- 1School of Microbiology, University College Cork, Cork, Ireland
- 2Teagasc Food Research Centre, Fermoy, Ireland
- 3APC Microbiome Institute, University College Cork, Cork, Ireland
Several examples of highly modified antimicrobial peptides have been described. While many such peptides are non-ribosomally synthesized, ribosomally synthesized equivalents are being discovered with increased frequency. Of the latter group, the lantibiotics continue to attract most attention. In the present review, we discuss the implementation of in vivo and in vitro engineering systems to alter, and even enhance, the antimicrobial activity, antibacterial spectrum and physico-chemical properties, including heat stability, solubility, diffusion and protease resistance, of these compounds. Additionally, we discuss the potential applications of these lantibiotics for use as therapeutics.
Introduction
Given that antibiotic resistance has now reached a crisis point, novel compounds and innovative methods are urgently required to arrest the spread and development of drug-resistant infections in both the nosocomial and community environments. Ideally, such novel substances should exhibit distinctly different mechanisms of action to currently used chemotherapeutics in order to decrease resistance development. Ribosomally synthesized antimicrobial peptides produced by bacteria (bacteriocins) constitute an emerging class of natural products that have attracted considerable interest as promising alternatives to existing antibiotics (Sahl and Bierbaum, 2008). Within this diverse group of peptides, the lantibiotics, i.e., class I bacteriocins which contain the post-translationally modified amino acids lanthionine and methyllanthionine, have become the focus of many biomedical and pharmaceutical research groups due to their demonstrable high potency in vitro, multiple modes of action and ability to destroy target cells rapidly (Cotter et al., 2005; Cavera et al., 2015). In general, lantibiotics exhibit activity against Gram positive bacteria. Importantly, this includes many drug resistant targets including methicillin resistant Staphylococcus aureus (MRSA), vancomycin intermediate S. aureus (VISA), vancomycin resistant enterococci (VRE), Streptococcus pneumoniae and Clostridium difficile, amongst others (Cotter et al., 2013). Furthermore, several lantibiotic peptides have demonstrated excellent in vivo activities and have progressed toward clinical evaluation for the treatment of life-threatening diseases (Dawson and Scott, 2012; Sandiford, 2015). Indeed, these and a range of other desirable features make them suitable for use in human and veterinary medicine and also in the pharmaceutical industry (Dischinger et al., 2014). However, despite these promising attributes, there are a number of limitations that has prevented their more widespread use, including instability and/or insolubility at physiological pH, low production levels and susceptibility to proteolytic digestion. The implementation of multiple technologies, including genome mining as well as high-throughput screening strategies in combination with in vivo and in vitro expression systems has provided a wealth of information relating to the widespread existence, structural diversity and functionality of lantibiotics while facilitating the identification of structural regions that can be targeted to enhance their biological and physicochemical properties. The present review will focus on recent developments with regard to these achievements.
Lantibiotics: The Case for Therapeutic Use (in vitro and in vivo Potency)
New antimicrobials that possess novel modes of action, particularly against drug resistant organisms so that they can be specifically targeted for clinical applications, are required as a matter of urgency. In this regard lantibiotics hold considerable potential as a consequence of their unusual structure, unique mechanisms of action and their potency against multi-drug resistant bacteria. Today, close to 100 of these bioactive peptides have been described, the majority of which are produced by Gram-positive bacteria (Dischinger et al., 2014). The common feature that links all lantibiotics is the presence of a number of distinctive amino acids which result from enzymatically mediated post-translational modifications, including dehydration and cyclisation, leading to the formation of the eponymous (methyl)lanthionine bridges. These bridges convert the linear peptide chain into a polycyclic form giving structure and function to the peptide. It should be noted that only those peptides that display antimicrobial activity within the larger family of lanthionine-containing peptides or lanthipeptides are termed lantibiotics.
Many lantibiotics exert their antimicrobial action through complexation with lipid II, an essential precursor of the bacterial cell wall, either by inhibiting cell wall synthesis through sequestration of lipid II and/or by disruption of membrane integrity and pore formation (Breukink and de Kruijff, 2006). Indeed, the prototypical and best studied lantibiotic nisin performs both of these functions as a consequence of two distinct structural domains located at the N- and C-termini (Figure 1). It has been established that the A, B, and C rings form a “cage-like” enclosure that facilitates binding of the pyrophosphate moiety of lipid II, thus inhibiting cell wall synthesis (Hsu et al., 2004) This binding enhances the ability of the C-terminal segment, containing rings D and E, to form pores in the cell membrane, resulting in the rapid efflux of ions and cytoplasmic solutes (Wiedemann et al., 2001). This mechanism of action is not common to all lantibiotics, and some of them lack the ability to elicit pores or to bind lipid II or both, but can still exhibit antimicrobial activity (Pag and Sahl, 2002). The poor activity of lantibiotics toward Gram negative bacteria is due to the outer membrane (OM) of the Gram negative cell wall which acts as a barrier for the cell, restricting the access of the peptides to the cytoplasmic membrane (Nikaido and Vaara, 1985).
