- 1Key Laboratory of Coastal Biology and Bioresource Utilization, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 2State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Several strains have been reported to grow on 3-methyl-4-nitrophenol (3M4NP), the primary breakdown product of the excessively used insecticide fenitrothion. However, the microbial degradation of 3M4NP at molecular and biochemical levels remains unknown. Here, methyl-1,4-benzoquinone (MBQ) and methylhydroquinone (MHQ), rather than catechol proposed previously, were identified as the intermediates before ring cleavage during 3M4NP degradation by Burkholderia sp. strain SJ98. Real-time quantitative PCR analysis indicated that the pnpABA1CDEF cluster involved in para-nitrophenol (PNP) and 2-chloro-4-nitrophenol (2C4NP) catabolism was also likely responsible for 3M4NP degradation in this strain. Purified PNP 4-monooxygenase (PnpA) is able to catalyze the monooxygenation of 3M4NP to MBQ and exhibited an apparent Km value of 20.3 ± 2.54 μM for 3M4NP, and pnpA is absolutely necessary for the catabolism of 3M4NP by gene knock-out and complementation. PnpB, a 1,4-benzoquinone reductase catalyzes the reduction of MBQ to MHQ, and also found to enhance PnpA activity in vitro in the conversion of 3M4NP to MBQ. By sequential catalysis assays, PnpCD, PnpE, and PnpF were likely involved in the lower pathway of 3M4NP catabolism. Although NpcCD, NpcE, and NpcF are able to catalyze the sequential conversion of MHQ in vitro, these enzymes are unlikely involved in 3M4NP catabolism because their coding genes were not upregulated by 3M4NP induction in vivo. These results revealed that the enzymes involved in PNP and 2C4NP catabolism were also responsible for 3M4NP degradation in strain SJ98. This fills a gap in our understanding of the microbial degradation of 3M4NP at molecular and biochemical levels and also provides another example to illustrate the adaptive flexibility in microbial catabolism for structurally similar compounds.
Introduction
As a representative among the organophosphorus pesticides, fenitrothion (O,O-dimethylO-p-nitro-m-tolyl phosphorothioate), is a highly toxic chemical and extensively used to control major insect pests (Hayatsu et al., 2000; Zhang et al., 2006; Itoh et al., 2014), especially in developing countries. Under aerobic environmental conditions, fenitrothion could be degraded rapidly, in days to weeks (Takimoto et al., 1976; Spillner et al., 1979; Mikami et al., 1985), with production of the persistent pollutant 3M4NP, which has serious health effects to humans and animals as an endocrine-disrupting chemical (Furuta et al., 2004; Kanaly et al., 2005; Li et al., 2008, 2009; Verma and Rana, 2009). Naturally occurring bacterial isolates capable of degrading 3M4NP have received considerable attention since they provide the possibility of both environmental and in situ detoxification. So far, the microbial degradation of 3M4NP was predominantly reported in Burkholderia spp., such as strains SJ98 (originally classified as a Ralstonia sp.; Bhushan et al., 2000), SH-1 (Kim et al., 2007), NF100 (Hayatsu et al., 2000), FDS-1 (Zhang et al., 2006), and RKJ800 (Arora and Jain, 2012).
Structurally, 3M4NP is a analog of the priority environmental pollutant PNP and 2C4NP. These nitrophenols compounds are widely used for manufacturing of drugs, dyes, pesticides, herbicides, and fungicides (Arora et al., 2012, 2014). The microbial degradation of PNP (Spain and Gibson, 1991; Jain et al., 1994; Kadiyala and Spain, 1998; Kitagawa et al., 2004; Takeo et al., 2008; Zhang et al., 2009; Liu et al., 2010; Shen et al., 2010; Wei et al., 2010) and 2C4NP (Ghosh et al., 2010; Arora and Jain, 2011, 2012; Pandey et al., 2011; Min et al., 2014, 2016) has been extensively investigated. For PNP degradation, the hydroquinone pathway was initiated by a single-component PNP monooxygenase (Zhang et al., 2009; Shen et al., 2010; Wei et al., 2010) and the hydroxyquinol pathway was initiated by a two-component PNP monooxygenase (Kadiyala and Spain, 1998; Kitagawa et al., 2004; Takeo et al., 2008; Liu et al., 2010). Recently, the enzymes encoded by pnpABCDEF were proved to be involved in the chlorohydroquinone pathway of 2C4NP catabolism in Gram-negative Burkholderia sp. SJ98 (Min et al., 2014) and the enzymes encoded by pnpA1A2BC were proved to be responsible for the hydroxyquinol pathway of 2C4NP catabolism in Gram-positive Rhodococcus imtechensis RKJ300 (Min et al., 2016).
In contrast to PNP and 2C4NP, the knowledge of the microbial degradation of 3M4NP is limited, with no genetic or biochemical investigation being reported. So far, two different pathways based on different intermediates present during 3M4NP degradation have been proposed in above five 3M4NP utilizers. Strains SJ98 (Bhushan et al., 2000) and SH-1 (Kim et al., 2007) were reported to degrade 3M4NP with catechol as an intermediate, revealing that the methyl group of 3M4NP was removed before ring cleavage. However, strains RKJ800 (Arora and Jain, 2012), NF100 (Hayatsu et al., 2000) and FDS-1 (Zhang et al., 2006) were reported to degrade 3M4NP with MHQ as the ring cleavage substrate, indicating that removal of the methyl group occurs after ring cleavage in these three strains. However, none of these two alternative pathways has been characterized at the genetic and enzymatic levels, either for the initial degardation or the ring cleavge reaction.
Burkholderia sp. strain SJ98 was previously reported to utilize 3M4NP with catechol as the intermediates (Bhushan et al., 2000), but with no genetic and enzymatic inverstigation. The goal of this study was to inverstigate the microbial degradation of 3M4NP by strain SJ98 at molecular and biochemical levels. To our surprise, MBQ and MHQ, rather than catechol were identified as the intermediates before ring cleavage during 3M4NP degradation by this strain. On the other hand, the enzymes encoded by the pnpABA1CDEF cluster were proved to be also responsible for 3M4NP degradation by this strain, in addition to PNP and 2C4NP degradation (Min et al., 2014). This study fills a gap in our understanding of the microbial degradation mechanism of 3M4NP at the biochemical and genetic levels and also provides another example to illustrate the adaptive flexibility in microbial catabolism for structurally similar compounds. Considering that strain SJ98 is capable of degrading PNP as well as its chloro- and methyl-substituted derivatives, it is reasonable to conclude that it is of potential in bioremediation of these toxicants-contaminated sites.
Materials and Methods
Bacterial Strains, Plasmids, Primers, Media, and Culture Conditions
The bacterial strains and plasmids used in this study are described in Table 1, and the primers used are listed in Table 2. Escherichia coli strains were grown in lysogeny broth (LB) at 37°C. Burkholderia strains were grown at 30°C in minimal medium (MM; Xiao et al., 2006) supplemented with 0.5 mM 3M4NP (2 mM glucose was added to enhance the biomass when cultures were prepared for biotransformation assays). When necessary, kanamycin at 50 mg/ml was added to the medium.
Biotransformation and Intermediates Identification
Biotransformation of 3M4NP by strain SJ98 was performed as previously described for PNP and 2C4NP (Min et al., 2014). In order to accumulate the intermediates before ring cleavage, 1 mM 2, 2′-dipyridyl, a known inhibitor for ferrous-dependent aromatic ring dioxygenases (Chapman and Hopper, 1968; Kadiyala and Spain, 1998; Ferreira et al., 2008), was added into the biotransformation mixtures. Quantitative analyses of 3M4NP and its catabolic intermediates were performed by high performance liquid chromatography (HPLC) analysis. One unit of activity was defined as the amount of cell (milligram of cell dry weight) required to transform 1 μmol of 3M4NP per min at 30°C. For gas chromatography-mass spectrometry (GC-MS) analysis of the intermediates, the supernatant was extracted with ether after acidification, and the extract was then dried over sodium sulfate.
Analytical Methods
High performance liquid chromatography analysis was performed with an Agilent 1200 system (Agilent Technologies, Palo Alto, CA, USA) equipped with a diode array detector and an Agilent ZORBAX Eclipse XDB-C18 column (250 mm × 4.6 mm, 5 μm particle size). The mobile phase consisted of solvents A (0.1% acetic acid in water) and B (methanol) with a gradient program started with 10% of B, followed by increasing to 90% B from 0 to 12 min, hold at 90% B from 12 to 15 min, then back to 10% B in 0.1 min and equilibrate for 2.9 min. The flow rate was 1.0 ml/min, and the injection volume was 10 μl. MBQ, MHQ, and 3M4NP were quantified at 254, 290, and 320 nm, respectively. Under these conditions, authentic MBQ, MHQ, and 3M4NP had retention times of 11.7, 8.9, and 13.5 min, respectively.
