- 1Laboratory of Microbiology, Wageningen University, Wageningen, Netherlands
- 2Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Hørsholm, Denmark
- 3National Institute for Public Health and the Environment, Bilthoven, Netherlands
Wide application of antibiotics has contributed to the evolution of multi-drug resistant human pathogens, resulting in poorer treatment outcomes for infections. In the marine environment, seawater samples have been investigated as a resistance reservoir; however, no studies have methodically examined sponges as a reservoir of antibiotic resistance. Sponges could be important in this respect because they often contain diverse microbial communities that have the capacity to produce bioactive metabolites. Here, we applied functional metagenomics to study the presence and diversity of functional resistance genes in the sponges Aplysina aerophoba, Petrosia ficiformis, and Corticium candelabrum. We obtained 37 insert sequences facilitating resistance to D-cycloserine (n = 6), gentamicin (n = 1), amikacin (n = 7), trimethoprim (n = 17), chloramphenicol (n = 1), rifampicin (n = 2) and ampicillin (n = 3). Fifteen of 37 inserts harbored resistance genes that shared <90% amino acid identity with known gene products, whereas on 13 inserts no resistance gene could be identified with high confidence, in which case we predicted resistance to be mainly mediated by antibiotic efflux. One marine-specific ampicillin-resistance-conferring β-lactamase was identified in the genus Pseudovibrio with 41% global amino acid identity to the closest β-lactamase with demonstrated functionality, and subsequently classified into a new family termed PSV. Taken together, our results show that sponge microbiota host diverse and novel resistance genes that may be harnessed by phylogenetically distinct bacteria.
Introduction
In the last decades the massive medical and veterinary use of antibiotics has contributed to the selection of multi-drug resistant human pathogens, resulting in poorer treatment outcomes upon infection (Arias and Murray, 2009). Bacterial pathogens can evolve resistance vertically (Lenski, 1998) but mainly acquire resistance through horizontal gene transfer (Högberg et al., 2010), where resistance genes are obtained either from the indigenous human microbiota or from environmental microorganisms to which an individual is exposed (e.g., via food, water or soil). The current complement of resistance genes encountered in the human microbiota is suggested to have originated from natural environments, likely in part selected by prior anthropogenic antibiotic pressure (Teuber et al., 1999; Davies and Davies, 2010; Forslund et al., 2013). However, microbiota from isolated human populations have also been found to carry functional antibiotic resistance (AR) genes (Clemente et al., 2015). Soil has been implicated to be a key environmental reservoir of AR genes, as evidenced by the fact that AR genes with perfect nucleotide identity to those in human pathogens have been described in soil-dwelling bacteria (Forsberg et al., 2012).
In addition to soil, resistance genes have been found in a broad range of environments that contain complex microbial communities such as activated sludge (Mori et al., 2008; Munck et al., 2015), caves (Bhullar et al., 2012), glaciers (Segawa et al., 2013), rivers (Amos et al., 2014) and animals (Alves et al., 2014; Wichmann et al., 2014). As a result, all these environments, and probably many more, can act as potential reservoirs for resistance gene dissemination. It has been shown that AR is a natural phenomenon that predates the modern selective pressure of clinical antibiotic use (D'Costa et al., 2011), although little is known about the exact roles of these genes in their native environments (Sengupta et al., 2013). The marine environment is a major genetic reservoir of AR, which despite its tremendous bacterial diversity has been little studied in the context of AR gene dissemination (Yang et al., 2013). A recent study has identified a range of unclassified resistance genes in ocean water, thereby highlighting its importance as an environmental reservoir (Hatosy and Martiny, 2015).
Marine sponges are an ancient lineage of sessile filter-feeding metazoans. Many sponges harbor a dense and diverse microbiota comprising up to 40% of the total sponge volume, and as such, house a complex ecosystem characterized by host-microbe and microbe-microbe interactions (Webster and Taylor, 2012). Much sponge research has focused on the sponge microbiota as a source of novel bioactive compounds (Zhang et al., 2005), whereas its role as a reservoir of AR genes has received limited attention. Still, the fact that sponge microbiota can produce diverse antimicrobials (Laport et al., 2009; Mehbub et al., 2014) would suggest the presence of AR genes. An example of antimicrobials produced by sponge bacteria are rifampicin antibiotics, which at first were only known to be produced by soil Actinobacteria (August et al., 1998; Schupp et al., 1998), but more recently have also been found to be produced by a sponge-associated Actinobacterium sp. (Kim et al., 2006).
To date, research on functional AR genes in sponge microbiota has been limited to a Bacillus sp. isolated from the sponge Haliclona simulans. The Bacillus sp. was found to contain two small plasmids, one of which harbored the tetL tetracycline resistance gene (Phelan et al., 2011) whereas the other harbored the ermT erythromycin resistance gene (Barbosa et al., 2014). The plasmid with the tetL gene was shown to be nearly identical to three other tetracycline resistance plasmids identified in the honey bee pathogen Paenibacilus larvae pMA67, in the cheese-resident Lactobacillus sakei Rits 9, and in Sporosarcina urea pSU1 isolated from soil beneath a chicken farm (Phelan et al., 2011). Furthermore, a TEM β-lactamase-encoding gene was detected in a metatranscriptome dataset from the sponge Crambe crambe (Versluis et al., 2015), and the mecA, mupA, qnrB, and tetL resistance genes were detected in the sponge Petromica citrina by PCR with gene-specific primers (Laport et al., 2016). Based on these results it is tempting to speculate that the sponge microbiota indeed might act as a reservoir of functional AR genes.
The aim of this study was to systematically assess sponge microbiota as a reservoir of functional AR genes. We screened for functional resistance genes against 14 clinically relevant antibiotics in three Mediterranean high-microbial-abundance sponges, namely Aplysina aerophoba, Petrosia ficiformis, and Corticium candelabrum. Small-insert libraries were prepared in Escherichia coli with DNA isolated from sponge tissue of these three sponges (environmental DNA libraries), as well as from pooled DNA isolated from 31 bacterial isolates that were obtained from these sponges (sponge isolates DNA library) The 31 bacterial isolates were obtained from agar media supplemented with antibiotics as part of a previous high-throughput cultivation study (Versluis et al., under review).
Materials and Methods
Resistance Profiling of Bacterial Isolates
One small-insert library was prepared from genomic DNA obtained from 31 bacterial strains (Table 1) isolated from the sponges A. aerophoba, C. candelabrum and P. ficiformis (Supplementary Materials and Methods). Resistance of these isolates to polymyxin B, erythromycin, ciprofloxacin, cefotaxime, tetracycline, chloramphenicol, rifampicin, ampicillin and imipenem was investigated in a previous study (Supplementary Materials and Methods) (Versluis et al., under review). Additionally, in this study, resistance of the isolates was tested to gentamicin (50 μg/ml), D-cycloserine (50 μg/ml), chlortetracycline (50 μg/ml), amikacin (100 μg/ml) and trimethoprim (50 μg/ml).
