- 1State Key Laboratory of Bioreactor Engineering and Institute of Applied Chemistry, East China University of Science and Technology, Shanghai, China
- 2Shanghai Collaborative Innovation Center for Biomanufacturing Technology, Shanghai, China
- 3School of Biological Sciences, The University of Hong Kong, Hong Kong, China
CO2 sequestration in deep-subsurface formations including oil reservoirs is a potential measure to reduce the CO2 concentration in the atmosphere. However, the fate of the CO2 and the ecological influences in carbon dioxide capture and storage (CDCS) facilities is not understood clearly. In the current study, the fate of CO2 (in bicarbonate form; 0∼90 mM) with 10 mM of formate as electron donor and carbon source was investigated with high-temperature production water from oilfield in China. The isotope data showed that bicarbonate could be reduced to methane by methanogens and major pathway of methanogenesis could be syntrophic formate oxidation coupled with CO2 reduction and formate methanogenesis under the anaerobic conditions. The bicarbonate addition induced the shift of microbial community. Addition of bicarbonate and formate was associated with a decrease of Methanosarcinales, but promotion of Methanobacteriales in all treatments. Thermodesulfovibrio was the major group in all the samples and Thermacetogenium dominated in the high bicarbonate treatments. The results indicated that CO2 from CDCS could be transformed to methane and the possibility of microbial CO2 conversion for enhanced microbial energy recovery in oil reservoirs.
Introduction
In recent years, increasing atmospheric CO2 and the resulting climate problem become the focus of global issues. A total of 3.2 gigatonnes of CO2 is released by the combustion of fossil fuels each year and has led to an increase of atmospheric CO2 concentrations from 280 ppm in 18th century to 383 ppm in Glueck et al. (2010). Three mainly strategies to reduce CO2 emission and building up are: reducing the CO2 production, expanding the CO2 utilization, and CO2 sequestration and storage (Schrag, 2007; Yang et al., 2008). The major approaches to decrease the CO2 production include improvement of energy efficiency or employment of cleaner technologies. Based on energy utilization and economic input, CO2 is chemically relatively stable and a non-attractive raw material. Carbon dioxide capture and storage (CDCS) is regarded as a potential and practical method to reduce the CO2 emission into atmosphere. The injection of CO2 into the oil reservoirs and geosystems may not only enhance the oil recovery (EOR) but also store about two-thirds of the CO2 in underground systems (Mikkelsen et al., 2010; Dressel et al., 2011; Momeni et al., 2012). Injected CO2 could be an important factor and affects the microbial metabolism and ecophysiology in storage environment (Delgado et al., 2012; Lin et al., 2013). Injected CO2 could alter the microbial community and the methanogenic pathways (Mayumi et al., 2013) and raise the bicarbonate concentration in oil reservoirs (Kirk, 2011).
The injection of CO2 from CDCS can be transformed into methane by microorganisms for energy recovery as value-added options, but electron donors are essential for the process (Hattori et al., 2001; De Bok et al., 2004). Formate and H2 are known as carriers for interspecies electron transfer and CO2 can be reduced with formate and H2 as electron donors through CO2-reducing microorganisms in anoxic environments (Dolfing et al., 2008). Interconversion of H2 and CO2 to formate by the microorganism at ambient conditions has been reported (Schuchmann and Müller, 2013). The Thermococcus sp. are capable of formate oxidation with H2 production alone at high-temperature (Kim et al., 2010). Many species of microorganisms can grow on formate as a sole methanogenic substrate (Wood et al., 2003; Oren, 2014). Formate can be used by some types of methanogens with formate dehydrogenase alone for H2 production and methane production (Lupa et al., 2008). Formate plays an important role as H2 storage compound for CO2 reduction in subsurface formation.
Formate is an important metabolite in anaerobic alkane oxidation in Desulfatibacillum alkenivorans AK-01 (Callaghan et al., 2012). Formate also plays an important role as intermediates in syntrophic butyrate or propionate oxidization (Hattori et al., 2001). Hydrogen production based on formate oxidation has been described before (Bagramyan and Trchounian, 2003; Meshulam-Simon et al., 2007). And both thermophilic and mesophilic communities with a formate-oxidizing bacterium and a hydrogenotrophic methanogen have been constructed for demonstration of syntrophic growth on formate (Dolfing et al., 2008). Formate serve as important degradation intermediate of alkanes and precursor for methanogenesis in oil reservoirs, but very little is known about formate metabolism before.
The whole oil reservoir can be regarded as an anaerobic bioreactor with variety of specialized microorganisms and has the potential for bioconversion of CO2 (Liu et al., 2015). However, very little is known about the fate of sequestrated CO2 and the anaerobic metabolic pathway of formate in oil reservoir. In this study, the treatments were constructed with production water of high-temperature oil reservoirs amended with formate and different concentrations of C-13 labeled bicarbonate. Microbial community was analyzed based on 16S rRNA gene after incubation and high levels of methane generated. In addition, stable isotope technique was introduced to detect the fate of injected CO2. The objectives of this study were to provide evidence on the possibility of microbial conversion of CO2 and metabolic pathways of formate.
Materials and Methods
Preparation of Inoculum and Enrichment Cultures
The inoculum for the culture experiments was collected from water-flooded oilfield production water of Ba 18 block of Baolige, Huabei Oilfield in China, and cultured under anaerobic conditions at 55°C in the dark. Physicochemical characteristics of the inoculum is shown in Supplementary Table S1. About 2 mL of inoculum was transferred aseptically into a serum bottle (120 ml internal volume) with 50 ml of basal medium containing (g/L): NaCl, 0.20; MgCl2.6H2O, 1.20; CaCl2.2H2O, 0.10; NH4Cl, 0.25; KH2PO4, 0.75; K2HPO4, 1.16; KCl, 1.30; rezasurin, 0.0001. Vitamin stock solution and trace element stock solution of 1.0 (mL/L) and Na2S.9H2O (0.50 g/L) were added into the medium and the final pH of the basal medium was adjusted to 7.2. The detailed composition of the vitamin stock solution and trace element stock solution was described before (Wang et al., 2012b).
13C-bicarbonate was introduced with a final concentration of 0, 30, 60, and 90 mM. Formate was added as the carbon source and electron donor at 10 mM. Basal medium with different concentrations of bicarbonate without added formate was used as blank. Treatments were abbreviated accordingly as S0, S30, S60, and S90 for different bicarbonate concentrations. All sets of experiments were conducted in triplicate. During the operation, the serum bottles were sealed with pure N2 gas and removed the O2 from the systems. All the microcosms were incubated at 55°C in the dark.
Chemical Analysis
Gas composition (CH4, CO2, and H2) of headspace was measured using a Gas Chromatograph (GC112A, Shanghai Precision and Scientific Instrument, CO., Ltd, China) with a thermal conductivity detector (TCD) and a flame ionization detector (FID). The detailed method was followed as described before (Mbadinga et al., 2012). Formate and acetate were detected and quantified by Ion Chromatograph (IC DX-600, Dionex, CO., USA) with IonPac AS11-HC analytical column (4 mm × 250 mm) and ASRS 300 suppressor. The mobile phase was 2 mM of NaOH.
DNA and 16S rRNA Gene Amplification
Five mL of the culture samples were taken and concentrated by centrifugation at 12000 × g for 20 min at 4°C after 180 days cultivation. Total community DNA was extracted from the microbial biomass using the genomic DNA Kit (AxygenBiosciences, Inc., USA). Partial 16S rRNA genes of bacteria and archaea were amplified as previously described with primer 8F/805R (Zhou et al., 2013) and 340F/1000R (Gantner et al., 2011), respectively. Polymerase chain reaction (PCR) was performed in a 25 μl reaction volume containing 12.5 μM of each primer (1 μl), 50 ng of template DNA (2 μl), 12.5 μl of 2 × PCR master mix (Lifefeng Biotechnology, Shanghai, China) and 8.5 μl ddH2O. PCR programs were as follows: an initial denaturation step at 95°C for 5 min, followed by 32 cycles of 94°C for 45 s, 59°C for 40 s, and 72°C for 50 s, with a final elongation step at 72°C for 10 min. Genes were amplified on a Peltier Thermal Cycler (Bio-Rad, CO., USA).
