- 1Key Lab of Urban Environment and Health, Institute of Urban Environment, Chinese Academy of Sciences, Xiamen, China
- 2State Key Lab of Urban and Regional Ecology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
- 3University of Chinese Academy of Sciences, Beijing, China
Arsenic (As) resistance determinant ars operon is present in many bacteria and has been demonstrated to enhance As(V) resistance of bacteria. However, whole molecular mechanism adaptations of bacteria in response to As(V) stress remain largely unknown. In this study, transcriptional profiles of Enterobacteriaceae strain LSJC7 responding to As(V) stress were analyzed using RNA-seq and qRT-PCR. As expected, genes involved in As(V) uptake were down-regulated, those involved in As(V) reduction and As(III) efflux were up-regulated, which avoided cellular As accumulation. Reactive oxygen species and nitric oxide (NO) were induced, which caused cellular damages including DNA, protein, and Fe–S cluster damage in LSJC7. The expression of specific genes encoding transcriptional regulators, such as nsrR and soxRS were also induced. NsrR and SoxRS modulated many critical metabolic activities in As(V) stressed LSJC7 cells, including reactive species scavenging and repairing damaged DNA, proteins, and Fe–S clusters. Therefore, besides As uptake, reduction, and efflux; oxidative stress defense and damage repair were the main cellular adaptive responses of LSJC7 to As(V) stress.
Introduction
Arsenic (As) is considered to be a ubiquitous toxic substance and carcinogen, released from both anthropogenic and natural sources, and has high environmental impact (Zhu et al., 2014). It ranks first on the US Priority List of Hazardous Substances by the US Environmental Protection Agency and Agency for Toxic Substances and Disease Registry1. Typical redox states of As in living organisms are pentavalent As(V) and trivalent As(III) (Cullen and Reimer, 1989). As a consequence of their pervasiveness, microorganisms have developed special adaptive, resistance, and colonization mechanisms to cope with the presence of As. Microorganisms usually avoid As toxicity through uptake selectivity, efflux of As, As(III) oxidation, As(V) reduction, and methylation and volatilization (Zhu et al., 2014). Typically, most bacteria can reduce As(V) to the more toxic and mobile As(III) then extrude, which is the most common As resistance mechanism depending on the presence of ars genes (Lin et al., 2006).
It has been reported that metal and metalloid ions can participate in redox reactions to induce toxicity (Harrison et al., 2009). The redox reactions induced by As stress can be divided into two types (Harrison et al., 2009): As(V) reduction–oxidation reaction with cellular thiols (RSH), and production of reactive oxygen species (ROS) via intermediate S radical chemistry (Lemire et al., 2013). The production of ROS, such as superoxide anion (), hydrogen peroxide (H2O2), and hydroxyl radical (HO•), serves as critical signaling events in cell proliferation and survival (Ray et al., 2012), and can also modify organic molecules to induce cytotoxicity. The dual functions of ROS play an important role in cells by causing cellular metabolic regulation. Moreover, a majority of up-regulated genes that are exposed to As(III) are involved in the elimination of ROS (Parvatiyar et al., 2005; Cleiss-Arnold et al., 2010). Bacterial mutants deficient in cellular antioxidants or ROS-scavenging enzymes are often more sensitive to As than wild-type bacteria (Parvatiyar et al., 2005).
In mammals and plants, As(III) exposure additionally produces reactive nitrogen species (RNS), including nitric oxide (NO) and its derivatives (Lynn et al., 1998; Shi et al., 2004; Rao et al., 2011; Leterrier et al., 2012). NO is known to play important roles in living organisms as a defense mechanism and as a messenger that can directly and reversibly activate latent, readily available oxidative stress defense system(s) (Gusarov and Nudler, 2005). However, as the important species of RNS, when NO is overproduced, it will increase nitration, which could deactivate and damage specific nucleic acids, proteins, and lipids, known as nitrosative stress (Leterrier et al., 2012). Nevertheless, little information is available about As induced RNS in bacteria.
Although bacteria have been shown to have special As detoxification pathways, enzymes, and transporters, their metabolic processes are still adversely affected. For example, a microarray analysis shows that approximately 14% of the whole Herminiimonas asoxydans genome is significantly affected by 15 min of As(III) exposure (Cleiss-Arnold et al., 2010). Though numerous studies have explored the profiling analysis of the whole organism’s metabolic pathways in response to As stress in plants and animals (Zheng et al., 2005; Yu et al., 2012; Lafuente et al., 2014); the biochemical and molecular mechanisms of As-induced cytotoxicity in bacteria and bacterial responses to that cytotoxicity remain largely unknown. However, understanding the As resistance of bacteria is crucial because of the increasing As contamination in the environment.
LSJC7 is a Gram-negative strain isolated from an antimony tailing, generally contained an As composition of 0.04% (Liu et al., 2010; Su et al., 2012). The genome analysis has revealed that it contains 4,428 protein-coding genes, in which there are two As resistant operons (arsRDABC and arsRBC; Su et al., 2012). Our previous studies have shown that it has high resistance to many heavy metals and metalloid, especially to As(V) (Chen et al., 2015). Therefore, further investigation is required to understand the toxicity and adaptive responses of LSJC7 to As(V) stress.
Genome-wide transcriptome is an effective method for studying the toxicity and adaptive response pathways by identifying and linking the resulting perturbations (Kaur et al., 2006; Pillai et al., 2014). In this study, we exploited the global transcriptomic profiling to understand whole molecular perturbations and cellular pathways in response to As(V) stress in LSJC7. This enables us to obtain a more detailed picture of the processes affected by As. This will open up for exploration the understanding of the molecular regulating mechanisms of other heavy metal resistant bacteria and to understand the As mediated antibiotic resistance in bacteria.
Materials and Methods
Bacterial Strain and Growth Medium
A single clone of Enterobacteriaceae strain LSJC7 was identified with 16S rRNA and grown with LB medium (pH 7.0) overnight at 30°C and 220 rpm (Su et al., 2012), then it was diluted 1:100 with 100 ml of LB medium with/without Na3AsO4 at 4 mM final concentration in a 250 ml triangular flask as As treatment/control. Meanwhile, a medium control, containing 100 ml of LB medium and Na3AsO4 but no bacterium, was set up. Each group had three replicates and was incubated in the same conditions overnight as described above. The concentration of LSJC7 was measured using optical density at 600 nm (OD600).
As Speciation Analysis
After LSJC7 was inoculated for 6, 12, and 24 h, 30 ml culture was taken from each triangular flask and centrifuged at 6000 rpm for 5 min. Then 2 ml of liquid supernatant were taken as As speciation samples of medium and bacterial cells were rinsed with deionized water and ice-cold phosphate buffer [1 mM K2HPO4, 5 mM MES, and 0.5 mM Ca(NO3)2] for 10 min to remove apoplastic As. Freeze-dried cells and supernatants from medium were kept in the freezer at -80°C until As speciation analysis. Cells were incubated with 10 ml of 1% HNO3 overnight, then extraction was performed using 95°C water bath for 60 min with shaking every 10 min (Raber et al., 2012). After extracts cooled to room temperature, aliquots of 1 ml each were filtered through 0.22 μm nylon filter and used directly for HPLC-ICP-MS (7500a; Agilent Technologies) analysis as described previously (Wang et al., 2014). Supernatants from medium were diluted about 2,000-fold with Milli-Q water and filtered through 0.22 μm nylon filter and used directly for HPLC-ICP-MS analysis.
Total RNA Extraction and RNA-Seq
Both arsenate exposure and control LSJC7 cells were collected at OD600 = 0.5 for RNA extraction. Samples (50 ml each) were collected by centrifugation (8,000 × g, 10 min). Then the cell pellets were resuspended in 1 ml TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). Two-hundred microliters of chloroform was added to homogenized cells and was mixed well for 30 s and then was incubated at room temperature for 10 min. The aqueous phase was recovered after centrifugation at 12, 000 × g for 20 min at 4°C. Twofold of cold ethanol was added to aqueous phase and inverted gently, then incubated at -80°C for 30 min. The precipitated RNA was pelleted by centrifugation for 20 min at 12 000 × g. The RNA pellet was washed twice with cold 75% ethanol and then left to air dry. The RNA pellet was resuspended in 30 μl DEPC water. The RNA quantity was assessed using RNA 6000 Nano LabChip Kit in microcapillary electrophoresis (Agilent 2100 Bioanalyzer, Agilent Technologies, Australia). The RNA integrity number (RIN) above 8.0 was used for sequencing. Amount of 3 μg total RNA for each sample was used as input material for the RNA sequencing sample preparations. Sequencing libraries were generated using NEBNext Ultra (NEB, USA). Directional RNA Library Prep Kit for Illumina (NEB) was used following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (NEB). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH-) and second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dNTPs with dTTP were replaced by dUTP. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptors with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150∼200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, MA, USA). Then, 3 μl USER Enzyme (NEB) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index Primer. At last, products were purified using AMPure XP system and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2000 platform and 100 bp paired-end reads were generated.
