- 1State Key Laboratory of Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Chinese Center for Disease Control and Prevention, National Institute for Communicable Disease Control and Prevention, Beijing, China
- 2Pathogenic Biology Institute, University of South China, Hengyang, China
- 3School of Tropical and Laboratory Medicine, Hainan Medical University, Haikou, China
Shigella and Salmonella are frequently isolated from various food samples and can cause human gastroenteritis. Here, a novel multiple endonuclease restriction real-time loop-mediated isothermal amplification technology (MERT-LAMP) were successfully established and validated for simultaneous detection of Shigella strains and Salmonella strains in only a single reaction. Two sets of MERT-LAMP primers for 2 kinds of pathogens were designed from ipaH gene of Shigella spp. and invA gene of Salmonella spp., respectively. Under the constant condition at 63°C, the positive results were yielded in as short as 12 min with the genomic DNA extracted from the 19 Shigella strains and 14 Salmonella strains, and the target pathogens present in a sample could be simultaneously identified based on distinct fluorescence curves in real-time format. Accordingly, the multiplex detection assay significantly reduced effort, materials and reagents used, and amplification and differentiation were conducted at the same time, obviating the use of postdetection procedures. The analytical sensitivity of MERT-LAMP was found to be 62.5 and 125 fg DNA/reaction with genomic templates of Shigella strains and Salmonella strains, which was consist with normal LAMP assay, and at least 10- and 100-fold more sensitive than that of qPCR and conventional PCR approaches. The limit of detection of MERT-LAMP for Shigella strains and Salmonella strains detection in artificially contaminated milk samples was 5.8 and 6.4 CFU per vessel. In conclusion, the MERT-LAMP methodology described here demonstrated a potential and valuable means for simultaneous screening of Shigella and Salmonella in a wide variety of samples.
Introduction
Shigella and Salmonella, recognized as important human pathogens worldwide, are responsible for food-borne gastroenteritis, which cause bacterial diarrhea represent a huge public health problem (Yang et al., 2007). It was estimated that in the word about 1.8 million cases died from diarrheal illnesses, a large proportion of which were attributed to Shigella spp. and Salmonella spp. (Shao et al., 2011). Previous studies have reported that Shigella app. were associated with the majority of cases of bacillary dysentery prevalent in developing nations, and Salmonella spp. were the most common cause of food-borne infection outbreaks in almost all countries (Iseki et al., 2007; Lin et al., 2010; Lauri et al., 2011). Therefore, for food industry and public health, rapid, specific and sensitive methodologies to detect the pathogens are continuously required.
The culture-based techniques were used as the gold standard for the detection of Shigella spp. and Salmonella spp. in various samples, but the conventional procedures required multiple subculture steps, biochemical and serological confirmation, which took about 7 days, and were time-consuming and laborious (Mokhtari et al., 2012; Soria et al., 2013). In recent years, molecular technologies, such as PCR and real-time PCR assays, have been successfully applied to detect Shigella spp. and Salmonella spp., while these techniques might not be available for resource-poor regions because of the requirement of specialized laboratories, trained personnel, complicated apparatus and expensive reagents (Okamura et al., 2009; Liew et al., 2014; Wang et al., 2015b). In addition, PCR-based technologies obtained poor performance in practical samples, which contained low numbers of bacterial cells, thus, these techniques required an amplification of bacterial numbers by culturing practical samples for 4–8 h (Song et al., 2005). Herein, it is extremely critical to establish a simple, rapid, sensitive, and specific approaches for detection of Shigella spp. and Salmonella spp. in medical, field and food diagnostic laboratories.
Loop-mediated isothermal amplification (LAMP), as a simple, rapid, highly specific, and sensitive detection methodology, has been employed to detect various pathogens, including parasites, fungi, bacteria, and viruses (Mori and Notomi, 2009; Law et al., 2014). Due to detect a single target sequence, the diagnostic assay has been limited its more flexible and wider applications (Fang et al., 2011; Wang et al., 2014). In order to achieve multiplex LAMP (mLAMP) detection, end point analysis, such as agarose gel and pyrosequencing methods, have been employed to differentiate multiple target sequences, while these strategies required further processing and instrumentation, and did not permit real-time detection (Yang et al., 2007; Liang et al., 2012). More recently, a novel technology presented in our previous study, multiple endonuclease restriction real-time loop-mediated isothermal amplification (MERT-LAMP), overcame the technical difficulties posed by current mLAMP approaches, which was demonstrated to simultaneously detect multiple targets in a single reaction (Wang et al., 2015a). Furthermore, the MERT-LAMP approaches allowed real-time detection in only one isothermal amplification step and the positive results could be obtains in as short as 12 min. As such, the novel MERT-LAMP methodology constitutes a potentially valuable tool for rapid detection of a variety of pathogens in either commercial or field labs.
In present study, our objective is the development and validation of a diagnostic MERT-LAMP-based methodology for simultaneous detection of the Shigella spp. and Salmonella spp. with high specificity and sensitivity, by targeting ipaH (GenBank accession no. M32063) and invA (GenBank accession no. NC.003197) genes, respectively. The ipaH gene, coding an invasion-associated plasmid antigen, was present in multiple copies in both the chromosomes and the plasmids of all Shigella species, which could be selected as mark gene for detection of all Shigella strains (Gaudio et al., 1997; Vu et al., 2004). The Salmonella invasion gene (invA), has been demonstrated to exist in all Salmonella strains but not in non- Salmonella bacteria, thus, the feature allowed to establish invA-based molecular detection methods for diagnosis of all Salmonella stains (Galan et al., 1992; Vantarakis et al., 2000; Hara-Kudo et al., 2005).
Materials and Methods
Design of the MERT-LAMP Primers
Based on the ipaH gene of Shigella spp. and invA gene of Salmonella spp., two sets of MERT-LAMP primers were designed by PrimerExplorer V4 (Eiken Chemical) according to the mechanism of MERT-LAMP technology. Blast analysis confirmed that two sets of MERT-LAMP primers were specific for Shigella and Salmonella. The dark quenchers used were Black Hole Quencher-1 and Black Hole Quencher-2, and the fluorophores used were HEX and Cy5, which can be monitored in real-time system that was used for carrying out the MERT-LAMP amplifications. All of the oligomers were synthesized and purified by Tian-Yihuiyuan (Beijing, China). The details of target sequences, primer design, primers sequences and locations were shown in Table 1 and Figure 1.

Figure 1. Sequence and location of Shigella (ipaH) and Salmonella (invA) genes used to design MERT-LAMP primers. The nucleotide sequences of the sense strands of ipaH (A) and invA (B) are listed. The sites of primer sequences were underlined. Left arrows and right arrows showed complementary and sense sequences that are used.
