- 1Division of Pediatric Gastroenterology and Hepatology, Department of Pediatrics, Shuang Ho Hospital, Taipei Medical University, Taipei, Taiwan
- 2Department of Pediatrics, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 3Graduate Institute of Biochemical and Biomedical Engineering, National Taipei University of Technology, Taipei, Taiwan
- 4Institute of Biomedical Engineering and Nanomedicine – National Health Research Institutes, Zhunan, Taiwan
- 5Department of Surgery, Shuang Ho Hospital, Taipei Medical University, Taipei, Taiwan
- 6Department of Surgery, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
The yqiC gene of Salmonella enterica serovar Typhimurium (S. Typhimurium) regulates bacterial growth at different temperatures and mice survival after infection. However, the role of yqiC in bacterial colonization and host immunity remains unknown. We infected human LS174T, Caco-2, HeLa, and THP-1 cells with S. Typhimurium wild-type SL1344, its yqiC mutant, and its complemented strain. Bacterial colonization and internalization in the four cell lines significantly reduced on yqiC depletion. Post-infection production of interleukin-8 and human β-defensin-3 in LS174T cells significantly reduced because of yqiC deleted in S. Typhimurium. The phenotype of yqiC mutant exhibited few and short flagella, fimbriae on the cell surface, enhanced biofilm formation, upregulated type-1 fimbriae expression, and reduced bacterial motility. Type-1 fimbriae, flagella, SPI-1, and SPI-2 gene expression was quantified using real-time PCR. The data show that deletion of yqiC upregulated fimA and fimZ expression and downregulated flhD, fliZ, invA, and sseB expression. Furthermore, thin-layer chromatography and high-performance liquid chromatography revealed the absence of menaquinone in the yqiC mutant, thus validating the importance of yqiC in the bacterial electron transport chain. Therefore, YqiC can negatively regulate FimZ for type-1 fimbriae expression and manipulate the functions of its downstream virulence factors including flagella, SPI-1, and SPI-2 effectors.
Introduction
Salmonella enterica serovar Typhimurium (i.e., S. Typhimurium) is a notorious food-borne pathogen that causes gastroenteritis in animals and humans worldwide. Salmonella exhibit classic virulence factors, such as plasmids, toxins, fimbriae, and flagella (van Asten and van Dijk, 2005). Most virulence genes are clustered in regions distributed throughout the chromosome called SPIs. Thus far, at least 11 SPIs have been identified. SPI-1 and SPI-2, the most commonly studied SPIs, encode T3SSs that translocate the effector proteins into host cells or secrete them into the extracellular environment, thus affecting host biochemistry and cell physiology (Sabbagh et al., 2010). SPI-1 genes are involved in the key events of early disease phases, such as membrane ruffling through cytoskeleton manipulation (Zhou et al., 2001), virulence regulation (Raffatellu et al., 2005), and innate immunity activation, including neutrophil recruitment across the epithelium and NF-κB signaling activation (Lee et al., 2000). By contrast, SPI-2 is responsible for intracellular transport and survival, particularly in the systemic phase of a disease (Coburn et al., 2007), and for cyclooxygenase 2 induction (Uchiya and Nikai, 2004), host cytokine modulation (Uchiya and Nikai, 2005), and cell death (Knodler et al., 2005).
In S. Typhimurium, yqiC is located outside SPIs, and most of its functions are unclear, except for its contribution to bacterial survival at various temperatures and to host survival in mice (Carrica et al., 2011). In our preliminary study, a yqiC transposon mutant of S. Typhimurium, created using transposon-directed insertion-site sequencing, could not colonize HEp-2 cells (Fang, 2011). Thus, yqiC, a non-SPI gene, is responsible for bacterial colonization and subsequent invasion during early bacteria–host interactions. However, this finding requires affirmation. In the process of bacterial colonization, fimbrial (Baumler et al., 1996) or adhesin-based (Lambert and Smith, 2008) adherence and subsequent bacterial invasion contribute to Salmonella virulence, accompanied by host neutrophil recruitment and transcellular signaling that orchestrate the pathogenicity (McCormick et al., 1993; Pace et al., 1993). In Salmonella, fimbrial adhesins include Fim, Pef, Bcf, Sti, Stf, Saf, Stb, Csg, Stc, Std, Lpf, Stj, and Sth (McClelland et al., 2001), whereas non-fimbrial adhesins include ShdA, MisL, SadA, SiiE, and BapA (Wagner and Hensel, 2011). However, YqiC is not classified in either of these categories. The role of yqiC in the early pathogenesis of Salmonella in host cells, a topic explored in few studies, remains unknown. In Escherichia coli, yqiC is involved in inducing oxidative stress response in host cell membranes (Al Mamun et al., 2012). We previously used Vector NTI v10 (Invitrogen, Carlsbad, CA, USA) to compare the amino acid sequences of the YqiC of Salmonella to those of IbpA and IbpB of E. coli (Matuszewska et al., 2008), which exhibited approximately 25% homology. IbpA and IbpB in E. coli have been reported to mediate resistance to oxidative stress (Kitagawa et al., 2000). Thus far, 18 putative membrane proteins of the Yqi family have been identified in Bacillus subtilis (Kunst et al., 1997). Among these, YqiD and YqiE are possibly involved in the biosynthesis of menaquinone, an electron transport chain mediator (Julsing et al., 2007). Within bacteria, menaquinone serves as an antioxidant (Nowicka and Kruk, 2010), and the electron transport chain is involved in E. coli flagellation (Hertz and Bar-Tana, 1977). Therefore, menaquinone can be potentially linked to flagellar biosynthesis for bacterial virulence. Thus, YqiC may be involved in the regulation of menaquinone and virulence in Salmonella.
Type-1 fimbrial genes (fim) are involved in Salmonella colonization in host cells and in the regulation of flagellin. When Salmonella reach the intestinal tract, the proteinaceous hair-like structures named fimbriae mediate bacterial adherence to host cells (Duguid and Campbell, 1969). Notably, type-1 fimbriae have a mannose-specific binding capacity, which is responsible for the bacteria–IEC association (Lindquist et al., 1987). The inhibition of Salmonella type-1 fimbrial gene expression can attenuate bacterial virulence in swine and mice (Aslanzadeh and Paulissen, 1990; Althouse et al., 2003). The activation of Salmonella type-1 fimbriae is regulated by FimZ, a member of the fim gene cluster (Yeh et al., 1995). The synthesis of flagella and fimbriae are oppositely controlled because increased expression of FimZ results in non-motility of hyperfimbriated S. Typhimurium downregulates the flhDC master flagellar operon (Clegg and Hughes, 2002). Flagella are essential for bacterial motility and dissemination during Salmonella infection (Duan et al., 2013). In addition, flagellin is the major and the most extensively studied flagellar protein in the pathogen-associated molecular pattern of Salmonella, and is also a key virulence factor in bacterial invasion and host response induction (Kawai and Akira, 2011). Salmonellosis is characterized by the recruitment of neutrophils from the microvasculature to the infected intestines (Cheminay et al., 2004), driven by IL-8 released from the IECs (McCormick et al., 1993) after Salmonella flagellin binds with toll-like receptor 5 of IECs, subsequently activating the cellular nuclear factor NF-κB and MAPK signaling pathways (Yu et al., 2003; Kawai and Akira, 2011). However, the association between flagella and the non-SPI yqiC in Salmonella has not been explored.
Defensins affecting the host innate immunity after Salmonella infection are small cationic AMPs that have a key role in host defense properties and are present in granulocytic leucocytes, mucosal surface, skin, and other epithelia (Selsted and Ouellette, 2005). AMP expression in the gastrointestinal tract is either constitutive or inducible. hBD-1 is the major constitutive AMP synthesized on the intestinal epithelial surface, whereas hBD-2, hBD-3, and a small amount of hBD-4 are the major inducible proteins expressed only during infection or inflammation (Wehkamp et al., 2007). In Salmonella-infected Caco-2 cells, hBD-1 and hBD-2 mRNA expression can be activated (O’Neil et al., 1999), and Salmonella flagellin is the primary ligand that induces hBD-2 expression through NF-κB activation (Ogushi et al., 2001; Takahashi et al., 2001). Although, host responses of AvBD-3 and gallinacin-3 can be enhanced in the gastrointestinal tissues of broiler chickens after Salmonella infection (Ramasamy et al., 2012), whether hBD-3 expression is affected by Salmonella infection in the human intestinal epithelium remains unknown.
To investigate the effects of yqiC on Salmonella–host interaction, we determined whether yqiC affects the expression of virulence factors in Salmonella and the IL-8- and hBD-3-related innate immune responses of the human intestinal epithelial LS174T cells. According to the findings of morphological studies, we performed yeast agglutination tests and qRT-PCR analyses to confirm our hypothesis that yqiC expression upregulates type-1 fimbrial expression and downregulates flagellar, SPI-1, and SPI-2 gene expression. We also reviewed relevant literature to determine the correlation between yqiC and the electron transport chain and performed two chromatography techniques to examine whether yqiC affects menaquinone expression in S. Typhimurium. Rotenone inhibition assays were performed to determine whether the effect of the NADH dehydrogenase inhibitor in the electron transport chain was similar to that of yqiC in regulating selected type-1 fimbrial, flagellar, SPI-1, and SPI-2 gene expression in S. Typhimurium.
Materials and Methods
Bacterial Strains and Culture Conditions
All bacterial strains—the wild-type S. Typhimurium strain SL1344 and its isogenic yqiC-deleted mutant strain (ΔyqiC), yqiC-complement ΔyqiC strain (ΔyqiC′), fliC-deleted mutant strain (ΔfliC), and spaS-deleted SPI-1 mutant strain (ΔspaS) were used in this study. SL1344 and ΔspaS were kindly provided by Duncan Maskell. S. Typhimurium ΔyqiC and ΔfliC were created using a previously described lambda (λ)-red recombinase method, with slight modifications (Gust et al., 2003). The kanamycin resistance gene cassette containing the Salmonella yqiC-flanking sequence was amplified using yqiC-specific primers (forward, 5′-AATAAGAGCTAACACTTACCAGTTCAGGGAAACCACAATGAACCGGAATTGCCAGCTG-3′, and reverse, 5′-ATCTTATATGAGCGGGCCGTCAGGCCCGTTCACGTTTTTATCAGAAGAACTCGTCAAG-3′) from the PCRII-TOPO vector. The apramycin resistance gene cassette with Salmonella fliC-flanking sequence was amplified using fliC-specific primers (forward, 5′-CCAATAACATCAAGTTGTAATTGATAAGGAAAAGATCATGATTCCGGGGATCCGTCGAC-3′, and reverse, 5′-TGATTGTGTACCACGTGTCGGTGAATCAATCGCCGGATTATGTAGGCTGGAGCTGCTTC-3′) from the pIJ773 vector. These chimeric sequences were transformed into S. Typhimurium SL1344, which were complemented with the pKD20 plasmid. Recombination was performed using the λ-red recombinase method, and the synthesized strains were cultured and selected on Luria-Bertani (LB) agar supplemented with the appropriate antibiotic (50 μg/mL kanamycin or apramycin). For generating the yqiC-complemented S. Typhimurium strain (ΔyqiC′), the yqiC-coding sequence was amplified using yqiC-specific primers (forward, 5′-GCCTGCTGGCAGTAAAGC-3′, and reverse, 5′-GCTTCCGCTTGCGATTGC-3′) and cloned into the pACYC184 vector for restoring yqiC expression. S. Typhimurium ΔyqiC′ was maintained in LB broth supplemented with 20 μg/mL chloramphenicol at 37°C.
For maintenance, bacteria were cultured in LB broth (Difco/Becton Dickinson) or on LB agar plates at 37°C with appropriate antibiotics, if necessary. For gentamicin protection assay and ELISA (for estimating in vitro IL-8 and hBD-3 secretion), mid-log cultures of the S. Typhimurium strains were incubated in LB broth by shaking at 225 rpm and 37°C for 3 h when their OD600 was approximately 0.8. For TLC and HPLC, S. Typhimurium strains were cultured in overnight cultures by shaking at 225 rpm and 37°C. For other assays, the overnight cultures of the S. Typhimurium strains were prepared by incubating single bacterial colonies from LB agar plates into LB broths and then grown at 37°C for approximately 18 h.
Cell Culture and Bacterial Infections
The human colon carcinoma cell line LS174T [American Tissue Culture Collection (ATCC), Rockville, MD, USA; ATCC-CL188], the colon carcinoma cell line Caco-2 (ATCC-TB37), the human cervical adenocarcinoma cell line HeLa (ATCC-CCL2), and the human monocyte cell line THP-1 (ATCC-TIB202) were cultured according to the ATCC instructions and maintained at 5% CO2 and 37°C. When the cells attained 90% confluency, the adherent cells were subcultured through trypsinization by using a trypsin-EDTA solution (Gibco).
