- 1Institute of Biological Environmental and Rural Sciences, Aberystwyth University, Aberystwyth, UK
- 2Departament de Producció Animal, Escola Tècnica Superior d’Enginyeria Agrària, Universitat de Lleida, Lleida, Spain
First described in 1843, Rumen protozoa with their striking appearance were assumed to be important for the welfare of their host. However, despite contributing up to 50% of the bio-mass in the rumen, the role of protozoa in rumen microbial ecosystem remains unclear. Phylogenetic analysis of 18S rDNA libraries generated from the rumen of cattle, sheep, and goats has revealed an unexpected diversity of ciliated protozoa although variation in gene copy number between species makes it difficult to obtain absolute quantification. Despite repeated attempts it has proven impossible to maintain rumen protozoa in axenic culture. Thus it has been difficult to establish conclusively a role of ciliate protozoa in rumen fiber degradation. The development of techniques to clone and express ciliate genes in λ phage, together with bioinformatic indices to confirm the ciliate origin of the genes has allowed the isolation and characterization of fibrolytic genes from rumen protozoa. Elimination of the ciliate protozoa increases microbial protein supply by up to 30% and reduces methane production by up to 11%. Our recent findings suggest that holotrich protozoa play a disproportionate role in supporting methanogenesis whilst the small Entodinium are responsible for much of the bacterial protein turnover. As yet no method to control protozoa in the rumen that is safe and practically applicable has been developed, however a range of plant extract capable of controlling if not completely eliminating rumen protozoa have been described.
Introduction
First described in 1843, Rumen protozoa with their striking appearance were assumed to be important for the welfare of their host. However, despite the fact that protozoa can contribute up to 50% of the bio-mass in the rumen, the role of protozoa in rumen microbial ecosystem remains unclear (Williams and Coleman, 1992). Here we evaluate recent information on the role of ciliate protozoa in the rumen microbial ecosystem.
Diversity and Phylogeny of Rumen Ciliate Protozoa
Since they were first discovered by Gruby and Delafond (1843), studies on rumen protozoa have relied on morphologic identification by optical microscopy. There are currently no culture collections of rumen ciliates, so researchers have to use photomicrographs for identification (Ogimoto and Imai, 1981; Williams and Coleman, 1992) or line drawings (Dogiel, 1927; Kofoid and MacLennan, 1930, 1932, 1933; Latteur, 1968, 1969, 1970; Dehority, 1993). It is widely accepted that microscopic identification and counting represents the gold standard for analyzing ciliate community structure in rumen studies (Williams and Coleman, 1992). However, the polymorphic nature of these microbes (Dehority, 1994, 2006) requires a high level of experience by the researcher to identify rumen ciliates at a genera or species level (Dehority, 2008). Thus, alternative ways to identify protozoa are needed since microscopic techniques are laborious and highly demanding.
Early molecular studies on rumen ciliate protozoa focussed on sequencing of 18S rRNA genes to clarify the internal phylogeny of the class Litostomatea (Embley et al., 1995; Wright et al., 1997). Embley et al. (1995) were the first to describe the monophyletic position of the anaerobic rumen ciliates Dasytricha ruminantium and Polyplastron multivesiculatum based on 18S rRNA genes and Wright et al. (1997), Wright and Lynn (1997a,b) provided molecular data for Diplodinium dentatum, Entodinium caudatum, Epidinium caudatum, Eudiplodinium maggii, Isotricha intestinalis, Ophryoscolex purkynjei, and Polyplastron multivesiculatum.
Numerous qualitative and quantitative studies focused on the diversity of ciliates have been performed since then; these studies have used a variety of techniques, including restriction fragment length polymorphism (RFLP; Regensbogenova et al., 2004a, Tymensen et al., 2012b), denaturing gradient gel electrophoresis (DGGE; Regensbogenova et al., 2004b, McEwan et al., 2005; de la Fuente et al., 2009; Belanche et al., 2010a, Kittelmann and Janssen, 2011), real-time PCR (qPCR; Sylvester et al., 2004; Skillman et al., 2006; Belanche et al., 2010a, Kittelmann and Janssen, 2011), fluorescence in situ hybridization (FISH; Xia et al., 2014) and next generation sequencing (Kittelmann et al., 2013, 2015; Ishaq and Wright, 2014; Moon-van der Staay et al., 2014).
Microscopy holds several advantages over PCR-based molecular methods for studying ciliate protozoa. First, while the vast majority of intestinal ciliates have been characterized morphologically, there is a lack of 18S rRNA gene reference sequences for many of the observed genera and species. Second, as discussed below, copy number variation of ribosomal RNA genes across the different genera or under different growth conditions may skew the observed proportions of these genera in a sample (Medinger et al., 2010). Studies using 18S rRNA gene surveys reveal an apparent higher diversity of ciliates than estimate by conventional morphological methods (Moon-van der Staay et al., 2014).
Like other ciliates, rumen protozoa contain two kinds of nuclei: a micronucleus and a macronucleus. The micronucleus possesses clearly visible chromosomes, is diploid, and synthesizes only a trace of RNA. The macronucleus contains no discernible chromosomes, has many times the diploid amount of DNA, divides amitotically, and provides virtually all of the RNA needed to run the vegetative life of the cell (Prescott and Murti, 1974). The extremely high copy number of rDNA in the macronuclear genome of ciliates, as previously found (Gong et al., 2013), is understandable considering the life history, large cell size and rapid growth of these organisms (Cavalier-Smith, 2005; Zhu et al., 2005; Godhe et al., 2008). In rumen ciliates, while the small protozoa such as Entodinium, tended to be under-represented, larger protozoa such as Epidinium or Polyplastron tended to be over-represented by a pyrosequencing approach compared to microscopic enumeration (Kittelmann et al., 2015). Similarly, Epidinium caudatum, which is approximately five times larger by volume than Entodinium caudatum (Dehority, 1993) also has approximately five times more 18S rRNA gene copies encoded in its genome (Sylvester et al., 2009). These results agree with the finding that PCR-based methods return lower estimates of abundance of small Entodinium spp. (Tymensen et al., 2012a) and overestimates of the abundance of, e.g., Polyplastron spp. (Ishaq and Wright, 2014). Knowing the copy numbers and the variations of rDNA sequences within individual eukaryotes is important both for interpreting rDNA-based diversity surveys and when 18s rDNA is used to quantify protozoal biomass. (Crosby and Criddle, 2003; Thornhill et al., 2007; Herrera et al., 2009; Amaral-Zettler et al., 2011). To date no studies addressing the number of copies per cell and variations of rDNA in rumen protozoa have been published. This is especially true as rDNA-based barcoding and microbial diversity studies using high-throughput sequencing are becoming more popular (Amaral-Zettler et al., 2009) and molecular tools based on marker gene surveys are now widely used to study the diversity of other microbes (Schlötterer, 2004; Case et al., 2007; Langille et al., 2013). Studies in non-rumen ciliates have shown that the rDNA copy number variation between and within ciliate species highlighting the difficulty of using the rDNA sequence number-based approach to infer the relative abundance of microbial eukaryotic cells in environmental samples (Wintzingerode et al., 1997; Medinger et al., 2010). Thus, latest methods based on 18S rRNA genes may be unreliable when estimating α-diversity or relative abundances of different genera and species in a given sample, although they can, reliably determine trends in relative abundances of genera and species between different samples (β-diversity). More research comparing molecular and traditional methods is needed (de la Fuente et al., 2009; Tymensen et al., 2012b; Kittelmann et al., 2015).
Functional Genes in Rumen Ciliate Protozoa
Although there are numerous copies of rDNA in ciliate macronucleus, it is likely that only a small portion of these genes are transcriptionally active in ciliates under any given growth condition, as previously shown for other eukaryotes (Reeder, 1999). Studies in Tetrahymena thermophila, Paramecium tetraurelia, and Oxytricha trifallax have pinpointed the important role of non-coding RNAs in genome rearrangement events (Feng and Guang, 2013). In spirotrichous ciliates, such as Euplotes, Stylonychia, and Oxytricha, more than 95% of the micronuclear DNA is eliminated to form the macronucleus during sexual reproduction (Swart et al., 2013) possible as a mechanism to allow the cell to adapt during times of stress. Moreover, in the macronucleus, the remaining genome is severely fragmented and these fragments are sorted and reordered under the guidance of transcripts from the parental macronucleus to produce protein-coding genes (Nowacki et al., 2011). None of these processes have been studied so far in rumen protozoa, due to the difficulties in getting full length genomes of rumen ciliates.
Despite repeated attempts it has proven impossible to maintain rumen protozoa in axenic culture. Thus it has been difficult to establish conclusively a role of ciliate protozoa in the rumen and specifically fiber degradation. Early studies sought to isolate and characterize cellulose, hemicellulase and xylanase enzymes from washed protozoal preparations (Howard et al., 1960; Bailey et al., 1962; Clayet et al., 1992), however the presence of both extra and intra cellular symbiotic bacteria in protozoal preparations made it difficult to be sure that the isolated activity was truly of protozoal origin (Delfosse-Debusscher et al., 1979; Thines-Sempoux et al., 1980). We developed techniques to clone and express ciliate genes in λ phage (Eschenlauer et al., 1998), using FISH to confirm the protozoal identity of the expressed genes (Newbold et al., 2005) and developing bioinformatic indices to confirm the ciliate origin of the genes (McEwan et al., 2000). Using these techniques we and others have been able to isolate and characterize genes from a range of rumen protozoa (McEwan et al., 1999; Newbold et al., 2005; Belzecki et al., 2007; Boxma et al., 2007). This includes a wide range of fibrolytic enzymes a number of which have been found to contain multiple domains with binding domains and putative chimeric constructs being observed suggesting a highly evolved fibrolytic capacity in the rumen ciliates (Devillard et al., 1999, 2003; Takenaka et al., 1999, 2004; Wereszka et al., 2004; Bera-Maillet et al., 2005). This observation has been confirmed by recent metagenomic screening of protozoal glucosidases and eukaryotic metatranscriptomes that have confirmed that a diverse range of diverse glycoside hydrolases are present in the rumen protozoa (Findley et al., 2011; Qi et al., 2011).
Based on large-scale construction and analysis of phylogenies of over 4000 Expressed Sequence Tag libraries from the rumen ciliates Entodinium caudatum, Eudiplodinium maggii, Metadinium medium, Diploplastron affine, Polyplastron multivesiculatum, Epidinium ecaudatum, Isotricha prostoma, Isotricha intestinalis, and Dasytricha ruminantium. Ricard et al. (2006) concluded there was extensive evidence of horizontal gene transfer (HGT; 148 out of 3563 non-redundant genes) from bacteria and archaea in rumen ciliate genomes. Among the HGT candidates, they reported an over-representation (>75%) of genes involved in metabolism, specifically in the catabolism of complex carbohydrates (Ricard et al., 2006), suggesting that HGT may have been important in allowing rumen ciliates to adapt to new niches within the rumen and that fibrolytic genes were acquired by protozoan from bacterial sources (Findley et al., 2011).