Reagents
The DNA extraction kits (QIAamp DNA Mini Kits) were purchased from Qiagen (Beijing, China), and the Nb.BsrDI was purchased from New England Biolabs (Beijing, China). The Loopamp kits and Loopamp™ Fluorescent Detection Reagent (FD) were purchase from Eiken Chemical (Tokyo, Japan, and Beijing, China).
Bacterial Strains
A total of 54 strains used in this study were shown in Table 2. All strains were stored in 10% (w/v) glycerol broth an −70°C. These strains were refreshed three times on nutrient agar plate at 37°C, and then were used for enrichment and genomic DNA extraction.
Genomic DNA Extraction
According to the manufacturer's instructions, the genomic DNA templates were extracted from all culture strains using DNA extraction kits (QIAamp DNA minikits; Qiagen, Hilden, Germany). The extracted templates were tested with ultraviolet spectrophotometer at A260/280 and stored under at −20°C before they were used.
The Normal LAMP Assay
To test the usability of two sets of LAMP primers, the LAMP approach either for Shigella strains or Salmonella strains was performed as the following description. In brief, the reaction was conducted with the Loopamp Kit in a final volume of 25 μl containing 1.6 μM each FIP and BIP primers, 0.8 μM each LF and LB primers, 0.4 μM each F3 and B3 primers, 12.5 μl 2 × reaction mix, 1 μl of Bst DNA polymerase (8 U), 1 μl FD, and 1 μl DNA template.
The reaction mixtures of normal LAMP were heated for 1 h at 63°C and then at 85°C for 5 min to stop the reaction. Three mainstream techniques were application to monitor the normal LAMP amplification. The positive amplifications could be directly observed color change by FD reagent, and the products were also detected by electrophoresis on 2% agarose gels with ethidium bromide staining. Moreover, real-time monitoring of normal LAMP reactions was conducted by recording the optical density (OD) at 650 nm every 6 s using the Loopamp Real-time Turbidimeter LA-320C (Eiken Chemical Co., Ltd, Japan). A positive result was defined as a threshold value of >0.1 within 60 min and analysis of each sample (dilution) was determined at least two times.
The Standard MERT-LAMP Assay
To evaluate the feasibility of two sets of MERT-LAMP primers, the amplification mixtures of MERT-LAMP were carried out in a final volume of 25 μl containing 0.8 μM EFIP and FIP primers, 1.6 μM BIP primers, 0.8 μM each LF and LB primers, 0.4 μM each F3 and B3 primers, 12.5 μl 2 × reaction mix, 1 μl (8 U) of Bst DNA polymerase, 1.5 μl (15 U) of Nb.BsrDI endonuclease, and 1 μl DNA template. The mixture was incubated at 63°C for 55 min and then at 85°C for 5 min to stop the amplification. Mixture without DNA template was used as a negative control. After amplification, the MERT-LAMP products were directly observed the color change by FD reagent or confirmed by electrophoresis on 2% agarose gels with ethidium bromide staining. Furthermore, the MERT-LAMP reactions were measured by real-time detection.
The Multiplex MERT-LAMP Reaction
For multiplex reactions, the MERT-LAMP approach was conducted as the following system: 25 μl containing 0.4 μM Shi-EFIP and Shi-FIP primers, 0.8 μM Shi-BIP primers, 0.4 μM each Shi-LF and Shi-LB primers, 0.4 μM each Shi-F3 and Shi-B3 primers, 0.6 μM Sal-EFIP and Sal-FIP primers, 1.2 μM Sal-BIP primers, 0.6 μM each Sal-LF and Sal-LB primers, 0.4 μM each Sal-F3 and Sal-B3 primers, 12.5 μl 2 × reaction mix, 1 μl (8 U) of Bst DNA polymerase, 1.5 μl (15 U) of Nb.BsrDI endonuclease and 1 μl DNA template DNA each of Shigella strains and Salmonella strains. The MERT-LAMP mixtures were carried out at 63°C for 55 min in a real-time system, and reaction mixtures without the DNA template were used as a negative control. The lowest detectable template amount were examined in triplicate
In order to determine the optimal amplification temperature of two sets of MERT-LAMP primes, the amplification mixtures of multiplex MERT-LAMP were heated at a constant temperature ranging from 60°C to 67°C for 55 min and then heated at 85°C for 5 min to stop the reaction. Mixtures without DNA template were used as a negative control.
Evaluation of the Sensitivity of the MERT-LAMP Assay
To make a comparative analysis of MERT-LAMP, LAMP, qPCR, and PCR assays by using pure culture, genomic DNA templates were serially diluted. The limit of detection (LoD) of MERT-LAMP, LAMP, qPCR, and PCR assays was confirmed by genomic DNA amount of the template. The Shigella-qPCR, Shigella-PCR, Salmonella-qPCR and Salmonella-PCR assays have been develop in previous studies, which were employed to verified the LoD of qPCR and PCR technologies (Novinscak et al., 2007; Ueda and Kuwabara, 2009; Mokhtari et al., 2013).
Evaluation of the Specificity of the MERT-LAMP Assay
To assess the specificity of the MERT-LAMP methodology, the multiplex MERT-LAMP reactions were performed under the conditions described above with the genomic DNA templates from 55 strains (Table 2). Analysis of each sample was carried out twice independently.
Practical Application of MERT-LAMP to Shigella and Salmonella Detection in Food Samples
In order to test the applicability of MERT-LAMP technology in food sample, Shigella flexneri serotype 1d (ICDC-NPS001) and Salmonella Enteritidis (ICDC-NPSa003) were simultaneously added to the pasteurized milk samples, which were purchased from a grocery store in Beijing. The milk samples were confirmed as being Shigella- and Salmonella-negative by traditional culture assay and PCR. Firstly, to test the minimal detectable colony forming units (CFUs), the cultures with Shigella flexneri or Salmonella Enteritidis strains were serially diluted (10−1–10−9), and the aliquots of 100 μl appropriate dilution (10−6) was plated in triplicate on brain heart infusion (BHI). The CFUs were counted after 24 h at 37°C. The following steps, the aliquots of 100 μl appropriate dilutions with Shigella flexneri and Salmonella Enteritidis was simultaneously inoculated into the milk samples, and the number of Shigella was adjusted to approximate 2.9 × 105, 2.9 × 104, 2.9 × 103, 2.9 × 102, 2.9 × 101, and 2.9 × 100 CFU/ml, Salmonella for 3.2 × 105, 3.2 × 104, 3.2 × 103, 3.2 × 102, 3.2 × 101, and 3.2 × 100 CFU/ml. Simultaneously, aliquots (100 μl) of the artificially contaminated milk was used for DNA extraction, and the supernatants (2 μl) were used for multiplex MERT-LAMP, LAMP, real-time PCR and PCR detection. Non-contaminated milk sample was selected as negative control. This performance was carried out in triplicate independently.
