- 1Department of Microbiology, Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, Wrocław, Poland
- 2Max Planck Institute for Molecular Genetics, Berlin, Germany
- 3Laboratory of Biopolymers Structure, Intercollegiate Faculty of Biotechnology, University of Gdańsk and Medical University of Gdańsk, Gdańsk, Poland
- 4Department of Life Science Engineering, Fachbereich 2, HTW Berlin, Berlin, Germany
In bacteria, chromosome replication is initiated by the interaction of the initiator protein DnaA with a defined region of a chromosome at which DNA replication starts (oriC). While DnaA proteins share significant homology regardless of phylogeny, oriC regions exhibit more variable structures. The general architecture of oriCs is universal, i.e., they are composed of a cluster of DnaA binding sites, a DNA-unwinding element, and sequences that bind regulatory proteins. However, detailed structures of oriCs are shared by related species while being significantly different in unrelated bacteria. In this work, we characterized Epsilonproteobacterial oriC regions. Helicobacter pylori was the only species of the class for which oriC was characterized. A few unique features were found such as bipartite oriC structure, not encountered in any other Gram-negative species, and topology-sensitive DnaA-DNA interactions, which have not been found in any other bacterium. These unusual H. pylori oriC features raised questions of whether oriC structure and DnaA-DNA interactions are unique to this bacterium or whether they are common to related species. By in silico and in vitro analyses we identified putative oriCs in three Epsilonproteobacterial species: pathogenic Arcobacter butzleri, symbiotic Wolinella succinogenes, and free-living Sulfurimonas denitrificans. We propose that oriCs typically co-localize with ruvC-dnaA-dnaN in Epsilonproteobacteria, with the exception of Helicobacteriaceae species. The clusters of DnaA boxes localize upstream (oriC1) and downstream (oriC2) of dnaA, and they likely constitute bipartite origins. In all cases, DNA unwinding was shown to occur in oriC2. Unlike the DnaA box pattern, which is not conserved in Epsilonproteobacterial oriCs, the consensus DnaA box sequences and the mode of DnaA-DnaA box interactions are common to the class. We propose that the typical Epsilonproteobacterial DnaA box consists of the core nucleotide sequence 5′-TTCAC-3′ (4–8 nt), which, together with the significant changes in the DNA-binding motif of corresponding DnaAs, determines the unique molecular mechanism of DnaA-DNA interaction. Our results will facilitate identification of oriCs and subsequent identification of factors which regulate chromosome replication in other Epsilonproteobacteria. Since replication is controlled at the initiation step, it will help to better characterize life cycles of these species, many of which are considered as emerging pathogens.
Introduction
Chromosome replication is tightly controlled and strictly dependent on cell cycle progression. It is primarily regulated at the first step, initiation (Zakrzewska-Czerwińska et al., 2007; Katayama et al., 2010; Skarstad and Katayama, 2013; Leonard and Grimwade, 2015). The basic mechanism of initiation is conserved in nearly all bacteria. First, the initiator protein DnaA recognizes and binds to a specific chromosomal region, the replication origin oriC (Ozaki and Katayama, 2009; Katayama et al., 2010; Duderstadt et al., 2011; Kaguni, 2011). This interaction leads to the formation of a highly ordered nucleoprotein complex (orisome) followed by DNA strand separation within a DNA unwinding element (DUE; Rozgaja et al., 2011; Ozaki et al., 2012; Duderstadt and Berger, 2013). The unwound DNA region provides the entry site for the assembly of a multiprotein apparatus (replisome) that synthesizes the nascent DNA strands (Beattie and Reyes-Lamothe, 2015). Most of the information on bacterial chromosome replication comes from studies in Escherichia coli, whose oriC, DnaA, and DnaA-DNA reciprocal interactions as well as the accessory and regulatory factors have been thoroughly characterized (reviewed in Katayama et al., 2010; Kaguni, 2011; Leonard and Grimwade, 2015). The initiation of chromosome replication has also been studied in a few other species (Bacillus subtilis, Caulobacter crescentus, Mycobacterium tuberculosis, Streptomyces coelicolor, and Helicobacter pylori). Comprehensive studies on these species as well as species related to E. coli, B. subtilis, C. crescentus, or Mycoplasma sp. (Harding et al., 1982; Lartigue et al., 2003; Shaheen et al., 2009; Briggs et al., 2012) suggest that the specific activities of DnaA proteins (Zawilak-Pawlik et al., 2005), structures of the oriC regions (Briggs et al., 2012; Rajewska et al., 2012), modes of orisome assembly (Zawilak-Pawlik et al., 2005; Madiraju et al., 2006; Ozaki and Katayama, 2011; Briggs et al., 2012; Scholefield et al., 2012; Donczew et al., 2014), accessory proteins and regulatory mechanisms are shared by related species while being significantly different in unrelated bacteria (Wolański et al., 2014). However, it should be noted that in most bacterial species the mechanistic details of orisome assembly are still largely unknown.
OriCs are usually located in the vicinity of the dnaA and dnaN genes, and they can be mono- or bi-partite (Wolański et al., 2014). OriC regions are composed of three functional modules: a cluster (or clusters) of DnaA binding sites (DnaA boxes), a DNA-unwinding element (DUE), and sequences that bind regulatory proteins. Typical DnaA boxes are 9-mers with sequences similar to the “perfect,” high-affinity R-type E. coli DnaA box TTATCCACA with some degree of degeneracy (allowed mismatches ≤ 2; Wolański et al., 2014). However, different classes of “imperfect” DnaA boxes (I sites and tau boxes in E. coli, W-boxes in C. crescentus), which differ in sequence and length from the “perfect” boxes, have been shown to play important roles in DnaA oligomer assembly (McGarry et al., 2004; Kawakami et al., 2005; Ozaki and Katayama, 2009; Taylor et al., 2011). The arrangement of DnaA boxes in oriC (number, spacing, orientation) is not stochastic. DnaA boxes provide a molecular scaffold for sequential DnaA binding and oligomerisation, which leads to DNA unwinding in the DUE region. However, there is no “perfect” or “model” scaffold. There are a variety of DnaA box arrangements in bacterial oriCs, and this phenomenon is still not explained in terms of structure or function (Wolański et al., 2014; Leonard and Grimwade, 2015). The second important module, the DUE, is located outside of the DnaA box cluster, adjacent (~2 helical turns) to the last DnaA box in the scaffold. The DUE region usually contains tens of base pairs (bps) and is rich in thymines and adenines (an AT-rich region), which lower the thermodynamic stability of the DUE compared to sequences of equal AT/GC or high GC content. It has been recently shown that the region of the DUE proximal to the DnaA-box encodes a motif, a DnaA-trio, required by B. subtilis DnaA to open DNA and to assemble on ssDNA (Richardson et al., 2016). The last oriC module, the sequences that bind regulatory proteins (oriBPs, origin binding proteins), is the most divergent of all three modules (Wolański et al., 2014; Marczynski et al., 2015). These sequences can overlap with DnaA boxes or be located within the DUE or elsewhere within oriC. They bind different classes of proteins, such as nucleoid associated proteins (NAPs) or response regulators of two component systems. Their primary role is to efficiently transmit feedback information (positive or negative) from the environment and/or the cell itself to the oriC to rapidly adjust the replication rate.
Our previous work on H. pylori oriC revealed that it is, unlike origins of most Gram-negative bacteria, composed of two DnaA box clusters (DnaA box consensus sequence TCATTCACN), oriC1 and oriC2, flanking the dnaA gene (Donczew et al., 2012). The DnaA protein binds to both oriC1 and oriC2, bridging them together and looping out dnaA, in which it resembles B. subtilis orisome (Krause et al., 1997). Surprisingly, oriC2–DnaA interaction was shown to depend on DNA topology, and we identified two DnaA boxes (ts1 and ts2) which were bound only in a supercoiled form (Donczew et al., 2014). The DNA-unwinding element region is located in the oriC2 sub-region downstream of dnaA. These unusual H. pylori oriC features raised questions of whether they are unique to this bacterium or they are also common to related species. Thus, this work was undertaken to identify and characterize oriC regions in bacterial species from selected Epsilonproteobacteria. Epsilonproteobacteria are found globally and inhabit a wide variety of ecological niches (Eppinger et al., 2004; Gupta, 2006). Two species of Epsilonproteobacteria, H. pylori, and Campylobacter jejuni, are undisputed human pathogens (Atherton, 2006; Epps et al., 2013). Others are proposed to be emerging pathogens connected with gastrointestinal diseases and/or reproductive disorders in animals (Helicobacter sp., Campylobacter sp., Arcobacter sp.). However, many Epsilonproteobacteria are non-pathogenic (symbiotic or free living species), recognized as an ecologically significant group of bacteria occurring dominantly in various redoxclines such as in deep-sea hydrothermal environments or oil fields (Nakagawa and Takaki, 2009). Such diverse life styles of Epsilonproteobacteria might be reflected by the diversity of the initiation or regulatory factors involved in the initiation of chromosome replication of the species inhabiting various ecological niches. Thus, to perform a reliable and comprehensive comparative analysis of Epsilonproteobacterial origins of chromosome replication and to compare it with H. pylori oriC it was reasonable to select species representing both H. pylori-related as well as relatively unrelated genera and lifestyles. By a two-step approach (in silico analysis followed by experimental in vitro work) we were able to precisely determine the position of oriC on chromosomes of pathogenic A. butzleri, commensal Wolinella succinogenes and free-living S. denitrificans and characterize the two most conserved modules of their oriC regions, namely the DnaA box clusters and the DUE. The in vitro bound clusters of DnaA boxes are located upstream (oriC1) and downstream (oriC2) of dnaA. Thus, the identified origins likely constitute bipartite origins as in H. pylori. The DNA-unwinding element region is located in the oriC2 sub-region downstream of dnaA. The detailed comparative analysis allowed us to propose Epsilonproteobacterial oriC features which are typical for many origins of unrelated bacteria as well as unique for this class.
