- 1Department of Laboratory Medicine, Shanghai Medical College, Huashan Hospital, Fudan University, Shanghai, China
- 2Department of Laboratory Medicine, Renji Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
Infections of the bloodstream, central nervous system, peritoneum, joints, and other sterile areas are associated with high morbidity and sequelae risk. Timely initiation of effective antimicrobial therapy is crucial to improving patient prognosis. However, standard final identification and antimicrobial susceptibility tests (ASTs) are reported 16–48 h after a positive alert. For a rapid, effective and low-cost diagnosis, we combined matrix-assisted laser desorption/ionization time of flight mass spectrometry with a Vitek AST system, and performed rapid microbial identification (RMI) and rapid multiple AST (RMAST) on non-duplicated positive body fluid cultures collected from a hospital in Shanghai, China. Sterile body fluid positive culture and blood positive culture caused by Gram negative (GN) or polymicrobial were applied to the MALDI–TOF measurement directly. When positive blood culture caused by Gram positive (GP) bacteria or yeasts, they were resuspended in 1 ml brain heart infusion for 2 or 4 h enrichment, respectively. Regardless of enrichment, the RMI (completed in 40 min per sample) accurately identified GN and GP bacteria (98.9 and 87.2%, respectively), fungi (75.7%), and anaerobes (94.7%). Dominant species in multiple cultures and bacteria that failed to grow on the routing plates were correctly identified in 81.2 and 100% of cases, respectively. The category agreements of RMAST results, determined in the presence of various antibiotics, were similarly to previous studies. The RMI and RMAST results not only reduce the turnaround time of the patient report by 18–36 h, but also indicate whether a patient's antibiotic treatment should be accelerated, ceased or de-escalated, and adjusted the essential drugs modification for an optimized therapy.
Introduction
Infections of the bloodstream, central nervous system, peritoneum, joints, and other sterile areas are associated with high morbidity and risk of sequelae (Thigpen et al., 2011; Goto and Al-Hasan, 2013; Chon et al., 2014; Ascione et al., 2015; Bagheri-Nesami et al., 2015). Among these, bloodstream infection (BSI) is most serious, because it can rapidly deteriorate into sepsis, severe sepsis, or septic shock. BSIs have become a major cause of death in European intensive care units, incurring a mortality rate of 30–50% (Vincent et al., 2006). To improve this prognosis, timely initiation of effective antimicrobial therapy is essential (Kumar et al., 2006; Vincent et al., 2006; Dellinger et al., 2013; Chon et al., 2014; Ascione et al., 2015; Bagheri-Nesami et al., 2015). Currently, culture remains the gold standard of infection diagnosis. However, standard final identification (ID) and ASTs are reported 16–48 h after a positive alert. During this delay, the clinician must administer an empirical antimicrobial therapy, typically a broad-spectrum antibiotic or an antibiotic cocktail to cover all likely pathogens. However, inappropriate antimicrobial therapy will worsen the outcome (Ibrahim et al., 2000; Tumbarello et al., 2010; Cain et al., 2015). Moreover, the long-term use of dispensable broad-spectrum antibiotics promotes antibiotic resistance and spread, increases cost and lengthens hospital stays (Blot et al., 2002; Tumbarello et al., 2010).
Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI–TOF MS) promises a revolutionary breakthrough in clinical microbiology. The technology identifies bacteria within 6 min (Seng et al., 2009), with a species-level accuracy of 84.1–93.6% (Bizzini and Greub, 2010) and high sensitivity (~105 CFU). Currently, MALDI-TOF MS has shown the ability to distinguish vanB-positive Enterococcus faecium from isolates that do not possess this resistance gene (Griffin et al., 2012) and demonstrate the carbapenemase activity by detection of meropenem and the relevant degradation products (Hrabak et al., 2011). Another study focused on a proteome analysis of ampicillin-resistant Fusobacterium nucleatum was reported (Al-Haroni et al., 2008). Moreover, MALDI-TOF MS fingerprinting has been used to distinguish the different expression levels of cell wall components between resistant isolates and sensitive isolates (Xu et al., 2006). Recently, RMI by MALDI–TOF MS has been adopted in various protocols, including the commercial Sepsityper kit (Bruker Daltonics, Bremen, Germany; Martiny et al., 2012; Hazelton et al., 2014; Idelevich et al., 2014; Martinez et al., 2014; Schieffer et al., 2014; Morgenthaler and Kostrzewa, 2015), serum separator tubes (Stevenson et al., 2010; Schubert et al., 2011) and in-house methods (Martiny et al., 2012). These techniques return the RMI from positive bottles within 30 min to a few hours. They also reliably identify species-level GN and GP bacteria (with accuracies of 90 and 76%, respectively) and yeasts (66%) (Morgenthaler and Kostrzewa, 2015). However, the Sepsityper kit is expensive, whereas the in-house method involves multi-step washing/centrifugation and yields relatively low RMI accuracy for GP bacteria and yeasts.
Rapid ASTs have been attempted in several studies. For instance, methicillin-resistant staphylococci and vancomycin-resistant enterococci have been detected in real-time PCR–melt curve analysis (Chan et al., 2015). MALDI–TOF MS can detect subtle differences in isogenic Staphylococcus aureus, which determine the organism's resistance to methicillin or teicoplanin (Majcherczyk et al., 2006), ampicillin susceptibility (Grundt et al., 2012), and carbapenemase (Hrabák et al., 2012). All of the above methods are efficient; however, molecular methods cannot identify the expression of resistance genes, and other methods are limited to one or a few specific antibiotic determination profiles.
To overcome these difficulties, we determined the RMI and RMAST by a combined MALDI–TOF MS/Vitek AST system. Bacterial cells were directly extracted from positive sterile body fluid cultures in serum separator tubes, and proliferated in brain heart infusion broth (BHI) for 2–4 h.
This study aims to evaluate the reliability and accuracy of this protocol in fast pathogen diagnosis without additional costs and efforts.
Materials and Methods
Location
This study was conducted in Huashan Hospital (affiliated with Fudan University), located in the center of Shanghai, China. Huashan Hospital is one of the largest (1300 beds) comprehensive teaching hospitals in China, handling ~8000 admissions per day.
Clinical Samples
Sterile body fluids, including blood, cerebrospinal fluid (CSF), pleural fluid, ascitic fluid, pericardial effusion, joint cavity fluid, and vitreous fluid, were injected into blood culture bottles to improve the positive rate of clinical samples. From September 2014 to August 2015, we enrolled the first positive culture from each patient. Thus, we collected 485 non-duplicated positive cultures.
Blood Culture (BC) Bottles and BC System
The BC bottles were BacT/Alert aerobic/SA and anaerobic/SN (bioMérieux, Marcy l'Etoile, France), and BACTEC Plus Aerobic/F, BACTEC Plus Anaerobic/F and BACTEC Mycosis-IC/F (Becton Dickinson, USA; Figure S1). All bottles were incubated in automated BC systems (BacT/Alert 3D, bioMérieux and BACTEC FX, Becton Dickinson; Figure S1) until they tested positive. Positive cultures were analyzed immediately after a positive alert during laboratory hours (8 a.m. to 5 p.m.). Cultures that became positive later than this period were stored in the BC systems and analyzed the next morning.
