- Binns Lab, Department of Biology, University of Pennsylvania, Philadelphia, PA, USA
The plant pathogen Agrobacterium tumefaciens carries a virulence gene system that is required for the initiation of crown gall tumors on susceptible plants. Expression of the vir genes is activated by the VirA/VirG two component regulatory system. VirA is a histidine kinase which signals the presence of certain chemicals found at the site of a plant wound. The receiver domain located at its carboxyl terminus defines VirA as a hybrid histidine kinase. Here, we show that the VirA receiver interacts with the DNA-binding domain of VirG. This finding supports the hypothesis that the receiver acts as a recruiting factor for VirG. In addition, we show that removal of the VirA receiver allowed vir gene expression in response to glucose in a dose dependent manner, indicating that the receiver controls VirG activation and suggesting that the supplementary ChvE-sugar signal increases this activity.
Introduction
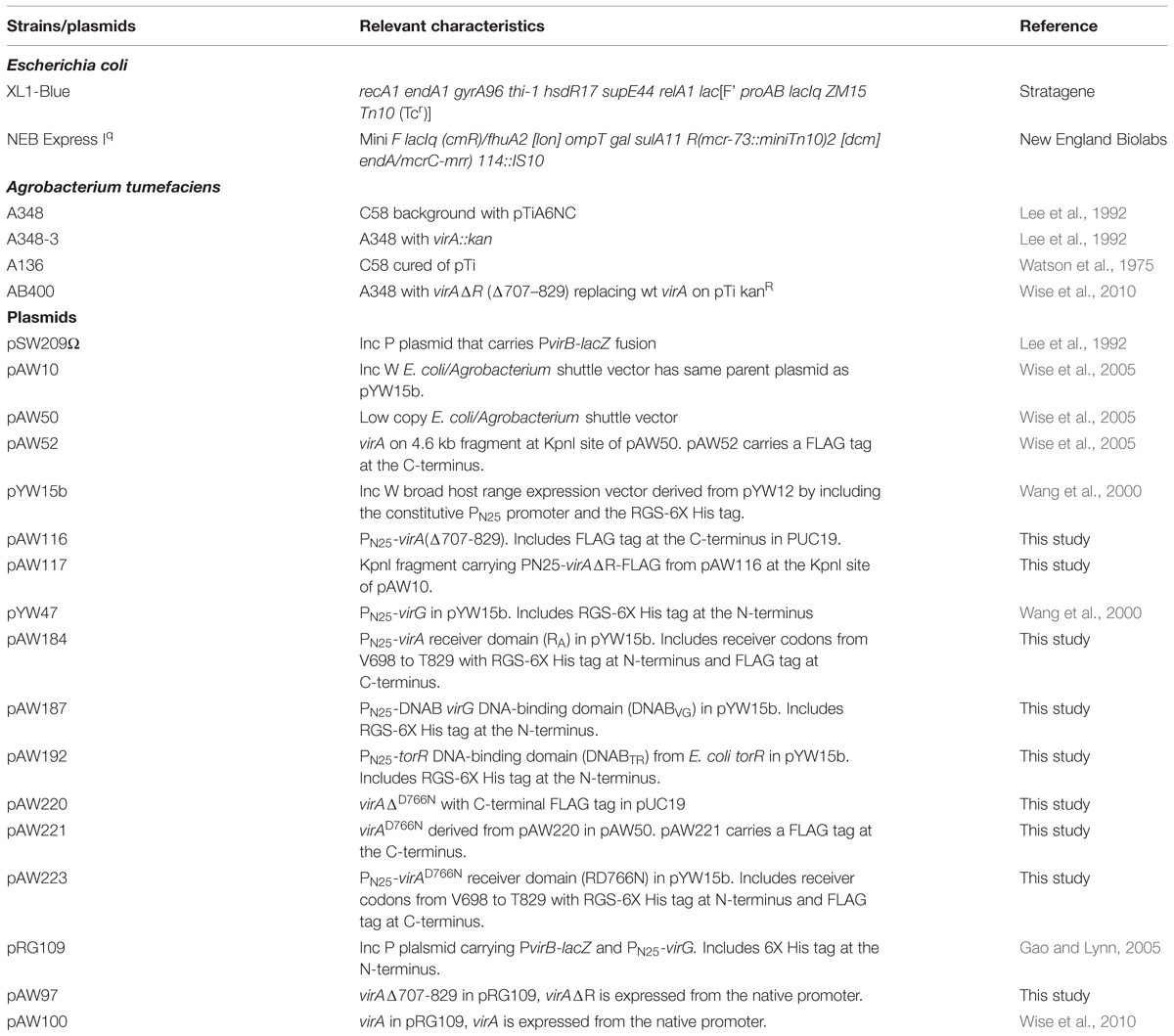
Agrobacterium tumefaciens is the causative agent of crown gall tumors on dicotyledonous plants. The virulence system that drives tumor formation depends on the ability of the VirA histidine kinase to signal the presence of certain chemical components found at the site of a plant wound. When the inducing signals (phenolics, monosaccharides, and low pH) are present, a phosphate group is transferred from the conserved histidine (H474) in VirA’s kinase region to a conserved aspartate (D52) in the receiver domain of the response regulator, VirG (see Figure 1). Phosphorylated VirG is able to bind the promoters of the virulence (vir) genes and activate their transcription (for reviews, see Gelvin, 2003; McCullen and Binns, 2006).
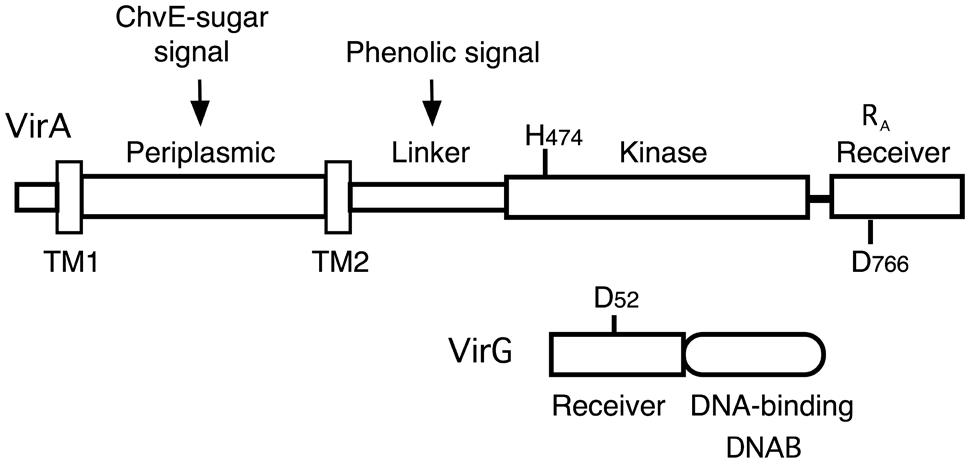
FIGURE 1. Diagram of VirA and VirG. VirA and VirG are arranged in functional domains. Key amino acids are indicated. H474 is the site of autophosphorylation in the VirA kinase domain. D52 is the aspartate in the VirG receiver domain that accepts the phosphate from VirA. D766N in the VirA receiver domain is analogous to VirG’s D52. RA denotes the receiver domain of VirA and DNAB represents the DNA-binding domain of VirG.
