- 1Biochimie, Biophysique et Génétique des Microorganismes, Institut des Sciences de la Vie, Université catholique de Louvain, Louvain-la-Neuve, Belgium
- 2Center for Systems Engineering and Applied Mechanics, ICTEAM, Université catholique de Louvain, Louvain-la-Neuve, Belgium
In streptococci, entry in competence is dictated by ComX abundance. In Streptococcus thermophilus, production of ComX is transient and tightly regulated during growth: it is positively regulated by the cell-cell communication system ComRS during the activation phase and negatively regulated during the shut-off phase by unidentified late competence gene(s). Interestingly, most S. thermophilus strains are not or weakly transformable in permissive growth conditions (i.e., chemically defined medium, CDM), suggesting that some players of the ComRS regulatory pathway are limiting. Here, we combined mathematical modeling and experimental approaches to identify the components of the ComRS system which are critical for both dynamics and amplitude of ComX production in S. thermophilus. We built a deterministic, population-scaled model of the time-course regulation of specific ComX production in CDM growth conditions. Strains LMD-9 and LMG18311 were respectively selected as representative of highly and weakly transformable strains. Results from in silico simulations and in vivo luciferase activities show that ComR concentration is the main limiting factor for the level of comX expression and controls the kinetics of spontaneous competence induction in strain LMD-9. In addition, the model predicts that the poor transformability of strain LMG18311 results from a 10-fold lower comR expression level compared to strain LMD-9. In agreement, comR overexpression in both strains was shown to induce higher competence levels with deregulated kinetics patterns during growth. In conclusion, we propose that the level of ComR production is one important factor that could explain competence heterogeneity among S. thermophilus strains.
Introduction
Genetic plasticity is at the core of the adaptation and evolution strategies in bacteria. Horizontal gene transfer (HGT) events allow individuals to rapidly acquire/lose phenotypic traits and have a major impact on bacterial evolution. Conjugation, transduction, and competence are the main HGT mechanisms, and define genetic exchange communities (Skippington and Ragan, 2011). Competence is defined as a transient physiological state that enables bacteria and archaea to take up exogenous naked DNA and to stably integrate it in their genome by homologous recombination (Johnsborg et al., 2007; Seitz and Blokesch, 2013). The transforming DNA can be passively or actively released from surrounding dead cells or siblings via a programmed cell lysis mechanism promoted by competent cells, respectively (Claverys et al., 2007; Berg et al., 2012; Wei and Havarstein, 2012; Borgeaud et al., 2015). Competence is increasingly viewed as a stress response that may increase adaptability and fitness in hostile conditions (Claverys et al., 2006; Perry et al., 2009; Charpentier et al., 2012; Dufour and Levesque, 2013).
DNA transformation is an energy-consuming process with possible deleterious effects on the population fitness due, for instance, to the acquisition of deleterious mutations and a slower growth rate of competent cells (Haijema et al., 2001; Johnsen et al., 2009; Moradigaravand and Engelstadter, 2013). To optimize the benefits vs. cost of natural transformation, bacteria have implemented complex and interconnected regulatory systems to turn on competence only when particular environmental and physiological parameters reach a critical point (for recent reviews, see Seitz and Blokesch, 2013; Johnston et al., 2014; Fontaine et al., 2015). For instance, streptococci use cell-cell communication systems to synchronize competence induction between individuals, allowing them to behave such as a multicellular organism. This coordination relies on the production and sensing of peptidic competence pheromones. Their production and secretion is generally initiated in response to specific environmental stresses (Claverys et al., 2006; Fontaine et al., 2015). At a critical extracellular concentration, competence pheromones activate the production of the master competence regulator in streptococci, ComX. ComX is a sigma factor (σX) that associates to the RNA polymerase to redirect transcription toward genes required for DNA transport and processing (also named late com genes). Competence pheromones also activate a positive feedback loop on their production that is thought to be necessary to coordinate and maintain competence induction in the population. Two main communication systems have been reported to directly control comX expression. The ComCDE three-component system is prevalent in species belonging to the mitis and anginosus taxonomic groups such as Streptococcus pneumoniae and Streptococcus anginosus, respectively. This system will not be extensively detailed here (For a recent review, see Fontaine et al., 2015). Briefly, the secreted competence pheromones (encoded by comC) are detected in the medium by the histidine kinase ComD, which transmits the information by phosphorelay to ComE. ComE~P then activates the transcription of comX and comCDE, thereby activating a positive feedback loop. The second system, named ComRS, is present in species belonging to the salivarius, mutans, pyogenic, suis, and bovis groups. Together with comX, the encoding genes of both ComCDE and ComRS communication systems belong to the so-called early com genes (Fontaine et al., 2015).
Growth in a chemically defined medium (CDM) devoid of complex oligopeptides seems to be optimal for ComRS activation (Mashburn-Warren et al., 2010; Desai et al., 2012; Son et al., 2012; Wenderska et al., 2012; Gardan et al., 2013; Morrison et al., 2013; Zaccaria et al., 2014). The best studied models are Streptococcus mutans (mutans group) and Streptococcus thermophilus (salivarius group) (Figure 1). In CDM, the ComS precursor is produced and secreted by as yet undiscovered transporter(s). During or after its secretion, ComS is matured by species-specific protease(s) (Khan et al., 2012; Gardan et al., 2013) to release the C-terminal pheromone domain named XIP (for σX inducing peptide; ComSext in Figure 1) (minimum active size is between 7 and 11 aa, see Fontaine et al., 2015). At a critical extracellular concentration, XIP peptides are re-imported into the cytoplasm through the Opp/Ami transporter (Gardan et al., 2009, 2013; Mashburn-Warren et al., 2010; Fontaine et al., 2010a). XIP then interacts with the transcriptional activator ComR and stimulates the binding of the ComR-XIP complex as a dimer to the ComR-box located in the promoter of comX and comS, thereby promoting their transcription. The ComR-XIP complex induces a positive feedback loop on ComS production, but not on ComR. In agreement, the comR promoter is devoid of any ComR-box (Fontaine et al., 2013). The control of ComR production is believed to be the target of distal regulatory systems in response to environmental cues (Mashburn-Warren et al., 2010; Fontaine et al., 2013, 2015). In S. mutans, distal comR regulators are the BsrRM-HdrRM relay in response to cell density (Okinaga et al., 2010; Xie et al., 2010) and the ScnRK two-component system involved in oxidative stress resistance (Kim et al., 2013).
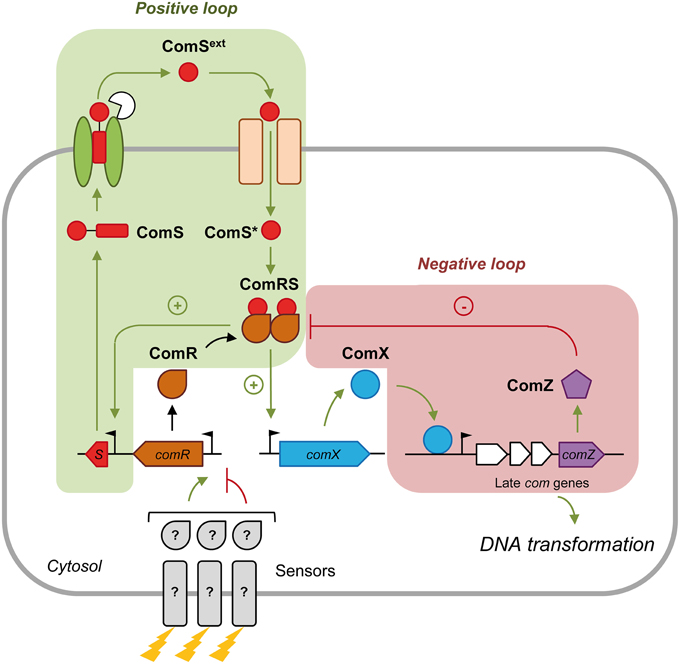
Figure 1. Regulation of comX expression by the ComRS signal transduction system in streptococci. Competence development is initiated by the production of the precursor of the competence pheromone ComS. ComS is then exported by an unknown transporter and matured by species-specific protease(s). At a critical concentration, the mature competence peptide ComSext (also named XIP) is sensed by a specific oligopeptide-binding protein OppA/AmiA and reimported through the Opp/Ami transporter. In the cytoplasm, the intracellular mature peptide ComS* interacts with the transcriptional regulator ComR. The resulting dimeric ComRS complex activates the transcription of early competence genes (green arrows) including comS (plus sign; positive feedback loop) and comX by binding to their promoter. Expression of comR is not activated by ComRS and is thought to be regulated by other regulators/sensors (?) in response to specific environmental cues (represented by lightning). ComX or σX induces transcription of late competence genes necessary for DNA transformation. Competence shut-off (red lines) in S. thermophilus involves at least one or more late gene product(s), named here ComZ. ComZ is thought to interact with and inactivate the ComRS complex (minus sign; negative feedback loop). The names of modeled peptidic/proteic species in system S1 are in black bold.
The shut-off mechanism of competence in ComRS-containing streptococci has been poorly investigated. In S. thermophilus, deletion of comX increases the level and duration of early com genes expression (e.g., comS) (Boutry et al., 2013), similarly to the case of S. pneumoniae (Mirouze et al., 2013; Weng et al., 2013). This suggests that some ComX-regulated genes are involved in this process. In S. mutans, active proteolysis of σX by the MecA-ClpCP machinery plays an important role in the kinetics of competence shut-off (Tian et al., 2013).
Kinetics of comX induction in CDM differs remarkably between species and strains (Desai et al., 2012; Gardan et al., 2013; Morrison et al., 2013; Zaccaria et al., 2014) and is greatly influenced by growth parameters such as pH (Guo et al., 2014; Son et al., 2015). For instance, the ComRS system of S. thermophilus is viewed as a timing device, rather than a true quorum-sensing system. Indeed, spontaneous expression of early com genes starts to increase at the early log phase, whatever the size of inocula, and lasts for about 1 h. In addition, induction of competence by XIP in S. thermophilus is less efficient in the lag and stationary phases (Gardan et al., 2013). This suggests that regulatory components need to be synthesized, or alternatively, degraded for optimal ComRS signaling (Fontaine et al., 2015).
As the ComRS system was recently discovered, many questions about its regulatory cascade still remain open. For instance, it is not clear how environmental parameters are integrated to fine-tune the activity of the ComRS complex. In this context, few ComRS-containing strains turn out to be spontaneously transformable in CDM, but most are responsive to artificial induction (Gardan et al., 2009; Fontaine et al., 2010a,b; Desai et al., 2012; Son et al., 2012; Morrison et al., 2013; Zaccaria et al., 2014). This suggests that some important steps of the regulatory cascade may either be limiting or missing in these growth conditions, which are far from those encountered in natural habitats.
The aim of this study is to deeply re-examine the ComRS regulatory cascade and identify the critical components for ComX production in species belonging to the salivarius group, using a combination of mathematical modeling and experimental molecular approaches. Based on the previous knowledge acquired on the ComRS regulatory cascade, S. thermophilus was chosen as a model bacterium. Strains LMD-9 and LMG18311 were respectively selected as representative of highly and weakly transformable strains (Fontaine et al., 2010b). A dynamic and mechanistic model of the ComRS system of strain LMD-9, partially calibrated by experimental data, was first built. This model, that integrates growth parameters, was then used to study qualitative and quantitative aspects (topology and dynamics) of the ComRS system in order to identify the regulatory determinants of competence development in S. thermophilus.
Materials and Methods
Experimental Procedures
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in the present study are listed in Supplementary Table S1. Plasmids derived from pGICB004 were specifically constructed in Escherichia coli EC1000. E. coli was grown in LB medium with shaking at 37°C (Sambrook et al., 1989). S. thermophilus was grown at 37°C in M17 broth (Difco Laboratories Inc., Detroit, MI) or in CDM, as described by Letort and Juillard (2001). All synthetic media contain 1% (wt vol−1) lactose (M17L and CDML broth, respectively). When required, spectinomycin (200 μg ml−1 for E. coli and S. thermophilus), erythromycin (250 μg ml−1 for E. coli, 2.5 μg ml−1 for S. thermophilus) or chloramphenicol (20 μg ml−1 for E. coli, 5 μg ml−1 for S. thermophilus) was added to the media. Solid agar plates were prepared by adding 2% (wt vol−1) agar to the medium. Solid plates inoculated with S. thermophilus cells are incubated anaerobically (BBL GasPak systems, Becton Dickinson, Franklin lakes, NJ) at 37°C.