RNA-Seq Reads Analysis and Differential Expression Analysis
Clean data were obtained using in-house perl scripts by removing reads containing adapter, reads containing ploy-N, and low quality reads from raw data of FASTQ format. RNA-Seq reads (clean data) were aligned to the LSJC7 reference genome2. Both building index of reference genome and aligning clean reads to reference genome were used in Bowtie2-2.0.6 (Langmead and Salzberg, 2012). HTSeq v0.5.4p33 was used to count the reads numbers mapped to each gene. And then Reads Per Kilobase of exon model per Million mapped reads (RPKM) of each gene was calculated based on the length of the gene and reads count was mapped to this gene (Mortazavi et al., 2008).
The read counts of each sequenced library were adjusted by edgeR program package (Robinson et al., 2010) through one scaling normalized factor. Differential expression analysis of two conditions was performed using the DEGSeq R package (1.12.0; Anders and Huber, 2010). The P-values were adjusted using the Benjamini and Hochberg (1995) method. Corrected P-value of 0.005 and log2 (Fold change) of 1 were set as the threshold for significantly differential expression.
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the GOseq R package, in which gene length bias was corrected (Young et al., 2010). GO terms with corrected P-value less than 0.05 were considered significantly enriched by differentially expressed genes.
Real Time Quantitative PCR (RT-qPCR)
Seventeen representative genes were selected and analyzed the expression levels via RT-qPCR to validate the RNA-Seq data. The primers were designed using Primer 3 and listed in Supplementary Table S4. One microgram of total RNA, with contamination DNA removed, was reverse-transcribed using random hexamers and SuperScript III (Invitrogen, USA) to synthesis cDNA according to kit instructions. RT-qPCR was performed with cDNA templates prepared from samples collected in RNA-seq experiment, using CFX96 Real-time PCR System (Bio-Rad, USA) with iQTM SYBR Green Supermix in 96-well plates. The reaction mixture was prepared following kit instructions. All reactions were carried out in three technical replicates. 16S rRNA was used as a reference gene to normalize the results. The relative abundance of gene transcripts among the treatment groups was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Endogenous ROS Detection
DCFH-DA (Beyotime, China) was diluted in phosphate buffer to make 100 μM working solution. Ten milliliters of LSJC7 culture (OD600 = 0.5) was pelleted by centrifugation and washed with phosphate buffer and resuspended in 5 ml of DCFH-DA working solution. Two-hundred microliters of the resultant cell suspension samples were transferred to a 96-well black plate and added 0.5 M As(V) to cell suspension to make final concentration of 0, 4, 16, 32 mM. Fluorescence was measured using a SpectraMax M5 Plate Reader (Molecular Devices, USA) with excitation/emission wavelengths of 488/525 nm.
Endogenous NO Detection
DAF-FM DA (Beyotime, China) was diluted in phosphate buffer to make 10 μM working solution. Ten milliliters of LSJC7 culture (OD600 = 0.5) was pelleted by centrifugation and washed with phosphate buffer and resuspended in 5 ml of DAF-FM DA working solution. Two-hundred microliters of the resultant cell suspension samples was transferred to a 96-well black plate and added 0.5 M As(V) to cell suspension to make final concentration of 0, 4, 16, 32 mM. Fluorescence was measured using a SpectraMax M5 Plate Reader (Molecular Devices, USA) with excitation/emission wavelengths of 494/517 nm.
Results and Discussion
Global Changes at Transcriptome Level
Global gene expression changes during LSJC7 growth with 4 mM As(V) or without As(V) (control) were analyzed using Illumina RNA deep sequencing (RNA-Seq) technology (van Vliet, 2010) to elucidate molecular mechanisms of As(V) tolerance. To avoid complete suppression of cellular metabolism, 4 mM solution, which was far below the EC50 (33 mM) for LSJC7 exposure to As(V) was chosen as the exposure concentration, and cultures at this concentration showed moderate growth inhibition. Cells in the two treatments were collected at the early exponential phase (OD600 was 0.5, Supplementary Figure S1). A total of 1.96 and 2.46 G clean bases were obtained from the control and As treatment, respectively. Over 98% of all clean reads aligned to the reference genome (Supplementary Table S2). The RNA-Seq raw reads were deposited to Sequence Read Archive (SRA) database of NCBI with accession numbers SRR3314666 and SRR3314667. Under As(V) exposure, 460 genes showed significant differential expression [log2 (Fold Change) > 1 and q-value < 0.005 or log2 (Fold Change) < -1 and q-value < 0.005], in which 285 genes were up-regulated and 175 genes were down-regulated, representing more than 10% of the genome and suggesting a major response of cellular physiology (Supplementary Table S3). Seventeen selected genes, including 11 up-regulated and 6 down-regulated genes on the list of RNA-Seq differentially expressed genes (Supplementary Table S1) were well validated by RT-qPCR (Figure 1). GO term enrichment differential expression genes were shown in Figure 2. Biological pathways with significant changes were those involved in protein metabolic process, protein folding, translation, amino acid metabolism, homeostatic process, nitrogen metabolism, and carbon metabolism. Molecular functions with significant changes were those involved in structural molecule activity, structural constituent of ribosome, molybdenum ion binding, electron carrier activity, iron–sulfur cluster binding, metal cluster binding, and heme binding.
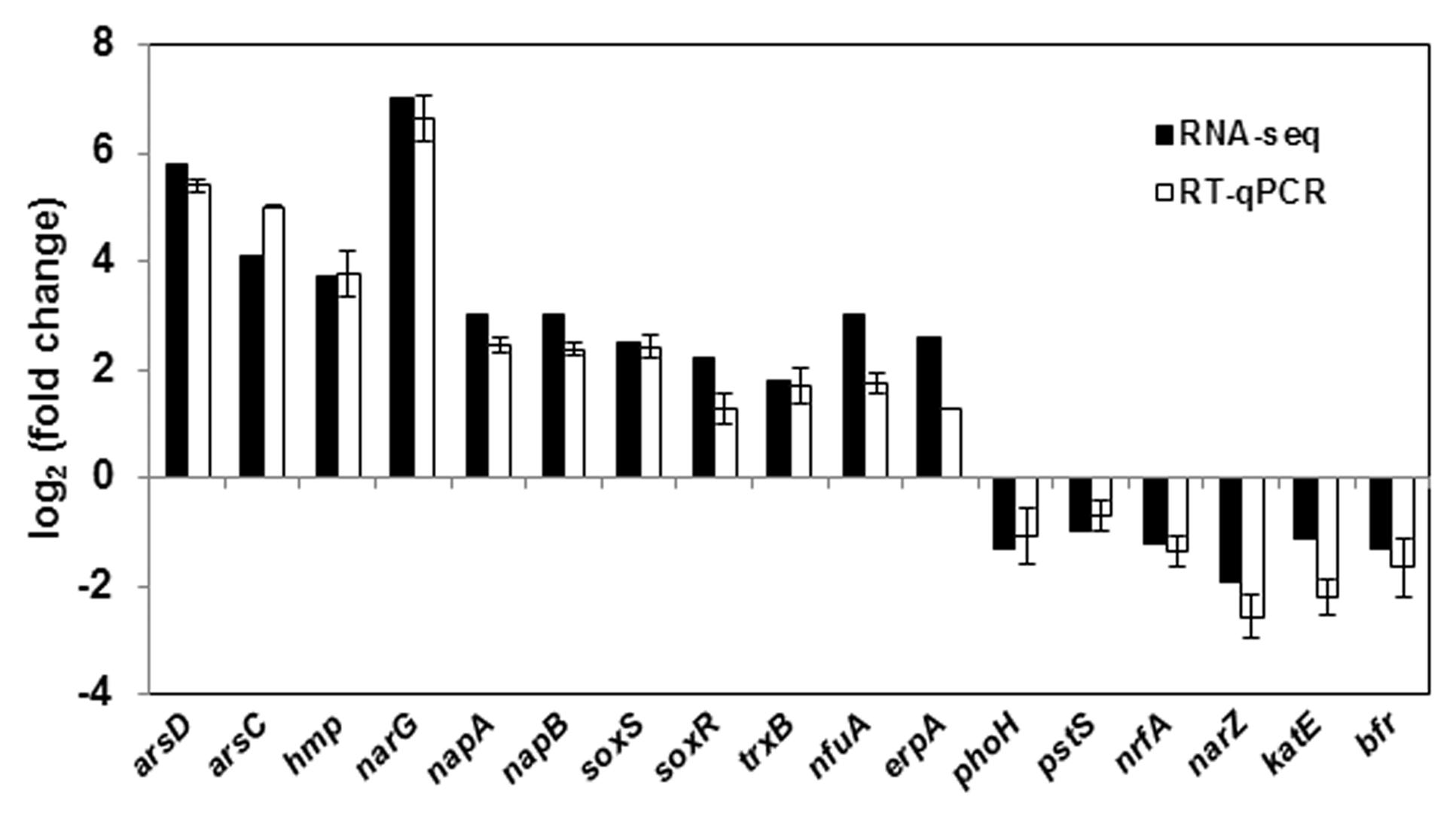
FIGURE 1. RT-qPCR validation of 17 genes with significant differential expression in As(V) stress. RT-qPCR data are mean ± SD from three biological replicates.
