- 1Department of Clinical Microbiology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 2Department of Critical Care Medicine and Respiratory Intensive Care Unit, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 3Department of Medical Microbiology and Parasitology, Institutes of Medical Sciences, Shanghai Jiaotong University School of Medicine, Shanghai, China
One hundred and two carbapenem-resistant Enterobacteriaceae (CRE) strains were isolated in a teaching hospital in Shanghai, China from 2012 to 2015. In a follow-up study, four New Delhi metallo-β-lactamase-5 (NDM-5)-producing strains were identified after screening these CRE strains, including 1 Klebsiella pneumoniae strain (RJ01), 1 Proteus mirabilis strain (RJ02), and 2 Escherichia coli strains (RJ03 and RJ04). All K. pneumoniae and E. coli isolates were resistant to carbapenems, third-generation cephalosporins, and piperacillin-tazobactam, but were susceptible to amikacin. No epidemiological links for either E. coli isolate were found by multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). However, MLST revealed a novel sequence type, ST2250, of the K. pneumoniae RJ01 strain. Inc types and sizes of blaNDM-5-carrying plasmids differed among the four isolates, although in P. mirabilis RJ02 and E. coli RJ03, blaNDM-5 was carried by conjugative IncX3 plasmids of nearly the same size (∼40 kb). Investigation of the genetic background of sequences flanking the blaNDM-5 gene showed that all four isolates shared the same genetic content (IS3000-ΔISAba125-IS5-blaNDM-5-ble-trpF-dsbC-IS26-ΔumuD), which was identical to that of the pNDM_MGR194 plasmid circulating in India. This is the first identification of blaNDM-5 in P. mirabilis, which suggests its further spread to Enterobacteriaceae, and indicates that IncX3 plasmids may play an important role in potentiating the spread of blaNDM.
Introduction
Carbapenemase-producing Enterobacteriaceae have become a challenge to clinical therapy because of the rapid worldwide dissemination of multi-drug resistance (Tangden and Giske, 2015). Among the newly emerged carbapenemases, New Delhi metallo-β-lactamase-1 (NDM-1)-producing strains, which are capable of hydrolyzing all β-lactams, but not monobactams, show high potential to cause a global health crisis (Moellering, 2010). In contrast to NDM-1, NDM-2 shows low affinity for penicillin, which implies that penicillin is perhaps a better option for treating Acinetobacter baumannii harboring NDM-2 (Tiwari and Moganty, 2013).
New Delhi metallo-β-lactamase-5, produced by an Escherichia coli strain, was first identified in the UK in 2011 from a patient with a recent history of hospitalization in India (Hornsey et al., 2011). The NDM-5 enzyme differed from NDM-1 by only two amino acid substitutions (Val88Leu and Met154Leu) and showed increased resistance to carbapenems and broad-spectrum cephalosporins. Since then, NDM-5-producing strains have also been identified in Algeria (Sassi et al., 2014), China (Yang et al., 2014), Japan (Nakano et al., 2014), Spain (Pitart et al., 2015), India (Krishnaraju et al., 2015), the United States (de Man et al., 2015), South Korea (Cho et al., 2015), and Australia (Wailan et al., 2015). In 2015, clones related to NDM-5-producing strains were reported in Denmark (five isolates) (Hammerum et al., 2015) and the Netherlands (Bathoorn et al., 2015). The widespread occurrence of NDM-5 in recent years highlights the need for international attention.
To date, the gene encoding NDM-5, blaNDM-5, has only been reported in E. coli and Klebsiella pneumoniae. To evaluate the potential transmission of blaNDM-5-harboring bacterial strains in Shanghai, we screened for NDM-5-producing Enterobacteriaceae, and blaNDM-5 was amplified from four isolates by PCR for a more comprehensive study of the gene.
