- 1Biotechnology Research Institute, Kenya Agricultural and Livestock Research Organization, Nairobi, Kenya
- 2Insect Pest Control Laboratory, Joint FAO/IAEA Programme of Nuclear Techniques in Food and Agriculture, International Atomic Energy Agency, Vienna, Austria
- 3Laboratory of Virology, Wageningen University, Wageningen, Netherlands
- 4Department of Medical Microbiology, Acıbadem University, İstanbul, Turkey
- 5Laboratory of Biochemistry, Wageningen University, Wageningen, Netherlands
- 6South African National Bioinformatics Institute, University of the Western Cape, Cape Town, South Africa
- 7Department of Biochemistry, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya
Glossina pallidipes salivary gland hypertrophy virus (GpSGHV; family Hytrosaviridae) is a dsDNA virus exclusively pathogenic to tsetse flies (Diptera; Glossinidae). The 190 kb GpSGHV genome contains 160 open reading frames and encodes more than 60 confirmed proteins. The asymptomatic GpSGHV infection in flies can convert to symptomatic infection that is characterized by overt salivary gland hypertrophy (SGH). Flies with SGH show reduced general fitness and reproductive dysfunction. Although the occurrence of SGH is an exception rather than the rule, G. pallidipes is thought to be the most susceptible to expression of overt SGH symptoms compared to other Glossina species that are largely asymptomatic. Although Glossina salivary glands (SGs) play an essential role in GpSGHV transmission, the functions of the salivary components during the virus infection are poorly understood. In this study, we used mass spectrometry to study SG proteomes of G. pallidipes and G. m. morsitans, two Glossina model species that exhibit differential GpSGHV pathologies (high and low incidence of SGH, respectively). A total of 540 host proteins were identified, of which 23 and 9 proteins were significantly up- and down-regulated, respectively, in G. pallidipes compared to G. m. morsitans. Whereas 58 GpSGHV proteins were detected in G. pallidipes F1 progenies, only 5 viral proteins were detected in G. m. morsitans. Unlike in G. pallidipes, qPCR assay did not show any significant increase in virus titers in G. m. morsitans F1 progenies, confirming that G. m. morsitans is less susceptible to GpSGHV infection and replication compared to G. pallidipes. Based on our results, we speculate that in the case of G. pallidipes, GpSGHV employs a repertoire of host intracellular signaling pathways for successful infection. In the case of G. m. morsitans, antiviral responses appeared to be dominant. These results are useful for designing additional tools to investigate the Glossina-GpSGHV interactions.
Introduction
Glossina pallidipes salivary gland hypertrophy virus (GpSGHV; family Hytrosaviridae) is a dsDNA virus whose 190 kb genome encodes more than 60 confirmed proteins (Abd-Alla et al., 2008, 2009b; Kariithi et al., 2013a). The Hytrosaviridae family consists of only one other member, the housefly Musca domestica (Diptera; Muscidae) hytrosavirus (MdSGHV; Coler et al., 1993). However, detection of hytrosavirus-like infection symptoms, i.e., the salivary gland hypertrophy syndrome (SGH) in the Narcissus bulb fly Merodon equestris (Diptera; Syrphidae; Amargier et al., 1979) and in male accessory gland filaments of the parasitic wasp Diachasmimorpha longicuadata (Hymenoptera; Braconidae; Luo and Zeng, 2010) implies that the Hytrosaviridae potentially contains other members. The intrinsic properties of hytrosaviruses, i.e., covert chronic infection of adult stages without expression of detectable SGH symptoms, have probably hindered the discovery of other Hytrosaviridae family members up until now. GpSGHV is exclusively pathogenic to the tsetse fly (Diptera; Glossinidae), the vector of a group of neglected tropical diseases called the African trypanosomiases (Mattioli et al., 2004). Research on GpSGHV pathobiology has been hindered by a lack of an in vitro cell culture system to support the virus replication (Abd-Alla et al., 2011a). Attempts to multiply GpSGHV in alternative insect hosts such as M. domestica have so far been unsuccessful. The only available method to multiply GpSGHV is via intra-hemocoelic injections of virus suspension in G. pallidipes (Kariithi et al., 2013b).
A mature GpSGHV virion contains four distinct structural components (nucleocapsid core, tegument, envelope, and helical surface projections) composed of 61 virally-encoded proteins (Kariithi et al., 2010). The GpSGHV virion also contains 51 host-derived cellular proteins: some are incorporated into the virus particles and may play roles in virus replication and transmission (Kariithi et al., 2013a,b). In G. pallidipes, GpSGHV is transmitted horizontally via saliva during feeding (Abd-Alla et al., 2010) and vertically (transovarial) via the fat body tracheal system and milk gland secretions (Boucias et al., 2013). GpSGHV infection in laboratory colonies of G. pallidipes can either be asymptomatic or symptomatic with the former being the most rampant in laboratory colonies of this tsetse species (Abd-Alla et al., 2010). However, the asymptomatic infection state can convert to a symptomatic state, leading to reproductive dysfunction and reduced fecundity in addition to SGH symptoms (Abd-Alla et al., 2007; Lietze et al., 2011; Boucias et al., 2013). More than 40% of salivary gland (SG) proteins appear to be specifically expressed in G. pallidipes flies with overt SGH symptoms but not in asymptomatic flies (Kariithi et al., 2011). Unlike in the laboratory tsetse fly colonies, GpSGHV infection is mainly covert (latent) in wild G. pallidipes populations. Occurrence of SGH symptoms have been reported in other Glossina species such as G. m. morsitans (Jura et al., 1993) and G. m. centralis (Sang et al., 1997). However, SGH symptoms are rare especially in species other than G. pallidipes. Notably, even in G. pallidipes the occurrence of SGH symptoms is an exception rather than the rule (Boucias et al., 2013). The pathobiology of GpSGHV in species other than G. pallidipes has not been so far investigated.
Whether naturally or artificially infected, the GpSGHV infection rate is low, but males are more susceptible to infections compared to females (Abd-Alla et al., 2007; Boucias et al., 2013). After acquisition through a blood meal, GpSGHV translocates to the SGs where it primarily replicates (Garcia-Maruniak et al., 2009). In G. pallidipes, intra-hemocoelic GpSGHV injection leads to significant increase in the viral titters in the whole fly, but the injected virus is not released via saliva during feeding and there is no development of overt SGH symptoms (Boucias et al., 2013). Rather, SGH symptoms are overt in the F1 progenies of the infected mothers. It is yet to be confirmed in which host tissues GpSGHV replicates after artificial injection. However, the current school of thought is that in naturally-infected G. pallidipes, the virus replicates in the male reproductive accessory glands (Sang et al., 1999) and the gut (Sang et al., 1997) but without any teratogenic effects.
The pathological, morphological and ultrastructural effects of GpSGHV infection in G. pallidipes SGs have been studied to considerable length (Kariithi et al., 2011, 2013a; Guerra et al., 2013). However, no such studies have been performed in other Glossina species. Further, the molecular basis for the differential GpSGHV pathology in different Glossina species is still unclear. Here, we investigated GpSGHV-induced modulation of total protein expression in the SGs of G. pallidipes and G. m. morsitans, with special emphasis on the host pathways that are potentially employed by the virus during infection. We hypothesized that GpSGHV infection in Glossina is under the control of host-and/or virus-encoded factors (proteins/peptides) whose interactions influence the expression or lack of overt SGH symptoms. We tested the hypothesis by comparing the SG proteomes of GpSGHV-infected vs. mock-infected G. pallidipes and G. m. morsitans flies. The host (and viral) proteins identified in this study are potential targets for control of GpSGHV infections in tsetse fly mass production facilities. For instance, antiviral strategies could be developed to block virus replication and egress (Esfandiarei et al., 2006; Cheshenko et al., 2010; Chen et al., 2011), prevent the establishment of virus replication complexes (Saxena et al., 2012) and prevent development of cellular proliferation (Guergnon et al., 2011). Such antiviral approaches are applicable in the control of virus infections in mass production of other insects.
Materials and Methods
Tsetse Flies
The G. m. morsitans and G. pallidipes flies used in this study were obtained from a colony maintained at the Joint FAO/IAEA Insect Pest Control Laboratories (IPCL), Seibersdorf, Austria. For each treatment described below, groups of experimental flies were kept in holding cages (diameter of 20 cm and height of 5 cm) at a density of 75 flies per cage and a mating ratio of 1:4 (male: female). The holding cages had netting on top and bottom for fly feeding and pupae collection, respectively. The experimental flies were reared at 23 ± 1°C, 75–80% relative humidity, 12 h scotophase and fed on defibrinated bovine blood meals (15–20 min; 3 times per week; Feldmann, 1994). The pupae from the sequential larviposition cycles were collected and incubated at 24°C until eclosion of the adult F1 progenies. For further analyses, male F1 progenies were selected from the fourth larviposition cycle (G4) based on available data that the incidence of SGH symptoms reaches 100% at the G4 (Boucias et al., 2013). It should be noted that males were used because they are significantly more susceptible to expression of SGH symptoms than the females (Abd-Alla et al., 2007). To allow for development of SGH symptoms, the selected F1 male progenies were reared for 4 weeks (equivalent to 12 blood meals) under the same insectaria conditions and handled as the parents. All the treatments described here were replicated at least three times.
Preparation of GpSGHV Inoculum and Injections of Tsetse Flies
To prepare the GpSGHV inoculum, one intact pair of SGs displaying overt SGH symptoms were dissected from an adult (10-day old) male G. pallidipes fly and stored in 1 ml of ice-cold sterile saline (pH 7.4). The SGs were then homogenized and clarified by brief centrifugation (500 × g; 3 min; 4°) to remove tissue debris. The supernatants were sterilized by passing through a 0.45-μm filter unit and the virus titters present in the filtrate were estimated by a quantitative polymerase chain reaction (qPCR) as described by Abd-Alla et al. (2009a). By this qPCR method, an average of 1 × 106 virus copies were estimated to be present in a 2 μl aliquot of the virus preparation and was used for tsetse fly injections. For infections, teneral (newly eclosed; non-fed) female G. m. morsitans and G. pallidipes flies were artificially (intra-hemocoelic) injected with the virus preparations using a protocol described by Boucias et al. (2013). Briefly, the female flies selected from the colony as described above were inoculated with either 2 μl of the virus inoculum or 2 μl of filter-sterilized PBS (mock infections). Following the injections, the females were mated with asymptomatic males; these females were then separated from the males and subsequently maintained in the insectary until they produced the F1 progenies as described above.
