- 1School of Molecular Biosciences, Washington State University, Pullman, WA, USA
- 2Institute of Biological Chemistry, Washington State University, Pullman, WA, USA
Bacteria have developed various stress response pathways to improve their assimilation and allocation of limited nutrients, such as nitrogen and phosphate. While both the nitrogen stress response (NSR) and phosphate stress response (PSR) have been studied individually, there are few experiments reported that characterize effects of multiple stresses on one or more pathways in Sinorhizobium meliloti, a facultatively symbiotic, nitrogen-fixing bacteria. The PII proteins, GlnB and GlnK, regulate the NSR activity, but analysis of global transcription changes in a PII deficient mutant suggest that the S. meliloti PII proteins may also regulate the PSR. PII double deletion mutants grow very slowly and pseudoreversion of the slow growth phenotype is common. To understand this phenomenon better, transposon mutants were isolated that had a faster growing phenotype. One mutation was in phoB, the response regulator for a two component regulatory system that is important in the PSR. phoB::Tn5 mutants had different phenotypes in the wild type compared to a PII deficient background. This led to the hypothesis that phosphate stress affects the NSR and conversely, that nitrogen stress affects the PSR. Our results show that phosphate availability affects glutamine synthetase activity and expression, which are often used as indicators of NSR activity, but that nitrogen availability did not affect alkaline phosphatase activity and expression, which are indicators of PSR activity. We conclude that the NSR is co-regulated by nitrogen and phosphate, whereas the PSR does not appear to be co-regulated by nitrogen in addition to its known phosphate regulation.
Introduction
Sinorhizobium meliloti are important bacteria due to their ability to fix atmospheric nitrogen during symbiosis with alfalfa and other legumes. Nitrogen availability is a common problem that farmers need to address in order to approach the yield potential of many crops. The application of nitrogen fertilizers to the soil is a common solution (Ladha et al., 2005; Hirsch and Mauchline, 2015). Using fertilizers is expensive and their use is also associated with significant environmental costs (Ladha et al., 2005; Spiertz, 2010; Hirsch and Mauchline, 2015). A significant fraction of the fertilizer typically runs off farmland and contaminates groundwater and river ecosystems. Microbial transformation of nitrogen fertilizers can lower soil pH, and affect crops that do not have the ability to compensate for the increased acidity. One way to reduce the need for nitrogen fertilizer applications is to grow legumes that can form symbiotic relationships with rhizobia, allowing biological nitrogen fixation (BNF) to occur in situ and on demand. In the process of establishing a symbiosis with a legume like alfalfa, nodules are formed on the roots after the bacteria infect emerging root hairs. In these nodules, bacteria invade plant cells and differentiate into an organelle-like state called a bacteroid. The bacteroids produce nitrogenase and associated proteins, which reduce atmospheric dinitrogen into ammonia, a process termed BNF. The plant can use the ammonia for incorporation into amino acids, nucleic acids, and other compounds necessary for plant survival (Patriarca et al., 2004; Oldroyd and Downie, 2008). In order to support nitrogen fixation, plants supply the bacteroids with carbon compounds and the nodule maintains a microaerobic environment that is compatible with bacterial respiration and nitrogen fixation. Establishing a symbiotic relationship is facultative for both the plant and bacteria and nodules are formed de novo in response to the plant’s need for nitrogen.
For BNF to occur and provide useable nitrogen to legumes, free-living rhizobia must be present in the soil and the bacteria therefore need to be able to survive in various soil conditions, adapting to nutrient limitations, soil acidity and moisture content. Soil nutrient availability can be dynamic, with the availability of nutrients shifting as the season and presence of other microorganisms changes (Vitousek et al., 2010; Fanin et al., 2016). Rhizobia have stress response pathways to deal with numerous environmental limitations and help them persist in the rhizosphere (Al-Niemi et al., 1997; Krol and Becker, 2004; Yurgel et al., 2012). Nutrient stress responses alter gene expression and protein activity to improve uptake and catabolism of substrates that contain phosphorus or nitrogen, more efficiently incorporate the compounds into metabolism and, in some situations, use biosynthetic pathways that are not as dependent on these nutrients (Battesti et al., 2011). The nitrogen and phosphate stress responses (PSRs) in S. meliloti have each been characterized. While certain aspects may not be entirely understood yet, a fair amount of detail has been elucidated for each pathway.
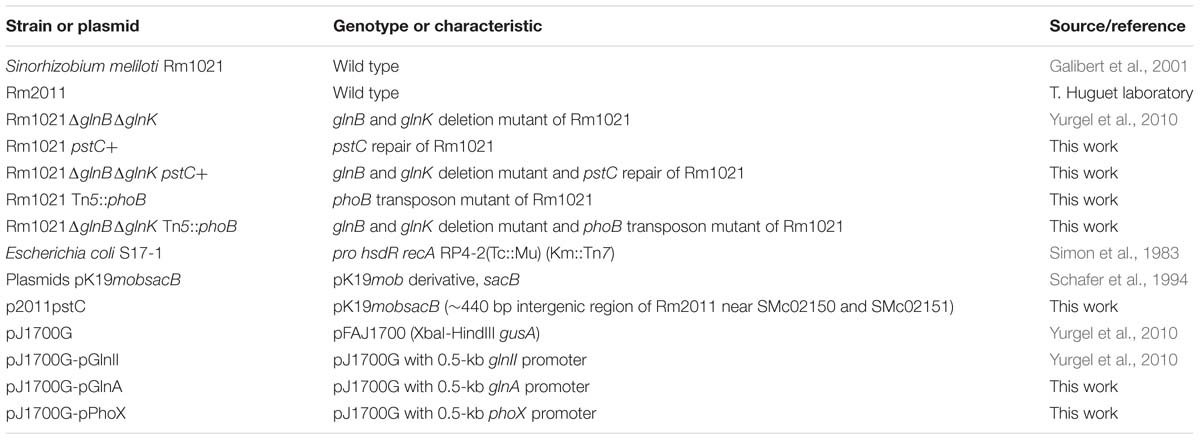
During nitrogen limitation, the nitrogen stress response (NSR) in S. meliloti is active (Figure 1A). The NSR increases uptake and mobilization of nitrogen while increasing the efficiency with which available nitrogen is used (Merrick and Edwards, 1995; Arcondeguy et al., 2001; Leigh and Dodsworth, 2007). Regulation of the core NSR in S. meliloti involves a sensor protein, GlnD, two PII proteins, GlnB and GlnK, and the NtrB/NtrC two component regulatory system. GlnD, a uridylyltransferase/uridylyl-cleavage enzyme, senses the ratio between glutamine and alpha-ketoglutarate, an index of nitrogen availability (Merrick and Edwards, 1995; Ikeda et al., 1996; Jiang et al., 2012). When alpha-ketoglutarate levels are high, indicating nitrogen deficiency, GlnD uridylylates GlnB and GlnK. GlnB-UMP and GlnK-UMP act on GlnE which regulates the adenylylation state of glutamine synthetase I (GSI) (Atkinson and Ninfa, 1998). GSI-AMP is inactive and when the adenylyl group is removed, GSI becomes activated allowing it to add ammonium to glutamate to form glutamine, a molecule which is used to donate nitrogen in many cellular biosynthetic reactions (Yurgel et al., 2010). In addition to influencing GlnE activity, GlnK regulates the AmtB transporter (Gruswitz et al., 2007). When nitrogen is low, GlnK-UMP acts on AmtB to increase uptake of nitrogen from the environment. GlnB-UMP acts on the two-component NtrB/NtrC system by allowing the NtrB sensor protein to be phosphorylated during low nitrogen conditions. NtrB-P then transfers the phosphate group to the response regulator, NtrC. NtrC-P increases transcription of target genes through DNA binding and interaction with RpoN, a sigma54 subunit of RNA polymerase. Genes activated by NtrC-P are primarily involved in nitrogen uptake and mobilization. One such target is glnII, coding for a second glutamine synthetase, GSII. glnII expression is often used as an indicator of NSR activity (Arcondeguy et al., 1997) due to its direct activation by NtrC-P.