The phenolic signal and slightly acidic pH are essential for vir gene induction. Certain monosaccharides (arabinose, glucose, galactose, glucuronic acid, and others), are not, by themselves, inducing agents. However, their presence greatly enhances vir gene expression when the concentration of phenolic inducer is low (Ankenbauer and Nester, 1990; Cangelosi et al., 1990). Reception of the phenolic signal requires the linker region of VirA (Chang and Winans, 1992) and likely involves a region that protein modeling indicates has structural similarity to a GAF domain (Gao and Lynn, 2007; Lin et al., 2014). The effect of sugar on the virulence system requires the periplasmic ChvE sugar-binding protein and is mediated via interaction of ChvE with the VirA periplasmic domain (Cangelosi et al., 1990; Chang and Winans, 1992; Shimoda et al., 1993; Hu et al., 2013).
VirA can be classified as a hybrid kinase because it carries a region with sequence similarity to the receiver domains of response regulators, including an aspartate at codon 766 that is analogous to the phosphorylatable aspartate (residue 52) in VirG’s receiver domain. We have previously suggested that VirA’s receiver domain functions differently than the receiver domains of hybrid kinases that are known to function as part of a phospho-relay (Wise et al., 2010). For example, while mutation of D766 reduces vir gene expression, it does not create a null mutation (Pazour et al., 1991). Furthermore, VirA does not include an HPT (histidine phospho-transfer) domain and in vitro experiments using purified proteins indicate that an HPT domain is not required for phosphate transfer between VirA and VirG (Jin et al., 1990). Most importantly, the VirA receiver is not essential for vir gene expression. Deletion of the receiver domain does change vir gene expression, but that effect varies significantly depending on how virG or virAΔR are expressed. As shown in earlier studies that used constitutively expressed virG, VirAΔR is capable of activating vir gene expression in the absence of the normally essential phenolic inducer, provided glucose or arabinose is available (Chang and Winans, 1992; Chang et al., 1996). These early results defined the receiver as an inhibitor of vir gene expression. However, replacing wild type virA on the tumor-inducing plasmid (pTi) with virAΔR demonstrated that the receiver domain of VirA is essential for vir gene expression if wild type virG is expressed solely from its natural position on pTi (Wise et al., 2010). Thus, the receiver has both activating and inhibitory effects on vir gene expression.
In this study, we examined the positive and negative roles of the VirA receiver (RA) in regulating vir gene expression and found the following: (1) RA, expressed in trans, did not correct the null phenotype of a virAΔR strain. In fact, RA inhibited vir gene expression in the wild type background more strongly than a mutant receiver carrying the D766N mutation (R766N). (2) The VirA receiver (RA) formed a stable complex with the DNA-binding domain of VirG. RA did not form a stable attachment to the DNA-binding domain of TorR, a response regulator which like VirG is a member of the OmpR/PhoB subclass of response regulators (Simon et al., 1994; Martinez-Hackert and Stock, 1997). (3) Glucose dose response assays in the presence or absence of the phenolic inducer acetosyringone (AS) indicated that VirAΔR and full-length VirA have similar dose responses to the ChvE-sugar signal, indicating that one function of the VirA receiver is to keep vir gene expression off when sugars are available, but no phenolic inducer is present.
Materials and Methods
Bacterial Strains and Plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. Standard cloning methods were used for DNA ligation, PCR, plasmid isolation, and DNA analysis. Details of plasmid construction and primer sequences are included in Supplementary Table S1. Escherichia coli XL1-blue was used for plasmid construction and amplification. E. coli strain NEB Express Iq was used for protein expression for pull-down experiments. Use of a particular Agrobacterium strain for an experiment is indicated in the associated legend.
β-Galactosidase Assays for Analysis of vir Gene Induction
vir gene induction was analyzed by triplicate measurements of β-galactosidase activity produced from a virB-lacZ fusion carried on the indicated plasmid. Overnight MG/L (Wise et al., 2006) cultures of Agrobacterium sp. were used to inoculate AB∗ (Wise et al., 2006) media containing 0.22% glycerol, 0.22% glucose, and/or AS as indicated. AB∗ culture samples were assayed according to the method of Miller (1972). Graphed values are the average of triplicate assays with the standard derivative indicated by error bars.
VirA Receiver Domain Pull-Down Assays
The plasmid constructs used in the pull-down experiments are diagramed in Figure 4. NEB Express Iq cells carrying pAW184, pAW187, or pAW192 were grown in LB medium supplemented with 70 μg/ml ampicillin at 37°C to an OD600 = 0.5. Isopropyl-β-D-thio-galactopyranoside (Sigma) was added to 1 mM and incubation was continued at 25°C for 5 h. Cells were precipitated, washed and re-suspended in a lysis buffer consisting of 50 mM NaPO4 pH 7.6, 50 mM NaCl, 100 μg/ml lysozyme (Affymetrix), EDTA-free protease inhibitor (Roche), 20 μg/ml DNAse I, and 10 μg/ml RNAse A. Cells were opened with sonication, and lysates were cleared with centrifugation at 10 krpm for 10 min at 4°C. When desired, Ni-NTA agarose (Qiagen) was used to obtain purified proteins, as per Qiagen instructions. For pull-down experiments, purified protein and/or cleared lysate was mixed with anti-FLAG M2 affinity gel (Sigma) in a TBS buffer consisting of 50 mM Tris-HCl pH 7.5, 75 mM NaCl, 1 mM EDTA, and 0.8% TritonX-100. Incubation with mixing was done in 2 ml micro-centrifuge tubes overnight at 4°C. Following incubation, samples were centrifuged to precipitate the FLAG affinity gel. A supernatant sample was reserved from this initial precipitation. The affinity gel with bound proteins was then washed three times with TBS. To avoid collecting unbound proteins that precipitated along with the affinity gel, the gel was transferred to Pierce Spin cups with paper filter (Thermo-Scientific) for three additional washes. Protein was eluted from the affinity gel according to the method recommended by Sigma using SDS-PAGE sample buffer without a reducing agent (60 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 0.002% bromophenol blue).
Immunoblot Analysis
β-mercaptoethanol was added to pull-down eluates to 5%. Samples were heated 20 min at 85°C and equivalent volumes were loaded on a 15% SDS-polyacrylamide gel. Proteins were visualized by Western blot analysis with anti-RGS-His antibody (Qiagen), ECL anti-mouse horseradish peroxidase-linked antibody (GE Healthcare) and ECL Western Blotting Detection reagent (Amersham).
Results
Constitutive Expression of virAΔR Corrects the Null Phenotype of virAΔR Expressed from the Ti Plasmid
Replacement of wild type virA on pTi with virAΔR (AB400) creates a null phenotype, even though the strain carries wild type virG at the native location (Wise et al., 2010). In this case because virA and virG are under the control of their own auto-inducible promoters, the levels of both VirA and VirG are expected to be extremely low (e.g., see Lee et al., 1992; Wise et al., 2010). As shown previously, constitutive expression of virG in AB400 (Wise et al., 2010) or A136 (Chang and Winans, 1992) allows VirAΔR to function. Figure 2 shows that the null phenotype of AB400 can also be corrected by constitutive expression of virAΔR (pAW117). In fact, in the glycerol medium, vir gene expression was significantly higher with constitutively expressed virAΔR than with constitutively expressed virG (pYW47). In medium containing glucose and AS, vir gene expression was similar whether the cells constitutively expressed virG or virAΔR, with the aforementioned exception of constitutively expressed virG bypassing the requirement for AS. The phenotype resulting from constitutive expression of virAΔR contrasted with the phenotype previously observed when virAΔR is expressed from its native promoter on the same multi-copy vector in the virA deletion strain, A348-3 (Wise et al., 2010). In that case, virAΔR was inactive unless the media included both sugar and a phenolic inducer. The variable results indicate that the initial concentrations of VirA and VirG are prime determinants of the ultimate vir gene induction phenotype. A summary table of various strains, their genotypes and vir gene expression phenotypes is presented in Supplementary Table S2.