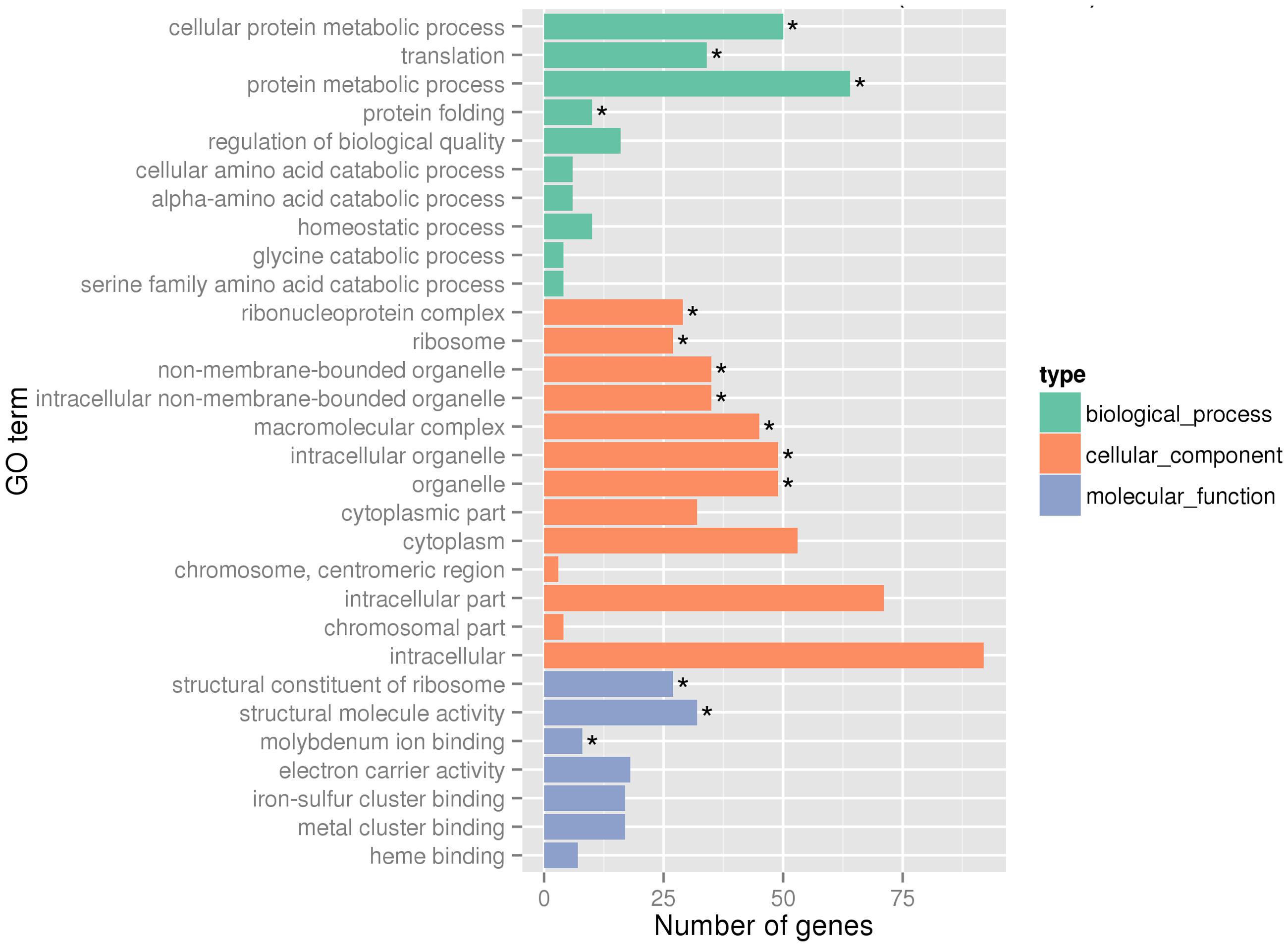
FIGURE 2. Gene Ontology (GO) terms enrichment at transcriptome level. Asterisks represent significantly enriched GO terms (P < 0.05).
Genes Related to As(V) Influx Pumps
Previous report showed that the uptake of arsenate into cells occurs via inorganic phosphate carriers, because As(V) is an analog of inorganic phosphate (Elias et al., 2012). This is consistent with our findings in this study. Our RNA-seq results showed that some genes related to inorganic phosphate transport, including pstS and phoH, were down-regulated (Supplementary Table S1) under As(V) exposure. The same expression profile was also shown by RT-qPCR (Figure 1). The pstS gene, encoding a high affinity phosphate binding protein, was down-regulated by 2.1-fold, which might decrease the efficiency of the whole gene cluster pstSCAB by limiting the uptake of arsenate (Dyhrman and Haley, 2006). Similarly, phoH, encoding protein PhoH which is also named phosphate starvation-inducible protein PsiH, was down-regulated by 2.4-fold. PhoH contains an ATP-binding motif and actually binds to ATP and is bound by PhoB protein, the transcriptional activator of the pho regulon, which regulates genes controlling phosphate mobilization, uptake, and metabolism, but its function in vivo is not known (Kim et al., 1993). It was suspected that the down-regulation of phoH resulted in the decrease of arsenate uptake by the regulation of the pho regulon. However, the expression of another phosphate transporter gene pitA (low-affinity inorganic phosphate transporter 1) did not change significantly in this study, indicating that PitA was either a constitutive expression or not the main arsenate transporter in LSJC7. These results suggest that reduced expression of influx pumps might be the strategy of LSJC7 to decrease the uptake of As(V).
As(V) Is Reduced and Extruded as As(III) by LSJC7
The concentration of intracellular As(III) increased significantly from exponential phase to stationary phase, while the concentration of As(V) stayed at about 20 mg/kg during the whole incubation time (Figure 3A). From sampling time point 6 to 24 h, the proportion of As(III) increased from 18 to 58% in the medium, In contrast, the concentration of As(V) in control sample set did not change during the whole process (Figure 3B). These results suggest that LSJC7 can reduce As(V) to As(III) quickly and extrude As(III) out of the cell. These reactions should be mediated by arsRDABC and arsRBC operons in LSJC7. Under As(V) exposure, the transcriptions of genes from the two ars operons were up-regulated by 4.6- to 56.4-fold (Supplementary Table S1 and Figure 1). The arsRBC operon encodes three proteins: ArsR, ArsB, and ArsC. ArsR regulates expression of the chromosomal ars operon (Lin et al., 2007). ArsC first converts As(V) to As(III) as an arsenate reductase and ArsB then extrudes As(III) as an As(OH)3/H+ antiporter, conferring resistance (Lin et al., 2007). ArsC can use reduced thioredoxin (Trx) or glutaredoxin (Grx) to convert As(V) to As(III), namely Trx-linked ArsC or Grx-linked ArsC. In our study, genes encoding the Trx system, including trxA, trxC, nrdH, and trxB were up-regulated by 2.8- to 21.3-fold in LSJC7 exposed to As(V), suggesting the Trx-linked ArsC in LSJC7. The Trx system, composed of thioredoxin reductase (TrxB encoded by trxB in our study), Trx (Thio encoded by trxA, Thio2 encoded by trxC and NrdH encoded by nrdH in our study), and NADPH is known to play critical roles in the redox reaction regulation (Lu et al., 2007), either through redox signaling or more directly as electron donors to the numerous members of the Prx family (Dietz, 2003). The arsRDABC operon encodes two additional proteins, ArsA forming a complex with ArsB that catalyzes ATP-driven As(III) efflux and ArsD which is a metallochaperone that delivers As(III) to ArsA ATPase. ArsA and ArsD increased As(III) efflux and resistance in the environment. The ars operon has been found in nearly every sequenced bacterial genome, of which more than 400 bacterial and archaeal ars operons containing arsA and arsD genes have been identified (Ajees et al., 2011). Specially, LSJC7, having two ars operons in its genome may remarkably enhance As(V) resistance.