Gas chromatography-mass spectrometry was performed using a Thermo Fisher Trace GC Ultra gas chromatograph equipped with an HP-5MS capillary column (30 m × 0.25 mm × 0.25 μm) and coupled to a Thermo Fisher ITQ 900 ion trap mass spectrometer. The conditions used for GC-MS analysis were the same as those described previously (Zhang et al., 2009). Under these conditions, authentic MBQ and MHQ had GC retention times of 10.06 and 16.05 min, respectively. The intermediates were identified based on comparisons of the mass spectra with those of authentic compounds and those available in an NIST98 MS data library, in addition to comparing the GC retention times of the intermediates with those of the authentic compounds.
High performance liquid chromatography-MS analysis was carried out on a Dionex UltiMate 3000 RS HPLC system coupled to an LCQ FleetTM ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Chromatographic separation was carried out on a Hypersil GOLD column (150 × 4.6 mm, 3 μm particle size) using gradient elution. The mobile phase consisted of solvents A (water) and B (acetonitrile) as follows: 10% B from 0 to 5 min; increasing to 60% B from 5 to 20 min, hold at 60% B from 20 to 25 min, then back to10% B in 0.1 min and equilibrate for 1.9 min. The flow rate was 0.8 ml/min. The mass spectrometer was operated in the negative ion mode and full scan (40–300 m/z). The sample solutions were nebulized and evaporated using nitrogen as the sheath and auxiliary gas at the flow rate of 35 and 10 arb (1 arb = 0.3 l/min), respectively. The ion spray voltage was set at 5 kV, and the transfer capillary temperature was maintained at 300°C.
Real-Time Quantitative PCR
The total RNA was isolated with an Easy Pure RNA Kit (TransGen Biotech, Beijing, China) and reverse transcribed into cDNA using a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen). Reverse transcription PCR was carried out with the primers described in Table 2. Transcriptional analysis was carried out in order to investigate whether the pnpABA1CDEF (Min et al., 2014) and pnpE2E1FD clusters (Vikram et al., 2012) involved in PNP catabolism in strain SJ98 were highly transcribed in response to 3M4NP. Real-time quantitative PCR (RT-qPCR) was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) using TransStart Tip Green qPCR SuperMix (TransGen) with primers in Table 2. All samples were run in triplicate in three independent experiments. Relative expression levels were estimated using the 2-ΔΔCT method (Livak and Schmittgen, 2001), and the 16S rRNA gene was used as a reference for normalization.
Protein Expression and Purification
N-terminal His-tagged Pnp proteins (H6-PnpA, H6-PnpB, H6-PnpCD, H6-PnpE, and PnpF) were expressed by E. coli Rosetta(DE3)pLysS carrying the corresponding expression plasmids constructed previously (Min et al., 2014). In order to express the N-terminal His-tagged Npc proteins, npcCD, npcE, and npcF were amplified using the primers listed in Table 2, digested with NdeI and XhoI, and inserted into pET-28a to obtain expression plasmids. The plasmids were then transformed into E. coli Rosetta(DE3)pLysS for protein expression and purification as described (Liu and Zhou, 2012).
Enzyme Assays
The activity assays of PNP 4-monooxygenase (PnpA) against 3M4NP and 1,4-benzoquinone reductase (PnpB) against MBQ were performed as previously described for these two enzymes against 2C4NP and 2-chloro-1,4-benzoquinone, respectively (Min et al., 2014). Identification of the products from H6-PnpA or H6-PnpB-catalyzed reactions was carried out by HPLC and GC-MS analysis as described previously (Zhang et al., 2009). Enzyme activity assays of hydroquinone dioxygenase (PnpCD and NpcCD), 4-hydroxymuconic semialdehyde dehydrogenase (PnpE and NpcE) and maleylacetate reductase (PnpF and NpcF) involved in the sequential transformation of MHQ were performed as previously described for sequential transformation of hydroquinone and chlorohydroquinone (Min et al., 2014).
In the case of the kinetics assays of H6-PnpA against 3M4NP, 7 concentrations of 3M4NP raging from 5 to 100 μM were used while the concentration of NADPH was fixed at 400 μM. Data from three independent sets of experiments were fitted with the Michaelis–Menten equation by OriginPro 8 software. The protein concentration was determined according to the Bradford method (Bradford, 1976) with bovine serum albumin as a standard. One unit of enzyme activity was defined as the amount of protein required to catalyze the conversion of 1 μmol of 3M4NP per min at 30°C. The nitrite concentration was determined as previously described (Lessner et al., 2002).
Results
Identification of MBQ and MHQ as Metabolites
The intermediates of 3M4NP catabolism were identified by HPLC analysis after the confirmation of strain SJ98 as a 3M4NP utilizer. Initially, no metabolite was captured during 3M4NP degradation. Therefore, 2, 2′-dipyridyl was added to the biotransformation system in order to accumulate the intermediates before ring cleavage. In this way, two intermediates with HPLC retention times of 11.7 and 8.9 min, respectively, were captured during 3M4NP degradation. The retention times of these two metabolites matched precisely to that of the standard MBQ and MHQ. Moreover, the identification of MBQ and MHQ was also confirmed by GC-MS analysis by comparison with mass spectra of the authentic compounds (Figure 1). In a time course assay of biotransformation, 3M4NP consumption (523 μM) was approximately equivalent to the total accumulation of both MBQ (55 μM) and MHQ (410 μM) (Figure 2), indicating a nearly stoichiometric formation of MBQ and MHQ from 3M4NP. Hence, the identification of metabolites clearly indicated that strain SJ98 degraded 3M4NP with MBQ and MHQ as the intermediates before ring cleavage (Figure 3A).
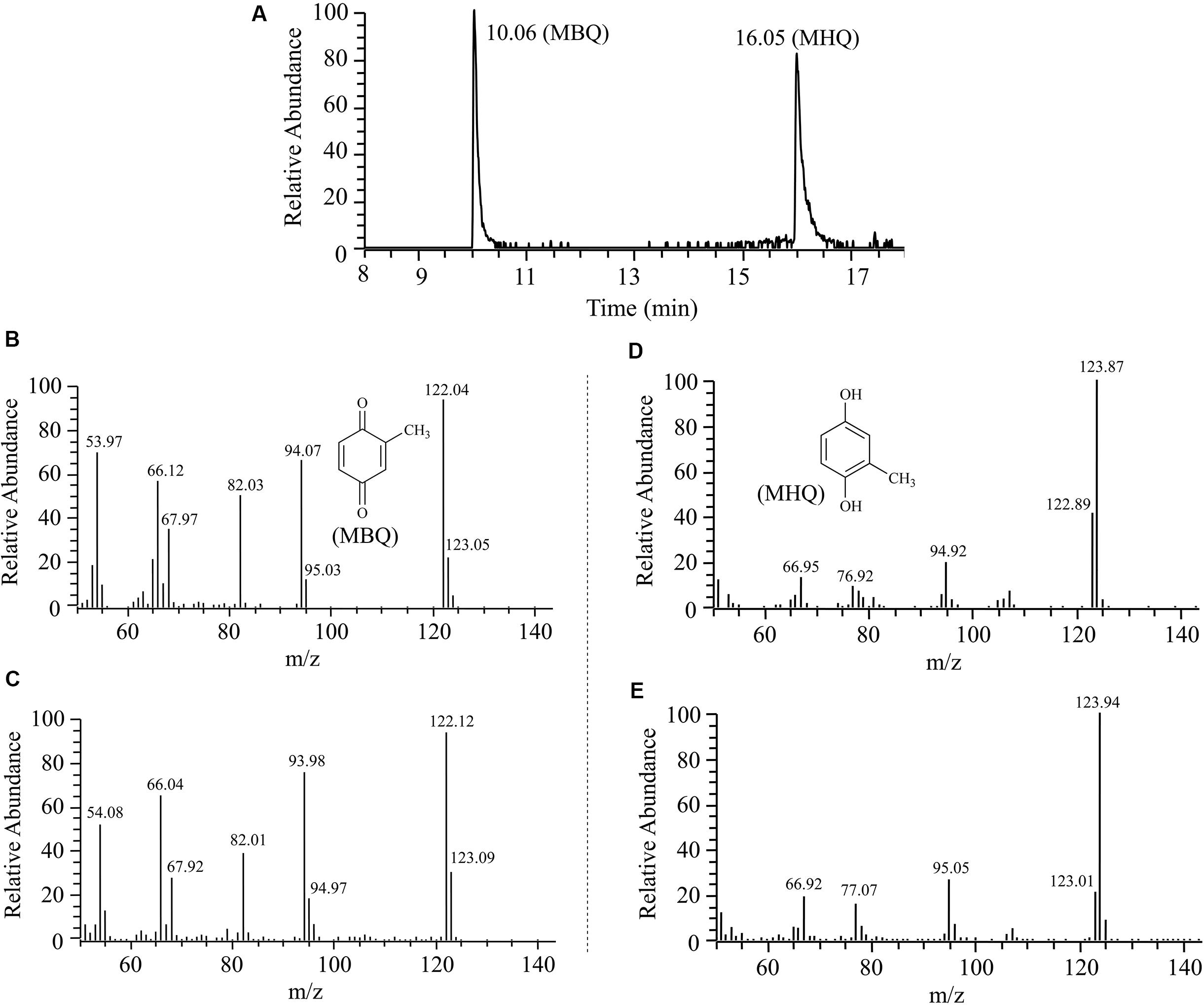
FIGURE 1. Gas chromatography-mass spectrometry (GC-MS) analysis of the intermediates captured during 3M4NP degradation by Burkholderia sp. strain SJ98. (A) The gas chromatogram is the extracted ion current chromatogram at m/z 122.00 ± 0.50 and 124.00 ± 0.50 from the total ion current chromatogram. (B) Mass spectrum of authentic methyl-1,4-benzoquinone (MBQ). (C) Mass spectrum of the GC peak at 10.06 min, identical to that of authentic MBQ. (D) Mass spectrum of authentic methylhydroquinone (MHQ). (E) Mass spectrum of the GC peak at 16.05 min, identical to that of authentic MHQ.