Role of Protozoa in the Rumen
Despite the fact that that protozoa make up a large portion of the rumen biomass, their role in ruminal fermentation and their contribution to the metabolism and nutrition of the host is still an area of substantial controversy (Williams and Coleman, 1992). In the last section of this paper we will review different strategies to manipulate the rumen protozoal density; however most of these dietary interventions also lead to modifications in rumen function making it difficult to assess the effect of the rumen protozoa per se. Rumen protozoa are not essential to the animal to survive and defaunation (the removal of protozoa from the rumen using a wide variety of chemicals and physical techniques) and protozoa-free animals have been used to study the role of ciliate protozoa in the rumen function without been affected by dietary interventions (Williams and Coleman, 1992). This paper does not aim to conduct a complete review on the effect of defaunation but compiles the most relevant publications since the excellent and complete review by Williams and Coleman (1992). Thus, a meta-analysis was conducted to study the main effects of defaunation based on 23 in vivo studies comprising 48 comparisons (Kreuzer et al., 1986; Vermorel and Jouany, 1989; Ushida et al., 1990; Frumholtz, 1991; Nagaraja et al., 1992; Hegarty et al., 1994, 2008; Faichney et al., 1999; Chandramoni et al., 2001; Machmuller et al., 2003; Eugène et al., 2004; Ozutsumi et al., 2005; Ohene-Adjei et al., 2007; Yáñez-Ruiz et al., 2007a,b; Bird et al., 2008; Morgavi et al., 2008, 2012; Belanche et al., 2011, 2012b, 2015; Mosoni et al., 2011; Zhou et al., 2011). Most of the studies were performed using sheep (87%), while the rest used cattle (13%). Isolation of new born animals from their mothers (40%), use of detergents and other chemicals (35%, using sodium lauryl sulfate, alkanes, synperonic NP9, calcium peroxide, copper sulfate etc.), and ruminal manipulation (25%, emptying and washing of the rumen), were used in order to achieve defaunation of the animals. Trials where the effect of additives, other than defaunation, were significant were discarded from the meta-analysis. Similarly, studies using monofaunated animals or selective defaunation were not included. The majority of the data (75%) were from trials in which animals were fed at maintenance and the remaining trials were from production trials. Animals were mainly fed mixed diets (90%) composed of forages supplemented with concentrate, while 10% of the diets were purely composed of forage. Rumen fermentation data and methane emissions were reported in most trials (69%), while information about digestion (31%), animal performances (12%) and rumen microbial populations (10%) was less abundant. Methane production was measured in chambers (75% in open chambers or respiratory calorimeters) or with the SF6 (sulfur hexafluoride) tracer method (25% of studies).
Multiple comparisons were included from an individual publication with multiple studies. For each comparison included in the analysis, the effect size was calculated as the natural logarithm of the response ratio (mean value in the defaunated treatment divided by mean value in control treatment) and variance of the ratio calculated based on the reported standard deviation or standard error of the mean for each comparison (Viechtbauer, 2010). All defaunation effects were weighted according to the number of observations (n) in each comparison. The meta-analysis was computed fitting a random-effect model with a DerSimonian-Laird estimator (Dersimonian and Laird, 1989) for assessing heterogeneity (τ2) in the Metafor package of R for each category separately as follow:
where θi = true effect size in the ith study, μ = overall true effect size and ei = random deviation from the overall effect size [ui ∼ N (0, τ2)] (Viechtbauer, 2010). For evaluating the response ratio, values below one indicated a negative, while values above one indicated a positive effect of defaunation on that particular parameter.
This meta-analysis confirmed many of the previously reported results (Williams and Coleman, 1992) and helped clarify number of areas in which results were still conflicting. Figure 1 shows the main effects of defaunation on the rumen function indicating that the physical and chemical characteristics of the rumen environment were changed by defaunation; although the nature of the observed change was not always consistent. Rumen volume and solid turnover rate were unaffected by defaunation, while the liquid turnover rate tended to decrease (–14%, P = 0.07). Elimination of protozoa from the rumen significantly decreased rumen OM digestibility (–7%, P = 0.008) and particularly NDF (–20%, P = 0.040) and ADF digestibility (–16%, P = 0.100), probably as a result of the loss of protozoal fibrolytic activity. This activity seems however to differ across the different protozoal groups: Large Ophyroscolecidae such as Epidinium, Polyplastron and Eudiplodinium have greater endoglucanase and xylanase activity (Williams and Coleman, 1992). On the other hand, Entodinium spp. have only weak activity (Williams and Coleman, 1992). Similarly, Dasytricha has glucosidase and cellobiosidase activity but negligible fibrolytic activity (Takenaka et al., 2004). The lower rumen digestibility in defaunated animals is partially compensated by a greater post ruminal digestion resulting in less pronounced differences in terms of total tract digestibility for OM (–4%, P = 0.089), CP (–3%, P = 0.034), NDF (–11%, P < 0.001), and ADF (–9%, P < 0.001; Figure 2). Another compensatory factor could be a shift toward more energetically efficient reactions in the rumen (less methane emissions) and less metabolic energy required by defaunated animals to eliminate the excess of urea as a result of the lower bacterial protein breakdown and ammonia levels in the rumen. Despite these compensatory mechanism, the decrease in feed digestibility is likely to be the main drawback of defaunation since it could limit the feed intake and efficiency of feed utilization at production levels of intake; this coupled to the lack of a commercially viable approach (see below) to defaunation, mean that defaunation is not recommended as a methane mitigation strategy under farm conditions (Hristov et al., 2013).
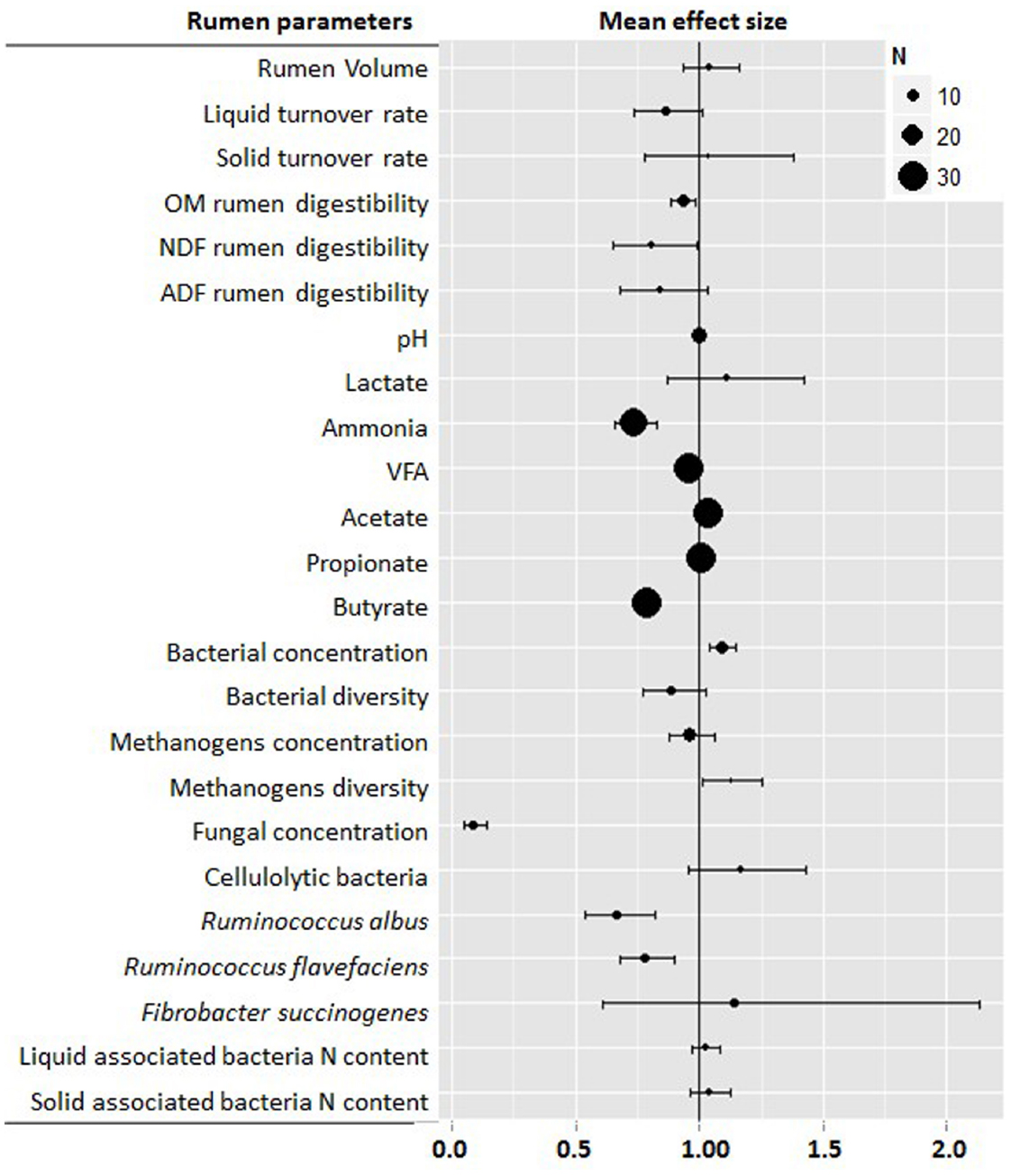
Figure 1. Meta-analysis describing the effects of defaunation on rumen function. For each parameter information is provided about the number of studies, observations (n), and range of values. Graph shows the mean effects and 95% confidential intervals. Values below one indicate a negative effect, while those above one indicate a positive effect of defaunation on that particular parameter.
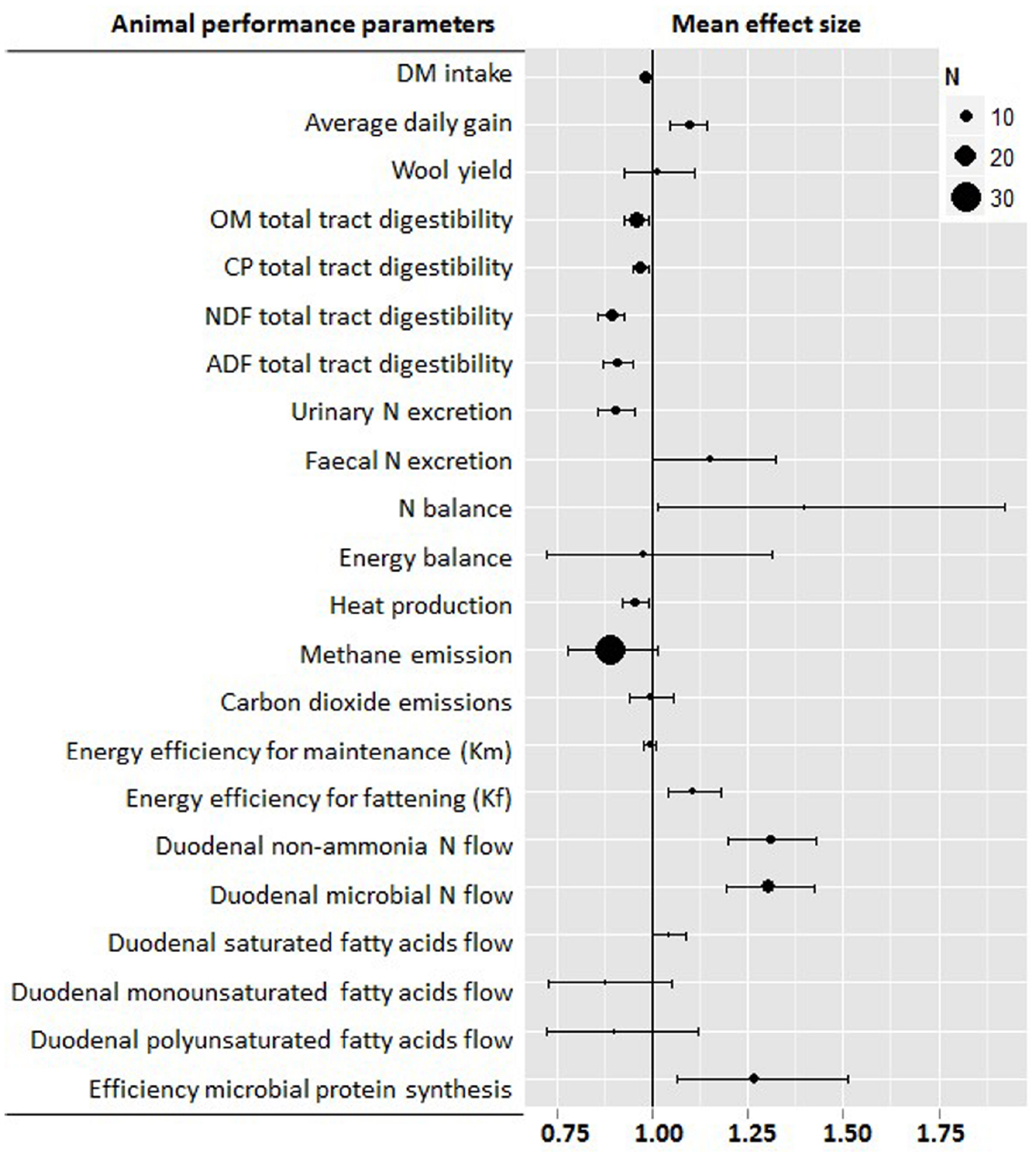
Figure 2. Meta-analysis describing the effects of defaunation animal performance. For each parameter information is provided about the number of studies, observations (n), and range of values. Graph shows the mean effects and 95% confidential intervals. Values below one indicate a negative effect, while those above one indicate a positive effect of defaunation on that particular parameter.