For bacterial colonization and invasion assays in triplicate, 5 × 105 LS174T, Caco-2, HeLa, or THP-1 cells were seeded per well onto 12-well plates and cultured for 4–5 days (final cell concentration, approximately 2 × 106 cells/well). Only THP-1 cells were treated with 10 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich) for cell attachment in wells before infection assays. The media of all four cell lines were replaced with FBS-free media before bacterial inoculation, and the cells were then infected with mid-log cultures of S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ (MOI = 5, i.e., approximately 1 × 107 CFU/well) for 2 h. Subsequently, the infected cells were washed with PBS three times to eliminate non-adherent non-invading bacteria, incubated in FBS-free plain media without gentamicin for 1 h, washed with PBS three times, and lysed using 1% Triton X-100 (Sigma-Aldrich) to generate an output pool A containing cell-associated bacteria. After the initial 2-h infection, the infected cells in the remaining wells were incubated in FBS-free media supplemented with 100 μg/mL gentamicin (Sigma-Aldrich) for 1 h to kill extracellular bacteria. Then, these cells were washed with PBS three times and lysed using 1% Triton X-100 to generate the output pool B containing intracellular bacteria. Serial dilutions of the bacterial lysates from output pools A and B were plated onto LB agar plates, and the CFUs were counted after overnight incubation to determine the number of viable bacteria in the CFUs in terms of the initial inoculum, expressed as CFU per initial inoculum of 2.5 × 106 cells.
Enzyme-Linked Immunosorbent Assay
For quantifying protein secretion from host cells after infection or stimulation, LS174T cells in individual wells were treated with LB broth containing S. Typhimurium SL1344, ΔyqiC, ΔyqiC′, or ΔfliC (MOI = 5); 50 ng/mL IL-1β (a proinflammatory cytokine; R&D System Europe); or 100 ng/mL Salmonella flagellin (InvivoGen) for 2 h by using the same protocol as earlier prescribed in the bacterial invasion assay to generate output pool B. Next, they were initially incubated in media supplemented with 100 μg/mL gentamicin for 1 h and then in media supplemented with 10 μg/mL gentamicin for an additional 15 h. After the above 18-h treatment, the media from the final 15-h incubation in these wells were harvested and centrifuged, and the supernatants were collected for performing ELISAs for IL-8 and hBD-3 in triplicate according to manufacturer’s instructions (human IL-8 ELISA development kit and human BD-3 ELISA development kit; PreproTech EC). IL-8 and hBD-3 concentrations in the media were determined at 405 nm on a SpectraMax reader (Molecular Devices, Sunnyvale, CA, USA) and calculated according to the standard curves plotted from serially diluted standard solutions using Microsoft Office Excel 2007.
Transmission Electron Microscopy
For morphological evaluation of S. Typhimurium strains through TEM, a loopful of bacteria was obtained from the colonies of each strain grown on LB agar plates and suspended in 300 μL of PBS. Resuspended bacterial solutions were centrifuged and washed with PBS two times. After another round of centrifugation, the supernatants were removed, and the bacterial pellets were then negatively stained using 2% phosphotungstic acid (Sigma-Aldrich). Finally, the stained bacterial suspensions were deposited on a carbon-coated grid, and all samples were observed under a Hitachi H-600 transmission electron microscope (Hitachi, Ltd, Tokyo, Japan).
Yeast Agglutination Test
Yeast agglutination tests were performed in triplicate as described previously (Wang et al., 2012). Each bacterial strain was streaked on LB agar plates and then incubated at 28 and 37°C for 18 h. A loopful of bacterial colonies was obtained from the LB agar plates and resuspended in 100 μL of PBS. Next, 30 μL of bacterial suspension was mixed with an equal amount of 3% (w/v) Saccharomyces cerevisiae cells (Sigma-Aldrich) on a glass slide. The formation of agglutination fragments indicated mannose-sensitive fimbriae adherence. In addition, the bacterial suspension was mixed with 3% (w/v) S. cerevisiae cells and additional 3% (w/v) D-mannose solution (Sigma-Aldrich) for confirming whether yeast agglutination was caused by mannose-specific binding mediated by type-1 fimbriae.
Biofilm Formation Assay
Biofilm formation assays were performed in triplicate as described previously (O’Toole, 2011). The overnight cultures of S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ were diluted in LB broth without salt, and 100 μL of each bacterial dilution was added into each well in triplicate in 96-well microplates for additional incubation at 37°C for 18 h. After incubation, the bacteria were removed and washed with PBS. Then, 125 μL of 0.1% crystal violet (Sigma-Aldrich) was added into each well, which was subsequently washed with PBS. Finally, 125 μL of 30% acetic acid (Sigma-Aldrich) was added into each well. The absorbance in each well was determined at 550 nm on the SpectraMax reader (Molecular Devices).
Bacterial Motility Assay
The bacterial motility assays were performed in triplicate. Semisolid LB agar plates containing 0.3% agar were used for the bacterial motility assay. Overnight-cultured bacteria (10 μL) were inoculated onto the centers of the semisolid LB agar plates, which were incubated at 37°C for 6 h. The diameters of the growth zones represent the strength of bacterial motility. S. Typhimurium ΔfliC and ΔspaS were used as the negative and positive controls, respectively.
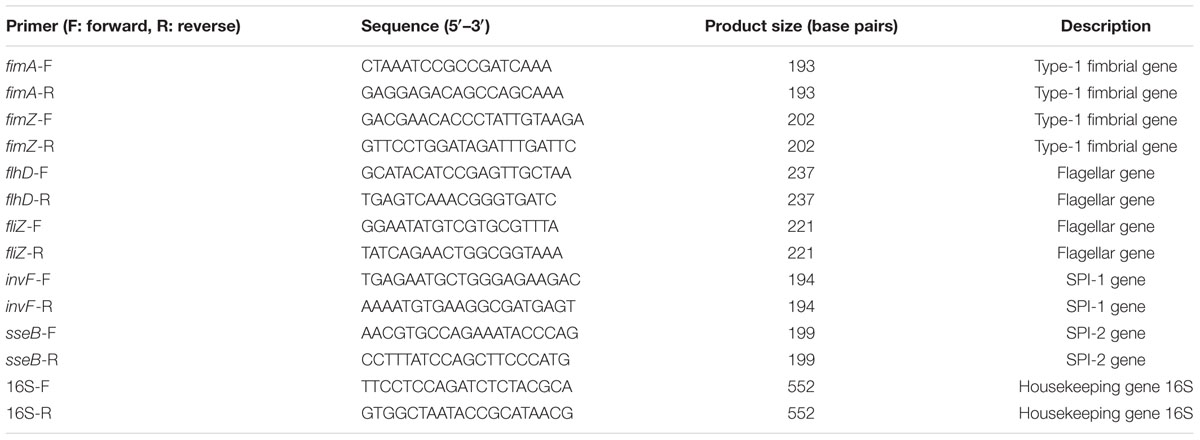
qRT-PCR Analysis
Total RNA of S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ was isolated from their overnight cultures by using the Total RNA Miniprep Purification Kit (Genemark, Taichung, Taiwan) according to manufacturer’s protocol. Total RNA samples were cleaned and purified using RNase-free DNase I (1 unit/1 μg RNA; NEB, Beverly, MA, USA). Next, 0.5 μg of RNA was reverse transcribed to cDNA using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer’s instruction. Oligonucleotide primer pairs specific to the target genes, namely fimA, fimZ, flhD, fliZ, invF, and sseB, and the housekeeping 16S ribosomal RNA gene (Table 1) were designed using Primer3 and BLAST1. By using the Bio-Rad C100 Real-Time PCR System, 0.1 μg of cDNA was amplified in a 20-μL reaction solution containing 0.25 μM of each primer and 10 μL of iQ SyBr green supermix (BioRad) with 40 cycles of enzyme activation at 95°C for 3 min, denaturation at 95°C for 15 s, annealing at 48°C for 30 s, and extension at 72°C for 30 s. The mRNA transcription levels were calculated using the ΔΔCt method as described previously (Livak and Schmittgen, 2001), and the expression levels of 16S ribosomal RNA were used for normalization. The mRNA expression levels of fimA, fimZ, flhD, fliZ, invF, and sseB were expressed as fold-change relative to S. Typhimurium SL1344.
NADH Dehydrogenase Inhibition Assay
To determine the involvement of the electron transport chain in Salmonella yqiC virulence, 100 μg/mL rotenone (Sigma-Aldrich), an NADH dehydrogenase inhibitor (Ueno et al., 1994), was added to the S. Typhimurium SL1344 cultures in LB broth and incubated at 37°C for 18 h. The total RNA of the prepared overnight cultures of S. Typhimurium SL1344 and ΔyqiC and S. Typhimurium SL1344 mixed with rotenone were isolated and processed in triplicate as described for the qRT-PCR analysis. The mRNA expression levels of fimA, fimZ, flhD, fliZ, invF, and sseB in S. Typhimurium SL1344 mixed with rotenone were compared with those in S. Typhimurium ΔyqiC.
Menaquinone Complementation Assay
To study the role of menaquinone in Salmonella yqiC virulence, 100 μg/mL menaquinone (Sigma-Aldrich) was added into the LB broth containing S. Typhimurium ΔyqiC and then coincubated at 37°C for 18 h. The total RNA of S. Typhimurium ΔyqiC was isolated and processed as described for the qRT-PCR analysis. The mRNA expression levels of fimA, fimZ, flhD, fliZ, invF, and sseB in the S. Typhimurium ΔyqiC complemented with menaquinone were compared with those in S. Typhimurium SL1344.
TLC for Menaquinone
We extracted menaquinone from S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ according to a protocol described previously (Kwon et al., 2000). All bacterial strains were cultured in 500 mL of 0.5% glucose and minimal salt medium (33.9 g/L Na2HPO4.7H2O, 15 g/L KH2PO4, 5 g/L NH4Cl, and 2.5 g/L NaCl) with vigorous shaking and incubated at 37°C overnight. The bacteria were harvested by centrifugation at 10,000 rpm, and the supernatants were discarded. The pellets were suspended in acetone and filtered through Whatman No. 1 filter papers (pore size, 11 μm; thickness, 0.18 mm). The filtrates were dried under reduced pressure. Two drops of acetone were added to the dried materials for resuspension. Finally, the suspensions were spotted on a Baker Si250F silica gel plate (J. T. Baker, Phillipsburg, NJ, USA) and separated using petroleum ether–ethyl ether (70:30) or chloroform.
HPLC for Menaquinone
For reconfirming the presence of menaquinone in S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′, the bacterial menaquinone was extracted using acetone, as described in the TLC procedure. The acetone extracts were centrifuged and filtered using Whatman No. 1 filter papers to remove cell debris. Subsequently, the filtered extracts were dried under decreasing pressure and treated with 1 mL of acetonitrile. The reconstituted mixtures were filtered using Sep-Pak C18 cartridges (Waters, Milford, MA, USA) and subjected to HPLC (Hitachi L-7400) on a Syncronis C18 HPLC column (Thermo Fisher Scientific, Waltham, MA, USA). Menaquinone was detected at 245 nm on a UV detector.
Statistical Analysis
All experiments were performed in triplicate, and data were analyzed using the Student’s t-test to compare the differences between the control and S. Typhimurium groups. A p-value of <0.05 was considered statistically significant.
Results
yqiC Deletion Inhibits S. Typhimurium Colonization and Invasion of Four Human Cell Lines
To investigate whether yqiC affects the colonizing and invading abilities of S. Typhimurium, three non-phagocytic human epithelial cell lines and one phagocytic cell line were infected with S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′. Our bacterial colonization and invasion assays demonstrated that approximately 4.8 × 106, 2.2 × 106, and 1.5 × 106 CFU of S. Typhimurium SL1344 cells colonized LS174T and THP-1 (at similar levels), HeLa, and Caco-2 cells, respectively (white bars in output pools A, Figure 1). The internalization rates of S. Typhimurium SL1344 were the highest in LS174T cells, followed by THP-1, HeLa, and Caco-2 cells (white bars in output pools B, Figure 1). Deletion of yqiC significantly inhibited the colonization and invasion of S. Typhimurium in all four cell types (striated bars, Figure 1). Furthermore, 20–50% inhibition of S. Typhimurium ΔyqiC in colonization and invasion was partially recovered by the complementation of yqiC in the output pool A or B (black bars, Figure 1), thus suggesting the significance of yqiC to the colonizing and invading abilities of S. Typhimurium. Therefore, yqiC is required in the early pathogenesis S. Typhimurium, thus clarifying its significance to the colonization and invasion of S. Typhimurium in various host cells, irrespective of the cell type.