Materials and Methods
Bacterial Strains, Detection of Carbapenemase-encoding Genes, and Antimicrobial Susceptibility Testing
From 2012 to 2015, 102 carbapenem-resistant Enterobacteriaceae (CRE) strains were isolated using the VITEK® two Compact system (bioMérieux, Durham, NC, USA) in the clinical microbiology laboratory of Ruijin Hospital in Shanghai, China. In a retrospective study, we amplified common carbapenemase genes (blaKPC, blaIMP, blaV IM, blaOXA-48, and blaNDM) (Nordmann et al., 2011) from all 102 CRE isolates and sequenced the positive products; four blaNDM-5-carrying strains were identified for further study. Among them, two strains were isolated in January 2014, and the other two strains were isolated in July and September of the same year, respectively. The minimum inhibitory concentrations (MICs) of amikacin, ciprofloxacin, ceftazidime, ceftriaxone, meropenem, imipenem, ertapenem, cefepime, piperacillin-tazobactam, and aztreonam were determined by the E-test (bioMérieux, France), according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (M100-S25) (Clinical and Laboratory Standards Institute [CLSI], 2015). E. coli ATCC25922 was used for quality control.
Determination of Genetic Relatedness
Multilocus sequence typing (MLST) was performed on the E. coli1 and K. pneumoniae2 isolates, respectively. Two E. coli isolates were further analyzed by pulsed-field gel electrophoresis (PFGE). Salmonella enterica serotype Braenderup H9812 was used as a size marker. Restriction patterns were compared visually and interpreted on the basis of previously defined criteria (Tenover et al., 1995).
Plasmid Conjugation and Incompatibility Typing
A plasmid-conjugation experiment was performed between the four blaNDM-5-positive isolates and sodium azide-resistant E. coli J53AzR as the recipient strain. Transconjugants were selected on MacConkey agar plates containing 125 mg/L sodium azide and 1.0 mg/L imipenem. Antimicrobial susceptibility testing and PCR amplification of the transconjugants were subsequently performed to confirm whether the plasmid was successfully transferred to the recipient. Plasmid incompatibility types of the isolates were identified by PCR-based replicon typing (Carattoli et al., 2005). The positive products were sequenced and used to make digoxigenin (DIG)-labeled specific probes to identify the blaNDM-5-carring plasmid.
S1-PFGE and Southern Blotting
Plasmid-harboring bacteria were digested with S1 nuclease after being embedded in 1% SeaKem Gold Agarose gels (Lonza, Rockland, ME, USA). PFGE was performed as described above. The plasmid DNA was then transferred to positively charged nylon membranes (Roche Diagnostics GmbH Mannheim, Germany) and hybridized against DIG-labeled blaNDM-5-specific probes. Incompatibility group identification for blaNDM-5-carrying plasmids was conducted by hybridizing against blaIncX3 and blaIncFII-specific probes in the same manner.
Analysis of the Genetic Background Flanking the blaNDM-5 Gene
Primers were designed based on the reported blaNDM-5-flanking sequences to determine the genetic background of the blaNDM-5-harboring strains (Supplementary Table S1). PCR experiments were performed using the same thermocycling conditions for all strains, as follows: one cycle of 94°C for 4 min; followed by 35 cycles of 94°C for 30 s, 58°C for 40 s, and 72°C for 1 min; and a final cycle at 72°C for 10 min. The amplification products were sequenced and compared with sequences deposited in the BLAST database3.
Results
Bacterial Strains and Antimicrobial Susceptibility Testing
In this study, four blaNDM-5-positive isolates (RJ01–RJ04) were recovered from three hospitalized patients and one outpatient in Ruijin Hospital. One hospitalized patient with blaNDM-5-carrying E. coli isolated from a rectal swab was found to be a carrier. In contrast, the other three patients from whom blaNDM-5-carrying strains were isolated from drainage fluid, a wound site, or mid-stream urine were symptomatic. Other patients in the same ward were also screened, but no CRE isolates were observed. None of the patients had ever been abroad. Among the isolates, RJ01 was a K. pneumoniae isolate, RJ02 was a Proteus mirabilis isolate, and RJ03 and RJ04 were E. coli isolates. All E. coli and K. pneumoniae isolates were resistant to carbapenems, third-generation cephalosporins, and piperacillin-tazobactam, but were susceptible to amikacin (Table 1). Notably, RJ04 was resistant to aztreonam, suggesting the coexistence of other resistance mechanisms. P. mirabilis RJ02 was highly resistant to imipenem, but was susceptible and showed intermediate resistance to meropenem and ertapenem, respectively.