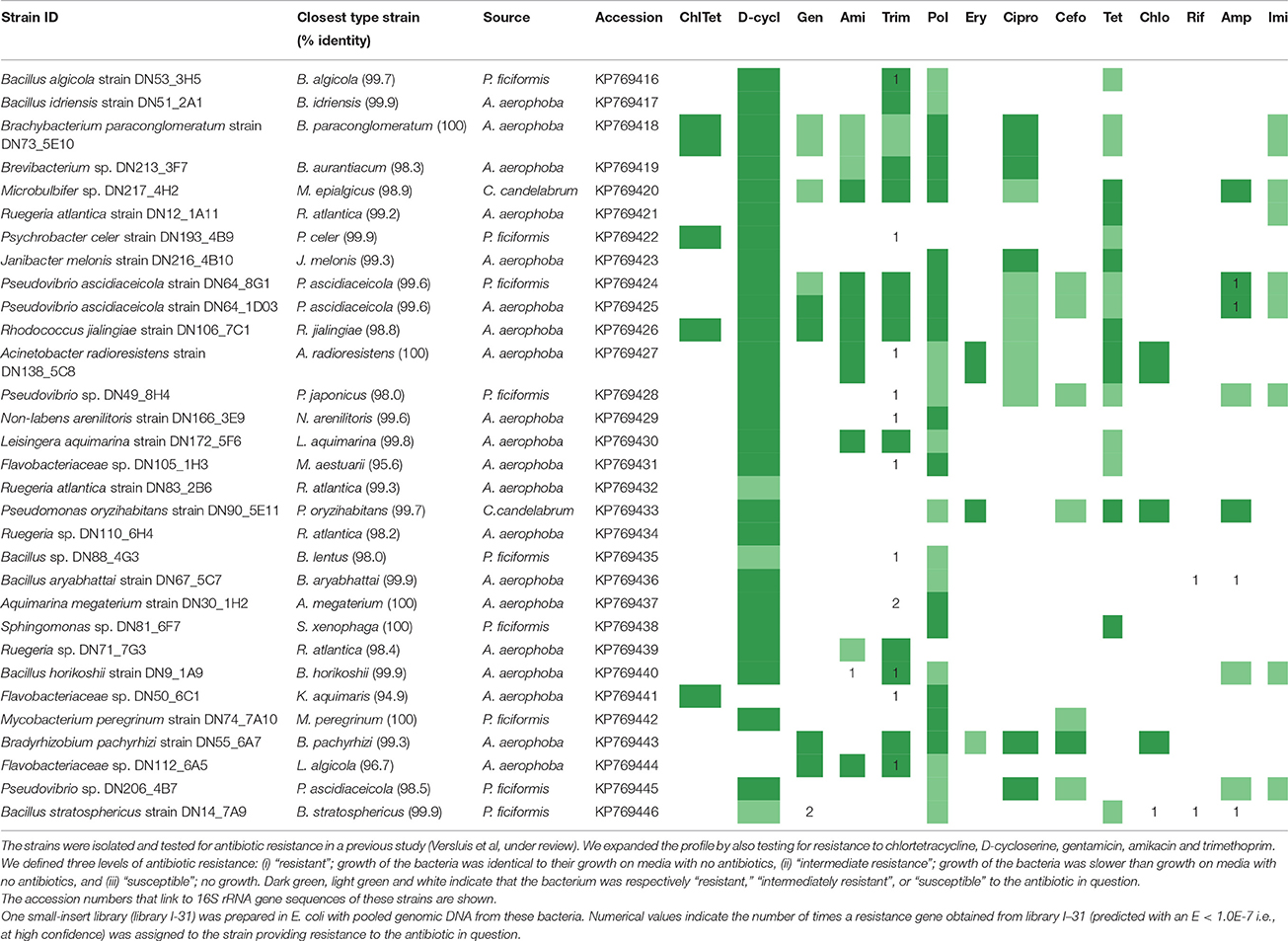
Table 1. Resistance profiles of bacterial strains isolated from the sponges A. aerophoba, C. candelabrum and P. ficiformis.
Preparation of Linearized Vector pZE21
An E. coli TOP10 strain harboring the plasmid pZE21 was inoculated into 10 ml LB broth with kanamycin (50 μg/ml). The culture was incubated at 37°C overnight. Plasmid isolation was performed with the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Waltham, United States). The DNA concentration was measured by Qubit®; (Thermo Fisher Scientific), and the isolated plasmid was electrophoresed on a 1% agarose gel for visual inspection. Next, the plasmid was cut by HincII #R0103C (NEB, Ipswich, Massachusetts) according to manufacturer's instructions. Subsequently, the plasmid ends were dephosphorylated according to the protocol for dephosphorylation of 5′-ends of DNA by rSAP (NEB). Finally, the linearized and dephosphorylated plasmid DNA was purified with the Clean-up Concentrator kit (A&A Biotechnology, Gdynia, Poland).
Preparation of Small-Insert Libraries
To prepare a small-insert library with a mixture of genomic DNA from 31 pure culture isolates (Table 1), we individually isolated DNA with the MasterPure™ DNA Purification Kit (Epicentre, Madison, Wisconsin). The DNA concentration was measured by Qubit® 2.0, and DNA from the different isolates was then pooled at equivalent mass. DNA from sponge tissue samples was isolated with the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). DNA was fragmented by the E210 sonicator (Covaris, Woburn, Massachusetts) where default operating conditions were used to achieve an average fragment size of 2 kbp. The fragmented DNA was electrophoresed on a 1% agarose gel, and DNA ranging from 0.7 to 5 kbp was purified by using the GeneJet Gel Extraction Kit (Thermo Fisher Scientific). In the final gel purification step, DNA was eluted with nuclease-free water instead of elution buffer. Subsequently the DNA ends were repaired with the NEBNext® End Repair Module (NEB) according to manufacturer's instructions. The end-repaired DNA was cleaned with the Clean-up Concentrator kit (A&A Biotechnology), and elution was performed with nuclease-free water. The TaKaRa MightyMix DNA ligation kit (Clontech Laboratories Inc., Mountain View, United States) was used for ligation of the end-repaired fragments into the linearized pZE21 vector. Each ligation mixture consisted of 2 μl Ligation Mix and 2 μl nuclease-free water containing 600 ng insert DNA and 120 ng linearized vector (5:1 ratio). The resulting product was used for two electroporation reactions at 2 μl each, thereby creating two distinct small-insert libraries. At electroporation, 2 μl ligation mixture was added to 50 μl One Shot® TOP10 Electrocomp® E. coli cells (Thermo Fisher Scientific). The sample was transferred to a 1 mm cuvet and subjected to an electric pulse at 1.8 kV, 2.5 μF, and 200 Ω. To recover the E. coli cells, 1 ml SOC medium was added, and the sample was incubated at 37°C for 1 h. After recovery, 2 μl of the cell suspension was ten-fold serially diluted, and plated on LB agar media containing kanamycin (50 μg/ml) in order to estimate library size based on CFU counts. For each library, twelve clones were picked and colony PCRs were performed with primers pZE21_81_104_57C and pZE21_151_174rc_58C (Sommer et al., 2009), which flank the insertion site. The average product size (nt), the CFU count (in no. of colonies per μl) and the total volume (in μl) were multiplied in order to estimate the library size. In order to achieve library amplification, the remainder of the cell suspension after recovery was transferred to 10 ml LB medium with kanamycin (50 μg/ml) and incubated overnight at 37°C. After library amplification, another CFU count was performed. The total number of CFUs pre- and post-amplification was divided in order to estimate the extent of library expansion. Libraries were mixed to contain 20% glycerol and stored in cryotubes at −80°C. We also verified the function of a β-lactamase resistance gene by cloning it into the pZE21 vector as a single gene using the methods described in this paragraph, starting from the ligation step. The insert DNA consisted of amplicon sequences that were obtained by gene-specific PCR.
