- 1Department of Basic Veterinary Medicine, College of Veterinary Medicine, Northwest A&F University, Yangling, China
- 2Department of Anatomy, Physiology and Pharmacology, College of Veterinary Medicine, Auburn University, Auburn, AL, USA
Objective: The aim of this study was to explore the occurrence and molecular characterization of extended-spectrum β-lactamases (ESBL), plasmid-mediated AmpC β-lactamase (pAmpC) and carbapenemases among ESBL-producing multidrug resistant (MDR) Escherichia coli from dogs and cats in the United States.
Methods: Of 2443 E.coli isolated from dogs and cats collected between August 2009 and January 2013, 68 isolates were confirmed as ESBL-producing MDR ones. PCR and sequencing were performed to identify β-lactamases and plasmid-mediated quinolone resistance (PMQR) genes, and shed light on the virulence gene profiles, phylogenetic groups and ST types.
Results: Phylogenic group D and B2 accounted for 69.1% of the isolates. 50 (73.5%) isolates carried CTX-M ESBL gene, and the most predominant specific CTX-M subtype identified was blaCTX−M−15 (n = 33), followed by blaCTX−M−1 (n = 32), blaCTX−M−123 (n = 27), blaCTX−M−9 (n = 19) and blaCTX−M−14 (n = 19), and blaCTX−M−123 was firstly reported in E. coli isolates in the United States alone or in association. Other β-lactamase genes blaTEM, blaSHV, blaOXA−48, and blaCMY−2 were detected in 41.2, 29.4, 19.1, and 17.6% of 68 ESBL-producing MDR isolates, respectively. The blaTEM and blaSHV genes were classfied as ESBLs with the exception of the blaTEM−1 gene. Additionally, 42.6% (29/68) of isolates co-expressed blaCTX−M−15 and PMQR gene aac(6′)-Ib-c. The overall occurrence of virulence genes ranged from 11.8 (ireA) to 88.2% (malX), and most of virulence genes were less frequent among CTX-M-producing isolates than non-CTX-M isolates with the exception of malX and iutA. The 68 isolates analyzed were assigned to 31 STs with six being novel. Three pandemic clonal lineages ST131 (n = 10), ST648 (n = 9), and ST405 (n = 9) accounted for more than 41% of the investigated isolates, and ST648 and ST405 of phylogenetic D were firstly reported in E. coli from dogs and cats in the United States.
Conclusion: blaCTX−M−123 of ESBLs and carbapenemase blaOXA−48 were firstly reported in ESBL-producing MDR E.coli from dogs and cats in the United States, and ST131, ST648, and ST405 were the predominant clonal groups.
Introduction
Extraintestinal pathogenic strains of Escherichia coli (ExPEC) are the most important dogs and cats bacterial pathogens associated with extraintestinal infections (Beutin, 1999). However, extended-spectrum β-lactamase (ESBL)-producing ExPEC are isolated worldwide with increasing frequency from human and animal clinical isolates (Pitout, 2012). The occurrence of β-lactamases, including ESBLs, plasmid-mediated AmpC β-lactamases (pAmpC) and carbapenemases among E. coli pose serious challenges to the use of penicillins, extended-spectrum cephalosporins (3rd and 4th generation cephalosporins), monobactams, and carbapenems (Karisik et al., 2008; Geser et al., 2012). Furthermore, ESBL-producing isolates are often cross-resistant to fluoroquinolones and other antimicrobial agents, thus expressed multidrug resistance (MDR). This combination of properties can significantly affect the course and outcomes of infections. β-lactamase genes commonly located on mobile genetic elements, such as plasmids, transposons, or integrons, and the resistance plasmids can easily be transferred between bacterial isolates by conjugation mechanism. Accordingly, transmission of β-lactamase genes between companion animals and owner has become a subject of active discussion as companion animals could be potential sources of ESBL-producing E. coli isolates causing community-acquired infections (Schmiedel et al., 2014).
Although the ESBLs, pAmpCs and carbapenemases in E. coli isolates from humans and animals have been characterized in various studies around the world, knowledge about the β-lactamases and population structure in MDR E. coli isolates from companion animals in the United States is limited. Prior to the current study only two studies have described the occurrence and the diversity of ESBLs in E. coli from dogs and cats in the United States (O'Keefe et al., 2010; Shaheen et al., 2011), and the isolates were collected from September 2004 to December 2007, and May 2008 to May 2009, respectively. However, the β-lactamases, particularly CTX-M-type ESBLs, are characterized by ongoing and complex evolution. Currently, greater than 150 variants have been identified, and several chimeras, e.g., blaCTX−M−64 and blaCTX−M−123 have been reported since 2009 (He et al., 2013). Moreover, several novel β-lactamases, e.g., blaKPC, blaNDM−1, and blaOXA−48 are emerging worldwide in E. coli isolated from humans or animals.
The aim of the present study was to (i) investigate the occurrence and molecular characterization of ESBL-producing MDR E. coli recovered from clinical cases of infection in dogs and cats in the United States, over a period of time ranging from August 2009 to January 2013, and (ii) characterize the association between β-lactamases, phylogenetic groups, virulence genes and the ST types.