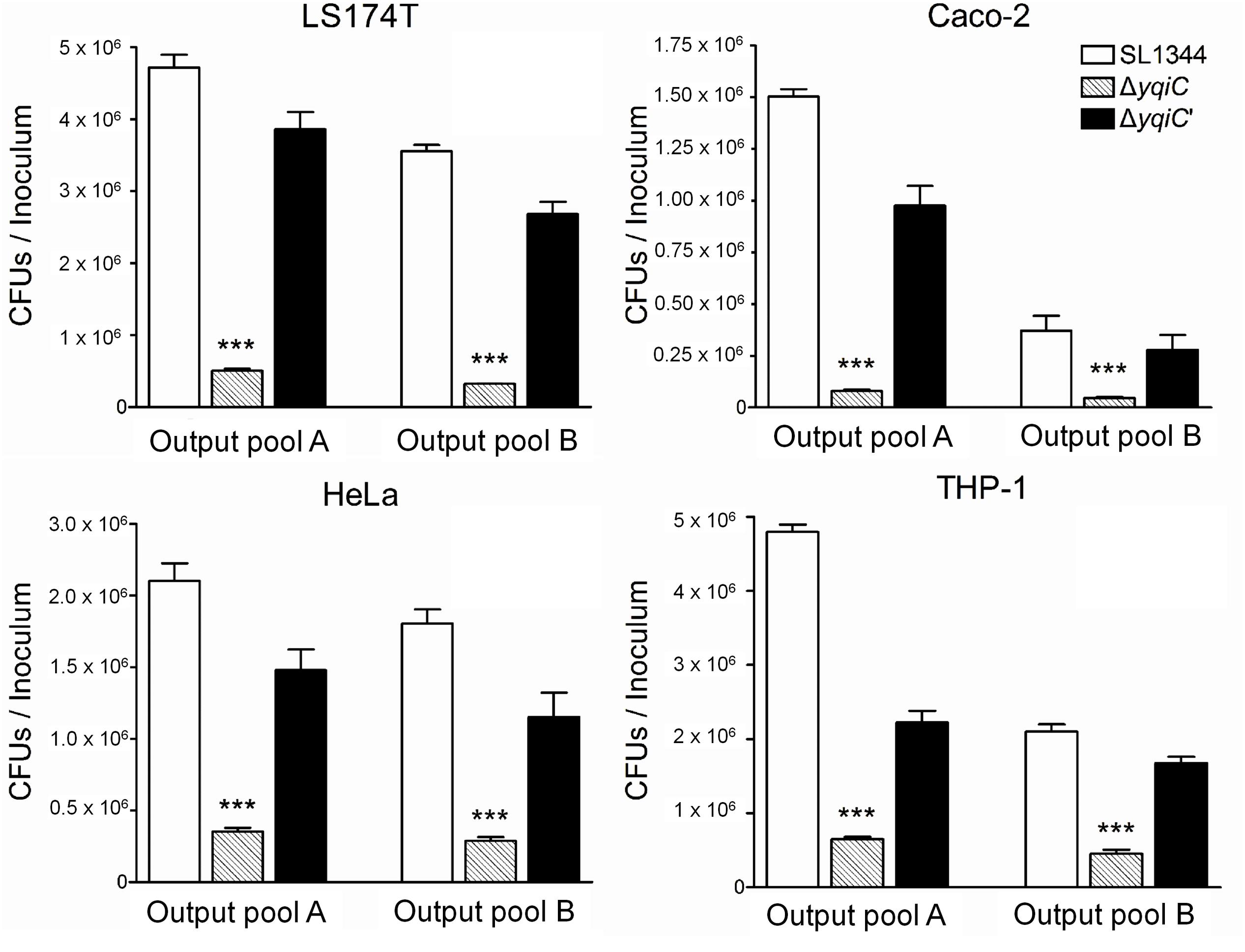
FIGURE 1. Colonization and invasion rates of three Salmonella Typhimurium strains in four human cell lines. LS174T, Caco-2, HeLa, and THP-1 cells were infected with S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′. The CFUs in output pools A and B represent the colonized and intracellular bacteria, respectively. The CFUs of the cell lysates containing bacteria were counted, and the bacterial counts of the colonizing and invading S. Typhimurium ΔyqiC and ΔyqiC′ CFUs were compared with those of the colonizing and invading S. Typhimurium SL1344 using the Student’s t-test. ∗∗∗p < 0.001.
yqiC and fliC Contribute to IL-8 Secretion of LS174T Cells after S. Typhimurium Infection
To determine whether Salmonella yqiC affects the induction of host inflammation, confluent LS174T cells were treated with S. Typhimurium SL1344, ΔyqiC, ΔyqiC′, and ΔfliC; IL-1β; and Salmonella flagellin for examining host IL-8 secretion. IL-8 secretion was significantly augmented in the LS174T cells treated with IL-1β and S. Typhimurium SL1344 compared with that in untreated cells (Figure 2A). Moreover, the secretion was higher in the cells treated with Salmonella flagellin than in those treated with IL-1β. In addition, IL-8 secretion significantly decreased in the LS174T cells infected with S. Typhimurium ΔyqiC and ΔfliC compared with those infected with S. Typhimurium SL1344. The IL-8 secretion levels in the S. Typhimurium ΔyqiC′- and SL1344-infected cells were similar (Figure 2A). Two positive controls, Salmonella flagellin and IL-1β (at the administered dosage) significantly increased IL-8 production compared with untreated controls (Figure 2A). Thus, disruption of either yqiC or fliC remarkably suppressed S. Typhimurium-induced IL-8 production in LS174T cells. Thus, Salmonella flagellin and YqiC have a crucial role in the activation of IL-8 release from LS174T cells after S. Typhimurium infection.
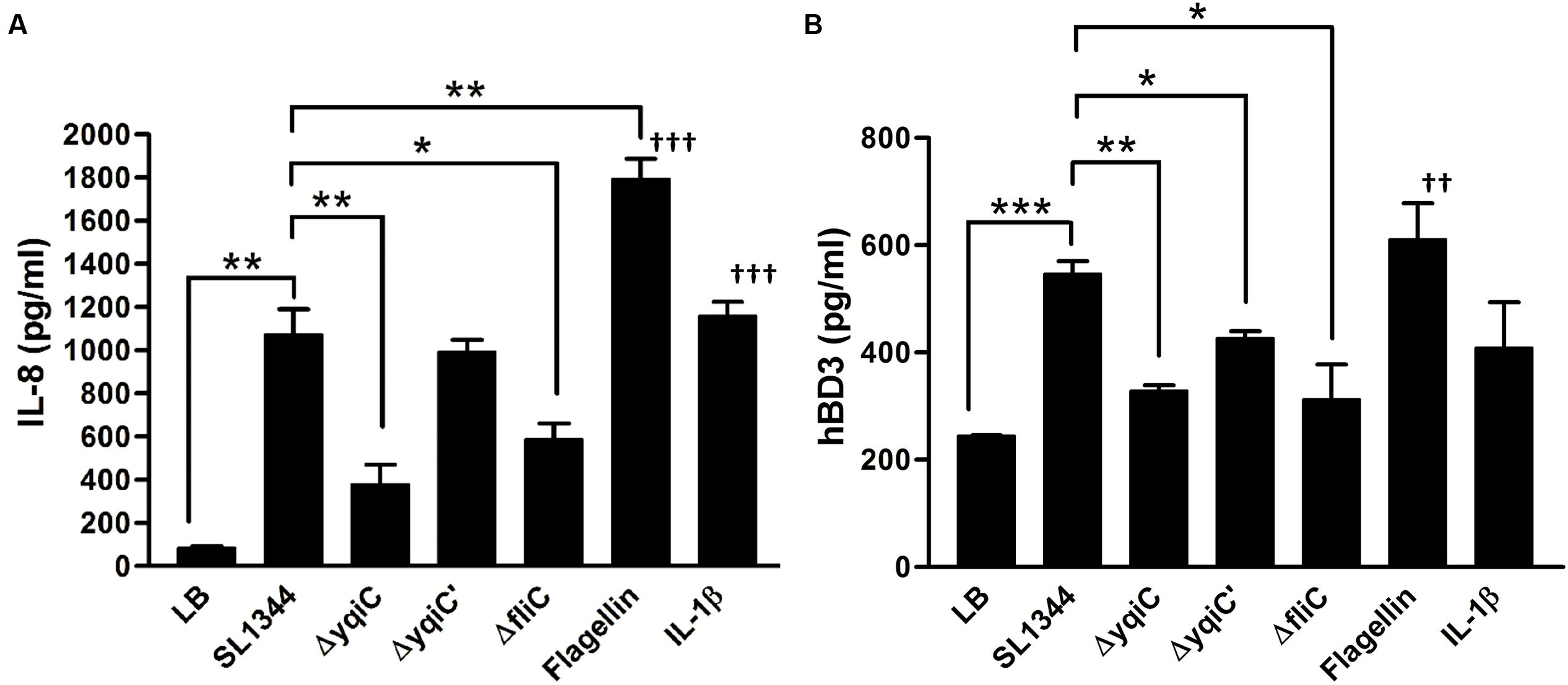
FIGURE 2. Secretion of IL-8 and hBD-3 in LS174T cells after treatment with S. Typhimurium SL1344, ΔyqiC, ΔyqiC′, ΔfliC, Salmonella flagellin, and IL-1β. LS174T cells were treated with S. Typhimurium SL1344 (MOI = 5), 100 ng/mL Salmonella flagellin, and 50 ng/mL IL-1β, according to the protocol in the bacterial invasion assay to generate output pool B. After 18-h treatment, the media from the final 15-h incubation in these wells were examined for quantification of IL-8 (A) and hBD-3 (B) secretion by using ELISA. Flagellin- and IL-1β-treated cells were the positive controls. The Student’s t-test was used for analyzing any significant differences in IL-8 and hBD-3 secretion levels between the S. Typhimurium SL1344-infected cells and the other groups (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001), and significant differences in IL-8 and hBD-3 secretion levels between negative control (LB-treated cells) and positive controls (††p < 0.01, †††p < 0.001).
yqiC and fliC Are Required for hBD-3 Production in LS174T Cells after S. Typhimurium Infection
Because hBD-3 can be expressed in LS174T cells (Fahlgren et al., 2004), but not in Caco-2 cells (Ou et al., 2009), we selected LS174T cells as an in vitro model for studying the involvement of yqiC and fliC in hBD-3-related host innate immunity in human IECs. Confluent LS174T cells were treated with S. Typhimurium SL1344, ΔyqiC, ΔyqiC′, and ΔfliC; IL-1β; and Salmonella flagellin for determining host hBD-3 production. S. Typhimurium significantly induced increased hBD-3 production in LS174T cells (Figure 2B). By contrast, hBD-3 secretion significantly decreased in the LS174T cells infected with S. Typhimurium ΔyqiC and ΔfliC compared with those infected with S. Typhimurium SL1344, and ΔyqiC′ partially restored hBD-3 secretion levels (Figure 2B). Thus, yqiC and fliC have a similar effect on hBD-3 production in LS174T cells after S. Typhimurium infection. Salmonella flagellin, but not IL-1β (at the administered dosage), significantly increased hBD-3 production compared with untreated controls (Figure 2B). The proinflammatory cytokine IL-1β appeared to be an inefficient positive control compared with Salmonella flagellin in terms of hBD-3 secretion in LS174T cells. Therefore, LS174T cells produce hBD-3 after S. Typhimurium infection, and yqiC and fliC of S. Typhimurium contribute to host hBD-3 expression. Thus, we hypothesized that yqiC of S. Typhimurium affects flagellation, flagella-related functions, and host responses.
TEM Revealed Fewer and Shorter Flagella and Presence of Fimbriae in S. Typhimurium ΔyqiC
To validate our hypothesis that yqiC is responsible for S. Typhimurium flagellation, cell surfaces of S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ were examined through TEM after negative staining. The results revealed remarkable differences in the bacterial surfaces of the wild-type and mutant S. Typhimurium strains. S. Typhimurium SL1344 exhibited numerous intact long flagella surrounding the bacterial cell (Figures 3A,B). By contrast, only a few of the fragmented flagella with short shafts surrounding the bacterial cells were observed in S. Typhimurium ΔyqiC, with identifiable type-1 fimbriae-like appendages on their cell surfaces (Figures 3C,D). S. Typhimurium ΔyqiC′ exhibited identical flagellar expression and the absence of fimbriae-mimicking structures, as observed in S. Typhimurium SL1344 (Figures 3E,F). Thus, yqiC deletion impairs flagella formation and activates the expression of type-1 fimbriae-like structures on the bacterial cell surface.