The total concentration and production of fermentation products also differs in faunated and defaunated animals. A decrease in rumen ammonia (–26%, P < 0.001) is probably the most consistent of the observed effects of protozoal elimination and seems to be due to decreased bacterial protein breakdown and feed protein degradability in the absence of rumen protozoa (Williams and Coleman, 1992). Moreover it has been demonstrated that although bacterial predation by rumen protozoa is dependent on the protozoal size, holotrich protozoa have a much lower predatory activity than entodiniomorphids (Belanche et al., 2012a) and ultimately a lower impact on rumen ammonia concentration (Belanche et al., 2015) and duodenal microbial protein flow (Ivan et al., 2000a; Ivan, 2009). However, this negative effect of entodiniomorphids on the duodenal protein flow may be overestimated due to the lack of a reliable marker to measure protozoal flow (Broderick and Merchen, 1992). In previous reviews higher lactate levels have been reported in defaunated animals (Williams and Coleman, 1992) because protozoa consume lactate more rapidly than bacteria (Newbold et al., 1986). Our meta-analysis showed a numerical increase in lactate concentration in defaunated animals (+11%) but this increase was non-significant possibly because the dataset used in this meta-analysis was mainly composed by high-forage diets which are less prone to promote these particular nutritional disorders.
Although rumen pH was unaffected, the lower VFA concentration observed in defaunated animals (–5%, P = 0.013) seems to highlight the role of protozoa in the synthesis of VFA and feed degradation. The ability of protozoa to engulf exogenous fatty acids (Karnati et al., 2009) may divert more carbon toward VFA production in preference to fatty acid synthesis and ultimately increase VFA production. More interestingly, our data suggested that defaunation substantially decreased butyrate (–22%, P < 0.001), slightly increased acetate (+3%, P < 0.001) and had no effect on propionate molar proportions. In a previous meta-analysis (Noziere et al., 2011) it has been reported that NDF digestibility positively correlates with acetate molar proportion (r = 0.95) but negative with propionate (r = –0.94) and butyrate (r = –0.91), therefore our observations might indicate that the shift in the molar proportions of VFA induced by defaunation seems to be mainly driven by a decrease in the fiber digestion. On the basis of stoichiometry, such a shift in rumen VFA production should result in a decrease in methane production as less metabolic H2 will be available as a substrate for methanogenesis (Demeyer et al., 1996). The effect of defaunation on methane production is however still not clear; in various reviews compiling in vivo and in vitro studies (Hegarty, 1999) or just in vivo (Morgavi et al., 2010) it was concluded that removal of protozoa from the rumen would result in a 13 and 10.5% decrease in methane production, which fully agree with our results based on more recent studies (–11%, P = 0.074). In line with these observations, in a meta-analysis containing 28 experiments and 91 treatments, it has been reported a significant linear relationship between methane emissions and protozoal concentration (r = 0.96) with a decrease in methane yield averaged 8.14 g/kg DMI per each log unit reduction in the protozoal concentration (Guyader et al., 2014), suggesting that protozoa played a catalytic role in rumen methanogenesis. The reasons for the lower methane emissions in defaunated animals are still controversial since the number of rumen protozoa explains only one part (approximately 47%) of the variability in methane emissions indicating that rumen methanogenesis is a complex process in which multiple microbes are involved (Morgavi et al., 2010). A number of mechanisms by which protozoa could enhance methanogenesis are possible based on their ability to produce H2 in their hydrogenosomes (a mitochondria-like organelle), their ability to host epi- and endo-symbiotic methanogens and to protect them from oxygen toxicity (Fenchel and Finlay, 2006). This interaction is a typical example of interspecies H2 transfer that favors both the methanogens and the protozoa (Ushida et al., 1997). One hypothesis is that defaunation leads to decreased methanogen numbers, which are the sole producers of methane in the rumen, but our meta-analysis reported that this decrease in methanogens levels was not significant (–3%, P = 0.48). An alternative hypothesis suggests that defaunation results in the elimination of protozoa-associated methanogens, which could be considered as one of the most active methanogens communities in the rumen (Finlay et al., 1994), however this later hypothesis based on the substitution of methanogen communities which differ in their methanogenic activity requires further investigation. Several authors have studied the endo-symbiotic archaeal population in protozoa (Sharp et al., 1998; Irbis and Ushida, 2004; Regensbogenova et al., 2004b; Tymensen et al., 2012a) and most of them agree that Methanobrevibacter sp. is the predominant genus, while the contribution of Methanomicrobium sp. and Methanomassiliicoccales to the endosymbiotic methanogen community is variable and could indicate differences among protozoal groups. The development of molecular techniques over the last decades has allowed colleagues to further explore these microbial interactions revealing that ciliate endosymbiotic methanogens differ from rumen free-living methanogens (Tokura et al., 1997; Sharp et al., 1998). In a recent publication it has also been demonstrated that holotrich protozoa have a different endosymbiotic methanogens than entodiniomorphids (Belanche et al., 2014) possibly because either holotrich protozoa have more active hydrogenosomes than entodiniomorphids (Paul et al., 1990) and/or rapid synthesis of glycogen by holotrich protozoa in the presence of excess carbohydrates generate more hydrogen (Hall, 2011; Denton et al., 2015). These differences may explain the greater impact of holotrich protozoa on rumen methanogenesis compared to entodiniomorphids (Belanche et al., 2015). This later observation is based on the successive inoculation of fauna-free sheep with various protozoal groups, thus it should be cautiously interpreted due to possible confounding effects of treatment and period.
The effect of defaunation on the rumen microbial ecosystem is not limited to only the methanogen population and it has been demonstrated that defaunated animals had significantly greater ruminal bacterial populations (+9%, P < 0.001) than control animals. The reason for this seems to be based on the availability of an ecological niche for the bacteria when protozoa are not present in the rumen combined with the removal of protozoal predation (Williams and Coleman, 1992). Recent studies using molecular techniques have demonstrated that defaunation also modified the structure of the bacterial community leading to a simplification in population structure and lower bacterial diversity (Belanche et al., 2012b, 2015). Our meta-analysis revealed a drop in the concentration of fibrolytic microbes such as anaerobic fungi (–92%, P < 0.001), Ruminococcus albus (–34%, P < 0.001) and Ruminococcus flavefaciens (–22%, P < 0.001) in the absence of protozoa. This observation suggests that fiber digestion in the rumen is a complicated task which requires the symbiotic collaboration of several fibrolytic microbes, including rumen protozoa, to carry out the initial stages of fiber colonization and digestion; therefore the absence of rumen protozoa seems to have a detrimental effect on this fibrolytic consortium and ultimately in fiber digestion. However, Hsu et al. (1991) reported an increase in ruminal fungal zoospores in defaunated animals possibly as a result of the removal of protozoal predation and competition for nutrients (protozoa vs. fungi), clearly more studies are needed to fully understand protozoa interactions with other rumen microbes under different dietary regimes.
The effect of the presence of rumen protozoa on pathogen’s survival in the rumen and pathogen shedding is another area of interest. As noted above rumen protozoa engulf and digest a wide range of bacteria (Williams and Coleman, 1992) and can reduce the shedding of potential pathogens from the animal, although the effect is highly dependent on the composition of the protozoal population present (Stanford et al., 2010). However, it has also been shown that rumen protozoa enhance the pathogenicity of certain pathogens leaving the rumen (Rasmussen et al., 2005; Carlson et al., 2007) suggesting that more work is needed in this area.
Figure 2 summarizes the effects of protozoa on animal productivity as the indirect effect of defaunation on the rumen function. Defaunation improved feed conversion rate as a result of the lower DM intake (–2%, P < 0.001) and greater average daily gain (+9%, P < 0.001). These positive effects of defaunation are particularly obvious with poor quality diets in which the average daily gain was low, possibly as a result of the lower availability of fermentable energy and rumen digestible protein for the rumen microbes (Williams and Coleman, 1992). In a previous meta-analysis the effect of defaunation appeared more pronounced when the ratio N/NDF was below 6 and the percentage of concentrate lower than 40% (Eugène et al., 2004). As defaunated animals have lower feed digestibility, absorbed energy is lower than in conventional animals. Thus better feed conversion rate after defaunation may be mainly attributed to the higher efficiency of utilization of absorbed nutrients. Indeed our meta-analysis showed that defaunation promotes a greater efficiency of energy utilization for fattening (+11%, P = 0.001), possibly as a result of a lower heat production (–5%, P = 0.010). Other hypothesis to explain the better performance of defaunated animals rely on a more efficient use of dietary protein. There is clear evidence that holotrich protozoa leave the rumen more slowly than bacteria (Abe et al., 1981). The amount and rate of protozoal flow to the lower gut is however the subject of much debate. Firkins et al. (2007), found that post-ruminal flow of protozoa is proportional to rumen protozoal biomass. Although, protozoal counts in the rumen and abomasal fluid indicated that abomasal counts were only 6–64% of rumen fluid counts (Punia et al., 1992), numbers of protozoa in free rumen fluid are unreliable indicators of protozoal biomass and outflow, because the majority of rumen protozoa (63–90%) are found either associated with feed particles or sequestered in the rumen wall (Hook et al., 2012). The use of specific markers of protozoa such as 2-aminoethylphosphonic acid has been questioned because it is present in bacteria and in feed (Ling and Buttery, 1978). Protozoal outflow measured by use of a general microbial markers (e.g., 15N and 35S and purine bases), microbial N minus specific bacterial markers (such as 2,6-diaminopimelic acid, DAPA) yielded variable estimates of protozoal outflow and sometimes even negative values due to methodological limitations for the markers used (Broderick and Merchen, 1992). Recent studies based on the use of 18S rRNA as a novel protozoal marker have reported that although protozoa can represent up to 60% of the rumen microbial biomass, they rarely exceeds 20% of the microbial protein flow into the small intestine (Sylvester et al., 2004, 2005; Yáñez-Ruiz et al., 2006; Belanche et al., 2011, 2012b). However, these new techniques are not free from their own limitations: Sylvester et al. (2005) observed that up to 48% of the protozoal N was actually N from contaminating bacteria which could led to inflate the protozoal N pool in the rumen, particularly when animals have low feed intakes and low particulate passage rate (Dijkstra, 1994). Similarly, Belanche et al. (2010b) observed a greater degradation of protozoal DNA compared to bacterial DNA during abomasal digestion which could result in an underestimation of the protozoal N outflow from the rumen. Despite these limitations, these new findings seem to support that rumen protozoal are partially sequestrated in the rumen. This rumen sequestration is however not equal for all protozoal groups; holotrich protozoa associate to the feed particles after feeding due to their strong chemotaxis toward sugars (Diaz et al., 2014), but rapidly migrate to the ventral reticulorumen to prevent being washed out of the rumen (Karnati et al., 2007), on the contrary Entodiniomorphids also associate to feed due to their moderate chemotaxis toward glucose and peptides but do not show the same affinity to the rumen wall (Diaz et al., 2014) and thus flow out the rumen with the particulate phase (Hook et al., 2012).
