Challenging muscle homeostasis uncovers novel chaperone interactions in Caenorhabditis elegans
- 1Department of Life Sciences and The National Institute for Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer Sheva, Israel
- 2Institute for Genetics and Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne, Germany
- 3Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases, CECAD Research Center, University of Cologne, Cologne, Germany
Proteome stability is central to cellular function and the lifespan of an organism. This is apparent in muscle cells, where incorrect folding and assembly of the sarcomere contributes to disease and aging. Apart from the myosin-assembly factor UNC-45, the complete network of chaperones involved in assembly and maintenance of muscle tissue is currently unknown. To identify additional factors required for sarcomere quality control, we performed genetic screens based on suppressed or synthetic motility defects in Caenorhabditis elegans. In addition to ethyl methyl sulfonate-based mutagenesis, we employed RNAi-mediated knockdown of candidate chaperones in unc-45 temperature-sensitive mutants and screened for impaired movement at permissive conditions. This approach confirmed the cooperation between UNC-45 and Hsp90. Moreover, the screens identified three novel co-chaperones, CeHop (STI-1), CeAha1 (C01G10.8) and Cep23 (ZC395.10), required for muscle integrity. The specific identification of Hsp90 and Hsp90 co-chaperones highlights the physiological role of Hsp90 in myosin folding. Our work thus provides a clear example of how a combination of mild perturbations to the proteostasis network can uncover specific quality control modules.
Introduction
The term molecular chaperone was first coined to describe the transient involvement of the nuclear protein nucleoplasmin in the assembly of the nucleosome subunits (Laskey et al., 1978). The concept of “assisting proteins” was then adopted to define a group of highly conserved proteins, some of which are heat shock proteins (HSPs), found to be transiently involved in polypeptide folding and in the assembly of various protein complexes, without being part of the final structure (Pelham, 1986; Ellis, 1987; Hemmingsen et al., 1988).
In vitro studies using purified components established a role for molecular chaperones in preventing aggregation and in the unfolding and reactivation of denatured proteins (Li et al., 2011; Bardwell and Jakob, 2012; Mattoo and Goloubinoff, 2014). For example, denatured model substrates, such as malate dehydrogenase, rhodanese and luciferase, were used in structure-function studies to elucidate chaperone activity, as a proxies for correct protein folding (Goloubinoff et al., 1989; Buchner et al., 1991; Martin et al., 1991; Schroder et al., 1993). In contrast, in vivo studies have been hampered by a lack of phenotypes that can be readily monitored. This challenge is further amplified when one considers that a single substrate is able to interact with many different chaperones within the cell.
Identifying specialized chaperones required for muscle assembly and maintenance offers a prime example of the difficulties associated with studying protein folding in vivo (Kim et al., 2008; Willis et al., 2009; Willis and Patterson, 2010). Although myosin subunits can spontaneously assemble into thick, filament-like structures in vitro, the myosin motor domain does not fold, assemble or function in an autonomous manner in vivo (Barral and Epstein, 1999; Srikakulam and Winkelmann, 2004). Thus far, screening for detectable phenotypes, such as those related to motility, muscle elongation and myofilament organization in Caenorhabditis elegans, identified only two chaperones, namely UNC-45 and UNC-23, among ~200 proteins required for sarcomere assembly, maintenance or function (Meissner et al., 2009; Benian and Epstein, 2011). While biochemical analysis of UNC-45-based interactions identified the molecular chaperone Hsp90 (Barral et al., 2002), knockdown or mutations of C. elegans Hsp90 (daf-21) results in only a minor effect on motility (Gaiser et al., 2011). This gap between phenotype and function might result from redundant functions of the proteostasis network, compensating for monogenic defects (Balch et al., 2008; Sawyer et al., 2011).
To study tissue-specific chaperone networks in a multicellular organism, we focused on the essential role of the myosin chaperone UNC-45, a member of the conserved UCS (UNC-45/CRO1/She4p) family, with the aim of uncovering novel chaperones involved in myosin folding. Temperature-sensitive (ts) mutants of unc-45 result in motility and sarcomere organization defects at restrictive conditions but have no apparent phenotype at the permissive growth temperature of 15°C. unc-45-associated phenotypes can be exposed under permissive conditions or masked under restrictive conditions by genetic or chemical remodeling of proteostasis (Gidalevitz et al., 2006, 2009; Alavez et al., 2011; Gazda et al., 2013; Melkani et al., 2013; Feldman et al., 2014). Thus, unc-45(ts) can be viewed as a sensitized background to screen for molecular chaperones specifically associated with muscle function.
Here, we used a non-biased mutagenesis screen to identify chaperones that ameliorate unc-45(ts) phenotypes at restrictive conditions and targeted RNAi synthetic interaction screens to identify chaperones that aggravate unc-45(ts) phenotypes under permissive conditions. Using an epistatic analysis approach, we genetically verified the interaction between UNC-45 and Hsp90. Moreover, we identified the Hsp90 co-chaperones CeHop (STI-1), CeAha1 (C01G10.8), and Cep23 (ZC395.10) as novel and specific modulators of UNC-45-Hsp90 myosin-associated functions.
Materials and Methods
Nematodes and Growth Conditions
C. elegans strains Bristol N2 (wild-type), DR94 (unc-45(m94)), CB286 (unc-45(e286)), RW1596 (myo-3(st386); stEx30[pmyo-3::MYO-3::GFP; pRF4(rol-6(su1006))]) and RB2525 (C01G10.8(ok3501)) were obtained from the Caenorhabditis Genetics Center. Animals were backcrossed to N2 at least four times. Strain ABZ100 [unc-45(m94); C01G10.8 (ok3501)] was generated by crossing DR94 (unc-45(m94)) males with RB2525 (C01G10.8 (ok3501)) hermaphrodites. The m94 allele was confirmed by incubating eggs at the restrictive temperature (25°C) and testing for the characteristic movement phenotype. The ok3501 allele was confirmed by genotyping PCR (primers used: left primer AGAAACGGCGATACGGCTAA; right primer: CAATGCAAGCTCGCCCTATT).
Nematodes were grown on NGM plates seeded with the Escherichia coli OP50-1 strain at 15°C. Synchronized L1 larvae were obtained as follows: Synchronized high density plates containing adult worms and L1 progeny were washed with M9 solution and collected into tubes. Then, L1 larvae were separated from the adult worm population by centrifugation. For all experiments, 5–20 μl of M9 containing 30–130 L1 larvae where transferred onto fresh plates seeded with E. coli HT115 (DE3) containing the indicated RNAi vectors and grown at 15 or 20°C for the duration of the experiment. The animals were then taken for phenotypic analysis, imaging and/or antibody staining. At 15°C, the first day of egg-laying was set at 120 h after L1 transfer, at 65 h for growth at 20°C and at 50 h for growth at 25°C. The first day of egg-laying was defined as the point when animals grown on HT115 (DE3) bacteria containing an empty vector control were able to lay eggs.
Ethyl Methanesulfonate (EMS) Screen
Thirteen thousand unc-45(m94) worms were grown at the permissive temperature (15°C) to young adulthood and mutagenized using 50 mM EMS, as described earlier (Brenner, 1974). To isolate both recessive and dominate suppressors, 88,000 F1 progeny were grown at the permissive temperature and F2 progeny were shifted to the restrictive temperature (22°C) at the L1 larvae stage, avoiding UNC-45-associated embryonic arrest. 18,000 viable F2 animals grown at the restrictive temperature were then gently transferred to new plates and placed distally from the food source. Seventy-seven animals that reached the food source and exhibited coordinated movement were individually transferred to new plates and phenotype transmission was determined at 20°C, with sterile lines being excluded. Next, the six remaining lines were assayed for motility (thrashing assay) and UNC-45 localization was imaged by immuno-staining with anti-UNC-45 antibodies (Gazda et al., 2013). Three lines that exhibited both normal motility and muscle organization, together with body wall muscle-localized UNC-45, were collected for sequencing analysis.