Positive Cultures Processing for RMI Analysis and RMAST
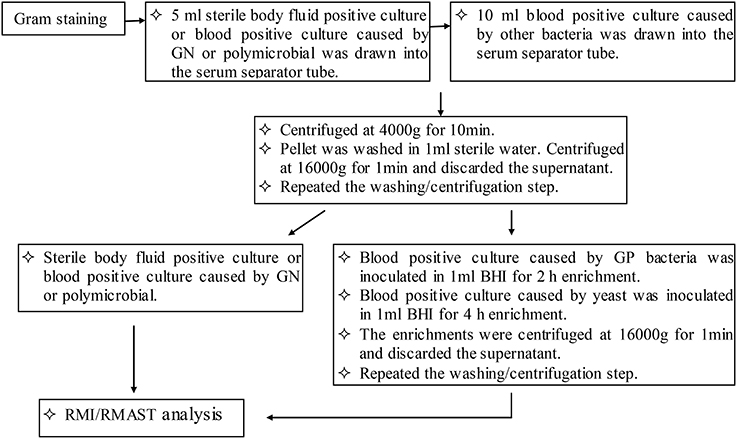
Based on the Gram staining results, 5 ml (sterile body fluid positive culture and blood positive culture caused by GN or polymicrobial) or 10 ml (blood positive culture caused by other bacteria) were drawn into one or two serum separator tubes (BD Vacutainer SSTII Advance, USA). Bacteria were pelleted by centrifugation at 4000 g for 10 min. The bacteria became aggregated at the surface of the polymeric gel, while the red-blood-cell component was sedimented beneath the gel layer. The supernatant was discarded and the bacterial pellet was gently resuspended in 1 ml sterile distilled water, without disrupting the gel layer. The suspension was transferred to a sterile Eppendorf tube, mixed thoroughly, and centrifuged at 16,000 g for 1 min. The supernatant was discarded and the washing/centrifugation steps were repeated once. The pellets were resuspended in 100 μl sterile distilled water. A portion of the resuspension was drawn and subjected to an ethanol/formic acid extraction procedure. The rest of the resuspension was prepared for 0.5 McFarland (McF) and subjected to RMAST as described for standard AST.
Prior to the ethanol/formic acid extraction, pellets of blood cultures caused by other bacteria (but not those caused by GN or polymicrobial) were resuspended in 1 ml BHI for enrichment. GP bacteria and yeasts were inoculated at 37°C with shaking at 200 rpm for 2 and 4 h, respectively. The enrichments were pelleted at 16,000 g for 1 min. After discarding the supernatant, the pellets were washed with 1 ml sterile distilled water, and re-centrifuged at 16,000 g for 1 min. Again, the supernatant was discarded.
In the ethanol/formic acid extraction procedure, a portion of the bacterial pellet was resuspended in 300 μl water by vortexing. The suspension was thoroughly mixed with 900 μl absolute ethanol and then centrifuged at 16,000 g for 1 min. The supernatant was discarded and the residual ethanol was removed after a repeat centrifugation. The cell pellet was air dried and dissolved in 30 μl of 70% formic acid by thorough vortexing. After adding 30 μl acetonitrile, the dissolved pellet was centrifuged at 16,000 g for 2 min and 1 μl supernatant was spotted onto a steel target plate for MALDI–TOF MS (Bruker Daltonics, Bremen, Germany) analysis (Figure 1).
Standard Identification and Antimicrobial Susceptibility Testing
The positive broths were sub-cultured onto routing plates of 5% sheep blood agar, chocolate, or anaerobic blood agar. Bacteria that failed to grow on the routing plates were inoculated on self-made plates composed of 20 ml sterile blood culture broth (bioMérieux, Marcy l'Etoile, France; Becton Dickinson, USA) and 3.9% agar powder (Oxoid, Thermo Fisher Scientific, England). Plates were grown in the incubator (Thermo Scientific Forma, USA) at 35°C in 5% CO2 or an anaerobic atmosphere until visible colonies appeared. A pure bacterial colony was smeared onto a steel target plate for identification by MALDI–TOF MS. For yeasts, 1 μl of formic acid was added to the plate and air dried for 5 min.
In the antimicrobial susceptibility testing, Vitek cards AST-GN13, AST-Gp67, and AST-Gp68 were used for GN bacteria, staphylococci/enterococci and Streptococcus pneumoniae, respectively. Other streptococci and yeasts were tested with ATB-STREP5 and ATB-FUNGUS3 (bioMérieux, Marcyl' Etoile, France), respectively. The GN bacteria, staphylococci, enterococci, streptococci, or yeast pellets were dissolved in 0.45% saline solution to prepare 0.5–0.63 McF or 2 McF suspension, respectively. The Vitek card AST-GN13 or AST-Gp67/AST-Gp68 were filled with suspension composed of 3 ml 0.45% saline solution and 145 or 280 μl 0.5–0.63 McF suspension, respectively. The ATB-STREP5 strip was filled with suspension composed of ATB S medium and 200 μl 0.5 McF suspension. The ATB-FUNGUS3 strip was filled with suspension composed of ATB F2 medium and 20 μl 2McF suspension.
Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry
The spot was overlaid with 1 μl MALDI matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile–2.5% trifluoroacetic acid). After drying at room temperature for 5 min, sample was subjected to analysis of the bacteria protein using MALDI-TOF MS system. The spectrum was obtained in linear positive-ion mode range from 2000 to 20 000 Da. Each spot was measured manually on five different positions by using 1000 laser shots at 25 Hz in groups of 40 shots. The MALDI Bruker Biotyper 3.0 software and library (Bruker Daltonics) were used for spectra analysis. According to the spectra matching and score criterion, MALDI Bruker Biotyper 3.0 software obtained appropriate scores and identification results. The workflow was shown in Figure 2A.
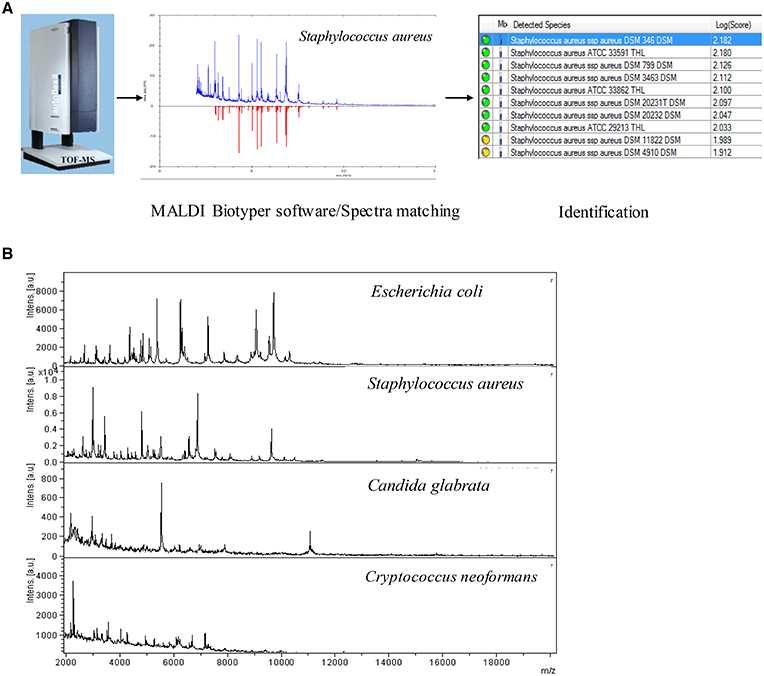
Figure 2. (A) The RMI workflow of MALDI-TOF MS system. (B) RMI spectra of representative bacterial species.