Materials and Methods
Bacterial Isolates
Between August 2009 and January 2013, a total of 2443 E. coli isolates from urine, wound, ear, genital tract, anal sac, nasal structure, and soft tissue samples of dogs and cats with presumed naturally-occurring infection in six geographical regions of the United States: West (California), South (North Carolina), Central (Missouri), Midwest (Ohio and Illinois), and Southeast (Alabama), Northeast (Massachusetts) were received from a nationally recognized veterinary diagnostic laboratory. Isolates were reconfirmed to be E. coli upon receipt by the Clinical Pharmacology Laboratory (CPL) at Auburn University based on reculture overnight on CHROMagar Orientation (BD Diagnostics, Franklin Lakes, NJ) at 37°C, and then the isolates were harvested and stored in tryptic soy broth containing 30% glycerol at −80°C until studied.
Susceptibility Testing and Initial ESBL Identification
Antimicrobial susceptibility testing was performed for all 2443 isolates using 96 well custom microdilution susceptibility plates according to the manufacturer's protocol (Trek Diagnostic Systems, Inc., Cleveland, OH). Susceptibility testing was performed using 16 antimicrobials representing six antimicrobial classes and classified into 12 antimicrobial categories: penicillins: ampicillin; penicillins + β-lactam inhibitor: amoxicillin-clavulanic acid; anti-pseudomonal + β-lactam inhibitor: ticarcillin-clavulanic acid; non-extended spectrum cephalosporins (1st generation cephalosporins): cephalothin; extended-spectrum cephalosporins (3rd and 4th generation cephalosporins): cefotaxime, ceftazidime, and cefpodoxime; cephamycins: cefoxitin; carbapenems: meropenem; tetracyclines: doxycycline; phenicols: chloramphenicol; fluoroquinolones: enrofloxacin and ciprofloxacin; aminoglycosides: gentamicin and amikacin; and folate pathway inhibitor: sulfamethoxazole-trimethoprim (Magiorakos et al., 2012; Thungrat et al., 2015). All MIC determinations were performed in triplicates and E. coli ATCC 25922 was used for quality control. The results were interpreted according to the guidelines of Clinical Laboratories Standards Institute (CLSI; CLSI, 2013). The MICs were recorded using the Sensititre Vizion system (Trek Diagnostic Systems), and each isolate was categorized in terms of its resistant phenotype as to: susceptible (S), non-multidrug resistance (DR) or MDR. DR was defined as resistance to 1 or 2 antimicrobial classes, and MDR was defined as resistance to three or more antimicrobial classes.
Additionally, all the 2443 E. coli isolates were screened for ESBL production using microdilution-based Sensititre (TREK diagnostic systems, Cleveland, Ohio) with ESBL Confirmatory MIC plates (ESB1F) as described previouly (Aly et al., 2012). Finally, the ESBL-producing isolates expressed MDR phenotype were used in the current study.
Phylogenetic Grouping and Virulence Genotyping
The distribution of phylogenetic groups amongest the ESBL-producing MDR isolates was determined by the new quadruplex PCR as recently described by Clermont et al. (Clermont et al., 2013). Genomic DNA were extracted from bacterial preparations using the PreMan® Ultra Preparation Reagent according to the manufacturer's protocol. The presence of 17 virulence genes [fimH, sfa/focDE, afa/draBC, papA, papC, papG alleles (I, II, III), hlyA, cnf1, kpsM II, fyuA, iutA, ireA, iroN, traT, and malX] known for their association with pathogenicity ExPEC isolates was ascertained in each isolate by use of established PCR assay as reported previously (Johnson and Stell, 2000; Liu et al., 2015).
Identification of β-Lactamase Genes and Other Resistance Genes
The occurrence of β-lactamase genes blaCTX−Ms, blaTEM, blaSHV, blaCMY−2, blaKPC, blaNDM−1, and blaOXA−48 were identified by PCR and subsequent sequencing using specific primers and conditions previously described (Yan et al., 2004; Poirel et al., 2011; Shaheen et al., 2011). Furthermore, the identification of plasmid-mediated quinolone resistance (PMQR) genes [qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr and qepA] was carried out as described previously (Liu et al., 2012).
Transfer of Resistance Genes by Conjugation
We tested whether the ESBL-producing E. coli isolates harboring blaCTX−Ms, blaTEM, blaSHV, or blaOXA−48 enzymes were transferable. Conjugation was performed by broth mating at 37°C on 10 ESBL-producing MDR isolates using plasmid-free sodium azide resistant E. coli J53 (J53 AZr) as recipient as described previously (Shaheen et al., 2011). Transconjugants were selected on tryptic soy agar plates supplemented with sodium azide (150 μg/ml) and cefotaxime (2 μg/ml). Antimicrobial susceptibility, confirmatory tests for ESBL production, and PCR detection of ESBL genes were performed on all transconjugants as described to confirm transfer of ESBL genes.
Multilocus Sequence Typing (MLST)
MLST was performed using seven conserved housekeeping genes of E. coli (adk, fumC, gyrB, icd, mdh, purA, and recA). A detailed scheme of the MLST procedure, including the primers, PCR conditions, allelic type and sequence type assignment methods, is available at MLST databases at the at the Warwick University website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
Statistical Analysis
Significance was determined by Pearson's Chi-squared test with Yates continuity correction using “R” software (version 3.0.1), and the level of significance was set at P < 0.05.
