- 1United States Department of Agriculture – Agricultural Research Service, Beltsville, MD, USA
- 2Instituto de Biología Molecular y Celular de Plantas, Universidad Politécnica de Valencia–Consejo Superior de Investigaciones Científicas, Valencia, Spain
- 3UMR 1332 Biologie du Fruit et Pathologie, Institut National de la Recherche Agronomique, Université de Bordeaux, Bordeaux, France
- 4Consiglio per la Ricerca in Agricoltura e l’analisi dell’Economia Agraria, Centro di Ricerca per la Patologia Vegetale, Rome, Italy
Next-generation sequencing (NGS) has been applied to plant virology since 2009. NGS provides highly efficient, rapid, low cost DNA, or RNA high-throughput sequencing of the genomes of plant viruses and viroids and of the specific small RNAs generated during the infection process. These small RNAs, which cover frequently the whole genome of the infectious agent, are 21–24 nt long and are known as vsRNAs for viruses and vd-sRNAs for viroids. NGS has been used in a number of studies in plant virology including, but not limited to, discovery of novel viruses and viroids as well as detection and identification of those pathogens already known, analysis of genome diversity and evolution, and study of pathogen epidemiology. The genome engineering editing method, clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system has been successfully used recently to engineer resistance to DNA geminiviruses (family, Geminiviridae) by targeting different viral genome sequences in infected Nicotiana benthamiana or Arabidopsis plants. The DNA viruses targeted include tomato yellow leaf curl virus and merremia mosaic virus (begomovirus); beet curly top virus and beet severe curly top virus (curtovirus); and bean yellow dwarf virus (mastrevirus). The technique has also been used against the RNA viruses zucchini yellow mosaic virus, papaya ringspot virus and turnip mosaic virus (potyvirus) and cucumber vein yellowing virus (ipomovirus, family, Potyviridae) by targeting the translation initiation genes eIF4E in cucumber or Arabidopsis plants. From these recent advances of major importance, it is expected that NGS and CRISPR-Cas technologies will play a significant role in the very near future in advancing the field of plant virology and connecting it with other related fields of biology.
Introduction
The field of virology was born in the late 1890s when it was found that the tobacco mosaic disease is caused by a novel form of infectious agent named “ultravirus” and referred to as ‘contagium vivum fluidum’ (soluble living germ or contagious living fluid; Hadidi and Barba, 2012). The virus, later named tobacco mosaic virus (TMV), was the first to be described and become an iconic one, especially in the first half of the 20th century. TMV was purified and crystallized by Wendell M. Stanley in 1935, who also described its structure, and Heinz Fraenkel-Conrat in 1950s was the first to show that its replication is directed by genetic information encoded within its RNA core (Hadidi and Barba, 2012), thus starting molecular virology. Molecular virology was also instrumental in the discovery, by Theodor O. Diener, of a novel class of infectious small RNA in 1971 as the causal agent of potato spindle tuber disease (Diener, 1971a,b), which he later named potato spindle tuber viroid (PSTVd; Diener, 1972). Viroids are very small circular, non-coding RNAs (Sänger et al., 1976; Owens et al., 1977). Unlike virus particles (virions), which are made of protein coats encapsidating nucleic acid genomes, viroids are naked single-stranded (ss) RNAs without protein coats, 247–401 nt in length and with a high degree of internal base pairing; they are the smallest known infectious agents (Hadidi et al., 2003). The 2012 Report of the International Committee for the Taxonomy of Viruses (ICTV) lists about 900 plant virus species (King et al., 2012). The current number of viroid species is 32 (Di Serio et al., 2014). Viruses and viroids infect vegetable, field, ornamental, and/or tree crops as well as wild plant species, in which they may induce significant effects on plant health such as a decrease of desired quality or loss of yield, causing a reduction in income for farmers, other crop producers, and distributors, and higher prices for consumers. Moreover, increased international movement, globalization of trade, propagation material, and seed supply systems all influence the introduction and proliferation of virus and viroid diseases across countries. Crop yield reductions attributed to specific virus or viroid infections in specific crops may vary from less than 10% to more than 80–90% for viruses (Waterworth and Hadidi, 1998; Hadidi et al., 2011; Barba and Hadidi, 2015) and from 17% to close to 100% for viroids (Singh et al., 1971; Zelazny et al., 1982; Hadidi et al., 2003). Some viruses and viroids can be largely latent (symptomsless) in some of their infected hosts (Hadidi et al., 1998, 2003, 2011). These latent agents, however, may be pathogenic in other hosts and their infections may result in yield reduction and general weakness of plants (Hadidi et al., 1998, 2003, 2011; Hadidi and Barba, 2012; Barba et al., 2015).
The very large variability among plant viruses and among viroids complicates their discovery, detection, quarantine, and certification as well as their etiological studies by standard methods, which could be overcome or made easier by next-generation sequencing (NGS) technology. Similarly, despite progress in understanding virus or viroid–plant interactions, obtaining plants resistant to these pathogens by conventional breeding or transgenic strategies are often complicated and faces the problem of resistance durability. Virus or viroid genome and/or plant gene editing is expected to play a significant role in developing transgene-free plants resistant to viruses or viroids. Advances in NGS capabilities and the rapid development and widespread adoption of a simple, inexpensive, easy to use and very effective genome engineering editing method known as “clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR-Cas)” are revolutionizing the fields of genetics, genomics, molecular biology and others. In this article we discuss the progress made in plant virology during the last 7 years by NGS and very recently by CRISPR-Cas systems.
NGS in Plant Virology
Next-generation sequencing, combined with informatics for de novo discovery and assembly of plant virus or viroid genome reads, has been used since 2009, first in discovering novel DNA and RNA viruses (Adams et al., 2009; Al Rwahnih et al., 2009; Kreuze et al., 2009), and in detecting and identifying RNA viruses (Al Rwahnih et al., 2009; Donaire et al., 2009; Kreuze et al., 2009), as well as viroids (Al Rwahnih et al., 2009). Moreover, it was used in the same year for sequencing viroid-derived small RNAs (vd-sRNAs) to study the role of RNA silencing in plant–viroid interactions, particularly for nuclear-replicating viroids (Navarro et al., 2009), and to investigate the genesis and possible pathogenesis of vd-sRNAs from a chloroplast-replicating viroid (Di Serio et al., 2009). Subsequent data support a role of vd-sRNAs in viroid pathogenesis (Navarro et al., 2012a; Adkar-Purushothama et al., 2015; Avina-Padilla et al., 2015), as previously shown for an sRNA derived from the satellite RNA Y of cucumber mosaic virus (CMV; Smith et al., 2011; Shimura et al., 2011). vd-sRNAs and virus small RNAs (vsRNAs), 21–24 nt in length, are generated in host plants by their silencing machinery in response to infection by these foreign replicons. RNA silencing is a cell surveillance system that recognizes double-stranded (ds) RNA and ssRNA with a compact secondary structure, and specifically inactivates viroids and RNA viruses (by post-transcriptional gene silencing) as well as DNA viruses (by transcriptional and/or post-transcriptional gene silencing), using small interfering RNAs as a guide (for review see: Barba and Hadidi, 2009; Sano et al., 2010; Hammann and Steger, 2012; Navarro et al., 2012b; Wang et al., 2012; Flores et al., 2015; Zhang et al., 2015).