Library Screening and Insert Sequencing
Libraries were plated at 25X (predicted) library coverage (i.e., every clone is expected to be present 25 times) on LB agar plates containing one of the following antibiotics: ampicillin (20 μg/ml), ciprofloxacin (1 μg/ml), tetracycline (20 μg/ml), chloramphenicol (20 μg/ml), polymyxin B (2 μg/ml), trimethoprim (2 μg/ml), erythromycin (200 μg/ml), cefotaxime (25 μg/ml), rifampicin (25 μg/ml), imipenem (20 μg/ml), gentamicin (20 μg/ml), D-cycloserine (100 μg/ml), chlortetracycline (20 μg/ml) and amikacin (50 μg/ml). E. coli TOP10 cells without a plasmid were used as negative control. Inserts from antibiotic resistant clones were Sanger sequenced from both the 5′ and 3′ flanks with respectively the pZE21_81_104_57C and pZE21_151_174rc_58C primers. Clones were assumed to contain identical inserts if sequences were more than 99% identical over a stretch of >400 bp. One clone per set of identical clones was selected for further analysis. The full-length inserts of these representative clones were sequenced by primer walking. Sanger sequencing was performed by flanking primers custom-designed with Primer3Plus (Untergasser et al., 2007) until GeneStudio version 2.2.0.0 could assemble a contig that spanned the entire insert. Finally, vector sequences were removed by DNA Baser version 3.5.4.2.
Analysis of Inserts Conferring Resistance
ORFs were identified by MetaGeneMark (Zhu et al., 2010), and the corresponding nucleotide and derived amino acid sequences were extracted. Amino acid sequences were used for BLASTp (Altschul et al., 1990) against the NCBI non-redundant protein database and the CARD database (McArthur et al., 2013). BLASTn of the sequences against the NCBI nr/nt and CARD databases were performed as well. Resistance functions were also predicted with HMMER (http://hmmer.org/) using the profile hidden Markov models (pHMMs) of the Resfams database as a reference (Gibson et al., 2014). We defined a resistance gene to be identified with high confidence if it was detected at an E-value of <1E-7 by either BLASTp or BLASTn against the Comprehensive Antibiotic Resistance Database (CARD), or by employing the pHMMs of the Resfams database. In addition, the resistance function needed to match the observed resistance phenotype. InterProScan (Mitchell et al., 2015) was used for classifying proteins into families and for predicting the presence of domains. Clustal Omega (Sievers et al., 2011) was used to globally align resistance genes with their best hit in NCBI's non-redundant protein database and to calculate a global amino acid identity value. ISfinder (Siguier et al., 2006) was used to identify IS elements. The ACLAME server (Leplae et al., 2004) was used to investigate if parts of the insert sequences were previously identified on mobile genetic elements.
Assignment of Inserts Conferring Resistance to Bacterial Isolates
Since one small-insert library was prepared with genomic DNA from 31 different sponge-associated bacterial isolates (as opposed to metagenomic DNA), we could use insert sequence-specific PCRs to assign inserts to the bacterium of origin. For this purpose, primers were designed with Primer3Plus (Untergasser et al., 2007). The reaction mixture of a given detection PCR consisted of: 10 μl 5X Green GoTaq Reaction Buffer (Promega, Fitchburg, Wisconsin), 0.2 mM dNTPs (Promega), 1 μM forward primer, 1 μM reverse primer, 0.5 μl GoTaq G2 DNA polymerase (5 U/μl, Promega), 1 μl genomic DNA (10–20 ng/μl), and 32.5 μl nuclease-free water (Promega). The PCR program consisted of: initial denaturation of 30 s at 98°C; 35 cycles of denaturation at 98°C for 10 s, annealing at 57°C for 20 s, and extension at 72°C for 20 s; and final extension at 72°C for 10 min. The PCR product was analyzed on a 1% agarose gel. If neither the initial PCR nor a repetition with a 1°C reduction in annealing temperature yielded a product, then new primers were designed. If a detection PCR yielded a product with genomic DNA from multiple isolates, then the amplification products were sequenced by Sanger sequencing. Inserts were assigned to an isolate if the Sanger sequence was 99–100% identical to the insert sequence.
Nucleotide Sequence Accession Numbers
The metagenomic insert sequences were deposited under accession numbers KU577908 to KU577944. The insert that contained blaPSV-1 as a single gene was deposited under accession KU926347.
Results
Composition and Screening of Metagenomic Libraries
In order to investigate the presence and diversity of functional AR genes in sponge microbiota, small-insert libraries were prepared in E. coli with a mixture of gDNA from 31 bacterial strains isolated from the sponges A. aerophoba, P. ficiformis, and C. candelabrum (library I-31; 0.8 Gb), and with DNA directly isolated from the same three sponges (libraries Aa; 0.2 Gb, Cc; 0.8 Gb, Pf; 1.7 Gb). 16S ribosomal RNA (rRNA) gene amplicon sequence analysis of the bacterial gDNA pool that was used to prepare library I-31 confirmed that DNA from all 31 isolates was present (data not shown). Amongst these 31 sponge-associated bacteria, resistance to 14 out of 16 antibiotics was observed, whereas merely intermediate resistance, signifying a reduced growth rate, to imipenem was observed. None of the isolates were resistant to rifampicin (Table 1). Screening of the four different libraries for AR yielded 37 clones with unique inserts conferring resistance to D-cycloserine (n = 6), gentamicin (n = 1), amikacin (n = 7), trimethoprim (n = 17), chloramphenicol (n = 1), rifampicin (n = 2) and ampicillin (n = 3) (Table 2). No clones were obtained that were resistant to chlortetracycline, polymyxin, erythromycin, ciprofloxacin, cefotaxime, tetracycline or imipenem. The majority of resistant clones (30 of 37) were derived from library I-31, whereas 2 clones, 0 clones and 5 clones were derived from libraries Aa, Cc, and Pf, respectively.
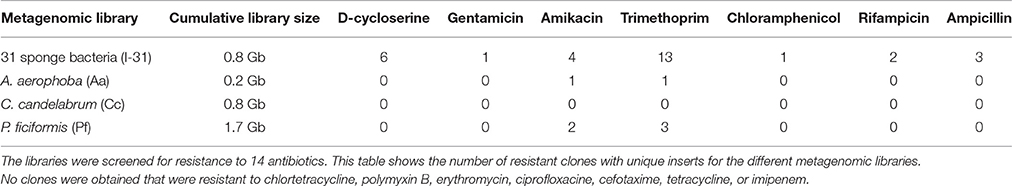
Table 2. Small-insert libraries in E. coli were made with DNA isolated from 31 sponge bacteria, and with DNA isolated from A. aerophoba, C. candelabrum and P. ficiformis tissue.
Resistance Gene Diversity and Uniqueness
The unique full-length inserts (mean insert size = 2974 bp ± 1627 [s.d.]) were sequenced by Sanger sequencing, which occasionally required multiple iterations of primer walking. We defined a resistance gene to be identified with high confidence if the resistance function was assigned at an E-value of <1E-7, and furthermore the resistance function needed to match the observed resistance phenotype.
In total, 26 resistance genes distributed over 24 different inserts were identified that met our confidence threshold (Table S1). The uniqueness of the resistance genes was evaluated by performing a global alignment at the amino acid level with the closest BLASTp hit in NCBI's non-redundant protein database. The majority of confidently identified resistance genes obtained from library I-31 (17 of 21) were predicted to code for proteins that had >70% identity at the amino acid level with known gene products, whereas proteins encoded by the genes (n = 5) obtained from the metagenomic libraries of environmental sponge DNA had <60% amino acid identity with known gene products (Figures 1A,B). Overall, most confidently identified resistance genes (15/26) were predicted to encode trimethoprim-resistance-conferring dihydrofolate reductases (Figure 1C). The confidently identified AR genes with the lowest similarity to known genes were predicted to code for a glycerol-3-phosphate acyltransferase and a GNAT family acetyltransferase with respectively 32 and 36% amino acid identity with the closest hit in NCBI's non-redundant protein database. Both of these amikacin-resistance-conferring genes were detected in libraries of environmental sponge DNA.