RNA Interference (RNAi)
RNAi knockdown treatments were performed as previously described (Ben-Zvi et al., 2009). Chaperone RNAi constructs were obtained from the “RNAi chaperone library” kindly provided by Prof. Richard Morimoto, Northwestern University. For all RNAi experiments, L1 larvae were obtained and transferred onto RNAi plates. Unless otherwise indicated, animals were maintained at 15°C as described above. Each RNAi treatment was performed at least three times, and >30 animals per experimental condition were scored. Animals fed on E. coli HT115 (DE3) containing the empty vector (pL4440) or unc-45 RNAi were included as negative and positive controls in each experiment, respectively (Figure S1). In addition, changes in mRNA and encoded protein levels of several genes were determined by Real-Time PCR or Western blot, respectively, to test the efficiency of the RNAi treatment.
Motility Assays
Age-synchronized populations of >30 animals were used for each assay. Animals were grown at 15°C for the duration of the experiment. On the first and second days of egg-laying, worms were tested for paralysis/slow movement by striking the plates gently against a table three times and monitoring movement. Worms that moved their heads but failed to move their bodies were scored as paralyzed. RNAi knockdown of unc-45 resulted in a strong paralyzed phenotype that was used as reference. To examine for small changes in motility, age-synchronized young adult animals (n = 20) movement was assessed by the thrashing assay, as previously described (Gazda et al., 2013).
RNA Levels
Total RNA was extracted from age-synchronized L4 animals using TRIzol reagent (Invitrogen). RNA was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad). mRNA levels were measured by quantitative PCR (qPCR), performed on a C1000 Thermal Cycler (Bio-Rad) with KAPA SYBRFAST qPCR Master Mix (KAPA Biosystems). The results were analyzed using CFX Manager software (Bio-Rad). Relative transcript levels were determined by averaging the CT of triplicate values for the genes examined and normalizing those to CT values obtained for 18S rRNA of the same sample using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Protein Levels
Age-synchronized populations were collected and lyzed in SDS sample buffer (98°C for 10 min). Samples were separated by SDS-SB gel electrophoresis and analyzed by western blot, using anti-tubulin (Sigma), anti-CeHop and anti-CeAha1 antibodies (kindly provided by Dr. Klaus Richter). Peroxidase-conjugated AffiniPure goat anti-rabbit antibodies served as secondary antibodies.
Co-Immunoprecipitation
An age-synchronized population was collected in M9 buffer, flash-frozen and lyzed using a hand drill. The lysate was maintained at 4°C. Samples were diluted in RIPA buffer (Sigma) supplemented with protease inhibitors (Roche) and centrifuged to remove debris. The supernatant was divided into equal aliquots. Two aliquots were incubated for 30 min at 4°C with either anti-CeHop or anti-CeAha1 antibodies and one aliquot was left untreated. Protein A/G PLUS-Agarose beads (Santa Cruz) were added to the samples, which were incubated for additional 30 min at 4°C. The samples were centrifuged (1000× g, 5 min) and supernatants and pellet were collected. Pellets containing the agarose beads were washed three times in RIPA buffer. Pellet and supernatant fractions were boiled (95°C for 10 min) in SDS sample buffer, resolved by SDS-SB gel electrophoresis and analyzed by western blot using anti-CeHop, anti-CeAha1, anti-CeHsp90 (kindly provided by Prof. Richard Morimoto) or anti-UNC-45 antibodies (Gazda et al., 2013). Peroxidase-conjugated AffiniPure goat anti-rabbit antibodies served as secondary antibodies.
Immunofluorescence Staining
Immunofluorescent staining of MYO-3, CeHop and CeAha1 was performed as previously described (Karady et al., 2013). Animals were stained with antibodies for MYO-3 (5-6-S, Hybridoma Bank), CeHop or CeAha1 (kindly provided by Dr. Klaus Richter). DyLight 488 goat anti-mouse, 549 goat anti-rabbit (Jackson Immuno-Research) or 633 goat anti-mouse (Thermo Scientific) antibodies served as secondary antibodies. Animals were imaged using a Leica DM5500 B confocal microscope. Fluorophores were excited using the 488, 549, and 633 nm laser lines, respectively. Immunofluorescent staining of UNC-45 was performed as described (Gazda et al., 2013).
Results
Using unc-45 Mutants to Identify Novel Myosin-Folding Modifiers
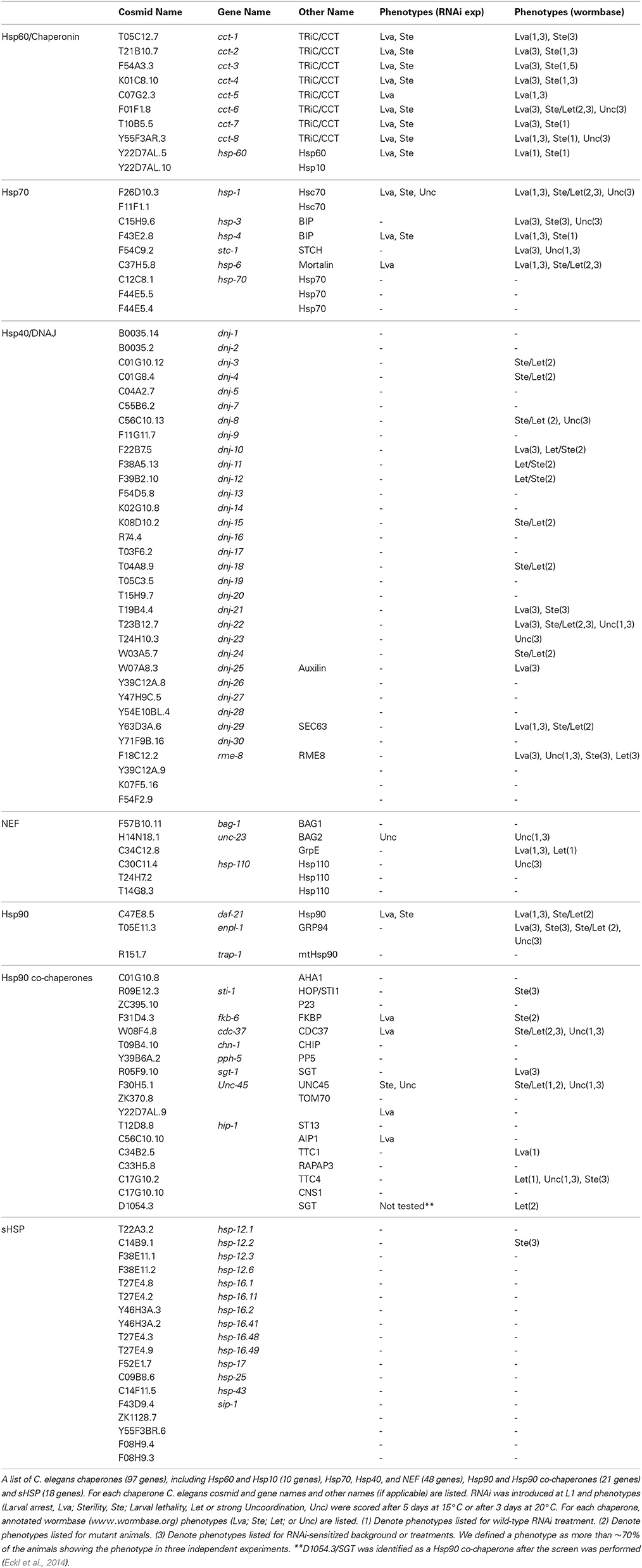
The gene encoding UNC-45, an essential muscle-specific chaperone, was used for our mutagenesis screening approach. A single amino acid substitute in the UCS domain of C. elegans UNC-45, E781K, or L822F (the m94 and e286 alleles, respectively), results in temperature-sensitive motility defects and myosin disorganization phenotypes when animals are grown at restrictive conditions (>22°C). However, animals show no phenotypes during development and early adulthood when grown at the permissive temperature (15°C). To identify chaperones that play a redundant role in myosin assembly and muscle maintenance, we screened for modifiers that can mask unc-45-associated phenotypes under restrictive conditions. Because RNAi knockdown of chaperone expression is expected to aggravate unc-45 phenotypes, we used mutagenesis to screen for compensatory factors. Specifically, EMS was used to induce mutations in the germline of unc-45(m94) hermaphrodite worms. F2 animals were shifted to the restrictive temperature (22°C) and screened for mutants with improved motility. Mutant animals, identified by their ability to reach a distant E. coli food source on the plate, were isolated (Figure 1A). The identified suppressor mutants with restored UNC-45 muscle organization were selected for further genetic analysis (Figures 1A–C). We first sequenced the unc-45 gene of the identified suppressors to search for possible intragenic changes. Of the three identified alleles (hh10, hh11, and hh12), all proved to be revertants, presenting the A2342G substitution that restores the m94 allele of the unc-45 gene to the wild-type version (Figure 1D). This observation suggests that introducing another change in the unc-45 gene or a mutation(s) elsewhere in the genome was unable to compensate unc-45(m94)-specific phenotypes under restrictive conditions. This indicates an essential role for UNC-45 that cannot be bypassed by other chaperones.