Detection of Absorbance and Luminescence
Small volumes (300 μl) of culture samples (OD600 of 0.05) were incubated in the wells of a sterile covered white microplate with a transparent bottom (Greiner, Alphen a/d Rijn, The Netherlands). Growth (OD600) and luciferase (Lux) activity (expressed in relative light unit; RLU) were monitored at 10-min intervals during 5 h in a Varioskan Flash multi-mode reader (ThermoFisher Scientific, Zellic, Belgium) as previously described (Fontaine et al., 2010a, 2013). In the supplementation experiments, different concentrations of synthetic forms of ComS17-24 (LPYFAGCL) (purity >95%) supplied by Peptide 2.0 (Chantilly, VA) were added to the 300 μl-culture samples after 1 h of growth at 37°C.
Natural Transformation Experiments
Experiments were performed as previously described (Fontaine et al., 2010a,b, 2013). The DNA, either pGIUD0855cat (1 μg), pGILFspec::PcomR (1 μg) or purified overlap PCR products (25 ng), were added to 300 μl culture samples. The transformation frequency was calculated as the number of antibiotic-resistant CFUs per ml divided by the total number of viable CFUs per ml. Antibiotic-resistant CFUs were selected on chloramphenicol in case the transforming DNA is pGIUD0855cat or a P32-cat-encompassing PCR product, or on spectinomycin when the transforming DNA encodes a Pspec-spec cassette (plasmid pGILFspec::PcomR). Integration of the antibiotic resistance cassette at the right location in the chromosome of transformants was checked by PCR (primer pairs used are listed in Supplementary Table S2).
DNA Techniques and Electrotransformation
General molecular biology techniques were performed according to the instructions given by Sambrook et al. (1989). S. thermophilus LMD-9 and S. thermophilus LMG18311 chromosomal DNA was prepared as described by Ferain et al. (1996). Electrotransformation of E. coli was performed as described by Dower et al. (1988). Electrocompetent S. thermophilus cells were prepared as previously described (Blomqvist et al., 2006). After transformation with 1 μg of plasmid DNA, cells were immediately resuspended in 1 ml M17L and incubated anaerobically during 6 h at 37°C (pMG36e derivatives) (van de Guchte et al., 1989) or 29°C (pGICB001) (Law et al., 1995). PCRs were performed with Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) in a GeneAmp PCR system 2400 (Applied Biosystems, Foster City, CA). The primers used in this study were purchased from Eurogentec (Seraing, Belgium) and are listed in Supplementary Table S2.
Deletion of DprA by Natural Transformation
The DprA− derivatives of LMD-9 reporter strains were constructed by exchanging the dprA open reading frame (ORF) (sequence between the start and stop codons) with the chloramphenicol resistance cassette lox66-P32-cat-lox71, as previously described (Fontaine et al., 2010b). Primers used are listed in Supplementary Table S2.
Construction of Reporter Strains
Luminescent LMG18311 reporter strains CB009 and LF146 were constructed by replacing part of the blp locus (from blpU to blpX) by the transcriptional fusion PcomX-luxAB and PcomR-luxAB-spec, respectively. This was achieved by transforming pGICB004 derivatives, respectively pGICB001 and pGILF::PcomR, which carries blp recombination fragments, as previously described (Fontaine et al., 2010a; Fleuchot et al., 2011). Plasmid pGILFspec::PcomR was obtained in two steps. First, the spectinomycin resistance cassette was cloned between luxAB and the blp recombination sequence of pGICB004 in order to easily select the desired transformants on spectinomycin. To this end, Pspec-spec was amplified by PCR from plasmid pR412 with primer pairs LoxSpec-SmaI-F/LoxSpec-Pvu-R, digested by SmaI and PvuII and cloned in pGICB004 linearized by SmaI, yielding pGILFspec. Second, a PCR product corresponding to the PcomR sequence from LMG18311 and digested by EcoRI and SpeI was cloned in similarly digested plasmid pGILFspec (see Supplementary Tables S1, S2).
EMSA Experiments
The double-stranded DNA fragment Cy3-PcomS used in the EMSA experiments (approximately 200 bp) was amplified by PCR from S. thermophilus LMD-9 (Supplementary Table S2). The 5′ end of the forward primer used was coupled to the Alexa 555 fluorophore. ComR-Strep from S. thermophilus LMD-9 was purified as previously described (Fontaine et al., 2013). Typically, a gel shift reaction (20 μl) was performed in a binding buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, 1 mg ml−1 BSA) and contained 150 ng labeled probe and 4 μM ComR-Strep proteins. When necessary, 0.2 or 2 μM of ComS17-24 peptides are added. The reaction is incubated at 37°C for 10 min prior to loading of the samples on a native TBE 5% gel. The gel is next subjected to 80 V for approximately 1 h in TBE buffer. DNA complexes were detected by fluorescence on the Ettan DIGE Imager with bandpass excitation filters (nm) of 540/25 (Cy3) and bandpass emission filters of 595/25 (Cy3) (GE Healthcare, Waukesha, WI).
Mathematical Modeling
Our model describes the mean behavior of a competent cell in a homogenous S. thermophilus LMD-9 population grown in CDML conditions. The model focuses on the timing device function of the ComRS system, solely. In this context, ComS exportation/maturation and importation are modeled to create a time delay in the signaling cascade. The choice of not modeling the cell-cell communication aspect of ComS in S. thermophilus is here justified by experimental evidences that (i) secreted mature ComS peptides (XIP) do not freely diffuse in CDML medium and do rather remain associated to the surface of secreting cells (Gardan et al., 2013) and that (ii) the competence cycle (on- and off-steps) takes place at low cell densities (at least in CDML growth conditions) (Gardan et al., 2009, 2013; Fontaine et al., 2010a).
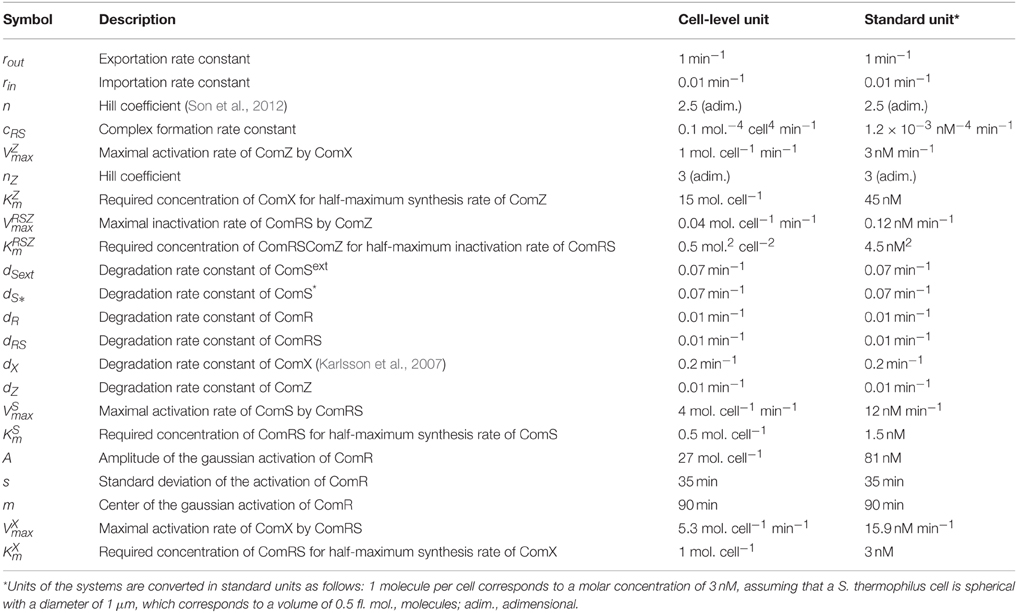
System S1 is composed of seven deterministic ordinary differential equations (ODE) describing the time-course evolution of the seven proteic/peptidic species involved in the (transcriptional) control of ComX production: precursor peptides ComS, secreted peptides ComSext, intracellular mature peptides ComS*, ComR, active complexes ComRS, ComX, and ComZ. These variables are expressed in number of molecules per cell (Table 1). In addition, an equation representing the time-evolution of the cell density X(t) is added (Equation 1, expressed in number of cells per ml), since the development of competence is tightly coupled to the growth process. ComZ is a hypothetical actor that represents the protein(s) of the late phase responsible for competence shut-off. The description of variables and parameters of all equations is presented in Tables 1, 2, respectively.
According to the structure of the regulatory network illustrated in Figure 1, the production of some variables (e.g., ComR, ComS, ComX) is controlled by a basal term b(t) and activation and/or repression terms. The time-course evolution of all variables is also negatively affected by a degradation term, which depends on a specific degradation rate d (we assume first-order degradation kinetics), and a dilution term which accounts for cell division and is thus directly related to the time-dependent growth rate μ(t).
ODE Equations of System S1
Equation (1) models the growth kinetics of LMD-9 cells in CDML conditions. It allows to model competence development during the different growth phases (lag, exponential, and stationary). The time-varying cell density X(t) (expressed in cell ml−1) was computed from a mean experimental growth curve. To convert experimental OD600 units in number of cells per ml, we assumed that a OD600 unit of 1 = 5 × 108 cells ml−1. This conversion was deduced from a plating and CFU (colony forming unit) counting experiment (data not shown). The varying growth rate μ(t) (expressed in min−1) was then computed from X(t) according to Equation (1) (see Supplementary Figure S1A)
Equation (2) describes how the specific number (i.e., per cell) of cytoplasmic precursor ComS peptides (Figure 1) varies with time. ComS is produced in the cell at a non-constant basal rate bS(t), and its production is activated by the ComRS complex according to a Michaelis-Menten function, as modeled in Son et al. (2012). This activation term represents the positive feedback loop which activates and maintains ComS production. The term “exportation” on the right side represents the secretion of ComS in the extracellular medium. The latter is modeled as a non-saturating step: it depends on a constant exportation rate rout and increases directly with ComS(t) concentration. ComS maturation was also assumed to be concomitant with exportation. The last term models ComS dilution due to cell division. The degradation rate of the cytoplasmic precursor ComS was assumed to be null, reflecting the hypothesis that ComS peptides are directly secreted into the extracellular medium upon synthesis.
Equation (3) describes the time-course evolution of number of ComSext, the secreted mature form of ComS, referred to as XIP (Mashburn-Warren et al., 2010). As noted above, ComSext peptides were assumed to remain closely associated to the cell surface, as evidenced in the literature (Gardan et al., 2013). This hypothesis is compatible with the timing-device mechanism of competence regulation in S. thermophilus proposed by Gardan et al. (2013). Expressing ComSext values in molecules cell−1 is thus relevant in this species. Nevertheless, it is important to note that the specific units (molecules cell−1) can be directly converted in volumetric units (molecules ml−1) at any time t based on the time-dependent value of X(t) (expressed in number of cells ml−1) (see Equation 1). In Equation 3, ComSext concentration at the cell surface is positively affected by the rate of ComS exportation (same term as above) and negatively affected by the rates of (i) mature ComSext re-importation in the cytoplasm, (ii) ComSext degradation in the extracellular medium and (iii) cell division (related to Equation 1). Re-importation of ComSext through the Opp system was also assumed to depend on a first-order importation constant rin. This simplification can be justified by the fact that in CDM conditions, the Opp transporter is not saturated by nutritive oligopeptides, in contrast to complex medium like M17 (Gardan et al., 2013; Son et al., 2012).
Equation (4) describes the evolution of number of mature intracellular ComS* peptides. In this equation, the importation flux increases linearly with specific ComSext concentration i.e., the number of ComSext peptides that are physically associated to one cell. The mechanistic effect of cell density on the importation flux is not modeled in Equation (4) as the cell-cell signaling function of ComS is not considered in the ComRS model. We fit the degradation constants in order to keep the concentrations of ComR and ComS inside the cells close to a 1:1 ratio. The negative term cRS(ComR(t)ComS*(t))n corresponds to the formation of the ComRS complex, according to the mass action law, as modeled in Son et al. (2012).
