- 1Department of Biology, University of Pisa, Pisa, Italy
- 2Department of Invertebrate Zoology, Saint Petersburg State University, Saint Petersburg, Russia
- 3Department of Veterinary Medicine, University of Milan, Milan, Italy
- 4Department of Zoology, Andhra University, Visakhapatnam, India
- 5Department of Biotechnology, Andhra University, Visakhapatnam, India
Holospora spp. and “Candidatus Gortzia infectiva”, known as Holospora-like bacteria (HLB), are commonly found as nuclear endosymbionts of ciliates, especially the Paramecium genus. HLB are related by phylogenetic relationships, morphological features, and life-cycles, which involve two alternating morphotypes: reproductive and infectious forms (RF, IF). In this paper we describe a novel species belonging to the “Ca. Gortzia” genus, detected in P. multimicronucleatum, a ciliate for which infection by an HLB has not been reported, discovered in India. This novel endosymbiont shows unusual and surprising features with respect to other HLB, such as large variations in IF morphology and the occasional ability to reproduce in the host cytoplasm. We propose the name of “Candidatus Gortzia shahrazadis” for this novel HLB. Moreover, we report two additional species of HLB from Indian Paramecium populations: “Ca. Gortzia infectiva” (from P. jenningsi), and H. obtusa (from P. caudatum); the latter is the first record of Holospora from a tropical country. Although tropical, we retrieved H. obtusa at an elevation of 706 m corresponding to a moderate climate not unlike conditions where Holospora are normally found, suggesting the genus Holospora does exist in tropical countries, but restricted to higher elevations.
Introduction
Ciliates (Alveolata, Ciliophora) have long been known to be potential hosts for bacteria (Preer et al., 1974; Görtz, 1983; Heckmann and Schmidt, 1987; reviewed in Görtz, 1996, 2006; Fokin, 2004a, 2012; Fujishima, 2009a). Molecular characterization shows a large biodiversity among these symbionts (Irbis and Ushida, 2004; Vannini et al., 2004, 2010, 2014; Rinke et al., 2006; Schrallhammer et al., 2006, 2011, 2013; Ferrantini et al., 2009; Boscaro et al., 2012, 2013a,b,c; Gong et al., 2014; Senra et al., 2015; Szokoli et al., 2016) and also the type of interaction with their hosts can range from mutualistic to parasitic (Kusch et al., 2002; Vannini et al., 2003, 2007; Fels and Kaltz, 2006; Fellous et al., 2011). In this work, we use De Bary's definition of “symbiosys” (de Bary, 1879), as “the living together of two differently named organisms”, independent of effects on the organisms involved, thereby including mutualism, parasitism, and commensalism. The terms “symbionts” and “endosymbionts” also use this definition.
Holospora is a well-known genus of endosymbiotic bacteria. The Holospora name, meaning “whole spore” was invoked by Hafkine (1890), to describe rod-shaped bacteria, similar to spores, infecting a French population of P. caudatum (found in an aquarium), including H. obtusa, H. undulata and H. elegans (Hafkine, 1890; redescribed in Gromov and Ossipov, 1981; Preer and Preer, 1982). Since then, many other Holospora species have been described (although most lack a proper description according to the bacterial taxonomic code), such as “H. acuminata”, “H. bacillata”, H. caryophila, “H. curvata”, “H. curviuscula”, “H. recta”, and Holospora spp. (Ossipov et al., 1980; Preer and Preer, 1982; Borchsenius et al., 1983; Fokin, 1991; Fokin and Sabaneyeva, 1993; Fokin et al., 1999, 2006; reviewed in Görtz and Schmidt, 2005; Ferrantini et al., 2007). The Holospora-like bacteria (HLB) group includes all known Holospora species and “Candidatus Gortzia infectiva”, recently found in a stable endosymbiosis in the P. jenningsi macronucleus (MA) (Boscaro et al., 2013a). These two genera of bacteria share similarities in morphology, in life-cycles and show a close phylogenetic relation (Boscaro et al., 2013a).
Holospora-like bacteria (HLB) are Gram-negative, non-motile Alphaproteobacteria, inhabiting either the MA or the micronucleus (MI) of Paramecium species (for review see Görtz and Schmidt, 2005; Fokin and Görtz, 2009), as well as the MA of Frontonia (Fokin et al., 2006; Ferrantini et al., 2007).
All HLB live in an obligate endosymbiosis with their hosts, and have variable degrees of nuclear and host specificity (Ossipov, 1973; Fujishima and Görtz, 1983; Görtz, 1983; Fokin, 2000; Boscaro et al., 2013a; for review on Holospora see Fujishima, 2009b). They undergo a distinctive life-cycle involving two different forms: the smaller and almost round reproductive form (1–3 μm) (RF), and the much more elongated, rod-like infectious form (4–20 μm) (IF), in which it is possible to recognize several cell parts: cytoplasm, periplasm, and an apical structure, the “recognition tip” (Görtz and Diekmann, 1980; Görtz et al., 1989, 1990; Fujishima et al., 1990; Boscaro et al., 2013a).
Holospora-like bacteria (HLB) can be spread by both horizontal and vertical transmission (reviewed in Fokin and Görtz, 2009). The IFs invade a new host cell via a phagocytotic pathway and, after acidosome fusion, are able to escape from digestive vacuoles and reach the nucleus of the ciliate (Görtz and Wiemann, 1989) via movement mediated by the host cytoskeleton (Fokin et al., 2003; Sabaneyeva et al., 2005, 2009). Inside the nucleus, IFs differentiate into reproductive forms, which divide by binary fission. During host cell division, RFs are shared between two daughter nuclei. In most Holospora species (i.e., “H. acuminata”, “H. curviuscula”, H. elegans, H. obtusa, “H. recta”, and H. undulata) the majority of infectious forms are concentrated in a central part of the dividing nucleus—called the “connecting piece”—that will be released into the environment (Wiemann and Görtz, 1989). The remaining Holospora species (i.e., “H. bacillata”, H. caryophila, “H. curvata”) and “Ca. Gortzia infectiva” do not show this particular localization (Fokin et al., 1996; Boscaro et al., 2013a; Fokin, 2015), and instead are released from the host by an inverted path of infection (Fokin and Sabaneyeva, 1997; Fokin, 2015): IFs are released singularly or in small groups from the nucleus into the cytoplasm, and later into the environment.
Holospora species have been widely recorded by protistologists, allowing researchers to produce a biogeography of these endosymbionts (Fokin and Görtz, 2009), although the infection rate in Paramecium populations is not always permanent and follows cyclic fluctuations (Fokin and Görtz, 2009; Duncan et al., 2015).
Although the genus Paramecium is nearly cosmopolitan (Fokin, 1997, 2010/11; Przyboś and Fokin, 2000), Holospora have only been isolated in cold to temperate areas, in the north of America, Asia and Europe (Hori and Fujishima, 2003; Fokin, 2004b; Fokin et al., 2006). However, this pattern is most likely biased by the lack of sampling in tropical countries, where sampling efforts have been limited (Fokin et al., 2004; Görtz, 2008; Fokin and Sera, 2014). An exception is “Ca. Gortzia infectiva”, which has only been found in a sample from Thailand, (Boscaro et al., 2013a), showing that HLB can exist in tropical climates.
In this paper, we report the retrieval of HLB from another tropical area, the south of India. We found and described a novel species of HLB, which is the first reported from P. multimicronucleatum. Moreover, we found “Ca. Gortzia infectiva” in the MA of an Indian population of P. jenningsi and, quite surprisingly, also H. obtusa infecting the MA of P. caudatum from India. Our records represent the first report of “Ca. Gortzia infectiva” from India and the first finding of the Holospora genus in a tropical country (although from a higher elevation with a moderate climate), at the lowest latitude ever reported. Thus, our study provides new and important information about HLB distribution, reshaping HLB biogeography.
Materials and Methods
Paramecium Collection and Identification
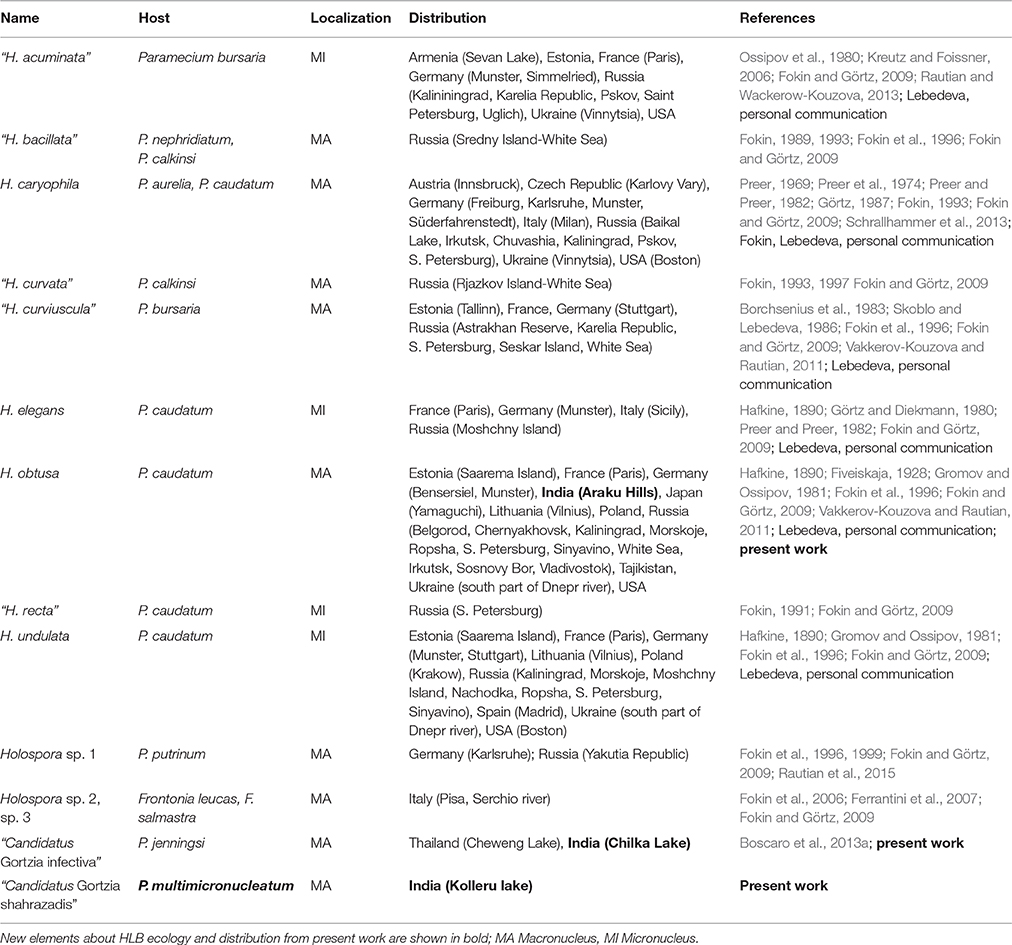
Sampling was carried out in India, during 2014: P. multimicronucleatum population TP2 was collected in freshwater Kolleru Lake, Andhra Pradesh (N 16°44′16.0″ E 81°24′18.0″; 28th September); P. multimicronucleatum PC6 strain was sampled in Pedda Cheruvu, the largest freshwater water body in the Vizianagaram district, Andhra Pradesh (N 18°06′36.0″ E 83°24′0.0″; 23rd November); P. jenningsi population BJ1 was collected in the brackish water Chilka Lake, Odisha (N 19°44′37.0″ E 85°12′44.4″; 3rd February); P. caudatum population SH2 was collected in a freshwater stream on Araku Hills, 706 m above the sea level, in Visakhapatnam district, Andhra Pradesh (N 18°16′32.3″ E 83°02′15.6″; 27th January). Where not indicated, samples were collected at sea level. The Araku hills have a moderate climate: the average maximum day-time temperature is 32.6°C and the average minimum temperature is 24.0°C. During the Indian summer season (March-June) the average maximum temperature is 35.2°C, the minimum is 26.2°C (http://www.yr.no/place/india/Andhra_Pradesh/Araku/statistics.html).