Detection of Viral DNA in Infected G. pallidipes and G. m. morsitans Flies
To confirm GpSGHV infections, the 4 week-old F1 male progenies produced by the mock- and virus-infected mothers were screened using a diagnostic PCR protocol described by Abd-Alla et al. (2007). For this, genomic DNA was extracted from one intermediate excised leg of individual flies using DNeasy Tissue Kit (QIAGEN Inc., Valencia, CA). PCR amplifications were performed using primers and conditions previously described (Abd-Alla et al., 2007, 2011b), and the PCR products analyzed on 1% agarose gels. Flies were considered to be non-infected, moderately-infected or highly infected if there were no visible bands or showed faint bands or thick bands, respectively, on agarose gels as previously described (Abd-Alla et al., 2010). The actual virus copy numbers and the virus density levels were determined using qPCR essentially as described by Abd-Alla et al. (2009b). For virus density levels, the qPCR data were normalized against the tsetse β-tubulin gene (Wang et al., 2013). The samples with high virus infections (based on the agarose gels) were subsequently used for mass spectrometry measurements as virus-infected samples. This cut off was especially used in the case of G. m. morsitans flies, which do not usually show overt SGH symptoms. For G. pallidipes, samples with overt SGH symptoms (corresponding to samples with high infections from the agarose gels) were selected for mass spectrometry measurements. For negative controls, samples from 4-week-old male flies were selected from the mock-infected fly groups (confirmed to be PCR-negative). Ten flies from each of the replicated treatments described above were selected for subsequent SG dissections.
Preparation of SG Protein Extracts
To prepare protein extracts, SGs from the F1 progenies described above were dissected 2 days after the flies had their last blood meals to allow for full digestions (Abd-Alla et al., 2009a). From each of the selected flies, intact pairs of SGs were individually dissected, during which the occurrence of SGH symptoms was assessed. The dissected SGs were preserved (at 4°C) in 150 μl sterile saline complemented with protease inhibitors (Roche Diagnostics, Germany). Then, each of the pool of 10 pairs of SGs were homogenized using a glass/Teflon homogenizer and ultra-sonicated (Sonifier cell disruptor, Branson, CT, USA) as described by Kariithi et al. (2013a). The homogenates were freeze-thawed and clarified three times by centrifugation (7500 × g; 10 min; 4°C). The supernatants were pooled and the proteins quantified using BCA Protein Assay (Bio-Rad) according to manufacturer's instructions. Then, equal quantities (600 ng) of the proteins (from each of the pooled 10 SGs per each of the three replicates) were electrophoresed using 12% SDS-PAGE gels (Invitrogen) as described by Green and Sambrook (2012). The gels were stained with colloidal CBB stain (NuPAGE Novex; Invitrogen). The middle sections of entire gel lanes were excised, and each of the gel lanes was divided into eight slices (equal portions) each of which was cut into approximately 1 mm3 pieces.
Mass Spectrometry and Identification of SG Proteins
To prepare peptides for mass spectrometry measurements, the gel slices containing the SG proteins were subjected to in-gel trypsin digestions as previously described (Kariithi et al., 2013a). Briefly, after washing the gel pieces with 50 mM ammonium bicarbonate (ABC) buffer and ABC buffer/50% (vol/vol) acetonitrile (ACN), proteins were reduced and alkylated using dithiothreitol and iodoacetamide, followed by washing with ABC/ABC-ACN buffers and trypsin digestions. Tryptic peptides were then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS; Lu et al., 2011). To identify the SG proteins, the MS/MS spectra obtained from the LC-MS/MS measurements were searched against a tsetse fly database, a GpSGHV database, a contaminant database containing sequences of common contaminants, and a decoy database constructed by reversing all protein sequences downloaded from UniProt. The MS searches were performed using MaxQuant v 1.3.0.5 (Cox and Mann, 2008) and Andromeda as the database search engine (Cox et al., 2011). A maximum false discovery rate (FDR) of less than 0.01 was set at the peptide and protein levels. MaxQuant search parameters included variable oxidation of M, fixed carboxamidomethylation of C, and extra variable modifications for de-amidation of N and Q. “Label-free quantification” (LFQ) and “match between runs” (set to 2 min) options were enabled. De-amidated peptides were allowed to be used for protein quantification.
Quantification and Characterization of SG Proteins
To quantify the SG proteins, the resulting MaxQuant protein list was filtered to show only those proteins with a minimum of two peptides matching the same protein, of which at least one peptide was unique and unmodified. All other quantification settings were set at default. Any hits to the decoy sequences and hits with modified peptides only were deleted from the list of protein/peptides groups. To easily compare abundances of the same proteins between the controls and virus-infected samples, logarithms (Log10) of normalized LFQs were used. Logarithms of the total intensity corrected for the number of measurable peptides (i.e., intensity based absolute quantization; iBAQ) were used to compare the levels of different proteins from the same sample (mocks vs. GpSGHV-infected; Schwanhausser et al., 2011). Proteins were considered to be up- or down-regulated when their Log10 protein abundance ratios were larger or smaller than zero, respectively. Proteins were considered significantly up-regulated when their Log10 protein abundance ratios were larger than six. Gene Ontology annotation of the identified proteins were created using Blast2GO v 3.0.4 (Conesa et al., 2005). Analyses of the protein motif/domain were performed using various bioinformatics softwares, including SMART (Schultz et al., 1998) and InterProScan (Zdobnov and Apweiler, 2001).
Results
Detection of Viral DNA in Infected G. pallidipes and G. m. morsitans Flies
We first analyzed GpSGHV replication in the F1 progenies from virus-infected mothers. We detected varying levels of GpSGHV infections in G. m. morsitans flies: non-infected, moderately infected and highly infected as evidenced by the absence of bands, faint bands and thick bands, respectively, in the agarose gels presented in the Figure 1A. The different GpSGHV infection levels we obtained for G. m. morsitans in Figure 1 were comparable to the results obtained for G. pallidipes, as well as results from our previous studies in G. pallidipes (compare with Figure 1 in Abd-Alla et al., 2010). qPCR analysis of the samples in the “highly infected” category revealed high virus genome copies in G. m. morsitans (Figure 1B). When dissected however, none of these F1 progenies from GpSGHV-infected G. m. morsitans mothers showed any SGH symptoms. This is unlike in the G. pallidipes F1 progenies, which revealed 100% prevalence of SGH symptoms (data not shown), a result which was in agreement with our recent report (Boucias et al., 2013). Notably, the GpSGHV density levels in F1 progenies of G. pallidipes were significantly higher (P = 0.0014) compared to the virus-infected G. m. morsitans F1 progenies (Figure 1C). Importantly, the virus density levels in both the mock- and the virus-infected G. m. morsitans F1 progenies were comparable to those of the mock-infected G. pallidipes (see Figure 1C).
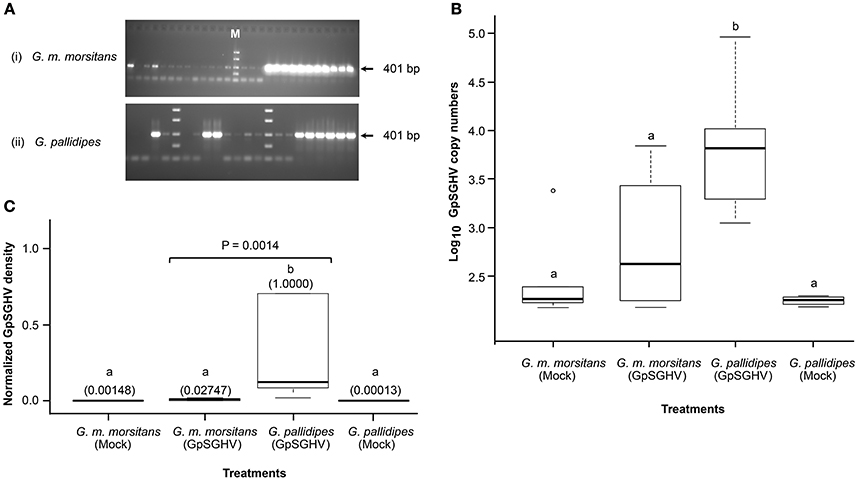
Figure 1. Detection of GpSGHV infections in experimental flies. (A) Sections of agarose gels used to analyze GpSGHV infections in virus-infected G. m. morsitans (i) and G. pallidipes flies (ii) using a diagnostic PCR protocol (Abd-Alla et al., 2007). Shown are non-infected samples (no bands), moderately-infected samples (faint bands) and highly infected samples (thick bands). M is molecular marker. The PCR amplifications were performed using genomic DNA extracted from single intermediate legs excised from 4-week old male F1 progenies produced by the control (Mock), or virus-infected (GpSGHV) flies. Determination of the GpSGHV copy numbers and the virus density levels by qPCR are shown in (B,C), respectively. For determination of virus copy numbers (B), 10-fold serially diluted viral DNA (targeting odv-e66 gene) were used as internal standards as described by Abd-Alla et al., 2009a. For determination of the virus expression levels, qPCR data were normalized using a tsetse fly housekeeping gene (β-tubulin). Viral density levels in the virus-infected G. pallidipes progenies were significantly higher (P = 0.0014) than the levels in the virus-infected G. m. morsitans flies. The values in the parentheses (C) indicate the virus density levels. Letters a and b represent significant differences between the samples (i.e., there was no significant difference between samples labeled a, while a and b were significantly different).
Determination and Characterization of Glossina SG Proteomes
We then performed mass spectrometry on the SG protein extracts from the F1 progenies of G. m. morsitans flies with high viral titters and the G. pallidipes flies with overt SGH symptoms. Analyses of the GpSGHV-infected G. m. morsitans and G. pallidipes proteomes compared to their mock-infected counterparts resulted in 3815 unique peptides that mapped to 863 non-redundant proteins. Of these proteins, 87.7% (n = 757) were host (Glossina)-specific, while 8.5% (n = 73) and 3.8% (n = 32) were from GpSGHV and bacterial endosymbionts (Wigglesworthia glossinidia and Sodalis glossinidius), respectively. We then filtered out proteins with single and modified peptides, which resulted in 606 proteins, of which 540, 58, and 9 proteins were from the host, GpSGHV and W. glossinidia, respectively. All the identified proteins are detailed in Supplementary Material (Tables S1–S8).