In Equation (5), ComR is produced at a time-dependent basal rate bR(t) and its expression is activated by unknown distal regulators whose actions are summed up in deterministic Gaussian function N(m, σ ,t): the effects of all the activators/inhibitors of ComR are embedded in this bell-shaped function:
Formation of the ComRS complex, as well as ComR degradation and cell division, negatively influences ComR(t) production.
Equation (6) describes the evolution of active ComRS complexes. A ½ stoichiometric coefficient was introduced to describe the formation rate of the active complex since 2 ComS and 2 ComR molecules are needed to produce 1 ComRS molecule (Fontaine et al., 2013). Production of ComRS is negatively influenced by its sequestration in an inactive complex, ComRSZ (for the role of ComZ, see Equation 8 below), which forms according to a Michaelis-Menten function. As mentioned above, this negative term models the negative feedback loop that operates during competence shut-off (see description of Equation 8 for detailed information).
Equation (7) has the same structure as Equation (2): ComX is produced at a basal rate bX(t), and its expression is activated by the ComRS complex.
Equation (8) describes the evolution over time of the ComZ protein, which represents the putative actor(s) of the late phase responsible for competence shut-off. We made the hypothesis of a null basal production rate for ComZ, which is assumed to be produced only in the presence of ComX. The model proposes that ComZ associates with ComRS to inactivate it (Figure 1). This hypothesis is based on the shut-off mechanism described in S. pneumoniae in which the late protein DprA was shown to interact with ComE~P to block ComE-driven transcription of comX (Mirouze et al., 2013).
These eight ODEs form the system S1. At any time t, the state s(t) of the system is determined by the cell density (X(t), see Equation 1) and the number of each molecular species (ξi(t), see Equations 2–8), and is defined by the following vector, evolving in the positive orthant (9):
Determination of Model Parameters
The determination of numerical values for the 22 parameters of system S1 was performed in a stepwise fashion, as described in details below. Briefly, system S1 was first simplified by replacing the production terms for ComR(t), ComS(t), and ComX(t) with lumped production terms derived from experimental measurements. This reduced system (termed S2) only contained 15 of the 22 parameters of system S1. In a second step, the 15 parameters of system S2 were determined, according to several mechanistic hypotheses and constraints. In a final step, the lumped, experiment-derived production terms for ComR(t), ComS(t), and ComX(t) were replaced with their mechanistic equivalents in system S1, and the remaining seven parameters were fitted.
Step 1—model simplification and inclusion of experimental data on ComS, ComR, and ComX production rates
To determine the time-dependent production rates (due to basal production rates and activation rates) prod(t) of variables ComR(t), ComS(t), and ComX(t), we used data on the specific luciferase activities driven from CMDL cultures of LMD-9 derivative strains containing PcomR-luxAB, PcomS-luxAB, or PcomX-luxAB reporter fusions (Fontaine et al., 2013; Supplementary Figure S1B), based on the assumption that the specific production rate of luciferase in a given reporter strain (prodLuxAB(t); molecules cell−1 min−1) is identical to the specific production rate of the peptide or protein under control of the same promoter (prod(t); molecules cell−1 min−1):
In order to determine prodLuxAB(t) from the experimentally observed light emission profiles (L(t); RLU OD) of the reporter strains, the following assumptions were made:
(i) the observed light emission L was assumed to be directly proportional to the amount of luciferase LuxAB:
(ii) the net production rate of luciferase dLuxAB(t)/dt was assumed to be the difference between its absolute production rate (prodLuxAB(t)), its degradation according to a constant degradation rate dLuxAB and its dilution due to cell growth:
Hence, by combining Equations (10–12), the production rates of ComS, ComR, and ComX can be reformulated as a function of the observed light emission as follows:
For the dilution term, μ(t) was calculated from the experimental growth curve of S. thermophilus LMD-9 in CDML, as described in Equation (1) (Supplementary Figure S1A). The degradation rate of luciferase dLuxAB was set to 0.015 min−1, as observed experimentally in S. thermophilus (data not shown). The value of the proportionality constant k (2.197 10−4 RLU molecule−1) was chosen so as to mimic the experimentally observed light emission profiles of the three reporter strains (LMD-9 derivatives containing PcomS-luxAB, PcomX-luxAB, and PcomR-luxAB). Parameter k was thus assumed to be an intrinsic property of the luciferase LuxAB, independent on the promoter driving its expression. For determining k, an additional constraint was set on the system, by imposing a maximal production rate for ComS (prodS(t)) of 20 molecules per minute, in accordance with the estimated maximal synthesis rate of small peptides (Bolouri and Davidson, 2003).
Step 2—determination of parameters for system S2
Since the seven parameters of the modeled production rates for ComS(t), ComS(t), and ComX(t) are hidden behind the experimental production rates, the number of parameters to estimate in system S2 is only m = 15, instead of 22 in system S1. Based on mechanistic assumptions and literature information, specific values were assigned to 2 of the 15 parameters:
(i) The Hill coefficient for formation of the ComRS complex was set to n = 2.5, according to Son et al. (2012), to capture the dimeric character of the complex (Fontaine et al., 2013).
(ii) The degradation rate of ComX (dX) was set to 0.2 min−1, according to Karlsson et al. (2007). This corresponds to a half-life of around 3 min, which is in the same range as the experimental half-life estimated for ComX in S. pneumoniae (~4 min) (Luo and Morrison, 2003).
In addition, the degradation rates of the other variables were related by the following inequality:
which is the mathematical expression of imposing (i) that the half-lives of the peptides/proteins should lie between around 1 min and 1 h, and (ii) that, due to the respective molecular weights of the molecules, the degradation of small peptides (ComSext and ComS*) is faster than the degradation of bigger proteins. Because of their similar size (~10 amino acids; Gardan et al., 2013), the degradation rates of ComSext and ComS* were assumed to be identical. The possible influence of ComS* on the stability of the ComRS complex was neglected, as previously assumed by Voigt et al. (2005) for a similar protein-peptide complex. Hence, the degradation rates of ComR and ComRS were assumed to be identical. Finally, since ComZ is a hypothetical protein, its degradation rate cannot be anticipated on rational grounds: by default, it was constrained to be identical to that of ComR and ComRS.
Numerical values for the remaining ten parameters of system S2 were set with the requirement that the resulting model adequately describes the following experimentally observed behaviors:
(a) Since we initially assumed that luciferase production in the reporter strains is proportional to the production of the corresponding peptides (see Step 1—Model Simplification and Inclusion of Experimental Data on ComS, ComR, and ComX production Rates), two qualitative requirements were set on the simulated profiles of intracellular concentrations of ComR, ComS, and ComX. First, the time sequence of the initial increase in concentration of the three peptides/proteins (Figure 2A) must qualitatively match the time sequence of the onset of light production (Supplementary Figure S1B), i.e., if luciferase activity is first detected in the PcomR-luxAB reporter strain, then the simulated ComR accumulation must start before that of ComS and ComX. The production of the late competence protein ComZ must be delayed compared to ComX, as observed experimentally between the activation of PcomX-luxAB and PcomGA-luxAB, another late com gene (data not shown). Second, the time sequence of the maximum intracellular concentration of the three peptides/proteins (Figure 2A) (null derivative, where the production term equals the consumption, degradation, and dilution terms) must qualitatively match the time sequence of experimentally observed maximum luciferase activity in the corresponding reporter strains (Supplementary Figure S1B). When we compare Figure 2A (modeled kinetics) and Supplementary Figure S1B (experimental kinetics), we see that the two qualitative requirements are globally satisfied. The shape and amplitudes of the simulated curves must however not necessarily match those of luciferase activity, since except for the dilution term, consumption terms for ComR, ComS, and ComX differ between each other and from the degradation term of LuxAB.
(b) It was required that in the modeled system, the concentration of ComX in simulated wild-type LMD-9 cells is not at saturation. In other words, we imposed that the maximum concentration of ComX in simulated cells can be further increased upon addition of extracellular ComS (i.e., non-zero initial values of the variable ComSext(t)), in agreement with experimental observations showing that natural transformation frequencies and the activity of PcomX-luxAB in wild-type LMD-9 can be further increased by addition of the synthetic mature ComS peptide ComS17-24 (Fontaine et al., 2010a and data not shown).
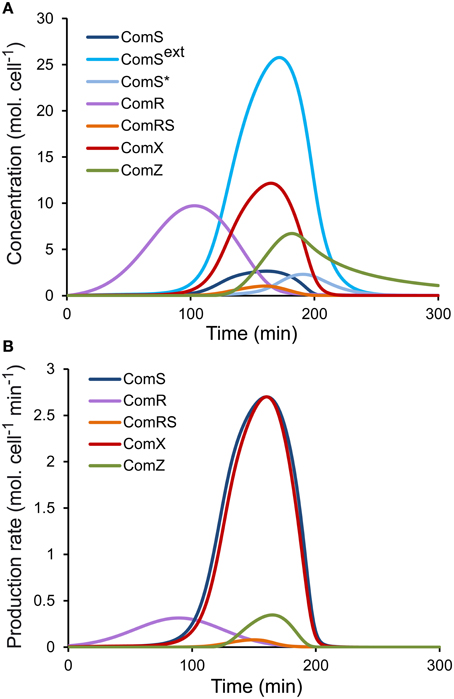
Figure 2. Simulated profiles of molecular species of the ComRS model. Time-course evolution of (A) the concentrations (molecules (mol.) cell−1) and (B) the production rates [molecules (mol.) cell−1 min−1] of the modeled molecular species in a WT LMD-9 strain in CDML growth conditions.
Step 3—determination of the remaining parameters for system S1
Using the 15 parameter values determined in the previous step, we then moved back to system S1, and replaced the experimentally-derived production rates prodR(t), prodS(t), and prodX(t) with their explicit mechanistic equivalents in Equations (5), (2), and (7) respectively. In all three cases, the mechanistic production terms are composed of a basal production rate term and an activation term.
The time-dependent basal expression rate of ComS (bS(t)) was derived from an experimental dataset of luciferase activity in a CDML culture of an LMD-9 ΔcomR strain containing a PcomS-luxAB reporter fusion (Supplementary Figure S1C). Indeed, since the deletion of comR prevents formation of the ComRS complex, it therefore suppresses the activation of ComS production by ComRS, and the remaining luciferase activity thus represents the basal activity of promoter PcomS (Fontaine et al., 2013). Specific luciferase activity (RLU OD) was converted into the basal production rate bS(t) (molecules cell−1 min−1) as described in step 1 for determination of the production rates of ComR, ComS, and ComX, using the formula:
where LΔcomR is the luciferase activity of the ΔcomR strain containing a PcomS-luxAB reporter fusion. We used the same value of 2.197 × 10−4 for the proportionality constant k for ComR, ComS, and ComX. For simplicity, we also assumed that the basal expression rates of ComR and ComX are identical to bS(t): bR(t) = bX(t) = bS(t). It is worth noting that these basal production functions are needed in the model to initiate the competence processus from zero initial concentrations. But their amplitudes remain negligible, with respect to the activation terms, all along the culture. Therefore, the convenient approximation of taking identical functions is not a really restrictive condition here.
Numerical values were then set for the seven additional parameters of system S1, which define the activation rates of ComS (K and V) and ComX (K and V) by ComRS and the activation rate of ComR by distal regulators (m, σ, A). Besides the qualitative requirements already used for system S2 (see Step 2—determination of Parameters for System S2), we also imposed the requirement that the simulated profiles of ComR, ComS, and ComX production rates (basal rate + activation rate) (Figure 2B) are similar in shape to the corresponding luciferase production rates (prodL(t)) deduced from experimental observations (Supplementary Figure S2). Constraints were also imposed on the parameter values for K, K, V, and V. Since our model does not explicitly deal with processes such as transcription and translation, Km and Vmax values cannot be directly compared to existing biological values of other systems. Nevertheless, we constrained those parameters to biologically relevant values, i.e., in the nM range as reported by Jabbari et al. (2011). We also imposed that K is lower than K because the binding affinity of ComR to its DNA motif (ComR-box) was previously predicted to be higher in PcomS compared to PcomX (Fontaine et al., 2013).
The resulting parameter values for the 22 parameters of system S1 are provided in Table 2.