Samples were grown at room temperature and fed Cerophyll medium inoculated with Raoultella planticola (see Boscaro et al., 2013a for details). Unfortunately the SH2 and BJ1 populations survived only a few weeks in laboratory conditions, but for P. multimicronucleatum population TP2 we were able to create a monoclonal culture, named TP2-2.
Living observations and images were made with an Axio Lab.A1 (Zeiss) microscope and/or by an Orthoplan Leitz microscope equipped with differential interference contrast (DIC), as well as a Leica DMR microscope at × 300–1250 magnifications. 18S rDNA sequencing was performed to confirm morphological identifications. About 20 cells for each population were isolated and stored in 70% Ethanol at −20°C until genomic extraction using a NucleoSpin™ Plant II kit (Macherey-Nagel, Germany). The polymerase chain reaction (PCR) was carried out with the following primers: 18S F9 (5′- CTG GTT GAT CCT GCC AG -3′), (Medlin et al., 1988), and 18S R1513Hypo (5′- TGA TCC TTC YGC AGG TTC -3′), (Petroni et al., 2002). All PCRs were performed in a 40 μl reaction volume with 0.25 μl primers (100 μM), TaKaRa PCR reagents and ExTaq (Takara Bio, Japan) using a C1000™ Thermal Cycler (Bio-Rad, Hercules, CA). The PCR program used was: denaturation at 94°C for 30 s, annealing at 55°C for 30 s, elongation at 72°C for 2 min, and final elongation step at 72° for 6 min. PCR products were cleaned with the EuroGOLD Cycle-Pure kit (EuroClone, Milano, Italy) and sent to GATC Biotech Company (Germany) for sequencing with the following internal primers: 18S R536 (5′-CTG GAA TTA CCG CGG CTG-3′), 18S R1052 (5′-AAC TAA GAA CGG CCA TGC A-3′) and 18S F783 (5′-GAC GAT CAG ATA CCG TC-3′), (Rosati et al., 2004).
Characterization of Endosymbionts
Examination of living cells show that SH2, BJ1, and TP2/TP2-2 paramecia exhibit MA bacterial infections. Transmission electron microscopy (TEM) analysis was performed on the TP2-2 monoclonal strain following the protocol in Boscaro et al. (2013a).
We carried out cross-infection experiments using infected TP2-2 cells as donor, and the endosymbiont-free P. multimicronucleatum strain PC6 as recipient. Experimental infection was carried out by preparing a homogenate of infected cells according to Preer (1969). Recipient cells were infected by mixing equal volumes of cell culture with the donor cell homogenate in a 3 mL depression slide, and incubating at room temperature. In order to check the infection status a set of living cells (n = 10) was observed by DIC after 2, 24 and 48 h after mixture with the homogenate. We could test the trans-infection with only one species of Paramecium as receiver, due to the small number of cells in the TP2 culture.
Endosymbiont 16S sequences were obtained for all the populations using the Alphaproteobacteria universal primers 16S Alfa F19b, 16S R1522b (Table 1); sequencing was initially carried out using bacterial universal primers (16S F343 ND, 16S F785 ND, 16S R515 ND); subsequently we designed specific sequencing primers for each species (for details see Table 1). For the TP2 endosymbiont, two additional PCR reactions were performed with two different sets of primers: 16S Alfa F19b together with 16S Gortz R659 (annealing temperature was changed to 51°C) and 16S F114HoloCaedi together with 16S R1488 Holo. The first PCR product was sequenced using R534 Gortzia, the second with F1008 Gortzia and 16S R1328HoloCaedi, and the sequencing results assembled into a single sequence. The 16S rRNA gene sequences were aligned using the ARB software package (Ludwig et al., 2004) and manually checked against more than 600,000 bacterial sequences from SSU rRNA SILVA 123 Ref NR 99 database (Quast et al., 2013). For phylogenetic analyses 58 sequences were employed: together with the 3 new sequences, 48 selected sequences belonging to order Rickettsiales and 7 sequences belonging to class Alphaproteobacteria as outgroup (sequences not shown in the tree are listed in Table S1). The alignment was reduced in length, producing a 1632 character matrix. Maximum likelihood (ML) analyses (PHYML 5.3.2) (Guindon and Gascuel, 2003) and Bayesian inference (BI) analyses (MrBayes 3.2) (Ronquist et al., 2012) were performed, with the GTR +I +G substitution model, as indicated by AIC (Akaike's information criterion), calculated by jModelTest 2.2 (Darriba et al., 2012). ML analysis was applied with 1000 pseudoreplicates, while for BI analysis, three different Markov Chain Monte Carlo runs were employed, with one cold chain and three heated chains each, running for 500,000 generations.
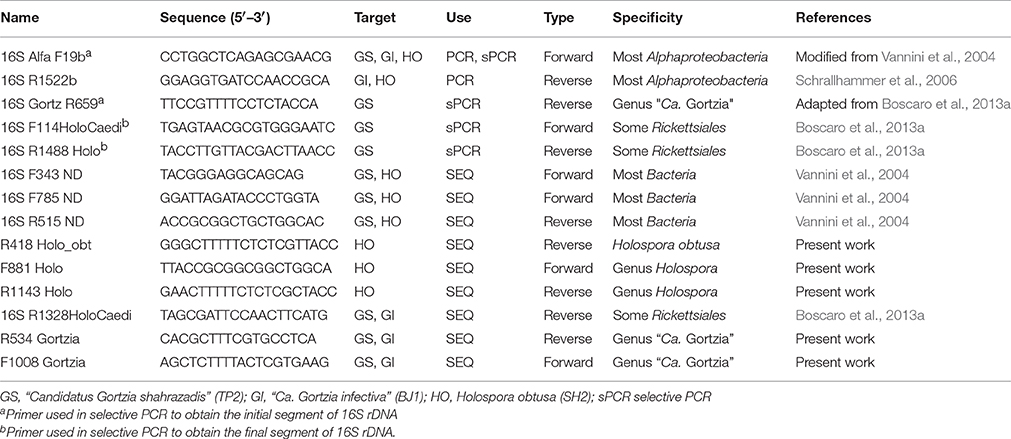
Table 1. List of primers used for 16S rRNA encoding gene sequencing of Holospora-like bacteria from Paramecium spp.
Fluorescence In Situ Hybridization (FISH) analyses was performed as described by Boscaro et al. (2013a). A first set of experiments was carried out with P. jenningsi BJ1 and P. multimicronucleatum TP2 using a probe designed for “Ca. Gortzia infectiva”, GortProb659 (5′-TTCCGTTTTCCTCTACCA-3′), (Boscaro et al., 2013a), labeled with a cyanine 3 (Cy3) fluorophore at the 5′ end, together with a Bacterial universal probe EUB338 (word5′-GCTGCCTCCCGTAGGAGT-3′) (Amann et al., 1990), labeled with fluorescein isothiocyanate (FITC) at the 5′ end. We then designed two new species-specific probes able to distinguish between the two endosymbionts from P. jenningsi and P. multimicronucleatum, since GortProb659 was found to label both. We designed GortzInf_1268 (word5′-TCCTGATTCGCTCAAGGTC-3′; FITC fluorophore in 5′ end), specific for “Ca. G. infectiva”, and GortzSha_1266 (word5′-TTTTGATTTGCTCAAGGTCGC-3′; Cy3 fluorophore in 5′ end), specific for the new HLB from P. multimicronucleatum. Both probes were tested in silico on the RDP (ribosomal database project) (Cole et al., 2009) and SILVA database using TestProbe 3.0 (Quast et al., 2013), allowing 0 mismatches. It was not possible to experimentally test GortzInf_1268 because P. jenningsi BJ1 infected with “Ca. Gortzia infectiva” was lost. We tested GortzSha_1266 (Cy3) in competition with GortzInf_1268 (FITC) or EUB338 (FITC) on the P. multimicronucleatum endosymbiont.
Results
Paramecium Species Identification
Paramecium TP2 and PC6 exhibited the typical cell morphology and size of P. multimicronucleatum morphospecies (Figure 1) (Fokin, 1997, 2010/11; Fokin and Chivilev, 2000), showing the presence of several vesicular-type MI (Figure 1J). We obtained two identical 1710 bp 18S rDNA sequences that matched more than 10 P. multimicronucleatum sequences (including HG315606, HE662762, and other) with 99.35–99.88% identity. For this reason, we identify our TP2 and PC6 organisms as P. multimicronucleatum. Nevertheless, our sequences reached only 98.49% identity if compared to another group of P. multimicronucleatum sequences, including the one for the morphospecies (AF255361) (Strüder-Kypke et al., 2000). Such variability in 18S rDNA sequences inside the taxon suggests the presence of cryptic species, a situation that will be addressed in forthcoming studies. Paramecium BJ1 was identified as P. jenningsi (Figure 2), due to the presence of two chromosomal-type MI (strangely, in some cases, more than two) (Fokin, 1997) (Figure 2B) and by 18S rDNA sequence (1711 bp) identity with P. jenningsi (HE662760, AF100311) of 99.88%. We identified Paramecium SH2 as P. caudatum (Figure 3) because it exhibited one compact-type MI (Figure 3C) and its 18S rDNA sequence (1710 bp) matched P. caudatum HE664171 with 99.00% identity, with 1 indel and 16 mismatches. This number of mismatches is unusual in 18S rDNA sequences of the same Paramecium species, suggesting again a possible cryptic P. caudatum species. These newly obtained sequences are available from the ENA database: LT549005 (P. multimicronucleatum TP2); LT549006 (P. multimicronucleatum PC6); LT549003 (P. jenningsi BJ1); LT549004 (P. caudatum SH2).