Interpretation of RMI and RMAST
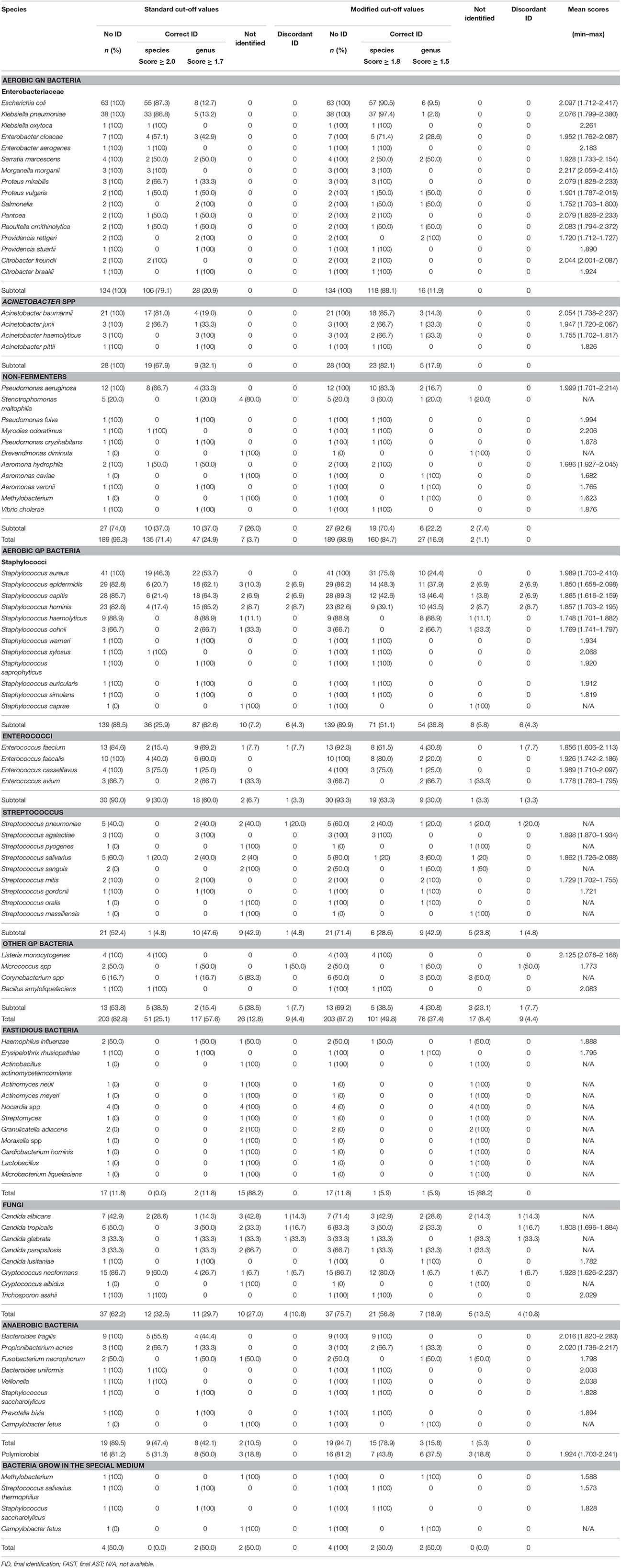
RMI results were scored by the manufacturer's standard criterion (cut-off values of 1.7 and 2.0 for acceptable identification to the genus and species levels, respectively) and by the modified criterion (cut-off values of 1.5 and 1.8 for at least three identical results in the list at the genus and species levels, respectively). These criteria have been described in previous studies (Schmidt et al., 2012; Machen et al., 2014; Martinez et al., 2014; Morgenthaler and Kostrzewa, 2015). The RMI results were compared with the final reports. Non-identifiable samples included correctly identified samples with scores below the genus level cut-off value and samples that presented no peak or a very weak signal. Discordant identification included samples that the RMI results were inconsistent with the final reports which proved correct. Discrepancies were resolved by 16S rRNA (for bacteria; primer F, 5′-AGAGTTTGATGATGGCTCAG-3′; primer R, 5′-ACCGCAACTGCTGGCAC-3′; expected PCR product size: ~800 bp) or 18S rRNA (for fungi; primer F, 5′-GATACCGTCGTAGTCTTA-3′; primer R, 5′-ATTCCTCGTTGAAGAGC-3′; expected PCR product size: ~800 bp) amplification. DNA was extracted from colonies sub-cultured for 24 h using Genomic DNA isolation kit (Sangon Biotech, Shanghai, China). Thermal cycler conditions were 94°C for 5 min, followed by 35 cycles of 94°C (30 s), 55°C (30 s), and 72°C (30 s), with a final extension at 72°C (7min).
RMAST results were compared with those obtained from the standard method. The minimum inhibitory concentrations (MIC) obtained by both methods were translated into clinical categories (susceptible, intermediate, resistant), following the CLSI recommendations. The comparison between the direct and standard inoculation methods was categorized as agreement, very major error (VME, false susceptibility), major error (ME, false resistance), or minor error (mE, susceptible/resistant versus intermediate susceptibility). Discrepancies in MICs were resolved by broth dilution methods according to the CLSI (2015) guidelines.
Evaluation of Clinical Relevance
To prevent the development of resistance, to reduce toxicity, and to reduce costs, the antimicrobial regimen should be ceased when detected typical contaminants (Monoculture grew Coagulase-negative staphylococcus among multiple cultures, Micrococcus spp, Corynebacterium spp, Bacillus spp, Propionibacterium acnes) and assessed for potential de-escalation (De-escalation refers to narrow the spectrum of antimicrobial coverage and choose the most appropriate single-agent therapy). Antibiotic therapy should be installed when detected pathogenic species that cannot be distinguished by Gram staining and modified when detected intrinsic resistant bacteria (Bacteria were intrinsic antibiotic-resistance. Stenotrophomonas maltophilia, Enterococcus casselifavus, and Candida glabrata are intrinsic resistant to carbapenem, vancomycin, and fluconazole, respectively) or acquired resistant bacteria (Antibiotic-resistance was mediated by plasmid, resistant enzymes, or other resistance mechanisms). RMI/RMAST results were reported to the clinician. To assess the impact of our approach on the management of sterile body infections, we determined whether the results can instructively recommend an optimal therapeutic scheme. Unreliable results were excluded. Indications leading to a treatment change are listed in Table 1.