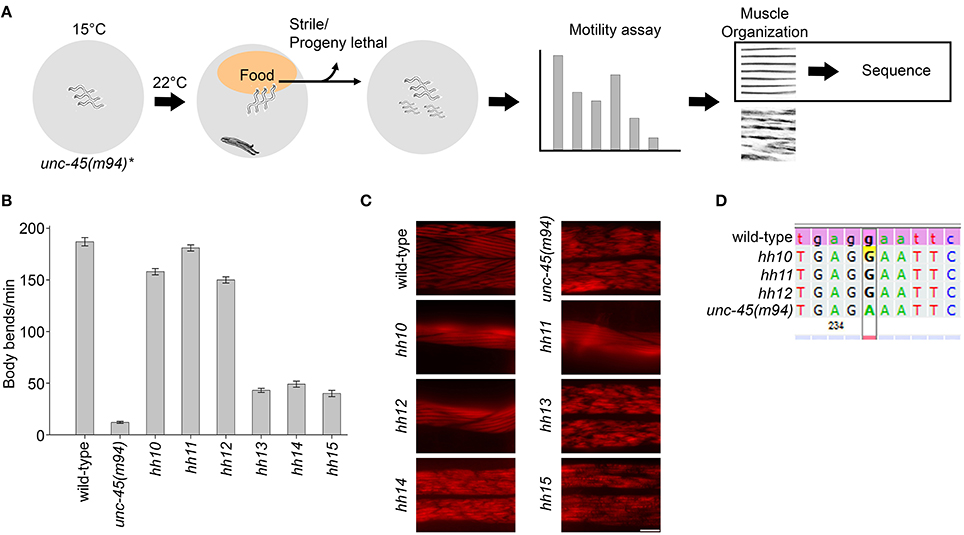
Figure 1. EMS screen for suppressors of the unc-45(m94) phenotype under restrictive conditions. (A) Schematic representation of the EMS-based screen used to identify suppressors of unc-45(m94) animals. (B) Age-synchronized wild-type, unc-45(m94) and isolated suppressor (hh10, hh11, hh12, hh13, hh14, and hh15) animals were grown at 22°C and scored for motility on day 1 of adulthood. Data are mean ± s.e.m. (C) Confocal images of body-wall muscle. Age-synchronized wild-type, unc-45(m94) and isolated suppressor (hh10, hh11, hh12, hh13, hh14, and hh15) animals were grown at 22°C, collected on day 1 of adulthood and stained with α-UNC-45 antibodies. Scale bar is 10 μm. (D) Sequence alignment of wild-type, unc-45(m94) and isolated suppressors (hh10, hh11, and hh12) highlighting base position 2341. Sequence analysis of selected suppressors (hh10, hh11, and hh12) revealed the A2341G mutation, which results in the synthesis of a wild-type version of the unc-45 gene.
A Targeted RNAi Screen for Chaperone Function
One possible interpretation of the mutagenesis screen results is that the lack of unc-45(ts) phenotypes at permissive conditions might result from the redundant but not compensatory functions of other chaperones in the muscle (Balch et al., 2008; Sawyer et al., 2011). We reasoned that affecting two chaperones that have no gross phenotypes on their own could counteract redundancy and uncover novel chaperone interactions. To test this approach, we first compiled a list of C. elegans chaperones (97 genes) (Table 1) and examined the effects of down-regulation of each chaperone in wild-type animals (Figure 2A). To draw up this list, we relied on gene annotations and biochemical and functional data. We focused our scope to specific chaperone families and their co-chaperones, including Hsp60 and Hsp10 (10 genes), Hsp70, Hsp40, and NEF (48 genes), Hsp90 and Hsp90 co-chaperones (21 genes) and sHSP (18 genes).