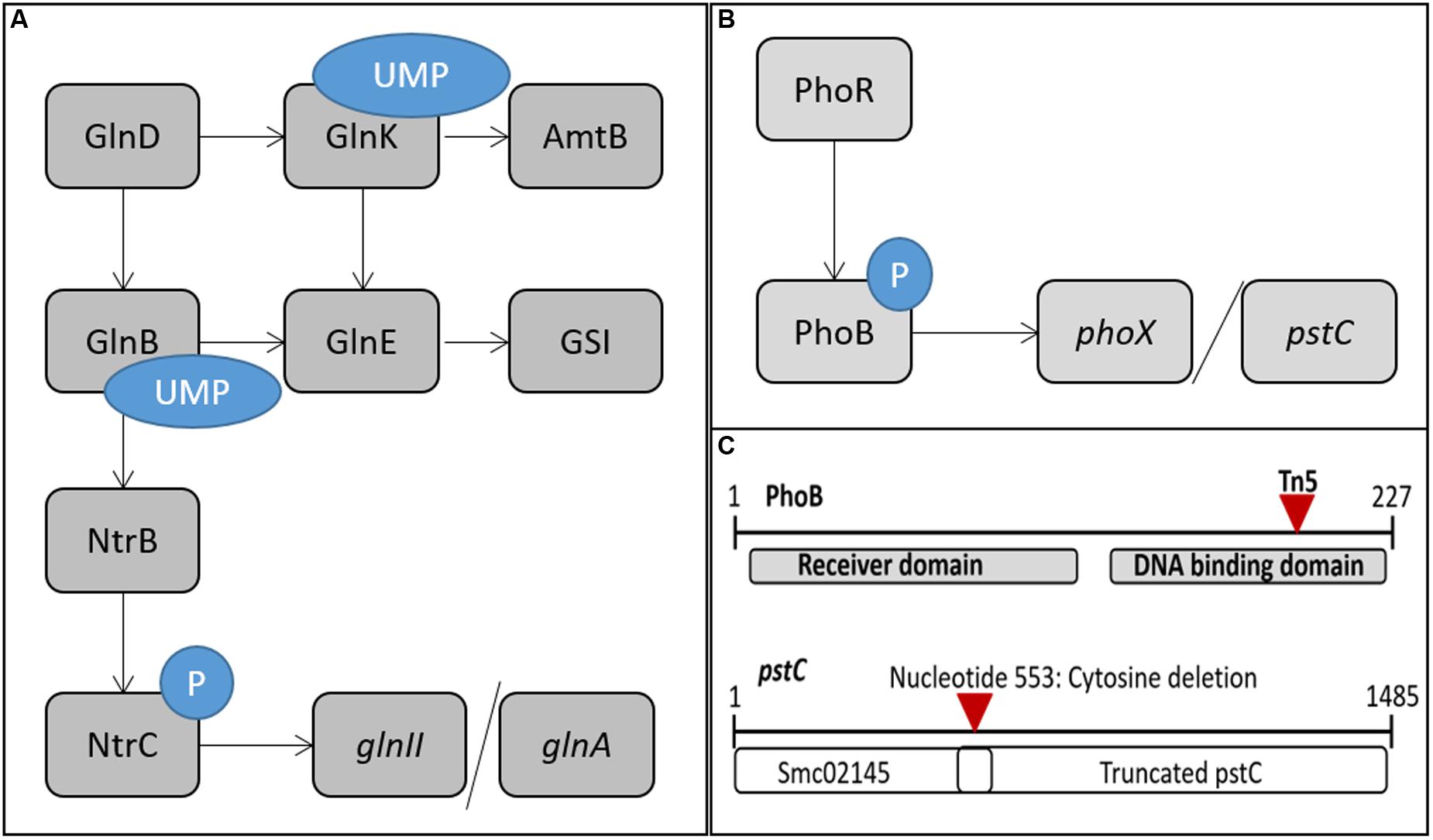
FIGURE 1. The NSR and PSR are used by Sinorhizobium meliloti to cope with nitrogen and phosphate limitations. Cartoon models of the regulatory circuits of the NSR and PSR are depicted here, as well as the phoB::Tn5 mutation, and pstC repair. (A) Model of the NSR during low nitrogen conditions. In response to low nitrogen, GlnD uridylylates GlnB and GlnK. GlnK-UMP activates the ammonium transporter AmtB. Both GlnB-UMP and GlnK-UMP act on GlnE to deadenylylate GSI-AMP and produce the active form of GSI. GlnB-UMP activates the NtrB/NtrC two-component system. NtrB phosphorylates NtrC. NtrC-P binds target promoters to activate a set of genes, including amtB, glnII, and glnA. (B) Model of the PSR during low phosphate conditions. In response to low levels of phosphate, PhoR phosphorylates PhoB. PhoB-P binds target promoters, such as phoX and pstC, to activate gene expression. (C) Cartoon depicting the site of Tn5 transposon insertion in PhoB and the mutation in pstC leading to the dysfunctional Pst transport system in S. meliloti Rm1021.
Deletion of the PII proteins results in slow growing, small colonies in S. meliloti (Yurgel et al., 2010). Poor growth in strains lacking the PII proteins is also observed in Rhodospirillum rubrum (Zhang et al., 2001), Escherichia coli (minimal media only) (Atkinson and Ninfa, 1998), Azospirillum brasilense (de Zamaroczy, 1998), and Azorhizobium caulinodans (Michel-Reydellet and Kaminski, 1999). Spontaneous reversion of the growth rate in these strains does occur, a phenomena known as pseudoreversion. In addition to poor growth, PII deletion strains consistently display increased GS activity. Suppressor mutations were identified in R. rubrum that restore growth of a PII deletion strain (Zhang et al., 2006). The mutations were found in five distinct genes and in each case, total GS activity was decreased compared to the parent strain, leading to the conclusion that excessive GS activity is leading to the slow growth phenotype of PII deletion strains. In S. meliloti, the Rm1021ΔglnBΔglnK deletion strain (hereafter referred to as BK) displays increased GS activity compared to wild type 1021 (Yurgel et al., 2010), but there are no published studies relating the growth rate to GS activity in BK pseudorevertants.
In the rhizosphere, both phosphate and nitrogen are often limiting nutrients. S. meliloti engage a PSR to cope with phosphate limitation (Figure 1B) (Al-Niemi et al., 1997). The PSR involves fewer regulatory and sensor proteins than the NSR, with the core regulation carried out by a two-component regulatory system, PhoR/PhoB. In this system, PhoR is the sensor kinase that detects the phosphate status of the cell. When phosphate supply is limited, PhoR becomes phosphorylated and then transfers the phosphate to PhoB, the response regulator. PhoB-P then regulates transcription of its target genes by binding in promoter regions where it interacts with RpoD, the sigma 70 subunit of RNA polymerase (Makino et al., 1993; Al-Niemi et al., 1997). PhoB regulated genes include those involved in phosphate uptake, transport and metabolism as well as exopolysaccharide production and cell protection genes (Mendrygal and Gonzalez, 2000; Yuan et al., 2006a). Alkaline phosphatases (AP) like PhoX have increased expression and activity during phosphate stress enabling the bacteria to release phosphate from environmental compounds and make it available for transport into the cell (Zaheer et al., 2009).
Transport of phosphate is complicated and involves several different transport systems that differ in their capacity and affinity. The high affinity, high velocity pst transporter is one of the primary transport systems for phosphate uptake in S. meliloti. Rm1021, a strain that is commonly used when conducting research on S. meliloti, has a defective pstC gene that produces a truncated protein (Figure 1C) (Yuan et al., 2006a). This leads to defective transport, resulting in constitutive activation of PhoB (Yuan et al., 2006a) and a moderate level of PSR activation at all times. In the course of this work, the pstC gene mutation was repaired and experiments were performed in both wild type Rm1021 and a strain of Rm1021 with the repaired allele, which has been designated pstC+. This allowed us to relate previous studies performed in Rm1021 (Yuan et al., 2006a; Yurgel et al., 2010, 2013) with studies done here and in other strains having a properly functioning PSR (Arcondeguy et al., 1997; Krol and Becker, 2004).
The NSR and PSR pathways have been characterized independently and only recently has data been presented indicating regulatory crosstalk (Krol and Becker, 2004; Martín et al., 2011; Yurgel et al., 2013). It is accepted that glnII is regulated by the NtrB/NtrC system, but the expression of glnII has been shown to be dependent on environmental phosphate conditions as well (Krol and Becker, 2004). Additionally, a change in expression of phosphate-related genes during nitrogen stress was described leading to more evidence of crosstalk between these two stress responses (Yurgel et al., 2013). We hypothesized that there was a coordination of the NSR and PSR and that phosphate conditions may also regulate aspects of the NSR, such as GS expression or activity. It was also hypothesized that nitrogen may affect aspects of the PSR, such as AP expression or activity. In this paper, we characterize a phoB::Tn5 mutant that alters the phenotypes of a PII deletion strain and lends support to crosstalk between the regulatory systems. We also demonstrate that phosphate conditions affect GS expression and activity, but nitrogen does not affect AP expression and activity in Rm1021 strains.
Materials and Methods
Strains, Plasmids, and Media
Strains and plasmids used are listed in Table 1. S. meliloti was grown at 30°C minimal mannitol NH4Cl or YMB liquid medium or medium that had been solidified with 1.5% agar (Sigma) (Somerville and Kahn, 1983). For assays using high or low nitrogen and/or phosphate, Min MOPS (Al-Niemi et al., 1997) liquid media containing 0.4% NH4Cl (nitrogen-sufficient) or 0.04% glutamate (nitrogen-deficient) was used. KH2PO4 was added at 2 mM for phosphate sufficient conditions and 50 μM for phosphate deficient conditions (Bardin et al., 1996). E. coli strains were grown at 37°C on LB media (Sambrook, 2001). Neomycin was added at 200 μg/mL and tetracycline at 10 μg/mL for S. meliloti. For E. coli, kanamycin was added at 40 μg/mL. LBMC was used for transduction (Finan et al., 1984). 5-Bromo-4-chloro-3-indolyl phosphate (X-Phos) was added to solid media at a concentration of 50 μg/mL.
Genetic Techniques
Standard procedures were used for DNA manipulations (Sambrook, 2001). Tn5 transposon mutants were generated as described by Yurgel and Kahn (2008) using the non-replicating suicide plasmid pSUP5011 to deliver the transposon to S. meliloti. Arbitrary PCR was performed to identify the location of the transposon insertions (Yurgel and Kahn, 2008). Plasmids pJ1700G-pGlnA and pJ1700G-pPhoX were constructed in a manner similar to pJ1700G-pGlnII as described by Yurgel et al. (2010).