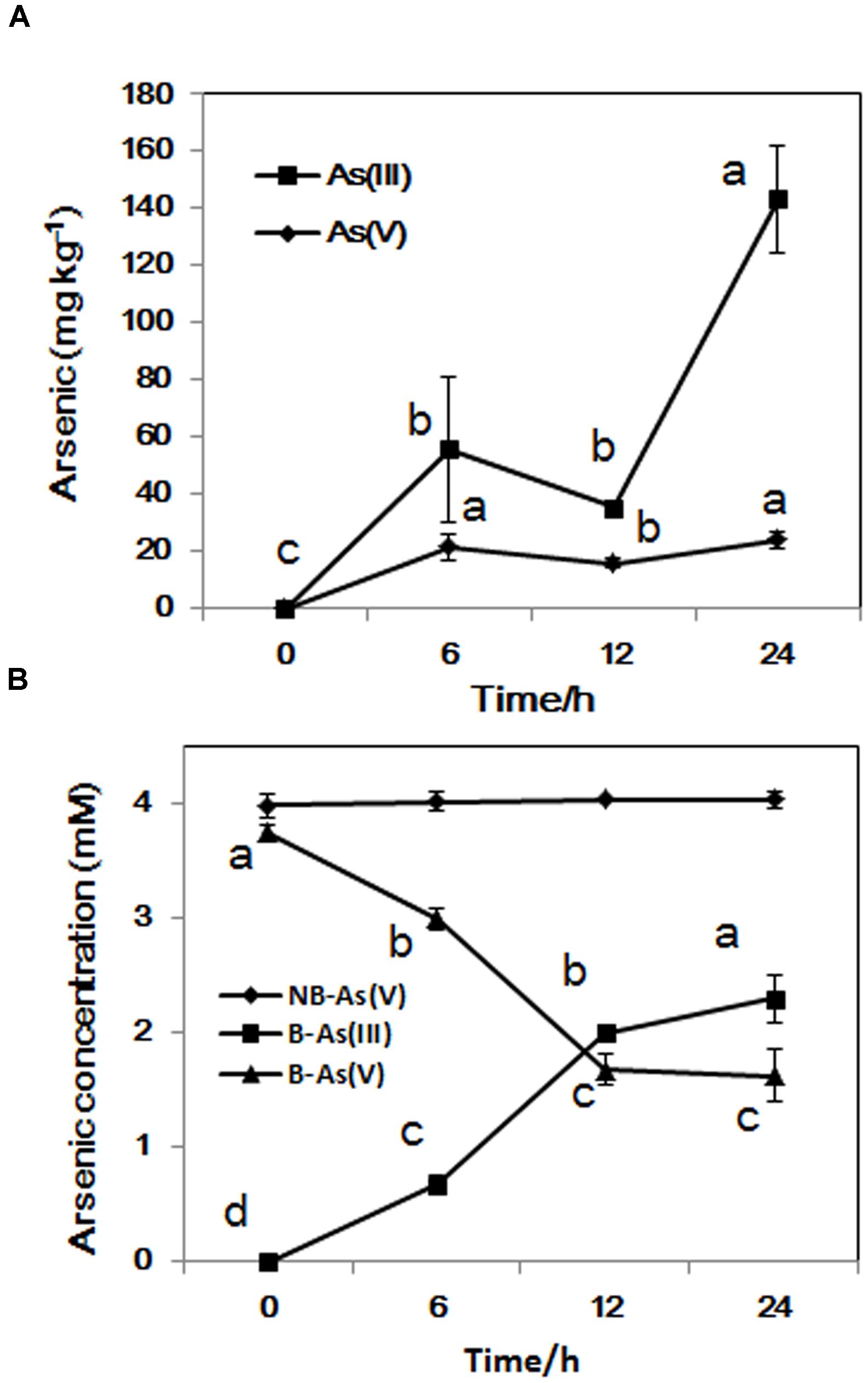
FIGURE 3. (A) As accumulated in LSJC7 cells; (B) Fresh LSJC7 cell culture (B) and medium control (NB, without LSJC7 cells) were exposed to 4 mM As(V) in LB medium to investigate As(V) reduction. Results are shown as means of three replicates. Different lowercase letters indicate statistically significant difference at p < 0.05.
Genes Related to Nitrate Respiration
Nitrate reductase activity in bacteria could be increased by many environmental stress factors, including hypoxia, NO, and silver nanoparticles, because nitrate acts as the effective alternate terminal electron acceptor after molecular oxygen (Du et al., 2012; Eoh and Rhee, 2013). LSJC7 possesses three nitrate reductases encoded by narLXKGHJI, narZYWV, and napFDAGHBC. Transcriptome results showed that genes in narLXKGHJI and napFDAGHBC operon were dramatically up-regulated 24.4 to 138.5-fold and 2.7 to 8.1-fold, respectively, narZ and narY were down-regulated about fourfold (Supplementary Table S1 and Figure 1). Meanwhile, many of the genes implicated in aerobic respiration (TCA cycle), such as sdhB and frdA, were down-regulated around twofold under As stress (Supplementary Table S1). From these results, we suspected that LSJC7 might prefer to use anaerobic nitrate respiration for energy production under As(V) stress.
On the other hand, nitrate respiration also could accumulate nitrite in vivo. In mycobacteria, export of the nitrite, a toxic by-product of nitrate respiration, by NarK3 and NarU is the main protection strategy (Tan et al., 2010). Under As stress, 138.5-fold up-regulated narK gene might indicate the export of nitrite by NarK was important part of the protection strategy. Also consistent with previous report that NirBD is required to reduce nitrite to ammonia in Enterobacteria (Cole, 1996), nirB was also up-regulated by 2.3-fold under As(V) stress in our study. It has been also demonstrated that low concentrations of NO are generated as a by-product of this metabolism (Corker and Poole, 2003; Spiro, 2007).
Endogenous NO Up-regulated nsrR and hmp
Intracellular NO production was detected in As(V) stressed LSJC7 cells using a DAF-FM DA probe. The concentration of NO increased with the As(V) concentration in the medium when LSJC7 was exposed to 0, 4, 16, and 32 mM As (Figure 4A). Consistently, genes encoding proteins of NO regulon (NsrR) and detoxification (Hmp and NrfA) were also regulated when LSJC7 was exposed to 4 mM As (Supplementary Table S1). NO can be generated via nitrite reduction by enzymatic or non-enzymatic mechanisms in Gram-negative bacteria (Puyaubert and Baudouin, 2014). NsrR is a global transcription repressor whose activity is responding to NO. Targets for NsrR regulation are at least 30 genes, including hmp, nrfA, ytfE, hcp-hcr, napF, napD, napH, and napB (Filenko et al., 2007). Flavohemoprotein (also named Flavohemoglobin, Hmp) is involved in conversion of NO to nitrate with substantial O2 consumption (Stevanin et al., 2000). The hmp genes were dramatically up-regulated by 13.3-fold, which suggested high concentration of NO was harmful to cells and strongly needed to be detoxified. Cytochrome c552 (NrfA), encoded by the nrfA gene, acts as a nitrite reductase and is also responsible for NO detoxification (Pittman et al., 2007). The nrfA gene is negatively regulated by NsrR and down-regulated by 2.1-fold (Supplementary Table S1 and Figure 1), suggesting NO detoxification regulated by hmp to generate nitrate may provide more benefits to LSJC7. Our data suggested that NO was generated under As(V) stress, while regulation system of NO was also activated to control its homeostasis and against nitrosative stress.