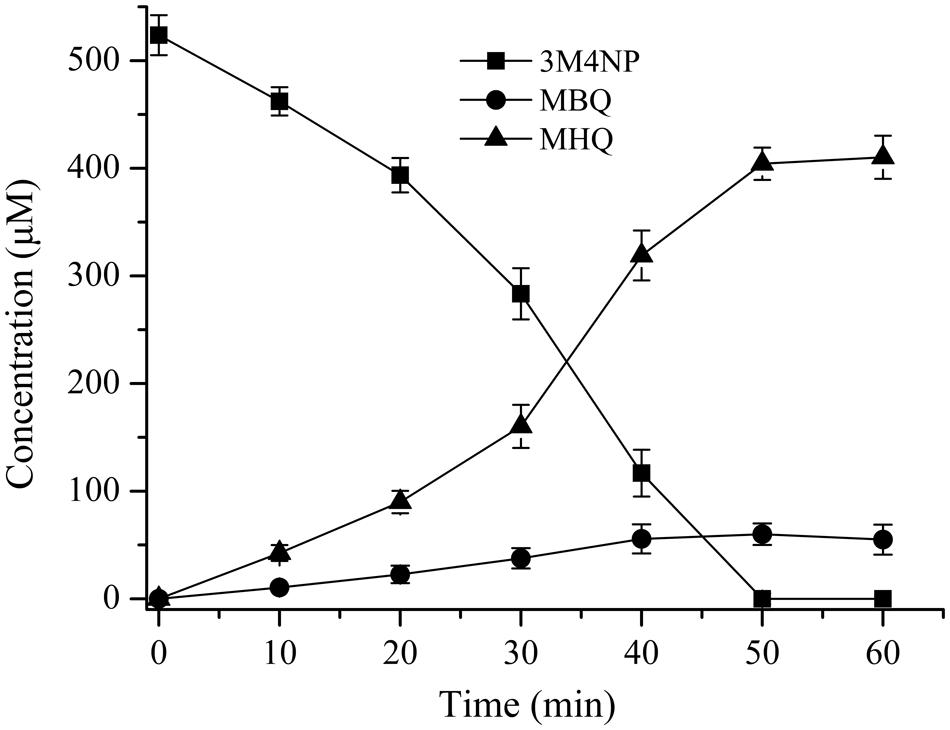
FIGURE 2. Time course of 3M4NP degradation by Burkholderia sp. strain SJ98 in whole cell biotransformation. In order to accumulate the intermediates before ring cleavage, 1 mM 2, 2′-dipyridyl was added to inhibit the ring cleavage enzyme. The disappearance of 3M4NP and the appearance of the products (MBQ and MHQ) were quantified by HPLC. The experiments were performed in triplicate; the results were the average of three independent experiments, and error bars indicate standard deviations.
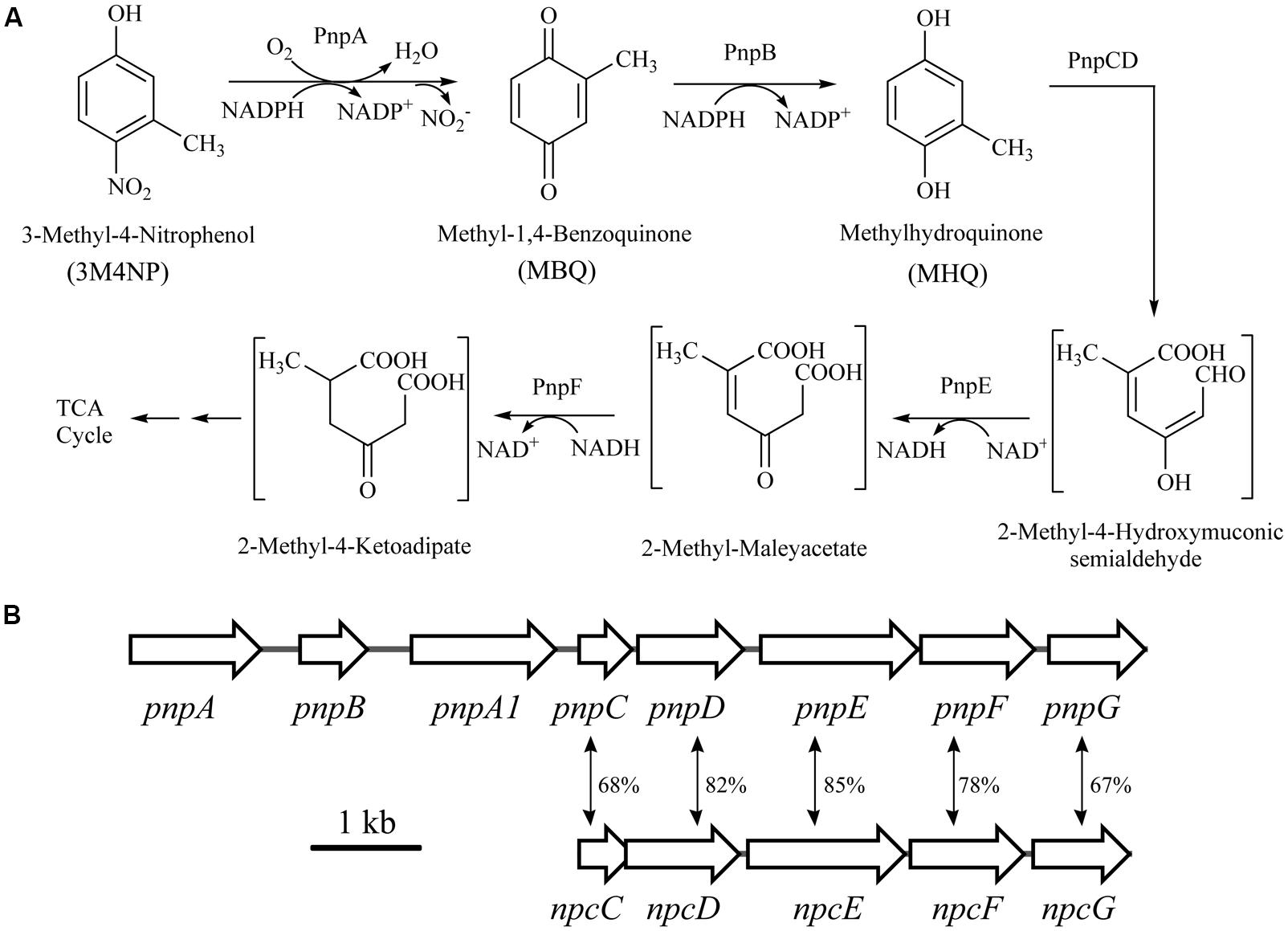
FIGURE 3. (A) Proposed pathways for 3M4NP catabolism in Burkholderia sp. strain SJ98, together with the catabolic reactions catalyzed by pnp gene products. (B) Organization of the pnp and npc gene clusters of strain SJ98. The large black arrows indicate the sizes and directions of transcription of each gene or ORF. The double-headed arrows demonstrated the alignment of the catabolic genes with significant similarities. Percentage identities at the amino acid level are indicated adjacent.
Biotransformation of 3M4NP by Strain SJ98
The uninduced cells of strain SJ98 exhibit negligible activity for 3M4NP. However, the 3M4NP-induced cells exhibited a specific activity of 3.27 ± 0.46 U mg-1 for 3M4NP, indicating that the enzymes involved in 3M4NP degradation in strain SJ98 are inducible. Interestingly, PNP-induced cells degrade 3M4NP (3.56 ± 0.64 U mg-1) while 3M4NP-induced strain SJ98 also have the ability to degrade PNP (5.56 ± 0.84 U mg-1). This finding indicated that the enzymes involved in PNP catabolism were likely also responsible for 3M4NP degradation by strain SJ98.