Construction of 16S rRNA Genes Libraries and Phylogenetic Analysis
Polymerase chain reaction products were cloned with a pMD 19-T simple vector kit (TaKaRa Bio, Inc., Japan) after checking by 1.8% (agarose) gel electrophoresis. Clones were randomly selected from plates and sequenced on an ABI 377 automated sequencer. The 16S rRNA sequences were checked and phylogenetic trees were generated with the protocol described before (Wang et al., 2012a). Operational taxonomic units (OTUs) were defined with the similarity of more than 97%. Phylogenetic trees of 16S rRNA genes retrieved from the original inoculum, S0, S30, S60, and S90 samples, were constructed. Sequences were performed by MEGA 5 with OTU identity of 97%. The topology of the tree was obtained with the neighbor-joining method. Bootstrap values (n = 1000 replicates) of ≥75% are showed.
Data Analysis of Stable Isotope
Carbon-13 isotopic compositions of CH4 and CO2 were determined with isotope ratio mass spectrometer (IRMS Delta V PLUS, Thermo Scientific, CO., USA). The samples were concentrated with PreCon connector to reduce dose. The standard international delta notation for reporting isotopic value ratios were relative to the VPDB standard. The references gas was CO2 and δ13 of CPDB was –23.73‰, which was calibrated by stable isotope of carbon in charcoal black. The system was performed with ISODATNT software.
Thermodynamic Calculations
Gibbs free energy data for thermodynamic calculations with different temperatures (Table 1) were taken from (Amend and Shock, 2001). ΔG°T is standard Gibbs free energy at temperature T and ΔG°′T is modified with the protons from ΔG°T. The change of Gibbs free energy (ΔG′T) for the reaction is calculated with the formula: ΔG′55 = ΔG°′55 – RTln([R]a/[P]b), R and P are the abbreviations of reactants and products, respectively; a and b are the stoichiometric numbers of each composition.
Nucleotide Sequence Accession Numbers
The partial gene sequences for bacteria and archaea obtained from the clone libraries were submitted to GenBank database under accession numbers KR049100–KR049155 and KR017718–KR017745, respectively.
Results
Methane and Hydrogen Production
Methane production increased after the initial 35 days of incubation under strictly anaerobic conditions, and an obvious lag period was observed in S90 compared with the other treatments (Figures 1A,B). The rate of methane production in S90 was about 1.95 pmol d-1 ml-1 during the culture of 35∼80 days, while it was about 25.5, 41.4, and 39.1 pmol d-1 ml-1 in S0, S30, and S60, respectively. After 180 days of incubation, methane production in all the treatments reached approximately 136 μmol/bottle, corresponding to almost exhaustion of the 500 μmol/bottle of formate introduced into the medium (Supplementary Table S2). Methane production was nearly equal to the formate consumed indicating that almost all of the methane produced was by formate reduction directly or indirectly. The ultimate methane production rate of all the samples was about 19.6 pmol d-1 ml-1. An obvious hydrogen production was found in S0 samples and subsequently depleted, and little hydrogen was detected in other samples amended with bicarbonate addition after 35 days and decreased with the further incubation of the cultures (Figure 1B), which showed the same trend with the S0 sample. The addition of bicarbonate decreased the amount of hydrogen production. There was no methane or hydrogen detected in all the blank treatments without added formate.
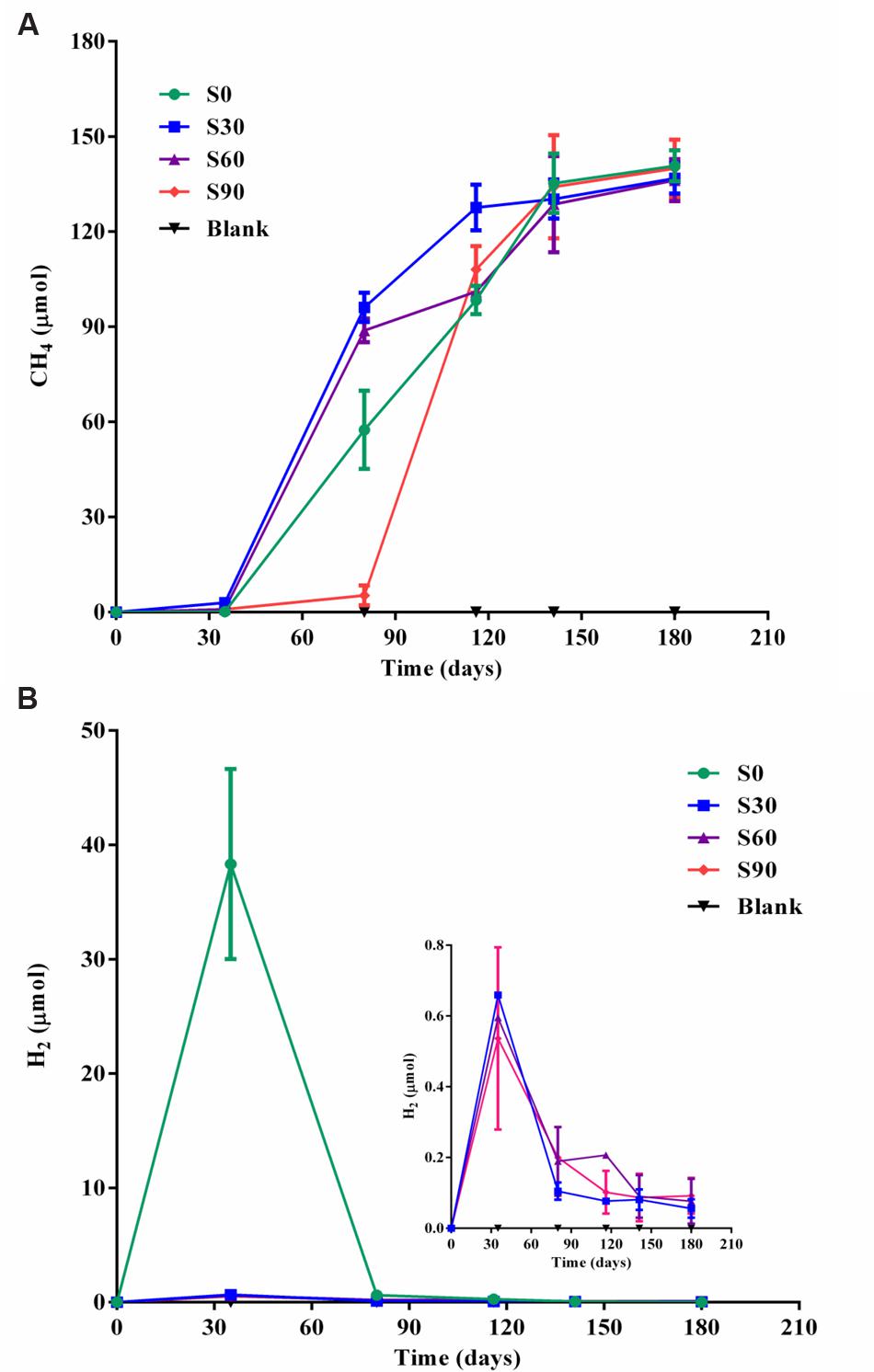
FIGURE 1. Methane and hydrogen production in the cultures. Methane production (A) and hydrogen production (B) in serum bottle maintained under anaerobic conditions. Blank are the cultures with different concentrations of bicarbonate without added formate. S0, S30, S60, and S90 treatments were the cultures with formate and different concentrations of bicarbonate (0, 30, 60, and 90 mM, respectively).
