- 1Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan
- 2Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, Tokyo, Japan
- 3Japan Science and Technology Agency, CREST, Saitama, Japan
- 4Japan Science and Technology Agency, PRESTO, Saitama, Japan
Phosphatidylglycerol (PG) and cardiolipin (CL) are two essential classes of phospholipid in plants and algae. Phosphatidylglycerophosphate synthase (PGPS) and cardiolipin synthase (CLS) involved in the biosynthesis of PG and CL belong to CDP-alcohol phosphotransferase and share overall amino acid sequence homology. However, it remains elusive whether PGPS and CLS are functionally distinct in vivo. Here, we report identification of a gene encoding CLS in Chlamydomonas reinhardtii, CrCLS1, and its functional compatibility. Whereas CrCLS1 did not complement the growth phenotype of a PGPS mutant of Synechocystis sp. PCC 6803, it rescued the temperature-sensitive growth phenotype, growth profile with different carbon sources, phospholipid composition and enzyme activity of Δcrd1, a CLS mutant of Saccharomyces cerevisiae. These results suggest that CrCLS1 encodes a functional CLS of C. reinhardtii as the first identified algal CLS, whose enzyme function is distinct from that of PGPSs from C. reinhardtii. Comparison of CDP-alcohol phosphotransferase motif between PGPS and CLS among different species revealed a possible additional motif that might define the substrate specificity of these closely related enzymes.
Introduction
Functional specificity of an enzyme is crucial in keeping metabolic reactions in order. This largely relies on the substrate specificity defined by the catalytic motif. Thus, enzymes are often categorized into groups according to the existence of common catalytic motif(s). In phospholipid metabolism, a number of important reaction steps are catalyzed by CDP-alcohol phosphotransferases (Li-Beisson et al., 2013, 2015). These include CLS, PGPS, phosphatidylinositol (PI) synthase, phosphatidylserine (PS) synthase, phosphatidylcholine (PC) synthase, and aminoalcohol phosphotransferase for the biosynthesis of CL, PG, PI, PS, PC, and PE, respectively. Because these are the major phospholipid classes found in diverse organisms from bacteria to mammals and seed plants, it can be stated that CDP-alcohol phosphotransferases are crucial in the entire phospholipid metabolism.
In plants and algae, PG is an indispensable phospholipid class in photosynthetic function (Hagio et al., 2000, 2002; Sato et al., 2000; Babiychuk et al., 2003; Yu and Benning, 2003). Moreover, CL, which is an anionic phospholipid class widely distributed in different kingdom and found exclusively at the inner membrane of mitochondria (Lewis and McElhaney, 2009), has an essential role in mitochondrial function and thus plant growth (Katayama et al., 2004; Pineau et al., 2013). The biosynthesis of these lipid classes begins with the conversion of phosphatidic acid (PA) into CDP-diacylglycerol (CDP-DAG) by CDP-DAG synthase (CDS; Sato et al., 2000; Haselier et al., 2010; Zhou et al., 2013). Next, PGPS coverts CDP-DAG to phosphatidylglycerol phosphate (PGP), which is dephosphorylated by PGP phosphatase (PGPP) to produce PG (Muller and Frentzen, 2001; Hagio et al., 2000, 2002; Wu et al., 2006; Osman et al., 2010; Hung et al., 2015b). Furthermore, PG is converted to CL by CLS in mitochondria (Kadenbach et al., 1982; Jiang et al., 1997, 1999). Initially, 3 PGPSs were proposed in Arabidopsis thaliana (PGP1, PGP2, and PGP3) based on the amino acid sequence similarity (Xu et al., 2002). However, the third isoform (PGP3) was later shown not to be a functional PGPS but instead functions as CLS (Katayama et al., 2004). Subsequent gene knockout studies defined distinct in vivo function of CLS associated with mitochondrial function (Pineau et al., 2013). Thus, PGPS and CLS are functionally independent, although they are homologous and belong to the same CDP-alcohol phosphotransferase family in A. thaliana. Recently, we identified and characterized genes for PGPS of Chlamydomonas reinhardtii (Hung et al., 2015a). We demonstrated two functional PGPS isoforms; however, genome-wide search identified an additional PGPS homolog, which is more homologous with CLS. This reminded us of the case in A. thaliana described above. Because reciprocal genetic complementation was not performed yet in A. thaliana or any other model organisms, it remains elusive whether PGPS and CLS are functionally distinct in vivo.
In this report, we identified the additional homolog of PGPS in C. reinhardtii, designated CrCLS1 (Cre13.g604700), and performed reciprocal functional complementation assay using pgsA, a PGPS mutant of Synechocystis sp. PCC 6803 and Δcrd1, a CLS mutant of Saccharomyces cerevisiae. The result of functional complementation in these mutants, along with phenotype observation, lipid analysis and enzyme activity assay, demonstrated that CrCLS1 encodes a functional CLS but not PGPS. We compared sequence similarity in detail between PGPS and CLS and noted some difference adjacent to the defined CDP-OH-P motif. Our results suggest non-overlapping function of PGPS and CLS, through the identification and characterization of CLS in C. reinhardtii as the first report of CLS in algae.
Materials and Methods
Strains
The strains produced in this work are listed in Supplementary Table S1.
Protein Sequence Analysis
The multiple alignment of protein sequences was performed by use of CLUSTALW1. The mitochondrial targeting sequence was predicted by use of the subcellular localization program MitoProtII2 (Claros and Vincens, 1996).
Cloning of Plasmid Vectors
CrCLS1 (Cre13.g604700): To construct pCH069, a 1,060-bp fragment was amplified from the cDNA template of C. reinhardtii strain CC-503 (cw92 mt+) with the primers CH227 and CH228, and cloned into pENTR/D-TOPO. Then, to construct pCH178, the open reading frame (ORF) of CrCLS1 was amplified from pCH069 with the primers CH831 and CH832, and inserted into XbaI and EcoRI sites of pCH078 (Hung et al., 2013). To construct pCH158, the ORF of CrCLS1 was amplified from pCH069 with the primers CH776 and CH777 and inserted into NdeI and HpaI sites of pTCP2031V (Satoh et al., 2001). The primers and plasmids used in this study are described in Supplementary Tables S2 and 3, respectively.