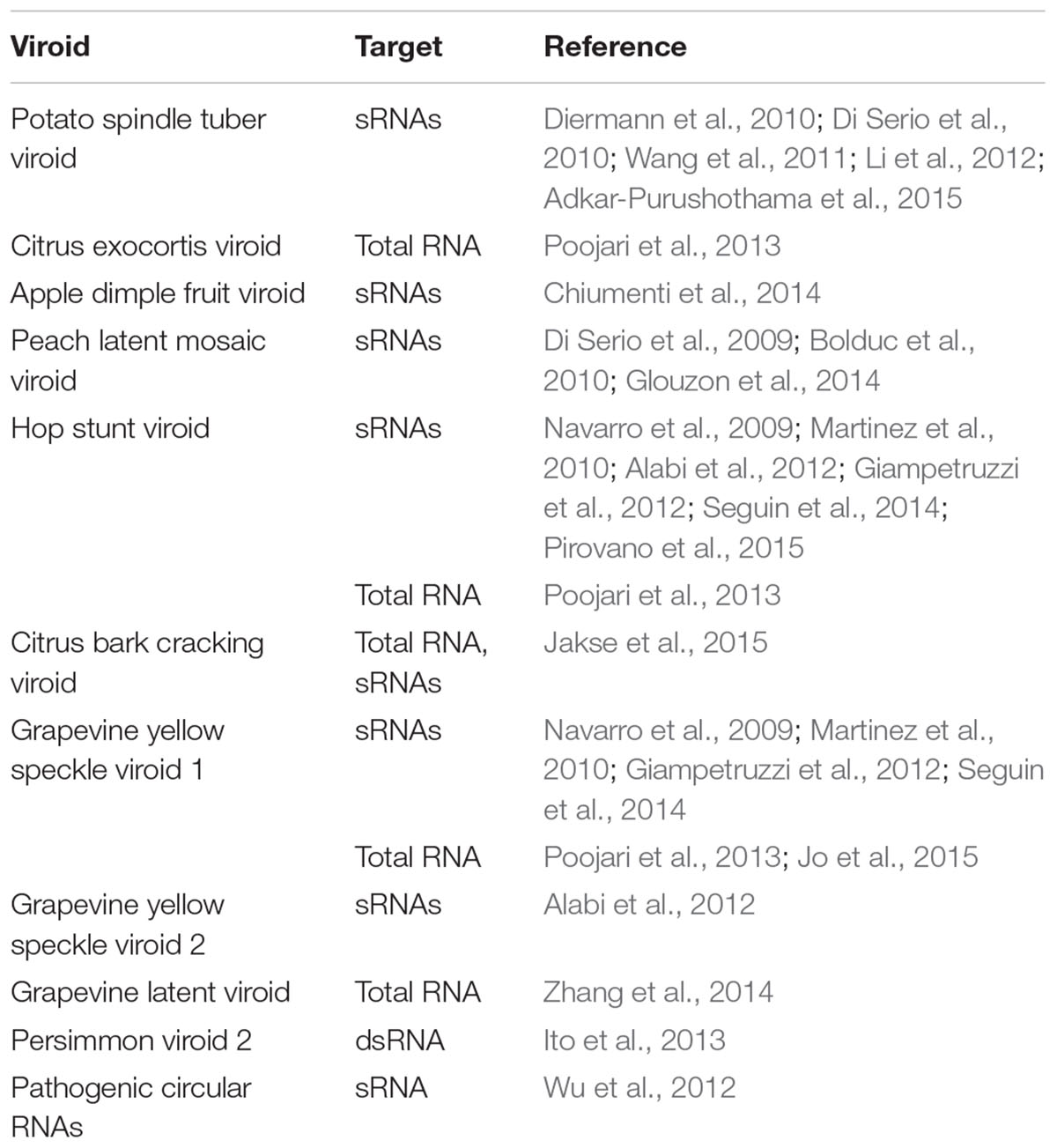
Next-generation sequencing of vsRNAs or vd-sRNAs has also been used in various studies on viruses or viroids, which include, but are not limited to, their characterization, profiling, distribution, accumulation, biogenesis, as well as their use in extending the known pathogen host range, and in virus strain differentiation, systemic movement, virus or viroid-host interaction, viroid evolution, and pathogenesis, mutation, mRNA targeting, and others (Tables 1 and 2).
Next-generation sequencing can in a single experiment determine the sequence of hundreds of thousands to millions of vsRNA or vd-sRNA, which can be re-assembled to obtain the genomic sequence of the virus or viroid genome(s) of interest but can also be compared to the host genome in an effort to identify genes that may be down-regulated upon infection as a consequence of their local homology with the infecting virus or viroid. In parallel, the identification in otherwise healthy plants of sRNAs with homology to viral satellite RNAs has provided recently a tentative scenario for their evolution from the host plant genome, offering a possible solution to the long lasting conundrum of the origin of these infectious agents (Zahid et al., 2015; Wang and Smith, 2016).
Discovery and Diagnostics of Plant Viruses and Viroids by NGS
Next-generation sequencing has significantly increased the number of novel plant viruses discovered and characterized both in host plants and in insect vectors. More than 100 novel DNA and RNA plant viruses from different genera and families have been reported in the recent years (Hadidi and Barba, 2012; Barba et al., 2014; Ho and Tzanetakis, 2014; Barba and Hadidi, 2015; Roossinck et al., 2015; Wu et al., 2015). Only two novel viroids, however, were discovered: persimmon viroid 2 (Ito et al., 2013) and grapevine latent viroid (Zhang et al., 2014). This trend has been observed in crop plants but also, to a very large extent, in wild plant species through the use of NGS in metagenomic approaches (Stobbe and Roossinck, 2014; Roossinck, 2015; Roossinck et al., 2015). Likewise, the sequences of many novel virus and viroid strains have been reported. In addition to discovering novel viruses, the complete nucleotide sequences of many known viruses were determined by NGS for complete virus characterization and/or virus identification in known and new hosts or for other reasons. For example, NGS has shown that the artichoke latent virus (ArLV) is a member of the genus Macluravirus, family Potyviridae, and that ranunculus latent virus should be considered as a strain of ArLV but not a distinct species (Minutillo et al., 2015); potato virus Y and potato virus S have been identified in Maori potato (Solanum tuberosum) and turnip mosaic virus in rengarenga (Arthropodium cirratum), which is a new host (Blouin et al., 2016). NGS analyses do not generally provide the final word on a new virus or viroid. The genome sequence generally has to be finalized using PCR-based approaches and Sanger sequencing and, as a general rule, the existence of the new virus or viroid should always be sought using an alternative technique. In addition, NGS librarary prepation methods with minimal bias should be used in order to obtain accurate and easy to interpret data (Van Dijk et al., 2014).
Another area where NGS has proven very valuable is in the detection of isolates, strains, or variants of known viruses that escape existing detection procedures and, particularly, PCR assays. The data obtained may afford a better knowledge of the polyvalence or specificity of existing assays and, if needed, facilitate the design of new detection primers of broader specificity for improving the classical detection assays. For example, the NGS discovery of a non-detectable isolate of plum bark necrotic and stem pitting associated virus led Marais et al. (2014) to develop a new PCR assay of broader specificity.
NGS has provided a very powerful alternative for detection and identification of plant viruses and viroids without a priori knowledge of pathogen sequence as required for PCR-based detection and identification methods. For this reason, NGS has become a universal approach for accurate detection and identification of many novel and known plant viruses. Viroids are also accurately and easily detected and identified by NGS (Table 3). Thus, NGS has the potential to be used as a primary diagnostic tool for plant viruses and viroids as the cost of sequencing platforms has become more competitive and affordable. Currently, the cost of NGS-based diagnostics is still high as compared to that of a PCR or serological assay, so that the technique is limited to situations where exhaustiveness is critical, such as quarantine or when trying to identify a causal agent, or to situations involving high value added samples, such as nuclear stock mother plants used for production of certified planting materials. It should be stressed that nucleic acids purifications and sequencing bank preparation protocols may have to be optimized and fine-tuned/adapted to particular plant species that may contain inhibitory substances that may otherwise interfere with the sensitivity of the detection procedure.
The volume and diversity of international movements of plant materials, including exchanges of germplasm or newly bred cultivars, have increased considerably during the last few decades, which have also created additional pathways for the introduction of plant viruses and viroids in new areas or for the emergence of novel virus or virus-like agents. Preventive measures for blocking the introduction and spread of these pathogens include the implementations of plant quarantine (phytosanitary regulation) and certification programs. These programs are tools that can be used to protect or improve the health status of cultivated plants (Foster and Hadidi, 1998; Barba, 1998; Barba et al., 2003; Singh et al., 2003; Reed and Foster, 2011; Roy, 2011). NGS has played a significant role in revealing new viruses and viroids of quarantine and/or certification importance. For example, imported sugarcane plants in quarantine in Montpellier, France, were found infected with a novel geminivirus, tentatively named sugarcane white streak virus, which had escaped detection by standard detection tests (Candresse et al., 2014). Similarly, NGS allowed the identification of a novel luteovirus in imported nectarine trees in the US (Bag et al., 2015; Villamor et al., 2016) and of a novel marafivirus (Villamor et al., 2016), suggesting that this technology could/should be adopted as a post-entry quarantine measure (see below). Also, very recently, NGS revealed that the causal agent of severe stunting and death of hop plants in Slovenia is citrus bark cracking viroid (CBCVd; Jakse et al., 2015), a pathogen reported previously in citrus plants wherein it induces minor damage (Duran-Vila and Semancik, 2003). Subsequently, CBCVd has been added to the certification list of hop planting material in Slovenia; in addition, the European and Mediterranean Plant Protection Organization (EPPO) has included the viroid to “The EPPO Alert List” (Jakse et al., 2015), so member countries and other countries may include it in their certification or quarantine programs or become aware of potential problems.