Dynamics of Microbial Community
16S rRNA clone libraries were constructed to analyze the microbial community under different bicarbonate concentrations compared with the original inoculum. A total of 460 sequences were obtained and then used to analyze the archaeal composition in inoculum, S0, S30, S60, and S90 samples and 9, 5, 4, 2, and 2 OTUs were resulted with 97% similarity, respectively (Figure 2). All the archaeal sequences belong to Methanomicrobiales, Methanobacteriales, Methanosarcinales, and Crenachaeota. Those affiliating with Methanomicrobiales and Methanobacteriales are CO2-reducing methanogens while Methanosarcinales have a variety of methanogenic biochemical pathways. Crenachaeota is known as ammonia-oxidizing bacteria (Hatzenpichler et al., 2008) and have no reported information about methanogenesis. The original inoculum contained the most varieties of archaea among all the five samples. With the addition of formate and increase in bicarbonate concentrations, the archaeal varieties decreased and only Methanobacteriales and Crenachaeota were detected in S60 and S90. In the original inoculum, Methanosarcinales were dominated (68.7%) with 3 OTUs (Inoculum-A-4, Inoculum-A-5, and Inoculum-A-7) and 3 OTUs (Inoculum-A-2, Inoculum-A-3 and Inoculum-A-6) with close identities to Methanobacteriales (22.4%), 2 OTUs (Inoculum-A-8 and Inoculum-A-9) belonged to Methanomicrobiales (7.5%) were also detected and OTU Inoculum-A-1 (1.4%) clustered into Crenachaeota. Methanobacteriales (72.4%∼83.8%) were dominated in all the culture treatments amended with formate. Methanomicrobiales, Methanobacteriales, Methanosarcinales and Crenachaeota were all detected in the S0 and S30 samples, but only Methanobacteriales and Crenachaeota were detected in the treatments (S60 and S90) with high bicarbonate concentrations.
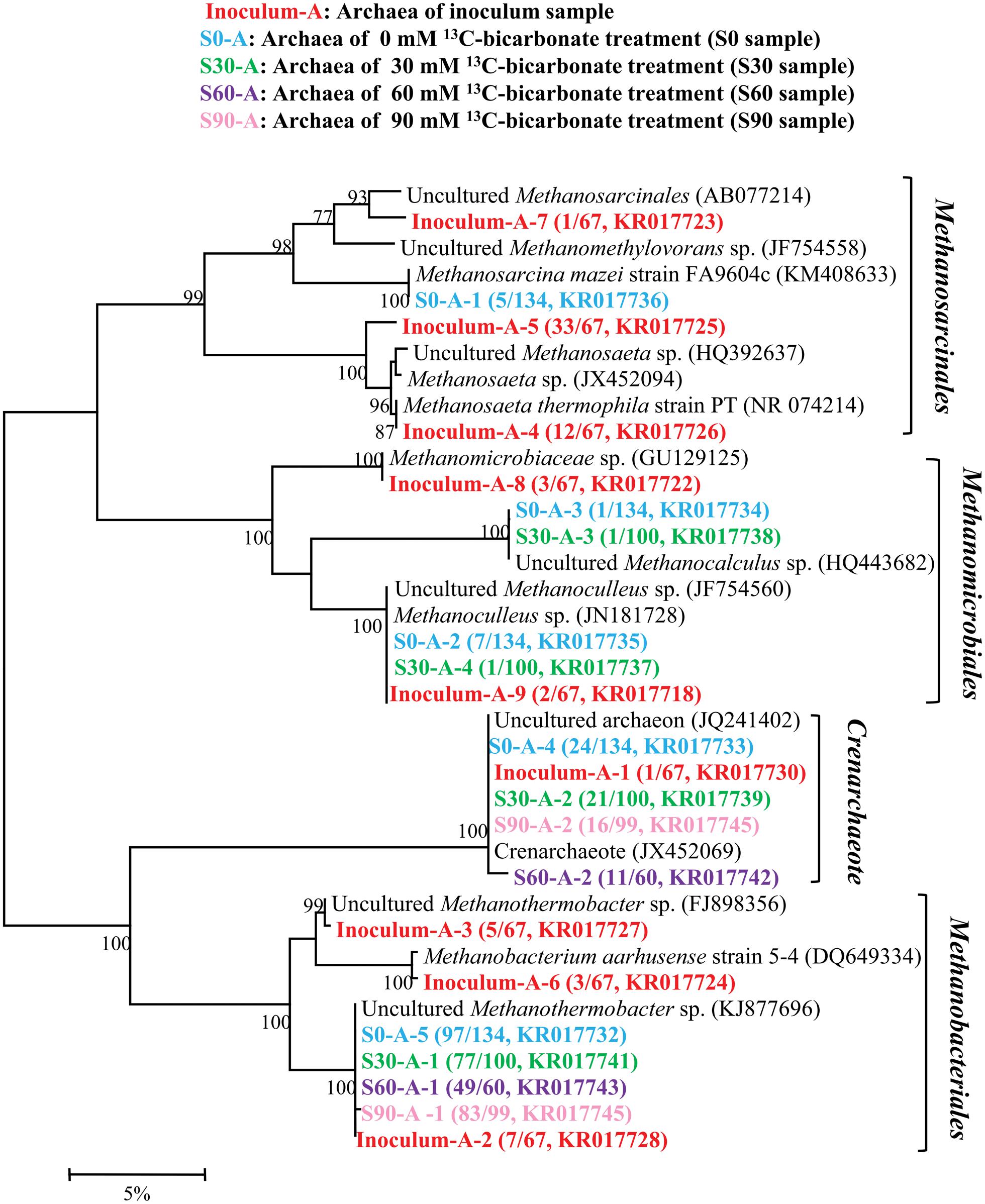
FIGURE 2. Phylogenetic tree of archaeal genes retrieved from five samples (shown with different color). Sequences were performed by MEGA 5 with OTU identity of 97%. The topology of the tree was obtained with the neighbor-joining method. Bootstrap values (n = 1000 replicates) of ≥75% are showed. Numbers in brackets are the appearance frequencies of the identical sequences in clones and the accession number.
By using bacterial gene specific primers, a total of 365 sequences were obtained and then analyzed with the 97% similarity to construct a phylogenetic tree about bacteria, and 7, 11, 10, 6, and 10 OTUs were found in the inoculum, S0, S30, S60, and S90 samples, respectively (Figure 3). Bacteria showed high varieties in all the five samples and most sequences belonged to Thermotogae, Synergistetes, Acetothermia, Nitrospirales, and Firmicutes. Thermodesulfovibrio belonged to Nitrospirales and Clostridia were found in all the five samples. Thermodesulfovibrio was dominated in the treatments (inoculum, S0, and S30) with low concentrations of bicarbonate. Thermacetogenium, a member of Clostridia, increased its presence in the treatments (S60 and S90) with high bicarbonate concentrations. Thermoanaerobacteriaceae was only detected in the treatments amended with bicarbonate addition (S30, S60, and S90).