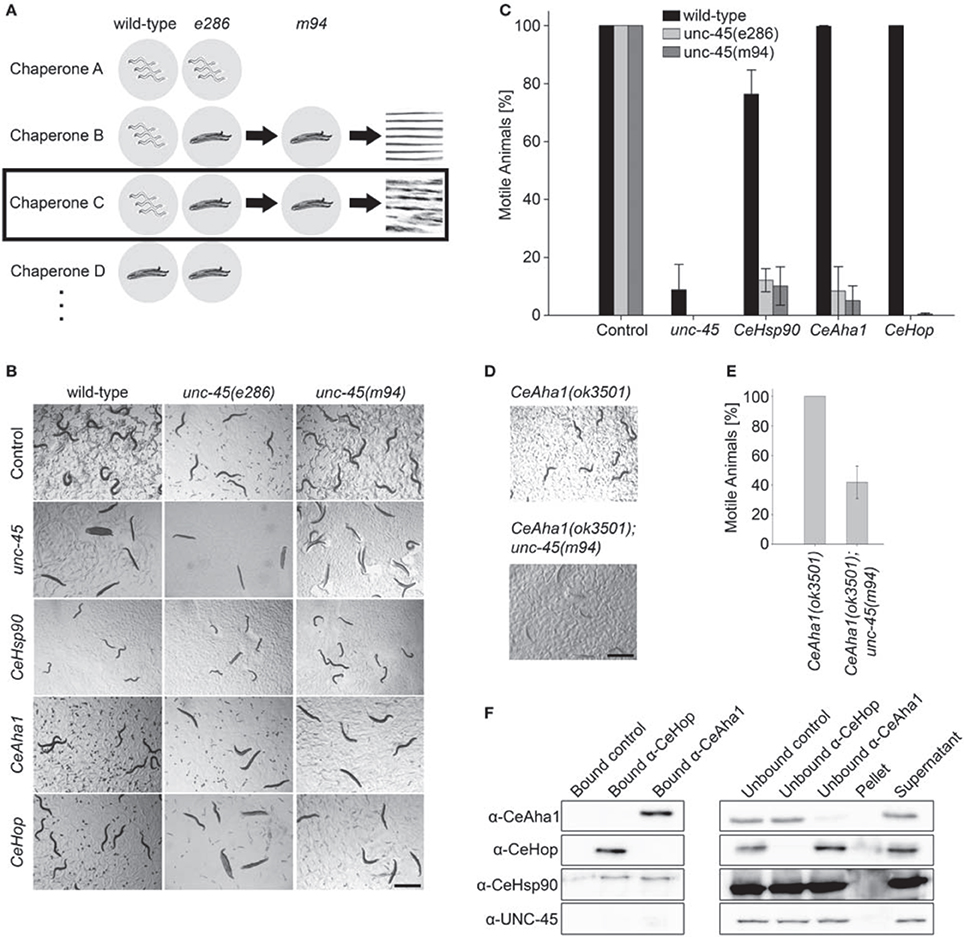
Figure 2. Using unc-45 as bait to identify novel myosin chaperones. (A) Schematic representation of the unc-45-targeted screen. (B) Age-synchronized (L1) wild-type, unc-45(e286) or unc-45(m94) animals grown at 15°C were transferred to plates containing control, unc-45, CeHsp90 (daf-21), CeHop (sti-1), or CeAha1 (C01G10.8) RNAi-expressing bacteria and images were taken on day 2 of adulthood. Scale bar is 1 mm. (C) Age-synchronized (L1) wild-type, unc-45(e286) or unc-45(m94) animals treated as in (B) were scored for motility on day 2 of adulthood. Data are mean ± s.e.m. (D) Age-synchronized RB2525 [C01G10.8(ok3501)] or ABZ100 (unc-45(m94); C01G10.8(ok3501)) animals were grown at 15°C on regular plates. Images were taken on day 2 of adulthood. Scale bar is 1 mm. (E) Age-synchronized (L1) RB2525 [C01G10.8(ok3501)] or ABZ100 [unc-45(m94); C01G10.8(ok3501)] animals treated as in (D) were scored for motility on day 2 of adulthood. Data are mean ± s.e.m. (F) CeAha1, CeHop, Hsp90, or UNC-45 were co-immunoprecipitated from total extracts of wild-type animals using anti-CeHop or anti-CeAha1 antibodies. Non-specific binding (bound control), unbound fractions (unbound control, unbound Hop, and unbound Aha1), and untreated fractions (pellet and supernatant) are included for reference.
RNAi was introduced at the first larval stage of the C. elegans life cycle (L1) and its effects were examined after 5 days at 15°C (or after 3 days at 20°C), until animals grown on control RNAi reached adulthood (Table 1). Down-regulation of genes encoding 10 chaperones (17 genes), including CTT, HSP-1 (Hsc70/HSPA8), HSP-4 (BIP), HSP-6 (Mortalin), and DAF-21 (Hsp90), resulted in severe organismal phenotypes, such as lethality, developmental arrest, or sterility (Table 1, Figure S2). In contrast, down-regulation of genes encoding most chaperones examined had no apparent phenotypes (Table 1). Our data are in agreement with published RNAi screens for wild-type RNAi treatment (Table 1, data curated by wormbase). When knockout mutants or data from enhanced RNAi efficacy experiments, (for example via dsDNA injection into the gonads) were considered, more acute phenotypes were observed for some chaperones (Table 1, data curated by wormbase). This further supports our conclusion that the proteostasis network may buffer mild perturbations in the folding capacity of the organism.
As expected, down-regulation of only two chaperones, unc-45 or unc-23, showed a marked movement defect. For example, based on their impact on muscular ultrastructure or their involvement in myopathies, molecular chaperones, such as TRIC (cct), αB-crystallin/HSPB5 (hsp-25), and DNAJB6 (dnj-24), were suggested to play a key role in sarcomere assembly and maintenance (Ding and Candido, 2000; Vang et al., 2005; Kim et al., 2008; Willis et al., 2009; Willis and Patterson, 2010; Garrido et al., 2012; Haslbeck et al., 2012; Sarparanta et al., 2012). Although these chaperones are associated with the sarcomere (Leroux and Candido, 1997; Ding and Candido, 2000; Meissner et al., 2011), knockdown of their genes by feeding RNAi did not result in notable motility defects (Table 1).
Screening for Chaperones that Genetically Interact with unc-45 Mutants
RNAi knockdown of chaperones was next performed with unc-45(ts) mutant strains under permissive conditions. Age-synchronized L1 unc-45(e286) mutant animals kept at permissive conditions (15°C) were treated with RNAi for different molecular chaperones and monitored for motility defects. We first sought chaperones that cause unc-45(e286)-specific phenotypes at permissive conditions when depleted (Figure 2A). Of the 96 RNAi constructs tested, five genes induced loss of coordination and/or an egg-laying defect, resulting in reduced motility in more than 70% of the unc-45(e286) mutant animals. As noted above, down-regulation of unc-45 or unc-23 also induced a loss of coordination in wild-type animals (Table 1); these were discarded from the candidate gene list (Figure 2A). The remaining chaperones identified in the screen are CeHsp90 (daf-21), CeHop (sti-1) and CeAha1 (C01G10.8) (Figures 2B,C). RNAi treatment for CeHsp90, encoding a known myosin chaperone, resulted in a strong larval arrest phenotype in both wild-type and unc-45(e286) animals but induced a strong motility defect only in unc-45(e286) animals (76.4 ± 8.3%, as compared to 12.1 ± 4% motile animals). CeHop and CeAha1 RNAi only affected unc-45(e286)-expressing animals, where complete paralysis resulted (Figures 2B,C).
Given that genetic background variations can confound the interpretation of genetic interactions, the three genes that affected unc-45(e286) were tested for their effects on animals carrying the unc-45(m94) allele (Figure 2A). Age-synchronized L1 unc-45(m94) mutant worms maintained under permissive conditions were treated with RNAi for CeHsp90, CeHop or CeAha1 and compared to animals that took up the empty vector control. Similar to the e286 allele, unc-45(m94) animals treated by RNAi for CeHsp90, CeHop, or CeAha1 were completely paralyzed on day 2 of adulthood under permissive conditions (Figures 2B,C). Although CeAha1 or CeHop RNAi treatment reduced the levels of the corresponding proteins to a similar extent in all strains examined, CeHop and CeAha1 RNAi only induced paralysis in unc-45(ts)-expressing animals (Figures 2B,C and Figure S3). Moreover, only 41.8 ± 11% of the double mutant of unc-45(m94) and a deletion allele of CeAha1 (RB2525 strain) were motile by the second day of adulthood, in contrast to the respective single mutant strains (Figures 2D,E). Thus, CeHop and CeAha1 are novel genetic modifiers of unc-45 and are associated with muscle function.
