- 1Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt
- 2Department of Pharmacology, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt
- 3Department of Poultry Diseases, Animal Health Research Institute, Cairo, Egypt
- 4Departments of Biotechnology, Animal Health Research Institute, Giza, Egypt
- 5Department of Microbiology, National Research Center, Cairo, Egypt
Methicillin-resistant Staphylococcus aureus (MRSA) have been found in various farm animal species throughout the world. Yet, methicillin-susceptible S. aureus (MSSA), methicillin-susceptible non-S. aureus (MS-NSA), and methicillin-resistant non-S. aureus (MR-NSA) were not investigated. Therefore, we persued to determine the diversity in their phenotypic virulence assay, phenotypic antimicrobial resistance profile and molecular characterization in one of the food chains in Egypt. Samples were collected during 2013 from beef meat at retail. Twenty seven isolates comprising five species (S. hyicus, S. aureus, S. schleiferi subsp. coagulans, S. intermedius, and S. lentus) were characterized for their antibiotic resistance phenotypic profile and antibiotic resistance genes (mecA, cfr, gyrA, gyrB, and grlA). Out of the 27 Staphylococcus isolates only one isolate was resistant to the 12 antibiotics representing nine classes. Raw beef meat sold across the Great Cairo zone, contains 66.7% of MRS, with highest prevalence was reported in S. aureus (66.7%), while the MRS non-S. aureus strains constituted 66.7% from which S. hyicus (60%), S. intermedius (33.3%), S. schleiferi subsp. coagulans (100%), and S. lentus (100%) were MRS. Seven S. aureus, six S. hyicus, four S. schleiferi subsp. coagulans, three S. intermedius, and one S. lentus isolates although being resistant to oxacillin yet, 11/27 (40.7%) carried the mecA gene. At the same time, the cfr gene was present in 2 of the nine S. aureus isolates, and totally undetectable in S. hyicus, S. schleiferi subsp. coagulans, S. intermedius, and S. lentus. Although, global researches largely focused into MRSA and MR-NSA in animals on pigs, the analysis of our results stipulates, that buffaloes and cattle could be MRSA dispersers and that this theme is not specific to pigs. Detection of MSSA virulence determinants is a must, as although oxacillin resistance may be absent yet, the MSSA may carry the virulence determinants which could be a source of perilous S. aureus for the human community.
Introduction
In Egypt, since more than three millennia dating from the Eighteenth Dynasty chronology, the domestic cattle are incorporated with all types of agricultural activities in addition to their provision of meat, internal organs (heart, kidneys, pancreas, rumen, intestine), milk, cheese, ghee, skin, bone, manure, horns, and tendons (Conférence publique de Mlle M-Christine Lavier Toulon, 2000; Benderitter, 2003).
Antimicrobial agents are widely spread and intensively used in the food production systems (FAO, 2015). The aftermath of this operation varies considerably globally as a result of the interaction between human (social structure) and animal demography (species, distribution, and density), farming systems, contaminated water sources, socio-economic strategies (manufacturing, commerce, availability of food, and animal fitness), and nation and world-wide trade. Although, antibiotic use in food animals has been concealed from the public scene yet, antibiotic application in meat producing animals have impacted on public health and has left no room for confusion after being scientifically documented (Conniff, 2015). In 2010, an announcement having a public consequential impact was made public by the FDA to the world in that 28.7 million pounds of antibiotics, which comprises 80% of the total amount of antibiotics utilized in the USA, were being poured down in the animal production industry not to medical care and that currently, two million people are annually sickened by resistant bacterial infections and that 23,000 die annually according to the CDC (Conniff, 2015).
Eventually, besides tracing the use of antibiotics, the effectual monitoring of antimicrobial resistance is the linchpin of any national and international successful achievement in their control policy. The pivotal role of monitoring was highlighted in the 2001 WHO Global Strategy for Containment of antimicrobial resistance and on the occasion of World Health Day 2011, wherein resistance, antimicrobial consumption, and disease stress are included as prime listings. The facts and statistics collected can be used to implement the government's rationale for introducing such radical legislation in the perfection of antibiotic use, to propose strategy and discern the prime concern for employment to provide public support regionally and globally.
Staphylococcus aureus is a hazard because of its detrimental effects on animal health and its capacity for transmission from animals to humans and vice-versa (Peton and Le Loir, 2014). The isolation universality of multidrug-resistant staphylococci from meat samples has been perceived by several researchers (Santos-Sanches et al., 2000; Khan et al., 2005; Huber et al., 2011; Bhargava and Zhang, 2014; Chajęcka-Wierzchowska et al., 2015; Guran and Kahya, 2015). This should be taken seriously, especially when the finding of Waters et al. (2011) documents that, close to 50 percent of meat samples tested from U.S. grocery stores are contaminated with the bacteria S. aureus and resistant to at least three classes of antibiotics. Methicillin-resistant strains of S. aureus (MRSA), which is linked to a wide range of human diseases (Köck et al., 2010; Fätkenheuer and Kaasch, 2014; Yaw et al., 2014), is notorious for its related-Nous to food contamination and its potentiality for its transmission to humans prevails. The pre-eminent disquiet in the European Union, has become the prevention and control of MRSA being ascertained as the most public health problem (Köck et al., 2010), an act that should be implemented in Egypt. Also, the high prevalence of MR-CNS found in livestock production (Huber et al., 2011) becomes a relevant consternation in view of the dispersing potentiality of mecA to MRSA as several coagulase negative staphylococcus (CNS) species, which are also non-S. aureus (NSA), can serve as reservoirs for mecA (Bhargava and Zhang, 2014). This consigned multiresistant CNS-NSA strains the intuitive capability to develop into an emerging problem for veterinary medicine.
Although CNS have emerged as important pathogens, little is known about the virulence factors of these bacteria (Stepanovic et al., 2001). The most important virulence factor of CNS is assumed to be the capacity for biofilm formation and, that testing for biofilm production could be a useful marker for the pathogenicity of CNS (Stepanovic et al., 2001).
In general, we have not come across any investigations involving biofilm formation, and the antibiotic resistant genes mecA, cfr, gyrA, gyrB, and grlA in MRSA in beef meat and in methicillin-resistant non-S. aureus (MR-NSA). Thereby, we aimed an endeavor to identify one of the main hazardous threat to public health, Staphylococcus species, to determine the generality of contamination of beef meat as a measurement for the exposure of consumers to MRSA and MR-NSA in retail beef meat in the Egyptian market, the rate of biofilm producers among the tested S. aureus comparable to non-S. aureus strains, their carriage to the antibiotic resistant genes mecA, cfr, gyrA, gyrB, and grlA and their consequence as a public health hazard.
Materials and Methods
Sample Collection
During the year 2013, 100 samples of retail whole beef were collected from several butchers and marts in the Great Cairo zone. Each sample weighing at least 500 g was aseptically collected with the minimum of cross-contamination and immediately conveyed to the laboratory to be microbiologically examined.
Isolation and Identification
To 50 g of meat samples, 100 ml of phosphate-buffered saline was added all in a sterile plastic bag which were vigorously shaken for 2 min to remove bacteria. Then 1 ml of each rinsate was used for isolation and detection of Staphylococcus isolates by the Double Enrichment method. The isolates were used for further culture to be consequently biochemically characterized as S. aureus (n = nine) and non-S. aureus (n = 18) according to Thorberg and Brandstrom (2000). A sub-culture of the 27 isolates were stored at −27°C in tryptic soy broth and fetal calf serum [Gibco/Invitrogen Carlsbad (CA) USA] containing 2% yeast extract (v/v) to be revitalized for phenotypic virulence assay, phenotypic antimicrobial resistance profile and molecular characterization.
Phenotypic Virulence Assays
For the 27 isolated staphylococci, four virulence factors were evaluated (alpha, beta and delta hemolysins, cytotoxicity, and biofilm formation) using phenotypic tests.