Recently, the US Department of Agriculture, Animal and Plant Inspection Service (APHIS), Plant Protection and Quarantine (PPQ) formed an internal working group to discuss the application of NGS to PPQ policy and operations. There are a number of challenges, however, that have to be addressed on the use of NGS in quarantine before a regulatory policy can be implemented. These include standardization of testing methods, interpretation of test results, biology of the discovered new pathogen, constructing a reliable database of whole genome sequence of pathogens of quarantine importance and others (E. V. Podleckis, personal communication 2016; M. K. Nakhla, personal communication 2016). It is expected that these challenges will be soon resolved due to advances in NGS capabilities and the rapid adaptation of this technology in plant pathogen diagnostics. NGS can become instrumental in releasing plants in quarantine and certification programs at a faster rate than current strategies while improving our ability to prevent the introduction of foreign plant viruses and/or viroids into new countries. Thus, NGS has the potential to be utilized in plant quarantine and certification programs standard assays in North America and Europe in the near future as, when compared with routine conventional assays, it could reduce significantly the number of non-detected viruses and viroids.
Relationship of DNA Green Algal Viruses and ssRNA Plant Viruses to Human Disorders/Diseases as Revealed by NGS
While no plant virus has been shown so far to be harmful to humans, the use of NGS approaches has yielded recently a few tantalizing results that may in time question this long held vision. Sequences homologous to the DNA of the green algal virus acanthocystis turfacea chlorella virus 1 (ATCV 1) were detected and identified in human oropharyngeal samples of healthy normal adults without any physical or mental disorder/illness (Yolken et al., 2014). Mice inoculated with ATCV 1, however, developed memory loss and other symptoms indicating a general decrease performance in several cognitive domains. On the other hand, DNA viruses that infect the green alga Phaeodactylum tricornutum have been associated with vaginitis (Stepanova et al., 2011). Women with this disease swam in the Black Sea two to three months before the symptoms appeared. Very recently, it was shown by NGS that the green alga DNA virus TsV-N1 that infects Tetraselmis striata has two genes with closest similarity to genes in parasites of the human urogenital system, Trichomonas vaginalis and Candida albicans (Pagarete et al., 2015).
Similarly, the observation that the RNA viral community of the human feces is dominated by plant viruses (Zhang et al., 2006) came as a surprise. It prompted further efforts that led to a report that the ssRNA pepper mild mottle virus (PMMV) was highly represented in a human population, that anti PMMV IgM antibodies could be detected in some persons, and that some relationship between PMMV detection and some clinical symptoms might exist (Colson et al., 2010). These findings might be also correlated with a report showing that the negative ssRNA tomato spotted wilt virus (TSWV) is able to replicate in human cell lines expressing the viral “polymerase-bound host factor” (de Medeiros et al., 2005).
More insights into the role of PMMV, TSWV, algal viruses and other higher plant and algal viruses in human diseases may be revealed by metagenomic studies using NGS of human gut viromes and other organs of patients (Balique et al., 2015). Thus, the relationship between these viruses and human disorders/diseases may need serious re-evaluation.
Possible Sequencing of Old/Ancient Viruses and Viroids by NGS
NGS technology advances, plus new sample preparation techniques, have allowed researchers to sequence complete ancient genomes from modern human ancestors and archaic humans (Gibson, 2015). NGS has also revealed the sequence of the ancient DNA of the 19th century late blight oomycetes pathogen, Phytophthora infestans, which caused the Irish potato famine of 1845–1847 (Martin et al., 2013). The high quality of the pathogen DNA in the 166–168 years old herbarium material suggests that DNA and RNA plant viruses as well as viroids in dried plant samples in herbaria, museums, or other places world-wide, could be used for studies of past epidemics and/or evolution of these pathogens by NGS. Studies using RT-PCR identified peach latent mosaic viroid in 50-year-old herbarium-preserved peach leaves showing symptoms of peach calico disease (Guy, 2013), and apple scar skin viroid in a 10-year-old air dried apple tree twig with no disease symptoms (A. Hadidi, unpuplished data). Plant viruses were also reported in 50- to 100-year-old herbarium samples using different traditional detection methods (for references, see Guy, 2013). In what may be the most remarkable result to date, Smith et al. (2014) were able to assemble the complete genome of a barley stripe mosaic virus (BSMV) isolate from small RNA sequences from barley grains that were approximately 750 years old. Interestingly, the sequence obtained does not fit well the phylogenetic reconstruction of the evolutionary timeline for BSMV, questioning the previously reconstructed history of BSMV and the hypothesis of a recent origin of the virus. Similarly, aged or older plant and soil samples could also be analyzed by NGS for plant viruses and viroids, as illustrated by the recent discovery of viral genomes in 700-years old caribou feces from a subarctic ice patch (Ng et al., 2014) and of a giant DNA virus named pithovirus sibericum in a 30,000 years old Siberian permafrost sample (Legendre et al., 2014). Such analyses would potentially allow to gain knowledge on the evolutionary history of plant viruses and viroids over the past few millennia, a time period hypothesized to have seen the emergence of several very important viral genera, as for example the evolutionary radiation of the potyviruses (Gibbs et al., 2008), the most important and numerous plant virus genus.
Genome Editing Using the CRISPR-Cas9 System in Plant Virology
CRISPR associated Cas proteins (CRISPR-Cas) systems act as archaea and bacteria immune systems against invading foreign DNAs (Mojica et al., 2005; Barrangour et al., 2007; Makarova et al., 2011; Sorek et al., 2013). The Cas proteins, such as Cas9, are RNA-directed endonucleases able to recognize and cleave nucleic acids on the basis of sequence complementarities (Brouns et al., 2008; Bhaya et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012; Westra et al., 2012; Sorek et al., 2013) and to modify the targeted sequences (Hsu et al., 2014). Cas9 can be targeted to specific DNA genomic sequences by engineering separately an encoded small guide RNA (sgRNA) with which it forms a complex (Doudna and Charpentier, 2014). Thus, only a short RNA sequence must therefore be synthesized to confer recognition of a new target. Recently, an alternative to the Cas9 enzyme, Cpf1, has been reported (Zetsche et al., 2015), which is smaller in size and makes genome editing easier and more precise.
RNA-guided cleavage paired with donor-guided repair allows easy introduction of any desired modification in a living cell. The possibility of re-directing the dsDNA targeting capability of CRISPR-Cas9 for RNA-guided ssRNA binding and/or cleavage (which is denoted RCas9, an RNA targeting Cas9) has been reported (O’Connell et al., 2014), and even more recently, that C2c2 effector functions as an RNA-guided RNA-targeting CRISPR (Abudayyeh et al., 2016).
The double-strand breaks created by Cas9 induce insertion or deletion (indel) mutations in the targeted gene or genome sequence of an organism (Ran et al., 2013), which are repaired by non-homologous end joining (NHEJ) and/or homology directed repair (HDR). NHEJ aligns one to few complementary bases for the re-ligation of two ends, whereas HDR uses longer stretches of sequence homology to repair DNA breaks. Screening, identification and frequency of mutations can be done efficiently and rapidly using NGS rather than the first-generation Sanger sequencing method (Bell et al., 2014).
During the last few years, the CRISPR-Cas based systems have become the method of choice for genome editing by introducing or correcting genetic mutations in a wide variety of biological contexts: cell lines, animals (including humans) and plants (Belhaj et al., 2015), as well as human RNA and DNA viruses (Price et al., 2015; Kennedy and Cullen, 2015). The advantages of the CRISPR-Cas genome editing over the other known genome editing systems are that it is faster, easier to use and applicable to many species, as opposed to being species-specific, as it has been used in organisms recalcitrant to previous attempts at genome engineering. It is also versatile as it can be used to introduce or delete a number of different genes at a time and does not require many manipulating tools.
Developing Plants Resistant to DNA Geminiviruses
Geminiviruses (family Geminiviridae) are circular ssDNA viruses with genomes of 2.3 to slightly above 5 kb, distributed worldwide and transmitted by insects, which cause serious damage to many economically important dicotyledonous and monocotyledonous crops (Moffat, 1999; Shamloul et al., 2001; Mansoor et al., 2003; Vanitharani et al., 2005; Czosnek et al., 2013; Hanley-Bowdoin et al., 2013). They replicate in plant nuclei through dsDNA intermediates that also serve as templates for transcription (Pilartz and Jeske, 1992). Current strategies for controlling geminiviruses vary from conventional breeding of resistant cultivars and insect vectors control to molecular methods based on transgenic plants expressing mutated viral proteins, RNA-mediated interference and others (Pilartz and Jeske, 1992; Vanderschuren et al., 2007; Aragao and Faria, 2009; Reyes et al., 2013; Yang C.-F. et al., 2014; Lapidot et al., 2015). All these strategies have met so far with marginal success.
Very recently, the application of the CRISPR-Cas systems targeting geminiviruses has been shown to enhance resistance to tomato yellow leaf curl virus (TYLCV, genus Begomovirus) and bean yellow dwarf virus (BeYDV, genus Mastrevirus) in Nicotiana benthamiana (Ali et al., 2015; Baltes et al., 2015) and to beet severe curly top virus (BSCTV, genus Curtovirus) in N. benthamiana and in Arabidopsis (Ji et al., 2015).