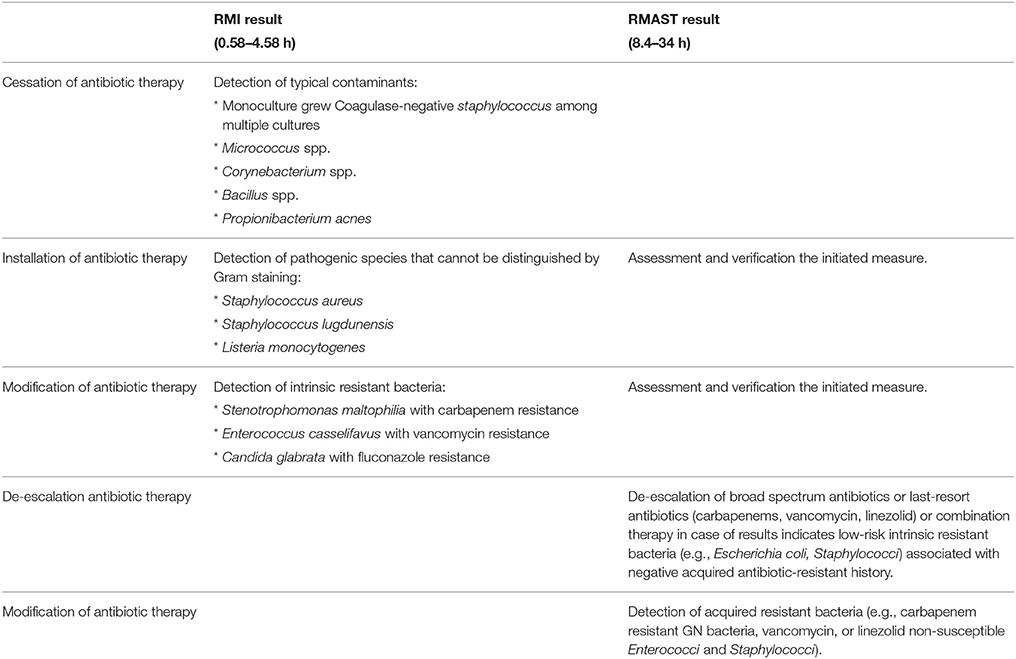
Table 1. Indications for optimized antibiotic therapy and recommendations based on RMI /RMAST results.
Statistical Analysis
Statistical time comparisons were conducted by a Wilcoxon signed-rank test (GraphPad Prism 5.0, CA, USA). P < 0.05 was considered statistically significant. All statistical tests were two-tailed.
Results
Rapid Microorganism Identification
Regardless of enrichment, the RMI test of each sample was completed in 40 min (15 min for the pellet collection/washing/centrifugation steps, 15 min for the extraction procedure, 5 min for the sample spotting/drying steps, 5 min for MALDI–TOF MS measurement). The RMI was evaluated in 485 non-duplicated positive cultures (Table 2). The compositions of the positive cultures are listed in Table S1.
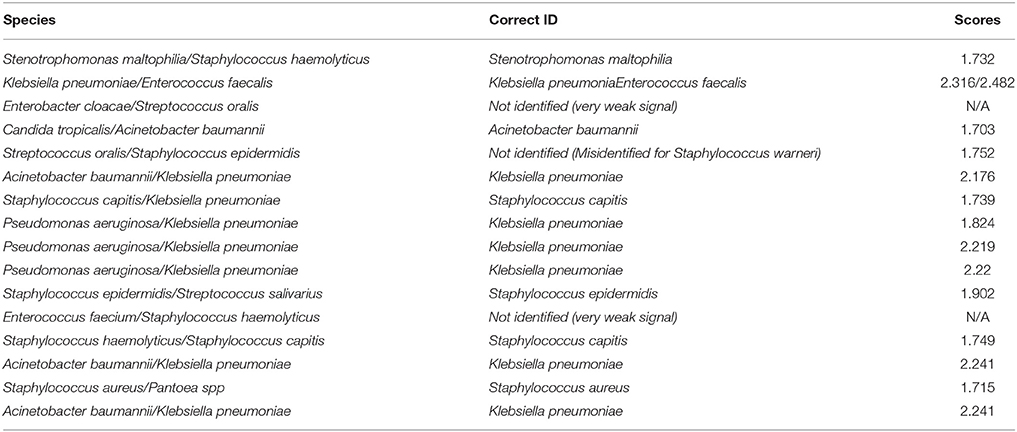
The RMI spectra of representative bacterial species were shown in Figure 2B. Scored by the standard criterion, the correct identification rates of RMI were 96.3% for GN bacteria (182/189), 82.8% for GP bacteria (168/203), 11.8% for fastidious bacteria (2/17), 62.2% for fungi (23/37), and 89.5% for anaerobic bacteria (17/19). In cultures of polymicrobial, the dominant bacterium was correctly identified in 81.2% (13/16) of cases and bacteria that failed to grow on the routing plates were identified with a success rate of 50.0% (2/4). Scored by the modified criterion, the correct identification rates of RMI were 98.9% (187/189) for GN bacteria, 87.2% (177/203) for GP bacteria, 11.8% (2/17) for fastidious bacteria, 75.7% (28/37) for fungi, and 94.7% (18/19) for anaerobic bacteria (18/19). In cultures containing polymicrobial and bacteria that failed to grow on the routing plates, the correct identification rates were 81.2% (13/16; dominant species) and 100.0% (4/4), respectively. The modified criterion provided more accurate RMI scores than the standard criterion, with no relevant misidentification at the genus level (Tables 2, 3). The RMI accuracy was lower in blood samples than in other sterile body fluids (Table S1). The mean RMI scores and the RMI results of polymicrobial are listed in Tables 2, 3, respectively.
Rapid Antimicrobial Susceptibility Test
Among the 320 non-duplicated positive cultures, 140 GN bacteria, 105 Staphylococcus spp., 28 Enterococcus spp., 14 Streptococcus spp., 33 fungi, and 3349 bacterial /antimicrobial combinations, were analyzed in the RMAST investigation (Table 4).