Results
Antimicrobial Susceptibility
Among the 2443 investigated E. coli isolates, 92 isolates (3.8%) were ESBL producers, including 68 (73.9%) MDR isolates, 20 (21.5%) DR isolates, and 4 (4.4%) S isolates. Among the 68 ESBL-producing MDR isolates (including 52 dog and 16 cat isolates), 97.1% (66/68) isolates were resistance to cephalothin, followed by cefotaxime (94.1%), ampicillin (92.6%), cefpodoxime (91.2%), amoxicillin-clavulanic acid (86.8%), ticarcillin-clavulanic acid (85.3%), ceftazidime (69.1%), cefoxitin (44.1%), and meropenem (17.6%). Moreover, some of the investigated ESBL-producing MDR isolates were also resistant to non-β-lactam agents, including ciprofloxacin (91.2%), doxycycline (88.2%), enrofloxacin (82.4%), sulfamethoxazole-trimethoprim (50%), chloramphenicol (44.1%), gentamicin (39.7 %), and amikacin (30.9%).
Phylogenetic Groups and the Virulence Genes Distribution
Phylogenetic analysis showed that the predominant phylogenetic groups were D (35.3%) and B2 (33.8%), followed by C (11.8%), A (10.3%), B1 (5.9%), E (1.5%), and F (1.5%). Fourteen of seventeen investigated virulence genes were detected, with the overall occurrence ranging from 11.8% (ireA) to 88.2% (malX) with the exception of papG I and papG II, which were not detected in any isolate. The isolates of phylogenic group B2 harbored more virulence genes (mean 7.3), and followed by group B1 (mean 5.8), group D (mean 5.6), group A (mean 4.7), and group C (mean 3.8). Furthermore, CTX-M-producing isolates possessed more virulence genes (mean 8.4) than did non-CTX-M isolates (mean 4.0; P < 0.0001). Several virulence genes, including sfa/focDE, afa/draBC, papA, papC, papG III, hlyA, cnf1, and iroN, were significantly more common or even exclusively present in non-CTX-M-producing isolates, whereas traT was significantly more common in CTX-M-producing isolates than in non-CTX-M isolates (72 vs. 22.2%, P = 0.0008). Additionally, the occurrence of virulence genes among ESBL-producing MDR E. coli was significantly lower than among non-ESBL isolates with the exception of malX (P < 0.01; Table 1).
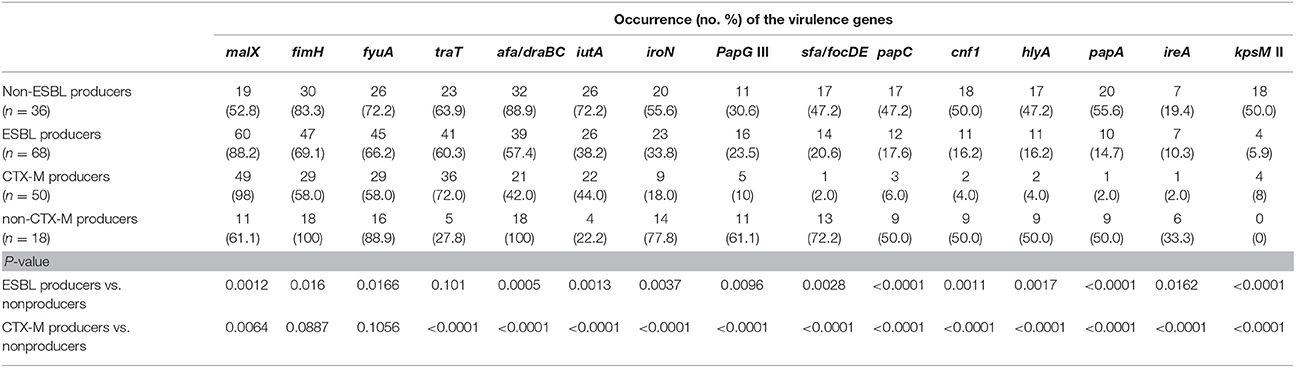
Table 1. The occurrence of virulence genes in the ESBL-producing and non-ESBL-producing MDR E. coli isolates.