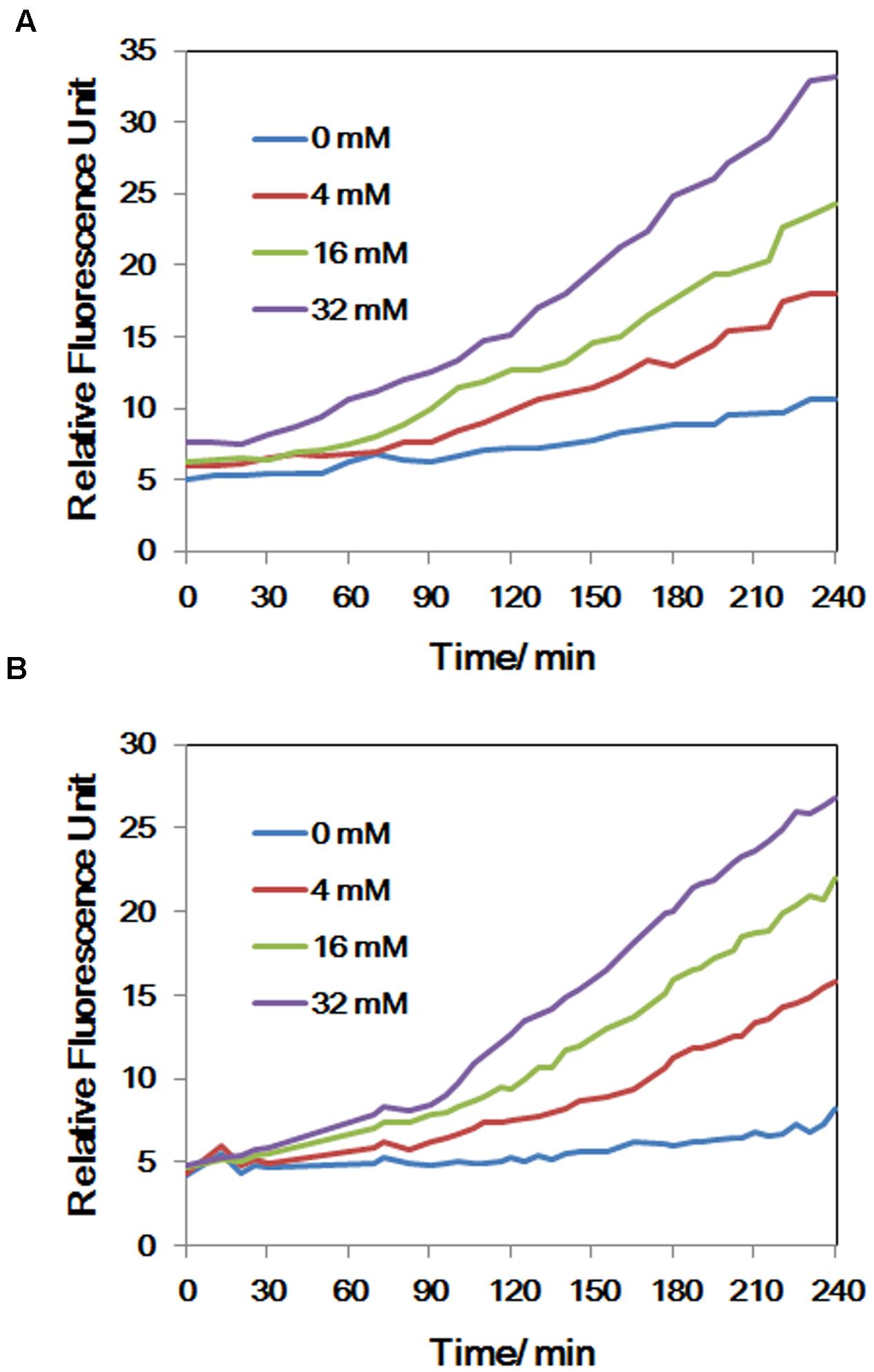
FIGURE 4. (A) The concentrations of NO of LSJC7 exposure to 0, 4, 16, and 32 mM As; (B) The concentrations of ROS of LSJC7 exposure to 0, 4, 16, and 32 mM As. Results are shown as means of three replicates.
ROS Generation under As(V) Stress
To demonstrate that ROS were produced in LSJC7 under As(V) stress, we examined the intracellular concentration of ROS of LSJC7 exposed to 0, 4, 16, and 32 mM As(V). As expected, the concentration of ROS increased with increasing As(V) concentrations in the medium and the treatment time (Figure 4B). It has been reported that ROS played critical roles in bacterial cell death from antibiotics, hydroxyurea, and silver stress (Davies et al., 2009; Morones-Ramirez et al., 2013; Dwyer et al., 2014). ROS mainly interacts with thiols, metal centers, nucleotide bases, protein tyrosines, and lipids (Fang, 2004) and these interactions result in damages to nucleic acids, proteins, and lipids, thus affecting normal cellular functions (Yang et al., 2007; Ray et al., 2012). In our transcriptome results, many repair genes were up-regulated robustly (Supplementary Table S1), which might indicate that ROS induced by As(V) would be an important reason for cellular damage. However, gene encoding superoxide dismutase (SOD), which directly nullifies ROS, was not found up-regulated in As(V) stressed cells. Moreover, genes encoding Prxs, including osmC, katG, katE, and cpo, were down-regulated by 2.1 to 3.1-fold in LSJC7 exposed to As(V) (Supplementary Table S1 and Figure 1). These results suggested that alternative antioxidant systems might be used against ROS under As(V) stress.
Genes Related to Fe–S Clusters Biogenesis and Iron Recruiting
Reactive oxygen species can directly affect IscR, which modulates a set of genes involved in iron–sulfur prosthetic groups (Fe–S clusters) biogenesis (namely, the Isc and Suf systems as well as ErpA and NfuA; Blanchard et al., 2007; Py and Barras, 2010). RNA-seq results showed that genes iscR, erpA, and nfuA were up-regulated by 3.6 to 8.2-fold in As(V) stressed cells (Supplementary Table S1). Meanwhile, the expression of erpA and nfuA was also validated by RT-qPCR (Figure 1). These results indicate increased Fe–S cluster biogenesis. Many genes encoding proteins containing Fe–S clusters (Fe–S proteins) were regulated in As(V) stressed cells; most of them were up-regulated. These proteins have prominent roles in multiple important cellular processes, including respiration (NapF, NapA, NapG, NapH, NarG, NarH encoded by napF, napA, napG, napH, narG, narH, and NarY, NarZ encoded by narY, narZ), central metabolism (FhuF, BfD, Hcp, Hcr encoded by fhuF, bfd, hcp, hcr), biosynthetic pathways (NfuA, ErpA encoded by nfuA, erpA), and gene regulation (NsrR, IscR, SoxR encoded by nsrR iscR, soxR). The robust regulation of these Fe–S proteins could be the reason of increased Fe–S clusters biogenesis. In As(V) stressed cells more iron should be recruited to alleviate the stress. Consistent with this, genes encoding proteins related to iron uptake were regulated under As(V) exposure (Supplementary Table S1); feoA encoding FeoA for ferrous iron uptake (Lau et al., 2013; Weaver et al., 2013), fhuA, fepD, and fhuF encoding FhuA, FepD, and FhuF for ferric iron (siderophore) uptake (Chenault and Earhart, 1992; Cleiss-Arnold et al., 2010; Miethke, 2013), were up-regulated by 3.0 to 7.7-fold; exbB encoding ExbB for the energy-transducing TonB–ExbB–ExbD complex of ferric iron transport (Andrews et al., 2003) was up-regulated 5.3-fold. Genes bfd and bfr, encoding iron storage protein bacterioferritin-associated ferredoxin (Bfd) and bacterioferritin (Bfr), are reciprocally regulated by iron availability via ferric uptake regulator (Quail et al., 1996). Bfd and Bfr can store iron as ferric iron to protect cells against ROS resulting from ferrous iron overload (Andrews et al., 2003). In RNA-seq results, bfd was up-regulated by 22.8-fold, while bfr was down-regulated by 2.4-fold (Supplementary Table S1). The regulations of these iron related genes indicated As(V) stressed cells robustly recruited iron to cells.
Genes Related to Iron–Sulfur Cluster Repair and Suppressed Fenton Reaction
The gene ytfE, known as one of NsrR’s targets (Filenko et al., 2007; Spiro, 2007), encoding Fe–S cluster repair protein YtfE, was significantly up-regulated, by 72.2-fold (Supplementary Table S1), suggesting that robust repair occurred in damaged Fe–S clusters in As(V) stressed cells. Fe–S clusters in dehydratases can be damaged by univalent oxidants, converting the clusters to inactive form and causing concomitant loss of Fe2+ which fuels Fenton reaction to produce the highly reactive HO• (Djaman et al., 2004; Jang and Imlay, 2007; Py and Barras, 2010). Fe2+ is recruited and integrated to Fe–S clusters by YtfE, which could decrease the cascade of the Fenton reaction (Py and Barras, 2010). The H2O2 scavenging system was another critical component to decrease Fenton reaction. In our study, the hcp–hcr genes, also modulated by NsrR, which encodes a hybrid cluster protein Hcp and its redox partner Hcr, were up-regulated under As(V) stress. Hcp–Hcr complex has been suggested to be a hydroxylamine reductase or peroxidase (Wolfe et al., 2002; Almeida et al., 2006). This result suggested that Hcp–Hcr might play an important role in scavenging H2O2 in As(V) stressed cells. Taken together, NO could induce Fe–S cluster repair and suppress Fenton reaction by modulating the expression of nsrR.
Genes Related to DNA and Protein Repair
In our study, the transcriptions of soxR and soxS were highly up-regulated under As(V) exposure (Supplementary Table S1 and Figure 1). These two genes encode two separate transcription activators SoxR and SoxS that participate in a two-step activation (Pomposiello and Demple, 2001). SoxR is a transcription factor containing [2Fe–2S] clusters, which coordinate the response to oxidative stress (Lushchak, 2011). SoxR can be distinctly activated by superoxide stress and thereby induce SOD in aerobic stress (Pomposiello and Demple, 2001; Gu and Imlay, 2011). However, in anaerobic cells, SoxR is able to be activated by redox drugs when alternative respiratory acceptors were provided (Gu and Imlay, 2011). SoxR also exhibits a mechanism of activation by NO, which is independent of the presence of oxygen (Pomposiello and Demple, 2001). In E. coli, SoxRS modulates more than 100 genes, encoding various proteins (Blanchard et al., 2007), which have diverse biological functions; including NADPH regeneration, superoxide scavenging, DNA repair, protein repair, recycling of damaged macromolecules, xenobiotic efflux, and carbon metabolism (Pomposiello and Demple, 2001; Blanchard et al., 2007). We suggest that the LSJC7 SoxRS gene, at least, could modulate DNA and protein repair in As(V) stressed cells, though more studies should be done to illuminate its targets. Because As(V) stressed cells were undergoing a hypoxia circumstance and the gene encoding SOD was not found significantly modulated, we propose that NO was the reason to activate SoxRS in LSJC7.