Protozoa predate on bacteria as their main protein source (Williams and Coleman, 1992) and as a result, defaunation makes the rumen more efficient in terms of proteosynthesis increasing the duodenal flow of microbial protein (+30%, P < 0.001) and total non-ammonia N flow (+31%, P < 0.001). Defaunation also increased the efficiency of microbial protein synthesis (+27%, P = 0.008) as a result of both a better microbial proteosynthesis and a lower OM digestion. Protozoal generation time is far higher than that of bacteria, thus the energetic requirements for maintenance are higher when expressed as a ratio of protein leaving the rumen (Williams and Coleman, 1992). As a result, the presence of protozoa has a negative impact on the overall energetic efficiency of the rumen ecosystem. In addition, defaunation can also modify the composition of the rumen bacteria (Belanche et al., 2012b) and ultimately the amino acid profile of the duodenal protein supply promoting an increase in specific amino acids such as leucine, threonine and arginine (Ivan, 2009), but not lysine which is considered the main limiting amino acid in high producing animals (Hristov and Jouany, 2005). After a series of in vivo experiments in which fauna-free lambs were progressively inoculated with protozoal species (i.e., Isotricha intestinalis, Dasytricha ruminantium, Polyplastron multivesiculatum, Epidinium ecaudatum, Eudiplodinium maggi, and Entodinium caudatum) in different sequential orders (Ivan et al., 2000a,b; Ivan, 2009), Ivan concluded that holotrich protozoa engulf only a very small number of rumen bacteria and have a small effect on the duodenal NAN flow and protein metabolism. Similarly, holotrich protozoa had no significant effect on the fiber digestion. This together, with the contribution of holotrich’s to rumen methanogenesis (Belanche et al., 2015), seems to indicate that presence of holotrich protozoa in the rumen is of little value to ruminant production, unless high-carbohydrate diets are used, in which case the presence of holotrich protozoa could be beneficial as a result of their ability to engulf and accumulate starch grains and soluble carbohydrates (Williams and Coleman, 1992). This engulfment of highly fermentable carbohydrates prevents alternative bacterial fermentation that would otherwise decrease pH and increase the onset of lactic acid acidosis (Mackie et al., 1978). On the contrary cellulolytic protozoa (Polyplastron, Epidinium, and Eudiplodinium) could be beneficial to animals fed with fibrous diets. However, these fibrolytic protozoa and Entodinium spp. have a substantial potential to engulf and degrade bacteria (Belanche et al., 2012a) and might be detrimental in terms of protein utilization by the ruminant host. Therefore their presence in the rumen may not be helpful in animals fed low protein diets because their fibrolytic beneficial effects would be counterbalanced by the sensitivity of fibrolytic bacteria and anaerobic fungi to N shortage in the rumen (Belanche et al., 2012c). Thus it seems that defaunation decreased urinary N losses here (–10%, P < 0.001) due to a combination of lower dietary CP degradation in the rumen and lower rumen bacterial breakdown. However, the great variability observed in terms of N balance highlights the fact that defaunation may have different effects on the overall efficiency of N utilization depending on the diet consumed by the animal in particular the level of intake and particle passage rate through the tract (Dijkstra, 1994).
The effects of defaunation on ruminal lipid metabolism are less defined. The protozoa contribute to the total microbial lipolytic activity but their role in bio-hydrogenation is less well-understood (Williams and Coleman, 1992) although they do contribute significantly to flow of unsaturated fatty acids leaving the rumen (Yáñez-Ruiz et al., 2006). Using steers fed diets with different chlorophyll levels, it was demonstrated that the high levels of polyunsaturated fatty acids in protozoal cells appears to be associates with ingestion of chloroplasts (Huws et al., 2009). This chloroplasts uptake seems to be specific of Entodiniomorphids since no engulfed chloroplasts have been found in holotrich protozoa (Huws et al., 2012). Thus protozoa appear to protect chloroplast unsaturated fatty acids from the rumen bio-hydrogenation increasing the duodenal flows of mono and polyunsaturated fatty acids. Our meta-analysis agrees with this observation, and defaunation promoted an increase of saturated fatty acids (+4%, P = 0.046) and a numerical decrease in mono and polyunsaturated fatty acids (–13 and –10%, respectively) which possibly did not reached significance as a result of diet-dependent effects.
The findings reported in our meta-analysis need to be carefully interpreted since most of the studies had methodological limitations due to the intrinsic difficulty of the defaunation process leading to:
(1) Confusion between the effect of defaunation and period when using the same experimental animals in time,
(2) Nearly all studies estimating protozoal pool size have failed to report duodenal flows of protozoal biomass and vice versa, moreover protozoal flow is often underestimated using traditional microbial markers (Broderick and Merchen, 1992),
(3) Studies assessing methane production have not accounted for other variable effects besides protozoal abundance (i.e., NDF digestibility),
(4) Animals are often fed at levels far below the production levels of intake,
(5) In some studies the adaptation period after defaunation/refaunation is too short for the microbial ecosystem to fully adapt,
(6) Elimination of rumen protozoa modifies the ecological structure in the rumen and ultimately alternative microbial groups can take over ecological niche previously filled by protozoa.
Thus, more studies are needed using state-of-the-art technologies to quantify protozoal activity (i.e., fibrolytic and proteolytic) as well as the ruminal protozoal pool size relative to protozoal N outflow for better understanding of the role of protozoa on the ruminant’s metabolism. These studies should be done at production levels of intake in order to truly assess the effect of defaunation per unit of product (milk or meat) produced.
Manipulation of Rumen Protozoa
Current concerns regarding the role of livestock in global warning has driven researchers to search for strategies to manipulate rumen protozoa to decrease methane production. As has been previously mentioned, there is a linear relationship between protozoal concentration and methane emissions (Guyader et al., 2014) and it has been estimated that between 9 and 37% of ruminal methane production can be attributed to methanogens associated with protozoa in the rumen (Finlay et al., 1994; Newbold et al., 1995; Machmuller et al., 2003).
Most approaches to defaunation rarely result in the total removal of protozoa from the rumen with their effectiveness largely dependent on diet composition (Hegarty, 1999). Treatments normally used to partially or completely defaunate the rumen include: chemicals that are toxic to protozoa (copper sulfate, dioctyl sodium sulfosuccinate, alcohol ethoxy-late or alkanates, calcium peroxide), ionophores, lipids, and saponins (Williams and Coleman, 1992; Jouany, 1996; Hook et al., 2010).
A recent meta-analysis by Guyader et al. (2014) has shown a concomitant reduction in protozoal numbers and methane emissions in 31% of 70 studies using different methane reduction strategies. Most of the studies used lipids as a protozoal control/methane mitigation strategy. The authors also reported that the antiprotozoal effect of lipids depends on the fatty acid composition with medium chain fatty acids more effective than polyunsaturated fatty acids in controlling protozoal numbers. Supplements rich in polyunsaturated fatty acids such as linoleic acid (C18:2 from soybean and sunflower) and linolenic acid (C18:3 from linseed) have been shown to have a negative effect on methane production (4.1 and 4.8% decrease per percentage unit of added lipids, respectively, Martin et al., 2010). The antimethanogenic effect of polyunsaturated fatty acids has been related to their toxic effect on cellulolytic bacteria (Nagaraja et al., 1997) and protozoa (Doreau and Ferlay, 1995). Although it was originally suggested that the biohydrogenation of polyunsaturated fatty acids in the rumen could represent an alternative H2 sink to methanogenesis (Lennarz, 1966), it is now believed that the significance of biohydrogenation to the overall H2 sink is small (Nagaraja et al., 1997). It has been shown that medium chain fatty acids have potent antiprotozoal effect. Several studies have reported decreased ruminal methanogenesis when supplementing lauric acid (C12:0) and myristic acid (C14:0), either in pure forms or in products rich in these fatty acids (coconut oil) under in vitro (Dohme et al., 2001; Soliva et al., 2004) or in vivo conditions (Machmuller et al., 2003; Jordan et al., 2006). Martin et al. (2010) reported that medium chain fatty acids, mainly provided by coconut oil, resulted in a decrease in methane of 7.3% per percentage unit of added lipid. However, the methane suppression effect observed was not always related to a decrease in protozoa concentration (Machmuller et al., 2003) which may be due to a direct effect of medium chain fatty acids on methanogens (Dohme et al., 1999; Panyakaew et al., 2013). A recent study (Faciola and Broderick, 2014), found that both coconut oil and lauric acid reduced the number of protozoa by 40% but whereas lauric acid altered fiber digestibility coconut oil did not. However, reductions in methane production and a concomitant decrease in dry matter intake when coconut oil and lauric acid were used as defaunating agents has been reported (Hristov et al., 2013), which would potentially limit their practical on-farm use. Clearly more studies are needed to work out how to use oils and fatty acids to control protozoa in the rumen
The literature suggests that saponins mitigate methanogenesis mainly by reducing the numbers of protozoa, whilst condensed tannins act by both reducing the number of protozoa and by a direct toxic effects on methanogens, whereas essential oils act mostly by a direct toxic effect on methanogens (Cieslak et al., 2013). In agreement with this information, a meta-analysis of the effect of phytochemicals on methanogenesis (Patra, 2010) has shown that changes in protozoa numbers followed a linear relationship with changes in methane production by saponins (r = 0.69) and tannins (r = 0.55), but this relationship was weaker (r = 0.45) with respect to essential oils. Methane inhibition by organosulfur compounds was not associated with changes in the protozoal population (Patra, 2010) as such compounds specifically inhibit methanogenic archaea. For tannin containing plants, the antimethanogenic activity has been attributed to the group of condensed tannins. It has been suggested that tannins have a direct effect on ruminal methanogens and an indirect effect on hydrogen production due to a reduction in fiber digestion (Tavendale et al., 2005). Regarding their effect on protozoa, some studies have reported no effect whereas others have shown a reduction in protozoa numbers in the presence of tannins (reviewed by Patra and Saxena, 2009). This inconsistency is probably due to differences in the structure and dose of the condensed tannin. Saponins, shows a more consistent inhibitory effect on rumen protozoa in the available literature. Goel and Makkar (2012) have suggested that the risk of impaired rumen function and thus reduced animal productivity is greater with tannins than with saponins and when used to decrease methane production, the effective concentration range for tannins is narrower than for saponins. Saponins are glycosylated triterpenes or steroids where the saponin is the aglycone while the glycone is a carbohydrate unit consisting of a monosaccharide or smaller oligosaccharide entity. The most commonly sources of saponins used in ruminant nutrition are Yucca schidigera, rich in sterol saponins (10%), and Quillaja saponaria which contains triterpene saponins. Lately, other sources of saponins such as tea saponins have being explored (Hu et al., 2005; Guo et al., 2008). The antiprotozoal effect of saponins is related to their interaction with the sterol moiety which is present in the membrane of protozoa (Patra and Saxena, 2009). It has been suggested that saponins with the same aglycone may have a different effect on protozoa depending on the sugar composition and arrangement (Wina et al., 2006). This effect seems to be transitory due to the cleavage of the glycosidic bond by rumen microbes (Newbold et al., 1997). The suppression of rumen protozoa by saponins or saponin-containing plants has been consistently observed in in vitro studies (Wina et al., 2005). However, in vivo studies have shown that the antiprotozoal effect of saponins tends to disappears after 7–14 days of administration (Wina et al., 2005; Patra and Saxena, 2009). It has been suggested that when saponins are deglycosylated to sapogenins by rumen microbes they become inactive (Newbold et al., 1997). Thus, it can be hypothesized that the combination of saponins with glycosidase inhibitors would avoid deglycosylation, maintaining the intact saponin and, therefore, the activity in the rumen. Preliminary in vitro studies (Ramos-Morales et al., 2014) using 2,5-Dihydroxymethyl-3,4-dihydroxypyrrolidine (DMDP) as a glycosidase inhibitor combined with a plant extract rich in saponins have shown the potential of this approach to maintain saponin activity over time. Another approach that is being explored by our research group is the synthesis of a chemically modified saponin without the natural glycoside bonds so the enzymatic cleavage would be structurally prohibited. The effects of saponins on rumen fermentation have not been found to be consistent. These discrepancies appear to be related to the chemical structure and dosage of saponins, diet composition, as well as adaptation of the microorganisms to saponins (Wina et al., 2005; Patra and Saxena, 2009).
