- 1Division of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA, USA
- 2Life and Environmental Sciences, School of Natural Science, University of California at Merced, Merced, CA, USA
Hydrogen atoms from water and food are incorporated into biomass during cellular metabolism and biosynthesis, fractionating the isotopes of hydrogen—protium and deuterium—that are recorded in biomolecules. While these fractionations are often relatively constant in plants, large variations in the magnitude of fractionation are observed for many heterotrophic microbes utilizing different central metabolic pathways. The correlation between metabolism and lipid δ2H provides a potential basis for reconstructing environmental and ecological parameters, but the calibration dataset has thus far been limited mainly to aerobes. Here we report on the hydrogen isotopic fractionations of lipids produced by nitrate-respiring and sulfate-reducing bacteria. We observe only small differences in fractionation between oxygen- and nitrate-respiring growth conditions, with a typical pattern of variation between substrates that is broadly consistent with previously described trends. In contrast, fractionation by sulfate-reducing bacteria does not vary significantly between different substrates, even when autotrophic and heterotrophic growth conditions are compared. This result is in marked contrast to previously published observations and has significant implications for the interpretation of environmental hydrogen isotope data. We evaluate these trends in light of metabolic gene content of each strain, growth rate, and potential flux and reservoir-size effects of cellular hydrogen, but find no single variable that can account for the differences between nitrate- and sulfate-respiring bacteria. The emerging picture of bacterial hydrogen isotope fractionation is therefore more complex than the simple correspondence between δ2H and metabolic pathway previously understood from aerobes. Despite the complexity, the large signals and rich variability of observed lipid δ2H suggest much potential as an environmental recorder of metabolism.
Introduction
The hydrogen isotopic compositions of organic molecules, primarily lipids, have frequently been used as indicators of biosynthetic processes (Chikaraishi and Naraoka, 2001; Chikaraishi et al., 2009), patterns of organic matter cycling (Chikaraishi and Naraoka, 2005; Jones et al., 2008), and as proxies for the isotopic composition of water in paleoenvironmental reconstructions (Sauer et al., 2001; Huang et al., 2002; Sachse et al., 2012). To first order, the hydrogen isotopic composition (expressed as a δ2H-value) of organic matter reflects that of growth water, offset by some (often large) fractionation associated with biosynthesis (Estep and Hoering, 1980; Sessions et al., 1999; Hayes, 2001). However, very large ranges of δ2H-values are observed in natural samples, even those occurring in a single species or environment, indicating that other processes are important as well (Jones et al., 2008; Li et al., 2009; Osburn et al., 2011). Known sources of variability include environmental conditions that may change the isotopic composition of intracellular water, such as salinity in planktonic organisms and aquatic macrophytes or humidity and transpiration in plants (Sachse and Sachs, 2008; Pu and Weiguo, 2011; Romero-Viana et al., 2013), or changes in biosynthetic fractionation such as between isoprenoid and n-alkyl lipids (Sessions et al., 1999). In addition, it has been shown that the central metabolic pathways used by microorganisms to process organic substrates are correlated with lipid δ2H (Zhang et al., 2009). These metabolism-associated isotope effects span up to a 500‰; range for a specific fatty acid in a single microbial strain, far exceeding most other forms of variability.
Patterns of lipid δ2H proposed by Zhang et al. (2009) include strong 2H-enrichment of lipids in aerobic heterotrophs, strong 2H-depletion in chemoautotrophs, and intermediate 2H-depletion by photoautotrophs. During heterotrophic growth, the level of 2H-enrichment is loosely correlated with the central metabolic pathway being activated by a particular substrate, with growth on tricarboxylic acid (TCA) cycle intermediates like acetate and succinate producing the most extreme 2H-enrichments (Zhang et al., 2009). Such trends have subsequently been replicated in other (aerobic) bacterial strains (Heinzelmann et al., 2015a,b), an archaeon (Dirghangi and Pagani, 2013b), and a heterotrophic eukaryote (Dirghangi and Pagani, 2013a). Isotope effects associated with the enzymatic protonation (reduction) of NADP+ during central metabolism, which can lead to either 2H depletions or enrichments in NADPH (Zhang et al., 2009), have been hypothesized to explain metabolic trends in hydrogen isotopes. This hydrogen is then transferred to lipids as an intact hydride donor in biosynthetic reactions (White, 2000; Zhang et al., 2009), facilitating transfer of the isotope label. Several recent culturing studies support this hypothesis (Dirghangi and Pagani, 2013b; Dawson et al., 2015; Heinzelmann et al., 2015a,b).
Using hydrogen isotopic fractionations as a proxy for microbial metabolism has considerable potential utility as an environmental indicator, extending the role of compound specific lipid δ2H beyond recording environmental water, to recording metabolic diversity. Because lipids have far greater preservation potential than nucleic acids, they may provide a coarser bur much longer-lasting record than do genomic approaches. Evaluation of these trends in modern environmental samples including hot springs (Naraoka et al., 2010; Osburn et al., 2011), marine particulate organic carbon (POC; Jones et al., 2008), and marine sediments (Li et al., 2009; Heinzelmann et al., 2015c) have revealed large ranges in lipid δ2H that are potentially attributable to heterotrophic 2H-enrichment and chemoautotrophic 2H-depletion. In addition, recent culturing studies have shown the metabolic effects to be larger in magnitude than those stemming from growth phase or salinity (Dirghangi and Pagani, 2013a,b; Chivall et al., 2014; Dawson et al., 2015; Heinzelmann et al., 2015a,b).
While these initial studies are promising, they include data primarily from aerobic heterotrophs and chemoautotrophs (Zhang et al., 2009; Heinzelmann et al., 2015b). Microbes with anaerobic metabolism are widespread in both marine and terrestrial environments and are often of great interest in reconstructing paleoenvironmental and paleoecological conditions. To date, published data for anaerobic growth includes the following: three photoheterotrophs (Zhang et al., 2009; Heinzelmann et al., 2015b), three sulfate reducers (Campbell et al., 2009; Dawson et al., 2015; Leavitt et al., 2016), and an acetogen (Valentine et al., 2004), primarily phototrophic or chemoautotrophic microbes. Dawson et al. (2015) analyzed the anaerobic heterotroph Desulfococcus multivorans and measured an 80‰; range in hydrogen isotope fractionation between lipids and growth water (εL–W) for pure cultures grown on a range of heterotrophic substrates. The growth substrates that lead to the most 2H-enriched lipids were acetate and succinate, consistent with previous findings; however, εL–W averaged −137 and −127‰;, significantly 2H-depleted relative to analogous aerobic cultures. This discrepancy, along with the general lack of published information on hydrogen isotope fractionations associated with anaerobic heterotrophs, arguably a dominant metabolism in sedimentary environments worldwide, is the primary motivation for our study.
Here we report on hydrogen isotopic fractionations in lipids produced by several facultative and obligate anaerobes. The organisms chosen for this study were selected in order to make direct comparisons of autotrophic vs. heterotrophic growth under both aerobic and anaerobic conditions. Escherichia coli, Paracoccus denitrificans, and Shewanella oneidensis can be grown under nearly identical conditions with either O2 or nitrate as an electron acceptor, providing a direct comparison between aerobic and anaerobic growth. P. denitrificans can also grow chemoautotrophically. We also studied five strains of strictly anaerobic sulfate reducing bacteria (SRB), all Deltaproteobacteria but differing significantly in both their specific metabolic function and known biochemical pathways (Muyzer and Stams, 2008). They include representatives with very broad heterotrophic specificity (D. autotrophicum, D. propionicus, and D. multivorans) as well as those capable of both autotrophic and heterotrophic growth (D. autotrophicum, D. hydrogenophilus, and D. multivorans). D. alaskensis provides a direct point of comparison to the recent work of Leavitt et al. (2016). Our results suggest that significant differences in εL–W exist between nitrate-reducing and sulfate-reducing organisms. Whether such patterns are widespread among anaerobes remains to be determined.
Materials and Methods
Bacterial Strains and Culturing Conditions
Strains for growth trials were selected based on their metabolic capability, sequenced genomes, and accessibility. Five SRBs, Desulfobacterium autotrophicum (DSM 3382), Desulfobacter hydrogenophilus (DSM-3380), Desulfobulbus propionicus (DSM 2032), Desulfovibrio alaskensis (DSM 16109), and D. multivorans (DSM 2059) were purchased from the DSMZ culture collection. E. coli K-12 MG1655 is the same strain as in Zhang et al. (2009). Paracoccus denitrificans (B-3785) was supplied by the ARS culture collection. The laboratory of Kenneth Nealson at the University of Southern California supplied S. oneidensis.
