- College of Veterinary Medicine, Shandong Agricultural University, Sino-German Cooperative Research Centre for Zoonosis of Animal Origin Shandong Province, Tai’an, China
In this study, differentially expressed proteins in A549 cells (human lung adenocarcinoma epithelial cell line) infected with H9N2 avian influenza virus (AIV) were investigated by two-dimensional electrophoresis (2-DE). Sixteen different spots between the groups (ratio > 2, p < 0.05) were identified with mass spectrometry identification. Proteins located in the downstream of the NF-κB and IFN transcription factor pathways were identified, e.g., ISG15. Actin and keratin were also identified, suggesting that the cytoskeleton may plays an important role in the AIV infection of mammalian cells. These findings could provide insights into the interaction between host and influenza viruses and might provide valuable information for clarifying the pathogenesis of viral infections as well.
Introduction
The avian influenza virus (AIV) H9N2 subtype has been circulating in domestic poultry in mainland China since 1994 (Tang et al., 1998). H9N2 influenza viruses occasionally transmit to humans, which have raised public concerns about a pandemic potential for this subtype of influenza viruses (Lin et al., 2000; Matrosovich et al., 2001). The infection of humans with avian H9N2 virus has been reported in China since the late 1990s (Peiris et al., 1999; Butt et al., 2005; Cheng et al., 2011). Some isolates of H9N2 viruses with the ability to binding SA-a2,6 receptors can infect humans (Ha et al., 2001; Matrosovich et al., 2001). The expanded receptor specificity of H9N2 AIVs has raised concerns about their pathogenicity in humans (Huang et al., 2014). Recently, increasing evidence has shown that avian H9N2 virus might act as a source of novel human influenza viruses (Jin et al., 2014). The internal genes of newly emerged human infections with AIV subtype H7N9 and H10N8 subtype AIV are derived from H9N2 AIV (Liu et al., 2013, 2015). There have been a lot of relevant reports about the mechanism underlying the pathogenicity and transmission of H9N2 AIV genes (Li et al., 2012, 2014; Zhong et al., 2014). However, there have been fewer proteomic studies on the host after infection (Liu et al., 2008). Therefore, there is a growing need to investigate host cells infected with the H9N2 virus to elucidate potential target proteins for viral infection and adaptation studies.
Researchers have applied proteomic approaches to study cellular proteins involved in the process of H5N1, H3N2, and H1N1 virus infection (Baas et al., 2006; Mayer et al., 2007; Coombs et al., 2010; Wu et al., 2013). However, alterations of cellular proteins in human airway epithelial cell lines infected by H9N2 influenza virus have not been reported. H9N2 AIV can infect the target cells of human lung tissue, namely type II alveolar cells and bronchial epithelial cells (Zhang et al., 2013). The A549 cell line, which originated from human airway epithelial cells, was found susceptible to a strain of H9N2 influenza viruses that we screened (unpublished data). In addition to this, A549 cell line has been used for H9N2 in vitro studies (Lee et al., 2010; Shahsavandi et al., 2013; Hamidreza et al., 2016). Therefore, the A549 cell line was selected for the present proteomic study.
To better understand the molecular and cellular basis of H9N2 infection and adaptation in human airway cells, we used proteomic approaches to study the patterns of cellular proteins with variable expression upon H9N2 virus infection. Our findings may assist with the investigations into the pathogenesis and adaptation of H9N2 virus in airway epithelial cells, and with the search for potential protein targets for further studies.
Materials and Methods
Cell Culture and Infection
Human lung epithelial A549 cells (ATCC CRL-185TM) were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) (GIBCO, Grand Island, NY, USA) at pH 7.2, supplemented with 10% fetal bovine serum (TransGen, Beijing, China) and penicillin (100 U/mL)/streptomycin (100 μg/mL) and were grown in an incubator at 37°C in a 5% CO2 humidified atmosphere.
In order to examine the difference of expressed proteins of the H9N2 virus in A549 cells effectively, specific pathogen free (SPF) embryonic chicken eggs (9-day-old) were used to amplify H9N2 virus - A/Chicken/Shandong/ch/2011(CK/SD/ch), which provided by the Center for Animal Disease Control Engineering of Shandong Province. Next, the amplified H9N2 virus was inoculated onto monolayers of the A549 cells line (1.6 × 106 cells/mL), a multiplicity of infection (MOI) of 1 was used in this study. Viral infection was carried out in DMEM with 2% FBS and pen/strep at 37°C in 5% CO2. The control groups were infected with the allantoic fluid of a healthy SPF chick embryo. The infected cells were harvested at 24 h post-infection (hpi) and 72 hpi by scraping followed by washing three times with 1 × PBS. In addition, in order to measure the titer of H9N2 virus, a standard hemagglutination (HA) titer assay was conducted.