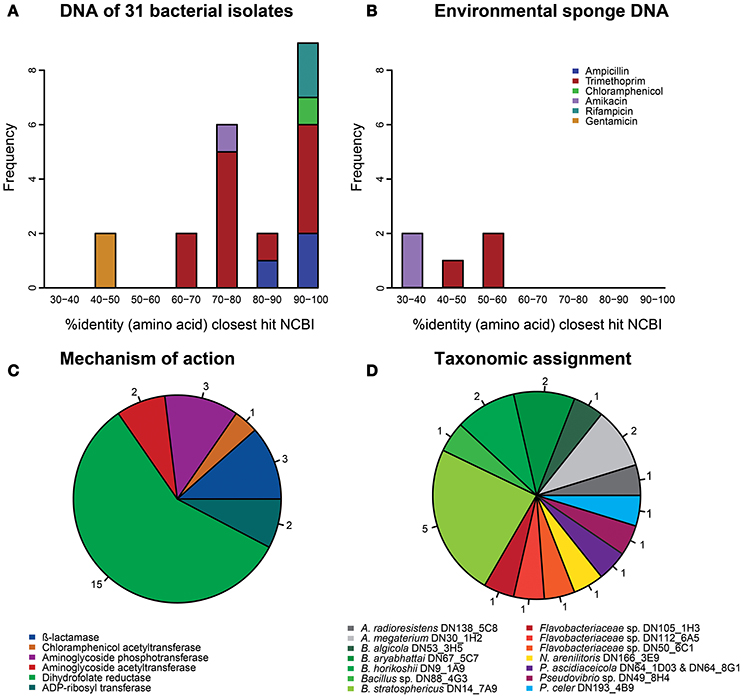
Figure 1. Characterization of the 26 antibiotic resistance genes that were identified with high confidence (E < 1.0E-7). (A) Amino acid identity distribution of the resistance genes that were obtained from the library of 31 sponge bacteria with their best hit (bitscore sorted) in NCBI's database. (B) Amino acid identity distribution of the resistance genes that were obtained from libraries based on DNA from sponge tissue with their best hit (bitscore sorted) in NCBI's database. (C) The mechanisms of action of all 26 resistance genes. (D) The taxonomic assignments of the resistance genes that were obtained from the library of 31 sponge bacteria.
For 13 insert sequences a resistance gene could not be identified meeting our threshold. Those inserts conferred resistance to amikacin (n = 4), D-cycloserine (n = 6), and trimethoprim (n = 3) (Table 3, Table S2). In these cases, besides BLAST searches against the CARD and NCBI nr/nt databases and motif detection by application of Resfams pHMMs, also protein domain and protein family predications by InterProScan were evaluated in order to manually predict AR gene presence. Most (12 of 13) inserts contained multiple open reading frames (ORFs), which prevented the unequivocal assignment of resistance functions to individual genes. For three of four inserts that conferred resistance to amikacin, we predicted the presence of genes encoding aminoglycoside-modifying enzymes. On one of these inserts (clone Env_Ami3) a gene was identified that we predicted to code for an enzyme that putatively modifies amikacin by phosphorylation. On the other two inserts, we predicted genes encoding an aminoglycoside acetyltransferase (clone Iso_Ami2) and an aminoglycoside methyltransferase (clone Iso_Ami3). The fourth insert conferring amikacin resistance (clone Iso_Ami4) contained a single gene that shared 93% amino acid identity with an amino acid transporter, and as such we predicted the gene to confer AR via antibiotic efflux. Six inserts conferred resistance to D-cycloserine. On all of these six inserts we predicted the presence of genes encoding proteins involved in transmembrane transport based on high similarity to known transporters. We predicted that three of the six D-cycloserine resistance-conferring inserts harbored genes encoding transporters that belong to the major facilitator superfamily. Three other inserts on which an AR gene could not be confidently identified conferred resistance to trimethoprim. On one of these inserts (clone Env_Trim4), we predicted that a thymidylate synthethase may be responsible for the resistance phenotype. The two other inserts conferring trimethoprim resistance (clones Iso_trim13 and Iso_trim14) putatively encoded oxidoreductases that could confer trimethoprim resistance.
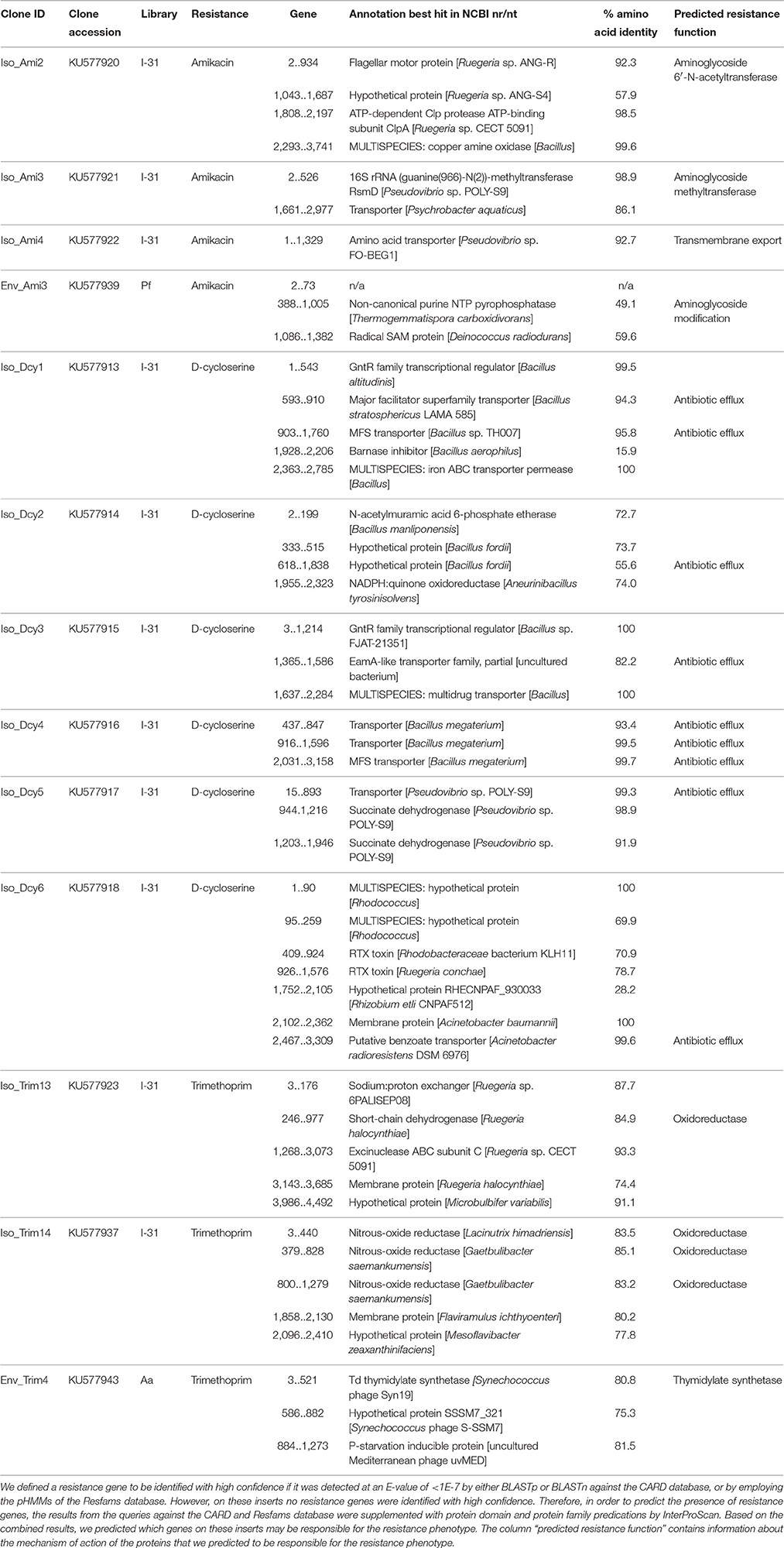
Table 3. Inserts that confer resistance to amikacin, D-cycloserine or trimethoprim on which no AR gene was identified with high confidence.
