- 1Department of Biochemistry and Molecular Biology, Graduate School of Science and Engineering, Saitama University, Saitama, Japan
- 2Biological Science Laboratories, KAO Corporation, Wakayama, Japan
- 3Core Research of Evolutional Science and Technology, Japan Science and Technology Agency, Saitama, Japan
LexA is a well-established transcriptional repressor of SOS genes induced by DNA damage in Escherichia coli and other bacterial species. However, LexA in the cyanobacterium Synechocystis sp. PCC 6803 has been suggested not to be involved in SOS response. In this study, we performed RNA-seq analysis of the wild-type strain and the lexA-disrupted mutant to obtain the comprehensive view of LexA-regulated genes in Synechocystis. Disruption of lexA positively or negatively affected expression of genes related to various cellular functions such as phototactic motility, accumulation of the major compatible solute glucosylglycerol and subunits of bidirectional hydrogenase, photosystem I, and phycobilisome complexes. We also observed increase in the expression level of genes related to iron and manganese uptake in the mutant at the later stage of cultivation. However, none of the genes related to DNA metabolism were affected by disruption of lexA. DNA gel mobility shift assay using the recombinant LexA protein suggested that LexA binds to the upstream region of pilA7, pilA9, ggpS, and slr1670 to directly regulate their expression, but changes in the expression level of photosystem I genes by disruption of lexA is likely a secondary effect.
Introduction
The LexA protein in Escherichia coli has been well-characterized as the key regulator of the SOS response induced by DNA damage (Butala et al., 2009). Under non-stress conditions, LexA binds to the promoter regions of more than 40 genes involved in the SOS response and represses their expression. When DNA is damaged, LexA undergoes autoproteolytic cleavage upon association with RecA protein activated through binding of single-stranded DNA fragments. As a consequence of auto-cleavage of the Ala84-Gly85 peptide bond carried out by Ser119 and Lys156, LexA loses DNA binding activity, thereby inducing the SOS response.
Genes encoding LexA homologs are highly conserved in bacterial genomes and LexA-dependent transcriptional regulation of genes involved in DNA repair has been reported in various bacterial species (Erill et al., 2007; Butala et al., 2009), indicating that the regulation of SOS regulon by LexA might be a universal adaptation strategy of bacteria to DNA damage. However, LexA homologs in several cyanobacterial species were suggested not to be involved in the typical E. coli-type SOS regulation. In Anabaena sp. PCC 7120, auto-cleavage of the Ala84-Gly85 bond of LexA does not occur at physiological pH even in the presence of activated RecA (Kumar et al., 2015). In the case of Synechocystis sp. PCC 6803 (S.6803), LexA lacks the conserved Ala-Gly auto-cleavage site and the serine of the Ser-Lys dyad required for auto-cleavage activity (Patterson-Fortin et al., 2006) and auto-cleavage of LexA in S.6803 has not been reported so far. DNA microarray analysis revealed that LexA depletion did not affect the expression level of genes involved in DNA metabolism (Domain et al., 2004).
The cellular processes regulated by LexA in S.6803 have been implied by studies reporting isolation of LexA as a binding factor to the promoter region of specific genes, such as the hoxEFUYH operon encoding bidirectional hydrogenase (Gutekunst et al., 2005; Oliveira and Lindblad, 2005), crhR encoding RNA helicase (Patterson-Fortin et al., 2006), and sbtA encoding sodium-dependent bicarbonate transporter (Lieman-Hurwitz et al., 2009). Domain et al. (2004) performed DNA microarray analysis of the LexA-depleted strain and found that most of genes affected were previously reported to be regulated by the availability of inorganic carbon (Wang et al., 2004). Kamei et al. (2001) reported that the lexA-disrupted mutant of the motile strain of S.6803 (denoted PCC strain) showed non-motile phenotype. DNA microarray analysis revealed that expression of the pilA genes encoding the subunits of the type IV pilus-like structure was lowered in the mutant. Although regulation of various cellular processes has been suggested, we currently have still a fragmentary understanding of the function of LexA in S.6803.
DNA microarray analysis has been the most popular methods of genome-wide transcriptome profiling. However, it has been supplanted by RNA-seq analysis in which isolated transcripts are converted into the complementary DNA (cDNA) followed by direct sequence in a massively parallel DNA sequencing-based approach. The advantages of RNA-seq over DNA microarray are its higher resolution and better dynamic range of detecting differential gene expression (Zhao et al., 2014). In order to obtain the comprehensive view of LexA-regulated genes in S.6803, here we performed RNA-seq analysis of the wild-type (WT) strain and the lexA-disrupted mutant. The results of RNA-seq analysis indicate that LexA in S.6803 regulates specific cellular functions such as phototactic motility, accumulation of the major compatible solute glucosylglycerol and subunits of bidirectional hydrogenase, and photosynthetic complexes, but not the SOS response. DNA gel mobility shift assay using the recombinant LexA protein suggested that LexA binds to the upstream region of pilA7, pilA9, ggpS, and slr1670 to directly regulate their expression.
Materials and Methods
Strains and Culture Conditions
A glucose-tolerant non-motile strain (GT strain) of Synechocystis sp. PCC 6803 was grown at 32°C in BG-11 medium containing 20 mM HEPES-NaOH, pH 7.0, under continuous illumination at 20 μmol photons m−2 s−1 with bubbling of air. The lexA (sll1626)-disrupted mutant (ΔlexA) was grown under the same conditions, except that 20 μg mL−1 kanamycin (Km) was added to the medium. Cell density was estimated by measuring OD730 using a spectrophotometer (model UV-160A, Shimadzu).
Generation of the lexA (sll1626)-Disrupted Mutant
The coding region of lexA (612 bp, from nucleotide 1319330 to 1318719 according to numbering in CyanoBase) was disrupted by insertion of a kanamycin resistance (Kmr) cassette. The upstream and downstream fragments including the lexA coding sequence were amplified by PCR from the genomic DNA of the WT strain using the primer sets lexA-F and Km-lexA-R (for amplification of 404 bp upstream fragment, from nucleotide 1319525 to 1319122) and Km-lexA-F and lexA-R (for amplification of 394 bp downstream fragment, from nucleotide 1318996 to 1318603; Table S1). Kmr cassette was PCR amplified from the pRL161 plasmid using the primer set Km-F and Km-R (Table S1). The amplified lexA fragments and Kmr cassette were fused together by the fusion PCR method (Wang et al., 2002) using the primer set lexA-F and lexA-R. The WT strain was transformed with the fusion PCR product and transformants (ΔlexA mutant) were selected in the presence of Km.
RNA Gel Blot Analysis
Isolation of total RNA by the hot phenol method and RNA gel blot analyses, using DIG RNA Labeling and Detection Kit (Roche), were performed as described previously (Muramatsu and Hihara, 2003). Template DNA fragments for in vitro transcription to generate RNA probes were prepared by PCR using the primers shown in Table S1.