Ali et al. (2015) engineered sgRNAs targeting coding and non-coding TYLCV sequences, including the conserved non-coding intergenic region (IR) of about 300 nt that can form a stem-loop structure containing the origin of replication and promoter sequences for RNA polymerase II (Czosnek et al., 2013; Mori et al., 2013; Yang X. et al., 2014). SgRNAs targeting the IR were the most efficient in reducing TYLCV DNA titer and attenuating disease symptoms. The sgRNAs-Cas9 system was also successful in targeting simultaneously the three geminiviruses TYLCV, beet curly top virus (genus Curtovirus) and merremia mosaic virus (genus Begomovirus) when sgRNAs specific for the IR sequence of each virus were used (Ali et al., 2015).
Similarly, Ji et al. (2015) targeted coding and non-coding sequences of BSCTV genome. SgRNA-Cas9 constructs inhibited the virus DNA replication at varying levels. Disease symptoms were also attenuated to different levels, which ranged from severe to mild leaf curly symptoms. Over-expressing sgRNA-Cas9 specifically targeting the viral DNA genome sequences resulted in virus-resistant plants.
It has been shown that two sgRNAs targeting the same BeYDV genome site in infected N. benthamiana plants can significantly increase resistance as compared to using only one single sgRNA (Baltes et al., 2015). Moreover, NGS analysis of indels within the viral genome suggested that Cas9 can bind to the viral genome and introduce the dsDNA breaks at the targeted sites, and also indicated that most mutations were 1–2 bp indels.
Developing Plants Resistant to RNA Viruses
Plant RNA viruses depend on host factors for their replication and infection as their coding capacity is limited (Sanfacon and Jovel, 2007; Nagy and Pogany, 2012; Wang, 2015). Recessive genes, including those coding for the translation initiation factors, confer plant resistance to infection by RNA viruses (Truniger and Aranda, 2009; Sanfaçon, 2015). More specifically, the eukaryotic translation initiation factor eIF4E and its isoform have been identified in cucumber (Cucumus sativus L.; Rodríguez-Hernández et al., 2012). In infected plant cells both proteins interact with the small VPg protein of some RNA viruses, such as members of the families Potyviridae and Secoviridae and of the genus Sobemovirus and alterations in such interactions result in a broad-spectrum resistance to viruses (Sanfaçon, 2015).
It had been shown that engineering mutations in the host plant eIF4E through the use of the Targeting-Induced Local Lesions in Genome (TILLING) strategy could provide resistance against potyviruses in a range of plants (Julio et al., 2015; Gauffier et al., 2016). The CRISPR-Cas9 sgRNA system has now been used to efficiently inactivate these genes and generate virus-resistant plants (Chandrasekaran et al., 2016; Pyott et al., 2016). For example, the targeting of two sites of eIF4E gene in cucumber allowed developing plants resistant to infection by five positive-strand RNA viruses (Chandrasekaran et al., 2016). The targeted gene sites were within the first and third exon sequences. Small indels and small nucleotide polymorphisms (SNPs) were observed in the T1 generation. Homozygous T3 progeny with 20 and four deletions in the eIF4E gene were immune to infection by cucumber vein yellowing virus (family Potyviridae, genus Ipomovirus), and resistant to papaya ringspot virus-W and zucchini yellow mosaic virus (family Potyviridae, genus Potyvirus). As expected, the plants remained susceptible to viruses that do not appear to highjack the host eIF4E to complete their cycle, such as cucumber mosaic virus (family Bromoviridae, genus Cucumovirus) or cucumber green mottle mosaic virus (family, Virgaviridae, genus Tobamovirus). Pyott et al. (2016) were also successful in engineering complete resistance in Arabidopsis to turnip mosaic virus, a potyvirus, using the CRISPR/Cas9 technology targeting the plant elF (iso) 4E gene.
Conclusion and Prospective
For the last seven years, NGS and bioinformatics have provided rapid and low cost DNA and/or RNA sequencing for plant viruses and viroids. Full genomes or virus- or viroid-specific small RNAs, which cover essentially the whole genome, have been sequenced for discovery of novel pathogens, as well as for pathogen detection and identification, replication, ecology, epidemiology, and pathogen-host interactions. Many novel plant RNA and DNA viruses were successfully discovered but only a couple of novel viroids were revealed. Known viruses and viroids were successfully deteted and identified by NGS. Prior knowledge of virus or viroid sequence was not needed, which has made NGS a universal rapid and accurate method for pathogen discovery and diagnostics. Using NGS in a quarantine program, a novel virus in imported sugarcane was discovered that previously escaped detection by standard detection methods. Similarly, using NGS in a certification program, the causal agent of severe hop stunt disease has been very recently discovered and now the disease is under control. In the very near future NGS may be utilized in plant quarantine and certification programs to significantly increase their capacities and reliabilities. NGS is playing a significant role in connecting plant virology with other related fields of biology such as genome editing. Very recently, CRISPR-Cas9 system has been used in developing plants resistant to five DNA geminiviruses and four RNA potyviruses. Since CRISPR/Cas9 system has the capabilities to work on DNA or RNA sequences, expanding the use of these capabilities in research on DNA/RNA viruses or viroids, in association with NGS, may open widely the door for studying control of diseases caused by these pathogens at the genomic level. These advances are propelled in part by synergies between two powerful technologies: NGS and genome engineering.
Author Contributions
AH conceived and wrote the manuscript, and RF, TC, and MB revised it and made useful suggestions. AH prepared the final version of the manuscript and all authors approved it.
Funding
Work in RF laboratory has been supported by the Spanish Ministerio de Economía y Competitividad (grant BFU2014-56812-P), and in MB laboratory by Consiglio per la Ricerca in Agricoltura e l’analisi dell’Economia Agraria (CREA).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer VP declared a shared affiliation, though no other collaboration, with one of the authors RF to the handling Editor, who ensured that the process nevertheless met the standards of a fair and objective review.
References
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B. T., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573
Adams, I. P., Glover, R. H., Monger, W. A., Mumford, R., Jackeviciene, E., Navalinskiene, M., et al. (2009). Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol. Plant Pathol. 10, 537–545. doi: 10.1111/j.1364-3703.2009.00545.x
Adkar-Purushothama, C. R., Brosseau, C., Giguère, T., Sano, T., Moffett, P., and Perreault, J. P. (2015). Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 27, 2178–2194. doi: 10.1105/tpc.15.00523
Al Rwahnih, M., Daubert, S., Golino, D., and Rowhani, A. (2009). Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 387, 395–401. doi: 10.1016/j.virol.2009.02.028
Alabi, O. J., Zheng, Y., Jagadeeswaran, G., Sunkar, R., and Naidu, R. (2012). “High-throughput sequence analysis of small RNAs in grapevine (Vitis vinifera L.) affected by grapevine leafroll disease,” in Proceedings of the17th Congress of ICVG, Davis, CA.
Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M., and Mahfouz, M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16:238. doi: 10.1186/s13059-015-0799-6
Aragao, F. J. L., and Faria, J. C. (2009). First transgenic geminivirus-resistant plant in the field. Nat. Biotechnol. 27, 1086–1088. doi: 10.1038/nbt1209-1086
Aregger, M., Borah, B. K., Seguin, J., Rajeswaran, R., Gubaeva, E. G., Zvereva, A. S., et al. (2012). Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 8:e1002941. doi: 10.1371/journal.ppat.1002941
Avina-Padilla, K., Martínez de la Vega, O., Rivera-Bustamante, R., Martínez-Soriano, J. P., Owens, R. A., Hammond, R. W., et al. (2015). In silico prediction and validation of potential gene targets for pospiviroid-derived small RNAs during tomato infection. Gene 564, 197–205. doi: 10.1016/j.gene.2015.03.076
Bag, S., Al Rwahnih, M., Li, A., Gonzalez, A., Rowhani, A., Uyemoto, J. K., et al. (2015). Detection of a new luteovirus in imported nectarine trees: a case study to propose adoption of metagenomics in post-entry quarantine. Phytopathology 105, 840–846. doi: 10.1094/PHYTO-09-14-0262-R
Balique, F., Lecoq, H., Raoult, D., and Colson, P. (2015). Can plant viruses cross the kingdom border and be pathogenic to humans? Viruses 7, 2074–2098. doi: 10.3390/v7042074
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 1:15145. doi: 10.1038/nplants.2015.145
Barba, A. (1998). “Virus certification of fruit tree propagative material in Western Europe,” in Plant Virus Disease Control, eds A. Hadidi, R. K. Khetarpal, and H. Kogenazawa (St. Paul, MN: APS Press), 288–293.
Barba, M., Czosnek, H., and Hadidi, A. (2014). Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 6, 106–136. doi: 10.3390/v6010106
Barba, M., Gumpf, D. J., and Hadidi, A. (2003). “Quarantine in imported germplasm,” in Viroids, eds A. Hadidi, R. Flores, J. W. Randles, and J. W. Semancik (Collingwood, VIC: CSIRO Publishing), 303–311.