Hemolysis Pattern on Blood Agar
The 27 tested isolates were subcultured onto 5% defibrinated sheep and rabbit blood (Merck, Darmstad, Germany) at 37°C for 24 h according to the procedure of Futagawa-Saito et al. (2006) to obtain isolated colonies. The type of hemolysis was recorded as α-, β-, and double (α + β). After incubation at 37°C for 24 h the clear zone around the colonies was registered as positive reaction. Some S. aureus strains proved the presence of CAMP-like factor, a special hemolysin, the activity of which is revealed by streaking the bacterial studied strain on 5% sheep blood agar, at 8 mm distance perpendicularly on the β-hemolytic S. aureus ATCC 25923 reference strain, and incubating the plate aerobically at 37°C for 24 h. The occurrence of synergic β-haemolysis at the confluence of the two spot areas indicates the presence of CAMP-like factor. The S. aureus (ATCC 25923 and ATCC 49775) and S. intermedius Hajek (ATCC strain 49052) were used as reference strains.
Cytotoxicity Assay
The cytotoxicity assay was carried out using Vero (African green monkey kidney) cells in 96-well microtiter trays according to the protocol of Tao et al. (1999). Suspensions of each of the 27 staphylococcal strains in distilled water were adjusted to 0.5 McFarland standard. Then, 20 μl of bacterial suspensions were added to 3.5 ml of brain heart infusion broth (BBL, Becton Dickinson Microbiology Systems). The tubes were incubated 2 days at 35°C, and thereafter for 2 days at room temperature. After centrifugation of bacterial broth cultures, 20 μl of supernatants were added in triplicate to 180 μl of cell culture medium. After incubation at 37°C, in a humid atmosphere and 5% CO2, the cytotoxic effect of each staphylococcal strain on the Vero monolayer morphology was microscopically perceived up to 5 days.
Biofilm Formation
Biofilm formation assay was determined by the Christensen's tube method and the Congo red agar assay (CRA) as proposed by Osman et al. (2015, 2016). The biofilm-producer ATCC 25923 and the biofilm negative ATCC 12228 were used as reference strains.
Phenotypic Characterization of Slime Production in CRA
This method is based on the characteristic cultural morphology of biofilm-forming bacteria on Congo red medium. The strains were cultured on CRA plates, composed of brain heart infusion broth (BHI) (Oxoid Ltd, Hampshire, England) 37 g/l, 10 g/l Congo red and 50 g/l sucrose and agar No 1 (Oxoid Ltd, Hampshire, England) 10 g/l. The plates were subsequently incubated for 24 h at 37°C and overnight at room temperature. The production of black colonies with a dry crystalline consistency by the organisms was taken to indicate slime production as non-slime-producing strains develop red colonies. ATCC 35982, a non-slime-producing strain (−), ATCC 35983, a moderate slime producing strain (++), and ATCC 35984, a strong slime producing strain (++), served as controls.
Investigation of Biofilm Production by Adherence to Borosilicate Test Tubes
A qualitative assessment of biofilm formation was determined by tube assay. An overnight culture of each of the 27 isolated staphylococci were grown in MHB and incubated for 24 h at 37°C. The tubes were removed and washed with phosphate buffer saline and dried. Tubes were stained with 0.1% crystal violet for 10 min. Excess stain was removed and tubes were washed with distilled water. Tubes were then dried and observed for biofilm formation. Biofilm formation was considered positive when a visible film lined the wall and bottom of the tube. Ring formation at the liquid-air interface was not indicative of biofilm formation. The scoring for tube method was done according to the results of the control strains. The isolates were classified into three categories: Biofilm formation was considered positive when a visible film lined the wall and the bottom of the tube. The amount of biofilm formed was scored as 1-weak/none, 2-moderate, and 3-high/strong.
Phenotypic Antibiotic Resistance Profile
Resistance of the 27 isolated staphylococci to 12 specific antibiotics (Table 3) were validated by culturing each isolate on Mueller-Hinton agar (MHA) supplemented with 0.5% NaCl and 2% for oxacillin, to obtain a fresh culture, to be determined by the disk diffusion method according to procedures of the CLSI (2012). Antimicrobials on the WHO's critically important antimicrobials list (2012) were selected for testing according to their importance to human and animal health (FAO/WHO/OIE, 2008; WHO, 2009). One loopful of colony material was subcultured in 5 ml Mueller-Hinton broth for an overnight at 35°C, vortexed well and adjusted to obtain a turbidity comparable to that of 0.5 McFarland opacity standard (bioMe'rieux, Marcy 1'Etoile, France). The bacterial suspensions were streaked on MHA plates with a cotton swab and with an antibiotic disc dispenser, the discs were placed on the agar surface, plates were incubated at 37°C, the standard temperature used for antimicrobial susceptibility testing, for 24 h, and the diameter of the inhibition zones was measured. The isolates were classified as sensitive, intermediate and resistant based on the diameter of the clearing zone according to CLSI (2012) guidelines. The antibiotic discs represented nine classes of antibiotics: ampicillin-sulbactam, methicillin, penicillin, and oxacillin (Penicillins); chloramphenicol (Chloramphenicols); ciprofloxacin (Florinated quinolones); clindamycin (Lincosamides); gentamycin (Aminoglycosides); erythromycine (Macrolides); sulphamethoxazole/trimethoprim (Sulphonamides); tetracycline (Tetracyclines); vancomycine (Glycopeptides). S. aureus ATCC 29213 (β-lactamase positive), and S. aureus ATCC 25923 (β-lactamase negative) were used as quality control. Multidrug resistance was reported as a single isolate resistant (intermediate or complete) to 3 or more antimicrobial classes (Waters et al., 2011). Bacteria strains that were resistant and intermediate were categorized as non- susceptible while the sensitive strains were categorized as susceptible.
DNA Extraction
Bacterial DNA from the 27 staphylococci were isolated from an overnight cultures using the boiling protocol previously described (Sowmya et al., 2012). DNA precipitates were resuspended in an appropriate volume of Tris-EDTA buffer solution.
PCR for Detection of Genes Encoding Antimicrobial Resistance
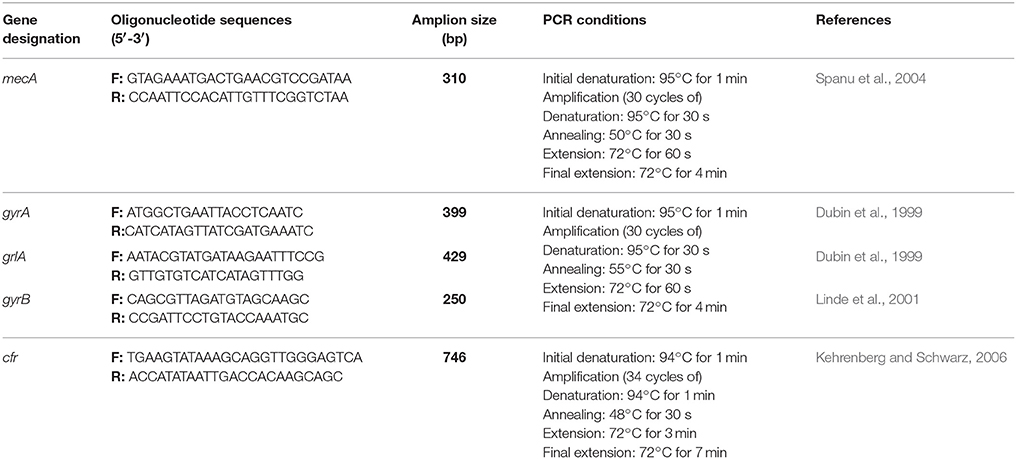
The 27 Staphylococcus species isolates were confirmed as MSSA, MRSA, methicillin-susceptible non-Staphylococcus aureus (non-MSSA), and methicillin-resistant non-Staphylococcus aureus (non-MRSA) molecularly to identify mecA (the gold-standard reference method for this analysis). In addition, four antimicrobial resistance genes responsible for quinolone resistance (gyrA, gyrB, and grlA genes) which are habitually found in S. aureus according resistance in the quinolone-resistance-determining-regions (QRDRs) and responsible for quinolone resistance (gyrA, gyrB, and grlA genes) and the cfr gene which confers resistance to five antibiotic classes (oxazolidinones, phenicols, streptogramin compounds, lincosamidins, and pleuromutilins usually referred to as PhLOPSA phenotype) were also amplified. Annealing temperatures and oligonucleotides used are presented in Table 1. The PCR runs included a reagent positive and negative control (without template DNA). Three independent PCR amplifications were carried out with a GeneAmp PCR System 2400 (Perkin-Elmer, Weiterstadt, Germany). Two microliters of template DNA was added to 23 mL of distilled sterile water, and finally 25 mL of reaction mixture containing 10mM Tris-HCl (pH8.3), 50mM KCl, 2.5mM MgCl2, 100 mM deoxynucleoside triphosphates, 1 mM of each primer and 3U of Taq polymerase were added. The amplicons were analyzed by electrophoresis on 1.5% (w/v) agarose gel in TBE buffer.