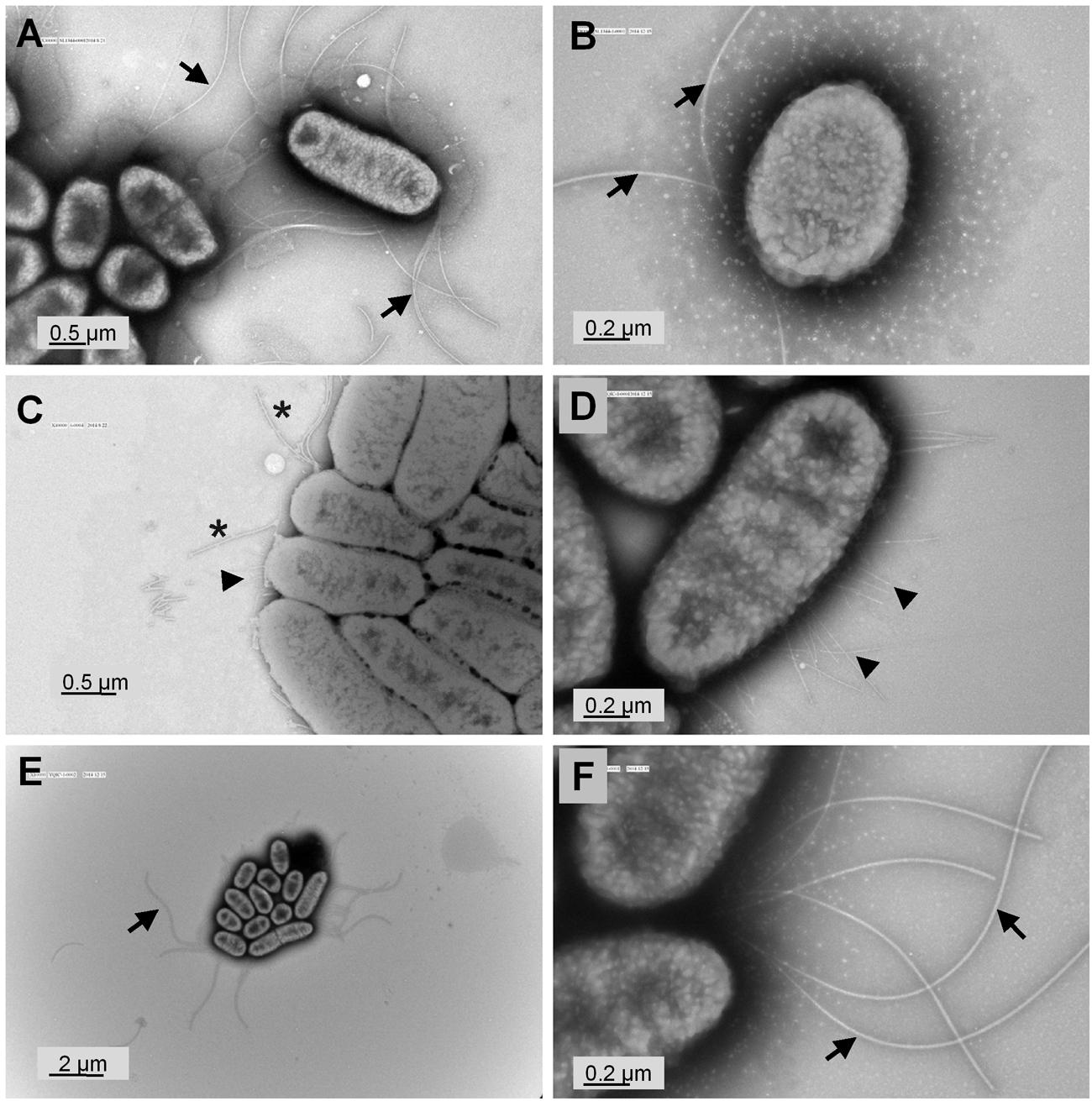
FIGURE 3. Transmission electron micrographs of S. Typhimurium SL1344 and ΔyqiC. Representative transmission electron micrographs after negative staining of at least four different fields of view from each bacterial strain reveal the morphology of S. Typhimurium SL1344 (A,B), ΔyqiC (C,D), and ΔyqiC′ (E,F). Numerous long flagella (arrows) were observed on the surfaces of S. Typhimurium SL1344 [magnification: (A) 30,000×; (B) 80,000×]. Type-1 fimbriae (arrowheads) and defective flagella (asterisks) were observed in S. Typhimurium ΔyqiC [magnification: (C) 40,000×; (D) 80,000×]. Similar to S. Typhimurium SL1344, numerous long flagella without fimbriae were observed in S. Typhimurium ΔyqiC′ [magnification: (E) 10,000×; (F): 80,000×].
yqiC Deletion Enhances Type-1 Fimbrial Expression in S. Typhimurium
To investigate whether yqiC deletion activates type-1 fimbrial expression, we performed yeast agglutination tests for determining the mannose-specific binding of type-1 fimbriae in S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′. No agglutination was observed in S. Typhimurium SL1344. However, significant yeast agglutination was observed in S. Typhimurium ΔyqiC, and small fragments disappeared in S. Typhimurium ΔyqiC′, respectively (left and middle panels, Figure 4A). Nonetheless, when D-mannose was added to the bacterial suspension of S. Typhimurium ΔyqiC to block the binding of its type-1 fimbriae with yeast, these fragments disappeared and this phenomenon appeared similar to the result in S. Typhimurium SL1344 (right, Figure 4A). Therefore, yqiC can suppress mannose-specific type-1 fimbrial expression in S. Typhimurium. Further, the biofilm formation assays demonstrated that that the biofilm formation was enhanced in S. Typhimurium ΔyqiC (Figure 4B), and it was restored in S. Typhimurium ΔyqiC′. According to these results, yqiC can prevent biofilm formation by inhibiting type-1 fimbrial expression in S. Typhimurium.
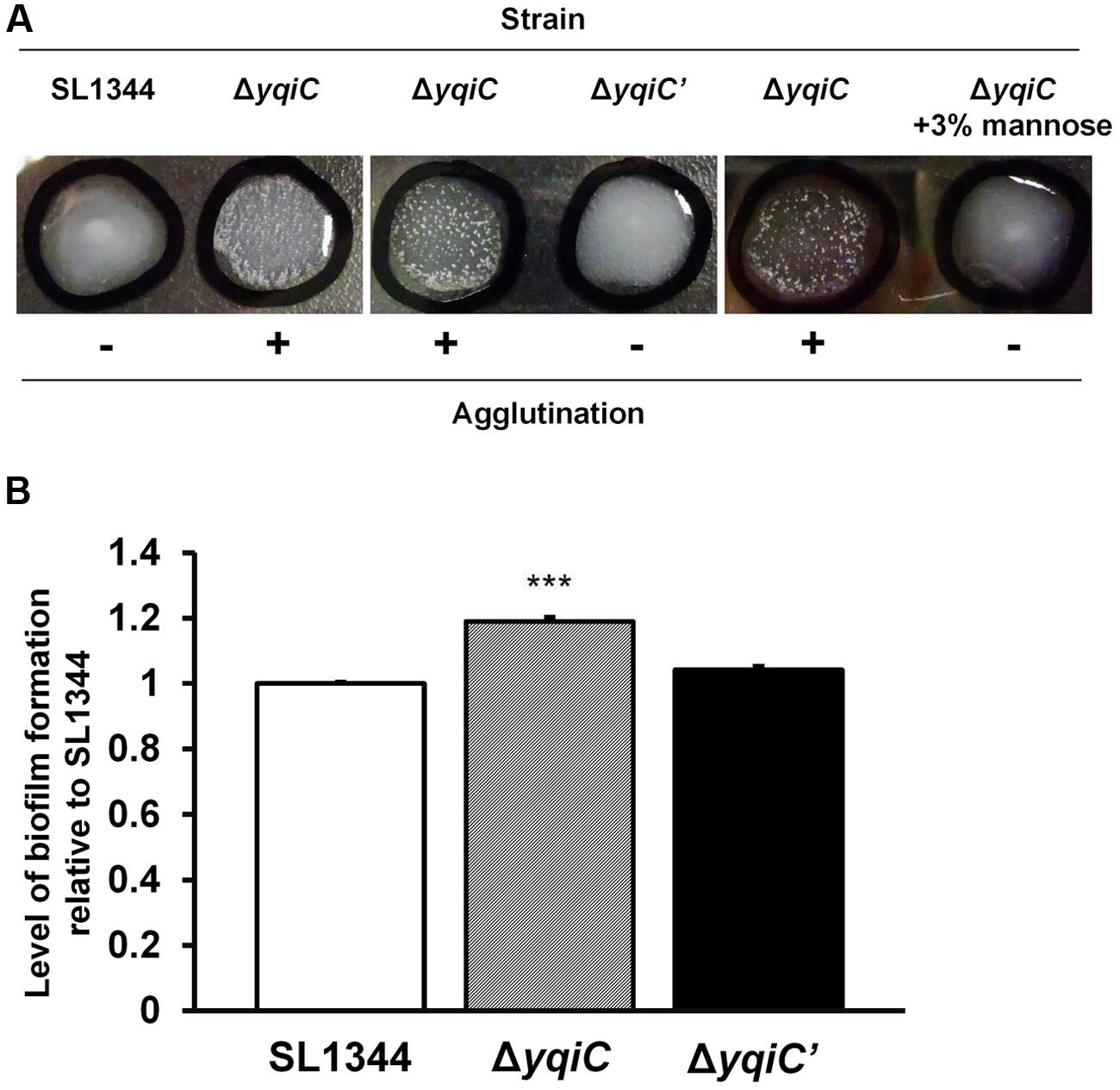
FIGURE 4. Phenotypic analysis of type-1 fimbrial expression in S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′. (A) Yeast agglutination tests for S. Typhimurium strains incubated at 37°C. Formation of small agglutination fragments in S. Typhimurium SL1344 ΔyqiC indicated that the bacteria can express type-1 fimbriae and agglutinate with the yeast cells. The addition of 3% D-mannose interfered with receptor binding on the surface of S. Typhimurium SL1344 and ΔyqiC, which could not agglutinate with yeast cells to form small fragments, thus confirming that the yqiC deletion activated the expression of type-1 fimbriae. (B) The biofilm formation assay. S. Typhimurium SL1344, ΔyqiC and ΔyqiC′ were cultured in 96-well microplates at 37°C for 18 h. Then, all wells were washed with PBS and stained using 0.1% crystal violet for subsequent measurement of the absorbance detected at OD550. The relative levels of biofilm formation in S. Typhimurium ΔyqiC and ΔyqiC′ were compared with those in S. Typhimurium SL1344 using the Student’s t-test. ∗∗∗p < 0.001.
yqiC Is Involved in S. Typhimurium Motility
yqiC is required for S. Typhimurium colonization and invasion and for inducing fliC-like IL-8 and hBD-3 secretion in host cells. Moreover, fliC is involved in bacterial motility. To examine whether yqiC contributes to bacterial motility, we compared the maximal diameters of the motility zones of five S. Typhimurium strains and observed that the diameter of the motility zone of the positive control S. Typhimurium ΔfliC was smaller than that of S. Typhimurium SL1344, whereas the diameters of the motility zones of S. Typhimurium SL1344 and its SPI-1 mutant ΔspaS were equal (upper, Figure 5). Moreover, the diameter of the motility zone of S. Typhimurium ΔyqiC was smaller than that of S. Typhimurium SL1344 and ΔfliC (Figure 5). The diameters of the motility zone of S. Typhimurium SL1344 was almost equal to that of S. Typhimurium ΔyqiC′ (Figure 5). Therefore, the loss of yqiC can downregulate S. Typhimurium motility, and its effect is more prominent than that of the loss of fliC.