Immunoblot Analysis
Total proteins were extracted from Synechocystis cells as described previously (Ishii and Hihara, 2008) and separated by 15% (w/v) SDS-PAGE, followed by electroblotting onto PVDF membranes (Immobilon-P; Millipore). Immunodetection was done using a rabbit polyclonal antibody raised against His-LexA recombinant protein. Goat anti-rabbit IgG conjugated to alkaline phosphatase was used as a secondary antibody.
Determination of Pigment Contents
In vivo absorption spectra of whole cells suspended in BG-11 medium were measured at room temperature using a spectrophotometer (V-650 Spectrometer, JASCO) with ISV-722 integrating sphare. Chlorophyll and phycocyanin contents were calculated from the peak heights of absorption spectra using the equations described in Arnon et al. (1974).
RNA-seq Analysis
RNA-seq analysis was carried out using cultures at OD730 = 0.5 and OD730 = 1.0 with three biological replicates. WT and ΔlexA were inoculated into new media at OD730 = 0.1 and incubated for 50 and 80 h, respectively, to be harvested at OD730 = 0.5. Similarly, WT and ΔlexA were inoculated at OD730 = 0.1 and incubated for 70 and 120 h, respectively, to be harvested at OD730 = 1.0. Isolation of total RNA by the hot phenol method was performed as described previously (Muramatsu and Hihara, 2003). To eliminate genomic DNA from total RNA samples, each sample was added with DNase I (TaKaRa) and incubated at 37°C for 3 h. Total RNA concentration was measured with Nanodrop 2000 (Thermo Fisher Scientific). The Ribo-Zero Magnetic Kit for Bacteria (Epicentre) was used to remove ribosomal RNA from each sample. Concentration and quality of mRNA samples were examined using an Agilent 2100 Bioanalyzer. TruSeq RNA Sample Prep Kit v2 (Illumina) was used for cDNA library construction, and the libraries were sequenced using the Illumina MiSeq system. 12 samples in total were analyzed using two cartridge of MiSeq Reagent Kit v3 (Illumina).
A total of 64 million reads data was obtained from 12 samples. To quantify expression level of each gene, nucleotide sequences of obtained reads were mapped to the genomic sequence of GT-I strain of S.6803 (Kanesaki et al., 2012) (NC_017038.1; http://www.ncbi.nlm.nih.gov/nuccore/NC_017038) using CLC Genomics Workbench 7.5.1 software (Qiagen). Raw read counts were divided by length of the transcripts and total number of million mapped reads in each sample to obtain reads per kilobase per million (RPKM) values (Mortazavi et al., 2008). TCC package of R software (Sun et al., 2013) was used to detect the differentially expressed genes between WT and ΔlexA. A false discovery rate of <0.01 was considered to be significant.
Overexpression and Purification of His-lexA
The coding region of the lexA gene was amplified by PCR using the primers lexA-NdeI-F and lexA-XhoI-R (Table S1), containing NdeI and XhoI sites at their 5′ end, respectively. The amplified lexA coding fragment was cloned into the pT7Blue T-vector (Novagen), digested with NdeI and XhoI and subcloned into the same restriction sites in pET28a vector (Novagen) to express the LexA protein with an N-terminal 6 × His-tag.
E. coli BL21(DE3) harboring the His-LexA expression construct was grown to an OD600 = 0.6 in 250 mL of 2 × yeast extract-tryptone (YT) medium containing 20 μg mL−1 Km at 37°C and induced with 0.013% of isopropyl β-D-thiogalactoside for 3 h. The cells were pelleted by centrifugation at 5800 g for 2 min, resuspended in 50 mM sodium phosphate buffer, pH 7.4, containing 0.5 M NaCl and 60 mM imidazole, and disrupted by three rounds of sonication with Sonifier 450 (Branson) for 2 min with interval of 1 min on ice. After the removal of whole cells and insoluble material by centrifugation, the soluble protein fraction was filtered through a 0.2 μm filter (DISMIC-25CS; ADVANTEC). His-LexA was purified by nickel-affinity column chromatography using a HisTrap FF crude (GE Healthcare). The soluble protein fraction was applied to the column equilibrated with 20 mM phosphate buffer, pH 7.4, containing 0.5 M NaCl and 60 mM imidazole, washed with 20 mM phosphate buffer, pH 7.4, containing 0.5 M NaCl and 80 mM imidazole, and eluted with 20 mM phosphate buffer, pH 7.4, containing 0.5 M NaCl and 300 mM imidazole. Purified His-LexA was desalted by a HiTrap Desalting column (GE Healthcare). Protein composition was examined by 15% (w/v) SDS-PAGE followed by staining with Coomassie Brilliant Blue R-250.
DNA Gel Mobility Shift Assay
Probes for DNA gel mobility shift assays were obtained by PCR amplification with primers shown in Table S1 using genomic DNA as a template. The 3′ end of the DNA fragment for each probe was labeled with digoxigenin (DIG)-ddUTP by using the terminal transferase method according to the manufacturer's instructions (DIG gel shift kit 2nd generation; Roche). Gel mobility shift assays were performed by using a DIG gel shift kit 2nd generation (Roche) according to the manufacturer's instruction except that 1 mM DTT was added to the reaction mixture.
Results
Characterization of the lexA (sll1626)-Disrupted Mutant
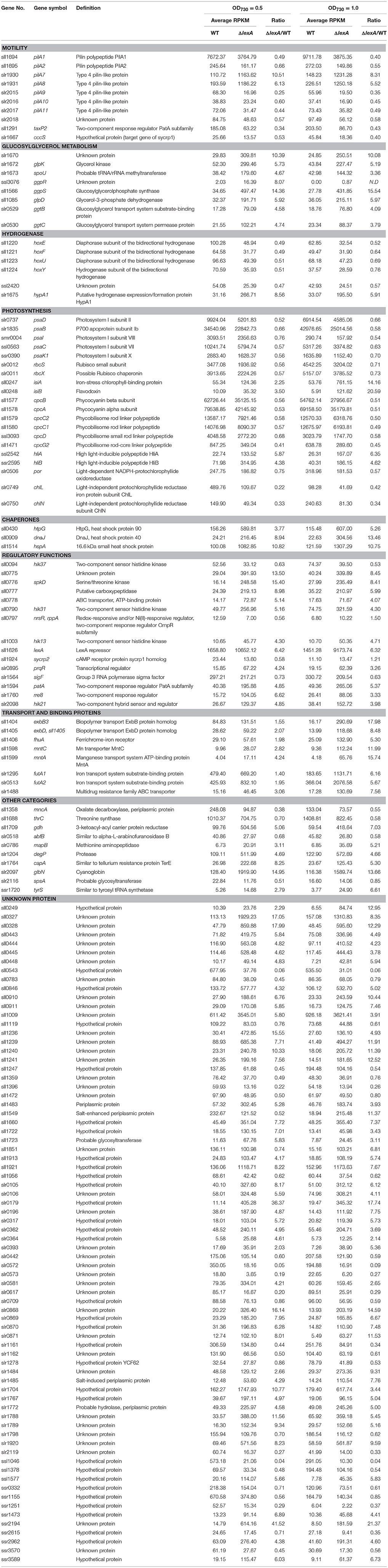
To reveal the function of LexA in GT strain of S.6803, we disrupted the lexA gene by inserting a Kmr cassette within the coding region (Figure 1A). Although a fully segregated mutant was not obtained (Figure 1B), RNA gel blot and immunoblot analyses revealed that both the lexA transcript (Figure 1C) and LexA protein (Figure 1D) levels were below the detection limit in the partially segregated mutant (ΔlexA) grown under normal growth conditions. Under the same conditions, ΔlexA displayed several abnormal phenotypes. The doubling time of ΔlexA was longer (31.4 h) than that of WT (19.5 h) at log phase, whereas the difference in growth rate between strains became smaller at stationary phase (Figure 1E). Amounts of chlorophyll and phycocyanin in ΔlexA calculated from the peak heights of cellular absorption spectra were 93 and 80% of WT levels, respectively (Figure 1F). Microscopic observation revealed that cell size of ΔlexA was heterogeneous and tended to be larger than that of WT (Figure 1G).