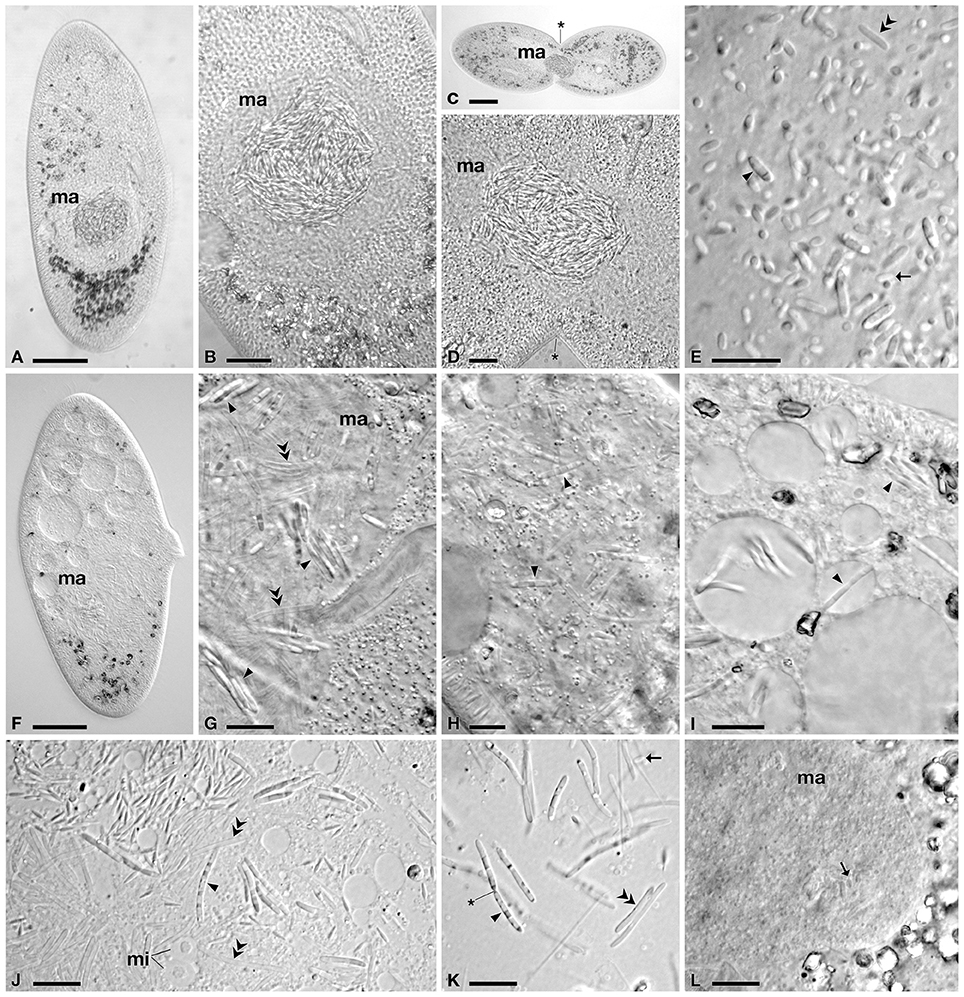
Figure 1. Light microscopy of infected cells of P. multimicronucleatum. (A–E) TP2 cells a few weeks after environmental sampling and (F–K) after 1 year in culture. (A) Whole cell with macronuclear infection. (B) Infected macronucleus (ma), with bacteria clustering inside. (C) Infected cell undergoing division and (D) detail of its ma, showing absence of connecting piece. (E) Bacterial forms released after ma squashing, showing classical size and morphology of reproductive (rf) and infectious (if) forms. (F) Whole cell with macronuclear infection after 1 year of cultivation, in which bacterial cells are less visible. (G) Detail of infected ma, in which if cluster together in small groups, among a large number of transient forms (tf). (H) if in host cytoplasm. (I) Detail of if in host cytoplasm inside and outside vacuoles. (J) Detail of host vesicular type micronuclei (mi) and if, showing some changes in morphology, such as size and unusual striated/dotted pattern. (K) if undergoing binary fission and tf after cell squashing. (L) ma of strain PC6, infected by bacteria during cross-infection experiments, showing classical rf morphology. Arrowheads indicate infectious forms, arrows reproductive forms and double arrowheads transient forms. The asterisk indicates the cleavage furrow. Bars stand for 30 μm (A,F), 20 μm (C,H) and 10 μm (B,D,E,G,I–L).
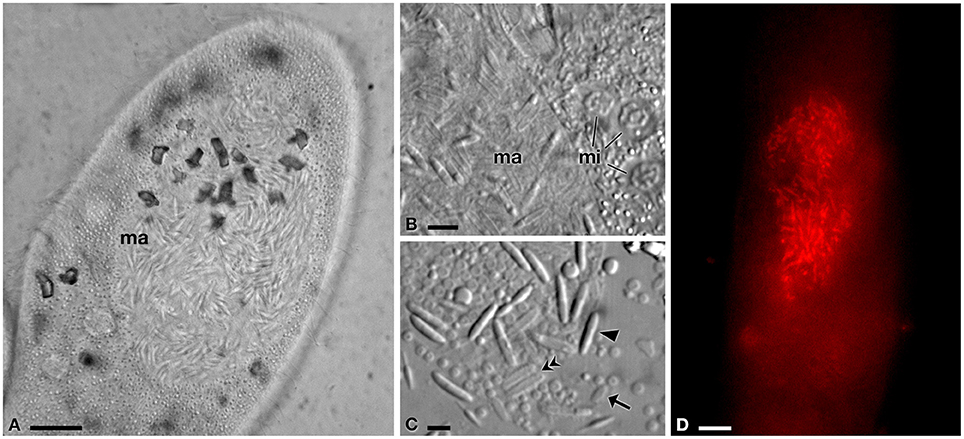
Figure 2. Light microscopy and FISH analyses of infected P. jenningsi BJ1. (A) Cell with hyperinfected macronucleus (ma). (B) Infected ma and three chromosomal type micronuclei (mi). (C) Detail of reproductive, infectious and transient forms, after ma crushing. (D) Positive signal of the probe GortProb659 (labeled with Cy3, emitting in red) inside host ma, during FISH experiment. Arrowheads indicate infectious forms, arrows reproductive forms and double arrowheads transient forms. Bars stand for 10 μm (A,D) and 5 μm (B,C).
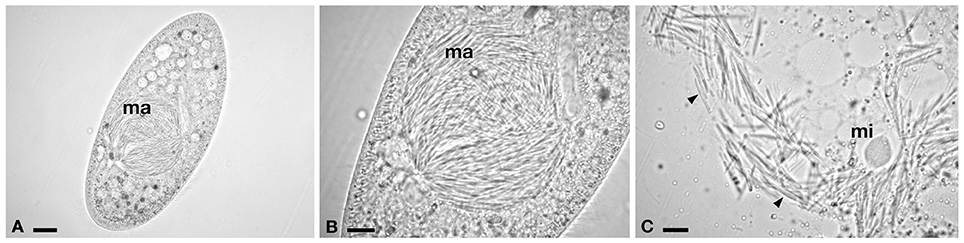
Figure 3. Light microscopy of infected P. caudatum SH2. (A) Whole cell with macronuclear infection. (B) Detail of hyperinfected macronucleus (ma). (C) Compact type micronucleus (mi) and rod-shaped bacteria, released after ma squashing. Arrowheads indicate infectious forms. Bars stand for 20 μm (A) and 10 μm (B,C).
Characterization of New HLB from P. multimicronucleatum TP2
Cells of P. multimicronucleatum TP2 exhibited 100% MA infection by rod shaped bacteria (Figure 1). A few weeks after isolation Paramecium cells manifested hyperinfected MAs, with dense clusters of bacteria (Figures 1A,B). The symbionts showed the typical morphology of HLB, with two different forms: the small RF (2.5–3.3 μm) and the larger, rod shaped IF, (6.9–10.7 μm) (Figure 1E). The IF were characterized by slightly tapered ends and by darker/reflecting parts in their bodies. In addition, some transient forms (TF)—large but not containing reflecting material—were present (Figure 1E). One year after isolation and subcloning, we repeated the analysis to check the infection status of TP2-2 and noticed that the distribution of the bacteria in the MA had changed: IF were clustered together in several small groups inside the nucleus (Figures 1F,G). IF and TF increased their length in comparison to previous observations, reaching 8.2–14.7 μm and 6.7–15.5 μm, respectively. In addition, the number of TF increased dramatically, whereas classical RF were fewer and/or less visible, compared to the first observations. We also observed IF dividing by binary fission (Figure 1K). Moreover, many IF showed an abnormal distribution of periplasmic parts inside the cell, forming a striated/dotted pattern (Figures 1J,K).
Quite surprisingly, we found these HLB not only in the MA but also evenly distributed in the cytoplasm, inside and outside food vacuoles (Figures 1H,I, 4G, 5D,F,G); using TEM and FISH techniques we detected large numbers of IFs and TFs, but also some RFs (Figures 4G, 5G). This phenomenon was observed during a random check 4 months after sampling and occurred in 10–20% of analyzed cells (n = 40). Due to the low number of cells in our culture, it was not possible to periodically check the status of infection and its progress, however, 1 year after sampling this unusual cytoplasmic distribution was present in 50% of the analyzed cells (n = 42).
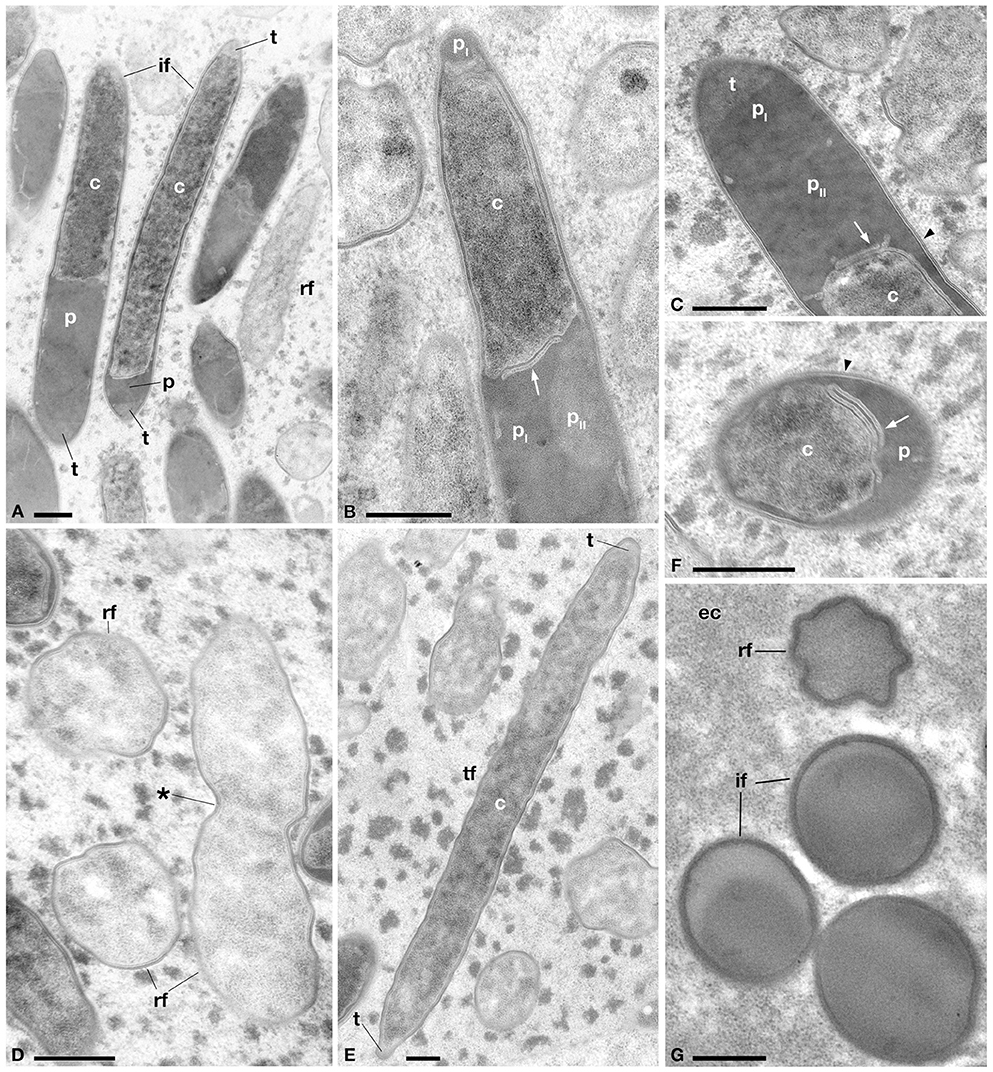
Figure 4. Ultrastructural morphology of the novel Holospora-like bacteria harbored by P. multimicronucleatum, after 1 year of cultivation. (A) Longitudinal section of reproductive (rf) and infectious (if) forms, the latter showing various degrees of cell compartmentalization in cytoplasm (c), periplasm (p) and recognition tip (t). (B) Detail of if showing cytoplasmic extrusion and periplasm with two electron-dense areas, a darker (pI) and a lighter (pII) one. (C) Detail of t, pI, and pII in if. (D) rf in transverse section and in longitudinal section during binary fission. (E) Transient form (tf) in longitudinal section with t at both ends of cellular body. (F) Transverse section of if showing irregular distribution of p around cytoplasmic part. (G) Transverse section of if and rf inside eukaryotic cytoplasm (ec) of host. Arrows indicate cytoplasmic extrusion in periplasmic space, arrowhead indicates the membrane layers surrounding IF. The asterisk indicates the cleavage furrow. Bars stand for 0.5 μm.