Simulation Protocols
All computations and simulations were done using the MATLAB (R2012a) software. In particular, the ode23 routine was used for the integrations over time, with a null initial condition (except for dose-response simulations). Our full MATLAB source code is available as Supplementary Data Sheet 1. Experimental data necessary to run the code are available as Supplementary Data Sheet 2.
The mean behavior of a wild-type cell was simulated in silico by integrating system S1 over time (from t = 0 to t = 400 min), with the parameters listed in Table 2. A null initial condition was used: at t = 0, all cells were assumed to be devoid of any molecule of the modeled variables: s(t0) = {X (t0), 0, 0, 0, 0, 0, 0,0}.
Simulation of knocked-out strains
To mimic a strain deleted for a gene of interest, the corresponding equation was set to zero in S1, and that modified S1 system was integrated over time with the same ode23 routine.
Simulation of dose-responses to ComS
To mimic the supplementation of ComS17-24 peptides in the growth medium, the variable ComSext(t) was given a non-zero initial value, between 0 and 2000 molecules per cell, at t = 0: s(t0) = {X (t0), 0, V0, 0, 0, 0, 0,0}.
To study the consequence of decreasing the affinity between ComR and ComS on dose-response, the complex formation constant cRS was decreased by a factor ranging from 101 to 105. To simulate supplementation of ComS(17-24) at different times T of growth, the initial condition of ComSext(t) was set to 0 until time T, then at time T (T = [30, 60, 90, 120, 150, 180, 210] in min), an initial condition of ComSext molecules (i.e., 150 molecules per cell) was set. For each condition (various ComSext concentrations or induction times), the system S1 was integrated until tend = 400 min with the ode23 routine and the maximal amount of ComX molecules per cell reached over time was computed.
Simulation of ComR or ComS overproduction
The different levels of ComR or ComS production were simulated by replacing their respective basal rate bR(t) and bS(t) by a constant value R: R = max(b(t)) IF.
When not otherwise stated, the induction factor (IF) was set to 50.
Sensitivity analysis
Our sensitivity analysis was performed by varying the value of each parameter within an interval ranging of 0.1–10 times its assigned value (listed in Table 2) unless otherwise stated. The maximal value of ComX obtained over time in this range was then computed.
Results
Modeling the Control of the Master Competence Regulator ComX By ComRS
In CDM conditions, the ComRS signaling system dominantly controls comX expression in S. thermophilus (Fontaine et al., 2013) and S. mutans (Mashburn-Warren et al., 2010; Son et al., 2012; Reck et al., 2015). We thus hypothesized that the modeling of the ComRS regulatory pathway would recapitulate regulation of ComX production in those growth conditions.
The LMD-9 strain of S. thermophilus was chosen as model strain since the competence regulating network of this strain has been extensively studied and a range of experimental data are available on the expression of key competence genes in wild type (WT) and competence-deficient backgrounds (Fontaine et al., 2010a, 2013; Gardan et al., 2013). Importantly, LMD-9 is also naturally competent in CDM, in contrast to most strains tested so far, where exogenous addition of micromolar concentrations of mature ComS peptides is necessary to reach high levels of competence (Fontaine et al., 2010b).
In CDM growth conditions, competence for natural transformation in S. thermophilus is a transient phenomenon. To take into account this expression dynamics, ordinary differential equations (ODEs) were used to model the time-course evolution of (known) principal molecular species involved in comX induction and shut-off (Figure 1). The ODE formalism is widely used in systems biology and presents several advantages: it allows to set up structural and dynamical models with an adjustable level of detail, allowing to capture various physical phenomena in a transparent way, to use experimental time-course data, and to test different hypotheses easily (de Jong, 2002).
The modeled peptide/proteins (expressed in number of molecules per cell) are: (i) cytoplasmic prepeptides ComS; (ii) mature secreted pheromones ComSext (also named XIP); (iii) reimported mature pheromones ComS*; (iv) unbound cytoplasmic ComR transcriptional regulators; (v) active ComRS complexes; (vi) competence sigma factor ComX and (vii) ComZ, which embeds the actor(s) controlled by ComX that are responsible for competence shut-off. ComZ is assumed to inhibit the activity of ComRS by forming a ternary ComRSZ complex (Figure 1). An equation modeling the population cell density over time (X(t), expressed in number of cells per ml) was also added to the system, to couple competence development to the growth process. At any time t, the state of the system consequently depends on the cell density.
The numerical values of the n (n = 22) parameters of the eight ODE equations were empirically determined on the basis of (i) literature; (ii) mechanistic hypotheses; and (iii) experimental data (see Materials and Methods for detailed information). The latter consist of the growth parameters of strain LMD-9 (Supplementary Figure S1A) and specific luciferase activities driven from promoters PcomX, PcomS, and PcomR in WT and ComR− reporter strains grown in CDM conditions (Supplementary Figures S1B,C).
Meta-Hypotheses
Three meta-hypotheses were assumed to build the mathematical model of the ComRS regulatory cascade. First, our model describes the behavior of a homogenous population of cells in which all the individuals are assumed to act exactly the same way, i.e., we made a mean-field approximation of individual cells. The modeled behavior can thus be seen as the mean behavior over a large sample of cells, neglecting the intercellular variations. This simplification was made because the experimental data used to calibrate the model parameters derive from luciferase activities measured at the population scale level (see Materials and Methods). This explains why the values of some parameters are not whole numbers in our simulations (Table 2). Second, we used the quasi-steady-state assumption for mRNA dynamics, hence focusing on the protein level only. This simplification is generally accepted because RNAs fluctuate much more rapidly, and hence, can be assumed to be in quasi-steady-state compared to the slower varying protein level. Third, we assumed that the transcription and translation steps are instantaneous: as previously discussed (Young and Bremer, 1975; Proshkin et al., 2010), delays can be neglected in prokaryotes due to the lack of compartmentalization.
Model Validation
To get confident in our parameter setting and in the predictive power of our model, the output of different sets of simulation were compared to existing or original experimental data. The chosen simulations aimed to test reactivity, dynamics and topology of the system.
Reactivity to ComS Induction
It was previously shown that the competence default of ComS− derivatives or poorly transformable strains may be supplemented by exogenous synthetic ComS17-24 peptides (Fontaine et al., 2010a,b, 2013). This behavior is typically observed in pheromone-based signaling systems (Sturme et al., 2002). One first set of validation consisted thus in testing if our model responded similarly to extracellular mature ComS (experimental synthetic ComS17-24 or simulated ComSext) in a ComS− background. To this end, the ComS dose-response curves of experimental and simulated ComS− backgrounds were compared. Experimental curves were obtained by measuring the maximum luciferase activity emitted by the ComS− PcomX-luxAB and PcomS-luxAB reporter strains in response to increasing synthetic ComS17-24 concentrations. The simulated curves were obtained by computing the maximum number of ComX molecules per cell obtained by integrating our system with an increasing initial concentration for ComSext. To mimic a ComS− background in our model, Equation (2), describing the production of ComS, was set to zero (see Materials and Methods). Experimental molar ComS17-24 concentrations were converted into molecules per cell to facilitate the comparison. As respectively illustrated in Figures 3A,B, experimental (blue) and simulated (red) curves are both qualitatively and quantitatively comparable. Indeed, the reactivity of the in silico and in vivo systems is in the same range: ComX concentrations were respectively induced ~280- and 560-fold by saturating peptide concentrations. Interestingly, the minimal pheromone concentration that is needed to experimentally turn on the system is ~104-fold higher than the concentration needed to activate the system in silico. This suggests that all the synthetic ComS17-24 peptides that are added to the cell culture are not effectively reimported inside the cells. This could be due to ComS degradation by cell surface proteases (e.g., HtrA), as described for other pheromones (Cassone et al., 2012), ComS aggregation in the growth medium or a limitation in the diffusion of ComS molecules through the cell wall.
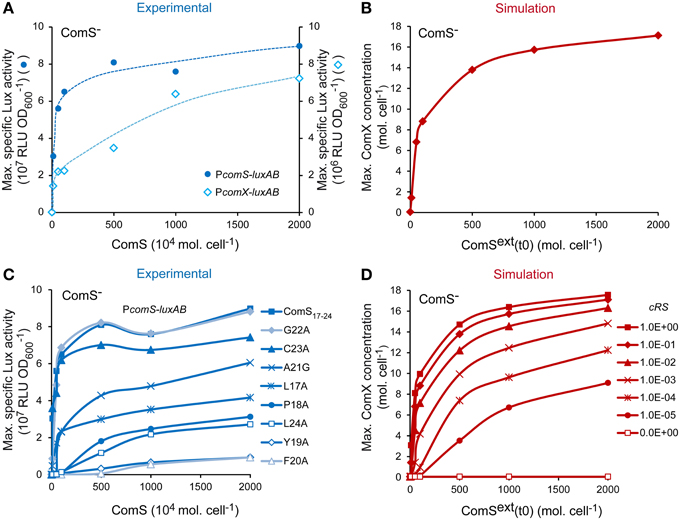
Figure 3. Validation of the reactivity of the ComRS model. (A) Maximum specific luciferase activity (RLU OD) driven by PcomX and PcomS in the ComS− strains LF118 and LF134, respectively, in response to increasing ComS17-24 concentrations. (B) In silico computation of the maximum ComX concentrations (molecules (mol.) cell−1) reached in response to increasing initial ComSext concentrations (mol. cell−1) in a simulated ComS− background. (C) Impact of the systematic replacement of each amino acid residue of ComS17-24 with Alanine on the reactivity of the PcomS-luxAB fusion in ComS− strain (LF134). Graph shows the maximum RLU OD measured after addition of increasing peptides concentrations (D) In silico impact of decreasing the affinity between ComR and ComS on the maximum ComX concentrations (mol. cell−1) reached. This was done by decreasing the value of the ComRS complex formation rate constant cRS by 10-fold steps. Concentrations of ComSext (mol. cell−1) and ComS17-24 (nM) used are 0, 10, 50, 100, 500, 1000, and 2000. To enable comparison between in silico and in vitro experiments, experimental nanomolar units of ComS(17-24) were converted in 104 mol. cell−1 by assuming a density of 5 × 107cells ml−1 at the moment of peptide addition (i.e., after 1 h of incubation in CDML). (A,C) The results of one representative experiment are shown.
Previous studies showed that the reactivity of streptococci to ComS induction is species-specific, suggesting that it is strongly dependent on the specificity of the ComR-ComS interaction. In S. thermophilus, some mutations in ComS17-24 were shown to decrease its biological activity, probably by affecting the formation of active ComRS complexes (Fontaine et al., 2013). Indeed, the efficiency of ComS17-24 variants to induce PcomS-luxAB in vivo (see the dose-response curves on Figure 3C) is strongly related to its ability to induce the formation of ternary PcomS-ComRS complexes in vitro (Supplementary Figure S3). A second set of simulations aimed thus to test whether ComX production was similarly dependent on the rate of ComRS complex formation in our model. The value of the ComRS complex formation rate constant cRS was decreased (see Materials and Methods) and dose-response curves were simulated by computing the maximum number of ComX molecules produced in a ComS− simulated cell (Figure 3D). As expected, our model was able to simulate the effect of ComS mutations that affect the reactivity of ComRS in vivo: the amplitudes and profiles of simulated dose-response curves resemble those observed with mutated ComS17-24 variants in the reporter strain (compare Figure 3C and Figure 3D).
Dynamics of Induction
In CDM medium, the timing of induction by mature ComS peptides (i.e., ComS(17-24)) was previously shown to be critical to reach optimal competence levels (Gardan et al., 2013; Figure 4A). To test the dynamics of ComX production over time in our model, we simulated the supplementation of mature ComS peptides (i.e., ComSext) at different moments during growth of a ComS− strain (see Materials and Methods). Maximum ComX concentrations computed at each induction time is in good accordance with experimental data (compare Figure 4A and Figure 4B) (Gardan et al., 2013). Indeed, ComX production is inducible at all times of growth but both the rate and amplitude are weaker at time 0 and reach an optimum between 60 and 90 min after inoculation. The apparent discrepancy between modeled and experimental output when cells reach the stationary phase (i.e., at induction times of 180 and 210 min) is probably due to the loss of luciferase activities resulting from a depletion of FMNH2 (data not shown and Gardan et al., 2013).