Complementation Assay of the Synechocystis sp. PCC 6803 pgsA Mutant by CrCLS1
Complementation assay of the Synechocystis sp. PCC 6803 pgsA mutant by CrCLS1 (pCH158) was performed as described previously (Hung et al., 2015a).
Complementation Assay of the S. cerevisiae Δcrd1 Mutant by CrCLS1
Complementation assay of the S. cerevisiae Δcrd1 mutant by CrCLS1 (pCH178) was performed as described previously (Hung et al., 2015a).
Lipid Extraction and Analysis
Lipid extraction and analysis were performed as previously described (Hung et al., 2013) except that 2D thin-layer chromatography (TLC) was used to separate phospholipid classes with the solvent system of chloroform/methanol/7 N ammonia 120:80:8 (by vol) for the first dimension and chloroform/methanol/acetic acid/water 170:20:15:3 (by vol) for the second dimension (Nakamura et al., 2003).
Radiolabeling Assay of CLS Activity
Logarithmically growing cells were resuspended in 5 ml SC-Ura medium at the cell density (OD600 of 5) with 30 μCi KH232PO4 (PerkinElmer). After shaking incubation for 8 h at room temperature, lipids were extracted from cells by the method of Bligh and Dyer (1959). Lipids spotted on a TLC plate (Silica gel 60G, Merck) were developed with chloroform/methanol/acetic acid (65:25:8, v/v/v) (Haselier et al., 2010) along with PG and CL (Sigma–Aldrich) as standards and radioactive spots were visualized by Imaging Plate (Fuji Film) and BAS-2500 (GE Healthcare). Unlabeled PG and CL were stained with 0.01% primuline in 80% (v/v) acetone and detected under UV light.
RNA Extraction and cDNA Synthesis
RNA extraction and cDNA synthesis were performed as previously described (Hung et al., 2013).
Quantitative RT-PCR
Quantitative RT-PCR analysis involved the ABI 7500 Real Time PCR System (Applied Biosystems) with the specific oligonucleotide primer sets, CH955 and CH956, CH957 and CH958, and CH531 and CH532, for CrCLS1, CRD1, and ACT1, respectively. Gene expression was normalized to that of ACT1. Data were averaged by three technical replicates in the same run and three biological replicates in separate runs. The primer sequences are described in Supplementary Table S2.
Results
Sequence Analysis of CrCLS1
To compare the amino acid sequence similarity of the putative CrCLS1 with other known CLSs in different organisms, the deduced amino acid sequence of CrCLS1 was compared with those of S. cerevisiae CRD1, Homo sapiens CLS1, A. thaliana CLS, and Drosophila melanogaster CLS, which are functionally characterized CLS (Figure 1) (Tuller et al., 1998; Katayama et al., 2004; Chen et al., 2006; Acehan et al., 2011). In Figure 1, the region containing the CDP-OH-P motif D(X)2DG(X)2AR(X)8-9G(X)3D(X)3D is underlined and asterisks indicate the conserved eight amino acid residues. All eight amino acids were conserved in CrCLS1, which suggests that CrCLS1 encodes a functional CLS. In addition, CrCLS1 contained a putative N-terminal mitochondrial targeting sequence predicted by the subcellular localization program MitoProtII, suggesting a possible localization of CrCLS1 in mitochondria, where CL is exclusively localized.

FIGURE 1. Multiple amino acid sequence alignment of Chlamydomonas reinhardtii CLS1 (CrCLS1) with other known CLSs, Saccharomyces cerevisiae CRD1 (ScCRD1), Homo sapiens CLS1 (HsCLS1), Arabidopsis thaliana CLS (AtCLS), and Drosophila melanogaster CLS (DmCLS). The region conserved among proteins containing a CDP-OH-P motif (PF01066.9) is underlined. Asterisks indicate the amino acid residues conserved in all sequences of proteins with the CDP-OH-P motif. Square frames indicate the terminal amino acid residues of the predicted cleavage site of putative N-terminal mitochondrial targeting sequence.
Complementation of pgsA by CrCLS1
To examine whether CrCLS1 functions as PGPS, we transformed CrCLS1 into the pgsA mutant of Synechocystis sp. PCC 6803, which abolishes PGPS activity and thus requires exogenous supplementation of PG for growth (Hagio et al., 2000). As shown in Figure 2, whereas the CrPGP1 and CrPGP2 functionally complemented the lethal phenotype of the pgsA mutant as reported previously (Hung et al., 2015a), CrCLS1 failed to complement the growth phenotype, showing the rescued growth only in the presence of PG. Therefore, CrCLS1 does not function as a PGPS in vivo in Synechocystis sp. PCC 6803.
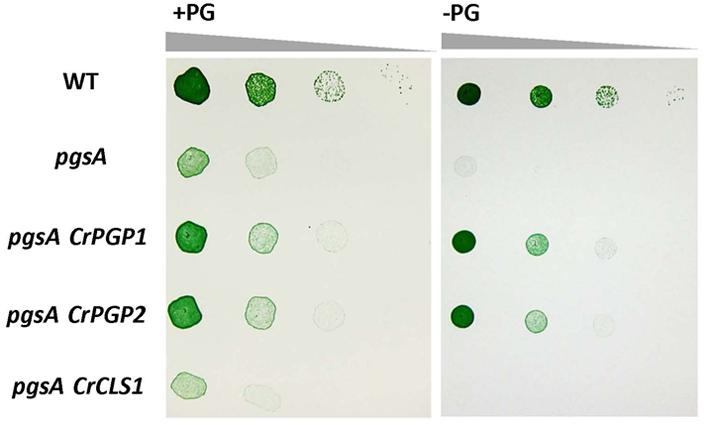
FIGURE 2. Heterologous complementation of Synechocystis sp. PCC 6803 pgsA mutant by CrPGP1, CrPGP2 and CrCLS1. Growth of the wild type, pgsA, pgsA CrPGP1, pgsA CrPGP2, and pgsA CrCLS1 were compared on solid BG-11 media with or without PG supplementation. Spotting involved serial 10-fold dilution from left to right starting at OD730 of 0.05, with 5 μl each spotted onto a BG-11 agar plate with or without 20 μM PG and incubated under 50∼60 μmol photons m-2 s-1 for 5 days at 30°C. Images are representative of three biological replicates.