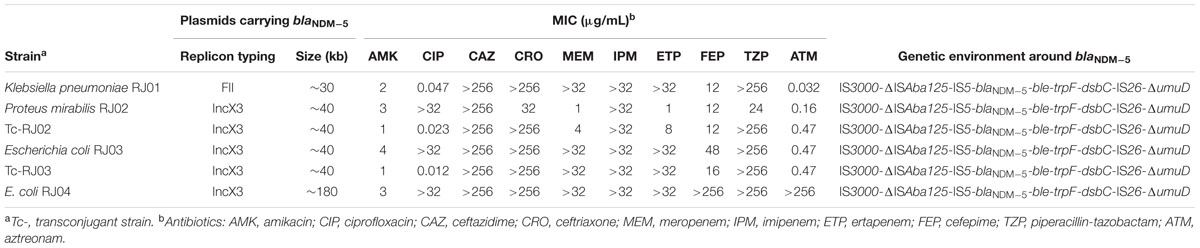
TABLE 1. Antimicrobial susceptibility of the four NDM-5-producing isolates and their transconjugants.
Genetic Relatedness of Four Isolates
According to the MLST results, E. coli RJ03 and E. coli RJ04 belonged to sequence type (ST) 354 and ST156, respectively. In accordance with the MLST results, the differences of the PFGE patterns confirmed that the two E. coli isolates are not clonally related. Because the sequence of K. pneumoniae RJ01 did not match any known STs, it was assigned a novel sequence type, ST2250 (10-20-2-1-9-11-25).
Characteristics of the blaNDM-5-Carrying Plasmids
In our study, RJ02 and RJ03 successfully transferred blaNDM-5 to the recipient, whereas RJ01 and RJ04 did not. An antimicrobial susceptibility test and PCR amplification were performed to confirm the presence of the gene and its phenotype in the transconjugants. S1-PFGE and subsequent Southern hybridization against DIG-labeled blaNDM-5 with incompatibility group-specific probes revealed that blaNDM-5 was carried by IncX3 plasmids of nearly the same size (∼40 kb) in P. mirabilis RJ02 and E. coli RJ03 (Figure 1), by a ∼180-kb IncX3 plasmid in the case of E. coli RJ04, and by a ∼30-kb IncFII plasmid in the case of K. pneumoniae RJ01.
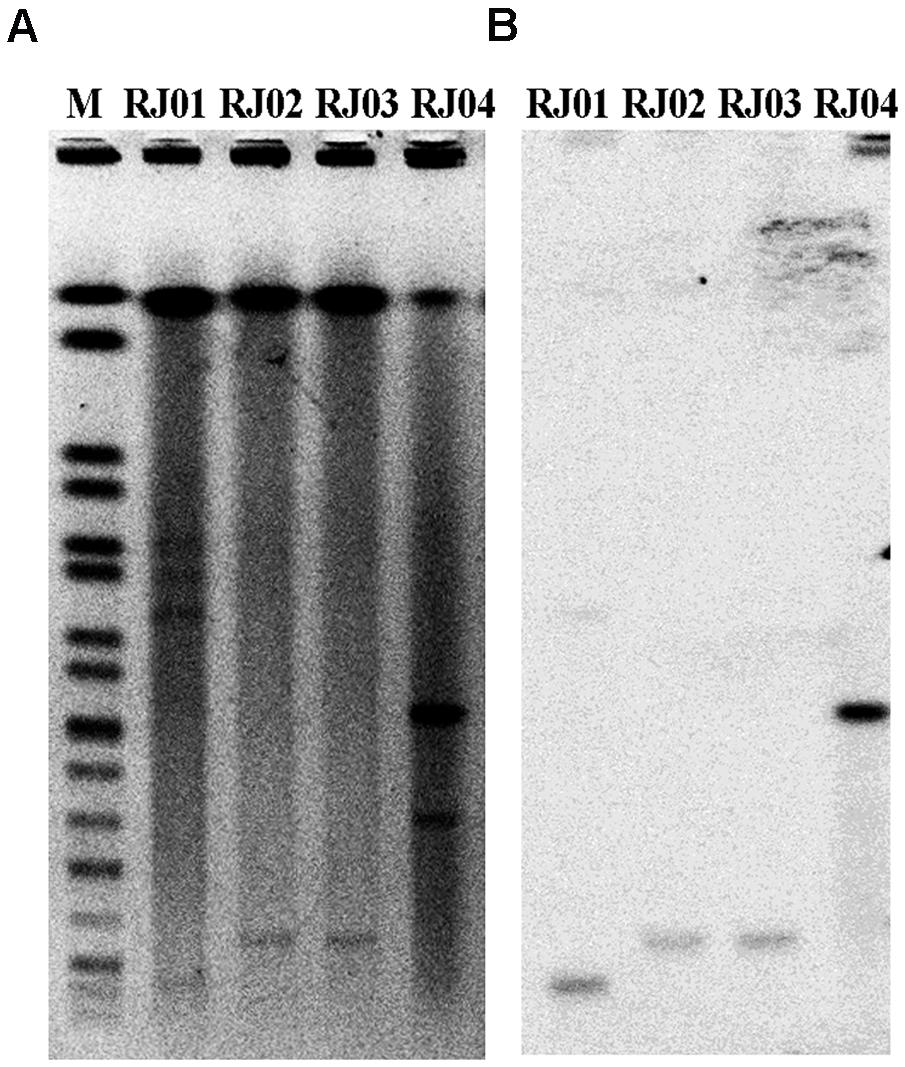
FIGURE 1. S1-pulsed-field gel electrophoresis (S1-PFGE) patterns of the four blaNDM-5-producing isolates (A) and Southern hybridization with a probe specific for blaNDM-5 (B). M, marker; Salmonella enterica serotype Braenderup H9812.
Genetic Surroundings of blaNDM-5