Effects of GpSGHV Infections on the Overall SG Protein Expression Patterns
When we compared the LC-MS/MS data sets obtained from the GpSGHV-infected to mock-infected SGs, we found clear differential protein expression patterns in response to the virus infections in G. pallidipes and G. m. morsitans vs. their respective mock-infected controls (compare Figure 2 and Figure 3). From these two figures, GpSGHV infection had more drastic effects in the protein expression in the SGs of G. pallidipes than that of the G. m. morsitans. In the Figure 2, GpSGHV infection in G. m. morsitans had little overall effects on the host's SG protein expression patterns, i.e., majority of the proteins (confidently identified by ≥2 unique peptides per protein) were aligned around the y-axis (circled). On the other hand, a cohort of SG proteins were detectable in the proteome of GpSGHV infected G. pallidipes, but were hardly detectable in the proteome of the mock-infected flies (see circled proteins in Figure 3). Notably, in contrast to the G. m. morsitans SG proteins (Figure 2), only few proteins were not significantly affected by GpSGHV-infection in G. pallidipes (see proteins along the y-axis in Figure 3). Overall, comparing the GpSGHV-infected flies to their mock-infected counterparts, the majority of the host's SG proteins in G. m. morsitans had less than 10-fold up- or down-regulation compared to the proteome of G. pallidipes (see dotted red lines in Figures 2, 3).
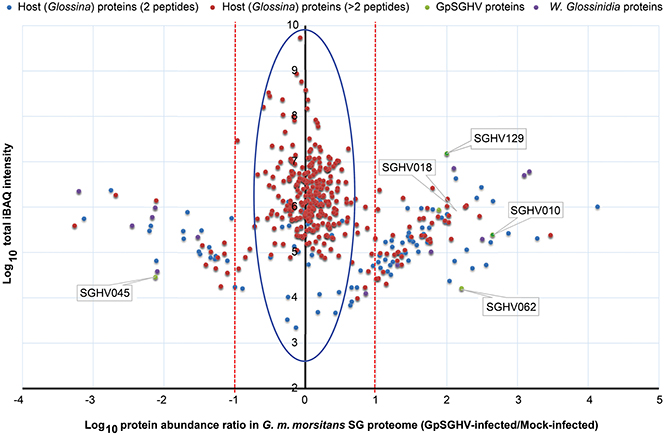
Figure 2. Abundance distribution ratios of G. m. morsitans SG proteins. The figure depicts the distribution of proteins detected in the SG proteome of G. m. morsitans infected by GpSGHV compared to the mock-infected controls. Shown are the host proteins that were detected by two (light blue) or more (red) peptides per protein. GpSGHV and Wigglesworthia glossinidia proteins are shown in green and purple, respectively. The proteins that were up-regulated and down-regulated in GpSGHV-infected SG are shown on the right and left sides of the Y-axis, respectively (in the blue large circle). The dotted red lines represent 10-fold protein regulation. iBAQ denotes intensity-based absolute quantification.
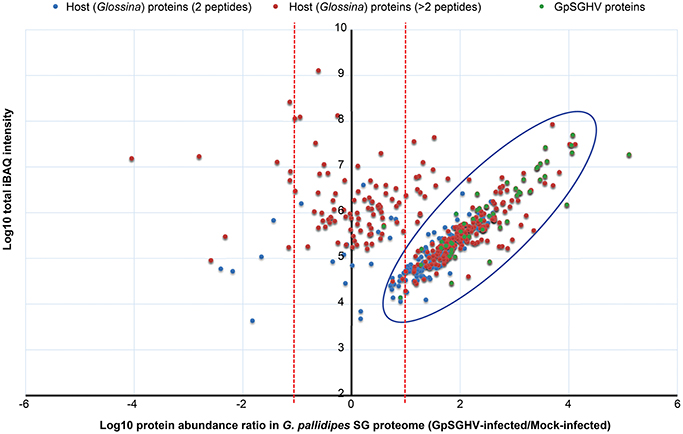
Figure 3. Abundance distribution ratios of G. pallidipes SG proteins. The figure depicts the distribution of proteins detected in the SG proteome of G. pallidipes infected by GpSGHV compared to the mock-infected controls. The host proteins detected by two or more peptides per protein are shown in blue and red, respectively, while the GpSGHV proteins are shown in green. The proteins that were up-regulated and down-regulated in GpSGHV-infected SG are shown on the right and left sides of the Y-axis, respectively. The large blue circle depicts proteins that were detectable in the SG proteome of GpSGHV-infected but not in the proteome of mock-infected G. pallidipes. Proteins which were not significantly modulated are depicted along the y-axis. The dotted red lines represent 10-fold protein regulation. iBAQ denotes intensity-based absolute quantification.
Identification of Differentially Expressed Proteins in Response to GpSGHV Infection
We then made a more comprehensive comparison of the GpSGHV-induced protein modulation by generating a log-log plot of the abundance distribution ratios of G. m. morsitans and G. pallidipes SG proteins. By combining the proteomics datasets obtained from the G. m. morsitans and G. pallidipes SG proteomes (Figures 2, 3, respectively), the identified host proteins fell into two broad categories. The first category consisted of proteins that were down-regulated or up-regulated in either or both G. m. morsitans and G. pallidipes proteomes. The second category consisted of the proteins that were detectable in one of the two Glossina species and in not the other. These categories are presented in the Figure 4.
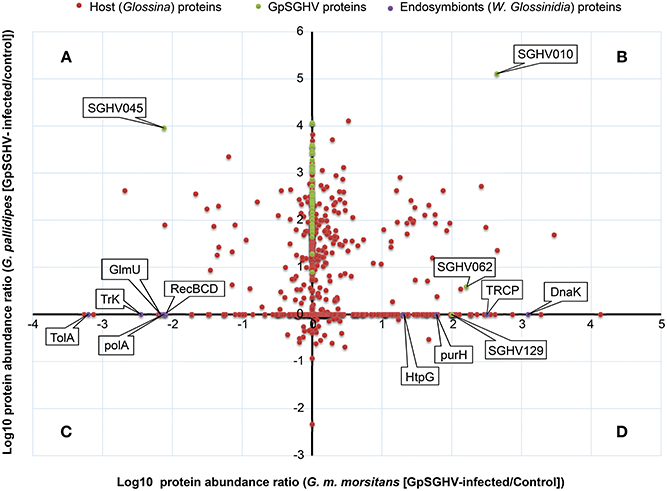
Figure 4. Abundance distribution ratios of GpSGHV-infected G. m. morsitans and G. pallidipes SG proteins. The figure shows a log-log PLOT of the host, viral and endosymbiont proteins (shown in red, green and purple, respectively). (A) Proteins down-regulated in G. m. morsitans but up-regulated in G. pallidipes. (B) Proteins up-regulated in both G. m. morsitans and G. pallidipes. (C) Proteins down-regulated in G. pallidipes but up-regulated in G. m. morsitans. (D) Proteins down-regulated in both G. m. morsitans and G. pallidipes. Proteins aligned along the Y-axis were detectable in G. pallidipes but were not detectable in G. m. morsitans, while the proteins aligned along the X-axis were detectable in G. m. morsitans but not detectable in G. pallidipes.
Proteins in the first category consisted of four groups: First, compared to mock-infected controls, a total of 57 proteins were up-regulated in the GpSGHV-infected G. pallidipes SG proteome but were down-regulated in G. m. morsitans (Figure 4A; Table S1). Of the 57 proteins, 23 showed more than 100-fold up-regulation in G. pallidipes proteome and they were all down-regulated in virus-infected G. m. morsitans (Table 1). Second, 134 proteins were up-regulated in both GpSGHV-infected G. m. morsitans and G. pallidipes SG proteomes compared to their mock-infected counterparts (Figure 4B; Table S2). Third, compared to the mock-infected controls, 18 proteins were down-regulated in the virus-infected G. pallidipes but up-regulated G. m. morsitans (Figure 4C; Table S3), nine of which were up-regulated ≥ 5-fold in G. m. morsitans compared to the G. pallidipes SG proteomes (Table 2). Lastly, nine proteins were down-regulated in both virus-infected G. m. morsitans and G. pallidipes as measured from their proteomes (Figure 4D; Table S4).

Table 1. Twenty-three host proteins that were more than 100-fold up-regulated in the GpSGHV-infected G. pallidipes but were down-regulated in the proteome of virus-infected G. m. morsitans.
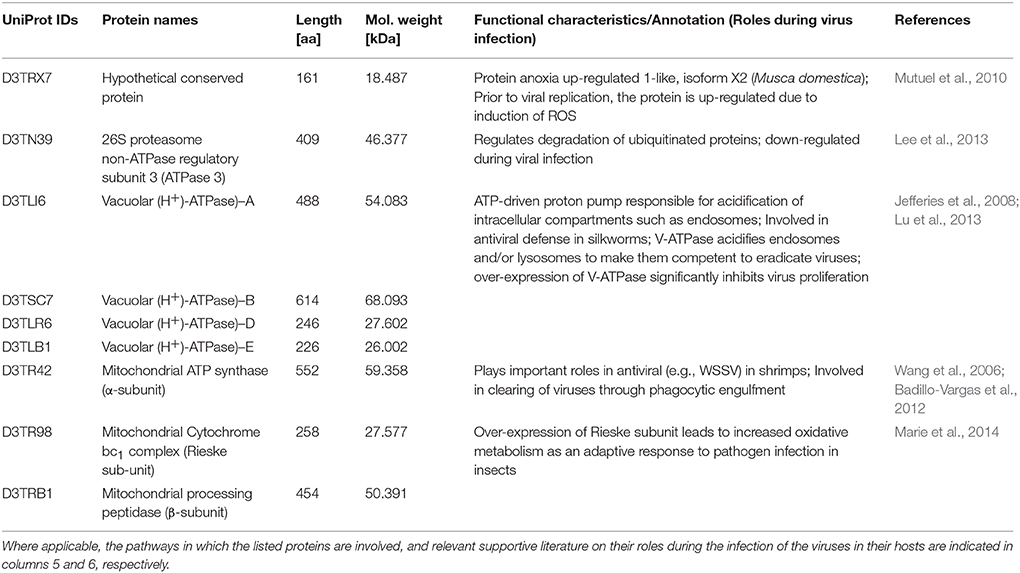
Table 2. Nine proteins that were more than 5-fold down-regulated in the GpSGHV-infected G. pallidipes but were up-regulated in the proteome of virus-infected G. m. morsitans.