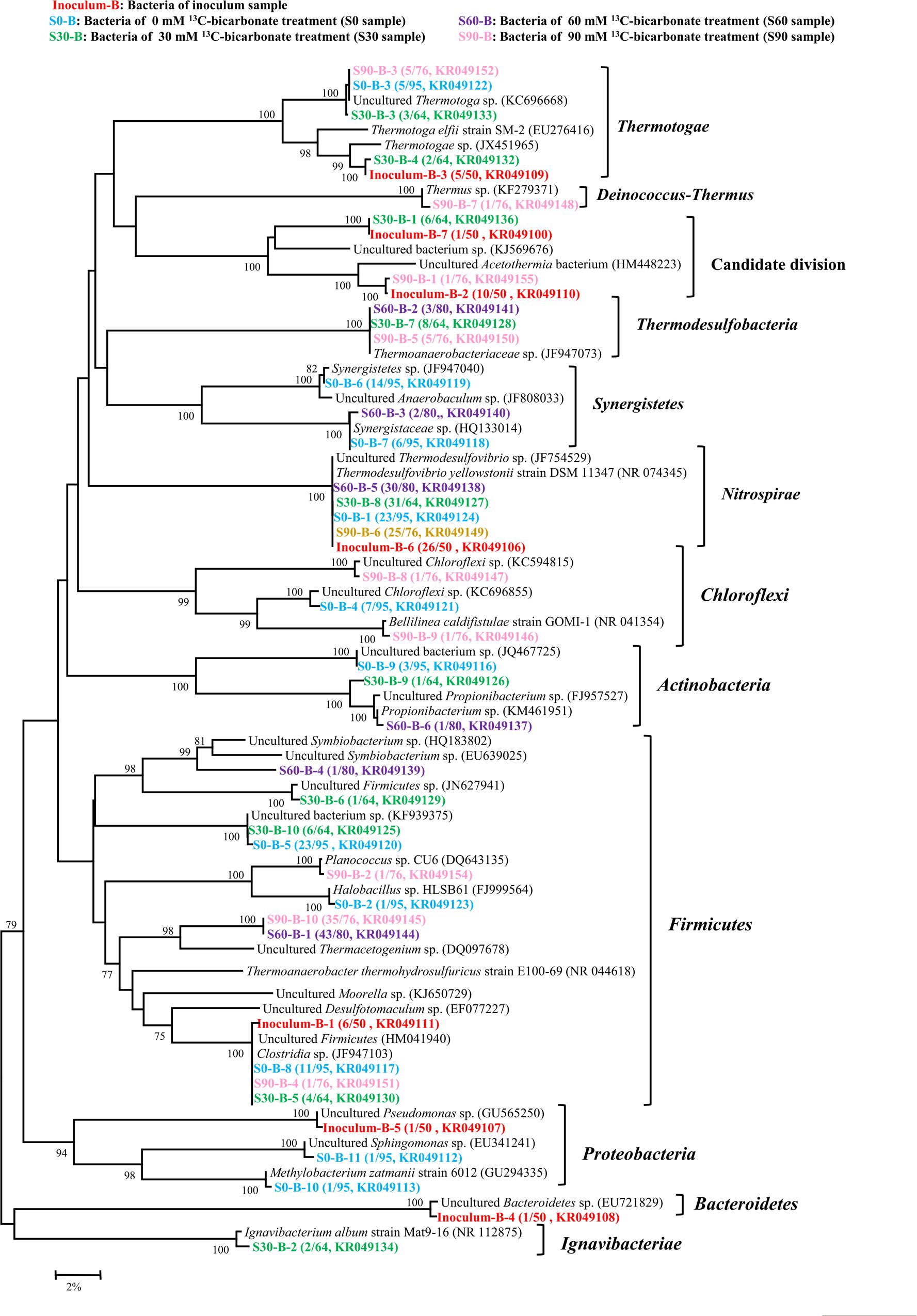
FIGURE 3. Phylogenetic tree of bacterial genes retrieved from five samples (shown with different color). Sequences were performed by MEGA 5 with OTU identity of 97%. The topology of the tree was obtained with the neighbor-joining method. Bootstrap values (n = 1000 replicates) of ≥75% are showed. Numbers in brackets are the appearance frequencies of the identical sequences in clones and the accession number.
Stable Carbon Isotope Analysis
Bicarbonate was added to mimic the highly buffered condition due to the injected CO2 and stable isotope C-13 was used to identify the fate of CO2 (bicarbonate form) in incubation with deep-subsurface fluid. The carbon isotopic compositions of methane and CO2 were detected after incubation for 180 days. In the treatment without 13C-bicarbonate addition, there was no obvious 13CH4 detected (Figure 4). But hydrogen production was detected and subsequently consumed (Figure 1B), indicating that H2 were produced from formate and then used for methane production. 13CH4 were detected in the treatments with 13C-bicarbonate addition (516.03∼675.46‰; Figure 4) and the production rate of 13CH4 increased as 13C-bicarbonate concentration increased, which suggested that C-13 labeled carbon dioxide (or 13C-bicarbonate) were reduced to 13CH4 and promoted the production rate of 13CH4. The ratio of labeled C-13 stayed as 13C-bicarbonate increased as injected 13C-bicarbonate concentration increased, while the amount of all detected C-13 and the percentage of labeled C-13 stayed as 13CO2 or 13CH4 decreased (Table 2). Carbon dioxide without any labeled 13C was detected (173.88∼383.48‰) in the culture treatments, which should be produced from formate. In the treatments amended with different concentrations of 13C-bicarbonate, formate was oxidized to CO2 and high concentration of CO2 with an increasing incorporation of 13CO2 and then a large fraction of 13CO2 with small fraction of CO2 were reduced to 13CH4 and CH4. (formate oxidation: HCOO- + H2O = HCO3- + H2; CO2 reduction: HCO3- + 4H2 + H+ = CH4 + 3H2O). CO2/13CO2 also could be involved in formate methanogenesis directly by methanogens and produced CH4/13CH4.
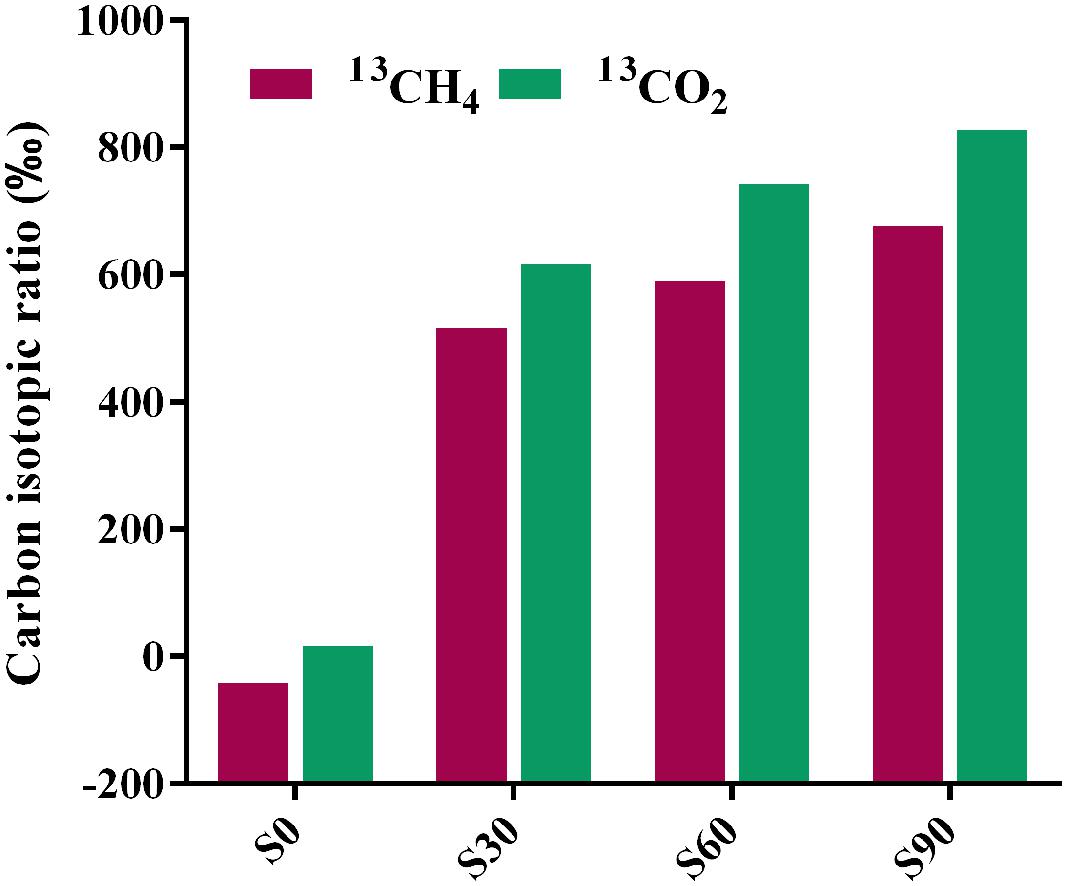
FIGURE 4. Carbon isotopic ratios of methane and CO2 after incubation. The relevant data are shown in Supplementary Table S3. S0, S30, S60, and S90 treatments were the cultures with formate and different concentrations of bicarbonate (0, 30, 60, and 90 mM, respectively).