Distribution of β-Lactamases and PMQR Genes
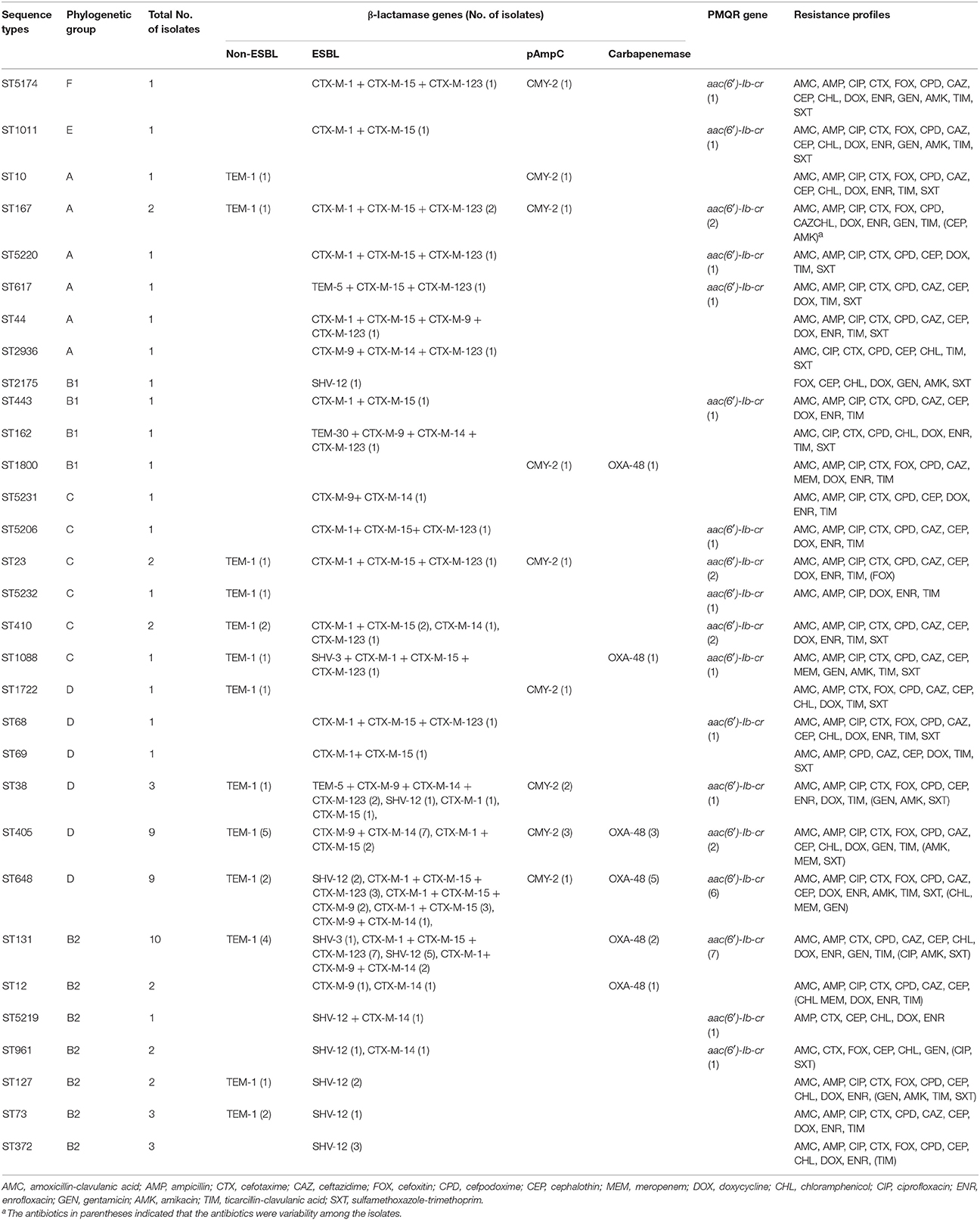
The distribution of β-lactamase and PMQR genes among the 68 ESBL-positive MDR E. coli isolates was shown in Table 2. The results showed that blaTEM, blaSHV, blaCTX−M, blaCMY−2, and blaOXA−48 were detected in 28 (41.2%), 20 (29.4%), 50 (73.5%), 12 (17.6%), and 13 (19.1%) isolates, respectively. 94.1% (64/68) of the isolates harbored two or more β-lactamase genes, and one isolate from dog with severe urinary tract infection co-harbored eight tested genes [blaTEM−5, blaSHV−12, blaCMY−2, blaCTX−M−15, blaCTX−M−1, blaCTX−M−14, blaCTX−M−123, and aac(6′)-Ib-cr; Table 2]. For the blaCTX−M positive isolates, CTX-M enzymes were clustered in CTX-M-1 (n = 35), CTX-M-9 (n = 22), and hybrid β-lactamases (n = 27) clusters. CTX-M-1 and CTX-M-9 double-positive group accounted for 10.3% of isolates, and three isolates co-harbored CTX-M-1, CTX-M-9 as well as hybrid β-lactamase. blaCTX−M−15 (n = 33) was the predominant genotype in blaCTX−M positive isolates, and followed by blaCTX−M−1 (n = 32), blaCTX−M−123 (n = 27), blaCTX−M−9 (n = 19), and blaCTX−M−14 (n = 19). Sequencing of blaTEM gene revealed 24 blaTEM−1, three blaTEM−5, and one blaTEM−30, whereas sequencing of blaSHV gene revealed 17 blaSHV−12 and two blaSHV−3. All blaTEM and blaSHV genes were classfied as ESBLs with the exception of the blaTEM−1 gene based on the sequencing. Moreover, 48.5% (33/68) of investigated isolates harbored aac(6′)-Ib-cr, while none of the isolates carried qnr and qepA genes. The vast majority of aac(6′)-Ib-cr-producing isolates were positive for blaCTX−M−15, blaCTX−M−1, and blaCTX−M−123, but negative for CTX-M 9 group enzymes despite blaCTX−M−14 and aac(6′)-Ib-cr coexisted in three isolates.