Results
Identification of Staphylococci
We identified 27 staphylococcal isolates belonging to five different staphylococcal species. These included S. hyicus (n = 10, 37%), S. aureus (n = 9, 33.3%), S. schleiferi subsp. coagulans (n = 4, 14.8%), S. intermedius (n = 3, 11.1%), and S. lentus (n = 1, 3.7%).
Phenotypic Virulence Assays
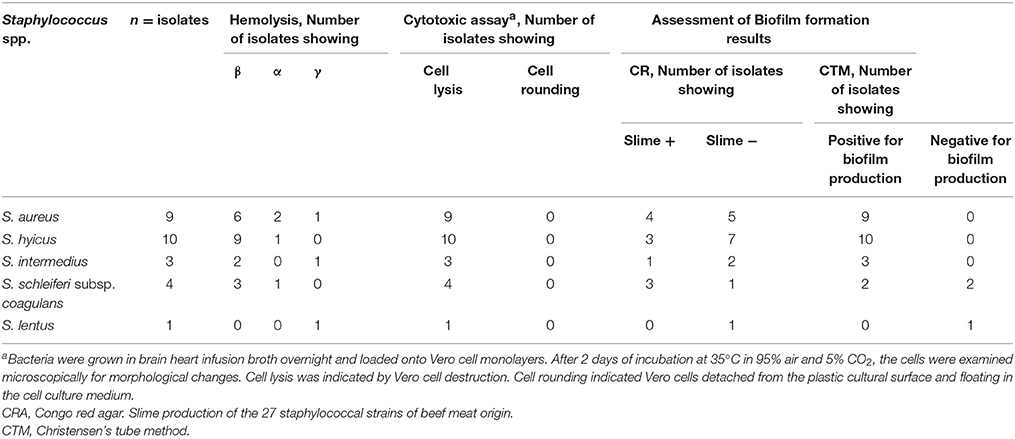
Summarized results of examining the 27 staphylococci isolates for factors that possibly contribute to their virulence are presented in Table 2.
Haemolysin Production
Haemolytic activity on plates with 5% defibrinated rabbit or sheep blood was established. In the strains showing hemolytic activity, the three patterns of haemolysis (β, α, and γ) were observed, 66.7% were demonstrated to be β-hemolytic, 22.2% were α-hemolytic, and 10.1% were γ-hemolytic.
Cytotoxicity Assay
All of the 27 Staphylococcus isolates were cytotoxic to Vero cells.
Biofilm Producing Phenotype
We found a highly resistant pattern among biofilm producers in comparison with non-biofilm producers. Among the 7/9 vancomycin resistant S. aureus isolates, they were also a biofilm producer (77.8%). Generally, the 20 vancomycin resistant S. aureus and non-S. aureus species were also biofilm producers. The affinity of the 27 Staphylococcus species to form a biofilm was found to be in favor of the glass surface by 88.9% (24/27).
Tube Method
The qualitative tube adherence test depends on the visual assessment of the degree of adherence of Staphylococcus isolates to the sides of borosilicate test tubes. The 27 Staphylococcus isolates were screened for biofilm formation by tube method. Results of the screening for biofilm by the tube method is shown in Table 2. Out of the 27 Staphylococcus isolates, 24 (88.9%) were strongly positive to the ability of biofilm formation, while 3 (11.1%) were negative to biofilm formation. Biofilm formation affinity was observed in the nine S. aureus isolates (100%) and 15/18 in the non-S. aureus isolates (83.3%).
Congo Red Agar Method
Among the 27 Staphylococcus isolates, 11 (40.7 %) showed black colonies with dry crystalline consistency which is indicative of slime production whereas 16 (59.3 %) isolates showed pink colored colonies with mucoid appearance on the CRA plates which were taken as negative for slime production (Table 2). Ability to produce slime was evident in 4/9 (44.4%) of the S. aureus and 7/18 (38.9%) of the non-S. aureus isolates. The studies on slime formation by the non-S. aureus species of medical importance revealed that S. hyicus, S. schleiferi subsp. Coagulans, and S. intermedius showed that up to 30% (3/10), 75% (3/4) and 33.3% (1/3), respectively, of these species produced slime, while S. lentus (1/1) was unable to produce slime.
Antibiotic Resistance Phenotypes for All Staphylococci
The phenotypic resistance pattern, prevalence and diversity of the 27 Staphylococcus species isolated from the beef meat are recorded in Table 3. The 27 Staphylococcus species were resistant to the 12 antibiotics with variable degrees as to each of the antibiotics used as indicated in Table 3 to reveal in ascending order: ampicillin-sulbactam 14.8% (4/27), chloramphenicol 29.6% (8/27), erythromycin 48.1% (13/27), ciprofloxacin 59.3% (16/27), methicillin 63% (17/27), tetracycline 66.7% (18/27), vancomycine 74.1% (20/27), oxacillin 77.8% (21/27), penicillin 92.6% (25/27), clindamycin 96.3% (26/27), gentamycin 96.3% (26/27), and sulphamethoxazole/trimethoprim 100% (27/27). The different Staphylococcus species also reflect a difference in their antibiotic resistance to the 12 antibiotics each species with a different capacity to resistance: S. lentus 91.7% (11/12), S. aureus 77.8% (84/108), S. intermedius 72.2% (26/36), S. schleiferi subsp. coagulans 70.8% (34/48), and S. hyicus 55% (66/120).
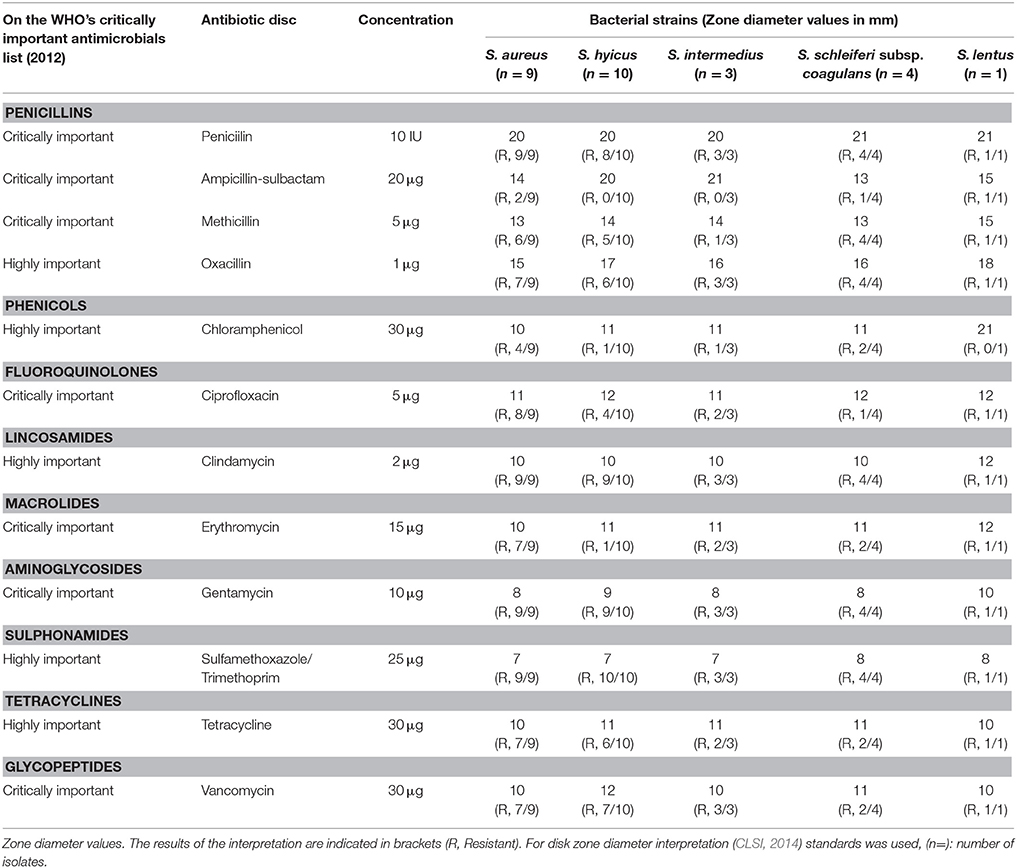
Table 3. Results of antimicrobial resistance tests by disk diffusion method for Staphylococcus species.