Similarly, the proteins that were detectable in the proteome of one of the two GpSGHV-infected tsetse species and not in the other fell into two main groups. First, compared to mock-infected flies, 189 proteins were detectable in the virus-infected G. m. morsitans SG proteome but not in that of G. pallidipes (Figure 4, x-axis), 65.1% (n = 123) of which were up-regulated (Table S5). Second, 133 proteins were detectable in G. pallidipes but not in G. m. morsitans (Figure 4, y-axis), 96.9% of which were up-regulated (Table S6).
A closer look at the proteomics data indicate that majority of the heavily modulated proteins in the SG proteome of G. pallidipes appear to be spread over a wide range of host pathways. These pathways included, among others: ATP/ubiquitin-dependent and 26S proteasome (UPS) pathways, integrin-liked kinase pathway, transketolase pathway, hippo signaling pathway and diverse signaling pathways (Table 1). In the case of G. m. morsitans, proteins potentially involved in the host's antiviral defenses appear to be quite dominant. Some of the antiviral defense-related systems included induction of innate immune response via reactive oxygen species (virus degradation), the ubiquitin/26S proteasome system, V-ATPase system (virus degradation via acidification of endosomes and or/lysosomes), the phagocytic engulfment system (to clear virus infection) and adaptive mitochondrial-mediated immune responses (interfere with production of progeny virus) (Table 2).
Detection of Virus-/Endosymbiont-Specific Proteins in Glossina SG Proteomes
Whereas only five GpSGHV proteins (encoded by ORFs SGHV018, SGHV010, SGHV045, SGHV062 and SGHV129) were detectable in G. m. morsitans, a total of 58 proteins were detected in the SG proteome of G. pallidipes (See Figures 2–4; Table S7). These proteomics results reflects the findings we obtained from the qPCR assays described above. It should be noted that SGHV010 is the most abundant of all GpSGHV proteins; SGHV062 is a high-molecular weight (512 kDa) viral protein, while SGHV045 is a one of the major viral envelop proteins (Kariithi et al., 2013a). The high abundance and large size, respectively, of these virion proteins possibly explains their detection in G. m. morsitans. Further, proteins encoded by the ORFs SGHV018 and SGHV129 have not been detected in our previous proteomic studies in G. pallidipes, and were in the current study only detectable in the G. m. morsitans proteome but not in the G. pallidipes proteome (Figure 2). The failure to detect other viral proteins in G. m. morsitans does not necessarily imply complete absence of these proteins. Rather, their abundances could have been too low or their detection could have been masked by the highly abundant host proteins.
All the nine Wigglesworthia proteins were in the detected in the SG proteome of G. m. morsitans (Table S8), probably because unlike the G. m. morsitans, the genome of G. pallidipes is not yet available. Whereas four of the nine Wigglesworthia proteins were up-regulated in the virus-infected compared to mock-infected flies, five were down-regulated (Figure 4). The up-regulated proteins included transcription repair coupling factor, chaperone protein DnaK, chaperone protein HtpG, and IMP-cyclohydrolase. Down-regulated proteins included protein TolA, DNA polymerase I, exonuclease V, bifunctional enzyme GlmU and a potassium transporter protein. Since Wigglesworthia is housed within the host's bacteriocytes (Pais et al., 2008), the detection of Wigglesworthia proteins in the G. m. morsitans SGs suggest that these proteins likely “leak” into the hemolymph, potentially during bacteriocytes turnover, and eventually move from the hemocoel to the SGs.
Discussion
G. m. morsitans is less Permissive to GpSGHV Infections than G. pallidipes
Hytrosaviruses replicate primarily in the SG tissue of their insects hosts (Garcia-Maruniak et al., 2009). As such, one would expect that the virus-induced modulation of the SG microenvironment (i.e., morphological and biochemical/functional features of the tissue) results in the expression of various proteins/peptides specifically to the advantage of viral replication and dissemination. Whereas GpSGHV in G. pallidipes occurs in both asymptomatic and symptomatic infection states, the virus infection in other Glossina species is almost always asymptomatic. We have previously demonstrated the dynamics of the development of virus-induced SGH symptoms (Abd-Alla et al., 2010, 2013) and trans-generational transmission of the virus in G. pallidipes (Boucias et al., 2013). However, the case of GpSGHV infection in other Glossina species has not been investigated. Importantly, we have so far not observed any overt SGH symptoms in the G. morsitans colony maintained at the IPCL Seibersdorf, which was used as the source of the experimental flies we have described here.
In the current study, the effects of GpSGHV infections in G. m. morsitans were remarkably different from previous studies. For instance, virus injection into newly larvipositioned third-instar larvae of two Morsitans groups resulted in varying prevalence of SGH-like symptoms in developed adults, which ranged from 1.1% (Jura et al., 1993) to 4.0% (Kokwaro et al., 1990) in G. m. morsitans, and up to 100% in G. m. centralis (Sang et al., 1997). In our case, we did not observe any SGH symptoms in G. m. morsitans, potentially because whereas we injected the virus into newly eclosed adults, the researchers in the previous studies injected the virus suspensions into larvae. We therefore conclude that the injected virus is capable of infecting and replicating during ontogeny on the SGs during pupation (as evidenced from the previous studies). Further, the injected virus appears incapable of infecting and inducing overt SGH symptoms in fully differentiated SG cells in adults (as evidenced from our study).
Notably, the observed high virus titters in G. m. morsitans could represent DNA replication but may not represent production of infectious viral particles. The comparable virus titters between the virus-infected G. m. morsitans and the mock-infected G. pallidipes suggest that the virus may be undergoing only partial replication in adult cells to maintain steady-state titters throughout the adulthood. This is in agreement with previous studies in G. pallidipes whereby, utilizing a diagnostic PCR, Abd-Alla et al. (2007) detected GpSGHV PCR positives in 100% of colonized G. pallidipes that did not exhibit overt SGH symptoms. Taken together, the analyses of GpSGHV loads (by agarose gels), virus density levels (by qPCR), and protein expression (by LC-MS/MS) imply that either G. m. morsitans, at least in the SGs, is far less permissive to virus replication or that the virus undergoes limited replication in G. m. morsitans (whereby only a subset of genes are expressed) compared to G. pallidipes. Potentially, unlike in G. pallidipes where SGH symptoms can be overt, we are of the opinion that GpSGHV infection is entirely latent in G. m. morsitans as previously proposed by Kariithi et al. (2013b). Potentially, the difference in GpSGHV replication, i.e., partial replication or latency in G. m. morsitans vs. active replication in G. pallidipes, explains the differences in the repertoire of proteins detected in the SG proteomes of the two Glossina species.
GpSGHV Potentially Modulates G. pallidipes Key Pathways for Efficient Infection
The clear GpSGHV-induced differential modulation of SG protein expression in Glossina raises the question of what host pathways are potentially globally regulated to facilitate successful virus infection. It is well known that for cellular entry and induction of pathogenesis, many viruses manipulate key host signaling pathways that globally regulate many cellular processes (Diehl and Schaal, 2013). So far, we have not been able to elucidate the precise mechanism(s) through which GpSGHV induces overt SGH symptoms, mainly because of a lack of cell culture system to support the virus multiplication (Arif and Pavlik, 2013). Therefore, the precise mechanisms of GpSGHV infection (cellular attachment, entry, intracellular trafficking, replication, maturation, and egress) in Glossina remains elusive. In an attempt to unravel the pathobiology of GpSGHV, we draw inferences from other virus-host systems that have been studied so far.
Of the proteins we identified in this study, proteins that showed significant differential expression patterns in virus-infected flies are particularly interesting since they potentially reflect involvement in GpSGHV pathogenesis. In this regard, the proteins that were significantly up-regulated in the SG proteome of G. pallidipes but down-regulated in that of G. morsitans (Table 1) are interesting to focus on. Also important were the nine proteins found to be up-regulated in proteome of G. morsitans but down-regulated in the proteome of G. pallidipes (Table 2). Notably, our annotation of the nine proteins revealed that these proteins may be involved in pathways related to the host's antiviral responses to virus infection (See Table 2 and the references therein). In the following sections, we briefly discuss the potential roles of the proteins stipulated in Tables 1, 2 with regard to viral entry into host cells, intracellular trafficking, and evasion of host's immune response, replication/translation and cellular proliferation.
Viral Entry and Intracellular Trafficking
For entry, some viruses attach to host cell receptors thus inducing conformational changes that cause fusion of the viral envelope with the host's plasma membrane (Thorley et al., 2010). This is followed by delivery of the viral nucleocapsids into the cellular cytoplasm and uncoating of the viral genome. Integrin-linked kinases (ILKs), which were up-regulated in G. pallidipes (D3TMU2; Table 1), have been implicated in viral cellular entry. For instance, Kaposi's sarcoma-associated herpesvirus (KSHV) envelop glycoprotein B (gB) hijacks ILKs to induce the FAK-Src-PI3K-RhoGTPase signaling pathway (Naranatt et al., 2003; Sharma-Walia et al., 2005). Similar to the KSHV gB envelope protein, the GpSGHV SGHV038 protein, which was detected in the current study (Table S1), contains an arginyl-glycyl-aspartic acid (RGD) motif that may interact with the host's ILKs. Pending experimental validations, GpSGHV potentially employs an entry mechanism similar to KSHV (Krishnan et al., 2005).