Results
Confirmation of Shigella- and Salmonella-LAMP Products
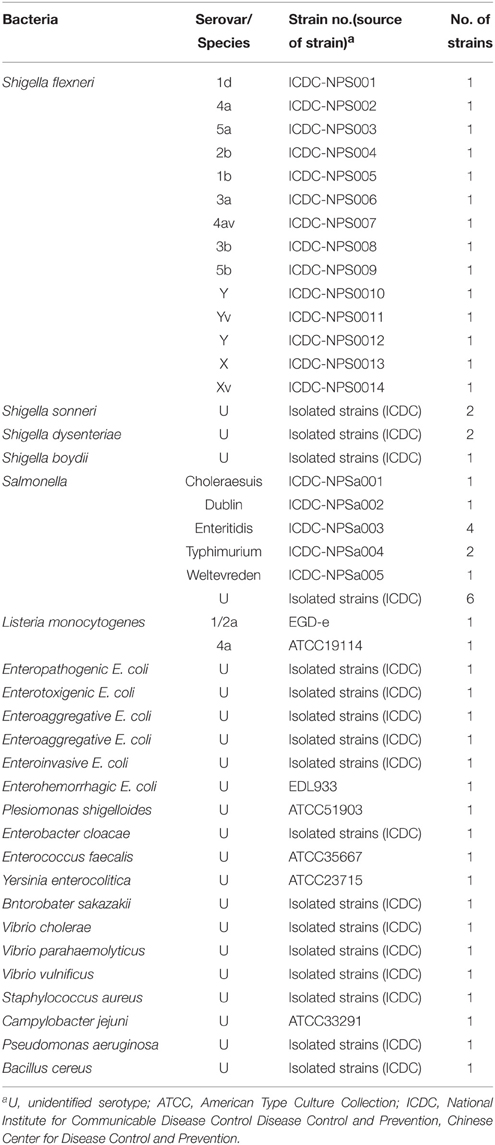
The color change of positive amplifications in Shigella- and Salmonella-LAMP tubes from light gray to green were directly observed by naked eyes within 1 h incubation periods (Figures 2A,B). The normal LAMP products were also detected by 2% agarose gel electrophoresis, and the typical ladder-like patterns were visual (Figures 2C,D). Thus, two sets of normal LAMP primers for Shigella and Salmonella detection were good candidates for establishing the MERT-LAMP methodologies.
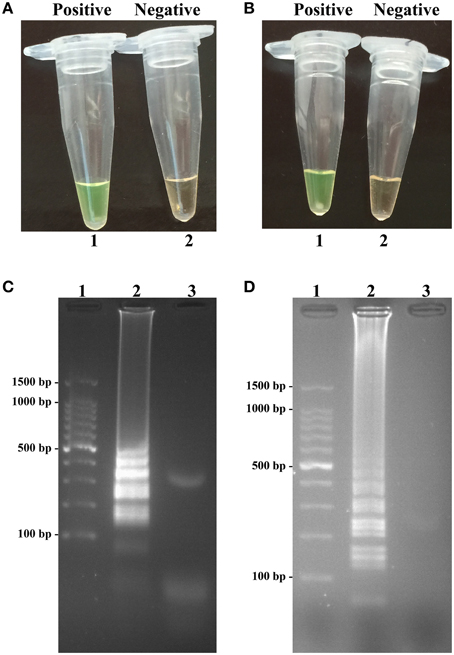
Figure 2. Confirmation and detection of Shigella- and Salmonella-LAMP products. (A,B) Color change of Shigella- and Salmonella-LAMP tubes; tube 1, positive amplification; tube 2, negative amplification. (C,D), 2% agarose gel electrophoresis applied to Shigella- and Salmonella-LAMP products; lane 1, DL 100-bp DNA marker; lane 2, positive LAMP reaction, lane 3, negative LAMP reaction.
Sensitivity of Shigella- and Salmonella-LAMP Assay in Pure Culture
Sensitivity of LAMP reactions on Shigella and Salmonella were examined by analyzing products generated from the serial dilutions (2.5 ng, 250 pg, 25 pg, 2.5 pg, 250 fg, 125 fg, 62.5 fg, and 31.25 fg per microliter) of the Shigella and Salmonella genomic DNA templates in triplicate. As shown in Figures 3A,B, the LAMP amplifications were monitored by real-time turbidity detection; the LoD of Shigella-LAMP approaches was 62.5 fg/reaction (Figure 3A), and the LoD of Salmonella-LAMP assays was 125 fg/reaction (Figure 3B). Moreover, the positive reactions by 2% agarose gel electrophoresis were observed as a ladder-like pattern, the LoD of the agarose gel electrophoresis detection for Shigella- and Salmonella-LAMP reactions was identical with turbidity measurement (Figures 3C,D).
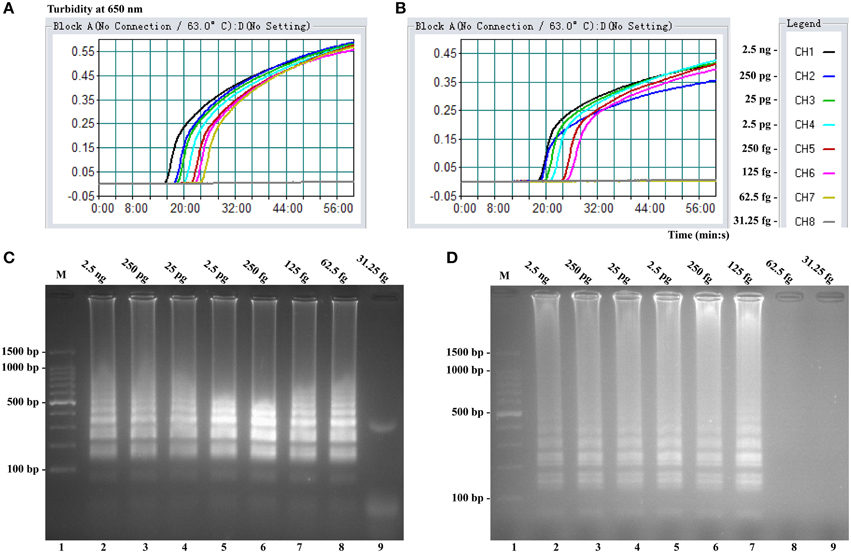
Figure 3. Sensitivity of the ipaH- and invA-LAMP assays using serially genomic DNA with Shigella strains and Salmonella strains as templates. Sensitivity of ipaH-LAMP (A) and invA-LAMP (B) for Shigella and Salmonella detection was analyzed by real-time measurement of turbidity and the corresponding curves of concentrations of genomic DNA were marked in the figure. The LoD of ipaH-LAMP assay was 62.5 fg per tube, and the invA-LAMP for 125 fg per reaction. Sensitivity of ipaH-LAMP (C) and invA-LAMP (D) for Shigella and Salmonella detection was monitored by 2% agarose gel electrophoresis, and the positive amplifications were seen as a ladder-like pattern on 2% agarose gel electrophoresis analysis. Lane 1, DL 100-bp DNA marker.