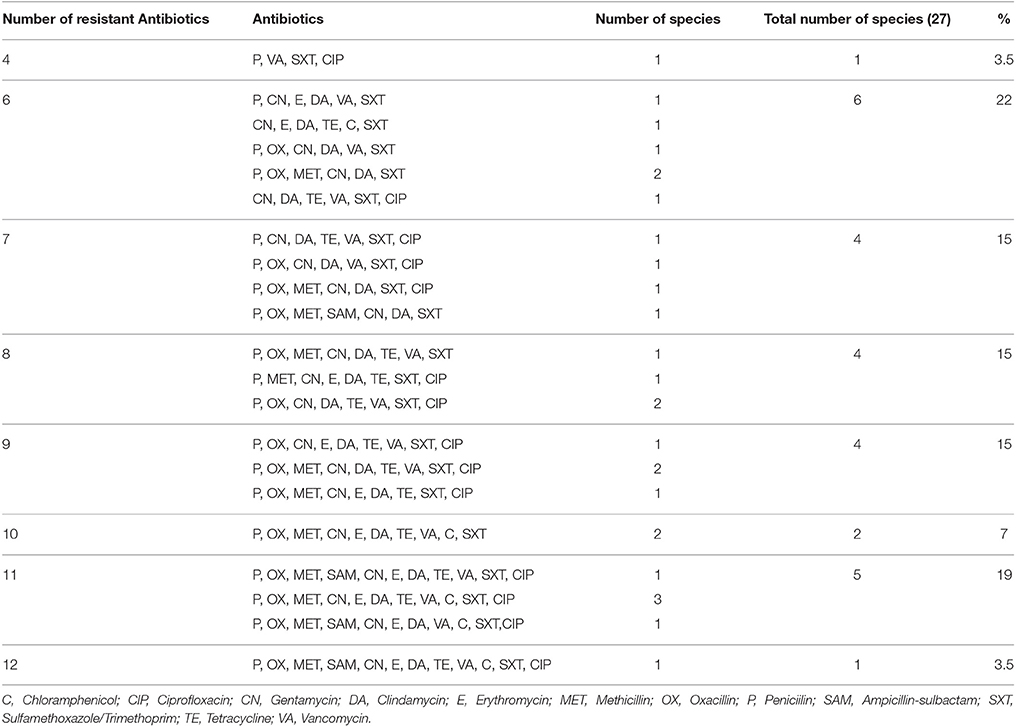
The several MDR combination patterns are recorded in Table 4. The Table reveals, that there were 21 different combination patterns ranging from four to twelve antibiotics in each combination. Out of the 27 Staphylococcus isolates only one isolate was resistant to the 12 antibiotics representing nine classes. This isolate was one of the nine S. aureus isolates (Table 5). The Table also reveals the wide dispersity in the number of antibiotic combinations in each isolate and species of the staphylococci isolates (S. aureus, five combinations; S. hyicus, five combinations; S. intermedius, three combinations; S. schleiferi subsp. coagulans two combinations; and S. lentus, one combination).
Widespread Distribution of Antibiotic Resistance Genes among Staphylococcal Isolates
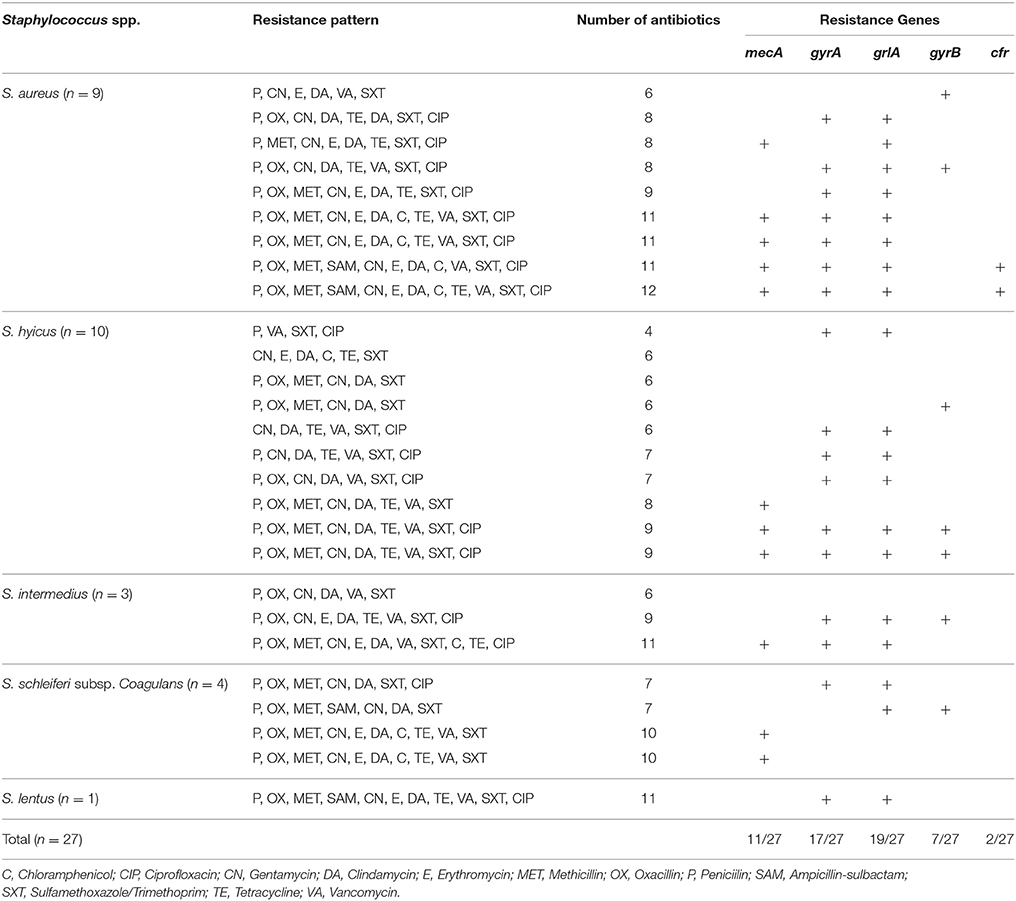
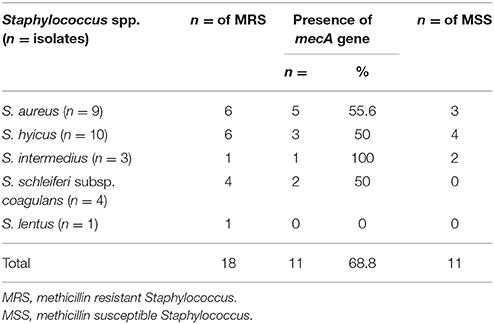
We screened the meat Staphylococcus isolates for the presence of the multiple resistance genes mecA, cfr, gyrA, gyrB, and grlA genes among Staphylococci isolated from beef meat in Egypt. The PCR amplification results are summarized in Table 5. All except for one staphylococcal isolate (S. lentus) beared > two antibiotic resistant genes and a noticeable number of these staphylococci (17/27, 63.7%) carried four different antibiotic resistant genes. In particular, the nine S. aureus isolates carried the five antibiotic resistant genes that confer resistance to the 12 different antibiotics tested in this investigation. Seven S. aureus, six S. hyicus, four S. schleiferi subsp. coagulans, three S. intermedius and one S. lentus isolates although being resistant to oxacillin yet, 11/27 (40.7%) carried the mecA gene. The association between the mecA gene and oxacillin resistance is outlined in Table 5. At the same time, the cfr gene was present in 2 of the nine S. aureus isolates, and totally undetectable in S. hyicus, S. schleiferi subsp. coagulans, S. intermedius and S. lentus. At the same time, the cfr gene was present in two of the nine S. aureus isolates, and totally undetectable in S. hyicus, S. schleiferi subsp. coagulans, S. intermedius, and S. lentus (Table 5).