Hop/STI1 (Hsp70-Hsp90 Organizing Protein/STress Inducible 1) and Aha1 (Activator of Hsp90 ATPase 1) are co-chaperones of Hsp90 that facilitate the maturation of Hsp90 substrates by modulating the Hsp90 ATPase cycle and thus driving Hsp90 conformational changes (Rohl et al., 2013). Hop/STI1 and Aha1 homologs in C. elegans were identified based on sequence analysis (Haslbeck et al., 2012). CeHop was shown to interact with Hsp90 by immunoprecipitation and to inhibit Hsp90 ATPase in vitro, similar to yeast Sti1 (Gaiser et al., 2009; Song et al., 2009). It is, therefore, likely that the interactions of CeHop and CeAha1 with unc-45 are mediated via CeHsp90. To test this directly, we performed immunoprecipitation and western blot analysis to monitor whether CeHop and CeAha1 form complexes with UNC-45 and CeHsp90. CeHop was found to be associated with CeHsp90 but not with CeAha1 or UNC-45 in agreement with previous mass spectrometry and co-immunoprecipitation studies (Song et al., 2009). Likewise, pull down of CeAha1 from wild-type total lysates captured CeHsp90 but not CeHop or UNC-45 (Figure 2F). Thus, as expected from their roles in other systems (Rohl et al., 2013), CeAha1 and CeHop form discrete complexes with CeHsp90 and do not directly interact with UNC-45.
The Hsp90 Co-Chaperones CeHop and CeAha1 are Linked to Muscle Homeostasis
CeHsp90 is a well-established myosin chaperone (Barral et al., 2002; Etard et al., 2007; Gazda et al., 2013), is localized to sarcomeric structures and affects myofilament organization in body wall muscles of C. elegans (Gaiser et al., 2011). To determine whether CeHop and CeAha1 also affect myosin folding, we first examined whether these proteins are also localized to the sarcomere. Immunofluorescent staining using specific antibodies to CeHop and CeAha1 revealed striated patterns, suggesting a role for these proteins in myofilament formation and/or maintenance (Figure 3A).
Figure 3. CeAha1 and CeHop are novel modifiers of unc-45 function. (A) Age-synchronized (L1) RW1596 animals, expressing MYO-3:GFP, were grown at 15°C, collected on day 2 of adulthood, stained with specific antibodies to CeAha1 or CeHop, and imaged by confocal microscopy. Scale bar is 10 μm. (B) Confocal images of body wall muscle. Age-synchronized wild-type, unc-45(e286) or unc-45(m94) animals were grown at 25°C, collected on day 2 of adulthood and stained with anti-MYO-3 antibodies. Scale bar is 10 μm. (C) Confocal images of body-wall muscle. Age-synchronized wild-type, unc-45(e286) or unc-45(m94) animals were grown at 15°C, transferred to plates containing control, CeAha1 (C01G10.8) or CeHop (sti-1) RNAi-expressing bacteria, collected on day 2 of adulthood and stained with anti-MYO-3 antibodies. Scale bar is 10 μm.
To directly test this prediction, we examined whether CeHop- and CeAha1-associated unc-45 phenotypes display disrupted myofilament organization. Therefore, we monitored the subcellular organization of myosin heavy chain A (MYO-3) using immuno-staining assays. Animals expressing unc-45(e286) or unc-45(m94) that were shifted to restrictive conditions after the first larval stage exhibited disruption of myofilament structure associated with mislocalization of MYO-3, while wild-type animals were unaffected (Figure 3B). MYO-3 organization in wild-type, unc-45(e286), or unc-45(m94) mutant animals under permissive conditions was mostly unaffected by the third day of adulthood, although myosin disorganization was observed at later time points (Ben-Zvi et al., 2009). Treatment of wild-type animals with CeHop and CeAha1 RNAi did not affect myofilament organization (monitored until the second day of adulthood). In contrast, treatment of unc-45(e286) mutant animals with CeHop and CeAha1 RNAi resulted in complete disruption of myofilament organization and MYO-3 mislocalization under permissive conditions (Figure 3C). Similar results were observed for unc-45(m94), although this mutant showed mild myosin disorganization in control animals. CeHop or CeAha1 RNAi-induced myofilament disruption was similar to the MYO-3 disorganization observed when unc-45 mutant animals were shifted to restrictive conditions (Figure 3B). Taken together, these data suggest that the Hsp90 co-chaperones CeHop and CeAha1 affect myosin organization when UNC-45 function is reduced. Moreover, the data suggest that Hsp90 and its co-chaperones are required to maintain unc-45(e286) and unc-45(m94) muscle organization.
The Hsp90 Co-Chaperone Cep23 (ZC395.10) is Required for Myosin Folding
The specific role of the Hsp90 system in modulating UNC-45-associated phenotypes suggested that other Hsp90 co-chaperones not identified in our screen might be required for unc-45(ts) motility. To re-examine the role of these co-chaperones in muscle function, wild-type and unc-45(e286) animals were treated with RNAi for known Hsp90 co-chaperones (Haslbeck et al., 2012, 2013) (17 genes; Table 1) and changes in motility were monitored. The detection threshold was increased in this secondary screen and animals were scored for any change in motility. Of the genes examined, only down-regulation of C. elegans p23 (Cep23; ZC395.10) resulted in motility defects of unc-45(e286) mutant animals but not wild-type animals. 39 ± 11.3% of the unc-45(e286) mutant animals examined were motile (Figures 4A,B and data not shown). Although Cep23 modulated unc-45(ts) motility to a lesser extent than CeHop or CeAha1 (Figures 2C, 4B), Cep23 effect on unc-45(ts) motility was highly significant (p < 0.005). Cep23 was considered as false negative of the screen and its role as an UNC-45 modifier was next examined.
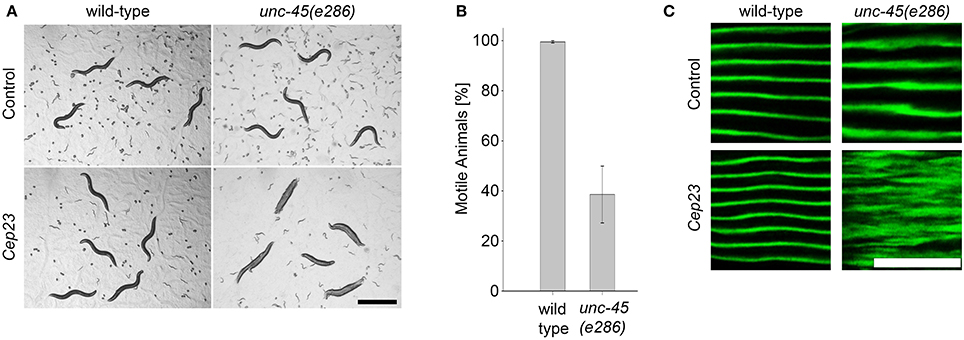
Figure 4. The Hsp90 co-chaperone Cep23 is required for myosin folding. (A) Age-synchronized (L1) wild-type or unc-45(e286) animals grown at 15°C were transferred to control or Cep23 RNAi-expressing bacteria and imaged on day 2 of adulthood. Scale bar is 1 mm. (B) Age-synchronized (L1) wild-type animals treated as in (A) were scored for motility on day 2 of adulthood. Data are mean ± s.e.m. (C) Confocal images of body wall muscle. Age-synchronized wild-type or unc-45(e286) mutant animals were grown at 15°C, transferred to plates containing control or Cep23 (ZC395.10) RNAi-expressing bacteria, collected on day 2 of adulthood and stained with anti-MYO-3 antibodies. Scale bar is 10 μm.
The Hsp90 co-chaperone p23 is an important component of the general Hsp90 chaperone cycle, forming a complex with CeHsp90 (Gaiser et al., 2010) that is localized to the sarcomere (Meissner et al., 2011). To test whether Cep23(RNAi) also affected myofilament organization, wild-type and unc-45(e286) mutant animals were monitored for changes in the subcellular organization of MYO-3 using immuno-staining assays. In agreement with the Cep23-mediated effects on motility, treatment of unc-45(e286) animals with Cep23(RNAi) resulted in severe MYO-3 disruption in affected animals (Figure 4C). Thus, Cep23 genetically interacts with unc-45, further supporting the specific role of the Hsp90 chaperone system in modulating unc-45 phenotypes.