Conjugation Experiments
We tested whether blaCTX−M genes or other β-lactamase genes in 10 selected isolates were transferable by conjugation experiments, and seven out of the 10 ESBL-producing isolates successfully transferred the β-lactamase genes to the recipient E. coli. PCR analysis showed the presence of respective blaCTX−M genes and other β-lactamase genes, including two blaOXA−48-carrying plasmids from all the transconjugants (Table 3). Meanwhile, PMQR gene aac-(6')-Ib-cr was co-transferred with β-lactamase genes. Generally, all donors and their transconjugants were resistant to amoxicillin-clavulanic acid, ampicillin, cefotaxime, cefoxitin, cefpodoxime, cephalothin, and ticarcillin-clavulanic acid, and all transconjugants exhibited an increase of at least eight-fold in MICs compared to the recipient, E. coli J53 AZr. The ciprofloxacin MICs for four transconjugants harboring aac-(6′)-Ib-cr ranged from 0.06 to 0.125 mg/L, representing an increase of two-fold to four-fold compared with the recipient (Table 3). Additionally, the transconjugants remained susceptible to meropenem, ciprofloxacin, gentamicin, chloramphenicol, and doxycycline, whereas one transconjugant harboring blaOXA−48 was resistant to sulfamethoxazole-trimethoprim and reduced the susceptibility to meropenem.
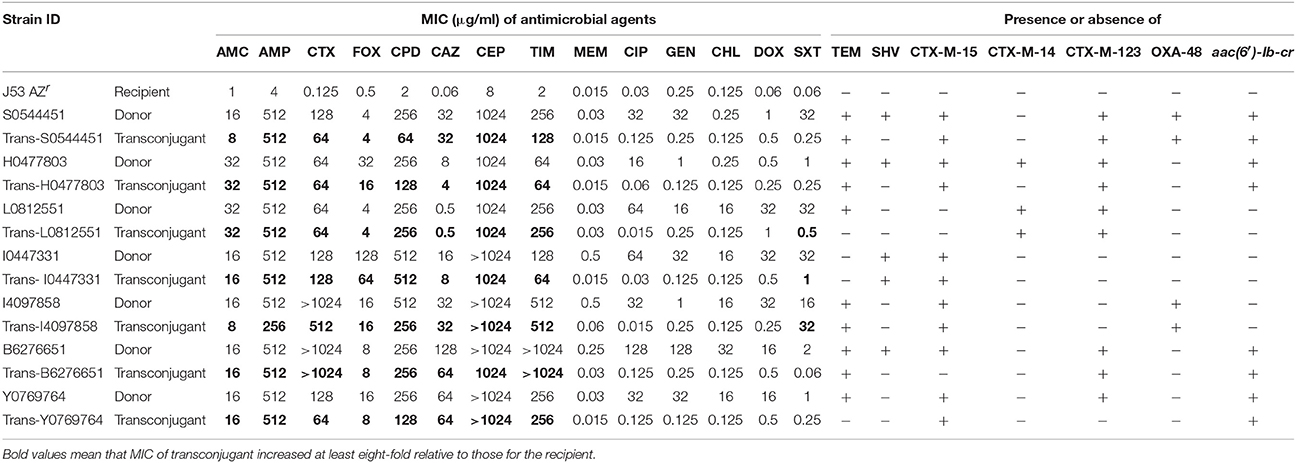
Table 3. Antibimicrobial susceptibility testing profiles of E. coli isolates used in the conjugation experiments.
MLST
The MLST investigation revealed that the 68 isolates were assigned to 31 STs, including six new STs (Table 2). Twelve STs were represented by more than two isolates, and other 19 STs contained a single isolate each. ST131 (n = 10), ST648 (n = 9), and ST405 (n = 9) accounted for more than 41% (28/68) of investigated isolates and 54% (27/50) of CTX-M-producing isolates, respectively. 74.2% (23/31) of STs, especially ST131, ST648, and ST405 were positively associated with CTX-M-producing isolates, while other STs, including ST10, ST5232, ST1722, ST2175, ST1800, ST73, ST372, and ST127 seem to have no relationship with CTX-Ms. Vast majority of ST131 and ST648 isolates were positively associated with blaCTX−M−15 and/or blaCTX−M−1 as well as blaCTX−M−123, whereas 77.8% of ST405 isolates were negatively associated with blaCTX−M−1, blaCTX−M−15, and blaCTX−M−123 genes. Moreover, 55.6% of ST648 isolates were positively associated with blaOXA−48, and 12 pAmpC genes blaCMY−2 were distributed in nine STs. Notably, all ST131, ST405, and ST648 isolates expressed resistance to ciprofloxacin and 3rd generation cephalosporins, whereas all ST131 isolates remained susceptible to cefoxitin. A strong correlation was revealed between the virulence gene profiles and STs, and the same STs showed the similar virulence gene profiles. Among the three most common STs, ST405 isolates harbored more virulence genes (mean 4.6), followed by ST131 (mean 4.4), and virulence genes were less abundant in ST648 isolates (mean 3.4). Almost all of the ST131 and ST405 isolates were positive for afa/draBC, traT, and malX genes, ST648 isolates were significantly associated with fimH, malX and traT, but negative for afa/draBC.
Discussion
ESBLs, pAmpC and carbapenemases are mostly responsible for the emerging resistance to the β-lactam antibiotics, especially the 3rd generation cephalosporins and carbapenems in E. coli (Pitout, 2012). In the present study, we conducted a molecular detection and characterization of the β-lactamase genes in ESBL-producing MDR E. coli isolates from dogs and cats in the United States over a period of time ranging from August 2009 to January 2013, and also revealed the association between the phylogenetic groups, virulence gene profiles, genetic backbones and β-lactamase types.