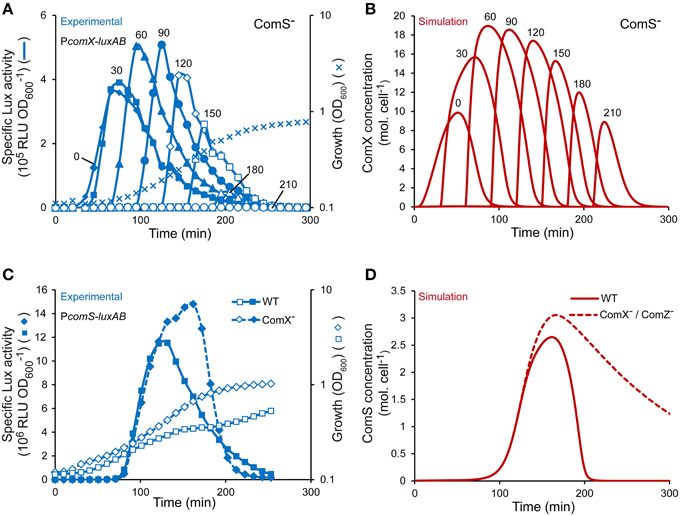
Figure 4. Validation of the dynamics and topology of the ComRS model. (A) Specific activity of PcomX upon ComS17-24 addition at different times of growth of a ComS− luminescent reporter strain (TIL1391). The curves show the specific luciferase activity (RLU OD) of the reporter strain measured directly after addition of 1 μM ComS17-24 at different times of growth. The growth curve (OD600) of the reporter strain grown in CDML in absence of ComS17-24 is shown. Data were adapted from Gardan et al. (2013). (B) Modeled kinetics of ComX concentrations (molecules (mol.) cell−1) in a ComS− background obtained by simulating the addition of ComSext (150 mol. cell−1) at different times of growth. The time T expressed in min (T = 0, 30, 60, 90, 120, 150, 180, and 210) at which ComS17-24 and ComSext were experimentally and in silico added, respectively, is indicated above each curve. (C) Comparison of the kinetics of the specific activity of PcomS between WT (LF121) and ComX− (LF122) reporter strains (RLU OD) in CDML growth conditions. (D) Comparison of modeled kinetics of ComS concentrations (mol. cell−1) between WT and ComX− or ComZ− strains. (A,C) The results of one representative experiment are shown.
Topology of the ComRS Model
The topology of model S1 was tested by simulating the behavior observed in a ComX− background, in which the activation of ComRS-controlled early gene comS is stronger and lasts longer, as a probable consequence of shut-off deregulation (Figure 4C) (Boutry et al., 2013). The ComX− background was simulated by setting equation describing the evolution of ComX(t) to zero in the model (Equation 7). As expected, the kinetics and amplitude of the simulated evolution of ComS concentrations in a ComX− background are similar to those of the luciferase activity reporting the activation of the PcomS promoter in the ComX− reporter strain LF122 (compare Figure 4C and Figure 4D). Competence shut-off is indeed delayed in both cases. It is noteworthy that this delay appears shorter in experiments compared to simulations. However, this difference is artefactual since the well-documented loss of luciferase activity in the stationary phase (Duncan et al., 1994; Koga et al., 2005) is largely responsible for the decrease in comS expression (Figure 4C). The delayed drop in simulated ComS concentration (Figure 4D) solely results from the negative terms modeled in the corresponding equation, which account for ComS exportation, degradation and cell division (see Materials and Methods).
ComR Abundance Limits the Competence Level of S. thermophilus LMD-9
The level of competence of S. thermophilus LMD-9 is directly dictated by the amount of ComX molecules produced (Fontaine et al., 2010a; Boutry et al., 2013). The spontaneous activation of comX expression in that strain is not at its maximum in CDM conditions since it may be further increased by adding saturating concentration of ComS (Fontaine et al., 2010a,b and data not shown). The ComRS model was used to determine which known actor limits comX expression in strain LMD-9 and, consequently, the competence level. To this end, a sensitivity analysis was exploited to identify the variable(s) and/or parameter(s) of the ComRS model having the most important impact on the amplitude of the model output. Parameters were grouped according to their role in equations (Supplementary Figure S4). Most sensitive parameters influence the specific amount of variables ComS (all peptidic forms: precursor, exported, and reimported mature peptide) and ComR. They account for their degradation rate (Supplementary Figure S4G), the rate of ComRS complex formation (Supplementary Figure S4B), and production rates (Supplementary Figures S4C,E,F,H). To further compare how ComS and ComR concentrations impact the competence level of LMD-9 cells, we tested in silico the effect of a ComS vs. ComR overproduction (respectively designated ComS+ and ComR+ in Figure 5A) on the maximum number of ComX molecules. Simulations were performed by multiplying the maximum basal production rate of ComR and ComS by a constant induction factor (IF) ranging from 1 to 100 (see Materials and Methods).
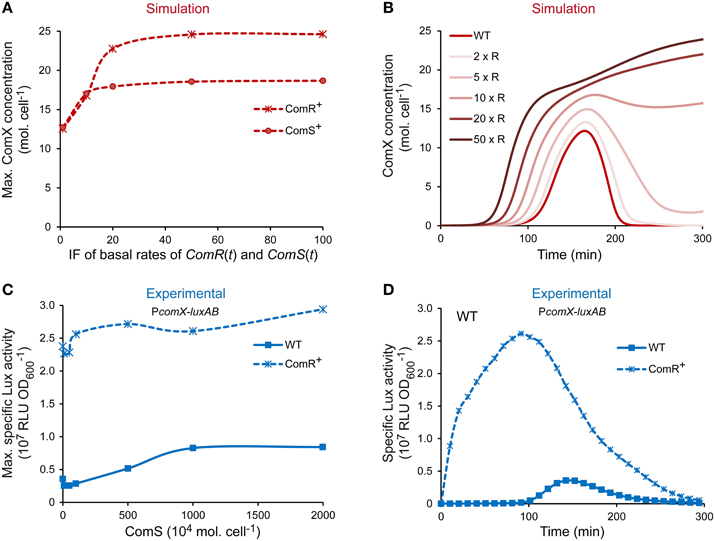
Figure 5. Effect of ComR and ComS overproduction on the level and timing of competence development in strain LMD-9. (A) Maximum ComX concentration [molecules (mol.) cell−1] computed by increasing the production rates of ComR and ComS. (B) Kinetics of ComX concentration (mol. cell−1) computed by increasing the production rate of ComR. Production rates were increased by multiplying the maximal basal production rate of variables ComR(t) and ComS(t) by a constant inducing factor (IF). IF used in (A,B) are [1, 10, 20, 50, 100] and [1, 2, 5, 10, 20, 50] (IF × R), respectively. (C) Comparison of the maximum specific luciferase activity of CB001 reporter strains (PcomX-luxAB) containing either an empty control vector (CB001 [pMG36eT]; WT) or the constitutive comR overexpression vector (CB001 [pMGcomRstrep]; ComR+) in response to increasing ComS17-24 concentration. ComS17-24 concentrations (nM) used are 0, 10, 50, 100, 500, 1000, and 2000. Nanomolar units were converted in 104 mol. cell−1 (see legend of Figure 3). (D) Comparison of the kinetics of specific PcomX activity in reporter strains containing either the comR overexpression vector (CB001 [pMGcomRstrep]; ComR+) or a control vector (CB001 [pMG36eT]; WT) in absence of ComS17-24. In all experiments, strains were inoculated in CDML at an OD600 of 0.05. Growth (OD600) and luciferase activity (RLU) were directly monitored until strains reached the stationary phase. (C,D) The results of one representative experiment are shown.
At IF ranging from 1 to 15, the maximum cellular ComX concentration increases similarly in both ComS+ and ComR+ simulated cells (Figure 5A). However, stronger induction resulted in a ~30% higher output in the case of ComR overproduction. At saturation, a 2- vs. 1.5-fold increase of ComX concentration is observed in ComR+ vs. ComS+ cells. Importantly, similar ouputs were obtained when the value of each parameter was varied separately within an interval ranging of 0.5–2 times its assigned value, indicating that this prediction is robust (Supplementary Figure S5).Those results suggest that increasing the cellular concentration of ComR vs. ComS will lead to stronger levels of comX expression in strain LMD-9. Interestingly, simulations also predict that the abundance of ComR controls the kinetics of competence in CDM. Indeed, the increased production rate of ComR is directly correlated to an earlier induction of ComX production (Figure 5B). To test those predictions experimentally, the competence level and kinetics of a WT and a ComR+ strain, which contains a comR expression vector (Boutry et al., 2013), were compared. The effect of increasing ComS abundance in both backgrounds was tested by monitoring PcomX in response to increasing ComS17-24 concentrations. Results obtained (Figures 5C,D) show that compared to WT cells, PcomX activity in ComR+ cells is (i) no more inducible by ComS17-24, indicating that ComS does not limit competence in those conditions; (ii) is stronger at all ComS17-24 concentrations tested (Figure 5C); and (iii) increases from the beginning of growth and is thus no more dependent on the growth phase (Figure 5D).
Altogether, our results strongly suggest that ComR abundance is the major limiting factor which controls the level of comX expression and the kinetics of competence induction in the naturally transformable strain LMD-9.
Low Competence of Strain LMG18311 is Due to Weak comR Expression
The ability of S. thermophilus to spontaneously turn on high competence levels under CDM growth conditions seems restricted to rare strains (Fontaine et al., 2010b). Since all of the key early competence genes to date are encoded in the genome of the poorly transformable strains, we assumed that differences in competence efficiency between S. thermophilus strains could be due to different production levels of actors of the ComRS signaling pathway. As ComR abundance was shown to limit competence in strain LMD-9, we studied the activity of PcomR and PcomX in the poorly transformable LMG18311 strain. As expected, maximum PcomX activity is very low in LMG18311 but might be increased 345-fold upon ComS17-24 supplementation (Figure 6A). Similar results were obtained for PcomS activity (data not shown), which confirms that similarly to other weak competent strain, ComS production is limiting in strain LMG18311. Interestingly, PcomR activity is also ~10-fold lower in strain LMG18311 compared to strain LMD-9 (Figure 6A).
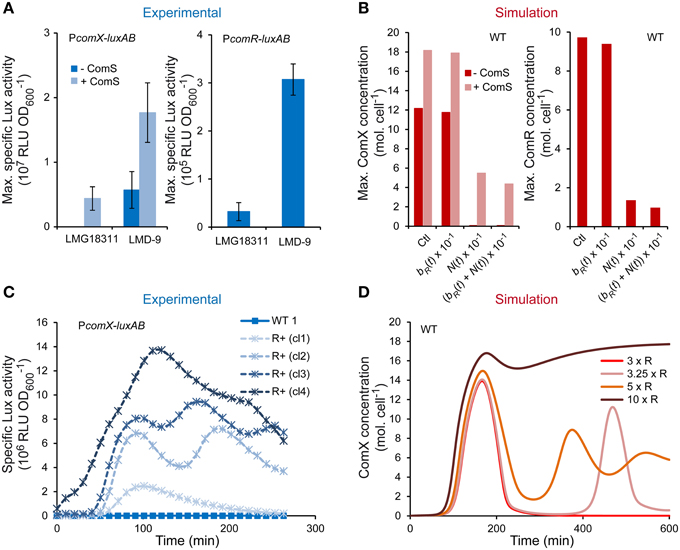
Figure 6. Role of ComR abundance on the competence level of S. thermophilus LMG18311. (A) Maximum specific luciferase activity driven by promoters PcomX (left panel) and PcomR (right panel) in respective reporter strains CB009 and LF146 grown in CDML. The symbols (− ComS) and (+ ComS) respectively indicate that no ComS17-24 and 1 μM ComS17-24 was added 1 h after inoculation. The values are means from triplicate experiments ± standard errors of the means (SEM). (B) Maximum concentrations [molecules (mol.) cell−1] of ComX (left panel) and ComR (right panel) computed by integrating the system with parameters values of ComR production rate, i.e., bR(t) + N(t) of system S1 (Ctl) (see Table 2), with a 10-fold lower ComR basal production rate (bR(t) × 10−1), with a 10-fold lower ComR activation rate (N(t) × 10−1) and with a 10-fold lower total ComR production rate ((bR(t) + N(t)) × 10−1). ComX molecules were computed by setting an initial condition of ComSext equal to zero (- ComS) or equal to 150 mol. cell−1 (+ ComS). (C) Kinetics of specific luciferase activity (RLU OD) by representative clones (cl1, cl2, cl3, and cl4) of the LMG18311 derivative reporter strains CB009 (PcomX-luxAB) containing an empty plasmid (CB009 [pMG36eT]; WT) or the constitutive comR overexpression vector (CB009 [pMGcomRstrep]; ComR+). (D) Consequences of increasing the maximum ComR basal production rate by a 3 (3 × R), 3.25 (3.25 × R), 5 (5 × R), and 10-fold (10 × R) factor on modeled kinetics of ComX concentrations (mol. cell−1). These induction factors were selected to illustrate the transition between mono-, bi-, and tri-phasic kinetics profiles.