All four blaNDM-5-positive isolates were found to have an identical genetic background, IS3000-ΔISAba125-IS5-blaNDM-5-ble-trpF-dsbC-IS26-ΔumuD, which is the same as that of isolate pNDM_MGR194 in India (GenBank Accession Number KF220657) (Krishnaraju et al., 2015).
Discussion
Since blaNDM-5 was first identified in the UK (Hornsey et al., 2011), strains harboring this gene have emerged in several countries (Sassi et al., 2014; Yang et al., 2014; de Man et al., 2015; Krishnaraju et al., 2015; Pitart et al., 2015; Wailan et al., 2015). Because NDM-5 has only been previously reported in E. coli and K. pneumoniae, the NDM-5 found in P. mirabilis RJ02 indicates the further spread of blaNDM-5 among different species of Enterobacteriaceae. Although NDM-5-producing strains are not as widespread as NDM-1-producing strains, their greater resistance to antimicrobial drugs make them a potential public health threat.
E. coli ST354 and ST156 were found in our study. Although ST648 has been reported both in the UK (Hornsey et al., 2011) and in Australia (Wailan et al., 2015), most reports have indicated a high ST diversity for blaNDM-5-positive E. coli (Sassi et al., 2014; Yang et al., 2014; Cho et al., 2015; Pitart et al., 2015). From 2014 to 2015, six NDM-5-producing ST16 K. pneumoniae isolates were identified among patients in Denmark (five isolates) (Hammerum et al., 2015) and the Netherlands (one isolate) (Bathoorn et al., 2015), who had not traveled recently. Before these reports were published, no other outbreaks of NDM-5-producing Gram-negative bacteria were reported. Although no evident genetic association was found between our blaNDM-5-positive isolates (ST2250) with other strains, the first report of an NDM-5-related outbreak in Europe showed an epidemic potential and deserved extensive attention.