Transcriptional Analysis of the pnpABA1CDEF and npcCDEF Clusters
In order to distinguish the pnpABA1CDEF and pnpE2E1FD clusters named originally in strain SJ98, the pnpE2E1FD cluster was tentatively renamed as npcCDEF in this study (Figure 3B). Considering that the genes of both clusters were in a single transcriptional operon, respectively, the transcriptional analyses of pnpA and npcC (each representing the operons they belong to) were carried out under various induction conditions by real-time quantitative PCR. The transcription level of pnpA under 3M4NP-induced condition was enhanced dramatically compared to that from the non-induced sample, with 54-fold increase (Figure 4), suggesting the involvement of Pnp proteins in 3M4NP catabolism. In contrast to pnpABA1CDEF cluster, the transcription level of the npcCDEF cluster was not increased by 3M4NP induction (Figure 4). On the other hand, no transcription increase of pnpA was observed by MHQ induction, a metabolite in 3M4NP catabolism.
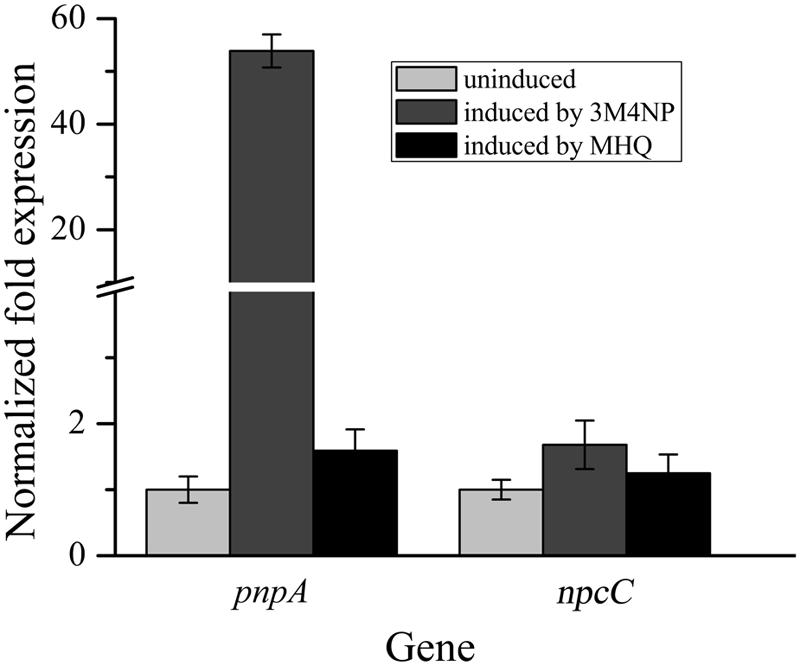
FIGURE 4. Transcriptional analyses of pnpA and npcC in Burkholderia sp. strain SJ98 under induced and uninduced conditions by RT-qPCR. The data are derived from three independent measurements, and error bars indicate standard deviations.
PnpA-Catalyzed Monooxygenation of 3M4NP to MBQ
Escherichia coli cells carrying pET-pnpA was found to be able to degrade 3M4NP by HPLC analysis, along with the stoichiometric release of nitrite ion. In contrast, neither 3M4NP consumption nor nitrite ion release was detected in the negative control when the cells harboring only pET-28a vector. The PnpA activity measured spectrophotometrically showed that the purified H6-PnpA catalyzed rapid degradation of 3M4NP (λmax, 396 nm) with a specific activity of 3.36 U mg-1, together with consumption of NADPH (λmax, 340 nm; Figure 5). By HPLC and GC-MS analyses, both MBQ and MHQ were detected as the products from 3M4NP monooxygenation catalyzed by the purified H6-PnpA. Kinetics assays revealed that the Km value of H6-PnpA for 3M4NP was 20.3 ± 2.54 mM.
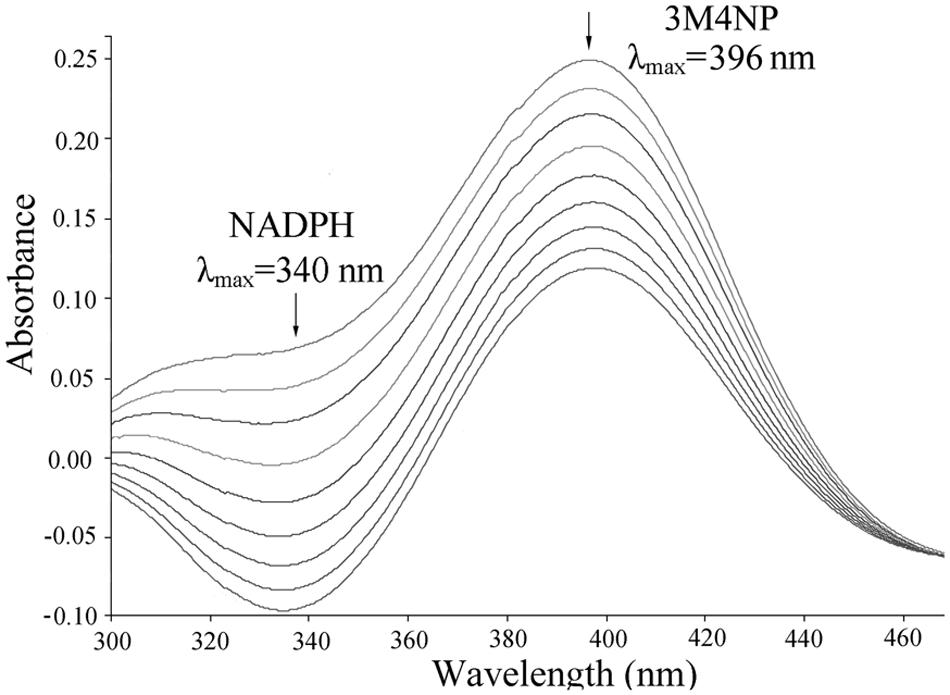
FIGURE 5. Spectral changes during the transformation of 3M4NP by H6-PnpA. Sample and reference cuvettes contained 0.1 mM NADPH, 0.03 mM FAD, 50 mM phosphate buffer (pH 7.4), and 20 μg H6-PnpA in a 0.5-ml mixture. The reaction was initiated by addition of 30 μM 3M4NP, and the spectra were recorded every minute after the addition of 3M4NP. The arrows indicate the directions of spectral changes.
PnpB-Catalyzed Reduction of MBQ to MHQ
The activity assay of PnpB against MBQ was carried out in order to confirm the presence of MBQ in the 3M4NP degradation. By HPLC analysis, the product of MBQ catalyzed by the purified H6-PnpB was identified as MHQ. Moreover, when H6-PnpB was added to a reaction mixture containing H6-PnpA, a clear increase in total 3M4NP monooxygenase activity was observed (Figure 6). A likely explanation for the enhanced activity was that PnpB reduced the formed MBQ to MHQ, probably preventing product inhibition of PnpA activity. Particularly, the increased 3M4NP monooxygenase activity with H6-PnpB further proves the formation of MBQ during 3M4NP catabolism in vivo, apart from the HPLC and GC-MS identification of MBQ as a metabolite.
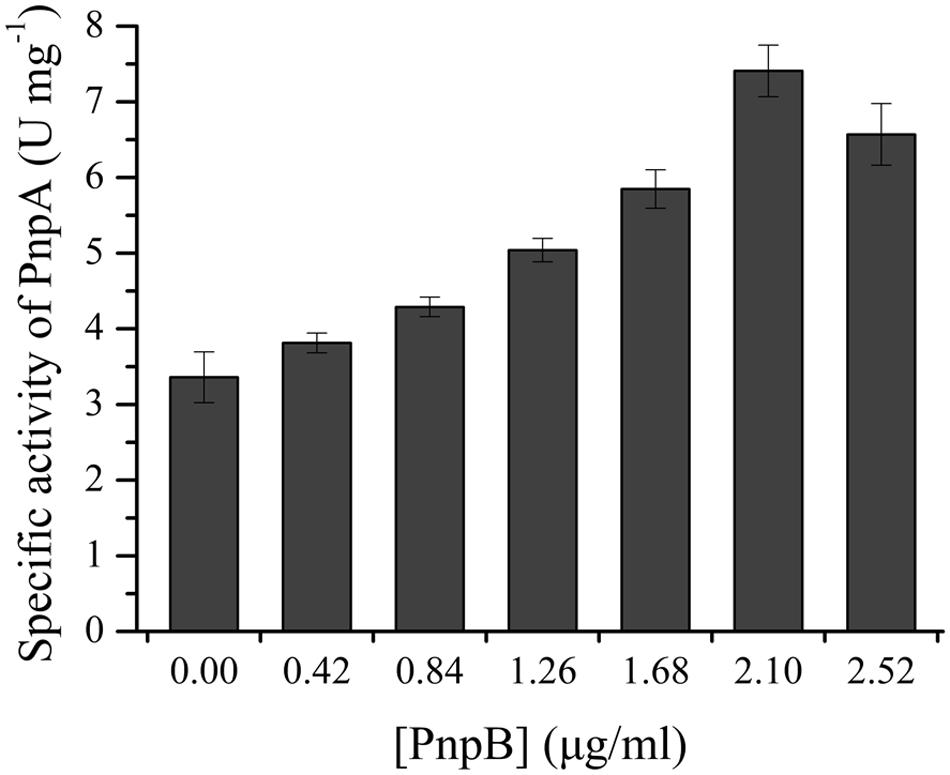
FIGURE 6. H6-PnpB enhanced the 3M4NP degradation by H6-PnpA. The reaction mixtures contained 0.1 mM NADPH, 0.03 mM FAD, 50 mM phosphate buffer (pH 7.4), 20 μg H6-PnpA and different amount of H6-PnpB (0–2.52 μg) in a 0.5 ml mixture. The reaction was initiated by addition of 30 μM 3M4NP.