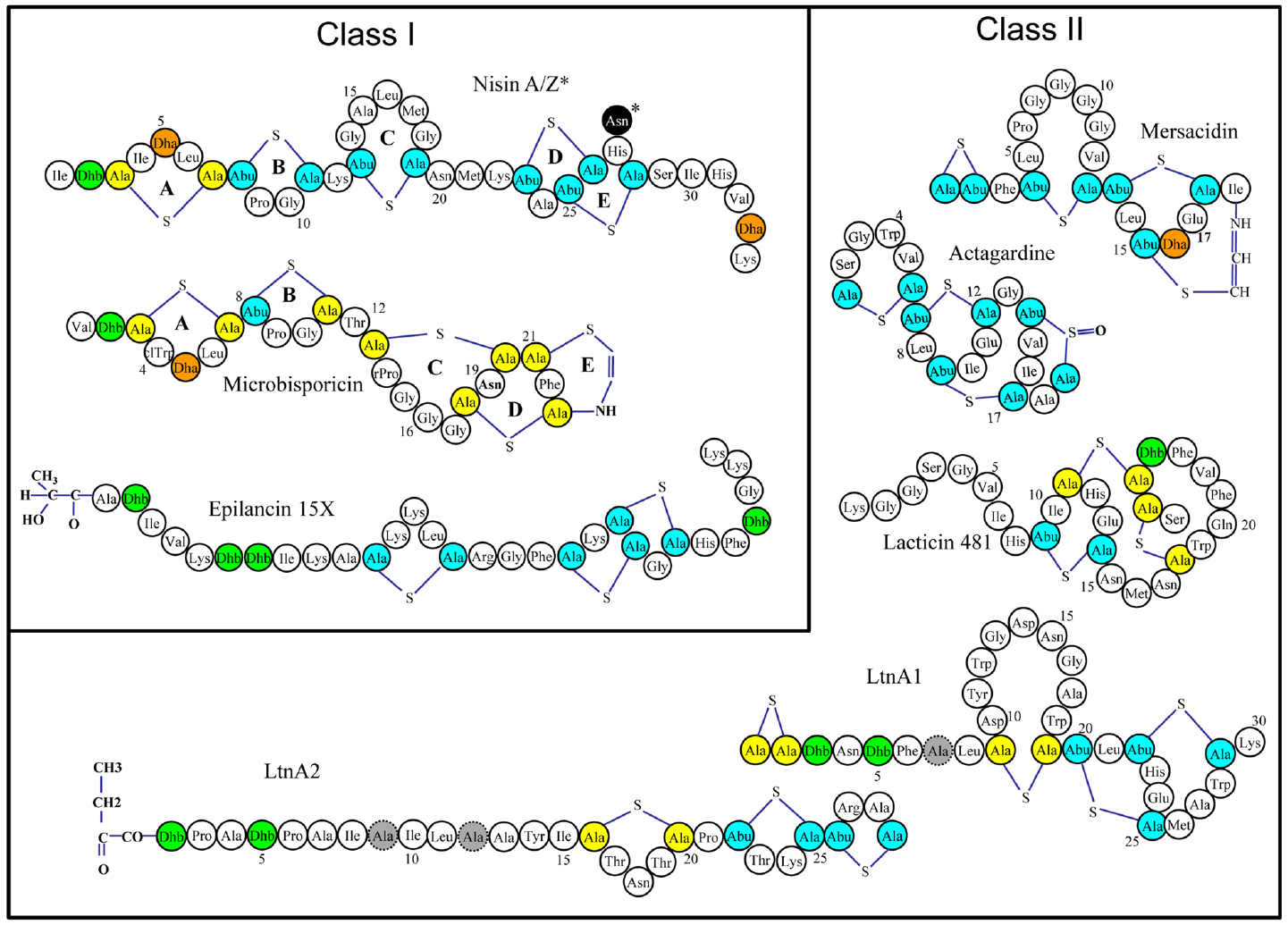
Figure 1. Representative structures of various single and two-component lantibiotics. Post translational modifications are indicated as follows: Abu: 2-aminobutyric acid, Ala-S-Ala: lanthionine, Abu-S-Ala: 3-methyllanthionine, Dha: dehydroalanine, Dhb: dehydrobutyrine. D-Ala residues (in lacticin 3147) are shaded gray.
Lantibiotics have been classified on the basis of their biosynthetic pathways (Willey and van der Donk, 2007). According to this scheme, class I lantibiotics are those modified by two separate enzymes, a LanB (dehydratase) and LanC (cyclase); class II are modified by a single LanM enzyme with both dehydratase and cyclase activity. The third and fourth classes of lanthipeptides are also modified by a single enzyme (general nomenclature LanKC for class III and LanL for class IV; van der Donk and Nair, 2014). Most of the class III lanthipeptides reported thus far have no or weak antimicrobial activities, but some have been shown to possess anti-allodynic/antinociceptive activity (Meindl et al., 2010; Iorio et al., 2014), antiviral activity (Férir et al., 2013) or morphogenetic activities (Willey and Gaskell, 2011). The designation Lan is used generically to refer to proteins associated with the biosynthesis of, or immunity to, lantibiotics. A typical lantibiotic operon will also contain genes encoding enzymes to carry out transport/processing (LanT), immunity (LanI and LanFEG), proteolytic processing (LanP) as well as the structural gene (LanA). Other enzymes, responsible for the formation of less common residues may also be present. Importantly, individual components of the lantibiotic biosynthetic machinery show even greater flexibility as demonstrated by their activity in vitro (Li et al., 2009).
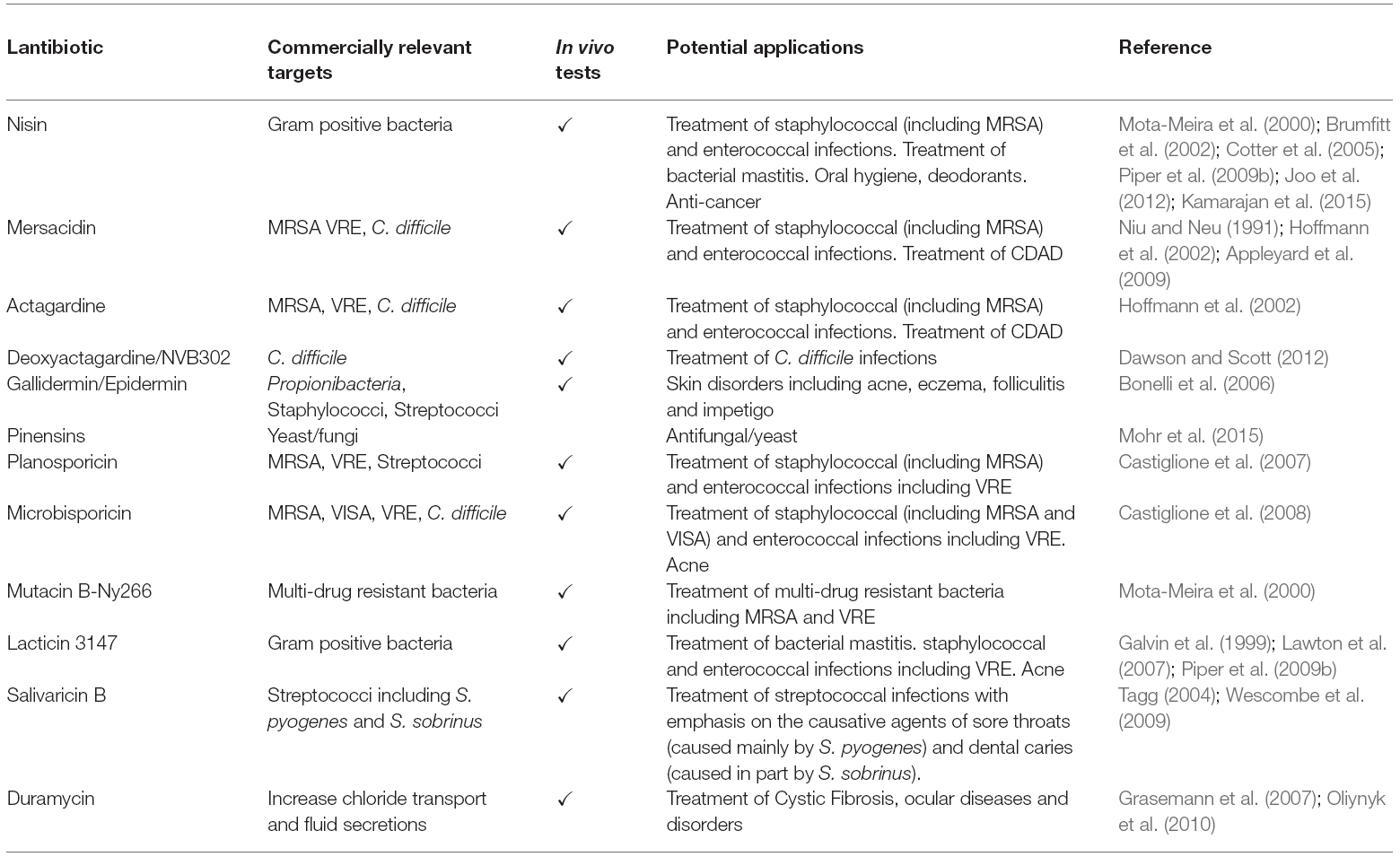
Activity of Lantibiotics in vitro
Although lantibiotics such as nisin have been in use for decades as safe and natural food preservatives (Delves-Broughton, 2005), the continued escalation of multi-drug resistant bacterial infections has led to a re-appraisal of their capacity for use against life-threatening infections. A multitude of studies have highlighted the in vitro potency of lantibiotics against nosocomial pathogens (the reader is directed to a comprehensive review: Piper et al., 2009a). Many lantibiotics, including lacticin 3147, mutacins B-Ny266 and 1140, nisin, mersacidin, epidermin, Pep5, and planosporicin exhibit activity against clinically-relevant targets (Table 1) such as MRSA, VRE, Propionibacterium acne, Streptococcus mutans, Streptococcus pyogenes, S. pneumoniae, C. difficile, Listeria, and Bacillus species (Severina et al., 1998; Galvin et al., 1999; Mota-Meira et al., 2000; Brumfitt et al., 2002; Rea et al., 2007; Ghobrial et al., 2009; Piper et al., 2009b). Notably, both Pep 5 and epidermin successfully inhibit the adhesion of staphylococcal cells to the surfaces of siliconized catheters (Fontana et al., 2006). Although it is the general view that lantibiotics exhibit less potential as chemotherapeutics to combat infections with Gram-negative organisms, lantibiotics including mutacin B-Ny266 are selectively active against a few strains of Neisseria and Helicobacter (Mota-Meira et al., 2000), while purified nisin displays activity against Escherichia coli (Kuwano et al., 2005).