Thermodynamics of Bicarbonate-Driven Methanogenic Reactions
Thermodynamic calculation was used to illustrate the energetically favorable metabolic pathways in the experiments with different concentrations of bicarbonate associated with microbial community. The theoretical constraints of archaea and bacteria were evaluated respectively with the data obtained in the present study with different concentration of bicarbonate. The thermodynamic calculation of different concentration of bicarbonate show the same trend. The calculations for the feasible reactions of hydrogen and formate in the sets of culture experiments amended with bicarbonate (S0, S30, S60, and S90) are as follows: CO2 reduction, acetate formation, formate methanogenesis, and formate oxidation (Table 1). The thermodynamics are shown in Figure 5. Acetate formation, formate methanogenesis, and formate oxidation were less energetically favorable with increasing bicarbonate concentrations and CO2 reduction became more energetically favorable. As other factors are fixed, CO2 reduction and formate oxidation are less sensitive to the change of bicarbonate concentration than formate methanogenesis and acetate formation because one mole of bicarbonate involves in per CO2 reduction and formate oxidation reaction. The change of Gibbs free energy (ΔG′55) calculated for the four reactions in all the treatments showed that all the reactions are exergonic under the experimental conditions and formate methanogenesis is the most favorable one. ΔG° 25°C of the formate oxidation is above the zero (1.12 kJ/mol) and ΔG° 55°C of the formate oxidation is only -0.92 kJ/mol, which are close to thermodynamic threshold (Figure 5 and Table 1). Both formate methanogenesis and formate oxidation are energetically favorable in the culture conditions.
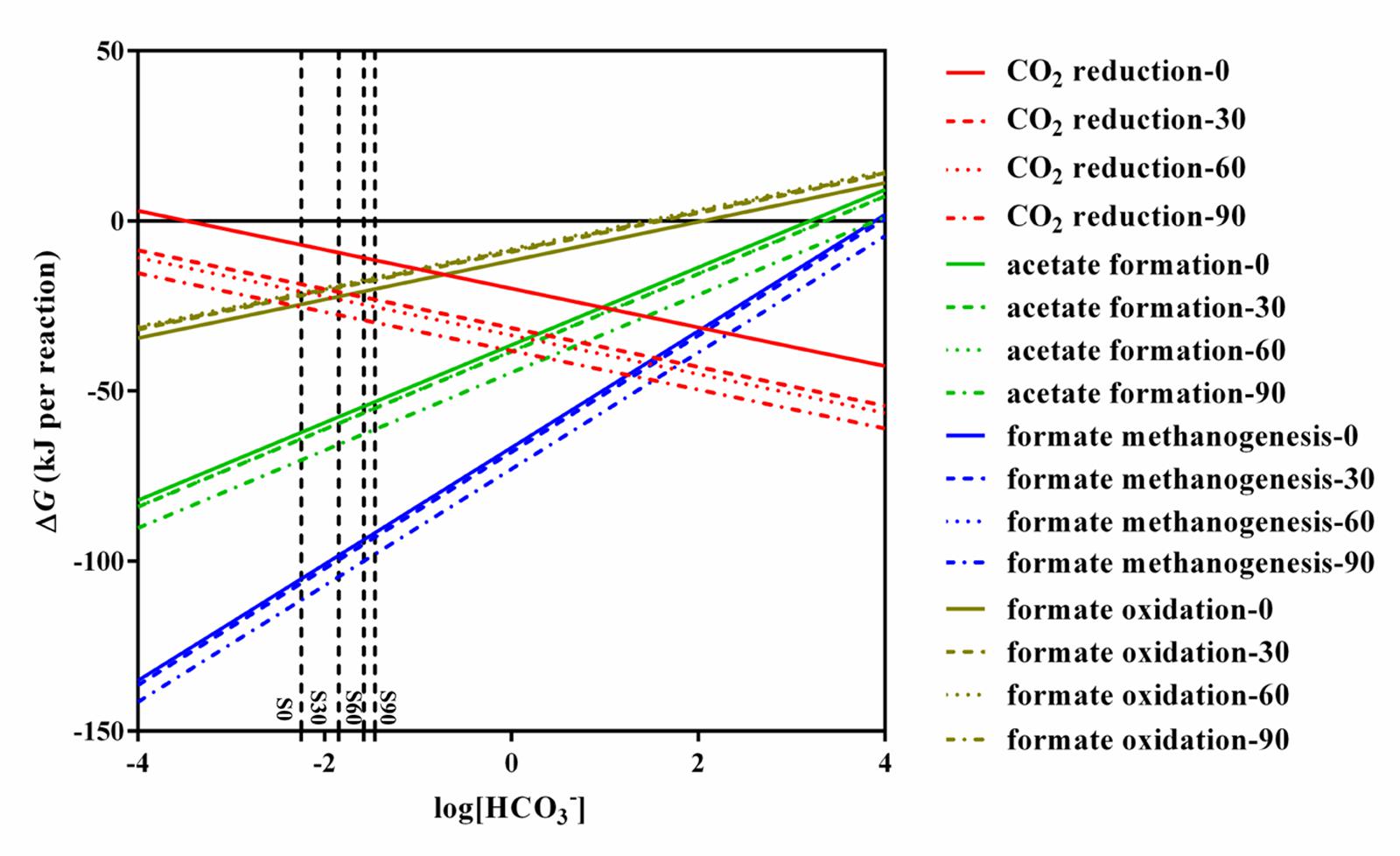
FIGURE 5. Effects of HCO3- on change in Gibbs free energy for CO2 reduction, acetate formation, formate methanogenesis, and formate oxidation. All lines in the figure were calculated using the conditions of S0, S30, S60, S90. The detail data were shown in Supplementary Table S2.
Discussion
Oil reservoir is an extreme environment with different conditions of temperatures, pH values, pressure, salinity, thermodynamic limits and barren habitat (Kobayashi et al., 2012; Mayer and Müller, 2014). These extreme conditions require microbes thriving in community to carry out biochemical reactions collectively. The injected CO2 from CDCS or MEER can alter the microbial community and the metabolic pathways in deep-subsurface environments, which may dictate the fate of CO2 (Mayumi et al., 2013; Ohtomo et al., 2013). The endogenous microorganisms in oil reservoirs can reduce CO2 with the electron derived from the electron donors, which define the oil reservoirs as a bioreactor for CO2 bioconversion to either fixation as acetate or as methane, a source of natural gas.
The Fate of the Injected CO2
Our studies showed that about 136 μmol methane produced after 180 days of incubation in each bottle and at least 516.03% was labeled in the treatments with 13C-bicarbonate addition, indicating that at least half of the methane produced were through the CO2 reduction pathway for methanogenesis. At the same time, part of injected bicarbonate were presented as 13C-bicarbonate in culture medium and 13CO2 of gas phase in headspace (Table 2). In original oil reservoir environments, residual CO2 from EOR could be converted to methane via methanogenesis, whereas most CO2 was dissolved in the formation fluids and appeared in gas phase after more than 30 years (Shelton et al., 2014). With the simulation reactor system in laboratory, injected CO2 could be transformed into acetate through homoacetogenesis, but methanogenesis was not detected under the condition simulating the in situ pressure and temperature (Ohtomo et al., 2013). Carbon dioxide can also be converted into formate through the carbon dioxide reductase or a whole cell system (Schuchmann and Müller, 2013).
High CO2 pressure invoke acetoclastic methanogenesis instead of syntrophic acetate oxidation coupled with CO2-reducing methanogenesis, when acetate and oil were added as substrates (Mayumi et al., 2013). Our results showed that when formate was added as substrate and electron donors, CO2 could be converted into methane through syntrophic formate oxidation coupled with CO2-reducing methanogenesis and formate methanogenesis. The bio-conversion rate of CO2 was about 19.6 pmol d-1 ml-1 in the treatments, and compared to original oil reservoir environments, the abundant supply of electron donor is essential to maintain and accelerate the process to take place. Naturally available H2 or electron resources for methanogenesis can be the primary requirements to improve the conversion rate of CO2 (Head et al., 2003; Cheng et al., 2009). And variety of enzymes (Yeates et al., 2008; Shekh et al., 2011; Alissandratos et al., 2014) and catalysts (Hull et al., 2012; Ziebart et al., 2012; Jeletic et al., 2013) can be used to convert the CO2 into reduced compounds under in situ and ex situ experimental conditions.