Sample Preparation
The cells were lysed with a lysis buffer containing 7 M urea, 2 M thiourea, 4% w/v CHAPS, 40 mM DTT, and 2% v/v IPG Buffer pH 4-7. After 60 min of gentle stirring at room temperature, the sample was centrifuged at 18 000 ×g at 4°C for 60 min. The supernatant was collected and the protein concentration was determined using the Bradford protein assay kit (TIANGEN, Beijing, China), according to the manufacturer’s instructions. The samples were then aliquoted and stored at -80°C until subsequent use.
Indirect Immunofluorescence
A549 cells were fixed with an acetone and ethanol solution (acetone: ethanol D 3:2) for 5 min, then washed with phosphate-buffered saline (PBS). Air-dried cells were incubated with an anti-hemagglutinin (HA) monoclonal antibody at a 1:500 dilution, at 37°C, for 1 h. After three washes with PBS, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG at a 1:500 dilution, at 37°, for 45 min. After three washes with PBS, H9N2 virus infected cells were air dried, and visualized and imaged with a Nikon inverted fluorescence microscope.
Two-Dimensional Gel Electrophoresis (2-DE)
Isoelectric focusing was carried out for a 1 h step-and-hold at 500 V, 1 h at a 1000 V gradient, 3 h at an 8000 V gradient, and a 2.5 h step-and-hold at 8000 V. After equilibration, strips were loaded on SDS–PAGE gels, and were electrophoresed for 45 min at a power of 5 W per gel, and then for approximately 6 h at a power of 12 W per gel until the dye reached the bottom of the gels. After electrophoresis, the gels were stained using silver staining methods.
Protein Identification and Database Search
The stained gels were scanned with Imagescanner III, and the gel images were processed by Image Master 2D Platinum 7.0 software. The differentially expressed protein spots (p < 0.05) with at least a two-fold difference in intensity were selected and subject to identification. Interesting protein spots were picked out from the stained gels, subjected to in-gel tryptic digestion and subsequently subjected to identification by Matrix-assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF/TOF) analysis. Combined mass spectrometry (MS) and tandem mass spectrometry (MS/MS) queries were performed using the Mascot search engine 2.2 in the NCBI-HUMAN database (268811, August 30, 2013) and uniprot-Homo sapiens database (134919, December 24, 2013).