Using Novel Chaperone Interactions in Muscle to Extend the Sarcomere Quality Control Network
To further extend the network of chaperones associated with muscle function, the chaperones identified in the screen can, in turn, be used to uncover novel chaperone interactions. As a proof of concept, the motility of CeAha1 mutant animals was used as bait to reveal interactions of this gene with CeHsp90 in muscle function. When CeAha1(ok3501) knockout animals were treated with CeHsp90(RNAi), only 13.9 ± 7% of the animals were motile by the first day of egg-laying, while no effect on motility was observed for CeAha1(ok3501) animals grown on the empty vector control (Figures 5A,B). This was specific to CeAha1(ok3501) animals, as most of the wild-type animals treated with CeHsp90 RNAi were motile (76.6±4%) (Figure 5B). Moreover, both wild-type and CeAha1(ok3501) animals treated with CeHsp90(RNAi) showed a complete larval arrest phenotype (Figure 5A). Thus, using CeAha1 as bait uncovered its interactions with CeHsp90 in muscle function and may serve to further uncover novel interactions with other molecular chaperones.
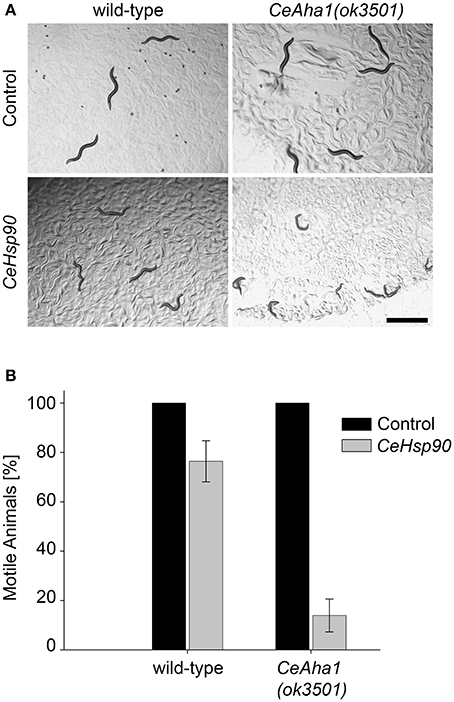
Figure 5. Extending the UNC-45 interaction network. (A) Age-synchronized (L1) wild-type or RB2525 [C01G10.8(ok3501)] animals grown at 15°C were transferred to plates containing control or CeHsp90 (daf-21) RNAi-expressing bacteria and images were taken on day 2 of adulthood. Scale bar is 1 mm. (B) Age-synchronized (L1) wild-type or RB2525 [C01G10.8(ok3501)] animals, treated as in (A), were scored for motility on day 2 of adulthood. Data are mean ± s.e.m.
Discussion
CeHop, CeAha1 and Cep23 are Novel Myosin Co-Chaperones
Incorrect folding and assembly of the sarcomere are prominent contributors to disease and aging (Herndon et al., 2002; Benian and Epstein, 2011). There is only limited understanding of the chaperone networks involved in sarcomere assembly and maintenance. Here, we used a genetic interaction approach as applied to a C. elegans body wall muscle model to identify novel chaperones that are required for sarcomere assembly and maintenance. To screen for novel sarcomere-associated chaperones, we specifically challenged the proteostasis network by studying animals with mutations affecting a known muscle chaperone, unc-45. These unbiased screens were able to genetically verify the known UNC-45-Hsp90 interaction (Barral et al., 2002; Gazda et al., 2013) and identified three Hsp90 co-chaperones, CeHop (STI-1), CeAha1 (C01G10.8) and Cep23 (ZC395.10), not previously associated with myosin folding (Gaiser et al., 2009, 2010, 2011; Song et al., 2009; Meissner et al., 2011; Haslbeck et al., 2012). Hsp90 binds numerous co-chaperones (17 in C. elegans; Table 1) (Haslbeck et al., 2012). Thus, our screen identified a specific set of co-chaperones required for Hsp90-myosin folding, including a client recruiter co-chaperone, CeHop, a remodeling co-chaperone, CeAha1, and a client maturation co-chaperone, Cep23. This set of co-chaperones forms a core chaperone folding cycle (Rohl et al., 2013). The screen, therefore, highlights the Hsp90 ATPase cycle as critical for Hsp90 function in myosin folding, specifically when UNC-45 function is compromised.
Mild Perturbations of the Proteostasis Network Uncover Specific Chaperone Interactions
Down-regulation of chaperone expression in an unc-45 mutant background did not lead to general proteostasis sensitivity as reflected by strong motility defects, regardless of the individual chaperone down-regulated. Rather, a specific set of chaperones was identified that function together to modulate Hsp90 substrate binding, remolding and releasing, and that buffer myosin folding when UNC-45 function is perturbed. This finding is strengthened by the unc-45(m94)-based mutagenesis screen, which only identified unc-45(m94) revertants as modifiers of UNC-45 function under restrictive conditions. In agreement, epistatic analyses of yeast genes required for folding in the endoplasmic reticulum identified specific aggravating and improving interactions between molecular chaperones (Jonikas et al., 2009), suggesting that mild perturbation of the proteostasis network can uncover specific chaperone interactions. Moreover, the discovery of a specific set of chaperones, such as unc-45 modulators, suggests that the proteostasis buffering capacity depends on modulating the function of chaperones that cooperate in the same process. In agreement, DnaK and trigger factor, chaperones that overlap in binding newly synthesized proteins, are redundant for bacterial growth under non-stress conditions but become essential when both are affected (Deuerling et al., 1999; Teter et al., 1999). Thus, a combination of mild perturbations of the proteostasis network, as when mildly affecting two chaperones, are required to uncover novel and specific chaperone interactions.
This specificity is somewhat surprising given that the chronic expression of misfolded proteins or aging unmask many unrelated metastable proteins (Gidalevitz et al., 2006; Ben-Zvi et al., 2009; Shemesh et al., 2013). The difference between the impact we observed for UNC-45 and the effect of aggregation-prone proteins could result from the impact of misfolded proteins and age-dependent modulation of the proteome on the proteostatic network (Shai et al., 2014). Indeed, expression of three distinct SOD1 mutants in C. elegans body wall muscle cells resulted in differential exposure of metastable mutant phenotypes, suggesting that each SOD1 mutant affects the proteostasis network differently (Gidalevitz et al., 2009). Moreover, expression of disease-related proteins was shown to inhibit calthris-mediated endocytosis in mammalian cells by engaging Hsc70. While over-expression of Hsc70 restored calthris-mediated endocytosis, it did not suppress aggregation (Yu et al., 2014). Given that other chaperones, such as Hsp90, DNJA, and Hsp110, were found to be associated with aggregates (Olzscha et al., 2011; Kirstein-Miles et al., 2013), many downstream functions may be affected by the expression of chronically misfolded proteins, resulting in a serious perturbation of the proteostatic network. This is in agreement with the organismal effects we noted upon down-regulating main folding chaperones, such as Hsc70 and mortalin (Figure S2). Indeed, more acute phenotypes were observed for many chaperones when knockout mutants are examined or when RNAi penetrance or efficacy was enhanced (Table 1). The enhanced penetrance of the phenotype could result from further reduction in the levels of expression or function of the chaperones, inducing more severe damage that can lead to different chaperonopathies, such as mutations in DNAJB6 that cause limb-girdle muscular dystrophy (Sarparanta et al., 2012). Thus, gross perturbations of the proteostatic network, as occur, for example, upon expression of aggregation-prone proteins, following down-regulation of a set of chaperones or with age, mask specific interactions and result in a failure of the quality control system to maintain proteostasis.
