- Departamento de Biología, Universidad Autónoma de Madrid, Madrid, Spain
The genomic sequence of Pseudomonas fluorescens F113 has shown the presence of a 41 kb cluster of genes that encode the production of a second flagellar apparatus. Among 2,535 pseudomonads strains with sequenced genomes, these genes are only present in the genomes of F113 and other six strains, all but one belonging to the P. fluorescens cluster of species, in the form of a genetic island. The genes are homologous to the flagellar genes of the soil bacterium Azotobacter vinelandii. Regulation of these genes is mediated by the flhDC master operon, instead of the typical regulation in pseudomonads, which is through fleQ. Under laboratory conditions, F113 does not produce this flagellum and the flhDC operon is not expressed. However, ectopic expression of the flhDC operon is enough for its production, resulting in a hypermotile strain. This flagellum is also produced under laboratory conditions by the kinB and algU mutants. Genetic analysis has shown that kinB strongly represses the expression of the flhDC operon. This operon is activated by the Vfr protein probably in a c-AMP dependent way. The strains producing this second flagellum are all hypermotile and present a tuft of polar flagella instead of the single polar flagellum produced by the wild-type strain. Phenotypic variants isolated from the rhizosphere produce this flagellum and mutation of the genes encoding it, results in a defect in competitive colonization, showing its importance for root colonization.
Introduction
Swimming and swarming bacterial motility are flagellum-dependent. In pathogenic bacteria, motility increases the virulence capacity of many bacterial species (Schmitt et al., 1994; Akerley et al., 1995). In plant-associated pseudomonads, motility is important to colonize the rhizosphere and full motility is a requisite for biocontrol and plant growth promotion (Chin-A-Woeng et al., 2000; Kamilova et al., 2005; Barahona et al., 2011).
Structure and function of the flagellum in Escherichia coli and Salmonella enterica, have been studied for many years. Both species show peritrichous flagella and the flagellar structure is built from inside out, requiring about 50 genes, whose expression is regulated in a hierarchy of several levels (Macnab, 2003; McCarter, 2006). At the highest level of this regulatory cascade is the master regulator flhDC operon, whose products, FlhC and FlhD are responsible for the flagellar regulon activation. In turn, the master regulator FlhDC is controlled by a cAMP-dependent system (Soutourina et al., 1999) which involves the CyaA adenylate cyclase and the c-AMP binding protein CRP (Botsford and Harman, 1992). A similar regulatory system has been shown for Azotobacter vinelandii, which also shows peritrichous flagella (Leon and Espin, 2008). The chromosome of this bacterium encodes both fleQ (see below) and flhDC, but only the later operon is required for flagella synthesis (Leon and Espin, 2008).
Within the pseudomonads, Pseudomonas aeruginosa and Pseudomonas fluorescens have been used as a model to study the synthesis and regulation of the flagellar apparatus. Conversely to enterobacteria, pseudomonads only produce one or two polar flagella. In the pseudomonads, FleQ is the master regulator (Arora et al., 1997; Dasgupta et al., 2003; Capdevila et al., 2004) and expression of most flagellar genes is controlled directly or indirectly by FleQ (Arora et al., 1997; Jyot et al., 2002; Dasgupta et al., 2003).
The sequencing of the genome of P. fluorescens F113 has shown that this strain encodes both types of flagella (Redondo-Nieto et al., 2012). The F113 genome contains all the genes required for the synthesis of a pseudomonads’ type flagellum, but also possesses 45 genes required for the synthesis of a second flagellum. Conversely to the operons encoding flagellar genes in pseudomonads, the region encoding the second F113 flagellum contains an flhDC operon (Redondo-Nieto et al., 2013). The 45 ORFs involved in the synthesis of this flagellum showed high homology to flagellar genes of A. vinelandii and enterobacteria. The region also showed synteny with the flagellar genes of A. vinelandii. The A. vinelandii chromosome harbors flagellar genes in two clusters, I and II. Genes in cluster I are conserved in the same order in the 41 kb region in the F113 chromosome. Cluster II in A. vinelandii is located 416 kb downstream of cluster I. A reduced version of this region, with its central part deleted and lacking 12 ORFs is located in an inverted orientation adjacent to cluster I in the F113 chromosome. All the genes present in A. vinelandii clusters, but absent in the F113 genome, encode chemotaxis proteins or proteins that are not essential for flagella synthesis (Redondo-Nieto et al., 2013).
Pseudomonas fluorescens F113 is able to colonize the rhizosphere of a wide variety of plants (Simons et al., 1996; Naseby and Lynch, 1999; Dekkers et al., 2000; Villacieros et al., 2003, 2005) and motility is a key trait for colonization. Hypermotile mutants (Barahona et al., 2010, 2011) or hypermotile phenotypic variants isolated from the rhizosphere (Martinez-Granero et al., 2006) are able to displace the wild-type strain in competitive colonization assays. For this reason, we have identified some genes that are part of independent regulatory pathways (Navazo et al., 2009) and regulate negatively motility, such as the two-component system GacA/S, the kinB and sadB genes, and the Wsp system. The two-component system GacA/S and the cytoplasmic protein SadB, repress the motility through fleQ (Navazo et al., 2009; Martinez-Granero et al., 2012). Swimming motility is also inhibited by kinB and the Wsp system, independently of FleQ.
In P. aeruginosa, KinB and AlgB form a two-component system involved in alginate synthesis (Ma et al., 1997) and also participate in the regulation of virulence and motility in this bacterium (Chand et al., 2011). A kinB mutant in F113 is more motile than the wild-type strain (Navazo et al., 2009). In this work, we show that the presence of the cryptic second flagellar apparatus in F113, which has homology to the A. vinelandii DJ, explains the hypermotility phenotype of the kinB mutant. The second flagellum absence in both F113 and kinB mutant, leads to a decrease in competitive root colonization ability.
Materials and Methods
Microorganisms, Growth Conditions, and Plasmids
Strains and plasmids used are listed in Supplementary Table 1. P. fluorescens F113 and derivatives were grown in SA medium (Scher and Baker, 1982), with shaking overnight at 28°C. Purified agar (1.5%; Pronadisa, Spain) was added for solid medium. Pseudomonas strains were grown in LB medium for RNA and protein extraction assays. When it was necessary, antibiotics were used at the following concentrations (μg ml-1): rifampicin 100; kanamycin 50; gentamicin 3; spectinomycin 100; and tetracycline 70. E. coli strains were grown in LB medium with shaking overnight at 37°C. In this case, antibiotics were used at the following concentrations (μg ml-1): kanamycin 25; gentamicin 10; ampicillin 100; spectinomycin 25; chloramphenicol 30; and tetracycline 10.
Swimming Motility Assays
Swimming was tested on SA medium with 0.3% purified agar. Bacteria were inoculated with a sterile toothpick in plates of 50 mm in diameter containing 12 ml of SA medium and incubated at 28°C. Swimming haloes were measured after 24 h. Every assay was performed three times with four replicates each time. Average swimming halo diameters (mm) and standard deviations were calculated and all dates were compared using statistic application software SPSS. Statistical significance was calculated by the Bonferroni test of variance analysis (ANOVA; P < 0.05).
Simple and Double Mutant Generation: Overexpression Assays
Mutants generation was performed by directed mutagenesis. Primers were designed (Supplementary Table 2) to amplify small fragments of flhDC, fliC2, cyaA, and vfr genes [300–500 nucleotides (nts); flhDC-F/R; fliC2-F/R; cyaA-F/R; vfr-F/R]. First, these internal fragments were cloned into cloning pGEM®T-easy vector and then, into the suicide vector pK18mobsacB (Schäfer et al., 1994) or pG18mob2 (Kirchner and Tauch, 2003; Supplementary Table 1). The constructs were introduced into F113 or F113-derived strains by triparental mating (in LB medium at 28°C during 16 h) using helper plasmid pRK600 (Finan et al., 1986; Supplementary Table 1) and selected for homologous recombination. The same method was used in the construction of double and triple mutants. All mutants were checked by Southern blotting and PCR. Primers were designed to amplify flhDC (flhDC-F2/R2; Supplementary Table 2). Overexpression of flhDC gene was achieved by cloning them under the control of the IPTG-inducible promoter present in the pVLT31 plasmid (de Lorenzo et al., 1993).