Following cellular entry, a critical phase in viral pathogenesis is intracellular trafficking of viral nucleocapsids, a process that requires intricate signaling. One of the key pathway components targeted by several viruses is the GTPase Rab2 protein. Notably, this protein was found to be up-regulated in G. pallidipes (D3TMA0; Table 1), unlike in G. m. morsitans. GTPases regulate membrane trafficking, particularly in the formation, motility and docking of vesicles (Zerial and McBride, 2001). Some viruses activate GTPase-mediated pathways to facilitate their intracellular trafficking (Chien et al., 2006). For instance, in the absence of Rab 1a/b, herpes simplex virus 1 (HSV-1) was unable to traffic from the ER to cytoplasmic viral assembly complexes, leading to a build-up of un-enveloped viral particles in the cell cytoplasm (Zenner et al., 2011). GTPases were indeed found to be up-regulated in shrimps infected with whispovirus (Wu and Zhang, 2007), another large invertebrate dsDNA virus like GpSGHV. It is tempting to postulate that GpSGHV up-regulates GTPases for intracellular trafficking in G. pallidipes, especially because the virus genome shares at least 28 putative homologs with the above-mentioned large dsDNA viruses, including HSV and whispoviruses (Abd-Alla et al., 2008).
Viral Replication and Dissemination in the Host
Apart from the knowledge that the host's SG is the primary replication organ for GpSGHV (Garcia-Maruniak et al., 2009), and that the virus is transmitted from the infected mother to the progeny via the milk gland secretions (Boucias et al., 2013), the precise virus replication and dissemination mechanisms are unknown. By comparing our data with the data available from other virus-host systems, it is possible to postulate theories on GpSGHV replication and dissemination in Glossina.
Some viruses modulate the mitochondrial transport machinery to provide energy necessary for replication, especially for the viruses whose genomes are A+T-rich (Ohta and Nishiyama, 2011; Anand and Tikoo, 2013). The GpSGHV genome is A+T-rich (72%; Abd-Alla et al., 2008), implying that virus-modulation of Glossina mitochondrial transport machinery is a good possibility. The mitochondrial oxoglutarate/malate carrier (OGC) protein is important for the tricarboxylic acid cycle (TCA), gluconeogenesis and nitrogen metabolism (Cappello et al., 2006). OGC is reportedly up-regulated as an adaptive response to prevent mitochondrial injury (Ripoli et al., 2010). Thus, the up-regulation of OGC (D3TM51) in G. pallidipes (Table 1) may be GpSGHV-induced when robust virus replication occurs, especially in the event of overt SGH symptoms. Another host protein targeted by viruses to facilitate replication is the transketolase (TKTL)-2 (D3TM09). We found TKTL protein to be up-regulated more than 100-fold in G. pallidipes (See Table 1) unlike in G. m. morsitans. TKTL provides a link between the glycolytic, pentose-phosphate, and nucleotide synthesis pathways (Brault et al., 2013). During active virus replication when rapid DNA synthesis is required, carbohydrate molecules are channeled to the DNA synthesis machinery through the TKTL pathway, a process of utmost importance in proliferating tissues (Chen et al., 2011). This is of particular interest in this case of induction of the SGH symptoms in G. pallidipes, especially because SGH is mainly due to cell proliferation (Guerra et al., 2013). Since the TKTL pathway allows synthesis of ribose without the need of oxygen, GpSGHV may highjack the TKTL pathway to circumvent the need for oxygen (Noch and Khalili, 2012), thus allowing rapid GpSGHV genome replication.
Another host protein involved in viral replication is proteasome α-4 (D3TP07; Table 1), a key protein in the ATP/ubiquitin-dependent non-lysosomal proteolytic pathway. For instance, the interaction of proteasome α-subunit PSMA7 with hepatitis C virus (HCV) led to an inhibition of host protease activity and thus stimulated transcription trans-activation by HCV (Krüger et al., 2001). Several other host proteins involved in viral replication that were detected in the current study included eIF3m (D3TMN6; Cheshenko et al., 2010), molecular chaperones (e.g., hsp90; D3TS03; Kariithi et al., 2011) and 26S proteasome regulatory complex proteins (D3TNJ0; Verchot, 2014; See Table 1). The hypothetical conserved protein (D3TRX7; Table 2) is 100% identical to the M. domestica anoxia up-regulated 1-like protein and its expression in our case is virus-induced. Mutuel et al. (2010) reported a significant induction of reactive oxygen species (ROS) in the tracheal and fat body systems of lepidopteran insects early in infection with Junonia coenia densovirus. The authors made this observation prior to viral replication before any detectable disease symptoms. Interestingly, decrease of ROS induction positively correlated with exponential phase of viral infection. It has been proposed that GpSGHV may replicate in the host's fat bodies, and that the host's tracheal system provides a conduit for the virus transmission (Kariithi, 2013). It is likely that this and perhaps other similar proteins play roles during GpSGHV replication. Taken together, our data provide potential targets for future investigations of how GpSGHV replicates and is disseminated in the host.
Viral Evasion of Host's Immune Responses
Upon successful cellular entry, viruses must evade the host's immune responses, a process for which the host's ubiquitin/proteasome system (UPS) has significant roles. In the current study, we detected the main components of the UPS, i.e., E3 ligase (D3TNJ0) and 26S proteasome (D3TRZ3) (Table 1). The UPS is essential for persistent infection of some viruses. For instance, plant RNA viruses in the family Luteoviridae (genera Poleroviruses and Enamoviruses) encode viral suppressors of RNA silencing (VSRs) that hijack the UPS components to promote degradation of key components of the host's RNA-interference (RNAi) system, thereby promoting virus replication (Verchot, 2014). DNA viruses are known to be under host RNAi surveillance. These include invertebrate iridoviruses (Bronkhorst et al., 2012; Kemp et al., 2013), baculoviruses (Jayachandran et al., 2012), densoviruses (Ma et al., 2011), whispoviruses (Huang and Zhang, 2013), and plant viruses (Blevins et al., 2006). As stated above, GpSGHV infections are frequently observed, but remain asymptomatic and seldom result in SGH symptoms both in nature and in laboratory-bred Glossina species. Although the questions of how GpSGHV infection progresses from a covert asymptomatic infection to an overt symptomatic infection are yet to be answered, we speculate that the virus is under host RNAi surveillance, hence components of the host's UPS systems form ideal candidates for further studies.
Another group of host antiviral defense proteins detected in this study were the V-ATPases (D3TLI6; D3TSC7; D3TLR6; and D3TLB1; Table 2), whose activity leads to acidification of intracellular compartments, necessary for multiple cellular processes (Jefferies et al., 2008). Recently, Lu et al. (2013) reported that over-expression of V-ATPase in Bombyx mori nucleopolyhedrovirus (BmNPV)-infected cells significantly inhibited viral proliferation. Potentially, the acidification of endosomes and lysosomes by V-ATPase renders these organelles competent for viral degradation. It is therefore not surprising that in the current study, V-ATPase were up-regulated in the SG proteome of G. m. morsitans as opposed to that of G. pallidipes (Table 2) as the former appear to be less permissive to GpSGHV replication compared to the latter. Similar to the V-ATPase, mitochondrial ATP synthase was down-regulated during white spot syndrome virus (WSSV) infection in shrimps (Wang et al., 2006).
The ubiquinol-cytochrome c reductase iron-sulfur subunit (Rieske subunit/bc1) detected in the current study (D3TR98; Table 2) was demonstrated to be up-regulated during the infection of Anopheles gambiae by Plasmodium falciparum (Marie et al., 2014). The up-regulation of bc1 in these two cases could have been due to the presence of the parasites in the mosquito, which could be a response involved in parasite resistance. The α- and β-subunits of mitochondrial processing peptidase (MPP; D3TRB1; Table 2) are homologous to the core 2 and core 1 proteins of the bc1 complex (Braun and Schmitz, 1995). MPP and bc1 complex appear to have similar pathogen-induced modulation patterns, and their up-regulation in G. m. morsitans may be an adaptive antiviral host resistant response.
Viral Persistent Infection and Induction of Cellular Pathology
It is still not clear how GpSGHV induces cellular proliferation in the host's SG tissue. However, other studies have demonstrated that some viruses induce cellular proliferation via modulation of specific signaling pathways. Protein phosphatase 2A (PP2A; D3TLN8; Table 1) is critical in the regulation of cell proliferation, signal transduction, cytoskeletal dynamics, and apoptosis (Seshacharyulu et al., 2013). Some viral proteins such as the small T antigen of SV40 specifically target and directly interact with and displace PP2A's scaffolding B subunit thereby inducing cellular proliferation (Guergnon et al., 2011). To activate intracellular signaling pathways, some viruses use various approaches to hijack the G-protein-coupled receptors (GPCRs; D3TPG7; Table 1), leading to enhancement of viral pathogenesis (Sodhi et al., 2004; Lin et al., 2005). Other viruses such as KSHV encode potent and constitutively active GPCR homologs that modulate cellular proliferation (Kirshner et al., 1999). Hypothetically, some GpSGHV envelop proteins (Table S1) could interact with host proteins to trigger signaling pathways resulting in hyperplasia as has been reported in other viruses such as the fowl poxvirus (Afonso et al., 2000). It is interesting to experimentally validate whether these virus and/or host proteins are actually involved in the development of SGH in Glossina.
Viral Assembly and Induction of SGH Symptoms
An important step during assembly of viruses is processing of viral mRNAs, which in some cases involve the host trans-acting splicing factors such as serine/arginine-rich proteins (SRps; Akopian et al., 2013). To ensure production of their own protein diversity, adenoviruses, HSV-1, influenza A viruses (IAV) and HIV manipulate mRNA splicing by phosphorylating SRps (Estmer-Nilsson et al., 2001; Sciabica et al., 2003; Fukuhara et al., 2006; Dubois et al., 2014). Therefore, it not surprising that in the current study, SRp 55 (D3TN12) was up-regulated in GpSGHV-infected G. pallidipes (active viral replication), but down-regulated in G. m. morsitans (less permissive to viral replication) (Table 1). However, it is currently unknown how GpSGHV mRNAs are processed. Another host protein involved in viral assembly is tailless-complex polypeptide protein-1 (TcP-1; D3TP27 and D3TMK9; Table 1). For instance, TcP-1 has been implicated in the assembly of hepatitis B/C virus capsids (Lingappa et al., 1994; Inoue et al., 2011), while annexins (D3TRS6 and D3TME1; Table 1) are involved in the HIV-1 assembly in lipid rafts (Harrist et al., 2009; Saxena et al., 2012). Other proteins that may be involved in viral assembly include the 26S proteasome non-ATPase regulatory subunit 3 (ATPase 3; D3TN39), which was down-regulated as seen in the SG proteome of G. pallidipes (Table 2). Potentially, this protein may regulate (by blocking) degradation of viral proteins. A gene similar to ATPase 3 was found to be down-regulated more than 10-fold in the rice stripe virus (RSV) infected small brown plant hopper, Laodelphax striatellus (Lee et al., 2013).