Materials and Methods
In silico Origin Predictions
The prediction of oriC-type replication origins in the genomes of A. butzleri RM4018 [GenBank entry CP000361.1], S. denitrificans DSM 1251 [GenBank entry CP000153.1.1], W. succinogenes DSM 1740 [GenBank entry BX571656.1] was performed in a stepwise procedure, similarly as described previously (Donczew et al., 2012). Briefly, it combined GC-skew analysis, prediction of superhelicity-dependent helically unstable DNA stretches (SIDDs) in intergenic regions in the vicinity of the inflection point (minimum) of the GC-skew, and DnaA box prediction. Details are described in Supplementary Materials.
Comparative Analysis of DnaA Amino Acid Sequences
Amino acids sequences of DnaA proteins from Proteobacteria and Actinobacteria were retrieved from UniProt amino acids sequence database (Boutet et al., 2016). To avoid sequence repetition, the search was performed on Ref90 subdatabase (Suzek et al., 2007). The Ref90 UniProt database was searched for term “chromosomal replication initiator protein DnaA” and results were further filtered to obtain sequences from Proteobacteria or Actinobacteria. Sequences of Proteobacterial DnaA proteins were further divided according to classification in the UniProt database into subfamilies: Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Delta/Epsilonproteobacteria according to classification proposed by Woese (1987). All amino acid sequences were subjected to multiple sequence alignment (MSA) using MAFFT algorithm (Katoh and Standley, 2013). MSAs were performed on the whole set of sequences, as well as on subsets related to Proteobacterial subfamilies. Analysis and visualization of the MSA results were performed using BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Materials and Culture Conditions
The strains, plasmids and proteins used in this work are listed in Table S1. The primer sequences used in this study are listed in Table S2. The genomic DNA of A. butzleri RM4018, S. denitrificans DSM 1251, and W. succinogenes DSM 1740 were used as templates to amplify DNA fragments for cloning. E. coli was grown at 30 or 37°C on solid or in liquid Luria-Bertani medium supplemented with 100 μg/ml ampicillin or 50 μg/ml kanamycin when necessary. Plasmids and DNA fragments were purified using a GeneJET Gel Extraction Kit, GeneJET Plasmid Miniprep Kit, GeneJET Plasmid Midiprep Kit (Thermo Scientific), or Plasmid Midi AX (A&A Biotechnology). DnaA proteins were purified as described in (Zawilak-Pawlik et al., 2006) with minor modifications (Supplementary Materials). In all subsequent analyses DnaA was supplemented with 3 mM ATP (electron microscopy) or 5 mM ATP (footprinting and P1 nuclease assay).
Footprinting, P1 Nuclease Assay, and Primer Extension (PE) Reactions
DMS and DNaseI footprinting was performed as described previously (Sasse-Dwight and Gralla, 1991; Krause et al., 1997; Zawilak et al., 2001; Donczew et al., 2014). The P1 nuclease assay was conducted similarly as described (Donczew et al., 2012). Details are described in Supplementary Materials.
Electron Microscopy
Electron microscopy was performed as described previously (Donczew et al., 2012, 2014). Details are described in Supplementary Materials.
Results
In silico Analysis Identifies oriCs at the Vicinity of dnaA
The in silico approach was similar to that previously applied to detect oriC in H. pylori (Donczew et al., 2012), namely a combination of GC-skew analysis, prediction of superhelicity-dependent helically unstable DNA stretches (SIDDs) in intergenic regions in the vicinity of the inflection point (minimum) of the GC-skew, and DnaA box prediction. We chose E. coli consensus DnaA box sequence [5′-TTWTNCACA allowing for 2 mismatches and 3 mismatches for closely-spaced DnaA boxes (Schaper and Messer, 1995)] in order not to bias the results by assuming that other Epsilonprotebacteria follow the H. pylori DnaA box consensus.
We identified putative origins of chromosome replication in three selected Epsilonproteobacterial species: A. butzleri, W. succinogenes, and S. denitrificans (Figure S1). In all three genomes, we obtained oriC predictions in the dnaA upstream region (Figure S2A), which we termed “oriC1” in analogy to the corresponding region in H. pylori (Donczew et al., 2012). The DoriC database predicts oriC at this position for A. butzleri RM4018 (DoriC entry ORI92240124) W. succinogenes DSM 1740 (DoriC entry ORI10010101) and S. denitrificans DSM 1251 (DoriC entry ORI10010173; Gao et al., 2013). We considered these oriC1 regions less likely to represent the regions where DNA unwinding would occur because no particular DnaA box could be assigned at a distance of ~2 helical turns to the SIDDs in the expected orientation. In all three genomes, we also obtained significant oriC predictions in the dnaA-dnaN intergenic region (Figures S1, S2B). These regions are characterized by the presence of significant SIDDs accompanied by the clusters of DnaA boxes with a SIDD-proximal DnaA box (or two closely spaced boxes) located at a distance of ~2 helical turns from the right border of the SIDD (Figure S2B). We assumed that these regions contain the DUEs and we termed them “oriC2” in analogy to the corresponding region in the H. pylori replication origin where unwinding occurs (Donczew et al., 2012). We obtained one additional oriC prediction for S. denitrificans, which we termed “oriC3” (Figure S2C). Due to a lack of DnaA box pattern conservation, we considered this prediction less likely than the oriC2 regions to represent the regions where unwinding would occur.
Epsilonproteobacterial DUEs Are Located in the dnaA-dnaN Intergenic Region
Next we analyzed the putative oriC regions in vitro. Because DnaA box clusters are also found outside of the origin sites, we focused on identification of the DUE as the most reliable feature of bacterial oriCs (Kitagawa et al., 1998; Okumura et al., 2012; Smith and Grossman, 2015). To experimentally identify the DUE position in predicted origins, P1 nuclease assay was applied. The method is widely used to identify helically unstable regions on a DNA strand, including DUEs. For the P1 nuclease assays, a series of plasmids containing in silico predicted single oriC regions was constructed, and cognate DnaA proteins were purified (Table S1 and Figure S3). The supercoiled plasmids were incubated with increasing amounts of DnaA protein, and the resulting single-stranded DNA regions were digested with P1 nuclease. Subsequently, site-specific digestion by PvuI or DrdI excised the DNA fragment from the plasmid, the size of which allowed us to approximately estimate the position of a region unwound by DnaA. The DnaA-dependent unwinding occurred exclusively in the predicted oriC2 regions for all analyzed Epsilonproteobacteria (Figure 1 and Figure S4). The relatively high concentration of DnaAs required for opening of the plasmid DNA at DUE suggest that there are unknown protein factors which facilitate DNA unwinding, similarly as HU enables unwinding of E. coli oriC (see Section Discussion). In the plasmids that were unwound in a DnaA-dependent manner (pori2 series plasmids), DNA fragments of ~550–650 bp were excised by P1/PvuI and P1/DrdI, indicating specific single-stranded DNA formation within the oriC2 regions (Figure 1). The plasmids were also unwound at a site within the vector sequence corresponding to the plasmid origin of replication regardless of DnaA presence or concentration (all lanes contained additional DNA fragments of 800 and 500 bp in PvuI and DrdI digestion, respectively). These results are consistent with the known phenomenon that the AT-rich regions present at the origins of replication or preceding transcription units are helically unstable and may undergo spontaneous transition to a single-stranded form (Kowalski et al., 1988).