PnpCD-, PnpE-, and PnpF-Catalyzed Sequential Reactions from MHQ in the Lower Pathway
When purified His6-PnpCD was incubated with MHQ, a spectral change from 290 to 320 nm occurred to form a new compound with a λmax of 320 nm (Figure 7A). In contrast, no spectral change was observed when His6-PnpCD was omitted from the mixture. By HPLC-MS analysis, the ring cleavage product of MHQ was found to have a retention time of 6.63 min and proposed as 2-methyl-4-hydroxymuconic semialdehyde, with the deprotonated ion peak at m/z 155.06 and its fragments at m/z 111.02 (loss of -COOH), at m/z 127.14 (loss of -CHO), and at m/z 141.07 (loss of –CH3) (Figure 7B).
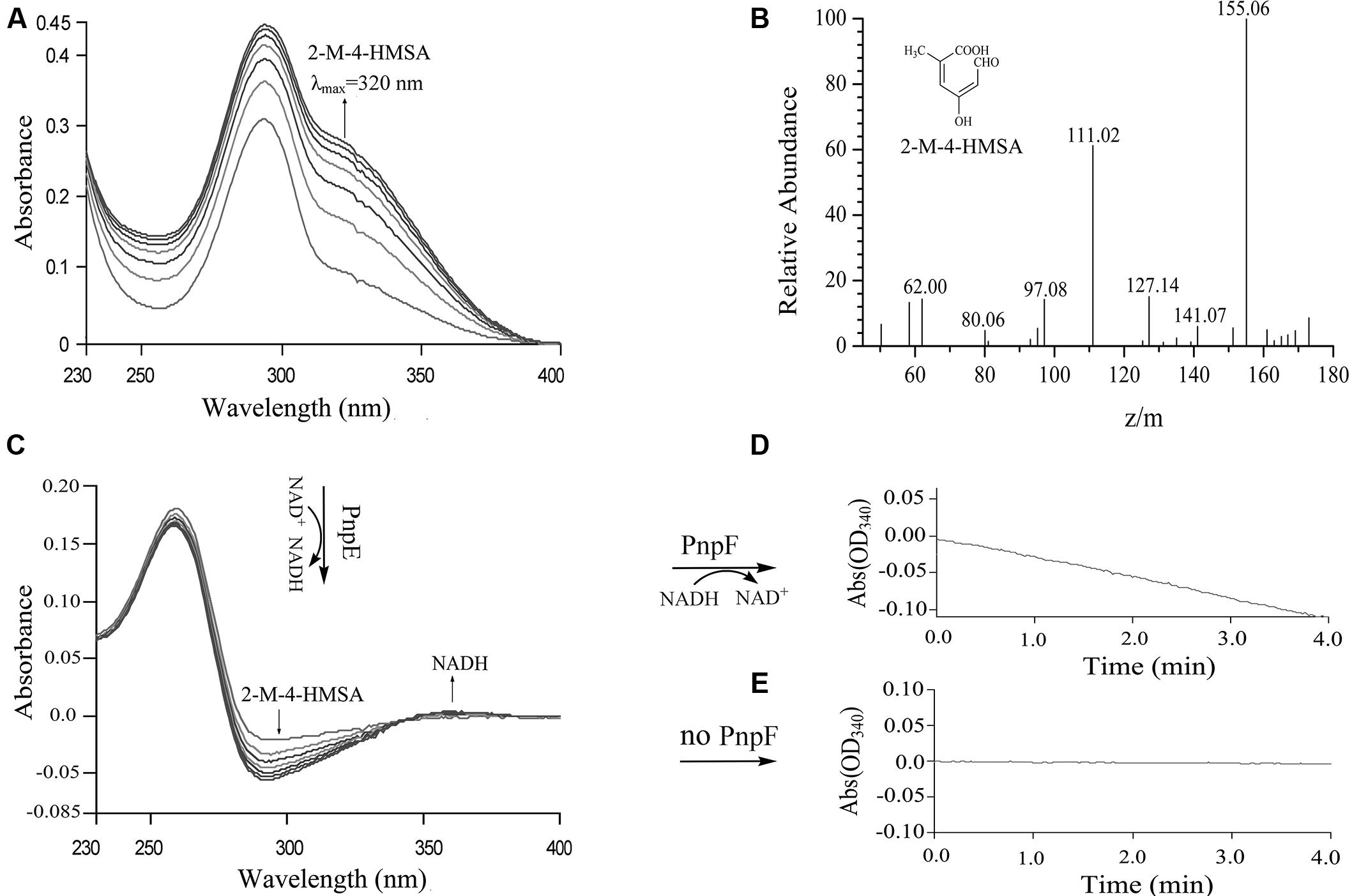
FIGURE 7. Enzyme activity assays of PnpCD, PnpE, and PnpF by sequential catalytic reactions with MHQ as the starting substrate. (A) Spectral changes during the transformation of MHQ by H6-PnpCD. Sample and reference cuvettes contained 0.04 mM Fe2+, 5 μg H6-PnpCD and 20 mM phosphate buffer (pH 7.4) in a 1 ml mixture. The reaction was initiated by the addition of 0.1 mM MHQ. (B) Mass spectrum of the product of MHQ catalyzed by purified PnpCD. The reaction mixture of MHQ catalyzed by H6-PnpCD was extracted with ethyl acetate, and the product was then identified by HPLC-MS analysis. (C) Enzyme activity assay of PnpE. Following the complete oxidation of MHQ by H6-PnpCD, the sample cuvette from the reaction in A was added by 50 μM NAD+, and then divided into two cuvettes (sample and reference). The spectra were recorded every minute after the addition of 20 μg H6-PnpE. (D,E) Enzyme activity assay of PnpF. Following the complete conversion of 2-methyl-4-hydroxymuconic semialdehyde (2-M-4-HMSA) to 2-methylmaleylacetate, the sample cuvette from the reaction in C was added by 50 μM NADH, and then divided into two cuvettes (sample and reference). H6-PnpF (20 μg; D) or buffer without H6-PnpF (E) was added to initiate the assay.
The activity assays of His6-PnpE and His6-PnpF were carried out by sequential catalyzes in order to investigate whether these two enzymes were also involved in 3M4NP degradation. The substrate of PnpE was 2-methyl-4-hydroxymuconic semialdehyde formed from the above PnpCD-catalyzed dioxygenation of MHQ. The absorbance of 2-methyl-4-hydroxymuconic semialdehyde (λmax, 320 nm) became progressively lower after addition of His6-PnpE, together with the production of NADH (Figure 7C), presumably forming 2-methylmaleylacetate. Subsequently, the reaction mixture of PnpE-catalyzed reaction was used to assay the activity of PnpF. As shown in Figure 7D, NADH was consumed gradually upon the addition of purified PnpF to the assay mixture, indicating that PnpF has 2-methylmaleylacetate reduction activity, presumably producing 2-methy-4-ketoadipate (Figure 3A). However, no oxidation of NADH was observed when H6-PnpF was omitted from the reaction mixture (Figure 7E). Although the transcription level of npc cluster has no apparent increase after 3M4NP induction, the purified His6-NpcCD, His6-NpcE, and His6-NpcF were also found to have the ability to catalyze the sequential conversion of MHQ, and the spectra changes are similar to those shown in Figure 7.