Confirmation and Detection of MERT-LAMP Products in Nonreal-Time Format
To validate the utility of two sets of MERT-LAMP primers, reaction systems were conducted in the presence or absence genomic DNA templates according to the standard MERT-LAMP condition. The color change of positive MERT-LAMP reactions in ipaH- and invA-MERT-LAMP tubes from light gray to green were directly seen by naked eyes within 1 h incubation periods (Figures 4A,B). Furthermore, the positive products were also detected by 2% agarose gel electrophoresis, and the typical ladder-like patterns were visible but not in negative control (Figures 4C,D). The results indicated the ipaH- and invA-MERT-LAMP primers were feasible for Shigella and Salmonella detection.
Figure 4. Confirmation and detection of ipaH- and invA-MERT-LAMP products. (A,B) Color change of ipaH- and invA-MERT-LAMP tubes; tube 1, positive amplification; tube 2, negative amplification. (C,D) 2% agarose gel electrophoresis applied to ipaH- and invA-MERT-LAMP products; lane 1, DL 100-bp DNA marker; lane 2, positive LAMP reaction, lane 3, negative LAMP reaction.
Analytical Sensitivity of MERT-LAMP Detection for a Single Target
In this study, we evaluated MERT-LAMP amplification in a single target format by using separate detection of ipaH (Shigella spp.-specific gene) and invA (Salmonella spp.-specific gene) from an Shigella and Salmonella genomic DNA templates with EFIP in each reaction. As expected, the release of quenching was observes as a robust increase of Cy5 and Hex signals, and positive results were obtained in ~12 min (Figures 5A,B). The LoD of MERT-LAMP methodology for independently detecting ipaH and invA genes was 62.5 and 125 fg of genomic DNA templates per reaction, respectively (Figures 5A,B). Furthermore, the positive amplifications by 2% agarose gel electrophoresis were seen as the typical ladder-like patterns were visible but not in negative control, and the LoD of the agarose gel electrophoresis detection for Shigella- and Salmonella-LAMP amplifications was identical with real-time measurement (Figures 5C,D).
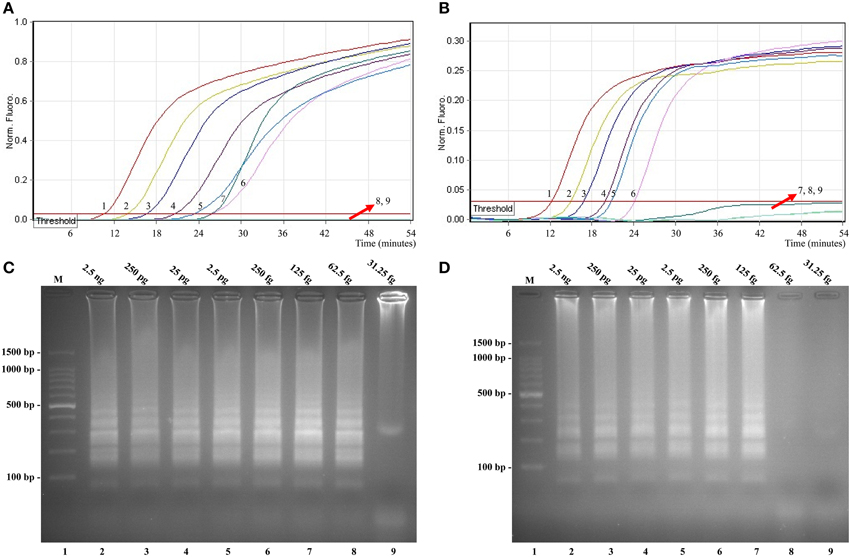
Figure 5. Sensitivity of the ipaH- and invA-MERT-LAMP assays using serially genomic DNA with Shigella strains and Salmonella strains as templates. Sensitivity of ipaH-MERT-LAMP (A) and invA-MERT-LAMP (B) for Shigella and Salmonella detection was monitored by real-time format, and signals 1, 2, 3, 4, 5, 6, 7, 8, and 9 represent DNA levels of 2.5 ng, 250 pg, 25 pg, 2.5 pg, 250 fg, 125 fg, 62.5 fg, and 31.25 fg per vessel and negative control. The LoD of ipaH-MERT-LAMP assay was 62.5 fg per tube, and the invA-MERT-LAMP for 125 fg per reaction. Sensitivity of ipaH-MERT-LAMP (C) and invA-MERT-LAMP (D) for Shigella and Salmonella detection was analyzed by 2% agarose gel electrophoresis, and the positive amplifications were observed as a ladder-like pattern on 2% agarose gel electrophoresis analysis. Lane 1, DL 100-bp DNA marker.
The LoD of MERT-LAMP, LAMP, qPCR, and PCR assays on Shigella was 62.5 fg, 62.5 fg, 2.5 pg and 25 pg, and on Salmonella was 125 fg, 125 fg, 2.5 pg, and 25 pg, respectively (Table 3, Figures 3, 5). The results indicated that the sensitivity of MERT-LAMP technique for detecting a single target was identical with normal LAMP approach, whereas was at least 10- and 100-fold more sensitive than that of qPCR and PCR assays, respectively.
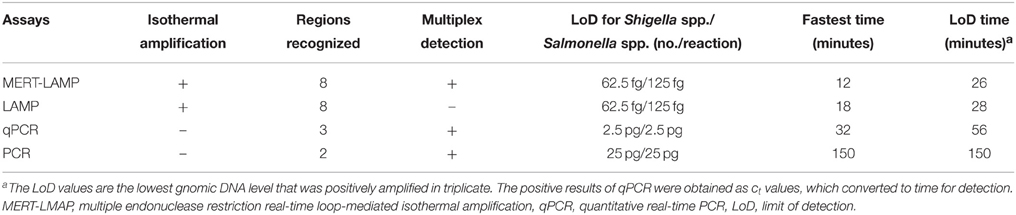
Table 3. The LoD and time for single MERT-LAMP targeting ipaH and invA genes compared with that of qPCR and conventional PCR approaches.
The Optimal Amplification Temperature of ipaH- and invA-MERT-LAMP Assays
In order to examine the optimal detection temperature of multiplex MERT-LMAP approach, the multiplex MERT-LMAP amplifications, which simultaneously contained the Shigella flexneri and Salmonella enteritidis genomic DNA templates at the level of 2.5 ng per tube, were performed at distinct temperatures (60°C–67°C) according to the multiplex MERT-LAMP reaction. The results were analyzed by means of real-time format, and the typical kinetics graphs were produced (Figure 6). Each detection temperature generated a robust signal corresponding to Cy5 and Hex channel, with the faster amplification observed for assay temperature of 62°C–65°C, which were considered as the standard temperature for multiplex MERT-LAMP detection. The detection temperature of 63°C was used for the rest of multiplex MERT-LAMP reaction conducted in this study.