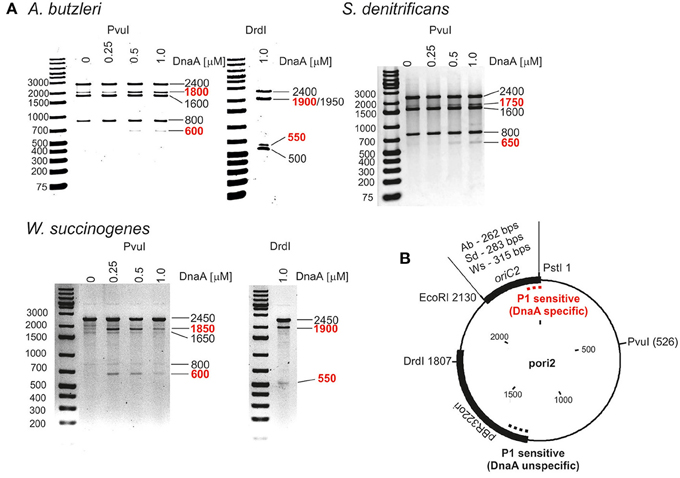
Figure 1. In vitro identification of DUEs in putative oriC regions of selected Epsilonproteobacteria. (A) P1 nuclease assay determining the DNA region susceptible to DnaA-dependent unwinding. Plasmids containing putative oriCs with DUE regions (pAbori2, pSdori2, pWsori2) were incubated with the indicated amounts of species-specific DnaA protein, digested by P1 nuclease, and restriction digested by PvuI or DrdI. The DNA fragments were visualized by separation on 1% agarose gels and ethidium bromide staining. DNA fragments produced in a DnaA-dependent manner are marked in red. (B) Schematic map of the plasmids used in the assay. The most important plasmid features are marked. The specific and nonspecific P1 sensitive regions are indicated by red and black dashed lines, respectively.
To precisely determine the unwound regions, PE reactions with 32P-labeled primers were performed on P1-digested oriC2 plasmid templates (Figure 2 and Figure S5; the primers are specified in Table S2). The primers hybridized to the template DNA ~40–80 bp away from the in silico predicted DUE region and were extended by Taq polymerase until it encountered the P1 nuclease digestion site. The detailed PE analysis confirmed that all oriC2 regions underwent DnaA-dependent unwinding. Thus, they all contained DUE sequences. The main part of each identified DUE region is an AT-rich region, which is a typical feature of bacterial origins (Figure 2 and Figure S5). In A. butzleri, it encompasses ~26 bps and contains ~4% GC residues (overall chromosomal GC content is 27.05%). The S. denitrificans DUE is 40 bps long and contains 20% GC (overall 34.46%). The W. succinogenes AT-rich region is 28 bps long and contains 14% GC (overall 48.46%). Analyses of the DUE sequences did not detect any repeats similar to 13-mer E. coli L, M, R repeats in the identified AT-rich regions.
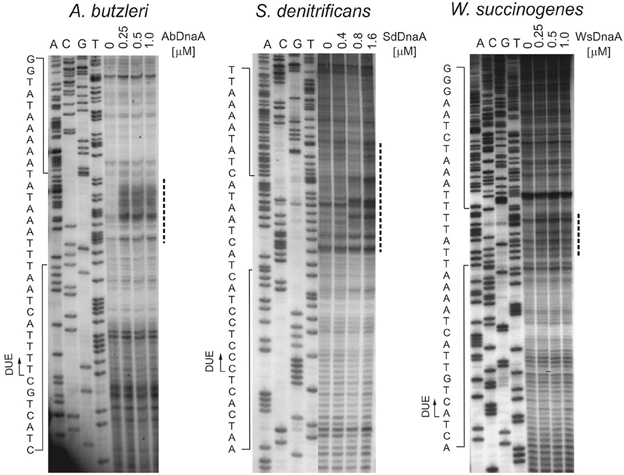
Figure 2. In vitro identification of the A. butzleri, S. denitrificans and W. succinogenes oriC sequence unwound by cognate DnaA proteins. Plasmids: pAbori2, pSdori2, and pWsori2 after incubation with the indicated amounts of DnaA, were digested by P1 nuclease and used as templates for PE reactions with 32P-labeled primers P2, P8, and P5, respectively. Dashed-lines indicate the nucleotides susceptible to P1 nuclease treatment. The boundaries of the DUE are marked with continuous-line arrows next to the presented sequences. Complementary results, with reverse primers are presented on Figure S5.
DnaA Box Clusters Are Located Upstream and Downstream of the dnaA Gene
The initial unwinding of DNA at the DUE site strictly depends on the DnaA interaction with oriC. Bacterial oriC regions usually contain one or two clusters of DnaA binding sites located in the vicinity of the DUE. They provide a platform for DnaA binding and proper oligomerisation, which leads to helix destabilization. Based on E. coli studies, the R-type DnaA box consensus sequence was proposed to be 5'-TTWTNCACA. However, the DnaA-binding sequences are variable, especially between distantly related species (Messer, 2002; Leonard and Méchali, 2013; Wolański et al., 2014). Thus, the precise determination of a DnaA box sequence in new species requires detailed in vitro analyses of the DnaA-DNA interaction. To determine the DnaA binding sites in the regions identified in silico, we used electrophoretic mobility shift assay (EMSA), electron microscopy (EM), and DMS footprinting.
Preliminary identification of DNA regions interacting with the DnaA protein was conducted using EMSA as described previously (Donczew et al., 2015). Fluorescently labeled PCR-amplified oriC sub-regions of A. butzleri, S. denitrificans, and W. succinogenes were incubated with increasing DnaA concentrations and subsequently resolved on a polyacrylamide gel (Figure S6). The EMSA indicated that in all analyzed origins, the DnaA protein was bound to oriC1 and oriC2 sub-regions; no binding of DnaA to the putative S. denitrificans oriC3 sub-region was observed. The binding of DnaA to oriC1 and oriC2 sub-regions was confirmed by electron microscopy (Figure 3). The pAbori1ori2, pSdori1ori2, and pWsoriori2 plasmids, containing oriC1 and oriC2 sub-regions separated by a dnaA gene, were incubated with corresponding DnaA proteins. The nucleoprotein complexes were subsequently stabilized by glutaraldehyde crosslinking and digested by ScaI to linearize plasmid molecules. The analysis revealed that the majority (70–90%) of the analyzed plasmid molecules were bound by DnaA (Figure 3). The incubation of DnaA with supercoiled plasmids led to formation of two predominant kinds of nucleoprotein complexes: 1/looped DNA structures (~25–30% of all bound molecules) with a single protein complex bound to two distant DNA regions (Figures 3A,C). The distance measurements between the plasmid ends and the protein core on ScaI digested nucleoprotein complexes confirmed the simultaneous binding of DnaA to oriC1 and oriC2 (Figures 3B,C); 2/plasmid molecules with a single protein complex bound to a single plasmid region, which constituted ~68–74% of all bound molecules (Figures 3A,C). The distance measurements confirmed the binding of DnaA to oriC1 or oriC2. 60–67% of the molecules were bound at oriC2 while 4–6% of the molecules were bound at oriC1. Approximately 3% of all the plasmid molecules were bound at unspecific regions (Figure 3C). This analysis suggested that DnaA exhibits higher affinity toward oriC2 than toward oriC1 or that the complexes formed at oriC2 are more stable than those formed on oriC1. The interaction between DnaA molecules bound to two suborigins apparently stabilized DnaA interactions with oriC1, because the majority of all oriC1 regions which were bound by DnaA were simultaneously joined to a protein complex interacting with oriC2. In summary, EMSA and EM confirmed DnaA binding to oriC1 and oriC2 and suggested that the origin organizations in A. butzleri, S. denitrificans, and W. succinogenes resembled that of H. pylori (Donczew et al., 2012).
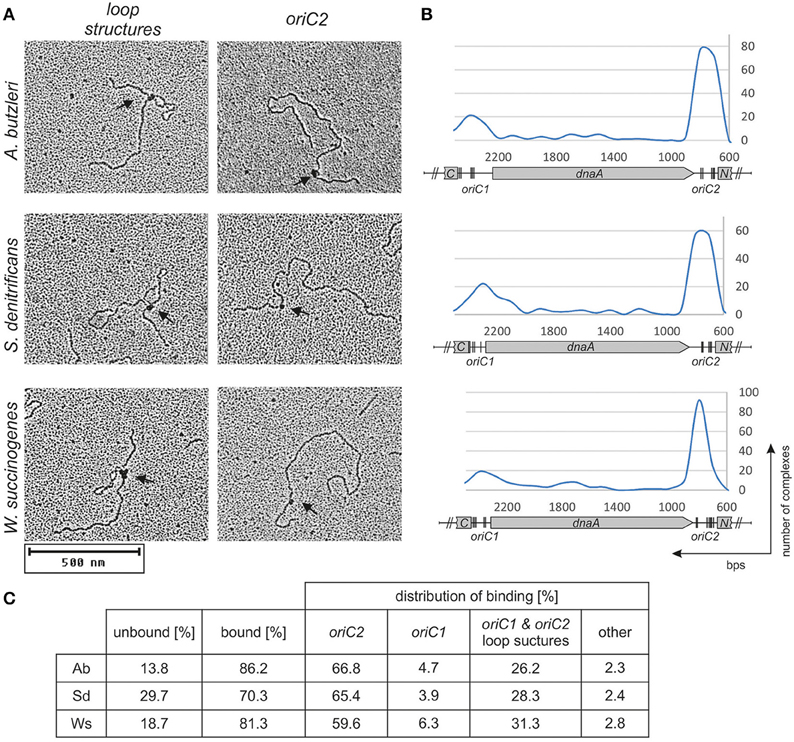
Figure 3. DnaA binding to supercoiled pori1ori2 plasmids. (A) Representative images from an EM analysis of DnaA interaction with the indicated plasmids are presented; only loop structures and DnaA-oriC2 complexes are shown. (B) Histograms of complexes of supercoiled oriC-plasmids with cognate DnaA proteins. Distribution of complexes were calculated based on an analysis of 200 molecules for each plasmid. The most characteristic features of each plasmid are shown below the histogram: oriC1, oriC2, dnaA, and fragments of ruvC (C) and dnaN (N). In silico predicted DnaA boxes (Figure S2) are indicated by vertical bars. (C) Statistical distribution and the level of DnaA binding is presented in the table below micrographs. The percentage of bound molecules and distribution of complexes were calculated based on an analysis of 200 molecules for each plasmid.