Replacement of a wild type allele of pstC was carried out through a cloning, conjugation, and transduction method (Labes et al., 1993). In Rm2011, a 438 base pair intergenic segment of DNA (chromosomal position 560651 to 561089) was amplified from an intergenic region upstream of pstC (SMc02145) between SMc02151 and SMc02150. The amplified DNA was ligated into plasmid pK19mobsacB to give p2011pstC. Using conjugation, the plasmid was integrated into the Rm2011 chromosome between SMc02151 and SMc02150, marking this region with a neomycin resistance gene contained within the plasmid. Using μ12 bacteriophage-mediated transduction, the integrated plasmid and nearby pst operon were delivered from Rm2011 to the Rm1021 strains of interest. Neomycin resistant colonies were selected and streaked for isolation from phage μ12. Visual confirmation of the repair of the pstC+ allele was observed on plates containing X-Phos, a substrate for AP that is an indirect method for observing PSR activation that results from the defect in pstC. Because of the phosphate transport defect caused by the pstC mutation, Rm1021 is normally blue on X-Phos media. White colonies were chosen after transduction. Phage-free colonies were then plated on YMB containing 5% sucrose to select for loss of the p2011pstC plasmid by recombination, leaving behind the pst operon and surrounding region from Rm2011. The repaired pstC+ gene was confirmed via DNA sequencing.
Transduction
Phage-mediated transduction was performed as described by Finan et al. (1984). Phage μ12 was used to deliver DNA into S. meliloti strains. Phage lysates were filtered with a 0.2 μ syringe filter to remove bacteria before infecting new cultures. Resulting colonies were selected on the appropriate antibiotic and streaked out until the phage was eliminated (Yurgel et al., 2012).
Electroporation of S. meliloti
Electroporation of S. meliloti was based on a protocol by Weigel and Glazebrook (2006) for transformation of Agrobacterium. Electrocompetent S. meliloti were made by growing 250 mL of cells in LB broth to an A600 of ∼0.2. Cells were spun down and washed with cold 10% glycerol four times, each time concentrating the cells in one fifth the volume of glycerol. Cells were resuspended in a final volume of 1 mL and flash frozen with liquid nitrogen before storage at -80°C. 50 μL of electrocompetent S. meliloti were incubated for 10 min with ∼2500 ng of plasmid before being electroporated at 2.0 kV in a 2 mm gap length cuvette. Cells were allowed to recover in 1 mL LB broth and grown overnight at 30°C with agitation. The recovered cells were plated on LB media containing the appropriate antibiotic for selection of S. meliloti containing the desired plasmid.
Replication Test
To test growth rate and colony size on Min MOPS media, replication tests were performed as described by Yurgel et al. (2010). Briefly, cells were resuspended to an A600 of 0.05. 100 μL of resuspended cells were added to one row of wells of a 96 well plate. Serial 10-fold dilutions were made ending with a 1:106 dilution. A sterile bolt replicator was used to transfer aliquots of each dilution to solid media. Plates were then incubated at 30°C and checked at various times to compare growth between the strains.
Western Blot
Western blot procedures were carried out according to Yurgel and Kahn (2008) with a few modifications. Cells were grown in high (containing 0.4% NH4Cl) or low (containing 0.04% glutamate) nitrogen media for 18 h before being spun down and resuspended in 1 M TE buffer. SDS loading dye was added and proteins were denatured by incubating at 95°C for 20 min. Proteins were separated on a 10% bis-tris acrylamide gel (Invitrogen) then transferred to a nitrocellulose membrane (Whatman) by electrophoresis. GSII was detected using a S. meliloti anti-GSII antibody (Shatters et al., 1989). The antibody/protein complex was detected using the Supersignal West Pico Chemiluminescence kit (Thermo Scientific).
GS Activity
Glutamine synthetase activity was measured by performing the γ-glutamyltransferase assay. Approximate GS activity was quantified based on the formation of γ-glutamylhydroxamate from glutamine and hydroxylamine. Cultures were grown on Minimal MOPS plates (Bardin et al., 1996) with 50 μM KH2PO4 (low Phosphate) or 2 mM KH2PO4 (high Phosphate) containing 0.4% NH4Cl (nitrogen-sufficient) or 0.04% glutamate (nitrogen-deficient) with succinate as a carbon source for 60 h and then colonies were resuspended in broth of the same composition. GS activity was then performed as described by Somerville and Kahn (1983). Activity units are calculated using A540/(mg protein∗min).
β-Glucuronidase Assay
Cultures were grown for 48 h in minimal MOPS media with 50 μM KH2PO4 (low phosphate) or 2 mM KH2PO4 (high phosphate) containing 0.4% NH4Cl (nitrogen-sufficient) or 0.04% glutamate (nitrogen-deficient). Cultures were diluted 1:40 and grown for 18 h. Expression reactions were run as described by Miller (1972) using 4-nitrophenyl-β-glucuronide (ONPGluc) as the colorimetric substrate. Culture absorbance at 600 nm was measured as well as absorbance at 415 nm, which was due to release of o-nitrophenol. Approximate β-D-glucuronidase activity was calculated using the formula A415/(A600∗min).
Alkaline Phosphatase Assay
Alkaline phosphatase activity was measured as described by Yurgel et al. (2000). Briefly, cultures were grown on minimal MOPS plates with sufficient or deficient nitrogen and/or phosphate for 72 h at 30°C. Cells were resuspended and washed in 1 M Tris HCl, pH 8 and resuspended again in 1 ml of 1 M Tris HCl. The reaction was started when 0.1 mL of p-nitrophenyl phosphate (pNPP) substrate (Sigma) was added to 0.9 mL of cells and incubated at 30°C for 60 min. Reactions were stopped by addition of 0.1 mL of 1 M KH2PO4. Absorbance of cell cultures was measured at 600 nm and absorbance of the reaction mix was measured at 420 nm. Activity units are A420/(A600∗min).
Results
Alleviation of BK Strain Growth Inhibition by a phoB::Tn5 Transposon Mutation
One of the most striking phenotypes of the BK deletion strain was its very slow growth rate and associated small colony size (Yurgel et al., 2010). S. meliloti BK was mutagenized with transposon Tn5, allowing the transposon to integrate randomly throughout the genome. Colonies were selected that were larger in size and seven distinct Tn5 insertion locations were identified in these colonies. Phage-mediated transduction was used to determine if the transposon was associated with faster growth by moving the neomycin resistant transposon into slow-growing BK cells. Of the seven identified locations, only one, containing a Tn5 insertion in phoB, was linked to improved growth of the BK strain, indicating that faster growth by the other six was due to unlinked mutations. The phoB::Tn5 mutation was in the DNA binding/sigma70 interacting C-terminal domain of PhoB, the response regulator in the PSR (Figure 1C) (Blanco et al., 2011).
PhoB regulates AP and AP activation is often used as a measure of the PSR and phosphate stress. In order to determine if the phoB mutant was able to activate the PSR, the mutant was placed on LB media containing X-Phos, a substrate for AP that produces a blue pigment when X-Phos is hydrolyzed (Long et al., 1988). Strains containing the phoB::Tn5 mutation produced white colonies on X-Phos media whereas strains with the unrepaired phoB were blue. This indicated that the phoB::Tn5 mutant lacked the ability to induce a PSR. The other six faster-growing transposon mutants were blue on X-Phos, suggesting that the spontaneous mutations that resulted in faster growth were not due to mutations inactivating AP or the PSR. This data indicates that mutations in other pathways, not just the PSR, can alter the growth rate of strains deficient in the PII proteins, GlnB and GlnK. Multiple attempts to delete phoB were unsuccessful and in the published literature, most phoB mutants in S. meliloti include a transposon or other mutation at the C-terminal end of the protein similar to the mutant isolated here (Krol and Becker, 2004). While the mutation in phoB was initially thought of as a knockout mutation, it may be a protein with altered function instead of a true knockout. The lack of a mutant described in literature as a complete gene deletion or disruption or mutation in the N-terminal region in S. meliloti suggests that this protein cannot be knocked out completely and that the truncated version may retain some sort of activity despite its inability to mediate the PSR.
In order to determine the impact of the phoB::Tn5 in a wild type background, the mutation was also transduced into Rm1021. A replication test of the various Rm1021 and BK strains discussed in this paper was performed on Min MOPS media with nitrogen and phosphate sufficient and deficient conditions. Figure 2 shows a representative plate on nitrogen and phosphate sufficient media after 3 days of growth. All media conditions showed similar growth patterns between strains, but it was most pronounced on the N and P sufficient media. BK grew much more slowly than Rm1021, as described previously (Yurgel et al., 2010). The BK strain containing the phoB::Tn5 mutation grew at a faster rate and produced larger colonies than the BK strain with wild type phoB. The BK strains had a smaller colony size than the Rm1021 strains for each pair of mutants (wild type, pstC+, phoB::Tn5), indicating that the absence of the PII genes affected colony size and growth rate in all backgrounds. However, adding either pstC+ or phoB::Tn5 to the BK background improved growth, suggesting that activation of the PSR was inhibitory.