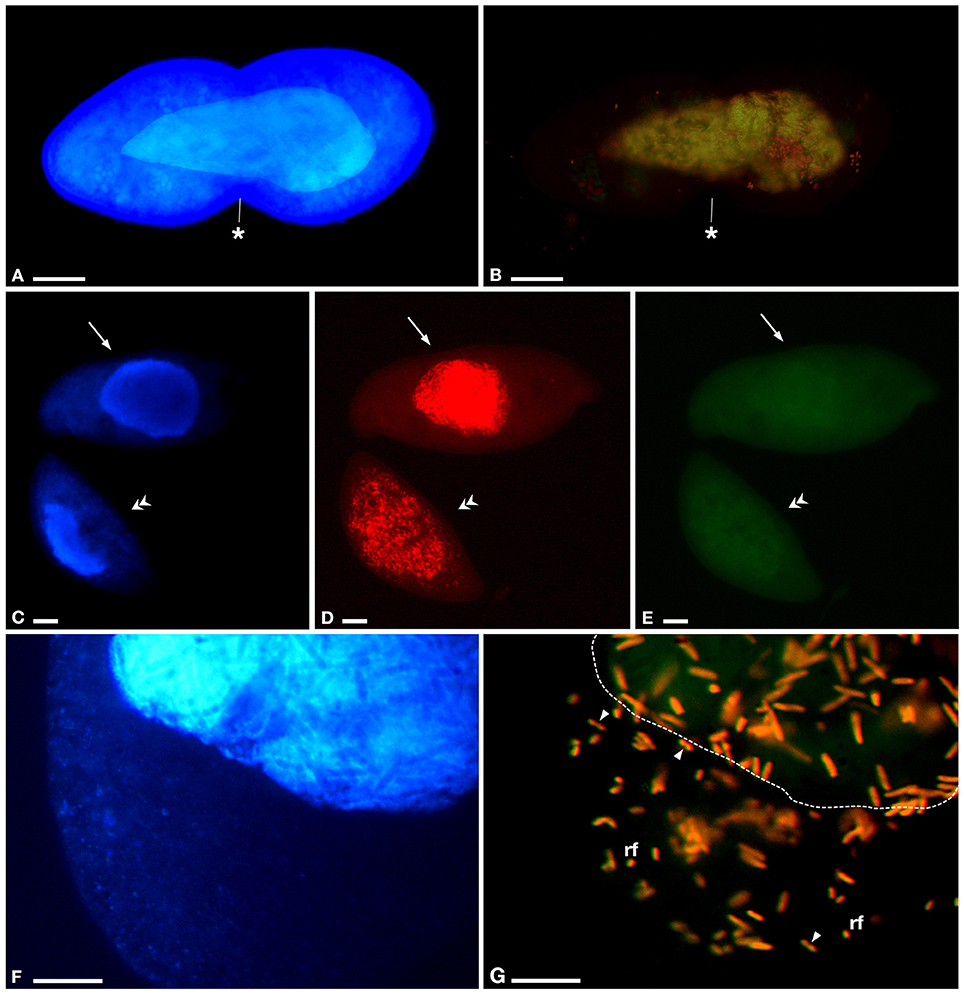
Figure 5. Results of fluorescence in situ hybridization (FISH) on P. multimicronucleatum TP2. Experiments were performed with different probe combinations: (A,B) GortProb659 (“Ca. Gortzia” genus-specific) and EUB338 (Bacteria), (C-E) GortzSha_1266 (“Ca. Gortzia shahrazadis” species-specific) and GortzInf_1268 (“Ca. Gortzia infectiva” species-specific), (F,G) GortzSha_1266 and EUB338. (A) Cell in division with infected macronucleus (ma), stained with 4,6-diamidino-2-phenylindole (DAPI), (B) positive to GortProb659 and EUB338 probes, completely overlapping. (C) Two cells fixed after 1year of culturing, showing intact ma stained with DAPI, (D) positive to GortzSha_1266, in ma (upper cell) and also in cytoplasm (lower cell), (E) same cells are negative to GortzInf_1268. (F) Closer view of cytoplasm and part of ma stained with DAPI of a P. multimicronucleatum cell infected by bacteria, (G) which are positive to GortzSha_1266 and EUB338 probes, with completely overlapped signals: reproductive forms (rf) and infectious forms well visible in host cytoplasm. Arrows indicate host cell with infection restricted to ma only, double arrowheads host cell with infection both in ma and in cytoplasm; arrowheads endosymbiont reproductive forms in cytoplasm during binary fission; dotted line position of ma inside host cell. The asterisks indicate the cleavage furrow. Bars stand for 20 μm (A–E) and 10 μm (F,G).
During host cell division, we never observed the “connecting piece” (Figures 1C,D, 5A,B). Endosymbionts were spread among daughter cells within MA pieces which, in many cases, were not equally shared in terms of size, suggesting that the bacteria interfere in host cell division.
During our cross-infection assay, we observed the presence of HLB inside the MA of P. multimicronucleatum PC6 48 h after the beginning of the experiment, showing the initial RF morphology observed in TP2 cells after a few weeks in cultivation (Figure 1L). From that moment, HLB cells in PC6 strain started to grow and differentiate, but after some days host cells started to decline and die.
Ultrastructural analysis of TP2 P. multimicronucleatum after 1 year of cultivation (Figure 4) supported live investigations using DIC microscope during the same period (Figures 1F–K). We were able to recognize different ultrastructural features characterizing the life-cycle stages described above: RF, IF, and TF. RF appeared short and roundish with the typical, homogeneous and relatively electron-transparent prokaryotic cytoplasm. The IF, longer and rod-shaped with slightly tapered ends, displayed differentiated cytoplasmic and periplasmic parts and a recognition tip-like structure in the apical part of the body (Figures 4A–C). In some cases, recognition tip-like structures were present at both ends of the cell (Figure 4A). The periplasmic regions of IF exhibited a very dense, osmiophilic pattern, while the recognition tip contained less osmiophilic material. The periplasm constituted a rather thin layer, often unevenly distributed beneath the outer cellular membrane of IF, sometimes forming outgrowths or sublayer invaginations in the cytoplasm (Figure 4F). We could discriminate at least two different periplasmic regions on the bases of their density: a darker, electron-dense part and a lighter one (Figures 4B,C). The IF cytoplasm was more heterogeneous and denser than the RF cytoplasm. Extensions of cytoplasm could be seen protruding into the periplasm in some cases (Figures 4B,C,F). Sometimes the same cell would manifest two or more stripes of dense periplasm, interposed with other cytoplasmic regions (probably forming the striped/dotted pattern previously observed via DIC microscopy). Another difference between IF and RF was their membrane composition: in the majority of IF the surface membrane was surrounded by fine fibrous material and, in some cases, manifests an additional membrane covering this fibrous-like layer (Figures 4C,F). TF appeared rod-shaped, with a size comparable to IF. The TF ultrastructure showed the presence of prokaryotic cytoplasm, without a distinctive periplasmic part (Figure 4E). The density of the TF cytoplasm resembles that observed in IF. Some TF manifest recognition tips at one or both ends of the body, as we observed in IF (Figure 4A). All forms showed a constant diameter of 0.7–0.8 μm.
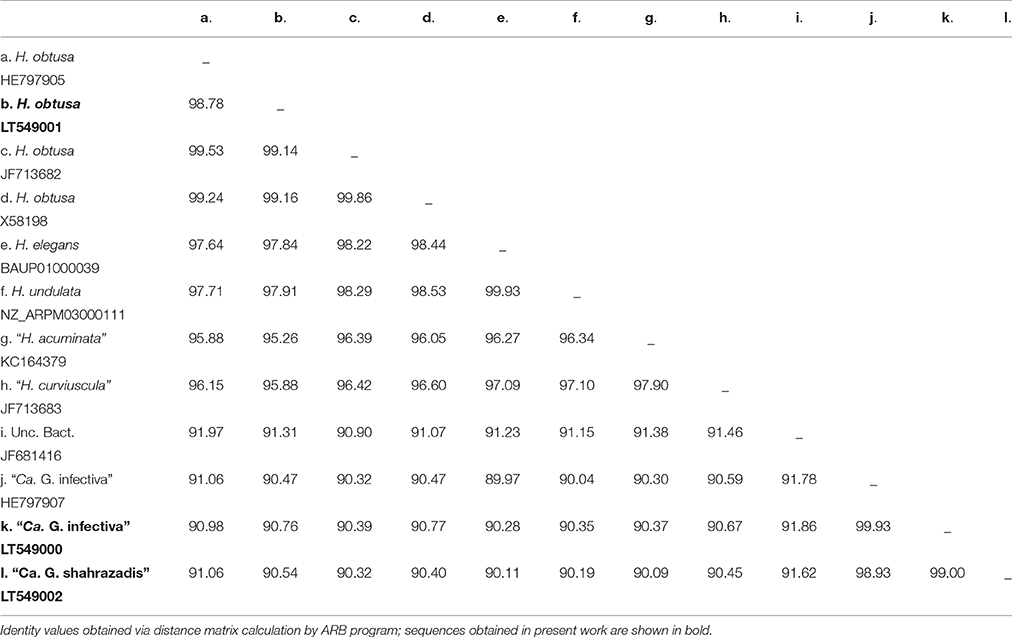
A 1398 bp long 16S rDNA sequence was obtained from this P. multimicronucleatum HLB and is available from ENA database under the accession number LT549002. From similarity matrix calculation we observed an identity value of 98.93% (15 mismatches) with “Ca. Gortzia infectiva” HE797907 and 90–91% with Holospora sequences (Table 2). In phylogenetic trees, this HLB species appeared strongly supported by statistical values as a member of the “Ca. Gortzia” clade (100/1.00), being a sister species of “Ca. Gortzia infectiva” (Figure 6).