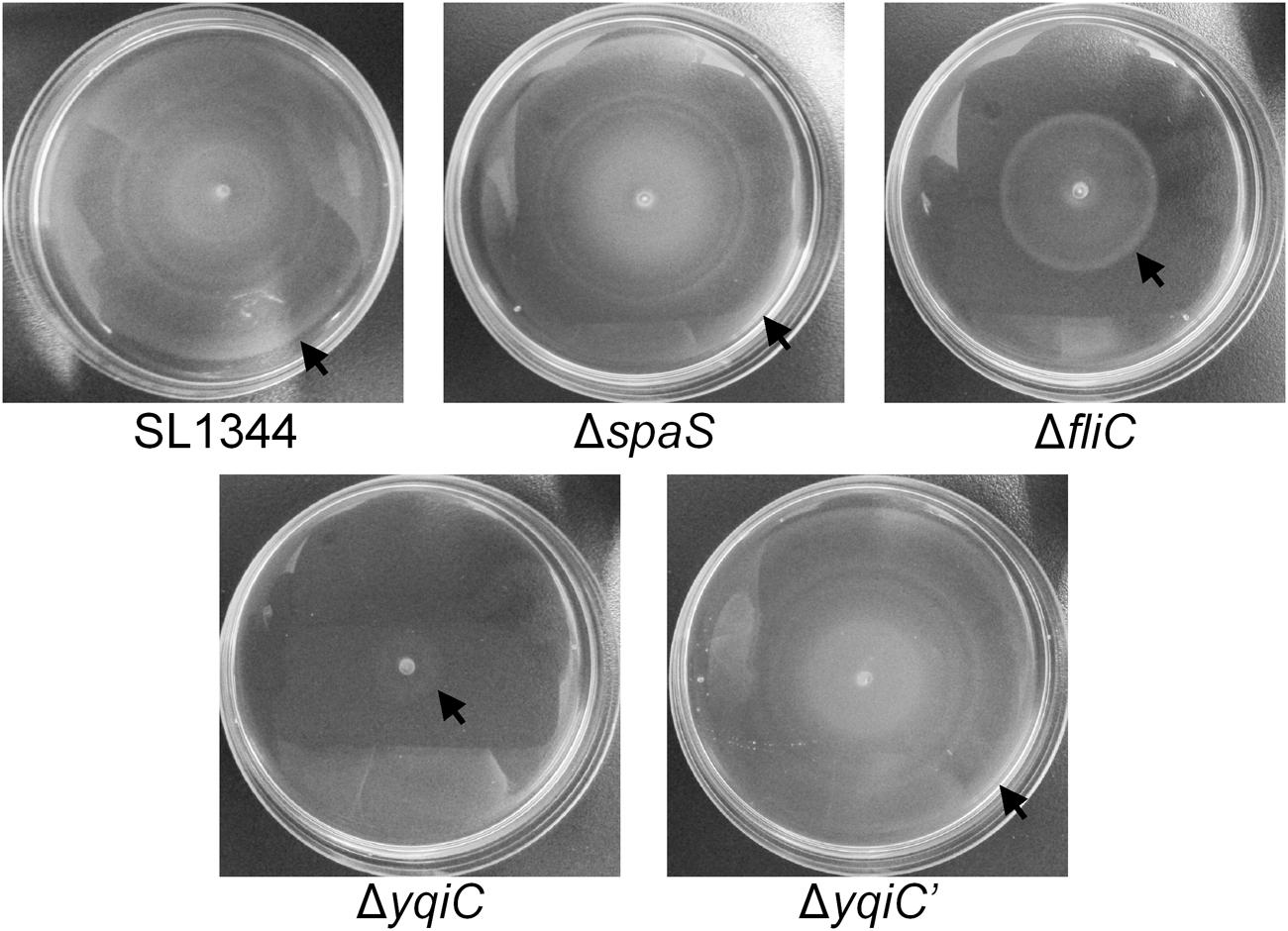
FIGURE 5. Bacterial motility assays for S. Typhimurium SL1344, ΔyqiC, ΔyqiC′, ΔfliC, and ΔspaS. The motilities of S. Typhimurium SL1344, ΔyqiC, ΔyqiC′, ΔfliC, and ΔspaS were examined through bacterial inoculation on semisolid LB agar plates, followed by a 6-h incubation at 37°C. The arrows indicate the outer rims of the motility zones.
yqiC Deletion Upregulates fim Gene Expression and Downregulates Flagella, SPI-1, and SPI-2 Gene Expression in S. Typhimurium
To further delineate the role of yqiC in regulating type-1 fimbrial, flagellar, SPI-1, and SPI-2 gene expression, we performed qRT-PCR for S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′. Compared with their expression in S. Typhimurium SL1344, fimA and fimZ expression was significantly upregulated in S. Typhimurium ΔyqiC (2.5–3 fold-change, p < 0.05), whereas their expression in S. Typhimurium ΔyqiC′ was similar to that in S. Typhimurium SL1344 (Figure 6A). By contrast, flhD, fliZ, invA, and sseB expression was significantly downregulated in S. Typhimurium ΔyqiC (-0.5 to -0.2 fold-change, p < 0.05; Figure 6A). In S. Typhimurium ΔyqiC′, the mRNA expression of the preceding genes was restored (Figure 6A). Our qRT-PCR results were consistent with the yqiC phenotypes obtained in TEM, yeast agglutination tests, and gentamicin protection and bacterial motility assays.
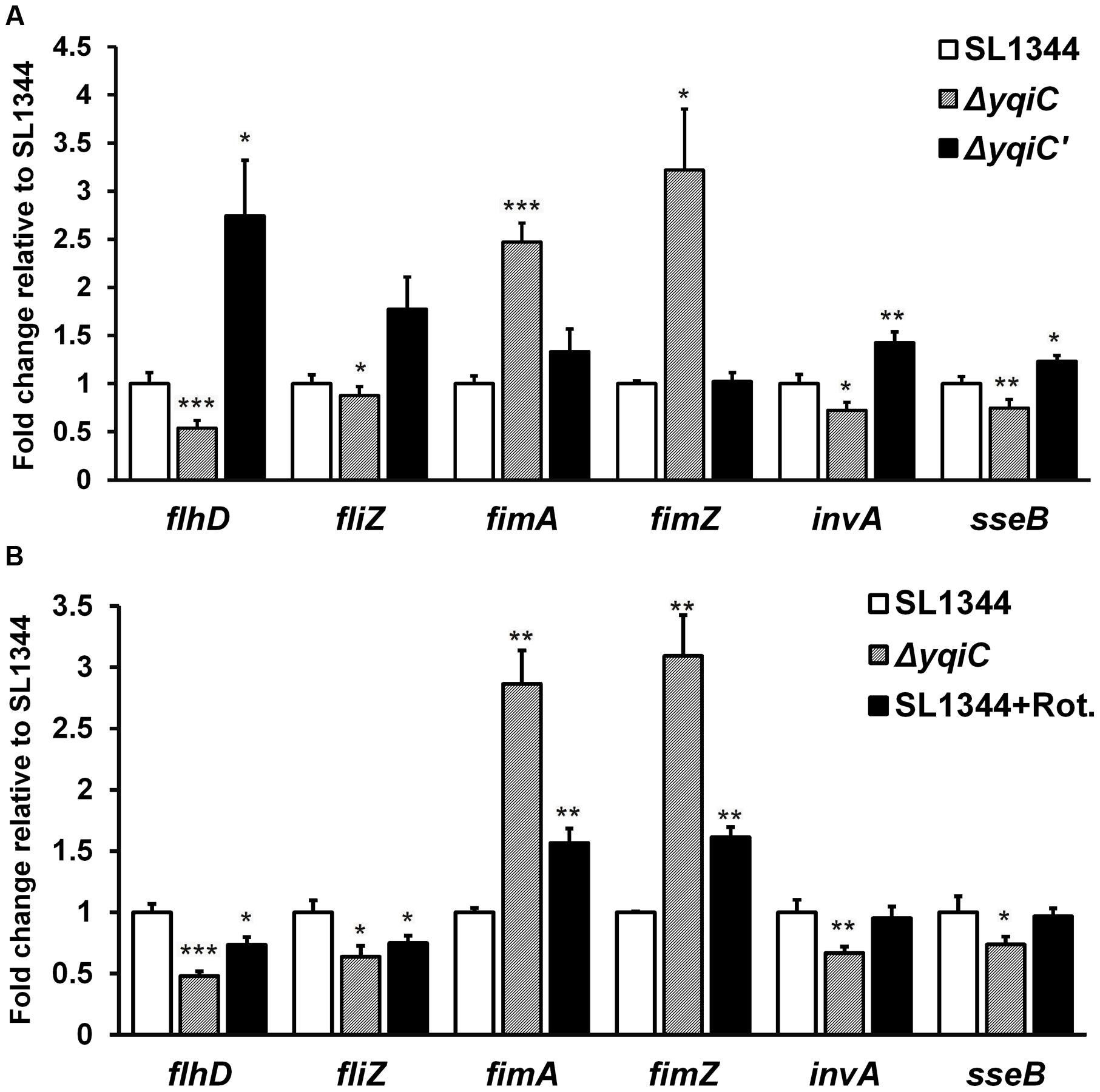
FIGURE 6. Gene expression analysis of flagella, type-1 fimbriae, SPI-1, and SPI-2 in S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′, and NADH dehydrogenase inhibition assays using qRT-PCR. (A) The mRNA expression levels of the type-1 fimbriae-related genes fimA and fimZ, the flagella-related genes flhD and fliZ, the SPI-1-related gene invA, and the SPI-2 gene sseB in S. Typhimurium ΔyqiC were compared with those of S. Typhimurium SL1344 and ΔyqiC′. (B) The NADH dehydrogenase inhibition assay revealed the effects of rotenone (Rot) and yqiC deletion on the regulation of the mRNA expression in flhD, fliZ, fimA, and fimZ, invA and sseB. The mRNA expression levels of the target genes are expressed as fold-change relative to the geometric means of their mRNA expression levels in S. Typhimurium SL1344. Any significant differences in the mRNA expression levels between S. Typhimurium SL1344 and other strains or conditions were analyzed using the Student’s t-test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 were considered statistically significant.
NADH Dehydrogenase Inhibition by Rotenone in the Electron Transport Chain Is Similar to Type-1 Fimbrial, Flagellar, SPI-1, and SPI-2 Gene Regulation in S. Typhimurium ΔyqiC
To study whether the electron transport chain is involved in regulating the phenotype of yqiC, we conducted inhibition assays by using rotenone, an NADH dehydrogenase inhibitor (Ueno et al., 1994), and investigated the effects of the electron transport chain on the yqiC-regulated expression of flagellar, type-1 fimbrial, SPI-1, and SPI-2 genes. The qRT-PCR results indicated that flhD and fliZ were significantly downregulated in rotenone-treated S. Typhimurium SL1344 and S. Typhimurium ΔyqiC (both p < 0.05; black and striated bars, respectively, Figure 6B). Rotenone significantly upregulated fimA and fimZ expression in S. Typhimurium SL1344 (both p < 0.01), although their expression levels were not as high as those in S. Typhimurium ΔyqiC (black and striated bars, respectively, Figure 6B). By contrast, rotenone had no significant effect on invA and sseB expression, in contrast to the downregulation induced by yqiC knockout in S. Typhimurium (Figure 6B). Therefore, rotenone inhibition resulted in a regulatory effect similar to that of yqiC deletion on flagella and type-1 fimbrial expression, suggesting that the electron transport chain is involved in yqiC phenotype expression. Whether the electron transport chain is linked to the yqiC-related modulation of flagellation and type-1 fimbriae activation in S. Typhimurium requires further studies for clarification. However, yqiC possibly regulates SPI-1 and SPI-2 gene expression in S. Typhimurium through pathways not associated with NADH dehydrogenase in the electron transport chain.
yqiC Is Indispensable for Menaquinone Biosynthesis in S. Typhimurium
Although, another two yqi genes yqiE and yqiD are involved in menaquinone biosynthesis in Bacillus subtilis (Julsing et al., 2007), it remains unknown whether yqiC is associated with the biosynthesis of the crucial electron transport chain mediator menaquinone in S. Typhimurium. We extracted and identified menaquinone contents from the cell membranes of S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ through TLC and HPLC. The spots eluted farthest from the start line on the TLC plate represented menaquinone (Rf 0.5-0.6, Figure 7A), whereas the secondary spots, also farther from the start line, were considered other quinone-like compounds or menaquinone intermediates (Rf 0.3-0.4, Figure 7A). TLC revealed that menaquinone was present in S. Typhimurium SL1344 and ΔyqiC′, but not in S. Typhimurium ΔyqiC. Furthermore, we performed HPLC for the membrane fraction components of the three S. Typhimurium strains to reconfirm our TLC results and used the commercial menaquinone K2 as a measurement standard (retention time 4.90 min). In HPLC, menaquinone peaks were only observed for S. Typhimurium SL1344 and ΔyqiC and not for S. Typhimurium ΔyqiC (Figure 7B).