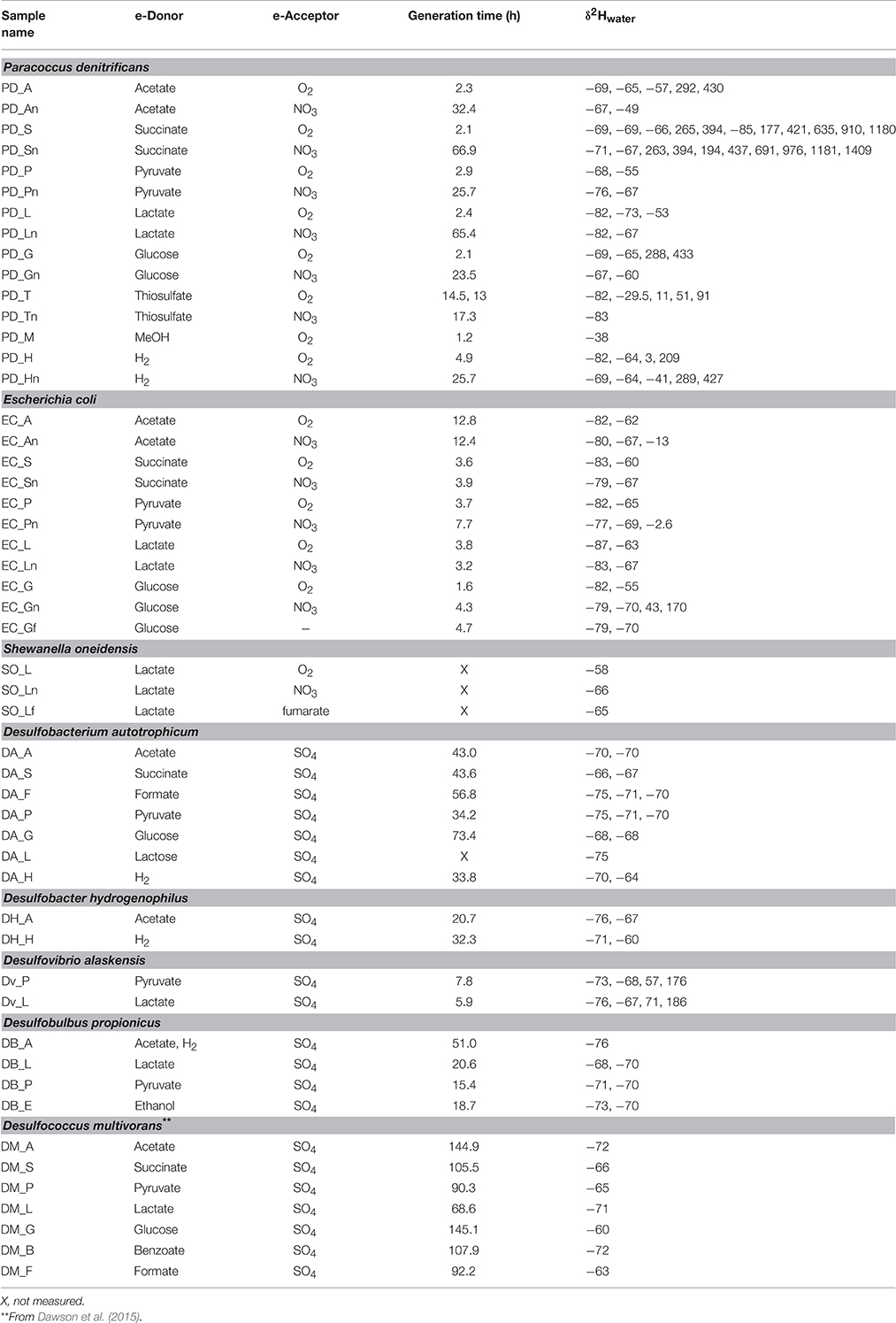
Strains were all grown in minimal media with single carbon sources (Table 1). Media were made up in deionized water (Milli-Q system), and where required the medium δ2H-value was adjusted by addition of filter-sterilized 2H2O. P. denitrificans and E. coli were grown in a low salt base (LWB) for all experiments. LWB contained per liter of water: 0.1 g NaCl, 0.5 g MgSO4·7H2O, 0.1 g CaCl2·2H2O, 0.05 g KCl, 0.03 g NH4Cl, 0.11 g KH2PO4·H2O, 1 ml vitamin solution, and 1 ml trace element solution (Breznak and Leadbetter, 2006). Sulfate-reducers were grown in Medium 383 (DSMZ) or Medium 194 (DSMZ; D. propionicus) supplementing the basal media with the trace element solution from the LWB medium, buffering with 1 g/L NaHCO3, and heterotrophic carbon source to 10 mM. Anaerobic media was cooled and prepared in an anaerobic chamber prior to flushing of the headspace with 80:20 N2:CO2 or H2:CO2 if H2 was the electron donor. Aerobic experiments were loosely capped and open to the atmosphere with the exception of cultures requiring H2 and O2, where media was degased under H2:CO2 and lab air was introduced by filter sterilization. Sulfate reducers were grown without shaking in the dark, whereas facultative anaerobes were shaken at ~120 RPM. Flasks, Balch tubes, and serum bottles for growth experiments were baked in a muffle furnace at 475°C prior to use. Cultures for lipid collection (generally 100 ml) were inoculated with 0.5–1 ml of actively growing culture before incubation at 25°C (E. coli, P. denitrificans, and S. oneidensis) or 28°C (SRBs). Growth curves were produced for each condition by monitoring optical density at 600 nm in separate smaller volumes with identical media with growth rates and doubling time calculated from regression of the exponential phase of growth curves after Widdel (2010). Cultures were harvested in late exponential or early stationary phase. Biological duplicates were produced for most growth conditions as indicated in Table 1 by multiple water isotope measurements.
Lipid Extraction and Identification
Harvested cultures were pelleted by centrifugation, frozen, lyophilized, and stored at −20°C until lipid extraction. Fatty acid methyl esters (FAMEs) were produced from cellular material in a combined extraction/derivatization procedure based on Rodríguez-Ruiz et al. (1998). For each sample, 1–100 mg of dry biomass was combined with 1 ml of hexane and 2 ml of 20:1 anhydrous methanol: acetyl chloride and reacted in a sealed glass culture tube at 100°C for 10 min. After cooling, 2 ml of water was added to the mixture followed by 3-fold extraction with 3 ml hexane. Where necessary, separation of saturated from unsaturated FAMEs was undertaken using Discovery®Ag-Ion solid phase extraction columns (Supelco), eluting the saturated, monounsaturated, and diunsaturated FAMEs in 96:4 hexane: acetone (6 ml), 90:10 hexane: acetone (4 ml), and acetone (4 ml), respectively. Fractions were evaporated to dryness under N2 and suspended in 1 ml hexane for analysis.
Fatty acids were identified by gas chromatography–mass spectrometry (GC-MS) and quantified via flame ionization detector (GC-FID). One microliter of each organic extract was injected into a ThermoFinnigan Trace GC with the effluent split ~9:1 between a DSQ mass spectrometer and FID. The sample was injected into a programmable temperature vaporization (PTV) injector operated in splitless mode and heated to 330°C in 24 s. The GC was equipped with a ZB-5 ms capillary column (30 m long, 0.25 mm I.D., 0.25 μm film) and was operated with a He carrier gas flow rate of 0.8 ml/min. The oven temperature was held for 1 min at 100°C, ramped at 20°C/min to 140°C, ramped at 3.0°C/min to 250°C and held for 1 min, then ramped at 20°C/min to 310°C and held for 10 min. Compounds were identified by comparison of mass spectra to the NIST 2004 library and/or by retention time to authentic standards. Samples were quantified relative to a known amount of either palmitic acid isobutyl ester (PAIBE) or a C23 n-alkane internal standard. The position and stereochemistry of double bonds was not determined.
Isotopic Analyses
Samples of culture medium for water hydrogen isotope analysis were collected either prior to incubation, or post-incubation by filtering through a 0.2 μm syringe filter or by collection of centrifuge supernatant after pelleting cells. Waters were analyzed using a Los Gatos DLT-100 spectroscopic Liquid Water Isotope Analyzer in eight-fold replicate against appropriate working standards (δ2H = −117.0‰;, −10.6, +287.3, and +457.6). Spectral interference was noted only for cultures containing MeOH, for which measurements of isotopic composition were made prior to adding the carbon substrate. Measured isotope ratios were converted to δ2H-values by comparison with the four standards, and normalized to the VSMOW-SLAP scale. Typical precision for these analyses was 1–2‰. All data reduction was performed using Visual Basic code written by us.
δ2H-values of FAMEs were measured using a ThermoFinnigan Trace GC coupled to a Delta+XP isotope ratio mass spectrometer (IRMS) via a pyrolysis interface (GC/TC) operated at 1391°C. External FAME standards were analyzed after every four samples. Eight to twenty-four microliters of each sample was injected using a PTV injector operated in splitless mode with solvent venting. A thick-film ZB-5 ms column (30 m long, 0.25 mm I.D., 1.00 μm film) was employed with He carrier gas flow rate at 1.4 ml/min. The GC oven temperature was held at 100°C for 1 min, ramped at 20°C/min to 205°C, ramped at 0.8°C/min to 220°C, ramped at 8°C/min to 320°C and held for 10 min. Peaks were identified by comparison of retention order and relative height to GC-MS chromatograms. Isotope ratios were calculated using ISODAT NT 2.5 software by comparison to methane reference gas peaks as described previously (Wang and Sessions, 2008) and are reported in the conventional δ2H notation (≡ Rsamp/Rstd −1) as permil (‰) deviations from the VSMOW standard. The root-mean-squared (RMS) error for external FAME standards run both before and between sample runs was 3.28 ± 0.8‰ (n = 190). Data were corrected for the calculated offset of bracketing external FAME standards. The standard deviation for replicate analyses of unknown analytes averaged 7.5‰ (n = 247). Samples were analyzed in triplicate. The factor averaged 3.48 ± 0.16 ppm/mV. Lipid isotope measurements were also corrected to exclude hydrogen contributed by the derivative methyl group. This was accomplished through determination of the MeOH δ2H by derivatizing a phthalic acid of known non-exchangable δ2H, then calculating non-exchangable lipid δ2H by isotopic mass balance. Fractionations between lipids and environmental water were calculated as εL–W = ((δ2Hl+1)/(δ2Hw+1) − 1) and are reported as permil (‰) variations.
Bulk biomass δ2H-values were measured in a Thermo Fisher thermo-chemolysis/elemental analyzer (TC/EA) interfaced through a Conflo IV into a Delta V Plus isotope ratio mass spectrometer at the University of California, Merced. Briefly, 0.1–0.4 mg of dried samples were weighed in duplicate into 3 × 5 mm silver capsules, sealed, then shipped to Merced, CA. Samples were held in ambient air for >1 month prior to final drying in a desiccator at room temperature. Silver boats with samples and standards were loaded into a Zero-blank autosampler (Costech Analytical) and purged with zero grade He for 10 min. Samples were introduced into a graphite and glassy carbon furnace at 1450°C. H2 was separated from CO via molecular sieve chromatography. The δ2H-values were determined relative to five standard compounds analyzed by the same methodology: stearic acid, mineral oil (Isoanalytical), pump oil (Estep and Hoering, 1980), ground chicken feather (Chris Romanek), and ground turkey feather (Chris Romanek). Stearic acid, mineral oil, and pump oil have hydrogen in non-exchangeable positions, whereas the chicken and turkey feathers potentially can exchange about 10% of their H at room temperature in air. Duplicate δ2H analysis of samples were typically within ±2‰ of an average value. These measurements include a small (~10%) portion of theoretically exchangeable hydrogen at room temperature. Because samples were of similar chemical composition, allowed to equilibrate with ambient atmospheric water vapor, and handled identically, the error associated with such exchangeable H is small. Two of the standards used (turkey and chicken feather) have similar percentages of “exchangeable hydrogen.” Part of our data processing includes comparing our measurements with δ2H-values of theoretically non-exchangeable hydrogen.