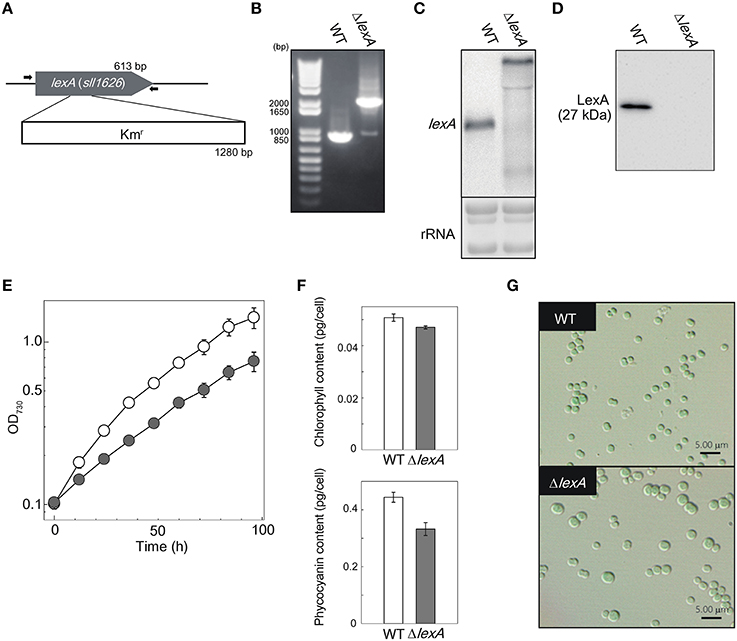
Figure 1. Generation and characterization of the ΔlexA mutant. (A) Scheme of the construct for disruption of lexA. The kanamycin resistance (Kmr) cartridge was inserted into the coding region. Arrows indicate the primers used for PCR amplification shown in (B). (B) PCR amplification of the lexA gene using genomic DNA from WT and the ΔlexA mutant as templates. (C) RNA gel blot analysis of the lexA transcripts detected by single-stranded RNA probe. 3 μg of total RNA were loaded per lane. Total RNA was stained with methylene blue to show the equal loading. (D) Immunoblot analysis of the LexA protein detected by anti-LexA antibody. 5 μg of total protein from cell lysate were loaded per lane. (E) Growth curves of WT (open circles) and the ΔlexA mutant (closed circles) under normal growth conditions. (F) Amounts of photosynthetic pigments calculated from the peak heights of cellular absorption spectra. (G) Observation of cell morphology by differential interference contrast microscopy.
RNA-seq Transcriptome Analysis
To investigate the difference in gene expression profile between WT and ΔlexA, total RNA was isolated from cultures incubated under normal growth conditions and RNA-seq analysis was performed. Figure 2 shows MA plots of the gene expression data obtained from cultures at OD730 = 0.5 and OD730 = 1.0. There were 1011 genes differentially expressed between strains at OD730 = 0.5 as shown in magenta (Figure 2A). Among them, expression levels of 315 genes were more than two-fold higher and those of 28 genes were more than two-fold lower in ΔlexA than in WT (Table S2). In the case of WT and ΔlexA cells at OD730 = 1.0, there were 447 genes differentially expressed between strains (Figure 2B). Among them, expression levels of 360 genes were more than two-fold higher and those of 21 genes were more than two-fold lower in ΔlexA than in WT (Table S3).
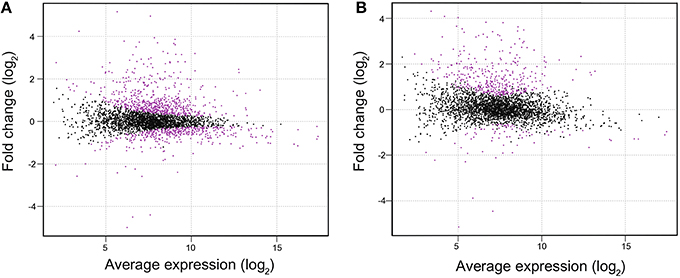
Figure 2. MA plots of RNA-seq data obtained from WT and ΔlexA cells at OD730 = 0.5 (A) and OD730 = 1.0 (B). The MA plot, a scatterplot of log2-fold-change (ΔlexA /WT) versus average expression in log2 scale for each gene, was produced using TCC package. Dots shown in magenta indicate differentially expressed genes with a false discovery rate < 0.01.
Table 1 shows the list of genes whose expression was affected by disruption of lexA. The higher resolution and better dynamic range of RNA-seq analysis compared to DNA microarray analysis enabled listing of small ORFs such as ssl1577, ggpR (ssl3076), ssr1251, ssr1473 and ssr3589, and genes with low expression level (low RPKM value) that cannot be detected by previous DNA microarray analyses. Differentially expressed genes can be categorized into several groups according to related cellular functions as mentioned below.
Motility-Related Genes
The motile strain of S.6803 exhibits phototactic motility dependent on the type IV-like thick pilus structure (Brahamsha and Bhaya, 2014). In S.6803 genome, there are multiple genes homologous to the pilA gene encoding the subunit of the type IV pilus-like structure. Among them, pilA1 was shown to be responsible for the thick pilus structure, motility, and transformation efficiency (Bhaya et al., 1999; Yoshihara et al., 2001), whereas functions of other pilA-like genes are unknown. We observed that their expression is positively or negatively affected by disruption of the lexA gene. Expression of pilA7-pilA8 was largely enhanced whereas that of pilA9-pilA10-pilA11 and pilA1-pilA2 decreased. The observed decrease in expression level of pilA1 and pilA9-pilA10-pilA11 was consistent with the results of DNA microarray analysis of the ΔlexA mutant in the motile PCC strain (Kamei et al., 2001).