Nisin has also been shown to effectively inhibit spore outgrowth including spores of Bacillus anthracis (Gut et al., 2008) and those of C. difficile (Nerandzic and Donskey, 2010). Additionally, studies have revealed that the lantibiotic gallidermin efficiently prevents biofilm formation in both S. aureus and S. epidermidis species (Saising et al., 2012).
Recently, an intriguing and novel (sub-)class of lantibiotics termed pinensins were found to be highly active against many filamentous fungi and yeasts but displayed only weak antibacterial activity. Not only do pinensin A and pinensin B represent the first examples of a lantibiotic fungicide, they are also the first lantibiotics to be isolated from a Gram-negative native producer (Mohr et al., 2015).
Lantibiotics Demonstrate in vivo Potency
While the in vitro success of a chemotherapeutic agent does not always necessarily translate to in vivo efficacy, there have been a number of encouraging studies to suggest that this may not be a major shortcoming of lantibiotics. For instance, mutacin B-Ny266 was shown to be as active as vancomycin against MRSA in vivo (Mota-Meira et al., 2005), mersacidin was able to effectively eradicate an MRSA infection in a mouse rhinitis model (Kruszewska et al., 2004) and Nisin F, a natural nisin variant, was also found to successfully control S. aureus infection in rats (De Kwaadsteniet et al., 2009). Similarly, microbsporicin (Figure 1) (NAI-107) was evaluated for its therapeutic potential in nosocomial infection and demonstrated efficacy against MRSA in a rat endocarditis model (Jabes et al., 2011). The efficacy of MU1140 (mutacin 1140) has also been investigated in vivo (Ghobrial et al., 2009) and is currently in pre-clinical development for the treatment of Gram positive infections. NVB302, a derivative of deoxyactagardine B, is currently undergoing phase I clinical trials as a therapeutic for the treatment of C. difficile infections due to its selective targeting of this organism over the predominantly Gram negative normal gut flora (Dawson and Scott, 2012). Investigations into the use of lantibiotics to control the microorganisms responsible for dental plaque, halitosis and “sore throat” infections have also yielded promising results (Hillman, 2002; Burton et al., 2006; Dierksen et al., 2007).
Some lantibiotics possess additional bioactivities that hold promise for therapeutic application. A smaller subcategory of lantibiotics, such as cinnamycin and duramycin, have been found to influence eukaryotic metabolic functions by binding phosphatidylethanolamine in cell membranes and, in turn, inhibiting the enzyme phospholipase A2 (Marki et al., 1991). In addition to this activity, duramycin demonstrated efficacy in the treatment of cystic fibrosis by inhalation (Grasemann et al., 2007) as a result of its ability to stimulate chloride secretion in bronchial epithelial cells (Oliynyk et al., 2010).