Australian researchers have demonstrated the potential of vaccination against methanogens as a method for mitigating methane emissions (Wright et al., 2004). Although this technology is still developing it provides many options for long term methane reduction. Similarly, protozoa, as providers of hydrogen to methanogens, or acetogens which compete for hydrogen with methanogens, could be possible vaccine targets for the reduction of methane emissions. An immunological approach has been explored for defaunation (Williams et al., 2008). Vaccination of sheep with entodinia or mixed protozoal antigens reduced protozoa numbers and IgG antibodies generated against rumen protozoa remained active and continued to bind target cells for up to 8 h. However, no in vivo effect on rumen protozoa has been observed. It has been suggested that the reasons for the lack of effect may be related to insufficient amount of specific Ig delivered in saliva, need of an adjuvant to optimize the production of salivary antibodies, target other antigens of protozoa to generate a greater immune response.
Conclusion
Since the landmark publication of the Rumen Protozoa by Williams and Coleman (1992), there has been steady but perhaps not spectacular progress in our understanding of rumen protozoa. The advent of molecular techniques has led to a raft of publications regarding protozoal diversity in the rumen and while as discussed above the techniques used have within themselves limitations in their ability to accurately quantify individual protozoal genera they have provided new insights into the diversity of ciliate protozoa in different ruminant species, in different geographies and under different dietary situations. There has been steady progress in the area of defaunation and whilst at this stage no commercially available defaunation technique has yet been marketed, it seems likely that plant extracts can be used to control protozoa in the rumen; if not completely eliminate them. Work on the consequence of elimination of protozoa has largely focused on their role in methanogenesis, reflecting current concerns regarding the role of ruminants in greenhouse gas production. However the use of meta-analysis of existing data combined with new defaunation studies have help clarify our understanding of the role of protozoa in the rumen as illustrated in Figures 1 and 2. It is however perhaps the development of molecular techniques to clone and characterize protozoal genes, originally from single species but more recently from metagenomic and transcriptomic sources, that seems to offer the greatest, but as yet largely unfulfilled, potential to help truly elucidate the role of rumen protozoa in the rumen and in the absence of progress in developing axenic culture of rumen protozoa more effort needs to be put into characterizing rumen protozoa activity through molecular methods.
Author Contributions
All authors have equally contributed to the preparation of this publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CN and AB acknowledge the support of the Biotechnology and Biological Sciences Research Council, UK via grant number BB/J0013/1.
References
Abe, M., Iriki, T., Tobe, N., and Shibui, H. (1981). Sequestration of holotrich protozoa in the reticulo-rumen of cattle. Appl. Environ. Microbiol. 41, 758–765.
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., and Huse, S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of v9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4:e6372. doi: 10.1371/journal.pone.0006372
Amaral-Zettler, L. A., Zettler, E. R., Theroux, S. M., Palacios, C., Aguilera, A., and Amils, R. (2011). Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J. 5, 42–50. doi: 10.1038/ismej.2010.101
Bailey, R. W., Clarke, R. T., and Wright, D. E. (1962). Carbohydrases of the rumen ciliate Epidinium ecaudatum (Crawley). Biochem. J. 83, 517–523. doi: 10.1042/bj0830517
Belanche, A., Abecia, L., Holtrop, G., Guada, J. A., Castrillo, C., de la Fuente, G., et al. (2011). Study of the effect of presence or absence of protozoa on rumen fermentation and microbial protein contribution to the chyme. J. Anim. Sci. 89, 4163–4174. doi: 10.2527/jas.2010-3703
Belanche, A., Balcells, J., De La Fuente, G., Yañez-Ruíz, D. R., Fondevila, M., and Calleja, L. (2010a). Description of development of rumen ecosystem by PCR assay in milk-fed, weaned and finished lambs in an intensive fattening system. J. Anim. Physiol. Anim. Nutr. 94, 648–658. doi: 10.1111/j.1439-0396.2009.00952.x
Belanche, A., Erroa, I. R., Balcells, J., and Calleja, L. (2010b). Use of quantitative real-time PCR to assess the in vitro survival of specific DNA gene sequences of rumen microbes under simulated abomasal conditions. J. Anim. Physiol. Anim. Nutr. 94, 204–211. doi: 10.1111/j.1439-0396.2008.00901.x
Belanche, A., de la Fuente, G., Moorby, J. M., and Newbold, C. J. (2012a). Bacterial protein degradation by different rumen protozoal groups. J. Anim. Sci. 90, 4495–4504. doi: 10.2527/jas.2012-5118
Belanche, A., de la Fuente, G., Pinloche, E., Newbold, C. J., and Balcells, J. (2012b). Effect of diet and absence of protozoa on the rumen microbial community and on the representativeness of bacterial fractions used in the determination of microbial protein synthesis. J. Anim. Sci. 90, 3924–3936. doi: 10.2527/jas.2011-4802
Belanche, A., Doreau, M., Edwards, J. E., Moorby, J. M., Pinloche, E., and Newbold, C. J. (2012c). Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. J. Nutr. 142, 1684–1692. doi: 10.3945/jn.112.159574
Belanche, A., de la Fuente, G., and Newbold, C. J. (2014). Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 90, 663–677. doi: 10.1111/1574-6941.12423
Belanche, A., de la Fuente, G., and Newbold, C. J. (2015). Effect of progressive inoculation of fauna-free sheep with holotrich protozoa and total-fauna on rumen fermentation, microbial diversity and methane emissions. FEMS Microbiol. Ecol. 362, 1–10. doi: 10.1093/femsec/fiu026
Belzecki, G., Newbold, C. J., McEwan, N. R., McIntosh, F. M., and Michalowski, T. (2007). Characterization of the amylolytic properties of the rumen ciliate protozoan Eudiplodinium maggii. J. Anim. Feed Sci. 16, 590–606.
Bera-Maillet, C., Devillard, E., Cezette, M., Jouany, J. P., and Forano, E. (2005). Xylanases and carboxymethylcellulases of the rumen protozoa Polyplastron multivesiculatum, Eudiplodinium maggii and Entodinium sp. FEMS Microbiol. Lett. 244, 149–156. doi: 10.1016/j.femsle.2005.01.035
Bird, S. H., Hegarty, R. S., and Woodgate, R. (2008). Persistence of defaunation effects on digestion and methane production in ewes. Aust. J. Exp. Agric. 48, 152–155. doi: 10.1071/EA07298
Boxma, B., Ricard, G., van Hoek, A. H., Severing, E., Moon-van der Staay, S. Y., van der Staay, G. W., et al. (2007). The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol. Biol. 7:230. doi: 10.1186/1471-2148-7-230
Broderick, G. A., and Merchen, N. R. (1992). Markers for quantifying microbial protein synthesis in the rumen. J. Dairy Sci. 75, 2618–2632.
Carlson, S. A., Sharma, V. K., McCuddin, Z. P., Rasmussen, M. A., and Franklin, S. K. (2007). Involvement of a Salmonella genomic island 1 gene in the rumen protozoan-mediated enhancement of invasion for multiple-antibiotic-resistant Salmonella enterica serovar Typhimurium. Infect. Immun. 75, 792–800. doi: 10.1128/IAI.00679-06
Case, R. J., Boucher, Y., Dahllöf, I., Holmström, C., Doolittle, W. F., and Kjelleberg, S. (2007). Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73, 278–288. doi: 10.1128/AEM.01177-06
Cavalier-Smith, T. (2005). Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 95, 147–175. doi: 10.1093/aob/mci010
Chandramoni, Jadhao, S. B., Tiwari, C. M., Haque, N., Murari Lal, and Khan, M. Y. (2001). Energy metabolism and methane production in faunated and defaunated sheep fed two diets with same concentrate to roughage ratio (70 : 30) but varying in composition. Asian-Australas. J. Anim. Sci. 14, 1238–1244. doi: 10.5713/ajas.2001.1238
Cieslak, A., Szumacher-Strabel, M., Stochmal, A., and Oleszek, W. (2013). Plant components with specific activities against rumen methanogens. Animal 7, 253–265. doi: 10.1017/S1751731113000852
Clayet, F., Senaud, J., and Bohatier, J. (1992). Chromatographic separation of some cell wall polysaccharide-degrading enzymes of the sheep rumen ciliate Epidinium caudatum. Ann. Zootech. 41, 81. doi: 10.1051/animres:19920141
Crosby, L. D., and Criddle, C. S. (2003). Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34, 790–803.
Dehority, B. A. (1993). Laboratory Manual for Classification and Morphology of Ruminal Ciliate Protozoa. Boca Raton, FL: CRC Press.
Dehority, B. A. (1994). Rumen ciliate protozoa of the blue duiker (Cephalophus monticola), with observations on morphological variation lines within the species Entodinium dubardi. J. Euk. Microbiol. 41, 103–111. doi: 10.1111/j.1550-7408.1994.tb01481.x
Dehority, B. A. (2006). Studies on the variation in caudal spination of Epidinium. Zootaxa 1305, 33–39.
Dehority, B. A. (2008). Improved in vitro procedure for maintaining stock cultures of three genera of rumen protozoa. J. Anim. Sci. 86, 1395–1401. doi: 10.2527/jas.2007-0238
de la Fuente, G., Belanche, A., Abecia, L., Dehority, B. A., and Fondevila, M. (2009). Rumen protozoal diversity in the Spanish ibex (Capra pyrenaica hispanica) as compared with domestic goats (Capra hircus). Eur. J. Protistol. 45, 112–120. doi: 10.1016/j.ejop.2008.07.001
Delfosse-Debusscher, J., Van Hoof, F., Hellings, P., and Thines-Sempoux, D. (1979). Hydrolytic activities of rumen ciliates. Ann. Rech. Vet. 10, 258–260.