The next step was to identify DnaA boxes at oriC1 and oriC2 by DMS footprinting. The DMS footprinting method is based on the specific methylation of guanine and, to a lesser degree, adenine residues by dimethyl sulfate. As a result of methylation, the proximate phosphodiester bond of the DNA backbone becomes susceptible to piperidine cleavage. Proteins bound to specific DNA regions hinder DMS modification and, consequently, nucleic acid fragmentation. A subsequent primer extension reaction allows the identification of the protein binding site, which becomes apparent as decreased intensity of DNA bands on a footprinting gel. Plasmids (Table S1) containing the investigated sub-regions of A. butzleri (pAbori1 and pAbori2), S. denitrificans (pSdori1 and pSdori2), and W. succinogenes (pWsori1 and pWsori2) were incubated with increasing concentrations of cognate DnaA protein, methylated by DMS and piperidine-cleaved. To determine protein binding sites, sets of primers that were complementary to the upstream regions of putative DnaA boxes (Table S2) were used in the PE reactions. We detected multiple G residues protected by DnaA protein in both origin sub-regions of S. denitrificans, A. butzleri, and W. succinogenes (Figure 4 and Figures S7–S9). The subsequent comparison of the DNA sequences in the vicinity of protected G residues identified 8–10 DnaA boxes at the oriC1-dnaA-oriC2 regions for each of the investigated Epsilonproteobacteria (Figure 5 and Figures S7–S9). It should be noted that although the DnaA concentrations required for detection of DnaA-DNA interactions were relatively high (between 0.4 and 1.6 μM), the specificity of the binding was maintained, because only G residues located within the sequence similar to E. coli 5′TTWTNCACA motif were protected from DMS modification. Other G residues, present elsewhere in the region, with the exception of a region that becomes hyper-methylated upon DnaA binding to oriC2 in A. butzleri and W. succinogenes (hs region, Donczew et al., 2014), were insensitive to DMS treatment (see Section Discussion). The exact localization of DnaA binding sites, orientation and number of boxes in oriC1 differs greatly among selected Epsilonproteobacteria (Figure 5 and Figures S7B, S8C, S9C), whereas the oriC2 region preserved the general features of a typical bacterial origin of replication. These features include the distance between the DUE and R1E. coli—type DnaA box (~8–18 bps, R1E. coli—type box is a DUE-proximal DnaA box in reverse orientation, as in the R1 box in E. coli oriC), the orientation of the R1E. coli—type box in respect to the DUE, and the opposite arrangement of the DUE-distal box. We were unable to confirm DnaA binding to the in silico predicted pairs of head-to-tail boxes that are essential for the formation of a functional orisome in H. pylori and that play a crucial role in its DUE unwinding (ts boxes; Donczew et al., 2014; see Section Discussion). The sequences of in vitro determined DnaA boxes were assembled to generate a consensus DnaA box sequence for each studied bacterium (Figure 5): A. butzleri 5′ HTWTTCACW, S. denitrificans 5′ HHATTCACA, and W. succinogenes 5′ TTWTTCACN. The comparison of DnaA box consensus sequences of the analyzed Epsilonproteobacteria, together with the consensus H. pylori DnaA box sequence 5′-TCATTCACN (Donczew et al., 2014 and Figure 5), revealed that between the species the boxes are relatively diverse in the first three and the very last nucleotide positions, similarly as was observed before in other bacteria (Tsodikov and Biswas, 2011; Wolański et al., 2014), but they are characterized by a conserved 5-nt core sequence 5′-TTCAC (4–8th residue of a 9-mer; Figure 5). We propose that this core sequence is a hallmark of DnaA boxes in Epsilonproteobacteria which distinguishes these boxes from other bacterial species.
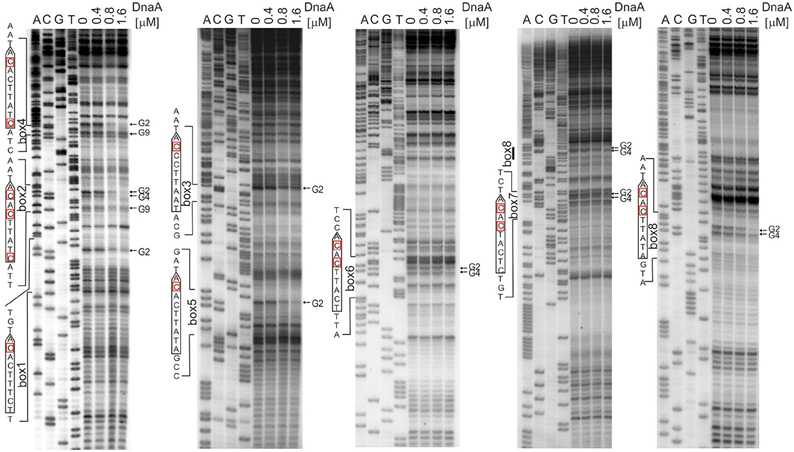
Figure 4. Identification of the DnaA boxes in the S. denitrificans oriC region. DMS footprinting analysis of DnaA-oriC interactions. Plasmid pSdori1ori2 was incubated with the indicated concentrations of the DnaA protein, methylated with DMS and used as a template for PE reactions; primers used to map DnaA boxes are specified in Table S2. Sequences of identified boxes are presented on the left of each panel; protected guanosine residues (G) are indicated with arrows. Densitometric plots are presented on Figure S7. Similar experiments were conducted for A. butzleri and W. succinogenes (Figures S8, S9, respectively).
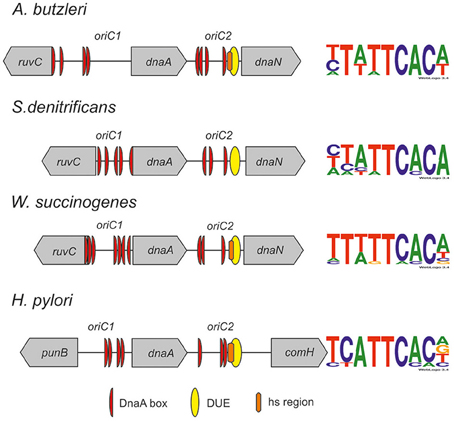
Figure 5. Schematic presentation of the most characteristic features identified by DMS and P1 nuclease assay in origins of replication of selected Epsilonproteobacteria. DnaA boxes, DUE and DMS hypermethylated regions (hs regions) similar to those identified by Donczew et al., 2014 in H. pylori are presented. The picture is not drawn to scale. The consensus Epsilonproteobacteria DnaA box sequences are based on the DMS analyses presented in this work and by Donczew et al., 2014. The logos were made with the WebLogo website (http://weblogo.threeplusone.com/). Detailed sequences of S. denitrificans, A. butzleri, and W. succinogenes oriC regions are presented on Figures S7–S9, respectively.
Epsilonproetoabcterial DnaA's Specificity toward DnaA Boxes is Different than That of E. coli and M. tuberculosis DnaAs
Experimentally identified Epsilonproteobacterial DnaA boxes follow the general DnaA box pattern. However, we observed two distinct features of these boxes: the conserved T residue at the 5th position of the DnaA box and the protection of two G residues by DnaA from DMS modification (G residues at the bottom strand of the DnaA box). In 33 DnaA boxes identified in Epsilonproteobacteria (26 in this work and 7 in H. pylori) the fifth position of the DnaA box was occupied by a T residue. In contrast, in E. coli and M. tuberculosis, the C residue is preserved at the 5th position of the DnaA box. However, it should be noted that this residue is not important for sequence-specific E. coli and M. tuberculosis DnaA binding to DnaA boxes (Schaper and Messer, 1995; Fujikawa et al., 2003; Tsodikov and Biswas, 2011). All of the identified Epsilonproteobacterial DnaA boxes were protected at both G residues from DMS modification upon DnaA binding. In similar DMS experiments, E. coli and M. tuberculosis DnaA proteins protect the 2nd guanine residue (G2) while expose the 4th guanine residue (G4) (Grimwade et al., 2000; Madiraju et al., 2006; Kaur et al., 2014). These two unique features prompted us to confirm the intrinsic ability of Epsilonproteobacterial DnaAs to interact with G4 of the DnaA box and the importance of the T5 residue for Epsilonproteobacteria DnaA-DNA interactions.