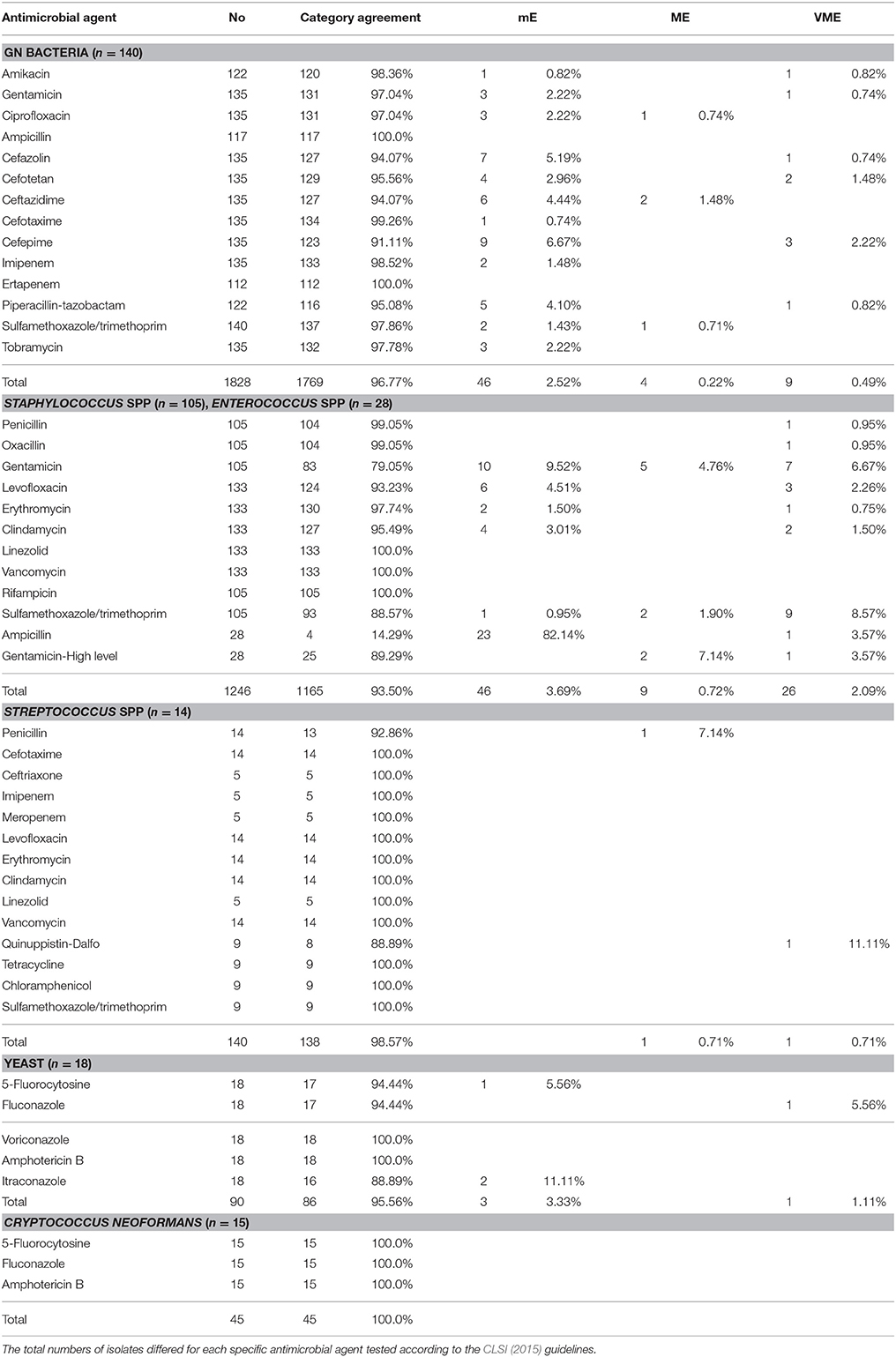
Table 4. Bacterial/antimicrobial combinations and errors in bacterial isolates from positive-culture sterile body fluids.
The RMAST was actually performed on 142 GN bacteria (109 Enterobacteriaceae, 14 Acinetobacter spp., and 19 non-fermentative bacteria), but two cultures (1 Acinetobacter junii, 1 Brevundimonas diminuta) grew poorly so were excluded from the analysis. In total, 1828 bacterial/antimicrobial combinations were analyzed with a category agreement of 96.77%, a mE rate of 2.52%, an ME rate of 0.22%, and a VME rate of 0.49%. Errors were found in Escherichia coli (1.09% mE, 0.05% ME, 0.27% VME), Klebsiella pneumoniae (0.66% mE, 0.05% ME, 0.71% VME), Enterobacter cloacae (0.22% mE), Enterobacter aerogenes (0.05% mE), Morganella morganii (0.16% mE), Raoultella ornithinolytica (0.11% mE), Providencia rettgeri (0.05% mE), Pseudomonas aeruginosa (0.22% mE, 0.05% ME, 0.11% VME), and other non-fermenters (0.22% mE, 0.11% ME, 0.05% VME).
Although, RMAST results were available for all 28 Enterococcus spp., they were available for only 105 of the 114 Staphylococcus spp.; 10 cases failed because of poor growth (4 Staphylococcus epidermidis, 2 S. hominis, 1 S. capitis, 1 S. auricularis, 1 S. simulans, and 1 S. caprae). In total, 1246 bacterial/antimicrobial combinations were analyzed with a category agreement of 93.50%, an mE rate of 3.69%, an ME rate of 0.72%, and a VME rate of 2.09%. Errors were found in S. aureus (0.32% mE, 0.16% VME), Coagulase-negative staphylococcus (1.28% mE, 0.56% ME, 1.76% VME), and Enterococci spp. (0.32% mE, 0.16% ME, 0.16% VME).
RMAST was originally performed on 21 Streptococcus spp., including 5 S. pneumoniae, 3 S. agalactiae, 1 S. pyogenes, and 12 S. viridians, but 7 of the S. viridans failed because of poor growth, yielding 14 RMAST results. In total, 140 bacterial/antimicrobial combinations were analyzed with a category agreement of 98.57%, an ME rate of 0.71%, and a VME rate of 0.71%.
RMAST results were available for 33 of the 37 fungal isolates (7 Candida albicans, 6 C. tropicalis, 3 C. parapsilosis, 3 C. glabrata, 1 C. lusitaniae, 1 Trichosporon asahii, 15 Cryptococcus neoformans, and 1 C. albidus). Four cases (1 C. albicans, 1 C. parapsilosis, 1 C. glabrata, and 1 Cryptococcus albidus) grew poorly and were thus excluded. In total, 90 bacterial/antimicrobial combinations were analyzed with a category agreement of 95.56%, an mE rate of 3.33% and a VME rate of 1.11% in the yeast group. Forty-five bacterial/antimicrobial combinations were also analyzed with a category agreement of 100% in the C. neoformans group.
Forty-three multi-drug resistant bacteria were detected in this study, including 1 vancomycin-intermediate Staphylococcus epidermidis, 1 vancomycin-resistant Enterococcus faecium, 1 linezolid-resistant S. aureus, 8 linezolid-resistant S. capitis, 16 carbapenem non-susceptible Enterobacteriaceae, 8 carbapenem non-susceptible Acinetobacter spp., and 6 carbapenem non-susceptible non-fermenting spp. The RMAST category agreement of multi-drug resistant bacteria was 97.68%, with 1 mE in carbapenem non-susceptible Enterobacteriaceae.
Time to Identification and Antimicrobial Susceptibility Testing
The average times of the RMI (vs. final identification report) were 0.58 h for GN, polymicrobial and sterile body fluid bacteria (vs. 18.1 h), 2.58 h for GP bacteria (vs. 18.1 h), 3.53 h for fungi (0.58 h in sterile body fluid and 4.58 h in blood, vs. 32.2 h), 1.36 h for anaerobic bacteria (0.58 h for GN and 2.58 h for GP bacteria, vs. 27.8 h), and 2.5 h for bacteria that failed to grow on the routing plates (vs. 64 h). The results are presented in Figure 3.
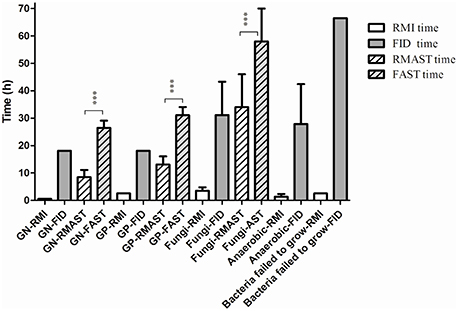
Figure 3. Comparison between the average times of RMI/RMAST and FID/FAST. The average times of RMAST vs. the conventional method were of great statistical significant (p < 0.0001). ***p < 0.0001. Bacteria failed to grow: bacteria failed to grow on the routing plates.