The Crucial Role of pnpA in 3M4NP Catabolism
Strains SJ98ΔpnpA (with pnpA deleted) and SJ98ΔpnpA1 (with pnpA1 deleted) described in Table 1 were, respectively, used to investigate the physiological roles of pnpA and pnpA1 in 3M4NP degradation in vivo. Strain SJ98ΔpnpA was no longer to grow with 3M4NP as sole carbon and energy source, and pnpA-complemented mutant SJ98ΔpnpA[pRK415-pnpA] regained its ability to utilize 3M4NP (Figure 8). This indicated that pnpA is absolutely essential for strain SJ98 to utilize 3M4NP. In contrast, although strain SJ98ΔpnpA1 still has the ability to grow on 3M4NP, its maximum specific growth rate (μm, 0.154 h-1) is approximately 28% lower than that of wild-type strain SJ98 (μm, 0.214 h-1), in addition to a slower rate of 3M4NP removal (9.3 μM h-1 for SJ98ΔpnpA1, compared with 13.0 μM h-1 for the wild type strain; Figure 8). This indicated that pnpA1 makes a partial contribution in 3M4NP catabolism in strain SJ98, but it is not necessary in 3M4NP catabolism and cannot maintain the cell growth alone.
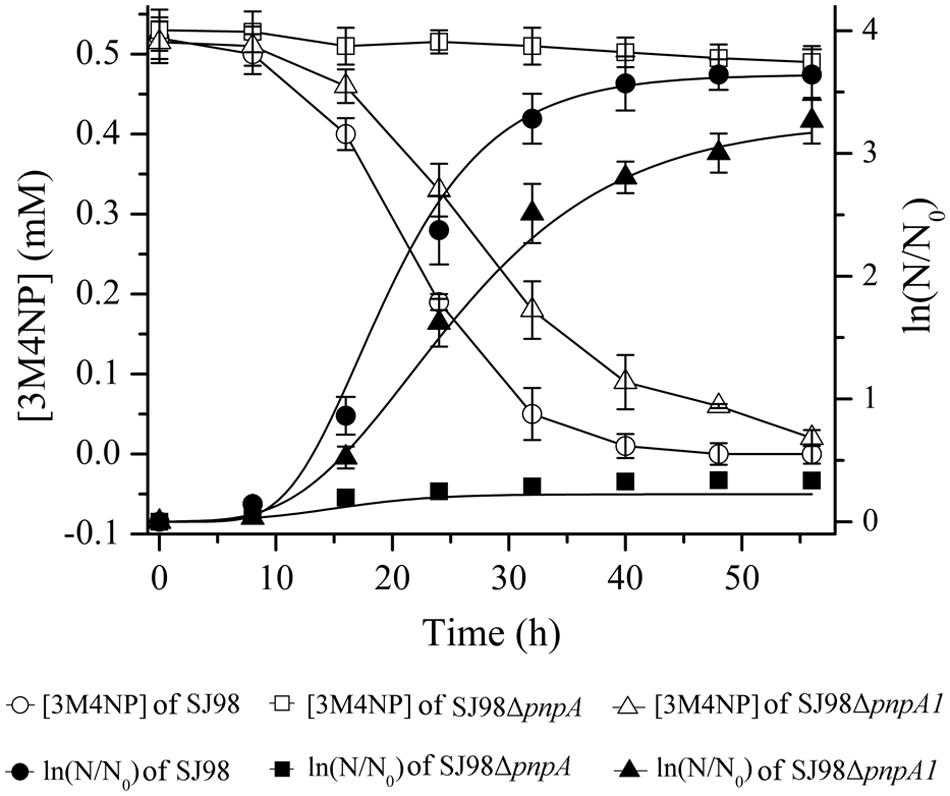
FIGURE 8. Time course of 3M4NP degradation and cell growth in cultures of Burkholderia sp. strains SJ98, SJ98ΔpnpA and SJ98ΔpnpA1. N, numberof cells; N0, initial number of cells. All of the experiments were performed in triplicate; the results were the average of three independent experiments, and error bars indicate standard deviations. The data of pnpA-complemented mutant SJ98ΔpnpA[pRK415-pnpA] were not shown but similar to those of Burkholderia sp. strain SJ98.
Discussion
Considering that fenitrothion could be degraded rapidly to 3M4NP in the environment (Takimoto et al., 1976; Spillner et al., 1979; Mikami et al., 1985), the contamination in the fenitrothion-treated environmentis is more likely due to the presence of 3M4NP rather than fenitrothion itself. In recent years, the microbial degradation of 3M4NP has been attracting considerable attentions (Bhushan et al., 2000; Hayatsu et al., 2000; Zhang et al., 2006; Kim et al., 2007; Arora and Jain, 2012). Burkholderia sp. strain SJ98, previously identified as Ralstonia sp. SJ98 was proposed to degrade 3M4NP with catechol as a metabolite (Bhushan et al., 2000), with no genetic and enzymatic evidences. However, catechol was not detected as the metabolites of 3M4NP degradation by either HPLC or GC-MS analyses. Furthermore, quantitative analysis of the intermediates have shown that strain SJ98 degrade 3M4NP with MBQ and MHQ as the metabolites before ring cleavage, the same as that reported in Burkholderia sp. strains NF100 (Hayatsu et al., 2000), FDS-1 (Zhang et al., 2006) and RKJ800 (Arora and Jain, 2012). Subsequent biochemical and genetic analyses fill a gap in our knowledge of the 3M4NP degradation mechanism at the molecular and biochemical levels in a bacterial strain.
Recently, the enzymes encoded by the pnpABA1CDEF cluster were reported to be responsible for the catabolism of PNP and 2C4NP in strain SJ98 (Min et al., 2014). In this study, the high transcription of the pnpABA1CDEF cluster in the 3M4NP-induced cell indicated that the enzymes encoded by the cluster were also likely involved in 3M4NP catabolism in this strain. Enzymatic assay has shown that PnpA has the ability to catalyze the monooxygenation of 3M4NP, and its encoding gene is absolutely necessary for strain SJ98 to grow on 3M4NP. Although both MBQ and MHQ were detected during 3M4NP degradation by purified PnpA, the direct product should be MBQ because it is generally accepted that monooxygenases produce a quinone after removal of an electron-withdrawn nitro group from a phenolic compound (Haigler et al., 1996; Perry and Zylstra, 2007; Liu et al., 2010; Min et al., 2014). The detection of MHQ is probably due to the non-enzymatic reaction of MBQ in the presence of NADPH, and the same explanations were also proposed previously for PNP (Perry and Zylstra, 2007; Zhang et al., 2009), 2C4NP (Min et al., 2014, 2016) and 2,4,6-trichlorophenol (Xun and Webster, 2004) monooxygenation. Moreover, the enhancement of 3M4NP monooxygenase activity of PnpA by PnpB also confirms the involvement of MBQ during 3M4NP degradation by strain SJ98. The enhanced activity of PnpA is likely due to the reduction of MBQ to MHQ by PnpB, possibly preventing product inhibition of PnpA. Previously, the analogous phenomena were also observed during monooxygenation of PNP (Zhang et al., 2009), 2C4NP (Min et al., 2014) and 2,4,6-trichlorophenol (Belchik and Xun, 2008) in the presence of corresponding quinone reductases. On the other hand, although PnpA catalyzes the monooxygenation of 3M4NP, in addition to PNP and 2C4NP (Min et al., 2014), its Km value for 2C4NP (6.2 ± 0.76 mM reported previously) is evidently lower than those for 3M4NP (20.3 ± 2.54 mM) and PNP (25.4 ± 3.63 mM reported previously), implying that 2C4NP is the probable physiological substrate for PnpA in strain SJ98.
Although Burkholderia sp. strains NF100 (Hayatsu et al., 2000), FDS-1 (Zhang et al., 2006), and RKJ800 (Arora and Jain, 2012), were also proposed to degrade 3M4NP with MHQ as the ring cleavage substrate, MBQ was not actually detected and the fate of MHQ was not revealed in these cases. In current study, PnpCD was found to be able to catalyze the dioxygenation of MHO, presumably forming 2-methyl-4-hydroxymuconic semialdehyde (Figure 3A). This is similar to previous reports in which the hydroquinone dioxygenase (HapCD) from Pseudomonas fluorescens ACB was also reported to be able to catalyze the dioxygenation of MHQ to methyl substituted 4-hydroxymuconic semialdehyde (Moonen et al., 2008). PnpE and PnpF were found to be able to catalyze the sequential conversion of 2-methyl-4-hydroxymuconic semialdehyde, presumably forming 2-methylmaleylacetate and 2-methy-4-ketoadipate, respectively. Previously, a maleylacetate reductase from Pseudomonas sp. strain B13 was also found to have the ability to catalyze the reduction of 2-methylmaleylacetate (Kaschabek and Reineke, 1993, 1995). Although NpcCD, NpcE, and NpcF also have the ability to catalyze the sequential transformation of MHQ in vitro, their coding genes were not upregulated by 3M4NP induction. Therefore, it is reasonable to conclude that NpcCD, NpcE, and NpcF are unlikely involved in the catabolism of 3M4NP in this strain.