Results
Each strain produced a characteristic suite of fatty acids (FA) whose identity and abundance varied substantially between strains, but were generally consistent in the same strain grown on different substrates (see Supplementary Table). The SRBs produced a more complex mixture of fatty acids often including branched and odd chain FA whereas E. coli, P. denitrificans, and S. oneidensis produced a more limited set (Figure S1), consistent with previous reports (Taylor and Parkes, 1983; Dowling et al., 1986). The range of δ2H-values measured for different FA structures from a single culture was often large, particularly for the SRB (Figure 1B and Figure S1). For example, D. autotrophicum grown autotrophically on H2 yielded a range of fatty acid δ2H-values from −210 to −315‰ (for 18:0 and 16:1 FA, respectively). This range shifted to −217 to −325‰ (for 16:0 and cyc-16:0 FA, respectively) when grown on succinate. The δ2H-values and relative ordering of individual compounds varied considerably across both substrates and strains. While there are tendencies for some structures (e.g., branched FA) to be relatively enriched or depleted, no robust inter- or intra-strain trends are apparent in this dataset (Figure S1). To allow for quantitative comparison between strains that produce different fatty acids, we calculated the abundance-weighted mean fractionation between lipids and water (εL–W) using all measured fatty acids (Figure 1A). Weighted-mean epsilon values for the D. autotrophicum trials discussed above are −247 and −250‰, respectively, indicating that the change in substrate does not significantly alter the isotopic composition of the overall fatty acid pool. In contrast, εL–W16:0 values are −235 and −278‰; for the two growth conditions, respectively. These results demonstrate the dynamic and complex nature of hydrogen isotope fractionation, particularly if individual molecular structures are considered in isolation. A summary of the observed fractionations for each organism, grouped by growth condition and substrate, is presented in Figure 1.
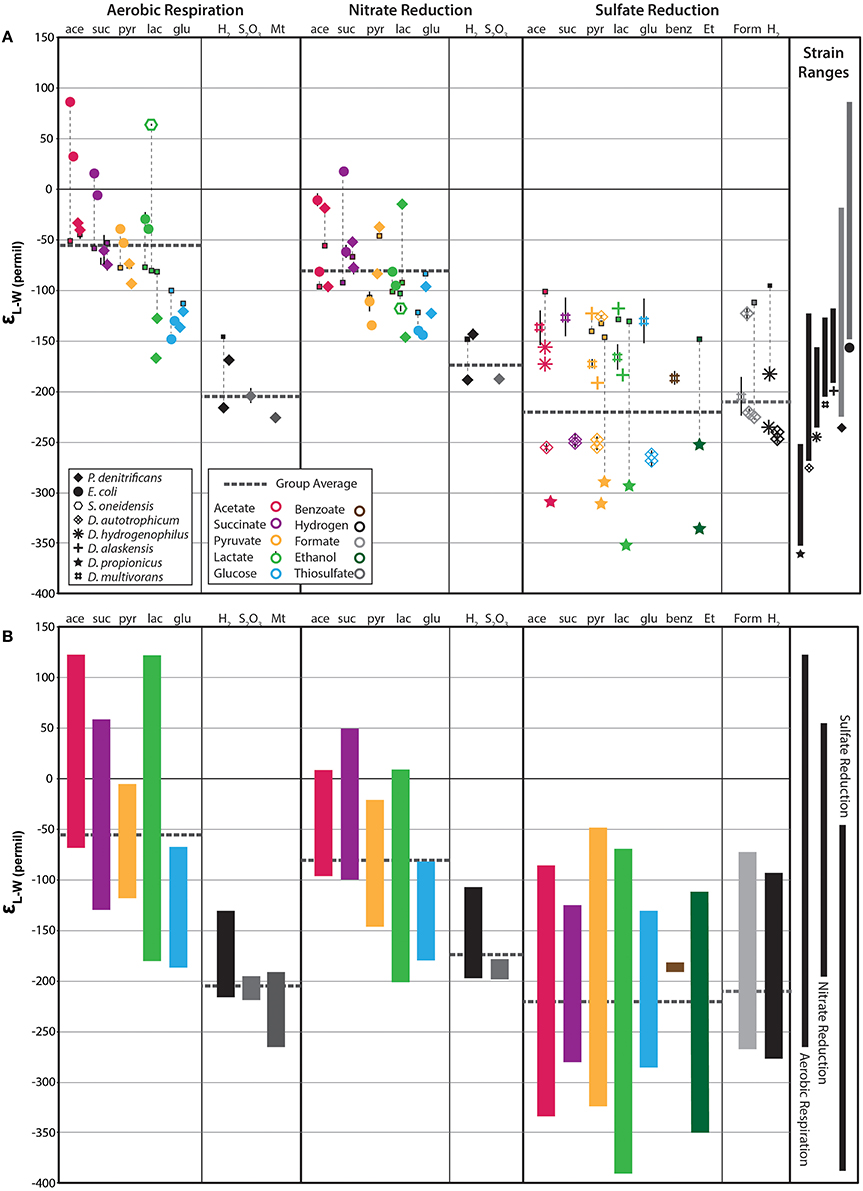
Figure 1. Fractionations between fatty acids and water (εL−W) for all non-labeled experiments. (A) Individual symbols represent the abundance-weighted average εL−W for all fatty acids measured in a given culture experiment. Bulk biomass δ2H-values are shown as small squares with thin vertical dashed lines extending to the corresponding εL−W-value. Thick, horizontal, dark gray, dashed lines indicate the group average (e.g., heterotrophic aerobic respiration). The range of abundance-weighted average εL−W for each strain is shown on the far right. (B) Vertical colored bars indicate the total compound-specific range for each substrate condition. Compound-specific data is plotted in Figure S1. Black bars on the far right illustrate the total range of compound specific data for aerobic respiration, nitrate reduction, and sulfate reduction, respectively.
We observe a large range of εL–W-values that vary between both strains and growth substrates. The fractionations exhibited by aerobes (left-hand group in Figure 1) vary strongly with substrate, with each strain exhibiting a large range of values (~250‰). The same organisms grown anaerobically under nitrate-reducing conditions (middle panel of Figure 1) yielded a generally similar, albeit slightly diminished, pattern of fractionation. Growth on lactate was a significant exception (see below). In contrast, fractionations measured for SRBs (right-hand panels of Figure 1) did not vary significantly between growth substrates, but did vary substantially between individual strains.
O2 Respiring Cultures
Cultures grown under aerobic conditions produced the broadest range of mean fractionation, ranging from −226 to +86‰ (average −80‰; Figure 1). The previously observed correspondence with metabolic substrate is apparent, with significant 2H-enrichments observed for lipids under heterotrophic conditions (−167 to +86‰, average −56‰) and 2H-depletions for autotrophic conditions (−226 to −169‰, average −204‰). Moreover, growth on acetate yielded the most positive values for εL–W (−40 to +80‰), followed by succinate (−74 to 15‰), pyruvate (−93 to −39‰), and glucose (−148 to −121‰), the same order as has previously been observed (Zhang et al., 2009). Cultures grown on lactate produced a very large range of fractionations (−167 to +63‰), with the four individual strains varying systematically within this range. For example, lipids from P. denitrificans cultures grown on lactate are relatively 2H-depleted (lower εL–W) and are similar to those grown on glucose, whereas lipids from E. coli cultures are more 2H-enriched (higher εL–W). S. oneidensis produced very 2H-enriched lipids while growing aerobically on lactate.
Lipid δ2H and bulk cellular biomass δ2H-values are strongly correlated, as would be expected (Figure 2A). However, the range of values in bulk δ2H (−50 to −150‰) is only about half that of the abundance-weighted mean lipid values (−80 to +100‰), with the two crossing at around −100‰. The greater range of lipid δ2H relative to that of bulk biomass cannot be attributed solely to exchangeable hydrogen, as this would require >50% exchangeable H. Possible origins of this pattern are discussed below in Section NADPH Isotope Exchange and Cellular Flux Effects.
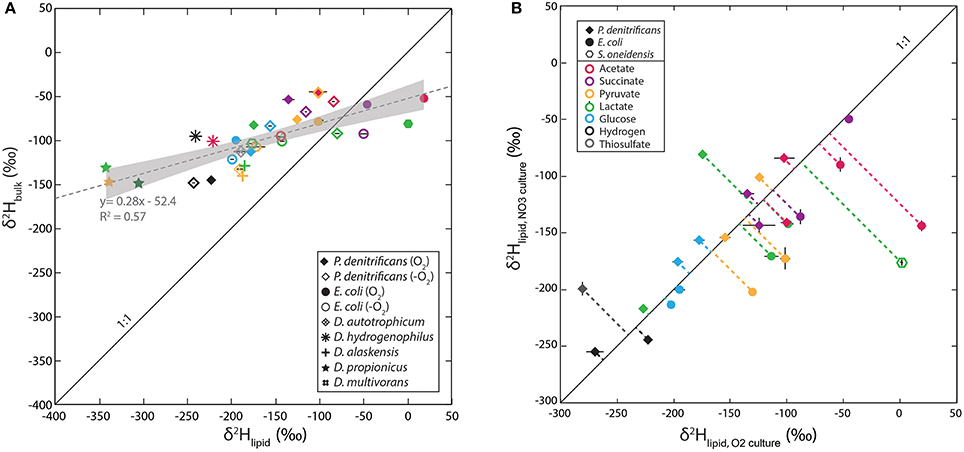
Figure 2. (A) Cross-plot of lipid vs. bulk cellular δ2H-values. The dotted gray line in (A) is a linear regression of the data and gray shaded envelope shows 95% confidence intervals on the regression. (B) Cross-plot of δ2H-values for aerobic and anaerobic culture conditions for facultative anaerobes. Symbols reflect strains type and color reflects substrate in both graphs. Solid diagonal lines are slopes of 1:1 plotted for reference.