A previous study conducted in Algeria showed that all three blaNDM-5-positive E. coli strains tested could transfer their resistant plasmids to the azide-resistant E. coli strain J53 (Sassi et al., 2014), which implied the possibility of horizontal transfer of the blaNDM-5 gene. In our study, two strains successfully transferred blaNDM-5 to the recipient, thereby reconfirming the horizontal transfer ability. IncX3 plasmids might have played an important role in mediating the horizontal transmission of blaNDM (Ho et al., 2012; Sonnevend et al., 2013), a possibility that has been supported by the results of several studies (Gottig et al., 2013; Yang et al., 2014; Krishnaraju et al., 2015). Here, blaNDM-5 was carried by IncX3 plasmids in three strains in our study. To date, IncX3 plasmids carrying blaNDM-5 have been reported in China (Yang et al., 2014), India (Krishnaraju et al., 2015), Australia (Wailan et al., 2015), and Denmark (Hammerum et al., 2015). Therefore, our current study complements these previous data. IncX-type plasmids have a narrow host range in Enterobacteriaceae. The fact that IncX-type plasmids have been shown to be conjugatable in most studies could explain the rapid spread of NDM-carrying isolates. Therefore, it is imperative that effective and feasible measures are taken immediately to control the dissemination of these resistant plasmids.
India is considered to be one the main reservoirs of blaNDM (Nordmann, 2014). Travel to endemic countries is generally associated with the global dissemination of carbapenemase-producing Enterobacteriaceae (Savard and Perl, 2014). Although the four isolates shared the same genetic background with isolate pNDM_MGR194 from India (GenBank Accession Number KF220657) (Krishnaraju et al., 2015), none of the four patients had ever been abroad. Previous reports of blaNDM-containing plasmids isolated from patients in China also showed little contact with the Indian subcontinent (Wang et al., 2012). However, as international travel is becoming increasingly common, some routes of transmission between people might be unrecognized. We further compared the genetic environments flanking the blaNDM-5 genes among isolates from China (Yang et al., 2014), India (Krishnaraju et al., 2015), and Japan (Nakano et al., 2014) and found that they were nearly identical, except for the number and position of the IS5 insertion. Therefore, we speculate that horizontal transference has played an important role in dissemination, although more data are needed to explain how the blaNDM-5 gene has spread among these countries.
In summary, our study provides evidence for the further spread of the blaNDM-5 gene in Enterobacteriaceae. These results thus expand previous data on NDM-5-producing strains, and support the speculation that the IncX3-type plasmids have played a major role in the global dissemination of NDM-producing Enterobacteriaceae. This work strongly highlights the urgent need for effective action to control the horizontal spread of blaNDM-5.
Author Contributions
FZ, LX, XW, LH, XG, YN, HQ, and JS: substantial contributions to the conception and design of the work; revising it critically for important intellectual content; final approval of the version to be published; agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved; analyzed the data; contributed materials.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81472010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00424
Footnotes
- ^ http://mlst.warwick.ac.uk/mlst/dbs/Ecoli
- ^ http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html
- ^ http://www.ncbi.nlm.nih.gov/BLAST
References
Bathoorn, E., Rossen, J. W., Lokate, M., Friedrich, A. W., and Hammerum, A. M. (2015). Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro. Surveill 20:41. doi: 10.2807/1560-7917.es.2015.20.41.30040
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Cho, S. Y., Huh, H. J., Baek, J. Y., Chung, N. Y., Ryu, J. G., Ki, C. S., et al. (2015). Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases. South Korea. Emerg. Infect. Dis. 21, 1088–1089. doi: 10.3201/eid2106.150048
Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial Susceptibility Testing[S]: Twenty-fifth Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