Barba, M., and Hadidi, A. (2015). An overview of plant pathology and application of next-generation sequencing technologies. CAB Rev. 10, 1–21. doi: 10.1079/PAVSNNR201510005
Barba, M., Ilardi, V., and Pasquini, G. (2015). Control of pome and stone fruit virus diseases. Adv. Virus Res. 91, 47–61. doi: 10.1016/bs.aivir.2014.11.001
Barrangour, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., and Nekrasov, V. (2015). Editng plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. doi: 10.1016/j.copbio.2014.11.007
Bell, C. C., Magor, G. W., Gillender, K. R., and Perkins, A. C. (2014). A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genomics 15:1002. doi: 10.1186/1471-2164-15-1002
Bhaya, D., Davison, M., and Barrangou, R. (2011). CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. doi: 10.1146/annurev-genet-110410-132430
Blevins, T., Rajeswaran, R., Aregger, M., Borah, B. K., Schepetilnikov, M., Baerlocher, L., et al. (2011). Massive production of small RNAs from a non-coding region of cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 39, 5003–5014. doi: 10.1093/nar/gkr119
Blouin, A. G., Ross, H. A., Hobson-Peters, J., O’Brien, C., Warren, B., and MacDiarmid, R. (2016). A new virus discovered by immunocapture of double-stranded RNA, a rapid method for virus enrichment in metagenomic studies. Mol. Ecol. Resour. doi: 10.1111/1755-0998.12525 [Epub ahead of print].
Bolduc, F., Hoareau, C., St-Pierre, P., and Perreault, J. P. (2010). In-depth sequencing of the siRNAs associated with peach latent mosaic viroid infection. BMC Mol. Biol. 11:16. doi: 10.1186/1471-2199-11-16
Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Candresse, T., Filloux, D., Muhire, B., Julian, C., Galzi, S., Fort, G., et al. (2014). Appearances can be deceptive: revealing a hidden viral infection with deep sequencing in a plant quarantine context. PLoS ONE 9:e102945. doi: 10.1371/journal.pone.0102945
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chiumenti, M., Torchetti, E. M., Di Serio, F., and Minafra, A. (2014). Identification and characterization of a viroid resembling apple dimple fruit viroid in fig (Ficus carica L.) by next generation sequencing of small RNAs. Virus Res. 188, 54–59. doi: 10.1016/j.virusres.2014.03.026
Colson, P., Richet, H., Desnues, C., Balique, F., Moal, V., Grob, J.-J., et al. (2010). Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS ONE 5:e10041. doi: 10.1371/journal.pone.0010041
Czosnek, H., Eybishtz, A., Sade, D., Gorovits, R., Sobol, I., Bejarano, E., et al. (2013). Discovering host genes involved in the infection by the tomato yellow leaf curl virus complex and in the establishment of resistance to the virus using tobacco-rattle virus-based post transcriptional gene silencing. Viruses 5, 998–1022. doi: 10.3390/v5030998
de Medeiros, R. B., Figueiredo, J., Resende Rde, O., and De Avila, A. C. (2005). Expression of a viral polymerase-bound host factor turns human cell lines permissive to a plant- and insect-infecting virus. Proc. Natl. Acad. Sci. U.S.A. 102, 1175–1180. doi: 10.1073/pnas.0406668102
Di Serio, F., Flores, R., Verhoeven, J. T. J., Li, S. F., Pallás, V., Randles, J. W., et al. (2014). Current status of viroid taxonomy. Arch. Virol. 159, 3467–3478. doi: 10.1007/s00705-014-2200-6
Di Serio, F., Gisel, A., Navarro, B., Delgado, S., Martínez de Alba, A. E., Donvito, G., et al. (2009). Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: implications, for their genesis and for pathogenesis. PLoS ONE 4:e7539. doi: 10.1371/journal.pone.0007539
Di Serio, F., Martínez de Alba, A. E., Navarro, B. A., Gisel, A., and Flores, R. (2010). RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J. Virol. 84, 2477–2489. doi: 10.1128/JVI.02336-09
Diener, T. O. (1971a). Potato spindle tuber virus with properties of a free nucleic acid. III. Subcellular location of PSTV-RNA and the question of whether virions exist in extracts or in situ. Virology 43, 75–89. doi: 10.1016/0042-6822(71)90226-1
Diener, T. O. (1971b). Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology 45, 411–428. doi: 10.1016/0042-6822(71)90342-4
Diener, T. O. (1972). Potato spindle tuber viroid. VII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 50, 606–609. doi: 10.1016/0042-6822(72)90412-6
Diermann, N., Matousek, J., Junge, M., Riesner, D., and Steger, G. (2010). Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 391, 1379–1390. doi: 10.1515/BC.2010.148
Donaire, L., Wang, Y., González-Ibeas, D., Mayer, K. F., Aranda, M. A., and Llave, C. (2009). Deep sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392, 203–214. doi: 10.1016/j.virol.2009.07.005
Doudna, J. A., and Charpentier, E. (2014). Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Dunham, J. P., Simmons, H. E., Holmes, E. C., and Stephenson, A. G. (2014). Molecular analysis of viral (zucchini yellow mosaic virus) genetic diversity during systemic movement through a Cucurbita pepo vine. Virus Res. 191, 172–179. doi: 10.1016/j.virusres.2014.07.030
Duran-Vila, N., and Semancik, J. S. (2003). “Citrus viroids,” in Viroids, eds A. Hadidi, R. Flores, J. W. Randles, and J. W. Semancik (Collingwood, VIC: CSIRO Publishing), 178–194.
Flores, R., Minoia, S., Carbonell, A., Gisel, A., Delgado, S., López-Carrasco, A., et al. (2015). Viroids, the simplest RNA replicons: how they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 209, 136–145. doi: 10.1016/j.virusres.2015.02.027
Foster, J. A., and Hadidi, A. (1998). “Exclusion of plant viruses,” in Plant Virus Disease Control, eds A. Hadidi, R. K. Khetarpal, and H. Kogenazawa (St. Paul, MN: APS Press), 208–229.
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–E2586. doi: 10.1073/pnas.1208507109
Gauffier, C., Lebaron, C., Moretti, A., Constant, C., Moquet, F., Bonnet, G., et al. (2016). A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J. 85, 717–729. doi: 10.1111/tpj.13136
Giampetruzzi, A., Roumi, V., Roberto, R., Malossini, U., Yoshikawa, N., La Notte, P., et al. (2012). A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small small RNAs in Cv Pinot gris. Virus Res. 163, 262–268. doi: 10.1016/j.virusres.2011.10.010
Gibbs, A. J., Ohshima, K., Phillips, M. J., and Gibbs, M. J. (2008). The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS ONE 3:e2523. doi: 10.1371/journal.pone.0002523
Gibson, A. (2015). Revolution in human evolution. Science 349, 362–366. doi: 10.1126/science.349.6246.362
Glouzon, J. P.-S., Bolduc, F., Wang, S., Najmanovich, R. J., and Perreault, J.-P. (2014). Deep-sequencing of the peach latent mosaic viroid reveals new aspects of population heterogeneity. PLoS ONE 9:e87297. doi: 10.1371/journal.pone.0087297
Guy, P. L. (2013). Ancient RNA? RT-PCR of 50-year-old RNA identifies peach latent mosaic viroid. Arch. Virol. 158, 691–694. doi: 10.1007/s00705-012-1527-0
Hadidi, A., and Barba, M. (2012). Next-generation sequencing: historical perspective and current applications in plant virology. Petria 22, 262–277.
Hadidi, A., Barba, M., Candresses, T., and Jelkmann, W. (eds). (2011). Virus and Virus-Like Diseases of Pome and Stone Fruits. St. Paul, MN: APS Press, 429.
Hadidi, A., Flores, R., Randles, J. W., and Semancik, J. S. (eds). (2003). Viroids. Collingwood, VIC: CSIRO Publishing, 370.
Hadidi, A., Khetarpal, R. K., and Kogenazawa, H. (eds). (1998). Plant Virus Disease Control. St. Paul, MN: APS Press, 684.