Induction of the SGH symptoms is possibly a reflection of active production of viable progeny virus particles. During active virus progeny production, enveloped viruses are known to depend on the endoplasmic reticulum (ER) for maturation of viral envelope glycoproteins, and proteins involved in the formation of replication complexes, assembly, envelopment and genome packaging (Medigeshi et al., 2007; Scheel and Rice, 2013). This imposes a tremendous protein load in the ER, leading to ER stress. Consequently, ER stress results in the induction of the unfolded protein response (UPR), an evolutionary conserved prosurvival pathway that signals the nucleus to induce the expression of various chaperones (Walter and Ron, 2011). In some cases, interaction between the induced chaperones and viral proteins is critical for processing of viral proteins and assembly of mature virions. During prolonged and overwhelming ER stress, UPR switches from being prosurvival to proaptotic (Szegezdi et al., 2006). Prolonged virus-induced ER stress/UPR responses modulate a variety of signaling pathways that contribute to viral pathogenesis (Fung and Liu, 2014), and may lead to cellular proliferation and hypertrophy. The up-regulation of UPR-associated proteins and several molecular chaperones in flies with SGH symptoms (see Table 1) implicates the UPR/ER stress machinery in the development of overt SGH symptoms in G. pallidipes. As discussed above, the TKTL pathway may also be involved in the expression of overt SGH symptoms in G. pallidipes. The current data provide potential targets for development of rationally designed antiviral strategies in large tsetse fly rearings for sterile insect technique.
Conclusions
The data presented in this study provide hints as to why G. m. morsitans is much less susceptible host to GpSGHV infection compared to G. pallidipes. The known and/or putative functions inferred from sequence similarity analyses revealed that the differentially modulated proteins we have identified are potentially involved in various aspects of GpSGHV pathogenesis. Specifically, and like in many other viruses, GpSGHV appears to deploy a repertoire of strategies to exploit the host intracellular signaling pathways for replication, especially in G. pallidipes. In the case of G. m. morsitans, host proteins involved in antiviral defense systems appeared to be dominant. Some of the pathways that appear to be targets of the virus include the UPR and TKTL pathways, implicating their involvement in expression of overt SGH symptoms in G. pallidipes. The proteins involved in these pathways deserve further functional (experimental) validations to understand the relevance of the differences in their expression patterns. The current study is a critical baseline data in a new Coordinated Research Project (CRP) initiated by IAEA, aimed at gaining a deeper knowledge of the Glossina/symbiont/GpSGHV tripartite interactions and how these interactions affect Trypanosoma parasite transmission (Van Den Abbeele et al., 2013). We have designed RNAi bioassays to further investigate how asymptomatic GpSGHV infection is maintained in Glossina. Candidate proteins experimentally validated as essential for efficient GpSGHV pathogenesis are ideal targets for developing rationally designed antiviral strategies to control the virus infections in tsetse mass rearing facilities. In a larger view, our data are important for future studies on molecular and biochemical routes employed by members of the new entrants into the family of insect viruses, the Hytrosaviridae.
Author Contributions
HK, İİ, JV, MV, and AA participated in the design of the study. AA and IM set up the bioassays. HK and İİ processed and quantified the salivary gland proteins. SB performed the LC-MS/MS measurements. SB and HK analyzed the proteomics data sets. HK and EM annotated/characterized the proteins. HK wrote the manuscript. JV, MV, AA, İİ, EM, EO, and SN contributed in writing the paper, providing critical comments and suggestions. All the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, International Atomic Energy Agency (CRP No.: D4.20.15) Vienna, Austria and the Netherlands Fellowship Program (Grant No.: CF7548/2011), The Netherlands. The authors would like to acknowledge Ms. Carmen Marin of the IPCL Seibersdorf for rearing the experimental flies.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00089
References
Abd-Alla, A., Bossin, H., Cousserans, F., Parker, A., Bergoin, M., and Robinson, A. (2007). Development of a non-destructive PCR method for detection of the salivary gland hypertrophy virus (SGHV) in tsetse flies. J. Virol. Meth. 139, 143–149. doi: 10.1016/j.jviromet.2006.09.018
Abd-Alla, A., Cousserans, F., Parker, A., Bergoin, M., Chiraz, J., and Robinson, A. (2009a). Quantitative PCR analysis of the salivary gland hypertrophy virus (GpSGHV) in a laboratory colony of Glossina pallidipes. Virus Res. 139, 48–53. doi: 10.1016/j.virusres.2008.10.006
Abd-Alla, A. M. M., Cousserans, F., Parker, A. G., Jehle, J. A., Parker, N. J., Vlak, J. M., et al. (2008). Genome analysis of a Glossina pallidipes salivary gland hypertrophy virus reveals a novel large double-stranded circular DNA virus. J. Virol. 82, 4595–4611. doi: 10.1128/JVI.02588-07
Abd-Alla, A. M. M., Kariithi, H. M., Mohamed, A. H., Lapiz, E., Parker, A. G., and Vreysen, M. J. B. (2013). Managing hytrosavirus infections in Glossina pallidipes colonies: feeding regime affects the prevalence of salivary gland hypertrophy syndrome. PLoS ONE 8:e61875. doi: 10.1371/journal.pone.0061875
Abd-Alla, A. M. M., Vlak, J. M., Bergoin, M., Maruniak, J. E., Parker, A. G., Burand, J. P., et al. (2009b). Hytrosaviridae: a proposal for classification and nomenclature of a new insect virus family. Arch. Virol. 154, 909–918. doi: 10.1007/s00705-009-0398-5
Abd-Alla, A. M. M., Kariithi, H. M., Parker, A. G., Robinson, A. S., Kiflom, M., Bergoin, M., et al. (2010). Dynamics of the salivary gland hypertrophy virus in laboratory colonies of Glossina pallidipes (Diptera: Glossinidae). Virus Res. 150, 103–110. doi: 10.1016/j.virusres.2010.03.001
Abd-Alla, A. M. M., Parker, A. G., Vreysen, M. J. B., and Bergoin, M. (2011a). Tsetse salivary gland hypertrophy virus: hope or hindrance for tsetse control? PLoS Negl. Trop. Dis. 5:e1220. doi: 10.1371/journal.pntd.0001220
Abd-Alla, A. M. M., Salem, T. Z., Parker, A. G., Wang, Y., Jehle, J. A., Vreysen, M. J. B., et al. (2011b). Universal primers for rapid detection of Hytrosaviruses. J. Virol. Meth. 171, 280–283. doi: 10.1016/j.jviromet.2010.09.025
Afonso, C. L., Tulman, E. R., Lu, Z., Zsak, L., Kutish, G. F., and Rock, D. L. (2000). The genome of fowlpox virus. J. Virol. 74, 3815–3831. doi: 10.1128/JVI.74.8.3815-3831.2000
Akopian, D., Shen, K., Zhang, X., and Shan, S. (2013). Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721. doi: 10.1146/annurev-biochem-072711-164732
Alcaide-Loridan, C., and Jupin, I. (2012). Ubiquitin and plant viruses, let's play together! Plant Physiol. 160, 72–82. doi: 10.1104/pp.112.201905
Amargier, A., Lyon, J. P., Vago, C., Meynadier, G., and Veyrunes, J. C. (1979). Mise en evidence et purification d'un virus dans la proliferation monstrueuse glandulaire d'insectes. Étude sur Merodon equestris F. (Diptere: Syrphidae). C. R. Acad. Sci. D. 289, 481–484.