The average times of RMAST (vs. the conventional method) were 8.4 ± 2.6 h for GN bacteria (vs. 26.4 ± 2.6 h; p < 0.0001), 13.1 ± 3.0 h for GP bacteria (vs. 31.1 ± 3.0 h; p < 0.0001), 34 ± 12 h for fungi (vs. 58 ± 12 h; p < 0.0001; Figure 3).
Analysis of Clinical Relevance
Among 485 RMI cases (Figure 4), 14.85% of the results indicated that clinicians should adjust their treatment. Specifically, antibiotic therapy should be ceased in 3.71% of patients, initiated in 9.28% of patients, and modified in 1.86% of patients. Among the 320 RMAST results (Figure 4), 65% indicated that clinicians could optimize the therapeutic regimen by de-escalating (51.6%) or modifying (13.44%) the antibiotic therapy.
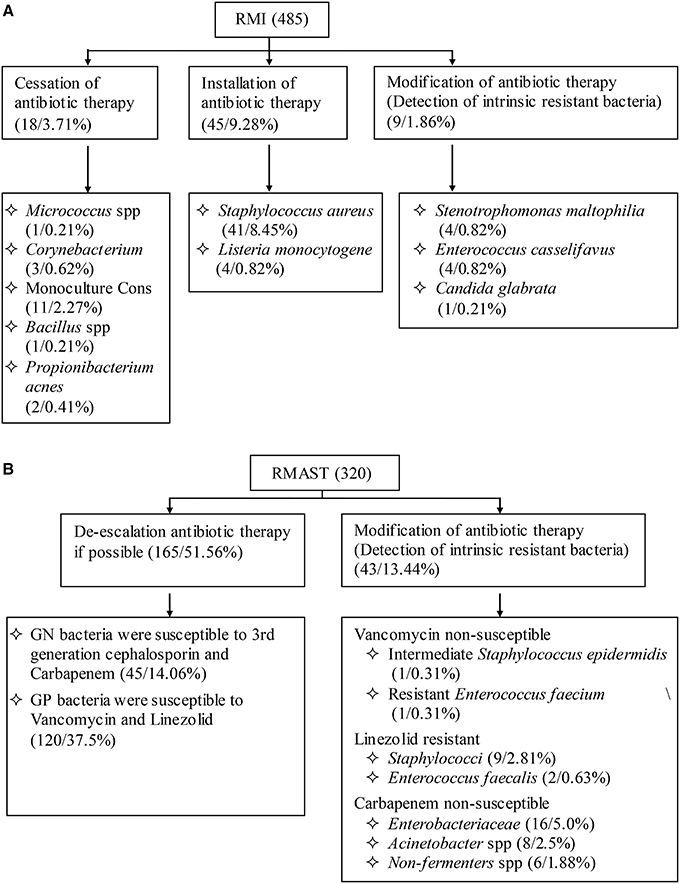
Figure 4. (A): Influence of RMI results on empirical antimicrobial therapy. (B): Influence of RMAST results on empirical antimicrobial therapy.
Discussion
Sterile body fluid infections tend to trigger systemic or local tissue damage. Because of their severity and morbidity, BSIs and acute bacterial meningitis have placed a great burden on health care settings (Adhikari et al., 2010; Thigpen et al., 2011; Goto and Al-Hasan, 2013; Portnoy et al., 2015). Early diagnosis and rapid intervention are critical to improving patient prognosis.
This study incorporated a combined MALDI–TOF MS/Vitek AST system into the clinical microbiology laboratory workflow. The aim was to provide clinicians with fast and accurate identification results and multiple precise MIC values for optimizing the therapeutic regimen.
Numerous strategies can identify RMI pathogens from positive blood samples by MALDI–TOF MS (Hazelton et al., 2014; Idelevich et al., 2014; Martinez et al., 2014; Schieffer et al., 2014; Chan et al., 2015; Morgenthaler and Kostrzewa, 2015). The RMI accuracy of GN and GP bacteria or yeasts identified by these strategies has been widely reported, but the investigated species were mostly single. Comprehensive analyses are relatively rare. In this study, we collected 485 non-duplicated positive cultures from blood, csf, ascitic fluid, vitreous fluid, and eye tissues samples, and initiated by GN and GP bacteria, streptococci, fungi, fastidious bacteria, anaerobic bacteria, polymicrobial, and bacteria that failed to grow on the routing plates, and systematically evaluated the accuracy and feasibility of RMI. Data showed the RMI accuracies of CSF, ascitic fluid, vitreous fluid, and eye tissues (almost 100%) were obviously higher than blood sample (57.1–100%), especially for CSF, ascitic fluid, vitreous fluid, and eye tissues samples which might attribute to the blood protein interference. Moreover, the correct RMI rate of GN and GP bacteria reached 98.9 and 87.2%, respectively, similar to the results of previous studies (Hazelton et al., 2014; Schieffer et al., 2014), whereas that of fungi (75.7%) was slightly higher than in previous studies (Idelevich et al., 2014). We attribute this improvement to the 4 h enrichment and constitution of the sample source. The RMI accuracy of csf infections caused by C. neoformans was especially high (100%, n = 9). Fastidious bacteria yielded the least accurate results because they grow poorly; this problem deserves further exploration. The dominant species in polymicrobial was correctly identified in 81.2% of cases. In addition, to the best of our knowledge, we were the first to characterize the RMIs of rare bacteria that failed to grow on routing plates (including Methylobacterium, Streptococcus salivarius thermophilus, Staphylococcus saccharolyticus, and Campylobacter fetus). The RMI accuracy reached 100% for these species (n = 4), although the sample size was small, and should be increased for an accurate evaluation. The accurate and early detection of salmonella is important for the control and prevention of hospital infections.
A variety of studies have focused on the resistance determinants and activities of resistant enzymes (Majcherczyk et al., 2006; Grundt et al., 2012; Hrabák et al., 2012; Chan et al., 2015), but the obtained data were relatively scarce and unable to provide accurate MIC values to clinicians (Idelevich et al., 2014; Machen et al., 2014). Unlike the former methods, the Vitek AST system is highly sensitive and well-standardized. In this study, the category agreements of GN bacteria, Staphylococcus and Enterococcus spp., Streptococcus spp., and fungi reached 96.77, 93.5, 98.57, and 95.56%, respectively, similarly to previous studies (Romero-Gómez et al., 2012; Wimmer et al., 2012). The percentage of major and very major errors was low among the 3349 bacteria/antimicrobial combinations. In a RMAST of GN bacteria, most of the VMEs (0.49%, n = 9) occurred in amikacin, gentamicin, cefazolin, cefotetan, cefepime, and piperacillin-tazobactam, as observed for E. coli and P. aeruginosa in a previous study (Wimmer et al., 2012). Moreover, the present study obtained VMEs from K. pneumoniae and other non-fermenters. In the RMAST of Staphylococcus spp., most of the VMEs (n = 26) occurred in the presence of gentamicin, levofloxacin, clindamycin, and sulfamethoxazole/trimethoprim. It is worth mentioning that the VITEK ATB AST system could read the data automatically or semi-automatically. The automatic interpretation of low-growth streptococcal and fungal species could be supplemented by manual assistance. Thus, the RMAST category agreement of fungi is obviously higher in our study than in previous study (Idelevich et al., 2014). Importantly, the RMAST category agreement of C. neoformans reached 100%.