Demeyer, D. I., Fiedler, D., and DeGraeve, K. G. (1996). Attempted induction of reductive acetogenesis into the rumen fermentation in vitro. Reprod. Nutr. Dev. 36, 233–240. doi: 10.1051/rnd:19960301
Denton, B. L., Diese, l. E., Firkins, J. L., and Hackmann, T. J. (2015). Accumulation of reserve carbohydrate by rumen protozoa and bacteria in competition for glucose. Appl. Environ. Microbiol. 81, 1832–1838. doi: 10.1128/AEM.03736-14
Dersimonian, R., and Laird, N. (1989). Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188. doi: 10.1016/0197-2456(86)90046-2
Devillard, E., Bera-Maillet, C., Flint, H. J., Scott, K. P., Newbold, C. J., Wallace, R. J., et al. (2003). Characterization of XYN10B, a modular xylanase from the ruminal protozoan Polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose. Biochem. J. 373, 495–503. doi: 10.1042/bj20021784
Devillard, E., Newbold, C. J., Scott, K. P., Forano, E., Wallace, R. J., Jouany, J. P., et al. (1999). A xylanase produced by the rumen ciliate protozoa Polyplastron multivesiculatum shows close sequence similarity with family 11 xylanases from gram positive bacteria. FEMS Microbiol. Lett. 181, 145–152. doi: 10.1111/j.1574-6968.1999.tb08837.x
Diaz, H. L., Knapp, J. R., Karnati, S. K. R., Dehority, B. A., and Firkins, J. L. (2014). Effects of wortmannin, sodium nitroprusside, insulin, genistein, and guanosine triphosphate on chemotaxis and cell growth of Entodinium caudatum, Epidinium caudatum, and mixed ruminal protozoa. J. Dairy Sci. 97, 2244–2253. doi: 10.3168/jds.2013-7429
Dijkstra, J. (1994). Simulation of the dynamics of protozoa in the rumen. Br. J. Nutr. 72, 679–699. doi: 10.1079/BJN19940071
Dohme, F., Machmüller, A., Estermann, B. L., Pfister, P., Wasserfallen, A., and Kreuzer, M. (1999). The role of the rumen ciliate protozoa for methane suppression caused by coconut oil. Lett. Appl. Microbiol. 29, 187–192. doi: 10.1046/j.1365-2672.1999.00614.x
Dohme, F., Machmüller, A., Wasserfallen, A., and Kreuzer, M. (2001). Ruminal methanogenesis as influenced by individual acids supplemented to complete ruminant diets. Lett. Appl. Microbiol. 32, 47–51. doi: 10.1046/j.1472-765x.2001.00863.x
Doreau, M., and Ferlay, A. (1995). Effect of dietary lipids on nitrogen metabolism in the rumen: a review. Livest. Prod. Sci. 43, 97–110. doi: 10.1016/0301-6226(95)00041-I
Embley, T. M., Finlay, B. J., Dyal, P. L., Hirt, R. P., and Wilkinson, M., Williams, A. G. (1995). Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 262, 87–93. doi: 10.1098/rspb.1995.0180
Eschenlauer, S. C., McEwan, N. R., Calza, R. E., Wallace, R. J., Onodera, R., and Newbold, C. J. (1998). Phylogenetic position and codon usage of two centrin genes from the rumen ciliate protozoan, Entodinium caudatum. FEMS Microbiol. Lett. 166, 147–154. doi: 10.1111/j.1574-6968.1998.tb13196.x
Eugène, M., Archimède, H., and Sauvant, D. (2004). Quantitative meta-analysis on the effects of defaunation of the rumen on growth, intake and digestion in ruminants. Livest. Prod. Sci. 85, 81–97. doi: 10.1016/S0301-6226(03)00117-9
Faciola, A. P., and Broderick, G. A. (2014). Effects of feeding lauric acid or coconut oil on ruminal protozoa numbers, fermentation pattern, digestion, omasal nutrient flow, and milk production in dairy cows. J. Dairy Sci. 97, 5088–5100. doi: 10.3168/jds.2013-7653
Faichney, G. J., Graham, N. M., and Walker, D. M. (1999). Rumen characteristics, methane emissions, and digestion in weaned lambs reared in isolation. Aust. J. Agric. Res. 50, 1083–1089. doi: 10.1071/AR98195
Fenchel, T., and Finlay, B. J. (2006). The diversity of microbes: resurgence of the phenotype. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361, 1965–1973. doi: 10.1098/rstb.2006.1924
Feng, X. Z., and Guang, S. H. (2013). Non-coding RNAs mediate the rearrangements of genomic DNA in ciliates. Sci. China Life Sci. 56, 937–943. doi: 10.1007/s11427-013-4539-4
Findley, S. D., Mormile, M. R., Sommer-Hurley, A., Zhang, X. C., Tipton, P., Arnett, K., et al. (2011). Activity-based metagenomic screening and biochemical characterization of bovine ruminal protozoan glycoside hydrolases. Appl. Environ. Microbiol. 77, 8106–8113. doi: 10.1128/AEM.05925-11
Finlay, B. J., Esteban, G., Clarke, K. J., Williams, A. G., Embley, T. M., and Hirt, R. P. (1994). Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol. Lett. 117, 157–162. doi: 10.1111/j.1574-6968.1994.tb06758.x
Firkins, J. L., Yu, Z., and Morrison, M. (2007). Ruminal nitrogen metabolism: perspectives for integration of microbiology and nutrition for dairy. J. Dairy Sci. 90(Suppl 1), E1–E16. doi: 10.3168/jds.2006-518
Frumholtz, P. P. (1991). Manipulation of the Rumen Fermentation and its Effect on Digestive Physiology. Ph.D. thesis, University of Aberdeen, Aberdeen.
Godhe, A., Asplund, M. E., Härnström, K., Saravanan, V., Tyagi, A., and Karunasagar, I. (2008). Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl. Environ. Microbiol. 74, 7174–7182. doi: 10.1128/AEM.01298-08
Goel, G., and Makkar, H. P. (2012). Methane mitigation from ruminants using tannins and saponins. Trop. Anim. Health Prod. 44, 729–739. doi: 10.1007/s11250-011-9966-2
Gong, J., Dong, J., Liu, X., and Massana, R. (2013). Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist 164, 369–379. doi: 10.1016/j.protis.2012.11.006
Gruby, D., and Delafond, H. M. O. (1843). Recherches sur des animalcules se développant en grand nombre dans l’estomac et dans les intestins, pendant la digestion des animaux herbivores et carnivores. Compt. Rend. Acad. Sci. 17, 1304–1308.
Guo, Y. Q., Liu, J. X., Lu, Y., Zhu, W. Y., Denman, S. E., and McSweeney, C. S. (2008). Effect of tea saponin on methanogenesis, microbial community structure and expression of mcrA gene, in cultures of rumen microorganisms. Lett. Appl. Microbiol. 47, 421–426. doi: 10.1111/j.1472-765X.2008.02459.x
Guyader, J., Eugene, M., Noziere, P., Morgavi, D. P., Doreau, M., and Martin, C. (2014). Influence of rumen protozoa on methane emission in ruminants: a meta-analysis approach. Animal 8, 1816–1825. doi: 10.1017/S1751731114001852
Hall, M. B. (2011). Isotrichid protozoa influence conversion of glucose to glycogen and other microbial products. J. Dairy Sci. 94, 4589–4602. doi: 10.3168/jds.2010-3878
Hegarty, R. S. (1999). Reducing rumen methane emissions through elimination of rumen protozoa. Aust. J. Agric. Res. 50, 1321–1327. doi: 10.1071/AR99008
Hegarty, R. S., Bird, S. H., Vanselow, B. A., and Woodgate, R. (2008). Effects of the absence of protozoa from birth or from weaning on the growth and methane production of lambs. Br. J. Nutr. 100, 1220–1227. doi: 10.1017/S0007114508981435
Hegarty, R. S., Nolan, J. V., and Leng, R. A. (1994). The effects of protozoa and of supplementation with nitrogen and sulfur on digestion and microbial-metabolism in the rumen of sheep. Aust. J. Agric. Res. 45, 1215–1227. doi: 10.1071/AR9941215
Herrera, M. L., Vallor, A. C., Gelfond, J. A., Patterson, T. F., and Wickes, B. L. (2009). Strain-dependent variation in 18S ribosomal DNA copy numbers in Aspergillus fumigatus. J. Clin. Microbiol. 47, 1325–1332. doi: 10.1128/JCM.02073-08
Hook, S. E., Dijkstra, J., Wright, A. D. G., McBride, B. W., and France, J. (2012). Modeling the distribution of ciliate protozoa in the reticulo-rumen using linear programming. J. Dairy Sci. 95, 255–265. doi: 10.3168/jds.2011-4352
Hook, S. E., Wright, A. G., and McBride, B. W. (2010). Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010, 945785. doi: 10.1155/2010/945785
Howard, B. H., Jones, G., and Purdom, M. R. (1960). The pentosanases of some rumen bacteria. Biochem. J. 74, 173–180. doi: 10.1042/bj0740173
Hristov, A. N., and Jouany, J. P. (2005). “Factors affecting the efficiency of nitrogen utilization in the rumen,” in Nitrogen and Phosphorus Nutrition of Cattle and Environment, eds A. N. Hristov and E. Pfeffer (Wallingford: CAB International), 177–166.
Hristov, A. N., Oh, J., Firkins, J. L., Dijkstra, J., Kebreab, E., Waghorn, G., et al. (2013). Mitigation of methane and nitrous oxide emissions from animal operations: a review of enteric methane mitigation options. J. Anim. Sci. 91, 5045–5069. doi: 10.2527/jas.2013-6583
Hsu, J. T., Fahey, G. C., Merchen, N. R., and Mackie, R. I. (1991). Effects of defaunation and various nitrogen supplementation regimens on microbial numbers and activity in the rumen of sheep. J. Anim. Sci. 69, 1279–1289.
Hu, W. L., Liu, J. X., Ye, J. A., Wu, Y. M., and Guo, Y. Q. (2005). Effect of tea saponin on rumen fermentation in vitro. Anim. Feed Sci. Technol. 120, 333–339. doi: 10.1016/j.anifeedsci.2005.02.029
Huws, S. A., Kim, E. J., Kingston-Smith, A. H., Lee, M. R. F., Muetzel, S. M., Cookson, A. R., et al. (2009). Rumen protozoa are rich in polyunsaturated fatty acids due to the ingestion of chloroplasts. FEMS Microbiol. Ecol. 69, 461–471. doi: 10.1111/j.1574-6941.2009.00717.x
Huws, S. A., Lee, M. R. F., Kingston-Smith, A. H., Kim, E. J., Scott, M. B., Tweed, J. K. S., et al. (2012). Ruminal protozoal contribution to the duodenal flow of fatty acids following feeding of steers on forages differing in chloroplast content. Br. J. Nutr. 108, 2207–2214. doi: 10.1017/S0007114512000335
Irbis, C., and Ushida, K. (2004). Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50, 203–212. doi: 10.2323/jgam.50.203
Ishaq, S. L., and Wright, A.-G. (2014). Design and validation of four new primers for next-generation sequencing to target the 18s rRNA genes of gastrointestinal ciliate protozoa. Appl. Environ. Microbiol. 80, 5515–5521. doi: 10.1128/AEM.01644-14
Ivan, M. (2009). Comparison of duodenal flow and digestibility in fauna-free sheep inoculated with Holotrich protozoa, Entodinium monofauna or total mixed protozoa population. Br. J. Nutr. 101, 34–40. doi: 10.1017/S0007114508984245
Ivan, M., Neill, L., and Entz, T. (2000a). Ruminal fermentation and duodenal flow following progressive inoculations of fauna-free wethers with major individual species of ciliate protozoa or total fauna. J. Anim. Sci. 78, 750–759.
Ivan, M., Neill, L., Forster, R., Alimon, R., Rode, L. M., and Entz, T. (2000b). Effects of Isotricha, Dasytricha, Entodinium, and total fauna on ruminal fermentation and duodenal flow in wethers fed different diets. J. Dairy Sci. 83, 776–787. doi: 10.3168/jds.S0022-0302(00)74940-X
Jordan, E., Lovett, D. K., Monahan, F. J., Callan, J., Flynn, B., and O’Mara, F. P. (2006). Effect of refined coconut oil or copra meal on methane output and on intake and performance of beef heifers. J. Anim. Sci. 84, 162–170. doi: 10.2527/jas.2005-354
Jouany, J. P. (1996). Effect of rumen protozoa on protein utilization by ruminants. J. Nutr. 126, 1335–1346.