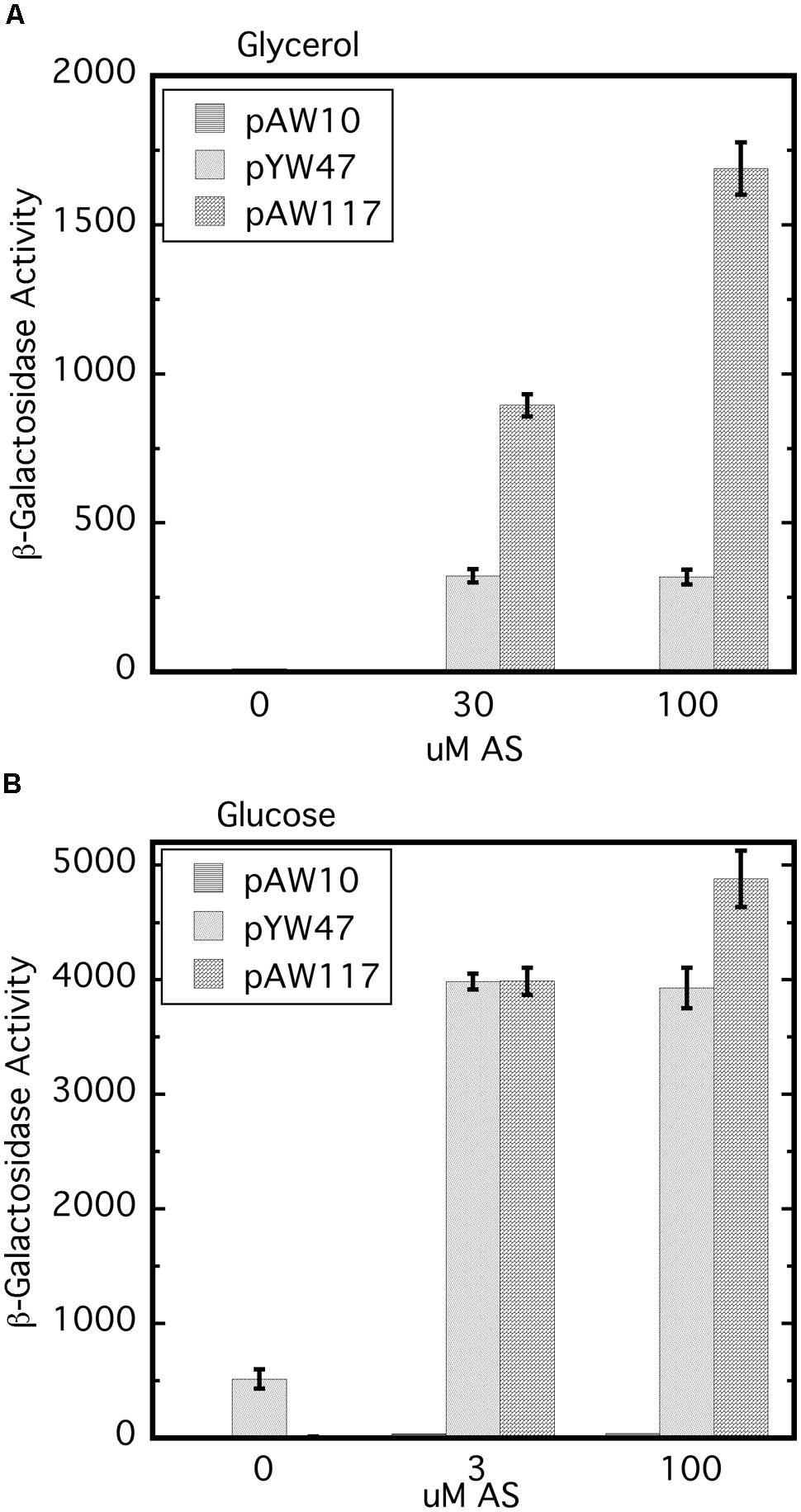
FIGURE 2. The null phenotype of AB400 (virAΔR on pTi) can be overcome by constitutive expression of either virAΔR or virG. AB400/pSW209Ω (virB-lacZ) carrying pAW10 (vector), pYW47 (PN25-virG) or pAW117 (PN25-virAΔR) were assayed for vir gene expression. (A) AB∗ induction medium included glycerol as the carbon source and 0, 30, or 100 μM AS. Student’s t-test indicates that pAW47 and pAW117 containing strains are different from one another (p > 0.95). (B) Induction medium included glucose in addition to glycerol and 0, 3, or 100 μM AS.
Expression of the VirA Receiver In Trans does not Correct the Null Phenotype of AB400
In vitro experiments have shown that the receiver domain of the Rhizobium leguminosarum hybrid kinase FixL is phosphorylated when expressed in trans to a version of FixL that carries an intact kinase, but no receiver (Boesten and Priefer, 2004). We reasoned that, if the receiver domain of VirA plays a role in phosphate relay to VirG, expression of the domain in trans could conceivably correct the null phenotype of AB400. However, constitutive expression of the receiver (pAW184) had no effect on vir gene expression in the AB400 background (Figure 3). The receiver domain expressed from pAW184 was shown to have biological activity through its ability to inhibit vir gene expression when included in the wild type background of A348 (see below). We concluded that the receiver must be attached to the remainder of the VirA protein in order to function as a positive factor for vir gene expression. This result does not disprove a role in phosphorylation for the receiver, but is consistent with a VirG recruiting function.