Taxonomic Assignment of Inserts from the Library of 31 Sponge Bacteria
Resistance genes that were confidently identified in library I-31 were assigned to their strain of origin by gene-specific PCRs, where the PCR amplicon covered at least part of the predicted resistance gene. Gene presence was checked in all 31 strains used for building the library by dedicated PCR reactions, which resulted in confidently identified AR genes being assigned to 15 of 31 bacterial strains in library I-31 (Table 1). Insert sequences for which no resistance gene could be identified with high confidence were assigned to a strain of origin at the loci given in Table S2. We discovered that several inserts may contain hybrid sequences, that is, the inserts consist of two different DNA fragments that were ligated into the vector. For example, the insert of clone Iso_Dcy6 was assigned to Ruegeria spp. in the region between 513 and 1372 bp and assigned to Acinetobacter radioresistens DN138_5C8 in the region between 2240 and 2851 (Table S2). We found that about half of the confidently identified resistance genes (11/21) were assigned to Bacillus spp., and Bacillus was the only taxon to which gentamicin, amikacin, chloramphenicol and rifampicin resistance genes were assigned (Table 1, Figure 1D). Even though gentamicin, amikacin, chloramphenicol, rifampicin and ampicillin resistance genes were assigned to Bacillus spp., the strains themselves were not found to be resistant to these antibiotics. Resistance genes that were assigned to the remaining taxa comprised mostly trimethoprim resistance genes, with an exception being an ampicillin resistance gene that was assigned to Pseudovibrio spp. (Table S1). Strains to which trimethoprim resistance genes were assigned were not trimethoprim resistant themselves in 8 out of 9 cases.
Novel β-Lactamase Family Identified in Pseudovibrio
The insert sequence of ampicillin-resistant clone Iso_Amp3 contained a gene predicted to encode a class A β-lactamase. The gene was assigned to Pseudovibrio ascidiaceicola DN64_1D03 and Pseudovibrio ascidiaceicola DN64_8G1, and the predicted protein shared 58% amino acid identity with the closest hit in the CBMAR database (Srivastava et al., 2014). Since this predicted protein displayed high divergence with members of known β-lactamase families it is a candidate to be classified into a new family (Jacoby, 2006). Therefore, we further tested the gene's function by cloning the gene-specific amplicon sequence that was generated with genomic DNA of Pseudovibrio ascidiaceicola DN64_1D03 as template into the pZE21 vector. The new clone, PSV1 (accession KU926347), was resistant to ampicillin, which confirmed the functionality of the β-lactamase gene. Remarkably, the coding sequence of the gene cloned using the genome-derived amplicon sequences was 75 bp longer than the coding sequence of the β-lactamase gene in the original metagenomic insert. Sanger sequencing of the genome-derived amplicon yielded a gene sequence that shared 100% nucleotide identity with the gene sequence in clone PSV1. Hence, we concluded that the longer β-lactamase gene in clone PSV1 is the exact gene that is present in the genome of Pseudovibrio ascidiaceicola DN64_1D03. We predict that even though a frameshift mutation yielded a shorter gene in case of the small-insert library, the encoded enzyme retained its function. We classified the genome-derived (longer) gene into a novel β-lactamase family named PSV after Pseudovibrio. The first member was designated blaPSV-1. BLASTp of the gene against the CBMAR database showed that LEN and SHV were the closest β-lactamase families. The best hit from the LEN and SHV families in both cases shared 50% amino acid identity with blaPSV-1. The best hit was blaLEN−22 (accession AM850912), with which blaPSV-1shared 41% global amino acid identity. The best hits in the NCBI non-redundant protein sequences database (≥76% amino acid identity) were exclusively genes in sequenced genomes of Pseudovibrio spp. (Figure 2). Additional resistance testing of clone PSV1 showed that blaPSV-1 conferred resistance to penicillin (50 μg/ml) but not against cefotaxime (20 μg/ml) and imipenem (20 μg/ml).
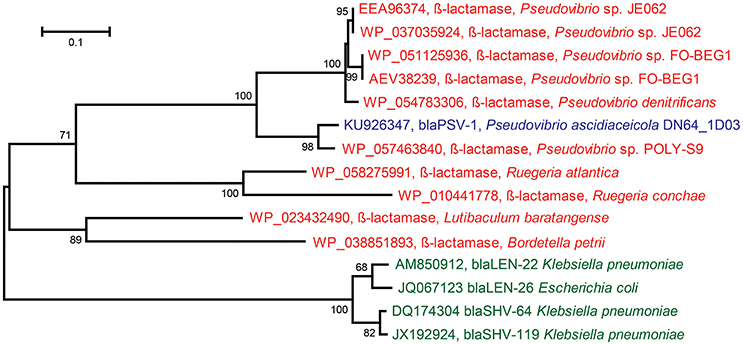
Figure 2. Maximum Likelihood Tree based on protein sequences of: the novel ß-lactamase (blaPSV-1) discovered in Pseudovibrio ascidiaceicola DN64_1D03 (blue), the closest homologs of the novel ß-lactamase in the NCBI non-redundant protein sequences database (red), and the closest homologs of the novel ß-lactamase in the CBMAR database (green) (the two closest homologs were picked for each of the two closest families: blaLEN and blaSHV). In this figure, only the proteins in green and the novel ß-lactamase (blaPSV-1) have demonstrated functionality. The tree was constructed in MEGA using 1000 iterations of bootstrapping. Bootstrap values <50 are not shown. The horizontal bar indicates the number of substitutions per site.
Resistance Gene Dissemination
Horizontal gene transfer is the main mechanism through which resistance genes are disseminated. Therefore, we investigated our sequences for evidence of gene mobilization. Comparison of the insert sequences with the ACLAME plasmid database showed that they were not previously identified on plasmids. Furthermore, ISfinder did not identify insertion sequences. We investigated whether the resistance genes identified in this study had been observed previously in a different genomic context by comparing our sequences with those in the NCBI nr/nt database (Table S3). A β-lactamase and an ADP-ribosyl transferase (those on clones Iso_Amp2 and Iso_Rif2, respectively), both of which were assigned to Bacillus aryabhattai DN67_5C7, shared very high (≥99%) identity with genes previously found in the non-marine Bacillus megaterium QM B1551, which suggests a relatively recent dissemination event. The insert of clone Iso_Amp2 shared 99% identity with the genome of Bacillus megaterium QM B1551 over the full-length insert (1352 bp). This insert produced four other significant alignments with sequences in the NCBI nr/nt database at >90% nucleotide identity, all of which belonged to Bacillus megaterium strains. The insert of clone Iso_Rif2 was 6717 bp in length but only the region from 5198 to 6717 bp aligned with the genome of Bacillus megaterium QM B1551. Besides the rifampicin-resistance-conferring gene, this region encodes a hypothetical protein and a gene that putatively encodes an enzyme involved in site specific recombination of plasmids. The gene putatively encoding a recombination enzyme was the only gene involved in gene mobilization that was predicted on the inserts.