Transmission Electron Microscopy
Pseudomonas fluorescens F113 and derivatives were grown in LB medium until optical density (OD600) of 0.8. Formvar-coated grids were placed on the top of a drop of bacterial cells for 30 s to allow bacterial adhesion. Grids were stained for 1 min 15 s with a 1% solution of potassium phosphotungstate and washed with a drop of sterile water (thrice for 30 s). In order to visualize the preparations, the microscope JEOL JEM 1010 (100 kV) was used (Laboratorio de Microscopía de Transmisión del Servicio Interdepartamental de Investigación, UAM) and the images were taken by the BioScan de Gatan camera. The images were analyzed through DigitalMicrograph 3.1 analyze system.
DNA Techniques
Standard methods (Sambrook et al., 1989) were used for DNA extraction, gene cloning, plasmid preparations, and agarose gel electrophoresis. Southern blots were performed with a non-radioactive detection kit, and a chemiluminescence method was used to detect hybridization signals according to the instructions of the manufacturer (Roche Boehringer Mannheim). PCR reactions were performed using the Tth enzyme (Biotools) under standard conditions. DNA sequencing was done by chain-termination method using DyeDeoxy terminator cycle sequencing kit protocol as described by the manufacturer (Applied Biosystems).
RNA Extraction, cDNA Synthesis, and Gene Expression Analysis
Total RNA was extracted using Trizol® according to manufacturer’s specifications (Invitrogen) from F113 wild-type and derivatives grown at 0.8 OD600 in LB medium. Genomic DNA was removed by RQ1 RNase-Free DNase treatment (Promega) for 30 min at 37°C. Later, RNA was purified using Trizol®. The concentration of RNA was spectrophotometrically determined in a Nanodrop® and integrity was verified in denaturing agarose gels. RT-PCRs were carried out using Illustra Ready-To-GoTM RT-PCR Beads kit from Amersham GE Healthcare. qRT-PCRs were performed in two steps: a first step of cDNA synthesis using the SuperScript®III First-Strand Synthesis System from Invitrogen and a second step of qPCR using the Power SYBR®Green PCR Master Mix from Applied Biosystems. Relative expression was estimated as 2-ΔΔCt using 16S DNA as housekeeping and wild-type ΔCt values as calibrator, as described by Livak and Schmittgen (2001). Every assay was performed three times with three replicates each time.
Protein Extraction and Western Blots
Proteins were extracted from 200 ml exponential phase grown cultures (OD600 = 0.8). In order to detach the flagella, the cultures were agitated by vortexing for 2 min and then centrifuged for 15 min at 7,500 rpm and 4°C. Extracellular proteins were extracted from the supernatant by precipitation for 10 min at RT with 10% (w/v) desoxycholic acid and then, 2 h at 4°C with 10% (w/v) trichloroacetic acid, followed by two washes with chilled acetone, and were finally resuspended in Laemmli buffer (Laemmli, 1970). Proteins were resolved by 12% SDS-PAGE and stained with Coomassie blue. The same electrophoretic conditions were used for Western blotting. Acrylamide gels were transferred onto nitrocellulose membranes for 1 h under standard conditions. The membranes were incubated with a 1:10,000 dilution of an anti-flagellin antiserum (de Weger et al., 1987) for 16 h at 4°C and then with a peroxidase-tagged secondary antibody (anti-rabbit immunoglobulin) for 1 h at RT. The enhanced chemiluminescence (ECL) method and Hyperfilm ECL (Amersham Biosciences) were used for development.
Root Competitive Colonization Assays
A modification of the “root-tip competitive colonization assay” (Simons et al., 1996) was used. Alfalfa seeds (Medicago sativa var. Resis) were surface disinfected in 70% ethanol for 2 min and then in diluted bleach (1:5; final sodium hypochlorite concentration, 1%) for 15 min and rinsed thoroughly with sterile distilled water. Seed vernalization was performed at 4°C for 16 h and was followed by incubation in darkness at 28°C for 1 day for germination. Germinated alfalfa seeds were sown in Leonard jar gnotobiotic systems (Villacieros et al., 2003) using ca. 3.5 l of sterile perlite as the solid substrate and 8 mM KNO3-supplemented FP (Fahraeus, 1957) as the mineral solution (500 ml/jar; replenished every 2 days). After 2 days, alfalfa seeds were inoculated with 108 cells of the appropriate strains. In competition experiments, strains were inoculated at a 1:1 ratio. In all cases 1 ml diluted culture was inoculated per plant. Plants were maintained under controlled conditions for 2 weeks: 16 h in the light at 25°C and 8 h in the dark at 18°C. Bacteria were recovered from the rhizosphere by vortexing the root tips (last centimeter of the main root) for 2 min in a tube containing FP medium and plating the appropriate dilutions on SA plates supplemented with appropriate antibiotics. Every experiment was performed three times with three replicates each time, and every replicate contained at least 20 plants.
Bioinformatic Analysis
Pseudomonas fluorescens F113 genomic sequence (NC_016830.1) from position 876600 to 961608 was used as query in Blastn v2.3.0+ (Camacho et al., 2009) to compare against complete and draft Pseudomonas genomes in NCBI database (nt and WGS on March 30, 2016). Own designed Python scripts were used to filter BLAST results. Genome annotations were obtained from NCBI database when possible; also RAST annotation pipeline (Aziz et al., 2008) was used. Synteny of the second flagellar region and its context was assessed from annotations and represented by using self-written Perl scripts. Protein homology was checked by Blastp (Camacho et al., 2009).
Phylogenomic distances between genomes harboring the flagellar island and close relative genomes was calculated with the genome-to-genome distance calculator (GGDC) 2.1 web service http://ggdc.dsmz.de (Meier-Kolthoff et al., 2013). Nucleotide sequence from each of the 45 ORFs from the flagellar island were retrieved from genomic annotation and aligned with Clustal Omega (Sievers et al., 2011). Resulting alignments were concatenated with own designed Python scripts following the order in P. fluorescens F113. The concatenated sequences were used to infer the phylogeny using maximum-likelihood method, Tamura–Nei model, and 1,000 bootstrap replicates using MEGA software (v7; Kumar et al., 2016).
Scripts are provided as Supplementary File 1. Statistical analyses were carried out with R software (R Core Team, 2013).
Results
The Pseudomonas fluorescens F113 Genome Encodes a Second Flagellar Apparatus in a Genomic Island
Analysis of the genome of P. fluorescens F113 showed that besides the genes required for the synthesis of the flagella, it harbors an additional 41 kb genetic region containing 45 ORFs that also showed homology with flagellar genes (Redondo-Nieto et al., 2012). This region is not present in other closely related strains belonging to the same phylogenomic group of P. fluorescens F113 (Redondo-Nieto et al., 2013; Garrido-Sanz et al., 2016). Syntenic comparison of the F113 genome and one of its closest sequenced relative, Pseudomonas brassicacearum NFM421 (Figure 1), showed that these 45 genes form a genetic island inserted in the intergenic region between genes PsF113_0737 and PsF113_0783 encoding a phosphatase and a peroxidase respectively, whose orthologs in P. brassicacearum are contiguous (PSEBRa709 and PSEBRa710). Abrupt loss of sequence homology between F113 and NFM421 indicated that the left border of the genetic island corresponds to F113 nucleotide 894337 and the right border to nucleotide 939056, the nucleotide before the stop codon of PsF113_0783. In the intergenic region between PSEBRa709 and PSEBRa710 there are 42 nts, which are present in other strains belonging to the F113 phylogenomic group but are not in the F113 genome. Those nucleotides could have been lost in F113 as result of the flagellar island insertion.