Recovery of Growth Defect in the Δcrd1 Mutant Complemented by CrCLS1
To investigate whether CrCLS1 encodes a functional CLS, we performed a heterologous complementation assay with the S. cerevisiae Δcrd1 mutant, because Synechocystis sp. PCC 6803 does not contain CL and no other CLS mutant is known in algae. As previously reported, Crd1p has CLS activity and Δcrd1 mutant cells show a temperature-sensitive growth defect, severe at 37°C but not at 30°C (Jiang et al., 1999). The temperature-sensitive phenotype of Δcrd1 mutant cells was rescued by heterologous complementation of HsCLS1 (Houtkooper et al., 2006), so we used this approach to investigate the function of CrCLS1. We cloned the ORF of CrCLS1 into a yeast shuttle vector and transformed it into Δcrd1 mutant cells. The Δcrd1 mutant harboring CrCLS1 fully recovered cell growth at 37°C, whereas the Δcrd1 mutant alone showed a growth defect at this temperature (Figure 3A). Therefore, CrCLS1 complemented the temperature-sensitive phenotype of Δcrd1, which suggests that CrCLS1 encodes a functional CLS of C. reinhardtii.
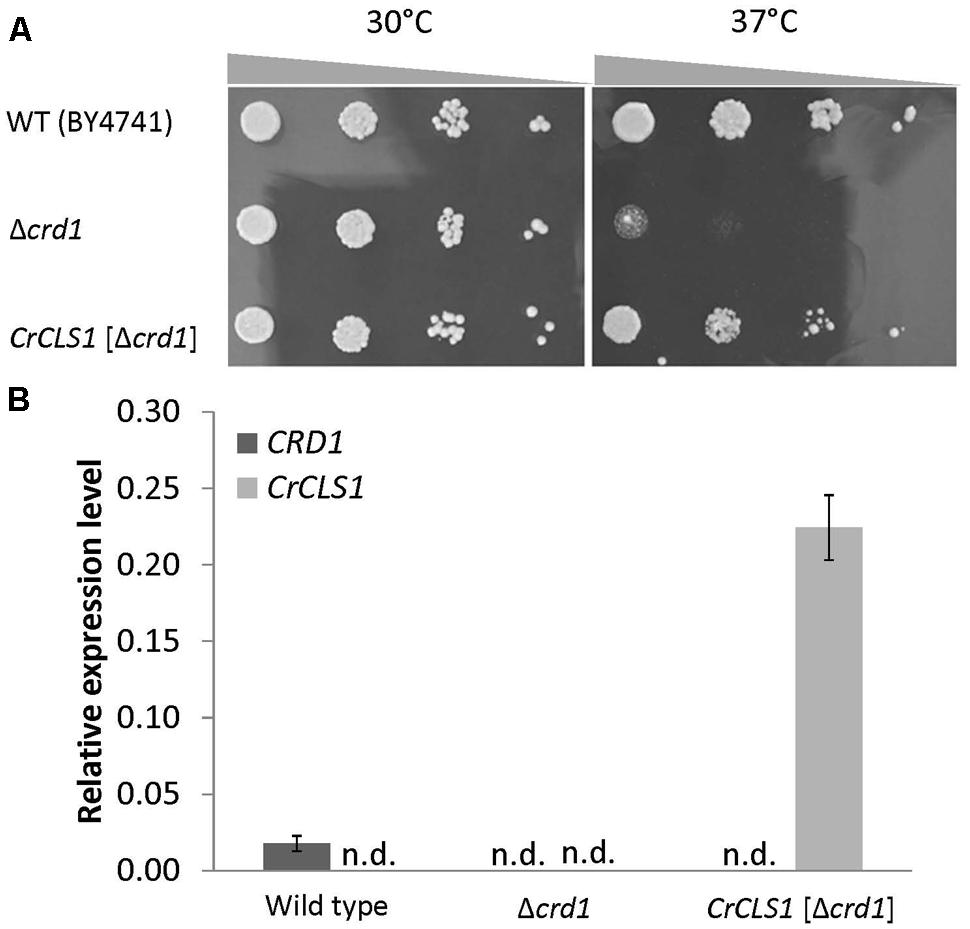
FIGURE 3. Heterologous complementation of temperature-sensitive growth defect in the S. cerevisiae Δcrd1 mutant with C. reinhardtii CLS1. (A) Culture of wild type, Δcrd1 mutant and Δcrd1 mutant harboring CrCLS1 involved serial 10-fold dilution from left to right starting at OD600 of 0.1; 5 μL each was spotted onto YPD media and incubated for 2 days at 30°C or 37°C. Images are representative of 3 replicates. (B) Relative expression of CRD1 and CrCLS1 in wild type, Δcrd1 and Δcrd1 CrCLS1. Levels are normalized to that of ACT1. Data are mean ± SD from three biological replicates. n.d., not detected.