Discussion
Functional metagenomic analysis of sponge-associated bacteria revealed diverse AR genes that conferred resistance to ampicillin, D-cycloserine, gentamicin, amikacin, trimethoprim, chloramphenicol and rifampicin. Seventeen of 26 confidently identified AR genes shared low homology (<90% amino acid identity) with known gene products. Therefore, these results show that sponges, like other environments that contain complex bacterial communities, appear to represent reservoirs of unique AR genes. In addition, we obtained 13 inserts that did not contain genes with significant similarity to known resistance genes, reinforcing the notion that sponges may act as reservoirs of yet unknown mechanisms of AR. Two resistance genes shared ≥99% nucleotide identity with AR genes detected outside the marine environment, and hence it is tempting to speculate that AR genes are being transmitted between sponges and non-marine environments and as such may be exploited by bacteria in other non-marine habitats. It has previously been shown that AR genes can be genetically linked, leading to co-selection (Galimand et al., 1997; Summers, 2006). None of our inserts contained AR genes with different resistance functions, which is likely in part due to the small insert size of the metagenomic libraries.
Out of 37 inserts harboring resistance genes, 30 inserts were obtained from the library of 31 sponge bacteria, and 7 inserts were obtained from the libraries of sponge tissue even though their cumulative library sizes were 0.8 and 2.7 Gbp, respectively. We expect that this discrepancy can be explained by libraries from sponge tissue DNA containing a substantial fraction of sponge DNA. No identical resistance genes were obtained from individual isolates and from sponge tissue in spite of the individual isolates being previously isolated from the tissue samples. This result, and taking into account the high AR gene diversity and limited scale of the experiment, suggests the presence of a substantial number of AR genes in sponges that are not yet identified. The number of AR genes that we identified was also restricted to those that are compatible with the expression system of the E. coli host, and to those that confer AR based on a DNA sequence of at most 5 kbp. From the sponge C. candelabrum no resistant clones were obtained, neither from the sponge tissue nor from the individual isolates. It should be noted, however, that the small-insert library of 31 bacteria contained only two isolates from C. candelabrum, which reduced the chance to identify a resistance gene from this sponge. Furthermore, the library of C. candelabrum tissue was 0.8 Gb in size and as such is expected to cover only a small fraction of the total unique DNA in this niche. Therefore, absence of resistance inserts for this sponge species should not be interpreted as an absence of AR genes.
Most confidently identified resistance genes (15/26) conferred resistance to trimethoprim by encoding the enzyme dihydrofolate reductase. Dihydrofolate reductase is a housekeeping enzyme that converts dihydrofolic acid to tetrahydrofolic acid. Tetrahydrofolic acid in turn is a required cofactor to convert dUMP to dTMP by thymidylate synthase, with dTMP being essential for DNA synthesis. Trimethoprim resistance due to additional dihydrofolate reductase genes can occur via a combination of two mechanisms: (i) the dihydrofolate reductase is overproduced leading to higher levels of the enzyme in the cell, and (ii) the heterologously introduced dihydrofolate reductase has decreased trimethoprim susceptibility due to mutations (Gibreel and Sköld, 1998; Coque et al., 1999; Rodrigues et al., 2016). The fact that strains to which dihydrofolate reductase genes were assigned were themselves in most cases (8/9) not trimethoprim resistant may have resulted from these strains having too little active dihydrofolate reductase to compensate for the inhibition by trimethoprim. Resistance in recombinant E. coli clones that host a plasmid-encoded dihydrofolate reductase may result from 50 to 70 copies of pZE21 plasmid being present per cell (Lutz and Bujard, 1997), thereby substantially increasing the concentration of active enzyme. Expression level differences of active enzyme could also explain other instances where the host to which an AR gene was assigned was itself not resistant. For example, Bacillus stratosphericus DN14_7A9 contained functional genes conferring resistance to gentamicin, chloramphenicol, rifampicin and ampicillin, but the strain itself was not resistant against these antibiotics. In the natural environment, these AR genes can potentially benefit their bacterial hosts in the presence of lower levels of antimicrobials than those applied in this study, or even if antimicrobials are present at sub-MIC levels (Andersson and Hughes, 2012; Sandegren, 2014). For example, an Escherichia coli strain harboring a single GyrA mutation was shown to outcompete the wild-type strain at a ciprofloxacin level of 1/230 the wild-type MIC (Gullberg et al., 2011).
The most unique confidently identified (predicted) resistance genes were those encoding a glycerol-3-phosphate acyltransferase and a GNAT family acetyltransferase with respectively 32 and 36% amino acid identity with the closest hit in NCBI's non-redundant protein database. These genes were derived from libraries Aa and Pf, respectively. Although aminoglycoside resistance genes might serve as defense against locally or self-produced antimicrobials (Walsh and Duffy, 2013), past studies have ascribed to these genes various other functional roles besides aminoglycoside modification. Aminoglycoside modification would then be accidental. For instance, a 2′-N-acetyltransferase was shown to be involved in the acetylation of peptidoglycan (Macinga et al., 1998). Reeves et al. (2013) suggested roles for aminoglycoside resistance genes in immune modulation and alleviation of cellular stress. In addition, aminoglycoside-3′-phosphotransferases are closely related to protein kinases (Davies, 2006). For one amikacin-resistance conferring insert, we predicted that resistance resulted from antibiotic efflux. The efflux pump showed high identity with an amino acid transporter that was not previously implicated in amikacin resistance. For another six metagenomic inserts from our libraries that conferred D-cycloserine resistance, we predicted the presence of efflux pumps that were also not previously linked to resistance. These results show that sponge-derived bacteria carry genes that provide AR by mechanisms that were not previously observed.
One novel β-lactamase (blaPSV-1) was discovered in P. ascidiaceicola DN64_1D03 that provided resistance to ampicillin and penicillin. Based on high amino acid sequence divergence with known β-lactamase families, the gene was placed in a novel family termed PSV, after Pseudovibrio. To date, all publicly available sequences that share ≥76% identity with blaPSV-1 at the amino acid level were found in Pseudovibrio spp. (Figure 2), a genus whose members are consistently being isolated from sponges and have never been isolated outside the marine environment (Lafi et al., 2005; Muscholl-Silberhorn et al., 2008; Menezes et al., 2010). The subsequent closest homologs of the novel β-lactamase were (predicted) β-lactamases from Ruegeria, a genus comprising marine bacteria (Wagner-Döbler and Biebl, 2006). Therefore, blaPSV-1 appears to be a marine-specific β-lactamase, and may correspondingly have a marine-specific role and/or substrate. In sponges, β-lactamases could serve as a defense molecule against β-lactam antimicrobials, and substances containing a β-lactam ring have been identified in sponges (Aviles and Rodriguez, 2010). On the other hand, β-lactamases have also been predicted to serve a role in disruption of cell signaling (Allen et al., 2009). Apart from the biological role of β-lactamases, their evolutionary origin is also still unclear (Hall and Barlow, 2004; Garau et al., 2005).
The majority of confidently identified AR genes (11/21) in library I-31 were assigned to Bacillus spp. including AR genes conferring resistance to ampicillin, chloramphenicol, gentamicin, amikacin and rifampicin. This is in line with the fact that, to date, the only sponge isolate that was shown to harbor functional resistance genes was Bacillus sp. strain HS24 (Phelan et al., 2011; Barbosa et al., 2014). These findings suggest that in sponges, Bacillus spp. are a reservoir of AR genes. However, sponges filter thousands of liters of water per day (Vogel, 1977), and as a result, sponge-associated microorganisms are not always permanent residents of the sponge holobiont. In fact, in all three investigated sponges Bacillus spp. represent less than 0.1% of the sponge microbiome (Versluis et al., under review). Therefore, we speculate that the diverse AR genes detected in the sponge-associated Bacillus spp. do not play a sponge-specific role but rather that these genes serve a more general role in the survival of these bacteria in the marine environment.