Hagen, C., Frizzi, A., Kao, J., Jia, L., Huang, M., Zhang, Y., et al. (2011). Using small RNA sequences to diagnose, sequence, and investigate the infectivity characteristics of vegetable-infecting viruses. Arch. Virol. 156, 1209–1216. doi: 10.1007/s00705-011-0979-y
Hammann, C., and Steger, G. (2012). Viroid-specific small RNA in plant disease. RNA Biol. 9, 809–819. doi: 10.4161/rna.19810
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., and Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
He, Y., Yang, Z., Hong, N., Wang, G., Ning, G., and Xu, W. (2015). Deep sequencing reveals a novel closterovirus associated with wild rose leaf rosette disease. Mol. Plant Pathol. 16, 449–458. doi: 10.1111/mpp.12202
Herranz, M. C., Navarro, J. A., Sommen, E., and Pallás, V. (2015). Comparative analysis among the small RNA populations of source, sink and conductive tissues in two different plant-virus pathosystems. BMC Genomics 16:117. doi: 10.1186/s12864-015-1327-5
Ho, T., and Tzanetakis, J. E. (2014). Development of a virus detection and discovery pipeline using next generation sequencing. Virology 47, 54–60. doi: 10.1016/j.virol.2014.09.019
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPRCas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, Q., Hollunder, J., Niehl, A., Korner, C. J., Gereige, D., Windels, D., et al. (2011). Specific impact of tobamavirus infection on the Arabidopsis small RNA profile. PLoS ONE 6:e19549. doi: 10.1371/journal.pone.0019549
Ito, T., Suzaki, K., Nakano, M., and Sato, A. (2013). Characterization of a new apscaviroid from American persimmon. Arch. Virol. 158, 2629–2631. doi: 10.1007/s00705-013-1772-x
Jakse, J., Radisek, S., Pokorn, T., Matousek, J., and Javornik, B. (2015). Deep-sequencing revealed Citrus bark cracking viroid (CBCVd) as a highly aggressive pathogen on hop. Plant Pathol. 64, 831–842. doi: 10.1111/ppa.12325
Ji, X., Zhang, H., Zhang, Y., Wang, Y., and Gao, C. (2015). Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1:15144. doi: 10.1038/nplants.2015.144
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. A. (2012). programmable dual-RNA guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jo, Y., Choi, H., Yoon, J.-Y., Choi, S.-K., and Cho, W. K. (2015). De novo genome assembly of grapevine yellow speckle viroid 1 from a grapevine transcriptome. Genome Announc. 3:e496-15. doi: 10.1128/genomeA.00496-15
Julio, E., Cotucheau, J., Decorps, C., Volpatti, R., Sentenac, C., Candresse, T., et al. (2015). A eukaryotic translation initiation factor 4E (eIF4E) is responsible for the “va” tobacco recessive resistance to potyviruses. Plant Mol. Biol. Rep. 33, 609–623. doi: 10.1007/s11105-014-0775-4
Kashif, M., Pietila, S., Artola, K., Tugume, A. K., Makinen, V., and Valkonen, J. P. T. (2012). Detection of viruses in sweetpotato from Honduras and Guatemala augmented by deep-sequencing of small-RNAs. Plant Dis. 96, 1430–1437. doi: 10.1094/PDIS-03-12-0268-RE
Kennedy, E. M., and Cullen, B. R. (2015). Bacterial CRISPR/Cas DNA endonuclease: a revolutionary technology that could dramatically impact viral research and treatment. Virology 47, 213–220. doi: 10.1016/j.virol.2015.02.024
King, A. M. Q., Adams, M. J., Carstens, E. B., and Lefkowitz, E. J. (eds). (2012). Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press, 1327.
Kreuze, J. F., Pérez, A., Untiveros, M., Quispe, D., Fuentes, S., Barker, I., et al. (2009). Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology 388, 1–7. doi: 10.1016/j.virol.2009.03.024
Kutnjak, D., Silvestre, R., Cuellar, W., Perez, W., Müller, G., Ravnikar, M., et al. (2014). Complete genome sequences of new divergent potato virus X isolates and discrimination between strains in a mixed infection using small RNAs sequencing approach. Virus Res. 191, 45–50. doi: 10.1016/j.virusres.2014.07.012
Lapidot, M., Karniel, U., Gelbart, D., Fogel, D., Evenor, D., Kutsher, Y., et al. (2015). A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genet. 11:e1005538. doi: 10.1371/journal.pgen.1005538
Legendre, M., Bartoli, J., Shmakova, L., Jeudy, S., Labadie, K., Adrait, A., et al. (2014). Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U.S.A. 111, 4274–4279. doi: 10.1073/pnas.1320670111
Li, R., Gao, S., Berendsen, S., Fei, Z., and Ling, K.-S. (2015). Complete genome sequence of a novel genotype of squash mosaic virus infecting squash in Spain. Genome Announc. 3, e1583-14. doi: 10.1128/genomeA.01583-14
Li, R., Gao, S., Fei, Z., and Ling, K.-S. (2013). Complete genome sequence of a new tobamovirus naturally infecting tomatoes in Mexico. Genome Announc. 1, e794-13. doi: 10.1128/genomeA.00794-13
Li, R., Gao, S., Hernandez, A. G., Wechter, W. P., Fei, Z., and Ling, K.-S. (2012). Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PLoS ONE 7:e37127. doi: 10.1371/journal.pone.0037127
Liang, P., Navarro, B., Zhang, Z., Wang, H., Lu, M., Xiao, H., et al. (2015). Identification and characterization of a novel geminivirus with a monopartite genome infecting apple trees. J. Gen. Virol. 96, 2411–2420. doi: 10.1099/vir.0.000173
Lin, K.-Y., Cheng, C.-P., Chang, B. C.-H., Wang, W.-C., Huang, Y.-W., Lee, Y.-S., et al. (2010). Global analysis of small interfering RNAs derived from Bamboo mosaic virus and its associated satellite RNAs in different plants. PLoS ONE 5:e11928. doi: 10.1371/journal.pone.0011928
Loconsole, G., Onelge, N., Potere, O., Giampetruzzi, A., Bozan, O., Satar, S., et al. (2012a). Identification and characterization of Citrus yellow vein clearing virus, a putative new member of the genus Mandarivirus. Phytopathology 102, 1168–1175. doi: 10.1094/PHYTO-06-12-0140-R
Loconsole, G., Saldarelli, P., Doddapaneni, H., Savino, V., Martelli, G. P., and Saponari, M. (2012b). Identification of a single-stranded DNA virus associated with citrus chlorotic dwarf disease, a new member of the family Geminiviridae. Virology 432, 162–172. doi: 10.1016/j.virol.2012.06.005
Ma, Y., Navarro, B., Zhang, Z., Lu, M., Zhou, X., Chi, S., et al. (2015). Identification and molecular characterization of a novel monopartite geminivirus associated with mulberry mosaic dwarf disease. J. Gen. Virol. 96, 2421–2434. doi: 10.1099/vir.0.000175
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Mansoor, S., Briddon, R. W., Zafar, Y., and Stanley, J. (2003). Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8, 128–134. doi: 10.1016/S1360-1385(03)00007-4
Marais, A., Faure, C., Couture, C., Bergey, B., Gentit, P., and Candresse, T. (2014). Characterization by deep sequencing of divergent plum bark necrosis stem pitting associated virus isolates and development of a broad-spectrum PBNSPaV-specific detection assay. Phytopathology 104, 660–666. doi: 10.1094/PHYTO-08-13-0229-R
Martin, M. D., Cappellini, E., Samaniego, J. A., Zepeda, M. L., Campos, P. F., Segium-Orlando, A., et al. (2013). Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Comm. 4:2172. doi: 10.1038/ncomms3172
Martinez, G., Donaire, L., Llave, C., Pallas, V., and Gómez, G. (2010). High-throughput sequencing of Hop stunt viroid-derived small RNAs from cucumber leaves and phloem. Mol. Plant Pathol. 11, 347–359. doi: 10.1111/j.1364-3703.2009.00608.x
Minutillo, S. A., Marais, A., Mascia, T., Faure, C., Svanella-Dumas, L., Theil, S., et al. (2015). Complete nucleotide sequence of artichoke latent virus shows it to be a member of the genus Macluravirus in the family Potyviridae. Phytopathology 105, 1155–1160. doi: 10.1094/PHYTO-01-15-0010-R
Mitter, N., Koundal, V., Williams, S., and Pappu, H. (2013). Differential expression of tomato spotted wilt virus-derived viral small RNAs in infected commercial and experimental host plants. PLoS ONE 8:e76276. doi: 10.1371/journal.pone.0076276
Moffat, A. S. (1999). Geminiviruses emerge as serious crop threat. Science 286, 1835. doi: 10.1126/science.286.5446.1835
Mojica, F. J. M., Diez-Villasenor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Mori, T., Takenaka, K., Domoto, F., Aoyama, Y., and Sera, T. (2013). Inhibition of binding of tomato yellow leaf curl virus rep to its replication origin by artificial zinc-finger protein. Mol. Biotechnol. 54, 198–203. doi: 10.1007/s12033-012-9552-5
Nagy, P. D., and Pogany, J. (2012). The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 10, 137–149. doi: 10.1038/nrmicro2692
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012a). Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 70, 991–1003. doi: 10.1111/j.1365-313X.2012.04940.x
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012b). Viroids: how to infect a host and cause disease without encoding proteins. Biochimie 94, 1474–1480. doi: 10.1016/j.biochi.2012.02.020
Navarro, B., Pantaleo, V., Gisel, A., Moxon, S., Dalmay, T., Bistray, G., et al. (2009). Deep sequencing of viroid-derived small RNAs from grapevine provides new insight on the role of RNA silencing in plant-viroid interaction. PLoS ONE 4:e7686. doi: 10.1371/journal.pone.0007686
Naveed, K., Mitter, N., Harper, A., Dhingra, A., and Pappua, H. R. (2014). Comparative analysis of virus-specific small RNA profiles of three biologically distinct strains of potato virus Y in infected potato (Solanum tuberosum) cv. Russet Burbank. Virus Res. 191, 153–160. doi: 10.1016/j.virusres.2014.07.005
Ng, T. F. F., Chen, L.-F., Zhou, Y., Shapiro, B., Stiller, M., Heintzman, P. D., et al. (2014). Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc. Natl. Acad. Sci. U.S.A. 111, 16106–16111. doi: 10.1073/pnas.1410429111
O’Connell, M. R., Oakes, B. L., Sternberg, S. H., East-Saletsky, A., Kaplan, M., and Doudna, J. A. (2014). Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266. doi: 10.1038/nature13769
Owens, R. A., Erbe, E., Hadidi, A., Steere, R. L., and Diener, T. O. (1977). Separation and infectivity of circular and linear forms of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U.S.A 74, 3859–3863. doi: 10.1073/pnas.74.9.3859
Pagarete, A., Grébert, T., Stepanova, O., Sandaa, R.-A., and Bratbak, G. (2015). Tsv-N1: a novel DNA algal virus that infects Tetraselmis striata. Viruses 7, 3937–3953. doi: 10.3390/v7072806
Pallett, D. W., Ho, T., Cooper, I., and Wang, H. (2010). Detection of cereal yellow dwarf virus using small interfering RNAs and enhanced infection rate with cocksfoot streak virus in wild cocksfoot grass (Dactylis glomerata). J. Virol. Methods 168, 223–227. doi: 10.1016/j.jviromet.2010.06.003
Pantaleo, V., Saldarelli, P., Miozzi, L., Giampetruzzi, A., Gisel, A., Moxon, S., et al. (2010). Deep sequencing analysis of viral short RNAs from an infected Pinot noir grapevine. Virology 408, 49–56. doi: 10.1016/j.virol.2010.09.001
Piernikarczyk, R., Matousek, J., Riesner, D., and Steger, G. (2013). “Potential mRNA targets of viroid-specific small RNA,” in Processeedings of the Abstracts International Workshop on Viroids and Satellite RNAs, Beijing, 23.