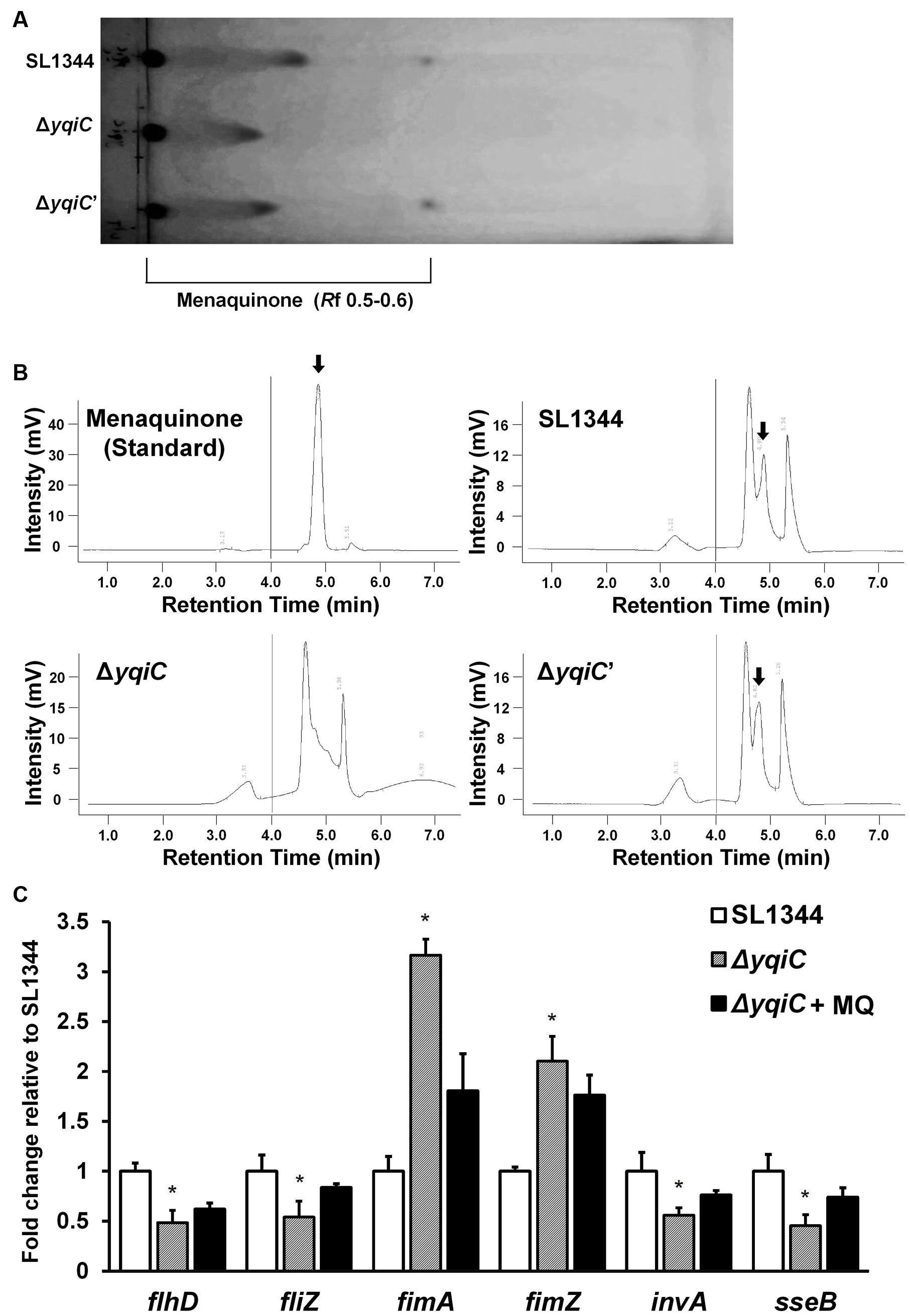
FIGURE 7. Thin-layer chromatography and HPLC for menaquinone in S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′, and menaquinone complementation assays using qRT-PCR. The menaquinone content within S. Typhimurium SL1344, ΔyqiC, and ΔyqiC′ was detected using TLC and HPLC. (A) TLC revealed the presence of menaquinone at an Rf of 0.5–0.6 in S. Typhimurium SL1344 and ΔyqiC′, but detectable menaquinone was absent in S. Typhimurium ΔyqiC. (B) HPLC revealed menaquinone peaks at a retention time of 4.90 min in S. Typhimurium SL1344 and ΔyqiC′ (arrows), but menaquinone peaks were absent in S. Typhimurium ΔyqiC. (C) In the menaquinone complementation assay, menaquinone (MQ) was added into the LB broth containing S. Typhimurium ΔyqiC for 18-h overnight culture. The mRNA expression levels of flhD, fliZ, fimA, fimZ, invA, and sseB of S. Typhimurium SL1344 ΔyqiC and menaquinone-complemented S. Typhimurium ΔyqiC were compared with those of S. Typhimurium SL1344 and are expressed as fold-change relative to the geometric means of their mRNA expression levels in S. Typhimurium SL1344. Any significant differences in the mRNA expression levels between S. Typhimurium SL1344 and other strains or conditions were analyzed using the Student’s t-test. ∗p < 0.05 was considered statistically significant.
Menaquinone Reverses the Effect of yqiC Deletion on Expression of Type-1 Fimbrial, Flagellar, SPI-1, and SPI-2 Genes in S. Typhimurium
To determine the role of menaquinone in Salmonella after yqiC deletion, menaquinone was added in S. Typhimurium ΔyqiC for 18-h incubation. The qRT-PCR results indicated that the downregulated expression of flhD, fliZ, invA, and sseB as well as the upregulated expression of fimA and fimZ were minimized after additional menaquinone treatment in S. Typhimurium ΔyqiC (Figure 7C). On simulating S. Typhimurium SL1344, complementation of menaquinone in the menaquinone-deficient S. Typhimurium ΔyqiC strain can recover the mRNA expression of type-1 fimbrial, flagellar, SPI-1, and SPI-2 genes.
Discussion
The phenotype of yqiC in Salmonella colonization and invasion of host cells might be controlled by environmental temperatures and bacterial cell-growth phases. The importance of yqiC in Salmonella virulence has been confirmed in mice in a previous study, in which all mice infected with the yqiC mutant survived for 30 days, whereas, all mice infected with the wild-type strain died (Carrica et al., 2011). However, the decisive stage of yqiC virulence during S. Typhimurium infection remains unknown. Although, the yqiC mutant completely lost its systemic virulence in mice, when cultured at 28°C, the mutant could invade murine macrophages and human epithelial HeLa cells for up to 24 h post in vitro infection as the wild-type (Carrica et al., 2011), hinting a late effect of yqiC on its virulence. By contrast, in our study, the yqiC mutant cultivated in the mid-log phase at 37°C lost its bacterial colonization and invasion abilities in the human intestinal epithelial LS174T cells during the first 18 h post-infection. The differences in the results of Carrica et al. (2011) and our study may be because of the bacterial virulence, which is influenced by temperature and cell-growth phases. Therefore, at 28°C, the bacteria did not exhibit the yqiC phenotype in bacterial colonization and invasion, but at 37°C, the yqiC effect was maximized for these two characteristics. Similarly, flagella formation can be enhanced at a low temperature by regulating clpP in Salmonella (Knudsen et al., 2014), and the Salmonella virulence can be affected by a temperature-sensing protein TlpA (Hurme and Rhen, 1998). In addition, cell growth phases can affect Salmonella virulence. A recent study in the flhDC promoter region of the Salmonella flagellar regulon reported that the flhDC transcription regulators are growth phase dependent, with the maximum expression in the mid-log cultures (Mouslim and Hughes, 2014). Thus, mid-log cultures of S. Typhimurium ΔyqiC used in our study might induce the phenotypic effect of yqiC on bacterial colonization and invasion, which was inhibited by overnight cultures and low temperature.
Salmonella employs yqiC to suppress type-1 fimbriae overexpression for manipulating other pathogenic factors, such as flagella and motility, in bacterial colonization and invasion in the human intestinal epithelium. In general, bacterial adherence and internalization are two major characteristics of Salmonella involved in bacterial colonization during the interactions between Salmonella and host cells. Type-1 fimbriae are major mediators during Salmonella biofilm formation (Boddicker et al., 2002). However, the deletion of yqiC activated type-1 fimbrial expression on the bacterial surface and biofilm formation, but led to loss of its colonization and invasion capability in all four human cell lines in our study. Thus, yqiC is required for Salmonella colonization and invasion in host cells through other pathogenic factors regulated by the type-1 fimbrial expression instead of the structural fimbrial expression on the bacterial surface and biofilm formation. Our data meant that the up-regulated type-1 fimbriae was not the crucial factor in yqiC mediated bacterial colonization. The colonization effect of type-1 fimbriae-like appendages on the S. Typhimurium surface and biofilm formation was probably overwhelmed by the augmented effect of flagella and SPI-1.
yqiC is involved in S. Typhimurium flagellation and motility, both of which are downregulated by the overexpression of the FimZ-centered type-1 fimbriae regulatory circuit. By contrast, yqiC can switch off this S. Typhimurium circuit for maintaining bacterial virulence factors, such as flagella, SPI-1, and SPI-2 effectors. Our results exhibited that yqiC deletion in Salmonella reduces the production of structural flagella on the bacterial outer membrane, thus reducing bacterial motility. Type-1 fimbrial expression in Salmonella is regulated by positive regulators FimZ and FimY (Yeh et al., 1995). fimZ controls type-1 fimbriae production and mediates bacterial motility. However, it is unclear whether fimY is correlated with bacterial motility. In addition, fimZ positively controls hilE, which negatively regulates hilD, an activator of hilA, to upregulate the expression of the SPI-1 gene invA (Baxter and Jones, 2005). FimZ appears to be the dominant activator of the PfimA promoter to regulate the expression of the fim structural genes. FimY activates FimZ expression and also activates its own expression slightly. FimZ alone can enhance the expression of hilE, a repressor of SPI-1 gene expression. Moreover, activated FimZ generates hyperfimbriated but non-motile S. Typhimurium in soft agar, and such non-motility correlates with the downregulation of the flhDC master flagellar operon (Clegg and Hughes, 2002). FliZ, a flagellar regulator, can inhibit the expression of the type-1 fimbrial gene through post-transcriptional regulation of FimZ (Saini et al., 2010). Therefore, FimZ can be considered a key molecule between flagellar and fimbrial formation in S. Typhimurium. In addition, hilD of Salmonella is involved in the regulation of SPI-2 gene expression (Martinez et al., 2011). The expression of FimZ and type-1 fimbriae in Salmonella gradually reduces after infecting murine macrophages, and the constitutive expression of FimZ and type-1 fimbriae can suppress the SPI-2 gene activation (Wang et al., 2013). Furthermore, the type-1 fimbrial regulator FimZ can control Salmonella motility by inactivating the flhDC flagellar operon (Clegg and Hughes, 2002). Our qRT-PCR analysis revealed that the expression of the SPI-1 gene invA, the SPI-2 gene sseB, and the flagellar gene flhD was downregulated in S. Typhimurium ΔyqiC. Bridging the existing knowledge, Salmonella yqiC can have a vital role in the orchestration of the type-1 fimbrial expression on flagella, SPI-1, and SPI-2, possibly through FimZ and FliZ (Figure 8).
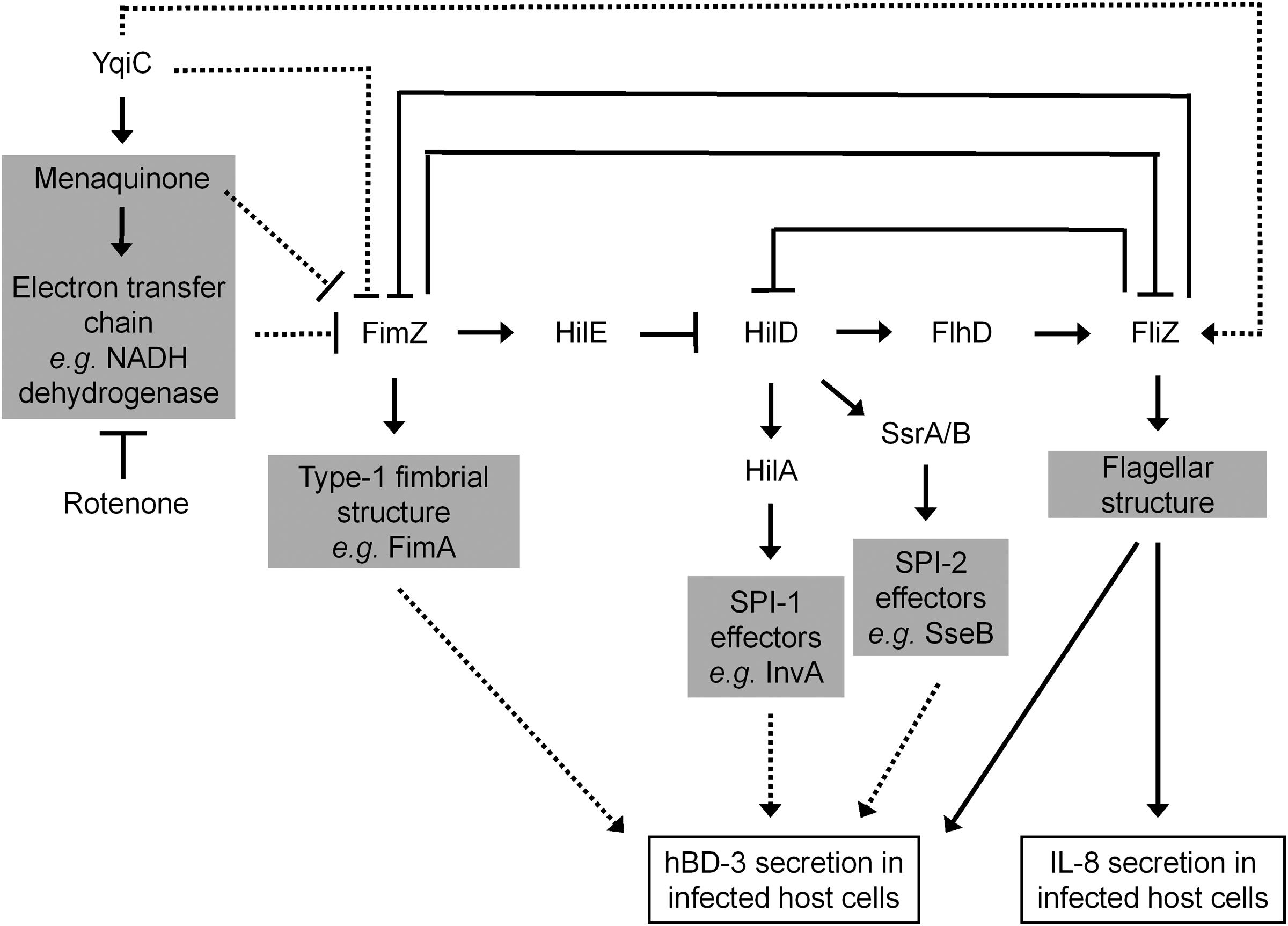
FIGURE 8. Scheme of the yqiC regulatory network in Salmonella. This scheme summarizes the role of yqiC in the regulation of type-1 fimbrial, SPI-1, SPI-2, and flagellar genes. YqiC enhances menaquinone biosynthesis and its related electron transport chain, negatively controls FimZ and its downstream regulatory network, and positively manipulates FliZ and its subsequent flagellation. Our data supported that YqiC can suppress the expression of type-1 fimbriae in Salmonella. The activation of the type-1 fimbrial regulator FimZ can inhibit the expression of flagellar, SPI-1, and SPI-2 genes by inhibiting the regulators HilD, HilA, SsrA/B, and FliZ, resulting in the suppression of the host immune responses, including the expression of IL-8 and hBD-3. The solid and dotted lines indicate well-documented regulations and putative connections in the regulator network, respectively.