Nitrate Reducing Cultures
Lipid-water fractionations observed for nitrate-reducing cultures were—with a few exceptions—broadly similar to those from aerobic cultures. The total range of εL–W-values for these cultures was −147 to + 18‰, averaging −81‰, which is 25‰ more 2H-depleted than the average for aerobic heterotrophy (dashed horizontal lines, Figure 1). Autotrophic nitrate-reducing cultures ranged from −188 to −143‰, averaging −173‰, 31‰ more 2H-enriched than their aerobic equivalents. However, neither of the paired differences between aerobic and anaerobic growth are significantly different than zero (p-values are 0.18 and 0.19, respectively). Figure 2B compares fractionation data between analogous experiments (same organism, substrate) respiring O2 vs. nitrate. Symbols plotting to the right of the diagonal are 2H-enriched in the aerobic condition whereas those plotting to the left are 2H-enriched in the anaerobic condition. Even at the level of individual substrates, there is no systematic pattern of variation between aerobic and anaerobic growth. For instance, acetate and lactate respiration yield lipid 2H-enrichment in the aerobic condition for E. coli and S. oneidensis but not P. denitrificans. Glucose respiration yields similar fractionations under both conditions for all strains, whereas H2 oxidation yields slight lipid 2H-enrichment for the anaerobic condition. The relationship between bulk biomass and lipid δ2H-values for nitrate reducing cultures is broadly similar to that observed in aerobes (Figure 2A).
Sulfate Reducing Cultures
In contrast to the cultures of aerobic and nitrate-respiring bacteria, fractionations for the sulfate-reducing cultures seemed to differ mainly between strains, rather than between growth substrates (Figure 1). The total range in in εL–W observed for heterotrophic sulfate-reducers is −352 to −117‰, but the combined strain average εL–W for acetate, succinate, pyruvate, lactate and glucose are all similar (−214, −208, −214, −222, and −220‰, respectively). The observed difference between the population mean εL–W-values of sulfate reducers and facultative anaerobes (O2 and NO3 respiring cultures) has strong statistical support (p < < 0.0001, student t-test) indicating that hydrogen isotopic fractionation patterns in lipids are controlled by different processes. The most divergent strain is D. propionicus whose entire range of εL–W-values are more 2H-depleted than those of the facultative anaerobes and only overlaps in one case with the next most 2H-depleted SRB D. autotrophicum. Notably, the lipids of the heterotrophic SRB are not particularly 2H-enriched, instead covering much of the range attributed previously to chemolithoautotrophs (Zhang et al., 2009; Heinzelmann et al., 2015b). In addition, the range of lipid εL–W-values for autotrophic sulfate reducers (including C1 formate oxidation) is −247 to −123‰, averaging −210‰, slightly 2H-enriched relative to the heterotrophic SRB cultures. These ranges include the data from Dawson et al. (2015) for D. multivorans (Figure 1, # symbols). Data from these cultures are generally similar to lipid εL–W-values for D. hydrogenophilus and D. alaskensis, but enriched relative to the lipid εL–W-values of D. propionicus and D. autotrophicum.
Discussion
Our data show that the δ2H-values of lipids can vary between strain, growth substrate (or electron donor), and electron acceptor. MANOVA (multivariate analysis of variance) tests on the whole data set reveal that strain (p = 0.0074) and electron acceptor (p = 0.02281) are significant in describing variation in εL–W but substrate is not (p = 0.956). The importance of electron acceptors in influencing lipid δ2H had not previously been appreciated. Differences in fractionation for both E. coli and P. denitrificans when respiring O2 vs. nitrate were relatively small. However, it is not obvious that they should differ at all, given that the relevant respiratory chains are nearly identical until the terminal reduction step and that nitrate anion carries no hydrogen atoms. Large differences in fractionation by sulfate reducers are perhaps less surprising, given that they are metabolically quite distinct. Nevertheless, SRB also conserve reducing equivalents (i.e., NADPH) via many of the same pathways as other bacteria, including the pentose phosphate pathway, TCA cycle, and transhydrogenases, so we expected fractionation to vary with growth substrate, which it did not. The lack of isotopic variation assigned to substrate-dependent fractionation in these organisms is perhaps an important clue toward understanding why εL–W is substrate dependent in aerobes and nitrate-reducers. In any event, a mechanistic understanding of these trends is critical to the future use of lipid δ2H as a recorder of microbial metabolism. Next we evaluate our data in light of known genetic components of each of organism and their measured growth rates, then explore a number of additional mechanisms that could contribute to such variability, highlighting key experiments that may help resolve this question in the future.
Central Metabolic Pathways
Current ideas about lipid δ2H variability assume that organic substrates provide only a small (<25%) proportion of lipid hydrogen, thus changes in the δ2H-values of substrates alone cannot explain the large changes observed in lipids (Zhang et al., 2009), although they may contribute to it. In contrast, more than half of FA hydrogen is attributed to direct reductions by NADPH, which is in turn produced during central carbon metabolism (Sessions et al., 1999; Zhang et al., 2009; Dirghangi and Pagani, 2013a,b; Heinzelmann et al., 2015b) with hydrogen ultimately derived from water. Moreover, the isotopic composition of growth substrates generally cannot be measured in natural environments. Thus, the measured fractionation between lipids and growth water serves as a convenient, albeit incomplete, description of the relevant cellular fractionations that we seek to understand. In the following discussion, we therefore focus on trying to explain values of εL–W.
Current data suggest modest 2H-enrichments of bacteria utilizing substrates that are processed within and through the TCA cycle (acetate, succinate, malate), modest 2H-depletion for those using glycolytic and pentose phosphate pathways, and significant 2H-depletions for those using chemoautotrophic metabolisms. Such patterns have generally been attributed to isotope effects accompanying the reduction of NADP+ by various enzymes (Zhang et al., 2009). One possible difference between aerobes and SRB is thus their compliment of NADP-reducing enzymes. As noted by Leavitt et al. (2016), other processes such as the consumption of NADPH by transhydrogenases may also impart measurable fractionations, and may also vary between aerobes and anaerobes. Reduction of NADP+ by still other pathways that are not present in aerobes, such as H2-consuming hydrogenase, offer still another possibility.
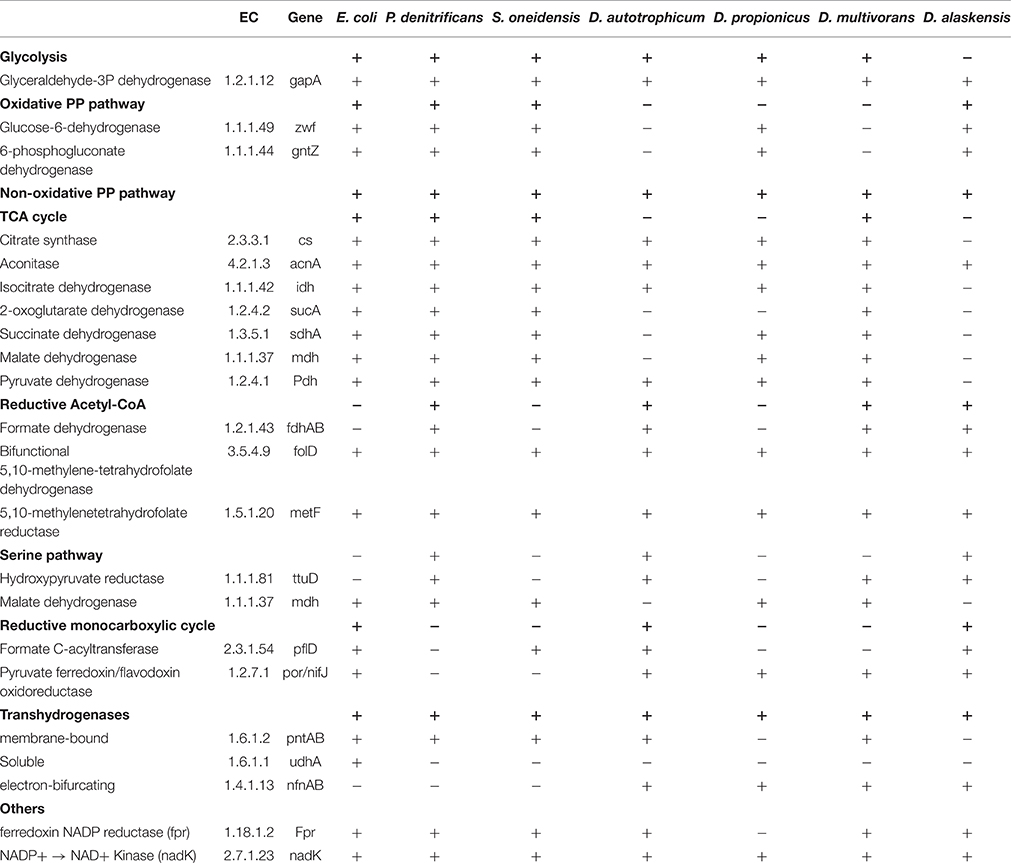
Table 2 documents the presence or absence of metabolic pathways (bold) and of genes for key enzymes (not bold) involved in NADPH processing, all based on the available annotated genomes of the studied strains. This analysis includes both those genes implicated in NADPH production or fractionation from Zhang et al. (2009) and others such as transhydrogenase variants highlighted in Price et al. (2014). All of the microbes in this study have complete sequenced genomes except D. hydrogenophilus, which is necessarily absent from this analysis. One important caveat is that we are looking at genome content and not gene expression, thus differences in expressed metabolic capacity under varying conditions cannot be assessed.
At the broadest level, the pangenome of the studied strains is extremely variable with only a limited number of relevant genes—and no complete pathways—present in all strains. The main exception to this pattern is the non-oxidative branch of the pentose phosphate pathway, which is present in all strains but does not produce NADPH. Major producers of NADPH during glucose metabolism in E. coli are isocitrate dehydrogenase (20–25%), membrane-bound transhydrogenase (35–45%), and the oxidative pentose phosphate pathway (PP pathway; 35–45%; Sauer et al., 2003). Isocitrate dehydrogenase is present in all strains but D. alaskensis, but is thought to produce minimal hydrogen isotope fractionation in product NADPH because the reaction is fully committed and acts on a lone substrate hydrogen (Zhang et al., 2009). All three facultative anaerobes possess both the PntAB transhydrogenase as well as a complete PP pathway.