Accurate RMI/RMAST results are useful for optimizing a therapeutic regimen. First, the RMI and RMAST results are returned at least 18–36 h earlier than the final reports. Notably, the RMI of rare bacteria that failed to grow on routing plates might be advanced by 66.5 h. Second, the RMI results may accelerate the installation antibiotic therapy of BSIs caused by S. aureus (8.45%), S. lugdunensis (not detected in the present study) and Listeria monocytogenes (0.82%), which due to morphology consistent with the contaminated bacteria and gram stain are indistinguishable. The RMI also hastens the modification of empirical antibiotic treatment in cases showing intrinsic resistance (1.86% of cultures in the present study). From another viewpoint, 3.71% of the cultures indicated cessation of antibiotic therapy. Early recognitions of these contaminants would avoid wastage of medical resources. Third, the RMAST results provide a variety of accurate MIC values, by which clinicians can choose drug multi-directionally and estimate therapeutic doses precisely. In the present RMAST results, 13.44% of the cases showed resistance to last-resort antibiotics (such as vancomycin, linezolid, and carbapenem), further alerting clinicians to adjust the treatment or initiate essential combination therapy. In contrast, 51.56% of the bacteria were susceptible to third-generation cephalosporin and carbapenem (GN bacteria) or vancomycin and linezolid (GP bacteria), suggesting that antibiotic therapy might be de-escalated in patients infected by these organisms.
In this study, we theoretically examined the utility of RMI/RMAST in optimizing therapeutic regimens. We did not conduct a practical investigation. Several studies (Martiny et al., 2013; Perez et al., 2013) prospectively demonstrated that rapid pathogen identification and antimicrobial stewardship reduces the hospital length of stay and total costs. However, these studies were limited to BSI caused by GN bacteria. Therefore, we tentatively propose a protocol that establishes the control and intervention groups and detects bacterial, yeast and fungal infections in diverse sterile body fluids. The impact of RMI/RMAST in decreasing the hospital length of stay, cost, and antibiotic selective pressure deserves further investigation.
Conclusions
The combined MALDI–TOF MS and Vitek AST system could obtain a rapid, accurate, reliable identification, and ASTs reports. The RMI and RMAST results not only reduce the turnaround time of the patient, but also guide clinicians whether a patient's antibiotic treatment should be accelerated, ceased or de-escalated, and adjusted the essential drugs modification for an optimized therapy.
Ethics Statement
This study was approved by the ethics committee of Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai, People's Republic of China (protocol HS-H-2014-0213). All subjects provided written informed consent before their inclusion in the study.
Authors Contributions
YT and BZ performed all experiments. BW assisted in antimicrobial susceptibility testing. ML, YL, YT, and BZ conceived the study and analyzed the results. ML and YL supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grants 81322025, 81171623 and 81371875), Outstanding Young Talent Plan of Shanghai (XYQ2011039), and Shanghai Shuguang Talent Project (12SG03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00523
Abbreviations
A list of abbreviations is shown in Table S2 in Supplementary Material.
References
Adhikari, N. K., Fowler, R. A., Bhagwanjee, S., and Rubenfeld, G. D. (2010). Critical care and the global burden of critical illness in adults. Lancet 376, 1339–1346. doi: 10.1016/S0140-6736(10)60446-1
Al-Haroni, M., Skaug, N., Bakken, V., and Cash, P. (2008). Proteomic analysis of ampicillin-resistant oral Fusobacterium nucleatum. Oral Microbiol. Immunol. 23, 36–42. doi: 10.1111/j.1399-302XX.2007.00387.x
Ascione, T., Pagliano, P., Mariconda, M., Rotondo, R., Balato, G., Toro, A., et al. (2015). Factors related to outcome of early and delayed prosthetic joint infections. J. Infect. 70, 30–36. doi: 10.1016/j.jinf.2014.07.008
Bagheri-Nesami, M., Babamahmoodi, F., and Nikkhah, A. (2015). Types, risk factors, clinical symptoms and diagnostic tests of acute adult meningitis in northern iran during 2006-2012. J. Clin. Diagn. Res. 9, IC01–IC05. doi: 10.7860/JCDR/2015/11991.5936
Bizzini, A., and Greub, G. (2010). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16, 1614–1619. doi: 10.1111/j.1469-0691.2010.03311.x
Blot, S., Vandewoude, K., De Bacquer, D., and Colardyn, F. (2002). Nosocomial bacteremia caused by antibiotic-resistant gram-negative bacteria in critically ill patients: clinical outcome and length of hospitalization. Clin. Infect. Dis. 34, 1600–1606. doi: 10.1086/340616
Cain, S. E., Kohn, J., Bookstaver, P. B., Albrecht, H., and Al-Hasan, M. N. (2015). Stratification of the impact of inappropriate empirical antimicrobial therapy for gram-negative bloodstream infections by predicted prognosis. Antimicrob. Agents Chemother. 59, 245–250. doi: 10.1128/AAC.03935-14
Chan, W. S., Chan, T. M., Lai, T. W., Chan, J. F., Lai, R. W., Lai, C. K., et al. (2015). Complementary use of MALDI-TOF MS and real-time PCR–melt curve analysis for rapid identification of methicillin-resistant staphylococci and VRE. J. Antimicrob. Chemother. 70, 441–447. doi: 10.1093/jac/dku411
Chon, Y. E., Kim, S. U., Lee, C. K., Park, J. Y., Kim, Y., Han, K. H., et al. (2014). Community-acquired vs. nosocomial spontaneous bacterial peritonitis in patients with liver cirrhosis. Hepatogastroenterology 61, 2283–2290.
Clinical Laboratory Standards Institute (CLSI) (2015). Performance Standards for Antimicrobial Susceptibility Testing; 25th Informational Supplement M100–S25. Wayne, PA: CLSI.