Transcriptome data showed that genes involved in both DNA and protein repair were modulated, indicating As(V) induced oxidative stress occurring. Nucleic acids could be damaged by Fenton chemistry, via the formation of either iron-oxo intermediates or HO• (Dwyer et al., 2014). The Gene smrA, encoding Smr protein (MutS2) which is a DNA repair enzyme (Fukui and Kuramitsu, 2011), was up-regulated by 2.1-fold under As stress (Supplementary Table S1). MutS2 has an endonuclease domain located in the C-terminal region also called the small MutS-related (Smr) domain (Moreira and Philippe, 1999). MutS is also found overexpressed in antibiotic-induced redox stress, which limits the antibiotic lethality (Dwyer et al., 2014). This result indicated that As(V) stress might induce DNA damage, however, cells could produce special proteins to resist or repair that damage.
Genes involved in protein turnover were also modulated in As(V) stress. Three out of the six genes encoding proteases were up-regulated, the other three were down-regulated, and 53 genes encoding proteins for protein synthesis and folding were particularly up-regulated (Supplementary Table S1). In our present study, genes of more than 10% of the genome showed significant differential expression under As stress, which must induce significant protein turnover. These results suggested vast scale protein synthesis and degradation processes under As(V) stress, which could repair the damaged proteins and produce new proteins for handling As(V) stress. Similar observations are also found with transition metals, including Mn(II), Fe(II), Co(II), Ni(II), Cu(II), and Zn(II) (Kaur et al., 2006), suggesting that protein turnover might be a general strategy to withstand metal(loid) stress.
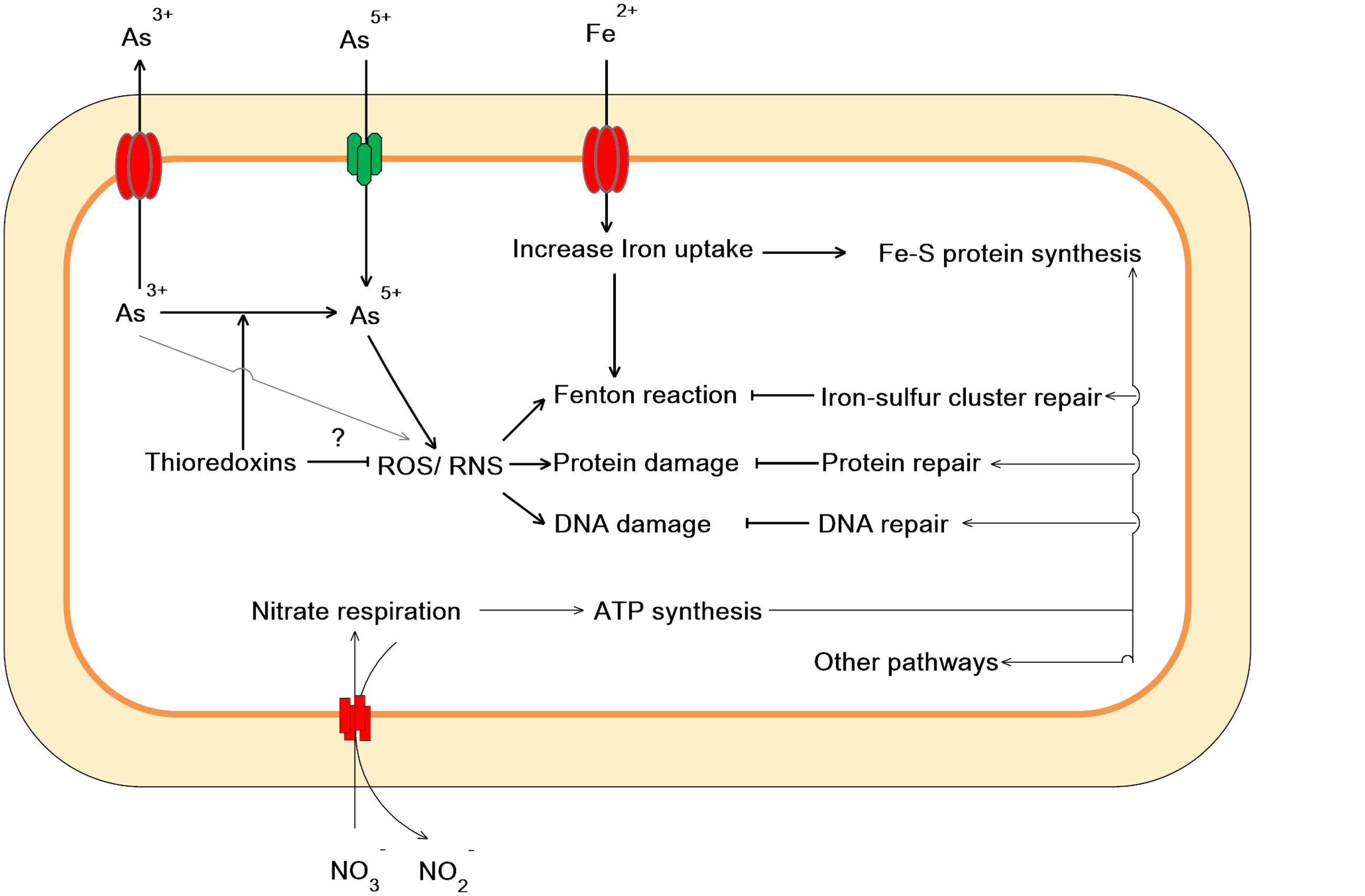
Taken together, our work showed that As(V) disturbed multiple cellular networks and processes of LSJC7 (Figures 5 and 6). To decrease the intracellular As accumulation, As(V) was reduced to As(III) and As(III) was expelled into medium. However, the transformation between As(V) and As(III) induced cascades of redox reactions in the cell and changed the cellular redox state and As(III) can strongly interact with vicinal thiols such as cysteine pairs in proteins (Hughes, 2002). Moreover, various types of reactive species, such as ROS and NO, could be involved in the damage of proteins and DNA. However, the expression of specific genes encoding transcriptional regulators, such as nsrR and soxRS, was increased under As stress. NsrR modulated the expression of several genes including hmp, nrfA, hcp–hcr, and ytfE, which were responsible for NO detoxification, H2O2 scavenging, and Fe–S clusters repair. Moreover, SoxRS in LSJC7 could modulate DNA and protein repair in As(V) stressed cells.