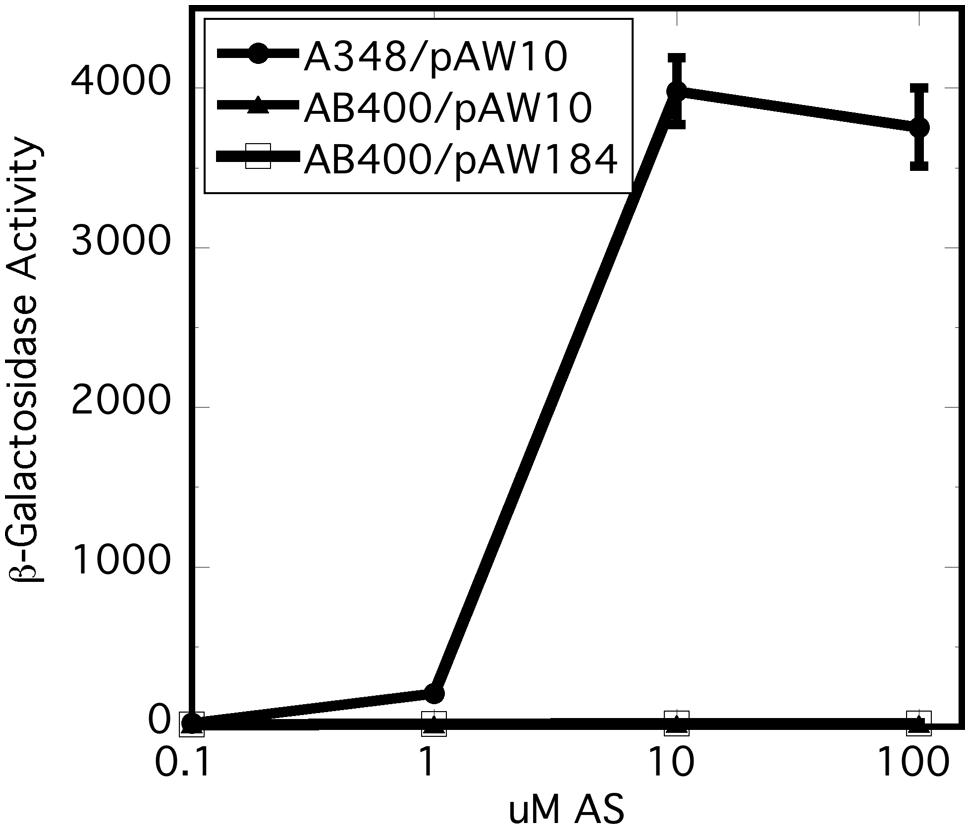
FIGURE 3. Expression of the virA receiver domain in trans did not correct the null phenotype of AB400. A348 carries wild type virG and wild type virA on pTi. AB400 carries wild type virG and virAΔR on pTi. pAW10 is an empty vector. pAW184 carries constitutively expressed virA receiver domain (see Figure 4). Cells were grown in AB∗ induction medium containing glucose and the indicated amounts of acetosyringone. Both AB400 and A348 carried the pSW209Ω (virB-lacZ) plasmid.
The VirA Receiver Domain Forms a Stable Complex with the VirG DNA-Binding Domain
virAΔR expressed from pTi created a situation where the proteins could not functionally interact unless the cellular content of one of the proteins was artificially increased (Figure 2). Several lines of evidence suggest that the receiver domain is not involved in a phospho-relay (Jin et al., 1990; Pazour et al., 1991; Mukhopadhyay et al., 2004; Wise et al., 2010). In addition, analysis indicates that hybrid histidine kinases have evolved independently through lateral recruitment (Zhang and Shi, 2005), suggesting possible functional variations. These considerations led to the hypothesis that the VirA receiver could act as a recruiting factor for VirG.
We tested this hypothesis with pull-down assays using the constructs shown in Figure 4. The three constructs, virA receiver domain (RA, pAW184), virG DNA-binding domain (DNABV G, pAW187), and the E. coli torR DNA-binding domain (DNABTR, pAW192) are all derivatives of pYW15b (Wang et al., 2000). TorR, like VirG, is included in the OmpR class of response regulators (Martinez-Hackert and Stock, 1997) and its DNA-binding domain was included as a control for non-specific binding. Each plasmid construct constitutively expresses the protein fragment with an RGS-6XHis tag at the N-terminus. In addition, the virA receiver construct is FLAG-tagged at its C-terminus. Cell lysates and, where indicated, Ni-NTA purified proteins were used in pull-down reactions that contained FLAG-affinity gel. (see Materials and Methods).
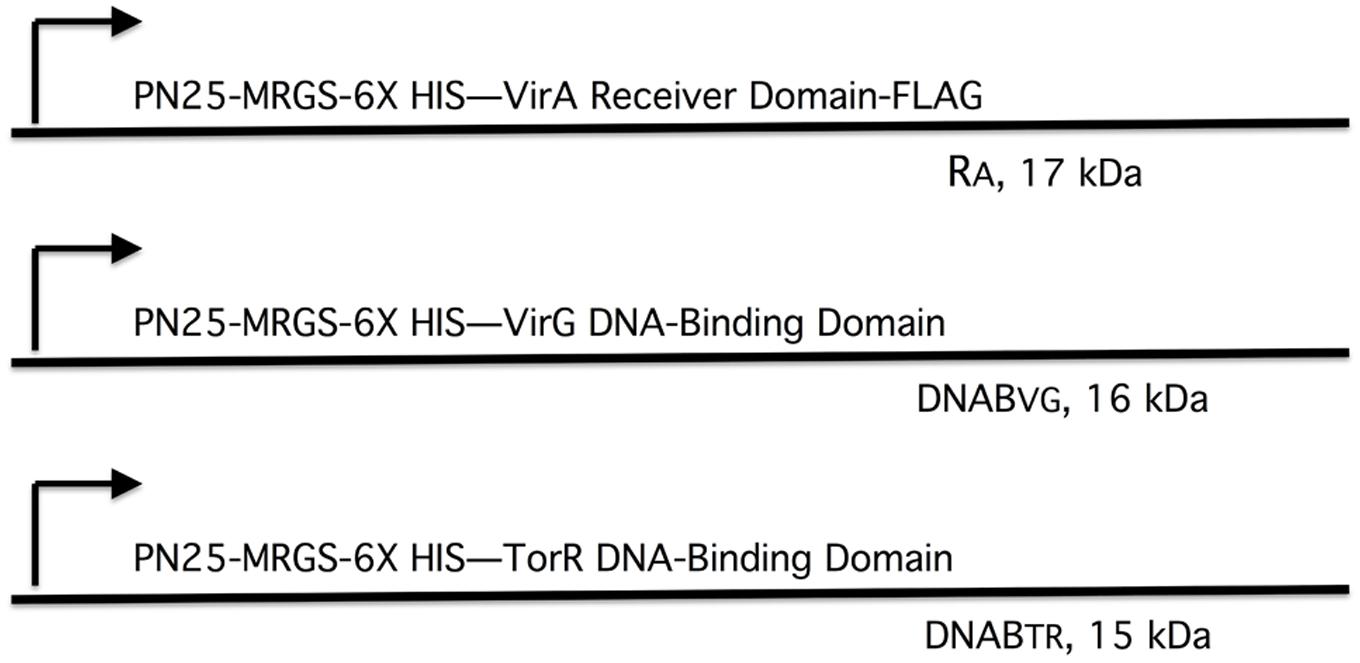
FIGURE 4. Plasmid constructs used in pull-down assays. The plasmids are derivatives of pYW15b (Wang et al., 2000). The VirA receiver domain (RA) is constitutively expressed from PN25 and includes a FLAG tag at the carboxyl end (pAW184). The VirG DNA-binding domain (DNABV G) and the TorR DNA-binding domain (DNABTR) are constitutively expressed from PN25 (pAW187 and pAW192, respectively). Each construct carries an RGS-6XHis tag near the amino terminus. The molecular weight, including epitope tags, is indicated for each protein fragment.
Figure 5 depicts the results of the pull-down experiments. In Figures 5A,B, the first three lanes (RA, DNABV G, and DNABTR) are purified proteins acting as size markers. The reaction loaded in lane 4 (Figure 5A) contained only lysate from cells that expressed the VirG DNA-binding domain (DNABV G). As expected, the FLAG affinity gel was unable to pull-down any protein from this reaction. Additional reactions, included in addition to lysate from cells expressing DNABV G, either lysate from cells expressing the VirA receiver domain (RA) or purified receiver domain (lanes 5 and 6, respectively). In both cases, the VirG DNA-binding domain was pulled down when the reaction mix included the VirA receiver domain. This was true despite extensive washing to remove proteins not bound to the affinity gel, indicating that the interaction between the VirA receiver and the VirG DNA-binding domain was stable under these conditions. A reaction that included lysate from cells expressing the TorR DNA-binding domain (DNABTR) along with the affinity gel (lane 7) pulled down no protein. The addition of VirA receiver domain lysate (lane 8) or purified receiver domain (lane 9) did not pull-down the TorR DNA-binding domain. Figure 5B shows supernatant from the initial precipitation (see Materials and Methods) of FLAG affinity gel following incubation. Supernatant loaded in lanes 7 and 9 indicate that purified RA was more limited in those reactions than RA present in whole lysates (lanes 5 and 8) and, thus only appears in the pull-down reaction, not the more dilute supernatants. Lanes 8, and 9 (Figure 5B) indicate that while DNABTR was included in the reactions containing RA, it remained in the supernatant. The fact that the VirA receiver did not bind to the DNA-binding domain of TorR indicates that the receiver has some specificity for the DNA-binding domain of its cognate response regulator VirG.