A Substrate-Centric Chaperone Interaction Map
One possible interpretation of our data is that different co-chaperones form functional modules in vivo that are specialized for a set of substrates to specifically impact their folding. This view of functional modules within the proteostasis network is supported by the roles of co-chaperones in directing chaperone function. For example, Hsp90 has different co-chaperones that perform similar functions for different substrates. Accordingly, kinases and steroid receptors interact with different subsets of Hsp90 co-chaperones (Rohl et al., 2013). Likewise, the combination of J proteins and NEF varies as a function of substrate and cellular function (Kampinga and Craig, 2010; Abrams et al., 2014). This view is further supported by our finding that C. elegans is more sensitive to down-regulation of genes encoding ATPase chaperone machines than those encoding their co-chaperones (compare 13/21 Hsp60/Hsp70/Hsp90 genes to 4/58 Hsp60/Hsp70/Hsp90 co-chaperone genes, Table 1). Co-chaperones that adjust chaperone function to a specific substrate demands may, therefore, give rise to a hierarchical structure of the proteostasis network with partially independent substrate-specific modules.
By directing our examination to how chaperones orchestrate sarcomere assembly and maintenance, we were able to identify key players in sarcomere folding that safeguard the functionality of muscle cells and are at risk in disease and aging. Any newly identified chaperone can be used to find additional chaperones and/or co-factors associated with muscle folding and assembly. Focusing on the substrate, rather than the chaperone, helps in the drawing of a genetic interaction map describing chaperone-substrate interactions in vivo.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The study was supported by an Israeli Council for Higher Education Alon Fellowship, a Marie Curie International Reintegration grant (FP7-268285), and an Israel Science Foundation grant (91/11) to Anat Ben-Zvi; by the Deutsche Forschungsgemeinschaft (CECAD, FOR885, SFB635, KFO 286, and DIP8 grant 2014376) and the European Research Council (consolidator grant 616499) to Thorsten Hoppe.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmolb.2014.00021/abstract
References
Abrams, J. L., Verghese, J., Gibney, P. A., and Morano, K. A. (2014). Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289, 13155–13167. doi: 10.1074/jbc.M113.530014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alavez, S., Vantipalli, M. C., Zucker, D. J., Klang, I. M., and Lithgow, G. J. (2011). Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature 472, 226–229. doi: 10.1038/nature09873
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Balch, W. E., Morimoto, R. I., Dillin, A., and Kelly, J. W. (2008). Adapting proteostasis for disease intervention. Science 319, 916–919. doi: 10.1126/science.1141448
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bardwell, J. C., and Jakob, U. (2012). Conditional disorder in chaperone action. Trends Biochem. Sci. 37, 517–525. doi: 10.1016/j.tibs.2012.08.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barral, J. M., and Epstein, H. F. (1999). Protein machines and self assembly in muscle organization. Bioessays 21, 813–823.
Barral, J. M., Hutagalung, A. H., Brinker, A., Hartl, F. U., and Epstein, H. F. (2002). Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295, 669–671. doi: 10.1126/science.1066648
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Benian, G. M., and Epstein, H. F. (2011). Caenorhabditis elegans muscle: a genetic and molecular model for protein interactions in the heart. Circ. Res. 109, 1082–1095. doi: 10.1161/CIRCRESAHA.110.237685
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ben-Zvi, A., Miller, E. A., and Morimoto, R. I. (2009). Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. U.S.A. 106, 14914–14919. doi: 10.1073/pnas.0902882106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buchner, J., Schmidt, M., Fuchs, M., Jaenicke, R., Rudolph, R., Schmid, F. X., et al. (1991). GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry 30, 1586–1591. doi: 10.1021/bi00220a020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deuerling, E., Schulze-Specking, A., Tomoyasu, T., Mogk, A., and Bukau, B. (1999). Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696.
Ding, L., and Candido, E. P. (2000). HSP25, a small heat shock protein associated with dense bodies and M-lines of body wall muscle in Caenorhabditis elegans. J. Biol. Chem. 275, 9510–9517. doi: 10.1074/jbc.275.13.9510
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eckl, J. M., Drazic, A., Rutz, D. A., and Richter, K. (2014). Nematode Sgt1-homologue D1054.3 binds open and closed conformations of Hsp90 via distinct binding sites. Biochemistry 53, 2505–2514. doi: 10.1021/bi5000542
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ellis, J. (1987). Proteins as molecular chaperones. Nature 328, 378–379. doi: 10.1038/328378a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Etard, C., Behra, M., Fischer, N., Hutcheson, D., Geisler, R., and Strahle, U. (2007). The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev. Biol. 308, 133–143. doi: 10.1016/j.ydbio.2007.05.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feldman, N., Kosolapov, L., and Ben-Zvi, A. (2014). Fluorodeoxyuridine improves Caenorhabditis elegans proteostasis independent of reproduction onset. PLoS ONE 9:e85964. doi: 10.1371/journal.pone.0085964
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaiser, A. M., Brandt, F., and Richter, K. (2009). The non-canonical Hop protein from Caenorhabditis elegans exerts essential functions and forms binary complexes with either Hsc70 or Hsp90. J. Mol. Biol. 391, 621–634. doi: 10.1016/j.jmb.2009.06.051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaiser, A. M., Kaiser, C. J., Haslbeck, V., and Richter, K. (2011). Downregulation of the Hsp90 system causes defects in muscle cells of Caenorhabditis elegans. PLoS ONE 6:e25485. doi: 10.1371/journal.pone.0025485
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaiser, A. M., Kretzschmar, A., and Richter, K. (2010). Cdc37-Hsp90 complexes are responsive to nucleotide-induced conformational changes and binding of further cofactors. J. Biol. Chem. 285, 40921–40932. doi: 10.1074/jbc.M110.131086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garrido, C., Paul, C., Seigneuric, R., and Kampinga, H. H. (2012). The small heat shock proteins family: the long forgotten chaperones. Int. J. Biochem. Cell Biol. 44, 1588–1592. doi: 10.1016/j.biocel.2012.02.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gazda, L., Pokrzywa, W., Hellerschmied, D., Lowe, T., Forne, I., Mueller-Planitz, F., et al. (2013). The myosin chaperone UNC-45 is organized in tandem modules to support myofilament formation in C. elegans. Cell 152, 183–195. doi: 10.1016/j.cell.2012.12.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R., and Morimoto, R. I. (2006). Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474. doi: 10.1126/science.1124514
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gidalevitz, T., Krupinski, T., Garcia, S. M., and Morimoto, R. I. (2009). Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genetics 5:e1000399. doi: 10.1371/journal.pgen.1000399
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goloubinoff, P., Christeller, J. T., Gatenby, A. A., and Lorimer, G. H. (1989). Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature 342, 884–889. doi: 10.1038/342884a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haslbeck, V., Eckl, J. M., Kaiser, C. J., Papsdorf, K., Hessling, M., and Richter, K. (2013). Chaperone-interacting TPR proteins in Caenorhabditis elegans. J. Mol. Biol. 425, 2922–2939. doi: 10.1016/j.jmb.2013.05.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haslbeck, V., Kaiser, C. J., and Richter, K. (2012). Hsp90 in non-mammalian metazoan model systems. Biochim. Biophys. Acta 1823, 712–721. doi: 10.1016/j.bbamcr.2011.09.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hemmingsen, S. M., Woolford, C., Van Der Vies, S. M., Tilly, K., Dennis, D. T., Georgopoulos, C. P., et al. (1988). Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333, 330–334. doi: 10.1038/333330a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herndon, L. A., Schmeissner, P. J., Dudaronek, J. M., Brown, P. A., Listner, K. M., Sakano, Y., et al. (2002). Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808–814. doi: 10.1038/nature01135