Expression of CrCLS1
To investigate whether CrCLS1 is appropriately expressed in the Δcrd1 mutant, we analyzed the gene expression of CrCLS1 in the Δcrd1 mutant harboring CrCLS1. The relative gene expression of CrCLS1 was 12.6-fold higher in the Δcrd1 mutant harboring CrCLS1 than CRD1 in the wild type (Figure 3B). Thus, CrCLS1 is sufficiently expressed in Δcrd1 mutant cells, which supports the functional complementation shown in Figure 3A.
Effect of Different Carbon Sources on the Growth of Δcrd1 Mutant Complemented by CrCLS1
A previous study showed that the growth of the Δcrd1 mutant under aerobic conditions was affected with ethanol used as the sole carbon source (Tuller et al., 1998). To investigate whether the Δcrd1 mutant harboring CrCLS1 rescued the growth defect under this condition, cells were grown in synthetic complete medium supplemented with 2% glucose or 2% ethanol as the sole carbon source. The growth rates of both the Δcrd1 mutant and Δcrd1 harboring CrCLS1 were indistinguishable from that of wild type in 2% glucose medium (Figure 4A). However, with 2% ethanol medium, the Δcrd1 mutant harboring CrCLS1 fully restored the growth phenotype to that of the wild type, whereas the Δcrd1 mutant showed growth retardation, as reported (Tuller et al., 1998) (Figure 4B). Therefore, CrCLS1 could complement the growth defect of the Δcrd1 mutant with ethanol supplementation as the carbon source.
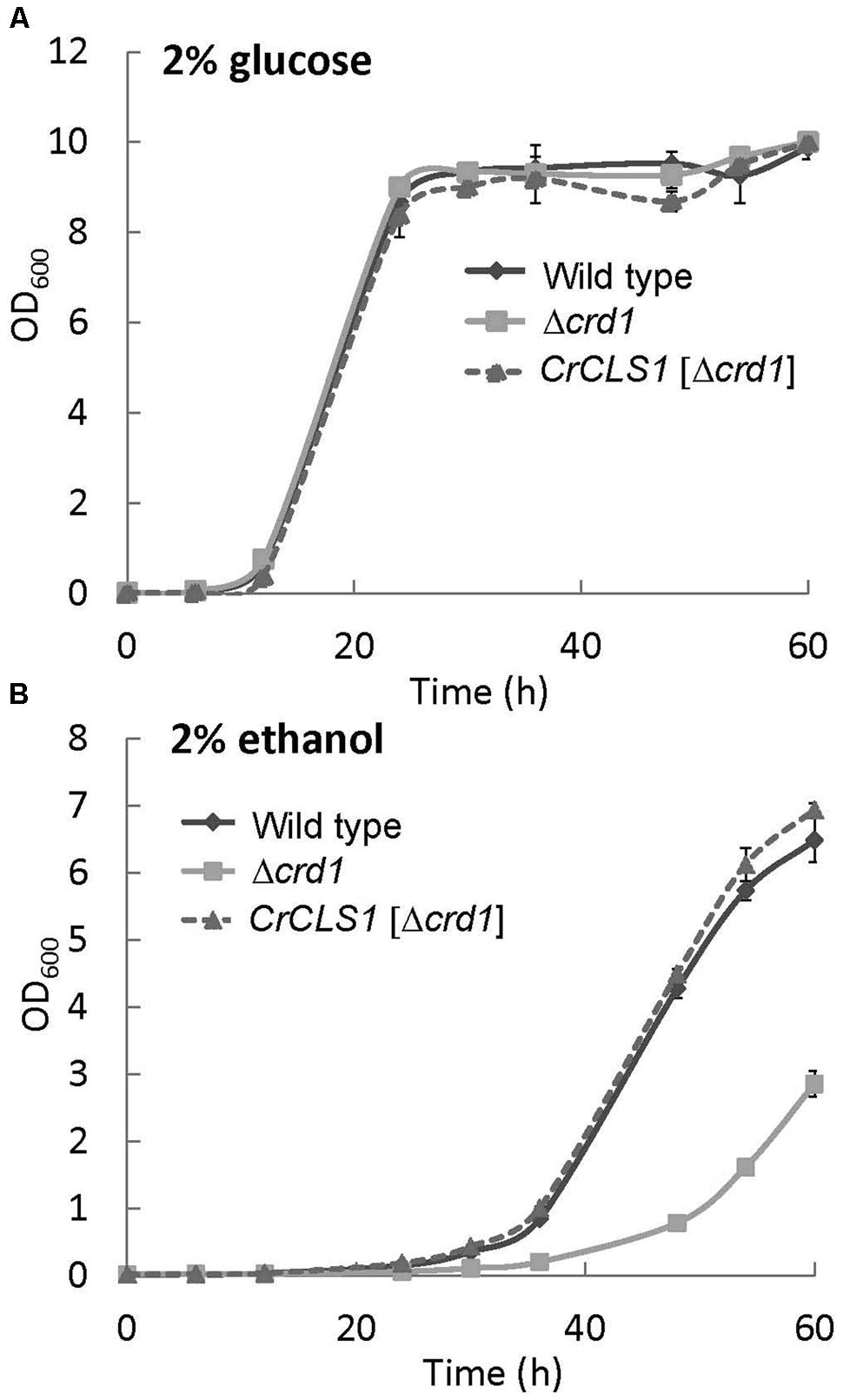
FIGURE 4. Growth of wild type, Δcrd1 and Δcrd1 CrCLS1 with different carbon sources. Cells were grown in synthetic complete medium supplemented with 2% glucose (A) or 2% ethanol (B). Cell growth was started at OD600 of 0.01 at 30°C. Data are mean ± SD from three biological replicates.