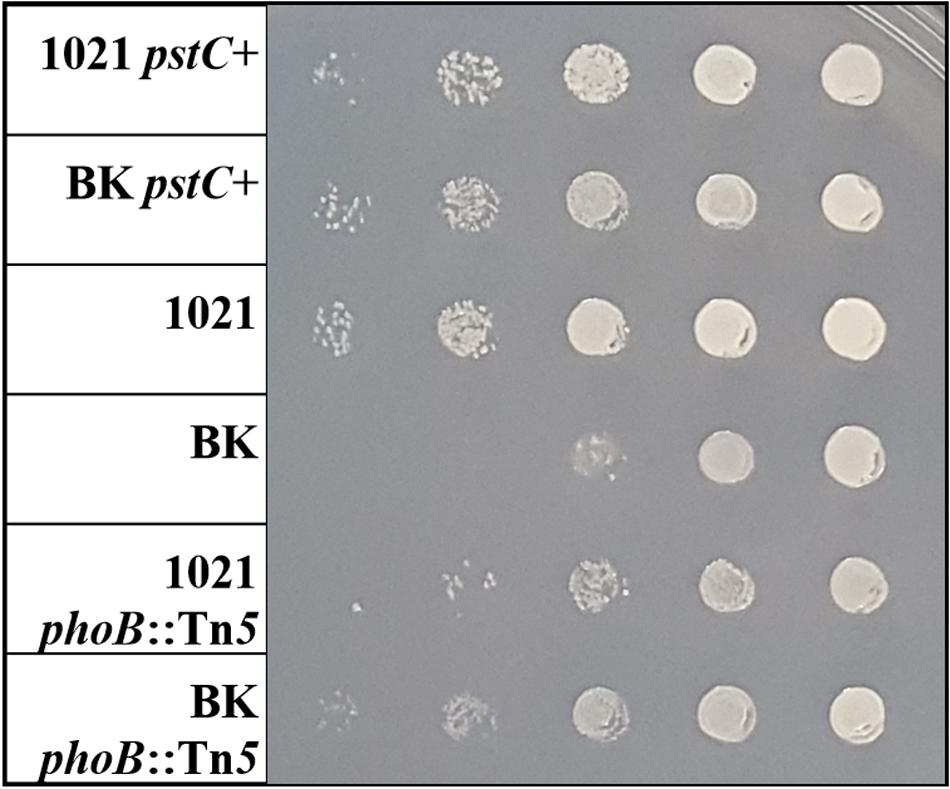
FIGURE 2. Replication test of various Rm1021 strains showing growth and colony size at 72 h on Min MOPS Glu with 50 μM KH2PO4 media. From left: Rm1021 pstC+, BK pstC+, Rm1021, BK, Rm1021 phoB::Tn5, BK phoB::Tn5. Columns are 1:10 dilutions from top to bottom.
GSII is Downregulated by the phoB Mutation
Another striking phenotype of the BK deletion strain observed previously was the altered abundance of GSII protein during both nitrogen sufficient and deficient conditions (Yurgel et al., 2010). Typically, GSII is only detected during nitrogen stress but its expression in the BK background was very high and actually higher when ammonium was present. To test whether GSII continued to be expressed in the BK strain containing the phoB::Tn5 mutation, the protein was visualized using Western blots (Figure 3A; Supplementary Figure S1). In Rm1021 phoB::Tn5, GSII was detected only during nitrogen stress, similar to the pattern of expression of GSII in Rm1021 without the phoB mutation. However, the BK phoB::Tn5 mutant did not have any detectable GSII protein when grown in either nitrogen condition, in contrast to what was observed in the BK strain and the wild type. GSII protein levels in pstC+ strains were similar to the parent strains (Figure 3B) in that GSII in Rm1021 and its pstC+ derivative were repressed by ammonium, while GSII was still expressed in BK and its pstC+ derivative in the presence of ammonium. These data suggest that GlnB/GlnK and PhoB are both involved in the expression of the GSII protein. Since the original expectation was that the ΔglnBΔglnK mutations in BK would block the NSR and expression of GSII, these results are consistent with the idea that expression of GSII in BK is anomalous and that the anomaly depends on PhoB.
FIGURE 3. Western blot of GSII protein abundance in various strains and growth conditions. Wild type (1021) or BK mutant strains with or without (A) the phoB transposon mutation or (B) a repaired pstC+ gene. In the nitrogen row the + or - indicates cultures grown in (+) MMNH4+ or (-) MM Glu, respectively.
The phenotypic differences in the phoB::Tn5 mutant between the Rm1021 and BK backgrounds suggest interactions between the nitrogen and PSRs. This hypothesis also suggests that phosphate availability may affect the NSR and nitrogen may affect the PSR. In addition to exploring the phosphate and nitrogen effects on the NSR and PSR, the phoB::Tn5 mutant strain was characterized further to better understand how GlnB/GlnK and PhoB may be interacting to jointly regulate the two stress responses.
Phosphate Availability Influences the NSR
To characterize the effects of nitrogen on the PSR, the PSR must be intact so that classical regulation and activation can be observed. Since the pstC mutation in Rm1021 partially activated the PSR, the strains were converted to pstC+ by transduction. Rm1021 without the repaired pstC gene was included in some experiments as a control (not shown) to ensure results were consistent with previously published data (Yurgel et al., 2010). The joint effects of nitrogen and phosphate availability on the NSR and PSR were explored in the pstC+ strains. Additionally, phoB::Tn5 mutant strains were included in some experiments to gather information on how PhoB may be involved in the regulation of the NSR or crosstalk with the PII proteins. The pstC+ allele also reverses the slow growth characteristic of the BK strain (Figure 2), an effect that is consistent with the observed downregulation of the PSR in this strain as the result of improved phosphate uptake. While this change does not disrupt PhoB, it does lead to less PhoB-activated transcription.
Total GS enzymatic activity was used to determine whether the NSR was active during nitrogen limitation in the newly constructed strains and if the level of activity was influenced by phosphate stress. Most rhizobia are unusual because they contain two distinct GS proteins that are regulated by the NSR using circuits that involve the PII proteins. The expression and overall activity of GSI and GSII are upregulated when nitrogen is limited and downregulated when nitrogen is sufficient. It was shown that, compared to wild type Rm1021, total GS activity increased in a strain missing the PII proteins because, while GSII protein was present in Rm1021 only during nitrogen limitation, GSII protein was present in the BK deletion strain at a very high level when ammonium was available and at a lower level when the nitrogen source was glutamate (Yurgel et al., 2012). The observed increase in GSII protein may be related to the observation that, regardless of nitrogen source, all of the detected GSI in the BK strain was present as GSI-AMP, the inactive form of GSI. In Rm1021, GSI, but not GSI-AMP, was detected under nitrogen stressed conditions, reflecting the need for active enzyme (Yurgel et al., 2010). These results are consistent with the need for PII proteins to transduce the signal generated by GlnD that nitrogen is limited.
Glutamine synthetase activity was affected by phosphate in a wild type background, but not in a PII deletion background. Figure 4 shows total GS activity in the pstC+ and phoB::Tn5 strains during high and low nitrogen and phosphate conditions. (The data used to generate Figures 4 and 5 are included in Supplementary Table S1 in order to permit other comparisons to be made.) When nitrogen was sufficient, phosphate stress did not have a significant effect on total GS activity in any strain. In contrast, when nitrogen was limited, phosphate deficient conditions repressed GS activity by ∼2-fold in Rm1021 pstC+ and Rm1021 phoB::Tn5 compared to phosphate sufficient conditions. This reduction in GS activity when phosphate availability was limited was not seen in the BK pstC+ strain. The BK phoB::Tn5 strain had a very low level of total GS activity regardless of the nitrogen or phosphate available, consistent with the idea that both the NSR and PSR were important in enzyme production or activation.