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jonikas, M. C., Collins, S. R., Denic, V., Oh, E., Quan, E. M., Schmid, V., et al. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697. doi: 10.1126/science.1167983
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kampinga, H. H., and Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. doi: 10.1038/nrm2941
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karady, I., Frumkin, A., Dror, S., Shemesh, N., Shai, N., and Ben-Zvi, A. (2013). Using Caenorhabditis elegans as a model system to study protein homeostasis in a multicellular organism. J. Vis. Exp. 18:e50840. doi: 10.3791/50840
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J., Lowe, T., and Hoppe, T. (2008). Protein quality control gets muscle into shape. Trends Cell Biol. 18, 264–272. doi: 10.1016/j.tcb.2008.03.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kirstein-Miles, J., Scior, A., Deuerling, E., and Morimoto, R. I. (2013). The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 32, 1451–1468. doi: 10.1038/emboj.2013.87
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laskey, R. A., Honda, B. M., Mills, A. D., and Finch, J. T. (1978). Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275, 416–420. doi: 10.1038/275416a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leroux, M. R., and Candido, E. P. (1997). Subunit characterization of the Caenorhabditis elegans chaperonin containing TCP-1 and expression pattern of the gene encoding CCT-1. Biochem. Biophys. Res. Commun. 241, 687–692. doi: 10.1006/bbrc.1997.7889
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X., Matilainen, O., Jin, C., Glover-Cutter, K. M., Holmberg, C. I., and Blackwell, T. K. (2011). Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet 7:e1002119. doi: 10.1371/journal.pgen.1002119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, J., Langer, T., Boteva, R., Schramel, A., Horwich, A. L., and Hartl, F. U. (1991). Chaperonin-mediated protein folding at the surface of groEL through a “molten globule”-like intermediate. Nature 352, 36–42. doi: 10.1038/352036a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mattoo, R. U., and Goloubinoff, P. (2014). Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins. Cell. Mol. Life Sci. 71, 3311–3325. doi: 10.1007/s00018-014-1627-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meissner, B., Rogalski, T., Viveiros, R., Warner, A., Plastino, L., Lorch, A., et al. (2011). Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS ONE 6:e19937. doi: 10.1371/journal.pone.0019937
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meissner, B., Warner, A., Wong, K., Dube, N., Lorch, A., Mckay, S. J., et al. (2009). An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet 5:e1000537. doi: 10.1371/journal.pgen.1000537
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Melkani, G. C., Trujillo, A. S., Ramos, R., Bodmer, R., Bernstein, S. I., and Ocorr, K. (2013). Huntington's disease induced cardiac amyloidosis is reversed by modulating protein folding and oxidative stress pathways in the Drosophila heart. PLoS Genet 9:e1004024. doi: 10.1371/journal.pgen.1004024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olzscha, H., Schermann, S. M., Woerner, A. C., Pinkert, S., Hecht, M. H., Tartaglia, G. G., et al. (2011). Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78. doi: 10.1016/j.cell.2010.11.050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pelham, H. R. (1986). Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell 46, 959–961. doi: 10.1016/0092-8674(86)90693-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohl, A., Rohrberg, J., and Buchner, J. (2013). The chaperone Hsp90: changing partners for demanding clients. Trends Biochem. Sci. 38, 253–262. doi: 10.1016/j.tibs.2013.02.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sarparanta, J., Jonson, P. H., Golzio, C., Sandell, S., Luque, H., Screen, M., et al. (2012). Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat. Genet. 44, 450–455, S1–S2. doi: 10.1038/ng.1103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sawyer, J. M., Glass, S., Li, T., Shemer, G., White, N. D., Starostina, N. G., et al. (2011). Overcoming redundancy: an RNAi enhancer screen for morphogenesis genes in Caenorhabditis elegans. Genetics 188, 549–564. doi: 10.1534/genetics.111.129486
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schroder, H., Langer, T., Hartl, F. U., and Bukau, B. (1993). DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144.
Shai, N., Shemesh, N., and Ben-Zvi, A. (2014). Remodeling of proteostasis upon transition to adulthood is linked to reproduction onset. Curr. Genomics 15, 122–129. doi: 10.2174/1389202915666140221005023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shemesh, N., Shai, N., and Ben-Zvi, A. (2013). Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell 12, 814–822. doi: 10.1111/acel.12110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, H. O., Lee, W., An, K., Lee, H. S., Cho, J. H., Park, Z. Y., et al. (2009). C. elegans STI-1, the homolog of Sti1/Hop, is involved in aging and stress response. J. Mol. Biol. 390, 604–617. doi: 10.1016/j.jmb.2009.05.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Srikakulam, R., and Winkelmann, D. A. (2004). Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 117, 641–652. doi: 10.1242/jcs.00899
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teter, S. A., Houry, W. A., Ang, D., Tradler, T., Rockabrand, D., Fischer, G., et al. (1999). Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97, 755–765. doi: 10.1016/S0092-8674(00)80787-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vang, S., Corydon, T. J., Borglum, A. D., Scott, M. D., Frydman, J., Mogensen, J., et al. (2005). Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. FEBS J. 272, 2037–2049. doi: 10.1111/j.1742-4658.2005.04630.x
Willis, M. S., and Patterson, C. (2010). Hold me tight: role of the heat shock protein family of chaperones in cardiac disease. Circulation 122, 1740–1751. doi: 10.1161/CIRCULATIONAHA.110.942250
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Willis, M. S., Schisler, J. C., Portbury, A. L., and Patterson, C. (2009). Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc. Res. 81, 439–448. doi: 10.1093/cvr/cvn289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, A., Shibata, Y., Shah, B., Calamini, B., Lo, D. C., and Morimoto, R. I. (2014). Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc. Natl. Acad. Sci. U.S.A. 111, E1481–E1490. doi: 10.1073/pnas.1321811111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Caenorhabditis elegans, chaperones, DAF-21, Hsp90, misfolding, myosin, proteostasis, UNC-45
Citation: Frumkin A, Dror S, Pokrzywa W, Bar-Lavan Y, Karady I, Hoppe T and Ben-Zvi A (2014) Challenging muscle homeostasis uncovers novel chaperone interactions in Caenorhabditis elegans. Front. Mol. Biosci. 1:21. doi: 10.3389/fmolb.2014.00021
Received: 03 September 2014; Accepted: 18 October 2014;
Published online: 06 November 2014.
Edited by:
Abdussalam Azem, Tel Aviv University, IsraelReviewed by:
Harm Kampinga, University Medical Center Groningen, NetherlandsDavid Vilchez, University of Cologne, Germany
Copyright © 2014 Frumkin, Dror, Pokrzywa, Bar-Lavan, Karady, Hoppe and Ben-Zvi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anat Ben-Zvi, Department of Life Sciences, Ben-Gurion University of the Negev, 1 Ben-Gurion Ave., PO Box 653, Beer Sheva 84105, Israel e-mail: anatbz@bgu.ac.il
†These authors have contributed equally to this work.
 Anna Frumkin
Anna Frumkin Shiran Dror
Shiran Dror Wojciech Pokrzywa
Wojciech Pokrzywa Yael Bar-Lavan
Yael Bar-Lavan Ido Karady
Ido Karady Thorsten Hoppe
Thorsten Hoppe Anat Ben-Zvi
Anat Ben-Zvi