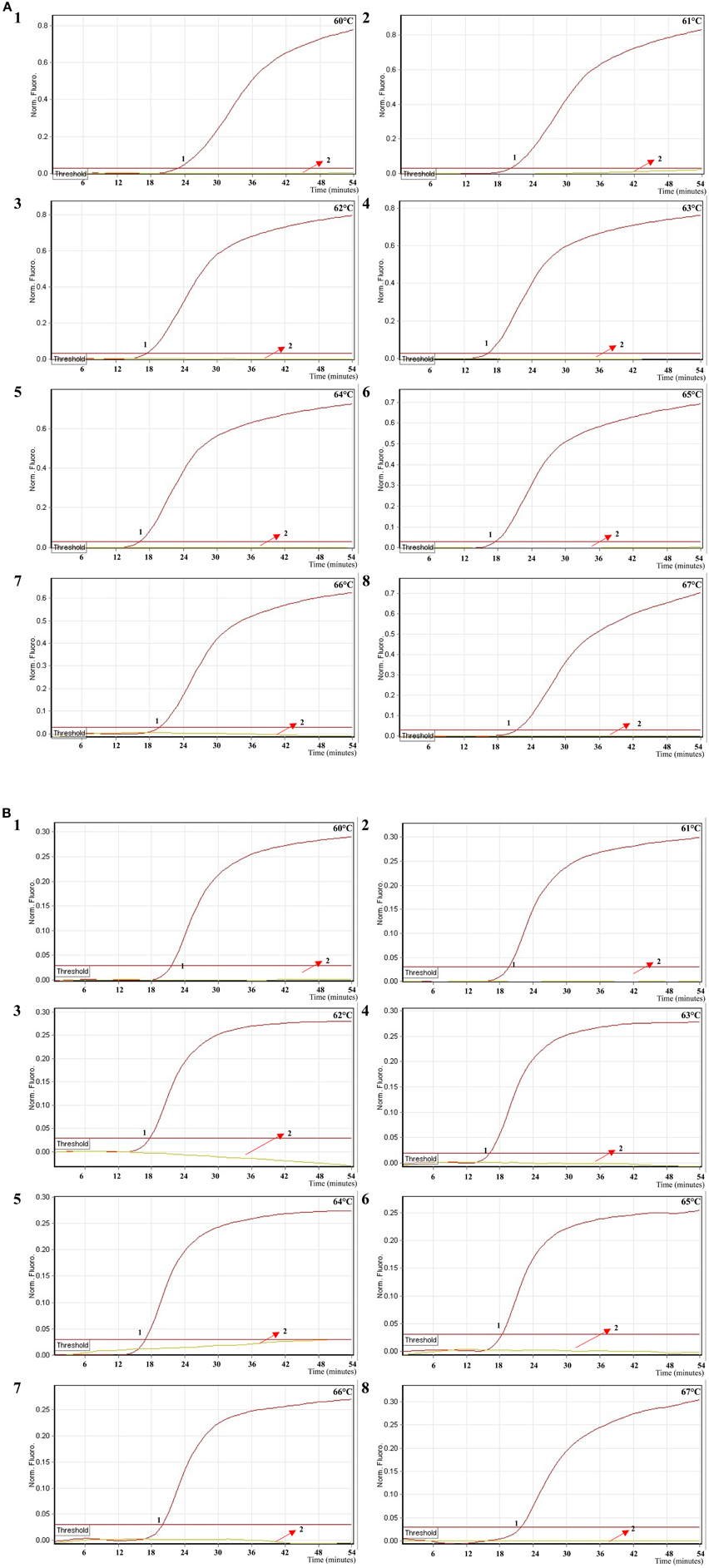
Figure 6. The optimal temperature for multiplex MERT-LAMP assay. Two sets of MERT-LMAP primers targeting ipaH and invA genes were used in the same reaction tube, (A,B) were simultaneously obtained from Cy5 (labeling Shi-EFIP of ipaH) and Hex (labeling Sal-EFIP of invA) channels, respectively. The multiplex MERT-LAMP amplifications were detected by means of real-time format, and the corresponding curves of DNA concentrations were listed. Signal 1 indicates Shigella flexneri strains of in Cy5 channel (A), Salmonella Enteritidis strain for Hex channel (B), and signal 2 indicates negative control. Eight kinetic graphs (1–8) were generated at different amplification temperature (60°C–67°C) with Shigella genomic DNA at the level of 2.5 ng in Cy5 channel (A); another eight kinetic graphs (1–8) were yielded at different detection temperature (60°C–67°C) with Salmonella genomic DNA at the level of 2.5 ng in Hex channel (B). The graphs from 62°C to 65°C show robust amplification.
Analytical Sensitivity of MERT-LAMP Detection for Multiple Targets in a Reaction
In order to simultaneously identify multiple targets in a single reaction, we slightly modified the amount of the primers on the base of standard MERT-LAMP reaction, and the multiplex MERT-LAMP systems were also performed at 63°C for 55 min. Two different fluorescence curves were simultaneously yielded from multiplex MERT-LAMP reactions containing two complete primer sets and their corresponding genomic DNA templates (Figure 7). The MERT-LAMP assay successfully detected Shigella and Salmonella in a single reaction, and simultaneously presented two sets of robust signals for two targets. The LoD of MERT-LAMP technology for simultaneously differentiating ipaH and invA genes was 62.5 and 125 fg of each genomic DAN per tube, respectively (Figure 7). No difference of sensitivity was seen between detecting multiple targets and a single target in a MERT-LAMP system.
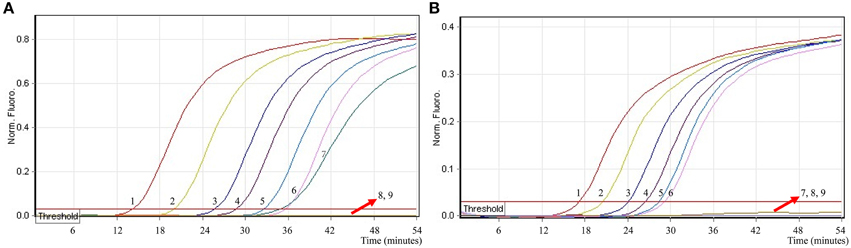
Figure 7. The sensitivity of multiplex MERT-LAMP assay for simultaneously detecting two target pathogens. Two sets of MERT-LMAP primers targeting ipaH and invA genes were simultaneously added to a reaction tube. (A,B) were simultaneously generated from Cy5 (labeling Shi-EFIP of ipaH) and Hex (labeling Sal-EFIP of invA) channels, respectively. Sensitivity of multiplex-MERT-LAMP for simultaneously detecting Shigella (A) and Salmonella (B) was monitored by real-time format, and signals 1, 2, 3, 4, 5, 6, 7, 8, and 9 represent DNA levels of 2.5 ng, 250 pg, 25 pg, 2.5 pg, 250 fg, 125 fg, 62.5 fg, and 31.25 fg per vessel and negative control. The LoD of multiplex MERT-LAMP assay for Shiglla detection was 62.5 fg per tube, and the LoD of multiplex MERT-LAMP for Salmonella detection was 125 fg per reaction.