Lipid Contents of the Δcrd1 Mutant Complemented by CrCLS1
The phospholipid profiles of the Δcrd1 mutant were previously analyzed by radiolabeling (Tuller et al., 1998) or mass spectrometry (Zhang et al., 2003). However, whether the Δcrd1 mutant alters the composition of major membrane phospholipid classes remained unclear. To investigate whether the complementation of the growth defect observed in Figures 3 and 4 is associated with lipid compositional change, we analyzed the major phospholipid composition of these strains. The Δcrd1 mutant showed an increase in PC content and decrease in PS and PI contents as compared with the wild type (Figure 5A). In the Δcrd1 mutant harboring CrCLS1, phospholipid composition was restored to a level similar to that of the wild type. The fatty acid composition of PE, PC, and PI was similar among the three strains (Figure 5B). Thus, CrCLS1 encodes a functional CLS that complements lipid compositional changes in the Δcrd1 mutant.
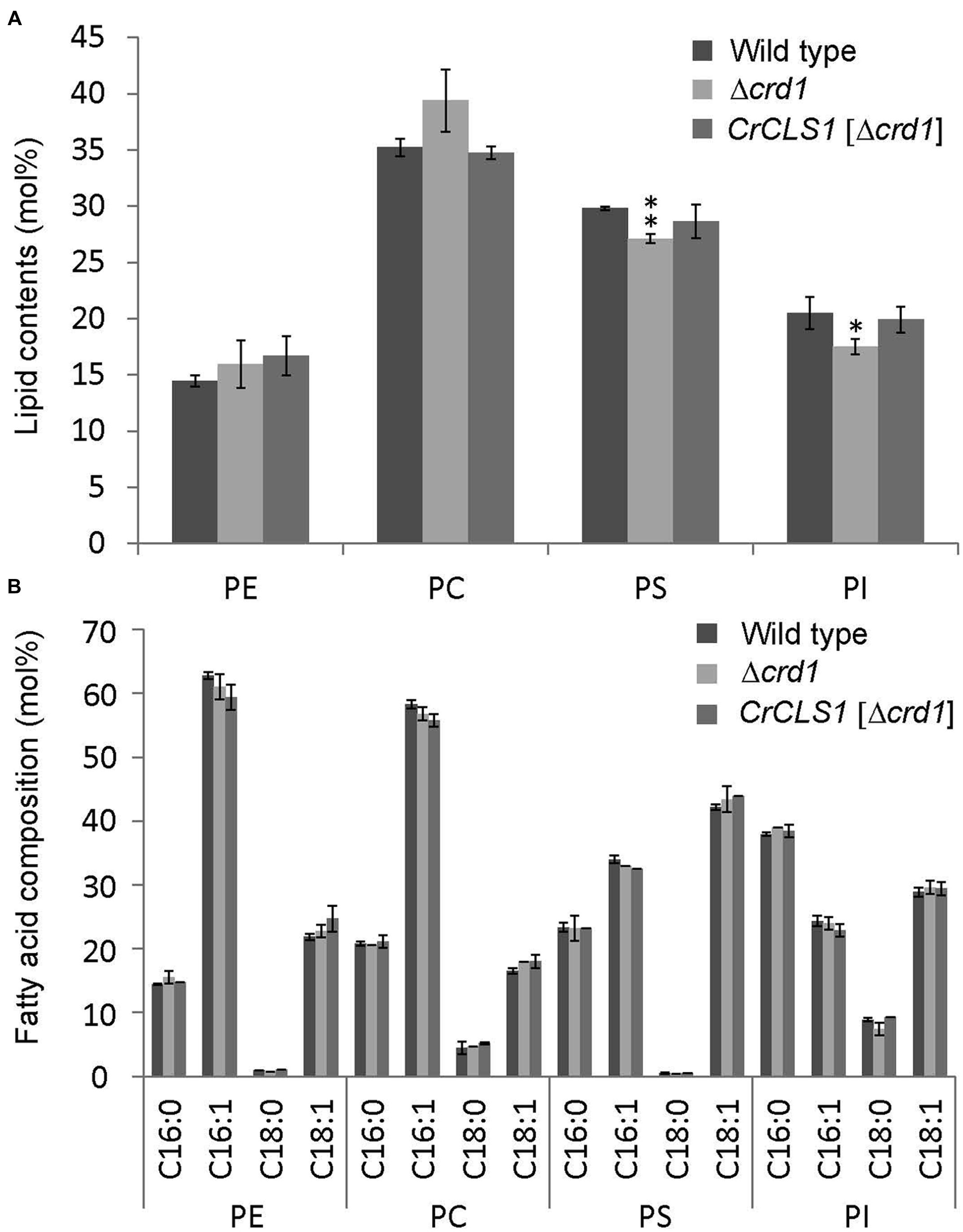
FIGURE 5. Phospholipid content (A) and fatty acid composition (B) of wild type, Δcrd1 and Δcrd1 CrCLS1. Total lipids were extracted from cells grown to stationary phase and separated by 2D thin-layer chromatography; content of phospholipids was quantified by gas chromatography. Data are mean ± SD from three biological replicates. Asterisks indicate statistical significance by Student’s t-test (∗P < 0.05, ∗∗P < 0.001). PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine.
Enzyme Activity of CrCLS1 Expressed in the Δcrd1
To investigate whether CrCLS1 encodes a functional CLS to restore the CL synthesis defect in the Δcrd1, we performed radiolabeling assay to analyze CLS activity. As shown in Figure 6, the Δcrd1 mutant harboring CrCLS1 recovered radiolabeled spot that co-migrates with the commercial standard of CL, which is present in wild type but absent in the Δcrd1 mutant, demonstrating that the activity of CLS was recovered in the mutant harboring CrCLS1. Thus, CrCLS1 encodes a functional CLS that complements CL synthesis defect of Δcrd1 mutant.