In contrast, only two of the five SRBs (D. propionicus and D. alaskensis) contain NADP+-reducing enzymes of the PP pathway, although all sulfate reducers have the NfnAB electron-bifurcating transhydrogenase. D. autotrophicum and D. multivorans also contain genes coding for the PntAB transhydrogenase. However, these different enzymatic compliments do not correspond to differences in isotope fractionation by the different SRBs, as the ranges of δ2H-values for the most metabolically similar organisms are essentially non-overlapping. The electron-bifurcating transhydrogenase NfnAB that is shown to be relevant to fractionations in D. alaskensis by Leavitt et al. (2016) is present all SRBs and so may also play a role here. However, strains with the same transhydrogenase gene content growing on the same substrates express different fractionations, complicating this correspondence.
The most extreme fractionations we observe for SRBs are the lipid 2H-depletions produced by D. propionicus when grown on any heterotrophic substrate. Notably this organism lacks a complete TCA-cycle and the PntAB transhydrogenase. The lack of a complete TCA-cycle is shared by D. autotrophicum, the SRB with the second most depleted lipids, which instead processes heterotrophic substrates through the reductive monocarboxylic pathway (Wood-Ljungahl Pathway; Strittmatter et al., 2009). Moreover, growth on substrates that activate the TCA cycle (acetate, succinate, malate) are broadly associated with 2H enrichments of lipids. While it is tempting to conclude that deficiency in TCA cycle genes leads to the observed isotopic depletions, the trend does not hold for D. alaskensis, which has a similar gene complement to D. autotrophicum but more 2H-enriched lipids. It remains possible that lack of TCA-cycle metabolism is broadly associated with 2H-depletions, but that D. alaskensis is unusually 2H-enriched for other reasons, such as operation of the NfnAB transhydrogenase (Leavitt et al., 2016).
In summary, the presence or absence of any single pathway or enzyme does not account for the observed range and distribution of isotope fractionations across the studied strains. There are, however, some correlations, suggesting that biochemical pathways exert partial control. Genes and pathways that we hypothesize to have some effect, but require further testing beyond the scope of this work include 2-oxoglutarate dehydrogenase, the oxidative PP pathway, and the transhydrogenase PntAB. The transhydrogenase NfnAB has been shown independently to have some effect (Leavitt et al., 2016).
Growth Rate
Growth rate is a potential physiological control on the hydrogen isotopic composition of lipids (Sachse et al., 2012). Working with transhydrogenase mutants of D. alaskensis, Leavitt et al. (2016) have shown that hydrogen isotope fractionation is strongly correlated with growth rate in that organism. Growth rate has the potential to impact the δ2H-value of the intracellular water pool, the availability of NADPH for anabolism, and cellular equilibration time of any other intracellular pool of hydrogen. It is also a variable that will plausibly change for facultative anaerobes going from oxic to anoxic conditions. To evaluate whether growth rate was a significant factor in our dataset we monitored the growth rate of each strain under each culture condition (Figure 3 and Figure S2). We hypothesized that a slower growth rate (i.e., longer generation time) would increase the extent of H-isotopic exchange between water and NADPH, decreasing observed fractionations in lipids and biomass, and producing a strongly positive slope on Figure 3.
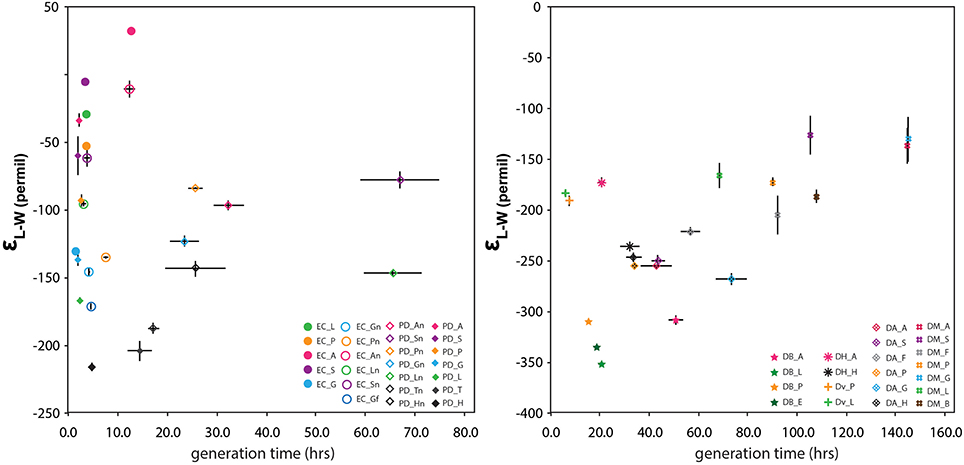
Figure 3. The relationship between generation time (h) and εL–W for facultative anaerobes (left) and sulfate-reducers (right). Error bars for fractionation reflect replicate measurements whereas those for generation time derive from standard error on growth curve regressions.
Generation time varied considerably between our cultures ranging from 1.6 h to just over 6 days, with aerobic heterotrophs growing the fastest and D. multivorans the slowest (Table 1, Figure 3). Generation time is not correlated with isotopic fractionation (R2 = 0.03, N = 46) and was not shown to be significant on the whole nor for individual metabolic groups via MANOVA analysis. Cultures that produced both the most 2H-enriched and 2H-depleted lipids have relatively fast generation times (<24 h). Linear regression of isotopic fractionations against growth rates for aerobic, nitrate-respiring, and sulfate-reducing cultures considered separately reveal possible differences between these groups. While the δ2H-values of lipids from factitive anaerobes are not correlated with generation time under either condition (R2 = 0.013 and N = 12 for O2, 0.02 for NO3 and N = 13), lipids from sulfate reducing cultures are positively correlated with generation time (0.96‰/h, R2 = 0.38, N = 21). This latter result has the same sign but half the magnitude and with a very different intercept than the correlation identified by Leavitt et al. (2016). In our data, this correlation is driven mainly by the relative 2H-depletion and rapid growth of D. propionicus and the 2H-enrichment and slow growth of D. multivorans. For example, if D. multivorans is excluded from the regression, this trend disappears (slope –0.38, R2 0.01, N = 14). It should also be noted that a correlation is not observed for data from individual strains, which might be expected if growth rate was a dominant controlling variable for lipid δ2H. Also, MANOVA tests for just SRBs found generation time to be insignificant for describing variations in fractionation. Given that we did not grow D. alaskensis under conditions leading to widely varying growth rate, we are unable to directly determine whether our results are compatible with those of Leavitt et al. (2016). Nevertheless, our conclusion is generally opposite theirs, that growth rate is not strongly correlated with δ2H across the SRB. Additional experiments under continuous culture conditions might establish a more plausible link between isotopic fractionation and growth rate for the sulfate reducing bacteria.
Other Possible Controls on Fractionation
Based on our understanding of cellular hydrogen fluxes and fractionations (Figure 4), a number of other possible controls on lipid hydrogen isotopes can be considered. Fatty acid biosynthesis is conserved throughout the bacterial domain (White, 2000), so changes in the biosynthetic enzymes are not therefore expected to be a major source of isotopic variability. During biosynthesis, chain elongation occurs by progressive addition of acetyl-CoA molecules to the primer molecule. A number of hydrogen additions occur during hydration of double bonds with either water or NADPH serving as the H donor (White et al., 2005). It follows then that sources of hydrogen on the final fatty acid are acetyl-CoA, water, and NADPH (Figure 4, FW, FA, FNP). The stoichiometry of these contributions depends slightly on the chain length and structure of the final lipid (e.g., unsaturated, saturated, or methyl branched), but essentially half of non-exchangeable hydrogen on fatty acids comes from NADPH (Sessions et al., 1999). The other half is sourced equally between intracellular water and the precursor acetyl-CoA. If we assume that the intrinsic biosynthetic fractionations are invariant in a single organism when growing on different substrates, then the isotopic composition of either water or the biosynthetic precursors acetyl-CoA or NADPH must change. In the following sections we explore the potential for intracellular water (δw), transhydrogenase activity (εT), and NADPH residence time and flux effects to affect major changes in lipid isotopic composition (δL).
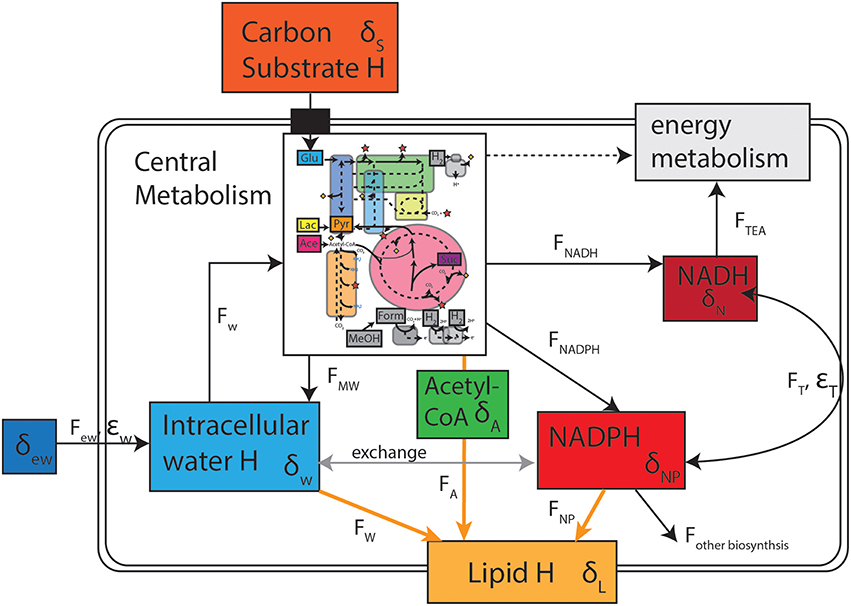
Figure 4. Conceptual model of the reservoirs of hydrogen as well as fluxes and fractionations that contribute to the ultimate isotopic composition of lipids. Inset of central metabolism is after that of Sauer et al. (2003) and Zhang et al. (2009).