Furthermore, we observed that several genes other than pilA involved in motility were affected by disruption of lexA. Expression of pixG-pixH-pixI-pixJ1-pixJ2-pixL (sll0038-0043) encoding regulatory factors involved in positive phototaxis increased in the mutant (Tables S2). It has been reported that motility is controlled by cAMP level in S.6803 and inactivation of cya1 encoding adenylate cyclase or sycrp1 encoding cAMP receptor protein results in loss of motility (Terauchi and Ohmori, 1999; Yoshimura et al., 2002a). Although expression of cya1 and sycrp1 itself was not so much affected by disruption of lexA, decrease in expression levels of five genes, pilA9-pilA10-pilA11-slr2018 and cccS (slr1667), out of six genes reported to be decreased by disruption of sycrp1(Yoshimura et al., 2002b) was observed (Table 1). cccS is also considered to be related to motility, since its disruption resulted in loss of the thick pili (Yoshimura et al., 2010). We observed expression level of sycrp2 is lower in ΔlexA (Table 1), although involvement of SYCRP2 in regulation of motility has not been reported.
Glucosylglycerol-Related Genes
In S.6803, glucosylglycerol (GG) is a major compatible solute to adapt to high-salt or high-osmotic pressure conditions (Klähn and Hagemann, 2011). A set of genes related to GG biosynthesis (ggp, glp) and uptake (ggt) are organized into several gene clusters such as ggtBCD (slr0529-0531), ggpS-glpD (sll1566-sll1085), ggpP-ggtA (slr0746-0747), and slr1670-glpK-spoU-slr1674-hypA1(slr1670-1675) in S.6803 genome (Mikkat and Hagemann, 2000; Klähn et al., 2010). RNA-seq analysis revealed that expression levels of these gene clusters were significantly higher in ΔlexA than WT (Table 1 and Table S2). Klähn et al. (2010) reported that a small ORF, ggpR (ssl3076), exists overlapping with the transcription initiation site of ggpS and its promoter region. Expression of ggpR was also induced by disruption of lexA (Table 1).
Hydogenase-Related Genes
Expression level of the hoxE-hoxF-hoxU-hoxY-hoxH operon encoding subunits of bidirectional NiFe-hydrogenase was lower in ΔlexA. This observation is consistent with the previous study reporting that LexA acts as a transcriptional activator for the hox operon (Gutekunst et al., 2005). On the other hand, the expression level of the hypA1 gene involved in hydrogenase maturation increased in ΔlexA.
Photosynthesis-Related Genes
In S.6803, photosystem (PS) I complex is comprised of 11 subunits and genes encoding these subunits (psa) are dispersed throughout the genome (Kaneko et al., 1996). We found that the expression level of every PSI gene was lower in ΔlexA than WT. The expression level of genes encoding subunits of phycobilisome (cpc, apc) was also lower in the mutant, whereas the expression level of genes encoding PSII subunits (psb) was not so much affected by disruption of lexA. Expression levels of chlL-chlN encoding subunits of light-independent protochlorophyllide reductase and that of por encoding light-dependent protochlorophyllide reductase were lower in ΔlexA. Both light-dependent and -independent enzymes catalyzing the last step of chlorophyll biosynthesis are likely to be under the control of LexA. On the other hand, expression level of hliA and hliB encoding high-light inducible proteins was higher in ΔlexA than WT.
SOS-Response Related Genes
Previous studies suggested that LexA in S.6803 is not involved in the SOS response. Neither lexA nor recA expression was induced upon UV-irradiation (Domain et al., 2004; Patterson-Fortin et al., 2006) and none of DNA metabolism-related genes was listed as genes induced or repressed by LexA depletion (Kamei et al., 2001; Domain et al., 2004). Similarly, induction or repression of DNA metabolism-related genes by disruption of lexA was not observed in our RNA-seq analysis.
Genes Differentially Expressed in ΔlexA at the Later Stage of Growth
Several genes expressed under iron-limiting conditions such as exbB-exbD-fhuA operon involved in inorganic iron uptake (Jiang et al., 2015), futA1 and futA2 encoding subunits of iron transporter (Katoh et al., 2001), and isiA-isiB operon encoding iron-stress inducible proteins (Vinnemeier et al., 1998) were highly induced in ΔlexA at OD730 = 1.0 but not in OD730 = 0.5. In the case of mntA and mntC encoding subunits of manganese transporter (Bartsevich and Pakrasi, 1995), their expression level was already higher in ΔlexA at OD730 = 0.5 and showed further increase at OD730 = 1.0.
DNA Gel Mobility Shift Assay
DNA gel mobility shift assay was performed to examine whether LexA directly regulates expression of putative target genes listed by RNA-seq analysis (Figure 3). We observed induction of the pilA7-pilA8 operon and repression of the pilA9-pilA10-pilA11 operon in ΔlexA (Table 1). Binding of His-LexA to the promoter regions of both operons (for the pilA7 operon from nucleotide 2222102 to 2222304 and for the pilA9 operon from nucleotide 755577 to 755778, according to numbering in CyanoBase) was observed, indicating that LexA directly activates or represses expression of these pilA operons. We also examined whether His-LexA binds to the upstream region of the two divergently transcribed operons, ggpS-glpD and slr1670-glpK-spoU-slr1674-hypA1, both of which are highly induced in ΔlexA. His-LexA bound to the promoter fragment of each operon (for the ggpS operon from nucleotide 1949371 to 1949186 and for the slr1670 operon from nucleotide 1949332 to 1949534). It is notable that LexA-binding site for the the ggpS operon is within the coding region of ggpR (nucleotide 1949372 to 1949100). Our results suggest that LexA binds to at least two binding site located in the intergenic region of the ggpS and slr1670 operons to repress their expression. Next, we examined the binding of LexA to the upstream region of PSI genes by using light-responsive promoter fragments containing the HLR1 sequence recognized by the response regulator RpaB (Seino et al., 2009). Binding of His-LexA to the promoter region of PSI genes was not observed (Figure 3) or much weaker than that to the pilA7, pilA9, ggpS, and slr1670 promoters and not reproducible. This indicates that decrease in expression levels of PSI genes in ΔlexA may be a secondary effect.
Figure 3. DNA gel mobility shift assay of the promoter segments of putative target genes with His-LexA. DIG-labeled promoter segments of pilA7, pilA9, ggpS, slr1670, and psaD were incubated for 25 min at room temperature with His-LexA added at indicated concentrations. five-fold and 50-fold excess amounts of the non-labeled promoter segments were added as a competitor. Samples were separated on a 6% polyacrylamide gel.
Discussion
Effects of Disruption of the lexA Gene in S.6803
In this study, we created the gene-disrupted mutant of lexA in GT strain of S.6803 to obtain the comprehensive view of LexA regulon by RNA-seq analysis. Although Kamei et al. (2001) successfully obtained the fully-segregated lexA mutant from the motile PCC strain, in most cases the ΔlexA mutant invariably retained the WT copy of the lexA gene (Domain et al., 2004; Gutekunst et al., 2005) and we also could not obtain fully-segregated mutant (Figure 1B). The heterogeneous appearance of the ΔlexA mutant cells (Figure 1G) may be caused by difference in the extent of segregation. However, despite the existence of the WT copy of lexA, immunoblot analysis revealed that LexA protein level was below the detection limit in our mutant (Figure 1D).