Remarkably, the first instance of a lantibiotic, or indeed any bacteriocin, to prevent the growth of cancer cells has been confirmed. In a study by Joo and coworkers, nisin Z was shown to be effective in the treatment of head and neck squamous cell carcinoma (HNSCC; Joo et al., 2012). In subsequent mouse trials involving a highly purified form of nisin Z, reduced tumorigenesis in vivo was observed and long-term treatment with nisin Z extended survival. In addition, nisin treated mice exhibited normal organ histology with no evidence of inflammation, fibrosis or necrosis (Kamarajan et al., 2015).
Bioengineering and Synthetic Biology- Generating More Effective Lantibiotics
Bioengineering (engineering within the cell) and the use of synthetic biology-based (in vitro engineering) approaches have been important for advancing our understanding of the fundamentals of bacteriocin activity and structure–function relationships (these approaches are the subject of a number of recent comprehensive reviews: Tabor, 2014; Escano and Smith, 2015). However, there is also a steadily growing number of engineered lantibiotic peptides that demonstrate enhanced functionalities (activity and/or stability) which make them more attractive from a clinical perspective (Cotter et al., 2013). The following provides some recent examples of bioengineered lantibiotics exhibiting enhanced pharmacological and physicochemical properties as well as developments in genetic systems to increase peptide yields.
Several bioengineered variants of the prototypical lantibiotic nisin have been generated that provide excellent examples of how lantibiotic functionality can be modulated by as little as one residue change. The nisin Z derivatives N20K, M21K, N27K, H31K generated by protein engineering displayed improved solubility, particularly at alkaline pH values where the solubility of the parent nisin is particularly reduced (Rollema et al., 1995; Yuan et al., 2004). Furthermore, the consequences of effecting single residue alterations at distinct locations in nisin has generated variants that exhibit not only improved antimicrobial activity against strains of clinical relevance (MRSA, VRE, VISA, MRSP, and C. difficile) but has also brought about the widening of its antimicrobial spectrum to include some Gram negative bacteria (Field et al., 2008, 2012, 2015; Molloy et al., 2013). More dramatic substitutions at the location of rings A and B at the N-terminal end of nisin A revealed that the various activities of nisin can be altered by changing the amino acid arrangement in this region of the peptide (Rink et al., 2007). The hinge-region of nisin has also been the subject of mutagenesis resulting in variants with enhanced antimicrobial activity (Field et al., 2008; Healy et al., 2013) as well as derivatives with an enhanced ability to diffuse through complex polymers (Rouse et al., 2012). In both mutacin 1140 and nukacin ISK-1 peptides, single residue changes brought about a significant increase in activity against several Gram positive strains(Islam et al., 2009; Chen et al., 2013). Similarly, mutagenesis of the mersacidin gene was ultimately successful in that several variants were identified which exhibited enhanced activity against a range of different targets including clinically relevant MRSA, VRE and S. pneumoniae (Appleyard et al., 2009).
Generating enhanced variants of two-component lantibiotics presents an even greater challenge given that two peptides are required to work jointly in synergy. However, a lacticin 3147 derivative with enhanced activity against a pathogenic strain of S. aureus was recently identified (Field et al., 2013), the first occasion such an increase in antibacterial properties has been observed for bioengineered two-component lantibiotics.
Synthetic biology approaches are another promising means to provide insights into structure-stability relationships and generate novel derivatives with improved function. Chemical synthesis enables the limitations of the modification machinery to be bypassed, extending the range of analogs that can be produced. For example, deoxyactagardine B is a single peptide lantibiotic that is rigid, compact and globular and differs from actagardine (Figure 1) by two amino acids and the absence of a sulfoxide bond (Boakes et al., 2010). A synthetically introduced C-terminal modification (1, 7 diaminoheptane) produced a variant, NVB302, that displayed greater solubility and activity compared to the parent molecule. NVB302 is now in phase I clinical trials for the treatment of C. difficile infections (Dawson and Scott, 2012). It has also been established that lantibiotics are susceptible to oxidation of the sulfur-containing lanthionine and this can lead to sharp decreases in antimicrobial activity. In the case of lactocin S, lanthionines were replaced with diaminopimelate to produce several analogs, one of which revealed greater stability whilst still retaining 100% biological activity (Ross et al., 2012).
Chemical synthesis methods were employed to produce enhanced analogs of the lantibiotic epilancin 15X (Knerr and van der Donk, 2012). A novel approach termed in vitro mutasynthesis has produced improved variants of the class II lantibiotic lacticin 481. Here, non-standard amino acids were introduced into the structural peptide by organic synthesis, and subsequently modified in vitro with purified LctM to generate derivatives with superior specific activity against a target strain (Levengood et al., 2009). Notably, synthetic chemistry approaches were employed to generate hybrids of nisin and vancomycin that demonstrated a 40-fold increase in potency compared to each of the components separately (Arnusch et al., 2008). Similarly, the nisin N-terminus (1–12) was synthetically modified by the coupling of simple membrane-active lipids to create biologically active and proteolytically stable hybrids (Koopmans et al., 2015).
Regardless of these bioengineering successes, one concern that remains to be tackled is that of production. Indeed, the discovery, study and application of lantibiotics is often compromised by limited, or the absence of, production of these peptides by the native producer, a problem which is further compounded when working with bioengineered derivatives. However, a number of instances have demonstrated that quite the opposite effect can be achieved in terms of production. In the case of mutacin 1140 and nukacin ISK-1, single residue alterations did not increase specific activity but instead increased peptide production by up to fourfold (Islam et al., 2009; Chen et al., 2013). Importantly, a recent study involving synthetic biology approaches describes the development of a genetic system that facilitates significant overproduction of nisin (Kong and Lu, 2014). Although heterologous expression of lantibiotic peptides (and their bioengineered derivatives) has been demonstrated in the Gram negative host E. coli on several occasions (Nagao et al., 2007; Caetano et al., 2011, 2014; Shi et al., 2012; Basi-Chipalu et al., 2015), a recent study describes a multigene assembly strategy for the overexpression of the two-component lantibiotic lichenicidin in E. coli (Kuthning et al., 2015). Such systems may also help in attaining higher yields to simplify isolation of and improve cost-efficiency of novel derivatives that are often compromised by limited production.