de Man, T. J., Perry, K. A., Avillan, J. J., Rasheed, J. K., and Limbago, B. M. (2015). Draft genome sequence of a New Delhi metallo-beta-lactamase-5 (NDM-5)-producing multidrug-resistant Escherichia coli isolate. Genome Announc. 3:e00017. doi: 10.1128/genomeA.00017-15
Gottig, S., Hamprecht, A. G., Christ, S., Kempf, V. A., and Wichelhaus, T. A. (2013). Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. J. Antimicrob. Chemother. 68, 1737–1740. doi: 10.1093/jac/dkt088
Hammerum, A. M., Hansen, F., Olesen, B., Struve, C., Holzknecht, B. J., Andersen, P. S., et al. (2015). Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J. Global Antimicrob. Resist. 3, 219–221. doi: 10.1016/j.jgar.2015.05.003
Ho, P. L., Li, Z., Lo, W. U., Cheung, Y. Y., Lin, C. H., Sham, P. C., et al. (2012). Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microbes Infect. 1:e39. doi: 10.1038/emi.2012.37
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Moellering, R. C. Jr. (2010). NDM-1–a cause for worldwide concern. N. Engl. J. Med. 363, 2377–2379. doi: 10.1056/NEJMp1011715
Nakano, R., Nakano, A., Hikosaka, K., Kawakami, S., Matsunaga, N., Asahara, M., et al. (2014). First report of metallo-beta-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob. Agents Chemother. 58, 7611–7612. doi: 10.1128/AAC.04265-14
Nordmann, P. (2014). Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med. Mal. Infect. 44, 51–56. doi: 10.1016/j.medmal.2013.11.007
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Pitart, C., Sole, M., Roca, I., Roman, A., Moreno, A., Vila, J., et al. (2015). Molecular characterization of blaNDM-5 carried on an IncFII plasmid in an Escherichia coli isolate from a nontraveler patient in Spain. Antimicrob. Agents Chemother. 59, 659–662. doi: 10.1128/AAC.04040-14
Sassi, A., Loucif, L., Gupta, S. K., Dekhil, M., Chettibi, H., and Rolain, J. M. (2014). NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob. Agents Chemother. 58, 5606–5608. doi: 10.1128/AAC.02818-13
Savard, P., and Perl, T. M. (2014). Combating the spread of carbapenemases in Enterobacteriaceae: a battle that infection prevention should not lose. Clin. Microbiol. Infect. 20, 854–861. doi: 10.1111/1469-0691.12748
Sonnevend, A., Al Baloushi, A., Ghazawi, A., Hashmey, R., Girgis, S., Hamadeh, M. B., et al. (2013). Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J. Med. Microbiol. 62, 1044–1050. doi: 10.1099/jmm.0.059014-0
Tangden, T., and Giske, C. G. (2015). Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J. Intern. Med. 277, 501–512. doi: 10.1111/joim.12342
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Tiwari, V., and Moganty, R. R. (2013). Structural studies on New Delhi Metallo-beta-lactamase (NDM-2) suggest old beta-lactam, penicillin to be better antibiotic for NDM-2-harbouring Acinetobacter baumanni. J. Biomol. Struct. Dyn. 31, 591–601. doi: 10.1080/07391102.2012.706075
Wailan, A. M., Paterson, D. L., Caffery, M., Sowden, D., and Sidjabat, H. E. (2015). Draft Genome Sequence of NDM-5-Producing Escherichia coli Sequence tType 648 and Genetic Context of blaNDM-5 in Australia. Genome Announc. 3:e00194. doi: 10.1128/genomeA.00194-15
Wang, Y., Wu, C., Zhang, Q., Qi, J., Liu, H., Wang, Y., et al. (2012). Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS ONE 7:e37152. doi: 10.1371/journal.pone.0037152
Keywords: Enterobacteriaceae, carbapenemase, NDM-5, IncX3 type plasmid, epidemiology
Citation: Zhang F, Xie L, Wang X, Han L, Guo X, Ni Y, Qu H and Sun J (2016) Further Spread of blaNDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front. Microbiol. 7:424. doi: 10.3389/fmicb.2016.00424
Received: 09 December 2015; Accepted: 16 March 2016;
Published: 30 March 2016.
Edited by:
John W. A. Rossen, University Medical Center Groningen, NetherlandsReviewed by:
Ruud H. Deurenberg, University Medical Center Groningen, NetherlandsErik Bathoorn, University Medical Center Groningen, Netherlands
Copyright © 2016 Zhang, Xie, Wang, Han, Guo, Ni, Qu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongping Qu, hongpingqu0412@hotmail.com; Jingyong Sun, 13671578899@126.com
 Fangfang Zhang1
Fangfang Zhang1 Jingyong Sun
Jingyong Sun