First, we directly compared the E. coli DnaA and A. butzleri DnaA interactions with H. pylori oriC regions. For both proteins the boxes were not optimal, neither in sequence of a single box nor in an overall organization of DnaA boxes in oriC. However, they should be recognized by both proteins since they contain the core sequence 5′-TTCAC (4–8 bps) important for Epsilonproteobacteria (represented by A. buztleri DnaA here), and recognizable by E. coli DnaA. We applied the DMS footprint assay to be able to observe interactions of DnaAs with guanines. Both proteins bound boxes located at H. pylori oriC1 (boxes 2–4; Donczew et al., 2014; Figure 6A). Both proteins recognized H. pylori DnaA boxes according to their intrinsic molecular pattern—A. butzleri DnaA protected both G residues (G2 and G4) while E. coli DnaA protected G2 and exposed G4 to DMS (Figure 6A). The differences in DnaA interaction with G residues between Epsilonproteobacterial DnaAs and EcDnaA were confirmed by DNaseI footprinting. The H. pylori GST-HpDnaA(IV) protein (Zawilak et al., 2001) interacted with boxes 2–3 and 4–5; this interaction almost entirely protected the boxes from DNaseI digestion (Figure S10). E. coli DnaA also interacted with these boxes, however, in contrast to H. pylori DnaA, it exposed DNA to DNaseI digestion at positions corresponding to G4 of each DnaA box.
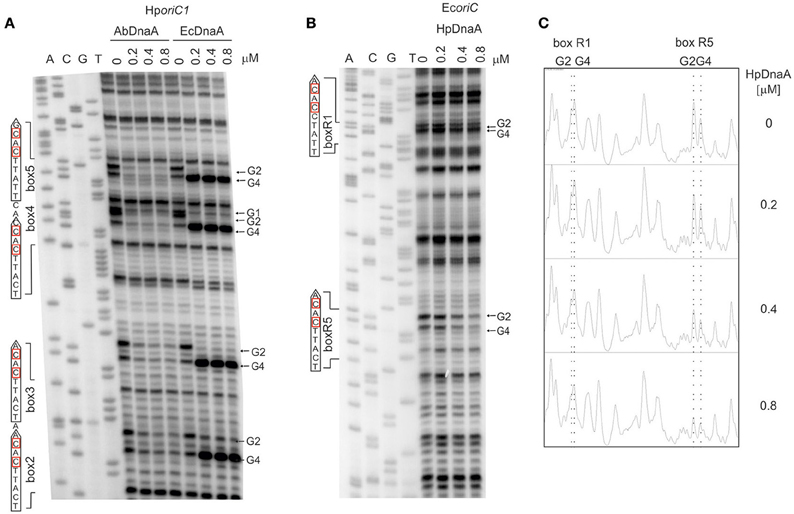
Figure 6. The specificity of interaction between Epsilonproteobacterial DnaAs and DnaA boxes. (A) DMS footprinting analysis of interactions of A. butzleri and E. coli DnaA with H. pylori oriC1. Plasmid pori1ori2 was incubated with the indicated DnaA protein concentrations, methylated with DMS and used as a template for PE reactions; primers used to map DnaA boxes are specified in Table S2. Sequences of identified boxes are presented on the left of each panel; protected guanosine residues (G) are indicated with arrows. (B) DMS footprinting analysis of interactions between H. pylori DnaA and E. coli oriC. Plasmid pOC170 was incubated with the indicated concentrations of the DnaA protein, methylated with DMS and used as a template for PE reactions; primers used to map DnaA boxes are listed in Table S2. Sequences of identified boxes are presented on the left of each panel; protected guanosine residues (G) are indicated with arrows. (C) Densitometric plots, which supplement the footprinting data. The plots were obtained for the lanes corresponding to the indicated amounts of DnaA protein. Protected guanosine residues (G) are indicated with dotted lines.
To verify the importance of the presence of T residue at 5th position of the DnaA box for the interaction with Epsilonproteobacterial DnaA we analyzed interactions of H. pylori DnaA with E. coli oriC by DMS footprinting. We were able to detect binding of H. pylori DnaA to the E. coli R5 DnaA box, which is identical to the strong H. pylori c2 and c3 DnaA boxes, i.e., it contains T at the 5th residue. We could also detect significantly weaker interaction of H. pylori DnaA with the R1 DnaA box which contains C at the 5th position (Figures 6B,C). This suggests that T residue at the 5th position in the DnaA box is important for H. pylori DnaA binding to DnaA boxes. Notably, H. pylori DnaA protected both G residues of E. coli DnaA boxes.
Further analyses are required to reveal the molecular interactions between DnaA and DNA which could explain the observed distinctions in DnaA box recognition between Epsilonproteobacterial DnaAs and E. coli DnaA. It has been previously shown that base-specific interactions with major and minor grooves of the DnaA box DNA are made by amino acid residues located at three regions of domain IV of DnaA: a basic loop between helix 2 and helix 3 (residue 399 in E. coli), helix 4 (residue 423 in E. coli), and helix 5 (residues 432–435, 438–439 in E. coli; Blaesing et al., 2000; Fujikawa et al., 2003; Tsodikov and Biswas, 2011). As it is shown in Figure 7, the arginine equivalent to E. coli R399 is maintained in all of the investigated Epsilonproteobacterial sequences. The DNA-protein interaction site located on helix 4 displays some more diversity - the position equivalent to P423 in E. coli DnaA is occupied by proline (e.g., A. butzleri DnaA) or leucine (e.g., H. pylori DnaA) residues in Epsilonproteobacteria. However, taking into account that A. butzleri DnaA and H. pylori DnaA display similar specificity of DnaA-DNA interactions, the diversity within helix 4 is probably not responsible for the observed DnaA box recognition pattern. Thus, most of the observed changes might arise from base-specific interactions between DNA and amino acids located at the N-terminus of helix 5 of DnaA's domain IV (Figure 7). This motif is about eight residues long and begins with positively charged residues corresponding to R432 in E. coli. Interestingly, for most of the Proteobacterial families arginine in this position is observed, however in the case of Epsilon/Delta Proteobacteria mostly a lysine residue occurs. The next, highly conservative HD dyad (positions 433–434 in E. coli) is followed by a threonine residue in most of the investigated sequences (position 435 E. coli). However, for Epsilon/Delta Proteobacteria this position is occupied mostly by serine residues or, less frequently, by threonine or alanine (see Figure 7). The next residue, that directly interacts with DNA, is located in position 438 (in E. coli). That position is usually occupied by hydrophobic residues (leucine, methionine), however, for many epsilon/delta proteobacterial DnaA sequences a polar serine residue is present at position 438. The last element of the helix 5 DNA binding motif is a position equivalent to E. coli H439, which is maintained in most of the analyzed sequences (Figure 7). In the case of epsilon/delta proteobacteria, histidine, and tyrosine was also observed (30 and 7%, respectively), but a lysine residue was the most frequently present (54%). In conclusion, the performed analysis of DnaA domain IV amino acid sequences of Proteobacteria and Actinobacteria reveals that especially positions equivalent to E. coli DnaA residues 435, 438–439 display some significant variation which could be responsible for the observed differences in DnaA box recognition between Epsilonproteobacterial DnaAs and DnaAs of other bacterial classes. However, more detailed studies are required to find molecular/structural features responsible for Epsilonproteobacterial DnaAs specificity toward their DnaA boxes.
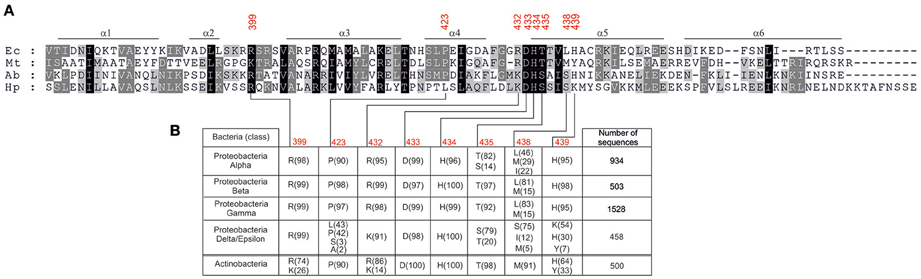
Figure 7. Amino acids occurrence frequency at interface between DnaA domain IV and DNA for various bacterial classes. (A) Multiple-sequence alignment of DnaA homologs from E. coli (Ec), M. tuberculosis (Mt), A. butzleri (Ab), and H. pylori (Hp). The red numbers indicate amino acids in E. coli DnaA involved in base-specific interactions. (B) The amino acid residues occurring most frequently in positions responsible for base-specific DNA-DnaA interactions are presented. Proteobacterial sequences are divided into classes, the numbers of analyzed sequences are shown in the last column, the frequency (in percent) of the amino acid occurrence at each position is shown in parenthesis.