It is generally thought that all the enzymes involved in the complete catabolic pathway of a compound are able to catalyze the initial transformation of this compound, as well as the subsequent reactions of all formed intermediates, to simple organic acids channeled into TCA cycle. Interestingly, the catabolism of PNP, 2C4NP, and 3M4NP by strain SJ98 of the current study are catalyzed by the same complete set of enzymes encode by the single pnpABA1CDEF cluster. This indicated that all of the Pnp enzymes exhibited extended substrate specificity for every single metabolites (chlorinated or methylated), resulting in its growth on all these three nitrophenols. In contrast, the Gram-positive PNP and 2C4NP utilizer R. imtechensis RKJ300, with a two-component PNP monooxygenase to initiate 2C4NP and PNP catabolism (Min et al., 2016), was found to be unable to grow on 3M4NP in this study. This could be due to several reasons, such as: (i) PnpA1A2 or other enzymes are inactive toward 3M4NP or methylated metabolites in the pathway; or (ii) inability of 3M4NP to induce the expression of the enzymes. During the investigation of ortho-nitrophenol (ONP) degradation by P. putida B2, the ONP monooxygenase from this strain has the ability to catalyze the monooxygenation of ONP, 4-chloro-2-nitrophenol and 4-methyl-2-nitrophenol to the ring-cleavage substrates catechol, 4-chloro-catechol and 4-methyl-catechol, respectively, but the ring-cleavage catechol dioxygenase in this strain virtually has no activity toward 4-chloro-catechol (Zeyer et al., 1986). However, the ring-cleavage enzyme PnpCD from strain SJ98 is able to efficiently catalyze the dioxygenation of hydroquinone, chlorohydroquinone (Min et al., 2014) and MHQ, the products of PNP, 2C4NP, and 3M4NP, respectively. Moreover, the enzymes involved in ONP catabolism in strain B2 only induced by unsubstituted ONP but not chloro- and methyl-substituted ONPs, whereas the enzymes involved in PNP catabolism in strain SJ98 is induced by PNP, 3M4NP (methyl-PNP) and 2C4NP (chloro-PNP).
Despite that the detailed genetic determinant of 3M4NP catabolism in Burkholderia sp. strain NF100 was unknown, previous plasmid curing experiments indicated that the genes involved in the conversion of 3M4NP to MHQ were located on the chromosome, whereas the genes responsible for MHQ degradation were on the plasmid pNF1 (Hayatsu et al., 2000). This suggested that the functional genes for the entire 3M4NP degradation in strain NF100 were separated into at least two non-contiguous gene clusters. However, the genes involved in 3M4NP catabolism in strain SJ98 were clustered together into single gene cluster/operon (pnpABA1CDEF). Moreover, the enzymes involved in PNP catabolism are likely not responsible for 3M4NP degradation in strain NF100 (Hayatsu et al., 2000), whereas the catabolism of 3M4NP, PNP, and 2C4NP share the same set of enzymes in strain SJ98 as demonstrated in a previous study and this study. These indicated that strains NF100 and SJ98 likely have different evolutionary patterns in acquiring 3M4NP and PNP catabolic ability.
Conclusion
Burkholderia sp. strain SJ98 degraded 3M4NP via MBQ and MHQ as the intermediates before ring cleavage. Purified PnpA catalyzes the monooxygenation of 3M4NP to MBQ, and pnpA is absolutely necessary for the catabolism of 3M4NP. PnpB catalyzes the reduction of MBQ to MHQ. PnpCD, PnpE, and PnpF were involved in the lower pathway of 3M4NP catabolism. These revealed that the enzymes involved in PNP and 2C4NP catabolism were also responsible for 3M4NP degradation in strain SJ98. The present study fills a gap in our understanding of the microbial degradation of 3M4NP at molecular and biochemical levels. Strain SJ98 is capable of degrading PNP and its chloro- and methyl-substituted derivatives, suggesting its potential role in bioremediation of these toxicants-contaminated sites.
Author Contributions
Conceived and designed the experiments: N-YZ, XH, and JM. Performed the experiments: JM and YL. Analyzed the data: JM, N-YZ, and XH. Wrote the paper: JM, XH, and N-YZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the Yantai Science and Technology Project (grant 2014ZH088), the National Key Basic Research Program of China (973 Program, grant 2012CB725202), and National Natural Science Foundation of China (grant 41376138).
Abbreviations
2C4NP, 2-chloro-4-nitrophenol; 3M4NP, 3-methyl-4-nitrophenol; MBQ, methyl-1,4-benzoquinone; MHQ, methylhydroquinone; PNP, para-nitrophenol.
References
Arora, P. K., and Jain, R. K. (2011). Pathway for degradation of 2-chloro-4-nitrophenol in Arthrobacter sp. SJCon. Curr. Microbiol. 63, 568–573. doi: 10.1007/s00284-011-0022-2
Arora, P. K., and Jain, R. K. (2012). Metabolism of 2-chloro-4-nitrophenol in a Gram negative bacterium, Burkholderia sp RKJ 800. PLoS ONE 7:e38676. doi: 10.1371/journal.pone.0038676
Arora, P. K., Sasikala, C., and Ramana, C. V. (2012). Degradation of chlorinated nitroaromatic compounds. Appl. Microbiol. Biotechnol. 93, 2265–2277. doi: 10.1007/s00253-012-3927-1
Arora, P. K., Srivastava, A., and Singh, V. P. (2014). Bacterial degradation of nitrophenols and their derivatives. J. Hazard. Mater. 266, 42–59. doi: 10.1016/j.jhazmat.2013.12.011
Belchik, S. M., and Xun, L. Y. (2008). Functions of flavin reductase and quinone reductase in 2,4,6-trichlorophenol degradation by Cupriavidus necator JMP134. J. Bacteriol. 190, 1615–1619. doi: 10.1128/Jb.01697-07
Bhushan, B., Samanta, S. K., Chauhan, A., Chakraborti, A. K., and Jain, R. K. (2000). Chemotaxis and biodegradation of 3-methyl- 4-nitrophenol by Ralstonia sp. SJ98. Biochem. Biophys. Res. Commun. 275, 129–133. doi: 10.1006/bbrc.2000.3216
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chapman, P. J., and Hopper, D. J. (1968). The bacterial metabolism of 2,4-xylenol. Biochem. J. 110, 491–498. doi: 10.1042/bj1100491
Ferreira, M. I., Marchesi, J. R., and Janssen, D. B. (2008). Degradation of 4-fluorophenol by Arthrobacter sp. strain IF1. Appl. Microbiol. Biotechnol. 78, 709–717. doi: 10.1007/s00253-008-1343-3
Furuta, C., Suzuki, A. K., Taneda, S., Kamata, K., Hayashi, H., Mori, Y., et al. (2004). Estrogenic activities of nitrophenols in diesel exhaust particles. Biol. Reprod. 70, 1527–1533. doi: 10.1095/biolreprod.103.024810
Ghosh, A., Khurana, M., Chauhan, A., Takeo, M., Chakraborti, A. K., and Jain, R. K. (2010). Degradation of 4-nitrophenol, 2-chloro-4-nitrophenol, and 2,4-dinitrophenol by Rhodococcus imtechensis strain RKJ300. Environ. Sci. Technol. 44, 1069–1077. doi: 10.1021/es9034123
Haigler, B. E., Suen, W. C., and Spain, J. C. (1996). Purification and sequence analysis of 4-methyl-5-nitrocatechol oxygenase from Burkholderia sp. strain DNT. J. Bacteriol. 178, 6019–6024.
Hayatsu, M., Hirano, M., and Tokuda, S. (2000). Involvement of two plasmids in fenitrothion degradation by Burkholderia sp. Strain NF100. Appl. Environ. Microbiol. 66, 1737–1740. doi: 10.1128/AEM.66.4.1737-1740.2000
Itoh, H., Navarro, R., Takeshita, K., Tago, K., Hayatsu, M., Hori, T., et al. (2014). Bacterial population succession and adaptation affected by insecticide application and soil spraying history. Front. Microbiol. 5:457. doi: 10.3389/fmicb.2014.00457
Jain, R. K., Dreisbach, J. H., and Spain, J. C. (1994). Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl. Environ. Microbiol. 60, 3030–3032.
Kadiyala, V., and Spain, J. C. (1998). A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl. Environ. Microbiol. 64, 2479–2484.
Kanaly, R. A., Kim, I. S., and Hur, H. G. (2005). Biotransformation of 3-methyl-4-nitrophenol, a main product of the insecticide fenitrothion, by Aspergillus niger. J. Agric. Food. Chem. 53, 6426–6431. doi: 10.1021/jf050679w
Kaschabek, S. R., and Reineke, W. (1993). Degradation of chloroaromatics- purification and characterization of maleylacetate reductase from Pseudomonas sp. strain B13. J. Bacteriol. 175, 6075–6081.