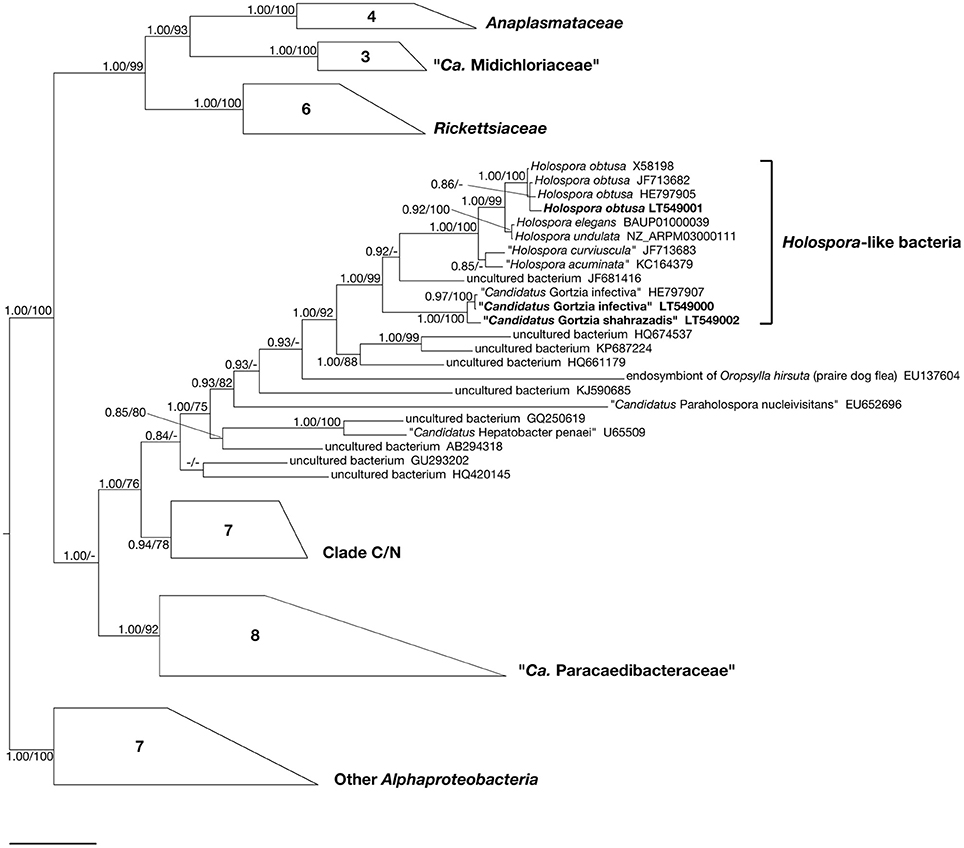
Figure 6. Bayesian inference tree of the order Rickettsiales based on 16S rDNA sequences. Numbers associated to nodes represent posterior probabilities and maximum likelihood bootstrap values, respectively (only values above 0.80–70 are shown). Numbers inside trapezoids correspond to sequences used to represent that clade. Sequences obtained in the present work are in bold characters. Bar stands for an estimated sequence divergence of 6%.
Preliminary FISH analyses suggested the presence of members of the “Ca. Gortzia” genus in P. multimicronucleatum TP2 (Figures 5A,B). In a double hybridization experiment with the newly designed species-specific probes GortzSha_1266 and GortzInf_1268, HLB from P. multimicronucleatum showed a positive signal only to GortzSha_1266 and not to the specific probe for “Ca. Gortzia infectiva” (Figures 5C–E). Moreover, FISH results were positive to GortzSha_1266 and EUB338 probes, with completely overlapping signals (Figures 5F,G), suggesting that the new HLB was the only bacterial species harbored by the TP2 cells. As detected from live observations, in many cases we found bacteria positive to the GortzSha_1266 probe not only in the MA but also in the cytoplasm (Figure 5G), with a homogeneous distribution. No broken MA were detected, which could have explained this unusual distribution.
These results confirmed both the presence of HLB corresponding to our 16S sequences inside the P. multimicronucleatum host and that it is possible to discriminate this bacterium from the already characterized “Ca. Gortzia infectiva”, using species-specific probes. The sequence of probe GortzSha_1266 matches no sequences in the RDP database, while GortzInf_1268 matches 4 sequences belonging to “Ca. Gortzia infectiva” clones. Sequences of these two new probes were deposited into probeBase database (Greuter et al., 2016).
“Ca. Gortzia infectiva” from India, Endosymbiont of P. jenningsi BJ1
Paramecium jenningsi BJ1 showed a “Ca. Gortzia infectiva”-related infection in the MA, from images taken of live specimens and from FISH results (Figure 2). The 16S rDNA sequence from this endosymbiont, 1432 bp long, showed 99.93% of identity (1 mismatch) with “Ca. Gortzia infectiva” HE797907 from Thailand, with which it forms a monophyletic species-level group, supported by strong statistical values (100/0.97) (Figure 6). The identity values with the other two newly characterized endosymbiont sequences, TP2 and SH2, were 99.00 and 90.76%, respectively (14 mismatches and 115 mismatches plus 18 indels, respectively) (Table 2). Our sequence is available from ENA database under the following accession number: LT549000, “Ca. Gortzia infectiva” BJ1.
Holospora obtusa from India, Endosymbiont of P. caudatum SH2
Live observations, supported by molecular analysis, confirmed the presence of H. obtusa in the MA of P. caudatum SH2 (Figure 3), from which we obtained a 16S rDNA sequence of 1434 bp, available on ENA database under the accession number LT549001, H. obtusa SH2. The identity was 98.78–99.16% with other H. obtusa sequences (HE797905, JF713682, X58198), (12 mismatches - 11 mismatches/2 indels) (Table 2). Although this H. obtusa from India showed a slightly higher divergence in comparison with its previously known conspecifics, from the phylogenetic analysis these four sequences clearly form a monophyletic clade (Figure 6), supported by high values of bootstrap and posterior probability (100/1.00).
Holospora obtusa was the sister group of the H. undulata (NZ_ARPM03000111) and H. elegans (BAUP01000039) clade. Together with “H. curviuscula” (JF713683) and “H. acuminata” (KC164379) and one uncultured bacterium (JF681416) they formed a monophyletic lineage, sister clade of the “Ca. Gortzia” genus. The sequences of H. undulata and H. elegans showed 99.93% identity, differing by only 1 bp.
Discussion
Novel HLB from P. multimicronucleatum TP2
In the present study, we describe a novel HLB, belonging to the “Ca. Gortzia” genus. It occurred in P. multimicronucletum, a Paramecium species in which HLB have never been documented (Fokin and Görtz, 2009). This endosymbiont shares many traits with the Holospora and “Ca. Gortzia” genera, such as the peculiar life-cycle involving two different stages (IF, RF), and ultrastructural internal compartmentalization of IF (Görtz and Wiemann, 1989; Wiemann and Görtz, 1989). Furthermore, it can be included in the group of “Ca. Gortzia infectiva”, H. caryophila, “H. bacillata”, and “H. curvata”, due to the absence of a “connecting piece” and the presence of several membrane layers surrounding IF. This particular feature, never observed in those Holospora species able to induce the “connecting piece”, is probably related to host-invasion or a releasing mechanism (Fokin et al., 1996; Fokin and Sabaneyeva, 1997; Fokin, 2015). Moreover, as observed in “H. bacillata” (Fokin, 1989), IF were able to divide by binary fission. On the other hand, we detected ultrastructural differences with other HLB (Schmidt et al., 1987; Görtz et al., 1990; Wiemann and Görtz, 1991): IF of the new bacterium showed a darker, less homogeneous cytoplasm, an unevenly distributed periplasm with two distinguishable regions, and a recognition tip that did not show the additional subdivisions detected in “Ca. Gortzia infectiva” (Boscaro et al., 2013a). The presence of two recognition tip-like structures in IF and TF has been interpreted as an early stage of division for IF (since IF can divide) or differentiation and division, in the case of TF.
Our analyses emphasizes additional and surprising features, such as the simultaneous and extensive presence of bacteria both in the host cytoplasm and MA: this pattern has never been reported for any species of Holospora nor for “Ca. Gortzia infectiva” (Fokin and Görtz, 2009; Boscaro et al., 2013a). The presence of IF in the cytoplasm has previously been reported and interpreted as a secondary infection of the host or as IF trafficking through the cytosol to exit the cell (Fokin and Sabaneyeva, 1997; Fokin, 2015). The presence of dividing RF in the cytoplasm (Figure 5G) suggest that in this case, unique in HLB, the symbiont is able to complete its life-cycle in the cytoplasm. Further investigation is needed to resolve this issue.
The only organism with a similar behavior among phylogenetically relatives is “Ca. Paraholospora nucleivisitans”, a cytoplasmatic endosymbiont of P. sexaurelia, but observed to enter the MA of its host (Eschbach et al., 2009); but it was rarely present in both cell compartments simultaneous. Furthermore, it showed different morphological features and low 16S rDNA identity (83.95%) with all HLB, indicating that this feature is not a shared derived character of the two organisms.
The other new characteristic of this novel HLB, as mentioned above, is the highly variable size and shape of IF and TF, observed after several months of culturing. IF, indeed, showed an abnormal pattern of growth (it almost doubled its length in some cases) and bacterial periplasmic distribution, when compared to the classical HLB morphology. We do not yet know the cause for these unusual variations in IF/TF shape. The overall supposition is that “aberrant” IFs faced some physiological problem in developing the classical morphology. Indeed, the increased number of TFs in the MA could be caused by an inability to exit this stage, with a consequent increase in size. When they were able to develop cellular differentiation, the periplasmic part has been produced in an unevenly pattern. In other words, these traits could be due to physiological stress, disease, or mutations that accumulate in vertically inherited bacteria, with a probable reduction in the efficiency of horizontal transfer, or due to changes in host/symbiont interactions occurred under laboratory growth conditions. The fact that the classical HLB morphology is observed in the experimentally infected PC6 strain could be due either to a positive selection for non-altered bacteria, still able to perform horizontal infection correctly, or to the capability of bacteria to restore the initial cell morphology in a different, unaltered, host environment. Although these are just hypothesis, the observed features can be used for taxonomic purposes. From a molecular point of view, this new HLB is phylogenetically close to “Ca. Gortzia infectiva” from P. jenningsi. Our results suggest that “Ca. Gortzia infectiva” and the new HLB coexist in the same geographical region and may have host specificity, a feature common in the sister genus Holospora.
To conclude, for this HLB we suggest the status of a novel species for the following reasons: 1) the different morphology and the striking phenotypic plasticity, never observed before in HLB; 2) the ability to infect both the MA and the cytoplasm; 3) the new host species in which it has been found, P. multimicronucleatum, given the fact that species-specificity between host and endosymbiont is well-known for most HLB; 4) the 16S rDNA distance from closely related species, which is compatible with threshold values proposed for new species description (Rosselló-Móra and Amann, 2015). For this new HLB species, belonging to the “Ca. Gortzia” genus, we propose the name “Candidatus Gortzia shahrazadis”, accordingly with Murray and colleagues (Murray and Schleifer, 1994; Murray and Stackebrandt, 1995). This specific name was chosen due to the Asian origin of this species and because one of the authors is known to say “I will finish this paper after one thousand and one nights.” A description of this new species is present at the end of the discussion.
“Ca. Gortzia infectiva” and Holospora obtusa from India
We detected HLB in an Indian Paramecium population. A first analysis of phenotypic characters and host specificity led to the species attributions “Ca. Gortzia infectiva” and H. obtusa, respectively, from P. jenningsi and P. caudatum. In both cases, the typical IF and RF were observed inside the host MA, with typical size and features described in the literature (Görtz et al., 1989; Boscaro et al., 2013a). Molecular analyses was performed on fixed material and, in both cases, results were congruent with live observations, confirming the species assignment, despite both Paramecium caudatum 18S rDNA sequence and its H. obusa 16S rDNA sequences showed some peculiarities when compared to sequences of conspecific organisms, suggesting a level of divergence. Phylogenetic analysis showed clear association of the Indian H. obtusa with its conspecifics sampled in other countries. Nevertheless, according to branch lengths, Indian H. obtusa SH2 showed a clear divergence from northern strains.