Detailed Analysis of Selected Isolates: Species Determination
The 27 isolates were chosen for further detailed examination on the basis of their ciprofloxacin and oxacillin from the disc diffusion screening. There was interspecies and intraspecies variation among the non-S. aureus species listed in Table 3. The four ciprofloxacin-susceptible, oxacillin -susceptible isolates examined, were S. hyicus. Again, four ciprofloxacin-resistant, oxacillin susceptible isolates were S. hyicus. Of the four ciprofloxacin-resistant, oxacillin -resistant isolates, four were S. hyicus, two were S. intermedius and one was S. schleiferi subsp. coagulans and S. lentus each. The study shows that, the occurrence of MRS from beef meat is not uncommon (Table 6). Raw beef meat sold across the Great Cairo zone, contains 66.7% of MRS (18/27), with highest prevalence was reported in S. aureus (6/9, 66.7%), while the MRS non-S. aureus strains 12/18 (66.7%) from which S. hyicus (6/10, 60%), S. intermedius (1/3, 33.3%), S. schleiferi subsp. coagulans (4/4, 100%), and S. lentus (1/1, 100%) were MRS.
Survey of Methicillin, Oxacillin, and Ciprofloxacin Resistance
As seen in Table 3, the distribution of oxacillin among the non-S. aureus (CNS) isolates was quite broad (14/18, 77.8%). The overall frequency of ciprofloxacin resistance among non-S. aureus (CNS) was 44.4% (8/18), while that among methicillin-resistant non-S. aureus (MRCNS) was 61.1% (11/18), higher, as expected, than that for methicillin-susceptible non-S. aureus (MSCNS) 33.3% (6/18). The distribution of S. aureus and non-S. aureus ciprofloxacin susceptibilities as a function of methicillin susceptibilities in addition to the mecA resistance determinant which is responsible for the methicillin resistance, is recorded in Table 6.
Discussion
The non-S. aureus species that have been isolated in the present investigation were recorded as relevant in their ability to produce human infections (S. lentus, Stepanovic et al., 2003; S. hyicus, Casanova et al., 2011; S. schleiferi subsp. coagulans, Davis et al., 2013; and S. intermedius, Koci et al., 2015).
Humans are at a risk of being infected with resistant food-borne bacteria via the food chain (Khan et al., 2000; Pesavento et al., 2007) from reservoir animals (de Jong et al., 2009; Ewers et al., 2009; O'Keefe et al., 2010; Marshall and Levy, 2011). A previous research (Waters et al., 2011) has suggested that, when buying meat at any butcher shop, supermarket or local grocery shop, is an equally likely chance of it being contaminated with drug-resistant bacteria. The WHO has revealed a report on the global serious threat of antibiotic resistant strains of bacterial pathogens occurring currently in every locality of the world with no limitations and to affect everybody, at any age and in any country (CDC, 2013).
In animals, comparison of resistance rates to antimicrobials can geographically vary among internationally and nationally according to the antimicrobial usage regime (Harada and Asai, 2010; Doyle et al., 2013) due to sample collection schemes, sample types, their minimum capability to implement AMR surveillance, rock-bottom financial and technical resources and differences in the implemented methodologies in the form of credibility, duplicability, enequitable significance, price, duration and simplicity (Strommenger et al., 2006; Cookson et al., 2007; Faria et al., 2008; Vindel et al., 2009; David et al., 2013) which makes any comparative effort to be unadvisable.
A wide dispersion of two categories of resistance has been revealed in the present investigation: 100% of our isolates carried the QRDRs responsible for quinolone resistance genes (gyrA and grlA) (17/27, 63% and 19/27, 70.4%) and the mecA resistance determinant (11/27, 40.7%), responsible for the methicillin and oxacillin resistance (Table 5). The mecA determinant has caused severe complications during the treatment MRSA in humans (Lee, 2003) and for S. epidermidis infections too (Kitao, 2003) when the hypothesis that, mecA is transferred from non-S. aureus staphylococci to S. aureus (Grundmann et al., 2006; Bhargava and Zhang, 2014) is taken into consideration.
The French National Monitoring Program (RESABO) established evidence that S. aureus is more susceptible to antimicrobial drugs than non-S. aureus species (Werckenthin et al., 2001). Conversely, our results had a commensurate regular and sequence discernible in the antibiotic resistance genes between S. aureus and non-S. aureus isolates. The results showed a congruity between phenotypic resistance and molecular genotyping for oxacillin, QRDRs, and PhLOPSA phenotypes. Generally, our results are in assonance with Chajęcka-Wierzchowska et al. (2015) who displayed that, all methicillin resistant staphylococci harbored mecA gene but in consonance with the investigation outcomes of Martineau et al. (2000), Lee et al. (2004), and Guran and Kahya (2015), who indicated the occurrence of staphylococci strains resistant to methicillin or/and oxacillin in the absence of the mecA gene. The reason behind this variable expression could be related to the hyperproduction of β-lactamase type A (Martineau et al., 2000) and/or cellular growth conditions according to Lee et al. (2004). Globally, there are considerable discrepancies between different countries in the MRSA prevalence rate on beef meat (Normanno et al., 2007; van Loo et al., 2007a,b; Chan et al., 2008; Pu et al., 2009; Lim et al., 2010; Weese, 2010; Weese et al., 2010; Bhargava et al., 2011; Chua et al., 2011; Hanson et al., 2011; Dalhoff, 2012; O'Brien et al., 2012; Buyukcangaz et al., 2013; Jackson et al., 2013; Nnachi et al., 2014; WHO, 2014).
The animals are the first reservoirs for CNS bearing the cfr resistance gene which endows resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics (PhLOPSA of phenotype) and some macrolides (Kehrenberg et al., 2005). An interesting and fortunate observation was that, we detected the gene cfr in 22.2% (2/9) of the S. aureus isolates, a rate which was lower than the detection rates of food animal samples in previous studies (Wang et al., 2012; Zeng et al., 2014) while the non-S. aureus isolates did not carry this gene. The cfr multiresistance gene is usually found in isolates of human origin (Chen et al., 2013; Cui et al., 2013) and persistence in retail meat (LaMarre et al., 2011). The insignificant detection rate found in this study (2/27, 7.4%), indicated the neglible opportunity of the cfr to be spread through contaminated meat with Staphylococci in the local market by consumers in Egypt, the diminished possibility of this gene to enter the food cycle, and the chance of additional distribution of cfr from an animal reservoir to S. aureus, MRSA or any other staphylococcal species associated with colonization and infection in humans (Witte and Cuny, 2011) is abolished. Timely detection and targeted prevention of further dissemination of MRSA present on retail beef meat and the cfr gene becomes justified in order to preserve efficacy of linezolid due to its importance and preference in the treatment of staphylococcal infections (Witte and Cuny, 2011).
Fluoroquinolones (FQs) are antibiotics effective against a wide range of organisms (Oliphant and Green, 2002) and having a role in the chemotherapy and postexposure prophylaxis for organisms, that could be used in biologic warfare. The extensive use of FQs has led to the springing up of FQ-resistant S. aureus, especially among MRSA strains (Oliphant and Green, 2002) in the QRDRs of the gyrA, gyrB, grlA, and grlB genes (Hooper, 2002; Oliphant and Green, 2002; Redgrave et al., 2014; Parkinson et al., 2015). Gade and Qazi (2013) signified that, ciprofloxacin can no longer be used as an empirical therapy against MRSA infections to be replaced by vancomycin as the alternative drug to which we are in dispute as we recorded a 77.8% resistance in the MRSA. It should be noted that, the access to any available literature on any information concerning the gyrA, gyrB, grlA, and grlB genes in meat was not available.