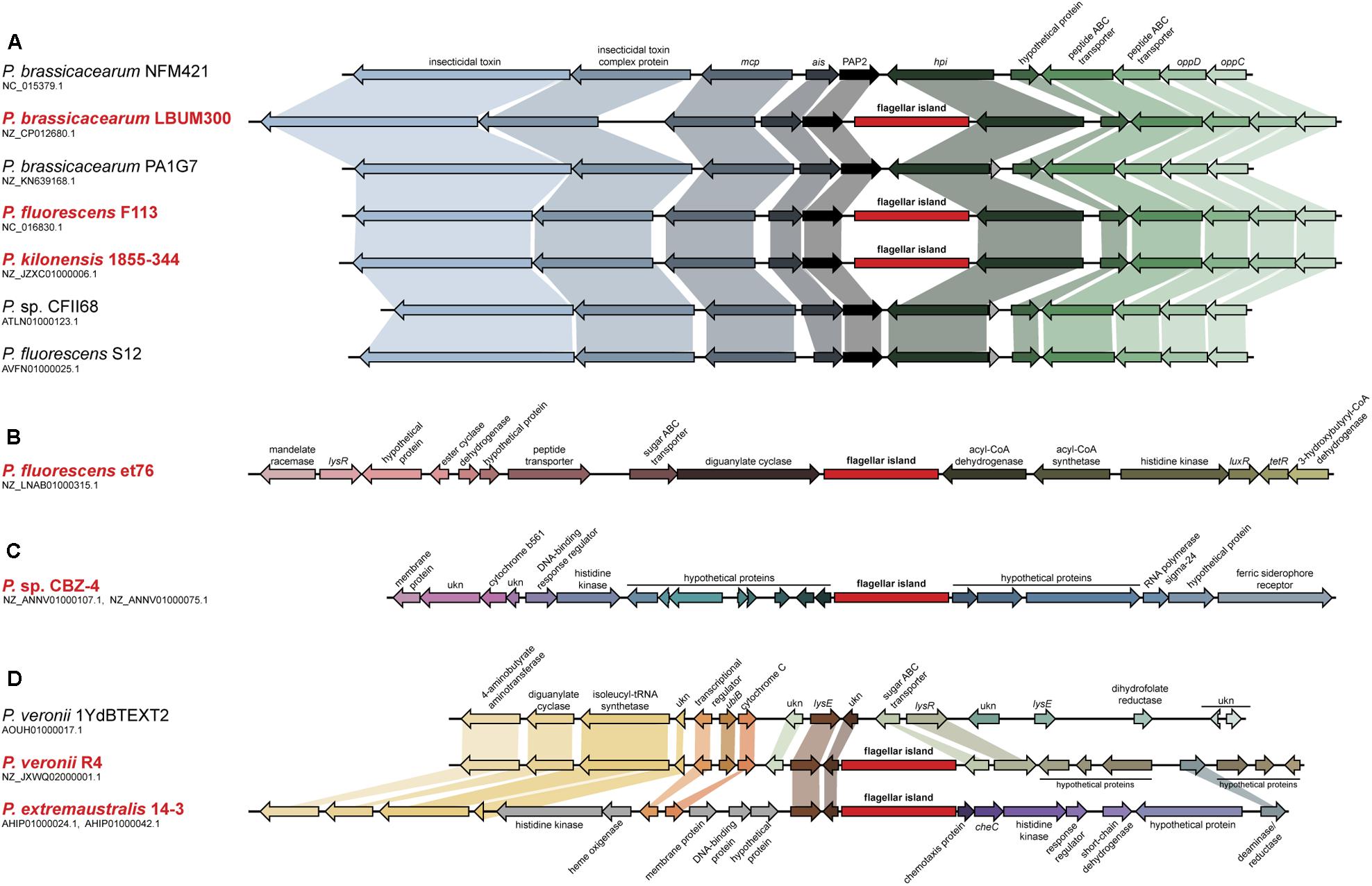
FIGURE 1. Analysis of the second flagellar island insertion events in strains belonging to the P. fluorescens complex. Syntenic context of genomes harboring the second flagellar island (red typing) and comparison with close relative genomes lacking the island (black typing). (A) Insertion event 1, affecting strains LBUM300, F113, and 1855-344. Close relative strains NFM421, PA1G7, CFII68, and S12, are shown for reference. (B) Insertion event 2, affecting strain et76. (C) Insertion event 3, affecting strain CBZ-4. (D) Insertion event 4, affecting strains R4 and 14-3. Close relative strain 1YdBTEXT2 is shown for reference. Second flagellar island is not in scale.
The analysis of pseudomonads genomes deposited both in the nr/nt and WGS NCBI databases, showed that besides F113, this flagellar island was also present in other seven strains, Pseudomonas extremaustralis 14-3b, Pseudomonas sp. CBZ-4, Pseudomonas veronii R4, P. brassicacearum LBUM300, Pseudomonas kilonensis 1855-344, P. fluorescens et76 and Pseudomonas putida ATH-43. Strains F113, 1855-344, LBUM300, and et76 are phylogenetically related and belong to the Pseudomonas corrugata subgroup, while strains 14-3b, CBZ-4, and R4 are also phylogenetically related and belong to the P. fluorescens subgroup (Figure 2A; Garrido-Sanz et al., 2016). Outside the P. fluorescens complex of species, the island was only found in a P. putida strain (ATH-43). The genomic location of the island is identical in strains F113, LBUM300, and 1855-344 (Figure 1) indicating that the presence of the island in their genomes originated from a single insertion event. The location of the island in strain et79 is in a different genomic context, indicating an independent insertion event (Figure 1). Similarly, the location of the flagellar island in strains R4 and 14-3 is also identical, albeit different that in the P. corrugata group strains (Figure 1). The insertion in the CBZ-4 strain is in another different region (Figure 1). These results indicate that within the P. fluorescens complex, the appearance of the flagellar island occurred in four independent insertion events. These events are also supported by a phylogenetic and phylogenomic analysis of the island (Figure 2). As shown in Figure 2B the evolution of the island is in accordance with the evolution of the genomes in which they are inserted. Furthermore, genetic organization within the island is highly conserved in all the analyzed strains (Supplementary Figure 1).
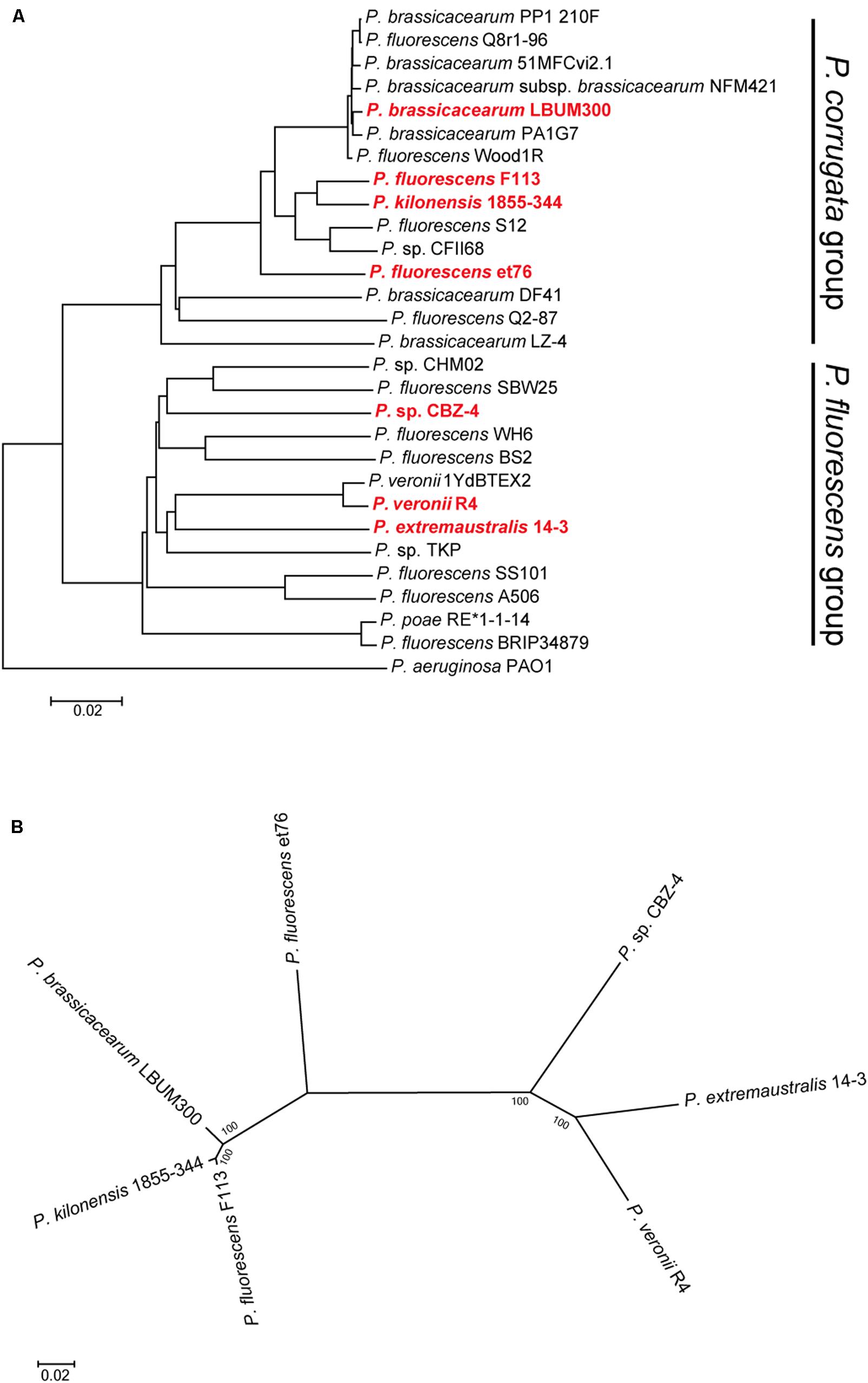
FIGURE 2. Phylogenetic distribution of the second flagellar island in strains belonging to the P. fluorescens complex. (A) Phylogenomic tree based on GGDC. Red bold typing indicates genomes harboring the second flagellar island. Insertion events 1 and 2, lie in strains belonging to the P. corrugata group, while insertion events 3 and 4, lie in strains belonging to the P. fluorescens group. (B) Co-evolution of the second flagellar island with the genomes harboring it. Multilocus sequence analysis of the 45 ORFs concatenated sequences from the second flagellar island shows agreement with the proposed four insertion events and the phylogenetic distance between the genomes harboring the island. Phylogenetic tree was built using maximum-likelihood method, Tamura–Nei model and 1,000 bootstrap replicates.