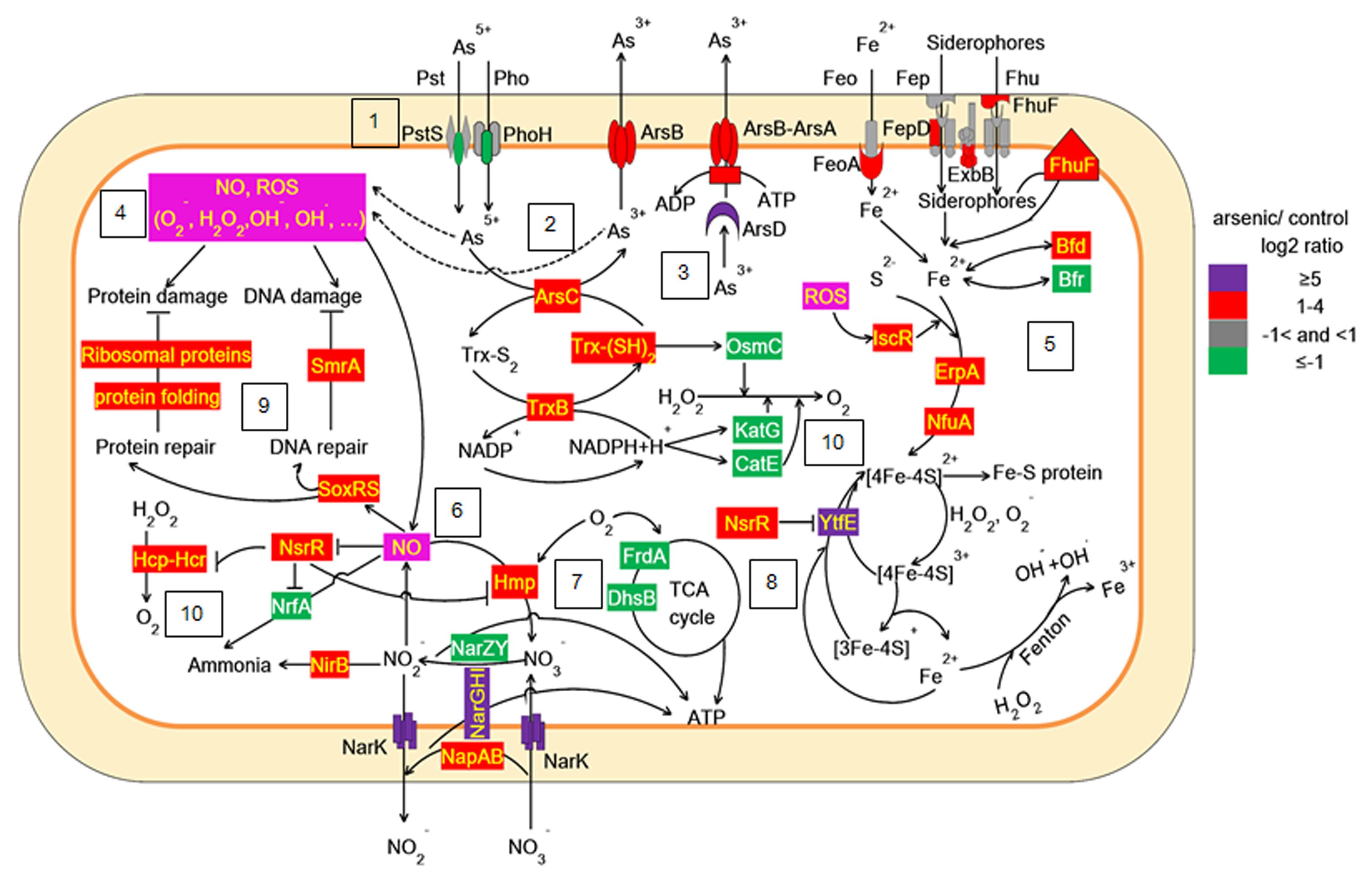
FIGURE 5. Proposed scheme illustrating the toxicity and adaptive-response pathways of LSJC7 to As(V) stress. (1) As(V) influx pumps was down-regulation; (2) ArsC converted As(V) to As(III); (3) ArsB extruded As(III); (4) Arsenic induced ROS and NO generation; (5) ROS induced Fe–S clusters biogenesis and iron recruiting; (6) Endogenous NO up-regulated nsrR and hmp; (7) Genes related to nitrate respiration and TCA cycle; (8) NO induced Iron–sulfur cluster repair and suppressed Fenton reaction; (9) SoxRS activated by NO might modulate DNA and protein repair; (10) Peroxiredoxins were modulated.
Conclusion
Enterobacteriaceae strain LSJC7 has evolved multiple strategies to survive As stress. It not only actively decreases cellular As concentration, but also launches an oxidative stress defense system to prevent cells from damages induced by As and reactive species, and repair some unavoidable cell damages. The same strategies could be employed by most Gram-negative bacteria.
Author Contributions
JY, Y-GZ, and J-QS conceived and designed the project. YZ, SC, and XH did the transcriptomic experiments. XX and YY performed the arsenic speciation analysis. YZ and XH analyzed the data. YZ, SC, Y-GZ and JY wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work is supported by the Natural Science Foundation of Fujian Province (2014J01141) and the National Natural Science Foundation of China (31270161 and 41571130063).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00636
FIGURE S1 | Growth rates of LSJC7 under 4 mM As(V) stress. The arrows represent the sampling time for transcriptome, and the boxes represent the sampling time for As species analysis.
Footnotes
- ^ http://www.atsdr.cdc.gov/SPL/index.html
- ^ http://www.ncbi.nlm.nih.gov/nuccore/AMFN00000000
- ^ https://pypi.python.org/pypi/HTSeq
References
Ajees, A. A., Yang, J., and Rosen, B. P. (2011). The ArsD As(III) metallochaperone. Biometals 24, 391–399. doi: 10.1007/s10534-010-9398-x
Almeida, C. C., Romao, C. V., Lindley, P. F., Teixeira, M., and Saraiva, L. M. (2006). The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 281, 32445–32450. doi: 10.1074/jbc.M605888200
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Andrews, S. C., Robinson, A. K., and Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate-a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Blanchard, J. L., Wholey, W. Y., Conlon, E. M., and Pomposiello, P. J. (2007). Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS ONE 2:e1186. doi: 10.1371/Journal.Pone.0001186
Chen, S., Li, X., Sun, G., Zhang, Y., Su, J., and Ye, J. (2015). Heavy metal induced antibiotic resistance in bacterium LSJC7. Int. J. Mol. Sci. 16, 23390–23404. doi: 10.3390/ijms161023390
Chenault, S. S., and Earhart, C. F. (1992). Identification of hydrophobic proteins FepD and FepG of the Escherichia coli ferrienterobactin permease. J. Gen. Microbiol. 138, 2167–2171. doi: 10.1099/00221287-138-10-2167
Cleiss-Arnold, J., Koechler, S., Proux, C., Fardeau, M. L., Dillies, M. A., Coppee, J. Y., et al. (2010). Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709. doi: 10.1186/1471-2164-11-709
Cole, J. (1996). Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136, 1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x
Corker, H., and Poole, R. K. (2003). Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 278, 31584–31592. doi: 10.1074/jbc.M303282200
Cullen, W. R., and Reimer, K. J. (1989). Arsenic speciation in the environment. Chem. Rev. 89, 713–764. doi: 10.1021/cr00094a002
Davies, B. W., Kohanski, M. A., Simmons, L. A., Winkler, J. A., Collins, J. J., and Walker, G. C. (2009). Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell 36, 845–860. doi: 10.1016/j.molcel.2009.11.024
Dietz, K. J. (2003). Plant peroxiredoxins. Annu. Rev. Plant Biol. 54, 93–107. doi: 10.1146/annurev.arplant.54.031902.134934
Djaman, O., Outten, F. W., and Imlay, J. A. (2004). Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 279, 44590–44599. doi: 10.1074/jbc.M406487200
Du, H., Lo, T. M., Sitompul, J., and Chang, M. W. (2012). Systems-level analysis of Escherichia coli response to silver nanoparticles: the roles of anaerobic respiration in microbial resistance. Biochem. Biophys. Res. Commun. 424, 657–662. doi: 10.1016/j.bbrc.2012.06.134
Dwyer, D. J., Belenky, P. A., Yang, J. H., MacDonald, I. C., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, E2100–E2109. doi: 10.1073/pnas.1401876111
Dyhrman, S. T., and Haley, S. T. (2006). Phosphorus scavenging in the unicellular marine diazotroph Crocosphaera watsonii. Appl. Environ. Microbiol. 72, 1452–1458. doi: 10.1128/AEM.72.2.1452-1458.2006
Elias, M., Wellner, A., Goldin-Azulay, K., Chabriere, E., Vorholt, J. A., Erb, T. J., et al. (2012). The molecular basis of phosphate discrimination in arsenate-rich environments. Nature 491, 134–137. doi: 10.1038/nature11517
Eoh, H., and Rhee, K. Y. (2013). Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 6554–6559. doi: 10.1073/pnas.1219375110
Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832. doi: 10.1038/nrmicro1004
Filenko, N., Spiro, S., Browning, D. F., Squire, D., Overton, T. W., Cole, J., et al. (2007). The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 189, 4410–4417. doi: 10.1128/JB.00080-07
Fukui, K., and Kuramitsu, S. (2011). Structure and function of the small MutS-related domain. Mol. Biol. Int. 2011, 691735. doi: 10.4061/2011/691735
Gu, M., and Imlay, J. A. (2011). The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 79, 1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x
Gusarov, I., and Nudler, E. (2005). NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 13855–13860. doi: 10.1073/pnas.0504307102
Harrison, J. J., Tremaroli, V., Stan, M. A., Chan, C. S., Vacchi-Suzzi, C., Heyne, B. J., et al. (2009). Chromosomal antioxidant genes have metal ion-specific roles as determinants of bacterial metal tolerance. Environ. Microbiol. 11, 2491–2509. doi: 10.1111/j.1462-2920.2009.01973.x
Hughes, M. F. (2002). Arsenic toxicity and potential mechanisms of action. Toxicol. Lett. 133, 1–16. doi: 10.1016/S0378-4274(02)00084-X
Jang, S., and Imlay, J. A. (2007). Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282, 929–937. doi: 10.1074/jbc.M607646200
Kaur, A., Pan, M., Meislin, M., Facciotti, M. T., El-Gewely, R., and Baliga, N. S. (2006). A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 16, 841–854. doi: 10.1101/gr.5189606
Kim, S. K., Makino, K., Amemura, M., Shinagawa, H., and Nakata, A. (1993). Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J. Bacteriol. 175, 1316–1324.