Karnati, S. K. R., Sylvester, J. T., Noftsger, S. M., Yu, Z., St-Pierre, N. R., and Firkins, J. L. (2007). Assessment of ruminal bacterial populations and protozoal generation time in cows fed different methionine sources. J. Dairy Sci. 90, 798–809. doi: 10.3168/jds.S0022-0302(07)71564-3
Karnati, S. K. R., Sylvester, J. T., Ribeiro, C. V. D. M., Gilligan, L. E., and Firkins, J. L. (2009). Investigating unsaturated fat, monensin, or bromoethanesulfonate in continuous cultures retaining ruminal protozoa. I. Fermentation, biohydrogenation, and microbial protein synthesis. J. Dairy Sci. 92, 3849–3860. doi: 10.3168/jds.2008-1436
Kittelmann, S., and Janssen, P. H. (2011). Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. FEMS Microbiol. Ecol. 75, 468–481. doi: 10.1111/j.1574-6941.2010.01022.x
Kittelmann, S., Devente, S. R., Kirk, M. R., Seedorf, H., Dehority, B. A., and Janssen, P. H. (2015). Phylogeny of the intestinal ciliates including first sequences from Charonina ventriculi and comparison of microscopy and 18S rRNA gene pyrosequencing for rumen ciliate community structure analysis. Appl. Environ. Microbiol. 81, 2433–2444. doi: 10.1128/AEM.03697-14
Kittelmann, S., Seedor, H., Walters, W. A., Clemente, J. C., Knight, R., Gordon, J. I., et al. (2013). Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS ONE 8:e47879. doi: 10.1371/journal.pone.0047879
Kofoid, C. A., and MacLennan, R. F. (1930). Ciliates from Bos indicus Linn. I. The genus Entodinium. Stein. Univ. Calif. Publ. Zool. 33, 471–544.
Kofoid, C. A., and MacLennan, R. F. (1932). Ciliates from Bos indicus Linn II. A revision of Diplodinium Schuberg. Univ. Calif. Publ. Zool. 37, 53–152.
Kofoid, C. A., and MacLennan, R. F. (1933). Ciliates from Bos indicus Linn III. Epidinium Crawley. Epiplastron gen. nov., and Ophryoscolex Stein. Univ. Calif. Publ. Zool. 39, 1–34.
Kreuzer, M., Kirchgessner, M., and Müller, H. L. (1986). Effect of defaunation on the loss of energy in wethers fed different quantities of cellulose and normal or steamflaked maize starch. Anim. Feed Sci. Technol. 16, 233–241. doi: 10.1016/0377-8401(86)90114-8
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Latteur, B. (1968). Revision systematique de la Famille des Ophryoscolescidae Stein, 1858: sous-famille Entodiniinae Lubinsky, 1957. Genre Entodinium Stein, 1858. Ann. Soc. R. Zool. de Belgique 98, 1–41.
Latteur, B. (1969). Revision systematique de la famille des Ophryoscolecidae Stein, 1858: sous-famille Entodiniinae Lubinsky, 1957. Genre Entodinium Stein, 1958. Ann. Soc. R. Zool. de Belgique 99, 3–25.
Latteur, B. (1970). Revision Systematique de la Famille des Ophryoscolecidae Stein, 1858: sous-famille des Diplodiniinae Lubinsky, 1957. Genre Diplodinium (Schuberg, 1888) sensu novo. Ann. Soc. R. Zool. de Belgique 100, 275–312.
Lennarz, W. J. (1966). Lipid metabolism in the bacteria. Adv. Lipid Res. 4, 175–225. doi: 10.1016/b978-1-4831-9940-5.50012-0
Ling, J. R., and Buttery, P. J. (1978). The simultaneous use of ribonucleic acid, 35S, 2,6-diaminopimelic acid and 2-aminoethylphosphonic acid as markers of microbial nitrogen entering the duodenum of sheep. Br. J. Nutr. 39, 165–179. doi: 10.1079/BJN19780023
Machmuller, A., Soliva, C. R., and Kreuzer, R. (2003). Effect of coconut oil and defaunation treatment on methanogenesis in sheep. Reprod. Nutr. Dev. 43, 41–55. doi: 10.1051/rnd:2003005
Mackie, R. I., Gilchrist, F. M. C., Robberts, A. M., Hannah, P. E., and Schwartz, H. M. (1978). Microbiological and chemical changes in the rumen during the stepwise adaptation of sheep to high concentrate diets. J. Agric. Sci. 90, 241–254. doi: 10.1017/S0021859600055313
Martin, C., Morgavi, D. P., and Doreau, M. (2010). Methane mitigation in ruminants: from microbe to the farm scale. Animal 4, 351–365. doi: 10.1017/S1751731109990620
McEwan, N. R., Abecia, L., Regensbogenova, M., Adam, C. L., Findlay, P. A., and Newbold, C. J. (2005). Rumen microbial population dynamics in response to photoperiod. Lett. Appl. Microbiol. 41, 97–101. doi: 10.1111/j.1472-765X.2005.01707.x
McEwan, N. R., Eschenlauer, S. C. P., Calza, R. E., Wallace, R. J., and Newbold, C. J. (1999). Protozoal sequences may reveal additional isoforms of the 14-3-3 protein family. Protist 150, 257–264. doi: 10.1016/S1434-4610(99)70028-3
McEwan, N. R., Gatherer, D., Eschenlauer, S. C. P., McIntosh, F. M., Calza, R. E., Wallace, R. J., et al. (2000). An unusual codon usage pattern in the ciliate family Ophryoscolecidae and its implications for determining the source of cloned DNA. Anaerobe 6, 21–28. doi: 10.1006/anae.1999.0310
Medinger, R., Nolte, V., Pandey, R. V., Jost, S., Ottenwaelder, B., Schloetterer, C., et al. (2010). Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol. Ecol. 19, 32–40. doi: 10.1111/j.1365-294X.2009.04478.x
Moon-van der Staay, S. Y., van der Staay, G. W. M., Michalowski, T., Jouany, J. P., Pristas, P., Javorský, P., et al. (2014). The symbiotic intestinal ciliates and the evolution of their hosts. Eur. J. Protistol. 50, 166–173. doi: 10.1016/j.ejop.2014.01.004
Morgavi, D. P., Forano, E., Martin, C., and Newbold, C. J. (2010). Microbial ecosystem and methanogenesis in ruminants. Animal 4, 1024–1036. doi: 10.1017/S1751731110000546
Morgavi, D. P., Jouany, J. P., and Martin, C. (2008). Changes in methane emission and rumen fermentation parameters induced by refaunation in sheep. Aust. J. Exp. Agric. 48, 69–72. doi: 10.1071/EA07236
Morgavi, D. P., Martin, C., Jouany, J. P., and Ranilla, M. J. (2012). Rumen protozoa and methanogenesis: not a simple cause-effect relationship. Br. J. Nutr. 107, 388–397. doi: 10.1017/S0007114511002935
Mosoni, P., Martin, C., Forano, E., and Morgavi, D. P. (2011). Long-term defaunation increases the abundance of cellulolytic ruminococci and methanogens but does not affect the bacterial and methanogen diversity in the rumen of sheep. J. Anim. Sci. 89, 783–791. doi: 10.2527/jas.2010-2947
Nagaraja, T. G., Newbold, C. J., Van Nevel, C. J., and Demeyer, D. I. (1997). “Manipulation of ruminal fermentation,” in The Rumen Microbial Ecosystem, eds P. N. Hobson and C. S. Stewart (London: Blackie Academic and Professional), 523–632.
Nagaraja, T. G., Towne, G., and Beharka, A. A. (1992). Moderation of ruminal fermentation by ciliated protozoa in cattle fed a high-grain diet. Appl. Environ. Microbiol. 58, 2410–2414.
Newbold, C. J., Chamberlain, D. G., and Williams, A. G. (1986). The effects of defaunation on the metabolism of lactic acid in the rumen. J. Sci. Food Agric. 37, 1083–1090. doi: 10.1002/jsfa.2740371105
Newbold, C. J., ElHassan, S. M., Wang, J., Ortega, M. E., and Wallace, R. J. (1997). Influence of foliage from African multipurpose trees on activity of rumen protozoa and bacteria. Br. J. Nutr. 78, 237–249. doi: 10.1079/BJN19970143
Newbold, C. J., Lassalas, B., and Jouany, J. P. (1995). The importance of methanogensis associated with ciliate protozoa in ruminal methane production in vitro. Letts. Appl. Micro. 21, 230–234.
Newbold, C. J., McEwan, N. R., Calza, R. E., Chareyron, E. N., Duval, S. M., Eschenlauer, S. C. P., et al. (2005). An NAD+-dependent glutamate dehydrogenase cloned from the ruminal ciliate protozoan, Entodinium caudatum. FEMS Microbiol. Lett. 247, 113–121. doi: 10.1016/j.femsle.2005.04.034
Nowacki, M., Shetty, K., and Landweber, L. F. (2011). RNA-mediated epigenetic programming of genome rearrangements. Ann. Rev. Genomics Hum. Genet. 12, 367–389. doi: 10.1146/annurev-genom-082410-101420
Noziere, P., Glasser, F., and Sauvant, D. (2011). In vivo production and molar percentages of volatile fatty acids in the rumen: a quantitative review by an empirical approach. Animal 5, 403–414. doi: 10.1017/S1751731110002016
Ohene-Adjei, S., Teather, R. M., Ivan, M., and Forster, R. J. (2007). Postinoculation protozoan establishment and association patterns of methanogenic archaea in the ovine rumen. Appl. Environ. Microbiol. 73, 4609–4618. doi: 10.1128/AEM.02687-06
Ozutsumi, Y., Tajima, K., Takenaka, A., and Itabashi, H. (2005). The effect of protozoa on the composition of rumen bacteria in cattle using 16S rRNA gene clone libraries. Biosci. Biotechnol. Biochem. 69, 499–506. doi: 10.1271/bbb.69.499
Panyakaew, P., Boon, N., Goel, G., Yuangklang, C., Schonewille, J. T., Hendriks, W. H., et al. (2013). Effect of supplementing coconut or krabok oil, rich in medium-chain fatty acids on ruminal fermentation, protozoa and archaeal population of bulls. Animal 7, 1950–1958. doi: 10.1017/S1751731113001766
Patra, K. A., and Saxena, J. (2009). Dietary phytochemicals as rumen modifiers: a review of the effects on microbial population. Antonie Van Leeuwenhoek 96, 363–375. doi: 10.1007/s10482-009-9364-1
Patra, A. K. (2010). Meta-analyses of effects of phytochemicals on digestibility and rumen fermentation characteristics associated with methanogenesis. J. Sci. Food Agric. 90, 2700–2708. doi: 10.1002/jsfa.4143
Paul, R. G., Williams, A. G., and Butler, R. D. (1990). Hydrogenosomes in the rumen entodiniomorphid ciliate Polyplastron multivesiculatum. J. Gen. Microbiol. 136, 1981–1989. doi: 10.1099/00221287-136-10-1981
Prescott, D. M., and Murti, K. G. (1974). Chromosome structure in ciliated protozoans. Cold. Spring Harb. Symp. Quant. Biol. 38, 609–618. doi: 10.1101/SQB.1974.038.01.065
Punia, B. S., Leibholz, J., and Faichney, G. J. (1992). Rate of production of protozoa in the rumen and the flow of protozoal nitrogen to the duodenum in sheep and cattle given a pelleted diet of lucerne hay and barley. J. Agric. Sci. 118, 229–236. doi: 10.1017/S0021859600068830
Qi, M., Wang, P., O’Toole, N., Barboza, P. S., Ungerfeld, E., Leigh, M. B., et al. (2011). Snapshot of the eukaryotic gene expression in muskoxen rumen—a metatranscriptomic approach. PLoS ONE 6:e20521. doi: 10.1371/journal.pone.0020521
Ramos-Morales, E., de la Fuente, G., Nash, R., Preskett, D., and Newbold, C. J. (2014). “In vitro effect of the combination of Ivy fruit refined extract and a glucosidase inhibitor on rumen fermentation and protozoa activity,” in Proceedings of Rowett-INRA 2014, Gut Microbiology: From Sequence To Function, Aberdeen, p 116.