A major drawback that has yet to be overcome with respect to therapeutic use is the sensitivity of lantibiotics to proteolytic cleavage by intestinal enzymes. For example, nisin, pep5 and epidermin have been shown to be susceptible to the proteases trypsin and chymotrypsin (Jarvis and Mahoney, 1969; Bierbaum et al., 1996). Bioengineering strategies could be employed to replace the residues that serve as recognition sites by these and other digestive enzymes and potentially overcome the issue of vulnerability to proteolytic breakdown in the gastrointestinal tract. Indeed, the recent discovery of the class II lantibiotic pseudomycoicidin (which was found to be resistant to trypsin) provides the perfect example for this approach. A trypsin cleavage site which is located in the conserved lipid II binding motif, is protected by the presence of at least one thioether ring structure. This was confirmed by experiments with site-directed mutant peptides where the removal of thioether forming Cys residues resulted in the establishment of protease sensitivity (Basi-Chipalu et al., 2015).
Lastly, it should be remarked that the efficacy of individual lantibiotics could be further boosted through combination with other antimicrobials or membrane-active substances. For example, nisin displayed synergistic activity with the antibiotics colistin and clarithromycin against Pseudomonas aeruginosa (Giacometti et al., 2000) and with ramoplanin and other non-β-lactam antibiotics against many strains of MRSA and VRE (Brumfitt et al., 2002). Similarly, nisin-ceftriaxone and nisin-cefotaxime were found to be highly synergistic against clinical isolates of Salmonella enterica serovar Typhimurium as evident by checkerboard test and time-kill assay (Rishi et al., 2014).
Conclusion
Lantibiotics possess many of the attributes essential for the treatment of infections caused by multi-drug resistant bacteria and their potential for use as alternatives to traditional antibiotic therapies has been mooted for decades. While greater than 100 lantibiotic peptides have been described, not all of these have been characterized in great depth and so many may possess traits of commercial value. Indeed, as the number of microbial genome sequences has increased dramatically, an even larger collection of new lantibiotic biosynthetic gene clusters has been revealed. These clusters can be applied directly or, the information gained from their analysis, can be used indirectly to guide the bioengineering of new and existing peptide structures.
Finally, although nisin remains the only lantibiotic that is extensively exploited, its full use as a therapeutic entity has not yet been fulfilled, in part due to its low solubility and stability at physiological pH. It is thus notable that a broad range of technologies have been developed for the engineering of lantibiotics and the past decade has seen several bioengineering studies describe the generation of peptide derivatives including nisin with enhanced functionality in terms of specific activity, spectrum of activity, solubility and/or temperature and pH stability. Critically, genetic systems are in continuous development to increase yields of peptide that may aid commercial viability. The further application of these systems to enhance nisin and other lantibiotics has the potential to lead to the development of novel derivatives for therapeutic use. Additionally, bioengineering in combination with semi-synthesis will expand structural diversity still further. It is thus likely that these peptides will be only the first of many generations of bioengineered lantibiotic and lantibiotic-like peptides. Given these recent developments and the fact that several lantibiotics are currently in clinical and preclinical trials reinforces our belief that bioengineered lantibiotics can contribute to a solution to antibiotic resistance across a broad range of bacterial pathogens.
Author Contributions
DF drafted the manuscript. PC, CH, and RR revised and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DF, CH, PC, RR are supported by the Irish Government under the National Development Plan, through a Science Foundation Ireland (SFI) Technology and Innovation Development Award (TIDA 14/TIDA/2286) to DF, a SFI Investigator awards to CH and RR (10/IN.1/B3027), SFI-PI funding (11/PI/1137) to PDC and the Alimentary Pharmabiotic Centre under Grant Number SFI/12/RC/2273.
References
Appleyard, A. N., Choi, S., Read, D. M., Lightfoot, A., Boakes, S., Hoffmann, A., et al. (2009). Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem. Biol. 16, 490–498. doi: 10.1016/j.chembiol.2009.03.011
Arnusch, C. J., Bonvin, A. M., Verel, A. M., Jansen, W. T., Liskamp, R. M., de Kruijff, B., et al. (2008). The vancomycin-nisin(1–12) hybrid restores activity against vancomycin resistant Enterococci. Biochemistry 47, 12661–12663. doi: 10.1021/bi801597b
Basi-Chipalu, S., Dischinger, J., Josten, M., Szekat, C., Zweynert, A., Sahl, H. G., et al. (2015). Pseudomycoicidin, a Class II Lantibiotic from Bacillus pseudomycoides. Appl. Environ. Microbiol. 81, 3419–3429. doi: 10.1128/AEM.00299-15
Bierbaum, G., Szekat, C., Josten, M., Heidrich, C., Kempter, C., Jung, G., et al. (1996). Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol. 62, 385–392.