In our trees all HLB cluster together: Holospora clade appeared as sister group of the “Ca. Gortzia” genus. In general, our phylogeny of the order Rickettsiales is in agreement with previously published analyses (Boscaro et al., 2013a; Hess et al., 2016). Additionally, we note that the sequences belonging to H. undulata and H. elegans are very similar. While past analyses included a short partial sequence (479 bp) from H. elegans (AB297813) (e.g., Boscaro et al., 2013a), to our knowledge this is the first study reporting the full-length gene sequence of this species, derived from its genomic assembly (Dohra et al., 2014). On the basis of the present data and given the fact that both H. undulata and H. elegans have been found in MI of P. caudatum, H. undulata—which has an unusual undulated shape in comparison to other Holospora—could be a morphotype of the species H. elegans. Nevertheless, further investigation is needed, including analysis of a greater number of strains and comparison at the whole-genome level. To our knowledge, the several strains of H. undulata molecularly characterized by our and other laboratories are all identical to the presently published 16S rDNA sequences (data not shown), while for H. elegans only a single complete sequence is available (Dohra et al., 2014). In any case, since these two species were described in 1890 by Hafkine (Hafkine, 1890), it seems particularly important to clarify this issue.
Reconsideration about HLBs Distribution
Our data confirm the presence of HLB endosymbionts in Paramecium species inhabiting different and distant locations in India. P. multimicronucleatum (TP2) was collected from freshwater Kolleru lake in Andhra Pradesh; P. jenningsi (BJ1) was sampled in Chilka Lake, a brackish lagoon in Odisha; and P. caudatum (SH2) was found in a freshwater stream at the top of Araku Hills (Andhra Pradesh). All these findings strongly suggest a relatively common presence of HLB in Indian ciliate populations, adapted to different types of tropical habitats.
To date Holospora has been considered an endosymbiont adapted to cold and temperate countries. It has been demonstrated that transmission efficiency of H. undulata, for example, is strongly affected by temperature, being more effective at 10°C than at 23 and 30°C (Fels and Kaltz, 2006). Moreover, Holospora was never found in tropical habitats. For all these reasons, our data introduce new and interesting information about HLB's distribution, being the lowest latitude record for the Holospora genus reported (Figure 7; Table 3).
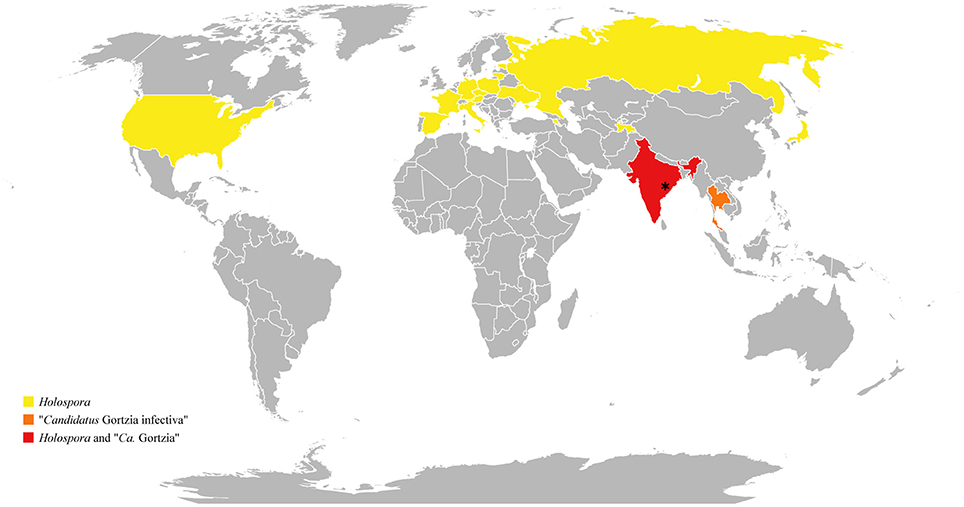
Figure 7. Holospora-like bacteria distribution. Yellow, orange, and red indicate countries in which Holospora-like bacteria have so far been detected, whereas a gray indicates those areas not yet investigated for their presence. Asterisk indicates sampling area of Holospora obtusa and “Ca. Gortzia” species characterized in the present work.
Nevertheless, the stream from which we sampled H. obtusa was located on the top of Araku hills, 700 m above the sea level, where climatic conditions were more temperate than those in the lowlands. We hypothesize that Holospora species are found in tropical countries, but in areas with more temperate environments, such as hills or mountains, generating a patchy distribution. The relatively high 16S rDNA divergence of this specific Holospora population could result from a small population size and possible genetic isolation due to patchy and unconnected distributions.
In contrast, members of “Ca. Gortzia” clade have been found exclusively in tropical areas at sea level (Boscaro et al., 2013a). These results lead to the conclusion that the “Ca. Gortzia” genus is more adapted to tropical climates than Holospora.
From an ecological perspective, our results address the possible function of HLB in Paramecium eco-physiology. Some have identified Holospora as true parasites (Görtz, 1988; Lohse et al., 2006), while others emphasize that those bacteria confer resistance to the host cell against challenging environmental conditions, such as salinity (Smurov and Fokin, 1998) or temperature variations (Hori and Fujishima, 2003). H. obtusa is able to enhance heat-shock gene expression of the host P. caudatum when exposed to increasing temperatures during laboratory experiments (Hori and Fujishima, 2003). Thus, it is possible that HLB confer a degree of benefit to their host if exposed to temperature stress, although this hypothesis is controversial (Duncan et al., 2010).
Obviously, another necessary consideration is that the biogeographic distribution of HLB is highly dependent on their hosts' distribution. The Paramecium genus is cosmopolitan, although some species seem to be adapted to certain climatic conditions. P. jenningsi, for example, has been found mostly in tropical countries, whereas P. caudatum is reported mainly from northern areas with temperate climates (Wichterman, 1986; Fokin, 2010/11). However, fewer sampling efforts in tropical countries strongly affects the present understanding of ciliate distribution, and thus the distribution of their endosymbionts (Görtz, 2008; Fokin and Sera, 2014).
To conclude, our record of H. obtusa from India reshapes the known Holospora distribution and underlines the need for further sampling and research efforts, to better clarify the ecological significance of HLB.
Emended Description of “Candidatus Gortzia” Genus (Boscaro et al., 2013a)
Gortzia (Gor'tzi.a; N.L. fem. n. Gortzia, in honor of Professor Emeritus Hans-Dieter Görtz). Gram-negative, Alphaproteobacteria, Rickettsiales, belonging to Holospora-like bacteria clade, together with Holospora genus and some other uncultured organisms. Macronuclear endosymbiont of Paramecium spp. (P. jenningsi, P. quadecaurelia, P. multimicronucleatum), and also cytoplasmatic in case of P. multimicronucleatum. “Ca. Gortzia”, has two different stages in its life-cycle: the short reproductive form (RF) and the elongated, rod-shaped infectious form (IF). IF showed cellular subcompartments: cytoplasm, periplasm, recognition tip. Periplasm and recognition tip could manifest parts with different electron-densities, according with species. No “connecting piece” or killer traits detected. The type species is “Ca. Gortzia infectiva” (Boscaro et al., 2013a). Only another species has been described: “Ca. Gortzia shahrazadis” (present work). Basis of assignment: positive matching with the 16S rRNA-targeting oligonucleotide genus-specific probe GortProb659 (5′-TTCCGTTTTCCTCTACCA-3′).
Description of “Candidatus Gortzia shahrazadis”
Gortzia shahrazadis (Gor'tzi.a shah.ra.za'dis; N.L. fem. n. Gortzia, in honor of Professor Emeritus Hans-Dieter Görtz; N.L. adj. shahrazadis, of Shahrazad, main character and charming story teller in the Arabian collection of tales, “The One Thousand and One Nights”). Mainly macronuclear endosymbiont which can sometimes be observed in the cytoplasm of P. multimicronucleatum, sampled from Kolleru, a freshwater lake in India. It has two life-cycle stages: a small, reproductive form (RF: 2.5–3.3 μm) and a rod-shaped infectious form (IF). The latter shows cellular compartmentalization: cytoplasm, periplasm (with two areas distinguishable by different density), and recognition tip. Two morphotypes of IF are detected during different periods of laboratory culturing: a shorter IF at the beginning (6.9–10.7 μm), and a longer one (8.2–14.7 μm) with irregular distribution of periplasm after 1 year of cultivation. The latter are found together with a high number of transient forms (TF), which are long (6.7–15.5 μm), rod-shaped and not yet differentiated into IF at an ultrastructural level. All forms showed a constant diameter of 0.7–0.8 μm. No production of a “connecting piece” during host cell division was observed. Several membranaceous layers surrounded fully differentiated IF. Basis of assignment: 16S rRNA gene sequence (ENA database, accession number LT549002) and positive matching with the 16S rRNA-targeting oligonucleotide probe GortzSha_1266 (5′-TTTTGATTTGCTCAAGGTCGC-3′).
Author Contributions
VS, SF, CB, and VN actively performed sampling in India. CB, VN, BS, CK organized and logistically supported sampling activity in India, VS carried out most of FISH experiments, molecular and phylogenetic analyses, probes and primers design. SF performed preliminary microscopic observation finding all the endosymbionts, morphologically identified the hosts, and performed TEM analysis. MC helped to realize the phylogenetic study. CB and VN performed part of the molecular characterization. SF, FV, BS, CK, and GP carefully supervised all the experiments and gave suggestions for data interpretation. VS wrote the manuscript. All the authors were involved in manuscript revision. GP coordinated the whole research activity.
Funding
This work was supported by the European Commission FP7-PEOPLE-2009-IRSES project CINAR PATHOBACTER (247658) and by the PRIN fellowship (protocol 2012A4F828_002) from the Italian Research Ministry (MIUR), and by the European Commission FP7 post grant Open Access Pilot Open AIRE.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the Marine Biology Laboratory, Andhra University, India for providing the research facilities. Prof. Raman Akkur and Prof. Prabhakara Rao Yallapragada are acknowledged for their valuable support. Special thanks to Ms. Natalia Lebedeva and to Jr. Prof. Martina Schrallhammer for their kind support in the reconstruction of Holospora distribution, to Margherita Tagliavia and Tom Doak for English revision. Simone Gabrielli is acknowledged for his patient and precious assistance in tree and photo editing.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01704/full#supplementary-material
References
Amann, R. I., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925.
Borchsenius, O. N., Skoblo, I. I., and Ossipov, D. V. (1983). Holospora curviuscula- A new species of macronuclear symbiotic bacteria of Paramecium bursaria. Tsitologiya 25, 91–97.