Analytical Specificity of the Multiplex MERT-LAMP Methodology
To evaluate the specificity of multiplex MERT-LAMP technique, a total of 54 strains, including 19 Shigella and 15 Salmonella and 20 non-Shigella and non-Salmonella, were subjected to MERT-LAMP reaction. By observation, positive results could be generated only when genomic DNAs of Shigella strains and Salmonella strains were used as templates in multiplex MERT-LAMP assay, and the target pathogens were correctly distinguished (Figure 8). In contrast, negative control, non-Shigella stains and non-Salmonella strains tested by multiplex MERT-LAMP technology were negative after 55-min incubation period. These results showed that the multiplex MERT-LAMP assay presented here was specific to target sequence identification.
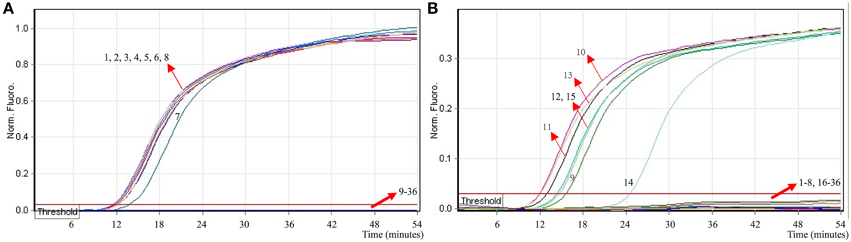
Figure 8. The specificity of multiplex MERT-LMAP detection for different strains. The multiplex MERT-LAMP reactions were conducted using different genomic DNA templates and were analyzed by means of real-time format. (A,B) were simultaneously yielded from Cy5 and Hex channels. Signals 1–8, Shigella flexneri strains of serovar 1d (ICDC-NPS001), 4a (ICDC-NPS002), 5a (ICDC-NPS003), 2b (ICDC-NPS004), 1b (ICDC-NPS005), Shigella boydii, Shigella sonneri and Shigella dysenteriae; signals 9–13, Salmonella Choleraesuis (ICDC-NPSa001), Salmonella Dublin (ICDC-NPSa002), Salmonella Enteritidis (ICDC-NPSa003), Salmonella Typhimurium (ICDC-NPSa004), Salmonella Weltevreden (ICDC-NPSa005); signals 14 and 15, two Salmonella strains of unidentified serotype; signals 16–35, Listeria monocytogenes stains of serovar 1/2a (EGD-e), Listeria monocytogenes stains of serovar 4a (ATCC19114), Enteropathogenic E. coli, Enterotoxigenic E. coli, Enteroaggregative E. coli, Enteroaggregative E. coli, Enteroinvasive E. coli, Enterohemorrhagic E. coli, Plesiomonas shigelloides, Enterobacter cloacae, Enterococcus faecalis, Yersinia enterocolitica, Bntorobater sakazakii, Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, Staphylococcus aureus, Campylobacter jejuni, Pseudomonas aeruginosa, and Bacillus cereus, signal 36, negative control.
Multiplex MERT-LAMP Technology for Artificially Contaminated Milk Sample
To ascertain the applicability the MERT-LAMP technology as a surveillance tool for Shigella and Salmonella in food, the MERT-LAMP assay was tested by the artificially inoculating Shigella stain and Salmonella strain into milk sample. The multiplex MERT-LAMP approach could yield positive amplification when the contaminated numbers of Shigella and Salmonella were more than 2.9 × 102 CFU/ml (~5.8 CFU/tube) and 3.2 × 102 CFU/ml (~6.4 CFU/tube), respectively, and the two target pathogens were simultaneously detected in a single MERT-LAMP reaction (Figure 9, Table 4). The non-contaminated milk sample was found to be negative.
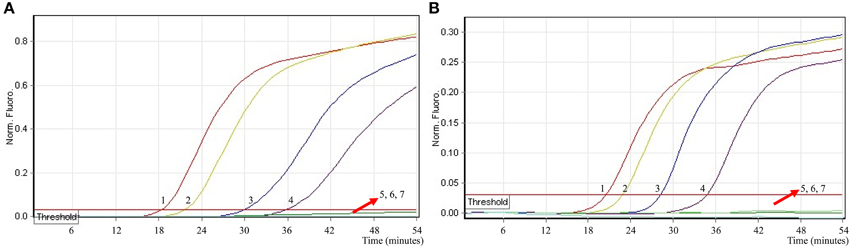
Figure 9. The sensitivity of multiplex MERT-LAMP assay for simultaneously detecting two target pathogens in artificially contaminated milk samples. Two sets of MERT-LMAP primers targeting ipaH and invA genes were added to a reaction tube. (A,B) were simultaneously generated from Cy5 (labeling Shi-EFIP of ipaH) and Hex (labeling Sal-EFIP of invA) channels, respectively. Sensitivity of multiplex-MERT-LAMP for simultaneously detecting Shigella (A) and Salmonella (B) in artificially contaminates milk samples was analyzed by real-time format, and signals 1, 2, 3, 4, 5, 6, and 7 represent Shigella DNA levels of 5800, 580, 58, 5.8, 0.58, and 0.058 CFU per reaction and negative control; Salmonella DNA levels for 6400, 640, 64, 6.4, 0.64, and 0.064 CFU per reaction and negative control. The LoD of multiplex MERT-LAMP assay for Shiglla detection in artificially contaminates milk samples was 5.8 CFU per tube, and the LoD of multiplex MERT-LAMP for Salmonella detection in artificially contaminates milk samples was 6.4 CFU per reaction.
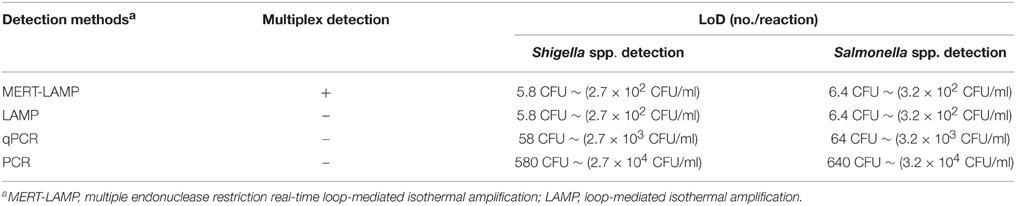
Table 4. Comparison of MERT-LMAP, LAMP, qPCR, and PCR assay for detection of Shigella spp. and Salmonella spp. in artificially contaminated milk samples.