Rasmussen, M. A., Carlson, S. A., Franklin, S. K., McCuddin, Z. P., Wu, M. T., and Sharma, V. K. (2005). Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant salmonella enterica bearing SGI1. Infect. Immun. 73, 4668–4675. doi: 10.1128/IAI.73.8.4668-4675.2005
Reeder, R. H. (1999). Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 62, 293–327. doi: 10.1016/S0079-6603(08)60511-5
Regensbogenova, M., Kisidayova, S., Michalowski, T., Javorsky, P., Moon-Van Der Staay, S. Y., Moon-Van Der Staay, G. W. M., et al. (2004a). Rapid identification of rumen protozoa by restriction analysis of amplified 18S rRNA gene. Acta Protozool. 43, 219–224.
Regensbogenova, M., McEwan, N. R., Javorsky, P., Kisidayova, S., Michalowski, T., Newbold, C. J., et al. (2004b). A re-appraisal of the diversity of the methanogens associated with the rumen ciliates. FEMS Microbiol. Lett. 238, 307–313. doi: 10.1111/j.1574-6968.2004.tb09771.x
Ricard, G., McEwan, N. R., Dutilh, B. E., Jouany, J. P., Macheboeuf, D., Mitsumori, M., et al. (2006). Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7:22. doi: 10.1186/1471-2164-7-22
Schlötterer, C. (2004). The evolution of molecular markers—just a matter of fashion? Nat. Rev. Genet. 5, 63–69. doi: 10.1038/nrg1249
Sharp, R., Ziemer, C. J., Stern, M. D., and Stahl, D. A. (1998). Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol. Ecol. 26, 71–78. doi: 10.1111/j.1574-6941.1998.tb01563.x
Skillman, L. C., Toovey, A. F., Williams, A. J., and Wright, A.-D. G. (2006). Development and validation of a real-time PCR method to quantify rumen protozoa and examination of variability between Entodinium populations in sheep offered a hay-based diet. Appl. Environ. Microbiol. 72, 200–206. doi: 10.1128/AEM.72.1.200-206.2006
Soliva, C. R., Meile, L., Hindrichsen, I. K., Kreuzer, M., and Machmüller, A. (2004). Myristic acid supports the immediate inhibitory effect of lauric acid on ruminal methanogens and methane release. Anaerobe 10, 269–276. doi: 10.1016/j.anaerobe.2004.06.003
Stanford, K., Bach, S. J., Stephens, T. P., and Mcallister, T. A. (2010). Effect of rumen protozoa on Escherichia coli O157:H7 in the rumen and feces of specifically faunated sheep. J. Food Prot. 73, 2197–2202.
Swart, E. C., Bracht, J. R., Magrini, V., Minx, P., Chen, X., Zhou, Y., et al. (2013). The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16000 tiny chromosomes. PLoS Biol. 11:e1001473. doi: 10.1371/journal.pbio.1001473
Sylvester, J. T., Karnati, S. K. R., Yu, Z. T., Morrison, M., and Firkins, J. L. (2004). Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 134, 3378–3384.
Sylvester, J. T., Karnati, S. K. R., Dehority, B. A., Morrison, M., Smith, G. L., St-Pierre, N. R., et al. (2009). Rumen ciliated protozoa decrease generation time and adjust 18S ribosomal DNA copies to adapt to decreased transfer interval, starvation, and monensin. J. Dairy Sci. 92, 256–269. doi: 10.3168/jds.2008-1417
Sylvester, J. T., Karnati, S. K. R., Yu, Z., Newbold, C. J., and Firkins, J. L. (2005). Evaluation of a real-time PCR assay quantifying the ruminal pool size and duodenal flow of protozoal nitrogen. J. Dairy Sci. 88, 2083–2095. doi: 10.3168/jds.S0022-0302(05)72885-X
Takenaka, A., D’Silva, C. G. D., Kudo, H., Itabashi, H., and Cheng, K. J. (1999). Molecular cloning, expression, and characterization of an endo-beta-1,4-glucanase cDNA from Epidinium caudatum. J. Gen. Appl. Microbiol. 45, 57–61. doi: 10.2323/jgam.45.57
Takenaka, A., Tajima, K., Mitsumori, M., and Kajikawa, H. (2004). Fiber digestion by rumen ciliate protozoa. Microbes Environ. 19, 203–210. doi: 10.1264/jsme2.19.203
Tavendale, M. H., Meagher, L. P., Pacheco, D., Walker, N., Attwood, G. T., and Sivakumaran, S. (2005). Methane production from in vitro rumen incubations with Lotus pedunculatus and Medicago sativa, and effects of extractable condensed tannin fractions on methanogenesis. Anim. Feed Sci. Technol. 123–124, 403–419. doi: 10.1016/j.anifeedsci.2005.04.037
Thines-Sempoux, D., Delfosse-Debusscher, J., and Latteur, D. (1980). Mechanism of cellulose degradation by the rumen ciliates. Arch. Int. Physiol. Biochim. 88, 102–104.
Thornhill, D. J., Lajeunesse, T. C., and Santos, S. R. (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol. Ecol. 16, 5326–5340. doi: 10.1111/j.1365-294X.2007.03576.x
Tokura, M., Ushida, K., Miyazaki, K., and Kojima, Y. (1997). Methanogens associated with rumen ciliates. FEMS Microbiol. Ecol. 22, 137–143. doi: 10.1111/j.1574-6941.1997.tb00365.x
Tymensen, L. D., Beauchemin, K. A., and McAllister, T. A. (2012a). Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology 158, 1808–1817. doi: 10.1099/mic.0.057984-0
Tymensen, L., Barkley, C., and McAllister, T. A. (2012b). Relative diversity and community structure analysis of rumen protozoa according to T-RFLP and microscopic methods. J. Microbiol. Methods 88, 1–6. doi: 10.1016/j.mimet.2011.09.005
Ushida, K., Kayouli, C., Smet, S. D., and Jouany, J. P. (1990). Effect of defaunation on protein and fibre digestion in sheep fed on ammonia-treated straw-based diets with or without maize. Br. J. Nutr. 64, 756–775. doi: 10.1079/BJN19900078
Ushida, K., Newbold, C. J., and Jouany, J. P. (1997). Interspecies hydrogen transfer between the rumen ciliate Polyplastron multivesiculatum and Methanosarcina barkeri. J. Gen. Appl. Microbiol. 43, 129–131. doi: 10.2323/jgam.43.129
Vermorel, M., and Jouany, J. P. (1989). Effects of rumen protozoa on energy utilization by wethers of two diets based on ammonia-treated straw supplemented or not with maize. Asian-Australas. J. Anim. Sci. 2, 7–17. doi: 10.5713/ajas.1989.475
Viechtbauer, W. (2010). Conducting meta-analyses in R with the metafor package. J. Statis. Soft. 36, 1–48. doi: 10.18637/jss.v036.i03
Wereszka, K., McIntosh, F. M., Michalowski, T., Jouany, J.-P., Nsabimana, E., Mache-boeuf, D., et al. (2004). A cellulase produced by the rumen protozoan Epidinium ecaudatum is of bacterial origin and has an unusual pH optimum. Endocytobiosis Cell Res. 15, 561–569.
Williams, Y. J., Rea, S. M., Popovski, S., Pimm, C. L., Williams, A. J., Toovey, A. F., et al. (2008). Reponses of sheep to a vaccination of entodinial or mixed rumen protozoal antigens to reduce rumen protozoal numbers. Br. J. Nutr. 99, 100–109. doi: 10.1017/S0007114507801553
Wina, E., Muetzel, S., and Becker, K. (2005). The impact of saponins or saponin-containing plant materials on ruminant production—a review. J. Sci. Food Agric. 53, 8093–8105. doi: 10.1021/jf048053d
Wina, E., Muetzel, S., and Becker, K. (2006). Effects of daily and interval feeding of Sapindus rarak saponins on protozoa, rumen fermentation parameters and digestibility in sheep. Asian-Australas. J. Anim. Sci. 19, 1580–1587. doi: 10.5713/ajas.2006.1580
Wintzingerode, F. V., Göbel, U. B., and Stackebrandt, E. (1997). Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21, 213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x
Wright, A.-D. G., Dehority, B. A., and Lynn, D. H. (1997). Phylogeny of the rumen ciliates Entodinium, Epidinium and Polyplastron (Litostomatea: Entodiniomorphida) inferred from small subunit ribosomal RNA sequences. J. Eukaryot. Microbiol. 44, 61–67. doi: 10.1111/j.1550-7408.1997.tb05693.x
Wright, A.-D. G., Kennedy, P., O’Neill, C., Toovey, A. F., Popovski, S., Rea, S. M., et al. (2004). Reducing methane emissions in sheep by immunization against rumen methanogens. Vaccine 22, 3976–3985. doi: 10.1016/j.vaccine.2004.03.053
Wright, A.-D. G., and Lynn, D. H. (1997a). Monophyly of the trichostome ciliates (Phylum Ciliophora: Class Litostomatea) tested using new 18S rRNA sequences from the Vestibuliferids, Isotricha intestinalis and Dasytricha ruminantium, and the Haptorian, Didinium nasutum. Eur. J. Protistol. 33, 305–315. doi: 10.1016/S0932-4739(97)80008-9
Wright, A.-D. G., and Lynn, D. H. (1997b). Phylogenetic analysis of the rumen ciliate family Ophryoscolecidae based on 18S ribosomal RNA sequences, with new sequences from Diplodinium, Eudiplodinium, and Ophryoscolex. Can. J. Zool. 75, 963–970. doi: 10.1139/z97-117
Xia, Y., Kong, Y. H., Seviour, R., Forster, R. J., Kisidayova, S., and McAllister, T. A. (2014). Fluorescence in situ hybridization probing of protozoal Entodinium spp. and their methanogenic colonizers in the rumen of cattle fed alfalfa hay or triticale straw. J. Appl. Microbiol. 116, 14–22. doi: 10.1111/jam.12356
Yáñez-Ruiz, D. R., Hart, K. J., Martin-Garcia, A. I., Belanche, A., and Newbold, C. J. (2007a). “The effect of absence of protozoa on methane emissions by lambs” in Proceedings of the British Society of Animal Science, Penicuik, p. 47.
Yáñez-Ruiz, D. R., Williams, S., and Newbold, C. J. (2007b). The effect of absence of protozoa on rumen biohydrogenation and the fatty acid composition of lamb muscle. Br. J. Nutr. 97, 938–948. doi: 10.1017/S0007114507675187
Yáñez-Ruiz, D. R., Scollan, N. D., Merry, R. J., and Newbold, C. J. (2006). Contribution of rumen protozoa to duodenal flow of nitrogen, conjugated linoleic acid and vaccenic acid in steers fed silages differing in their water-soluble carbohydrate content. Br. J. Nutr. 96, 861–869. doi: 10.1017/BJN20061927
Zhou, Y. Y., Mao, H. L., Jiang, F., Wang, J. K., Liu, J. X., and McSweeney, C. S. (2011). Inhibition of rumen methanogenesis by tea saponins with reference to fermentation pattern and microbial communities in Hu sheep. Anim. Feed Sci. Technol. 166, 93–100. doi: 10.1016/j.anifeedsci.2011.04.007
Keywords: rumen, protozoa, defaunation, methane, microbial diversity
Citation: Newbold CJ, de la Fuente G, Belanche A, Ramos-Morales E and McEwan NR (2015) The Role of Ciliate Protozoa in the Rumen. Front. Microbiol. 6:1313. doi: 10.3389/fmicb.2015.01313
Received: 04 August 2015; Accepted: 09 November 2015;
Published: 26 November 2015.
Edited by:
Garret Suen, University of Wisconsin-Madison, USAReviewed by:
Diego P. Morgavi, Institute National de la Recherche Agronomique, FranceJeffrey Firkins, The Ohio State University, USA
Copyright © 2015 Newbold, de la Fuente, Belanche, Ramos-Morales and McEwan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles J. Newbold, cjn@aber.ac.uk
 Charles J. Newbold
Charles J. Newbold