Anand, S. K., and Tikoo, S. K. (2013). Viruses as modulators of mitochondrial functions. Adv. Virol. 2013, 17. doi: 10.1155/2013/738794
Arif, B., and Pavlik, L. (2013). Insect cell culture: virus replication and applications in biotechnology. J. Invertebr. Pathol. 112 (Suppl. 1), S138–S141. doi: 10.1016/j.jip.2012.07.011
Badillo-Vargas, I. E., Rotenberg, D., Schneweis, D. J., Hiromasa, Y., Tomich, J. M., and Whitfield, A. E. (2012). Proteomic analysis of Frankliniella occidentalis and differentially expressed proteins in response to tomato spotted wilt virus infection. J. Virol. 86, 8793–8809. doi: 10.1128/JVI.00285-12
Beaton, A. R., Rodriguez, J., Reddy, Y. K., and Roy, P. (2002). The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc. Natl. Acad. Sci U.S.A. 99, 13154–13159. doi: 10.1073/pnas.192432299
Bishop-Lilly, K. A., Turell, M. J., Willner, K. M., Butani, A., Nolan, N. M., Lentz, S. M., et al. (2010). Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl. Trop. Dis. 4:e878. doi: 10.1371/journal.pntd.0000878
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H. S., et al. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Boucias, D. G., Kariithi, H. M., Bourtzis, K., Schneider, D. I., Kelley, K., Miller, W. J., et al. (2013). Trans-generational transmission of the Glossina pallidipes hytrosavirus depends on the presence of a functional symbiome. PLoS ONE 8:e61150. doi: 10.1371/journal.pone.0061150
Brault, C., Levy, P. L., and Bartosch, B. (2013). Hepatitis C virus-induced mitochondrial dysfunctions. Viruses 5, 954–980. doi: 10.3390/v5030954
Braun, H. P., and Schmitz, U. K. (1995). Are the ‘core’ proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease? Trends Biochem. Sci. 20, 171–175. doi: 10.1016/S0968-0004(00)88999-9
Bronkhorst, A. W., van Cleef, K. W., Vodovar, N., İnce, İ. A., Blanc, H., Vlak, J. M., et al. (2012). The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. U.S.A. 109, E3604–E3613. doi: 10.1073/pnas.1207213109
Cappello, A. R., Curcio, R., Valeria, M. D., Stipani, I., Robinson, A. J., Kunji, E. R., et al. (2006). Functional and structural role of amino acid residues in the even-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J. Mol. Biol. 363, 51–62. doi: 10.1016/j.jmb.2006.08.041
Chen, A. J., Gao, L., Wang, X. W., Zhao, X. F., and Wang, J. X. (2013). SUMO-conjugating enzyme E2 UBC9 mediates viral immediate-early protein SUMOylation in crayfish to facilitate reproduction of white spot syndrome virus. J. Virol. 87, 636–647. doi: 10.1128/JVI.01671-12
Chen, I. T., Aoki, T., Huang, Y. T., Hirono, I., Chen, T. C., Huang, J. Y., et al. (2011). White spot syndrome virus induces metabolic changes resembling the warburg effect in shrimp hemocytes in the early stage of infection. J. Virol. 85, 12919–12928. doi: 10.1128/JVI.05385-11
Cheshenko, N., Trepanier, J. B., Segarra, T. J., Fuller, A. O., and Herold, B. C. (2010). HSV usurps eukaryotic initiation factor 3 subunit M for viral protein translation: novel prevention target. PLoS ONE 5:e11829. doi: 10.1371/journal.pone.0011829
Chien, Y., Kim, S., Bumeister, R., Loo, Y. M., Kwon, S. W., Johnson, C. L., et al. (2006). RalB GTPase-mediated activation of the IkB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127, 157–170. doi: 10.1016/j.cell.2006.08.034
Coler, R. R., Boucias, D. G., Frank, J. H., Maruniak, J. E., Garcia-Canedo, A., and Pendland, J. C. (1993). Characterization and description of a virus causing salivary gland hyperplasia in the housefly, Musca domestica. Med. Vet. Entomol. 7, 275–282. doi: 10.1111/j.1365-2915.1993.tb00688.x
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
Diehl, N., and Schaal, H. (2013). Make yourself at home: viral hijacking of the PI3K/Akt signaling pathway. Viruses, 5, 3192–3212. doi: 10.3390/v5123192
Dubois, J., Terrier, O., and Rosa-Calatrava, M. (2014). Influenza viruses and mRNA splicing: doing more with less. mBio 5:e00070-14. doi: 10.1128/mBio.00070-14
Edwards, L. A., Shabbits, J. A., Bally, M., and Dedhar, S. (2004). Integrin-linked kinase (ILK) in combination molecular targeting. Cancer Treat. Res. 119, 59–75. doi: 10.1007/1-4020-7847-1_4
Eldin, P., Papon, L., Oteiza, A., Brocchi, E., Lawson, T. G., and Mechti, N. (2009). TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol. 90, 536–545. doi: 10.1099/vir.0.006288-0
Esfandiarei, M., Suarez, A., Amaral, A., Si, X., Rahmani, M., Dedhar, S., et al. (2006). Novel role for integrin-linked kinase in modulation of coxsackievirus B3 replication and virus-induced cardiomyocyte injury. Circ. Res. 99, 354–361. doi: 10.1161/01.RES.0000237022.72726.01
Estmer-Nilsson, C., Petersen-Mahrt, S., Durot, C., Shtrichman, R., Krainer, A. R., Kleinberger, T., et al. (2001). The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20, 864–871. doi: 10.1093/emboj/20.4.864
Feldmann, U. (1994). “Guidelines for the rearing of tsetse flies using the membrane feeding technique,” in Techniques of Insect Rearing for the Development of Integrated Pest and Vector Management Strategies, ed J. P. R. Ochieng-Odero (Nairobi: ICIPE Science Press), 449–471.
Fukuhara, T., Hosoya, T., Shimizu, S., Sumi, K., Oshiro, T., Yoshinaka, Y., et al. (2006). Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U.S.A. 103, 11329–11333. doi: 10.1073/pnas.0604616103
Fung, T. S., and Liu, D. X. (2014). Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 5:296. doi: 10.3389/fmicb.2014.00296
Garcia-Maruniak, A., Abd-Alla, A. M. M., Salem, T. Z., Parker, A. G., van Oers, M. M., Maruniak, J. E., et al. (2009). Two viruses that cause salivary gland hypertrophy in Glossina pallidipes and Musca domestica are related and form a distinct phylogenetic clade. J. Gen. Virol. 90, 334–346. doi: 10.1099/vir.0.006783-0
Golebiewski, L., Liu, H., Javier, R. T., and Rice, A. P. (2011). The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions. J. Virol. 85, 10639–10648. doi: 10.1128/JVI.05070-11
Green, M. R., and Sambrook, J. (eds.). (2012). “Analysis of DNA,” in Molecular Cloning: A Laboratory Manual, 4 Edn. (New York, NY: Cold Spring Harbor Laboratory Press), 81–156.
Grove, J., and Marsh, M. (2011). The cell biology of receptor-mediated virus entry. J. Cell. Biol. 195, 1071–1082. doi: 10.1083/jcb.201108131
Guergnon, J., Godet, A. N., Galioot, A., Falanga, P. B., Colle, J. H., Cayla, X., et al. (2011). PP2A targeting by viral proteins: a widespread biological strategy from DNA/RNA tumor viruses to HIV-1. Biochim. Biophys. Acta. 1812, 1498–1507. doi: 10.1016/j.bbadis.2011.07.001
Guerra, L., Stoffolano, J. G. Jr., Gambellini, G., Masci, V. L., Belardinelli, M. C., and Fausto, A. M. (2013). Ultrastructure of the salivary glands of non-infected and infected glands in Glossina pallidipes by the salivary glands hypertrophy virus. J. Invertebr. Pathol. 112 (Suppl. 1), S53–S61. doi: 10.1016/j.jip.2012.04.003
Harrist, A. V., Ryzhova, E. V., Harvey, T., and Gonzalez-Scarano, F. (2009). Anx2 interacts with HIV-1 Gag at phosphatidylinositol (4,5) bisphosphate-containing lipid rafts and increases viral production in 293T cells. PLoS ONE 4:e5020. doi: 10.1371/journal.pone.0005020
Huang, T., and Zhang, X. (2013). Host defense against DNA virus infection in shrimp is mediated by the siRNA pathway. Eur. J. Immunol. 43, 137–146. doi: 10.1002/eji.201242806
Inoue, Y., Aizaki, H., Hara, H., Matsuda, M., Ando, T., Shimoji, T., et al. (2011). Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology 410, 38–47. doi: 10.1016/j.virol.2010.10.026
Javier, R. T., and Rice, A. P. (2011). Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 85, 11544–11556. doi: 10.1128/JVI.05410-11
Jayachandran, B., Hussain, M., and Asgari, S. (2012). RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 86, 13729–13734. doi: 10.1128/JVI.02041-12
Jefferies, K. C., Cipriano, D. J., and Forgac, M. (2008). Function, structure and regulation of the vacuolar (H+)-ATPases. Arch. Biochem. Biophys. 476, 33–42. doi: 10.1016/j.abb.2008.03.025
Jura, W. G. Z. O., Zdarek, J., and Otieno, L. H. (1993). A simple method for artificial infection of tsetse, Glossina morsitans morsitans larvae with the DNA virus of G. pallidipes. Int. J. Trop. Insect Sci. 14, 383–387. doi: 10.1017/S1742758400014909
Kariithi, H. M. (2013). Glossina Hytrosavirus Control Strategies in Tsetse Fly Factories: Application of Infectomics in Virus Management. Ph.d. thesis, Wageningen University.
Kariithi, H. M., van Lent, J. W., Boeren, S., Abd-Alla, A. M., Ýnce, Ý. A., van Oers, M. M., et al. (2013a). Correlation between structure, protein composition, morphogenesis and cytopathology of Glossina pallidipes salivary gland hypertrophy virus. J. Gen. Virol. 94, 193–208. doi: 10.1099/vir.0.047423-0
Kariithi, H. M., İnce, İ. A., Boeren, S., Abd-Alla, A. M. M., Parker, A. G., Aksoy, S., et al. (2011). The salivary secretome of the tsetse fly, Glossina pallidipes (Diptera: Glossinidae) infected by salivary gland hypertrophy virus. PLoS Negl. Trop. Dis. 5:e1371. doi: 10.1371/journal.pntd.0001371
Kariithi, H. M., Ìnce, İ. A., Boeren, S., Vervoort, J., Bergoin, M., van Oers, M. M., et al. (2010). Proteomic analysis of Glossina pallidipes salivary gland hypertrophy virus virions for immune intervention in tsetse fly colonies. J. Gen. Virol. 91, 3065–3074. doi: 10.1099/vir.0.023671-0
Kariithi, H. M., van Oers, M. M., Vlak, J. M., Vreysen, M. J. B., Parker, A. G., and Abd-Alla, A. M. M. (2013b). Virology, epidemiology and pathology of Glossina hytrosavirus, and its control prospects in laboratory colonies of the tsetse fly, Glossina pallidipes (Diptera; Glossinidae). Insects 4, 287–319. doi: 10.3390/insects4030287
Kemp, C., Mueller, S., Goto, A., Barbier, V., Paro, S., Bonnay, F., et al. (2013). Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658. doi: 10.4049/jimmunol.1102486
Kirshner, J. R., Staskus, K., Haase, A., Lagunoff, M., and Ganem, D. (1999). Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 73, 6006–6014.