Both yqiC and fliC are required for host IL-8 production in human IECs after S. Typhimurium infection. Bacterial flagellin is considered the main ligand of Salmonella that elicits inflammatory responses, such as IL-8 production, in host cells (McCormick et al., 1993; Hayashi et al., 2001). In this study, IL-8 and hBD-3 secretion from LS174T cells reduced after S. Typhimurium ΔyqiC infection compared with that of S. Typhimurium SL1344, and the reduced IL-8 secretion was non-significantly higher on S. Typhimurium ΔyqiC infection than on S. Typhimurium ΔfliC infection (Figure 2A). These findings suggest that the non-SPI gene yqiC is a Salmonella virulence genetic factor for induction of host cell inflammation and is at least as influential as fliC. The significantly attenuated colonization and invasion of S. Typhimurium ΔyqiC, resulting in fewer intracellular bacteria compared with the wild-type strain, can partly clarify the decreased IL-8 secretion from the infected host cells. Meanwhile, such attenuated host inflammation can be attributed to the effects of other virulence factors affected by yqiC deletion, including downregulation of the SPI-1 and SPI-2 genes invA and sseB through upregulation of fimZ in S. Typhimurium ΔyqiC (Figure 6). Furthermore, the reduced expression of structural flagella in S. Typhimurium ΔyqiC (Figure 3), observed in this study, further addresses the contribution of yqiC to Salmonella virulence.
Both yqiC and fliC are responsible for hBD-3 production in LS174T cells after S. Typhimurium infection. This is the first study to report that S. Typhimurium can trigger hBD-3 secretion in human IECs. hBD-3 expression is significantly higher in normal tonsils and skin, but absent in the small intestine (Harder et al., 2001). Thus far, avian studies have reported contrasting results associated with β-defensin-3 expression. AvBD-3 expression is enhanced in uninfected broiler chicken guts (Ramasamy et al., 2012), and its gene expression is upregulated in the bone marrow and spleen after Salmonella infection (Ma et al., 2013). By contrast, AvBD-3 expression is significantly upregulated in the uninfected chicken ovaries, but not in the Salmonella-infected chicken ovaries (Ebers et al., 2009; Michailidis et al., 2012). Our study by using hBD-3-expressing LS174T cells presented further evidence that hBD-3 is constitutively secreted in the uninfected human intestinal epithelium and that the expression can be significantly increased by S. Typhimurium infection. Similar to the induction of human hBD-2 expression in Caco-2 cells (Ogushi et al., 2001; Takahashi et al., 2001), our study demonstrated that hBD-3 production can be induced by Salmonella flagellin, and yqiC has a role in the regulatory network involving type-1 fimbriae, SPI-1, SPI-2, and flagella (Figure 8). We hypothesize that in addition to flagellar genes, the expression of type-1 fimbrial, SPI-1, or SPI-2 genes is involved in the regulation of hBD-3 expression in host cells, and additional studies are warranted for clarification.
For the first time, we demonstrated that Salmonella yqiC accounts for menaquinone biosynthesis within bacterial cells and has a key role in flagellation and suppression of type-1 fimbrial expression, similar to the effects of NADH dehydrogenase in the electron transport chain. Menaquinone is one of the isoprenoid quinines that are located in the inner membrane of bacteria and function as electron and proton carriers in the respiratory electron transport chain and as antioxidants (Nowicka and Kruk, 2010). Most gram-negative facultative anaerobic rods, such as E. coli and Klebsiella pneumoniae, contain mixtures of menaquinones, demethylmenaquinones, and ubiquinones (electron carriers in the electron transport chain), with the menaquinones and demethylmenaquinones being the major quinone types (Collins and Jones, 1981). However, the composition of quinone pools in Salmonella remains unclear. In bacteria, menaquinone-carried electrons can be bound with the oxidized reductants such as NADH, succinate, formate, and hydrogen, whereas menaquinones can be oxidized by coupling them with reduction of oxidants such as oxygen, nitrogen dioxide, nitrite, and sulfate. Thus, the redox loop mechanism or the respiratory chain drives the proton pumps for ATP synthesis (Lancaster and Simon, 2002). In addition, the flagellar export apparatus of Salmonella comprising FlhA, FlhB, FliO, FliP, FliQ, and FliR exerts its ATPase activity at the cytoplasmic side, where ATP is hydrolyzed as the energy source to drive flagellin exportation for the assembly of each flagellum (Minamino et al., 2014). In E. coli, the respiratory electron transport system correlates with flagella formation (Hertz and Bar-Tana, 1977). In Salmonella, the ubiquinone synthetic pathway is required for flagellar biosynthesis (Bar-Tana et al., 1980). However, the mechanism underlying menaquinone-controlled flagellation in Salmonella remains unclear. ubiB, is probably involved in menaquinone or ubiquinone biosynthesis in Salmonella, and can suppress type-1 fimbrial expression because ubiB deletion in Salmonella enhances type-1 fimbrial expression (Chuang et al., 2008). These findings are consistent with our result that absence of menaquinone activated the expression of type-1 fimbriae, and defective flagellation coexisted in S. Typhimurium ΔyqiC. In this study, complementation with menaquinone in S. Typhimurium ΔyqiC confirmed that menaquinone is required for type-1 fimbrial, flagellar, SPI-1, and SPI-2 gene expression when yqiC is disrupted in S. Typhimurium. Therefore, menaquinone is a critical mediator within Salmonella affected by yqiC to negatively control fimZ expression. Meanwhile, activated fimZ expression can reduce flagellar biosynthesis and SPI-1 and SPI-2 gene expression (Figure 8). Because of the lack of menaquinone inhibitors, we could not confirm whether menaquinone downregulates fimZ expression directly or through the electron transport chain. Menaquinone analogs can be used as inhibitors to inhibit the growth of Staphylococcus aureus, B. anthracis, Streptococcus pyogenes, and Streptococcus agalactiae (Schlievert et al., 2013). In this study, we used an NADH dehydrogenase inhibitor, rotenone, in S. Typhimurium, and its effects on the regulation of type-1 fimbrial and flagellar gene expression, but not on the regulation of SPI-1 and SPI-2 gene expression, were similar to those of yqiC deletion. Rotenone only affects NADH dehydrogenase activity in complex I of the electron transport chain (Ueno et al., 1994), but succinate dehydrogenase and nitrate reductase complexes are also responsible for electron transport in bacteria (Hurdle et al., 2011). Therefore, the regulation of SPI-1 and SPI-2 gene expression by Salmonella yqiC correlates with other enzymes, rather than NADH dehydrogenase, in the electron transport chain. Furthermore, YqiC of S. Typhimurium shares approximately 25% amino acid homology with IbpA and IbpB of E. coli, as per our preliminary alignments. IbpA and IbpB, the heat shock proteins of E. coli, are correlated with E. coli resistance to oxidative stress induced by hydrogen peroxide and copper (Kitagawa et al., 2000; Matuszewska et al., 2008). Thus, yqiC might facilitate cell damage repair under oxidative stress through menaquinone and its related processes in the electron transport chain. This claim warrants further investigation.
Conclusion
yqiC significantly contributes to Salmonella colonization and invasion as well as host inflammation and innate immunity after infection. Our study provides additional novel knowledge regarding the presence of an upstream virulence gene manipulating fimZ-dominated type-1 fimbriae regulation and its downstream virulence factors, flagella, SPI-1, and SPI-2. We observed that the electron transport chain, including its functioning compound menaquinone, is possibly involved in regulation of these crucial Salmonella virulence factors. Therefore, the evidence suggests that yqiC is a promising target gene for developing novel antimicrobials and immunomodulators against salmonellosis.
Author Contributions
K-CW performed the research and wrote the article; C-HH, S-MD, and C-KC analyzed data; H-WF and M-TH performed the technique of molecular biology, and S-BF designed the research and assisted correction of the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the Career Development Grant from the National Health Research Institutes, Taiwan (NHRI-EX105-10234SC), and the National Taipei University of Technology-Taipei Medical University Joint Research Program (NTUT-TMU-101-18).
Abbreviations
AMP, antimicrobial peptide; AvBD, avian β-defensin; CFU, colony-forming unit; E. coli, Escherichia coli; ELISA, enzyme-linked immunosorbent assay; FBS, fetal bovine serum; fim, Type-1 fimbriae; hBD, human β-defensin; HPLC, high-performance liquid chromatography; IEC, intestinal epithelial cell; IL, interleukin; MAPK, mitogen-activated protein kinase; MQ, menaquinone; NADH, nicotinamide adenine dinucleotide (NAD) + hydrogen (H); NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; PBS, phosphate buffered saline; qRT-PCR, quantitative real-time polymerase chain reaction; S. Typhimurium, Salmonella enterica serovar Typhimurium; SPI, Salmonella pathogenicity island; T1SSs, type I secretion systems; T3SSs, type III secretion systems; TEM, transmission electron microscopy; TLC, thin-layer chromatography.
Footnotes
References
Al Mamun, A. A., Lombardo, M. J., Shee, C., Lisewski, A. M., Gonzalez, C., Lin, D., et al. (2012). Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338, 1344–1348. doi: 10.1126/science.1226683
Althouse, C., Patterson, S., Fedorka-Cray, P., and Isaacson, R. E. (2003). Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71, 6446–6452. doi: 10.1128/IAI.71.11.6446-6452.2003
Aslanzadeh, J., and Paulissen, L. J. (1990). Adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol. Immunol. 34, 885–893. doi: 10.1111/j.1348-0421.1990.tb01067.x
Bar-Tana, J., Howlett, B. J., and Hertz, R. (1980). Ubiquinone synthetic pathway in flagellation of Salmonella typhimurium. J. Bacteriol. 143, 637–643.
Baumler, A. J., Tsolis, R. M., and Heffron, F. (1996). Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64, 1862–1865.
Baxter, M. A., and Jones, B. D. (2005). The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 73, 1377–1385. doi: 10.1128/IAI.73.3.1377-1385.2005
Boddicker, J. D., Ledeboer, N. A., Jagnow, J., Jones, B. D., and Clegg, S. (2002). Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45, 1255–1265. doi: 10.1046/j.1365-2958.2002.03121.x
Carrica, M. C., Craig, P. O., Garcia-Angulo, V. A., Aguirre, A., Garcia-Vescovi, E., Goldbaum, F. A., et al. (2011). YqiC of Salmonella enterica serovar typhimurium is a membrane fusogenic protein required for mice colonization. BMC Microbiol. 11:95. doi: 10.1186/1471-2180-11-95
Cheminay, C., Chakravortty, D., and Hensel, M. (2004). Role of neutrophils in murine salmonellosis. Infect. Immun. 72, 468–477. doi: 10.1128/IAI.72.1.468-477.2004
Chuang, Y. C., Wang, K. C., Chen, Y. T., Yang, C. H., Men, S. C., Fan, C. C., et al. (2008). Identification of the genetic determinants of Salmonella enterica serotype Typhimurium that may regulate the expression of the type 1 fimbriae in response to solid agar and static broth culture conditions. BMC Microbiol. 8:126. doi: 10.1186/1471-2180-8-126
Clegg, S., and Hughes, K. T. (2002). FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 1209–1213. doi: 10.1128/jb.184.4.1209-1213.2002
Coburn, B., Sekirov, I., and Finlay, B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549. doi: 10.1128/CMR.00013-07
Collins, M. D., and Jones, D. (1981). Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45, 316–354.