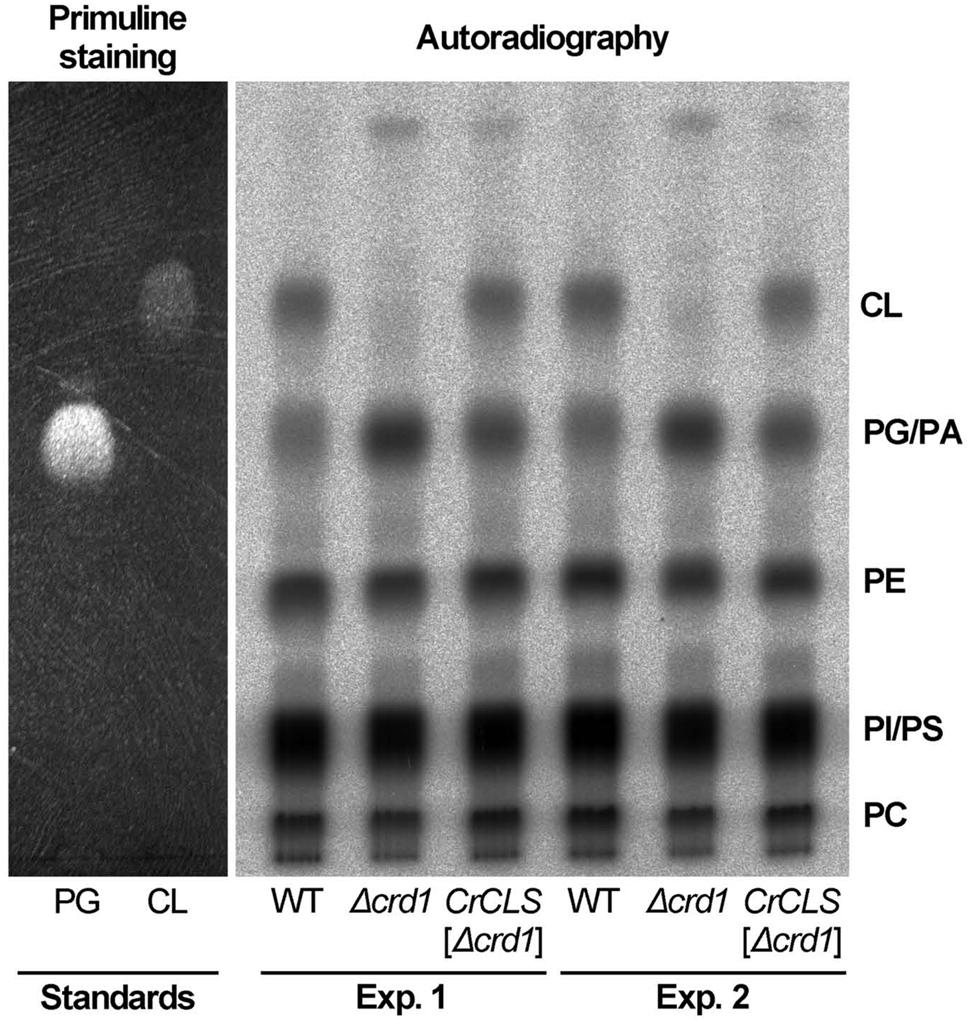
FIGURE 6. CrCLS complemented cardiolipin synthesis in Δcrd1 cells. Wild type (WT), Δcrd1 or Δcrd1 CrCLS cells were grown in the presence of [32P]-phosphate for 8 h at room temperature. Radioactivity incorporated into phospholipids was determined by TLC analysis and autoradiography, with unlabeled PG and CL visualized by primuline staining as standards. Two biologically independent experiments (Experiments 1 and 2) were shown. PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol; PS, phosphatidylserine; PA, phosphatidic acid.
Discussion
Present study reported identification of a CLS gene in C. reinhardtii, CrCLS1, and examined its in vivo function by heterologous complementation of pgsA, a PGPS mutant of Synechocystis sp. PCC 6803, and Δcrd1, a CLS mutant of S. cerevisiae. Whereas CrCLS1 did not complement the growth phenotype of pgsA, it rescued the temperature-sensitive growth phenotype, growth profile with different carbon sources, phospholipid composition and enzyme activity of Δcrd1 of S. cerevisiae. These results suggest that CrCLS1 is a functional gene for CLS of C. reinhardtii as the first identified algal CLS, which is functionally incompatible with PGPS despite their sequence homology.
Physiological roles of CrCLS1 in C. reinhardtii are not reported yet; however, several transcriptomic studies have shown gene expression profiles in response to environmental stresses. For example, expression of CrCLS1 is down-regulated in response to the deprivation of iron (Urzica et al., 2013) and nitrogen (Goodenough et al., 2014). Conversely, an upregulation is seen by copper deficiency (Castruita et al., 2011) and singlet oxygen stress (Wakao et al., 2014). These data suggest possible roles of CrCLS1 in adaptation to circumvent environmental stresses.
Given that both PGPS and CLS belong to the CDP-alcohol phosphotransferase family and the relevant CDP-OH-P motifs are closely related (Katayama et al., 2004), what defines substrate specificity of these enzymes?
Recently, structural basis for catalysis in a CDP-alcohol phosphotransferase was revealed by crystallographic analysis (Sciara et al., 2014). According to this structure, conserved amino acid residues in the CDP-OH-P motif are associated with CDP-DAG. Since CDP-DAG is the common substrate between CLS and PGPS, this study suggests that an additional motif recognizes the other substrate (PG for CLS; glycerol 3-phosphate for PGPS). We aligned the amino acid sequences of core CDP-OH-P motif among three CLSs (C. reinhardtii cardiolipin synthase 1, CrCLS1; S. cerevisiae CRD1, ScCRD1; A. thaliana CLS, AtCLS) and five PGPSs (C. reinhardtii PGP1, CrPGP1; C. reinhardtii PGP2, CrPGP2; A. thaliana PGP1, AtPGP1; A. thaliana PGP2, AtPGP2; Synechocystis sp. PCC 6803 PgsA, SynPgsA) (Figure 7). While the eight amino acid residues of the core CDP-OH-P motif D(X)2DG(X)2AR(X)8-9G(X)3D(X)3D indicated by asterisks in Figure 7 were conserved between the PGPS and CLS, we noted that seven amino acids (FxxAxxT) immediately before the core CDP-OH-P motif were highly conserved among PGPSs but not CLSs (underlined in Figure 7). In addition, we found additional four amino acid residues that were conserved among PGPS but not in CLS (indicated by dots in Figure 7). It is possible that these additional residues may define the substrate specificity between PGPS and CLS. Detailed structural analysis as well as enzymatic characterization of these residues are anticipated to experimentally validate this proposal.