Methanogenic Pathway Analysis in the Experiments
Biological CO2 fixation is a common phenomenon in biology. Six autotrophic carbon fixation pathways have been reported (Berg et al., 2010) and most key enzymes of these six biochemical pathways have been detected in oil reservoirs (Liu et al., 2015). There are three main methanogenic pathways in oil reservoirs: CO2-reducing, acetoclastic and methylotrophic with different substrates: CO2, acetate and methyl group containing compounds, respectively (Nazaries et al., 2013) and direct interspecies electron transfer as a new model for methanogenesis has been confirmed (Rotaru et al., 2014). In this study, formate was amended into the treatments as the carbon source and electron donor. The proposed pathways of anaerobic formate transformation and related CO2 conversion were shown in Figure 6. Formate can be involved in CO2 conversion and methanogenesis through many kinds of pathways.
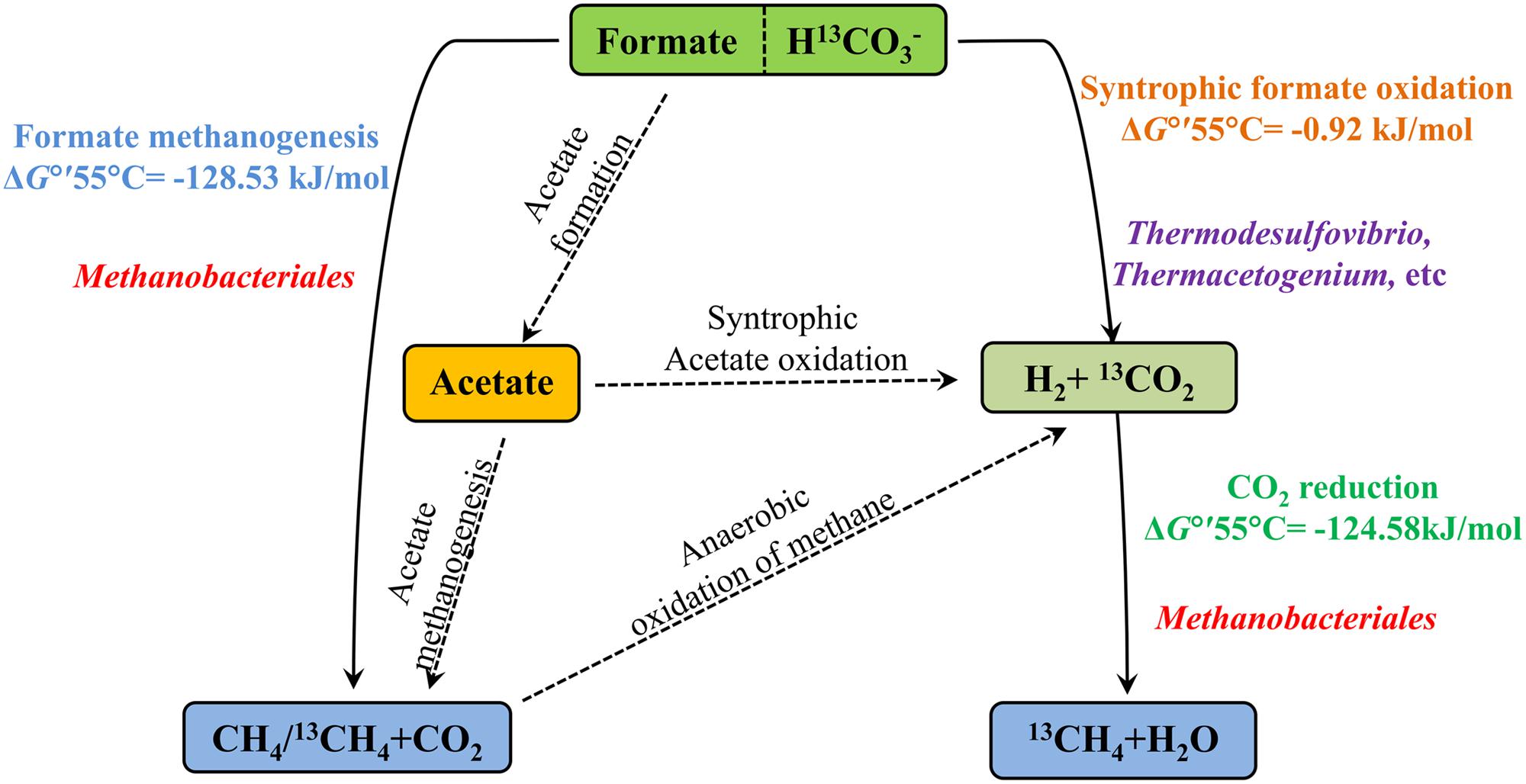
FIGURE 6. The proposed pathways of anaerobic formate transformation and related CO2 conversion. Solid lines mean the pathways were detected in the treatments and dotted lines mean not. ΔG°′55°C is standard Gibbs free energy at temperature 55°C and modified with protons (Table 1). Microorganisms are positioned according their probable role in the treatments.
Methane production was nearly equal to the formate consumed and consistent with stoichiometric formate oxidation reaction, indicating syntrophic formate oxidation and formate methanogenesis would be involved. Hydrogen was detected in all the samples at 35 days and decreased with the further incubation of the cultures (Figure 1B), suggesting that hydrogen was produced and then used for the CO2 reduction, which also indicated that syntrophic formate oxidation coupled with CO2 reduction and formate methanogenesis would be responsible for methanogenesis. The hydrogen could be generated through syntrophic formate oxidation or as intermediate during the formate methanogenesis in the samples. The obvious hydrogen production was found in S0 treatments and little hydrogen was detected in other culture samples amended with bicarbonate addition, because a high concentration of hydrogen could not be accumulated in these treatments with the limitation of Gibbs free energy threshold (ΔG′ = 0, Table 1). Meanwhile, CO2 reduction, formate oxidation, and formate methanogenesis were also below the threshold values, indicating these reactions were energetically favorable and spontaneous in the treatments. The production and subsequent consumption of hydrogen suggest that CO2 reduction dominated in the systems.
Formate was once known as nutritional substrate but not for methanogenesis directly by methanogens (Tanner et al., 1989), but some genera of Methanobacteriaceae, which can use formate directly for methanogenesis were found (Oren, 2014). Methane with C-13 labeled was detected in the treatments with amendment of 13C-bicarbonate (516.03∼675.46‰), suggesting that 13C-bicarbonate (or 13CO2) was reduced to 13CH4 (Figure 4), indicating the CO2-reducing pathway was dominant in the experiments. Carbon dioxide without any labeled 13CO2 was detected (173.88∼383.48‰) in the treatments (Figure 4), which should be produced by syntrophic formate oxidation. Formate is known as a potential energy source and important degradation intermediate in ecosystem. Based on our results, syntrophic formate oxidation coupled with CO2 reduction and formate methanogenesis could be the possible methanogenic pathway in the experiments.
CO2-Induced the Shift of Microbial Community
Oil reservoir is a typical anaerobic environment and the endogenous microorganisms in oil reservoir can degrade alkanes and produce methane (Mbadinga et al., 2011). In this study, the archaea in the inoculum had a high diversity and the ability to produce methane through CO2-reducing, acetoclastic and methylotrophic methanogenic pathways (Figure 2). However, some types of archaea were not detected in the treatments with the addition of formate and increase of bicarbonate concentrations, Methanosarcinales disappeared in the sample amended with 30 mM of bicarbonate, and only Methanobacteriales and Crenachaeota were detected in S60 and S90 treatments. The results indicated that Methanomicrobiales and Methanosarcinales were sensitive to the high bicarbonate concentrations. Compared to the original inoculum, Methanobacteriales was dominated in all the treatment cultures in the presence of formate, suggesting formate promoted the growth of the Methanobacteriales. Methanobacteriales is a kind of strict CO2-reducing methanogens, it also possesses formate dehydrogenase and could be candidate for formate-dependent H2 production and methanogenesis directly (Lupa et al., 2008), which is in agreement with the mentioned methanogenic pathway.