The prevalence of 3.8% ESBL-producing E. coli found in this study is similar to that recorded in a recent study (3%; Shaheen et al., 2011) but higher than the first survey (1%; O'Keefe et al., 2010) among E. coli from dogs and cats in the United States. Surprisingly, 73.9% (68/92) of the ESBL-producing E. coli exhibited MDR phenotype, and 75% of MDR isolates were resistant to more than 10 antimicrobial agents tested. Phylogenetic groups D and B2 were the main phylogenetic groups in this study, and it was similar to the phylogenetic subtype distribution of the ESBL-producing isolates from human patients (Hu et al., 2013), which further demonstrated that isolates in phylogenetic groups D and B2 were associated with extraintestinal infections. Among the 68 ESBL-producing MDR isolates, blaCTX−M was prominent and detected in 73.5% (50/68) of isolates, whereas two previous similar surveys carried out in different states in the United States showed that the corresponding prevalence of blaCTX−M were 16.7% and 89.7%, respectively (O'Keefe et al., 2010; Shaheen et al., 2011). It is indicated that the geographical regions, time, resistant phenotype and the history of antimicrobial treatment of the animals can affect the prevalence of blaCTX−M gene. The high prevalence of blaCTX−M strongly suggests a significant role for E. coli isolates from companion animals as ESBL gene reservoirs, which poses an additional risk to humans. Therefore, monitoring of the spread of blaCTX−M genes in E. coli isolates in dogs and cats is urgently needed. Althouth blaCTX−M−15 was still the most frequently encountered gene, the specific genotype of blaCTX−M is undergoing changes, which was supported by available evidence from the occurrence of CTX-M-9 group as well as the occurrence of a novel hybrid β-lactamase gene blaCTX−M−123. blaCTX−M−123 was firstly discovered in E. coli from pig feces in China in 2013 (He et al., 2013), and afterward in human specimen (Hu et al., 2013). It is interesting to note that blaCTX−M−15 is also the most widely distributed ESBL gene among human-associated Enterobacteriaceae (Cantón and Coque, 2006). These finding revealed the possibility of cross-transmission between animals and humans. Moreover, several isolates appear only with blaTEM−1, blaCMY−2, or blaOXA−48, suggesting that these isolates perhaps carry other ESBL genes, which will require further studies.
blaCMY−2 was the most prevalent pAmpC, and it not only confer resistance to a wide range of extended-spectrum cephalosporins but also are not affected by β-lactamase inhibitors. blaCMY−2 was dected in 17.6% of the isolates in our study, and it was significantly lower than the occurance of blaCMY−2 (89%) in E. coli from companion animals in a previous study in the United States (Shaheen et al., 2011). We supposed that the occurance of blaCMY−2 might be underestimated since only the ESBL-producing MDR isolates were characterized in this study. Meanwhile, our results showed that majority (58.3%) of CMY-2-producing isolates belonged to phylogenetic group D, consistent with a previous study in E. coli from human in Australia (Sidjabat et al., 2014). This similar distribution of phylogenetic group further certified that blaCMY−2 can also be transferred between different bacterial species and between animals and humans (Li et al., 2007; Shaheen et al., 2011). blaOXA−48 was initially reported in Klebsiella pneumoniae isolates in Turkey in 2001(Poirel et al., 2004) and afterward in other Mediterranean countries (Spain, France, Italy, Egypt, and Lebanon Turkey) (Girlich et al., 2014). In 2013, it was firstly discovered in E. coli from dogs in Germany (Stolle et al., 2013). blaOXA−48 can hydrolyze carbapenems and β-lactamase inhibitors but has no activity toward broad-spectrum cephalosporins (Mathers et al., 2013). Our data showed that about 19% of the isolates carried the blaOXA−48, and they were mostly associated with meropenem resistance, sequence types ST648, ST405, and ST131 as well as different combinations of β-lactamase genes. To our knowledge, blaOXA−48 was firstly reported in the United States in 2012 (Poirel et al., 2012), and the present study is the first report of blaOXA−48 in E. coli from dogs and cats in the United States. Moreover, blaOXA−48 can transfer with other β-lactamases and aac(6′)-Ib-cr. This finding also revealed possibility of the transfer between humans and companion animals appears highly probable through multiple potential pathways although blaOXA−48 is still sporadic occurrence in animals.
aac(6′)-Ib-cr was the exclusive PMQR gene in this study, and CTX-M-producing isolates (particularly blaCTX−M−15 positive isolates) showed significantly higher occurrence of aac(6′)-Ib-cr compared to non-CTX-M or non-ESBL isolates (62 vs. 11.1 vs. 10%, P < 0.001). The frequent combination of blaCTX−M−15 and aac(6′)-Ib-cr in this study further supported the previous studies that coproduction of β-lactamases and PMQR genes could conduce to the dissemination of MDR isolates, and also reflect the fact that genes encoding resistance to β-lactams and quinolones are located on the same plasmid. Although it was not the primary focus of this study, our results coincided with a previous study (Qin et al., 2013) that ESBL-producing isolates presented a lower occurrence of studied virulence genes compared with non-ESBL isolates (the data from another study in our laboratory) with the exception of malX gene, and CTX-M-producing E. coli harbored fewer virulence genes than non-CTX-M isolates (P < 0.0001). A possible reason why individual virulence gene increased among ESBL-producing is that might be a fitness trade-off for the ESBL to survive antibiotics exposure (Qin et al., 2013) and the difference source of E. coli. The exact explanation needs additional study in the future.