A phenomenon we noticed is that in our study, several inserts may contain hybrid sequences, that is, the inserts consist of two different DNA fragments that were ligated into the same vector. As a consequence, the assignment of inserts to a strain of origin can depend on the sequence locus that is amplified by detection PCR. We suspect that these hybrid inserts are an artifact of the cloning strategy involving ligation of blunt end fragments. Because in the present study, one of the libraries was prepared from DNA extracted from 31 bacterial isolates, and thus a defined pool of starting DNA was used, detection of hybrid inserts was possible. However, based on this finding, for all studies using similar cloning methods conclusions about the genetic context of AR genes on metagenomic inserts should be drawn carefully. Here, to ensure that all confidently identified resistance genes were assigned to the correct strain, we performed gene-specific PCRs (Table S1).
In conclusion, we show that sponges constitute a reservoir of diverse functional AR genes. We detected functional AR genes that show little similarity to known AR genes, as well as functional AR genes that have no similarity to known AR genes. One ampicillin resistance gene, blaPSV-1, was placed into a novel family of marine-specific β-lactamases. These results raise questions as to the roles of these genes in bacteria residing in arguably the oldest lineage of metazoans (Hentschel et al., 2006), the sponges. Furthermore, the functionality of our observed AR genes in E. coli shows that they can potentially be harnessed by phylogenetically distinct bacteria in other environments.
Author Contributions
DV was involved in experimental design, performed the experiments and the analysis, wrote the manuscript, and prepared the figures. MV, HS, and DS were involved in experimental design and jointly supervised the work. MR carried out part of the experimental work. All authors revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the European Union through the EvoTAR project (Grant agreement no. 282004) and by a grant of BE-Basic-FES funds from the Dutch Ministry of Economic Affairs (project 7.1.5). MS and MR additionally acknowledge the Lundbeck Foundation for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01848/full#supplementary-material
References
Allen, H. K., Moe, L. A., Rodbumrer, J., Gaarder, A., and Handelsman, J. (2009). Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 3, 243–251. doi: 10.1038/ismej.2008.86
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alves, M. S., Pereira, A., Araújo, S. M., Castro, B. B., Correia, A. C., and Henriques, I. (2014). Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 5:426. doi: 10.3389/fmicb.2014.00426
Amos, G. C., Zhang, L., Hawkey, P. M., Gaze, W. H., and Wellington, E. M. (2014). Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Vet. Microbiol. 171, 441–447. doi: 10.1016/j.vetmic.2014.02.017
Andersson, D. I., and Hughes, D. (2012). Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist. Updat. 15, 162–172. doi: 10.1016/j.drup.2012.03.005
Arias, C. A., and Murray, B. E. (2009). Antibiotic-resistant bugs in the 21st century–a clinical super-challenge. N. Engl. J. Med. 360, 439–443. doi: 10.1056/NEJMp0804651
August, P. R., Tang, L., Yoon, Y. J., Ning, S., Müller, R., Yu, T. W., et al. (1998). Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 5, 69–79. doi: 10.1016/S1074-5521(98)90141-7
Aviles, E., and Rodriguez, A. D. (2010). Monamphilectine A, a potent antimalarial β-lactam from marine sponge Hymeniacidon sp: isolation, structure, semisynthesis, and bioactivity. Org. Lett. 12, 5290–5293. doi: 10.1021/ol102351z
Barbosa, T. M., Phelan, R. W., Leong, D., Morrissey, J. P., Adams, C., Dobson, A. D., et al. (2014). A novel erythromycin resistance plasmid from Bacillus sp. strain HS24, isolated from the marine sponge Haliclona simulans. PLoS ONE 9:e115583. doi: 10.1371/journal.pone.0115583
Bhullar, K., Waglechner, N., Pawlowski, A., Koteva, K., Banks, E. D., Johnston, M. D., et al. (2012). Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 7:e34953. doi: 10.1371/journal.pone.0034953
Clemente, J. C., Pehrsson, E. C., Blaser, M. J., Sandhu, K., Gao, Z., Wang, B., et al. (2015). The microbiome of uncontacted Amerindians. Sci. Adv. 1:e1500183. doi: 10.1126/sciadv.1500183
Coque, T. M., Singh, K. V., Weinstock, G. M., and Murray, B. E. (1999). Characterization of dihydrofolate reductase genes from trimethoprim-susceptible and trimethoprim-resistant strains of Enterococcus faecalis. Antimicrob. Agents Chemother. 43, 141–147.
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Davies, J. E. (2006). Aminoglycosides: ancient and modern. J. Antibiot. 59, 529–532. doi: 10.1038/ja.2006.73
D'Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W., Schwarz, C., et al. (2011). Antibiotic resistance is ancient. Nature 477, 457–461. doi: 10.1038/nature10388
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Forslund, K., Sunagawa, S., Kultima, J. R., Mende, D. R., Arumugam, M., Typas, A., et al. (2013). Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23, 1163–1169. doi: 10.1101/gr.155465.113
Galimand, M., Guiyoule, A., Gerbaud, G., Rasoamanana, B., Chanteau, S., Carniel, E., et al. (1997). Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 337, 677–680. doi: 10.1056/NEJM199709043371004
Garau, G., Di Guilmi, A. M., and Hall, B. G. (2005). Structure-based phylogeny of the metallo-β-lactamases. Antimicrob. Agents Chemother. 49, 2778–2784. doi: 10.1128/AAC.49.7.2778-2784.2005
Gibreel, A., and Sköld, O. (1998). High-level resistance to trimethoprim in clinical isolates of Campylobacter jejuni by acquisition of foreign genes (dfr1 and dfr9) expressing drug-insensitive dihydrofolate reductases. Antimicrob. Agents Chemother. 42, 3059–3064.
Gibson, M. K., Forsberg, K. J., and Dantas, G. (2014). Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216. doi: 10.1038/ismej.2014.106
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. doi: 10.1371/journal.ppat.1002158
Hall, B. G., and Barlow, M. (2004). Evolution of the serine β-lactamases: past, present and future. Drug Resist. Updat. 7, 111–123. doi: 10.1016/j.drup.2004.02.003
Hatosy, S. M., and Martiny, A. C. (2015). The Ocean as a global reservoir of antibiotic resistance genes. Appl. Environ. Microbiol. 81, 7593–7599. doi: 10.1128/AEM.00736-15
Hentschel, U., Usher, K. M., and Taylor, M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55, 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Högberg, L. D., Heddini, A., and Cars, O. (2010). The global need for effective antibiotics: challenges and recent advances. Trends Pharmacol. Sci. 31, 509–515. doi: 10.1016/j.tips.2010.08.002
Jacoby, G. A. (2006). β-lactamase nomenclature. Antimicrob. Agents Chemother. 50, 1123–1129. doi: 10.1128/AAC.50.4.1123-1129.2006
Kim, T. K., Hewavitharana, A. K., Shaw, P. N., and Fuerst, J. A. (2006). Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol. 72, 2118–2125. doi: 10.1128/AEM.72.3.2118-2125.2006
Lafi, F. F., Garson, M. J., and Fuerst, J. A. (2005). Culturable bacterial symbionts isolated from two distinct sponge species (Pseudoceratina clavata and Rhabdastrella globostellata) from the Great Barrier Reef display similar phylogenetic diversity. Microb. Ecol. 50, 213–220. doi: 10.1007/s00248-004-0202-8
Laport, M. S., Pontes, P. V. M., dos Santos, D. S., Santos-Gandelman, J. d. F., Muricy, G., Bauwens, M., et al. (2016). Antibiotic resistance genes detected in the marine sponge Petromica citrina from Brazilian coast. Braz. J. Microbiol. 47, 617–620. doi: 10.1016/j.bjm.2016.04.016
Laport, M. S., Santos, O. C., and Muricy, G. (2009). Marine Sponges: potential sources of new antimicrobial drugs. Curr. Pharm. Biotechnol. 10, 86–105. doi: 10.2174/138920109787048625
Lenski, R. E. (1998). Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1, 265–270.