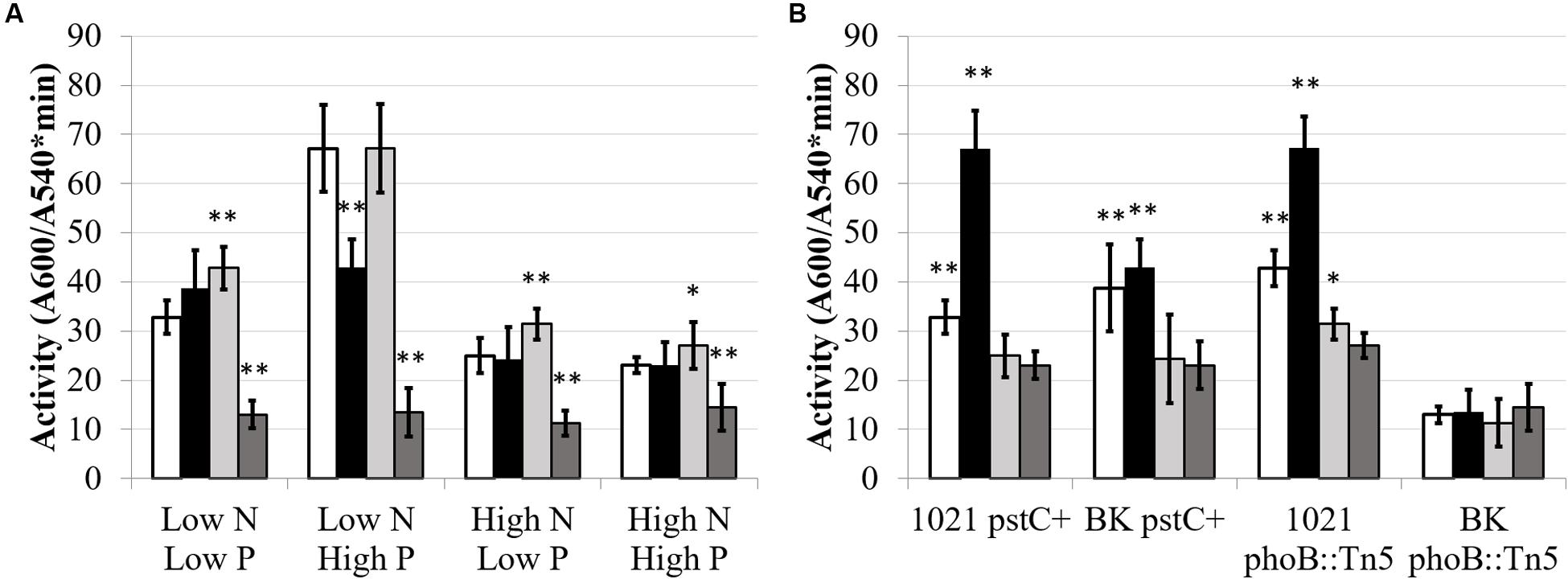
FIGURE 4. Total glutamine synthetase activity under different media conditions. Each bar represents three replicates each from three experiments for a total n = 9. Error bars are ±1 SD. Activity units are A540/(mg protein∗minute). (A) Comparison showing differences between strains for each media type on the X-axis. Asterisks are significant differences from Rm1021 pstC+ for each media condition. ∗∗P < 0.01; ∗P < 0.05 as determined by a Student’s t-test. White bar is 1021 pstC+, black bar is BK pstC+, light gray bar is 1021 phoB::Tn5, dark gray bar is BK phoB::Tn5. (B) Comparison showing differences between media types for each strain on the X-axis. Asterisks are significant differences from High N High P media conditions for each strain. ∗∗P < 0.01; ∗P < 0.05 as determined by a Student’s t-test. White bar is Low N Low P media, black bar is Low N High P media, light gray bar is High N Low P media, dark gray bar is High N High P media.
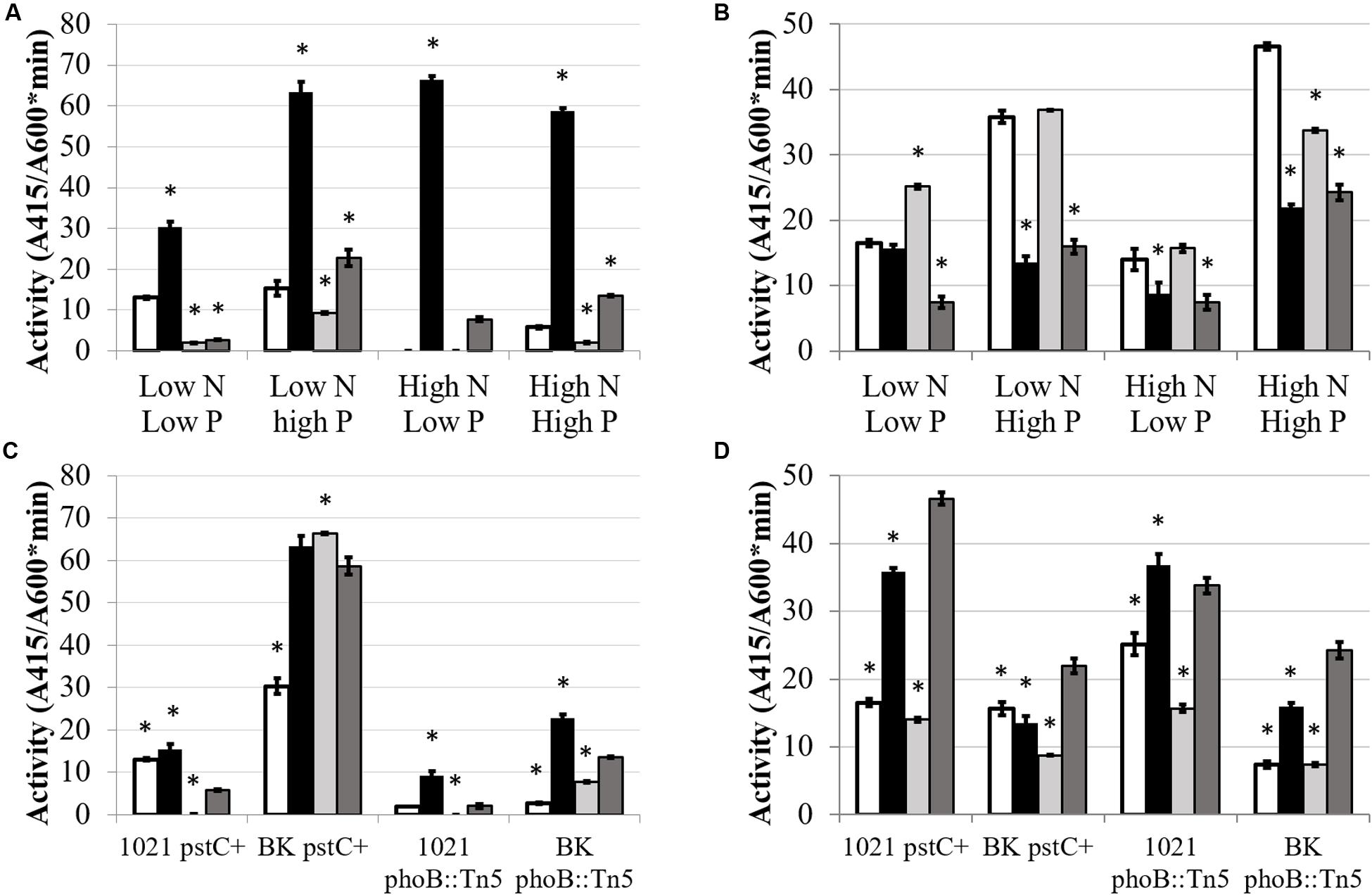
FIGURE 5. GusA fusion expression of glnII (A,C) and glnA (B,D) as a function of media and genetic background. Each bar represents 3 replicates for a total n = 3. Error bars are ±1 SD. Activity units are A415/(A600∗minute). (A,B) Comparison showing differences between strains for each media type on the X-axis. Asterisks are significant differences from Rm1021 pstC+ for each media condition as determined by a Student’s t-test at P < 0.01. White bar is 1021 pstC+, black bar is BK pstC+, light gray bar is 1021 phoB::Tn5, dark gray bar is BK phoB::Tn5. (C,D) Comparison showing differences between media type for each strain on the X-axis. Asterisks are significant differences from High N High P for each strain as determined by a Student’s t-test at P < 0.01. White bar is Low N Low P media, black bar is Low N High P media, light gray bar is High N Low P media, dark gray bar is High N High P media.
Glutamine synthetase I activity is primarily regulated through adenylylation whereas GSII is primarily regulated through transcription and subsequent translation of the glnII gene. GSI and GSII are the protein products of the glnA and glnII genes. Activity of the glnA and glnII promoters was examined to see if they responded to phosphate limitation in parallel with GS activity. If phosphate conditions had an effect on the GS activities by influencing transcription, this would be most obvious in the effect on glnII expression. Expression of glnA and glnII were assayed by measuring β-glucuronidase activity generated by the activity of the glnA and glnII promoters fused to a gusA reporter gene in plasmids pJ1700G-pGlnA and pJ1700G-pGlnII.
As expected, glnII expression was activated by nitrogen limitation and repressed by ammonium (Figure 5). However, the level of this response was regulated by phosphate availability. In Rm1021 pstC+, changing phosphate concentration did not lead to a significant change in glnII expression when nitrogen was limited, but adding phosphate significantly stimulated glnII expression during nitrogen sufficient conditions from an undetectable level to a low but measurable β-glucuronidase activity.
Expression of the glnII-gusA fusion was strikingly higher in the BK pstC+ background than in the Rm1021 pstC+ background, an observation consistent with GSII protein abundance. The level of expression in BK pstC+ ranged from ∼3-fold higher than in Rm1021 pstC+ under the “inducing” condition of low N and nearly 100-fold higher in high N where glnII expression in Rm1021 pstC+ was almost completely repressed. The BK pstC+ strain had reduced activity during phosphate limitation compared to sufficiency when nitrogen was limited, demonstrating a response to phosphate levels under conditions when glnII expression is typically induced. Introducing the phoB::Tn5 mutation substantially lowered glnII-gusA expression in both the wild type and BK backgrounds under each of the media conditions tested. The addition of phosphate raised the level of expression of glnII-gusA in both Rm1021 and BK with the phoB::Tn5 mutation indicating that GSII expression was induced by phosphate in the absence of a proper PhoB regulatory protein.
Unlike GSII, GSI is regulated post-translationally by adenylylation. In rhizobia, glnA transcription is not as significant in controlling the amount of GSI activity present and the effect of nitrogen concentration on glnA-gusA expression observed here was not as dramatic as the effect on glnII-gusA (Figure 5). Raising phosphate increased the expression of glnA-gusA for both Rm1021 pstC+ and BK pstC+ when nitrogen was available but only Rm1021 pstC+ had significantly higher expression when nitrogen was limiting. The presence or absence of the phoB::Tn5 mutation did not have a strong effect on the expression of glnA-gusA under most conditions but, for both 1021 phoB::Tn5 and BK phoB::Tn5, phosphate increased glnA-gusA expression by about twofold when phosphate was higher under similar nitrogen conditions.