Results
In current study, we examined the HA titer of H9N2 before infection, and the titer was 27. However, the titers in supernatant of infected A549 cells were 28 (24 hpi) and 24 (72 hpi), respectively. The changes characteristic to the type of cytopathic effect of A549 cells following 24 hpi and 72 hpi with light microscope. The cells had minimal changes, mainly in cell shrinkage, rounding and suspension of a few of them at 24 hpi and about half lost the adhesion to the surface of the cell culture plastic at 72 hpi.
Next, indirect immunofluorescence assay (IFA) was used to verify the infection of A549 cells by H9N2 AIV. Mouse monoclonal antibody against viral nucleoprotein was used as the primary antibody and goat antimouse IgG/FITC was the secondary antibody. The result showed that about 45% of the A549 cells were infected by CK/SD/ch at 24 hpi (Figure 1).
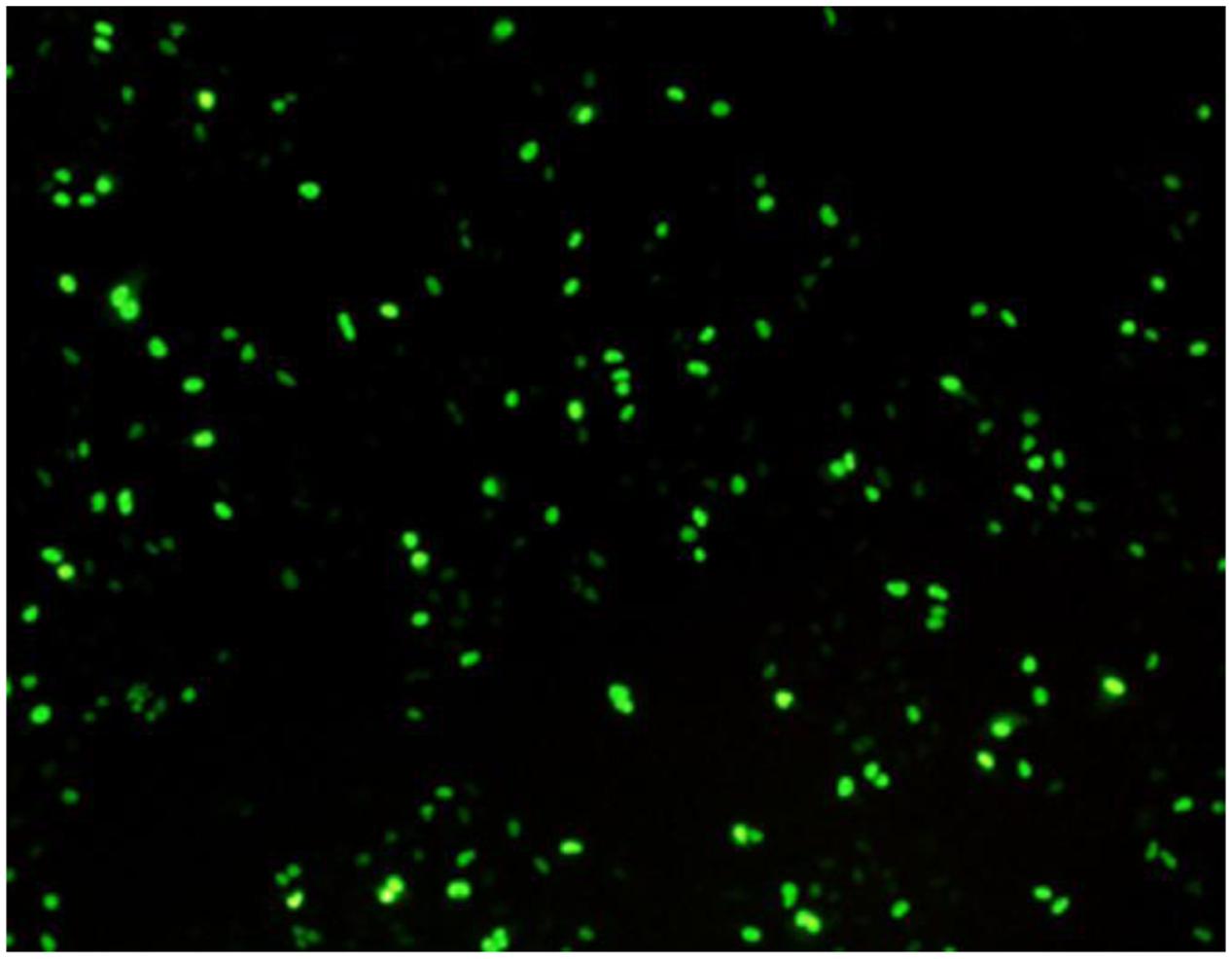
FIGURE 1. Indirect immunofluorescence analysis of A549 cells infected with CK/SD/ch confirmed infection. (Approximately 45% of the cells were infected by H9N2 at 24 hpi).
Protein extracts (300 μg) from A549 cells at different time post infection were loaded on to 2-DE gels. For each time point three replicate gels were run. A total of 481 proteins were detected in every gel. Differentially expressed proteins were judged by the criterion of an increase or decrease of spot intensities by at least two fold (Figures 2–3). The majority of the differentially expressed protein spots were also illustrated in enlarged formats (Figure 4). After searching NCBI-HUMAN and uniprot-Homo sapiens databases using the Mascot search engine, sixteen differentially expressed proteins were identified at different time points post-infection. Six protein spots of them (spots 319, 194, 167, 166, 160, and 9) were found to be up-regulated at both 24 hpi and 72 hpi (Tables 1 and 2). Only three protein spots (spots 169, 92, and 90) were found to be down-regulated at above two time points (Tables 3 and 4).
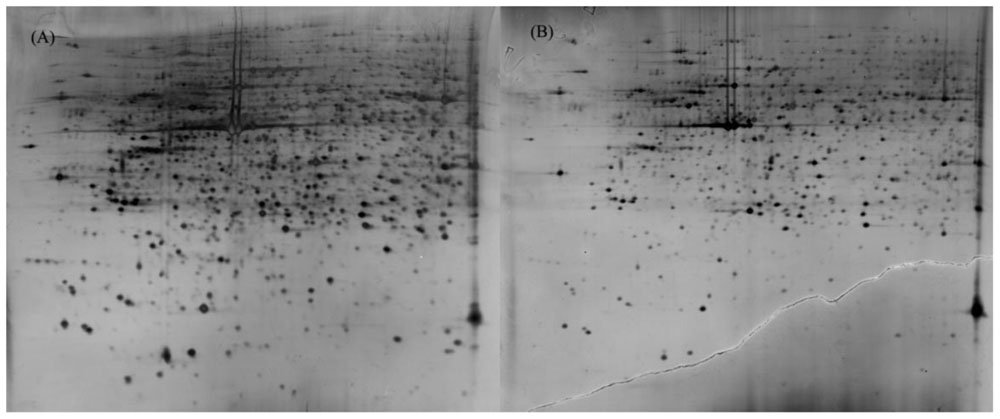
FIGURE 2. 2-DE gel images of AIV-infected and control groups of A549 cells at 24 hpi. (A): 2-DE gel of the AIV-infected group. (B): 2-DE gel of the control group.