Pilartz, M., and Jeske, H. (1992). Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 189, 800–802. doi: 10.1016/0042-6822(92)90610-2
Pirovano, W., Miozzi, L., Boetzer, M., and Pantaleo, V. (2015). Bioinformatics approaches for viral metagenomics in plants using short RNAs: model case of study and application to a Cicer arietinum population. Front. Microbiol. 5:790. doi: 10.3389/fmicb.2014.00790
Poojari, S., Alabi, O. J., Fofanov, V. Y., and Naidu, R. A. (2013). A leafhopper-transmissible DNA virus with novel evolutionary lineage in the family Geminiviridae implicated in grapevine red leaf disease by next-generation sequencing. PLoS ONE 8:e64194. doi: 10.1371/journal.pone.0064194
Price, A. A., Sampson, T. R., Ratner, H. K., Grakoui, A., and Weiss, D. S. (2015). Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 112, 6164–6169. doi: 10.1073/pnas.1422340112
Pyott, D. E., Sheehan, E., and Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. doi: 10.1111/mpp.12417 [Epub ahead of print].
Qi, X., Bao, F. S., and Xie, Z. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4:e4971. doi: 10.1371/journal.pone.0004971
Rajeswaran, R., Aregger, M., Zvereva, A. S., Borah, B. K., Gubaeva, E. G., and Pooggin, M. M. (2012). Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 40, 6241–6254. doi: 10.1093/nar/gks242
Ran, F. A., Hsu, P. D., Wright, J., Agrawala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas system. Nat. Protoc. 153, 910–918.
Reed, P. J., and Foster, J. A. (2011). “Exclusion of pome and stone fruit viruses, viroids and phytoplasmas by certification and quarantine,” in Virus and Virus-Like Diseases of Pome and Stone Fruits, eds A. Hadidi, M. Barba, T. Candresses, and W. Jelkmann (St. Paul, MN: APS Press), 381–388.
Reyes, M. I., Nash, T. E., Dallas, M. M., Ascencio-Ibanez, J. T., and Hanley-Bowdoin, L. (2013). Peptide aptamers that bind to geminivirus replication proteins confer a resistance phenotype to tomato yellow leaf curl virus and tomato mottle virus infection in tomato. J. Virol. 87, 9691–9706. doi: 10.1128/JVI.01095-13
Rodríguez-Hernández, A. M., Gosalvez, B., Sempere, R. N., Burgos, L., Aranda, M. A., and Truniger, V. (2012). Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 13, 755–763. doi: 10.1111/j.1364-3703.2012.00785.x
Roossinck, M. J. (2015). Metagenomics of plant and fungal viruses reveals an abundance of persistent lifestyles. Front. Microbiol. 5:767. doi: 10.3389/fmicb.2014.00767
Roossinck, M. J., Martin, D. P., and Roumagnac, P. (2015). Plant virus metagenomics: advances in virus discovery. Phytopathology 105, 716–727. doi: 10.1094/PHYTO-12-14-0356-RVW
Roy, A., Choudhary, N., Guillermo, L. M., Shao, J., Govindarajulu, A., Achor, D., et al. (2013). A novel virus of the Genus Cilevirus causing symptoms similar to citrus leprosis. Phytopathology 103, 488–500. doi: 10.1094/PHYTO-07-12-0177-R
Roy, A.-S. (2011). “Control measures of pome and stone fruit viruses, viroids, and phytoplasmas: role of international organizations,” in Virus and Virus-Like Diseases of Pome and Stone Fruits, eds A. Hadidi, M. Barba, T. Candresses, and W. Jelkmann (St. Paul, MN: APS Press), 407–413.
Ruiz-Ruiz, S., Navarro, B., Gisel, A., Pena, L., Navarro, L., Moreno, P., et al. (2011). Citrus tristeza virus infection induces the accumulation of viral small RNAs (21–24 nt) mapping preferentially at the 3’-terminal region of the genomic RNA and affects the host small RNA profile. Plant Mol. Biol. 75, 607–619. doi: 10.1007/s11103-011-9754-4
Sanfaçon, H. (2015). Plant translation factors and virus resistance. Viruses 7, 3392–3419. doi: 10.3390/v7072778
Sanfacon, H., and Jovel, J. (2007). “Interactions between plant and virus proteomes in susceptible hosts: identification of new targets for antiviral strategies,” in Biotechnology and Plant Disease Management, eds Z. K. Punja, S. H. De Boer, and H. Sanfacon (Wallingford: CAB International), 87–108.
Sänger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856. doi: 10.1073/pnas.73.11.3852
Sano, T., Barba, M., Li, S.-F., and Hadidi, A. (2010). Viroids and RNA silencing: mechanism, role in viroid pathogenicity and development of viroid-resistant plants. GM Crops 1, 1–7. doi: 10.4161/gmcr.1.2.11871
Seguin, J., Rajeswaran, R., Malpica-López, N., Martin, R. R., Kasschau, K., Dolja, V. V., et al. (2014). De novo reconstruction of consensus master genomes of plant RNA and DNA viruses from siRNAs. PLoS ONE 9:e88513. doi: 10.1371/journal.pone.0088513
Shamloul, A. M., Abdallah, N. A., Madkour, M. A., and Hadidi, A. (2001). Sensitive detection of the Egyptian species of sugarcane streak virus by PCR-probe capture hybridization (PCR-ELISA) and its complete nucleotide sequence. J. Virol. Methods 92, 45–54. doi: 10.1016/S0166-0934(00)00272-X
Shimura, H., Pantaleo, V., Ishihara, T., Myojo, N., Inaba, J. I., Sueda, K., et al. (2011). Viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. doi: 10.1371/journal.ppat.1002021
Silva, T. F., Romanel, E. A. C., Andrade, R. R. S., Farinelli, L., Osteras, M., Deluen, C., et al. (2011). Profile of small interfering RNAa from cotton plants infected with the polerovirus Cotton leafroll dwarf virus. BMC Mol. Biol. 12:40. doi: 10.1186/1471-2199-12-40
Singh, R. P., Finnie, R. E., and Bagnal, R. H. (1971). Losses due to the potato spindle virus. Am. Potato J. 48, 266–267. doi: 10.1007/BF02861592
Singh, R. P., Randles, J. W., and Hadidi, A. (2003). “Strategies for the control of viroid diseases,” in Viroids, eds A. Hadidi, R. Flores, J. W. Randles, and J. W. Semancik (Collingwood, VIC: CSIRO Publishing), 295–302.