Intracellular Water
Intracellular water directly contributes (as H+) at least 25% of the carbon-bound hydrogen on a fatty acid. The isotopic composition of this water (δw) is often assumed to be equal to or in static equilibrium with extracellular water (δew) due to the rapid movement of water in and out of cells through aquaporin channels (Kreuzer-Martin et al., 2006). This assumption is challenged for high-salinity environments where restricted membrane permeability may contribute to the trend of decreasing lipid-water fractionation (Sachse and Sachs, 2008). Intracellular water also derives from metabolic processes (FMW), mainly respiration, and the balance between this source and inward diffusion of extracellular water has been shown to change with growth rate and phase (Kreuzer-Martin et al., 2006; Romero-Viana et al., 2013). Kreuzer-Martin et al. (2005, 2006) quantitatively evaluated the sources of intracellular water in E. coli and found that during very rapid growth (doubling time of ~0.5 h), a majority of intracellular water was derived from metabolic rather than medium water. This respiratory contribution was reduced during stationary phase and at slower growth rates, thus introducing a mechanism for growth phase related hydrogen isotope effects (Kreuzer-Martin et al., 2005, 2006). Although our cultures were all harvested at a similar growth phase, it is possible that the relative offset between intra- and extra-cellular water could vary between aerobic and anaerobic growth of a single strain or between strains on different substrates. Testing this hypothesis will require measurements of δw from cultures, which produce different δL. Notably, our cultures were growing more slowly than those shown to produce this effect in Kreuzer-Martin et al. (2006) and we observed similar fractionations for cultures harvested intentionally in stationary phase (see Supplementary data PD_L(SP)_-82), neither of which support a role for changes in metabolic water to lipid δ2H.
We tested extracellular water incorporation in growth trials with P. denitrificans and D. alaskensis using variable δ2Hew and a range of metabolic substrates. For these experiments lipid δ2H is strongly correlated with growth medium water δ2H in all cases with regression slopes ranging from 0.48 to 0.68 (Figure 5). The slope of linear regressions between δ2Hwater and δ2Hlipid in Figure 5 are driven by a combination of fractional water incorporation (Xw), fractionation between lipid and water (εl∕w), and fractionation between lipid and substrate (εl∕s) (See Sessions and Hayes, 2005; Zhang et al., 2009 for discussion). This complexity prevents the direct interpretation of the correlation slope as fractional water incorporation, but comparisons can be made following the logic discussed in Zhang et al. (2009) (Figure S3).
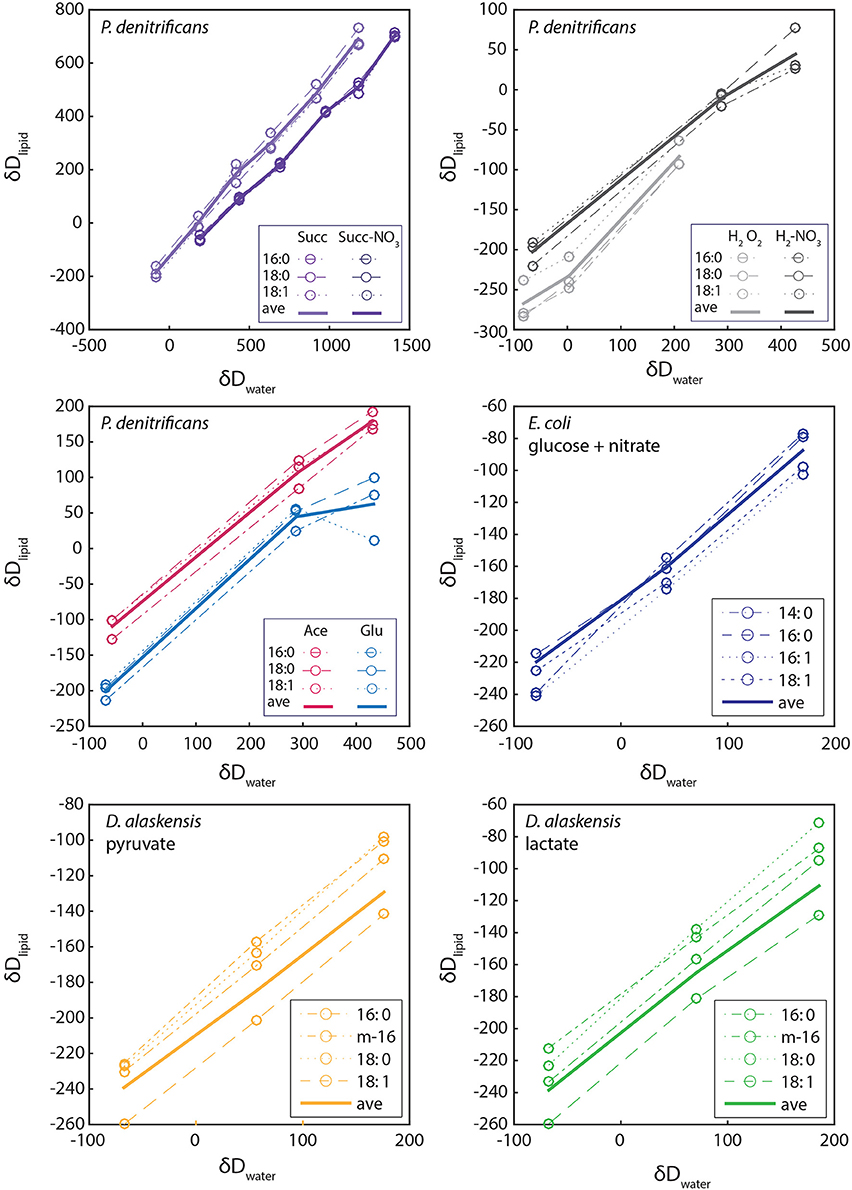
Figure 5. The relationship between hydrogen isotopic composition of growth water and lipids for P. denitrificans, E. coli, and D. alaskensis. Linear regressions (not shown) were performed on the points for weighted average lipid values (bold lines), rather than data for individual lipids (dotted and dashed lines, see legend).
We directly compared δ2H of lipids produced during aerobic and anaerobic growth of P. denitrificans on succinate and hydrogen for a large range of δ2Hwater. We observe a decrease in slope for the anaerobic condition for both substrates, although with an opposite trend in intercept values (Figure 5). Assuming biosynthetic fractionations remain constant, this change in slope could be interpreted as a decrease in fractional water incorporation (or reduced equilibration of other pools with water) under the anaerobic condition.
An alternative interpretation is an increase in isotope fractionation between lipid and substrate. Slopes were identical and intermediate in value for growth on acetate and glucose. We do not see significant change in slope between facultative anaerobes and SRBs with similar results from D. alaskensis cultures grown on lactate and pyruvate (slopes 0.48 – 0.60). Our results are entirely within the range of those from Zhang et al. (2009) precluding an obvious mechanistic link between growth water incorporation and the unusual fractionation patterns observed in anaerobes. This approach may however yield more conclusive results with testing of additional substrates and culture conditions.
Transhydrogenase Activity
Transhydrogenase enzymes moderate the balance between NADPH and NADH in the cell (Griffiths and Roberton, 1966; White, 2000), vary in their activity based on metabolic conditions (Sauer et al., 2003), and are associated with large isotope effects (Jackson et al., 1999). There is thus ample reason to think these enzymes are key determinants in large hydrogen isotopic shifts. Different central metabolic pathways produce varying amounts of NADH vs. NADPH, leading to a potential imbalance between supply and demand of NADPH for anabolism. Transhydrogenase enzymes modulate the transfer of a hydride (H−) moiety from NADH to NADP+, or from NADPH to NAD+. In E. coli these two processes are accomplished via two different transhydrogenase enzymes: an energy-dependent, membrane-bound version (PntAB, Table 2) that produces NADPH, and a soluble version (UdhA) that catalyzes the opposite reaction. Isotope labeling experiments in E. coli show that growth on glucose results in 35–45% of the NADPH pool being produced via the PntAB transhydrogenase, i.e. NADPH is underproduced relative to anabolic demand. In contrast, overproduction of NADPH is observed during growth on acetate, and this surplus is reduced using the soluble UdhA enzyme (Sauer et al., 2003). Given a normal isotope effect for both classes of enzymes (typically 800–3500‰; Jackson et al., 1999), we might expect that underproduction of NADPH leads to 2H-depletion (NADPH is the enzymatic product) and overproduction leads to 2H-enrichment (NADPH is the reactant), a pattern that is broadly similar to that observed in E. coli.
In addition to these two transhydrogenases found in E. coli, a third, electron bifurcating transhydrogenase (NfnAB) has been shown to be important in the growth of D. alaskensis (Price et al., 2014). This enzyme is reversible depending on substrate and allows for ATP production despite the high activation energy of sulfate (Price et al., 2014). Mutants deficient in NfnAB are shown to produce lipids that are 2H-enriched by up to 75‰relative to the wild type under conditions of NADPH consumption, whereas they are nearly identical under conditions of NADPH production by NfnAB (Leavitt et al., 2016). This is the opposite direction of fractionation than predicted by a normal isotope effect, i.e. consumption of NADPH by NfnAB should lead to 2H enrichment, and eliminating that activity should correspond to 2H depletion.
The strains studied here are all genetically capable of expressing at least one transhydrogenase variant with the exception of D. propionicus (Table 2), although they differ significantly in how many and which versions they possess. E. coli is the only organism to possess genes for both the membrane-bound (PntAB) and the soluble (UdhA) transhydrogenases as described in Sauer et al. (2003). P. denitrificans, S. oneidensis, and D. multivorans contain only PntAB coding genes, D. alaskensis has only those for NfnAB, whereas D. autotrophicum contains genes for both PntAB and NfnAB. While genetic potential does not necessarily indicate enzymatic activity, the inverse is true: organisms lacking a specific gene lack the corresponding transhydrogenase. For instance, expression of transhydrogenases by E. coli and P. denitrificans when switching between growth on glucose to acetate, must vary as E. coli has been demonstrated to upregulate the expression of the UdhA encoding transhydrogenase that P. denitrificans lacks (Sauer et al., 2003). Interestingly, we do observe large 2H-enrichment of E. coli lipids relative to those of P. denitrificans when growing on acetate but not on glucose, consistent with this prediction (Figure 1). Direct demonstration of differential transhydrogenase expression between these cultures and conditions will be required to prove a mechanistic link.