Boscaro, V., Fokin, S. I., Schrallhammer, M., Schweikert, M., and Petroni, G. (2013a). Revised systematics of Holospora-like bacteria and characterization of “Candidatus Gortzia infectiva,” a novel macronuclear symbiont of Paramecium jenningsi. Microb. Ecol. 65, 255–267. doi: 10.1007/s00248-012-0110-2
Boscaro, V., Petroni, G., Ristori, A., Verni, F., and Vannini, C. (2013b). “Candidatus Defluviella procrastinata” and “Candidatus Cyrtobacter zanobii,” two novel ciliate endosymbionts belonging to the “Midichloria clade.” Microb. Ecol. 65, 302–310. doi: 10.1007/s00248-012-0170-3
Boscaro, V., Schrallhammer, M., Benken, K. A., Krenek, S., Szokoli, F., Berendonk, T. U., et al. (2013c). Rediscovering the genus Lyticum, multiflagellated symbionts of the order Rickettsiales. Sci. Rep. 3:3305. doi: 10.1038/srep03305
Boscaro, V., Vannini, C., Fokin, S. I., Verni, F., and Petroni, G. (2012). Characterization of “Candidatus Nebulobacter yamunensis” from the cytoplasm of Euplotes aediculatus (Ciliophora, Spirotrichea) and emended description of the family Francisellaceae. Syst. Appl. Microbiol. 35, 432–440. doi: 10.1016/j.syapm.2012.07.003
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic. Acids. Res. 37, 141–145. doi: 10.1093/nar/gkn879
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
Dohra, H., Tanaka, K., Suzuki, T., Fujishima, M., and Suzuki, H. (2014). Draft genome sequences of three Holospora species (Holospora obtusa, Holospora undulata, and Holospora elegans), endonuclear symbiotic bacteria of the ciliate Paramecium caudatum. FEMS Microbiol. Lett. 359, 16–18. doi: 10.1111/1574-6968.12577
Duncan, A. B., Fellous, S., Accot, R., Alart, M., Sobandi, K. C., Cosiaux, A., et al. (2010). Parasite-mediated protection against osmotic stress for Paramecium caudatum infected by Holospora undulata is host genotype specific. FEMS Microbiol. Ecol. 74, 353–360. doi: 10.1111/j.1574-6941.2010.00952.x
Duncan, A. B., Gonzalez, A., and Kaltz, O. (2015). Dispersal, environmental forcing, and parasites combine to affect metapopulation synchrony and stability. Ecology 96, 284–290. doi: 10.1890/14-0137.1
Eschbach, E., Pfannkuchen, M., Schweikert, M., Drutschmann, D., Brümmer, F., Fokin, S. I., et al. (2009). “Candidatus Paraholospora nucleivisitans,” an intracellular bacterium in Paramecium sexaurelia shuttles between the cytoplasm and the nucleus of its host. Syst. Appl. Microbiol. 32, 490–500. doi: 10.1016/j.syapm.2009.07.004
Fellous, S., Quillery, E., Duncan, A. B., and Kaltz, O. (2011). Parasitic infection reduces dispersal of ciliate host. Biol. Lett. 7, 327–329. doi: 10.1098/rsbl.2010.0862
Fels, D., and Kaltz, O. (2006). Temperature-dependent transmission and latency of Holospora undulata, a micronucleus-specific parasite of the ciliate Paramecium caudatum. Proc. Biol. Sci. 273, 1031–1038. doi: 10.1098/rspb.2005.3404
Ferrantini, F., Fokin, S. I., Modeo, L., Andreoli, I., Dini, F., Görtz, H. D., et al. (2009). “Candidatus Cryptoprodotis polytropus,” a novel Rickettsia-like organism in the ciliated protist Pseudomicrothorax dubius (Ciliophora, Nassophorea). J. Eukaryot. Microbiol. 56, 119–129. doi: 10.1111/j.1550-7408.2008.00377.x
Ferrantini, F., Fokin, S. I., Vannini, C., Verni, F., and Petroni, G. (2007). Characterization of a novel Holospora-like symbiont from Frontonia (Ciliophora, Oligohymenophorea). J. Eukaryot. Microbiol. 54:75a.
Fiveiskaja, A. (1928). Einfluss der Kernparasiten der Infusorien auf den Stoffwechsel. Arch. Protistenkd. 65, 75–298.
Fokin, S. I. (1989). Bacterial endobionts of the ciliate Paramecium woodruffi. I. Endobionts of the macronucleus. Tsitologiya 31, 839–844.
Fokin, S. I. (1991). Holospora recta sp. nov. a micronucleus-specific endobiont of the ciliate Paramecium caudatum. Tsitologiya 33, 135–141.
Fokin, S. I. (1993). Bacterial endobionts of ciliates and their employment in experimental protozoology. Tsitologiya 35, 59–91.
Fokin, S. I. (1997). Morphological diversity of the micronuclei in Paramecium. Arch. Protistenkd. 148, 375–387. doi: 10.1016/S0003-9365(97)80017-6
Fokin, S. I. (2000). Host specificity of Holospora and its relationships with Paramecium phylogeny. Jpn. J. Protozool. 33, 94.
Fokin, S. I. (2004a). Bacterial endocytobionts of Ciliophora and their interactions with the host cell. Int. Rev. Cytol. 236, 181–249. doi: 10.1016/S0074-7696(04)36005-5
Fokin, S. I. (2004b). Some bacterial infections of Paramecium. Occurrence and dynamics. Abstr 23 An Meet DGP. Ber. Nat. Med. Ver. 27.
Fokin, S. I. (2010/11). Paramecium genus: biodiversity, some morphological features the key to the main morphospecies discrimination. Protistology 6, 227–235.
Fokin, S. I. (2012). Frequency and biodiversity of symbionts in representatives of the main classes of Ciliophora. Eur. J. Protistol. 48, 138–148. doi: 10.1016/j.ejop.2011.12.001
Fokin, S. I. (2015). “Release of Holospora-like bacteria in different ciliate species,” in 7th ECOP-ISOP Joint Meeting (Sevilla), 279.
Fokin, S. I., Brigge, T., Brenner, J., and Görtz, H. D. (1996). Holospora species infecting the nuclei of Paramecium appear to belong into two groups of bacteria. Eur. J. Protistol. 32, 19–24. doi: 10.1016/S0932-4739(96)80072-1
Fokin, S. I., Brigge, T., and Görtz, H. D. (1999). An infectious bacterium inhabiting the macronucleus of Paramecium putrinum. J. Euk. Microbiol. 46, 11a.
Fokin, S. I., and Chivilev, S. M. (2000). Paramecium morphometric analysis and taxonomy. Acta Protozool. 39, 1–14.
Fokin, S. I., and Görtz, H. D. (2009). “Diversity of Holospora bacteria in Paramecium and their characterization,” in Endosymbionts in Paramecium, ed M. Fujishima (Münster: Microbiology Monograph), 161–199.
Fokin, S. I., Przyboś, E., Chivilev, S. M., Beier, C. L., Horn, M., Skotarczak, D., et al. (2004). Morphological and molecular investigations of Paramecium schewiakoffi sp. nov. (Ciliophora, Oligohymenophorea) and current status of taxonomy. Eur. J. Protistol. 40, 225–243. doi: 10.1016/j.ejop.2004.02.001
Fokin, S. I., and Sabaneyeva, E. V. (1997). Release of endonucleobiotic bacteria Holospora bacillata and Holospora curvata from the macronucleus of their host cells Paramecium woodruffi and Paramecium calkinsi. Endocyt. Cell. Res. 12, 49–55.
Fokin, S. I., Schrallhammer, M., Vanini, C., Ferrantini, F., Petroni, G., and Görtz, H. D. (2006). “New Holospora endocytobionts in some common ciliates,” in 25th Wiss Tag Deut Gesel Protozool Liebenwalde (Berlin), 28.
Fokin, S. I., and Sera, V. (2014). The hidden biodiversity of ciliate-endosymbionts systems. JSM Microbiol. 2, 1015.
Fokin, S. I., Skovorodkin, I. N., and Sabaneyeva, E. V. (2003). Initial steps of infection of the ciliate Paramecium by bacteria Holospora spp. Tsitologiya 45, 1127–1233.
Fokin, S., and Sabaneyeva, E. V. (1993). Bacterial endocytobionts of the ciliate Paramecium calkinsi. Eur. J. Protistol. 29, 390–395. doi: 10.1016/S0932-4739(11)80401-3
Fujishima, M. (2009a). Endosymbionts in Paramecium, ed M. Fujishima (Münster: Microbiology Monograph).
Fujishima, M. (2009b). “Infection and maintenance of Holospora species in Paramecium caudatum,” in Endosymbionts in Paramecium, ed M. Fujishima (Münster: Microbiology Monograph), 201–225.
Fujishima, M., and Görtz, H. D. (1983). Infection of macronuclear anlagen of Paramecium caudatum with the macronucleus-specific symbiont Holospora obtusa. J. Cell Sci. 64, 137–146.
Fujishima, M., Sawabe, H., and Iwatsuki, K. (1990). Scanning electron microscopic observation of differentiation from the reproductive short form to the infectious long form of Holospora obtusa. J. Protozool. 37, 123–128. doi: 10.1111/j.1550-7408.1990.tb05881.x
Gong, J., Qing, Y., Guo, X., and Warren, A. (2014). “Candidatus Sonnebornia yantaiensis,” a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea). Syst. Apll. Microbiol. 37, 35–41. doi: 10.1016/j.syapm.2013.08.007
Görtz, H. D. (1987). Different endocytobionts simultaneously colonizing ciliate cells. Ann. N. Y. Acad. Sci. 503, 261–268. doi: 10.1111/j.1749-6632.1987.tb40613.x
Görtz, H. D. (1988). “Endocytobiosis,” in Paramecium, ed H. D. Görtz (Berlin, Heidelberg: Springer Verlag), 393–405.
Görtz, H. D. (1996). “Symbiosis in ciliates,” in Ciliates. Cells as organisms, eds K. Hausmann and P. S. Bradbury (Stuttgart: Fisher), 441–462.
Görtz, H. D. (2006). “Symbiotic associations between ciliates and prokaryotes,” in The Prokaryotes, eds M. Dworki, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 364–402.
Görtz, H. D. (2008). Towards an understanding of the distribution, dynamics and ecological significance of bacterial symbioses in protists. Denisia 23, 307–311.
Görtz, H. D., Ahlers, N., and Robenek, H. (1989). Ultrastructure of the infectious and reproductive forms of Holospora obtusa, a bacterium infecting the macronucleus of Paramecium caudatum. J. Gen. Microbiol. 135, 3079–3085. doi: 10.1099/00221287-135-11-3079
Görtz, H. D., and Diekmann, J. (1980). life-cycle and infectivity of Holospora elegans Hafkine, a micronucleus-specific symbiont of Paramecium caudatum (Ehrenberg). Protistologica 16, 591–603.
Görtz, H. D., Lellig, S., Miosga, O., and Wiemann, M. (1990). Changes in fine structure and polypeptide pattern during development of Holospora obtusa, a bacterium infecting the macronucleus of Paramecium caudatum. J. Bacteriol. 172, 5664–5669.
Görtz, H. D., and Schmidt, H. J. (2005). “Holosporaceae fam. nov.,” in Bergey's Manual of Systematic Bacteriology, Vol. 2, Part C, 2nd Edn., eds G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (New York, NY: Springer), 146–160.
Görtz, H. D., and Wiemann, M. (1989). Route of infection of the bacteria Holospora elegans and Holospora obtusa into the nuclei of Paramecium caudatum. Eur. J. Protistol. 24, 101–109. doi: 10.1016/S0932-4739(89)80037-9
Greuter, D., Loy, A., Horn, M., and Rattei, T. (2016). probeBase—an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res. 44, D586–D589. doi: 10.1093/nar/gkv1232
Gromov, B. V., and Ossipov, D. V. (1981). Holospora (ex Hafkine 1890) nom. rev., a genus of bacteria inhabiting the nuclei of paramecia. Int. J. Syst. Bacteriol. 31, 348–352. doi: 10.1099/00207713-31-3-348
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Heckmann, K., and Schmidt, H. J. (1987). Polynucleobacter necessarius gen nov, sp nov, an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int. J. Syst. Bacteriol. 37, 456–457. doi: 10.1099/00207713-37-4-456
Hess, S., Suthaus, A., and Melkonian, M. (2016). Characterisation of “Candidatus Finniella” (Rickettsiales, Alphaproteobacteria), novel endosymbionts of Viridiraptorid amoeboflagellates (Cercozoa, Rhizaria). Appl. Environ. Microbiol. 82, 659–670. doi: 10.1128/AEM.02680-15
Hori, M., and Fujishima, M. (2003). The endosymbiotic bacterium Holospora obtusa enhances heat-shock gene expression of the Paramecium caudatum. J. Eukaryot. Microbiol. 50, 293–298. doi: 10.1111/j.1550-7408.2003.tb00137.x
Irbis, C., and Ushida, K. (2004). Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50, 203–212. doi: 10.2323/jgam.50.203
Kreutz, M., and Foissner, W. (2006). The Sphagnum ponds of Simmelried in Germany: a biodiversity hot-spot for microscopic organisms. Protozool. Monogr. 3, 1–267.