Lafuente, A., Perez-Palacios, P., Doukkali, B., Molina-Sanchez, M. D., Jimenez-Zurdo, J. I., Caviedes, M. A., et al. (2014). Unraveling the effect of arsenic on the model Medicago-Ensifer interaction: a transcriptomic meta-analysis. New Phytol. 205, 255–272. doi: 10.1111/nph.13009
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lau, C. K., Ishida, H., Liu, Z., and Vogel, H. J. (2013). Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J. Bacteriol. 195, 46–55. doi: 10.1128/JB.01121-12
Lemire, J. A., Harrison, J. J., and Turner, R. J. (2013). Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384. doi: 10.1038/nrmicro3028
Leterrier, M., Airaki, M., Palma, J. M., Chaki, M., Barroso, J. B., and Corpas, F. J. (2012). Arsenic triggers the nitric oxide (NO) and S-nitrosoglutathione (GSNO) metabolism in Arabidopsis. Environ. Pollut. 166, 136–143. doi: 10.1016/j.envpol.2012.03.012
Lin, Y. F., Walmsley, A. R., and Rosen, B. P. (2006). An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. U.S.A. 103, 15617–15622. doi: 10.1073/pnas.0603974103
Lin, Y. F., Yang, J. B., and Rosen, B. P. (2007). ArsD: an As(III) metallochaperone for the ArsAB As(III)-translocating ATPase. J. Bioenerg. Biomembr. 39, 453–458. doi: 10.1007/s10863-007-9113-y
Liu, F., Le, X. C., McKnight-Whitford, A., Xia, Y., Wu, F., Elswick, E., et al. (2010). Antimony speciation and contamination of waters in the Xikuangshan antimony mining and smelting area, China. Environ. Geochem. Health 32, 401–413. doi: 10.1007/s10653-010-9284-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J., Chew, E. H., and Holmgren, A. (2007). Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc. Natl. Acad. Sci. U.S.A. 104, 12288–12293. doi: 10.1073/pnas.0701549104
Lushchak, V. I. (2011). Adaptive response to oxidative stress: bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 153, 175–190. doi: 10.1016/j.cbpc.2010.10.004
Lynn, S., Shiung, J. N., Gurr, J. R., and Jan, K. Y. (1998). Arsenite stimulates poly(ADP-ribosylation) by generation of nitric oxide. Free Radic. Biol. Med. 24, 442–449. doi: 10.1016/S0891-5849(97)00279-7
Miethke, M. (2013). Molecular strategies of microbial iron assimilation: from high-affinity complexes to cofactor assembly systems. Metallomics 5, 15–28. doi: 10.1039/c2mt20193c
Moreira, D., and Philippe, H. (1999). Smr: a bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem. Sci. 24, 298–300. doi: 10.1016/S0968-0004(99)01419-X
Morones-Ramirez, J. R., Winkler, J. A., Spina, C. S., and Collins, J. J. (2013). Silver enhances antibiotic activity against gram-negative bacteria. Sci. Transl. Med. 5:190ra181. doi: 10.1126/scitranslmed.3006276
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Parvatiyar, K., Alsabbagh, E. M., Ochsner, U. A., Stegemeyer, M. A., Smulian, A. G., Hwang, S. H., et al. (2005). Global analysis of cellular factors and responses involved in Pseudomonas aeruginosa resistance to arsenite. J. Bacteriol. 187, 4853–4864. doi: 10.1128/JB.187.14.4853-4864.2005
Pillai, S., Behra, R., Nestler, H., Suter, M. J., Sigg, L., and Schirmer, K. (2014). Linking toxicity and adaptive responses across the transcriptome, proteome, and phenotype of Chlamydomonas reinhardtii exposed to silver. Proc. Natl. Acad. Sci. U.S.A. 111, 3490–3495. doi: 10.1073/pnas.1319388111
Pittman, M. S., Elvers, K. T., Lee, L., Jones, M. A., Poole, R. K., Park, S. F., et al. (2007). Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 63, 575–590. doi: 10.1111/j.1365-2958.2006.05532.x
Pomposiello, P. J., and Demple, B. (2001). Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19, 109–114. doi: 10.1016/S0167-7799(00)01542-0
Puyaubert, J., and Baudouin, E. (2014). New clues for a cold case: nitric oxide response to low temperature. Plant Cell Environ. 37, 2623–2630. doi: 10.1111/pce.12329
Py, B., and Barras, F. (2010). Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8, 436–446. doi: 10.1038/nrmicro2356
Quail, M. A., Jordan, P., Grogan, J. M., Butt, J. N., Lutz, M., Thomson, A. J., et al. (1996). Spectroscopic and voltammetric characterisation of the bacterioferritin-associated ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 229, 635–642. doi: 10.1006/bbrc.1996.1856
Raber, G., Stock, N., Hanel, P., Murko, M., Navratilova, J., and Francesconi, K. A. (2012). An improved HPLC-ICPMS method for determining inorganic arsenic in food: application to rice, wheat and tuna fish. Food Chem. 134, 524–532. doi: 10.1016/j.foodchem.2012.02.113
Rao, K. P., Vani, G., Kumar, K., Wankhede, D. P., Misra, M., Gupta, M., et al. (2011). Arsenic stress activates MAP kinase in rice roots and leaves. Arch. Biochem. Biophys. 506, 73–82. doi: 10.1016/j.abb.2010.11.006
Ray, P. D., Huang, B. W., and Tsuji, Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 24, 981–990. doi: 10.1016/j.cellsig.2012.01.008
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Shi, H., Shi, X., and Liu, K. J. (2004). Oxidative mechanism of arsenic toxicity and carcinogenesis. Mol. Cell. Biochem. 255, 67–78. doi: 10.1023/B:MCBI.0000007262.26044.e8
Spiro, S. (2007). Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31, 193–211. doi: 10.1111/j.1574-6976.2006.00061.x
Stevanin, T. M., Ioannidis, N., Mills, C. E., Kim, S. O., Hughes, M. N., and Poole, R. K. (2000). Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo’ or bd, from nitric oxide. J. Biol. Chem. 275, 35868–35875. doi: 10.1074/jbc.M002471200
Su, J., Ye, J., and Zhu, Y. (2012). Draft genome sequence of a novel bacterial strain, LSJC7, belonging to the family Enterobacteriaceae with dual resistance to arsenic and tetracycline. J. Bacteriol. 194, 7005–7006. doi: 10.1128/JB.01652-12
Tan, M. P., Sequeira, P., Lin, W. W., Phong, W. Y., Cliff, P., Ng, S. H., et al. (2010). Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE 5:e13356. doi: 10.1371/journal.pone.0013356
van Vliet, A. H. (2010). Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 302, 1–7. doi: 10.1111/j.1574-6968.2009.01767.x
Wang, F. H., Qiao, M., Lv, Z. E., Guo, G. X., Jia, Y., Su, Y. H., et al. (2014). Impact of reclaimed water irrigation on antibiotic resistance in public parks, Beijing, China. Environ. Pollut. 184, 247–253. doi: 10.1016/j.envpol.2013.08.038
Weaver, E. A., Wyckoff, E. E., Mey, A. R., Morrison, R., and Payne, S. M. (2013). FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J. Bacteriol. 195, 4826–4835. doi: 10.1128/JB.00738-13
Wolfe, M. T., Heo, J., Garavelli, J. S., and Ludden, P. W. (2002). Hydroxylamine reductase activity of the hybrid cluster protein from Escherichia coli. J. Bacteriol. 184, 5898–5902. doi: 10.1128/JB.184.21.5898-5902.2002
Yang, P., He, X. Q., Peng, L., Li, A. P., Wang, X. R., Zhou, J. W., et al. (2007). The role of oxidative stress in hormesis induced by sodium arsenite in human embryo lung fibroblast (HELF) cellular proliferation model. J. Toxicol. Environ. Health A 70, 976–983. doi: 10.1080/15287390701290832
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yu, L. J., Luo, Y. F., Liao, B., Xie, L. J., Chen, L., Xiao, S., et al. (2012). Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol. 195, 97–112. doi: 10.1111/j.1469-8137.2012.04154.x
Zheng, P. Z., Wang, K. K., Zhang, Q. Y., Huang, Q. H., Du, Y. Z., Zhang, Q. H., et al. (2005). Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 102, 7653–7658. doi: 10.1073/pnas.0502825102
Keywords: transcriptome, arsenic, RNA-seq, reactive oxygen species, nitric oxide, Enterobacteriaceae
Citation: Zhang Y, Chen S, Hao X, Su J-Q, Xue X, Yan Y, Zhu Y-G and Ye J (2016) Transcriptomic Analysis Reveals Adaptive Responses of an Enterobacteriaceae Strain LSJC7 to Arsenic Exposure. Front. Microbiol. 7:636. doi: 10.3389/fmicb.2016.00636
Received: 24 January 2016; Accepted: 18 April 2016;
Published: 02 May 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Sebastian Guenther, Freie Universität Berlin, GermanyPing Li, China University of Geosciences, China
Copyright © 2016 Zhang, Chen, Hao, Su, Xue, Yan, Zhu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ye, jye@iue.ac.cn
 Yingjiao Zhang
Yingjiao Zhang Songcan Chen2,3
Songcan Chen2,3 Jian-Qiang Su
Jian-Qiang Su Jun Ye
Jun Ye