The contamination of carcasses with MRSA can occur via several means: (i) improper agricultural practices, (ii) use of impotable water for agricultural irrigation, (iii) the use of improperly disinfected appliancies used in the food industry, (iv) unsuitable temperatures during storage and transportation, (v) poor food handling procedures, vi) lack of rodent and/or insect control (FDA, 2012; WHO, 2012), (vii) through the butchers tools which are suitable vehicles for transmission (de Boer et al., 2009), (viii) on the meat display table, to buyers and vice versa (Emele et al., 1995). The environmental contamination of the slaughter-house, processing appliances or operative hands in these facilities with MRSA could result in a cross-contagion process to the meat (Wertheim et al., 2004; Alexandra et al., 2011), and consequently conceal the importance of the association between on-farm antibiotic use and contamination of the meat with MRSA. The most important outcome of our investigation was the alarming finding that, most of the MRSA and NSA-MR isolated from retail meat in the Egyptian market were >50% resistant to clinically important antimicrobials (sulfamethoxazole/trimethoprim, clindamycin, gentamycin, peniciilin, oxacillin, vancomycin, tetracycline, methicillin, ciprofloxacin, and erythromycin in descending order) imposing a severe threat to public health of the Egyptians. In addition, the prevalence rate of the MRSA and NSA-MR isolated from retail meat collected from sale outlets reflected an indication to the level of exposure of the consumers to these pathogens (Wagenaar et al., 2007).
Conclusions
To the best of our knowledge, the extent of a similar research topic has not been tackled in Egypt. In addition, the strength of our study lies in the fact that, previous researches on S. aureus isolated from foodstuffs were merely screened for the mecA gene, while in our investigation a selective enrichment of MRSA was adopted to be followed by a PCR technique for the identification for the presence of mecA gene. Also, we included in our investigation samples collected from three different geographic areas involving both municipal and agricultural sectors. In some of our strains, we found that although resistance was recorded, yet the responsible gene was not expressed which could be due to, the small number of resistance genes tested or/and the gene responsible for resistance was not included in our protocol. There are no existing programs for surveillance which allows us to recommend a large scale of research to be conducted to cover the whole country with specific zone system, specifying large number of sample size to be included covering most type of meat consumed in Egypt.
Author Contributions
KO developed the concept, designed experiments, collected, and analyzed data and prepared the manuscript; AA provided reagents and mice; JB and NH performed PCR assays; RE gave technical support and conceptual advice; AO performed PCR assays; MB gave technical support and conceptual advice; AS performed the experiments. All authors discussed the results and implications and commented on the manuscript at all stages.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alexandra, F., Britta, K., Gladys, K., Beatriz, G. R., Katja, A., André, H. J., et al. (2011). Methicillin Susceptible and Resistant Staphylococcus aureus from Farm to Fork Impact on Food Safety. Beograd: Tehnologija Mesa Osnivač i izdavač: Institut za higijenu i tehnologiju mesa.
Benderitter, T. (2003). Cows, Oxen and Bulls in the Ancient Egypt. English translation by Jon J. Hirst. The Osirisnet Project
Bhargava, K., Wang, X., Donabedian, S., Zervos, M., de Rocha, L., and Zhang, Y. (2011). Methicillin-resistant Staphylococcus aureus in retail meat, Detroit, Michigan, USA Emerg. Infect. Dis. 17, 1135–1137. doi: 10.3201/eid1706.101905
Bhargava, K., and Zhang, Y. (2014). Characterization of methicillin-resistant coagulase-negative staphylococci (MRCoNS) in retail meat. Food Microbiol. 42:56e60. doi: 10.1016/j.fm.2014.02.019
Buyukcangaz, E., Velasco, V., Sherwood, J. S., Stepan, R. M., Koslofsky, R. J., and Logue, C. M. (2013). Molecular typing of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) isolated from animals and retail meat in North Dakota, United States. Foodborne Pathog. Dis. 10, 608–617. doi: 10.1089/fpd.2012.1427
Casanova, C., Iselin, L., von Steiger, N., Droz, S., and Sendi, P. (2011). Staphylococcus hyicus bacteremia in a farmer. J. Clin. Microbiol. 49, 4377–4378. doi: 10.1128/JCM.05645-11
CDC (2013). Get Smart: Know When Antibiotics Work. Atlanta, GA: Centers for Disease Control and Prevention.
Chajęcka-Wierzchowska, W., Zadernowska, A., Nalepa, B., Sierpiñska, M., and Łsaniewska-Trokenheim, Ł. (2015). Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin–phenotypic and genotypic antibiotic resistance. Food Microbiol. 46, 222–226. doi: 10.1016/j.fm.2014.08.001
Chan, P. A., Wakeman, S. E., Angelone, A., and Mermel, L. A. (2008). Investigation of multi-drug resistant microbes in retail meats. J. Food Agric. Environ. 6, 71–75.
Chen, H., Wu, W., Ni, M., Liu, Y., Zhang, J., Xia, F., et al. (2013). Linezolid-resistant clinical isolates of enterococci and Staphylococcus cohnii from a multicentre study in China: molecular epidemiology and resistance mechanisms. Int. J. Antimicrob. Agents 42, 317–321. doi: 10.1016/j.ijantimicag.2013.06.008
Chua, K., Laurent, F., Coombs, G., Grayson, M. L., and Howden, B. P. (2011). Not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician's guide to community MRSA - its evolving antimicrobial resistance and implications for therapy. Clin. Infect. Dis. 52, 99–114. doi: 10.1093/cid/ciq067
CLSI (2012). Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. Wayne, PA: CLSI.
Conférence publique de Mlle M-Christine Lavier Toulon (2000). Vandier, J: Manuel d'Archéologie Égyptienne T IV, Picard.
Conniff, R. (2015). Antibiotic Resistance Threatens to Become an “Apocalyptic Scenario.” Available online at: http://www.takepart.com/feature/2014/06/06/antibiotic-resistance-from-meat
Cookson, B. D., Robinson, D. A., Monk, A. B., Murchan, S., Deplano, A., de Ryck, R., et al. (2007). Evaluation of molecular typing methods in characterizing a european collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 45, 1830–1837. doi: 10.1128/JCM.02402-06
Cui, L., Wang, Y., Li, Y., He, T., Schwarz, S., Ding, Y., et al. (2013). Cfr-mediated linezolid-resistance among methicillin-resistant coagulase-negative staphylococci from infections of humans. PLoS ONE 8:e57096. doi: 10.1371/journal.pone.0057096
Dalhoff, A. (2012). Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscipl. Perspect. Infect. Dis. 2012, 37.
David, M. Z., Taylor, A., Lynfield, R., Boxrud, D. J., Short, G., Zychowski, D., et al. (2013). Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and pcr for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. Medical Center. J. Clin. Microbiol. 51, 814–819. doi: 10.1128/JCM.02429-12
Davis, M. F., Cain, C. L., Brazil, A. M., and Rankin, S. C. (2013). Two coagulase-negative staphylococci emerging as potential zoonotic pathogens: wolves in sheep's clothing? Front. Microbiol. Antimicrob. Resis. Chemoth. 4:123. doi: 10.3389/fmicb.2013.00123
de Boer, E., Zwartkruis-Nahuis, J. T., Wit, B., Huijsdens, X. W., de Neeling, A. J., Bosch, T., et al. (2009). Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int. J. Food Microbiol. 134, 52–56. doi: 10.1016/j.ijfoodmicro.2008.12.007
de Jong, A., Bywater, R., Butty, P., Deroover, E., Godinho, K., Klein, U., et al. (2009). A pan-European survey of antimicrobial susceptibility towards human-use antimicrobial drugs among zoonotic and commensal enteric bacteria isolated from healthy food-producing animals. J. Antimicrob. Chemother. 63, 733–744. doi: 10.1093/jac/dkp012
Doyle, M. P., Loneragan, G. H., Scott, H. M., and Singer, R. S. (2013). Antimicrobial resistance: challenges and perspectives. Compr. Rev. Food Sci. Food Saf. 12, 234–248. doi: 10.1111/1541-4337.12008
Dubin, D. T., Fitzgibbon, J. E., Nahvi, M. D., and John, J. F. (1999). Topoisomerase sequences of coagulase-negative staphylococcal isolates resistant to ciprofloxacin or trovafloxacin. Antimicrob. Agents Chemother. 43, 1631–1637.
Emele, F. E., Pasculle, W., Glew, R. H., and Mbrey, N. (1995). Incidence and prevalence of Aeromonas hydrophila in meat sold in Enugu Main Market. Nigerian J. Microbiol. 10, 77–80.