Boakes, S., Appleyard, A. N., Cortes, J., and Dawson, M. J. (2010). Organization of the biosynthetic genes encoding deoxyactagardine B (DAB), a new lantibiotic produced by Actinoplanes liguriae NCIMB41362. J. Antibiot (Tokyo) 63, 351–358. doi: 10.1038/ja.2010.48
Bonelli, R. R., Schneider, T., Sahl, H. G., and Wiedemann, I. (2006). Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob. Agents Chemother. 50, 1449–1457. doi: 10.1128/AAC.50.4.1449-1457.2006
Breukink, E., and de Kruijff, B. (2006). Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–332. doi: 10.1038/nrd2004
Brumfitt, W., Salton, M. R., and Hamilton-Miller, J. M. (2002). Nisin, alone and combined with peptidoglycan-modulating antibiotics: activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. J. Antimicrob. Chemother. 50, 731–734. doi: 10.1093/jac/dkf190
Burton, J. P., Chilcott, C. N., Moore, C. J., Speiser, G., and Tagg, J. R. (2006). A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 100, 754–764. doi: 10.1111/j.1365-2672.2006.02837.x
Caetano, T., Barbosa, J., Moesker, E., Sussmuth, R. D., and Mendo, S. (2014). Bioengineering of lanthipeptides in Escherichia coli: assessing the specificity of lichenicidin and haloduracin biosynthetic machinery. Res. Microbiol. 165, 600–604. doi: 10.1016/j.resmic.2014.07.006
Caetano, T., Krawczyk, J. M., Mosker, E., Sussmuth, R. D., and Mendo, S. (2011). Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem. Biol. 18, 90–100. doi: 10.1016/j.chembiol.2010.11.010
Castiglione, F., Cavaletti, L., Losi, D., Lazzarini, A., Carrano, L., Feroggio, M., et al. (2007). A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry 46, 5884–5895. doi: 10.1021/bi700131x
Castiglione, F., Lazzarini, A., Carrano, L., Corti, E., Ciciliato, I., Gastaldo, L., et al. (2008). Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem. Biol. 15, 22–31. doi: 10.1016/j.chembiol.2007.11.009
Cavera, V. L., Arthur, T. D., Kashtanov, D., and Chikindas, M. L. (2015). Bacteriocins and their position in the next wave of conventional antibiotics. Int. J. Antimicrob. Agents 46, 494–501. doi: 10.1016/j.ijantimicag.2015.07.011
Chen, S., Wilson-Stanford, S., Cromwell, W., Hillman, J. D., Guerrero, A., Allen, C. A., et al. (2013). Site-directed mutations in the lanthipeptide mutacin 1140. Appl. Environ. Microbiol. 79, 4015–4023. doi: 10.1128/AEM.00704-13
Cotter, P. D., Hill, C., and Ross, R. P. (2005). Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Pept. Sci. 6, 61–75. doi: 10.2174/1389203053027584
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
Dawson, M. J., and Scott, R. W. (2012). New horizons for host defense peptides and lantibiotics. Curr. Opin. Pharmacol. 12, 545–550. doi: 10.1016/j.coph.2012.06.006
De Kwaadsteniet, M., Doeschate, K. T., and Dicks, L. M. (2009). Nisin F in the treatment of respiratory tract infections caused by Staphylococcus aureus. Lett. Appl. Microbiol. 48, 65–70. doi: 10.1111/j.1472-765X.2008.02488.x
Dierksen, K. P., Moore, C. J., Inglis, M., Wescombe, P. A., and Tagg, J. R. (2007). The effect of ingestion of milk supplemented with salivaricin A-producing Streptococcus salivarius on the bacteriocin-like inhibitory activity of streptococcal populations on the tongue. FEMS Microbiol. Ecol. 59, 584–591. doi: 10.1111/j.1574-6941.2006.00228.x
Dischinger, J., Basi Chipalu, S., and Bierbaum, G. (2014). Lantibiotics: promising candidates for future applications in health care. Int. J. Med. Microbiol. 304, 51–62. doi: 10.1016/j.ijmm.2013.09.003
Escano, J., and Smith, L. (2015). Multipronged approach for engineering novel peptide analogues of existing lantibiotics. Expert. Opin. Drug Discov. 10, 857–870. doi: 10.1517/17460441.2015.1049527
Férir, G., Petrova, M. I., Andrei, G., Huskens, D., Hoorelbeke, B., Snoeck, R., et al. (2013). The Lantibiotic peptide labyrinthopeptin A1 demonstrates broad Anti-HIV and Anti-HSV activity with potential for microbicidal applications. PLoS ONE 8:e64010. doi: 10.1371/journal.pone.0064010
Field, D., Begley, M., O’Connor, P. M., Daly, K. M., Hugenholtz, F., Cotter, P. D., et al. (2012). Bioengineered nisin a derivatives with enhanced activity against both gram positive and gram negative pathogens. PLoS ONE 7:e46884. doi: 10.1371/journal.pone.0046884
Field, D., Gaudin, N., Lyons, F., O’Connor, P. M., Cotter, P. D., Hill, C., et al. (2015). A bioengineered nisin derivative to control biofilms of Staphylococcus pseudintermedius. PLoS ONE 10:e0119684. doi: 10.1371/journal.pone.0119684
Field, D., Molloy, E. M., Iancu, C., Draper, L. A., PM, O. C., Cotter, P. D., et al. (2013). Saturation mutagenesis of selected residues of the alpha-peptide of the lantibiotic lacticin 3147 yields a derivative with enhanced antimicrobial activity. Microb. Biotechnol. 6, 564–575. doi: 10.1111/1751-7915.12041
Field, D., O’Connor, P. M., Cotter, P. D., Hill, C., and Ross, R. P. (2008). The generation of nisin variants with enhanced activity against specific gram-positive pathogens. Mol. Microbiol. 69, 218–230. doi: 10.1111/j.1365-2958.2008.06279.x
Fontana, M. B., de Bastos Mdo, C., and Brandelli, A. (2006). Bacteriocins Pep5 and epidermin inhibit Staphylococcus epidermidis adhesion to catheters. Curr. Microbiol. 52, 350–353. doi: 10.1007/s00284-005-0152-5
Galvin, M., Hill, C., and Ross, R. P. (1999). Lacticin 3147 displays activity in buffer against gram-positive bacterial pathogens which appear insensitive in standard plate assays. Lett. Appl. Microbiol. 28, 355–358. doi: 10.1046/j.1365-2672.1999.00550.x
Ghobrial, O. G., Derendorf, H., and Hillman, J. D. (2009). Pharmacodynamic activity of the lantibiotic MU1140. Int. J. Antimicrob. Agents 33, 70–74. doi: 10.1016/j.ijantimicag.2008.07.028
Giacometti, A., Cirioni, O., Barchiesi, F., and Scalise, G. (2000). In vitro activity and killing effect of polycationic peptides on methicillin-resistant Staphylococcus aureus and interactions with clinically used antibiotics. Diagn. Microbiol. Infect. Dis. 38, 115–118. doi: 10.1016/S0732-8893(00)00175-9
Grasemann, H., Stehling, F., Brunar, H., Widmann, R., Laliberte, T. W., Molina, L., et al. (2007). Inhalation of Moli1901 in patients with cystic fibrosis. Chest 131, 1461–1466. doi: 10.1378/chest.06-2085