Discussion
It has been recently proposed that bacterial oriC regions are central management systems controlling DNA replication as well as responsible for coordination of replication with other cellular processes (Marczynski et al., 2015). Thus, identification of the origin of chromosome replication is the first step in characterization of initiation of chromosome replication at the level of molecular mechanism as well as in the context of the cell cycle in individual bacterial species.
In this work, we identified and characterized experimentally the putative oriC regions of three Epsilonproteobacteria, namely A. butzleri, S. denitrificans, and W. succinogenes. We determined the overall structures of these regions as well as the sequences of individual DnaA boxes and DUEs. These analyses allowed us to propose features which are specific to the Epsilonproteobacteria as well as those which are common to bacteria in general. It should be noted that due to the lack of molecular biology techniques available for studies on the three Epsilonproteobacterial species the functionality of these origins has not been characterized in vivo. However, despite being “putative” origins they should be considered as reliably identified because all of the characterized modules (DnaA boxes, bipartite structure, and DUE) follow either the general or H. pylori specific oriC schemes.
General Structure of Epsilonproteobacterial oriCs
Similarly as previously shown for most other bacteria including H. pylori, the putative origins of chromosome replication of A. butzleri, S. denitrificans, and W. succinogenes are located in the vicinity of dnaA. Two clusters of DnaA boxes flank the dnaA gene; the DUE is located in the dnaA and dnaN intergenic region (Figure 5). Interestingly, in all Epsilonproteobacteria, with the exception of several species of the Helicobacter genus, the ruvC-dnaA-dnaN-gyrB locus is highly conserved (Figure S11A). The so far presented experimental results together with the in silico oriC predictions conducted for a few randomly chosen species of the same class suggest that Epsilonproteobacterial origins might be bipartite, with oriC1 and oriC2 conservatively located between ruvC-dnaA and dnaA-dnaN, respectively (Figure 5 and Figure S11B; data for oriC1 are not shown). The manner of DnaA interaction with the entire oriC1-dnaA-oriC2 region resembles that presented previously for oriCs of H. pylori and B. subtilis, and also for the E. coli oriC-mioC region and S. coelicolor oriC (Krause et al., 1997; Jakimowicz et al., 2000; Donczew et al., 2012). As observed on electron micrographs, ~1/3 of DNA molecules adopt the looped structure, in which two oriC sub-regions (or DnaA-DNA subcomplexes in E. coli and S. coelicolor) are joined together by the protein-protein interactions between DnaA molecules bound to each sub-region. This indicates that the DnaA protein of these species is characterized by intrinsic ability to join DnaA-DNA subcomplexes, provided that such subcomplexes are located on the same molecule, because no specific intermolecular interactions were observed between subcomplexes located on separate plasmid molecules. The clusters of DnaA boxes can also be bound independently by Epsilonproteobacterial DnaA, with predominant binding to oriC2. This indicates a higher affinity of Epsilonproteobacterial DnaA toward DUE proximal oriC2 or increased stability of DnaA-oriC2 complexes over DnaA-oriC1 complexes. In fact, the DnaA-oriC1 complexes were rarely observed in Epsilonproteobacterial oriC1-dnaA-oriC2 plasmids (Figure 3C and Donczew et al., 2014). However, as was shown for H. pylori, when oriC1 is detached from oriC1-dnaA-oriC2 context, it is efficiently bound by H. pylori DnaA as linear or supercoiled DNA (Donczew et al., 2012). This further supports the hypothesis of a complex interplay between oriC1 and oriC2 sub-regions. The role of such interplay is still not explained. For bipartite chromosomal and plasmid origins the regulatory role of such interaction is proposed (Krause et al., 1997; Moriya et al., 1999 and references herein). It is still not known what is the role of DnaA-mediated interaction between oriC and DnaA boxes at the mioC promoter in E. coli, but it might be related to an interplay between oriC activity and mioC transcription (Løbner-Olesen and Boye, 1992; Bates et al., 1997; Su'etsugu et al., 2003; Lies et al., 2015).
The DUEs of the three identified Epsilonproteobacterial origins, similarly as in H. pylori and B. subtilis, are located in oriC2. They are composed of the AT-rich sequence with no tandem repeats similar to E. coli L, M or R 13-mers. However, in all three species the DnaA-trio motif is found (Richardson et al., 2016). In A. butzleri and W. succinogenes a hypersensitivity to DMS of the region between R1E. coli—type DnaA box and DUE was observed, which resembles H. pylori hs region (Donczew et al., 2014). Although this phenomenon is not fully explained, it further confirms the correct assignment of Epsilonproteobacterial DUEs. Similarly to H. pylori, the interaction between oriC2 and DnaA is sufficient to unwind the DUE, which suggests that oriC1 plays additional role(s) in orisome assembly and/or regulation of chromosome replication. The oriC1 sub-region might be particularly important in vivo since its deletion is lethal in H. pylori and B. subtilis. It should be noted that, although DNA unwinding was driven by a relatively high DnaA concentration, it was localized exclusively in oriC2, 8-18 bps downstream of the R1-type DnaA box. Thus, it can be considered as highly specific DnaA dependent unwinding. However, it can't be excluded that another protein facilitates DnaA-dependent DUE unwinding in Epsilonproteobacteria, such as E. coli HU is indispensable for E. coli DUE opening in vitro (Hwang and Kornberg, 1992). Further studies are required to identify such protein(s) involved in assembly and/or regulation of Epsilonproteobacterial orisomes.
By detailed DMS footprinting we identified all the boxes bound in vitro by DnaA at oriC1 and oriC2 (Figure 5). The oriC boxes of the three species analyzed in this work and of H. pylori differ in number, orientation, and consensus sequence (see also below). Thus, we did not find any particular pattern of DnaA box arrangement conserved among the four Epsilonproteobacterial species. However, the orientation of the last box in a cluster, the DUE-proximal R1E. coli—type box, is conserved in all four species. It should be noted that on the contrary to in silico predictions, this box is a single not a double-box as ts1 and ts2 DnaA boxes of H. pylori. However, ts DnaA boxes, especially the H. pylori ts1 DnaA box, are very weak, thus they can be easily missed in analyses conducted under sub-optimal conditions such as in vitro studies. Nonetheless the identified R1E. coli—type DnaA boxes are oriented toward the DUE, as in all bacterial origins characterized so far (Rajewska et al., 2012; Wolański et al., 2014). The distance between the DUE and the adjacent in vitro bound DnaA box varies between 8 and 18 bps and is typical for the majority of known origins (Figures S7–S9). Interestingly, in the three analyzed species but not in H. pylori, the DUE-distal DnaA box in oriC2 is oriented outward from the DUE-proximal DnaA box. This feature is in agreement with other data showing that the clusters of DnaA boxes in many bacteria are characterized by similar tail-to-tail (outward) orientation of the distal boxes (Rajewska et al., 2012; Wolański et al., 2014). In E. coli, this orientation of the distal boxes as well as the asymmetrical orientation of left and right DnaA box clusters was proposed to be required for the formation of two oppositely-oriented DnaA subcomplexes (Rozgaja et al., 2011; Noguchi et al., 2015). The significance of this DnaA box orientation beyond the E. coli initiation complex is not known, but it is possible that the oppositely polarized DnaA oligomers are important for different orisome functions, such as unwinding of DNA and loading of other replisome proteins. It has been proposed that similar oppositely polarized and functionally divided DnaA oligomers might be formed on bipartite origins such as in H. pylori, but, taking into account the loop formation between suborigins, the uniform orientation of the boxes would be then required (Noguchi et al., 2015). However, the orientation of the DnaA boxes in three other Epsilonproteobacteria is not uniform. In all cases oriC2 DnaA binding sites are organized in two oppositely-directed (tail to tail) arrays of boxes. The in vitro bound DnaA boxes in A. butzleri oriC1 are oriented in the same direction, while DnaA boxes at oriC1 of W. succinogenes and S. denitrificans are oppositely-directed. Interestingly, the terminal boxes at W. succinogenes and S. denitrificans oriC1 are oriented inward (head-to-head) while those at oriC2 are oriented outward. This raises interesting questions of whether polarized DnaA oligomers are formed on bipartite (sub)origins, and what is the role of individual suborigin-DnaA complexes on orisome function.
The Specificity of DnaA-DnaA Box Interactions in Epsilonproteobacteria
In this work, we performed a detailed analysis of the DnaA boxes at oriCs bound in vitro by DnaA proteins of the analyzed Epsilonproteobacteria. By comparing the localization and orientation of DnaA boxes at oriC we concluded that there is no common DnaA box pattern in Epsilonproteobacteria (Figure 5). However, we noticed that the consensus sequence of Epsilonproteobacterial DnaA box is strictly conserved within the 5-nucleotide core 5′-TTCAC-3′ (4–8th position). The importance of nucleotides at other positions is species-dependent (Figure 5). Interestingly, the S. denitrificans DnaA box sequence is highly degenerated at the first three positions, while positions 4–9 are well-conserved. This may suggest that S. denitrificans boxes are not 9-mers but 6-mers. This is similar to the 5-mer W-boxes in C. crescentus, 7-mer boxes of M. tuberculosis, or 6-mer τ-sites of E. coli oriC (Kawakami et al., 2005; Taylor et al., 2011; Tsodikov and Biswas, 2011). Alternatively, DMS footprinting, a very sensitive method, does not discriminate between low- and high-affinity DnaA boxes thus, the identified DnaA binding sites might belong to different classes. Various classes of boxes might be partially responsible for regulating DnaA assembly during orisome formation such as ATP- and ADP-DnaA boxes in E. coli or G and W boxes in C. crescentus (Ozaki and Katayama, 2009; Taylor et al., 2011).