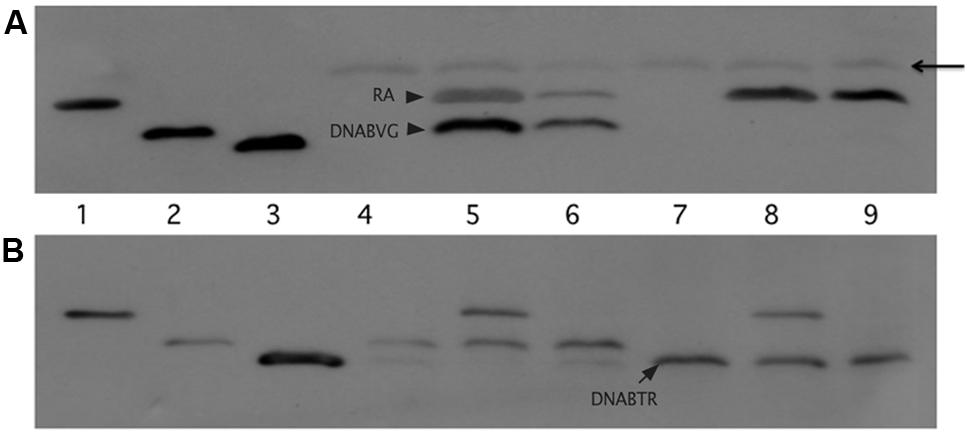
FIGURE 5. The VirA receiver domain binds to the VirG DNA-binding domain, but not to the DNA-binding domain of TorR. The first three lanes of both immunoblots show Ni-NTA purified proteins: lane 1, VirA receiver domain; lane 2 VirG DNA-binding domain; lane 3 TorR DNA-binding domain. (A) Top blot, lanes 4 through 9 were loaded with the eluates of the pull-down assays. Each reaction included equivalent amounts of FLAG-affinity gel and lysate from cells that produced the VirG DNA-binding domain from pAW187 (lanes 4, 5, and 6) or lysate from cells that produced the TorR DNA-binding domain from pAW192 (lanes 7, 8 and 9). Lanes 5 and 8 included lysate from cells that produced the VirA receiver domain (pAW184). Lanes 6 and 9 included lesser amounts of purified VirA receiver domain. The arrowhead indicates light chain antibody derived from the anti-FLAG affinity gel. (B) The lanes in the lower blot correspond to the lanes in blot (A), but lanes 4 through 9 were loaded with samples of supernatant recovered after the first precipitation of the affinity gel from the pull-down reactions, before washing. The supernatant samples show that significant amounts of DNABTR had been added to the pull-down reactions (A, lanes 7, 8 and 9) and remained in the supernatant (B, lanes 7, 8, and9).
Inhibition of vir Gene Expression by the Wild Type VirA Receiver is Stronger than Inhibition by the D766N Mutant Receiver in the A348 Wild Type Background
It has been reported previously that the D766N receiver mutation reduces vir gene expression by approximately 80% (Pazour et al., 1991). In our hands, VirAD766N expressed from a low copy vector (pAW221) in A348-3 (ΔvirA) reduced vir gene expression by 20–40% relative to wild type VirA (Figure 6A). The different results may be due to differences in plasmid copy number and strain variation. A348-3 carries wild type virG in the native position on the Ti plasmid whereas in the earlier work both virG and virA were expressed from a multi-copy plasmid in A136.
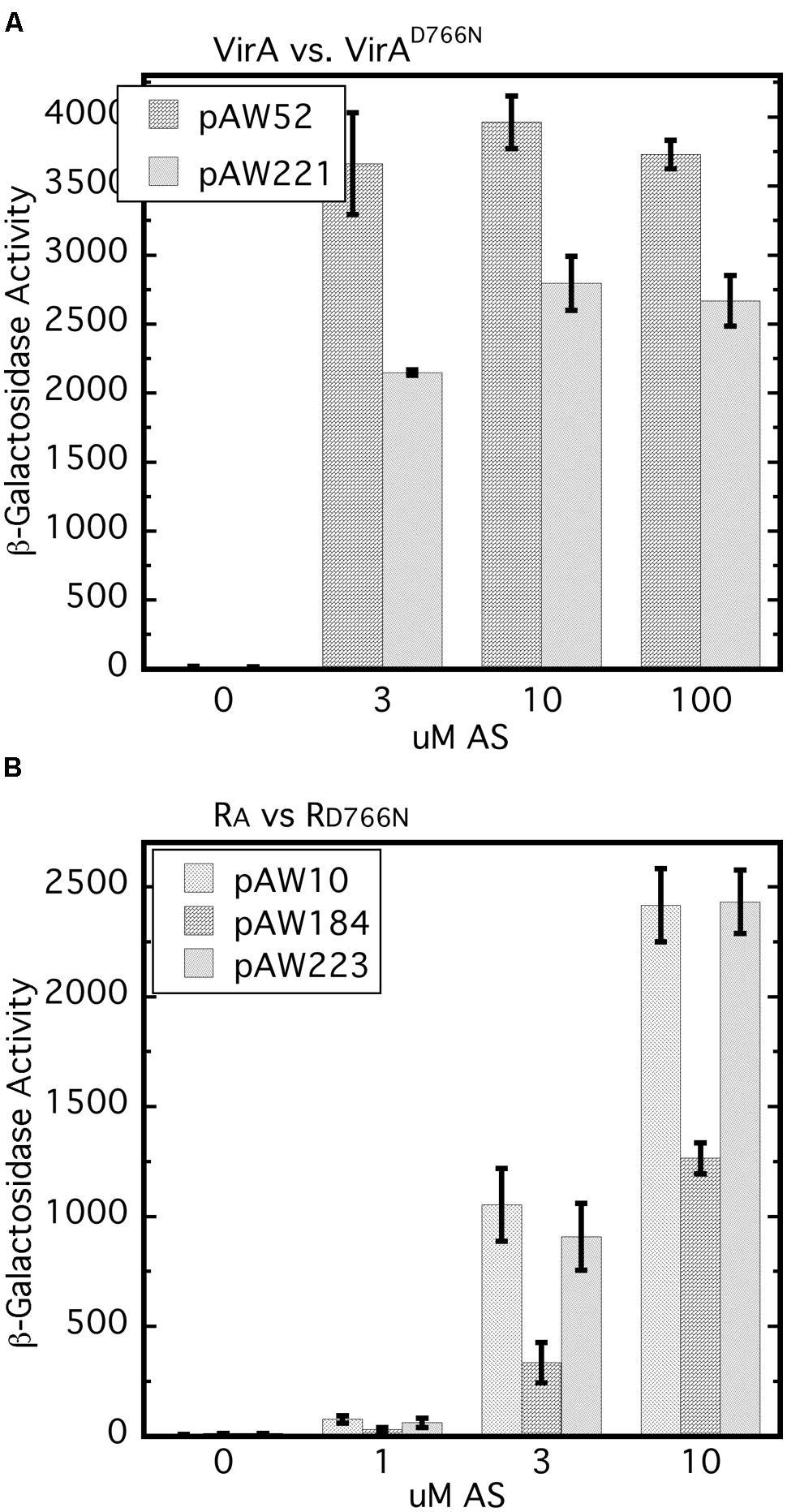
FIGURE 6. The D766N receiver mutation reduces vir gene expression and is less inhibitory than the wild type receiver. (A) vir gene expression that relies on VirAD766N is compared with that which depends on wild type VirA. A348-3 (ΔvirA) carrying pSW290Ω (virB-lacZ) and pAW52 (virA) or pAW221 (virAD766N) were grown in AB∗ induction medium with 0.22% glucose and the indicated amounts of AS. Student’s t-test showed that at 3, 10, and 30 μM AS VirAD766N is different than VirA (p > 0.95). (B) A348/pSW209Ω carrying pAW10 (vector), pAW184 (PN25-RA), or pAW223 (PN25-RD766N) was grown in AB∗ medium containing 0.22% glucose and AS. The Tukey test for these three strains indicated that A348/pSW209Ω/pAW184 is different than same strain with pAW10 or pAW223 (p > 0.95) and that pAW223 and pAW10 are not different (p > 0.95).