The ComRS model was next used to test whether the observed difference in comR expression accounts for low expression of comX. To mimic strain LMG18311, the production rate of ComR(t) was divided by 10. The latter is modeled as the sum of rates due to ComR basal production (bR(t)) and activation/repression by -as yet- unidentified regulator(s) (N(t)). As shown on Figure 6B, simulations done after dividing both terms reproduced the behavior of strain LMG18311: maximum cellular ComX concentration reaches high levels only in presence of initial ComSext concentrations. In addition, a similar four-fold difference in simulated and experimental outputs (respectively maximum ComX concentration and PcomX activity) was observed between strains LMG18311 and LMD-9 in ComS-induced conditions. Similar results were obtained when the rate of ComR(t) activation/repression solely was divided in the ComRS model, while it was not the case when only the basal rate was divided by 10. This suggests that the contribution of ComR basal production to ComX production is minor in S. thermophilus (Figure 6B).
In conclusion, the weak transformability of strain LMG18311 results from a low expression of comR, solely. This may prevent the activation of the positive feedback loop acting on ComS production.
Increasing ComR Concentration Induces Competence in the Weakly Transformable Strain LMG18311
To further confirm the key role of ComR concentration on the transformability of strain LMG18311, we tested the consequence of comR overexpression on this phenotype. As expected, both the mean transformation frequency (4.4 × 10−4 ± 4.0 × 10−4 vs. 2.1 × 10−6 ± 5.0 × 10−5) and the mean maximum specific luciferase activity (RLU OD) of PcomX-luxAB fusions (6.7 × 106 ± 3.5 × 106 vs. 4.4 × 104 ± 1.2 × 103) increased significantly in the reporter derivative strain carrying the comR overexpression vector compared to those carrying the empty vector. Interestingly, besides a high variability in the amplitude of PcomX activity (Figure 6C and data not shown), the various LMG18311 ComR+ clones also display variable (but reproducible) kinetics patterns, which is either monophasic (4/10 clones), biphasic (5/10 clones), or in rare instances triphasic (1/10 clone) (the specific luciferase activity of representative clones are presented in Figure 6C). To rationally interpret these results, we simulated increasing levels of ComR production by multiplying its maximum basal rate by a range of increasing values (1.5–20). For IF ranging from 3.25 to 7 (Figure 6D and data not shown), the time-course evolution of ComX concentration displays an oscillating behavior. The number of phases is directly proportional to the level of ComR production. At higher IF (Figure 6D), the phases merge while maximum amplitude continues to increase until saturation. Those predictions suggest that the different behaviors observed among LMG18311 ComR+ clones result from different levels of comR expression, which could for instance be due to different copy numbers of the corresponding expression vector.
Discussion
Competence for natural transformation is a major driving force in bacterial evolution. Hence, it is of key importance to understand how this transient phenomenon takes place and is regulated. In Gram-positive bacteria, the best documented competence regulation systems are those of the model species S. pneumoniae (ComCDE-ComX relay system) (Claverys et al., 2006) and Bacillus subtilis (ComK master regulator) (Hamoen et al., 2003). During the last decade, the regulatory cascades governing competence in these two species have been mathematically modeled for helping to solve some important aspects such as the entry in the competence state (Leisner et al., 2009; Liebal et al., 2010), competence shut-off (Karlsson et al., 2007; Schultz et al., 2007), competence bistability (Maamar and Dubnau, 2005; Dubnau and Losick, 2006; Son et al., 2012; Xi et al., 2013), decision-making process between developmental programs (competence vs. sporulation) (Schultz et al., 2009), or benefits and cost of competence development (Moradigaravand and Engelstadter, 2013). These modeling approaches were very useful to decipher these processes and to obtain in some cases counter-intuitive explanations.
The ComRS-ComX relay system, which regulates competence in a majority of streptococcal groups, was recently discovered (for a review, see Fontaine et al., 2015). Even if the available experimental data begin to accumulate, a series of questions remain open on its functioning such as the topology of the regulatory cascade or the critical components of this system responsible for the on-off state of competence development. Although a simple mathematical model integrating ComRS was built to study environmental inputs on competence bistability in S. mutans (Son et al., 2012), a kinetic model that integrates all known steps of comX regulation by ComRS was missing until now.
The ComRS Dynamic Model: Limitations and Future Developments
At this stage, our model presents some limitations partially due to mechanistic or meta-hypotheses that were done to construct the model. First, the model neglects the putative cell-to-cell communication role of ComS. However, although “self”-communication dominates over “others”-communication in peptide diffusion-limiting conditions (such as modeled here), the combination of specific parameters may influence the output of social behaviors mediated by pheromone-based regulatory systems, such as the rate of peptide secretion and importation, the strength of the positive feedback loop (Youk and Lim, 2014) and/or the possibility of communication mediated by physical contacts between cells. In the future, both aspects (egoist and altruistic) of ComS signaling will be integrated in our model in order to study their relative contribution in competence regulation in various growth conditions.
Other limitations of our current model are that (i) the simulated cell growth is always assumed to be the same between WT and mutants while altered growth profiles were experimentally observe in mutant strains, (ii) the model has been calibrated and evaluated only for CDML growth conditions (iii) the model only considers the effect of proximal actors on comX expression while the effects of all distal regulators are summed up in the temporal gaussian term (N(t)) of ComR due to a lack of knowledge on distal regulation, and (iv) the model is not calibrated on the true abundance of ComR, ComS, and ComX proteins, which is unknown in ComRS-containing streptococci, but on expression data based on the activity of the luciferase reporter system. This last point implicates that the model remains qualitative and could be used to predict behaviors and to compare quantitative effects, but not to directly infer quantitative data. So, future improvements of the model will aim to solve some of these limitations such as integrating detailed distal regulations (transcriptional and post-transcriptional), improving quantitative predictions by measuring the time-course production of key players of the system during growth, or taking into account both RNA and protein levels.
Finally, the question of the portability of the model developed for S. thermophilus to other ComRS-containing streptococci is an important aspect. Although differences exist between species regarding for instance the diffusion properties of ComS and the mechanism of competence shut-off (see below), the topology of the ComX induction cascade as well as growth conditions allowing its activation (i.e., CDM conditions) are strongly conserved among ComRS-containing streptococci (for a review, see Fontaine et al., 2015). This suggests that the adaptation of the model to other phylogenetically close streptococci (e.g., S. mutans, S. salivarius) could be achieved without major adjustments.
ComR Abundance is Critical for Competence Activation
One of the interesting outputs of the use of the model is that, besides the ComS pheromone, ComR is a limiting factor for maximum competence development of S. thermophilus in CDM conditions (Figure 5A). This was experimentally validated: the comR overexpression in strain LMD-9, which develops competence spontaneously, and strain LMG18311, which is weakly competent, resulted in a mean 10- and 100-fold higher level of comX expression, respectively (Figures 5C, 6C). It is noteworthy that increasing ComS abundance in strains LMD-9 and LMG18311 (through in silico or in vivo exogenous peptide supplementation) (Figures 6A,B) induces a similar positive effect on competence development but the amplitude of the response is correlated to the number of ComR molecules produced in each strain. Indeed, the induction of comX by saturating ComS concentrations remains stronger in strain LMD-9 vs. LMG18311 (Figures 6A,B). In regard to the topology of the competence induction cascade, ComR is indeed pivotal as its amount will determine the number of ComRS complexes which will be formed. A critical amount of ComRS complexes is probably required to activate the positive feedback loop acting on comS expression and hence induce competence in the population. Our results strongly suggest that this minimal amount is not reached in strain LMG18311 because of a too low comR expression level. The model predicts that increasing the ComR concentration in this strain shifts the balance toward formation of enough ComRS complexes to spontaneously induce competence. Interestingly, competence also turns on much earlier during growth of ComR+ vs. WT cells because of the immediate production of high amount of ComRS (data not shown), indicating that the timing device is constrained by the kinetics and amplitude of ComR production, solely. Such a critical role of ComR abundance does not seem to be restricted to the S. thermophilus species since it was previously reported that ComR overexpression in S. mutans UA159 resulted in a 10-fold increase in transformability (Mashburn-Warren et al., 2010). Increasing ComR production could thus provide a simple way to render weakly or non-competent strains highly competent without the requirement of adding synthetic ComS.
The role of ComR as a limiting factor of competence may be related to the structure of the ComRS signaling cascade in which ComR production is independent of the positive feedback loop (Figure 1). Indeed, this contrasts with the topology of most cell-cell communication systems in which productions of master regulator and pheromone are auto-regulated and coordinated at the transcriptional level, as for instance in the ComCDE system controlling competence in S. pneumoniae (Fontaine et al., 2015) and the streptococcal Rgg/Shp systems (Pérez-Pascual et al., 2015). This ensures that the same amount of regulator and associated pheromone are produced in these systems. The evolutionary consequence of uncoupling ComR and ComS production may be advantageous to fine-tune both kinetics and amplitude of competence activation in response to various environmental inputs. Currently, the modulation of comR transcription by distal regulatory systems (e.g., BsrRM-HdrRM relay and ScnRK) has only been shown in S. mutans (Okinaga et al., 2010; Xie et al., 2010; Kim et al., 2013). In addition, other signaling systems (i.e., S. pyogenes Rgg/Shp, and S. pneumoniae TprA/PhrA) of the RNPP family (the latter includes ComR) have also been shown as transcriptionally regulated by distal systems sensing either metal availability (Chang et al., 2015) or carbon source (Chang et al., 2015; Hoover et al., 2015). In S. thermophilus, no distal regulation acting on comR expression has been reported so far.
The comparative study of strains LMD-9 and LMG18311 indicate that competence heterogeneity between S. thermophilus strains (Fontaine et al., 2010b) could partially be due to differences in the comR expression level. The origin of these differences (i.e., LMD-9 vs. LMG18311) is not clear. Our model predicts that the activation or de-repression of comR expression by distal regulators, rather than the basal expression, is affected in strain LMG18311 compared to strain LMD-9 (Figure 6B). It may either be due to sequence polymorphism of the promoter region that affects binding of regulator(s), or to the presence of mutations in some comR distal regulators in poorly competent strains. Alternatively, in regard to the exceptional transformability of strain LMD-9 within the S. thermophilus species, it is possible that LMD-9 harbors specific mutations in comR regulator(s) allowing competence induction in otherwise non-permissive growth conditions. Consistent with this hypothesis of competence deregulation, strains of Bacillus licheniformis displaying efficient transformability carry mutations in competence regulator genes as a probable consequence of domestication (Jakobs and Meinhardt, 2015). Swapping comR promoter regions and determination of the sub-regions important for comR expression could help to begin answering this question.
Insights into Competence Shut-Off
In 2007, the use of an ODE mathematical model of the ComCDE system has suggested that the competence shut-off mechanism in S. pneumoniae was likely to involve a late phase repressor protein (Karlsson et al., 2007). A couple of years later, the shut-off mechanism in S. pneumoniae was experimentally unraveled and takes place at multiple levels: (i) competition between ComE and ComE~P (Martin et al., 2013), (ii) inhibition of ComX activity (Weng et al., 2013), and (iii) direct inhibition of ComE~P by DprA (Mirouze et al., 2013). The latter is a late phase protein that was initially proven as involved in DNA integration by recruiting RecA (Bergé et al., 2003; Mortier-Barrière et al., 2007).