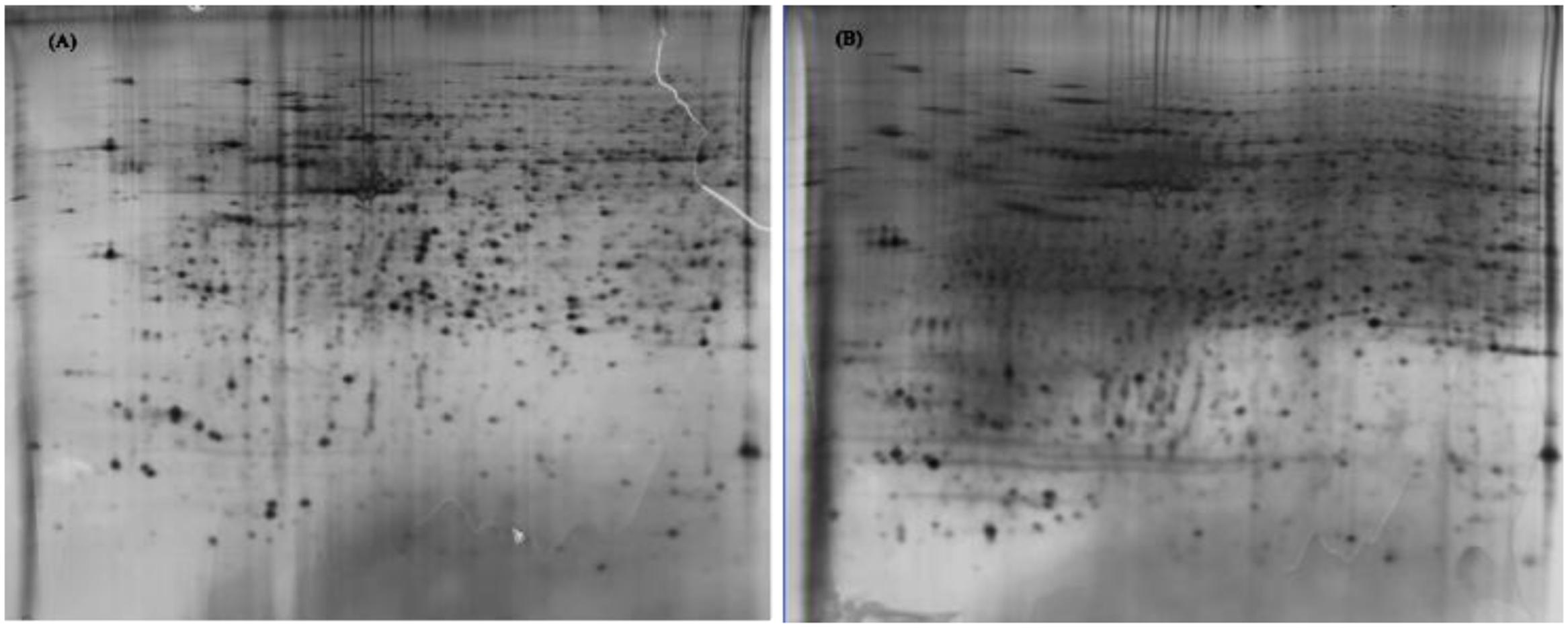
FIGURE 3. 2-DE gel images of AIV-infected and control groups of A549 cells at 72 hpi. (A): 2-DE gel of the AIV-infected group. (B): 2-DE gel of the control group.
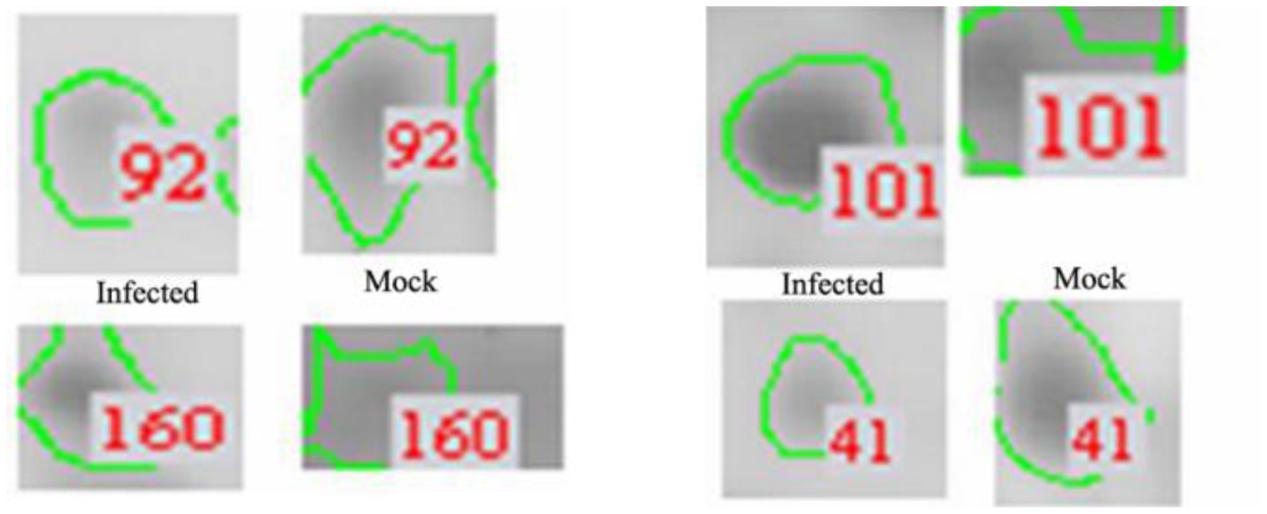
FIGURE 4. Enlarged regions of several differentially expressed protein spots. Differentially expressed protein spots are indicated by numbers and circles.