Nonetheless we observed two distinct features connected with the core consensus sequence of Epsilonproteobacterial DnaA boxes and DnaA-DnaA box interactions: strict conservation of thymine at the 5th position and the binding of Epsilonproteobacterial DnaA to guanine G4 of a DnaA box. So far the 5th positions of the E. coli consensus DnaA box (TTWTNCACA) and the M. tuberculosis DnaA box (YWRTCCACA) were considered to be variable without influencing the affinity toward cognate DnaAs (Schaper and Messer, 1995; Fujikawa et al., 2003; Tsodikov and Biswas, 2011). However, it should be noted that in both species, the 5th position of the DnaA box is preferentially occupied by the C residue. All other bases of the sequence, either of the upper or the lower strand, interact with DnaA, and any deviation from the most stringent TTATNCACA consensus sequence results in reduced DnaA affinity toward the less perfect boxes. Surprisingly, all Epsilonproteobacterial DnaA boxes are strictly conserved at the 5th position, which is occupied by the T residue. The other nucleotides within a core sequence are also highly conserved. The 8th position is occupied by the C residue, with the sole exception of the H. pylori ts1 DnaA box, in which C at the 8th position is substituted with A. The ts1 and ts2 DnaA boxes constitute a double DnaA box, which might require special sequence adjustment for proper and/or efficient DnaA binding. In addition, the ts1 DnaA box is bound with lower affinity than the ts2 box (Donczew et al., 2014), indicating that substitution at this position of a DnaA box negatively affects DnaA binding. Relatively rare C to A and vice versa substitutions at 6th and 7th positions, respectively, are tolerated. Other substitutions are not tolerated. For example, the DnaA box c1 (TTATAGACA), in which T5 is substituted by an A residue while C6 by a G residue, is not bound by DnaA neither in DMS nor DNaseI footprinting (data not shown) and should not be considered as a DnaA box any longer. The C residue at the 6th position of a DnaA box, which corresponds to G at the 4th position of the DnaA box in reverse orientation (for example H. pylori 5′-NGTGAATGA), has been shown to be protected from DMS modification upon interactions with DnaA. Such protection indicates that, in contrast to DnaAs from other phyla, the Epsilonproteobacterial DnaA proteins directly interact with this nucleotide residue. Altogether these data suggest that the molecular interactions between Epsilonproteobacterial DnaA and cognate DnaA boxes differ from those of E. coli and M. tuberculosis DnaA with cognate DnaA boxes and our preliminary analyses suggest that these differences arise from amino acid substitutions in helix 5 of domain IV of Epsilonproteobacterial DnaAs.
In summary, the identified origins of A. butzleri, S. denitrificans, and W. succinogenes are organized in a similar manner as previously characterized bacterial origins. The in silico and in vitro analyses of the origins of four bacteria from this class followed by global chromosome sequence analysis of the available Epsilonproteobacteria species allowed us to propose oriC features characteristic for the class, including the typical ruvC-dnaA-dnaN localization of oriC (with the exception of Helicobacteriaceae species), the bipartite oriC structure, and the core 5′-TTCAC (5–8th nucleotides of a 9-mer) consensus DnaA box sequence. We present evidence that the molecular interaction between Epsilonproteobacteria DnaA and DnaA box is significantly different from the interactions described for other bacteria, in particular E. coli and M. tuberculosis. Our comprehensive analysis of Epsilonproteobacteria opens possibilities for more precise and considerably quicker identification of origins in other bacteria of the class as well as further identification and characterization of factors involved in regulation of replication of Epsilonproteobacterial chromosomes. Taking into account that some of the known Epsilonproteobacteria are pathogenic (Campylobacter sp.) or are considered to be emerging pathogens (Arcobacter sp.) further studies on initiation of chromosome replication, the key step in the bacterial cell cycle, might help to better characterize life cycles of these species.
Author Contributions
PJ, RD, and AZ planned the experiments. PJ, RD, and AZ performed the experiments. TM provided electron microscopy facility. CW, MT, and SO planned and performed in silico analyses. PJ, RD, CW, and AZ analyzed data. PJ, RD, CW, SO, and AZ wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. Wösten and J. Simon for A. butzleri and W. succinogenes genomic DNA, respectively. This research was supported by the Foundation for Polish Science as part of the PARENT/BRIDGE programme (POMOST/2012-6/9) co-financed by the European Union under the European Regional Development Fund under the Operational Programme Innovative Economy and by a research grant SONATA BIS3 from the National Science Centre (DEC-2013/10/E/NZ1/00718). Computational resources were provided by the Informatics Center of the Metropolitan Academic Network (IC MAN–TASK) in Gdańsk, Poland. Publication supported by Wroclaw Centre of Biotechnology, programme The Leading National Research Centre (KNOW) for years 2014–2018.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01555
References
Atherton, J. C. (2006). The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 1, 63–96. doi: 10.1146/annurev.pathol.1.110304.100125
Bates, D. B., Boye, E., Asai, T., and Kogoma, T. (1997). The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 94, 12497–12502.
Beattie, T. R., and Reyes-Lamothe, R. (2015). A Replisome's journey through the bacterial chromosome. Front. Microbiol. 6:562. doi: 10.3389/fmicb.2015.00562
Blaesing, F., Weigel, C., Welzeck, M., and Messer, W. (2000). Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol. Microbiol. 36, 557–569. doi: 10.1046/j.1365-2958.2000.01881.x
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the UniProt knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Briggs, G. S., Smits, W. K., and Soultanas, P. (2012). Chromosomal replication initiation machinery of low-g+c-content firmicutes. J. Bacteriol. 194, 5162–5170. doi: 10.1128/JB.00865-12
Donczew, R., Makowski, L., Jaworski, P., Bezulska, M., Nowaczyk, M., Zakrzewska-Czerwińska, J., et al. (2015). The atypical response regulator HP1021 controls formation of the Helicobacter pylori replication initiation complex. Mol. Microbiol. 95, 297–312. doi: 10.1111/mmi.12866
Donczew, R., Mielke, T., Jaworski, P., Zakrzewska-Czerwińska, J., and Zawilak-Pawlik, A. (2014). Assembly of Helicobacter pylori initiation complex is determined by sequence-specific and topology-sensitive DnaA-oriC interactions. J. Mol. Biol. 426, 2769–2782. doi: 10.1016/j.jmb.2014.05.018
Donczew, R., Weigel, C., Lurz, R., Zakrzewska-Czerwinska, J., and Zawilak-Pawlik, A. (2012). Helicobacter pylori oriC-the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. doi: 10.1093/nar/gks742
Duderstadt, K. E., and Berger, J. M. (2013). A structural framework for replication origin opening by AAA+ initiation factors. Curr. Opin. Struct. Biol. 23, 144–153. doi: 10.1016/j.sbi.2012.11.012
Duderstadt, K. E., Chuang, K., and Berger, J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. doi: 10.1038/nature10455
Eppinger, M., Baar, C., Raddatz, G., Huson, D. H., and Schuster, S. C. (2004). Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2, 872–885. doi: 10.1038/nrmicro1024
Epps, S. V. R., Harvey, R. B., Hume, M. E., Phillips, T. D., Anderson, R. C., and Nisbet, D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health 10, 6292–6304. doi: 10.3390/ijerph10126292
Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. doi: 10.1093/nar/gkg309
Gao, F., Luo, H., and Zhang, C.-T. (2013). DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41, D90–D93. doi: 10.1093/nar/gks990
Grimwade, J. E., Ryan, V. T., and Leonard, A. C. (2000). IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35, 835–844. doi: 10.1046/j.1365-2958.2000.01755.x
Gupta, R. S. (2006). Molecular signatures (unique proteins and conserved indels) that are specific for the epsilon proteobacteria (Campylobacterales). BMC Genomics 7:167. doi: 10.1186/1471-2164-7-167
Harding, N. E., Cleary, J. M., Smith, D. W., Michon, J. J., Brusilow, W. S., and Zyskind, J. W. (1982). Chromosomal replication origins (oriC) of Enterobacter aerogenes and Klebsiella pneumoniae are functional in Escherichia coli. J. Bacteriol. 152, 983–993.
Hwang, D. S., and Kornberg, A. (1992). Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J. Biol. Chem. 267, 23087–23091.