The detection limit of multiplex MERT-LAMP technology was consistent with that of normal LAMP detection only for Shigella or Salmonella, respectively (Table 4). Comparatively, the qPCR and PCR assays produced positive results when the contaminate numbers of Shigella amounted to more than 2.9 × 103 CFU/ml (~58 CFU/tube) and 2.9 × 104 CFU/ml (~580 CFU/tube), Salmonella for 3.2 × 103 CFU/ml (~64 CFU/tube), and 3.2 × 104 CFU/ml (~640 CFU/tube), respectively. The results revealed that the sensitivity of multiplex MERT-LAMP assays was 10- and 100-fold more sensitive than that of qPCR and PCR methodologies (Table 4).
Discussion
Previous studies have reported that Shigella and Salmonella could contaminate similar types of samples, thus it would be significant to simultaneously identify the two pathogens (Li and Mustapha, 2004; MacRitchie et al., 2014). Culture-based techniques and biochemical are the most common detection assays, while require a long enrichment time and subsequent identification (Mokhtari et al., 2012; Soria et al., 2013). PCR-based technologies have been successfully developed to detect Shigella and Salmonella from various food products and environment samples (Okamura et al., 2009; Liew et al., 2014). However, these techniques require a sophisticated apparatus to denature the template DNA, which significantly limit its wider application in under-resourced settings and in field laboratories (Wang et al., 2014). In recent years, the LAMP methodology was widely applied to detect Shigella and Salmonella due to the simplicity and rapidity, while it was restricted to identify only a single target in a vessel.
As a novel isothermal detection technology, MERT-LAMP assay was recently devised and the working principle was successfully validated (Wang et al., 2015a). Under the isothermal condition, the MERT-LAMP methodology permitted real-time detection of multiple targets in a single reaction, and the amplification and detection were conducted simultaneously within 55 min. In this study, the MERT-LAMP technique for the simultaneous differentiation of Shigella and Salmonella was successfully developed and evaluated. The MERT-LAMP amplifications were carried out at only one isothermal step, obviating the use of a thermal cycling equipment, and did not further pyrosequencing and agarose gel detection. Therefore, the multiplex MERT-LAMP approach significantly reduced reagents, materials and effort used, and identification and amplification were performed at the same time, alleviating the use of further procedures. Moreover, the reaction vessels were kept closed in the whole course of the experiment, which effectively eliminated any carryover contamination.
Two species-specific genes (ipaH and invA) present in both Shigella and Salmonella were used as target sequences for designing MERT-LAMP primers (Vantarakis et al., 2000; Hara-Kudo et al., 2005). The specificity of multiplex MERT-LAMP reactions was successful examined, and the positive results were generated in the assay of Shigella strains and Salmonella strains but not for non-Shigella strains and non-Salmonella strains. Furthermore, the MERT-LAMP established here can simultaneously detect and correctly distinguish the target pathogens in a sample, which can be completed in only one isothermal detection step with easily interpretable results. Therefore, the MERT-LAMP assay combined with hand-held diagnostic devices can be as a potential tool for detecting target pathogens, which can be widely applied to various fields, such as point-of-care diagnosis, field testing, and more.
The established multiplex MERT-LAMP assay detected as little as 62.5 and 125 fg of Shigella and Salmonella DNA per tube in the pure cultures, which was consistent with that of normal LAMP and singleplex MERT-LAMP reactions. However, the normal LAMP technology was limited to detect only one target template (Yan et al., 2014). The analytical sensitivity of multiplex MERT-LMAP assay indicated that it could screen marginal amounts of target sequences and was at least 10- and 100-fold more sensitive than that of qPCR and PCR approaches, respectively. With regard to assay speed and sensitivity level obtained in our study, the MERT-LAMP methodology performed better than qPCR and PCR techniques, thus significantly shortening the total detection time (Table 3). Therefore, the MERT-LAMP assay presented here was recommended as a potential diagnostic tool for screening Shigella strains and Salmonella strains in a variety of samples.
To test the practical application of MERT-LAMP detection of Shigella strains and Salmonella strains in food products, artificially contaminated milk samples were analyzed by conventional PCR, qPCR, LAMP, and multiplex MERT-LAMP assays. The detection limit of multiplex MERT-LAMP for Shigella strains and Salmonella strains detection in artificially contaminated milk samples was 5.8 and 6.4 CFU per tube, which was also identical with normal LAMP analysis. The two target pathogens could be simultaneously screened within 40 min with only isothermal incubation step and the positive results were presented in real-time format (Figure 9). Moreover, the LoD of multiplex MERT-LAMP technology for artificially contaminated milk samples was 10- and 100-fold more sensitive than qPCR and conventional PCR assays. Herein, the multiplex MERT-LAMP approach was more suitable than LAMP, qPCR, and PCR techniques for multiplex, rapid, specific and sensitive for detection of Shigella strains and Salmonella strains in food samples.
In conclusion, a rapid, multiplex, specific, and sensitive MERT-LAMP assay for simultaneous detection of Shigella strains and Salmonella strains was established by targeting ipaH and invA genes, and the specificity, sensitivity and practical application were successfully evaluated. Thus, the novel MERT-LAMP assay could be used as a potential tool for facilitating simultaneous monitoring of Shigella strains and Salmonella strains in variety of samples.
Disclosures
Yi Wang, Yan Wang and Changyun Ye have filed for a patent from the State Intellectual Property Office of China, which covers the novel method and sequences included in this manuscript (application number CN201510792208.6).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the financial supports of the grants (Mega Project of Research on the Prevention and Control of HIV/AIDS, Viral Hepatitis Infectious Diseases 2011ZX10004-001, 2013ZX10004-101 to CY) from the Ministry of Science and Technology, People's Republic of China, and grant (2015SKLID507 to CY) from State Key Laboratory of Infectious Disease Prevention and Control, China CDC.
References
Fang, X., Chen, H., Yu, S., Jiang, X., and Kong, J. (2011). Predicting viruses accurately by a multiplex microfluidic loop-mediated isothermal amplification chip. Anal. Chem. 83, 690–695. doi: 10.1021/ac102858j
Galán, J. E., Ginocchio, C., and Costeas, P. (1992). Molecular and functional characterization of the Salmonella invasion gene invA: Homology of InvA to members of a new protein family. J. Bacteriol. 174, 4338–4349.