Gut, I. M., Prouty, A. M., Ballard, J. D., van der Donk, W. A., and Blanke, S. R. (2008). Inhibition of Bacillus anthracis spore outgrowth by nisin. Antimicrob. Agents Chemother. 52, 4281–4288. doi: 10.1128/AAC.00625-08
Healy, B., Field, D., O’Connor, P. M., Hill, C., Cotter, P. D., and Ross, R. P. (2013). Intensive mutagenesis of the nisin hinge leads to the rational design of enhanced derivatives. PLoS ONE 8:e79563. doi: 10.1371/journal.pone.0079563
Hillman, J. D. (2002). Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie Van Leeuwenhoek 82, 361–366. doi: 10.1023/A:1020695902160
Hoffmann, A., Pag, U., Wiedemann, I., and Sahl, H. G. (2002). Combination of antibiotic mechanisms in lantibiotics. Farmaco 57, 685–691. doi: 10.1016/S0014-827X(02)01208-9
Hsu, S. T., Breukink, E., Tischenko, E., Lutters, M. A., de Kruijff, B., Kaptein, R., et al. (2004). The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967. doi: 10.1038/nsmb830
Iorio, M., Sasso, O., Maffioli, S. I., Bertorelli, R., Monciardini, P., Sosio, M., et al. (2014). A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem. Biol. 9, 398–404. doi: 10.1021/cb400692w
Islam, M. R., Shioya, K., Nagao, J., Nishie, M., Jikuya, H., Zendo, T., et al. (2009). Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol. Microbiol. 72, 1438–1447. doi: 10.1111/j.1365-2958.2009.06733.x
Jabes, D., Brunati, C., Candiani, G., Riva, S., Romano’, G., and Donadio, S. (2011). Efficacy of the new lantibiotic NAI-107 in experimental infections induced by MDR Gram positive pathogens. Antimicrob. Agents Chemother. 55, 1671–1676. doi: 10.1128/AAC.01288-10
Jarvis, B., and Mahoney, R. R. (1969). Inactivation of nisin by alpha-chymotrypsin. J. Dairy Sci. 52, 1448–1449. doi: 10.3168/jds.S0022-0302(69)86771-8
Joo, N. E., Ritchie, K., Kamarajan, P., Miao, D., and Kapila, Y. L. (2012). Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med. 1, 295–305. doi: 10.1002/cam4.35
Kamarajan, P., Hayami, T., Matte, B., Liu, Y., Danciu, T., Ramamoorthy, A., et al. (2015). Nisin ZP, a bacteriocin and food preservative, inhibits head and neck cancer tumorigenesis and prolongs survival. PLoS ONE 10:e0131008. doi: 10.1371/journal.pone.0131008
Knerr, P. J., and van der Donk, W. A. (2012). Chemical synthesis and biological activity of analogues of the lantibiotic epilancin 15X. J. Am. Chem. Soc. 134, 7648–7651. doi: 10.1021/ja302435y
Kong, W., and Lu, T. (2014). Cloning and optimization of a nisin biosynthesis pathway for bacteriocin harvest. ACS Synth. Biol. 3, 439–445. doi: 10.1021/sb500225r
Koopmans, T., Wood, T. M., t Hart, P., Kleijn, L. H., Hendrickx, A. P., Willems, R. J., et al. (2015). Semisynthetic lipopeptides derived from nisin display antibacterial activity and lipid II binding on par with that of the parent compound. J. Am. Chem. Soc. 137, 9382–9389. doi: 10.1021/jacs.5b04501
Kruszewska, D., Sahl, H. G., Bierbaum, G., Pag, U., Hynes, S. O., and Ljungh, A. (2004). Mersacidin eradicates methicillin-resistant Staphylococcus aureus (MRSA) in a mouse rhinitis model. J. Antimicrob. Chemother. 54, 648–653. doi: 10.1093/jac/dkh387
Kuthning, A., Mosker, E., and Sussmuth, R. D. (2015). Engineering the heterologous expression of lanthipeptides in Escherichia coli by multigene assembly. Appl. Microbiol. Biotechnol. 99, 6351–6361. doi: 10.1007/s00253-015-6557-6
Kuwano, K., Tanaka, N., Shimizu, T., Nagatoshi, K., Nou, S., and Sonomoto, K. (2005). Dual antibacterial mechanisms of nisin Z against Gram-positive and Gram-negative bacteria. Int. J. Antimicrob. Agents 26, 396–402. doi: 10.1016/j.ijantimicag.2005.08.010
Lawton, E. M., Ross, R. P., Hill, C., and Cotter, P. D. (2007). Two-peptide lantibiotics: a medical perspective. Mini Rev. Med. Chem. 7, 1236–1247. doi: 10.2174/138955707782795638
Levengood, M. R., Knerr, P. J., Oman, T. J., and van der Donk, W. A. (2009). In vitro mutasynthesis of lantibiotic analogues containing non-proteinogenic amino acids. J. Am. Chem. Soc. 131, 12024–12025. doi: 10.1021/ja903239s
Li, B., Cooper, L. E., and van der Donk, W. A. (2009). In vitro studies of lantibiotic biosynthesis. Methods Enzymol. 458, 533–558. doi: 10.1016/S0076-6879(09)04821-6
Marki, F., Hanni, E., Fredenhagen, A., and van Oostrum, J. (1991). Mode of action of the lanthionine-containing peptide antibiotics duramycin, duramycin B and C, and cinnamycin as indirect inhibitors of phospholipase A2. Biochem. Pharmacol. 42, 2027–2035. doi: 10.1016/0006-2952(91)90604-4
Meindl, K., Schmiederer, T., Schneider, K., Reicke, A., Butz, D., Keller, S., et al. (2010). Labyrinthopeptins: a new class of carbacyclic lantibiotics. Angew. Chem. Int. Ed. Engl. 49, 1151–1154. doi: 10.1002/anie.200905773
Mohr, K. I., Volz, C., Jansen, R., Wray, V., Hoffmann, J., Bernecker, S., et al. (2015). Pinensins: the first antifungal lantibiotics. Angew. Chem. Int. Ed. 54, 11254–11258. doi: 10.1002/anie.201500927
Molloy, E. M., Field, D., Connor, P. M. O., Cotter, P. D., Hill, C., and Ross, R. P. (2013). Saturation mutagenesis of lysine 12 leads to the identification of derivatives of nisin a with enhanced antimicrobial activity. PLoS ONE 8:e58530. doi: 10.1371/journal.pone.0058530
Mota-Meira, M., LaPointe, G., Lacroix, C., and Lavoie, M. C. (2000). MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob. Agents Chemother. 44, 24–29. doi: 10.1128/AAC.44.1.24-29.2000
Mota-Meira, M., Morency, H., and Lavoie, M. C. (2005). In vivo activity of mutacin B-Ny266. J. Antimicrob. Chemother. 56, 869–871. doi: 10.1093/jac/dki295
Nagao, J., Aso, Y., Shioya, K., Nakayama, J., and Sonomoto, K. (2007). Lantibiotic engineering: molecular characterization and exploitation of lantibiotic-synthesizing enzymes for peptide engineering. J. Mol. Microbiol. Biotechnol. 13, 235–242. doi: 10.1159/000104749
Nerandzic, M. M., and Donskey, C. J. (2010). Triggering germination represents a novel strategy to enhance killing of Clostridium difficile Spores. PLoS ONE 5:e12285. doi: 10.1371/journal.pone.0012285
Nikaido, H., and Vaara, M. (1985). Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49, 1–32.