FIGURE 7. Multiple amino acid sequence alignment of the core motif of CDP-OH-P between CLSs and the PGPSs. Asterisks indicate the amino acid residues conserved among proteins with the CDP-OH-P motif, and dots indicate those conserved only among PGPSs. The amino acid residues present among PGPS but not among CLS are underlined. CrCLS1, C. reinhardtii CLS1; ScCRD1, S. cerevisiae CRD1; AtCLS, A. thaliana CLS; CrPGP1, C. reinhardtii PGP1; CrPGP2, C. reinhardtii PGP2; AtPGP1, A. thaliana PGP1; AtPGP2, A. thaliana PGP2; SynPgsA, Synechocystis sp. PCC 6803 PgsA.
Conclusion
We suggest functional specificity of CLS by the identification and characterization of a CLS, CrCLS1, in C. reinhardtii.
Author Contributions
KK, HW, and YN conceived research. KK and C-HH performed experiments and analyzed data. All authors wrote and commented on the manuscript and approved the contents.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Masahiro Ikeuchi (the University of Tokyo, Tokyo, Japan) for the supply of pTCP2031V. This research was supported by CREST (to HW) and PRESTO (to YN), Japan Science and Technology Agency, and the core budget of Institute of Plant and Microbial Biology, Academia Sinica (to YN).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01542
Abbreviations
CDP, cytidine 5′-diphosphate; CL, cardiolipin; CLS, cardiolipin synthase; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PGP, phosphatidylglycerol phosphate; PGPP, phosphatidylglycerophosphate phosphatase; PGPS, phosphatidylglycerophosphate synthase; PI, phosphatidylinositol; PS, phosphatidylserine.
Footnotes
References
Acehan, D., Malhotra, A., Xu, Y., Ren, M., Stokes, D. L., and Schlame, M. (2011). Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 100, 2184–2192. doi: 10.1016/j.bpj.2011.03.031
Babiychuk, E., Muller, F., Eubel, H., Braun, H. P., Frentzen, M., and Kushnir, S. (2003). Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J. 33, 899–909. doi: 10.1046/j.1365-313X.2003.01680.x
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Castruita, M., Casero, D., Karpowicz, S. J., Kropat, J., Vieler, A., Hsieh, S. I., et al. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292. doi: 10.1105/tpc.111.084400
Chen, D., Zhang, X. Y., and Shi, Y. (2006). Identification and functional characterization of hCLS1, a human cardiolipin synthase localized in mitochondria. Biochem. J. 398, 169–176. doi: 10.1042/BJ20060303
Claros, M. G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. doi: 10.1111/j.1432-1033.1996.00779.x
Goodenough, U., Blaby, I., Casero, D., Gallaher, S. D., Goodson, C., Johnson, S., et al. (2014). The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell 13, 591–613. doi: 10.1128/EC.00013-14
Hagio, M., Gombos, Z., Varkonyi, Z., Masamoto, K., Sato, N., Tsuzuki, M., et al. (2000). Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol. 124, 795–804. doi: 10.1104/pp.124.2.795
Hagio, M., Sakurai, I., Sato, S., Kato, T., Tabata, S., and Wada, H. (2002). Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol. 43, 1456–1464. doi: 10.1093/pcp/pcf185
Haselier, A., Akbari, H., Weth, A., Baumgartner, W., and Frentzen, M. (2010). Two closely related genes of Arabidopsis encode plastidial cytidinediphosphate diacylglycerol synthases essential for photoautotrophic growth. Plant Physiol. 153, 1372–1384. doi: 10.1104/pp.110.156422
Houtkooper, R. H., Akbari, H., van Lenthe, H., Kulik, W., Wanders, R. J., Frentzen, M., et al. (2006). Identification and characterization of human cardiolipin synthase. FEBS Lett. 580, 3059–3064. doi: 10.1016/j.febslet.2006.04.054
Hung, C. H., Endo, K., Kobayashi, K., Nakamura, Y., and Wada, H. (2015a). Characterization of Chlamydomonas reinhardtii phosphatidylglycerophosphate synthase in Synechocystis sp. PCC 6803. Front. Microbiol. 6:842. doi: 10.3389/fmicb.2015.00842
Hung, C. H., Kobayashi, K., Wada, H., and Nakamura, Y. (2015b). Isolation and characterization of a phosphatidylglycerophosphate phosphatase1. PGPP1, in Chlamydomonas reinhardtii. Plant Physiol. Biochem. 92, 56–61. doi: 10.1016/j.plaphy.2015.04.002
Hung, C. H., Ho, M. Y., Kanehara, K., and Nakamura, Y. (2013). Functional study of diacylglycerol acyltransferase type 2 family in Chlamydomonas reinhardtii. FEBS Lett. 587, 2364–2370. doi: 10.1016/j.febslet.2013.06.002
Jiang, F., Gu, Z., Granger, J. M., and Greenberg, M. L. (1999). Cardiolipin synthase expression is essential for growth at elevated temperature and is regulated by factors affecting mitochondrial development. Mol. Microbiol. 31, 373–379. doi: 10.1046/j.1365-2958.1999.01181.x
Jiang, F., Rizavi, H. S., and Greenberg, M. L. (1997). Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26, 481–491. doi: 10.1046/j.1365-2958.1997.5841950.x
Kadenbach, B., Mende, P., Kolbe, H. V., Stipani, I., and Palmieri, F. (1982). The mitochondrial phosphate carrier has an essential requirement for cardiolipin. FEBS Lett. 139, 109–112. doi: 10.1016/0014-5793(82)80498-5