Comparison of the aminoacid sequences of the ORFs in the island present in F113 confirmed that for most of them, closest relatives outside the pseudomonads were present in A. vinelandii genomes. However, a few ORFs showed higher homology with ORFs present in other Gamma Proteobacteria (Supplementary Table 3).
Genes for the Second Flagellum Are Not Expressed in P. fluorescens F113, but Are Expressed in a kinB Mutant Background
We have previously shown that ectopic expression of the flhCD genes in a wild-type F113 background resulted in hypermotility (Redondo-Nieto et al., 2013) indicating that the expression of the master regulatory operon is sufficient to trigger the regulatory cascade resulting in the formation of the second flagellar apparatus. However, the introduction of the plasmid was unable to restore motility to non-motile mutants such as fliC (data not shown), indicating that production of the first flagellum is necessary for the production of the second one or that a non-functional flagellum is produced under these conditions.
In order to test the functionality of the second flagellar apparatus, we checked the expression of the fliC2 gene encoding flagellin in F113 and isogenic mutants affected in motility (Figure 3A). RT-PCR experiments showed no expression of the gene in the wild-type strain, suggesting that a second flagellar apparatus is not produced by F113 under laboratory conditions. No expression was observed in the gacS, sadB, or wspR mutants. However, a kinB mutant, that had been isolated as a hypermotile mutant in a screening for motility repressors (Navazo et al., 2009) and an algU mutant, showed significant expression of the fliC2 gene (Figure 3A). Quantitative RT-PCR was used to determine the expression of flhDC and fliC2 in both mutant backgrounds compared to F113. As shown in Figure 3B, high level of expression was observed for both genes in both mutants. flhDC expression was six times higher in the kinB mutant and eight times higher in the algU mutant related to the wild-type strain. Furthermore, fliC2 expression was 4.5 times higher in kinB mutant and 11 times higher in algU than in F113.
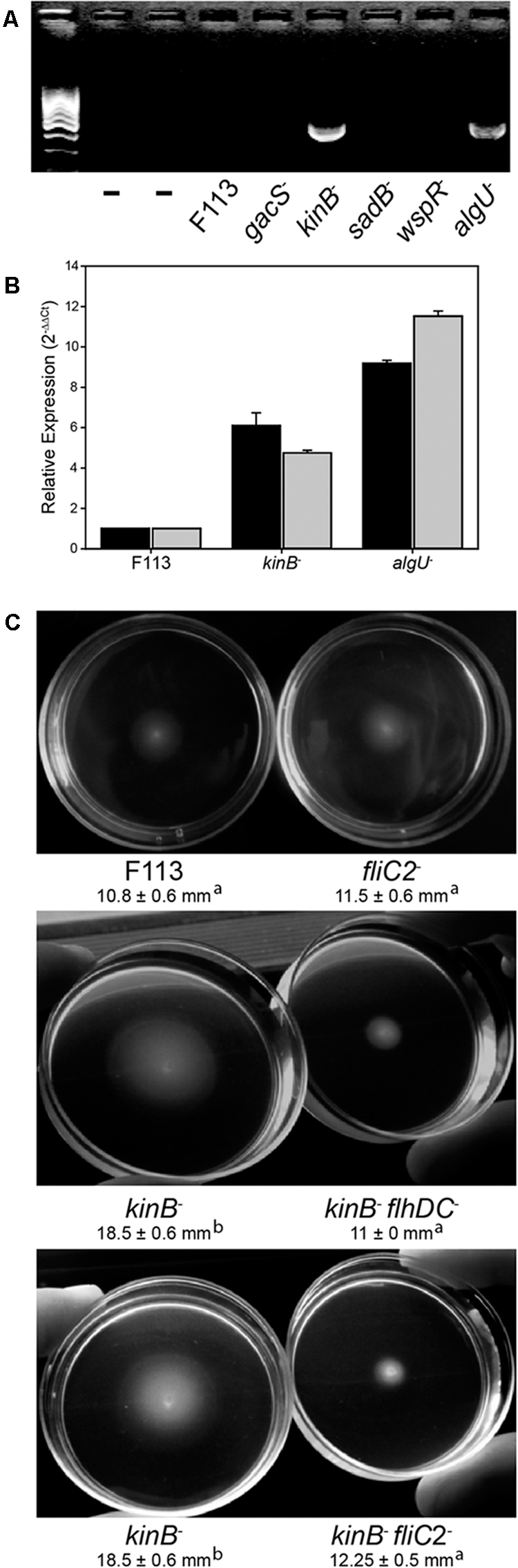
FIGURE 3. The second flagellar apparatus is not produced in P. fluorescens F113, but is produced in kinB and algU mutant backgrounds. (A) RT-PCR of the fliC2 gene in P. fluorescens F113 and different isogenic mutants. Expression was only observed in the kinB and algU mutant backgrounds. First two lanes are negative controls (without retrotranscription) of the kinB and algU RNA preparations. (B) Quantitative RT-PCR analysis of the flhDC (black) and fliC2 (gray) in P. fluorescens F113 and isogenic kinB and algU mutants. Differences between the wild-type strain and the two mutants, were statistically significant (p < 0.05) for both genes. (C) Swimming motility phenotype of P. fluorescens F113 and isogenic mutants affected in fliC2, kinB, kinBflhDC, and kinBfliC2, after 18 h of incubation. Different letters indicate statistically significant differences (p < 0.05). Motility of the strain ectopically overexpressing flhDC was two times (23 mm) the motility of the wild-type strain. Motility experiments were performed three times in triplicate.
To test whether the hypermotility phenotype of the kinB mutant was due to the production of a second flagellar apparatus, we generated flhDC and fliC2 mutations in a kinB mutant background. As shown in Figure 3C, both mutations suppressed the hypermotility phenotype of the kinB mutant, restoring wild-type motility. These results indicate that production of the second flagellar apparatus is causing the hypermotility of the kinB mutant. Furthermore, fliC2 mutation in a F113 background produces no change in the motility phenotype, confirming that the second flagellum is not expressed in the wild-type strain under the conditions tested (Figure 3C).
flhDC Are Positively Regulated by Vfr
It has been shown that in A. vinelandii, flhDC expression is activated through c-AMP by the Vfr protein (a CRP ortholog). In order to test whether this regulation was also functioning for the second flagellar apparatus in P. fluorescens F113, we generated vfr mutants in F113 and the kinB mutant. As shown in Figure 4, the vfr mutation had no effect in the swimming phenotype of F113. However, this mutation suppressed the hypermotile phenotype of the kinB mutant, restoring motility to wild-type levels. We also tested the effect of a mutation on the cyaA gene, encoding an adenylate cyclase, in the kinB background. This mutation also restored the wild-type motility phenotype to the kinB mutant (Figure 4). These results indicate that cyaA and vfr activate flhDC expression and therefore the production of the second flagellar apparatus and that this activation is strongly repressed by the kinB gene.
