920.2009.01865.x
Techtmann, S. M., Lebedinsky, A. V., Colman, A. S., Sokolova, T. G., Woyke, T., Goodwin, L., et al. (2012). Evidence for horizontal gene transfer of anaerobic carbon monoxide dehydrogenases. Front. Microbiol. 3:132. doi: 10.3389/fmicb.2012.00132
Terlesky, K., and Ferry, J. (1988). Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J. Biol. Chem. 263, 4075–4079.
Thauer, R. K., Kaster, A.-K., Goenrich, M., Schick, M., Hiromoto, T., and Shima, S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536. doi: 10.1146/annurev.biochem.030508.152103
Thauer, R. K., Kaster, A.-K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Tiquia-Arashiro, S. M. (2014). Thermophilic Carboxydotrophs and Their Applications in Biotechnology. New York City, NY: Springer. doi: 10.1007/978-3-319-11873-4
Tschech, A., and Pfennig, N. (1984). Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137, 163–167. doi: 10.1007/BF00414460
Uffen, R. L. (1976). Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate. Proc. Natl. Acad. Sci. U.S.A. 73, 3298–3302. doi: 10.1073/pnas.73.9.3298
Wang, S., Huang, H., Kahnt, J., Mueller, A. P., Köpke, M., and Thauer, R. K. (2013). NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386. doi: 10.1128/JB.00678-13
Wasserfallen, A., Nölling, J., Pfister, P., Reeve, J., and De Macario, E. C. (2000). Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int. J. Syst. Evol. Microbiol. 50, 43–53. doi: 10.1099/00207713-50-1-43
Weinstock, B., and Niki, H. (1972). Carbon monoxide balance in nature. Science 176, 290–292. doi: 10.1126/science.176.4032.290
Welte, C., and Deppenmeier, U. (2011). Re-evaluation of the function of the F420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J. 278, 1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x
Wohlfarth, G., Geerligs, G., and Diekert, G. (1990). Purification and properties of a NADH-dependent 5, 10-methylenetetrahydrofolate reductase from Peptostreptococcus productus. Eur. J. Biochem. 192, 411–417. doi: 10.1111/j.1432-1033.1990.tb19242.x
Wu, M., Ren, Q., Durkin, A. S., Daugherty, S. C., Brinkac, L. M., Dodson, R. J., et al. (2005). Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1:e65. doi: 10.1371/journal.pgen.0010065
Xie, B.-T., Liu, Z.-Y., Tian, L., Li, F.-L., and Chen, X.-H. (2015). Physiological response of Clostridium ljungdahlii DSM 13528 of ethanol production under different fermentation conditions. Bioresour. Technol. 177, 302–307. doi: 10.1016/j.biortech.2014.11.101
Yoneda, Y., Yoshida, T., Kawaichi, S., Daifuku, T., Takabe, K., and Sako, Y. (2012). Carboxydothermus pertinax sp. nov., a thermophilic, hydrogenogenic, Fe (III)-reducing, sulfur-reducing carboxydotrophic bacterium from an acidic hot spring. Int. J. Syst. Evol. Microbiol. 62, 1692–1697. doi: 10.1099/ijs.0.031583-0
Younesi, H., Najafpour, G., and Mohamed, A. R. (2005). Ethanol and acetate production from synthesis gas via fermentation processes using anaerobic bacterium, Clostridium ljungdahlii. Biochem. Eng. J. 27, 110–119. doi: 10.1016/j.bej.2005.08.015
Zavarzina, D. G., Sokolova, T. G., Tourova, T. P., Chernyh, N. A., Kostrikina, N. A., and Bonch-Osmolovskaya, E. A. (2007). Thermincola ferriacetica sp. nov., a new anaerobic, thermophilic, facultatively chemolithoautotrophic bacterium capable of dissimilatory Fe (III) reduction. Extremophiles 11, 1–7. doi: 10.1007/s00792-006-0004-7
Zhu, J., Zheng, H., Ai, G., Zhang, G., Liu, D., Liu, X., et al. (2012). The genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS ONE 7:e36756. doi: 10.1371/journal.pone.0036756
Zinder, S., and Anguish, T. (1992). Carbon monoxide, hydrogen, and formate metabolism during methanogenesis from acetate by thermophilic cultures of Methanosarcina and Methanothrix strains. Appl. Environ. Microbiol. 58, 3323–3329.
Keywords: water–gas shift reaction, syngas, carboxydotrophic, hydrogenogenesis, methanogenesis, acetogenesis
Citation: Diender M, Stams AJM and Sousa DZ (2015) Pathways and Bioenergetics of Anaerobic Carbon Monoxide Fermentation. Front. Microbiol. 6:1275. doi: 10.3389/fmicb.2015.01275
Received: 07 September 2015; Accepted: 31 October 2015;
Published: 19 November 2015.
Edited by:
Biswarup Mukhopadhyay, Virginia Polytechnic Institute and State University, USAReviewed by:
Michael Rother, Technische Universität Dresden, GermanyFrank T. Robb, University of Maryland School of Medicine, USA
Serge R. Guiot, National Research Council Canada, Canada
Copyright © 2015 Diender, Stams and Sousa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martijn Diender, martijn.diender@wur.nl
 Martijn Diender
Martijn Diender Alfons J. M. Stams
Alfons J. M. Stams Diana Z. Sousa
Diana Z. Sousa