A previous review suggested that attention should be paid to the rising of E. coli ST131, ST648, ST405, and ST38 isolates as they can play an important role in the worldwide distribution of CTX-M-producing E. coli (Pitout, 2012). It was further confirmed by our results since ST131, ST648, and ST405 accounted for 54% of the CTX-M-producing MDR isolates. ST131 was the predominant clone in this study, and all ST131 isolates remained susceptible to cefoxitin, which has been recently suggested as an alternative carbapenems for the treatment of infections by ESBL-producing E. coli (Guet-Revillet et al., 2014). It is noteworthy that nine ST648 isolates were strongly associated with blaCTX−M−15 (88.9%, 8/9), blaOXA−48 (55.6%, 5/9), and severe clinical signs. The zoonotic potential of ST648 ESBL-producing isolates has been indicated in the isolates from humans, domestic and wild animals in previous studies (Nicolas-Chanoine et al., 2008; Cortes et al., 2010), and two recent studies in Europe further suggested that ST648 clone may represent a novel genotype that combines MDR phenotype, extraintestinal virulence and zoonotic potential in companion animals (Huber et al., 2013; Ewers et al., 2014). Furthermore, ST131, ST648, and ST405 isolates have the similar β-lactamase gene combinations and resistance profiles, respectively. While it is alarming that other STs have various β-lactamase gene combinations, especially one ST38 isolate, which was associated with the highest frequency of β-lactamases and aac(6′)-Ib-cr, high level cephalosporins resistance (MICs ≥ 32 μg/ml), lowest frequency of virulence genes and severe clinical signs. Nevertheless, constant attention and further investigations for ST648 and ST38 isolates in companion animals are necessary as they are now rapidly and globally disseminated as well as the companion animals are more and more considered an important source of human infections as the physical closeness.
Conclusion
CTX-M-producing E. coli tend to have less virulent properties compared with the non-CTX-M isolates. CTX-Ms represented by blaCTX−M−1, blaCTX−M−15, and blaCTX−M−123 have spread rapidly. The occurrence of blaCTX−M−123 of ESBLs and blaOXA−48 carbapenemase were particularly striking, being reported here for the first time in E. coli from dogs and cats in the United States. ST131, ST648, and ST405 were the predominant clonal groups among the ESBL-producing E. coli, and all ST131 isolates remained susceptible to the cefoxitin. This information will be useful for assessing the epidemiological risk factors and appropriate use of antimicrobials for ESBL-producing E. coli infections of dogs and cats.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge the assistance of laboratory staff in the Clinical Pharmacology Laboratory of Auburn University. This study was supported by the Fundamental Research Funds for the Central Universities (no. 2452016044), National Science Foundation of Shaanxi province (no. 2014JM3071), and partially supported by from Morris Animal Foundation (no. D07-MS006).
References
Aly, S. A., Debavalya, N., Suh, S. J., Oryazabal, O. A., and Boothe, D. M. (2012). Molecular mechanisms of antimicrobial resistance in fecal Escherichia coli of healthy dogs after enrofloxacin or amoxicillin administration. Can. J. Microbiol. 58, 1288–1294. doi: 10.1139/w2012-105
Cantón, R., and Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
CLSI (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacterial Isolated from Animals; Approved Standard. 4th Edn and Supplement, CLSI Document VET01-A4 (Standard) and VET01-S2 (Supplement). Wayne, PA: Clinical and Laboratory Standards Institute.
Cortés, P., Blanc, V., Mora, A., Dahbi, G., Blanco, J. E., Blanco, M., et al. (2010). Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76, 2799–2805. doi: 10.1128/AEM.02421-09
Ewers, C., Bethe, A., Stamm, I., Grobbel, M., Kopp, P. A., Guerra, B., et al. (2014). CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 69, 1224–1230. doi: 10.1093/jac/dkt516
Geser, N., Stephan, R., and Hächler, H. (2012). Occurrence and characteristics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8:21. doi: 10.1186/1746-6148-8-21
Girlich, D., Bouihat, N., Poirel, L., Benouda, A., and Nordmann, P. (2014). High rate of faecal carriage of extended-spectrum beta-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin. Microbiol. Infect. 20, 350–354. doi: 10.1111/1469-0691.12325
Guet-Revillet, H., Emirian, A., Groh, M., Nebbad-Lechani, B., Weiss, E., Join-Lambert, O., et al. (2014). Pharmacological study of cefoxitin as an alternative antibiotic therapy to carbapenems in treatment of urinary tract infections due to extended-spectrum-beta-lactamase-producing Escherichia coli. Antimicrob. Agents Chemother. 58, 4899–4901. doi: 10.1128/AAC.02509-14
He, D., Partridge, S. R., Shen, J., Zeng, Z., Liu, L., Rao, L., et al. (2013). CTX-M-123, a novel hybrid of the CTX-M-1 and CTX-M-9 Group beta-lactamases recovered from Escherichia coli isolates in China. Antimicrob. Agents Chemother. 57, 4068–4071. doi: 10.1128/AAC.00541-13