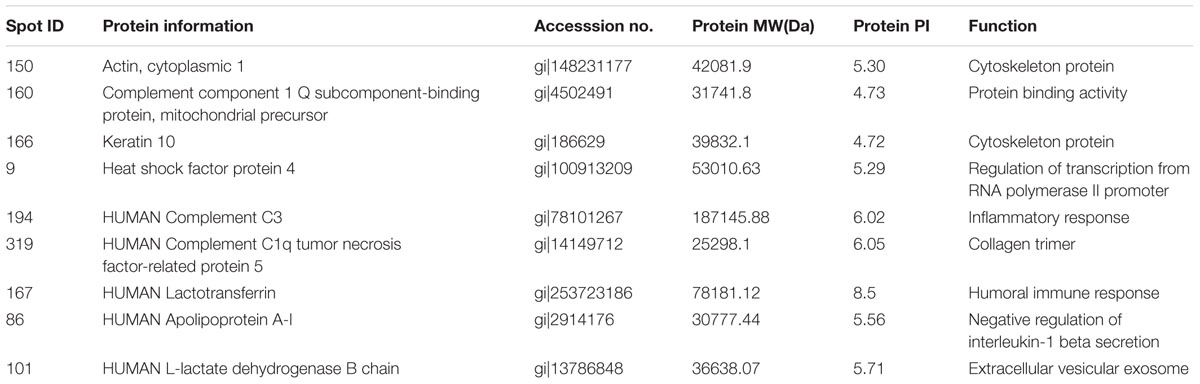
TABLE 1. Summary of upregulated proteins in A549 cells infected with influenza A H9N2 virus at 24 hpi (r > 2, p < 0.05).
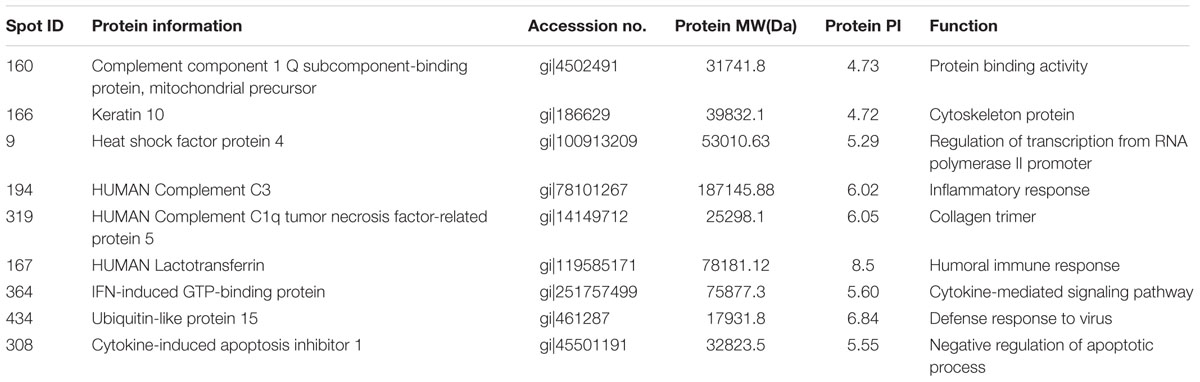
TABLE 2. Summary of upregulated proteins in A549 cells infected with influenza A H9N2 virus at 72 hpi (r > 2, p < 0.05).
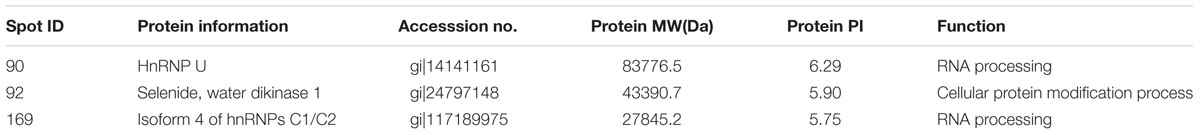
TABLE 3. Summary of downregulated proteins in A549 cells infected with influenza A H9N2 virus at 24 hpi (r > 2, p < 0.05).