In ComRS-containing streptococci, the shut-off mechanism is poorly understood and seems to differ among species (Fontaine et al., 2015). In S. mutans, it was shown that this mechanism was at least due to a post-translational control of ComX stability by the concerted action of the MecA adaptor protein and the ClpCP proteolytic system (Tian et al., 2013; Dong et al., 2014). In contrast, it has been shown that the MecA-ClpCP machinery does not contribute to the shut-off in S. thermophilus (Boutry et al., 2012; Wahl et al., 2014). However, it has been observed that ComX-negative strains exhibit a longer and stronger activation of genes belonging to the ComRS core regulon (i.e., comS and comX), without affecting comR expression (Boutry et al., 2013). This behavior could be simulated with our predictive model (Figures 4C,D), confirming that at least one late gene product is partially responsible for competence shut-off and acts by negatively affecting ComRS activity.
In our model, the feedback inhibition loop via the putative effector of shut-off ComZ is directed on the ComRS complex in order to form a ternary complex ComRSZ (Figure 1). By analogy with S. pneumoniae, the orthologue of DprA (which interacts and inhibits ComE activity in the pneumococcus) was an obvious putative candidate for this function. However, the early competence phase of S. thermophilus LMD-9 is not affected in a dprA deletion mutant compared to the WT (see Supplementary Figure S6), suggesting that the DprA orthologue is not involved in competence exit in this species. Although other late competence gene(s) could substitute DprA for sequestering active ComRS complexes in S. thermophilus, we cannot rule out alternative inactivation mechanisms such as (i) degradation of ComR and/or ComS, (ii) inactivation by post-translational modification of ComR and/or ComS (e.g., ComR phosphorylation), or (iii) inhibition of formation or activity of the ComRS complex (e.g., inhibitory peptide, competition for DNA binding). Changing the target of the negative loop in the model combined with modifications of parameters affecting the abundance of ComR and/or ComS could be useful to define at which level(s) the shut-off is taking place.
Oscillatory Behavior of Competence Activation
Besides the critical role of ComR in the strength of competence activation, ComR abundance also regulates an oscillatory behavior to competence activation. Indeed, an oscillating profile of comX expression was observed in silico and experimentally in the comR-overexpressing strain LMG18311 (Figures 6C,D). This provides an additional validation of the topological organization and the dynamical calibration of our model. Indeed, Novák and Tyson (2008) showed that the four minimal requirements for oscillations are: (i) a negative feedback loop, which is made in our setting of ComRS → ComX → ComZ⊣ComRS, (ii) a delay in this negative loop, implemented here by the positive loop made of ComS → ComSext → ComS* → ComRS → ComS, (iii) sufficient non-linearity, which arises here from our non-linear activation/repression functions and from the mass action law of the ComRS assembly with n = 2.5, and (iv) appropriate time-scales of the opposite forces, confirming that our parameter set is coherent. Moreover, according to the notation of Lenz and Sogaard-Andersen (2011), our system can be classified as a relaxation oscillator due to its structure (a time-delayed negative feedback loop coupled to a positive feedback loop), hence operating in a broad frequency range. Indeed, both amplitude and frequency of oscillations can be modulated by varying the ComR production level (Figures 6C,D).
Temporal oscillations have been reported in a wide range of biological networks and organisms: from bacterial gene circuits (Schultz et al., 2009, 2013) to eukaryotic circadian rhythms (Lenz and Sogaard-Andersen, 2011) and quorum-sensing systems (Sgro et al., 2015). In addition, synthetic oscillators have been successfully built (Elowitz and Leibler, 2000). Interestingly, cycles in the competence state have already been reported for S. pneumoniae (Chen and Morrison, 1987) and Helicobacter pylori (Baltrus and Guillemin, 2006). In S. pneumoniae, the activation of these multiple competence cycles (up to three during growth in batch cultures) in a WT strain was observed at high initial pH values (>7.25) while competence is inhibited in acidic conditions (Chen and Morrison, 1987). In WT strains of Helicobacter pylori, multiple competence phases occurred spontaneously but appeared to be strain-dependent (Baltrus and Guillemin, 2006). Although a similar cycling behavior is observed here in an artificial genetic background of S. thermophilus, oscillation in competence gene activation was also seen in response to ComS addition in the closely related species S. salivarius (data not shown), indicating that competence oscillation could be an intrinsic property of the ComRS signaling system. It has been previously shown that oscillations are synchronized in the whole population through quorum-sensing inducing molecules (McMillen et al., 2002; Schwab et al., 2012; Singh and Parmananda, 2013). However, it remains to be determined here if oscillations are taking place at the whole population level or if successive subpopulations are activating a single competence episode. Future time-lapse experiments at the single cell level with a GFP-reporter strain of competence activation will be needed to answer this question.
Finally, the question of the biological advantage conferred by an oscillating competence state has not yet been addressed. We can speculate that the benefit could be the lengthening of the transformation process in conditions where competence is highly beneficial for the population. Oscillations would then allow recovery phases that could be needed to relieve the inhibition of cell division resulting from competence activation, hence implementing a compromise between competence and growth, two processes that could be mutually exclusive (Haijema et al., 2001; Qi et al., 2005). However, one must keep in mind that the current observations were obtained in artificial laboratory conditions in which bacteria could exhibit exotic behaviors that would not or rarely be seen in natural conditions.
Concluding Remarks
To conclude, we designed an original approach combining mathematical modeling with molecular biology tools to build a robust data-based model of the ComRS signaling system, which helped us to confirm the topology of the competence regulatory network and to identify its critical components in salivarius streptococci. We demonstrated the key hierarchical importance of ComR in transformation efficiency and timing of competence activation, compatible with its putative role of master integration-sensor of distal regulations by environmental signals. In addition, our model shed light on the competence shut-off mechanism and proposed that the topology of the regulatory network based on ComRS allows an oscillatory behavior of competence activation. Future work will consist in exploiting our model to identify more finely at which level the shut-off is taking place in order to identify putative key player(s) involved in this mechanism. A possible oscillating competence state is also an exciting aspect that will require further investigation such as its occurrence at the single cell level and its biological relevance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We warmly thank P. Soumillon and P. Goffin for fruitful discussions regarding mathematical modeling and critically reading of the manuscript. We gratefully thank R. Gardan for providing the raw data of Figure 4A. This research has been funded by FNRS and the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office. LH holds a doctoral fellowship from FRIA. LF is a postdoctoral researcher at FNRS. PH is Senior Research Associate at FNRS.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01413
References
Baltrus, D. A., and Guillemin, K. (2006). Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol. Lett. 255, 148–155. doi: 10.1111/j.1574-6968.2005.00066.x
Berg, K. H., Biørnstad, T. J., Johnsborg, O., and Håvarstein, L. S. (2012). Properties and biological role of streptococcal fratricins. Appl. Environ. Microbiol. 78, 3515–3522. doi: 10.1128/AEM.00098-12
Bergé, M., Mortier-Barrière, I., Martin, B., and Claverys, J. P. (2003). Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50, 527–536. doi: 10.1046/j.1365-2958.2003.03702.x
Blomqvist, T., Steinmoen, H., and Håvarstein, L. S. (2006). Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 72, 6751–6756. doi: 10.1128/AEM.01156-06
Bolouri, H., and Davidson, E. H. (2003). Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc. Natl. Acad. Sci. U.S.A. 100, 9371–9376. doi: 10.1073/pnas.1533293100
Borgeaud, S., Metzger, L. C., Scrignari, T., and Blokesch, M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67. doi: 10.1126/science.1260064
Boutry, C., Delplace, B., Clippe, A., Fontaine, L., and Hols, P. (2013). SOS response activation and competence development are antagonistic mechanisms in Streptococcus thermophilus. J. Bacteriol. 195, 696–707. doi: 10.1128/JB.01605-12
Boutry, C., Wahl, A., Delplace, B., Clippe, A., Fontaine, L., and Hols, P. (2012). Adaptor protein MecA is a negative regulator of the expression of late competence genes in Streptococcus thermophilus. J. Bacteriol. 194, 1777–1788. doi: 10.1128/JB.06800-11
Cassone, M., Gagne, A. L., Spruce, L. A., Seeholzer, S. H., and Sebert, M. E. (2012). The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chem. 287, 38449–38459. doi: 10.1074/jbc.M112.391482
Chang, J. C., Jimenez, J. C., and Federle, M. J. (2015). Induction of a quorum sensing pathway by environmental signals enhances group A streptococcal resistance to lysozyme. Mol. Microbiol. 97, 1097–1113. doi: 10.1111/mmi.13088
Charpentier, X., Polard, P., and Claverys, J. P. (2012). Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS? Curr. Opin. Microbiol. 15, 570–576. doi: 10.1016/j.mib.2012.08.001
Chen, J. D., and Morrison, D. A. (1987). Modulation of competence for genetic transformation in Streptococcus pneumoniae. J. Gen. Microbiol. 133, 1959–1967. doi: 10.1099/00221287-133-7-1959
Claverys, J. P., Martin, B., and Håvarstein, L. S. (2007). Competence-induced fratricide in streptococci. Mol. Microbiol. 64, 1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x
Claverys, J. P., Prudhomme, M., and Martin, B. (2006). Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60, 451–475. doi: 10.1146/annurev.micro.60.080805.142139
de Jong, H. (2002). Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 9, 67–103. doi: 10.1089/10665270252833208
Desai, K., Mashburn-Warren, L., Federle, M. J., and Morrison, D. A. (2012). Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J. Bacteriol. 194, 3774–3780. doi: 10.1128/JB.00337-12
Dong, G., Tian, X. L., Gomez, Z. A., and Li, Y. H. (2014). Regulated proteolysis of the alternative sigma factor SigX in Streptococcus mutans: implication in the escape from competence. BMC. Microbiol. 14:183. doi: 10.1186/1471-2180-14-183
Dower, W. J., Miller, J. F., and Ragsdale, C. W. (1988). High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145. doi: 10.1093/nar/16.13.6127
Dubnau, D., and Losick, R. (2006). Bistability in bacteria. Mol. Microbiol. 61, 564–572. doi: 10.1111/j.1365-2958.2006.05249.x
Dufour, D., and Lévesque, C. M. (2013). Bacterial behaviors associated with the quorum-sensing peptide pheromone ('alarmone') in streptococci. Future Microbiol. 8, 593–605. doi: 10.2217/fmb.13.23
Duncan, S., Glover, L. A., Killham, K., and Prosser, J. I. (1994). Luminescence-based detection of activity of starved and viable but nonculturable bacteria. Appl. Environ. Microbiol. 60, 1308–1316.
Elowitz, M. B., and Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338. doi: 10.1038/35002125
Ferain, T., Hobbs, J. N. Jr., Richardson, J., Bernard, N., Garmyn, D., Hols, P., et al. (1996). Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J. Bacteriol. 178, 5431–5437.