An earlier study showed that constitutive expression of the virA receiver domain in trans resulted in restoration of repression of vir gene expression in a strain carrying virAΔR and constitutively expressed virG (Chang et al., 1996). Moreover, this repressive trans activity of the receiver domain was observed when it carried the D766N mutation. Because our work has shown the importance of the initial concentration of VirG we compared vir gene expression in the wild type background (A348) for cells that constitutively expressed either the wild type receiver (pAW184) or RD766N (pAW223), or carried the empty vector pAW10. The wild type receiver consistently repressed vir gene expression in the wild type background (Figure 6B). In several repetitions of this assay, RD766N had modest but not significant repressive activity while being expressed at a similar level as the RA construct (Supplementary Figure S1). Thus the inhibitory effect, which presumably depends on the ability of RA to sequester VirG through interaction with its DNA-binding domain, appears to be dependent on D766N. Interestingly, and consistent with Chang et al. (1996), RD766N seemed almost as inhibitory as RA when A348 also carried PN25-virG (Supplementary Figure S2). Taken together these results further emphasize that the initial VirG concentration affects its interactions with the VirA receiver domain and the effects of that domain on vir gene expression.
Glucose Dose Response Assays Show that the VirA Receiver Domain Regulates Phosphate Transfer to VirG and Suggests that the ChvE-Dependent Sugar Signal Increases Auto-Phosphorylation
Removal of the receiver domain of VirA increases activity of that protein, provided the cellular content of VirG is increased (Chang and Winans, 1992; Chang et al., 1996; Brencic et al., 2004; Wise et al., 2010). We investigated the repressive nature of the receiver domain by comparing the activity of full-length wild type VirA with that of VirAΔR in the presence of constitutively expressed virG in a glucose dose response assay. Figure 7 represents the results of glucose dose response assays done with 0, 1, and 10 μM AS. For this experiment, A136 carried either pAW97 (PN25-virG, virAΔR, virB-lacZ) or pAW100 (PN25-virG, virA, virB-lacZ). In Figure 7A, cells were grown with increasing amounts of glucose, but no phenolic inducer. Predictably, full-length VirA was inactive, while VirAΔR-dependent vir gene expression increased with the concentration of glucose. When 1 μM AS was included in the growth medium, VirAΔR had some activity in the absence of sugar (0 mM glucose) and full-length VirA became functional as the concentration of glucose was increased (Figure 7B). Increasing the AS concentration to 10 μM further increased vir gene expression for both strains (Figure 7C). Although VirAΔR could respond to either the phenolic inducer or glucose, full-length VirA required sugar for activity at these relatively low concentrations of AS (1 or 10 μM). However, half maximal expression occurred between 1 and 3 mM glucose for VirAΔR, regardless of the presence of AS. Half maximal activity was also between 1 and 3 mM glucose for full-length VirA if AS was present in the assay medium. Thus, the half-maximal response to glucose appeared to be independent of AS concentration and the presence of the receiver, while the primary effect of increased AS concentration for both strains appeared to be an increase in maximum vir gene expression at a particular glucose concentration. These observations suggest that the ChvE-dependent sugar signal and the phenolic inducer affect two functions of the protein that are largely separate.
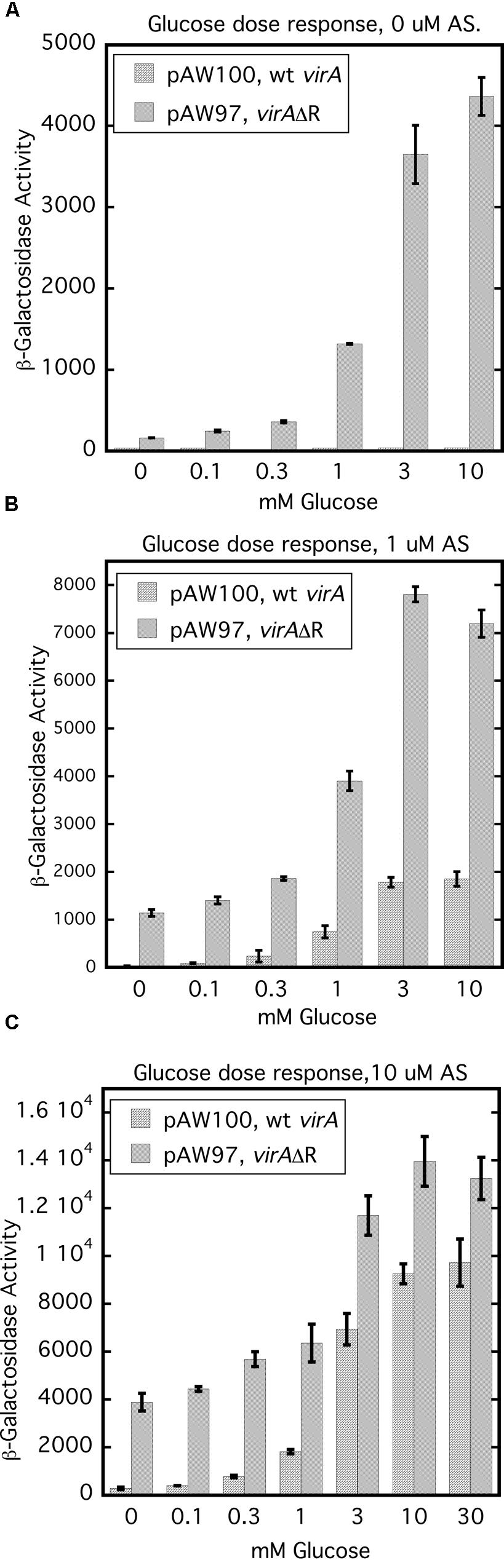
FIGURE 7. Glucose dose response assays indicate that the ChvE-dependent sugar signal affects a function of VirA that is partly independent from the effect of the phenolic inducer. A136 (no pTi) carrying either pAW100 (wt virA, PN25-virG, virB-lacZ) or pAW97 (virAΔR, PN25-virG, virB-lacZ) were assayed in AB∗ induction medium with increasing concentrations of glucose. (A) Induction media did not contain AS. (B) Induction media contained 1 μM AS. (C) Induction media contained 10 μM AS.