Some SRBs in this study have the genes for both PntAB and NfnAB, some have only PntAB, and still others have neither. However, the hydrogen isotope fractionations by SRBs we measured do not support significant differences in transhydrogenase fractionations. D. multivorans and D. alaskensis produce similar, small ranges of fractionations despite having genes for PntAB and NfnAB, respectively. D. autotrophicum has strongly 2H-depleted lipids with little variation between growth substrates, despite having the genetic potential to produce both pntAB and nfnAB. The strong 2H-depletion observed for D. propionicus despite a lack of transhydrogenase genes is interesting, but inconclusive at this point. In summary, the potential for transhydrogenase enzymes to influence the isotopic composition of NADPH (δNP) and thus lipids is clear, and there is experimental evidence that this is so in some particular organisms. However, it appears unlikely to be a single, over-riding control that could explain all hydrogen isotope variations for all of the studied organisms.
NADPH Isotope Exchange and Cellular Flux Effects
NADPH contributes up to half of the carbon-bound hydrogen on fatty acids and thus has significant leverage to influence lipid isotopic composition. Variation in the isotopic composition of the H− donated by NADPH (δNP) is hypothesized to be the dominant mechanism by which central metabolism influences δL(Zhang et al., 2009; Dirghangi and Pagani, 2013a,b; Heinzelmann et al., 2015b). Previously, control of δNP has been ascribed mainly to NADPH-reducing pathways (“supply”), although both the demand for NADPH and the process of isotopic exchange could also influence its δ2H-value. The latter, in particular, might vary with the cellular residence time of reduced NADPH, which in turn might plausibly vary between aerobic/anaerobic metabolism.
It is well-established that the cellular quotient of NADP(H) varies between strains and across growth conditions (London and Knight, 1966; Wimpenny and Firth, 1972; Matin and Gottschal, 1976; Karl, 1980). London and Knight (1966) specifically noted differences in the relative abundance of NAD+ between strict anaerobes, facultative anaerobes, and aerobes. Focusing just on NADPH, if we assume that a larger cellular quotient of reduced NADPH is correlated with a longer residence time for each molecule, then greater hydrogen isotope exchange with cellular water could result in damping the isotopic signals associated with NADPH supply. Existing reports are conflicting about the precise rate of isotopic exchange of NADPH in vivo, but at least one study (Saito et al., 1980) has observed rapid exchange in vitro. The extent to which either the size or the oxidation state of the NADPH reservoir influences lipid δ2H is not known, but this appears to be a promising avenue for further investigation.
It also likely that the various downstream biosynthetic processes that constitute the demand for reduced NADPH fractionate hydrogen isotopes. Even if individual isotope biosynthetic effects remain constant, a change in the relative proportions of downstream products (e.g. more or less lipids relative to other biochemical pools) could change their individual isotopic compositions even while the bulk isotopic composition changes very little (Hayes, 2001). Comparison of the bulk isotopic data to that of fatty acids (Figure 2A) does not favor this hypothesis. The δ2H-values of both lipids and bulk biomass change with growth substrate, and—more importantly—the order of relative 2H-depletion reverses (e.g., lipids are 2H-depleted relative to biomass in some cultures, and 2H-enriched in others). The latter pattern could not arise simply from changing the relative proportions of anabolic products with constant isotope effects.
Instead, the co-varying isotopic compositions of bulk biomass and lipids are more consistent with them receiving different amounts of H from a common source, whose δ2H-value varies between experiments. For example, our data (Figure 2A) can be reasonably fit by a 2-component mixing model, with one component (presumably water + substrate) having a constant δ2H of −64‰ and the other (presumably NADPH) having δ2H that varies between 0 and −700‰, with 50% of lipid hydrogen and only 13.5% of bulk biomass hydrogen coming from NADPH (Figure S4). This simple model is certainly an oversimplification, and is not a unique solution, but is at least consistent with what is known about the relative contributions of NADPH to lipids vs. other biomolecules such as proteins and sugars.
Conclusions
Our study reveals additional complexity of hydrogen isotope fractionation in microbial species. Two distinct patterns of fractionation were measured among heterotrophic bacteria: one in facultative nitrate-reducers that is qualitatively similar to that seen previously in aerobes, and a second in sulfate-reducing bacteria that is entirely distinct. Our data confirms that growth substrate is the dominant controlling variable for lipid δ2H-values of facultative anaerobes, consistent with previous reports. In contrast, SRB produce 2H-depleted lipids, apparently regardless of metabolic pathway or substrate, and instead converge on strain-specific values. Generally speaking, lipids produced during nitrate reduction and aerobic respiration tend to be more enriched, whereas those produced during sulfate reduction tend to be more depleted and in a very similar range as photoautotrophy. The distinction between autotrophic and heterotrophic growth observed by Zhang et al. (2009) and emphasized subsequently (Valentine, 2009; Heinzelmann et al., 2015b), was replicated in nitrate reducers but not sulfate reducers. In SRB, utilization of heterotrophic substrates results in the production of lipids that match or exceed 2H-depletions observed for autotrophic growth. Whether this result represents an exception or the rule for other obligate anaerobes is still unknown.
Author Contributions
This manuscript is part of the thesis work of MO who completed the bulk of the experiments, measurements, and writing of this manuscript. KD cultured one strain of bacteria and contributed significantly to the genetic components of the manuscript. MF contributed measurements of bulk microbial biomass that significantly shaped the arguments within the manuscript. The majority of experiments and measurements were completed in the laboratories of AS who also contributed significantly to the intellectual development of the project, advisement of MO, and formulation of the manuscript. All authors contributed to writing and editing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jared Leadbetter and Victoria Orphan for initial guidance and use of laboratory facilities, the ARS culture collection for strain access, Lichun Zhang and Fenfang Wu for laboratory assistance, Sebastian Kopf for helpful discussions, and Pratixa Savalia for help culturing S. oneidensis. This research was supported by an NSF GRFP award to MO, a Penn State Astrobiology Research Center [through the NASA Astrobiology Institute (NNA09DA76A)] postdoctoral fellowship to KD, and by EAR-1529120 to AS. We thank Christina Bradley and David Araiza (UC Merced) for assistance in measuring the bulk δ2H measurements of cultures.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01166
References
Breznak, J. A., and Leadbetter, J. R. (2006). “Termite gut spirochetes,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (Springer: New York), 318–329.
Campbell, B. J., Li, C., Sessions, A. L., and Valentine, D. L. (2009). Hydrogen isotopic fractionation in lipid biosynthesis by H2-consuming Desulfobacterium autotrophicum. Geochim. Cosmochim. Acta 73, 2744–2757. doi: 10.1016/j.gca.2009.02.034
Chikaraishi, Y., and Naraoka, H. (2001). Organic hydrogen-carbon isotope signatures of terrestrial higher plants during biosynthesis for distinctive photosynthetic pathways. Geochem. J. 35, 451–458. doi: 10.2343/geochemj.35.451
Chikaraishi, Y., and Naraoka, H. (2005). δ13C and δD identification of sources of lipid biomarkers in sediments of Lake Haruna (Japan). Geochim. Cosmochim. Acta 69, 3285–3297. doi: 10.1016/j.gca.2005.02.023
Chikaraishi, Y., Tanaka, R., Tanaka, A., and Ohkouchi, N. (2009). Fractionation of hydrogen isotopes during phytol biosynthesis. Org. Geochem. 40, 569–573. doi: 10.1016/j.orggeochem.2009.02.007
Chivall, D., M'Boule, D., Sinke-Schoen, D., Damsté, J. S. S., Schouten, S., and van der Meer, M. T. J. (2014). The effects of growth phase and salinity on the hydrogen isotopic composition of alkenones producedby coastal haptophyte algae. Geochim. Cosmochim. Acta 140, 381–390. doi: 10.1016/j.gca.2014.05.043
Dawson, K. S., Osburn, M. R., Sessions, A. L., and Orphan, V. J. (2015). Metabolic associations with archaea drive shifts in hydrogen isotope fractionation in sulfate-reducing bacterial lipids in cocultures and methane seeps. Geobiology 13, 462–477. doi: 10.1111/gbi.12140
Dirghangi, S. S., and Pagani, M. (2013a). Hydrogen isotope fractionation during lipid biosynthesis by Tetrahymena thermophila. Org. Geochem. 64, 105–111. doi: 10.1016/j.orggeochem.2013.09.007
Dirghangi, S. S., and Pagani, M. (2013b). Hydrogen isotope fractionation during lipid biosynthesis byHaloarcula marismortui. Geochim. Cosmochim. Acta 119, 381–390. doi: 10.1016/j.gca.2013.05.023
Dowling, N. J., Widdel, F., and White, D. (1986). Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulphate-reducers and other sulphide-forming bacteria. J. Gen. Microbiol. 132, 1815–1825.
Estep, M. F., and Hoering, T. C. (1980). Biogeochemistry of the stable hydrogen isotopes. Geochim. Cosmochim. Acta 44, 1197–1206. doi: 10.1016/0016-7037(80)90073-3
Griffiths, D. E. D., and Roberton, A. M. A. (1966). Energy-linked reactions in mitochondria: studies on the mechanism of the energy-linked transhydrogenase reaction. Acta Bioenerg. 118, 453–464. doi: 10.1016/s0926-6593(66)80089-9
Hayes, J. M. (2001). Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev. Mineral. Geochem. 43, 225–277. doi: 10.2138/gsrmg.43.1.225
Heinzelmann, S. M., Chivall, D., M'Boule, D., Sinke-Schoen, D., Villanueva, L., Damsté, J. S. S., et al. (2015a). Comparison of the effect of salinity on the D/H ratio of fatty acids of heterotrophic and photoautotrophic microorganisms. FEMS Microbiol. Lett. 362, fnv065. doi: 10.1093/femsle/fnv065
Heinzelmann, S. M., Villanueva, L., Lipsewers, Y., Sinke-Schoen, D., Sinninghe Damste, J., Schouten, S., et al. (2015c). “Assessing the metabolism of sedimentary microbial communities using the hydrogen isotopic composition of fatty acids,” in Metabolism of Microbial Communities in the Environment A Compound-Specific Stable Hydrogen Isotope Approach (Royal Netherlands Institute of Sea Research), 103–145. Ph.D. Thesis, Royal Netherlands Institute of Sea Research.