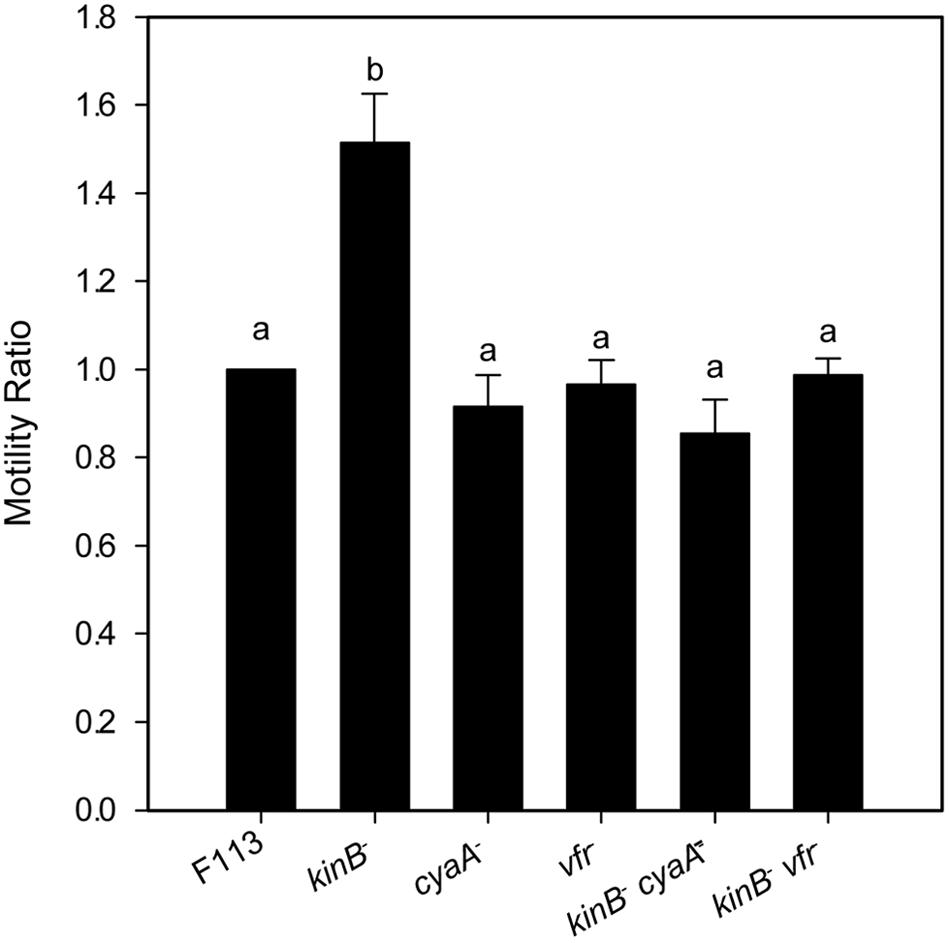
FIGURE 4. flhDC are positively regulated by Vfr. Swimming motility phenotype of Pseudomonas fluorescens F113 and isogenic mutants affected in kinB, cyaA, vfr, kinBcyaA, and kinBvfr. Each strain was tested four times and means and standard deviations of measures have been represented. Only the kinB mutant showed significant differences (p < 0.05) in swimming motility compared to wild-type strain. Same letters indicate no statistical differences, P < 0.05.
Production of the Second Flagellar Apparatus Results in Polar Hyperflagellated Bacteria
Transmission electron microscopy of negatively stained cells was used to monitor the flagellar number and location in different derivatives of P. fluorescens F113 (Figure 5). The wild-type strain (Figure 5A) presents a single polar flagellum, as has been previously described. Conversely, all the strains producing the second flagellar apparatus, such as the kinB mutant (Figure 5C) and the F113 (pflhDC; Figure 5B) presented a tuft of flagella in one of the poles of the cell. The same was true for the phenotypic variant S (Sánchez-Contreras et al., 2002; Figure 5D), a hypermotile variant isolated from the rhizosphere. A kinB mutant, harboring a mutation in the fliC2 gene (Figure 5E) showed the same flagellation that the wild-type strain.
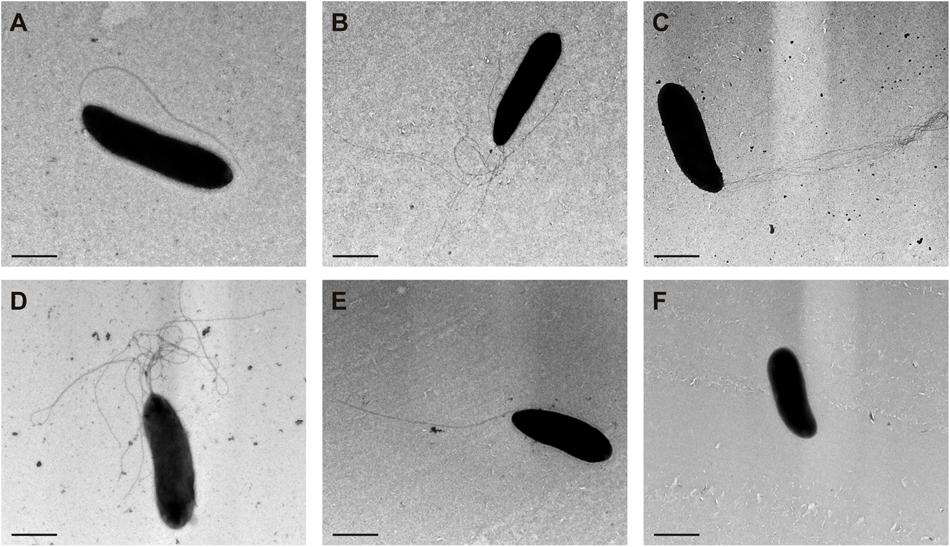
FIGURE 5. Production of the second flagellar apparatus results in polar hyperflagellated bacteria. Transmission electron microscopy image of (A) P. fluorescens F113; (B) F113 harboring a plasmid expressing flhDC; (C) kinB mutant; (D) phenotypic variant S; (E) kinBfliC2 mutant; (F) fliC mutant. All the strains producing the second flagellar apparatus showed a polar tuft of flagella.
In order to confirm that hyperflagellation resulted from production of the second flagellar apparatus and not by hyperproduction of the first flagellum, we extracted extracellular proteins from cultures of the wild-type strain, the kinB mutant and a sadB mutant, that has been previously shown to overproduce the FliC flagellin. As shown in Supplementary Figure 2, an anti-FliC antiserum that does not recognize FliC2, showed FliC overproduction only for the sadB mutant.
Production of the Second Flagellar Apparatus Is Important for Root Colonization
Since the phenotypic variant S appeared to be producing the second flagellar apparatus, we decided to test fliC2 expression in a battery of hypermotile phenotypic variants isolated from alfalfa rhizosphere (Martinez-Granero et al., 2006). All the phenotypic variants expressed the fliC2 gene (Figure 6A), suggesting that the rhizosphere environment selects for the production of the second flagellar apparatus. It is important to note that phenotypic variants arise at a much lower frequency from liquid medium cultures than in the rhizosphere (Martínez-Granero et al., 2005). We also tested the competitive colonization ability of the wild-type strain against a fliC2 mutant, unable to produce the second flagella. As shown in Figure 6B, the fliC2 mutant was displaced by the wild-type strain from the root tip. Furthermore, we analyzed the competitive colonization capacity of the kinB mutant that synthesizes both flagellum, and a kinBfliC2 double mutant, which can only produce the first flagellar apparatus. As shown in Figure 6B, the kinB mutant was able to displace the kinBfliC2 double mutant. Taken together, these results show that the production of the second flagella in the rhizosphere environment is an important trait for root competitive colonization.