Discussion
Agrobacterium tightly controls expression of the virulence genes to conserve energy and resources until an appropriate host plant is detected. VirA and VirG are both essential for virulence, but their interdependent and self-regulated expression (Winans et al., 1988) means that these proteins are present in miniscule amounts in the early stages of induction (Lee et al., 1992; Wise et al., 2010). Removing the receiver domain from VirA, as in AB400, appears to create a situation where productive encounters between VirA and VirG are too infrequent to up-regulate the vir gene control system. The null phenotype of AB400 could be overcome by increasing the cellular content of either VirAΔR or VirG (Figure 2). This finding likely reflects a situation whereby increasing the concentration of either of the proteins permits them to interact with sufficient frequency to further induce expression of the virAΔR and virG genes located on the Ti plasmid. Early experiments identified the receiver domain of VirA as an inhibitory factor: the normally essential phenolic inducer was unnecessary for gene expression mediated by VirAΔR. Those experiments were done with cells that constitutively expressed virG (Chang and Winans, 1992). In contrast, vir gene expression that depended on PN25-virAΔR in the AB400 background (Figure 2) did require the phenolic inducer even with glucose in the medium. This difference may reflect the fact that overexpressed VirAΔR is membrane-bound with limited mobility compared to the cytoplasmic VirG protein.
Our pull-down assays indicated that the receiver domain of VirA forms a stable complex with the DNA-binding domain of VirG. This complex formation is the probable determinant of both the receiver’s positive effect on vir gene expression and the explanation for the inhibitory effect found when the receiver is expressed in trans. Mechanisms that contribute specifically to the interaction of cognate regulatory proteins have been more studied in eukaryotes (Pawson and Nash, 2003), but a few have been identified in prokaryotes. Prokaryotic kinases that carry domains that function specifically as recruiting or docking factors for their regulatory partners include RcsD (Schmöe et al., 2011), BglF (Lopian et al., 2003), CikA (Zhang et al., 2006), and CheA (McEnvoy et al., 1996).
The interaction between the VirA receiver domain and the VirG DNA-binding domain has similarities in the Rcs regulatory system. The RcsD protein transfers a phosphate from an Hpt domain to RcsB, the system’s response regulator. Efficient phosphate transfer is contingent on stabilization of the RcsD/RcsB interaction and that stabilization is achieved through the interaction of an RcsB structural domain (ABL) with the DNA-binding domain of the response regulator (Schmöe et al., 2011). Perhaps, the role of response regulator recruitment should be considered for other hybrid kinases.
Brencic et al. (2004) examined inhibition of vir gene expression in merodiploids that expressed full-length or VirAΔR alleles in strains that also carried versions of VirA that were constitutively active in the absence of a phenolic inducer. The authors concluded that inhibitory activity depended on the receiver domain. Here, we have looked at vir gene inhibition in the wild type background of A348. Expression of RA in trans had a distinct inhibitory effect in the wild type background of A348. We suspect that the inhibition seen by Brencic et al. (2004) and in Figure 6B is due to detached receiver protein interacting with the VirG DNA-binding domain, thereby reducing the availability of VirG for phosphorylation by an intact VirA protein. We hypothesize that transfer of a phosphate group from the VirA kinase to the VirG receiver domain would release full-length VirA from VirG and allow activated VirG to bind the vir gene promoters.
Interestingly, RD766N was less inhibitory than the wild type receiver in the wild type background of A348, but nearly equivalent when virG was expressed from the constitutive PN25 promoter, the latter result being consistent with those of Chang et al. (1996). In our assays the D766N mutation in full-length VirA expressed from a low copy plasmid has a consistent negative effect on vir gene expression in induction medium containing glucose. It may be that both the reduced vir gene expression mediated by VirAD766N and the reduced inhibition by RD766N in A348 stem from a slightly reduced affinity of those proteins for the VirG DNA-binding domain. However, we were unable to show a clear difference in the affinity of RA and RD766N for the DNA-binding domain of VirG using pull-down assays. The basis for the observation that the inhibitory capacity of RD766N was nearly equivalent to RA in the presence of constitutively expressed virG is, at this point, not clear. Perhaps the high levels of VirG may drive the formation of VirG/RD766N complexes thereby reducing the level of VirG available for phosphorylation by VirA, but further exploration of various models is needed.
The glucose dose response experiments (Figure 7) demonstrated that the concentration of AS had no effect on the half-maximum values for vir gene expression which was consistently between 1 and 3 mM glucose. These results imply that the ChvE-sugar signal influences a function of the VirA protein that is independent from the effect of the phenolic inducer. VirAΔR’s ability to activate VirG when phenolics are absent may reflect increased auto-phosphorylation stimulated by the sugar signal. Provided the cellular concentration of VirG is sufficiently elevated, the absence of the VirA receiver allows the phosphate group to be easily transferred to VirG. A ChvE-sugar dependent increase in auto-phosphorylation is also consistent with the strong increase in vir gene expression driven by wild type VirA in medium that contains both a phenolic inducer and an enhancing monosaccharide relative to the significantly lower activity observed if glycerol replaces the sugar as carbon source. In the case of full-length VirA, a phenolic inducer is required, apparently, to counter the obstruction of phosphate transfer that defines an inhibitory function of the receiver. An alternative model is that the phenolic signaling alters the ‘residence’ time of VirG on VirA, thereby affecting activation of VirG.
As a soil bacterium, Agrobacterium competes with numerous other microbes for nutrients. Soil can be a rich source of carbohydrates depending on regional micro-biota and the availability of root exudates (Kögel-Knabner, 2002). Agrobacterium carries redundant sugar transport systems that help in the competition for resources (Wood et al., 2001; Zhao and Binns, 2014). A variety of phenolics, derived from decaying lignin, are potential inducers of virulence and may also exist in soil. However, phenolics in the environment tend to be less stable than plant derived sugars (Spielvogel et al., 2007). As more than 20 genes are involved in the virulence process, it is in the best interest of the bacterium to strictly limit their induction to the plant environment where a productive infection may result in a nutritional source. Thus, the VirA receiver domain appears to have a role, not only in recruiting VirG, but also in limiting VirG activation to conditions that mimic those found at the site of a plant wound.
Funding
This work was funded by grants from the National Science Foundation IOS# 11211019 and the University of Pennsylvania.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank David Lynn, Mark Goulian, and their associated lab members for interesting and helpful discussions regarding VirA, VirG, and other two-component regulatory systems. We appreciate the technical support provided by Cush El.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01546
References
Ankenbauer, R. G., and Nester, E. W. (1990). Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccarides. J. Bacteriol. 172, 6442–6446.
Boesten, B., and Priefer, U. B. (2004). The C-terminal receiver domain of the Rhizobium leguminosarum bv. viciae FixL protein is required for free-living microaerobic induction of the fnrN promoter. Microbiology 150, 3703–3713. doi: 10.1099/mic.0.27323-0
Brencic, A., Xia, Q., and Winans, S. C. (2004). VirA of Agrobacterium tumefaciens is an intradimer transphosphorylase and can actively block vir gene expression in the absence of phenolic signals. Mol. Microbiol. 52, 1349–1362. doi: 10.1111/j.1365-2958.2004.04057.x
Cangelosi, G. A., Ankenbauer, R. G., and Nester, E. W. (1990). Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. U.S.A. 87, 6708–6712. doi: 10.1073/pnas.87.17.6708
Chang, C. H., and Winans, S. C. (1992). Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 174, 7033–7039.