Bicarbonate concentration showed little impact on bacterial diversity. Thermodesulfovibrio belonged to Nitrospirales was a major group in the inoculum and all the treatments (Figure 3), especially in the low concentration treatments (inoculum, S0, and S30). Thermodesulfovibrio is a typical sulfate-reducing bacterium and grew through the reduction of sulfate with electron donors like hydrogen and formate. Thermodesulfovibrio and the CO2-reducing methanogen in co-culture experiments can produce methane syntrophically (Sekiguchi et al., 2008). In this study, Thermodesulfovibrio may use formate and co-work with methanogens for methane production. Clostridia were the most widely studied as hydrogen producer and acetogens in anaerobic environments (Drake et al., 2008; Calusinska et al., 2010), and Thermacetogenium belonged to Clostridia increased in the treatments with high bicarbonate concentrations (S60 and S90), known as a syntrophic acetate-oxidizing bacterium and acetogens (Hattori et al., 2000). However, there was no acetate accumulation (Supplementary Table S2) or acetoclastic methaogens (Figure 2) in the treatments with high bicarbonate concentrations (S60 and S90), suggesting that acetoclastic methanogenesis was not involved in the methane production of this study.
The microbial community in the experiments was capable of converting CO2 into methane through syntrophic formate oxidation coupled with CO2-reducing methanogenesis and formate methanogenesis. Thermodesulfovibrio and Methanobacteriales may play important role for the process in the system. The addition of bicarbonate can induce the shift of microbial community, especially in archaea.
In summary, under the microcosm study, most methane was produced by reducing the amended CO2 through the syntrophic formate oxidation coupled to CO2-reducing methanogenesis and formate methanogenesis with addition of bicarbonate. The results indicated that the microbial conversion of CO2 to methane is feasible with the microbial community in production water of high-temperature oil reservoirs. Thermodesulfovibrio and Methanobacteriales may be responsible for formate utilization and CO2 conversion. The results suggested that syntrophic formate oxidation coupled to CO2 reduction and formate methanogenesis could bealternative methanogenic pathway and gave some knowledge on formate metabolism in subsurface environment.
Author Contributions
J-DG and B-ZM designed the experiments, G-CY did the experiments, G-CY, LZ and SM carried out the microbial analysis. J-FL and S-ZY gave the suggestion for the experiments and results analysis. G-CY prepared the manuscript with contributions from all co-authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 41530318, 41273084, 41403066), the NSFC/RGC Joint Research Fund (Grant No. 41161160560) and the Fundamental Research Funds for the Central Universities of China (Grant No. WK1414029).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00365
References
Alissandratos, A., Kim, H. K., and Easton, C. J. (2014). Formate production through carbon dioxide hydrogenation with recombinant whole cell biocatalysts. Bioresour. Technol. 164, 7–11. doi: 10.1016/j.biortech.2014.04.064
Amend, J. P., and Shock, E. L. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25, 175–243. doi: 10.1111/j.1574-6976.2001.tb00576.x
Bagramyan, K., and Trchounian, A. (2003). Structural and functional features of formate hydrogen lyase, an enzyme of mixed-acid fermentation from Escherichia coli. Biochemistry (Moscow) 68, 1159–1170. doi: 10.1023/B:BIRY.0000009129.18714.a4
Berg, I. A., Kockelkorn, D., Ramos-Vera, W. H., Say, R. F., Zarzycki, J., Hügler, M., et al. (2010). Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 8, 447–460. doi: 10.1038/nrmicro2365
Callaghan, A. V., Morris, B. E. L., Pereira, I. A. C., McInerney, M. J., Austin, R. N., Groves, J. T., et al. (2012). The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation. Environ. Microbiol. 14, 101–113. doi: 10.1111/j.1462-2920.2011.02516.x
Calusinska, M., Happe, T., Joris, B., and Wilmotte, A. (2010). The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156, 1575–1588. doi: 10.1099/mic.0.032771–32770
Cheng, S., Xing, D., Call, D. F., and Logan, B. E. (2009). Direct biological conversion of electrical current into methane by electromethanogenesis. Environ. Sci. Technol. 43, 3953–3958. doi: 10.1021/es803531g
De Bok, F. A. M., Plugge, C. M., and Stams, A. J. M. (2004). Interspecies electron transfer in methanogenic propionate degrading consortia. Water Res. 38, 1368–1375. doi: 10.1016/j.watres.2003.11.028
Delgado, A. G., Parameswaran, P., Fajardo-Williams, D., Halden, R. U., and Krajmalnik-Brown, R. (2012). Role of bicarbonate as a pH buffer and electron sink in microbial dechlorination of chloroethenes. Microb. Cell Fact. 11:128. doi: 10.1186/1475-2859-11-128
Dolfing, J., Jiang, B., Henstra, A. M., Stams, A. J. M., and Plugge, C. M. (2008). Syntrophic growth on formate: a new microbial niche in anoxic environments. Appl. Environ. Microbiol. 74, 6126–6131. doi: 10.1128/aem.01428-1428
Drake, H. L., Gößner, A. S., and Daniel, S. L. (2008). Old acetogens, new light. Ann. N. Y. Acad. Sci. 1125, 100–128. doi: 10.1196/annals.1419.016
Dressel, B., Deel, D., Rodosta, T., Plasynski, S., Litynski, J., and Myer, L. (2011). CCS activities being performed by the U.S. DOE. Int. J. Environ. Res. Public Health 8, 300–320. doi: 10.3390/ijerph8020300
Gantner, S., Andersson, A. F., Alonso-Sáez, L., and Bertilsson, S. (2011). Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84, 12–18. doi: 10.1016/j.mimet.2010.10.001
Glueck, S. M., Gumus, S., Fabian, W. M. F., and Faber, K. (2010). Biocatalytic carboxylation. Chem. Soc. Rev. 39, 313–328. doi: 10.1039/B807875K
Hattori, S., Kamagata, Y., Hanada, S., and Shoun, H. (2000). Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 50, 1601–1609. doi: 10.1099/00207713-50-4-1601
Hattori, S., Luo, H., Shoun, H., and Kamagata, Y. (2001). Involvement of formate as an interspecies electron carrier in a syntrophic acetate-oxidizing anaerobic microorganism in coculture with methanogens. J. Biosci. Bioeng. 91, 294–298. doi: 10.1016/S1389-1723(01)80137-7
Hatzenpichler, R., Lebedeva, E. V., Spieck, E., Stoecker, K., Richter, A., Daims, H., et al. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U.S.A. 105, 2134–2139. doi: 10.1073/pnas.0708857105
Head, I. M., Jones, D. M., and Larter, S. R. (2003). Biological activity in the deep subsurface and the origin of heavy oil. Nature 426, 344–352. doi: 10.1038/nature02134
Hull, J. F., Himeda, Y., Wang, W.-H., Hashiguchi, B., Periana, R., Szalda, D. J., et al. (2012). Reversible hydrogen storage using CO2 and a proton-switchable iridium catalyst in aqueous media under mild temperatures and pressures. Nat. Chem. 4, 383–388. doi: 10.1038/nchem.1295
Jeletic, M. S., Mock, M. T., Appel, A. M., and Linehan, J. C. (2013). A cobalt-based catalyst for the hydrogenation of co2 under ambient conditions. J. Am. Chem. Soc. 135, 11533–11536. doi: 10.1021/ja406601v