To date, LexA in S.6803 has been reported to be involved in transcriptional regulation of genes related to various cellular functions. Our RNA-seq data are consistent with some of these reports, e.g., positive regulation of the hox operon reported by Gutekunst et al. (2005) and positive regulation of the pilA genes reported by Kamei et al. (2001). However, we could not observe the large effect of LexA depletion on carbon metabolism-related genes reported by Domain et al. (2004). Domain et al. isolated RNA for DNA microarray analysis from concentrated cultures incubated on plates for 2 h. The growth condition must be largely different from our liquid culture, which may cause the difference in gene expression profile. in vitro transcription/translation assay performed by Patterson-Fortin et al. (2006) showed that CrhR protein accumulation decreased in response to increasing LexA concentration. However, in our data, expression level of crhR was not affected by disruption of lexA.
RNA-seq data in this study suggested involvement of LexA in regulation of (1) phototactic motility, (2) accumulation of GG, (3) bidirectional hydrogenase, and (4) photosystem I and phycobilisome complexes. We also observed increase in expression level of genes related to iron and manganese uptake in ΔlexA at OD730 = 1.0. LexA may be involved in stage specific repression of these genes, but it is also possible that these genes were upregulated as a consequence of iron and manganese limitation in the mutant culture during prolonged incubation. We will discuss regulation of cellular processes (1)–(4) by LexA in the following sections.
Cellular Processes Regulated by LexA in S.6803
Phototactic Motility
Kamei et al. (2001) reported that disruption of the lexA gene in the motile PCC strain resulted in decrease in expression level of pilA genes and loss of thick pili and motility. Our RNA-seq analysis showed that expression levels of genes related to phototactic motility are largely affected by disruption of lexA also in the non-motile strain. In addition to the decrease in expression level of pilA1 and pilA9-pilA10-pilA11 reported in Kamei et al. (2001), we observed significant induction of pilA7-pilA8. Furthermore, expression levels of several genes related to positive phototaxis and cAMP signaling were affected. Although many non-motile mutants were so far isolated from the PCC strain, information on the mechanism of transcriptional regulation of motility-related genes is limited. Bhaya et al. (1999) reported decrease in expression level of pilA1 and pilA2 by disruption of the sigF gene encoding an alternative sigma factor. Yoshimura et al. (2002b) and Dienst et al. (2008) reported decrease in expression level of pilA9-pilA10-pilA11-slr2018 and cccS-cccP by disruption of sycrp1 encoding cAMP receptor protein and hfq encoding RNA chaperone homolog, respectively. Panichkin et al. (2006) reported decrease in expression level of pilA9-pilA10-pilA11-slr2018 and increase in that of pilA5-pilA6 and pilA1-pilA2 by disruption of spkA encoding Serine/threonine protein kinase. None of these reports showed the direct interaction of these regulatory factors with pilA genes and LexA in this study is the first report of binding of transcriptional regulator to their upstream region (Figure 3). Involvement of SYCRP1 in transcriptional regulation of pilA genes through the direct regulation of LexA is not likely, since no SYCRP1 binding sequence has been detected in the upstream region of the lexA gene (Omagari et al., 2008; Xu and Su, 2009). Further examination of relationship between LexA and previously identified regulatory factors which affect motility may be a key to understanding of signal transduction mechanism regulating phototactic motility.
Accumulation of GG
In order to acclimate to high-salt or high-osmotic pressure conditions, S.6803 accumulates the compatible solute GG. Upon a salt shock, genes related to both GG biosynthesis (ggp, glp) and uptake (ggt) are induced (Kanesaki et al., 2002; Marin et al., 2004). GG is synthesized by a two-step reaction in S.6803. First, condensation of ADP-glucose and glycerol 3-phosphate is catalyzed by GG-phosphate synthase (GgpS) and then the intermediate is dephosphorylated by GG-phosphate phosphatase (GgpP) (Hagemann and Erdmann, 1994). Glycerol-3-phosphate dehydrogenase (GlpD) and glycerol kinase (GlpK) are involved in the metabolism of glycerol-3-phosphate, a precursor of GG. Uptake of GG from the environment is performed by ABC transporter consisting of an ATP-binding protein (GgtA), a substrate-binding protein (GgtB) and two integral membrane proteins (GgtC and GgtD) in S.6803 (Mikkat and Hagemann, 2000). All of these genes are induced by the disruption of lexA (Table 1 and Table S2). DNA gel mobility shift assay revealed that His-LexA protein binds to the upstream region of two divergently transcribed operons, ggpS-glpD and slr1670-glpK-spoU-slr1674-hypA1 (Figure 3). To date, sigma factors SigF (Marin et al., 2002) and SigB (Nikkinen et al., 2012), a small protein GgpR (Klähn et al., 2010) and a response regulator Slr1588 (Chen et al., 2014) were reported to be involved in transcriptional regulation of the ggpS-glpD operon. Our result suggests the existence of the additional regulatory mechanism, namely, repression of the divergent ggpS and slr1670 operons by LexA. Expression of ggpR may also be repressed by LexA, judging from the fact its expression was induced by the disruption of lexA (Table 1). Salt-stress inducible genes such as hliA, hliB, hspA, prqR, degP, sll1723, sll0846, sll1483, slr1704, sll1236, slr0581, and ssr2194, reported in the previous DNA microarray studies (Kanesaki et al., 2002; Marin et al., 2004), were also induced by the disruption of lexA (Table 1). There is possibility that LexA acts as a repressor for multiple salt-stress inducible genes as well as the ggpS and slr1670 operons.
Bidirectional Hydogenase
Regulation of the hox operon by LexA has been extensively studied in S.6803 (Oliveira and Lindblad, 2009). LexA was shown to bind to two distinct regions of the hox promoter, −198 to −338 and −592 to −690, relative to the start codon of hoxE (Gutekunst et al., 2005; Oliveira and Lindblad, 2005) and work for positive regulation of hydrogenase activity (Gutekunst et al., 2005). Regulation of hydrogenase-related genes by LexA may be common among cyanobacterial species, judging from the reports on LexA homologs in Anabaena sp. PCC 7120 (Sjöholm et al., 2007) and Lyngbya majuscula CCAP 1446/4 (Ferreira et al., 2007).