Katayama, K., Sakurai, I., and Wada, H. (2004). Identification of an Arabidopsis thaliana gene for cardiolipin synthase located in mitochondria. FEBS Lett. 577, 193–198. doi: 10.1016/j.febslet.2004.10.009
Lewis, R. N., and McElhaney, R. N. (2009). The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim. Biophys. Acta 1788, 2069–2079. doi: 10.1016/j.bbamem.2009.03.014
Li-Beisson, Y., Beisson, F., and Riekhof, W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82, 504–522. doi: 10.1111/tpj.12787
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11:e0161. doi: 10.1199/tab.0161
Muller, F., and Frentzen, M. (2001). Phosphatidylglycerophosphate synthases from Arabidopsis thaliana. FEBS Lett. 509, 298–302. doi: 10.1016/S0014-5793(01)03163-5
Nakamura, Y., Arimitsu, H., Yamaryo, Y., Awai, K., Masuda, T., Shimada, H., et al. (2003). Digalactosyldiacylglycerol is a major glycolipid in floral organs of Petunia hybrida. Lipids 38, 1107–1112. doi: 10.1007/s11745-006-1166-x
Osman, C., Haag, M., Wieland, F. T., Brugger, B., and Langer, T. (2010). A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29, 1976–1987. doi: 10.1038/emboj.2010.98
Pineau, B., Bourge, M., Marion, J., Mauve, C., Gilard, F., Maneta-Peyret, L., et al. (2013). The importance of cardiolipin synthase for mitochondrial ultrastructure, respiratory function, plant development, and stress responses in Arabidopsis. Plant Cell 25, 4195–4208. doi: 10.1105/tpc.113.118018
Sato, N., Hagio, M., Wada, H., and Tsuzuki, M. (2000). Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 97, 10655–10660. doi: 10.1073/pnas.97.19.10655
Satoh, S., Ikeuchi, M., Mimuro, M., and Tanaka, A. (2001). Chlorophyll b expressed in Cyanobacteria functions as a light-harvesting antenna in photosystem I through flexibility of the proteins. J. Biol. Chem. 276, 4293–4297. doi: 10.1074/jbc.M008238200
Sciara, G., Clarke, O. B., Tomasek, D., Kloss, B., Tabuso, S., Byfield, R., et al. (2014). Structural basis for catalysis in a CDP-alcohol phosphotransferase. Nat. Commun. 5:4068. doi: 10.1038/ncomms5068
Tuller, G., Hrastnik, C., Achleitner, G., Schiefthaler, U., Klein, F., and Daum, G. (1998). YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421, 15–18. doi: 10.1016/S0014-5793(97)01525-1
Urzica, E. I., Vieler, A., Hong-Hermesdorf, A., Page, M. D., Casero, D., Gallaher, S. D., et al. (2013). Remodeling of membrane lipids in iron-starved Chlamydomonas. J. Biol. Chem. 288, 30246–30258. doi: 10.1074/jbc.M113.490425
Wakao, S., Chin, B. L., Ledford, H. K., Dent, R. M., Casero, D., Pellegrini, M., et al. (2014). Phosphoprotein SAK1 is a regulator of acclimation to singlet oxygen in Chlamydomonas reinhardtii. Elife 3:e02286. doi: 10.7554/eLife.02286
Wu, F., Yang, Z., and Kuang, T. (2006). Impaired photosynthesis in phosphatidylglycerol-deficient mutant of cyanobacterium Anabaena sp. PCC7120 with a disrupted gene encoding a putative phosphatidylglycerophosphatase. Plant Physiol. 141, 1274–1283. doi: 10.1104/.pp106.083451
Xu, C., Hartel, H., Wada, H., Hagio, M., Yu, B., Eakin, C., et al. (2002). The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 129, 594–604. doi: 10.1104/pp.002725
Yu, B., and Benning, C. (2003). Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J. 36, 762–770. doi: 10.1046/j.1365-313X.2003.01918.x
Zhang, M., Su, X., Mileykovskaya, E., Amoscato, A. A., and Dowhan, W. (2003). Cardiolipin is not required to maintain mitochondrial DNA stability or cell viability for Saccharomyces cerevisiae grown at elevated temperatures. J. Biol. Chem. 278, 35204–35210. doi: 10.1074/jbc.M306729200
Keywords: CDP-alcohol phosphotransferase, cardiolipin, cardiolipin synthase, phosphatidylglycerol, PGPS, Chlamydomonas reinhardtii Synechocystis sp. PCC 6803, Saccharomyces cerevisiae
Citation: Hung C-H, Kobayashi K, Wada H and Nakamura Y (2016) Functional Specificity of Cardiolipin Synthase Revealed by the Identification of a Cardiolipin Synthase CrCLS1 in Chlamydomonas reinhardtii. Front. Microbiol. 6:1542. doi: 10.3389/fmicb.2015.01542
Received: 15 October 2015; Accepted: 21 December 2015;
Published: 12 January 2016.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Alberto A. Iglesias, Instituto de Agrobiotecnología del Litoral – Consejo Nacional de Investigaciones Científicas y Tecnológicas, ArgentinaTakashi Osanai, Meiji University, Japan
Song Xue, Marine Bioengineering Group Department of Biotechnology Dalian Institute of Chemical Physics Chinese Academy of Sciences, China
Copyright © 2016 Hung, Kobayashi, Wada and Nakamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuki Nakamura, nakamura@gate.sinica.edu.tw
 Chun-Hsien Hung
Chun-Hsien Hung Koichi Kobayashi
Koichi Kobayashi Hajime Wada
Hajime Wada Yuki Nakamura
Yuki Nakamura