Fleuchot, B., Gitton, C., Guillot, A., Vidic, J., Nicolas, P., Besset, C., et al. (2011). Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80, 1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x
Fontaine, L., Boutry, C., de Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2010a). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Fontaine, L., Dandoy, D., Boutry, C., Delplace, B., de Frahan, M. H., Fremaux, C., et al. (2010b). Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl. Environ. Microbiol. 76, 7870–7877. doi: 10.1128/AEM.01671-10
Fontaine, L., Goffin, P., Dubout, H., Delplace, B., Baulard, A., Lecat-Guillet, N., et al. (2013). Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87, 1113–1132. doi: 10.1111/mmi.12157
Fontaine, L., Wahl, A., Fléchard, M., Mignolet, J., and Hols, P. (2015). Regulation of competence for natural transformation in streptococci. Infect. Genet. Evol. 33, 343–360. doi: 10.1016/j.meegid.2014.09.010
Gardan, R., Besset, C., Gitton, C., Guillot, A., Fontaine, L., Hols, P., et al. (2013). Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J. Bacteriol. 195, 1845–1855. doi: 10.1128/JB.02196-12
Gardan, R., Besset, C., Guillot, A., Gitton, C., and Monnet, V. (2009). The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191, 4647–4655. doi: 10.1128/JB.00257-09
Guo, Q., Ahn, S. J., Kaspar, J., Zhou, X., and Burne, R. A. (2014). Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J. Bacteriol. 196, 227–236. doi: 10.1128/JB.00995-13
Haijema, B. J., Hahn, J., Haynes, J., and Dubnau, D. (2001). A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40, 52–64. doi: 10.1046/j.1365-2958.2001.02363.x
Hamoen, L. W., Venema, G., and Kuipers, O. P. (2003). Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149, 9–17. doi: 10.1099/mic.0.26003-0
Hoover, S. E., Perez, A. J., Tsui, H. C., Sinha, D., Smiley, D. L., DiMarchi, R. D., et al. (2015). A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol. Microbiol. 97, 229–243. doi: 10.1111/mmi.13029
Jabbari, S., Heap, J. T., and King, J. R. (2011). Mathematical modelling of the sporulation-initiation network in Bacillus subtilis revealing the dual role of the putative quorum-sensing signal molecule PhrA. Bull. Math. Biol. 73, 181–211. doi: 10.1007/s11538-010-9530-7
Jakobs, M., and Meinhardt, F. (2015). What renders Bacilli genetically competent? A gaze beyond the model organism. Appl. Microbiol. Biotechnol. 99, 1557–1570. doi: 10.1007/s00253-014-6316-0
Johnsborg, O., Eldholm, V., and Håvarstein, L. S. (2007). Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158, 767–778. doi: 10.1016/j.resmic.2007.09.004
Johnsen, P. J., Dubnau, D., and Levin, B. R. (2009). Episodic selection and the maintenance of competence and natural transformation in Bacillus subtilis. Genetics 181, 1521–1533. doi: 10.1534/genetics.108.099523
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Karlsson, D., Karlsson, S., Gustafsson, E., Normark, B. H., and Nilsson, P. (2007). Modeling the regulation of the competence-evoking quorum sensing network in Streptococcus pneumoniae. Biosystems 90, 211–223. doi: 10.1016/j.biosystems.2006.08.005
Khan, R., Rukke, H. V., Ricomini Filho, A. P., Fimland, G., Arntzen, M. Ø., Thiede, B., et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194, 3781–3788. doi: 10.1128/JB.00624-12
Kim, J. N., Stanhope, M. J., and Burne, R. A. (2013). Core-gene-encoded peptide regulating virulence-associated traits in Streptococcus mutans. J. Bacteriol. 195, 2912–2920. doi: 10.1128/JB.00189-13
Koga, K., Harada, T., Shimizu, H., and Tanaka, K. (2005). Bacterial luciferase activity and the intracellular redox pool in Escherichia coli. Mol. Genet. Genomics 274, 180–188. doi: 10.1007/s00438-005-0008-5
Law, J., Buist, G., Haandrikman, A., Kok, J., Venema, G., and Leenhouts, K. (1995). A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177, 7011–7018.
Leisner, M., Kuhr, J. T., Rädler, J. O., Frey, E., and Maier, B. (2009). Kinetics of genetic switching into the state of bacterial competence. Biophys. J. 96, 1178–1188. doi: 10.1016/j.bpj.2008.10.034
Lenz, P., and Søgaard-Andersen, L. (2011). Temporal and spatial oscillations in bacteria. Nat. Rev. Microbiol. 9, 565–577. doi: 10.1038/nrmicro2612
Letort, C., and Juillard, V. (2001). Development of a minimal chemically-defined medium for the exponential growth of Streptococcus thermophilus. J. Appl. Microbiol. 91, 1023–1029. doi: 10.1046/j.1365-2672.2001.01469.x
Liebal, U. W., Millat, T., de Jong, I. G., Kuipers, O. P., Völker, U., and Wolkenhauer, O. (2010). How mathematical modelling elucidates signalling in Bacillus subtilis. Mol. Microbiol. 77, 1083–1095. doi: 10.1111/j.1365-2958.2010.07283.x
Luo, P., and Morrison, D. A. (2003). Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185, 349–358. doi: 10.1128/JB.185.1.349-358.2003
Maamar, H., and Dubnau, D. (2005). Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56, 615–624. doi: 10.1111/j.1365-2958.2005.04592.x
Martin, B., Soulet, A. L., Mirouze, N., Prudhomme, M., Mortier-Barrière, I., Granadel, C., et al. (2013). ComE/ComE~P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87, 394–411. doi: 10.1111/mmi.12104
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78, 589–606. doi: 10.1111/j.1365-2958.2010.07361.x
McMillen, D., Kopell, N., Hasty, J., and Collins, J. J. (2002). Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 679–684. doi: 10.1073/pnas.022642299
Mirouze, N., Bergé, M. A., Soulet, A. L., Mortier-Barrière, I., Quentin, Y., Fichant, G., et al. (2013). Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc. Natl. Acad. Sci. U.S.A. 110, E1035–E1044. doi: 10.1073/pnas.1219868110
Moradigaravand, D., and Engelstädter, J. (2013). The evolution of natural competence: disentangling costs and benefits of sex in bacteria. Am. Nat. 182, E112–E126. doi: 10.1086/671909
Morrison, D. A., Guédon, E., and Renault, P. (2013). Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus. J. Bacteriol. 195, 2612–2620. doi: 10.1128/JB.00230-13
Mortier-Barrière, I., Velten, M., Dupaigne, P., Mirouze, N., Piétrement, O., McGovern, S., et al. (2007). A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130, 824–836. doi: 10.1016/j.cell.2007.07.038
Novák, B., and Tyson, J. J. (2008). Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9, 981–991. doi: 10.1038/nrm2530
Okinaga, T., Xie, Z., Niu, G., Qi, F., and Merritt, J. (2010). Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol. Oral Microbiol. 25, 165–177. doi: 10.1111/j.2041-1014.2010.00574.x
Pérez-Pascual, D., Gaudu, P., Fleuchot, B., Besset, C., Rosinski-Chupin, I., Guillot, A., et al. (2015). RovS and its associated signaling peptide form a cell-to-cell communication system required for Streptococcus agalactiae pathogenesis. MBio 6:e02306-14. doi: 10.1128/mBio.02306-14
Perry, J. A., Jones, M. B., Peterson, S. N., Cvitkovitch, D. G., and Lévesque, C. M. (2009). Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72, 905–917. doi: 10.1111/j.1365-2958.2009.06693.x
Proshkin, S., Rahmouni, A. R., Mironov, A., and Nudler, E. (2010). Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508. doi: 10.1126/science.1184939
Qi, F., Kreth, J., Lévesque, C. M., Kay, O., Mair, R. W., Shi, W., et al. (2005). Peptide pheromone induced cell death of Streptococcus mutans. FEMS Microbiol. Lett. 251, 321–326. doi: 10.1016/j.femsle.2005.08.018
Reck, M., Tomasch, J., and Wagner-Döbler, I. (2015). The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS. Genet. 11:e1005353. doi: 10.1371/journal.pgen.1005353
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring, NY; Harbor, NY: Cold Spring Harbor Laboratory.
Schultz, D., Ben, J. E., Onuchic, J. N., and Wolynes, P. G. (2007). Molecular level stochastic model for competence cycles in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 104, 17582–17587. doi: 10.1073/pnas.0707965104
Schultz, D., Lu, M., Stavropoulos, T., Onuchic, J., and Ben-Jacob, E. (2013). Turning oscillations into opportunities: lessons from a bacterial decision gate. Sci. Rep. 3:1668. doi: 10.1038/srep01668
Schultz, D., Wolynes, P. G., Ben, J. E., and Onuchic, J. N. (2009). Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 106, 21027–21034. doi: 10.1073/pnas.0912185106
Schwab, D. J., Baetica, A., and Mehta, P. (2012). Dynamical quorum-sensing in oscillators coupled through an external medium. Physica D 241, 1782–1788. doi: 10.1016/j.physd.2012.08.005
Seitz, P., and Blokesch, M. (2013). Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 37, 336–363. doi: 10.1111/j.1574-6976.2012.00353.x
Sgro, A. E., Schwab, D. J., Noorbakhsh, J., Mestler, T., Mehta, P., and Gregor, T. (2015). From intracellular signaling to population oscillations: bridging size- and time-scales in collective behavior. Mol. Syst. Biol. 11, 779. doi: 10.15252/msb.20145352
Singh, H., and Parmananda, P. (2013). Quorum sensing via static coupling demonstrated by Chua's circuits. Phys. Rev. E. Stat. Nonlin. Soft. Matter Phys. 88:040903. doi: 10.1103/PhysRevE.88.040903
Skippington, E., and Ragan, M. A. (2011). Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol. Rev. 35, 707–735. doi: 10.1111/j.1574-6976.2010.00261.x
Son, M., Ahn, S. J., Guo, Q., Burne, R. A., and Hagen, S. J. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86, 258–272. doi: 10.1111/j.1365-2958.2012.08187.x
Son, M., Ghoreishi, D., Ahn, S. J., Burne, R. A., and Hagen, S. J. (2015). Sharply tuned pH Response of genetic competence regulation in Streptococcus mutans: a microfluidic study of the environmental sensitivity of comX. Appl. Environ. Microbiol. 81, 5622–5631. doi: 10.1128/AEM.01421-15
Sturme, M. H., Kleerebezem, M., Nakayama, J., Akkermans, A. D., Vaugha, E. E., and de Vos, W. M. (2002). Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek. 81, 233–243. doi: 10.1023/A:1020522919555
Tian, X. L., Dong, G., Liu, T., Gomez, Z. A., Wahl, A., Hols, P., et al. (2013). MecA protein acts as a negative regulator of genetic competence in Streptococcus mutans. J. Bacteriol. 195, 5196–5206. doi: 10.1128/JB.00821-13
van de Guchte, M., van der Vossen, J. M., Kok, J., and Venema, G. (1989). Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55, 224–228.
Voigt, C. A., Wolf, D. M., and Arkin, A. P. (2005). The Bacillus subtilis sin operon: an evolvable network motif. Genetics 169, 1187–1202. doi: 10.1534/genetics.104.031955
Wahl, A., Servais, F., Drucbert, A. S., Foulon, C., Fontaine, L., and Hols, P. (2014). Control of natural transformation in salivarius Streptococci through specific degradation of sigmaX by the MecA-ClpCP protease complex. J. Bacteriol. 196, 2807–2816. doi: 10.1128/JB.01758-14
Wei, H., and Håvarstein, L. S. (2012). Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl. Environ. Microbiol. 78, 5897–5905. doi: 10.1128/AEM.01343-12
Wenderska, I. B., Lukenda, N., Cordova, M., Magarvey, N., Cvitkovitch, D. G., and Senadheera, D. B. (2012). A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol. Lett. 336, 104–112. doi: 10.1111/j.1574-6968.2012.02660.x
Weng, L., Piotrowski, A., and Morrison, D. A. (2013). Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS ONE 8:e64197. doi: 10.1371/journal.pone.0064197
Xi, H., Duan, L., and Turcotte, M. (2013). Point-cycle bistability and stochasticity in a regulatory circuit for Bacillus subtilis competence. Math. Biosci. 244, 135–147. doi: 10.1016/j.mbs.2013.05.002
Xie, Z., Okinaga, T., Niu, G., Qi, F., and Merritt, J. (2010). Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol. Microbiol. 78, 1431–1447. doi: 10.1111/j.1365-2958.2010.07417.x
Youk, H., and Lim, W. A. (2014). Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science 343:1242782. doi: 10.1126/science.1242782
Young, R., and Bremer, H. (1975). Analysis of enzyme induction in bacteria. Biochem. J. 152, 243–254. doi: 10.1042/bj1520243
Keywords: timing device, feedback regulation, competence shut-off, ordinary differential equations, kinetics model, oscillation
Citation: Haustenne L, Bastin G, Hols P and Fontaine L (2015) Modeling of the ComRS Signaling Pathway Reveals the Limiting Factors Controlling Competence in Streptococcus thermophilus. Front. Microbiol. 6:1413. doi: 10.3389/fmicb.2015.01413
Received: 21 August 2015; Accepted: 27 November 2015;
Published: 22 December 2015.
Edited by:
Eugenie Huillet, Institut National de la Recherche Agronomique, FranceReviewed by:
Lilia Macovei, The Forsyth Institute, USAMichael J. Federle, University of Illinois at Chicago, USA
Pierre Nicolas, Institut National de la Recherche Agronomique, France
Copyright © 2015 Haustenne, Bastin, Hols and Fontaine. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laetitia Fontaine, laetitia.fontaine@uclouvain.be
†These authors have contributed equally to this work.
 Laurie Haustenne
Laurie Haustenne Georges Bastin
Georges Bastin Pascal Hols
Pascal Hols Laetitia Fontaine
Laetitia Fontaine