Dellinger, R. P., Levy, M. M., Rhodes, A., Annane, D., Gerlach, H., Opal, S. M., et al. (2013). Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 39, 165–228. doi: 10.1007/s00134-012-2769-8
Goto, M., and Al-Hasan, M. N. (2013). Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin. Microbiol. Infect. 19, 501–509. doi: 10.1111/1469-0691.12195
Griffin, P. M., Price, G. R., Schooneveldt, J. M., Schlebusch, S., Tilse, M. H., Urbanski, T., et al. (2012). Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 50, 2918–2931. doi: 10.1128/JCM.01000-12
Grundt, A., Findeisen, P., Miethke, T., Jäger, E., Ahmad-Nejad, P., and Neumaier, M. (2012). Rapid detection of ampicillin resistance in Escherichia coli by quantitative mass spectrometry. J. Clin. Microbiol. 50, 1727–1729. doi: 10.1128/JCM.00047-12
Hazelton, B., Thomas, L. C., Olma, T., Kok, J., O'Sullivan, M., Chen, S. C., et al. (2014). Rapid and accurate direct antibiotic susceptibility testing of blood culture broths using MALDI Sepsityper combined with the BD Phoenix automated system. J. Med. Microbiol. 63, 1590–1594. doi: 10.1099/jmm.0.075580-0
Hrabák, J., Studentová, V., Walková, R., Zemlicková, H., Jakubu, V., Chudácková, E., et al. (2012). Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-as sisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50, 2441–2443. doi: 10.1128/JCM.01002-12
Hrabak, J., Walkova, R., Studentova, V., Chudackova, E., and Bergerova, T. (2011). Carbapenemase activity detection by matrix-assisted laser desorption-ionization time of flight mass spectrometry. J. Clin. Microbiol. 49, 3222–3227. doi: 10.1128/JCM.00984-11
Ibrahim, E. H., Sherman, G., Ward, S., Fraser, V. J., and Kollef, M. H. (2000). The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the intensive care unit setting. CHEST 118, 146–155. doi: 10.1378/chest.118.1.146
Idelevich, E. A., Grunewald, C. M., Wüllenweber, J., and Becker, K. (2014). Rapid identification and susceptibility testing of Candida spp. from positive blood cultures by combination of direct MALDI-TOF mass spectrometry and direct inoculation of Vitek 2. PLoS ONE 9:e114834. doi: 10.1371/journal.pone.0114834
Kumar, A., Roberts, D., Wood, K. E., Light, B., Parrillo, J. E., Sharma, S., et al. (2006). Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34, 1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9
Machen, A., Drake, T., and Wang, Y. F. (2014). Same day identification and full panel antimicrobial susceptibility testing of bacteria from positive blood culture bottles made possible by a combined lysis-filtration method with MALDI-TOF VITEK mass spectrometry and the VITEK2 system. PLoS ONE 9:e87870. doi: 10.1371/journal.pone.0087870
Majcherczyk, P. A., McKenna, T., Moreillon, P., and Vaudaux, P. (2006). The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 255, 233–239. doi: 10.1111/j.1574-6968.2005.00060.x
Martinez, R. M., Bauerle, E. R., Fang, F. C., and Butler-Wu, S. M. (2014). Evaluation of three rapid diagnostic methods for direct identification of microorganisms in positive blood cultures. J. Clin. Microbiol. 52, 2521–2529. doi: 10.1128/JCM.00529-14
Martiny, D., Debaugnies, F., Gateff, D., Gérard, M., Aoun, M., Martin, C., Konopnicki, D., et al. (2013). Impact of rapid microbial identification directly from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry on patient management. Clin. Microbiol. Infect. 19, E568–E581. doi: 10.1111/1469-0691.12282
Martiny, D., Dediste, A., and Vandenberg, O. (2012). Comparison of an in-house method and the commercial Sepsityper™ kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2269–2281. doi: 10.1007/s10096-012-1566-1
Morgenthaler, N. G., and Kostrzewa, M. (2015). Rapid identification of pathogens in positive blood culture of patients with sepsis: review and meta-analysis of the performance of the sepsityper kit. Int. J. Microbiol. 2015:827416. doi: 10.1155/2015/827416
Perez, K. K., Olsen, R. J., Musick, W. L., Cernoch, P. L., Davis, J. R., Land, G. A., et al. (2013). Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch. Pathol. Lab. Med. 137, 1247–1254. doi: 10.5858/arpa.2012-0651-OA
Portnoy, A., Jit, M., Lauer, J., Blommaert, A., Ozawa, S., Stack, M., et al. (2015). Estimating costs of care for meningitis infections in low- and middle-income countries. Vaccine 33, A240–A247. doi: 10.1016/j.vaccine.2014.11.061
Romero-Gómez, M. P., Gómez-Gil, R., Paño-Pardo, J. R., and Mingorance, J. (2012). Identification and susceptibility testing of microorganism by direct inoculation from positive blood culture bottles by combining MALDI-TOF and Vitek-2 Compact is rapid and effective. J Infect. 65, 513–520. doi: 10.1016/j.jinf.2012.08.013
Schieffer, K. M., Tan, K. E., Stamper, P. D., Somogyi, A., Andrea, S. B., Wakefield, T., et al. (2014). Multicenter evaluation of the Sepsityper extraction kit andMALDI-TOF MS for direct identification of positive blood culture isolates using the BD BACTEC FX and VersaTREK diagnostic blood culture systems. J. Appl. Microbiol. 116, 934–941. doi: 10.1111/jam.12434
Schmidt, V., Jarosch, A., März, P., Sander, C., Vacata, V., and Kalka-Moll, W. (2012). Rapid identification of bacteria in positive blood culture by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31, 311ee7. doi: 10.1007/s10096-011-1312-0
Schubert, S., Weinert, K., Wagner, C., Gunzl, B., Wieser, A., Maier, T., et al. (2011). Noviel, improved sample preparation for rapid, direct identification from positive blood cultures using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) Mass Spectrometry. J. Mol. Diagn. 13, 701–706. doi: 10.1016/j.jmoldx.2011.07.004
Seng, P., Drancourt, M., Gouriet, F., La Scola, B., Fournier, P. E., Rolain, J. M., et al. (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49, 543–551. doi: 10.1086/600885
Stevenson, L. G., Drake, S. K., and Murray, P. R. (2010). Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48, 444–447. doi: 10.1128/JCM.01541-09
Thigpen, M. C., Whitney, C. G., Messonnier, N. E., Zell, E. R., Lynfield, R., Hadler, J. L., et al. (2011). Bacterial meningitis in the United States, 1998-2007. N. Engl. J. Med. 364, 2016–2025. doi: 10.1056/NEJMoa1005384
Tumbarello, M., Spanu, T. D., Bidino, R., Marchetti, M., Ruggeri, M., Trecarichi, E. M., et al. (2010). Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum-β- lactamase production and inadequate initial antibiotic therapy. Antimicrob. Agents Chemother. 54, 4085–4091. doi: 10.1128/AAC.00143-10
Vincent, J. L., Sakr, Y., Sprung, C. L., Ranieri, V. M., Reinhart, K., Gerlach, H., et al. (2006). Sepsis in European intensive care units: results of the SOAP study. Crit. Care Med. 34, 344–353. doi: 10.1097/01.CCM.0000194725.48928.3A
Wimmer, J. L., Long, S. W., Cernoch, P., Land, G. A., Davis, J. R., Musser, J. M., et al. (2012). Strategy for rapid identification and antibiotic susceptibility testing of Gram-negative bacteria directly recovered from positive blood cultures using the Bruker MALDI Biotyper and the BD Phoenix system. J. Clin. Microbiol. 50, 2452–2454. doi: 10.1128/JCM.00409-12
Keywords: MALDI-TOF mass spectrometry, Vitek AST system, sterile body fluids positive culture, rapid diagnosis, clinical impact
Citation: Tian Y, Zheng B, Wang B, Lin Y and Li M (2016) Rapid Identification and Multiple Susceptibility Testing of Pathogens from Positive-Culture Sterile Body Fluids by a Combined MALDI-TOF Mass Spectrometry and Vitek Susceptibility System. Front. Microbiol. 7:523. doi: 10.3389/fmicb.2016.00523
Received: 02 January 2016; Accepted: 30 March 2016;
Published: 20 April 2016.
Edited by:
Andres M. Perez, University of Minnesota, USAReviewed by:
Biswapriya Biswavas Misra, University of Florida, USASanti M. Mandal, Vidyasagar University, India
Copyright © 2016 Tian, Zheng, Wang, Lin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Lin, linyong7007@126.com;
Min Li, ruth_limin01@126.com
†These authors have contributed equally to this work.
 Yueru Tian
Yueru Tian Bing Zheng
Bing Zheng Bei Wang
Bei Wang Yong Lin
Yong Lin Min Li
Min Li