Photosystem I and Phycobilisome
In the ΔlexA mutant, chlorophyll and phycocyanin contents were lower than those in WT (Figure 1F). This may be caused by decreased expression level of genes encoding subunits of PSI (psa), subunits of phycobilisome (cpc, apc) and both light-dependent and -independent protochlorophyllide reductase (chlL, chlB, por). It is known that these photosynthesis-related genes show the quite similar response to the changing light environment (Muramatsu and Hihara, 2012). The response regulator RpaB regulates high-light response of photosynthesis-related genes by binding to their promoter regions under low-light conditions (Wilde and Hihara, 2016). PSI genes and hli genes are positively- and negatively-regulated target genes of RpaB, respectively (Kappell and van Waasbergen, 2007; Seki et al., 2007; Seino et al., 2009). Repression of PSI genes and induction of hli genes by disruption of LexA (Table 1) seem to suggest overlapping roles of RpaB and LexA in regulation of photosynthetic gene expression. However, clear and reproducible band shift was not observed when binding of His-LexA to the promoter regions of PSI genes was examined (Figure 3). It is possible that changes in expression levels of photosynthesis-related genes in ΔlexA are not the consequence of loss of regulation by LexA but a secondary effect.
Search for LexA Binding Sites in the Target Promoters
Our results of DNA gel mobility shift assay suggest that LexA binds to the upstream region of piA7, pilA9, ggpS and slr1670 to directly regulate their expression (Figure 3). To date, several nucleotide sequences for LexA binding site have been identified by DNA gel mobility shift assay, for example, 5′-TTTATTTGA ACTATTTTT-3′, 5′-TTTTTCGTTGTCTAA ATT-3′ (Oliveira and Lindblad, 2005), 5′-CTA-N9(AT-rich)-CTA-3′ (Patterson-Fortin and Owttrim, 2008), and 5′-AGT AACTAGTTCG-3′ (Gutekunst et al., 2005) in S.6803 and 5′-TAG TACTAATGTTCTA-3′ in A.7120. (Mazón et al., 2004). However, these LexA binding sequences could not be found in the promoter fragments to which His-LexA bound. Instead, we found that a 5′-TTTTG(A/T)TNAC-3′ sequence commonly exists in these promoter fragments (Figure S1). The sequence is located around the putative transcription start site in the case of the negatively-regulated target genes, ggpS, piA7, and slr1670, whereas it is located further upstream region in the case of the positively-regulated pilA9 gene. It has been reported that a certain global transcriptional regulator, such as NtcA and RpaB in S.6803, can act as both repressor and activator dependent on the location of the binding site (García-Domínguez et al., 2000; Seino et al., 2009). Binding of the transcriptional regulator causes repression when its binding site overlaps the RNA polymerase-binding site, whereas activating effect is observed when the binding site is located further upstream. The location of 5′-TTTTG(A/T)TNAC-3′ sequence in four LexA-target promoters seems consistent with the scheme.
Physiological Roles of Cyanobacterial LexA
Results of RNA-seq analysis (Table 1) together with DNA gel mobility shift assay (Figure 3) suggest LexA in S.6803 can positively or negatively regulate various cellular processes such as phototactic motility, GG accumulation and hydogenase activity. Regulation of such a wide range of cellular processes by LexA was reported in other bacterial species. For example, the lexA mutant of Clostridium difficile showed pleiotrophic phenotypes such as filamentous structure due to inhibition of cell division, decreased sporulation, decrease in swimming motility and increased biofilm formation (Walter et al., 2015). In this case, LexA acts as a regulator of DNA damage in addition to the above mentioned biological functions. In contrast, DNA microarray data from different research groups (Kamei et al., 2001; Domain et al., 2004) and our RNA-seq data suggest LexA in S.6803 is not involved in regulation of SOS genes. In S.6803, expression of lexA and recA was not induced upon UV-irradiation (Domain et al., 2004; Patterson-Fortin et al., 2006). Similarly, in Anabaena sp. PCC 7120, expression of lexA was not induced upon UV-B exposure or treatment with a DNA damaging agent mitomycin C (Kumar et al., 2015). In these freshwater species, LexA-independent protection mechanism for DNA damage may have evolved and LexA may have become devoted to regulating other cellular processes. Then, what is the physiological meaning of the coordinated regulation of phototactic motility, GG accumulation, and hydogenase activity by LexA in S.6803? We searched for environmental conditions where LexA-target genes are coordinately regulated using CyanoEXpress gene expression database (http://cyanoexpress.sysbiolab.eu/) and found that salt stress causes induction of GG metabolism-related genes and repression of hox operon and pilA genes in WT (Shoumskaya et al., 2005; Dickson et al., 2012). The expression profile is similar to that observed by disruption of lexA (Table 1), indicating the possibility that transcriptional regulation by LexA is temporarily inactivated under salt stress conditions.
Recently, it has been suggested that the SOS response in the marine Synechococcus is regulated by LexA like E. coli (Blot et al., 2011; Tetu et al., 2013). Cyanobacterial LexA genes can be clustered into three groups, Clade A containing Gloeobacter violaceus PCC 7421, Clade C containing marine picocyanobacteria and Clade B containing most remaining species (Li et al., 2010). There may exist high degree of variation of LexA regulons among species belonging to these three clades. By examination of what kind of cellular processes LexA regulates, we will be able to know decision of each species about how to use the transcriptional regulator LexA for better adaptation to changing environment.
Author Contributions
The study was conceived by AYK and YH, with design input from AKK. Experiments were performed by AYK and AKK. Data analysis and interpretation was done by all authors. The manuscript was prepared by AYK and YH, and reviewed by all authors.
Funding
This work was financially supported by the Core Research of Evolutional Science & Technology (CREST) programs from the Japan Science and Technology Agency (JST).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00193
Table S1. Oligonucleotides used in this study.
Table S2. RNA-seq data of WT and ΔlexA at OD730 = 0.5.
Table S3. RNA-seq data of WT and ΔlexA at OD730 = 1.0.
Figure S1. Consensus sequences for LexA binding site in four LexA-target promoters identified with MEME.
References
Arnon, D. I., McSwain, B. D., Tsujimoto, H. Y., and Wada, K. (1974). Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim. Biophys. Acta 357, 231–245. doi: 10.1016/0005-2728(74)90063-2
Bartsevich, V. V., and Pakrasi, H. B. (1995). Molecular identification of an ABC transporter complex for manganese: analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 14, 1845–1853.
Bhaya, D., Watanabe, N., Ogawa, T., and Grossman, A. R. (1999). The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc. Natl. Acad. Sci. U.S.A. 96, 3188–3193. doi: 10.1073/pnas.96.6.3188
Blot, N., Mella-Flores, D., Six, C., Le Corguille, G., Boutte, C., Peyrat, A., et al. (2011). Light history influences the response of the marine cyanobacterium Synechococcus sp. WH7803 to oxidative stress. Plant Physiol. 156, 1934–1954. doi: 10.1104/pp.111.174714
Brahamsha, B., and Bhaya, D. (2014). “Motility in unicellular and filamentous Cyanobacteria,” in The Cell Biology of Cyanobacteria, eds E. Flores and A. Herrero (Dorset: Caister Academic Press), 233–262.