Chang, C. H., Zhu, J., and Winans, S. C. (1996). Pleotropic phenotypes caused by genetic ablation of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 178, 4710–4716.
Gao, R., and Lynn, D. G. (2005). Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J. Bacteriol. 187, 2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005
Gao, R., and Lynn, D. G. (2007). Integration of rotation and piston motions in coiled-coil signal transduction. J. Bacteriol. 189, 6048–6056. doi: 10.1128/JB.00459-07
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Hu, X. Z., Zhao, J. L., DeGrado, W. F., and Binns, A. N. (2013). Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc. Natl. Acad. Sci. U.S.A. 110, 678–683. doi: 10.1073/pnas.1215033110
Jin, S., Prusti, R. K., Roitsch, T., Ankenbauer, R. G., and Nester, E. W. (1990). Phosphorylation of the VirG protein of Agrobacterium tumefaciens by the autophosphorylated VirA protein: essential role in biological activity of VirG. J. Bacteriol. 172, 4945–4950.
Kögel-Knabner, I. (2002). The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 34, 139–162. doi: 10.1016/S0038-0717(01)00158-4
Lee, K., Dudley, M. W., Hess, K. M., Lynn, D. G., Joerger, R. D., and Binns, A. N. (1992). Mechanism of activation of Agrobacterium virulence genes: identification of phenol binding proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 8666–8670. doi: 10.1073/pnas.89.18.8666
Lin, Y. H., Pierce, B. D., Fang, F., Wise, A. A., Binns, A. N., and Lynn, D. G. (2014). Role of the VirA histidine autokinase of Agrobacterium tumefaciens in the initial steps of pathogenesis. Front. Plant Sci. 5:195. doi: 10.3389/fpls.2014.00195
Lopian, L., Nussbaum-Shochat, A., O’Day-Kerstein, K., Wright, A., and Amster-Choder, O. (2003). The BglF sensor recruits the BglG transcription regulator to the membrane and releases it on stimulation. Proc. Natl. Acad. Sci. U.S.A. 100, 7099–7104. doi: 10.1073/pnas.1037608100
Martinez-Hackert, E., and Stock, A. M. (1997). Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269, 301–312. doi: 10.1006/jmbi.1997.1065
McCullen, C. A., and Binns, A. N. (2006). Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22, 101–127. doi: 10.1146/annurev.cellbio.22.011105.102022
McEnvoy, M. M., Muhandiram, D. R., Kay, L. E., and Dahlquist, F. W. (1996). Structure and dynamics of a CheY-binding domain of the chemotaxis kinase CheA determined by nuclear magnetic resonance spectroscopy. Biochemistry 35, 5633–5640. doi: 10.1021/bi952707h
Miller, J. H. (1972). Experiments in Molecular Genetics. (Cold Springs Harbor, NY: Cold Springs Harbor Laboratory Press), 352–355.
Mukhopadhyay, A., Gao, R., and Lynn, D. G. (2004). Integrating input from multiple signals: the VirA/VirG two-component systen of Agrobacterium tumefaciens. ChemBioChem 5, 1535–1542. doi: 10.1002/cbic.200300828
Pawson, T., and Nash, P. (2003). Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452. doi: 10.1126/science.1083653
Pazour, G. J., Ta, C. N., and Das, A. (1991). Mutants of Agrobacterium tumefaciens with elevated vir gene expression. Proc. Natl. Acad. Sci. U.S.A. 88, 6941–6945. doi: 10.1073/pnas.88.16.6941
Schmöe, K., Rogov, V. V., Rogova, N. Y., Löhr, F., Göhr, F., Gütert, P., et al. (2011). Structural insights into Rcs phosphotransfer: the newly identified RcsD-ABL domain enhances interaction with the response regulator RcsB. Structure 19, 557–587. doi: 10.1016/j.str.2011.01.012
Shimoda, N., Toyoda-Yamamoto, A., Aoki, S., and Machida, Y. (1993). Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J. Biol. Chem. 268, 26552–26558.
Simon, G., Méjean, V., Jourlin, C., Chippaux, M., and Pascal, M. C. (1994). The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J. Bacteriol. 176, 5601–5606.
Spielvogel, S., Prietzel, J., and Kögel-Knabner, I. (2007). Changes of lignin phenols and neutral sugars in different soil types of a high-elevation forest ecosystem 25 years after forest dieback. Soil Biol. Biochem. 39, 655–668. doi: 10.1016/j.soilbio.2006.09.018
Wang, Y., Mukhophadyhyay, A., Horvitz, V. R., Binns, A. N., and Lynn, D. N. (2000). Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene 242, 105–114. doi: 10.1016/S0378-1119(99)00541-7
Watson, B., Currier, T. C., Gordon, M. P., Chilton, M. D., and Nester, E. W. (1975). Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 123, 255–264.
Winans, S. C., Kerstetter, R. A., and Nester, E. W. (1988). Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J. Bacteriol. 170, 4047–4054.
Wise, A. A., Fang, F., Lin, Y. H., He, F., Lynn, D. G., and Binns, A. N. (2010). The receiver domain of hybrid histidine kinase VirA: an enhancing factor for vir gene expression in Agrobacterium tumefaciens. J. Bacteriol. 192, 1534–1542. doi: 10.1128/JB.01007-09
Wise, A. A., Liu, Z., and Binns, A. N. (2006). “Culture and Maintenance,” in Agrobacterium Protocols, 2nd Edn, Vol. 1, ed. K. Wang (Totowa, NJ: Humana Press), 3–13.
Wise, A. A., Voinov, L., and Binns, A. N. (2005). Intersubunit complementation of sugar signal transduction in VirA heterodimers and posttranslational regulation of VirA activity in Agrobacterium tumefaciens. J. Bacteriol. 187, 213–223. doi: 10.1128/JB.187.1.213-223.2005
Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323. doi: 10.1126/science.1066804
Zhang, W., and Shi, L. (2005). Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid histidine kinases. Microbiology 151, 2159–2173. doi: 10.1099/mic.0.27987-0
Zhang, X., Dong, G., and Golden, S. S. (2006). The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol. Microbiol. 60, 658–668. doi: 10.1111/j.1365-2958.2006.05138.x
Keywords: hybrid histidine kinase, response regulator, receiver domain, signal transduction, Agrobacterium, plant pathogenesis
Citation: Wise AA and Binns AN (2016) The Receiver of the Agrobacterium tumefaciens VirA Histidine Kinase Forms a Stable Interaction with VirG to Activate Virulence Gene Expression. Front. Microbiol. 6:1546. doi: 10.3389/fmicb.2015.01546
Received: 28 August 2015; Accepted: 21 December 2015;
Published: 08 January 2016.
Edited by:
Mark Findlay Belmonte, University of Manitoba, CanadaReviewed by:
Melanie J. Filiatrault, United States Department of Agriculture – Agricultural Research Service, USALois Banta, Williams College, USA
Copyright © 2016 Wise and Binns. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew N. Binns, abinns@sas.upenn.edu
 Arlene A. Wise
Arlene A. Wise Andrew N. Binns
Andrew N. Binns