Gaudio, P. A., Sethabutr, O., Echeverria, P., and Hoge, C. W. (1997). Utility of a polymerase chain reaction diagnostic system in a study of the epidemiology of shigellosis among dysentery patients, family contacts, and well controls living in a shigellosis-endemic area. J. Infect. Dis. 176, 1013–1018. doi: 10.1086/516531
Hara-Kudo, Y., Yoshino, M., Kojima, T., and Ikedo, M. (2005). Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253, 155–161. doi: 10.1016/j.femsle.2005.09.032
Iseki, H., Alhassan, A., Ohta, N., Thekisoe, O. M., Yokoyama, N., Inoue, N., et al. (2007). Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J. Microbiol. Methods 71, 281–287. doi: 10.1016/j.mimet.2007.09.019
Lauri, A., Castiglioni, B., and Mariani, P. (2011). Comprehensive analysis of Salmonella sequence polymorphisms and development of a LDR-UA assay for the detection and characterization of selected serotypes. Appl. Microbiol. Biotechnol. 91, 189–210. doi: 10.1007/s00253-011-3308-1
Law, J. W., Ab Mutalib, N. S., Chan, K. G., and Lee, L. H. (2014). Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Li, Y., and Mustapha, A. (2004). Simultaneous detection of Escherichia coli O157:H7, Salmonella, and Shigella in apple cider and produce by a multiplex PCR. J. Food Prot. 67, 27–33.
Liang, C., Chu, Y., Cheng, S., Wu, H., Kajiyama, T., Kambara, H., et al. (2012). Multiplex loop-mediated isothermal amplification detection by sequence-based barcodes coupled with nicking endonuclease-mediated pyrosequencing. Anal. Chem. 84, 3758–3763. doi: 10.1021/ac3003825
Liew, P. S., Teh, C. S., Lau, Y. L., and Thong, K. L. (2014). A real-time loop-mediated isothermal amplification assay for rapid detection of Shigella species. Trop. Biomed. 31, 709–720.
Lin, W. S., Cheng, C. M., and Van, K. T. (2010). A quantitative PCR assay for rapid detection of Shigella species in fresh produce. J. Food Prot. 73, 221–233.
MacRitchie, L. A., Hunter, C. J., and Strachan, N. J. (2014). Consumer acceptability of interventions to reduce in the poultry food chain. Food Control 35, 260–266. doi: 10.1016/j.foodcont.2013.06.005
Mokhtari, W., Nsaibia, S., Gharbi, A., and Aouni, M. (2013). Real-time PCR using SYBR Green for the detection of Shigella spp. in food and stool samples. Mol. Cell Probes 27, 53–59. doi: 10.1016/j.mcp.2012.09.002
Mokhtari, W., Nsaibia, S., Majouri, D., Ben Hassen, A., Gharbi, A., and Aouni, M. (2012). Detection and characterization of Shigella species isolated from food and human stool samples in Nabeul, Tunisia, by molecular methods and culture techniques. J. Appl. Microbiol. 113, 209–222. doi: 10.1111/j.1365-2672.2012.05324.x
Mori, Y., and Notomi, T. (2009). Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15, 62–69. doi: 10.1007/s10156-009-0669-9
Novinscak, A., Surette, C., and Filion, M. (2007). Quantification of Salmonella spp. in composted biosolids using a TaqMan qPCR assay. J. Microbiol. Methods 70, 119–126. doi: 10.1016/j.mimet.2007.03.019
Okamura, M., Ohba, Y., Kikuchi, S., Takehara, K., Ikedo, M., Kojima, T., et al. (2009). Rapid, sensitive, and specific detection of the O4 group of Salmonella enterica by loop-mediated isothermal amplification. Avian Dis. 53, 216–221. doi: 10.1637/8450-081808-Reg.1
Shao, Y., Zhu, S., Jin, C., and Chen, F. (2011). Development of multiplex loop-mediated isothermal amplification-RFLP (mLAMP-RFLP) to detect Salmonella spp. and Shigella spp. in milk. Int. J. Food Microbiol. 148, 75–79. doi: 10.1016/j.ijfoodmicro.2011.05.004
Song, T., Toma, C., Nakasone, N., and Iwanaga, M. (2005). Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243, 259–263. doi: 10.1016/j.femsle.2004.12.014
Soria, M. C., Soria, M. A., Bueno, D. J., and Terzolo, H. R. (2013). Comparison of 3 culture methods and PCR assays for Salmonella gallinarum and Salmonella pullorum detection in poultry feed. Poult. Sci. 92, 1505–1515. doi: 10.3382/ps.2012-02926
Ueda, S., and Kuwabara, Y. (2009). The rapid detection of Salmonella from food samples by loop-mediated isothermal amplification (LAMP). Biocontrol Sci. 14, 73–76. doi: 10.4265/bio.14.73
Vantarakis, A., Komninou, G., Venieri, D., and Papapetropoulou, M. (2000). Development of a multiplex PCR detection of Salmonella spp. and Shigella spp. in mussels. Lett. Appl. Microbiol. 31, 105–109. doi: 10.1046/j.1365-2672.2000.00797.x
Vu, D. T., Sethabutr, O., Von Seidlein, L., Tran, V. T., Do, G. C., Bui, T. C., et al. (2004). Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J. Clin. Microbiol. 42, 2031–2035. doi: 10.1128/JCM.42.5.2031-2035.2004
Wang, Y., Wang, Y., Lan, R., Xu, H., Ma, A., Li, D., et al. (2015a). Multiple endonuclease restriction real-time loop-mediated isothermal amplification: A novel analytically rapid, sensitive, multiplex loop-mediated isothermal amplification detection technique. J. Mol. Diagn. 17, 392–401. doi: 10.1016/j.jmoldx.2015.03.002
Wang, Y., Wang, Y., Ma, A. J., Li, D. X., Luo, L. J., Liu, D. X., et al. (2015b). Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci. Rep. 5:11902. doi: 10.1038/srep11902
Wang, Y., Wang, Y., Xu, H., Dai, H., Meng, S., and Ye, C. (2014). Rapid and sensitive detection of listeria ivanovii by loop-mediated isothermal amplification of the smcL gene. PLoS ONE 9:e115868. doi: 10.1371/journal.pone.0115868
Yan, L., Zhou, J., Zheng, Y., Gamson, A. S., Roembke, B. T., Nakayama, S., et al. (2014). Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 10, 970–1003. doi: 10.1039/c3mb70304e
Keywords: Shigella spp., Salmonella spp., MERT-LAMP, LAMP, LoD
Citation: Wang Y, Wang Y, Luo L, Liu D, Luo X, Xu Y, Hu S, Niu L, Xu J and Ye C (2015) Rapid and Sensitive Detection of Shigella spp. and Salmonella spp. by Multiple Endonuclease Restriction Real-Time Loop-Mediated Isothermal Amplification Technique. Front. Microbiol. 6:1400. doi: 10.3389/fmicb.2015.01400
Received: 08 September 2015; Accepted: 24 November 2015;
Published: 14 December 2015.
Edited by:
Andrea Gomez-Zavaglia, Center for Research and Development in Food Cryotechnology (CONICET), ArgentinaCopyright © 2015 Wang, Wang, Luo, Liu, Luo, Xu, Hu, Niu, Xu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changyun Ye, yechangyun@icdc.cn
 Yi Wang
Yi Wang Yan Wang1
Yan Wang1 Dongxin Liu
Dongxin Liu Changyun Ye
Changyun Ye