Butala, M., Žgur-Bertok, D., and Busby, S. J. W. (2009). The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 66, 82–93. doi: 10.1007/s00018-008-8378-6
Chen, L., Wu, L., Zhu, Y., Song, Z., Wang, J., and Zhang, W. (2014). An orphan two-component response regulator Slr1588 involves salt tolerance by directly regulating synthesis of compatible solutes in photosynthetic Synechocystis sp. PCC 6803. Mol. BioSyst. 10, 1765–1774. doi: 10.1039/c4mb00095a
Dickson, D. J., Luterra, M. D., and Ely, R. L. (2012). Transcriptomic responses of Synechocystis sp. PCC 6803 encapsulated in silica gel. Appl. Microbiol. Biotechnol. 96, 183–196. doi: 10.1007/s00253-012-4307-6
Dienst, D., Duhring, U., Mollenkopf, H.-J., Vogel, J., Golecki, J., Hess, W. R., et al. (2008). The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 154, 3134–3143. doi: 10.1099/mic.0.2008/020222-0
Domain, F., Houot, L., Chauvat, F., and Cassier-Chauvat, C. (2004). Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53, 65–80. doi: 10.1111/j.1365-2958.2004.04100.x
Erill, I., Campoy, S., and Barbé, J. (2007). Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31, 637–656. doi: 10.1111/j.1574-6976.2007.00082.x
Ferreira, D., Leitão, E., Sjöholm, J., Oliveira, P., Lindblad, P., Moradas-Ferreira, P., et al. (2007). Transcription and regulation of the hydrogenase(s) accessory genes, hypFCDEAB, in the cyanobacterium Lyngbya majuscula CCAP 1446/4. Arch. Microbiol. 188, 609–617. doi: 10.1007/s00203-007-0281-2
García-Domínguez, M., Reyes, J. C., and Florencio, F. J. (2000). NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 35, 1192–1201. doi: 10.1046/j.1365-2958.2000.01789.x
Gutekunst, K., Phunpruch, S., Schwarz, C., Schuchardt, S., Schulz-Friedrich, R., and Appel, J. (2005). LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58, 810–823. doi: 10.1111/j.1365-2958.2005.04867.x
Hagemann, M., and Erdmann, N. (1994). Activation and pathway of glucosylglycerol synthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 140, 1427–1431. doi: 10.1099/00221287-140-6-1427
Ishii, A., and Hihara, Y. (2008). An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803. Plant Physiol. 148, 660–670. doi: 10.1104/pp.108.123505
Jiang, H.-B., Lou, W.-J., Ke, W.-T., Song, W.-Y., Price, N. M., and Qiu, B.-S. (2015). New insights into iron acquisition by cyanobacteria: an essential role for ExbB-ExbD complex in inorganic iron uptake. ISME J. 9, 297–309. doi: 10.1038/ismej.2014.123
Kamei, A., Hihara, Y., Yoshihara, S., Geng, X., Kanehisa, M., and Ikuechi, M. (2001). “Functional Analysis of lexA-like gene, sll1626 in Synechocystis sp. PCC 6803 using DNA microarray,” in PS2001 Proceedings of 12th International Congress on Photosynthesis (Melbourne, VIC: CSIRO Publishing), S41–013.
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. doi: 10.1093/dnares/3.3.109
Kanesaki, Y., Shiwa, Y., Tajima, N., Suzuki, M., Watanabe, S., Sato, N., et al. (2012). Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 19, 67–79. doi: 10.1093/dnares/dsr042
Kanesaki, Y., Suzuki, I., Allakhverdiev, S. I., Mikami, K., and Murata, N. (2002). Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290, 339–348. doi: 10.1006/bbrc.2001.6201
Kappell, A. D., and van Waasbergen, L. G. (2007). The response regulator RpaB binds the high light regulatory 1 sequence upstream of the high-light-inducible hliB gene from the cyanobacterium Synechocystis PCC 6803. Arch. Microbiol. 187, 337–342. doi: 10.1007/s00203-007-0213-1
Katoh, H., Hagino, N., and Grossman, A. R. (2001). Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183, 2779–2784. doi: 10.1128/JB.183.9.2779-2784.2001
Klähn, S., and Hagemann, M. (2011). Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 13, 551–562. doi: 10.1111/j.1462-2920.2010.02366.x
Klähn, S., Höhne, A., Simon, E., and Hagemann, M. (2010). The gene ssl3076 encodes a protein mediating the salt-induced expression of ggpS for the biosynthesis of the compatible solute glucosylglycerol in Synechocystis sp. strain PCC 6803. J. Bacteriol. 192, 4403–4412. doi: 10.1128/JB.00481-10
Kumar, A., Kirti, A., and Rajaram, H. (2015). LexA protein of cyanobacterium Anabaena sp. strain PCC7120 exhibits in vitro pH-dependent and RecA-independent autoproteolytic activity. Int. J. Biochem. Cell Biol. 59, 84–93. doi: 10.1016/j.biocel.2014.12.003
Li, S., Xu, M., and Su, Z. (2010). Computational analysis of LexA regulons in cyanobacteria. BMC Genomics 11:527. doi: 10.1186/1471-2164-11-527
Lieman-Hurwitz, J., Haimovich, M., Shalev-Malul, G., Ishii, A., Hihara, Y., Gaathon, A., et al. (2009). A cyanobacterial AbrB-like protein affects the apparent photosynthetic affinity for CO2 by modulating low-CO2-induced gene expression. Environ. Microbiol. 11, 927–936. doi: 10.1111/j.1462-2920.2008.01818.x
Marin, K., Huckauf, J., Fulda, S., and Hagemann, M. (2002). Salt-dependent expression of glucosylglycerol-phosphate synthase, involved in osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 184, 2870–2877. doi: 10.1128/JB.184.11.2870-2877.2002
Marin, K., Kanesaki, Y., Los, D. A., Murata, N., Suzuki, I., and Hagemann, M. (2004). Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 136, 300–329. doi: 10.1104/pp.104.045047
Mazón, G., Lucena, J. M., Campoy, S., Fernández de Henestrosa, A. R., Candau, P., and Barbé, J. (2004). LexA-binding sequences in Gram-positive and cyanobacteria are closely related. Mol. Genet. Genomics 271, 40–49. doi: 10.1007/s00438-003-0952-x
Mikkat, S., and Hagemann, M. (2000). Molecular analysis of the ggtBCD gene cluster of Synechocystis sp. strain PCC6803 encoding subunits of an ABC transporter for osmoprotective compounds. Arch. Microbiol. 174, 273–282. doi: 10.1007/s002030000201
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Muramatsu, M., and Hihara, Y. (2003). Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216, 446–453. doi: 10.1007/s00425-002-0859-5
Muramatsu, M., and Hihara, Y. (2012). Acclimation to high-light conditions in cyanobacteria: from gene expression to physiological responses. J. Plant Res. 125, 11–39. doi: 10.1007/s10265-011-0454-6