Effects of Nitrogen on PSR
The GS activity and expression results indicated that phosphate status affected aspects of the NSR. The hypothesis that nitrogen conditions would affect the PSR was also explored. As discussed previously, AP activity is an index of the PSR that is typically induced by low phosphate conditions. AP expression and activity were measured to determine if nitrogen limitation would influence the production of AP. The classical activation pattern was observed and displayed a very definite on/off activity pattern in the Rm1021 pstC+ strain when grown in low or high phosphate, respectively. When the cells were pstC+ in either wild type Rm1021 or the BK deletion strain, nitrogen limitation had no significant effect on the activity of AP (Figure 6). phoX, which codes for the major AP in Rm1021, is upregulated during phosphate limitation in a PhoB-dependent manner (Zaheer et al., 2009). Since phosphatases are typically inhibited by phosphate (Fernley and Walker, 1967), a gusA reporter fusion to phoX was constructed and introduced into the mutant strains to measure a reporter activity based on expression of the phoX promoter. The activity of β-glucuronidase from the phoX-gusA fusion mimicked the activity levels of AP for each strain and condition (Figure 7). The pstC+ strains showed increased expression during phosphate limitation and very minimal expression when the phosphate level was sufficient, but changing the nitrogen availability had no effect. The phoB::Tn5 mutant strains had minimal expression of the phoX-gusA, regardless of phosphate or nitrogen levels. These data show nitrogen status was not having an effect on AP expression or activity in addition to the known regulation by phosphate availability. When any of these strains carried the phoB::Tn5 transposon insertion, they had very low AP or phoX-gusA derived β-glucuronidase activity, regardless of nitrogen or phosphate conditions. This pattern of expression would be expected if the phoB gene was mutated, phoX was regulated by PhoB and the phoB::Tn5 mutation was not leaky in its control of the PSR. This implies that the phosphate responsiveness of both glnA and glnII expression was not due to some residual PhoB activity and that another level of phosphate regulation in S. meliloti appeared to be involved.
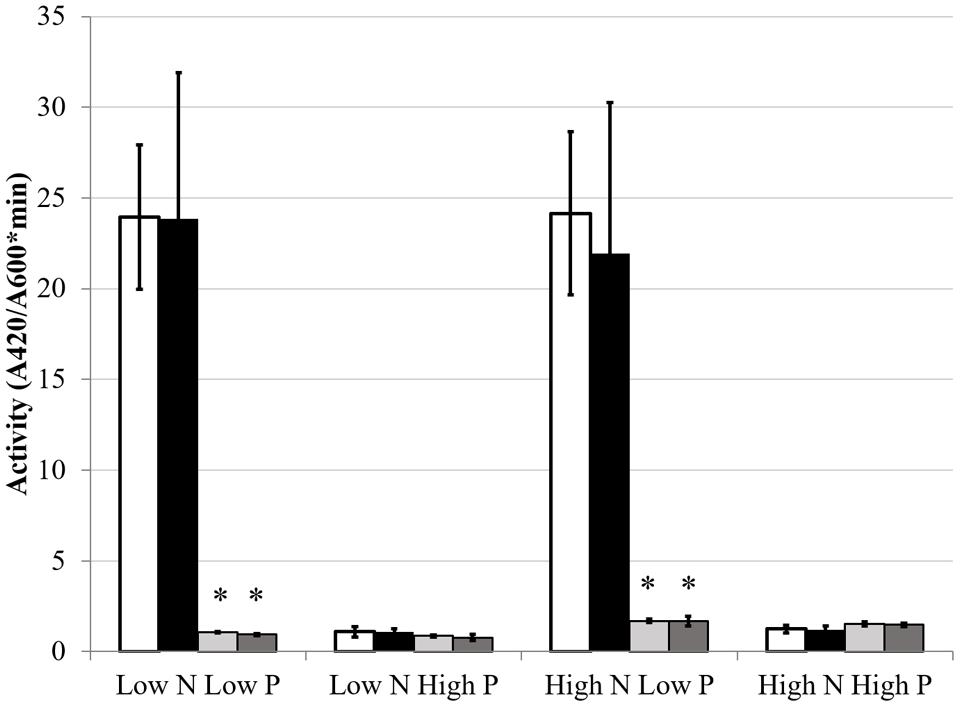
FIGURE 6. Alkaline phosphatase activity under different media conditions. Each bar represents three replicates each from three independent experiments for a total n = 9. Error bars are ±1 SD. Activity units are A420/(A600∗minute). In all cases, asterisks are significant differences from Rm1021 pstC+ for that media condition as determined by a Student’s t-test at P < 0.01. White bar is 1021 pstC+, black bar is BK pstC+, light gray bar is 1021 phoB::Tn5, dark gray bar is BK phoB::Tn5.
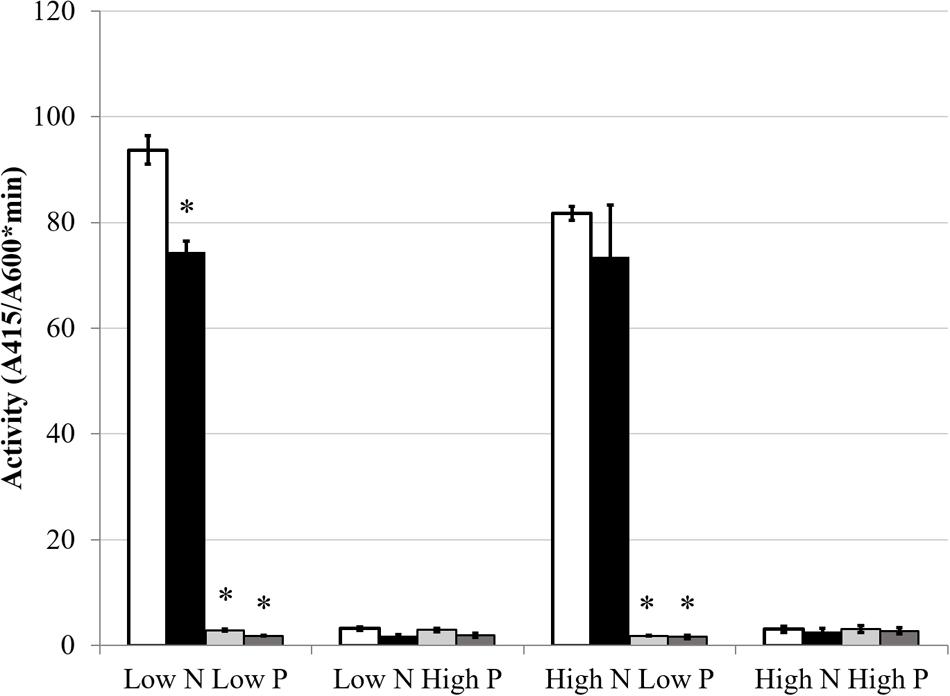
FIGURE 7. GusA fusion expression of phoX. Each bar represents three replicates each from two independent experiments for a total n = 6. Error bars are ±1 SD. Activity units are A415/(A600∗minute). In all cases, asterisks are significant differences from Rm1021 pstC+ for that media condition as determined by a Student’s t-test at P < 0.01. White bar is 1021 pstC+, black bar is BK pstC+, light gray bar is 1021 phoB::Tn5, dark gray bar is BK phoB::Tn5.
Discussion
The persistence of rhizobia in various soil types is crucial for BNF to occur during symbiosis with plants. Responding to various stresses and nutrient limitations in soil has led to the development of stress response regulatory pathways by bacteria. The nitrogen and PSRs of S. meliloti have been characterized independently, and recent studies have indicated a coordination of the stress responses. It had been shown that a mutant lacking the GlnB and GlnK PII proteins, which mediate the NSR, has a striking slow growth phenotype. Faster growing pseudorevertants occur at a significant rate in the BK strain and in order to understand which genes and proteins are involved in the pseudoreversion, transposon mutagenesis was performed. The mutagenesis screening led to the identification of a phoB::Tn5 mutation that could generate faster growing colonies.
In R. rubrum, the slow growth phenotype of a PII deletion strain was alleviated by additional mutations that each led to a decrease in overall GS activity (Zhang et al., 2006). Consistent with this, GSII protein was no longer detected in either nitrogen condition in the BK phoB::Tn5 mutant strain, in contrast to the elevated level of GSII found in the BK mutant. This aspect of the phoB phenotype was specific to the mutant that lacked PII proteins—the phoB::Tn5 mutation did not affect the pattern of GSII protein expression in Rm1021. It was previously shown that a group of PSR-regulated genes had altered expression in the absence of the PII proteins (Yurgel et al., 2013). The finding that a phoB mutation alleviated the growth inhibition in the BK strains led us to explore the hypothesis that the NSR may be affected by phosphate as well as nitrogen, and that nitrogen may be affecting the PSR in addition to phosphate. The effect of phosphate on the NSR was investigated by evaluation of GS genes expression and enzymatic activity and the effect of nitrogen on the PSR was investigated through the AP gene and protein activity.