Hu, Y. Y., Cai, J. C., Zhou, H. W., Chi, D., Zhang, X. F., Chen, W. L., et al. (2013). Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 79, 5988–5996. doi: 10.1128/AEM.01740-13
Huber, H., Zweifel, C., Wittenbrink, M. M., and Stephan, R. (2013). ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet. Microbiol. 162, 992–996. doi: 10.1016/j.vetmic.2012.10.029
Johnson, J. R., and Stell, A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181, 261–272. doi: 10.1086/315217
Karisik, E., Ellington, M. J., Livermore, D. M., and Woodford, N. (2008). Virulence factors in Escherichia coli with CTX-M-15 and other extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 61, 54–58. doi: 10.1093/jac/dkm401
Li, X. Z., Mehrotra, M., Ghimire, S., and Adewoye, L. (2007). beta-Lactam resistance and beta-lactamases in bacteria of animal origin. Vet. Microbiol. 121, 197–214. doi: 10.1016/j.vetmic.2007.01.015
Liu, X., Boothe, D. M., Thungrat, K., and Aly, S. (2012). Mechanisms accounting for fluoroquinolone multidrug resistance Escherichia coli isolated from companion animals. Vet. Microbiol. 161, 159–168. doi: 10.1016/j.vetmic.2012.07.019
Liu, X., Thungrat, K., and Boothe, D. M. (2015). Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PLoS ONE 10:e0143335. doi: 10.1371/journal.pone.0143335
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mathers, A. J., Hazen, K. C., Carroll, J., Yeh, A. J., Cox, H. L., Bonomo, R. A., et al. (2013). First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the new world. J. Clin. Microbiol. 51, 680–683. doi: 10.1128/JCM.02580-12
Nicolas-Chanoine, M. H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Canica̧, M. M., et al. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281. doi: 10.1093/jac/dkm464
O'Keefe, A., Hutton, T. A., Schifferli, D. M., and Rankin, S. C. (2010). First detection of CTX-M and SHV extended-spectrum beta-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob. Agents Chemother. 54, 3489–3492. doi: 10.1128/AAC.01701-09
Pitout, J. D. (2012). Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front. Microbiol. 3:9. doi: 10.3389/fmicb.2012.00009
Poirel, L., Héritier, C., Tolun, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Poirel, L., Potron, A., and Nordmann, P. (2012). OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67, 1597–1606. doi: 10.1093/jac/dks121
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Qin, X., Hu, F., Wu, S., Ye, X., Zhu, D., Zhang, Y., et al. (2013). Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS ONE 8:e61169. doi: 10.1371/journal.pone.0061169
Schmiedel, J., Falgenhauer, L., Domann, E., Bauerfeind, R., Prenger-Berninghoff, E., Imirzalioglu, C., et al. (2014). Multiresistant extended-spectrum beta-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 14:187. doi: 10.1186/1471-2180-14-187
Shaheen, B. W., Nayak, R., Foley, S. L., Kweon, O., Deck, J., Park, M., et al. (2011). Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob. Agents Chemother. 55, 5666–5675. doi: 10.1128/AAC.00656-11
Sidjabat, H. E., Seah, K. Y., Coleman, L., Sartor, A., Derrington, P., Heney, C., et al. (2014). Expansive spread of IncI1 plasmids carrying blaCMY−2 amongst Escherichia coli. Int. J. Antimicrob. Agents 44, 203–208. doi: 10.1016/j.ijantimicag.2014.04.016
Stolle, I., Prenger-Berninghoff, E., Stamm, I., Scheufen, S., Hassdenteufel, E., Guenther, S., et al. (2013). Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 68, 2802–2808. doi: 10.1093/jac/dkt259
Thungrat, K., Price, S. B., Carpenter, D. M., and Boothe, D. M. (2015). Antimicrobial susceptibility patterns of clinical Escherichia coli isolates from dogs and cats in the United States: January 2008 through January 2013. Vet. Microbiol. 179, 287–295. doi: 10.1016/j.vetmic.2015.06.012
Yan, J. J., Hong, C. Y., Ko, W. C., Chen, Y. J., Tsai, S. H., Chuang, C. L., et al. (2004). Dissemination of blaCMY-2 among Escherichia coli isolates from food animals, retail ground meats, and humans in southern Taiwan. Antimicrob. Agents Chemother. 48, 1353–1356. doi: 10.1128/AAC.48.4.1353-1356.2004
Keywords: Escherichia coli, ESBL, OXA-48 carbapenemase, multidrug resistance, companion animals
Citation: Liu X, Thungrat K and Boothe DM (2016) Occurrence of OXA-48 Carbapenemase and Other β-Lactamase Genes in ESBL-Producing Multidrug Resistant Escherichia coli from Dogs and Cats in the United States, 2009–2013. Front. Microbiol. 7:1057. doi: 10.3389/fmicb.2016.01057
Received: 15 March 2016; Accepted: 23 June 2016;
Published: 11 July 2016.
Edited by:
Benoit Doublet, National Institute for Agricultural Research, FranceReviewed by:
Jean-Yves Madec, ANSES-French Agency for Food, Environmental and Occupational Health & Safety, FranceAngela Novais, UCIBIO-Research Unit on Applied Molecular Biosciences, Portugal
Copyright © 2016 Liu, Thungrat and Boothe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqiang Liu, liuxiaoqiang142@163.com
 Xiaoqiang Liu
Xiaoqiang Liu Kamoltip Thungrat2
Kamoltip Thungrat2