Nikkinen, H.-L., Hakkila, K., Gunnelius, L., Huokko, T., Pollari, M., and Tyystjarvi, T. (2012). The SigB factor regulates multiple salt acclimation responses of the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 158, 514–523. doi: 10.1104/pp.111.190058
Oliveira, P., and Lindblad, P. (2005). LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 251, 59–66. doi: 10.1016/j.femsle.2005.07.024
Oliveira, P., and Lindblad, P. (2009). Transcriptional regulation of the cyanobacterial bidirectional Hox-hydrogenase. Dalton Trans. 7, 9990–9996. doi: 10.1039/b908593a
Omagari, K., Yoshimura, H., Suzuki, T., Takano, M., Ohmori, M., and Sarai, A. (2008). Delta G-based prediction and experimental confirmation of SYCRP1-binding sites on the Synechocystis genome. FEBS J. 275, 4786–4795. doi: 10.1111/j.1742-4658.2008.06618.x
Panichkin, V. B., Arakawa-Kobayashi, S., Kanaseki, T., Suzuki, I., Los, D. A., Shestakov, S. V., et al. (2006). Serine/threonine protein kinase SpkA in Synechocystis sp. strain PCC 6803 is a regulator of expression of three putative pilA operons, formation of thick pili, and cell motility. J. Bacteriol. 188, 7696–7699. doi: 10.1128/JB.00838-06
Patterson-Fortin, L. M., Colvin, K. R., and Owttrim, G. W. (2006). A LexA-related protein regulates redox-sensitive expression of the cyanobacterial RNA helicase, crhR. Nucleic Acids Res. 34, 3446–3454. doi: 10.1093/nar/gkl426
Patterson-Fortin, L. M., and Owttrim, G. W. (2008). A Synechocystis LexA-orthologue binds direct repeats in target genes. FEBS Lett. 582, 2424–2430. doi: 10.1016/j.febslet.2008.06.009
Seino, Y., Takahashi, T., and Hihara, Y. (2009). The response regulator RpaB binds to the upstream element of photosystem I genes to work for positive regulation under low-light conditions in Synechocystis sp. strain PCC 6803. J. Bacteriol. 191, 1581–1586. doi: 10.1128/JB.01588-08
Seki, A., Hanaoka, M., Akimoto, Y., Masuda, S., Iwasaki, H., and Tanaka, K. (2007). Induction of a group 2 σ factor, RPOD3, by high light and the underlying mechanism in Synechococcus elongatus PCC 7942. J. Biol. Chem. 282, 36887–36894. doi: 10.1074/jbc.M707582200
Shoumskaya, M. A., Paithoonrangsarid, K., Kanesaki, Y., Los, D. A., Zinchenko, V. V., Tanticharoen, M., et al. (2005). Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J. Biol. Chem. 280, 21531–21538. doi: 10.1074/jbc.M412174200
Sjöholm, J., Oliveira, P., and Lindblad, P. (2007). Transcription and regulation of the bidirectional hydrogenase in the cyanobacterium Nostoc sp. strain PCC 7120. Appl. Environ. Microbiol. 73, 5435–5446. doi: 10.1128/AEM.00756-07
Sun, J., Nishiyama, T., Shimizu, K., and Kadota, K. (2013). TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14:219. doi: 10.1186/1471-2105-14-219
Terauchi, K., and Ohmori, M. (1999). An adenylate cyclase, cya1, regulates cell motility in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 40, 248–251. doi: 10.1093/oxfordjournals.pcp.a029534
Tetu, S. G., Johnson, D. A., Varkey, D., Phillippy, K., Stuart, R. K., Dupont, C. L., et al. (2013). Impact of DNA damaging agents on genome-wide transcriptional profiles in two marine Synechococcus species. Front. Microbiol. 4:232. doi: 10.3389/fmicb.2013.00232
Vinnemeier, J., Kunert, A., and Hagemann, M. (1998). Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 169, 323–330. doi: 10.1111/j.1574-6968.1998.tb13336.x
Walter, B. M., Cartman, S. T., Minton, N. P., Butala, M., and Rupnik, M. (2015). The SOS response master regulator LexA is associated with sporulation, motility and biofilm formation in Clostridium difficile. PLoS ONE 10:e0144763. doi: 10.1371/journal.pone.0144763
Wang, H.-L., Postier, B. L., and Burnap, R. L. (2002). Polymerase chain reaction-based mutagenesis identify key transporters belonging to multigene families involved in Na+ and pH homeostasis of Synechocystis sp. PCC 6803. Mol. Microbiol. 44, 1493–1506. doi: 10.1046/j.1365-2958.2002.02983.x
Wang, H.-L., Postier, B. L., and Burnap, R. L. (2004). Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 279, 5739–5751. doi: 10.1074/jbc.M311336200
Wilde, A., and Hihara, Y. (2016). Transcriptional and posttranscriptional regulation of cyanobacterial photosynthesis. Biochim. Biophys. Acta 1857, 296–308. doi: 10.1016/j.bbabio.2015.11.002
Xu, M., and Su, Z. (2009). Computational prediction of cAMP receptor protein (CRP) binding sites in cyanobacterial genomes. BMC Genomics 10:23. doi: 10.1186/1471-2164-10-23
Yoshihara, S., Geng, X., Okamoto, S., Yura, K., Murata, T., Go, M., et al. (2001). Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 42, 63–73. doi: 10.1093/pcp/pce007
Yoshimura, H., Kaneko, Y., Ehira, S., Yoshihara, S., Ikeuchi, M., and Ohmori, M. (2010). CccS and CccP are involved in construction of cell surface components in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 51, 1163–1172. doi: 10.1093/pcp/pcq081
Yoshimura, H., Yanagisawa, S., Kanehisa, M., and Ohmori, M. (2002b). Screening for the target gene of cyanobacterial cAMP receptor protein SYCRP1. Mol. Microbiol. 43, 843–853. doi: 10.1046/j.1365-2958.2002.02790.x
Yoshimura, H., Yoshihara, S., Okamoto, S., Ikeuchi, M., and Ohmori, M. (2002a). A cAMP receptor protein, SYCRP1, is responsible for the cell motility of Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 460–463. doi: 10.1093/pcp/pcf050
Keywords: cyanobacteria, LexA, RNA-seq, Synechocystis, transcriptome
Citation: Kizawa A, Kawahara A, Takimura Y, Nishiyama Y and Hihara Y (2016) RNA-seq Profiling Reveals Novel Target Genes of LexA in the Cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 7:193. doi: 10.3389/fmicb.2016.00193
Received: 07 January 2016; Accepted: 04 February 2016;
Published: 19 February 2016.
Edited by:
Takashi Osanai, Meiji University, JapanCopyright © 2016 Kizawa, Kawahara, Takimura, Nishiyama and Hihara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yukako Hihara, hihara@molbiol.saitama-u.ac.jp
 Ayumi Kizawa
Ayumi Kizawa Akihito Kawahara
Akihito Kawahara Yasushi Takimura
Yasushi Takimura Yoshitaka Nishiyama
Yoshitaka Nishiyama Yukako Hihara
Yukako Hihara