Smith, N. A., Eamens, A. L., and Wang, M. B. (2011). Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022. doi: 10.1371/journal.ppat.1002022
Smith, O., Clapham, A., Rose, P., Liu, Y., Wang, J., and Allaby, R. G. (2014). A complete ancient RNA genome: identification, reconstruction and evolutionary history of archaeological barley stripe mosaic virus. Sci. Rep. 4:4003. doi: 10.1038/srep04003
Sorek, R., Lawrence, C. M., and Wiedenheft, B. (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266. doi: 10.1146/annurev-biochem-072911-172315
Stepanova, O. A., Solovyova, Y. V., and Solovyov, A. V. (2011). Results of algae viruses search in human clinical material. Ukr. Bioorg. Acta 9, 53–56.
Stobbe, A. H., and Roossinck, M. J. (2014). Plant virus metagenomics: what we know and why we need to know more. Front. Plant Sci. 5:150. doi: 10.3389/fpls.2014.00150
Szittya, G., Moxon, S., Pantaleo, V., Toth, G., Rusholme, P. R. L., Moulton, V., et al. (2010). Structural and functional analysis of viral siRNAs. PLoS Pathog. 6:e1000838. doi: 10.1371/journal.ppat.1000838
Truniger, V., and Aranda, M. A. (2009). “Recessive resistance to plant viruses,” in Advances in Virus Research, eds G. Loebenstein and J. P. Carr (Norfolk, VA: Caister Academic Press), 119–231.
Van Dijk, E., Jaszczyszyn, Y., and Thermes, C. (2014). Library preparation methods for next- generation sequencing: tone down the bias. Exp. Cell Res. 322, 12–20. doi: 10.1016/j.yexcr.2014.01.008
Vanderschuren, H., Stupak, M., Futterer, J., Gruissem, W., and Zhang, P. (2007). Engineering resistance to geminiviruses–review and perspectives. Plant Biotechnol. J. 5, 207–220. doi: 10.1111/j.1467-7652.2006.00217.x
Vanitharani, R., Chellappan, P., and Fauquet, C. M. (2005). Geminiviruses and RNA silencing. Trends Plant Sci. 10, 144–151. doi: 10.1016/j.tplants.2005.01.005
Villamor, D. E. V., Mekuria, T. A., and Eastwell, K. C. (2016). High-Throughput sequencing identifies novel viruses in nectarine: insights to the etiology of stem-pitting disease. Phytopathology 106, 519–527. doi: 10.1094/PHYTO-07-15-0168-R
Visser, M., Maree, H. J., Jasper, D. G., Rees, G., and Burger, J. T. (2014). High-throughput sequencing reveals small RNAs involved in ASGV infection. BMC Genomics 15:568. doi: 10.1186/1471-2164-15-568
Vives, M. C., Velazquez, K., Pina, J. A., Moreno, P., Guerri, J., and Navarro, L. (2013). Identification of a new enamovirus associated with citrus vein enation disease by deep sequencing of small RNAs. Phytopathology 103, 1077–1086. doi: 10.1094/PHYTO-03-13-0068-R
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Wang, M. B., Masuta, C., Smith, N. A., and Shimura, H. (2012). RNA silencing and plant viral diseases. Mol. Plant Microbe Interact. 25, 1275–1285. doi: 10.1094/MPMI-04-12-0093-CR
Wang, M. B., and Smith, N. A. (2016). Satellite RNA pathogens of plants: impacts and origins-an RNA silencing perspective. Wiley Interdiscip. Rev. RNA 7, 5–16. doi: 10.1002/wrna.1311
Wang, Y., Shibuya, M., Taneda, A., Kurauchi, T., Senda, M., Owens, R. A., et al. (2011). Accumulation of potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 413, 72–83. doi: 10.1016/j.virol.2011.01.021
Waterworth, H., and Hadidi, A. (1998). “Economic losses due to plant viruses,” in Plant Virus Disease Control, eds A. Hadidi, R. K. Khetarpal, and H. Kogenazawa (St. Paul, MN: APS Press), 1–13.
Westra, E. R., van Erp, P. B., Künne, T., Wong, S. P., Staals, R. H., Seegers, C. L., et al. (2012). CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605. doi: 10.1016/j.molcel.2012.03.018
Wu, Q., Ding, S.-W., Zhang, Y., and Zhu, S. (2015). Identification of viruses and viroids by next-generation sequencing and homology-dependent and homology-independent algorithms. Annu. Rev. Phytopathol. 53, 425–444. doi: 10.1146/annurev-phyto-080614-120030
Wu, Q., Wang, Y., Cao, M., Pantaleo, V., Burgyan, J., Li, W. X., et al. (2012). Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. U.S.A. 109, 3938–3943. doi: 10.1073/pnas.1117815109
Xu, Y., Huang, L., Fu, S., Wu, J., and Zhou, X. (2012). Population diversity of Rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphax striatellus. PLoS ONE 7:e46238. doi: 10.1371/journal.pone.0046238
Yan, F., Zhang, H., Adams, M., Yang, J., Peng, J., Antoniw, J., et al. (2010). Characterization of siRNAs derived from rice stripe virus in infected rice plants by deep sequencing. Arch. Virol. 155, 935–940. doi: 10.1007/s00705-010-0670-8
Yang, C.-F., Chen, K.-C., Cheng, Y.-H., Raja, J. A. J., Huang, Y.-L., Chien, W.-C., et al. (2014). Generation of marker-free transgenic plants concurrently resistant to a DNA geminivirus and a RNA tospovirus. Sci. Rep. 4:5717. doi: 10.1038/srep05717
Yang, X., Caro, M., Hutton, S. F., Scott, J. W., Guo, Y., Wang, X., et al. (2014). Fine mapping of the tomato yellow leaf curl virus resistance gene-on chromosome 11 of tomato. Mol. Breed. 34, 749–760.
Yang, X., Wang, Y., Guo, W., Xie, Y., Xie, Q., Fan, L., et al. (2011). Characterization of small interfering RNAs derived from the geminivirus/betasatellite complex using deep sequencing. PLoS ONE 6:e16928. doi: 10.1371/journal.pone.0016928
Yolken, R. H., Jones-Brando, L., Dunigan, D. D., Kannan, G., Dickerson, F., Severance, E., et al. (2014). Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice. Proc. Natl. Acad. Sci. U.S.A. 111, 16106–16111. doi: 10.1073/pnas.1418895111
Yoshikawa, N., Yamagishi, N., Yaegashi, H., and Ito, T. (2012). Deep sequence analysis of viral small RNAs from a green crinkle-diseased apple tree. Petria 22, 292–297.
Zahid, K., Zhao, J. H., Smith, N. A., Schumann, U., Fang, Y. Y., Dennis, E. S., et al. (2015). Nicotiana small RNA sequences support a host genome origin of cucumber mosaic virus satellite RNA. PLoS Genet. 11:e1004906. doi: 10.1371/journal.pgen.1004906
Zelazny, B., Randles, J. W., Boccardo, G., and Imperial, J. S. (1982). The viroid nature of the cadang-cadang disease of coconut palm. Sci. Filipinas 2, 45–63.
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, C., Wu, Z., Li, Y., and Wu, J. (2015). Biogenesis, function, and applications of virus-derived small RNAs in plants. Front. Microbiol. 6:1237. doi: 10.3389/fmicb.2015.01237
Zhang, T., Breitbart, M., Lee, W. H., Run, J., Wei, C. L., Soh, S. W. L., et al. (2006). RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:e3. doi: 10.1371/journal.pbio.0040003
Zhang, Y., Singh, K., Kaur, R., and Qiu, W. (2011). Association of a novel DNA virus with the grapevine vein-clearing and decline syndrome. Phytopathology 101, 1081–1090. doi: 10.1094/PHYTO-02-11-0034
Keywords: next-generation sequencing, plant virology, plant viruses, viroids, resistance to plant viruses by CRISPR-Cas9
Citation: Hadidi A, Flores R, Candresse T and Barba M (2016) Next-Generation Sequencing and Genome Editing in Plant Virology. Front. Microbiol. 7:1325. doi: 10.3389/fmicb.2016.01325
Received: 01 July 2016; Accepted: 11 August 2016;
Published: 26 August 2016.
Edited by:
Nobuhiro Suzuki, Okayama University, JapanReviewed by:
Carmen Hernandez, Spanish National Research Council, SpainVicente Pallas, Instituto de Biología Molecular y Celular de Plantas, Spain
Copyright © 2016 Hadidi, Flores, Candresse and Barba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Hadidi, ahadidi@yahoo.com
†Lead Scientist Emeritus
 Ahmed Hadidi
Ahmed Hadidi Ricardo Flores
Ricardo Flores Thierry Candresse
Thierry Candresse Marina Barba
Marina Barba