Heinzelmann, S. M., Villanueva, L., Sinke-Schoen, D., Sinninghe Damsté, J. S., Schouten, S., and van der Meer, M. T. J. (2015b). Impact of metabolism and growth phase on the hydrogen isotopic composition of microbial fatty acids. Front. Microbiol. 6:408. doi: 10.3389/fmicb.2015.00408
Huang, Y., Shuman, B., Wang, Y., and Webb, T. III (2002). Hydrogen isotope ratios of palmitic acid in lacustrine sediments record late Quaternary climate variations. Geology 30, 1103–1106. doi: 10.1130/0091-7613(2002)030<1103:HIROPA>2.0.CO;2
Jackson, J. B., Peake, S. J., and White, S. A. (1999). Structure and mechanism of proton-translocating transhydrogenase. FEBS Lett. 464, 1–8. doi: 10.1016/S0014-5793(99)01644-0
Jones, A. A., Sessions, A. L., Campbell, B. J., Li, C., and Valentine, D. L. (2008). D/H ratios of fatty acids from marine particulate organic matter in the California Borderland Basins. Org. Geochem. 39, 485–500. doi: 10.1016/j.orggeochem.2007.11.001
Karl, D. M. (1980). Cellular nucleotide measurements and applications in microbial ecology. Microbiol. Rev. 44, 739–796.
Kreuzer-Martin, H. W., Ehleringer, J. R., and Hegg, E. L. (2005). Oxygen isotopes indicate most intracellular water in log-phase Escherichia coli is derived from metabolism. Proc. Natl. Acad. Sci. U.S.A. 102, 17337–17341. doi: 10.1073/pnas.0506531102
Kreuzer-Martin, H. W., Lott, M. J., Ehleringer, J. R., and Hegg, E. L. (2006). Metabolic processes account for the majority of the intracellular water in log-phase Escherichia coli cells as revealed by hydrogen isotopes. Biochemistry 45, 13622–13630. doi: 10.1021/bi0609164
Leavitt, W. D., Flynn, T. M., Suess, M. K., and Bradley, A. S. (2016). Transhydrogenase and growth substrate influence lipid hydrogen isotope ratios in Desulfovibrio alaskensis G20. Front. Microbiol. 7:918. doi: 10.3389/fmicb.2016.00918
Li, C., Sessions, A. L., Kinnaman, F., and Valentine, D. L. (2009). Hydrogen-isotopic variability in lipids from Santa Barbara Basin sediments. Geochim. Cosmochim. Acta 73, 4803–4823. doi: 10.1016/j.gca.2009.05.056
London, J., and Knight, M. (1966). Concentrations of nicotinamide nucleotide coenzymes in micro-organisms. J. Gen. Microbiol. 44, 241–254. doi: 10.1099/00221287-44-2-241
Matin, A., and Gottschal, J. C. (1976). Influence of dilution rate on NAD(P) and NAD(P)H concentrations and ratios in a Pseudomonas sp. grown in continuous culture. J. Gen. Microbiol. 94, 333–341. doi: 10.1099/00221287-94-2-333
Muyzer, G., and Stams, A. J. M. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Naraoka, H., Uehara, T., Hanada, S., and Kakegawa, T. (2010). d13C-dD distribution in lipid biomarkers in a bacterial mat from a hot spring in Miyagi Prefecture, NE Japan. Org. Geochem. 41, 398–403. doi: 10.1016/j.orggeochem2009.11.008
Osburn, M., Sessions, A., and Spear, J. (2011). Hydrogen-isotopic variability in fatty acids from Yellowstone National Park hot spring microbial communities. Geochim. Cosmochim. Acta 75, 4830–4845. doi: 10.1016/j.gca.2011.05.038
Price, M. N., Ray, J., Wetmore, K. M., Kuehl, J. V., Bauer, S., Deutschbauer, A. M., et al. (2014). The genetic basis of energy conservation in the sulfate-reducing bacterium Desulfovibrio alaskensis G20. Front. Microbiol. 5:577. doi: 10.3389/fmicb.2014.00577
Pu, Z., and Weiguo, L. (2011). Effect of plant life form on relationship between dD values of leaf wax n-alkanes and altitude along Mount Taibai, China. Org. Geochem. 42, 100–107. doi: 10.1016/j.orggeochem.2010.11.005
Rodríguez-Ruiz, J., Belarbi, E.-H., Sánchez, J. L. G., and Alonso, D. L. (1998). Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 12, 689–691. doi: 10.1023/A:1008812904017
Romero-Viana, L., Kienel, U., Wilkes, H., and Sachse, D. (2013). Growth-dependent hydrogen isotopic fractionation of algal lipid biomarkers in hypersaline Isabel Lake (Mexico). Geochim. Cosmochim. Acta 106, 490–500. doi: 10.1016/j.gca.2012.12.017
Sachse, D., Billault, I., Bowen, G. J., Chikaraishi, Y., Dawson, T. E., Feakins, S. J., et al. (2012). Molecular paleohydrology: interpreting the hydrogen-isotopic composition of lipid biomarkers from photosynthesizing organisms. Annu. Rev. Earth Planet. Sci. 40, 221–249. doi: 10.1146/annurev-earth-042711-105535
Sachse, D., and Sachs, J. (2008). Inverse relationship between D/H fractionation in cyanobacterial lipids and salinity in Christmas Island saline ponds. Geochim. Cosmochim. Acta 72, 793–806. doi: 10.1016/j.gca.2007.11.022
Saito, K., Kawaguchi, A., Okuda, S., Seyama, Y., and Yamakawa, T. (1980). Incorporation of hydrogen-atoms from deuterated water and stereospecifically deuterium-labeled nicotinamide nucleotides into fatty-acids with the Escherichia-coli fatty-acid synthetase system. Biochim. Biophys. Acta 618, 202–213. doi: 10.1016/0005-2760(80)90026-0
Sauer, P. E., Eglinton, T. I., Hayes, J. M., Schimmelmann, A., and Sessions, A. L. (2001). Compound-specific D/H ratios of lipid biomarkers from sediments as a proxy for environmental and climatic conditions. Geochim. Cosmochim. Acta 65, 213–222. doi: 10.1016/S0016-7037(00)00520-2
Sauer, U., Canonaco, F., Heri, S., Perrenoud, A., and Fischer, E. (2003). The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279, 6613–6619. doi: 10.1074/jbc.M311657200
Sessions, A., and Hayes, J. (2005). Calculation of hydrogen isotopic fractionations in biogeochemical systems. Geochim. Cosmochim. Acta 69, 593–597. doi: 10.1016/j.gca.2004.08.005
Sessions, A. L., Burgoyne, T. W., Schimmelmann, A., and Hayes, J. M. (1999). Fractionation of hydrogen isotopes in lipid biosynthesis. Org. Geochem. 30, 1193–1200. doi: 10.1016/S0146-6380(99)00094-7
Strittmatter, A. W., Liesegang, H., Rabus, R., Decker, I., Amann, J., Andres, S., et al. (2009). Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 11, 1038–1055. doi: 10.1111/j.1462-2920.2008.01825.x
Taylor, J., and Parkes, R. J. (1983). The cellular fatty acids of the sulphate-reducing Bacteria, Desulfobacter sp., Desulfobulbus sp. and Desulfovibrio desulfuricans. Microbiology 129, 3303–3309. doi: 10.1099/00221287-129-11-3303
Valentine, D. L. (2009). Isotopic remembrance of metabolisms past. Proc. Natl. Acad. Sci. U.S.A. 106, 12565–12566. doi: 10.1073/pnas.0906428106
Valentine, D., Sessions, A., Tyler, S., and Chidthaisong, A. (2004). Hydrogen isotope fractionation during H2/CO2 acetogenesis: hydrogen utilization efficiency and the origin of lipid-bound hydrogen. Geobiology 2, 179–188. doi: 10.1111/j.1472-4677.2004.00030.x
Wang, Y., and Sessions, A. L. (2008). Memory effects in compound-specific D/H analysis by gas chromatography/pyrolysis/isotope-ratio mass spectrometry. Anal. Chem. 80, 9162–9170. doi: 10.1021/ac801170v
White, S. W., Zheng, J., Zhang, Y.-M., and Rock, C. O. (2005). The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831. doi: 10.1146/annurev.biochem.74.082803.133524
Wimpenny, J. W., and Firth, A. (1972). Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J. Bacteriol. 111, 24–32.
Keywords: fatty acids, hydrogen isotopes, sulfate-reducing bacteria, anaerobic microbial metabolism, NAD(P)H, transhydrogenase
Citation: Osburn MR, Dawson KS, Fogel ML and Sessions AL (2016) Fractionation of Hydrogen Isotopes by Sulfate- and Nitrate-Reducing Bacteria. Front. Microbiol. 7:1166. doi: 10.3389/fmicb.2016.01166
Received: 10 March 2016; Accepted: 13 July 2016;
Published: 02 August 2016.
Edited by:
Shuhei Ono, Massachusetts Institute of Technology, USAReviewed by:
William D. Leavitt, Washington University in St. Louis, USAYongsong Huang, Brown University, USA
Copyright © 2016 Osburn, Dawson, Fogel and Sessions. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magdalena R. Osburn, maggie@earth.northwestern.edu
†Present Address: Magdalena R. Osburn, Department of Earth and Planetary Science, Northwestern University, Evanston, IL, USA
 Magdalena R. Osburn
Magdalena R. Osburn Katherine S. Dawson
Katherine S. Dawson Marilyn L. Fogel2
Marilyn L. Fogel2 Alex L. Sessions
Alex L. Sessions