Ewers, C., Antão, E. M., Diehl, I., Philipp, H. C., and Wieler, L. H. (2009). Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 75, 184–192. doi: 10.1128/AEM.01324-08
FAO/WHO/OIE (2008). “Joint FAO/WHO/OIE expert meeting on critically important antimicrobials,” in Report of a Meeting Held in FAO (Rome; Geneva: WHO).
Faria, N. A., Carrico, J. A., Oliveira, D. C., Ramirez, M., and de Lencastre, H. (2008). Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 46, 136–144. doi: 10.1128/JCM.01684-07
Fätkenheuer, G., and Kaasch, A. J. (2014). How deadly is methicillin-resistant Staphylococcus aureus? Lancet Infect. Dis. 14, 905–907. doi: 10.1016/S1473-3099(14)70904-1
FDA (2012). Food and Drug Administration. Staphylococcus aureus: Foodborne Pathogenic Microorganisms and Natural Toxins Handbook, 2nd Edn. Washington, DC: US Food and Drug Administration.
Futagawa-Saito, K., Ba-Thein, W., Sakurai, N., and Fukuyasu, T. (2006). Prevalence of virulence factors in Staphylococcus intermedius isolates from dogs and pigeons. BMC Vet. Res. 2:4. doi: 10.1186/1746-6148-2-4
Gade, N. D., and Qazi, M. S. (2013). Fluoroquinolone therapy in Staphylococcus aureus infections: where do we stand? J. Lab. Phys. 5, 109–112. doi: 10.4103/0974-2727.119862
Grundmann, H., Aires-de-Sousa, M., Boyce, J., and Tiemersma, E. (2006). Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368, 874–885. doi: 10.1016/S0140-6736(06)68853-3
Guran, H. S., and Kahya, S. (2015). Species diversity and pheno- and genotypic antibiotic resistance patterns of staphylococci isolated from retail ground meats. J. Food Sci. 80, M1291–M1298. doi: 10.1111/1750-3841.12893
Hanson, B. M., Dressler, A. E., Harper, A. L., Scheibel, R. P., Wardyn, S. E., Roberts, L. K., et al. (2011). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) on retail meat in Iowa. J. Infect. Public Health 4, 169–174. doi: 10.1016/j.jiph.2011.06.001
Harada, K., and Asai, T. (2010). Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J. Biomed. Biotechnol. 2010, 12. doi: 10.1155/2010/180682
Hooper, D. C. (2002). Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2, 530–538. doi: 10.1016/S1473-3099(02)00369-9
Huber, H., Ziegler, D., Pflüger, V., Vogel, G., Zweifel, C., and Stephan, R. (2011). Prevalence and characteristics of methicillin-resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet. Res. 7:6. doi: 10.1186/1746-6148-7-6
Jackson, C. R., Davis, J. A., and Barrett, J. B. (2013). Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. J. Clin. Microbiol. 51, 1199–1207. doi: 10.1128/JCM.03166-12
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance Genes fexA and cfr among chloramphenicol-resistant staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L. H., and Vester, B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x
Khan, S. A., Nawaz, M. S., Khan, A. A., and Cerniglia, C. E. (2000). Transfer of erythromycin resistance from poultry to human clinical strains of Staphylococcus aureus. J. Clin. Microbiol. 38, 1832–1838.
Khan, S. A., Nawaz, M. S., Khan, A. A., Hopper, S. L., Jones, R. A., and Cerniglia, C. E. (2005). Molecular characterization of multidrug-resistant Enterococcus spp. from poultry and dairy farms: detection of virulence and vancomycin resistance gene markers by PCR. Mol. Cell Probes 19, 27–34. doi: 10.1016/j.mcp.2004.09.001
Kitao, T. (2003). Survey of methicillin-resistant coagulase-negative staphylococci isolated from the fingers of nursing students. J. Infect. Chemother. 9, 30–34. doi: 10.1007/s10156-002-0203-4
Koci, F., Sekar, A., Pacifico, L., and Esposito, A. (2015). A canine bug in a human heart. Q. J. Med. 108, 337–338. doi: 10.1093/qjmed/hcu123
Köck, R., Becker, K., Cookson, B., van Gemert-Pijnen, J. E., Harbarth, S., Kluytmans, J., et al. (2010). Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro. Surveill. 15, 19688.
LaMarre, J. M., Locke, J. B., Shaw, K. J., and Mankin, A. S. (2011). Low fitness cost of the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 55, 3714–3719. doi: 10.1128/AAC.00153-11
Lee, J. H. (2003). Methicillin (Oxacillin)-Resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69, 6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003
Lee, J. H., Jeong, J. M., Park, Y. H., Choi, S. S., Kim, Y. H., Chae, J. S., et al. (2004). Evaluation of the methicillin-resistant Staphylococcus aureus (MRSA)-screen latex agglutination test for detection of MRSA of animal origin. J. Clin. Microbiol. 42, 2780–2782. doi: 10.1128/JCM.42.6.2780-2782.2004
Lim, S. K., Nam, H. M., Park, H. J., Lee, H. S., Choi, M. J., Jung, S. C., et al. (2010). Prevalence and characterization of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J. Microbiol. Biotechnol. 20, 775–778. doi: 10.4014/jmb.0912.12022
Linde, H. J., Schmidt, M., Fuchs, E., Reischl, U., Niller, H. H., and Lehn, N. (2001). In vitro activities of six Quinolones and mechanisms of resistance in Staphylococcus aureus and coagulase-negative Staphylococci. Antimicrob. Agents Chemother. 45, 1553–1557. doi: 10.1128/AAC.45.5.1553-1557.2001
Marshall, B. M., and Levy, S. B. (2011). Food Animals and Antimicrobials: impacts on Human Health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Martineau, F., Picard, F. J., Lansac, N., Ménard, C., Roy, P. H., Ouellette, M., et al. (2000). Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. J. Clin. Microbiol. 44, 231–238. doi: 10.1128/aac.44.2.231-238.2000
Nnachi, A. U., Emele, F. E., Ukaegbu, C. O., Agah, M. V., Udu-Ibiam, O. E., Chukwu, O. S., et al. (2014). Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in raw meat and meat handlers in Onitsha, Nigeria. Europ. J. Prev. Med. 2, 9–15. doi: 10.11648/j.ejpm.20140201.12
Normanno, G., Corrente, M., La Salandra, G., Dambrosio, A., Quaglia, N. C., Parisi, A., et al. (2007). Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol. 117, 219–222. doi: 10.1016/j.ijfoodmicro.2007.04.006
O'Brien, A. M., Hanson, B. M., Farina, S. A., Wu, J. Y., Simmering, J. E., Wardyn, S. E., et al. (2012). MRSA in conventional and alternative retail pork products. PLoS ONE 7:e30092. doi: 10.1371/journal.pone.0030092
O'Keefe, A., Hutton, T. A., Schifferli, D. M., and Rankin, S. C. (2010). First detection of CTX-M and SHV extended-spectrum beta-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob. Agents Chemother. 54, 3489–3492. doi: 10.1128/AAC.01701-09
Oliphant, C. M., and Green, G. M. (2002). Quinolones: a comprehensive review. Am. Family Phys. 65, 455–464.