Kim, Y. J., Lee, H. S., Kim, E. S., Bae, S. S., Lim, J. K., Matsumi, R., et al. (2010). Formate-driven growth coupled with H2 production. Nature 467, 352–355. doi: 10.1038/nature09375
Kirk, M. F. (2011). Variation in energy available to populations of subsurface anaerobes in response to geological carbon storage. Environ. Sci. Technol. 45, 6676–6682. doi: 10.1021/es201279e
Kobayashi, H., Kawaguchi, H., Endo, K., Mayumi, D., Sakata, S., Ikarashi, M., et al. (2012). Analysis of methane production by microorganisms indigenous to a depleted oil reservoir for application in Microbial Enhanced Oil Recovery. J. Biosci. Bioeng. 113, 84–87. doi: 10.1016/j.jbiosc.2011.09.003
Lin, Y., Lü, F., Shao, L., and He, P. (2013). Influence of bicarbonate buffer on the methanogenetic pathway during thermophilic anaerobic digestion. Bioresour. Technol. 137, 245–253. doi: 10.1016/j.biortech.2013.03.093
Liu, J.-F., Sun, X.-B., Yang, G.-C., Mbadinga, S. M., Gu, J.-D., and Mu, B. (2015). Analysis of microbial communities in the oil reservoir subjected to CO2-flooding by using functional genes as molecular biomarkers for microbial CO2 Sequestration. Front. Microbiol. 6:236. doi: 10.3389/fmicb.2015.00236
Lupa, B., Hendrickson, E. L., Leigh, J. A., and Whitman, W. B. (2008). Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl. Environ. Microbiol. 74, 6584–6590. doi: 10.1128/AEM.01455-1458
Mayer, F., and Müller, V. (2014). Adaptations of anaerobic archaea to life under extreme energy limitation. FEMS Microbiol. Rev. 38, 449–472. doi: 10.1111/1574-6976.12043
Mayumi, D., Dolfing, J., Sakata, S., Maeda, H., Miyagawa, Y., Ikarashi, M., et al. (2013). Carbon dioxide concentration dictates alternative methanogenic pathways in oil reservoirs. Nat. Commun. 4, 1998–1998. doi: 10.1038/ncomms2998
Mbadinga, S., Li, K.-P., Zhou, L., Wang, L.-Y., Yang, S.-Z., Liu, J.-F., et al. (2012). Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl. Microbiol. Biotechnol. 96, 531–542. doi: 10.1007/s00253-011-3828-3828
Mbadinga, S. M., Wang, L.-Y., Zhou, L., Liu, J.-F., Gu, J.-D., and Mu, B.-Z. (2011). Microbial communities involved in anaerobic degradation of alkanes. Int. Biodeterior. Biodegrad. 65, 1–13. doi: 10.1016/j.ibiod.2010.11.009
Meshulam-Simon, G., Behrens, S., Choo, A. D., and Spormann, A. M. (2007). Hydrogen metabolism in Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73, 1153–1165. doi: 10.1128/aem.01588-1586
Mikkelsen, M., Jorgensen, M., and Krebs, F. C. (2010). The teraton challenge. A review of fixation and transformation of carbon dioxide. Energy Environ. Sci. 3, 43–81. doi: 10.1039/B912904A
Momeni, A., Aghajani, M., and Zargar, G. (2012). A Simulation study of carbon dioxide sequestration in a depleted oil reservoir. Pet. Sci. Technol. 30, 751–765. doi: 10.1080/10916466.2010.490809
Nazaries, L., Murrell, J. C., Millard, P., Baggs, L., and Singh, B. K. (2013). Methane, microbes and models: fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 15, 2395–2417. doi: 10.1111/1462-2920.12149
Ohtomo, Y., Ijiri, A., Ikegawa, Y., Tsutsumi, M., Imachi, H., Uramoto, G.-I., et al. (2013). Biological CO2 conversion to acetate in subsurface coal-sand formation using a high-pressure reactor system. Front. Microbiol. 4:361. doi: 10.3389/fmicb.2013.00361
Oren, A. (2014). “The family methanobacteriaceae,” in The Prokaryotes, eds E. Rosenberg, E. DeLong, S. Lory, E. Stackebrandt and F. Thompson (Berlin: Springer), 165–193.
Rotaru, A.-E., Shrestha, P. M., Liu, F., Shrestha, M., Shrestha, D., Embree, M., et al. (2014). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415. doi: 10.1039/C3EE42189A
Schrag, D. P. (2007). Preparing to capture carbon. Science 315, 812–813. doi: 10.1126/science.1137632
Schuchmann, K., and Müller, V. (2013). Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342, 1382–1385. doi: 10.1126/science.1244758
Sekiguchi, Y., Muramatsu, M., Imachi, H., Narihiro, T., Ohashi, A., Harada, H., et al. (2008). Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int. J. Syst. Evol. Microbiol. 58, 2541–2548. doi: 10.1099/ijs.0.2008/000893-890
Shekh, A. Y., Krishnamurthi, K., Mudliar, S. N., Yadav, R. R., Fulke, A. B., Devi, S. S., et al. (2011). Recent advancements in carbonic anhydrase–driven processes for CO2 sequestration: minireview. Crit. Rev. Environ. Sci. Technol. 42, 1419–1440. doi: 10.1080/10643389.2011.556884
Shelton, J. L., McIntosh, J. C., Warwick, P. D., and Yi, A. L. Z. (2014). Fate of injected CO2 in the wilcox group, louisiana, gulf coast basin: chemical and isotopic tracers of microbial–brine–rock–CO2 interactions. Appl. Geochem. 51, 155–169. doi: 10.1016/j.apgeochem.2014.09.015
Tanner, R. S., Mcinerney, M. J., and Nagle, D. (1989). Formate auxotroph of Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 171, 6534–6538.
Wang, L.-Y., Duan, R.-Y., Liu, J.-F., Yang, S.-Z., Gu, J.-D., and Mu, B.-Z. (2012a). Molecular analysis of the microbial community structures in water-flooding petroleum reservoirs with different temperatures. Biogeosciences 9, 4645–4659. doi: 10.5194/bg-9-4645-2012
Wang, L.-Y., Li, W., Mbadinga, S. M., Liu, J.-F., Gu, J.-D., and Mu, B.-Z. (2012b). Methanogenic microbial community composition of oily sludge and its enrichment amended with alkanes incubated for over 500 days. Geomicrobiol. J. 29, 716–726. doi: 10.1080/01490451.2011.619634
Wood, G. E., Haydock, A. K., and Leigh, J. A. (2003). Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 185, 2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003
Yang, H., Xu, Z., Fan, M., Gupta, R., Slimane, R. B., Bland, A. E., et al. (2008). Progress in carbon dioxide separation and capture: a review. J. Environ. Sci. 20, 14–27. doi: 10.1016/S1001-0742(08)60002-9
Yeates, T. O., Kerfeld, C. A., Heinhorst, S., Cannon, G. C., and Shively, J. M. (2008). Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691. doi: 10.1038/nrmicro1913
Zhou, F., Mbadinga, S. M., Liu, J.-F., Gu, J.-D., and Mu, B.-Z. (2013). Evaluation of microbial community composition in thermophilic methane-producing incubation of production water from a high-temperature oil reservoir. Environ. Technol. 34, 2681–2689. doi: 10.1080/09593330.2013.786135
Keywords: bicarbonate, oil reservoirs, stable isotope technique, CDCS, methanogenesis, CO2 conversion
Citation: Yang G-C, Zhou L, Mbadinga SM, Liu J-F, Yang S-Z, Gu J-D and Mu B-Z (2016) Formate-Dependent Microbial Conversion of CO2 and the Dominant Pathways of Methanogenesis in Production Water of High-temperature Oil Reservoirs Amended with Bicarbonate. Front. Microbiol. 7:365. doi: 10.3389/fmicb.2016.00365
Received: 23 January 2016; Accepted: 07 March 2016;
Published: 22 March 2016.
Edited by:
Fernando G. Fermoso, Instituto de la Grasa – Consejo Superior de Investigaciones Científicas, SpainReviewed by:
Uwe Strotmann, Westfälische Hochschule, GermanyPetra Worm, Wageningen University, Netherlands
Copyright © 2016 Yang, Zhou, Mbadinga, Liu, Yang, Gu and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo-Zhong Mu, bzmu@ecust.edu.cn
 Guang-Chao Yang
Guang-Chao Yang Lei Zhou
Lei Zhou Serge M. Mbadinga
Serge M. Mbadinga Jin-Feng Liu
Jin-Feng Liu Shi-Zhong Yang
Shi-Zhong Yang Ji-Dong Gu
Ji-Dong Gu Bo-Zhong Mu
Bo-Zhong Mu