Niu, W. W., and Neu, H. C. (1991). Activity of mersacidin, a novel peptide, compared with that of vancomycin, teicoplanin, and daptomycin. Antimicrob. Agents Chemother. 35, 998–1000. doi: 10.1128/AAC.35.5.998
Oliynyk, I., Varelogianni, G., Roomans, G. M., and Johannesson, M. (2010). Effect of duramycin on chloride transport and intracellular calcium concentration in cystic fibrosis and non-cystic fibrosis epithelia. APMIS 118, 982–990. doi: 10.1111/j.1600-0463.2010.02680.x
Pag, U., and Sahl, H. G. (2002). Multiple activities in lantibiotics–models for the design of novel antibiotics? Curr. Pharm Des. 8, 815–833. doi: 10.2174/1381612023395439
Piper, C., Cotter, P. D., Ross, R. P., and Hill, C. (2009a). Discovery of medically significant lantibiotics. Curr. Drug Discov. Technol. 6, 1–18. doi: 10.2174/157016309787581075
Piper, C., Draper, L. A., Cotter, P. D., Ross, R. P., and Hill, C. (2009b). A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J. Antimicrob. Chemother. 64, 546–551. doi: 10.1093/jac/dkp221
Rea, M. C., Clayton, E., O’Connor, P. M., Shanahan, F., Kiely, B., Ross, R. P., et al. (2007). Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56, 940–946. doi: 10.1099/jmm.0.47085-0
Rink, R., Wierenga, J., Kuipers, A., Kluskens, L. D., Driessen, A. J., Kuipers, O. P., et al. (2007). Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl. Environ. Microbiol. 73, 5809–5816. doi: 10.1128/AEM.01104-07
Rishi, P., Preet Singh, A., Garg, N., and Rishi, M. (2014). Evaluation of nisin-beta-lactam antibiotics against clinical strains of Salmonella enterica serovar Typhi. J Antibiot (Tokyo) 67, 807–811. doi: 10.1038/ja.2014.75
Rollema, H. S., Kuipers, O. P., Both, P., de Vos, W. M., and Siezen, R. J. (1995). Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environ. Microbiol. 61, 2873–2878.
Ross, A. C., McKinnie, S. M., and Vederas, J. C. (2012). The synthesis of active and stable diaminopimelate analogues of the lantibiotic peptide lactocin S. J. Am. Chem. Soc. 134, 2008–2011. doi: 10.1021/ja211088m
Rouse, S., Field, D., Daly, K. M., O’Connor, P. M., Cotter, P. D., Hill, C., et al. (2012). Bioengineered nisin derivatives with enhanced activity in complex matrices. Microb. Biotechnol. 5, 501–508. doi: 10.1111/j.1751-7915.2011.00324.x
Sahl, H., and Bierbaum, G. (2008). Multiple activities of natural agents such as defensins and bacteriocins suggest a change in strategy when developing new antimicrobials. Microbe 3, 467–473.
Saising, J., Dube, L., Ziebandt, A.-K., Voravuthikunchai, S. P., Nega, M., and Götz, F. (2012). Activity of Gallidermin on Staphylococcus aureus and Staphylococcus epidermidis Biofilms. Antimicrob. Agents Chemother. 56, 5804–5810. doi: 10.1128/AAC.01296-12
Sandiford, S. K. (2015). Perspectives on lantibiotic discovery—where have we failed and what improvements are required? Expert. Opin. Drug Discov. 10, 315–320. doi: 10.1517/17460441.2015.1016496
Severina, E., Severin, A., and Tomasz, A. (1998). Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J. Antimicrob. Chemother. 41, 341–347. doi: 10.1093/jac/41.3.341
Shi, Y., Bueno, A., and van der Donk, W. A. (2012). Heterologous production of the lantibiotic Ala(0)actagardine in Escherichia coli. Chem. Commun. (Camb) 48, 10966–10968. doi: 10.1039/c2cc36336d
Tabor, A. B. (2014). Recent advances in synthetic analogues of lantibiotics: what can we learn from these? Bioorg. Chem. 55, 39–50. doi: 10.1016/j.bioorg.2014.04.004
Tagg, J. R. (2004). Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. Indian J. Med. Res. 119(Suppl.), 13–16.
van der Donk, W. A., and Nair, S. K. (2014). Structure and mechanism of lanthipeptide biosynthetic enzymes. Curr. Opin. Struct. Biol. 29, 58–66. doi: 10.1016/j.sbi.2014.09.006
Wescombe, P. A., Heng, N. C. K., Burton, J. P., Chilcott, C. N., and Tagg, J. R. (2009). Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future Microbiol. 4, 819–835. doi: 10.2217/fmb.09.61
Wiedemann, I., Breukink, E., van Kraaij, C., Kuipers, O. P., Bierbaum, G., de Kruijff, B., et al. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. doi: 10.1074/jbc.M006770200
Willey, J. M., and Gaskell, A. A. (2011). Morphogenetic signaling molecules of the streptomycetes. Chem. Rev. 111, 174–187. doi: 10.1021/cr1000404
Willey, J. M., and van der Donk, W. A. (2007). Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501. doi: 10.1146/annurev.micro.61.080706.093501
Keywords: antimicrobial peptide, nisin, mutagenesis, lantibiotic, post-translational modification, bacteriocin, bacterial resistance
Citation: Field D, Cotter PD, Hill C and Ross RP (2015) Bioengineering Lantibiotics for Therapeutic Success. Front. Microbiol. 6:1363. doi: 10.3389/fmicb.2015.01363
Received: 29 September 2015; Accepted: 17 November 2015;
Published: 27 November 2015.
Edited by:
Enrica Pessione, University of Torino, ItalyReviewed by:
Dimitris Tsaltas, Cyprus University of Technology, CyprusShuji Tani, Osaka Prefecture University, Japan
Copyright © 2015 Field, Cotter, Hill and Ross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Des Field, des.field@ucc.ie
 Des Field
Des Field