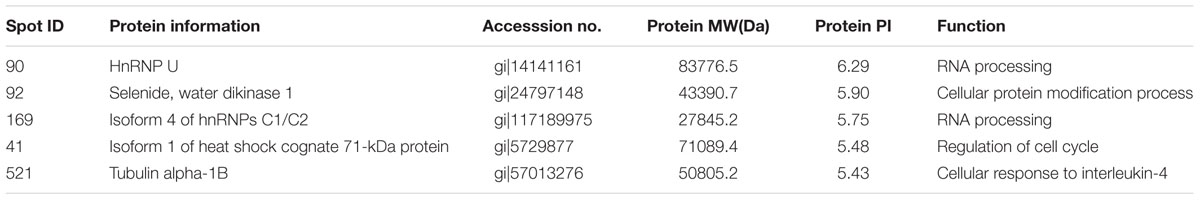
TABLE 4. Summary of downregulated proteins in A549 cells infected with influenza A H9N2 virus at 72 hpi (r > 2, p < 0.05).
Discussion
In this study, 24 h was selected as the first time point to carry out the statistics of differentially expressed proteins, for host cell morphology changes were not obvious at this time point. The differential expression proteins could affect response of antiviral immune of the host cells at this time point (Huang et al., 2014). 72 h was selected as the final stage of the infection to carry out statistics of differentially expressed proteins. Differential expression of proteins induced by human cells infected by AIV at different time points mainly involved cytoskeleton, cytokine mediated signaling pathways, mRNA transcription and expression of regulation, etc. (Liu et al., 2008; Sutejo et al., 2012).
In the current study, an overview of the differentially expressed proteins in response to infection by the H9N2 AIV was obtained by comparing differences in the abundance of proteins isolated from AIV-infected and mock-infected A549 cells.
Spot 150 (upregulated) was identified as actin, which is a major component of the cytoskeleton, and plays an important role in signaling pathways activated by virus infections (Liu et al., 2008). It is also thought to act as a regulator of transcription, for example, serve as a scaffold for the transport and/or anchorage of mRNA. The important roles of actin in gene transcription have been well described (Miralles and Visa, 2006). Actin can be used as a available transcription factor during virus synthetic protein using materials of host system. The actin cytoskeleton is also in favor of the release of progeny virus (Miralles and Visa, 2006). Besides, actin may also participate in endocytosis, by which the actin rearrangements contribute to virus particle internalization. This is achieved either by increasing endocytic activity (Pelkmans et al., 2002) or by bringing cell surface-bound virus particles to sites of high endocytic activity (Pollard and Borisy, 2003). The role of actin in H9N2 virus particle internalization in A549 cells needs further research.
Another component of cytoskeleton which displays considerable alteration in A549 cells infected with H9N2 influenza virus is cytokeretin, which forms intermediate filament in the cell. In this study, spot 166 (upregulated) was identified as keratin 10. In the cytoplasm, the network of cytokeratin filaments extending from the cell membrane to the nuclear membrane, play a role in the communication between the cell membrane and nuclear membrane. In addition, cytokeratin interacts with small nuclear ribonucleoprotein bodies containing small ribosomal protein and RNA.
In this study, actin and keratin in A549 cells infected with H9N2 AIV were identified, which may suggest that the cytoskeleton plays an important role in the AIV infection of mammalian cells with AIV.
Proteins located in the downstream of the NF-κB and IFN transcription factor pathways were identified, e.g., ISG15 (upregulated), which plays an important role in the antiviral process, especially that of the RNA virus. The ubiquitin-like modifier, ISG15, has been reported many times in the process of resisting influenza virus (Wu et al., 2013), and can be induced under virus and type I interferon stimulation. ISG15 plays an important role in regulating the antiviral innate immune response, and keeping interferon levels within a certain range, so that it can exert antiviral functions without causing immunologic damage. In addition, ISG15 may have antiviral effects by directly interacting with viral proteins (Zhao et al., 2010; Guan et al., 2011). In this study, it is possible that ISG15 was upregulated to play a role in the defense response to virus.
Spot 9 (downregulated) was HnRNP U (participating in pre-mRNA treatment) after identification, which interacts with NS1 protein during infection of H3N2 AIV (Coombs et al., 2010). NS1 protein can regulate cellular mRNA and viral mRNA translation. NS1 protein may hinder gene expression in normal cells by inhibiting ribonucleoprotein U-mediated mRNA expression. HnRNP U was downregulated in this study, which was consistent with previous findings during infections with H1N1 and H3N2 subtype viruses (Emmott et al., 2010). In our study, HnRNP U was also identified, which might suggest that HnRNP U can interact with NS1 during infection of H9N2 AIV.