Duan, Q., Zhou, M., Zhu, L., and Zhu, G. (2013). Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8. doi: 10.1002/jobm.201100335
Duguid, J. P., and Campbell, I. (1969). Antigens of the type-1 fimbriae of salmonellae and other enterobacteria. J. Med. Microbiol. 2, 535–553. doi: 10.1099/00222615-2-4-535
Ebers, K. L., Zhang, C. Y., Zhang, M. Z., Bailey, R. H., and Zhang, S. (2009). Transcriptional profiling avian β-defensins in chicken oviduct epithelial cells before and after infection with Salmonella enterica serovar Enteritidis. BMC Microbiol. 9:153. doi: 10.1186/1471-2180-9-153
Fahlgren, A., Hammarstrom, S., Danielsson, A., and Hammarstrom, M. L. (2004). β-Defensin-3 and -4 in intestinal epithelial cells display increased mRNA expression in ulcerative colitis. Clin. Exp. Immunol 137, 379–385. doi: 10.1111/j.1365-2249.2004.02543.x
Fang, S.-B. (2011). Early Interactions of non-Typhoidal Salmonella with Human Epithelium. Doctoral thesis, UCL University College London, London.
Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546. doi: 10.1073/pnas.0337542100
Harder, J., Bartels, J., Christophers, E., and Schroder, J. M. (2001). Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276, 5707–5713. doi: 10.1074/jbc.M008557200
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. doi: 10.1038/35074106
Hertz, R., and Bar-Tana, J. (1977). Anaerobic electron transport in anaerobic flagellum formation in Escherichia coli. J. Bacteriol. 132, 1034–1035.
Hurdle, J. G., O’Neill, A. J., Chopra, I., and Lee, R. E. (2011). Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9, 62–75. doi: 10.1038/nrmicro2474
Hurme, R., and Rhen, M. (1998). Temperature sensing in bacterial gene regulation–what it all boils down to. Mol. Microbiol. 30, 1–6. doi: 10.1046/j.1365-2958.1998.01049.x
Julsing, M. K., Rijpkema, M., Woerdenbag, H. J., Quax, W. J., and Kayser, O. (2007). Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis. Appl. Microbiol. Biotechnol. 75, 1377–1384. doi: 10.1007/s00253-007-0953-5
Kawai, T., and Akira, S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650. doi: 10.1016/j.immuni.2011.05.006
Kitagawa, M., Matsumura, Y., and Tsuchido, T. (2000). Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol. Lett. 184, 165–171. doi: 10.1111/j.1574-6968.2000.tb09009.x
Knodler, L. A., Finlay, B. B., and Steele-Mortimer, O. (2005). The Salmonella effector protein SopB protects epithelial cells from apoptosis by sustained activation of Akt. J. Biol. Chem. 280, 9058–9064. doi: 10.1074/jbc.M412588200
Knudsen, G. M., Nielsen, M. B., Thomsen, L. E., Aabo, S., Rychlik, I., and Olsen, J. E. (2014). The role of ClpP, RpoS and CsrA in growth and filament formation of Salmonella enterica serovar Typhimurium at low temperature. BMC Microbiol. 14:208. doi: 10.1186/s12866-014-0208-4
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., et al. (1997). The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256. doi: 10.1038/36786
Kwon, O., Kotsakis, A., and Meganathan, R. (2000). Ubiquinone (coenzyme Q) biosynthesis in Escherichia coli: identification of the ubiF gene. FEMS Microbiol. Lett. 186, 157–161. doi: 10.1111/j.1574-6968.2000.tb09097.x
Lambert, M. A., and Smith, S. G. (2008). The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol. 8:142. doi: 10.1186/1471-2180-8-142
Lancaster, C. R., and Simon, J. (2002). Succinate:quinone oxidoreductases from epsilon-proteobacteria. Biochim. Biophys. Acta 1553, 84–101. doi: 10.1016/S0005-2728(01)00230-4
Lee, C. A., Silva, M., Siber, A. M., Kelly, A. J., Galyov, E., and McCormick, B. A. (2000). A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. U.S.A. 97, 12283–12288. doi: 10.1073/pnas.97.22.12283
Lindquist, B. L., Lebenthal, E., Lee, P. C., Stinson, M. W., and Merrick, J. M. (1987). Adherence of Salmonella typhimurium to small-intestinal enterocytes of the rat. Infect. Immun. 55, 3044–3050.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, D., Zhang, M., Zhang, K., Liu, X., Han, Z., Shao, Y., et al. (2013). Identification of three novel avian β-defensins from goose and their significance in the pathogenesis of Salmonella. Mol. Immunol. 56, 521–529. doi: 10.1016/j.molimm.2013.05.227
Martinez, L. C., Yakhnin, H., Camacho, M. I., Georgellis, D., Babitzke, P., Puente, J. L., et al. (2011). Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 80, 1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x
Matuszewska, E., Kwiatkowska, J., Kuczynska-Wisnik, D., and Laskowska, E. (2008). Escherichia coli heat-shock proteins IbpA/B are involved in resistance to oxidative stress induced by copper. Microbiology 154(Pt 6), 1739–1747. doi: 10.1099/mic.0.2007/014696-0
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., et al. (2001). Complete genome sequence of Salmonella enterica serovar typhimurium LT2. Nature 413, 852–856. doi: 10.1038/35101614
McCormick, B. A., Colgan, S. P., Delp-Archer, C., Miller, S. I., and Madara, J. L. (1993). Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J. Cell Biol. 123, 895–907. doi: 10.1083/jcb.123.4.895
Michailidis, G., Avdi, M., and Argiriou, A. (2012). Transcriptional profiling of antimicrobial peptides avian β-defensins in the chicken ovary during sexual maturation and in response to Salmonella enteritidis infection. Res. Vet. Sci. 92, 60–65. doi: 10.1016/j.rvsc.2010.10.010
Minamino, T., Morimoto, Y. V., Kinoshita, M., Aldridge, P. D., and Namba, K. (2014). The bacterial flagellar protein export apparatus processively transports flagellar proteins even with extremely infrequent ATP hydrolysis. Sci. Rep. 4, 7579. doi: 10.1038/srep07579
Mouslim, C., and Hughes, K. T. (2014). The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 10:e1003987. doi: 10.1371/journal.ppat.1003987
Nowicka, B., and Kruk, J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797, 1587–1605. doi: 10.1016/j.bbabio.2010.06.007
Ogushi, K., Wada, A., Niidome, T., Mori, N., Oishi, K., Nagatake, T., et al. (2001). Salmonella enteritidis FliC (flagella filament protein) induces human β-defensin-2 mRNA production by Caco-2 cells. J. Biol. Chem. 276, 30521–30526. doi: 10.1074/jbc.M011618200
O’Neil, D. A., Porter, E. M., Elewaut, D., Anderson, G. M., Eckmann, L., Ganz, T., et al. (1999). Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163, 6718–6724.
O’Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 30, e2437. doi: 10.3791/2437
Ou, G., Baranov, V., Lundmark, E., Hammarstrom, S., and Hammarstrom, M. L. (2009). Contribution of intestinal epithelial cells to innate immunity of the human gut–studies on polarized monolayers of colon carcinoma cells. Scand. J. Immunol. 69, 150–161. doi: 10.1111/j.1365-3083.2008.02208.x
Pace, J., Hayman, M. J., and Galan, J. E. (1993). Signal transduction and invasion of epithelial cells by S. typhimurium. Cell 72, 505–514. doi: 10.1016/0092-8674(93)90070-7
Raffatellu, M., Wilson, R. P., Chessa, D., Andrews-Polymenis, H., Tran, Q. T., Lawhon, S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73, 146–154. doi: 10.1128/IAI.73.1.146-154.2005
Ramasamy, K. T., Verma, P., and Reddy, M. R. (2012). Differential gene expression of antimicrobial peptides β defensins in the gastrointestinal tract of Salmonella serovar pullorum infected broiler chickens. Vet. Res. Commun. 36, 57–62. doi: 10.1007/s11259-011-9512-8
Sabbagh, S. C., Forest, C. G., Lepage, C., Leclerc, J. M., and Daigle, F. (2010). So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305, 1–13. doi: 10.1111/j.1574-6968.2010.01904.x
Saini, S., Slauch, J. M., Aldridge, P. D., and Rao, C. V. (2010). Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J. Bacteriol. 192, 5767–5777. doi: 10.1128/JB.00624-10
Schlievert, P. M., Merriman, J. A., Salgado-Pabon, W., Mueller, E. A., Spaulding, A. R., Vu, B. G., et al. (2013). Menaquinone analogs inhibit growth of bacterial pathogens. Antimicrob. Agents Chemother. 57, 5432–5437. doi: 10.1128/AAC.01279-13
Selsted, M. E., and Ouellette, A. J. (2005). Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557. doi: 10.1038/ni1206
Takahashi, A., Wada, A., Ogushi, K., Maeda, K., Kawahara, T., Mawatari, K., et al. (2001). Production of β-defensin-2 by human colonic epithelial cells induced by Salmonella enteritidis flagella filament structural protein. FEBS Lett. 508, 484–488. doi: 10.1016/S0014-5793(01)03088-5
Uchiya, K., and Nikai, T. (2004). Salmonella enterica serovar Typhimurium infection induces cyclooxygenase 2 expression in macrophages: involvement of Salmonella pathogenicity island 2. Infect. Immun. 72, 6860–6869. doi: 10.1128/IAI.72.12.6860-6869.2004
Uchiya, K., and Nikai, T. (2005). Salmonella pathogenicity island 2-dependent expression of suppressor of cytokine signaling 3 in macrophages. Infect. Immun. 73, 5587–5594. doi: 10.1128/IAI.73.9.5587-5594.2005
Ueno, H., Miyoshi, H., Ebisui, K., and Iwamura, H. (1994). Comparison of the inhibitory action of natural rotenone and its stereoisomers with various NADH-ubiquinone reductases. Eur. J. Biochem. 225, 411–417. doi: 10.1111/j.1432-1033.1994.00411.x
van Asten, A. J., and van Dijk, J. E. (2005). Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol. Med. Microbiol. 44, 251–259. doi: 10.1016/j.femsim.2005.02.002
Wagner, C., and Hensel, M. (2011). Adhesive mechanisms of Salmonella enterica. Adv. Exp. Med. Biol. 715, 17–34. doi: 10.1007/978-94-007-0940-9_2
Wang, K. C., Hsu, Y. H., Huang, Y. N., Chen, T. H., Lin, J. H., Hsuan, S. L., et al. (2013). A low-pH medium in vitro or the environment within a macrophage decreases the transcriptional levels of fimA, fimZ and lrp in Salmonella enterica serovar Typhimurium. J. Biosci. 38, 499–507. doi: 10.1007/s12038-013-9347-2
Wang, K. C., Hsu, Y. H., Huang, Y. N., and Yeh, K. S. (2012). A previously uncharacterized gene stm0551 plays a repressive role in the regulation of type 1 fimbriae in Salmonella enterica serotype Typhimurium. BMC Microbiol. 12:111. doi: 10.1186/1471-2180-12-111
Wehkamp, J., Schauber, J., and Stange, E. F. (2007). Defensins and cathelicidins in gastrointestinal infections. Curr. Opin. Gastroenterol. 23, 32–38. doi: 10.1097/MOG.0b013e32801182c2
Yeh, K. S., Hancox, L. S., and Clegg, S. (1995). Construction and characterization of a fimZ mutant of Salmonella typhimurium. J. Bacteriol. 177, 6861–6865.
Yu, Y., Zeng, H., Lyons, S., Carlson, A., Merlin, D., Neish, A. S., et al. (2003). TLR5-mediated activation of p38 MAPK regulates epithelial IL-8 expression via posttranscriptional mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 285, G282–G290. doi: 10.1152/ajpgi.00503.2002
Zhou, D., Chen, L. M., Hernandez, L., Shears, S. B., and Galan, J. E. (2001). A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39, 248–259. doi: 10.1046/j.1365-2958.2001.02230.x
Keywords: yqiC, bacterial colonization, menaquinone, Salmonella Typhimurium, interleukin-8, human β-defensin-3, flagella, type-1 fimbriae
Citation: Wang K-C, Huang C-H, Ding S-M, Chen C-K, Fang H - W, Huang M - T and Fang S-B (2016) Role of yqiC in the Pathogenicity of Salmonella and Innate Immune Responses of Human Intestinal Epithelium. Front. Microbiol. 7:1614. doi: 10.3389/fmicb.2016.01614
Received: 29 July 2016; Accepted: 27 September 2016;
Published: 10 October 2016.
Edited by:
Fabrice Merien, Auckland University of Technology, New ZealandReviewed by:
Lydia Bogomolnaya, Texas A&M University Health Science Center, USAMasahiro Eguchi, National Institute of Animal Health (NARO), Japan
Copyright © 2016 Wang, Huang, Ding, Chen, Fang, Huang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiuh-Bin Fang, sbfang@tmu.edu.tw
 Ke-Chuan Wang
Ke-Chuan Wang Chih-Hung Huang3
Chih-Hung Huang3 Shiuh-Bin Fang
Shiuh-Bin Fang