Kusch, J., Czubatinsky, L., Wegmann, S., Hübner, M., Alter, M., and Albrecht, P. (2002). Competitive advantages of Caedibacter infected paramecia. Protist 153, 47–58. doi: 10.1078/1434-4610-00082
Lohse, K., Gutierrez, A., and Kaltz, O. (2006). Experimental evolution of resistance in Paramecium caudatum against the bacterial parasite Holospora undulata. Evolution 60, 1177–1186. doi: 10.1111/j.0014-3820.2006.tb01196.x
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Kumar, Y., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Murray, R. G., and Schleifer, K. H. (1994). A proposal for recording the properties of putative taxa of procaryotes. Int. J. Syst. Bacteriol. 44, 174–176. doi: 10.1099/00207713-44-1-174
Murray, R. G. E., and Stackebrandt, E. (1995). Implementation of the provisional status Candidatus for incompletely described prokaryotes. Int. J. Syst. Bacteriol. 45, 186–187. doi: 10.1099/00207713-45-1-186
Ossipov, D. V. (1973). Specific infectious specificity of the omega-particles, micronuclear symbiotic bacteria of Paramecium caudatum. Cytology 15, 211–217.
Ossipov, D. V., Skoblo, I. I., Borchsenius, O. N., and Rautian, M. S. (1980). Holospora acuminata – a new species of symbiotic bacterium from the micronucleus of the ciliate Paramecium bursaria Focke. Tsitologiya 22, 922–929.
Petroni, G., Dini, F., Verni, F., and Rosati, G. (2002). A molecular approach to the tangled intrageneric relationships underlying phylogeny in Euplotes (Ciliophora, Spirotrichea). Mol. Phylogenet. Evol. 22, 118–130. doi: 10.1006/mpev.2001.1030
Preer, J. R., and Preer, L. B. (1982). Revival of names of protozoan endosymbionts and proposal of Holospora caryophila nom. nov. Int. J. Syst. Bacteriol. 32, 140–141. doi: 10.1099/00207713-32-1-140
Preer, J. R., Preer, L. B., and Jurand, A. (1974). Kappa and other endosymbionts in Paramecium aurelia. Bacteriol. Rev. 38, 113.
Preer, L. B. (1969). Alpha, an infectious macronuclear symbiont of Paramecium aurelia. J. Protozool. 16, 570–578. doi: 10.1111/j.1550-7408.1969.tb02315.x
Przyboś, E., and Fokin, S. I. (2000). Data on the occurrence of species of the Paramecium aurelia complex world-wide. protistology 1, 179–184.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rautian, M. S., Beliavskaia, A., Kiselev, A., Logacheva, M., Garushyants, S., and Gelfand, M. (2015). “New intracellular symbionts of paramecia,” in 7th ECOP-ISOP Joint Meeting (Sevilla), 184.
Rautian, M. S., and Wackerow-Kouzova, N. D. (2013). Phylogenetic placement of two previously described intranuclear bacteria from the ciliate Paramecium bursaria (Protozoa, Ciliophora): “Holospora acuminata” and “Holospora curviuscula.” Int. J. Syst. Evol. Microbiol. 63, 1930–1933. doi: 10.1099/ijs.0.046631-0
Rinke, C., Schmitz-Esser, S., Stoecker, K., Nussbaumer, A. D., Molnár, D. A., Vanura, K., et al. (2006). “Candidatus Thiobios zoothamnicoli,” an ectosymbiotic bacterium covering the giant marine ciliate Zoothamnium niveum. Appl. Environ. Microbiol. 72, 2014–2021. doi: 10.1128/AEM.72.3.2014-2021.2006
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, D., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rosati, G., Modeo, L., Melai, M, Petroni, G., and Verni, F. (2004). A multidisciplinary approach to describe protists: a morphological, ultrastructural, and molecular study on Peritromus kahli Villeneuve-Brachon, 1940 (Ciliophora, Heterotrichea). J. Eukaryot. Microbiol. 51, 49–59. doi: 10.1111/j.1550-7408.2004.tb00160.x
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Sabaneyeva, E. V., Derkacheva, M. E., Benken, K. A., Fokin, S. I., Vainio, S., and Skovorodkin, I. N. (2009). Actin-based mechanism of Holospora obtusa trafficking in Paramecium caudatum. Protist 160, 205–219. doi: 10.1016/j.protis.2008.11.006
Sabaneyeva, E. V., Fokin, S. I., and Gavrilova, E. V, Kornilova, E. S. (2005). Nocodazole inhibits macronuclear infection with Holospora obtusa in Paramecium caudatum. Protoplasma 226, 147–153. doi: 10.1007/s00709-005-0121-7
Schmidt, H. J., Freiburg, M., and Görtz, H. D. (1987). Comparison of the infectious forms of two bacterial endonucleobionts, Holospora elegans and H. obtusa, from the ciliate Paramecium caudatum. Microbios 49, 189–197.
Schrallhammer, M., Ferrantini, F., Vannini, C., Galati, S., Schweikert, M., Görtz, H. D., et al. (2013). “Candidatus Megaira polyxenophila” gen nov, sp nov: considerations on evolutionary history, host range and shift of early divergent Rickettsiae. PLoS ONE 8:e72581. doi: 10.1371/journal.pone.0072581
Schrallhammer, M., Fokin, S. I., Schleifer, K. H., and Petroni, G. (2006). Molecular characterization of the obligate endosymbiont “Caedibacter macronucleorum” Fokin and Görtz, 1993 and of its Host Paramecium duboscqui Strain Ku4-8. J. Eukaryot. Microbiol. 53, 499–506. doi: 10.1111/j.1550-7408.2006.00133.x
Schrallhammer, M., Schweikert, M., Vallesi, A., Verni, F., and Petroni, G. (2011). Detection of a novel subspecies of Francisella noatunensis as endosymbiont of the ciliate Euplotes raikovi. Microb. Ecol. 61, 455–464. doi: 10.1007/s00248-010-9772-9
Senra, M. V., Dias, R. J., Castelli, M., Silva-Neto, I. D., Verni, F., Soares, C. A., et al. (2015). A house for two - double bacterial infection in Euplotes woodruffi Sq1 (Ciliophora, Euplotia) sampled in southeastern Brazil. Microb. Ecol. 71, 505–517. doi: 10.1007/s00248-015-0668-6
Skoblo, I. I., and Lebedeva, N. A. (1986). Infection of the nuclear apparatus of Paramecium bursaria (Ciliata) by the symbiotic bacterium Holospora curviuscula. Tsitologiya 28, 367–372.
Smurov, A. O., and Fokin, S. I. (1998). Resistance of Paramecium caudatum infected with endonuclear bacteria Holospora against salinity impact. Proc. Zool. Inst. RAN 276, 175–178.
Strüder-Kypke, M. C., Wright, A. D. G., Fokin, S. I., and Lynn, D. H. (2000). Phylogenetic relationships of the subclass Peniculia (Oligohymenophorea, Ciliophora) inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 47, 419–429. doi: 10.1111/j.1550-7408.2000.tb00069.x
Szokoli, F., Sabaneyeva, E. V., Castelli, M., Krenek, S., Schrallhammer, M., Soares, C. A., et al. (2016). “Candidatus Fokinia solitaria,” a novel “stand-alone” symbiotic lineage of Midichloriaceae (Rickettsiales). PLoS ONE 11:e0145743. doi: 10.1371/journal.pone.0145743
Vakkerov-Kouzova, N. D., and Rautian, M. S. (2011). Obtaining and characterization of “Holospora curviuscula” and Holospora obtusa, bacterial symbionts of the macronuclei of Paramecium bursaria and Paramecium caudatum. Microbiology 80, 728–732. doi: 10.1134/S0026261711050171
Vannini, C., Boscaro, V., Ferrantini, F., Benken, K. A., Mironov, T. I., Schweikert, M., et al. (2014). Flagellar movement in two bacteria of the family Rickettsiaceae: a re-evaluation of motility in an evolutionary perspective. PLoS ONE 9:e87718. doi: 10.1371/journal.pone.0087718
Vannini, C., Ferrantini, F., Schleifer, K. H., Ludwig, W., Verni, F., and Petroni, G. (2010). “Candidatus Anadelfobacter veles” and “Candidatus Cyrtobacter comes,” two new Rickettsiales species hosted by the protist ciliate Euplotes harpa (Ciliophora, Spirotrichea). Appl. Environ. Microbiol. 76, 4047–4054. doi: 10.1128/AEM.03105-09
Vannini, C., Lucchesi, S., and Rosati, G. (2007). Polynucleobacter: symbiotic bacteria in ciliates compensate for a genetic disorder in glycogenolysis. Symbiosis 44, 85–91.
Vannini, C., Petroni, G., Schena, A., Verni, F., and Rosati, G. (2003). Well-established mutualistic associations between ciliates and prokaryotes might be more widespread and diversified than so far supposed. Europ. J. Protistol. 39, 481–485. doi: 10.1078/0932-4739-00024
Vannini, C., Rosati, G., Verni, F., and Petroni, G. (2004). Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of “Candidatus Devosia euplotis.” Int. J. Syst. Evol. Microbiol. 54, 1151–1156. doi: 10.1099/ijs.0.02759-0
Wiemann, M., and Görtz, H. D. (1989). Release of the endonucleobiotic bacterium Holospora elegans from its host cell Paramecium caudatum. Europ. J. Protistol. 25, 100–108. doi: 10.1016/S0932-4739(89)80021-5
Keywords: Gortzia, Holospora, bacteria, Paramecium, endosymbiosis, distribution, India
Citation: Serra V, Fokin SI, Castelli M, Basuri CK, Nitla V, Verni F, Sandeep BV, Kalavati C and Petroni G (2016) “Candidatus Gortzia shahrazadis”, a Novel Endosymbiont of Paramecium multimicronucleatum and a Revision of the Biogeographical Distribution of Holospora-Like Bacteria. Front. Microbiol. 7:1704. doi: 10.3389/fmicb.2016.01704
Received: 13 May 2016; Accepted: 12 October 2016;
Published: 04 November 2016.
Edited by:
David Berry, University of Vienna, AustriaReviewed by:
Joan M. Bernhard, Woods Hole Oceanographic Institution, USAIlya Skovorodkin, Biocenter Oulu, Finland
Copyright © 2016 Serra, Fokin, Castelli, Basuri, Nitla, Verni, Sandeep, Kalavati and Petroni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulio Petroni, giulio.petroni@unipi.it
 Valentina Serra
Valentina Serra Sergei I. Fokin1,2
Sergei I. Fokin1,2 Michele Castelli
Michele Castelli Charan K. Basuri
Charan K. Basuri Giulio Petroni
Giulio Petroni