To date, the antiviral role of some proteins involved in the anti-infection process of the host cell is ambiguous, therefore, we discussed the main differential proteins that were found in the current study. From the discussion above, actin, keratin, ISG15 and HnRNP U played an important role in the process of the A549 cells infected with H9N2 AIV. These findings could provide insights into the interaction between host and influenza viruses and may provide valuable information for clarifying the pathogenesis of viral infections.
Author Contributions
GY and WL designed the experiment and completed most of the works. JL, DM, and LW analyzed some test results and collected materials. TC and YC gave experiment instruction. Thank all the authors’ contribution to the experiment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the project of National Natural Science Foundation of China “Molecular mechanism affecting airborne transmission and interspecies infection of H9N2 avian influenza virus” [31470258] and the State Key Laboratory of Pathogen and Biosecurity (Academy of Military Medical Science) “Research of aerosol formation of H9N2 AIV and the mechanism of infection in mammals” [SKLPBS1449].
References
Baas, T., Baskin, C. R., Diamond, D. L., Garcia-Sastre, A., Bielefeldt-Ohmann, H., and Tumpey, T. M. (2006). Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 80, 10813–10828. doi: 10.1128/JVI.00851-06
Butt, K. M., Smith, G. J., Chen, H., Zhang, L. J., Leung, Y. H., and Xu, K. M. (2005). Human infection with an avian H9N2 influenza a virus in Hong Kong in 2003. J. Clin. Microbiol. 43, 5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005
Cheng, V. C., Chan, J. F., Wen, X., Wu, W. L., and Que, T. L. (2011). Infection of immunocompromised patients by avian H9N2 influenza a virus. J. Infect. 62, 394–399. doi: 10.1016/j.jinf.2011.02.007
Coombs, K. M., Berard, A., Xu, W., Krokhin, O., Meng, X., and Cortens, J. P. (2010). Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 84, 10888–10906. doi: 10.1128/JVI.00431-10
Emmott, E., Wise, H., Loucaides, E. M., Matthews, D. A., Digard, P., and Hiscox, J. A. (2010). Quantitative proteomics using SILAC coupled to LC-MS/MS reveals changes in the nucleolar proteome in influenza A virus-infected cells. J. Proteome Res. 9, 5335–5345. doi: 10.1021/pr100593g
Guan, R., Ma, L. C., Leonard, P. G., Amer, B. R., Sridharan, H., and Zhao, C. (2011). Structural basis for the sequence-specific recognition of human tSG15 by the NSl protein of influenza B virus. Proc. Natl. Acad. Sci. U.S.A. 108, 13468–13473. doi: 10.1073/pnas.1107032108
Ha, Y., Stevens, D. J., Skehel, J. J., and Wiley, D. C. (2001). X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. U.S.A. 98, 11181–11186. doi: 10.1073/pnas.201401198
Hamidreza, F., Reza, T., and Alireza, H. (2016). Up-regulation of pro-inflammatory cytokines and chemokine production in avian influenza H9N2 virus-infected human lung epithelial cell line (A549). Immunol. Invest. 45, 116–129. doi: 10.3109/08820139.2015.1099663
Huang, R., Liu, J. Y., Liang, W., Wang, A. R., Liu, Z. H., and Yang, Y. (2014). Response profiles of cytokines and chemokines against avian H9N2 influenza virus within the mouse lung. Med. Microbiol. Immunol. 203, 109–114. doi: 10.1007/s00430-013-0317-y
Jin, Y., Yu, D., Ren, H. G., Yin, Z. Q., Huang, Z. S., and Hu, M. D. (2014). Phylogeography of Avian Influenza A H9N2 in China. BMC Genomics 15:1110. doi: 10.1186/1471-2164-15-1110
Lee, D. C., Mok, C. K., Law, A. H., Peiris, M., and Lau, A. S. (2010). Differential replication of avian influenza H9N2 viruses in human alveolar epithelial A549 cells. Virol. J. 7, 1–5. doi: 10.1186/1743-422X-7-71
Li, X. K., Qi, W. B., He, J., Ning, Z. Y., Hu, Y., and Tian, J. (2012). Molecular basis of efficient replication and pathogenicity of H9N2 avian influenza viruses in mice. PLoS ONE 7:e40118. doi: 10.1371/journal.pone.0040118
Li, X. Y., Shi, J. Z., Guo, J., Deng, G. H., Zhang, Q. Y., and Wang, J. L. (2014). Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 Avian Influenza viruses. PLoS Pathog. 10:e1004508. doi: 10.1371/journal.ppat.1004508