Jakimowicz, D., Majkadagger, J., Konopa, G., Wegrzyn, G., Messer, W., Schrempf, H., et al. (2000). Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 298, 351–364. doi: 10.1006/jmbi.2000.3686
Kaguni, J. M. (2011). Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 15, 606–613. doi: 10.1016/j.cbpa.2011.07.016
Katayama, T., Ozaki, S., Keyamura, K., and Fujimitsu, K. (2010). Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8, 163–170. doi: 10.1038/nrmicro2314
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kaur, G., Vora, M. P., Czerwonka, C. A., Rozgaja, T. A., Grimwade, J. E., and Leonard, A. C. (2014). Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 91, 1148–1163. doi: 10.1111/mmi.12525
Kawakami, H., Keyamura, K., and Katayama, T. (2005). Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 280, 27420–27430. doi: 10.1074/jbc.M502764200
Kitagawa, R., Ozaki, T., Moriya, S., and Ogawa, T. (1998). Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12, 3032–3043. doi: 10.1101/gad.12.19.3032
Kowalski, D., Natale, D. A., and Eddy, M. J. (1988). Stable DNA unwinding, not “breathing,” accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc. Natl. Acad. Sci. U.S.A. 85, 9464–9468. doi: 10.1073/pnas.85.24.9464
Krause, M., Rückert, B., Lurz, R., and Messer, W. (1997). Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274, 365–380. doi: 10.1006/jmbi.1997.1404
Lartigue, C., Blanchard, A., Renaudin, J., Thiaucourt, F., and Sirand-Pugnet, P. (2003). Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res. 31, 6610–6618. doi: 10.1093/nar/gkg848
Leonard, A. C., and Grimwade, J. E. (2015). The orisome: structure and function. Front. Microbiol. 6:545. doi: 10.3389/fmicb.2015.00545
Leonard, A. C., and Méchali, M. (2013). DNA replication origins. Cold Spring Harb. Perspect. Biol. 5:a010116. doi: 10.1101/cshperspect.a010116
Lies, M., Visser, B. J., Joshi, M. C., Magnan, D., and Bates, D. (2015). MioC and GidA proteins promote cell division in E. coli. Front. Microbiol. 6:516. doi: 10.3389/fmicb.2015.00516
Løbner-Olesen, A., and Boye, E. (1992). Different effects of mioC transcription on initiation of chromosomal and minichromosomal replication in Escherichia coli. Nucleic Acids Res. 20, 3029–3036.
Madiraju, M. V., Moomey, M., Neuenschwander, P. F., Muniruzzaman, S., Yamamoto, K., Grimwade, J. E., et al. (2006). The intrinsic ATPase activity of Mycobacterium tuberculosis DnaA promotes rapid oligomerization of DnaA on oriC. Mol. Microbiol. 59, 1876–1890. doi: 10.1111/j.1365-2958.2006.05068.x
Marczynski, G. T., Rolain, T., and Taylor, J. A. (2015). Redefining bacterial origins of replication as centralized information processors. Front. Microbiol. 6:610. doi: 10.3389/fmicb.2015.00610
McGarry, K. C., Ryan, V. T., Grimwade, J. E., and Leonard, A. C. (2004). Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816. doi: 10.1073/pnas.0400340101
Messer, W. (2002). The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26, 355–374. Available online at: http://www.ncbi.nlm.nih.gov/pubmed/12413665
Moriya, S., Imai, Y., Hassan, A. K., and Ogasawara, N. (1999). Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid 41, 17–29. doi: 10.1006/plas.1998.1381
Nakagawa, S., and Takaki, Y. (2009). “Nonpathogenic epsilonproteobacteria,” in Encyclopedia of Life Sciences (Chichester: John Wiley & Sons, Ltd.). doi: 10.1002/9780470015902.a0021895
Noguchi, Y., Sakiyama, Y., Kawakami, H., and Katayama, T. (2015). The Arg Fingers of Key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 290, 20295–20312. doi: 10.1074/jbc.M115.662601
Okumura, H., Yoshimura, M., Ueki, M., Oshima, T., Ogasawara, N., and Ishikawa, S. (2012). Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 40, 220–234. doi: 10.1093/nar/gkr716
Ozaki, S., and Katayama, T. (2009). DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62, 71–82. doi: 10.1016/j.plasmid.2009.06.003
Ozaki, S., and Katayama, T. (2011). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. doi: 10.1093/nar/gkr832
Ozaki, S., Noguchi, Y., Hayashi, Y., Miyazaki, E., and Katayama, T. (2012). Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 287, 37458–37471. doi: 10.1074/jbc.M112.372052
Rajewska, M., Wegrzyn, K., and Konieczny, I. (2012). AT-rich region and repeated sequences - the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 36, 408–434. doi: 10.1111/j.1574-6976.2011.00300.x
Richardson, T. T., Harran, O., and Murray, H. (2016). The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534, 412–416. doi: 10.1038/nature17962
Rozgaja, T. A., Grimwade, J. E., Iqbal, M., Czerwonka, C., Vora, M., and Leonard, A. C. (2011). Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 82, 475–488. doi: 10.1111/j.1365-2958.2011.07827.x
Sasse-Dwight, S., and Gralla, J. D. (1991). Footprinting protein-DNA complexes in vivo. Meth. Enzymol. 208, 146–168. doi: 10.1016/0076-6879(91)08012-7
Schaper, S., and Messer, W. (1995). Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270, 17622–17626. doi: 10.1074/jbc.270.29.17622
Scholefield, G., Errington, J., and Murray, H. (2012). Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31, 1542–1555. doi: 10.1038/emboj.2012.6
Shaheen, S. M., Ouimet, M.-C., and Marczynski, G. T. (2009). Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155, 1215–1225. doi: 10.1099/mic.0.025528-0
Skarstad, K., and Katayama, T. (2013). Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5:a012922. doi: 10.1101/cshperspect.a012922
Smith, J. L., and Grossman, A. D. (2015). In vitro whole genome DNA binding analysis of the bacterial replication initiator and transcription factor DnaA. PLoS Genet. 11:e1005258. doi: 10.1371/journal.pgen.1005258
Su'etsugu, M., Emoto, A., Fujimitsu, K., Keyamura, K., and Katayama, T. (2003). Transcriptional control for initiation of chromosomal replication in Escherichia coli: fluctuation of the level of origin transcription ensures timely initiation. Genes Cells 8, 731–745. doi: 10.1046/j.1365-2443.2003.00671.x
Suzek, B. E., Huang, H., McGarvey, P., Mazumder, R., and Wu, C. H. (2007). UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinforma. Oxf. Engl. 23, 1282–1288. doi: 10.1093/bioinformatics/btm098
Taylor, J. A., Ouimet, M.-C., Wargachuk, R., and Marczynski, G. T. (2011). The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol. Microbiol. 82, 312–326. doi: 10.1111/j.1365-2958.2011.07785.x
Tsodikov, O. V., and Biswas, T. (2011). Structural and thermodynamic signatures of DNA recognition by Mycobacterium tuberculosis DnaA. J. Mol. Biol. 410, 461–476. doi: 10.1016/j.jmb.2011.05.007
Wolański, M., Donczew, R., Zawilak-Pawlik, A., and Zakrzewska-Czerwińska, J. (2014). oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 5:735. doi: 10.3389/fmicb.2014.00735
Zakrzewska-Czerwińska, J., Jakimowicz, D., Zawilak-Pawlik, A., and Messer, W. (2007). Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31, 378–387. doi: 10.1111/j.1574-6976.2007.00070.x
Zawilak, A., Cebrat, S., Mackiewicz, P., Król-Hulewicz, A., Jakimowicz, D., Messer, W., et al. (2001). Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res. 29, 2251–2259. doi: 10.1093/nar/29.11.2251
Zawilak-Pawlik, A., Kois, A., Majka, J., Jakimowicz, D., Smulczyk-Krawczyszyn, A., Messer, W., et al. (2005). Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem. J. 389, 471–481. doi: 10.1042/BJ20050143
Keywords: Epsilonproteobacteria, initiation of chromosome replication, oriC, DnaA, DnaA box, orisome
Citation: Jaworski P, Donczew R, Mielke T, Thiel M, Oldziej S, Weigel C and Zawilak-Pawlik A (2016) Unique and Universal Features of Epsilonproteobacterial Origins of Chromosome Replication and DnaA-DnaA Box Interactions. Front. Microbiol. 7:1555. doi: 10.3389/fmicb.2016.01555
Received: 18 July 2016; Accepted: 16 September 2016;
Published: 30 September 2016.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Alan Leonard, Florida Institute of Technology, USAGregory Marczynski, McGill University, Canada
Copyright © 2016 Jaworski, Donczew, Mielke, Thiel, Oldziej, Weigel and Zawilak-Pawlik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Zawilak-Pawlik, zawilak@iitd.pan.wroc.pl
†Present Address: Rafal Donczew, Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA
 Pawel Jaworski
Pawel Jaworski Rafal Donczew1†
Rafal Donczew1† Thorsten Mielke
Thorsten Mielke Marcel Thiel
Marcel Thiel Stanislaw Oldziej
Stanislaw Oldziej Anna Zawilak-Pawlik
Anna Zawilak-Pawlik