Osman, K. M., Abd El-Razik, K. A., Marie, H. S., and Arafa, A. (2015). Relevance of biofilm formation and virulence of different species of coagulase-negative staphylococci to public health. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2009–2016. doi: 10.1007/s10096-015-2445-3
Osman, K. M., Abd El-Razik, K. A., Marie, H. S. H., and Arafa, A. (2016). Coagulase-negative Staphylococci collected from bovine milk: species and antimicrobial gene diversity. J. Food Saf. 36, 89–99. doi: 10.1111/jfs.12216
Parkinson, E. I., Bair, J. S., Nakamura, B. A., Lee, H. Y., Kuttab, H. I., Southgate, E. H., et al. (2015). Deoxynybomycins inhibit mutant DNA gyrase and rescue mice infected with fluoroquinolone-resistant bacteria. Nat. Commun. 6, 6947. doi: 10.1038/ncomms7947
Pesavento, G., Ducci, B., Comodo, N., and Nostro, A. L. (2007). Antimicrobial resistance profile of Staphylococcus aureus isolated from raw meat: a research for methicillin resistant Staphylococcus aureus (MRSA). Food Control 18, 196–200. doi: 10.1016/j.foodcont.2005.09.013
Peton, V., and Le Loir, Y. (2014). Staphylococcus aureus in veterinary medicine. Infect. Gen. Evol. 21, 602–615. doi: 10.1016/j.meegid.2013.08.011
Pu, S., Han, F., and Ge, B. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus strains from Louisiana retail meats. Appl. Environ. Microbiol. 75, 265–267. doi: 10.1128/AEM.01110-08
Redgrave, L. S., Sutton, S. B., Webber, M. A., and Piddock, L. J. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22, 438–445. doi: 10.1016/j.tim.2014.04.007
Santos-Sanches, I., Mato, R., de Lencastre, H., Tomasz, A., and CEM/NET Collaborators the International Collaborators (2000). Patterns of multidrug resistance among methicillin-resistant hospital isolates of coagulase-positive and coagulase-negative staphylococci collected in the international multicenter study RESIST in 1997 and 1998. Microb. Drug Resist. 6, 199–211. doi: 10.1089/mdr.2000.6.199
Sowmya, N., Thakur, M. S., and Manonmani, H. K. (2012). Rapid and simple DNA extraction method for the detection of enterotoxigenic Staphylococcus aureus directly from food samples: comparison of PCR and LAMP methods. J. Appl. Microbiol. 113, 106–113. doi: 10.1111/j.1365-2672.2012.05315.x
Stepanovic, S., Ježek, P., Vukovic, D., Dakic, I., and Petráš, P. (2003). Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J. Clin. Microbiol. 41, 5262–5264. doi: 10.1128/JCM.41.11.5262-5264.2003
Stepanovic, S., Vukovic, D., Trajkovic, V., Samardzic, T., Cupic, M., and Svabic-Vlahovic, M. (2001). Possible virulence factors of Staphylococcus sciuri. FEMS Microbiol. Lett. 199, 47–53. doi: 10.1128/JCM.41.11.5262-5264.2003
Strommenger, B., Kettlitz, C., Weniger, T., Harmsen, D., Friedrich, A. W., and Witte, W. (2006). Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44, 2533–2540. doi: 10.1128/JCM.00420-06
Tao, M., Yamashita, H., Watanabe, K., and Nagatake, T. (1999). Possible virulence factors of Staphylococcus aureus in a mouse septic model. FEMS Immunol. Med. Microbiol. 23, 135–146. doi: 10.1111/j.1574-695X.1999.tb01232.x
Spanu, T., Sanguinetti, M., D'Inzeo, T., Ciccaglione, D., Romano, L., Leone, F., et al. (2004). Identification of methicillin-resistant isolates of Staphylococcus aureus and coagulase-negative Staphylococci responsible for bloodstream infections with the Phoenix™ system. Diag. Microbiol. Infect. Dis. 48, 221–227. doi: 10.1016/j.diagmicrobio.2003.11.004
Thorberg, B. M., and Brandstrom, B. (2000). Evaluation of two commercial systems and a new identification scheme based on solid substrates for identifying coagulase-negative staphylococci from bovine mastitis. J. Vet. Med. B Infect. Dis. Vet. Public Health 47, 683–691. doi: 10.1046/j.1439-0450.2000.00399.x
van Loo, I., Huijsdens, X., Tiemersma, E., De Neeling, A., Van De Sande -Bruinsma, N., Beaujean, D., et al. (2007a). Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerging Infect. Dis. 13, 1834–1839. doi: 10.3201/eid1312.070384
van Loo, I., Huijsdens, X., Tiemersma, E., de Neeling, A., van de Sande-Bruinsma, N., and Beaujean, D. (2007b). Methicillin-Resistant Staphylococcus aureus in meat products, the Netherlands. Emerging Infect. Dis. 13, 1753–1755. doi: 10.3201/eid1311.070358
Vindel, A., Cuevas, O., Cercenado, E., Marcos, C., Bautista, V., Castellares, C., et al. (2009). Methicillin-resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. J. Clin. Microbiol. 47, 1620–1627. doi: 10.1128/JCM.01579-08
Wagenaar, J., Van Duijkeren, E., Troelstra, A., van de Giessen, A., Kluytmans, J., Mevius, D., et al. (2007). Vragen en antwoorden over MRSA in landbouwhuisdieren. (Questions and answers about MRSA in farm animals). Tijdschr. Diergeneeskd 132, 558–560.
Wang, Y., Zhang, W., Wang, J., Wu, C., Shen, Z., Fu, X., et al. (2012). Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56, 1485–1490. doi: 10.1128/AAC.05827-11
Waters, A. E., Contente-Cuomo, T., Buchhagen, J., Liu, C. M., Watson, L., Pearce, K., et al. (2011). Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 52, 1227–1230. doi: 10.1093/cid/cir181
Weese, J. S. (2010). Methicillin-resistant Staphylococcus aureus in animals. ILAR J. 51, 233–244. doi: 10.1093/ilar.51.3.233
Weese, J. S., Avery, B. P., and Reid-Smith, R. J. (2010). Detection and quantification of methicillin-resistant Staphylococcus aureus (MRSA) clones in retail meat products. Lett. Appl. Microbiol. 51, 338–342. doi: 10.1111/j.1472-765X.2010.02901.x
Werckenthin, C., Cardoso, M., Martel, J. L., and Schwarz, S. (2001). Antimicrobial resistance in staphylococci from animals with particular reference to bovine Staphylococcus aureus, porcine Staphylococcus hyicus, and canine Staphylococcus intermedius. Vet. Res. 32, 341–362. doi: 10.1051/vetres:2001129
Wertheim, H. F., Vos, M. C., Boelens, H. A., Voss, A., Vandenbroucke-Grauls, C. M., Meester, M. H., et al. (2004). Low prevalence of methicillin-resistant Staphylococcus aureus (MRSA) at hospital admission in the Netherlands: the value of search and destroy and restrictive antibiotic use. J. Hosp. Infect. 56, 321–325. doi: 10.1016/j.jhin.2004.01.026
WHO (2009). World Health Organization Critically Important Antimicrobials for Human Medicine, 2nd Revision. Geneva: Department of Food Safety and Zoonoses, WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR).
WHO (2012). World Health Organization: Food Production to Consumption. Geneva, World Health Organization.
WHO (2014). World Health Organization WHO's First Global Report on Antibiotic Resistance Reveals Serious, Worldwide Threat to Public Health. Geneva.
Witte, W., and Cuny, C. (2011). Emergence and spread of cfr-mediated multiresistance in staphylococci: an interdisciplinary challenge. Future Microbiol. 6, 925–931. doi: 10.2217/FMB.11.69
Yaw, L. K., Robinson, J. O., and Ho, K. M. (2014). A comparison of long-term outcomes after meticillin-resistant and meticillin-sensitive Staphylococcus aureus bacteraemia: an observational cohort study. Lancet Infect. Dis. 14, 967–975. doi: 10.1016/S1473-3099(14)70876-X
Keywords: methicillin- resistant-susceptible S. aureus, methicillin- resistant-susceptible non-S. aureus, beef meat, antibiotic resistance genes, biofilm
Citation: Osman KM, Amer AM, Badr JM, Helmy NM, Elhelw RA, Orabi A, Bakry M and Saad ASA (2016) Antimicrobial Resistance, Biofilm Formation and mecA Characterization of Methicillin-Susceptible S. aureus and Non-S. aureus of Beef Meat Origin in Egypt. Front. Microbiol. 7:222. doi: 10.3389/fmicb.2016.00222
Received: 06 December 2015; Accepted: 11 February 2016;
Published: 29 February 2016.
Edited by:
Octavio Luiz Franco, Universidade Catolica de Brasilia, BrazilReviewed by:
Marco Rinaldo Oggioni, University of Leicester, UKAnselmo Otero-Gonzalez, University of Havana, Cuba
Copyright © 2016 Osman, Amer, Badr, Helmy, Elhelw, Orabi, Bakry and Saad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kamelia M. Osman, kamelia-osman@hotmail.com
 Kamelia M. Osman
Kamelia M. Osman Aziza M. Amer2
Aziza M. Amer2 Nashwa M. Helmy
Nashwa M. Helmy