Lin, Y. P., Shaw, M., Gregory, V., Cameron, K., Lim, W., and Klimov, A. (2000). Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. U.S.A. 97, 9654–9658. doi: 10.1073/pnas.160270697
Liu, D., Shi, W., Shi, Y., Wang, D. Y., Xiao, H. X., and Li, W. (2013). Origin and diversity of novel avain influenza A H7N9 viruses causing human infection: phylogenetic, structual, and coalescent analyses. Lancet 381, 1926–1932. doi: 10.1016/S0140-6736(13)60938-1
Liu, M. B., Li, X. D., Yuan, H., Zhou, J. F., Wu, J. W., and Bo, H. (2015). Genetic diversity of avian influenza A (H10N8) virus in live poultry markets and its association with human infections in China. Sci. Rep. 5:7362. doi: 10.1038/srep07632
Liu, N., Song, W. J., Wang, P., Lee, K. C., Chan, W., Chen, H. L., et al. (2008). Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics 8, 1851–1858. doi: 10.1002/pmic.200700757
Matrosovich, M. N., Krauss, S., and Webster, R. G. (2001). H9N2 influenza a viruses from poultry in Asia have human virus-like receptor specificity. Virology 281, 156–162. doi: 10.1006/viro.2000.0799
Mayer, D., Molawi, K., Martinez-Sobrido, L., Ghanem, A., Thomas, S., and Baginsky, S. (2007). Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6, 672–682. doi: 10.1021/pr060432u
Miralles, F., and Visa, N. (2006). Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 18, 261–266. doi: 10.1016/j.ceb.2006.04.009
Peiris, M., Yuen, K. Y., Leung, C. W., Chan, K. H., Ip, P. L., and Lai, R. W. (1999). Human infection with influenza H9N2. Lancet 354, 916–917. doi: 10.1016/S0140-6736(99)03311-5
Pelkmans, L., Puntener, D., and Helenius, A. (2002). Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296, 535–539. doi: 10.1126/science.1069784
Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. doi: 10.1016/S0092-8674(03)00120-X
Shahsavandi, S., Ebrahimi, M. M., Sadeghi, K., Mosavi, S. Z., and Mohammadi, A. (2013). Dose- and time-dependent apoptosis induced by avian H9N2 influenza virus in human cells. Biomed. Res. Int. 2013, 1–7. doi: 10.1155/2013/524165
Sutejo, R., Yeo, D. S., Myaing, M. Z., Hui, C., Xia, J., Ko, D., et al. (2012). Activation of type I and III interferon signalling pathways occurs in lung epithelial cells infected with low pathogenic avian influenza viruses. PLoS ONE 7:e33732. doi: 10.1371/journal.pone.0033732
Tang, X. Y., Tian, G. B., Zhao, C. S., Zhou, J. F., and Yu, K. Z. (1998). Isolation and characterization of prevalent strains of avian influenza viruses in China. Chin. J. Anim. Poult. Infect. Dis. 20, 1–5.
Wu, X. P., Wang, S. Y., Yu, Y., Zhang, J. Y., Sun, Z. Y., and Yan, Y. (2013). Subcellular proteomic analysis of human host cells infected with H3N2 swine influenza virus. Proteomics 13, 3309–3326. doi: 10.1002/pmic.201300180
Zhang, Z. F., Fan, X. H., Chen, X. Y., Feng, A. L., and Yang, L. (2013). Avain influenza virus subtype H9N2 replicates in human lung tissues. Chin. J. Virol. 29, 206–210.
Zhao, C., Hsiang, T. Y., Kuo, R. L., and Krug, R. M. (2010). ISG15 conjugation system targets the viral NSl protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 107, 2253–2258. doi: 10.1073/pnas.0909144107
Keywords: H9N2 AIV, two-dimensional electrophoresis, A549 cells, differentially expressed proteins, proteomics
Citation: Yu G, Liang W, Liu J, Meng D, Wei L, Chai T and Cai Y (2016) Proteomic Analysis of Differential Expression of Cellular Proteins in Response to Avian H9N2 Virus Infection of A549 Cells. Front. Microbiol. 7:1962. doi: 10.3389/fmicb.2016.01962
Received: 10 May 2016; Accepted: 23 November 2016;
Published: 15 December 2016.
Edited by:
Leonard Peruski, US Centers for Disease Control and Prevention, USAReviewed by:
Haider Abdul-Lateef Mousa, University of Basrah, IraqShailesh D. Pawar, National Institute of Virology, India
Copyright © 2016 Yu, Liang, Liu, Meng, Wei, Chai and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tongjie Chai, chaitj117@163.com Yumei Cai, caiyum@163.com
†These authors have contributed equally to this work.
 Guanliu Yu
Guanliu Yu Wei Liang
Wei Liang Jiyuan Liu
Jiyuan Liu Dan Meng
Dan Meng Liangmeng Wei
Liangmeng Wei