In addition to the well-characterized nitrogen regulation of the GS enzymes, phosphate availability also affected S. meliloti GS expression and activity. The importance of the PII proteins in proper regulation of total GS activity was reinforced as well. In the wild type Rm1021 background, phosphate limitation repressed GS activity in both the pstC+ and phoB::Tn5 strains when nitrogen was limited. This indicated that, while GS activity depended on nitrogen conditions and regulation involving the PII proteins, phosphate stress further regulated GS activity and suggested the involvement of the PII proteins with the PSR, perhaps through a phosphate-responsive transcription factor. Since Rm1021 pstC+ and Rm1021 phoB::Tn5 show similar repression patterns of total GS activity, PhoB is unlikely to be involved in the low phosphate repression of GS activity—if it were, this repression should be abolished in the phoB::Tn5 strain. The BK pstC+ strain displayed the unusual increased GS activity during limited nitrogen, but not during nitrogen sufficient conditions. However, it was not responsive to changing phosphate levels. The consistent low GS activity in the BK phoB::Tn5 strain suggested that both BK and PhoB were required for the proper activation of GS. This phenotype coincided with the changes observed in GSII protein as detected via Western blot indicating some sort of interaction or coordinated regulation between GlnB/GlnK and PhoB.
PhoB is the response regulator of the two component PhoR/PhoB system (Figure 1). PhoR phosphorylates PhoB in response to low phosphate conditions, converting PhoB into the more transcriptionally active PhoB-P state. A mutation in PhoR would decrease PhoB-P. A small amount of PhoB could potentially be phosphorylated by small molecules (Wanner and Wilmes-Riesenberg, 1992), but the low level of phosphorylation would likely not be sufficient to induce the necessary response to phosphate stress. We have not investigated whether the lack of PhoB activation due to a PhoR knockout would produce similar phenotypes as the phoB::Tn5 mutant in regard to its regulation of the PSR. However, it is possible that regulatory crosstalk between two component systems is occurring, where PhoR-P is activating NtrC in the absence of PhoB, its cognate response regulator (Verhamme et al., 2002).
In wild type Rm1021, phosphate concentration affected glnII expression when nitrogen was available—low phosphate nearly abolished glnII expression when nitrogen was sufficient. One potential rationale is that the cell was allocating resources to the PSR when nitrogen was present and the cell did not need to increase the level of GS activity to assimilate sufficient nitrogen. When nitrogen and phosphate are both limited, cells must cope with these stresses simultaneously and may prioritize its response. It is likely the cell continues to commit resources to both stress responses instead of allowing one to function at a high level while the other is severely inhibited by a lack of resources. Another potential explanation for the dual regulation of GS by both nitrogen and phosphate could be that the cell was trying to balance the nitrogen and phosphate pools within the cell. Nucleic acid synthesis requires inputs of both nitrogen and phosphate and an imbalance in the resource pools could be inhibitory. Protein synthesis is less affected by phosphate availability since, while ATP and phosphate are needed at several steps in the process, phosphate is not directed into a sink by the process. The lack of a recognizable Pho box (Yuan et al., 2006b) upstream of glnII suggests that PhoB is not directly binding to the glnII promoter region. This idea is supported by the increased expression in response to additional phosphate that occurs even when PhoB is absent. The ntrB/ntrC promoter also lacks a recognizable Pho box, suggesting that PhoB is not binding in this region and affecting glnII expression by directly regulating ntrB/ntrC expression. In Streptomyces coelicolor, there is direct interaction between PhoP, the response regulator in a two component PSR system, and some promoters involved in its NSR through binding of PhoP at partial Pho box sequences (Rodríguez-García et al., 2009).
Previous studies demonstrated that glnII expression was increased in BK deletion strains. This result was explained by the need for a PII protein to dephosphorylate NtrC-P (Yurgel et al., 2010) so that NtrC remains phosphorylated and induces glnII expression. We found that glnII expression is elevated in the BK pstC+ strain in all media conditions, including in the presence of ammonium. One explanation for increased GSII in BK during nitrogen limitation is that the increased GSII compensates for the adenylylation and inactivation of GSI (Yurgel et al., 2010). Another potential explanation is that GSI is always modified in the BK deletion as a result of the very high levels of GSII protein. The unexpected connection reported here is that when PhoB is mutated in the BK strains, GSII expression and protein levels are drastically reduced. This observation supports the idea that GlnB/GlnK and PhoB mediate a coordinated response to some nutrient conditions.
The data show that the PSR, as measured by AP induction or activity, was not affected by nitrogen availability. This suggests that the cell prioritizes phosphate acquisition during phosphate limitation and concurrent nitrogen deficiency was not affecting activation of the PSR. Due to the low level of AP activity and expression when phosphate was available, there was essentially no opportunity to observe an impact of nitrogen deficiency on expression or activity of AP as was observed when glnII expression was altered by phosphate limitation. Yurgel et al. (2013) found that phoX was induced in the BK deletion strain and by nitrogen limitation in high phosphate media. In the phoX-gusA reporter fusion experiments described above, expression levels were low in high phosphate medium. The sensitivity of the Yurgel et al. (2013) transcriptome study may have allowed the detection of a threefold change in mRNA that was not observable as a comparable change in the level of the PhoX-GusA fusion protein. The lack of activity and expression of AP in the phoB::Tn5 strains demonstrated that the mutation was eliminating the ability of PhoB to induce PSR.
This coordinated response of GS to both nitrogen and phosphate is leading to the exploration of the interaction and dual regulation of the NSR by both nitrogen and phosphate at a deeper level. Global transcription changes in response to nitrogen and phosphate nutrient deficiencies are being characterized to determine how S. meliloti gene expression is regulated by multiple stresses and which cellular processes are impacted the most. A closer examination of PSR and NSR transcript changes is being conducted to determine if there are genes that are regulated by nitrogen or phosphate, respectively. Additionally, transcript changes in the BK background are being examined during one or both nutrient stresses to help understand the interaction between the PII genes and the PSR. Further research might include examining how post-translationally modified protein abundance may be altered by the various media conditions since signal transduction and enzyme activity are known to be important in the NSR. While the data presented here focus on the NSR and PSR, an important question highlighted by this research is whether there are interactions between other nutritional stress responses and, if so, whether these might be mediated by a well-defined integrative mechanism. Understanding how multiple stresses affect each pathway and ultimately gaining insight into how stress responses are coordinately regulated in bacteria may be crucial to understanding which strains are best suited to various soil conditions. The limitation of nitrogen, phosphate, or both in the soil influences the microbial community and can have significant effects on relative levels of bacteria and fungi present (Vitousek et al., 2010; Leff et al., 2015; Fanin et al., 2016). As nutrient levels change and may shift from one limitation to co-limitation, the microbial populations shift as well. A better understanding of how nitrogen and phosphate availability affects both the NSR and PSR can serve to facilitate the selection of strains that may be best suited for certain soil types and the change in nutrients across seasons. In the context of symbiotic nitrogen fixation, response to limiting factors in the soil may determine how rapidly a strain can respond to the opportunity presented by a legume root and be important in competitiveness and an increased crop yield.
Author Contributions
Study conception and design: KH, MK, SY; acquisition of data: KH, MM; analysis and interpretation of data: KH, MK, SY; drafting of manuscript: KH, MK; critical revision: KH, MK, SY.
Funding
This research was supported by the Office of Research in the College for Agricultural, Human and Natural Resource Sciences at Washington State University, and by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-FG03- 96ER20225.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank the WSU Laboratory for Biotechnology and Bioanalysis for DNA sequencing support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01928/full#supplementary-material
FIGURE S1 | (A) GSII Western blots from Figure 3 with Coomassie stained protein gel. Ladder on far right of stained gel, followed by lanes corresponding to the GSII Western conditions and strains as indicated. (B) GSII Western blots from Figure 3 with Coomassie stained membranes to show loaded protein.
References
Al-Niemi, T. S., Summers, M. L., Elkins, J. G., Kahn, M. L., and McDermott, T. R. (1997). Regulation of the phosphate stress response in Rhizobium meliloti by PhoB. Appl. Environ. Microbiol. 63, 4978–4981.
Arcondeguy, T., Huez, I., Tillard, P., Gangneux, C., de Billy, F., Gojon, A., et al. (1997). The Rhizobium meliloti PII protein, which controls bacterial nitrogen metabolism, affects alfalfa nodule development. Genes Dev. 11, 1194–1206. doi: 10.1101/gad.11.9.1194
Arcondeguy, T., Jack, R., and Merrick, M. (2001). PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65, 80–105. doi: 10.1128/MMBR.65.1.80-105.2001
Atkinson, M. R., and Ninfa, A. J. (1998). Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 29, 431–447. doi: 10.1046/j.1365-2958.1998.00932.x
Bardin, S., Dan, S., Osteras, M., and Finan, T. M. (1996). A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178, 4540–4547. doi: 10.1128/jb.178.15.4540-4547.1996
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Blanco, A. G., Canals, A., Bernués, J., Solà, M., and Coll, M. (2011). The structure of a transcription activation subcomplex reveals how σ70 is recruited to PhoB promoters. EMBO J. 30, 3776–3785. doi: 10.1038/emboj.2011.271
de Zamaroczy, M. (1998). Structural homologues PII and Pz of Azospirillum brasilense provide intracellular signalling for selective regulation of various nitrogen-dependent functions. Mol. Microbiol. 29, 449–463. doi: 10.1046/j.1365-2958.1998.00938.x
Fanin, N., Hättenschwiler, S., Chavez Soria, P. F., and Fromin, N. (2016). (A)synchronous availabilities of N and P regulate the activity and structure of the microbial decomposer community. Front. Microbiol. 6:1507.