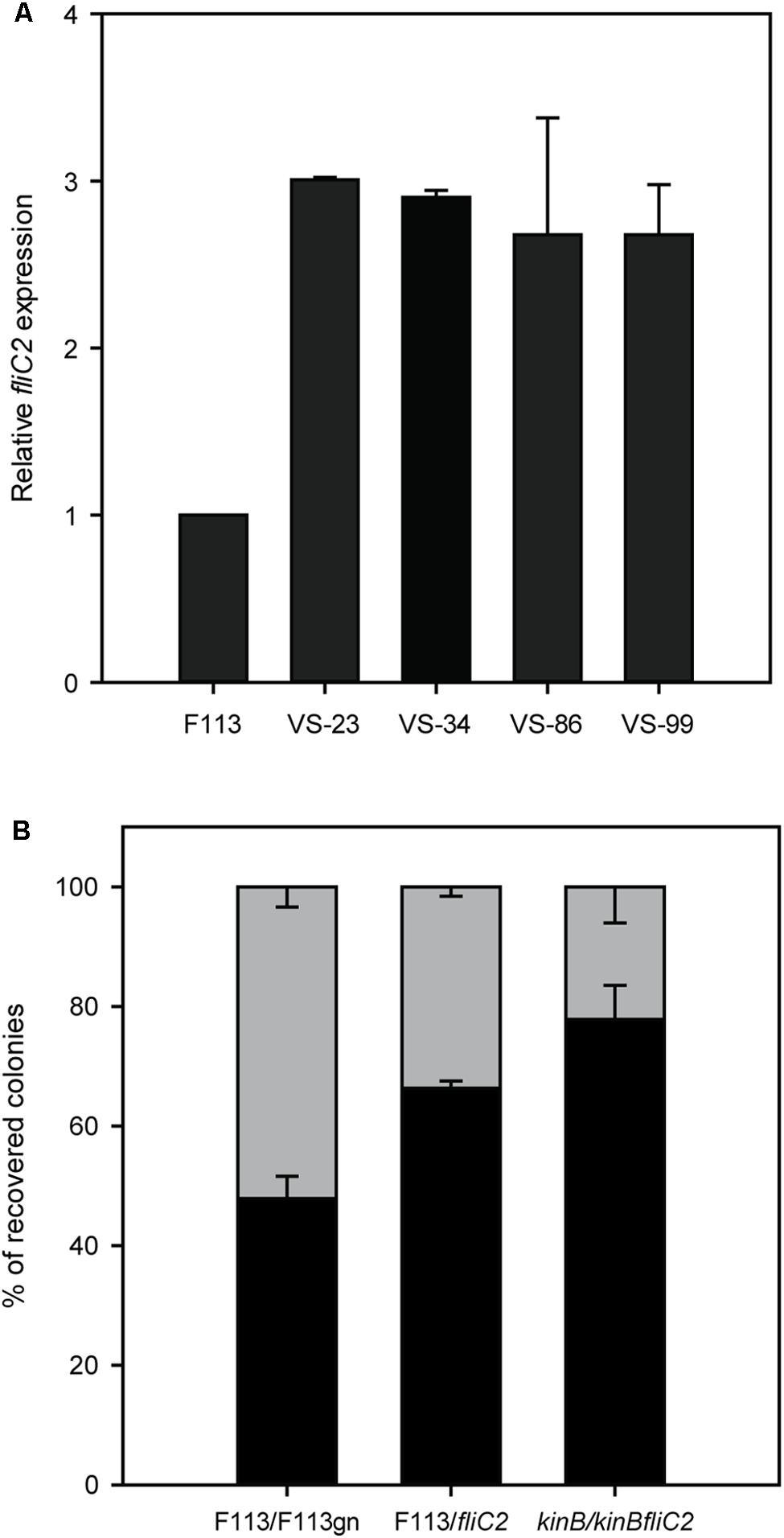
FIGURE 6. Production of the second flagellar apparatus is important for root colonization. (A) Quantitative RT-PCR analysis of the expression of fliC2 in Pseudomonas fluorescens F113 and its phenotypic variants isolated from the alfalfa rhizosphere. All the variants showed statistically significant (p < 0.05) differences with the wild-type strain. (B) Competitive root colonization assays of Pseudomonas fluorescens F113 and derivatives. Bacterial pairs were inoculated at a 1:1 proportion and colonies were recovered after 2 weeks of inoculation, in culture medium plates supplemented with the appropriate antibiotics. Every experiment was performed three times with three replicates each time, and every replicate contained at least 20 plants. Means and standard deviations are represented. Gray bars represent the percentage of colonies recovered from the tested strain (F113gn, fliC2 mutant and kinBfliC2 double mutant). Black bars represent the percentage of colonies recovered from the competitor (wild-type strains on first and second case, and kinB mutant on third case).
Discussion
Production of a second flagellar apparatus is an unusual trait in fluorescent pseudomonads. A genomic analysis has shown that within the genus Pseudomonas only a few strains, most belonging to the P. fluorescens complex of species encode the genes for its synthesis. Out of 212 sequenced strains within the complex, only seven strains contained this genetic region. Only one other sequenced pseudomonad, outside the P. fluorescens complex contained the genes for its synthesis. The genes encoding the synthesis of the second flagellar apparatus form a genetic island, since they are located in different genomic positions in the genomes of different strains. In the case of F113, the availability of a reference genome of a closely relative that does not contain the island, P. brassicacearum NFM421, has allowed to determine the precise location and boundaries of the genetic island.
Phylogenomic analysis, using whole-genome distance calculations (Meier-Kolthoff et al., 2013) showed that the seven strains belong to two of the eight subgroups that form the P. fluorescens complex of species (Garrido-Sanz et al., 2016): P. corrugata and P. fluorescens subgroups. The seven strains also had different origins, ranging from the plant-rhizosphere to Antarctic soil and water samples. Phylogenomic and phylogenetic analysis has shown that there have been at least five insertion events in the evolutionary story of this genetic island. It is likely that sequencing the genomes of new strains will result in finding new strains containing the flagellar island either at one of the identified locations or at a different one, representing novel insertion events. In this sense, we have recently isolated a strain RMP9, which is closely related to F113 and has the island inserted in the same position. Outside from the pseudomonads, closer homologs of the genes encoded by the genomic islands are in the A. vinelandii genome, although a few of them have closer relatives among different Gamma Proteobacteria. Therefore, the origin of the island is unclear.
Pseudomonas fluorescens F113 does not produce the second flagellar apparatus under laboratory conditions. However, the master regulator of flagella, flhDC, and the flagellin encoding gene, fliC2, are expressed in kinB and algU mutant backgrounds, indicating that both genes are strongly repressing the expression of the second flagellar genes. It has been shown that in A. vinelandii, motility is controlled by AlgU, which represses the flhDC operon (Leon and Espin, 2008). In pseudomonads, kinB expression requires the AlgU sigma factor (Damron et al., 2009). The results presented here in P. fluorescens F113 are in agreement with this data and indicate that in F113 the flhDC operon is under strict repression by both genes. Furthermore, we have previously shown that AmrZ, a transcriptional regulator which also requires AlgU for its transcription (Tart et al., 2005), downregulates the expression of the flhDC master operon (Martinez-Granero et al., 2014). Since in F113, AlgU also regulates fleQ transcription through repression by AmrZ (Martinez-Granero et al., 2012) it is clear that AlgU simultaneously regulates the synthesis of both flagellar apparatus in P. fluorescens F113, which is congruent with the proposed role of AlgU as a master regulatory gene for environmental adaption (Martinez-Granero et al., 2014).
Ectopic expression of the flhDC operon in F113 is enough for production of the second flagellum. When flhDC is expressed from a vector with a kinB and algU independent promoter, there is an increase in swimming motility, indicating the production of the second flagellar apparatus. Surprisingly, ectopic expression of flhDC in isogenic backgrounds unable to produce the first flagellum, did not restore motility and were aflagelated.
An F113 kinB mutant is hypermotile (Barahona et al., 2011). Here we show that wild-type motility is restored by mutating either fliC2 or flhDC in a kinB- background. These results justify the phenotype of the kinB mutant, showing that its hypermotility is due to the production of the second flagellar apparatus, which is repressed in the wild-type background. In enterobacteria, the flhDC expression is under c-AMP control through the CRP protein (Stella et al., 2008). Here we have shown a similar regulatory mechanism for the biosynthesis of the second flagellar apparatus: mutation of the vfr gene, encoding a CRP ortholog, in a kinB- background, resulted in suppression of the hypermotility phenotype, consistent with lack of second flagellar apparatus production. Identical result was obtained when mutating cyaA, which encodes an adenylate cyclase in the F113 genome. These results indicate that production of the second flagellar apparatus is dependent on c-AMP through the Vfr protein. Although in other pseudomonads it has been shown that Vfr might regulate the expression of fleQ (Dasgupta et al., 2002) and therefore the production of the canonical flagellar apparatus, this is not the case in F113. Figure 7 shows a model for the regulation of the biosynthesis of the second flagellar apparatus in P. fluorescens F113. According to this model, the master regulatory operon for the synthesis of this flagellum, flhDC, is subjected to positive and negative regulation. Positive regulation is exerted through CyaA and Vfr. The flhDC operon is strongly repressed by KinB and downregulated by AmrZ, which also downregulates the expression of the flagellar structural genes for the synthesis of the second flagellar apparatus (Martinez-Granero et al., 2014). Both kinB and amrZ require the AlgU sigma factor for expression. Production of the second flagellar system would therefore require the relief of the KinB repression and the activation of the flhDC operon by Vfr and c-AMP. It should be noted that both AlgU and AmrZ are implicated in the regulation of both flagellar apparatus, indicating that these sigma factor and transcriptional regulator are key nodes in the regulation of motility in F113.