Leplae, R., Hebrant, A., Wodak, S. J., and Toussaint, A. (2004). ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 32(Database issue), D45–D49. doi: 10.1093/nar/gkh084
Lutz, R., and Bujard, H. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210. doi: 10.1093/nar/25.6.1203
Macinga, D. R., Cook, G. M., Poole, R. K., and Rather, P. N. (1998). Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2′-N-acetyltransferase in P. stuartii. J. Bacteriol. 180, 128–135.
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Mehbub, M. F., Lei, J., Franco, C., and Zhang, W. (2014). Marine Sponge derived natural products between 2001 and 2010: trends and opportunities for discovery of bioactives. Mar. Drugs 12, 4539–4577. doi: 10.3390/md12084539
Menezes, C. B., Bonugli-Santos, R. C., Miqueletto, P. B., Passarini, M. R., Silva, C. H., Justo, M. R., et al. (2010). Microbial diversity associated with algae, ascidians and sponges from the north coast of Sao Paulo state, Brazil. Microbiol. Res. 165, 466–482. doi: 10.1016/j.micres.2009.09.005
Mitchell, A., Chang, H. Y., Daugherty, L., Fraser, M., Hunter, S., Lopez, R., et al. (2015). The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43(Database issue), D213–D221. doi: 10.1093/nar/gku1243
Mori, T., Mizuta, S., Suenaga, H., and Miyazaki, K. (2008). Metagenomic screening for bleomycin resistance genes. Appl. Environ. Microbiol. 74, 6803–6805. doi: 10.1128/AEM.00873-08
Munck, C., Albertsen, M., Telke, A., Ellabaan, M., Nielsen, P. H., and Sommer, M. O. (2015). Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 6, 8452. doi: 10.1038/ncomms9452
Muscholl-Silberhorn, A., Thiel, V., and Imhoff, J. F. (2008). Abundance and bioactivity of cultured sponge-associated bacteria from the Mediterranean sea. Microb. Ecol. 55, 94–106. doi: 10.1007/s00248-007-9255-9
Phelan, R. W., Clarke, C., Morrissey, J. P., Dobson, A. D., O'Gara, F., and Barbosa, T. M. (2011). Tetracycline resistance-encoding plasmid from Bacillus sp. strain #24, isolated from the marine sponge Haliclona simulans. Appl. Environ. Microbiol. 77, 327–329. doi: 10.1128/Aem.01239-10
Reeves, A. Z., Campbell, P. J., Sultana, R., Malik, S., Murray, M., Plikaytis, B. B., et al. (2013). Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob. Agents Chemother. 57, 1857–1865. doi: 10.1128/AAC.02191-12
Rodrigues, J. V., Bershtein, S., Li, A., Lozovsky, E. R., Hartl, D. L., and Shakhnovich, E. I. (2016). Biophysical principles predict fitness landscapes of drug resistance. Proc. Natl. Acad. Sci. U.S.A. 113, E1470–E1478. doi: 10.1073/pnas.1601441113
Sandegren, L. (2014). Selection of antibiotic resistance at very low antibiotic concentrations. Ups. J. Med. Sci. 119, 103–107. doi: 10.3109/03009734.2014.904457
Schupp, T., Toupet, C., Engel, N., and Goff, S. (1998). Cloning and sequence analysis of the putative rifamycin polyketide synthase gene cluster from Amycolatopsis mediterranei. FEMS Microbiol. Lett. 159, 201–207. doi: 10.1111/j.1574-6968.1998.tb12861.x
Segawa, T., Takeuchi, N., Rivera, A., Yamada, A., Yoshimura, Y., Barcaza, G., et al. (2013). Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 5, 127–134. doi: 10.1111/1758-2229.12011
Sengupta, S., Chattopadhyay, M. K., and Grossart, H. P. (2013). The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4:47. doi: 10.3389/fmicb.2013.00047
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(Database issue), D32–D36. doi: 10.1093/nar/gkj014
Sommer, M. O., Dantas, G., and Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131. doi: 10.1126/science.1176950
Srivastava, A., Singhal, N., Goel, M., Virdi, J. S., and Kumar, M. (2014). CBMAR: a comprehensive beta-lactamase molecular annotation resource. Database (Oxford). 2014:bau111. doi: 10.1093/database/bau111
Summers, A. O. (2006). Genetic linkage and horizontal gene transfer, the roots of the antibiotic multi-resistance problem. Anim. Biotechnol. 17, 125–135. doi: 10.1080/10495390600957217
Teuber, M., Meile, L., and Schwarz, F. (1999). Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek 76, 115–137. doi: 10.1023/A:1002035622988
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35(Web Server issue), W71–W74. doi: 10.1093/nar/gkm306
Versluis, D., D'Andrea, M. M., Ramiro Garcia, J., Leimena, M. M., Hugenholtz, F., Zhang, J., et al. (2015). Mining microbial metatranscriptomes for expression of antibiotic resistance genes under natural conditions. Sci. Rep. 5:11981. doi: 10.1038/srep11981
Vogel, S. (1977). Current-induced flow through living sponges in nature. Proc. Natl. Acad. Sci. U.S.A. 74, 2069–2071. doi: 10.1073/pnas.74.5.2069
Wagner-Döbler, I., and Biebl, H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280. doi: 10.1146/annurev.micro.60.080805.142115
Walsh, F., and Duffy, B. (2013). The culturable soil antibiotic resistome: a community of multi-drug resistant bacteria. PLoS ONE 8:e65567. doi: 10.1371/journal.pone.0065567
Webster, N. S., and Taylor, M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Wichmann, F., Udikovic-Kolic, N., Andrew, S., and Handelsman, J. (2014). Diverse antibiotic resistance genes in dairy cow manure. mBio 5:e01017–13. doi: 10.1128/mBio.01017-13
Yang, J., Wang, C., Shu, C., Liu, L., Geng, J., Hu, S., et al. (2013). Marine sediment bacteria harbor antibiotic resistance genes highly similar to those found in human pathogens. Microb. Ecol. 65, 975–981. doi: 10.1007/s00248-013-0187-2
Zhang, L., An, R., Wang, J., Sun, N., Zhang, S., Hu, J., et al. (2005). Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 8, 276–281. doi: 10.1016/j.mib.2005.04.008
Keywords: antibiotic resistance, sponge, microbiota, resistance gene, functional metagenomics
Citation: Versluis D, Rodriguez de Evgrafov M, Sommer MOA, Sipkema D, Smidt H and van Passel MWJ (2016) Sponge Microbiota Are a Reservoir of Functional Antibiotic Resistance Genes. Front. Microbiol. 7:1848. doi: 10.3389/fmicb.2016.01848
Received: 10 August 2016; Accepted: 03 November 2016;
Published: 17 November 2016.
Edited by:
Heather K. Allen, National Animal Disease Center, USAReviewed by:
Pierre Cornelis, Vrije Universiteit Brussel, BelgiumMaria Jose Gosalbes, University of Valencia, Spain
Copyright © 2016 Versluis, Rodriguez de Evgrafov, Sommer, Sipkema, Smidt and van Passel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dennis Versluis, dversluis@hotmail.com
Mark W. J. Van Passel, mark.van.passel@rivm.nl
 Dennis Versluis
Dennis Versluis Mari Rodriguez de Evgrafov
Mari Rodriguez de Evgrafov Morten O. A. Sommer
Morten O. A. Sommer Detmer Sipkema
Detmer Sipkema Hauke Smidt
Hauke Smidt Mark W. J. van Passel
Mark W. J. van Passel