Fernley, H. N., and Walker, P. G. (1967). Studies on alkaline phosphatase. Inhibition by phosphate derivatives and the substrate specificity. Biochem. J. 104, 1011–1018. doi: 10.1042/bj1041011
Finan, T. M., Hartweig, E., LeMieux, K., Bergman, K., Walker, G. C., and Signer, E. R. (1984). General transduction in Rhizobium meliloti. J. Bacteriol. 159, 120–124.
Galibert, F., Finan, T. M., Long, S. R., Puhler, A., Abola, P., Ampe, F., et al. (2001). The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293, 668–672. doi: 10.1126/science.1060966
Gruswitz, F., O’Connell, J., and Stroud, R. M. (2007). Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc. Natl. Acad. Sci. U.S.A. 104, 42–47. doi: 10.1073/pnas.0609796104
Hirsch, P. R., and Mauchline, T. H. (2015). Chapter two - the importance of the microbial N cycle in soil for crop plant nutrition. Adv. Appl. Microbiol. 93, 45–71. doi: 10.1016/bs.aambs.2015.09.001
Ikeda, T. P., Shauger, A. E., and Kustu, S. (1996). Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259, 589–607. doi: 10.1006/jmbi.1996.0342
Jiang, P., Ventura, A. C., and Ninfa, A. J. (2012). Characterization of the reconstituted UTase/UR-PII-NRII-NRI bicyclic signal transduction system that controls the transcription of nitrogen-regulated (Ntr) genes in Escherichia coli. Biochemistry 51, 9045–9057. doi: 10.1021/bi300575j
Krol, E., and Becker, A. (2004). Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics MGG 272, 1–17.
Labes, M., Rastogi, V., Watson, R., and Finan, T. M. (1993). Symbiotic nitrogen fixation by a nifA deletion mutant of Rhizobium meliloti: the role of an unusual ntrC allele. J. Bacteriol. 175, 2662–2673. doi: 10.1128/jb.175.9.2662-2673.1993
Ladha, J. K., Pathak, H., Krupnik, T. J., Six, J., and van Kessel, C. (2005). Efficiency of fertilizer nitrogen in cereal production: retrospects and prospects. B.-A. Adv. Agron. 87, 85–156.
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Leigh, J. A., and Dodsworth, J. A. (2007). Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61, 349–377. doi: 10.1146/annurev.micro.61.080706.093409
Long, S., McCUNE, S., and Walker, G. C. (1988). Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J. Bacteriol. 170, 4257–4265. doi: 10.1128/jb.170.9.4257-4265.1988
Makino, K., Amemura, M., Kim, S. K., Nakata, A., and Shinagawa, H. (1993). Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 7, 149–160. doi: 10.1101/gad.7.1.149
Martín, J. F., Sola-Landa, A., Santos-Beneit, F., Fernández-Martínez, L. T., Prieto, C., and Rodríguez-García, A. (2011). Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 4, 165–174. doi: 10.1111/j.1751-7915.2010.00235.x
Mendrygal, K. E., and Gonzalez, J. E. (2000). Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182, 599–606. doi: 10.1128/JB.182.3.599-606.2000
Merrick, M. J., and Edwards, R. A. (1995). Nitrogen control in bacteria. Microbiol. Rev. 59, 604–622.
Michel-Reydellet, N., and Kaminski, P. A. (1999). Azorhizobium caulinodans PIIand GlnK proteins control nitrogen fixation and ammonia assimilation. J. Bacteriol. 181, 2655–2658.
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Oldroyd, G. E. D., and Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
Patriarca, E. J., Tate, R., Ferraioli, S., and Iaccarino, M. (2004). Organogenesis of legume root nodules. Int. Rev. Cytol. 234, 201–262. doi: 10.1016/S0074-7696(04)34005-2
Rodríguez-García, A., Sola-Landa, A., Apel, K., Santos-Beneit, F., and Martín, J. F. (2009). Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 37, 3230–3242. doi: 10.1093/nar/gkp162
Sambrook, J. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Shatters, R. G., Somerville, J. E., and Kahn, M. L. (1989). Regulation of glutamine synthetase II activity in Rhizobium meliloti 104A14. J. Bacteriol. 171, 5087–5094. doi: 10.1128/jb.171.9.5087-5094.1989
Simon, R., Priefer, U., and Puhler, A. (1983). “Vector plasmids for in-vivo and in-vitro manipulations of gram-negative bacteria,” in Molecular Genetics of the Bacteria-Plant Interaction: The Rhiizobium Meliloti-Medicago Sativa System, ed. A. Pühler (Berlin: Springer-Verlag), 98–106.
Somerville, J. E., and Kahn, M. L. (1983). Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J. Bacteriol. 156, 168–176.
Spiertz, J. H. J. (2010). Nitrogen, sustainable agriculture and food security. A review. Agron. Sustain. Dev. 30, 43–55. doi: 10.1051/agro:2008064
Verhamme, D. T., Arents, J. C., Postma, P. W., Crielaard, W., and Hellingwerf, K. J. (2002). Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 148, 69–78. doi: 10.1099/00221287-148-1-69
Vitousek, P. M., Porder, S., Houlton, B. Z., and Chadwick, O. A. (2010). Terrestrial phosphorus limitation: mechanisms, implications, and nitrogen-phosphorus interactions. Ecol. Appl. Publ. Ecol. Soc. Am. 20, 5–15.
Wanner, B. L., and Wilmes-Riesenberg, M. R. (1992). Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174, 2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992
Weigel, D., and Glazebrook, J. (2006). Transformation of agrobacterium using electroporation. CSH Protoc. 2006:db.rot4665.
Yuan, Z.-C., Zaheer, R., and Finan, T. M. (2006a). Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188, 1089–1102. doi: 10.1128/JB.188.3.1089-1102.2006
Yuan, Z.-C., Zaheer, R., Morton, R., and Finan, T. M. (2006b). Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 34, 2686–2697. doi: 10.1093/nar/gkl365
Yurgel, S., Mortimer, M. W., Rogers, K. N., and Kahn, M. L. (2000). New substrates for the dicarboxylate transport system of Sinorhizobium meliloti. J. Bacteriol. 182, 4216–4221. doi: 10.1128/JB.182.15.4216-4221.2000
Yurgel, S. N., and Kahn, M. L. (2008). A mutant GlnD nitrogen sensor protein leads to a nitrogen-fixing but ineffective Sinorhizobium meliloti symbiosis with alfalfa. Proc. Natl. Acad. Sci. U.S.A. 105, 18958–18963. doi: 10.1073/pnas.0808048105
Yurgel, S. N., Rice, J., and Kahn, M. L. (2012). Nitrogen metabolism in Sinorhizobium meliloti-alfalfa symbiosis: dissecting the role of GlnD and PII proteins. Mol. Plant. Microbe Interact. 25, 355–362. doi: 10.1094/MPMI-09-11-0249
Yurgel, S. N., Rice, J., and Kahn, M. L. (2013). Transcriptome analysis of the role of GlnD/GlnBK in nitrogen stress adaptation by Sinorhizobium meliloti Rm1021. PLoS ONE 8:e58028. doi: 10.1371/journal.pone.0058028
Yurgel, S. N., Rice, J., Mulder, M., and Kahn, M. L. (2010). GlnB/GlnK PII proteins and regulation of the Sinorhizobium meliloti Rm1021 nitrogen stress response and symbiotic function. J. Bacteriol. 192, 2473–2481. doi: 10.1128/JB.01657-09
Zaheer, R., Morton, R., Proudfoot, M., Yakunin, A., and Finan, T. M. (2009). Genetic and biochemical properties of an alkaline phosphatase PhoX family protein found in many bacteria. Environ. Microbiol. 11, 1572–1587. doi: 10.1111/j.1462-2920.2009.01885.x
Zhang, Y., Pohlmann, E. L., Conrad, M. C., and Roberts, G. P. (2006). The poor growth of Rhodospirillum rubrum mutants lacking PII proteins is due to an excess of glutamine synthetase activity. Mol. Microbiol. 61, 497–510. doi: 10.1111/j.1365-2958.2006.05251.x
Keywords: nitrogen stress, phosphate stress, glutamine synthetase, alkaline phosphatase, PII nitrogen regulatory proteins, Phob
Citation: Hagberg KL, Yurgel SN, Mulder M and Kahn ML (2016) Interaction between Nitrogen and Phosphate Stress Responses in Sinorhizobium meliloti. Front. Microbiol. 7:1928. doi: 10.3389/fmicb.2016.01928
Received: 19 September 2016; Accepted: 16 November 2016;
Published: 30 November 2016.
Edited by:
Christian Sohlenkamp, National Autonomous University of Mexico, MexicoReviewed by:
Ernesto Ormeño-Orrillo, National Agrarian University, PeruDavid Salvador Zamorano Sanchez, University of California, Santa Cruz, USA
Copyright © 2016 Hagberg, Yurgel, Mulder and Kahn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael L. Kahn, kahn@wsu.edu
†Present address: Svetlana N. Yurgel, Department of Environmental Sciences, Dalhousie University, Truro, NS, Canada
 Kelly L. Hagberg
Kelly L. Hagberg Svetlana N. Yurgel
Svetlana N. Yurgel Monika Mulder
Monika Mulder Michael L. Kahn
Michael L. Kahn