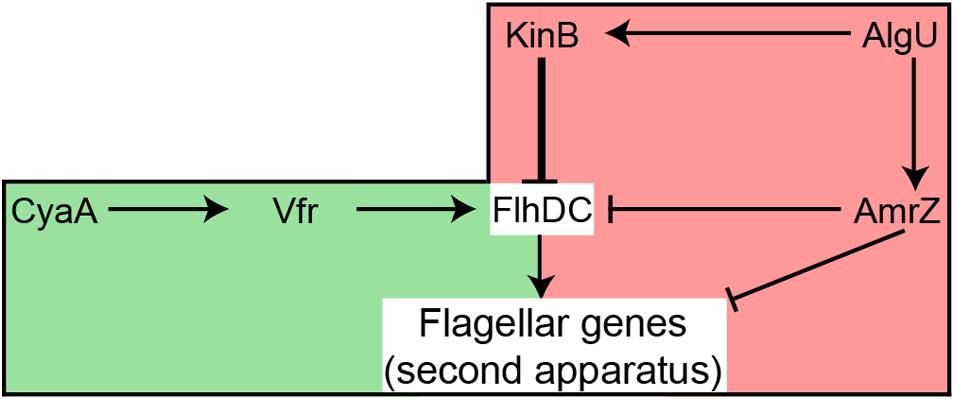
FIGURE 7. Hypothetical model showing the regulation of the production of the second flagellar apparatus in P. fluorescens F113. The flagellar master operon flhDC is positively regulated by Vfr, likely in response to c-AMP produced by CyaA. The master operon is strongly repressed (thick line) by KinB. AmrZ downregulates the expression of both the master operon and the flagellar structural genes. AlgU is required for the expression of kinB and amrZ. Green background indicates positive regulation, while red background indicates negative regulation of flhDC. Note that although AlgU and AmrZ regulate genes required for the synthesis of both flagellar apparatus, FlhDC only regulate the genes required for the second flagellar system.
We have also shown that the second flagellar apparatus is produced as a polar tuft of flagella. This tuft was observed in the wild-type strain, which otherwise produced a single polar flagellum, when ectopically expressing the flhDC operon. The tuft was also observed in the kinB mutant. Mutation of fliC2 in the kinB- background restored the formation of a single polar flagellum, showing that the tuft represents the second flagellar apparatus. Furthermore, by using an anti FliC1 antiserum, we have shown that the kinB mutant does not produce more FliC1 than the wild-type strain. Overproduction of FliC1 was, however, observed in a sadB mutant, that we have previously shown that overproduces this flagellin (Navazo et al., 2009). Flagella regulated by the flhDC master operon are usually peritrichous, as is the case for enterobacteria and A. vinelandii (Leon and Espin, 2008), however, a tuft of polar flagella controlled through flhDC is the normal flagellation of Burkholderia glumae (Jang et al., 2014), indicating that flagellar location does not depend on the master regulatory genes.
The tuft of flagella was also observed in a phenotypic variant isolated from the alfalfa rhizosphere (Sánchez-Contreras et al., 2002). This observation prompted us to determine the production of the second flagellar apparatus in this environment, observing that a number of previously isolated hypermotile phenotypic variants also expressed the fliC2 gene. These results suggested that the second flagellar apparatus might play a role in root colonization. Others and we, have previously shown that motility is one of the most important traits for competitive colonization of the rhizosphere (Barahona et al., 2011), since non-motile, non-chemotactic or mutants with reduced motility are displaced from the rhizosphere (de Weert et al., 2002; Capdevila et al., 2004). We have also observed that the rhizosphere selects for hypermotile phenotypic variants (Martinez-Granero et al., 2006). Therefore, it is likely that the hypermotility conferred by production of the second flagellar apparatus could be an important trait for competitive colonization of the root. This hypothesis was confirmed for strain F113, since blocking the production of the second flagellar apparatus by mutating the fliC2 gene, reduced the competitive colonization ability both in a wild-type and in a kinB mutant background. The fact that several of the strains shown to encode the second flagellar apparatus do not come from plant environments, make it difficult to assess the ecological importance of this system in the plant-root environment. Since colonization depends in multiple traits and is highly variable even in closely related strains, experiments comparing a number of rhizosphere isolates containing the island with isogenic or nearly isogenic strains that do not contain it would be necessary to generalize the ecological importance of this flagellar system.
Conclusion
Pseudomonas fluorescens F113 can produce two flagellar systems, the canonical, regulated by FleQ and a second system regulated by the flhDC master operon, similarly to enterobacteria and A. vinelandii. Genes for the synthesis of the second flagellar system are carried by a genetic island found only in a limited set of strains within the genus Pseudomonas. The second flagella is cryptic, but is produced by mutants in the kinB gene or by the wild-type strain when ectopically expressing the flhDC operon. Both flagella are coordinately regulated through AlgU, KinB, Vfr, and AmrZ. Production of the second flagellar apparatus in F113 results in a tuft of polar flagella, which increases motility, and gives a competitive advantage for root colonization.
Author Contributions
EB, AN, FM-G, and CM performed experiments and analyzed results. DG-S performed bioinformatic analysis. MR-N, MM, and RR conceived and supervised the study and wrote the manuscript.
Funding
Research was funded by Grant BIO2015-64480R (MINECO/FEDER EU).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank Centro de Computación Científica at UAM for giving us access to their computational facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01471
SUPPLEMENTARY FILE 1 | Scripts used in the bioinformatics analysis.
References
Akerley, B. J., Cotter, P. A., and Miller, J. F. (1995). Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80, 611–620. doi: 10.1016/0092-8674(95)90515-4
Arora, S. K., Ritchings, B. W., Almira, E. C., Lory, S., and Ramphal, R. (1997). A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179, 5574–5581.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Barahona, E., Navazo, A., Martinez-Granero, F., Zea-Bonilla, T., Perez-Jimenez, R. M., Martin, M., et al. (2011). Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 77, 5412–5419. doi: 10.1128/AEM.00320-11
Barahona, E., Navazo, A., Yousef-Coronado, F., Aguirre de Carcer, D., Martinez-Granero, F., Espinosa-Urgel, M., et al. (2010). Efficient rhizosphere colonization by Pseudomonas fluorescens F113 mutants unable to form biofilms on abiotic surfaces. Environ. Microbiol. 12, 3185–3195. doi: 10.1111/j.1462-2920.2010.02291.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST + : architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Capdevila, S., Martinez-Granero, F. M., Sanchez-Contreras, M., Rivilla, R., and Martin, M. (2004). Analysis of Pseudomonas fluorescens F113 genes implicated in flagellar filament synthesis and their role in competitive root colonization. Microbiology 150(Pt 11), 3889–3897. doi: 10.1099/mic.0.27362-0
Chand, N. S., Lee, J. S., Clatworthy, A. E., Golas, A. J., Smith, R. S., and Hung, D. T. (2011). The sensor kinase KinB regulates virulence in acute Pseudomonas aeruginosa infection. J. Bacteriol. 193, 2989–2999. doi: 10.1128/JB.01546-10
Chin-A-Woeng, T. F., Bloemberg, G. V., Mulders, I. H., Dekkers, L. C., and Lugtenberg, B. J. (2000). Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant Microbe Interact. 13, 1340–1345. doi: 10.1094/MPMI.2000.13.12.1340
Damron, F. H., Qiu, D., and Yu, H. D. (2009). The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 191, 2285–2295. doi: 10.1128/JB.01490-08
Dasgupta, N., Ferrell, E. P., Kanack, K. J., West, S. E., and Ramphal, R. (2002). fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184, 5240–5250. doi: 10.1128/JB.184.19.5240-5250.2002
Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S., et al. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824. doi: 10.1046/j.1365-2958.2003.03740.x
de Lorenzo, V., Eltisb, L., Kesslerb, B., and Timmisb, K. N. (1993). Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123, 17–24. doi: 10.1016/0378-1119(93)90533-9
de Weert, S., Vermeiren, H., Mulders, I. H., Kuiper, I., Hendrickx, N., Bloemberg, G. V., et al. (2002). Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 15, 1173–1180. doi: 10.1094/MPMI.2002.15.11.1173
de Weger, L. A., van der vlugt, C. I. M., Wijfjes, A. H. M., Bakker, P. A. H. M., and Schippers, B. (1987). Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 169, 2769–2773.