Kokwaro, E. D., Nyindo, M., and Chimtawi, M. (1990). Ultrastructural changes in salivary glands of tsetse, Glossina morsitans morsitans, infected with virus and rickettsia-like organisms. J. Invertebr. Pathol. 56, 337–346. doi: 10.1016/0022-2011(90)90120-U
Krishnan, H. H., Sharma-Walia, N., Zeng, L., Gao, S. J., and Chandran, B. (2005). Envelope glycoprotein gB of Kaposi's sarcoma-associated herpesvirus is essential for egress from infected cells. J. Virol. 79, 10952–10967. doi: 10.1128/JVI.79.17.10952-10967.2005
Krüger, M., Beger, C., Welch, P. J., Barber, J. R., Manns, M. P., and Wong-Staal, F. (2001). Involvement of proteasome a-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation. Mol. Cell. Biol. 21, 8357–8364. doi: 10.1128/MCB.21.24.8357-8364.2001
Lallemand-Breitenbach, V., and de Thé, H. (2010). PML nuclear bodies. Cold Spring Harb. Symp. Quant. Biol. 2:a000661. doi: 10.1101/cshperspect.a000661
Lee, J. H., Choi, J. Y., Tao, X. Y., Kim, J. S., Kim, W., and Je, Y. H. (2013). Transcriptome analysis of the small brown plant hopper, Laodelphax striatellus carrying rice stripe virus. Plant Pathol. J. 29, 330–337. doi: 10.5423/PPJ.NT.01.2013.0001
Lietze, V.-U., Abd-Alla, A., Vreysen, M., Geden, C. C., and Boucias, D. G. (2011). Salivary gland hypertrophy viruses: a novel group of insect pathogenic viruses. Ann. Rev. Entomol. 56, 63–80. doi: 10.1146/annurev-ento-120709-144841
Lin, Y. L., Mettling, C., Portales, P., Reant, B., Clot, J., and Corbeau, P. (2005). G-protein signaling triggered by R5 human immunodeficiency virus type 1 increases virus replication efficiency in primary T lymphocytes. J. Virol. 79, 7938–7941. doi: 10.1128/JVI.79.12.7938-7941.2005
Lingappa, J. R., Martin, R. L., Wong, M. L., Ganem, D., Welch, W. J., and Lingappa, V. R. (1994). A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J. Cell Biol. 125, 99–111. doi: 10.1083/jcb.125.1.99
Lloyd, R. E. (2013). Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscipl. Rev. 4, 317–331. doi: 10.1002/wrna.1162
Lu, J., Boeren, S., de Vries, S. C., van Valenberg, H. J., Vervoort, J., and Hettinga, K. (2011). Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J. Proteomics 75, 34–43. doi: 10.1016/j.jprot.2011.07.031
Lu, P., Xia, H., Gao, L., Pan, Y., Wang, Y., Cheng, X., et al. (2013). V-ATPase is involved in silkworm defense response against Bombyx mori nucleopolyhedrovirus. PLoS ONE 8:e64962. doi: 10.1371/journal.pone.0064962
Luo, L., and Zeng, L. (2010). A new rod-shaped virus from parasitic wasp Diachasmimorpha longicaudata (Hymenoptera: Braconidae). J. Invertebr. Pathol. 103, 165–169. doi: 10.1016/j.jip.2009.08.008
Ma, M., Huang, Y., Gong, Z., Zhuang, L., Li, C., Yang, H., et al. (2011). Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS ONE 6:e24758. doi: 10.1371/journal.pone.0024758
Marie, A., Holzmuller, P., Tchioffo, M., Rossignol, M., Demettre, E., Seveno, M., et al. (2014). Anopheles gambiae salivary protein expression modulated by wild Plasmodium falciparum infection: highlighting of new antigenic peptides as candidates of An. gambiae bites. Parasites Vectors 7:599. doi: 10.1186/s13071-014-0599-y
Mattioli, R. C., Feldmann, U., Hendrickx, G., Wint, W., Jannin, J., and Slingenbergh, J. (2004). Tsetse and trypanosomiasis intervention policies supporting sustainable animal-agricultural development. J. Food Agr. Environ. 2, 310–314.
Medigeshi, G. R., Lancaster, A. M., Hirsch, A. J., Briese, T., Lipkin, W. I., Defilippis, V., et al. (2007). West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J. Virol. 81, 10849–10860. doi: 10.1128/JVI.01151-07
Mutuel, D., Ravallec, M., Chabi, B., Multeau, C., Salmon, J. M., Fournier, P., et al. (2010). Pathogenesis of Junonia coenia densovirus in Spodoptera frugiperda: a route of infection that leads to hypoxia. Virology 403, 137–144. doi: 10.1016/j.virol.2010.04.003
Naranatt, P. P., Akula, S. M., Zien, C. A., Krishnan, H. H., and Chandran, B. (2003). Kaposi's sarcoma-associated herpesvirus induces the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling pathway in target cells early during infection: implications for infectivity. J. Virol. 77, 1524–1539. doi: 10.1128/JVI.77.2.1524-1539.2003
Noch, E., and Khalili, K. (2012). Oncogenic viruses and tumor glucose metabolism: like kids in a candy store. Mol. Cancer Ther. 11, 14–23. doi: 10.1158/1535-7163.MCT-11-0517
Ohta, A., and Nishiyama, Y. (2011). Mitochondria and viruses. Mitochondrion 11, 1–12. doi: 10.1016/j.mito.2010.08.006
Pais, R., Lohs, C., Wu, Y., Wang, J. W., and Aksoy, S. (2008). The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74, 5965–5974. doi: 10.1128/AEM.00741-08
Reineke, L. C., and Lloyd, R. E. (2013). Diversion of stress granules and P-bodies during viral infection. Virology 436, 255–267. doi: 10.1016/j.virol.2012.11.017
Ripoli, M., D'Aprile, A., Quarato, G., Sarasin-Filipowicz, M., Gouttenoire, J., Scrima, R., et al. (2010). Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1a-mediated glycolytic adaptation. J. Virol. 84, 647–660. doi: 10.1128/JVI.00769-09
Sang, R. C., Jura, W. G. Z. O., Otieno, L. H., Mwangi, R. W., and Ogaja, P. (1999). The effects of a tsetse DNA virus infection on the functions of the male accessory reproductive gland in the host fly Glossina morsitans centralis (Diptera: Glossinidae). Curr. Microbiol. 38, 349–354. doi: 10.1007/PL00006815
Sang, R. C., Jura, W. G. Z. O., Otieno, L. H., Tukei, P. M., and Mwangi, R. W. (1997). Effects of tsetse DNA virus infection on the survival of a host fly Glossina morsitans centralis (Diptera: Glossinidae). J. Invertebr. Pathol. 69, 253–260. doi: 10.1006/jipa.1996.4629
Saxena, V., Lai, C. K., Chao, T. C., Jeng, K. S., and Lai, M. M. (2012). Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J. Virol. 86, 4139–4150. doi: 10.1128/JVI.06327-11
Scheel, T. K., and Rice, C. M. (2013). Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19, 837–849. doi: 10.1038/nm.3248
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Schwanhausser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. doi: 10.1038/nature10098
Sciabica, K. S., Dai, Q. J., and Sandri-Goldin, R. M. (2003). ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22, 1608–1619. doi: 10.1093/emboj/cdg166
Seshacharyulu, P., Pandey, P., Datta, K., and Batra, S. K. (2013). Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 335, 9–18. doi: 10.1016/j.canlet.2013.02.036
Sharma-Walia, N., Krishnan, H. H., Naranatt, P. P., Zeng, L., Smith, M. S., and Chandran, B. (2005). ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection. J. Virol. 79, 10308–10329. doi: 10.1128/JVI.79.16.10308-10329.2005
Sodhi, A., Montaner, S., and Gutkind, J. S. (2004). Viral hijacking of G-protein-coupled-receptor signaling networks. Nat. Rev. Mol. Cell Biol. 5, 998–1012. doi: 10.1038/nrm1529
Sontag, E. (2001). Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal. 13, 7–16. doi: 10.1016/S0898-6568(00)00123-6
Szegezdi, E., Logue, S. E., Gorman, A. M., and Samali, A. (2006). Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885. doi: 10.1038/sj.embor.7400779
Thorley, J. A., McKeating, J. A., and Rappoport, J. Z. (2010). Mechanisms of viral entry: sneaking in the front door. Protoplasma, 244, 15–24. doi: 10.1007/s00709-010-0152-6
Van Den Abbeele, J., Bourtzis, K., Weiss, B., Cordön-Rosales, C., Miller, W., Abd-Alla, A. M. M., et al. (2013). Enhancing tsetse fly refractoriness to trypanosome infection—A new IAEA coordinated research project. J. Invertebr. Pathol. 112 (Suppl. 1), S142–S147. doi: 10.1016/j.jip.2012.07.020
Verchot, J. (2014). The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 5:66. doi: 10.3389/fpls.2014.00066
Walter, P., and Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. doi: 10.1126/science.1209038
Wang, B., Li, F., Dong, B., Zhang, X., Zhang, C., and Xiang, J. (2006). Discovery of the genes in response to white spot syndrome virus (WSSV) infection in Fenneropenaeus chinensis through cDNA microarray. Mar. Biotechnol. 8, 491–500. doi: 10.1007/s10126-005-6136-4
Wang, J., Brelsfoard, C., Wu, Y., and Aksoy, S. (2013). Intercommunity effects on microbiome and GpSGHV density regulation in tsetse flies. J. Invertebr. Pathol. 112, S32–S39. doi: 10.1016/j.jip.2012.03.028
Wu, W., and Zhang, X. (2007). Characterization of a Rab GTPase up-regulated in the shrimp Peneaus japonicus by virus infection. Fish. Shellfish Immunol. 23, 438–445. doi: 10.1016/j.fsi.2007.01.001
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Zenner, H. L., Yoshimura, S., Barr, F. A., and Crump, C. M. (2011). Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J. Virol. 85, 8012–8021. doi: 10.1128/JVI.00500-11
Keywords: SGH syndrome, asymptomatic infection, LC-MS/MS, pathogenesis, Hytrosaviridae, hypertrophy, unfolded protein response
Citation: Kariithi HM, Ince İA, Boeren S, Murungi EK, Meki IK, Otieno EA, Nyanjom SRG, van Oers MM, Vlak JM and Abd-Alla AMM (2016) Comparative Analysis of Salivary Gland Proteomes of Two Glossina Species that Exhibit Differential Hytrosavirus Pathologies. Front. Microbiol. 7:89. doi: 10.3389/fmicb.2016.00089
Received: 09 September 2015; Accepted: 18 January 2016;
Published: 09 February 2016.
Edited by:
Slobodan Paessler, University of Texas Medical Branch, USAReviewed by:
Jianwei Wang, Chinese Academy of Medical Sciences, ChinaCheng Huang, University of Texas Medical Branch, USA
Copyright © 2016 Kariithi, Ince, Boeren, Murungi, Meki, Otieno, Nyanjom, van Oers, Vlak and Abd-Alla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henry M. Kariithi, henry.kariithi@kalro.org;
Adly M. M. Abd-Alla, a.m.m.abd-alla@iaea.org
†These authors have contributed equally to this work.
 Henry M. Kariithi
Henry M. Kariithi İkbal Agah İnce
İkbal Agah İnce Sjef Boeren
Sjef Boeren Edwin K. Murungi
Edwin K. Murungi Irene K. Meki
Irene K. Meki Everlyne A. Otieno
Everlyne A. Otieno Steven R. G. Nyanjom
Steven R. G. Nyanjom Monique M. van Oers
Monique M. van Oers Just M. Vlak
Just M. Vlak Adly M. M. Abd-Alla
Adly M. M. Abd-Alla