Kaschabek, S. R., and Reineke, W. (1995). Maleylacetate reductase of Pseudomonas sp. strain B13-specificity of substrate conversion and halide elimination. J. Bacteriol. 177, 320–325.
Kim, S. H., Park, M. R., Han, S. I., Whang, K. S., Shim, J. H., and Kim, I. S. (2007). Degradation of 3-methyl-4-nitrophenol, a main product of the insecticide fenitrothion, by Burkholderia sp. SH-1 isolated from earthworm (Eisenia fetida) intestine. J. Appl. Biolog. Chem. 50, 281–287.
Kitagawa, W., Kimura, N., and Kamagata, Y. (2004). A novel p-nitrophenol degradation gene cluster from a Gram-positive bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 186, 4894–4902. doi: 10.1128/JB.186.15.4894-4902.2004
Lessner, D. J., Johnson, G. R., Parales, R. E., Spain, J. C., and Gibson, D. T. (2002). Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp strain JS765. Appl. Environ. Microbiol. 68, 634–641. doi: 10.1128/Aem.68.2.634-641.2002
Li, C. M., Suzuki, A. K., Takahashi, S., Taneda, S., Watanabe, G., and Taya, K. (2008). Effects of 3-methyl-4-nitrophenol on the reproductive toxicity in female Japanese quail (Coturnix japonica). Biol. Pharm. Bull, 31, 2158–2161. doi: 10.1248/BPB.31.2158
Li, X. Z., Li, C. M., Suzuki, A. K., Watanabe, G., Taneda, S., and Taya, K. (2009). Endocrine disruptive effect of 3-methyl-4-nitrophenol isolated from diesel exhaust particles in hershberger assay using castrated immature rats. Biosci. Biotechnol. Biochem. 73, 2018–2021. doi: 10.1271/bbb.90204
Liu, P. P., Zhang, J. J., and Zhou, N. Y. (2010). Characterization and mutagenesis of a two-component monooxygenase involved in para-nitrophenol degradation by an Arthrobacter strain. Int. Biodeterior. Biodegr. 64, 293–299. doi: 10.1016/j.ibiod.2010.03.001
Liu, T. T., and Zhou, N. Y. (2012). Novel L-cysteine-dependent maleylpyruvate isomerase in the gentisate pathway of Paenibacillus sp. strain NyZ101. J. Bacteriol. 194, 3987–3994. doi: 10.1128/JB.00050-12
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mikami, N., Sakata, S., Yamada, H., and Miyamoto, J. (1985). Further-studies on degradation of fenitrothion in soils. J. Pestic. Sci. 10, 491–500. doi: 10.1584/jpestics.10.491
Min, J., Zhang, J. J., and Zhou, N. Y. (2014). The gene cluster for para-nitrophenol catabolism is responsible for 2-chloro-4-nitrophenol degradation in Burkholderia sp. strain SJ98. Appl. Environ. Microbiol. 80, 6212–6222. doi: 10.1128/AEM.02093-14
Min, J., Zhang, J. J., and Zhou, N. Y. (2016). A two-component para-nitrophenol monooxygenase initiates a novel 2-chloro-4-nitrophenol catabolic pathway in Rhodococcus imtechensis RKJ300. Appl. Environ. Microbiol. 82, 714–723. doi: 10.1128/AEM.03042-15
Moonen, M. J., Synowsky, S. A., van den Berg, W. A., Westphal, A. H., Heck, A. J., van den Heuvel, R. H., et al. (2008). Hydroquinone dioxygenase from Pseudomonas fluorescens ACB: a novel member of the family of nonheme-iron(II)-dependent dioxygenases. J. Bacteriol. 190, 5199–5209. doi: 10.1128/JB.01945-07
Pandey, J., Heipieper, H. J., Chauhan, A., Arora, P. K., Prakash, D., Takeo, M., et al. (2011). Reductive dehalogenation mediated initiation of aerobic degradation of 2-chloro-4-nitrophenol (2C4NP) by Burkholderia sp. strain SJ98. Appl. Microbiol. Biotechnol. 92, 597–607. doi: 10.1007/s00253-011-3254-y
Perry, L. L., and Zylstra, G. J. (2007). Cloning of a gene cluster involved in the catabolism of p-nitrophenol by Arthrobacter sp. strain JS443 and characterization of the p-nitrophenol monooxygenase. J. Bacteriol. 189, 7563–7572. doi: 10.1128/JB.01849-06
Shen, W., Liu, W., Zhang, J., Tao, J., Deng, H., Cao, H., et al. (2010). Cloning and characterization of a gene cluster involved in the catabolism of p-nitrophenol from Pseudomonas putida DLL-E4. Bioresour. Technol. 101, 7516–7522. doi: 10.1016/j.biortech.2010.04.052
Spain, J. C., and Gibson, D. T. (1991). Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl. Environ. Microbiol. 57, 812–819.
Spillner, C. J., Jack, R., DeBaun, and Menn, J. J. (1979). Degradation of fenitrothion in forest soil and effects on forest soil microbes. J. Agric. Food Chem. 27, 1054–1060. doi: 10.1021/jf60225a009
Takeo, M., Murakami, M., Niihara, S., Yamamoto, K., Nishimura, M., Kato, D., et al. (2008). Mechanism of 4-nitrophenol oxidation in Rhodococcus sp. strain PN1: characterization of the two-component 4-nitrophenol hydroxylase and regulation of its expression. J. Bacteriol. 190, 7367–7374. doi: 10.1128/JB.00742-08
Takimoto, Y., Hirota, M. I., Inui, H., and Miyamoto, J. (1976). Decomposition and leaching of radioactive sumithion in 4 different soils under laboratory conditions. J. Pestic. Sci. 1, 131–143. doi: 10.1584/jpestics.1.131
Verma, Y., and Rana, S. V. S. (2009). Endocrinal toxicity of industrial solvents- A mini review. Indian J. Exp. Biol. 47, 537–549.
Vikram, S., Pandey, J., Bhalla, N., Pandey, G., Ghosh, A., Khan, F., et al. (2012). Branching of the p-nitrophenol (PNP) degradation pathway in Burkholderia sp. strain SJ98: evidences from genetic characterization of PNP gene cluster. AMB Express 2:30. doi: 10.1186/2191-0855-2-30
Wei, Q., Liu, H., Zhang, J. J., Wang, S. H., Xiao, Y., and Zhou, N. Y. (2010). Characterization of a para-nitrophenol catabolic cluster in Pseudomonas sp. strain NyZ402 and construction of an engineered strain capable of simultaneously mineralizing both para- and ortho-nitrophenols. Biodegradation 21, 575–584. doi: 10.1007/s10532-009-9325-4
Xiao, Y., Wu, J. F., Liu, H., Wang, S. J., Liu, S. J., and Zhou, N. Y. (2006). Characterization of genes involved in the initial reactions of 4-chloronitrobenzene degradation in Pseudomonas putida ZWL73. Appl. Microbiol. Biotechnol. 73, 166–171. doi: 10.1007/s00253-006-0441-3
Xun, L., and Webster, C. M. (2004). A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J. Biol. Chem. 279, 6696–6700. doi: 10.1074/JBC.M312072200
Zeyer, J., Kocher, H. P., and Timmis, K. N. (1986). Influence of para-substituents on the oxidative metabolism of o-nitrophenols by Pseudomonas putida B2. Appl. Environ. Microbiol. 52, 334–339.
Zhang, J. J., Liu, H., Xiao, Y., Zhang, X. E., and Zhou, N. Y. (2009). Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J. Bacteriol. 191, 2703–2710. doi: 10.1128/JB.01566-08
Keywords: 2-chloro-4-nitrophenol, 3-methyl-4-nitrophenol, Burkholderia sp. strain SJ98, catabolism, fenitrothion, para-nitrophenol
Citation: Min J, Lu Y, Hu X and Zhou N-Y (2016) Biochemical Characterization of 3-Methyl-4-nitrophenol Degradation in Burkholderia sp. Strain SJ98. Front. Microbiol. 7:791. doi: 10.3389/fmicb.2016.00791
Received: 21 December 2015; Accepted: 09 May 2016;
Published: 25 May 2016.
Edited by:
Pankaj Kumar Arora, Mahatma Jyotiba Phule Rohilkhand University, IndiaReviewed by:
Tapan Kumar Adhya, KIIT University, IndiaJay Prakash Verma, Banaras Hindu University, India
Copyright © 2016 Min, Lu, Hu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoke Hu, xkhu@yic.ac.cn; Ning-Yi Zhou, ningyi.zhou@sjtu.edu.cn
 Jun Min
Jun Min Yang Lu
Yang Lu Xiaoke Hu
Xiaoke Hu Ning-Yi Zhou
Ning-Yi Zhou