Dekkers, L. C., Mulders, I. H., Phoelich, C. C., Chin, A. W. T. F., Wijfjes, A. H., and Lugtenberg, B. J. (2000). The sss colonization gene of the tomato-Fusarium oxysporum f. sp. radicis-lycopersici biocontrol strain Pseudomonas fluorescens WCS365 can improve root colonization of other wild-type Pseudomonas spp.bacteria. Mol Plant Microbe Interact. 13, 1177–1183. doi: 10.1094/MPMI.2000.13.11.1177
Fahraeus, G. (1957). The infection of clover root hairs by nodule bacteria studied by simple glass technique. J. Genet. Microbiol. Rev. 16, 374–381.
Finan, T. M., Kunkel, B., De Vos, G. F., and Signer, E. R. (1986). Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167, 66–72.
Garrido-Sanz, D., Meier-Kolthoff, J. P., Göker, M., Martin, M., Rivilla, R., and Redondo-Nieto, M. (2016). Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE 11:e0150183. doi: 10.1371/journal.pone.0150183
Jang, M. S., Goo, E., An, J. H., Kim, J., and Hwang, I. (2014). Quorum sensing controls flagellar morphogenesis in Burkholderia glumae. PLoS ONE 9:e84831. doi: 10.1371/journal.pone.0084831
Jyot, J., Dasgupta, N., and Ramphal, R. (2002). FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 184, 5251–5260. doi: 10.1128/JB.184.19.5251-5260.2002
Kamilova, F., Validov, S., Azarova, T., Mulders, I., and Lugtenberg, B. (2005). Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 7, 1809–1817. doi: 10.1111/j.1462-2920.2005.00889.x
Kirchner, O., and Tauch, A. (2003). Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104, 287–299. doi: 10.1016/S0168-1656(03)00148-2
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laemmli, U. K. (1970). Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Leon, R., and Espin, G. (2008). flhDC, but not fleQ, regulates flagella biogenesis in Azotobacter vinelandii, and is under AlgU and CydR negative control. Microbiology 154(Pt 6), 1719–1728. doi: 10.1099/mic.0.2008/017665-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of gene expression data using real-time quantitative PCR and the 2-DDCTmethod. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, S., Wozniak, D. J., and Ohman, D. E. (1997). Identification of the histidine protein kinase KinB in Pseudomonas aeruginosa and its phosphorylation of the alginate regulator algB. J. Biol. Chem. 272, 17952–17960. doi: 10.1074/jbc.272.29.17952
Macnab, R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832
Martínez-Granero, F., Capdevila, S., Sánchez-Contreras, M., Martín, M., and Rivilla, R. (2005). Two site-specific recombinases are implicated in phenotypic variation and rhizosphere colonization in Pseudomonas fluorescens. Microbiology 151(Pt 3), 975–983. doi: 10.1099/mic.0.27583-0
Martinez-Granero, F., Navazo, A., Barahona, E., Redondo-Nieto, M., Rivilla, R., and Martin, M. (2012). The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS ONE 7:e31765. doi: 10.1371/journal.pone.0031765
Martinez-Granero, F., Redondo-Nieto, M., Vesga, P., Martin, M., and Rivilla, R. (2014). AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P. fluorescens F113. BMC Genomics 15:237. doi: 10.1186/1471-2164-15-237
Martinez-Granero, F., Rivilla, R., and Martin, M. (2006). Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 72, 3429–3434. doi: 10.1128/AEM.72.5.3429-3434.2006
McCarter, L. L. (2006). Regulation of flagella. Curr. Opin. Microbiol. 9, 180–186. doi: 10.1016/j.mib.2006.02.001
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Naseby, D. C., and Lynch, J. M. (1999). Effects of Pseudomonas fluorescens F113 on ecological functions in the pea rhizosphere are dependent on pH. Microb. Ecol. 37, 248–256. doi: 10.1007/s002489900147
Navazo, A., Barahona, E., Redondo-Nieto, M., Martinez-Granero, F., Rivilla, R., and Martin, M. (2009). Three independent signalling pathways repress motility in Pseudomonas fluorescens F113. Microb. Biotechnol. 2, 489–498. doi: 10.1111/j.1751-7915.2009.00103.x
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Redondo-Nieto, M., Barret, M., Morrisey, J. P., Germaine, K., Martinez-Granero, F., Barahona, E., et al. (2012). Genome sequence of the biocontrol strain Pseudomonas fluorescens F113. J. Bacteriol. 194, 1273–1274. doi: 10.1128/JB.06601-11
Redondo-Nieto, M., Barret, M., Morrissey, J., Germaine, K., Martinez-Granero, F., Barahona, E., et al. (2013). Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics 14:54. doi: 10.1186/1471-2164-14-54
Sambrook, J., Fritsch, E. F., and Maniatics, T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sánchez-Contreras, M., Martín, M., Villacieros, M., O’Gara, F., Bonilla, I., and Rivilla, R. (2002). Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 184, 1587–1596. doi: 10.1128/JB.184.6.1587-1596.2002
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Scher, F. M., and Baker, R. (1982). Effects of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium wilt pathogens. Phytopathology 72, 1567–1573. doi: 10.1094/Phyto-72-1567
Schmitt, C. K., Darnell, S. C., Tesh, V. L., Stocker, B. A., and O’Brien, A. D. (1994). Mutation of flgM attenuates virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J. Bacteriol. 176, 368–377.
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Simons, M., van der Bij, A. J., Brand, I., de Weger, L. A., Wijffelman, C. A., and Lugtenberg, B. J. (1996). Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant Microbe Interact. 9, 600–607. doi: 10.1094/MPMI-9-0600
Soutourina, O., Kolb, A., Krin, E., Laurent-Winter, C., Rimsky, S., Danchin, A., et al. (1999). Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181, 7500–7508.
Stella, N. A., Kalivoda, E. J., O’Dee, D. M., Nau, G. J., and Shanks, R. M. (2008). Catabolite repression control of flagellum production by Serratia marcescens. Res. Microbiol. 159, 562–568. doi: 10.1016/j.resmic.2008.07.003
Tart, A. H., Wolfgang, M. C., and Wozniak, D. J. (2005). The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 187, 7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005
Villacieros, M., Power, B., Sanchez-Contreras, M., Lloret, J., Oruezabal, R. I., Martín, M., et al. (2003). Colonization behaviour of Pseudomonas fluorescens and Sinorhizobium meliloti in the alfalfa (Medicago sativa) rhizosphere. Plant Soil 251, 47–54. doi: 10.1023/A:1022943708794
Villacieros, M., Whelan, C., Mackova, M., Molgaard, J., Sanchez-Contreras, M., Lloret, J., et al. (2005). Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol. 71, 2687–2694. doi: 10.1128/AEM.71.5.2687-2694.2005
Keywords: motility, pseudomonads, FlhDC, genetic island, rhizosphere
Citation: Barahona E, Navazo A, Garrido-Sanz D, Muriel C, Martínez-Granero F, Redondo-Nieto M, Martín M and Rivilla R (2016) Pseudomonas fluorescens F113 Can Produce a Second Flagellar Apparatus, Which Is Important for Plant Root Colonization. Front. Microbiol. 7:1471. doi: 10.3389/fmicb.2016.01471
Received: 05 May 2016; Accepted: 02 September 2016;
Published: 22 September 2016.
Edited by:
Erh-Min Lai, Academia Sinica, TaiwanReviewed by:
Stijn Spaepen, Max Planck Institute for Plant Breeding Research (MPG), GermanyMark Silby, University of Massachusetts Dartmouth, USA
Copyright © 2016 Barahona, Navazo, Garrido-Sanz, Muriel, Martínez-Granero, Redondo-Nieto, Martín and Rivilla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rafael Rivilla, rafael.rivilla@uam.es
†These authors have contributed equally to this work.
 Emma Barahona
Emma Barahona Ana Navazo†
Ana Navazo† Daniel Garrido-Sanz
Daniel Garrido-Sanz Candela Muriel
Candela Muriel Francisco Martínez-Granero
Francisco Martínez-Granero Miguel Redondo-Nieto
Miguel Redondo-Nieto Marta Martín
Marta Martín Rafael Rivilla
Rafael Rivilla